Mutant c-Jun and c-Fos proteins selectively activate expression of EBV lytic genes, including a subgroup of viral late genes, in the absence of viral DNA replication. These findings indicate that newly synthesized viral DNA is not invariably required for viral late gene expression. While viral DNA replication may be obligatory for late gene expression driven by viral transcription factors, it does not limit the ability of cellular transcription factors to activate expression of some viral late genes. Our results show that expression of all late genes may not be strictly dependent on viral lytic DNA replication. The c-Fos A151S mutation has been identified in a human cancer. c-Fos A151S in combination with wild-type c-Jun activates the EBV lytic cycle. Our data provide proof of principle that mutant cellular transcription factors could cause aberrant regulation of viral lytic cycle gene expression and play important roles in EBV-associated diseases.
KEYWORDS: AP-1, BZLF1, Epstein-Barr virus, late gene expression, viral gene regulation
ABSTRACT
Epstein-Barr virus (EBV) ZEBRA protein activates the EBV lytic cycle. Cellular AP-1 proteins with alanine-to-serine [AP-1(A/S)] substitutions homologous to ZEBRA(S186) assume some functions of EBV ZEBRA. These AP-1(A/S) mutants bind methylated EBV DNA and activate expression of some EBV genes. Here, we compare expression of 67 viral genes induced by ZEBRA versus expression induced by AP-1(A/S) proteins. AP-1(A/S) activated 24 genes to high levels and 15 genes to intermediate levels; activation of 28 genes by AP-1(A/S) was severely impaired. We show that AP-1(A/S) proteins are defective at stimulating viral lytic DNA replication. The impairment of expression of many late genes compared to that of ZEBRA is likely due to the inability of AP-1(A/S) proteins to promote viral DNA replication. However, even in the absence of detectable viral DNA replication, AP-1(A/S) proteins stimulated expression of a subgroup of late genes that encode viral structural proteins and immune modulators. In response to ZEBRA, expression of this subgroup of late genes was inhibited by phosphonoacetic acid (PAA), which is a potent viral replication inhibitor. However, when the lytic cycle was activated by AP-1(A/S), PAA did not reduce expression of this subgroup of late genes. We also provide genetic evidence, using the BMRF1 knockout bacmid, that these genes are true late genes in response to ZEBRA. AP-1(A/S) binds to the promoter region of at least one of these late genes, BDLF3, encoding an immune modulator.
IMPORTANCE Mutant c-Jun and c-Fos proteins selectively activate expression of EBV lytic genes, including a subgroup of viral late genes, in the absence of viral DNA replication. These findings indicate that newly synthesized viral DNA is not invariably required for viral late gene expression. While viral DNA replication may be obligatory for late gene expression driven by viral transcription factors, it does not limit the ability of cellular transcription factors to activate expression of some viral late genes. Our results show that expression of all late genes may not be strictly dependent on viral lytic DNA replication. The c-Fos A151S mutation has been identified in a human cancer. c-Fos A151S in combination with wild-type c-Jun activates the EBV lytic cycle. Our data provide proof of principle that mutant cellular transcription factors could cause aberrant regulation of viral lytic cycle gene expression and play important roles in EBV-associated diseases.
INTRODUCTION
The viral transcription factor ZEBRA (BZLF1), is responsible for reactivation of Epstein-Barr virus (EBV) from latency into the lytic cycle (1–3). During reactivation, the EBV lytic cycle is separable into four temporal stages: very early gene expression (also termed immediate early [IE]), early gene expression, viral DNA replication, and late gene expression. ZEBRA plays an essential role in each stage of lytic gene expression. ZEBRA induces expression of the very early protein Rta (4, 5); it synergizes with Rta to activate early genes whose products are required for viral DNA replication (6–11); it binds to the origin of lytic replication (oriLyt) and assembles the replication complex (12–14). ZEBRA suppresses aberrant expression of some late genes in the absence of viral DNA replication (15).
ZEBRA regulates viral gene transcription and DNA replication through binding directly to three different types of DNA motifs: canonical AP-1 sites (also known as TPA response sites, where TPA is 12-O-tetradecanoylphorbol-13-acetate) (16–18), unmethylated ZEBRA response elements (ZREs) (17, 19), and methylated ZREs (19, 20). The amino acid sequence of the basic DNA recognition domain of ZEBRA, a bZIP transcription factor, is similar to that of other bZIP family members such as the cellular AP-1 proteins c-Jun and c-Fos (16, 21–26). Crystal structures of c-Jun and c-Fos identified five amino acids that interact with DNA bases in the AP-1 binding site by either hydrogen bonding or van der Waals interactions (27). Of these five amino acids, four share positional homology with ZEBRA (22). The exception is ZEBRA S186; homologous AP-1 locations are c-Jun (A266) and c-Fos (A151) (22, 27).
The essential role of residue S186 in ZEBRA's ability to regulate gene transcription was demonstrated in a ZEBRA mutant with a single serine-to-alanine change at amino acid S186 [Z(S186A)]. The S186A mutation makes the DNA binding domain of ZEBRA structurally more similar to the DNA binding domain of human c-Jun and c-Fos (22, 28). Z(S186A) is deficient in activating the EBV lytic cycle (29–31). This defect was correlated to its inability to bind to methylated ZREs (20).
To further explore the inability of Z(S186A) to activate the viral lytic cycle, we made reciprocal mutations in the homologous amino acids in cellular AP-1 proteins, i.e., c-Jun(A266S) and c-Fos(A151S). These mutant AP-1 proteins with alanine-to-serine [AP-1(A/S)] changes could bind to ZRE2 and ZRE3, two methylated EBV DNA sites in the BRLF1 promoter, in a manner similar to ZEBRA and activate expression of some early lytic cycle genes (32). For example, AP-1(A/S) strongly activated expression of the Rta transcription factor (encoded by BRLF1) as well as the DNA polymerase processivity factor EA-D (encoded by BMRF1). However, AP-1(A/S) did not activate expression of either mRNA or protein of the canonical viral late gene FR3 (encoded by BFRF3), the minor capsid protein. The extent of viral gene expression in response to AP-1(A/S) was limited in the previous study. The ability of AP-1(A/S) to activate expression of BRLF1, five early RNAs, two late viral RNAs, and the viral long noncoding RNA BHLF1 was analyzed by Northern blotting. The inability of AP-1(A/S) to perform DNA replication was inferred from the indirect observation of its inability to activate expression of BFRF3.
Here, we extensively characterize the ability of AP-1(A/S) proteins to activate different phases of the viral lytic cycle. Since in the previous study AP-1(A/S) did not activate expression of FR3, a late gene, we asked whether AP-1(A/S) was capable of activating viral DNA replication. We demonstrate that AP-1(A/S) is unable to activate viral DNA replication even though it binds to oriLyt. In order to understand the failure of AP-1(A/S) to activate viral DNA replication, we undertook a quantitative PCR (qPCR)-based array of viral gene expression. We found that AP-1(A/S) was capable of selectively activating expression of 24 viral genes to levels as high as or higher than those activated by ZEBRA. Examining these 24 viral genes, we made the unexpected observation that, despite being unable to stimulate viral DNA replication, AP-1(A/S) did induce expression of some viral late gene mRNAs. AP-1(A/S) binds to the promoter region of one of these late genes, the immunomodulator BDLF3. This observation provides a plausible mechanism for the ability of AP-1(A/S) to activate late gene expression. Our results challenge the dogma that expression of all late genes is strictly dependent on viral lytic DNA replication (33–35). With the recent identification of the c-Fos(A151S) mutation in liver cancer by the Catalogue of Somatic Mutations in Cancer (COSMIC) (http://cancer.sanger.ac.uk/cosmic, 36), we provide a proof of principle that a naturally occurring mutant cellular transcription factor could cause aberrant regulation of viral lytic cycle gene expression, including viral late genes encoding viral immune modulators, in the absence of viral DNA replication.
RESULTS
Mutant AP-1(A/S) proteins do not induce expression of a classical viral late protein or promote viral DNA replication.
BZKO cells, which are HEK293 cells that contain an EBV bacmid lacking a functional ZEBRA gene (8), are incapable of spontaneously entering the EBV lytic cycle. However, EBV in BZKO cells can be activated into the lytic cycle by transfection with a plasmid expressing ZEBRA. Transfection of ZEBRA in BZKO cells led to expression of the very early protein Rta, the early protein EA-D, and the late protein FR3; these genes were used as markers for different phases of the lytic cycle. These results indicated that ZEBRA induced the full lytic cycle (Fig. 1A, lane 2). We compared the functions of ZEBRA in lytic cycle activation to those of mutant Z(S186A), wild-type AP-1, and mutant AP-1(A/S) proteins. Z(S186A) and AP-1 did not substitute for ZEBRA's functions in activating the EBV lytic cycle; they did not lead to expression of any viral proteins assayed (Fig. 1A, lanes 3 and 4). Transfection of BZKO cells with plasmids expressing mutant AP-1(A/S) proteins promoted expression of Rta protein to levels 4.4-fold higher than levels induced by ZEBRA (Fig. 1A, compare lanes 2 and 5) and expression of EA-D protein to levels similar to those induced by ZEBRA (Fig. 1A, compare lanes 2 and 5). However, AP-1(A/S) proteins failed to cause the viral late protein FR3 to be expressed (Fig. 1A, lane 5).
FIG 1.
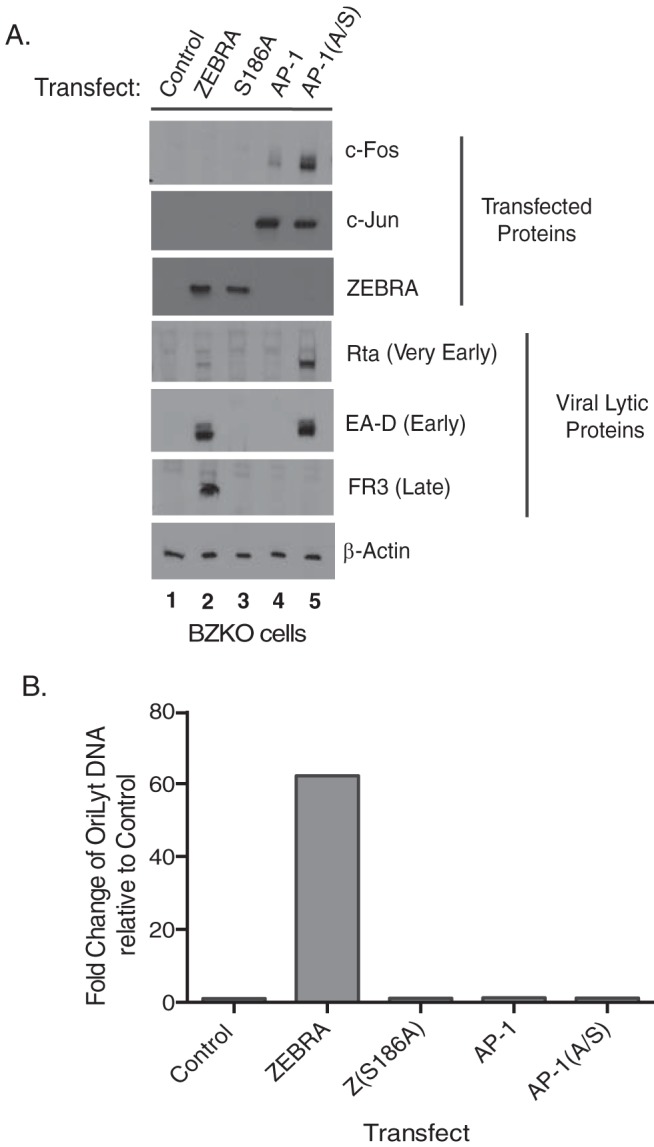
AP-1(A/S) activates expression of BMRF1 (EA-D), an EBV marker early protein, but does not induce expression of BFRF3, an EBV marker late protein, or DNA replication. (A) Western blot of BZKO cells harvested 48 h after transfection with control CMV plasmid vector, wild-type ZEBRA, Z(S186A), wild-type AP-1, or mutant AP-1(A/S), probed with the antibodies indicated. (B) qPCR for the expression of viral DNA performed on the same cells used for Western blot analysis in panel A. Western blotting and qPCR were performed once on cells used for the array shown in Fig. 3.
A viral DNA replication assay was performed on the same cells assayed for viral proteins. Transfection of a plasmid encoding ZEBRA led to a 62-fold increase in amplification of the viral genome. Transfection of plasmids expressing either Z(S186A), AP-1, or AP-1(A/S) showed no viral genome amplification compared to the level with the vector control (Fig. 1B). These results indicated that while AP-1(A/S) substituted for ZEBRA in inducing expression of Rta and EA-D, early viral lytic cycle genes, AP-1(A/S) was defective in activating viral DNA replication and expression of the viral late protein FR3.
Comparing binding of ZEBRA and AP-1(A/S) to that of oriLyt and the BRLF1 promoter.
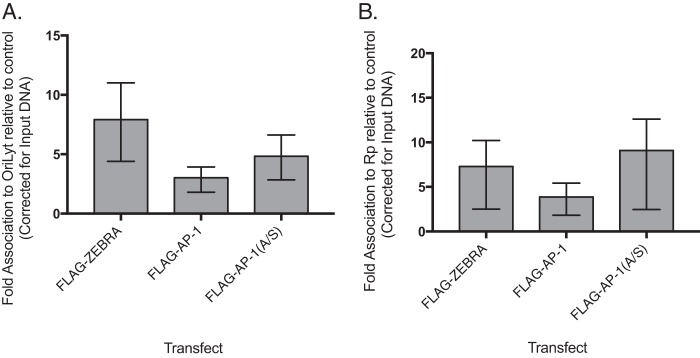
To determine whether AP-1(A/S) was unable to perform viral DNA replication due to an inability to bind to oriLyt, we performed chromatin immunoprecipitation (ChIP) on BZKO cells 48 h after transfection with a vector control, FLAG-tagged ZEBRA, FLAG-tagged AP-1, or FLAG-tagged AP-1(A/S) vector. The anti-FLAG antibody was used to immunoprecipitate all proteins assayed. ZEBRA bound to the oriLyt region containing ZREs 1 to 4 (Fig. 2A) and to the BRLF1 promoter (Rp) region containing ZREs 2 and 3 (Fig. 2 B). AP-1(A/S) also bound to oriLyt region containing ZREs 1 to 4. The binding of AP-1(A/S) to the oriLyt region containing ZREs 1 to 4 and the Rp region containing ZREs 2 and 3 was not significantly different from that of ZEBRA (Fig. 2B).
FIG 2.
AP-1(A/S) binds to oriLyt and to Rp of ZRE2 and -3. Chromatin immunoprecipitation was performed in BZKO cells harvested 48 h after transfection with a control vector, wild-type FLAG-ZEBRA, FLAG-Z(S186A), wild-type FLAG-AP-1, or mutant FLAG-AP-1(A/S). The FLAG antibody was used to immunoprecipitate all target proteins. Association to oriLyt (A) or Rp of ZRE2 and -3 (B) was determined using qPCR. Bar height indicates the mean of three experiments. Error bars show 95% confidence intervals determined using bootstrapping. ZEBRA, AP-1, and AP-1(A/S) all bind significantly to oriLyt and Rp, with no statistically significant differences among them.
Patterns of EBV gene expression induced by ZEBRA versus AP-1(A/S).
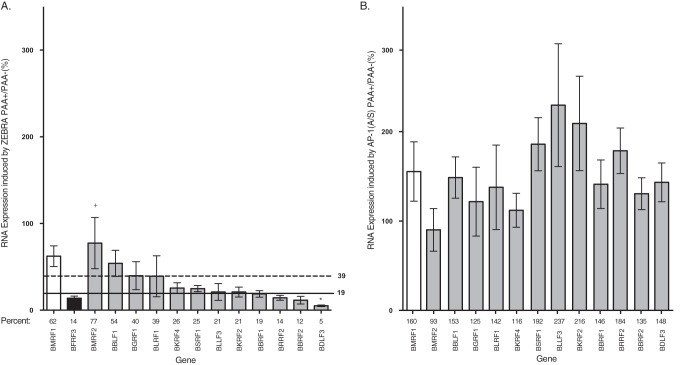
We characterized overall patterns of viral gene expression using two qPCR-based arrays of EBV genes. The arrays used cDNAs from cells under conditions described in the experiments shown in Fig. 1 (see Table S1 in the supplemental material). Transfection of ZEBRA led to increased expression of all of the EBV genes (Fig. 3). Transfection of the mutant Z(S186A) failed to activate any viral genes relative to the level of the vector control. This result indicates that Z(S186A) is completely defective at activation of the viral lytic cycle. Wild-type AP-1 proteins promoted expression of a small number of viral lytic genes at levels lower than when the same genes were induced by ZEBRA. Mutant AP-1(A/S) proteins activated the majority of the 67 EBV genes assayed. AP-1(A/S) promoted expression of 24 genes to high levels, comparable to those with ZEBRA, and 15 to intermediate levels, and 28 genes were expressed at levels less than 20% of the level observed when ZEBRA induced the lytic cycle (Fig. 3 and 4).
FIG 3.
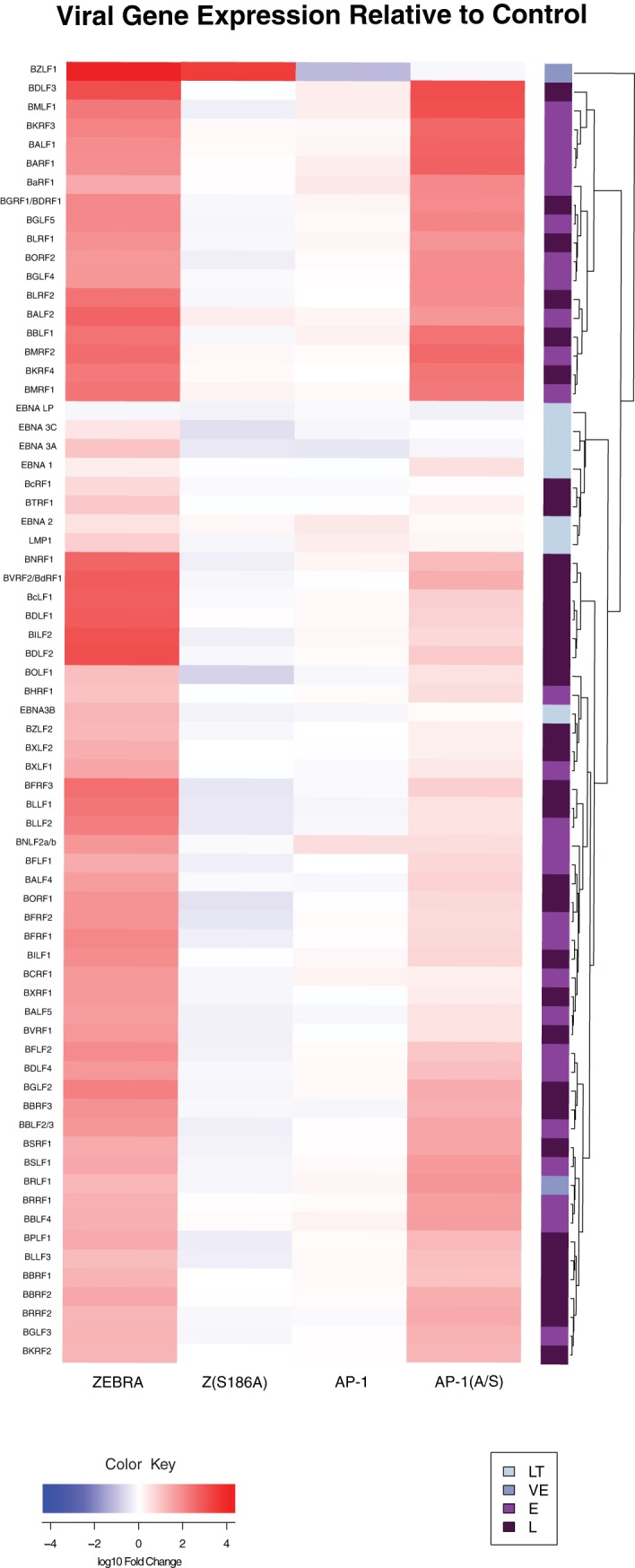
EBV gene expression induced by ZEBRA, Z(S186A), AP-1, and AP-1(A/S). Heat map of viral gene expression based on a qPCR array of cDNA from cells transfected with a control vector, ZEBRA, Z(S186A), wild-type AP-1, or AP-1(A/S) harvested at 48 h posttransfection (n = 1). Data are the average fold change of expression of 67 genes relative to expression of the vector control. Samples were standardized to GAPDH expression. Genes are displayed using hierarchical clustering under complete linkage with a Euclidean distance measure. cDNA was prepared from same samples as those illustrated in Fig. 1. LT, latent; L, late; E, early; VE, very early.
FIG 4.
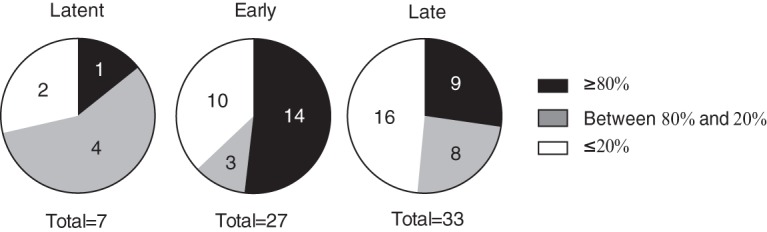
Extent of expression of different EBV gene temporal classes with activation by AP-1(A/S). Pie charts show the categories of expression of viral genes activated by AP-1(A/S) relative to levels with ZEBRA based on the qPCR cDNA array illustrated in Fig. 3. Viral genes are partitioned into the following sets in comparison to expression levels with ZEBRA activation: black, greater than or equal to 80%; gray, less than 80% and greater than 20%; white, less than or equal to 20%.
ZEBRA activated expression of all of the latency genes. Expression of six of the eight latency genes assayed, EBNA LP, EBNA3A, EBNA3C, EBNA1, EBNA2, and LMP1, clustered together in the array data, along with the late gene regulator BcRF1 and the late gene BTRF1 (37). Wild-type AP-1 proteins promoted expression of two latency genes, EBNA2 and LMP1, which were expressed at 82% and 32%, respectively, of the expression levels resulting from activation by ZEBRA. AP-1 proteins decreased the expression of the latent EBNA3A, -B, and -C transcripts compared to the level with the vector control. AP-1(A/S) led to expression of EBNA1 at high levels; four genes, EBNA2, EBNA3C, LMP1, and EBNA LP, were expressed at intermediate levels. AP-1(A/S) was deficient, compared to ZEBRA, in promoting expression of EBNA3A and EBNA3B (Fig. 3 and 4).
ZEBRA activated expression of all of the viral lytic cycle early genes. Z(S186A) and wild-type AP-1 did not activate expression of any viral early genes. AP-1(A/S) promoted expression of many viral early genes. Fourteen early genes were expressed to high levels, comparable to the levels with expression by ZEBRA, and three were expressed to intermediate levels. Ten early lytic genes were expressed at less than 20% of the level measured when ZEBRA induced the lytic cycle (Fig. 3 and 4; also Table S1).
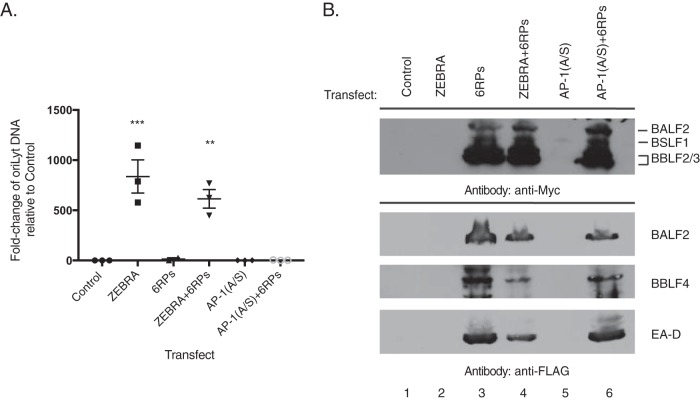
The inability of AP-1(A/S) to induce viral DNA replication could be explained by very low levels of expression of genes encoding two essential replication proteins, BALF5, the viral DNA polymerase, and/or BALF2, the single-stranded DNA binding protein. However, exogenous expression of six EBV-encoded replication proteins, BALF5, BSLF1, BBLF4, BBLF2/3, BALF2, and BMRF1, did not rescue the inability of AP-1(A/S) to drive viral DNA amplification (Fig. 5A).
FIG 5.
Exogenous expression of six viral replication proteins does not rescue EBV DNA replication by AP-1(A/S). (A) qPCR for relative abundance of viral DNA in BZKO cells harvested 48 h after transfection with control vector, wild-type ZEBRA, or mutant AP-1(A/S) without or with plasmids encoding six viral replication proteins (RPs), BALF5, BSLF1, BBLF4, BBLF2/3, BALF2, and BMRF1. In each experiment qPCR was performed on the same cells used for Western blot analysis. Each point shows one replicate. The horizontal line denotes the mean. Error bars show one standard error of the mean. P values were determined using pairwise Student's t tests with Dunnett's correction for multiple comparisons compared to results with the vector control. **, P ≤ 0.0021; ***, P ≤ 0.0002. (B) Western blotting performed on the same cells used for qPCR probed with the antibodies indicated. The blot is representative of three experiments.
AP-1(A/S) activates viral late gene expression.
Viral late gene expression is considered to be dependent upon replication of the viral genome (33, 34). Since AP-1(A/S) proteins were unable to stimulate viral DNA replication (Fig. 1B), we expected that these mutants would be unable to activate expression of any viral late genes. However, AP-1(A/S) proteins activated expression of a subset of genes that have been previously characterized as late genes based on their sensitivity to phosphonoacetic acid (PAA) in Akata cells treated with anti-IgG for 48 h to induce the lytic cycle (38). AP-1(A/S) activated nine late genes to high levels and eight to intermediate levels and was defective at expressing 16 late genes compared to expression by ZEBRA (Fig. 3, Fig. 4).
Using individual reverse transcription-quantitative PCR (RT-qPCR) assays, we validated expression of 12 viral late genes activated by AP-1(A/S) in BZKO cells. We selected the genes that were activated to greater than 80% relative to the level activated by ZEBRA in at least one array and a minimum of 50% relative to activation by ZEBRA in the other array (Fig. 3 and Table S1). The 12 genes chosen based on these criteria were BLRF1, BBLF1, BGRF1, BKRF1, BBFR1, BKRF2, BRRF2, BBRF1, BMRF2, BSRF1, BLLF3, and BDLF3 (Tables 1 and S1 and Fig. 3). Eight genes, BMRF2, BBLF1, BGRF1, BKRF4, BSRF1, BKRF2, BBRF1, and BRRF2, were activated by AP-1(A/S) to high levels, that is, above 70% of the levels activated by ZEBRA in RT-qPCR assays. The four remaining genes, BLRF1, BLLF3, BBRF2, and BDLF3, were activated at intermediate levels, with expression at 59.4%, 28.8%, 48.0%, and 35.7%, respectively, compared to the levels in response to ZEBRA (Fig. 6A). Thus, the individual RT-qPCRs confirmed results from the arrays demonstrating that a subset of late genes are expressed to high or intermediate levels by AP-1(A/S).
TABLE 1.
Late genes regulated by AP-1(A/S)
| Genea | Function | % activation by qPCRb |
% activation by RT-qPCRb | |
|---|---|---|---|---|
| Array 1 | Array 2 | |||
| BBLF1 | MyrP | 347 | 100 | 124 |
| BBRF1 | Portal protein, U16 homolog | 420 | 56 | 86 |
| BBRF2 | Egress protein, UL7 homolog | 222 | 73 | 86 |
| BDLF3 | gp150, immune evasion | 70 | 104 | 47 |
| BGRF1 | Tripartite terminase subunit UL15 homolog | 109 | 90 | 158 |
| BKRF2 | Envelope glycoprotein L | ND | 97 | 89 |
| BKRF4 | Tegument protein | 400 | 113 | 84 |
| BLLF3 | Deoxyuridine 5′ triphosphate nucleotidohydrolase | ND | 84 | 73 |
| BLRF1 | Glycoprotein N | 426 | 88 | 119 |
| BMRF2 | Viral attachment | 298 | 115 | 60 |
| BRRF2 | Tegument protein | 700 | 164 | 104 |
| BSRF1 | Tegument protein | 1176 | 169 | 72 |
All of these genes have previously been characterized as late genes in Akata cells treated with anti-IgG (38).
Activation was determined in BZKO cells. Values represent the ratio of activation by AP-1(A/S)/activation by ZEBRA. ND, not determined.
FIG 6.
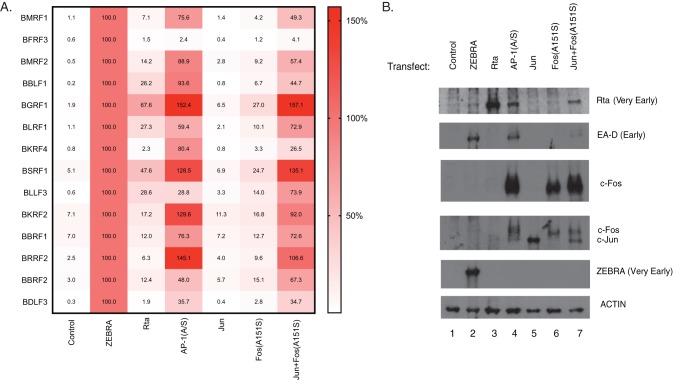
Expression of 12 putative late genes activated by AP-1(A/S), ZEBRA, Rta, AP-1(A/S), Jun, Fos(A151S), and Jun plus Fos(A151S). (A) Heat map of viral gene expression based on RT-qPCR RNA in BZKO cells transfected with a control vector, ZEBRA, Rta, AP-1(A/S), wild-type c-Jun, Fos(A151S), or wild-type c-Jun and Fos(A151S). Cells were harvested 48 h after transfection. Data are the average fold change relative to ZEBRA expression of three experiments. (B) Western blotting performed on the same cells used for RT-qPCR probed with the antibodies indicated. Blot is representative of three experiments.
Rta alone does not activate viral late gene expression to levels activated by AP-1(A/S).
Due to the ability of AP-1(A/S) to activate high-level expression of Rta protein, we hypothesized that it was possible that expression of viral late genes in the absence of viral DNA replication resulted from activation by Rta. To address this question, we transfected BZKO cells with plasmids encoding Rta or AP-1(A/S) and compared the expression levels of the 12 putative late genes. The transfection of Rta led to high levels of expression of Rta protein, with levels 3.5-fold higher than the levels of Rta protein induced by transfection of AP-1(A/S) (Fig. 6B). Six late genes, BKRF4, BKRF2, BBRF1, BRRF2, BBRF2, and BDLF3, were induced to be expressed at low levels by Rta and high levels by AP-1(A/S). Rta transfection resulted in higher levels of expression of five late genes, BBLF1, BGRF1, BLRF1, BSRF1, and BLLF3, with expression at 26.2%, 67.6%, 27.3%, 47.6%, and 28.6%, respectively, relative to the levels with ZEBRA. Four of these genes still were induced to be expressed to higher levels by AP-1(A/S) than by Rta. BLLF3 was the only late gene that was expressed to similar levels when cells were transfected with AP-1(A/S) or Rta. Analysis of the distribution of gene expression levels between AP-1(A/S) and Rta showed that gene expression when activated by Rta was significantly lower than that by AP-1(A/S) (P = 0.001). These results indicate that Rta alone is only partially capable of activating expression of 5 of the 12 viral late genes. Thus, the induction of expression of Rta by AP-1(A/S) does not account for the expression patterns of the viral late genes induced by AP-1(A/S).
Fos(A151S) mutant activates expression of viral late genes in the presence of wild-type c-Jun.
The recent identification of the Fos(A151S) mutation in human cancer provoked the question of whether the Fos(A151S) mutant was able to activate expression of viral late genes in the absence of Jun(A266S). Transfection of Fos(A151S) alone into BZKO cells activated trace amounts of Rta protein (Fig. 6B). Fos(A151S) induced expression of two of the viral late genes, BGRF1 and BSRF1, to 27.0% and 24.7%, respectively, relative to the levels of activation induced by ZEBRA (Fig. 6A).
Due to the inability of c-Fos to homodimerize, we hypothesized that the limited transcriptional activity of Fos(A151S) by itself might be due to lack of binding partners expressed in the cell. In order to address this possibility, we transfected Fos(A151S) with wild-type Jun into BZKO cells. Wild-type Jun alone did not activate any of the viral genes (Fig. 6A). Jun plus Fos(A151S) activated Rta protein to levels similar to those with activation by AP-1(A/S) (Fig. 6B). Jun plus Fos(A151S) activated all 12 putative late genes to levels similar to their levels of expression with activation by AP-1(A/S) in which both partners are mutants. Analysis of the distribution of gene expression levels identified no significant difference in the patterns of expression induced by AP-1(A/S) and Jun plus Fos(A151S) (P = 0.325). The studies demonstrate that in an AP-1 heterodimer only the Fos(A151S) mutant is needed to trigger lytic reactivation.
Sensitivity of expression of the subgroup of late genes activated by AP-1(A/S) to an inhibitor of viral replication.
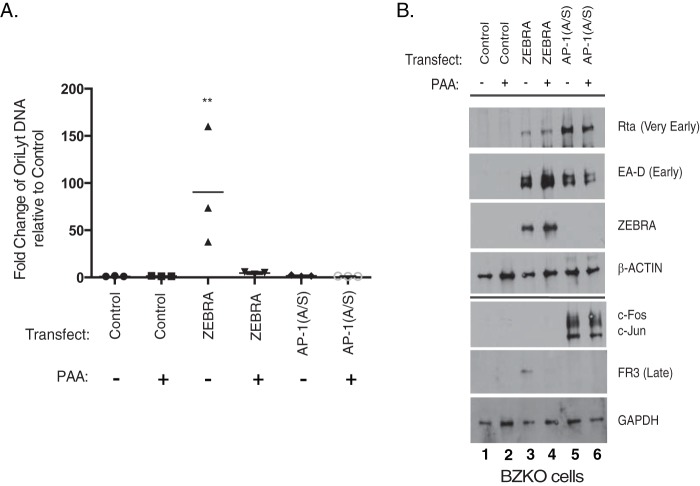
We used phosphonoacetic acid (PAA), a well-characterized inhibitor of the EBV DNA polymerase, to determine whether the 12 selected genes behaved as classical viral late genes when the lytic cycle was activated by ZEBRA or by AP-1(A/S). Transfection of BZKO cells with ZEBRA, but not AP-1(A/S), stimulated viral DNA replication (Fig. 7A). Treatment of ZEBRA-transfected BZKO cells with PAA dramatically reduced DNA replication to levels similar to those seen in cells transfected with the control vector (Fig. 7A). In BZKO cells transfected with ZEBRA, treatment with PAA completely inhibited expression of the late capsid protein FR3 to levels similar to those seen in cells transfected with the vector control (Fig. 7B, lanes 4 versus 3 and 1). Treatment with PAA had no effect on the expression of the very early protein Rta or the early protein EA-D in samples transfected with either ZEBRA or AP-1(A/S) (Fig. 7B, lane 3 versus 4 and lane 5 versus 6).
FIG 7.
PAA inhibits viral DNA replication and late protein expression induced by ZEBRA. (A) qPCR for relative abundance of viral DNA. In each experiment qPCR was performed on the same cells used for Western blot analysis. Each triangle represents one replicate. The horizontal line denotes the mean. P values were determined using pairwise Student's t tests with Dunnett's correction for multiple comparisons compared to results with the untreated control. **, P ≤ 0.0021. (B) Western blot of BZKO cells harvested 48 h after transfection with control vector, ZEBRA, or AP-1(A/S) and treated with PAA (500 μM) immediately after transfection. The blot is representative of three experiments.
RNA harvested from BZKO cells (Fig. 7A) was used to compare the relative sensitivities of the 12 candidate late genes to PAA when the genes were expressed under the influence of ZEBRA (Fig. 8A) or AP-1(A/S) (Fig. 8B). When the viral lytic cycle was induced by transfection of ZEBRA into BZKO cells, the 12 candidate late genes could be grouped into three categories with respect to their sensitivities to PAA (Table 2): four genes, BBLF1, BMRF2, BBLF1, and BBRF2, behaved as classical early genes falling within two standard errors of the mean (SEM) of the marker early gene BMRF1; four genes, BDLF3, BRRF2, BBRF1, and BBRF2, behaved as late genes, falling within two SEM of the marker late gene BFRF3. Four genes with intermediate sensitivity to PAA fell between two SEM of either BMRF1 or BFRF3. These four genes were BKRF4, BSRF1, BKRF2, and BLLF3 (Table 2). From the analysis of their sensitivities to PAA, we concluded that at least eight of the genes belong to the late or intermediate/late temporal class when the lytic cycle was activated by transfection of ZEBRA into BZKO cells. (Fig. 8A and Table 2).
FIG 8.
Expression of a group of late genes when activated by ZEBRA or AP-1(A/S) in response to PAA treatment. Fold change in expression was determined in BZKO cells treated with PAA relative to expression in untreated cells of 12 putative late genes following transfection with ZEBRA (A) or AP-1(A/S) (B). Bar height indicates the mean of three experiments. Error bars show one standard error of the mean. A marker early gene, BMRF1, is shown in white. A marker late gene, BFRF3, is shown in black. The 12 late genes activated by AP-1(A/S) in BZKO cells are shown in gray. In panel A the solid line represents 2 SEM of the expression level of BFRF3 in PAA-treated versus expression in untreated cells. The dashed line represents 2 SEM of expression of BMRF1 in PAA-treated versus expression in untreated cells. Genes with expression values below the solid line are considered late genes, and those with expression values above the dotted line are considered early genes. P values were determined using pairwise Student's t tests with Dunnett's correction for multiple comparisons compared to results with the control early gene BMRF1 or control late gene BFRF3. *, P ≤ 0.0332, compared to results with the BMRF1 control; +, P ≤ 0.0332 compared to results with the BFRF3 control. In panel B, BRFR3 was not shown because it was expressed at very low levels (<5% of the level expressed in cells transfected with ZEBRA).
TABLE 2.
Classification of late genes activated by AP-1(A/S) based on response to PAA and dependence on viral DNA replication in BMRF1 KO cells
| Gene | Classification in BZKO cellsa |
Classification in HH514-16 cellsa |
% activation by AP-1(A/S) in BZKO cellsa | Classification in BMRF1 KO cellsb |
|||
|---|---|---|---|---|---|---|---|
| % activation by ZEBRA | Temporal class | % activation by sodium butyrate | Temporal class | % activation by ZEBRA/ZEBRA + BMRF1 | Temporal class | ||
| BBLF1 | 54 | Early | 59 | Intermediate/late | 153 | 63 | Early |
| BBRF1 | 19 | Late | 37 | Late | 145 | 33 | Late |
| BBRF2 | 12 | Late | 30 | Late | 134 | 25 | Late |
| BDLF3 | 5 | Late | 14 | Late | 147 | 14 | Late |
| BGRF1 | 40 | Early | 34 | Late | 125 | 38 | Late |
| BKRF2 | 21 | Intermediate/late | 33 | Late | 215 | 12 | Late |
| BKRF4 | 25 | Intermediate/late | 59 | Intermediate/late | 115 | 71 | Early |
| BLLF3 | 21 | Intermediate/late | 57 | Intermediate/late | 237 | 101 | Early |
| BLRF1 | 39 | Early | 12 | Late | 142 | 11 | Late |
| BMRF2 | 77 | Early | 260 | Early | 92 | 88 | Early |
| BRRF2 | 14 | Late | 108 | Intermediate/early | 184 | 7 | Late |
| BSRF1 | 25 | Intermediate/late | 209 | Early | 191 | 15 | Late |
In response to PAA genes are classified as early genes if they fall within two standard errors of the mean (SEM) of BMRF1, a classical early gene. Genes are classified as late genes if they fall within two SEM of BFRF3, a classical late gene. In BZKO cells the cutoff for early genes is greater than or equal to 39% and the cutoff for late genes is less than or equal to 19%. In HH514-16 cells the cutoff for early genes is greater than or equal to 126% and the cutoff for late genes is less than or equal to 53%. Genes that fall between these two cutoffs are referred to as intermediate. Intermediate genes that fall close to a cutoff for early or late genes are referred to as intermediate/early or intermediate/late respectively. Activation levels were determined by RT-qPCR. Values are the ratio of activation with PAA/activation without PAA.
In response to viral DNA replication in BMRF1 KO cells genes are classified as early genes if their expression is significantly different from the expression of BFRF3 and as late genes if their expression is significantly different from the expression of BGLF4. Activation levels were determined by RT-qPCR. Values are the ratio of activation with ZEBRA/activation with ZEBRA plus BMRF1.
Remarkably, expression of all 12 putative late genes was completely resistant to the action of PAA when the lytic cycle was induced by AP-1(A/S) (Fig. 8B). In fact, expression of 11 of them was enhanced in the presence of PAA (Fig. 8B and Table 2). Comparison of the distribution of expression levels of the viral late genes by AP-1(A/S) and ZEBRA in the presence of PAA showed that expression of the 12 late genes was significantly higher with activation by AP-1(A/S) (P = 0.0001). Since transfection of AP-1(A/S) did not lead to amplification of viral lytic DNA in the absence or presence of PAA (Fig. 7A), we conclude from this experiment that 12 EBV genes, previously classified as belonging to the late temporal class in Akata cells treated with anti-IgG, are expressed to high levels in the absence of detectable viral DNA replication when the lytic cycle is induced by AP-1(A/S) in BZKO cells.
AP-1(A/S) selectively enhances expression of some late and early gene transcripts.
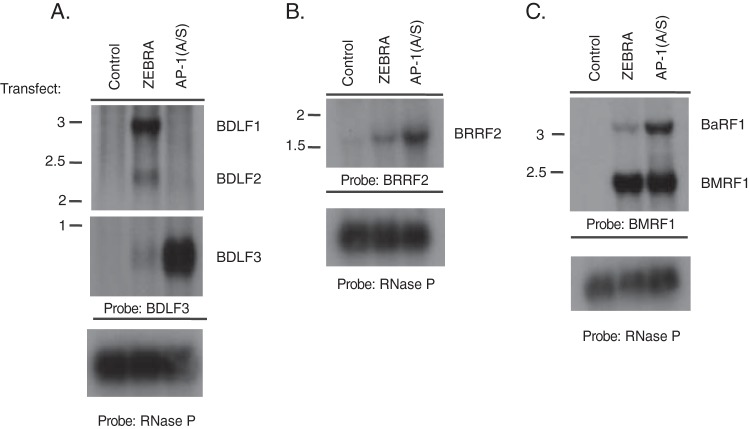
We sought to determine whether late transcripts activated by AP-1(A/S) are comparable in size to native transcripts induced to be expressed during the viral lytic cycle by ZEBRA. Northern blotting was performed in BZKO cells harvested 48 h after transfection with a control plasmid vector or with plasmids expressing either ZEBRA or AP-1(A/S). This allowed us to determine if RT-qPCR analysis was detecting native viral late transcripts or aberrant read-through from other transcripts. Two late genes induced by AP-1(A/S), BDLF3 and BRRF2, were chosen for this experiment based on their sensitivities to PAA in BZKO cells (Table 2) and on their high amplification in RT-qPCR analysis (Table 1). Transfection of ZEBRA led to the expression of three late viral transcripts, BDLF1, BDLF2, and BDLF3, all of which are coterminal with BDLF3; these three transcripts were observed by probing for BDLF3 (Fig. 9A, second lane, and 10A). In contrast, transfection of AP-1(A/S) led to expression of only the BDLF3 transcript to levels approximately 21 times higher than the level induced by transfection of ZEBRA (Fig. 9A, third versus second lane). BDLF3 was expressed at levels approximately 50% of the level with activation by ZEBRA when assayed by RT-qPCR (Fig. 5). However, RT-qPCR with BDLF3 primers also amplified BDLF1 and BDLF2 that are expressed only in response to ZEBRA. Therefore, expression of the BDLF3 gene by AP-1(A/S) appears lower than that with ZEBRA when assessed by RT-qPCR and higher than that with ZEBRA when assessed by Northern blotting. This was not unexpected because, in the case of BDLF3, the RT-qPCR measures the sum total of the three 3′ coterminal transcripts, while Northern blotting distinguishes between them.
FIG 9.
AP-1(A/S) selectively activates one component of a group of coterminal late lytic transcripts. Northern blots of BZKO cells harvested 48 h after transfection with control vector, wild-type ZEBRA, or mutant AP-1(A/S) were probed with radioactive double-stranded BDLF3 (A), BRRF2 (B), BMRF1 (C), or RNase P DNA probes as indicated. Predicted approximate RNA sizes are as follows: BDLF1, 3,032 bp; BDLF2, 2,122 bp; BDLF3, 750 bp; BRRF2, 1,672 bp; BaRF1, 3,381 bp; BMRF1, 2345 bp. Predicted RNA sizes are based on annotated TATA box and poly(A) tail sites available in the human herpesvirus 4 complete wild-type genome (GenBank accession number AJ507799.2).
FIG 10.
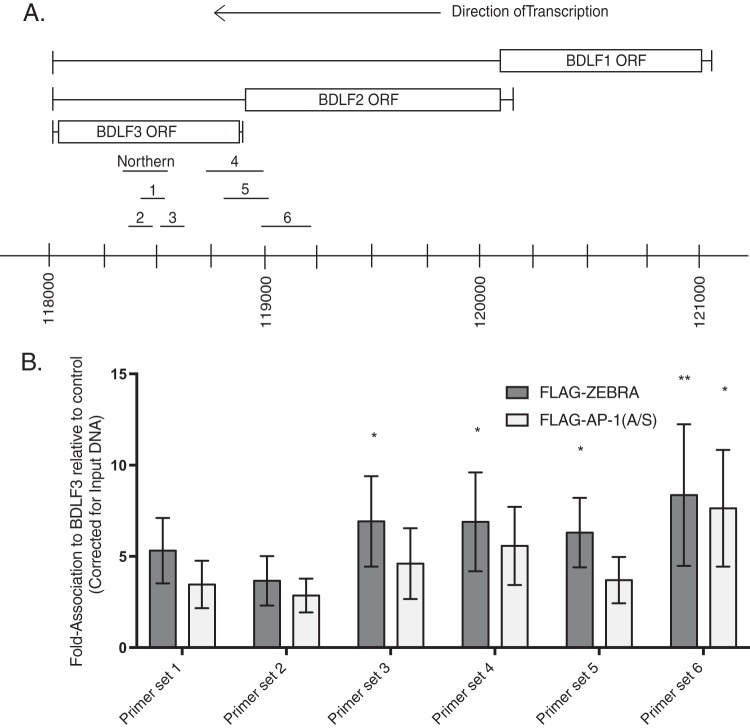
Both ZEBRA and AP-1(A/S) bind to the BDLF3 promoter. (A) Schematic drawing of the locus of BDLF1 to BDLF3 showing the locations of the six primer sets used for ChIP qPCR, indicated by 1 to 6, and the probe used for Northern blotting. The TATA box is used to denote the start of transcripts, and the poly(A) signal is used to denote the end of transcripts. The human herpesvirus 4 complete wild-type genome (GenBank accession number AJ507799.2) sequence was used to demarcate locations of genes. ORF, open reading frame. (B) Chromatin immunoprecipitation was performed in BZKO cells harvested 48 h after transfection with control FLAG vector, wild-type FLAG-ZEBRA, or mutant FLAG-AP-1(A/S). The FLAG antibody was used to immunoprecipitate all target proteins. Association to BDLF3 was determined using qPCR. Bar height indicates the mean of three experiments. Error bars show one standard error of the mean. P values were determined using pairwise Student's t tests with Dunnett's correction for multiple comparisons compared to results with the control FLAG vector. *, P ≤ 0.0332; **, P ≤ 0.0021.
Transcripts of the BRRF2 late gene were of similar sizes following transfection with either ZEBRA or AP-1 (A/S) (Fig. 9B). However, expression of BRRF2 mRNA was approximately 3.8 times higher when induced by AP-1(A/S) than when induced by ZEBRA (Fig. 9B). The marker early gene BMRF1 was induced by AP-1(A/S) to levels 76.5% of those induced by ZEBRA (Fig. 9C, lane 3 versus 2). However, the BMRF1 probe detected a coterminal early transcript, BaRF1, that was 3.4-fold more abundant when induced by AP-1(A/S) than when induced by ZEBRA (Fig. 9C). These results indicate that AP-1(A/S) activates EBV lytic transcripts in a gene-specific manner; the abundances of some early and late gene transcripts are enhanced when induced by AP-1(A/S).
ZEBRA and AP-1(A/S) bind to the BDLF3 promoter.
We asked whether the enhanced ability of AP-1(A/S) to promote expression of the BDLF3 transcript, compared to that of ZEBRA, correlated with enhanced ability of AP-1(A/S) to bind directly to the BDLF3 promoter. We compared binding of ZEBRA and AP-1(A/S) to the BDLF3 promoter region using ChIP under the same conditions used to measure their binding to oriLyt and Rp (Fig. 2). Ramasubramanyan et al. previously predicted possible ZEBRA binding in the BDLF3 region using ChIP with high-throughput sequencing (ChIP-seq) (39). We generated six primer sets spanning the region predicted to bind ZEBRA (Fig. 10A). Both ZEBRA and AP-1(A/S) bound to the BDLF3 promoter region. AP-1(A/S) did not bind better than ZEBRA to any of the six regions tested by qPCR. Binding levels of ZEBRA and AP-1(A/S) were equivalent using the upstream primer set 6 (Fig. 10B). These results provide a plausible mechanism by which AP-1(A/S) activates expression of BDLF3 by directly binding to the promoter. However, the experiment shows that the considerably higher activation of BDLF3 expression by AP-1(A/S) cannot be explained by higher DNA binding affinity to the BDLF3 promoter.
Temporal class of the 12 candidate late genes under conditions when the lytic cycle was activated in BL cells.
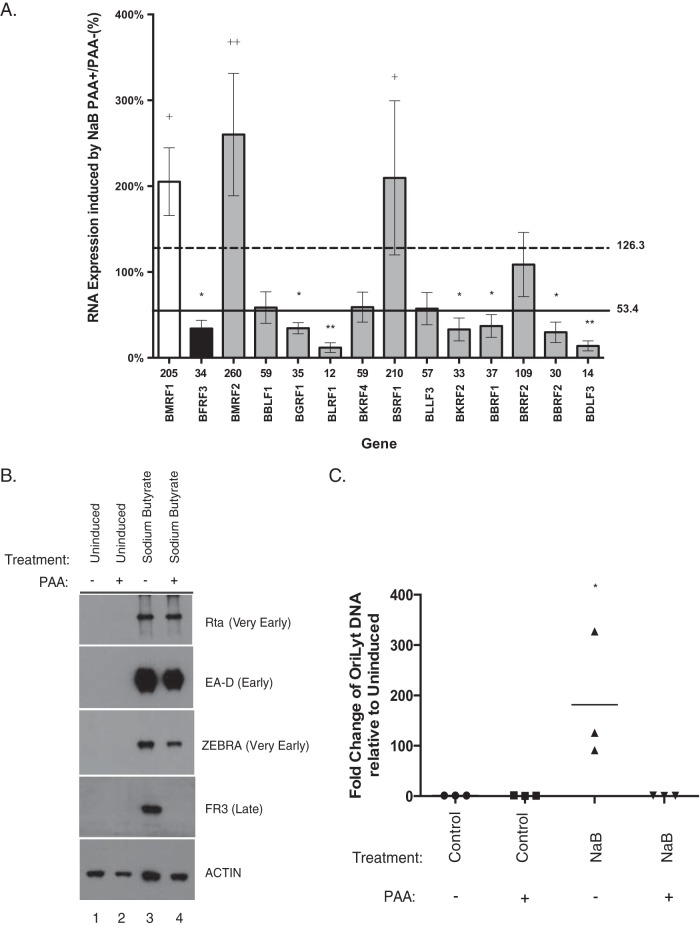
We used HH514-16 Burkitt lymphoma (BL) cells, in which the lytic cycle can be triggered by treatment with sodium butyrate, to characterize the sensitivity of the 12 candidate genes to PAA (Fig. 11A and Table 2). In HH514-16 cells treated with sodium butyrate, PAA did not affect the levels of Rta, ZEBRA, or EA-D proteins (Fig. 11B), but, as expected, it did inhibit production of the FR3 late protein (Fig. 11B). DNA replication assays performed on the same cells used for Western blot analysis showed that DNA abundance was 181-fold higher in butyrate-treated cells than in untreated cells (Fig. 11C). Treatment with PAA inhibited activation of DNA replication by sodium butyrate to levels similar to the level in untreated cells (Fig. 11C). In HH514-16 cells, treatment with both sodium butyrate and PAA enhanced expression of the marker early gene BMRF1 and reduced expression of the marker viral late gene BFRF3 compared to levels in cells treated with sodium butyrate alone (Fig. 11A). In HH514-16 cells activated by sodium butyrate and treated with PAA, 6 of the 12 AP-1(A/S)-responsive candidate late genes, BDLF3, BKRF2, BLRF1, BBRF2, BBRF1, and BGRF1, also behaved as late genes and fell within two SEM of expression of the marker late gene BFRF3. Two genes, BMRF2 and BSRF1, behaved similarly to the traditional early gene BMRF1 and were resistant to the activity of PAA. Four AP-1(A/S)-induced genes, BRRF2, BKRF4, BLLF3, and BBLF1, had intermediate sensitivity to PAA. Of these, BRRF2 was close to the cutoff for early genes. The three other genes, BBLF1, BKRF4, and BLLF3, had intermediate sensitivity to PAA, with 58.6%, 59.1%, and 57.3% of the expression level with sodium butyrate without PAA, respectively (Fig. 11A and Table 2). In the same experiments, expression of the marker early gene BMRF1 was enhanced by PAA. These results in a completely different biological system, consisting of different cells and a different inducing agent, confirmed that at least six of the candidate late genes that were activated by AP-1(A/S) in the absence of DNA replication in BZKO cells manifested late behavior when the lytic cycle was triggered in HH514-16 by butyrate (Table 2).
FIG 11.
Effect of PAA on the expression of the 12 putative late genes activated by sodium butyrate in HH514-16 cells. (A) Fold change in gene expression levels in H514-16 cells treated with sodium butyrate plus PAA relative to levels in cells treated with sodium butyrate alone at 72 h. Bar height indicates the mean of three experiments. Error bars show one standard error of the mean. A marker early gene, BMRF1, is shown in white. A marker late gene, BFRF3, is shown in black. The 12 late genes activated by AP-1(A/S) in BZKO cells are shown in gray. The solid line represents two SEM of the expression level of BFRF3 in PAA-treated cells versus the level in untreated cells. Values below this line represent late genes. The dashed line represents two SEM above the expression of BMRF1 in PAA-treated versus untreated cells. Values above this line represent early genes. P values were determined using pairwise Student's t tests with Dunnett's correction for multiple comparisons compared to results with the control. *, P ≤ 0.0332; **, P ≤ 0.0021 (both, for results compared to those with the BMRF1 control); +, P ≤ 0.0332; ++, P ≤ 0.0021 (both, for results compared to those with the BFRF3 control). (B) Western blotting was performed on HH514-16 cells harvested 72 h after lytic induction with sodium butyrate. The cells were untreated or treated with PAA (500 μM) immediately after induction. Blot is representative of three experiments. (C) Fold change in expression of viral DNA measured by qPCR, relative to expression with the vector control, was determined on the same cells used for Western blot analysis. Each point indicates one replicate. The horizontal line denotes the mean. P values were determined using pairwise Student's t tests with Dunnett's correction for multiple comparisons compared to results with an untreated control. *, P ≤ 0.0332.
Activation of 12 candidate late genes in replication-deficient Raji cells.
In order to determine the temporal class of the 12 candidate late genes in a genetic system where viral DNA replication cannot occur, we studied Raji cells, a Burkitt lymphoma cell line infected with a replication-deficient virus. Raji cells can be induced into the lytic cycle using 12-O-tetradecanoylphorbol-13-acetate (TPA). However, they lack a functional BALF2 (40), the single-stranded DNA binding protein (41), and are unable to perform viral DNA replication. In Raji cells treated with TPA, no detectable changes in viral DNA content were observed (Fig. 12B). As expected, treatment with TPA did induce RNA expression of the viral very early gene BZLF1 and the early gene BMRF1 in Raji cells (Fig. 12A). Unexpectedly, TPA treatment led to induction of mRNAs of all of the viral late genes tested, including the canonical viral late gene BFRF3 (Fig. 12A). These results in a genetically replication-deficient system are unexpected. They show that RNA expression of viral late genes in Raji cells is not dependent on viral DNA replication.
FIG 12.
Induction of Raji cells with TPA leads to expression of viral late RNAs in the absence of viral DNA replication. (A) Fold change in gene expression in Raji cells treated with TPA relative to levels in untreated cells at 48 h. Each point indicates one replicate. The horizontal lines denote the means. (B) Fold change in expression of viral DNA measured by qPCR, relative to levels in uninduced cells, was performed on the same cells used for RT-qPCR analysis. Each point indicates one replicate. The horizontal lines denote the means. (C) Western blotting was performed on Raji cells harvested 48 h after lytic induction with TPA. Lanes 1 to 3, biological replicates treated with TPA; lanes 4 to 6, untreated biological replicates; lane 7, positive control for FR3 in HH514-16 cells treated with sodium butyrate at 72 h as described in the legend of Fig. 11.
Activation of the 12 candidate late genes by ZEBRA in BMRF1 KO cells.
To determine the kinetic class of the 12 putative late genes expressed by AP-1(A/S), we used BMRF1 knockout (KO) cells, HEK293 cells containing an EBV bacmid lacking a functional BMRF1 gene (42). BMRF1 encodes the viral DNA processivity factor, a protein essential for viral DNA replication and late gene expression. Transfection of ZEBRA in BMRF1 KO cells failed to activate viral DNA replication and failed to induce expression of the canonical late FR3 protein (Fig. 13C). Cotransfection of ZEBRA and BMRF1 in BMRF1 KO cells restored viral DNA replication and activated expression of the late FR3 protein (Fig. 13C) (42). Using RT-qPCR, we compared expression levels of the 12 putative late genes with expression of BGLF4, a canonical early transcript, and BFRF3, a canonical late transcript. In this system, 8 of the 12 genes studied, BGRF1, BLRF1, BSRF1, BKRF2, BBRF1, BRRF2, BBRF2, and BDLF3, had significantly different levels of expression than the canonical early gene BGLF4. Expression levels of these same eight genes were not significantly different from the level of the canonical late gene BFRF3. These data show that in response to ZEBRA expression of 8 of the 12 putative late genes is dependent on replication of the viral genome. These results show that these genes are true late genes.
FIG 13.
Expression of 12 putative late genes activated by ZEBRA in the presence and absence of BMRF1 in BMRF1 KO cells. Fold change in expression levels in BMRF1 KO of 12 putative late genes following transfection with ZEBRA or ZEBRA plus BMRF1. (A) Fold change in gene expression levels following transfection of ZEBRA relative to the level with ZEBRA plus BMRF1 at 48 h. Bar height indicates the mean of three experiments. Error bars show one standard error of the mean. A marker early gene, BGLF4, is shown in white. A marker late gene, BFRF3, is shown in black. The 12 late genes activated by AP-1(A/S) in BZKO cells are shown in gray. P values were determined using pairwise Student's t tests with Dunnett's correction for multiple comparisons compared to results with the control early gene BGLF4 or control late gene BFRF3. *, P ≤ 0.0332; **, P ≤ 0.0021, ***, P ≤ 0.0002 (all, for results compared to the result with the BGLF4 control); +, P ≤ 0.0332; ++, P ≤ 0.0021; +++, P ≤ 0.0002 (all, for results compared to the result with the BFRF3 control). (B) Western blotting was performed on the same cells used for RT-qPCR probed with indicated antibodies. Blot is representative of three experiments. (C) The fold change in expression levels of viral DNA measured by qPCR relative to the level in the vector control was determined on one representative biological replicate used for RT-qPCR analysis.
DISCUSSION
Here, we describe novel and unexpected observations about transcriptional control of the EBV lytic cycle. Mutant cellular AP-1 proteins are capable of substituting for ZEBRA to drive expression of a majority of EBV genes (Fig. 3 and 4; see also Table S1 in the supplemental material). Although the mutant AP-1 proteins are unable to promote lytic viral DNA replication, they are nonetheless competent to induce expression of a subset of viral genes that are classified as late genes (38) (Fig. 3, 4, and 6 and Table 2). Thus, expression of EBV late genes may not be invariably dependent on lytic viral DNA replication (34, 35, 37).
AP-1(A/S) activates expression of a subset of viral lytic genes.
AP-1(A/S) strongly activates expression of mRNAs encoded by more than one-third (24 of 67 genes on the array) of viral genes assayed to levels that are similar or higher than levels with activation by ZEBRA (Fig. 3 and 4). One remarkable example is the BRLF1 gene which is activated 4.1-fold higher (Fig. 3) and its product Rta, which is activated 4.4-fold higher (Fig. 1A) by AP-1(A/S) than by ZEBRA. Expression of downstream genes that are more strongly enhanced by AP-1(A/S) than by ZEBRA could be influenced by the higher levels of the Rta transcription factor acting in synergy with AP-1(A/S) (32). Rta is known to have an extremely potent transcriptional activation domain (43). However, Rta itself is not responsible for activation of expression of EBV late genes (Fig. 6).
Approximately 40% of EBV genes (28 of 67) are activated to very low levels by AP-1(A/S) by comparison to levels with ZEBRA (Fig. 3 and 4). This group includes both early and late lytic cycle genes. Although our data indicate that AP-1(A/S) is capable of activation of a subset of viral late genes, the deficiency of AP-1(A/S) in promoting expression of many other viral late genes may be linked to its deficiency in promoting viral lytic DNA replication (Fig. 1B). AP-1(A/S) binds efficiently to methylated ZREs (32); therefore, binding to methylated DNA is not sufficient to activate high levels of expression of all EBV lytic genes. Differences between AP-1(A/S) and ZEBRA in the extents of viral gene expression must be influenced by differences in the transcriptional activation domains, DNA binding domains, and dimerization domains of the two transcription factors (21–23). These would influence interacting partners, chromatin reorganization, DNA binding specificity, and affinity for different DNA response elements. Thus, ZEBRA and AP-1(A/S) would be expected to differ functionally as activators or repressors of viral and cellular gene expression.
AP-1(A/S) does not promote viral lytic DNA replication.
Although AP-1(A/S) functions well as a viral transcription factor, it is unable to perform viral DNA replication. The binding of AP-1(A/S) to oriLyt is reduced compared to binding by ZEBRA (Fig. 2A). Previous work by El-Guindy et al. showed that the inability of a ZEBRA mutant, Z(S167A/S173A), which eliminates constitutive CK2 phosphorylation sites, to stimulate viral DNA replication was correlated to a 2.9-fold reduction in oriLyt binding as assessed by ChIP (44). AP-1(A/S) binding to oriLyt was reduced to a similar extent. Conversely, AP-1(A/S) binds as well as ZEBRA to Rp. These differences in binding to Rp and oriLyt may explain why AP-1(A/S) functions well as a transcription factor but poorly as a replication protein. Our results support the conclusion of El-Guindy et al. (44) that higher binding affinity is required for DNA replication than for viral transcription.
Another possible explanation for the replication defect is that AP-1(A/S) fails to stimulate expression of viral or cellular genes whose products mediate replication. AP-1(A/S) is defective at promoting expression of 10 viral early genes, two of which, BALF2 and BALF5, are known to be required for viral DNA replication (Fig. 3 and 4; Table S1). However, exogenous expression of the six known viral DNA replication proteins did not rescue viral DNA replication (Fig. 5). One or more of the EBV early genes, other than BALF5 and BALF2, which are expressed at low levels, may play a previously unknown role in viral DNA replication.
Additional hypotheses need to be explored in further experiments directed at unraveling the defect of AP-1(A/S) in replication. Even in the presence of all necessary replication proteins, AP-1(A/S) may be unable to substitute for ZEBRA's functions as an oriLyt binding protein that assembles the replication complex. Schepers et al. showed that the c-Jun transactivation domain fused to the GAL4 DNA binding domain and tethered to oriLyt via GAL4 binding sites was unable to perform viral lytic replication (13). Further experiments comparing the interaction of AP-1(A/S) and ZEBRA with all the components of the viral replication machinery will be necessary to fully explore this hypothesis.
AP-1(A/S) promotes late gene expression in the absence of viral lytic DNA replication.
The 12 genes, previously classified as late genes in the Akata cell system (38), that are induced by AP-1(A/S) even in the absence of viral lytic DNA replication have various functions in the viral life cycle, including capsid proteins, glycoproteins, tegument proteins, the viral dUTPase, and immune modulators (Table 1). We examined the possibilities that these genes were previously misclassified or that their temporal control was influenced by cell background. These genes were initially classified as late genes in the Akata cell system based on their PAA sensitivities and their expression kinetics (38). We analyzed their expression levels in BZKO cells in the absence or presence of the potent and specific viral lytic DNA replication inhibitor PAA (Fig. 8 and Table 2). When the viral lytic cycle was initiated in BZKO cells by transfection of ZEBRA in the presence of PAA, 8 of the 12 genes, BKRF4, BSRF1, BLLF3, BKRF2, BBRF1, BDLF3, BRRF2, and BBRF2, behaved similarly to the marker late gene BFRF3 and were very sensitive to PAA. In the same cell system, these same eight genes were completely resistant to PAA when the lytic cycle was activated by AP-1(A/S). In fact, their expression was enhanced compared to expression by AP-1(A/S) in the absence of PAA. To ascertain whether these genes behave as late genes in another biological system, we performed similar experiments in Burkitt lymphoma cells activated by sodium butyrate in the absence or presence of PAA (Fig. 8 and Table 2). In this cell background, expression of 9 of the 12 genes, BBLF1, BKRF4, BLLF3, BDLF3, BKRF2, BLRF1, BBRF2, BBRF1, and BGRF1, was inhibited by PAA. To confirm that the response of these 12 genes was not simply an effect of using PAA to inhibit viral DNA synthesis, we assessed the classification of these genes using the genetic system of BMRF1 KO cells. In this system 8 of the 12 genes, BGRF1, BLRF1, BSRF1, BKRF2, BBRF1, BRRF2, BBRF2, and BDLF3, were expressed in a manner dependent on viral DNA replication, similar to other canonical late genes (Fig. 13). We conclude that a subset of bona fide late genes are activated by AP-1(A/S) in the absence of viral lytic DNA replication. In the presence of ZEBRA, viral DNA replication is required for expression of these same genes.
Fos(A151S) mutant activates expression of 12 late genes activated by AP-1(A/S) in the absence of Jun(A266S) mutant.
The Fos(A151S) mutation was recently identified in the COSMIC database in a patient with hepatocellular cancer. Therefore, it was important to determine if this mutation by itself was capable of activating expression of the 12 putative late genes. In the presence of wild-type Jun, which does not activate viral lytic gene expression, Fos(A151S) activated the expression of the 12 putative late genes to an extent similar to AP-1(A/S) (Fig. 6). This result demonstrates that a single missense mutation in only one member of the AP-1 heterodimer is sufficient to drive viral late gene expression in the absence of viral DNA replication. In a more general way, a single mutant cellular protein that naturally occurs in tumors is capable of bypassing the canonical viral control mechanisms.
Possible role of late gene regulators.
AP-1(A/S) did not activate expression of four of six known late gene regulators assayed in the EBV gene expression array. These genes are BcRF1, the unique TATT binding protein, BDLF4, BFRF2, and BVLF1. AP-1(A/S) did activate expression of two late gene regulators, BGLF3 and the kinase BGLF4. The seventh late gene regulator, BGLF3.5, was not assayed in the arrays (37, 45). These results suggest that the expression of the subgroup of late genes induced by AP-1(A/S) may not be dependent on the full complement of canonical viral late gene regulators. When the same group of late genes responsive to AP-1(A/S) is stimulated by ZEBRA, all of these genes, except BSRF1, require all of the known late gene regulators (46). Most likely, EBV late genes that were not induced by AP-1(A/S) require both lytic viral DNA replication and the complete late gene initiation complex. However, recent work by McKenzie et al. showed that four viral late genes were expressed even in the absence of the canonical viral late preinitiation complex. These genes are transcribed in a method distinct from other late genes (46). BSRF1, one of the late genes activated by AP-1(A/S) in BZKO cells, is an example of a late gene whose expression is dependent on the BGLF4 kinase but independent of BGLF3 and other late gene regulators (46).
Not all viral late genes are regulated by the same mechanism in different cells.
Previous assignment of viral kinetic classes assumed that temporal expression of viral genes during reactivation was the same in all cell systems. However, our experiments show that cell background and lytic cycle-inducing stimulus influence the kinetics of viral gene expression. We examined the response of 12 candidate late genes to activation by ZEBRA in the presence of the potent viral DNA replication inhibitor PAA in two cell lines of different tissue origins, namely, BZKO cells, which are derived from the 293 human embryonic kidney cell line, and HH514-16 cells, a subclone of a lymphoid cell line derived from Burkitt lymphoma (Fig. 8 and 10). The responses of six genes to PAA were concordant in both cell lines; five genes were classified as late, and one, BMRF2, was classified as early. The responses of six genes were discordant in the two cell systems. Two genes, BSRF1 and BRRF2, displayed PAA sensitivity characteristic of late genes in BZKO cells but were resistant to PAA, a property of early genes, in HH514-16 cells. Three genes were classified as early in BZKO cells but displayed late characteristics in HH514-16 (Table 2). In the BMRF1 KO genetic system, the classification of the 12 genes was different from classification in both BZKO and HH514-16 cells. Among the three cell systems four genes, BBRF1, BBRF2, BDLF3, and BKRF2, were consistently classified as late, while one gene, BMRF2, behaved as an early gene in all cell systems. Moreover, there was a gradient of responses to PAA by late genes in the same cell line. Variable inhibition of late genes by PAA may indicate that, even in the same cell line, not all late genes are regulated by the same mechanisms (Fig. 8A and 10A). One possible explanation is that different extents of viral replication may license expression of different late genes. A potential biological reason for these phenomena is that EBV may require mechanisms to vary the temporal regulation of certain viral genes in response to cell or tissue type, differentiation, or physiologic conditions.
Lytic induction of Raji cells by TPA led to induction of all of the viral late genes assayed, including the canonical late gene BFRF3 (Fig. 12A). This result suggests that even canonical late genes may be expressed at low levels in the absence of detectable viral DNA replication. Treatment of BZKO cells or HH514-16 cells with PAA inhibits viral late gene expression. However, PAA does not completely abolish expression of viral late genes. This may be due to incomplete inhibition of viral DNA replication by PAA or to low levels of expression of late genes in the absence of viral DNA replication. All viral late genes may have a “leaky” component that can be expressed in the absence of viral DNA replication.
AP-1(A/S) selectively activates one member of a late transcription unit with coterminal transcripts.
Our observations (Fig. 9) of different behaviors of individual members of coterminal transcription units provides a mechanistic insight into selective action of AP-1(A/S) in regulation of expression of EBV lytic cycle genes. In one such transcription unit, AP-1(A/S) strongly and selectively activates expression of BDLF3 (Fig. 9A) but fails to activate expression of transcripts of two upstream late genes, BDLF1 and BDLF2, that are coterminal with BDLF3. Conversely, ZEBRA strongly activates BDLF1 and BDLF2 but only weakly activates BDLF3. In another coterminal early gene transcription unit, ZEBRA and AP-1(A/S) equally activate expression of BMRF1, but AP-1 (A/S) more potently activates transcription of BaRF1, which encodes a subunit of ribonucleotide reductase (Fig. 9C).
Our ChIP data show that AP-1(A/S) and ZEBRA bind with similar affinities to the promoters of BDLF3 (Fig. 10). The results of the BDLF3 Northern blot analysis and ChIP taken together suggest that, while binding of the viral and cellular transcription factors to the promoters of responsive genes may be necessary, binding alone is not sufficient to account for intensity or specificity of activation. These facets of transcriptional control might be attributed to specific protein-protein interactions by the two transcription factors or subtle differences in their interactions with DNA.
ZEBRA may link viral DNA replication and late gene expression.
The prevailing model for regulation of viral late genes postulates that they are regulated by a common mechanism (37, 45, 47–55). Conventional thinking holds that viral DNA replication in cis is an essential prerequisite to enable viral late gene transcription (33–35). We show that a subset of late genes that are activated by AP-1(A/S) in the absence of replication absolutely require DNA replication when the lytic cycle is induced by ZEBRA, but AP-1 (A/S) seems to bypass the requirement for replication for this group of genes. While AP-1(A/S) is capable of functioning as a viral transcription factor, it may lack certain activities needed for temporal regulation of the viral life cycle. Temporal regulation of late genes may consist of two distinct components: repression of these genes before replication and licensing of expression after replication. One function of ZEBRA itself may be to repress expression of late genes until after viral DNA replication has occurred. AP-1(A/S) may be unable to assume this role.
Under one other previously described experimental condition, ZEBRA assumed a repressive role that ensured proper temporal regulation of late gene transcription. The viral late gene BLRF2, encoding a tegument protein, was expressed in the absence of DNA replication when Rta was expressed in Raji cells (30). EBV DNA is incapable of undergoing DNA replication in the Raji cell line due to an inactivating mutation in BALF2, which is an essential replication protein (40). In the Raji cell background Rta does not cross-stimulate expression of ZEBRA (30). Introduction of either wild-type ZEBRA or some ZEBRA mutants into Raji cells synthesizing high levels of Rta prevented expression of BLRF2 (15). Mindful of this example, we considered the possibility that the 12 late genes were aberrantly induced because the AP-1(A/S) mutant promotes high levels of Rta in BZKO cells in the absence of ZEBRA. Experiments expressing Rta in the absence of ZEBRA showed that while Rta was capable of activating 5 of the 12 late genes induced by AP-1(A/S) (BBLF1, BGRF1, BLRF1, BSRF1, and BLLF3), all except BLLF3 were expressed to levels lower than when they were expressed by AP-1(A/S) (Fig. 6A). Another group of putative late genes were activated strongly by AP-1(A/S) but weakly by Rta. It is possible that Rta and AP-1(A/S) act in synergy to activate viral gene expression, in a manner similar to the relationship between Rta and ZEBRA. Moreover, in the EBV gene expression array from BZKO cells expressing AP-1(A/S) and high levels of Rta (Fig. 3 and Table S1), BLRF2 was not among the late genes shown to be expressed. These results make it less likely that the unopposed action of Rta accounts for the aberrant behavior of the late genes we have identified. An alternative hypothesis that needs to be explored in further experiments is that ZEBRA, but not AP-1(A/S), regulates transcription of cellular genes that link viral late gene expression and DNA replication.
In summary, AP-1(A/S) mutants shed light on poorly understood nuances of EBV lytic gene expression. AP-1(A/S) induces expression of some viral genes more strongly than ZEBRA but fails to induce expression of other viral genes. If viral lytic cycle gene promoters were exclusively controlled by some combinatorial action of ZEBRA and Rta and if AP-1(A/S) were to substitute for the transcriptional function of ZEBRA, then one would expect the pattern of gene expression promoted by AP-1(A/S) to closely resemble the pattern induced by ZEBRA. Moreover, since AP-1(A/S) does not invariably link viral late gene expression to viral DNA replication, the mutant cellular proteins provide valuable experimental tools with which to dissect ways in which replication does or does not facilitate late viral gene expression.
MATERIALS AND METHODS
Cell lines and transfection.
Human embryonic kidney 293 cells containing EBV/B95-8 bacmids with insertional inactivation of the EBV BZLF1 gene (BZKO) (8) or BMRF1 (BMRF1 KO) (42) were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% (vol/vol) fetal bovine serum (FBS), penicillin (50 U/ml), streptomycin (50 U/ml), and hygromycin (100 μg/ml). The HH514-16 subclone Burkitt lymphoma cells (56, 57) and Raji cells (40) were grown in RPMI 1640 medium with 8% FBS, penicillin (50 U/ml), and streptomycin (50 U/ml). BZKO cells were transfected with the DMRIE-C (1,2-dimyristyloxypropyl-3-dimethyl-hydroxy ethyl ammonium bromide and cholesterol) reagent (Invitrogen). HH514-16 cells were treated with 3 mM sodium butyrate to induce the EBV lytic cycle. Cells were harvested for analysis at 48 h after transfection (BZKO) or after treatment with sodium butyrate (HH514-16).
Expression vectors.
Expression vectors for the wild-type EBV BZLF1 gene and the Z(S186A) mutant containing EBV genomic DNA driven by the cytomegalovirus (CMV) immediate early (IE) promoter in pHD1013 have been previously described (29, 31). Expression vectors for wild-type human c-Jun, wild-type human c-Fos, c-Jun(A266S), and c-Fos(A151S) cloned with a FLAG tag in pCMV/Flag2 (Sigma-Aldrich) or untagged in pCMV6-XL5 vector (OriGene) were previously described (32). Expression vectors for the six replication proteins BALF5, BBLF2/3, BBLF4, BSLF1, and BMRF1 have been previously described (12, 58).
Gene expression qPCR arrays.
Methods for harvesting total RNA, selecting for poly(A) RNA, generating cDNA, and conducting the qPCR array have been described previously (59–62). Two arrays were used to analyze the data. Array 2 had more primer sets per gene than array 1. Data represent average fold change compared to the level with the vector control of each gene standardized to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The data are arranged using hierarchical clustering under complete linkage with a Euclidean distance measure (see the legend for Fig. 2 Table S1 in the supplemental material).
Western blot analysis.
Electrophoresis of total cell extracts was performed in sodium dodecyl sulfate (SDS) on 12% TGX (Tris-glycine extended) polyacrylamide gels (Bio-Rad). Gels were transferred to nitrocellulose membranes (Bio-Rad). Antibodies for the following were used: c-Jun (ab31419; Abcam), c-Fos (sc-52; Santa Cruz), ZEBRA (BZ1) (63), EA-D (R3.1) (64), Rta (31), FR3 (65), beta-actin (ab8227; Abcam), and GAPDH (ab8245; Abcam). Protein levels were determined by densitometry of radiographs and were normalized to beta-actin or GAPDH. Data shown are representative of the number of biological replicates indicated in the figure legends.
Northern blotting.
Preparation of total cellular RNA, radioactive probes, gel conditions, and transfer have been previously described (18, 66). The probes were double-stranded DNA prepared by PCR. The probes were purified by agarose gel electrophoresis. The positions of the probes on the sequence of the complete EBV genome (GenBank accession number NC_007605) are available upon request. Northern blots were also probed with the H1 component of RNase P to control for RNA loading (4).
DNA replication qPCR assay.
A DNA replication qPCR assay was performed as previously described (44). Primer sequences are available in Table 3.
TABLE 3.
Sequences of primers used for RT-qPCR and qPCR
| Primer target or name | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| BMRF1 | CAA CAC CGC ACT GGA GAG | GCC TGC TTC ACT TTC TTG G |
| 18S | GTA ACC CGT TGA ACC CCA TT | CCA TCC AAT CGG TAG TAG CG |
| oriLyt | TCC TCT TTT TGG GGT CTC TG | CCC TCC TCC TCT CGT TAT |
| Rp ZRE2-ZRE3 | GGG CAT TCC ATA AAG CAA ACA GG | CAT CTG GCT GGG GCA TTA AC |
| BRRF2 | GTT TTC GCA CGC ACC TTA T | TCG TCG GGA GAT AAA AGT GA |
| BKRF4 | GAC GAG GAG ATT GAT TTG GA | GTT GGG TAG ATG GCG AGA CT |
| BBLF1 | GGT GCC CTC TGG TCT CTT T | AAC AGC AAT TCA TCA TTT TCA GA |
| BBRF1 | CGT GGT GGA GGA GTT CAT AA | CATGGAGCGGATCTTCTTTAG |
| BBRF2 | CGT CCC TGA GTT CTA CAA TGC | GCG CAC ATC TCC ATA AGG TT |
| BDLF3 | GCC CCT ATA ACC ACA ACT GC | TAG TCA CAC CCG TGG AGG T |
| BGRF1 | GCA ACC ACC AAC CTC TTT CT | ATA GGC CGG ATC AAC GTA CA |
| BSRF1 | ATG GCC TTC TAT CTC CCA GA | CCA TCG TCT GCT CAT TTC TT |
| BFRF3 | GCC ATA GAC AAG AGG CAG AG | CGG AGG CTG CTA ATA GAT GA |
| BKRF2 | TGG CCT CCT TAA ATT CAC CT | CCA AAC AGG TCT TCC ACT GA |
| BLRF1 | GTC CTA AGA AAG CCG TTT GC | TCA AAG GCG AGA ATG TG |
| BMRF2 | GGC CAT TCC AGC ATC ATT | GGG TCC GTT CAC AAA CTT CT |
| BLLF3 | ACC ACA ACT GTG GAC CTA G | AGC CAG GTG GAT TTT GAG |
| BGLF4 | AAA AGA GGT TCA AGG AGA GCT AC | AGT CGT CTG CCA AGA GTT CA |
| BDLF3 ChIP 1 | TTG ATG GTG GAT GCG TTG GA | CGA CTA GCG CCC CTA TAA CC |
| BDLF3 ChIP 2 | GTG GCA GTG ATG TTC TGT GC | CGG TGA CAT TCA CTG GGA CA |
| BDLF3 ChIP 3 | GTT GTG GTT ATA GGG GCG CT | CAA CGG CTG TGA CTA CAC CA |
| BDLF3 ChIP 4 | CCG ACG GTG ATG GTG TAG TC | GAC CGC TCA TTC CCT GCA TA |
| BDLF3 ChIP 5 | GTA TGC AGG GAA TGA GCG GT | CAT TCC TAC GCC CTG CTT CA |
| BDLF3 ChIP 6 | GGT CAC CAA TGA CGG CCT AA | GGT GCC CAA ATC TAC AGG CT |
Reverse transcription-quantitative PCR.
Total RNA was isolated using a QIAshredder followed by an RNeasy Plus kit (Qiagen). The cells that were the source of isolated RNAs were the same cells that were used for protein in immunoblot experiments and for DNA replication qPCR assay. The relative transcript levels were determined with gene-specific primers, using an iScript SYBR green RT-PCR kit (Bio-Rad) or iTac SYBR green RT-PCR kit (Bio-Rad). Primer sequences are available in Table 3. Relative expression levels were calculated using the ΔΔCq (where Cq is quantification cycle) method and normalized to 18S RNA. RNA samples were assayed in triplicate.
ChIP.
Immunoprecipitation of viral DNA was performed by chemically cross-linking DNA-protein complexes formed in BZKO cells using 1% formaldehyde. The cells were incubated for 10 min at 37°C and then washed once in phosphate-buffered saline (Thermo Scientific) containing protease inhibitors (Millipore). Cells were resuspended in SDS lysis buffer (50m M Tris-HCl [pH 8.1], 1% SDS, and 10 mM EDTA) and sonicated four times on ice, for 10s each time, using a Sonifier 450 apparatus (Branson). Cell lysates were cleared by centrifugation, and the collected supernatants were diluted 10-fold in chromatin immunoprecipitation (ChIP) dilution buffer (16.7 mM Tris-HCl [pH 8.1], 0.01% SDS, 1.1% Triton X-100, 167 mM NaCl, 1.2 mM EDTA). FLAG-tagged ZEBRA-, AP-1-, and AP-1(A/S)-associated DNAs were immunoprecipitated using anti-FLAG mouse monoclonal antibody (F1805; Sigma). The immune complexes were collected on protein A/G agarose beads (Millipore). Binding to viral DNA was assessed by qPCR.
Gene classification.
We generated margins of noninferiority to classify genes as either early or late in the cell systems studied. Genes are classified as early genes if the means of the expression levels of the gene fall within two standard errors of the mean (SEM) of expression of BMRF1, a classical early gene, in cells treated with PAA relative to the level in untreated cells. Genes are classified as late genes if the means of the expression level of the gene fall within two SEM of expression BFRF3, a classical late gene, in cells treated with PAA relative to the level in untreated cells. Genes with expression levels that fall between these two cutoffs are referred to as intermediate. In HH514-16 cells treated with butyrate and PAA, the cutoff for early genes is greater than or equal to 126% relative to the expression level of BMRF1, and the cutoff for late genes is less than or equal to 53% of the level of BFRF3. In BZKO cells treated with PAA, the cutoff for early genes is greater than or equal to 39% of the level of BMRF1, and the cutoff for late genes is less than or equal to 19% of the level of BFRF3.
Statistical analysis.
Significance tests for the comparison of distributions in gene expression arrays were performed using a two-tailed Wilcoxon matched-pairs signed rank test, with an alpha of 0.05. Statistical tests comparing single gene expression to the level of a control were performed using a Student t test with Dunnett's correction for multiple comparisons; significant P values are indicated in the figure legends. Bootstrapping was used to estimate the 95% confidence interval on ChIP experiments shown in Fig. 3; binding is considered significantly different from the control vector if the confidence intervals do not overlap 1.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Institute (CA16038 and CA12055 to G.M. and CA019014 to D.P.D.) and from the National Institute of Allergy and Infectious Diseases (2T32AI007640 and 2T32AI055403 to D.E.L.).
We thank Daniel DiMaio for helpful and critical comments on the manuscript. We thank H.-J. Delecluse for providing us with BZKO cells. We thank Veronika Shabanova for assistance with the statistical analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01062-18.
REFERENCES
- 1.Grogan E, Jenson H, Countryman J, Heston L, Gradoville L, Miller G. 1987. Transfection of a rearranged viral DNA fragment, WZhet, stably converts latent Epstein-Barr viral infection to productive infection in lymphoid cells. Proc Natl Acad Sci U S A 84:1332–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Countryman J, Jenson H, Seibl R, Wolf H, Miller G. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J Virol 61:3672–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller G, El-Guindy A, Countryman J, Ye J, Gradoville L. 2007. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv Cancer Res 97:81–109. doi: 10.1016/S0065-230X(06)97004-3. [DOI] [PubMed] [Google Scholar]
- 4.Kolman JL, Taylor N, Gradoville L, Countryman J, Miller G. 1996. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol 70:1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P, Speck SH. 2003. Synergistic autoactivation of the Epstein-Barr virus immediate-early BRLF1 promoter by Rta and Zta. Virology 310:199–206. doi: 10.1016/S0042-6822(03)00145-4. [DOI] [PubMed] [Google Scholar]
- 6.Quinlivan EB, Holley-Guthrie EA, Norris M, Gutsch D, Bachenheimer SL, Kenney SC. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res 21:1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holley-Guthrie EA, Quinlivan EB, Mar EC, Kenney S. 1990. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol 64:3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse HJ. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J 19:3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J 5:3243–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rooney CM, Rowe DT, Ragot T, Farrell PJ. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J Virol 63:3109–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick JM, Lieberman PM, Hayward SD. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol 62:2274–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fixman ED, Hayward GS, Hayward SD. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol 69:2998–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schepers A, Pich D, Hammerschmidt W. 1993. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J 12:3921–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schepers A, Pich D, Hammerschmidt W. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367–376. doi: 10.1006/viro.1996.0325. [DOI] [PubMed] [Google Scholar]
- 15.El-Guindy AS, Miller G. 2004. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J Virol 78:7634–7644. doi: 10.1128/JVI.78.14.7634-7644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrell PJ, Rowe DT, Rooney CM, Kouzarides T. 1989. Epstein Barr virus BZLF 1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J 8:127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman PM, Hardwick JM, Sample J, Hayward GS, Hayward SD. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol 64:1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehman AM, Ellwood KB, Middleton BE, Carey M. 1998. Compensatory energetic relationships between upstream activators and the RNA polymerase II general transcription machinery. J Biol Chem 273:932–939. doi: 10.1074/jbc.273.2.932. [DOI] [PubMed] [Google Scholar]
- 19.Bhende PM, Seaman WT, Delecluse H-J, Kenney SC. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat Genet 36:1099–1104. doi: 10.1038/ng1424. [DOI] [PubMed] [Google Scholar]
- 20.Bhende PM, Seaman WT, Delecluse H-JJ, Kenney SC. 2005. BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue 186. J Virol 79:7338–7348. doi: 10.1128/JVI.79.12.7338-7348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouzarides T, Packham G, Cook A, Farrell PJ. 1991. The BZLF1 protein of EBV has a coiled coil dimerisation domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene 6:195–204. [PubMed] [Google Scholar]
- 22.Petosa C, Morand P, Baudin F, Moulin M, Artero J-B, Müller CW. 2006. Structural basis of lytic cycle activation by the Epstein-Barr virus ZEBRA protein. Mol Cell 21:565–572. doi: 10.1016/j.molcel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Flemington E, Speck SH. 1990. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc Natl Acad Sci U S A 87:9459–9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landschulz WH, Johnson PF, McKnight SL. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 25.Bohmann D, Bos T, Admon A, Nishimura T, Vogt P, Tjian R. 1987. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 238:1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- 26.Vogt PK, Bos TJ, Doolittle RF. 1987. Homology between the DNA-binding domain of the GCN4 regulatory protein of yeast and the carboxyl-terminal region of a protein coded for by the oncogene jun. Proc Natl Acad Sci U S A 84:3316–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glover JN, Harrison SC. 1995. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 28.Hong S, Wang D, Horton JR, Zhang X, Speck SH, Blumenthal RM, Cheng X. 2017. Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta. Nucleic Acids Res 45:2503–2515. doi: 10.1093/nar/gkx057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francis A, Gradoville L, Miller G. 1997. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol 71:3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragoczy T, Miller G. 1999. Role of the Epstein-Barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J Virol 73:9858–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heston L, El-Guindy A, Countryman J, Dela Cruz C, Delecluse H-J, Miller G. 2006. Amino acids in the basic domain of Epstein-Barr virus ZEBRA protein play distinct roles in DNA binding, activation of early lytic gene expression, and promotion of viral DNA replication. J Virol 80:9115–9133. doi: 10.1128/JVI.00909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu K-P, Heston L, Park R, Ding Z, Wang'ondu R, Delecluse H, Miller G. 2013. Latency of Epstein-Barr virus is disrupted by gain-of-function mutant cellular AP-1 proteins that preferentially bind methylated DNA. Proc Natl Acad Sci U S A 110:8176–8181. doi: 10.1073/pnas.1301577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serio TR, Kolman JL, Miller G. 1997. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J Virol 71:8726–8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djavadian R, Chiu YF, Johannsen E. 2016. An Epstein-Barr virus-encoded protein complex requires an origin of lytic replication in cis to mediate late gene transcription. PLoS Pathog 12:e1005718. doi: 10.1371/journal.ppat.1005718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honess RW, Roizman B. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol 14:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, YinKok C, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ. 2017. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aubry V, Mure F, Mariamé B, Deschamps T, Wyrwicz LS, Manet E, Gruffat H. 2014. Epstein-barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex. J Virol 88:12825–12838. doi: 10.1128/JVI.02139-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan J, Cahir-McFarland E, Zhao B, Kieff E. 2006. Virus and cell RNAs expressed during Epstein-Barr virus replication. J Virol 80:2548–2565. doi: 10.1128/JVI.80.5.2548-2565.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramasubramanyan S, Kanhere A, Osborn K, Flower K, Jenner RG, Sinclair AJ. 2012. Genome-wide analyses of Zta binding to the Epstein-Barr virus genome reveals interactions in both early and late lytic cycles and an epigenetic switch leading to an altered binding profile. J Virol 86:12494–12502. doi: 10.1128/JVI.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decaussin G, Leclerc V, Ooka T. 1995. The lytic cycle of Epstein-Barr virus in the nonproducer Raji line can be rescued by the expression of a 135-kilodalton protein encoded by the BALF2 open reading frame. J Virol 69:7309–7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng Y, Middeldorp J, Madjar J-J, Ooka T. 1997. A major DNA binding protein encoded by BALF2 open reading frame of Epstein-Barr virus (EBV) forms a complex with other EBV DNA-binding proteins: DNAase, EA-D, and DNA polymerase. Virology 239:285–295. doi: 10.1006/viro.1997.8891. [DOI] [PubMed] [Google Scholar]
- 42.Neuhierl B, Delecluse H-J. 2006. The Epstein-Barr virus BMRF1 gene is essential for lytic virus replication. J Virol 80:5078–5081. doi: 10.1128/JVI.80.10.5078-5081.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardwick JM, Tse L, Applegren N, Nicholas J, Veliuona MA. 1992. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol 66:5500–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Guindy A, Heston L, Delecluse H-J, Miller G. 2007. Phosphoacceptor site S173 in the regulatory domain of Epstein-Barr Virus ZEBRA protein is required for lytic DNA replication but not for activation of viral early genes. J Virol 81:3303–3316. doi: 10.1128/JVI.02445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Guindy A, Lopez-Giraldez F, Delecluse HJ, McKenzie J, Miller G. 2014. A locus encompassing the Epstein-Barr virus bglf4 kinase regulates expression of genes encoding viral structural proteins. PLoS Pathog 10:e1004307. doi: 10.1371/journal.ppat.1004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenzie J, Lopez-Giraldez F, Delecluse H-J, Walsh A, El-Guindy A. 2016. The Epstein-Barr virus immunoevasins BCRF1 and BPLF1 are expressed by a mechanism independent of the canonical late pre-initiation complex. PLoS Pathog 12:e1006008. doi: 10.1371/journal.ppat.1006008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruffat H, Kadjouf F, Mariame B, Manet E. 2012. The Epstein-Barr virus BcRF1 gene product is a TBP-like protein with an essential role in late Gene expression. J Virol 86:6023–6032. doi: 10.1128/JVI.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, VonDollen J, Maher MC, Johnson T, Newton W, Jager S, Shales M, Horner J, Hernandez RD, Krogan NJ, Glaunsinger BA. 2015. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell 57:349–360. doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis ZH, Hesser CR, Park J, Glaunsinger BA. 2016. Interaction between ORF24 and ORF34 in the Kaposi's sarcoma-associated herpesvirus late gene transcription factor complex is essential for viral late gene expression. J Virol 90:599–604. doi: 10.1128/JVI.02157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perng Y-C, Campbell JA, Lenschow DJ, Yu D. 2014. Human cytomegalovirus pUL79 is an elongation factor of RNA polymerase II for viral gene transcription. PLoS Pathog 10:e1004350. doi: 10.1371/journal.ppat.1004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Omoto S, Mocarski ES. 2014. Transcription of true late (γ2) cytomegalovirus genes requires UL92 function that is conserved among beta- and gammaherpesviruses. J Virol 88:120–130. doi: 10.1128/JVI.02983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arumugaswami V, Wu T-T, Martinez-Guzman D, Jia Q, Deng H, Reyes N, Sun R. 2006. ORF18 is a transfactor that is essential for late gene transcription of a gammaherpesvirus. J Virol 80:9730–9740. doi: 10.1128/JVI.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong E, Wu T-T, Reyes N, Deng H, Sun R. 2007. Murine gammaherpesvirus 68 open reading frame 24 is required for late gene expression after DNA replication. J Virol 81:6761–6764. doi: 10.1128/JVI.02726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perng Y-C, Qian Z, Fehr AR, Xuan B, Yu D. 2011. The human cytomegalovirus gene UL79 is required for the accumulation of late viral transcripts. J Virol 85:4841–4852. doi: 10.1128/JVI.02344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Omoto S, Mocarski ES. 2013. Cytomegalovirus UL91 is essential for transcription of viral true late (γ2) genes. J Virol 87:8651–8664. doi: 10.1128/JVI.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heston L, Rabson M, Brown N, Miller G. 1982. New Epstein-Barr virus variants from cellular subclones of P3J-HR1 Burkitt lymphoma. Nature 295:160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- 57.Taylor N, Countryman J, Rooney C, Katz D, Miller G. 1989. Expression of the BZLF1 latency-disrupting gene differs in standard and defective Epstein-Barr viruses. J Virol 63:1721–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El-Guindy A, Heston L, Miller G. 2010. A subset of replication proteins enhances origin recognition and lytic replication by the Epstein-Barr virus ZEBRA protein. PLoS Pathog 6:e1001054. doi: 10.1371/journal.ppat.1001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hilscher C, Vahrson W, Dittmer DP. 2005. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res 33:e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hosseinipour MC, Sweet KM, Xiong J, Namarika D, Mwafongo A, Nyirenda M, Chiwoko L, Kamwendo D, Hoffman I, Lee J, Phiri S, Vahrson W, Damania B, Dittmer DP. 2014. Viral profiling identifies multiple subtypes of Kaposi's sarcoma. mBio 5:e01633-14. doi: 10.1128/mBio.01633-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lock EF, Ziemiecke R, Marron J, Dittmer DP. 2010. Efficiency clustering for low-density microarrays and its application to QPCR. BMC Bioinformatics 11:386. doi: 10.1186/1471-2105-11-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dittmer DP, Hilscher CJ, Gulley ML, Yang EV, Chen M, Glaser R. 2008. Multiple pathways for Epstein-Barr virus episome loss from nasopharyngeal carcinoma. Int J Cancer 123:2105–2112. doi: 10.1002/ijc.23685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young LS, Lau R, Rowe M, Niedobitek G, Packham G, Shanahan F, Rowe DT, Greenspan D, Greenspan JS, Rickinson AB. 1991. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J Virol 65:2868–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pearson GR, Vroman B, Chase B, Sculley T, Hummel M, Kieff E. 1983. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J Virol 47:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Serio TR, Angeloni A, Kolman JL, Gradoville L, Sun R, Katz DA, Van Grunsven W, Middeldorp J, Miller G. 1996. Two 21-kilodalton components of the Epstein-Barr virus capsid antigen complex and their relationship to ZEBRA-associated protein p21 (ZAP21). J Virol 70:8047–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bishop JM. 1981. Enemies within: the genesis of retrovirus oncogenes. Cell 23:5–6. doi: 10.1016/0092-8674(81)90263-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.