Highly pathogenic influenza A virus (HPAIV) continues to pose a pandemic threat globally. From 2003 to 2017, H5N1 HPAIV caused 453 human deaths, giving it a high mortality rate (52.74%). This work shows that miR-324-5p suppresses H5N1 HPAIV replication by directly targeting the viral genome (thereby inhibiting viral gene expression) and cellular CUEDC2 gene, the negative regulator of the interferon pathway (thereby enhancing antiviral genes). Our study enhances the knowledge of the role of microRNAs in the cellular response to viral infection. Also, the study provides help in understanding how the host cells utilize small RNAs in controlling the viral burden.
KEYWORDS: antiviral immunity, avian viruses, innate immunity, viral RNA polymerase, miRNA
ABSTRACT
MicroRNAs (miRNAs) are small noncoding RNAs that are crucial posttranscriptional regulators for host mRNAs. Recent studies indicate that miRNAs may modulate host response during RNA virus infection. However, the role of miRNAs in immune response against H5N1 infection is not clearly understood. In this study, we showed that expression of cellular miRNA miR-324-5p was downregulated in A549 cells in response to infection with RNA viruses H5N1, A/PR8/H1N1, and Newcastle disease virus (NDV) and transfection with poly(I·C). We found that miR-324-5p inhibited H5N1 replication by targeting the PB1 viral RNA of H5N1 in host cells. In addition, transcriptome analysis revealed that miR-324-5p enhanced the expression of type I interferon, type III interferon, and interferon-inducible genes (ISGs) by targeting CUEDC2, the negative regulator of the JAK1-STAT3 pathway. Together, these findings highlight that the miR-324-5p plays a crucial role in host defense against H5N1 by targeting viral PB1 and host CUEDC2 to inhibit H5N1 replication.
IMPORTANCE Highly pathogenic influenza A virus (HPAIV) continues to pose a pandemic threat globally. From 2003 to 2017, H5N1 HPAIV caused 453 human deaths, giving it a high mortality rate (52.74%). This work shows that miR-324-5p suppresses H5N1 HPAIV replication by directly targeting the viral genome (thereby inhibiting viral gene expression) and cellular CUEDC2 gene, the negative regulator of the interferon pathway (thereby enhancing antiviral genes). Our study enhances the knowledge of the role of microRNAs in the cellular response to viral infection. Also, the study provides help in understanding how the host cells utilize small RNAs in controlling the viral burden.
INTRODUCTION
The highly pathogenic avian influenza A virus (HPAIV) continues to pose a pandemic threat due to zoonotic transmission to humans. The high degree of genetic shift and reassortment has evolved the influenza A virus in such a way that the virus acquired resistance against various antiviral drugs and therapeutic antibodies (1–3). HPAIV infection causes severe respiratory illness by rapid progression of pneumonia in avian and mammalian hosts, including humans. From 2003 to 2017, H5N1 HPAIV caused 453 human deaths among 859 cases reported, giving it a high mortality rate (52.74% [http://www.who.int/influenza/human_animal_interface/2017_06_15_tableH5N1-corrected.pdf?ua=1]). Recently, there has been increasing evidence that host-encoded small noncoding RNAs interact with the genomes of RNA viruses and inhibit viral replication. Thus, these RNAs could prove to be a potential candidate for controlling viral replication via interaction with viral transcripts or genomes (4–6).
MicroRNAs (miRNAs) are posttranscriptional gene regulators involved in fine tuning of gene expression, subsequent synthesis of protein, and regulation of cellular signaling pathways through interaction with the 3′ untranslated regions (3′ UTR) of target genes (7–9). Various studies have shown that expression of cellular miRNAs is profoundly influenced by viral infection and that altered miRNA expression leads to enhanced or suppressed antiviral responses (10–12). These responses may help in viral evasion or restriction during infection as shown previously for influenza virus, hepatitis C virus (HCV), and HIV-1 (13–15).
In this study, we showed that H5N1 infection suppresses miR-324-5p expression in lungs of C57BL/6 mice in vivo and in human primary small airway epithelial cells (SAECs) and the human lung carcinoma cell line A549 in vitro. Moreover, a similar pattern was observed in swine flu (influenza) patients. Ectopic expression of miR-324-5p in SAECs and A549 cells inhibited viral replication. The inhibition of viral replication was through direct interaction of miR-324-5p with PB1 transcript (PB1 encodes the subunit of viral RNA polymerase) via formation of the active RNA-induced silencing complex (RISC), resulting in less viral RNA polymerase and leading to decreased viral loads in various cell types.
Influenza A virus is sensed by Toll-like receptor 3 (TLR3), TLR7, TLR8, and RIG-I and induces an antiviral state (16–18). Uncontrolled activation of sensor-mediated antiviral signaling pathways can lead to a hyperinflammatory state or immunopathology (19). To develop appropriate antiviral immune responses, maintain immune homeostasis, and ensure viral clearance, the signaling pathways are tightly regulated by various negative regulators, such as SOCS1 to -7 and CUEDC2 (20, 21). Previously, it has been reported that several viruses promote expression of host negative regulators to dampen the antiviral immune state, thereby enhancing viral replication (22).
In our study, we found that ectopic expression of miR-324-5p enhanced the induction of antiviral genes, such as the type I (IFN-α4 and IFN-β) and type III (IFN-λ1, -λ2, and -λ3) interferon genes and interferon-inducible genes following H5N1 infection. To identify the mechanism, we performed high-throughput transcriptome sequencing (RNA-Seq) analysis in A549 cells overexpressing miR-324-5p and identified CUEDC2. CUEDC2 is a negative regulator of the JAK1-STAT3 pathway, and the 3′ UTR is targeted by miR-324-5p. Together, the results of our study show dual roles of miR-324-5p in regulating host antiviral immunity by targeting viral PB1 transcripts and the 3′ UTR of CUEDC2 to inhibit H5N1 replication.
RESULTS
miR-324-5p is downregulated during RNA virus infection and predicted to target the H5N1 genome.
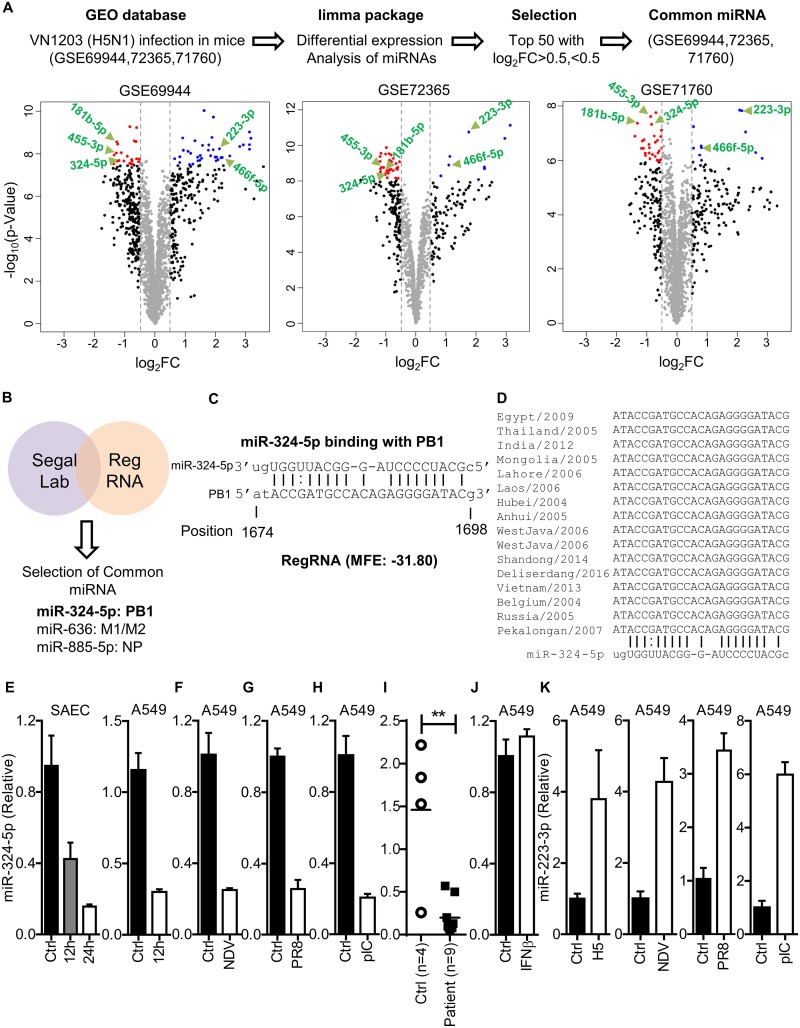
To identify the miRNAs which are significantly dysregulated during H5N1 (RNA virus) infection, the Gene Expression Omnibus (GEO) database was searched for experiments involving mouse infection with H5N1. Three data sets (GSE69944, GSE72365, and GSE71760) generated from three independent experiments were identified in which C57BL/6 mice were infected with A/Vietnam/1203/2004 (H5N1) and miRNA microarray analysis was performed using RNA from lung tissue at 4 days postinfection (p.i.). We reanalyzed these data sets using the limma package for differential expression, and on day 4 p.i., expression of a large number of miRNAs was found to be either upregulated or downregulated, as shown by volcano plot (Fig. 1A). The top 50 miRNAs from each of three data sets were selected with log2 fold changes (FC) of >0.5 or <0.5. Next, common miRNAs among these data sets were selected. miR-466f-5p and miR-223-3p were found to be upregulated, whereas miR-455-3p, miR-324-5p, and miR-181b-5p were downregulated (Fig. 1A).
FIG 1.
RNA virus infection downregulates the expression of miR-324-5p. (A) Schematic representation of work flow for data analysis. The volcano plot represents dysregulated miRNAs identified in lung tissue of C57BL/c mice infected with A/Vietnam/1203/2004 (H5N1) (176 PFU/mouse) at day 4 postinfection in three independent microarray data sets using the indicated screening algorithm (upper portion). Common miRNAs that were increased or decreased more than log2 FC (0.5) among the top 50 miRNAs in three independent studies were selected and are shown by the green arrows (lower portion). (B) List of candidate miRNAs targeting the H5N1 genome identified using Segal Lab and RegRNA1.0. The cutoffs used for Segal Lab and RegRNA1.0 were −20 kcal/mol and −25 kcal/mol, respectively. (C) Prediction of the miR-324-5p binding site in the PB1 gene of H5N1 and minimum free energy (MFE) of miRNA-PB1 duplex. The indicated position is relative to the first nucleotide of the PB1 gene. (D) Sequence alignment of the PB1 gene (target sequence) across different clades of H5N1 aligned using ClustalW. (E to H) Quantification of relative expression of hsa-miR-324-5p in SAECs and A549 cells with H5N1 infection (MOI, 1) at the indicated time points (E); A549 cells with NDV infection (MOI, 1) at 24 h (F); A549 cells with A/PR8/H1N1 (PR8) infection (MOI, 1) at 24 h (G); A549 cells with poly(I·C) transfection (1 μg/ml) at 24 h (H). (I) Quantification of relative expression of miR-324-5p in swine flu patients (n = 9) compared to healthy controls (n = 4). (J) Quantification of relative abundance of miR-324-5p in A549 cells in response to stimulation with IFN-β (100 IU/ml) after 12 h. (K) Quantification of relative expression of miR-223-3p in A549 cells infected with H5N1 (MOI, 1), NDV (MOI, 1), and PR8 (MOI, 1) and transfected with poly(I·C) (pIC; 1 μg/ml) at 24 h. Ctrl, control (uninfected); H5, H5N1 virus. Data are means ± SEMs from triplicate samples of a single experiment and are representative of results from three independent experiments (E to K).
Simultaneously, the binding prediction for all known miRNAs with the segmented genome of H5N1 was performed through an in silico approach using Segal Lab and RegRNA1.0 software, which predicts the binding by computing energy gained from miRNA-target formation and identifying homologs of regulatory RNA motifs, respectively (23, 24). To screen miRNA, minimum cutoff free energy (MFE) was kept at −20 kcal/mol and −25 kcal/mol for Segal Lab and RegRNA1.0, respectively, and the common miRNAs were selected. Both software packages predicted miR-324-5p, miR-885-5p, and miR-636 having binding sites in PB1, NP, and M1, respectively (Fig. 1B). We found that miR-324-5p is downregulated in mouse lung tissue in response to H5N1 infection, and it is predicted to have the binding site in PB1 (Fig. 1C), one of the essential genes of the H5N1 virus required for viral transcription and replication. Further analysis revealed that the binding site for miR-324-5p in the PB1 gene is highly conserved among various subtypes of H5N1 (Fig. 1D).
These in vivo observations were validated in vitro by infecting the primary human cells derived from lung known as small airway epithelial cells (SAECs) with H5N1, and it was found that miR-324-5p was downregulated at 12 and 24 h postinfection (Fig. 1E, left graph). Similar results were obtained when human alveolar basal epithelial (A549) cells were infected with H5N1 virus (Fig. 1E, right graph). Further, to test whether the downregulation of miR-324-5p is specific to H5N1 influenza virus, we infected A549 cells with Newcastle disease virus (NDV) or A/PR8/H1N1 (PR8) or transfected them with poly(I·C). We found that the expression of miR-324-5p was downregulated in response to infection with NDV or A/PR8/H1N1 virus and poly(I·C) transfection, indicating that downregulation of miR-324-5p might be a regulatory response of the host during RNA virus infection or to RNA virus pathogen-associated molecular patterns (PAMPs) (Fig. 1F to H). Also, we analyzed miR-324-5p expression in nasopharyngeal swab samples collected from H1N1 patients. miR-324-5p exhibited a pattern similar to that observed in vivo and in vitro (Fig. 1I). However, stimulation with recombinant IFN-β (rIFN-β) did not change the expression levels of miR-324-5p (Fig. 1J), indicating that the interferon signaling pathway has no role in downregulation of miR-324-5p. Further, to confirm that in vivo microarray results correlate with in vitro results, we tested the expression of miR-223-3p. We observed that expression of miR-223-3p increased in response to infection with H5N1 (as shown in Fig. 1A), NDV, and A/PR8/H1N1 and to transfection with poly(I·C), indicating that bioinformatics analysis of in vivo microarray is consistent with in vitro results (Fig. 1K). All together, these results indicate that expression of miR-324-5p is decreased in response to RNA virus infection.
miR-324-5p specifically targets the PB1 gene of H5N1.
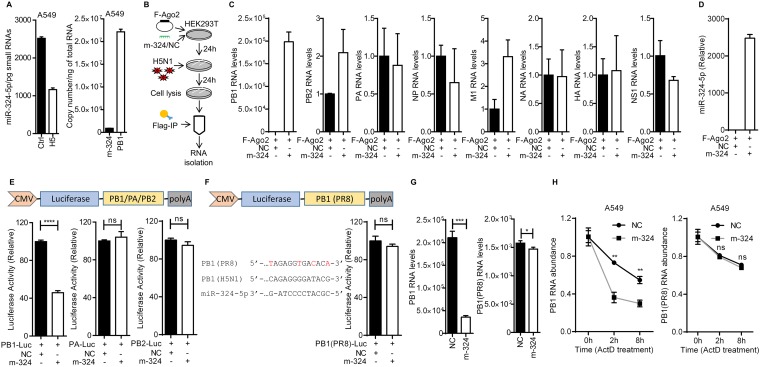
The observation that miR-324-5p is reduced in response to infection with H5N1 led us to investigate the copy number of miR-324-5p. Studies have shown that almost 60% of the microRNA expressed has no observable effect on the target transcript (25). microRNA expressed at >100 copies/pg of small RNA functions as a physiologically relevant candidate for suppressing transcript expression (26). We employed the same strategy to estimate the functionality of miR-324-5p in infected cells. To find out the copy number of miR-324-5p/pg small RNA, we infected A549 cells with H5N1 virus under the same conditions as for Fig. 1E (right graph), isolated the small RNAs, and calculated the copy number of miR-324-5p. The copy number of miR-324-5p in the H5N1-infected cells was found to be manifold higher than 100 copies/pg of small RNA (Fig. 2A, left graph), indicating that miR-324-5p is physiologically active for suppressing transcript expression in the host cell. Also, we calculated the copy number of miR-324-5p and PB1 RNAs in H5N1-infected cells. The copy numbers of miR-324-5p and PB1 in H5N1-infected cells were found to be (9.11 to 9.21) × 105 copies/ng of total RNA and (2.17 to 2.39) × 107 copies/ng of total RNA, respectively (Fig. 2A, right graph).
FIG 2.
miR-324-5p interacts with the PB1 gene of H5N1. (A) Quantification of copy number per picogram of small RNA of miR-324-5p in uninfected and H5N1-infected (MOI, 1) A549 cells (left) and quantification of copy number per nanogram of total RNA of miR-324-5p and PB1 in A549 cells infected with H5N1 at an MOI of 1 (right). The copy number of miR-324-5p and PB1 was calculated using miR-324-5p mimic and PB1 RNA as a standard. (B) Schematic representation of work flow for RNA immunoprecipitation (RNA-IP) assay with H5N1 virus. (C) HEK293T cells were transfected with Flag-tagged Ago2 (F-Ago2) along with m-324 (miR-324-5p mimic) or NC (negative-control mimic) and then infected with H5N1 for 24 h at an MOI of 1. Cells were subjected to RNA-IP with anti-Flag antibody-tagged beads, and the abundances of PB1, PB2, PA, NP, M1, NA, HA, and NS1 viral RNAs were quantified using qRT-PCR analysis. (D) Quantification of relative abundance of miR-324-5p in RNA-IP performed for panel B. (E) The PB1, PA, and PB2 genes were cloned into the pMIR-Report vector as shown. HEK293T cells were transfected with PB1-Luc, PA-Luc, or PB2-Luc (100 ng) along with 25 nM m-324 or NC, lysed after 24 h, and subjected to luciferase assay. CMV, cytomegalovirus. (F) The PB1 gene of A/PR8/H1N1 was cloned into the pMIR-Report vector as shown. PB1 of PR8 influenza virus was aligned with PB1 of H5N1 influenza. Mismatches between the two are indicated in red. HEK293T cells were transfected with PB1(PR8)-Luc along with 25 nM m-324 or NC, lysed after 24 h, and subjected to luciferase assay. (G) A549 cells were transfected with miR-324 or NC, followed by infection with H5N1 (left) or A/PR8/H1N1 (right) at an MOI of 1 after 24 h, and subjected to qRT-PCR analysis. (H) Quantification of the relative amounts of PB1 RNA in A549 cells that were transfected with NC or m-324, infected with H5N1 (left) or A/PR8/H1N1 (right) for 24 h at an MOI of 1, and then treated with actinomycin D (ActD; 10 μg/ml) for the indicated times. Data are means ± SEMs from triplicate samples of a single experiment and are representative of results from two independent experiments (A, C, D, and H) and three independent experiments (E to G). ****, P < 0.0001, ***, P < 0.001, **, P < 0.01, and *, P < 0.05, by two-tailed unpaired t test (E to H). ns, nonsignificant.
The observation that miR-324-5p has a binding site in the PB1 gene of H5N1 prompted us to validate the interaction. To this end, RNA immunoprecipitation (RNA-IP) was performed with HEK293T cells as shown in the schematic in Fig. 2B. For RNA-IP, we cotransfected cells with miR-324-5p mimic or the microRNA negative control (NC) and Flag-tagged Ago2 (major protein involved in RNA-induced silencing complex), and then cells were infected with H5N1. The Ago2 was pulled down using Flag-specific antibody, and the abundances of polymerase basic 1 (PB1), PB2, polymerase acidic (PA), nucleoprotein (NP), matrix protein 1 (M1), neuraminidase (NA), hemagglutinin (HA), and nonstructural protein 1 (NS1) RNAs associated with Ago2 were quantified. We found a high abundance of PB1 RNA in cells overexpressing miR-324-5p compared to NC (Fig. 2C), suggesting that Ago2 facilitates the binding of miR-324-5p with PB1. However, other viral RNAs were not found to be associated with Ago2 (Fig. 2C). The presence of miR-324-5p in the pulldown complex was confirmed by TaqMan assay using reverse transcription-quantitative PCR (qRT-PCR) (Fig. 2D). The results suggest that miR-324-5p interacts with the PB1 gene of H5N1. Furthermore, to test the functional significance of miR-324-5p and PB1 interaction, we constructed three luciferase reporter plasmids containing the PB1, PA, and PB2 genes, named PB1-Luc, PA-Luc, and PB2-Luc, respectively. HEK293T cells were cotransfected with miR-324-5p or NC and luciferase reporter plasmids, and then luciferase activity was measured. Luciferase activity of PB1-Luc was significantly reduced with miR-324-5p compared to NC, whereas no reduction was observed in the case of PA-Luc and PB2-Luc (Fig. 2E), indicating that miR-324-5p specifically interacts and inhibits PB1 expression upon binding. We also found that the miR-324-5p binding site in the PB1 gene of A/PR8/H1N1 influenza virus has several nucleotide mismatches when aligned with PB1 of H5N1 (Fig. 2F, left side). We cloned the PB1 gene of A/PR8/H1N1 into the luciferase reporter vector [called PB1(PR8)-Luc]. HEK293T cells were cotransfected with miR-324-5p or NC and luciferase reporter plasmid, and again luciferase activity was measured. The inhibitory effect of miR-324-5p was completely abolished due to mismatches in the miR-324-5p target site in PB1 derived from A/PR8/H1N1 (Fig. 2F, right side). Furthermore, A549 cells transfected with miR-324-5p or NC and subsequently infected with H5N1 or H1N1 were tested for the levels of PB1. Cells with miR-324-5p showed high levels of reduction in H5N1 PB1 transcripts (Fig. 2G, left graph), whereas a low level of reduction was observed for A/PR8/H1N1 PB1 transcript levels (Fig. 2G, right graph) in comparison to that in cells transfected with NC. To understand the mechanism of miR-324-5p-mediated suppression of PB1 expression, we transfected A549 cells with the miR-324-5p or NC mimic, infected the cells with H5N1 for 24 h, and then treated the cells with actinomycin D, an inhibitor of transcription. We observed that the degradation of PB1 RNA was higher in the presence of miR-324-5p than with NC (Fig. 2H, left graph), indicating that interaction of miR-324-5p with PB1 leads to degradation of PB1 RNA. Similarly, we performed the experiment after infection with A/PR8/H1N1. We observed that the levels of degradation of PB1 RNA were comparable in miR-324-5p- and NC-transfected cells (Fig. 2H, right graph), indicating that miR-324-5p does not degrade the PB1 RNA of A/PR8/H1N1. Together, the results suggest that miR-324-5p specifically interacts with the PB1 gene of H5N1 and suppresses PB1 expression by degrading its RNA.
miR-324-5p inhibits H5N1 viral replication.
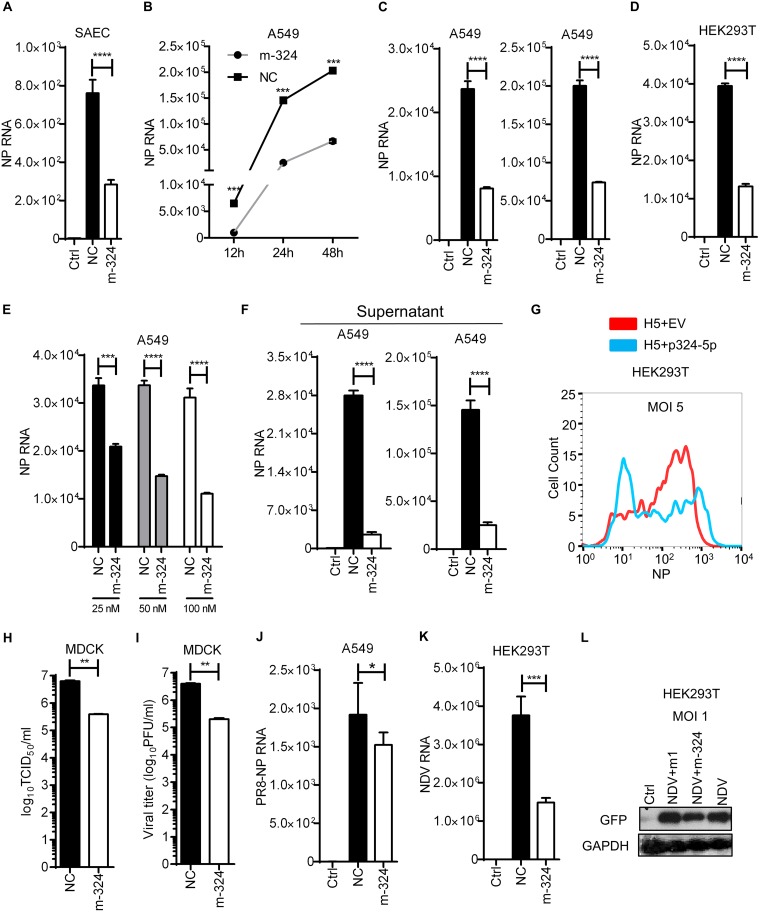
As miR-324-5p interacts with PB1 of H5N1, we sought to investigate the regulatory role of miR-324-5p in H5N1 replication. To this end, SAECs were transfected with miR-324-5p or NC, followed by infection with H5N1 at a multiplicity of infection (MOI) of 1, and then viral loads were quantified by measuring the expression of the H5N1 NP gene. The expression level of NP RNA, a signature of viral replication, was significantly decreased in miR-324-5p-overexpressing cells compared to that in NC-transfected cells (Fig. 3A). Next we examined the growth kinetics of the H5N1 in the presence of miR-324-5p or NC; we observed a significant reduction in viral load at 12, 24, and 48 h postinfection (Fig. 3B). Additionally, we observed that the viral load was significantly reduced at MOIs of 1 and 5 in A549 cells overexpressing miR-324-5p compared to those with NC (Fig. 3C). Next HEK293T cells were transfected with the miR-324-5p mimic or NC and subsequently infected with H5N1 at an MOI of 1. The miR-324-5p mimic-overexpressing cells showed a significant reduction of virus after 24 h of infection (Fig. 3D). Moreover, A549 cells transfected with different amounts of miR-324-5p showed reduction of viral load in a dose-dependent manner (Fig. 3E). As influenza virus progeny is released into the culture supernatant, viral production in the culture supernatant of A549 cells infected at MOIs of 5 and 10 were quantified. Consistent with previous results, the viral titer was significantly reduced in the supernatant of cells transfected with miR-324-5p mimic (Fig. 3F). Furthermore, the reduction in viral replication was confirmed by testing viral protein in infected cells by flow cytometry using H5N1 NP-specific antibody (Fig. 3G). To further confirm these results, we transfected A549 cells with the miR-324-5p or NC mimic, followed by infection with H5N1 at an MOI of 0.5. The supernatants containing released virus particles were collected after 24 h and used for the measurement of viral load by standard assays, such as 50% tissue culture infective dose (TCID50) and plaque assays, by infecting MDCK cells. miR-324-5p markedly reduced H5N1 viral titer as determined by TCID50 assay (Fig. 3H) and plaque assay (Fig. 3I). As shown in Fig. 2E, miR-324-5p does not bind with the PB1 gene of A/PR8/H1N1 due to a lack of a functional binding site; this prompted us to investigate the effect of miR-324-5p on viral replication of A/PR8/H1N1. We transfected A459 cells with miR-324-5p or NC and infected them with A/PR8/H1N1. We observed that miR-324-5p does not bind to the PB1 gene of A/PR8/H1N1 but still inhibited the replication of A/PR8/H1N1 (Fig. 3J), although not to same extent as observed with H5N1. To verify that inhibition of viral replication is associated only with influenza virus or other RNA viruses, we used NDV, a single-stranded RNA (ssRNA) virus which does not have an miR-324-5p target site in the genome. miR-324-5p inhibited NDV replication, as determined by quantifying RNA and viral protein by qRT-PCR (Fig. 3K) and Western blot analysis (Fig. 3L), respectively. Together, these results indicate that miR-324-5p inhibits H5N1 replication by interacting with H5N1 PB1 RNA, but results with A/PR8/H1N1 and NDV also indicate the possibility of simultaneous existence of another axis for viral inhibition.
FIG 3.
miR-324-5p inhibits H5N1 in various cell types. (A) SAECs were transfected with m-324 or NC at 100 nM, followed by infection with H5 at an MOI of 1. After 24 h, relative levels of NP RNA were measured by qRT-PCR. (B) A549 cells were transfected with 100 nM m-324 or NC, followed by infection with H5 at an MOI of 5. NP relative RNA was quantified by qRT-PCR at 12, 24, and 48 h after infection. (C) A549 cells were transfected with 100 nM m-324 or NC, followed by infection with H5 at MOIs of 1 (left) and 5 (right). NP relative RNA was quantified by qRT-PCR after 24 h of infection. (D) HEK293T cells were transfected with miR-324-5p mimic or NC for 24 h and subjected to infection with H5 at an MOI of 1 for 24 h, and NP was detected using qRT-PCR. (E) A549 cells were transfected with increasing amounts (25, 50, and 100 nM) of m-324 or NC and subjected to infection with H5N1 at an MOI of 5. NP relative RNA was quantified by qRT-PCR after 24 h. (F) A549 cells were transfected with 100 nM m-324 or NC, followed by infection with H5N1 at MOIs of 5 (left) and 10 (right) for 24 h. Relative levels of NP RNA were quantified in cell culture supernatant. (G) HEK293T cells were transfected with plasmid encoding miR-324-5p (p324-5p) or empty vector (EV; used as a negative control) for 24 h and subjected to infection with H5 at an MOI of 1 for 12 h, and NP was detected using flow cytometer analysis with anti-NP antibody. (H and I) Effect of miR-324-5p on H5N1 replication by TCID50 (H) and plaque (I) assays. A549 cells were transfected with m-324 or NC, followed by infection with H5N1 at an MOI of 0.5. After 24 h, supernatant containing viral particles was used for infecting MDCK cells. At 96 h after H5N1 infection, the viral load was determined by TCID50 assay (H) and plaque assay (I) in a 96-well plate. (J) A549 cells were transfected with 100 nM m-324 or NC, followed by infection with A/PR8/H1N1 (PR8) at an MOI of 1. NP relative RNA was quantified by qRT-PCR after 24 h of infection. (K) HEK293T cells were transfected with m-324 or NC, followed by infection at an MOI of 5. NDV RNA was quantified after 24 h using qRT-PCR analysis. (L) HEK293T cells were transfected with either m-324 or miR-1, followed by infection with GFP-expressing NDV (NDV-GFP) at an MOI of 1, and subjected to Western blotting after 24 h using anti-GFP antibody. Data are means ± SEMs from triplicate samples of a single experiment and are representative of results from three independent experiments. (A, C, D, F, J, and K) ****, P < 0.001, and *, P < 0.01, by one-way ANOVA. (B, E, H, and I) ****, P < 0.0001, ***, P < 0.001, and **, P < 0.01, by two-tailed unpaired t test.
miR-324-5p promotes antiviral innate immunity during H5N1 infection.
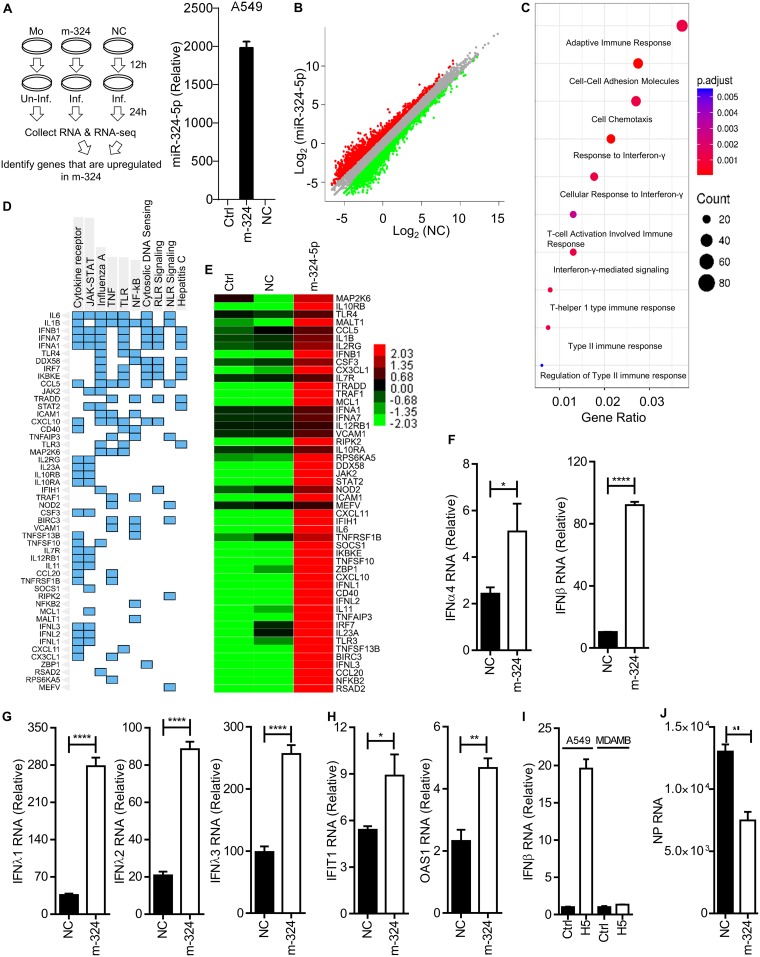
To investigate the possible existence of another axis for viral inhibition, A549 cells were either mock transfected or transfected with miR-324-5p or NC, followed by infection with H5N1 for 24 h, and subjected to transcriptome analysis using an Illumina next-generation sequencer (NGS) as shown in the schematic in Fig. 4A. Notably, the transfection efficiency of miR-324-5p was confirmed by qRT-PCR using miR-324-5p TaqMan (Fig. 4A, right side). Transcriptome analysis showed that 763 genes were upregulated more than 2-fold upon miR-324-5p transfection in comparison to findings with NC (Fig. 4B; see also Table S1 in the supplemental material). Further, Gene ontology (GO) analysis unveiled that the majority of genes are associated with the adaptive immune response, cell adhesion molecules, and responses to type I, II, and III interferons (Fig. 4C). Additionally, KEGG pathway analysis of upregulated genes indicated enrichment of genes for cytokine signaling, JAK-STAT, influenza A virus, tumor necrosis factor (TNF) signaling, Toll-like receptor (TLR) signaling, NF-κB signaling, cytosolic DNA sensing, RIG-I-like receptor (RLR) signaling, NOD-like receptor (NLR) signaling, and hepatitis C virus signaling pathways (Fig. 4D). Interestingly, we found that genes related to type I interferons (IFN-α1, IFN-α7, and IFN-β1), type III interferons (IFN-λ1 to -λ4), and interferon-stimulated genes (DDX58, IFIT1, IFIH1, etc.) were predominantly upregulated (Fig. 4E). Furthermore, the NGS results were verified independently by testing the expression of type I interferons (IFN-α4 and IFN-β), type III interferons (IFN-λ1, IFN-λ2, and IFN-λ3), and interferon-stimulated genes (IFIT1 and OAS1) using qRT-PCR after H5N1 infection. Consistent with NGS results, cells expressing miR-324 showed enhanced expression of IFN-α4, IFN-β, IFN-λ1, IFN-λ2, IFN-λ3, IFIT1, and OAS1 (Fig. 4F to H). These results indicate that miR-324-5p inhibits H5N1 replication by targeting the viral PB1 gene and enhancing antiviral genes. To confirm dual mechanisms for inhibition of H5N1 replication by miR-324-5p, we used MDAMB-231 cells, which have compromised IFNAR1 expression (27–29). Notably, IFNAR1 is required for the activation of the JAK-STAT pathway for production of antiviral genes, such as type I and type III interferon genes and ISGs. To this end, A549 and MDAMB-231 cells were either left uninfected or infected with H5N1 at an MOI of 1 and the IFN-β RNA level was quantified. As expected, IFN-β was induced in A549 cells; however, it was not induced in MDAMB-231 cells in response to H5N1 infection (Fig. 4I), confirming that MDAMB-231 cells are deficient in the interferon pathway. Next, we transfected MDAMB-231 cells with NC or miR-324-5p followed by infection with H5N1 and quantified the viral replication. We observed that miR-324-5p inhibited the replication of H5N1 (Fig. 4J) even in the absence of an interferon response, indicating that interaction of miR-324-5p with PB1 is one of the two factors involved in inhibition of H5N1 replication.
FIG 4.
miR-324-5p enhances ISGs and type I and type III interferons. (A) Schematic outline of transfection, infection (H5N1), RNA isolation, sequencing, and RNA-Seq analysis. Mo, mock; Inf., infected; Un-inf., uninfected. (B) Global expression analysis during m-324 overexpression in H5N1-infected A549 cells using R package. NC is plotted versus m-324 in H5N1-infected samples. Genes significantly changed (>2-fold) are colored in red and green for upregulated and downregulated, respectively. (C) Gene ontologies represented as dot plots. The circles are colored based on the significance (P value) of enrichment, while the sizes represent the number of genes that are enriched for that particular category. (D) Clustergram representing enriched KEGG pathways analysis as columns and input genes as rows; cells in the matrix (blue color) indicate if a gene is associated with the pathway. (E) Heat map analysis of upregulated genes associated with innate immune responses. (F to H) A549 cells were transfected with either m-324 or NC, followed by infection with H5N1 at an MOI of 5, and subjected to qRT-PCR analysis after 24 h to determine the relative expression of IFN-α4 and IFN-β (F), IFN-λ1, IFN-λ2, and IFN-λ3 (G), and IFIT1 and OAS1 (H) RNAs. (I) Equal numbers of A549 and MDAMB-231 cells were either left uninfected (Ctrl) or infected with H5N1 at an MOI of 1 for 24 h. IFN-β RNA levels were quantified by qRT-PCR. (J) MDAMB-231 cells were transfected with 100 nM m-324 or NC, followed by infection with H5N1 at an MOI of 1 for 24 h. Relative levels of NP RNA were quantified by qRT-PCR. Data are means ± SEMs of triplicate samples of a single experiment and are representative of results from three independent experiments (F to J). ****, P < 0.0001, ***, P < 0.001, **, P < 0.01, and *, P < 0.05, by two-tailed unpaired t test.
miR-324-5p enhances antiviral immune responses by targeting the 3′ UTR of the CUEDC2 transcript.
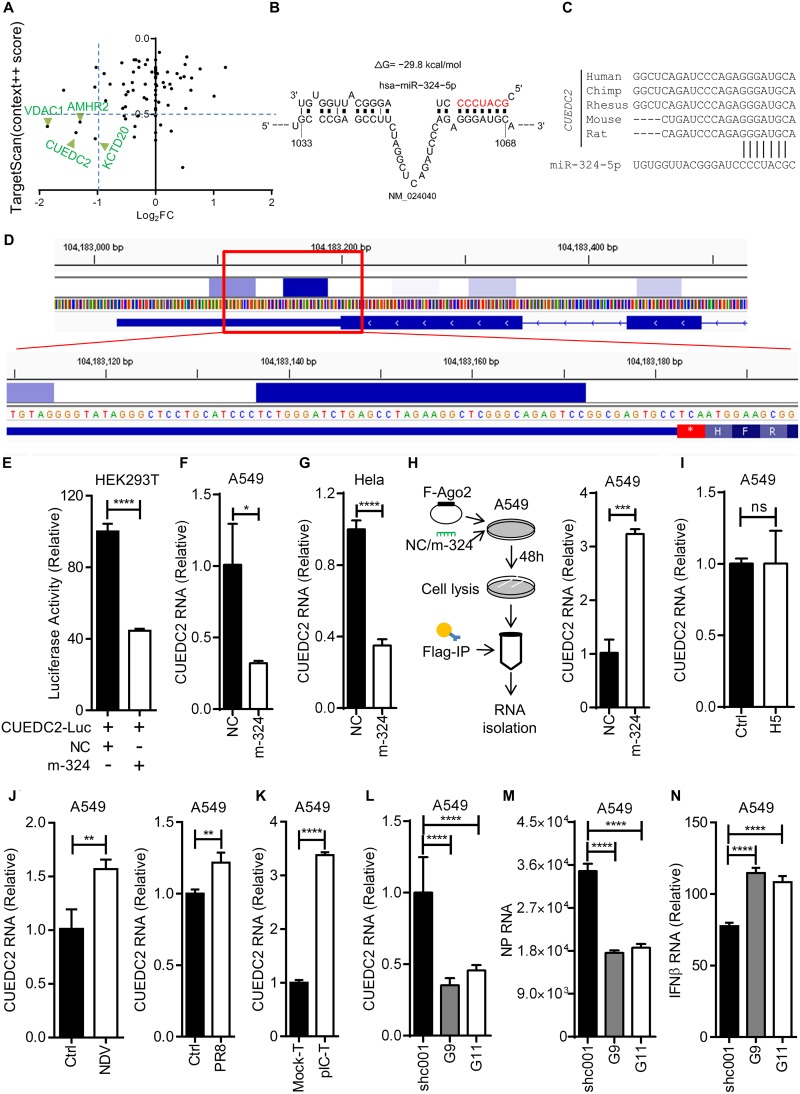
To understand underlying molecular mechanism of miR-324-5p-mediated enhanced production of type I and type III interferons and ISGs during H5N1 infection, the previously discussed transcriptome data were reanalyzed for the downregulated genes. Data analysis showed that 635 genes were downregulated in the miR-324-5p-transfected sample compared to the NC (Table S2). Next the downregulated genes were analyzed using target prediction algorithms, such as Targetscan and Miranda, for identification of miRNA targets in transcripts. A total of 4 out of 635 genes were commonly predicted by the different target prediction algorithms (Fig. 5A and B). Among all 4 genes, CUEDC2 has been previously reported to be involved as a negative regulator of the JAK1-STAT3 signaling pathway, whereas other genes had not been reported to be associated with immunity (21). Notably, the target site for miR-324-5p is conserved among the chimpanzee, rhesus monkey, mouse, and rat (Fig. 5C). CUEDC2 has also been shown to interact with Ago2 in CLIP-Seq (cross-linking immunoprecipitation followed by sequencing) analysis (30). However, to verify the binding of miR-324-5p with CUEDC2, we reanalyzed the CLIP of Ago2 (RISC complex) followed by high-throughput sequencing (CLIP-Seq; GEO accession no. GSE44404) and observed that the large number of Ago2 interacting reads aligned to the genomic region with the predicted miR-324-5p binding site (Fig. 5D). To further strengthen our findings, we cloned the CUEDC2 3′ UTR downstream of the luciferase gene in the pMIR-Report vector (named CUEDC2-Luc). HEK293T cells were cotransfected with miR-324-5p or NC and CUEDC2-Luc, and then luciferase activity was measured. Luciferase activity of CUEDC2-luc was significantly reduced with miR-324-5p compared to that with NC (Fig. 5E), indicating that miR-324-5p directly interacts with and inhibits CUEDC2 expression after binding to the seed sequence. Similar results were obtained at the RNA level by real-time PCR in A549 and HeLa cells (Fig. 5F and G). To further confirm the direct interaction between CUEDC2 and miR-324-5p, RNA-IP was performed with Flag-Ago2 in A549 cells as shown in the schematic in Fig. 5H. When F-Ago2 was pulled down using antibody specific for Flag, CUEDC2 was found to directly interact with the Ago2 (Fig. 5H). Next, expression of CUEDC2 during RNA virus infection was investigated in A549 cells using H5N1, NDV, and poly(I·C). Expression of CUEDC2 was not affected by H5N1 (Fig. 5I), whereas NDV and PR8 modestly induced the expression of CUEDC2 (Fig. 5J). On the other hand, poly(I·C) transfection in A549 cells enhanced CUEDC2 expression (Fig. 5K), suggesting that CUEDC2 induction may depend on the pathogenicity of the virus or ligand. Together, the results showed that miR-324-5p enhances the antiviral immune response by directly targeting the 3′ UTR of CUEDC2 transcripts. In order to confirm that CUEDC2 acts as a negative regulator of JAK1-STAT3 signaling to inhibit the antiviral response, we knocked down CUEDC2 in A549 cells. Knockdown was confirmed at the RNA level (Fig. 5L). Reduced expression of CUEDC2 resulted in an enhanced antiviral response and subsequently reduced H5N1 replication (Fig. 5M and N).
FIG 5.
miR-324-5p targets the 3′ UTR of the CUEDC2 gene. (A) Identification of genes targeted by miR-324-5p. Common genes between Targetscan (Context++ score > 0.5) and genes downregulated in m-324 RNA sequencing (log2 FC < −1) were selected and are shown with green arrows. (B) Prediction of miR-324-5p binding with the 3′ UTR of the CUEDC2 gene. NM_024040 is the RefSeq CUEDC2 transcript identifier, and “ΔG” represents energy of miR-324-5p-CUEDC2 complex. (C) Conserved miR-324-5p binding sites in the 3′ UTR of CUEDC2. (D) Ago2 interaction loci in the 3′ UTR of the CUEDC2 gene in 293S cells as visualized in Integrative Genomics Viewer (IGV). CLIP tags are represented by a heat map. Numbers represent the genomic location, and an asterisk represents the stop codon. (E) HEK293T cells were transfected with CUEDC2-Luc (CUEDC2) in the presence of m-324-5p or NC, lysed after 24 h, and subjected to luciferase assay. (F and G) A549 (F) and HeLa (G) cells were transfected with m-324 (miR-324-5p) or NC, followed by quantification of CUEDC2 RNA after 24 h. (H) Schematic of RNA-IP assay. A549 cells were transfected with Flag-tagged Ago2 (F-Ago2) in the presence of either m-324 or NC. After 48 h, cells were subjected to RNA-IP with anti-Flag antibody and CUEDC2 mRNA was quantified using qRT-PCR analysis. (I to K) Quantification of relative expression of CUEDC2 in A549 cells at 24 h after infection with H5 (MOI, 1) (I) and NDV (MOI, 1) or PR8 (MOI, 1) (J) and poly(I·C) transfection (pIC-T) (1 μg/ml) (K). (L to N) A549 cells were transfected with 1.5 μg control short hairpin RNA (shRNA) (shc001) or CUEDC2-specific shRNA (G9 or G11) for 36 h, followed by H5N1 infection (MOI, 1) for 24 h. qRT-PCR was used for quantification of CUEDC2 (L), NP RNA (M), and IFN-β expression (N). Data are means ± SEMs from triplicate samples of a single experiment and are representative of results from three independent experiments (E to N). (E to K) ****, P < 0.0001, ***, P < 0.001, **, P < 0.01, and *, P < 0.05, by two-tailed unpaired t test. (L and M) ***, P < 0.001, and **, P < 0.01, by one-way ANOVA.
Taken together, our data indicate that miR-324-5p suppresses H5N1 replication by directly targeting viral PB1 and host CUEDC2.
DISCUSSION
Innate immunity against viruses is initiated through sensing of viral components by a variety of sensors, resulting the development of an antiviral state (31, 32). Influenza virus has a segmented RNA genome, and it infects mammalian and avian hosts to establish disease via triggering of numerous signaling pathways (33). These biochemical pathways subsequently change the transcript levels of immune and nonimmune mediators and regulatory noncoding (small and long) RNAs (10, 16, 34). The differential expression of noncoding small RNAs during viral infection may provide an opportunity to interact with viral transcripts generated during viral replication and determines the disease outcome. In this study, we found such a microRNA, miR-324-5p, after analyzing miRNA microarray data for in vivo mouse infection with H5N1 and in silico prediction software. We showed that miR-324-5p targets the H5N1 genome and the host transcript CUEDC2, a negative regulator for induction of host antiviral responses, and that this bispecific interaction results in reduction of viral replication.
Sensing of viruses by cell surface and cytosolic sensors induces type I and III interferons and interferon-inducible genes to promote expression of wide array of genes involved particularly in inducing apoptosis in virally infected cells and to protect uninfected cells from viral infection (35, 36). The viruses have developed a variety of molecular strategies: some viruses express miRNA (37, 38), enhance the host miRNA targeting viral sensors and signal transducers, or suppress miRNA targeting negative regulators of antiviral immunity to overcome host immunity and establish infection (39, 40).
In this study, miR-324-5p was identified through in silico analysis of in vivo and in vitro data, and it targets the highly conserved PB1 genes of different subtypes of H5N1. The PB1 gene encodes a subunit of RNA polymerase essential for H5N1 transcription and replication during infection. Ectopic expression of miR-324-5p in SAECs and A549 cells significantly inhibited the replication of H5N1. Surprisingly, A/PR8/H1N1 and NDV (lacking miR-324-5p seed sequence) also showed modest reductions in replication, suggesting that miR-324-5p might target host genes involved in antiviral innate immunity. Notably, the PB1 genes of A/PR8/H1N1, H1N1, H3N2, and H7N9 have several mismatches in the miR-324-5p target site; however, all these viruses encode the similar amino acids at the miR-324-5p target site, indicating that changes in the nucleotide sequence could be one strategy of viruses to avoid miRNA-mediated inhibition of viral replication and promote infection (Fig. 6A). The genome-wide transcriptome analysis of A549 cells expressing miR-324-5p revealed that miR-324-5p also targets the 3′ UTR of CUEDC2 conserved in mammals, including mice and humans. CUEDC2 is a negative regulator of the JAK-STAT pathway for induction of type I and type III interferons. These findings describe the two-way restriction mechanism of H5N1 replication by miR-324-5p by targeting a viral gene (PB1) and host gene (CUEDC2).
FIG 6.
H5N1 downregulates miR-324-5p expression to promote viral evasion. (A) Multiple-sequence alignment of the target site of miR-324-5p in the PB1 gene across different strains of influenza A virus. Mismatches are shown by red crosses. H, R, G, D, and T are amino acids encoded by the target site nucleotides. (B) Model showing that miR-324-5p binds to the 3′ UTR of host CUEDC2 and viral PB1 (H5N1) and subsequently inhibits the replication of H5N1.
Recent studies have shown that cellular miRNA expression is profoundly influenced by pathogenic insults and that altered miRNA expression can affect the miRNA-mediated silencing of the host mRNA and RNA virus replication. Given the single-strand segmented genome of influenza virus, the viral genome can interact with host miRNA and make a silencing complex through host miRNA-viral RNA interaction. Previously, it has been reported that honeysuckle (HS)-encoded atypical miR-2911 directly targets influenza A virus genes and significantly inhibited viral replication (41). Similarly, our previous study also showed that host miR-485-5p restricted the replication of influenza A virus by targeting PB1 of influenza virus and enhancing RIG-I-mediated antiviral signaling at high viral titers (42). In contrast, another report showed that H5N1 downregulated miRNAs, such as miR-584-5p and miR-1249, that target the PB2 gene of the virus and inhibit viral replication (6). In another study, miR-3145 inhibited influenza virus replication by targeting and silencing the PB1 gene (43). All together, these studies along with our results indicate that the host utilizes miRNAs to restrict influenza virus via targeting the most essential genes encoding subunits (PB1 and PB2) of the viral RNA polymerase and that are indispensable for viral replication, suggesting that miRNAs are pivotal in innate immune defense against influenza virus and may be involved in restriction of other RNA viruses. Our results showed that infection of RNA viruses such as H5N1, H1N1, and NDV and viral PAMPs such as poly(I·C) downregulates the expression of miR-324-5p in response to infection with RNA viruses. The downregulation of miR-324-5p might suppress heightened antiviral immune-pathogenesis to maintain homeostasis. In this study, we showed that H5N1 inhibited the expression of host miR-324-5p in vivo and in vitro, resulting in less targeting of PB1 and CUEDC2 to promote H5N1 replication. Previously, miR-324-5p was reported to regulate expression of CUEDC2, which is crucial for modulating macrophage function (44). Additionally, miR-324-5p downregulates the potassium channel Kv4.2 protein, contributing to seizure onset (45).
Collectively, our results suggest that miR-324-5p acts as a two-way restriction factor for HPAIV H5N1. It directly targets the genome of H5N1, suppressing PB1 expression and subsequent viral replication. In addition, miR-324-5p also targets CUEDC2, a negative regulator of the JAK-STAT pathway, reducing its expression and thereby enhancing the expression of antiviral genes (Fig. 6B). Together, our findings provide insight into the role of miR-324-5p in host defense against H5N1 influenza virus.
MATERIALS AND METHODS
Ethics statement.
Experiments were performed after approval from the Institutional Ethical Committee (IEC), Indian Institute of Science Education and Research (IISER) Bhopal (IISERB/IEC/Certificate/20l7-II/01), and the Institute Biosafety Committee (IBSC), National Institute of High Security Animal Diseases (NIHSAD) Bhopal (HSADL/IBSC/2014-2/128).
Patient samples.
Nasopharyngeal swabs were collected from four healthy persons and nine patients with swine influenza (H1N1) in viral transport medium (VTM) vials at All India Institute of Medical Sciences (AIIMS) Bhopal. Viral RNA and total RNA were isolated using a QIAamp viral RNA minikit (Qiagen) and TRIzol LS reagent (Invitrogen), respectively. The extracted RNA was subjected to a CDC-approved real-time RT-PCR (rRT-PCR) assay for the qualitative detection of swine influenza viruses. RNA samples were collected from AIIMS Bhopal after approval from the IEC, IISER Bhopal.
Analysis of microarray data from the GEO database.
miRNA microarray data were obtained from the GEO database (GSE69944, GSE71760, and GSE72365). Identification of miRNAs differentially expressed in the above-mentioned data sets between uninfected and H5N1-infected samples was conducted with the GEO2R online tool (46). Various R packages were used for visualization of differentially expressed miRNAs.
RNA-Seq analysis.
Total RNA isolated using TRIzol reagent was processed to prepare cDNA libraries using TruSeq technology according to the manufacturer's instructions (Illumina, San Diego, CA). Libraries were sequenced using Illumina HiSeq2500, with a read length of 101 bp, by Bencos Research Solutions Pvt. Ltd., Mumbai, India. Assessment of read quality of row data was done using FastQC (0.11.5) (47). Trimmomatic was used to remove Illumina adaptors, and quality filtering of reads was done by the sliding-window approach (48). Approximately 20 million cleaned pair-end sequencing reads from each sample were uploaded to the Galaxy web platform and were analyzed at https://usegalaxy.org (49). Reads were mapped to the reference human genome (hg38) using TOPHAT2. Cufflinks was used to assemble the aligned RNA-Seq reads into transcripts and estimate the normalized abundance of the assembled transcripts as fragments per kilobase per million (FPKM) (50). Cuffmerge was used to merge together several Cufflinks assemblies. A merged gtf file produced by cuffmerge was provided as an input to Cuffdiff along with alignment files produced by TOPHAT2 for differential analysis between two samples.
Visualization of expression and differential expression results was done using various R packages. Gene ontology (GO) analysis was done using the web-based GEne SeT AnaLysis toolkit (51), and analysis of upregulated KEGG pathways was done using Enrichr (52). Cluster 3.0 and TreeView 1.1.6 (53) were used for making heat maps.
Reanalysis of CLIP-Seq data.
Raw Ago2 CLIP-Seq data associated with untreated 293S cells from data set GSE44404 were imported to the Galaxy web platform and were analyzed using the public server at https://usegalaxy.org. The quality of raw data was assessed, reads were trimmed, and cleaned reads were mapped to hg19 using Bowtie v2.3.2.2. Visualization was done using Integrative Genomics Viewer (IGV).
Cells, transfection, viruses, and reagents.
A549 human alveolar basal epithelial cells (Cell Repository, NCCS, India), HEK293T human embryonic kidney cells (ATCC CRL-3216), and HeLa cervical cancer cells (Cell Repository, NCCS, India) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. MDAMB-231 and MDCK cells were cultured in L-15 and Eagle's minimum essential medium (EMEM), respectively, supplemented with 10% FBS and 1% penicillin-streptomycin. Small airway epithelial cells (SAECs; Lonza) were cultured and maintained as per the manufacturer's instruction. Transfection of DNA, mimics, and poly(I·C) was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. All experiments related to HPAIV A/duck/India/02CA10/2011 (H5N1) and NDV LaSota were performed using single viral stocks in biosafety level 3 (BSL3) laboratories at the National Institute of High Security Animal Diseases (NIHSAD) and in BSL2 laboratories, respectively. The cells were infected in serum-free DMEM with HPAIV and NDV and washed with phosphate-buffered saline (PBS) after 1 h, and then PBS was replaced with DMEM supplemented with 1% FBS. DMEM, FBS, and penicillin-streptomycin were purchased from Invitrogen.
Generation of A/PR8/H1N1 virus.
A/PR8/H1N1 virus was generated using the eight-plasmid reverse genetics system. HEK293T (5 × 105) cells were seeded in a 6-well plate in DMEM supplemented with 10% FBS. The eight plasmids (500 ng each) were transfected using Lipofectamine 2000. After 24 h, medium was removed, cells were washed with PBS, and medium was replaced with 3 ml DMEM containing bovine serum albumin (BSA; 0.3%, wt/vol). l-1-Tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (1 μg/ml) was added into the medium after 48 h. After 96 h, medium was collected and filtered through a 0.2-μm filter. A/PR8/H1N1 virus was further amplified in embryonated chicken eggs.
Luciferase reporter assay.
HEK293T cells were seeded at 60 to 70% confluency into a 24-well plate and transfected with 50 ng transfection pRL-TK plasmid (control) and 100 ng luciferase reporter plasmid along with 25 nM miRNA mimics or negative-control mimic (NC). The cells were lysed at 24 h posttransfection, and luciferase activity was measured in total cell lysate by using Glomax (Promega).
Quantitative real-time reverse transcription-PCR.
Total RNA was extracted using TRIzol reagent (Invitrogen) and was used to synthesize cDNA with the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's protocol. Gene expression was measured by quantitative real-time PCR using gene-specific primers and SYBR green chemistry (Bio-Rad). Real-time PCR analysis was performed with TaqMan universal PCR master mix (Applied Biosystems) and TaqMan miRNA assays specific for miR-324-5p and U6 for quantification of respective miRNAs.
Fluorescence-activated cell sorting.
Cells were fixed using paraformaldehyde (PFA; 4%), permeabilized with Triton X-100 (0.1%), and incubated with anti-NP antibody (H5N1), followed by incubation with Alexa Fluor 488-conjugated donkey anti-mouse secondary antibody. The cells were then analyzed with a FACSAria III flow cytometer (Becton Dickinson), and the data were analyzed with FlowJo software (FlowJo).
RNA immunoprecipitation.
RNA immunoprecipitation was performed as described previously (54). HEK293T cells were lysed in lysis buffer (0.5% NP-40, 150 mM KCl, 25 mM Tris-glycine [pH 7.5]). Lysate was incubated with Flag M2 affinity beads (Sigma) for 6 h. Thereafter, the lysate was washed 5 times with washing buffer (300 mM NaCl, 50 mM Tris-glycine [pH 7.5], 5 mM MgCl2, 0.05% NP-40). The RNA was extracted from the immunoprecipitated ribonucleoproteins (RNPs) using the TRIzol reagent.
Measurement of viral titer.
MDCK cells were infected in the serum-free MEM medium with the cell culture supernatant containing the virus and washed with PBS after 1 h of infection. For plaque assay, cells were overlaid with 1.2% Avicel (Sigma-Aldrich; equivalent to Avicel RC-581 from FMC Corp.) in 2 ml maintenance medium and incubated at 37°C in 5% CO2. After 96 h, the cell monolayer was fixed with 4% paraformaldehyde for 20 min at 4°C. Cells were stained with 0.5% crystal violet solution and plaques were scored under microscope in transmitted light. For the 50% tissue culture infective dose (TCID50) assay, cell monolayers were fixed and stained with 1% crystal violet after 96 h of infection and the TCID50 was calculated based on the Reed and Muench method as described previously (55).
Statistical analysis.
All experiments were performed with appropriate control samples or mock-transfected samples. Experiments were performed twice or thrice independently of each other. All the data were analyzed using GraphPad Prism software for statistical significance. Student's two-tailed unpaired t test was performed for two groups, and analysis of variance (ANOVA) was used for three groups.
Data availability.
RNA-Seq data have been submitted to the Gene Expression Omnibus under accession number GSE108906.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by research grants BT/PR6009/GBD/27/382/2012 and BT/IN/Indo-UK/FADH/48/AM/2013 from the Department of Biotechnology, Government of India, to H.K. and A.M., respectively. We thank DBT—Advanced Level State Biotech Hub, Mizoram University, for financial assistance. This study was also partly supported by the Grant Research Program of the Institute for Genetic Medicine, Hokkaido University, Japan.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The pIRESneo-Flag/HA Ago2 plasmid was a gift from T. Tuschl (Addgene; plasmid 10822). The following reagent was obtained through BEI Resources, NIAID, NIH: human interferon beta (HuIFN-beta), NR-3080. We thank R. Fouchier for providing the A/PR8/H1N1 reverse genetics system, P. Palese for providing green fluorescent protein-expressing NDV (NDV-GFP), and Nagarjun Vijay for helping in bioinformatics analysis. We also thank Sanjana Bhattacharya and Athira S. Raj for technical support. We thank IISER Bhopal for providing the Central Instrumentation Facility and the director of ICAR-NIHSAD for providing the advanced biosafety level 3 facility.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01057-18.
REFERENCES
- 1.Barik S. 2012. New treatments for influenza. BMC Med 10:104. doi: 10.1186/1741-7015-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacob A, Sood R, Chanu KV, Bhatia S, Khandia R, Pateriya AK, Nagarajan S, Dimri U, Kulkarni DD. 2016. Amantadine resistance among highly pathogenic avian influenza viruses (H5N1) isolated from India. Microb Pathog 91:35–40. doi: 10.1016/j.micpath.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Maurer-Stroh S, Li Y, Bastien N, Gunalan V, Lee RTC, Eisenhaber F, Booth TF. 2014. Potential human adaptation mutation of influenza A(H5N1) virus, Canada. Emerg Infect Dis 20:1580–1582. doi: 10.3201/eid2009.140240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Y-J, Yang J, Fan X-L, Zhao H-B, Hu W, Li Z-P, Yu G-C, Ding X-R, Wang J-Z, Bo X-C, Zheng X-F, Zhou Z, Wang S-Q. 2012. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J Cell Mol Med 16:2539–2546. doi: 10.1111/j.1582-4934.2012.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol 84:8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, Zhang YY, Lu JS, Xia BH, Yang ZX, Zhu XD, Zhou XW, Huang PT. 2017. The highly pathogenic H5N1 influenza A virus down-regulated several cellular microRNAs which target viral genome. J Cell Mol Med 21:3076–3086. doi: 10.1111/jcmm.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Flynt AS, Lai EC. 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Buggele WA, Johnson KE, Horvath CM. 2012. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J Biol Chem 287:31027–31040. doi: 10.1074/jbc.M112.387670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. 2010. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol 185:6226. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 12.Yarbrough ML, Zhang K, Sakthivel R, Forst CV, Posner BA, Barber GN, White MA, Fontoura BMA. 2014. Primate-specific miR-576-3p sets host defense signaling threshold. Nat Commun 5:4963. doi: 10.1038/ncomms5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adoro S, Cubillos-Ruiz JR, Chen X, Deruaz M, Vrbanac VD, Song M, Park S, Murooka TT, Dudek TE, Luster AD, Tager AM, Streeck H, Bowman B, Walker BD, Kwon DS, Lazarevic V, Glimcher LH. 2015. IL-21 induces antiviral microRNA-29 in CD4 T cells to limit HIV-1 infection. Nat Commun 6:7562. doi: 10.1038/ncomms8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, Liu Y, Wu J. 2013. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog 9:e1003248. doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura S, Horie M, Daidoji T, Honda T, Yasugi M, Kuno A, Komori T, Okuzaki D, Narimatsu H, Nakaya T, Tomonaga K. 2016. Influenza A virus-induced expression of a GalNAc transferase, GALNT3, via microRNAs is required for enhanced viral replication. J Virol 90:1788–1801. doi: 10.1128/JVI.02246-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos I, Fernandez-Sesma A. 2012. Innate immunity to H5N1 influenza viruses in humans. Viruses 4:3363–3388. doi: 10.3390/v4123363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo J-S, Kato H, Fujita T. 2014. Sensing viral invasion by RIG-I like receptors. Curr Opin Microbiol 20:131–138. doi: 10.1016/j.mib.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Aittomäki S, Pesu M. 2014. Therapeutic targeting of the JAK/STAT pathway. Basic Clin Pharmacol Toxicol 114:18–23. [DOI] [PubMed] [Google Scholar]
- 20.Yoshimura A, Suzuki M, Sakaguchi R, Hanada T, Yasukawa H. 2012. SOCS, inflammation, and autoimmunity. Front Immunol 3:20. doi: 10.3389/fimmu.2012.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W-N, Wang L, Wang Q, Luo X, Fang D-F, Chen Y, Pan X, Man J-H, Xia Q, Jin B-F, Li W-H, Li T, Liang B, Chen L, Gong W-L, Yu M, Li A-L, Zhou T, Li H-Y. 2012. CUEDC2 (CUE domain-containing 2) and SOCS3 (suppressors of cytokine signaling 3) cooperate to negatively regulate Janus kinase 1/signal transducers and activators of transcription 3 signaling. J Biol Chem 287:382–392. doi: 10.1074/jbc.M111.276832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akhtar LN, Benveniste EN. 2011. Viral exploitation of host SOCS protein functions. J Virol 85:1912–1921. doi: 10.1128/JVI.01857-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39:1278. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 24.Huang H-Y, Chien C-H, Jen K-H, Huang H-D. 2006. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res 34:W429–W434. doi: 10.1093/nar/gkl333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. 2012. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. 2007. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol 25:1457. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 27.Wagner TC, Velichko S, Chesney Steven K, Biroc S, Harde D, Vogel D, Croze E. 2004. Interferon receptor expression regulates the antiproliferative effects of interferons on cancer cells and solid tumors. Int J Cancer 111:32–42. doi: 10.1002/ijc.20236. [DOI] [PubMed] [Google Scholar]
- 28.Matsushima-Miyagi T, Hatano K, Nomura M, Li-Wen L, Nishikawa T, Saga K, Shimbo T, Kaneda Y. 2012. TRAIL and Noxa are selectively upregulated in prostate cancer cells downstream of the RIG-I/MAVS signaling pathway by nonreplicating Sendai virus particles. Clin Cancer Res 18:6271. doi: 10.1158/1078-0432.CCR-12-1595. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Ingle H, Mishra S, Mahla RS, Kumar A, Kawai T, Akira S, Takaoka A, Raut AA, Kumar H. 2015. IPS-1 differentially induces TRAIL, BCL2, BIRC3 and PRKCE in type I interferons-dependent and -independent anticancer activity. Cell Death Dis 6:e1758. doi: 10.1038/cddis.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karginov FV, Hannon GJ. 2013. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev 27:1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev 243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 32.Goubau D, Deddouche S, Reis e Sousa C. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph U, Su YCF, Vijaykrishna D, Smith GJD. 2017. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir Viruses 11:74–84. doi: 10.1111/irv.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landeras-Bueno S, Ortín J. 2016. Regulation of influenza virus infection by long non-coding RNAs. Virus Res 212:78–84. doi: 10.1016/j.virusres.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Barber GN. 2001. Host defense, viruses and apoptosis. Cell Death Differ 8:113. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- 36.Stetson DB, Medzhitov R. 2006. Type I interferons in host defense. Immunity 25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Nukui M, Mori Y, Murphy EA. 2015. A human herpesvirus 6A-encoded microRNA: role in viral lytic replication. J Virol 89:2615–2627. doi: 10.1128/JVI.02007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. 2009. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol 83:10684–10693. doi: 10.1128/JVI.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Q, Fu S, Wang J. 2014. Hepatitis C virus infection decreases the expression of Toll-like receptors 3 and 7 via upregulation of miR-758. Arch Virol 159:2997–3003. doi: 10.1007/s00705-014-2167-3. [DOI] [PubMed] [Google Scholar]
- 40.Hu X, Ye J, Qin A, Zou H, Shao H, Qian K. 2015. Both microRNA-155 and virus-encoded MiR-155 ortholog regulate TLR3 expression. PLoS One 10:e0126012. doi: 10.1371/journal.pone.0126012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu J, Kong H, Zhang Q, Qi X, Hou D, Zhang L, Zhang G, Liu Y, Zhang Y, Li J, Wang J, Chen X, Wang H, Zhang J, Chen H, Zen K, Zhang C-Y. 2015. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res 25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingle H, Kumar S, Raut AA, Mishra A, Kulkarni DD, Kameyama T, Takaoka A, Akira S, Kumar H. 2015. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci Signal 8:ra126. doi: 10.1126/scisignal.aab3183. [DOI] [PubMed] [Google Scholar]
- 43.Khongnomnan K, Makkoch J, Poomipak W, Poovorawan Y, Payungporn S. 2015. Human miR-3145 inhibits influenza A viruses replication by targeting and silencing viral PB1 gene. Exp Biol Med 240:1630–1639. doi: 10.1177/1535370215589051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Wang S- X, Mu R, Luo X, Liu Z-S, Liang B, Zhuo H-L, Hao X-P, Wang Q, Fang D-F, Bai Z-F, Wang Q-Y, Wang H-M, Jin B-F, Gong W-L, Zhou T, Zhang X-M, Xia Q, Li T. 2014. Dysregulation of the miR-324-5p-CUEDC2 axis leads to macrophage dysfunction and is associated with colon cancer. Cell Rep 7:1982–1993. doi: 10.1016/j.celrep.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Gross C, Yao X, Engel T, Tiwari D, Xing L, Rowley S, Danielson SW, Thomas KT, Jimenez-Mateos EM, Schroeder LM, Pun RYK, Danzer SC, Henshall DC, Bassell GJ. 2016. MicroRNA-mediated downregulation of the potassium channel Kv4.2 contributes to seizure onset. Cell Rep 17:37–45. doi: 10.1016/j.celrep.2016.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis S, Meltzer PS. 2007. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 47.Andrews S. FastQC. A quality control tool for high throughput sequence data. Babraham Institute, Cambridge, United Kingdom: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- 48.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. 2017. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 54.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data have been submitted to the Gene Expression Omnibus under accession number GSE108906.