Structural analyses highlight the constraint of virus evolution to a limited number of capsid protein folds and assembly strategies that result in a functional virion. We report the cryo-EM and cryo-electron tomography structures and the results of atomic force microscopy studies of the infectious bursal disease virus (IBDV), a double-stranded RNA virus with an icosahedral capsid. We found evidence of a new inner shell that might act as an internal scaffold during IBDV assembly. The use of an internal scaffold is reminiscent of tailed dsDNA viruses, which constitute the most successful self-replicating system on Earth. The IBDV scaffold protein is multifunctional and, after capsid maturation, is genome bound to form ribonucleoprotein complexes. IBDV encompasses numerous functional and structural characteristics of RNA and DNA viruses; we suggest that IBDV is a modern descendant of ancestral viruses and comprises different features of current viral lineages.
KEYWORDS: cryo-EM and cryo-ET, dsRNA-binding protein, IBDV, scaffolding protein, virus assembly
ABSTRACT
Infectious bursal disease virus (IBDV), a nonenveloped, double-stranded RNA (dsRNA) virus with a T=13 icosahedral capsid, has a virion assembly strategy that initiates with a precursor particle based on an internal scaffold shell similar to that of tailed double-stranded DNA (dsDNA) viruses. In IBDV-infected cells, the assembly pathway results mainly in mature virions that package four dsRNA segments, although minor viral populations ranging from zero to three dsRNA segments also form. We used cryo-electron microscopy (cryo-EM), cryo-electron tomography, and atomic force microscopy to characterize these IBDV populations. The VP3 protein was found to act as a scaffold protein by building an irregular, ∼40-Å-thick internal shell without icosahedral symmetry, which facilitates formation of a precursor particle, the procapsid. Analysis of IBDV procapsid mechanical properties indicated a VP3 layer beneath the icosahedral shell, which increased the effective capsid thickness. Whereas scaffolding proteins are discharged in tailed dsDNA viruses, VP3 is a multifunctional protein. In mature virions, VP3 is bound to the dsRNA genome, which is organized as ribonucleoprotein complexes. IBDV is an amalgam of dsRNA viral ancestors and traits from dsDNA and single-stranded RNA (ssRNA) viruses.
IMPORTANCE Structural analyses highlight the constraint of virus evolution to a limited number of capsid protein folds and assembly strategies that result in a functional virion. We report the cryo-EM and cryo-electron tomography structures and the results of atomic force microscopy studies of the infectious bursal disease virus (IBDV), a double-stranded RNA virus with an icosahedral capsid. We found evidence of a new inner shell that might act as an internal scaffold during IBDV assembly. The use of an internal scaffold is reminiscent of tailed dsDNA viruses, which constitute the most successful self-replicating system on Earth. The IBDV scaffold protein is multifunctional and, after capsid maturation, is genome bound to form ribonucleoprotein complexes. IBDV encompasses numerous functional and structural characteristics of RNA and DNA viruses; we suggest that IBDV is a modern descendant of ancestral viruses and comprises different features of current viral lineages.
INTRODUCTION
Structural information related to the capsid protein (CP) has become crucial for identifying phylogenetic relationships among viruses (1). Based on their CP fold, viruses are grouped into a limited number of structure-based lineages (2). Four lineages have been established to date; these are (i) the double-stranded DNA (dsDNA) viruses, with an upright double β-barrel CP (prototypes are phage PRD1 and adenovirus), (ii) the tailed dsDNA phages, tailed haloarchaeal viruses, and herpesviruses, which all share the Hong Kong 97 (HK97)-like CP fold, (iii) the picornavirus-like superfamily, with a single horizontal β-barrel as the CP fold (3), and (iv) the double-stranded RNA (dsRNA) or bluetongue virus (BTV)-like viruses. Most of these lineages include icosahedral viruses, although poxviruses are linked to the PRD1-like lineage (4, 5).
The similarity of the CP fold and virion architecture within the same viral lineage entails the additional resemblance of structural components related to virion assembly (6) and genome packaging (7). Minor or transient participants, such as cementing proteins, portal or genome (dsDNA)-packaging proteins, and scaffold proteins (SP), function with the CP as a structural module encoded by a fixed group of genes (8). Viruses of the HK97-like lineage, although they have variety of forms of replication, share the same virion assembly principles. HK97-like virions assemble as a precursor particle, the procapsid, which subsequently expands to a mature capsid during genome packaging (9). CP assembly requires SP or its functional equivalent; viral expansion is initiated by genome packaging and is associated in some cases with the action of a protease in the capsid interior that cleaves SP. The portal complex, which is incorporated at the onset of capsid assembly, is responsible for viral genome encapsidation and includes an ATP-driven nanomotor; it is also found in the PRD1-adenovirus lineage (10). In the picornavirus-like superfamily that includes single-stranded RNA (ssRNA) viruses, CP assembly takes place around the viral genome in a coupled assembly-packaging process.
Most dsRNA viruses share a multishelled icosahedral capsid, in which the innermost T=1 icosahedral capsid provides a platform for RNA transcription and replication (11–13). The T=1 capsid has 120 copies of a plate-like protein, in which the CP fold is the hallmark of the BTV lineage, that nucleate around genomic ssRNA copies (14, 15). The inner shell acts as a template to prime the assembly of the T=13 surrounding capsids.
We analyzed the capsid assembly of the birnavirus infectious bursal disease virus (IBDV), a nonenveloped, icosahedral dsRNA virus (16, 17). Birnaviruses are exceptions, as they lack the 120-subunit T=1 core (16, 18); instead, they have a single ∼70-nm-diameter T=13l icosahedral capsid that encapsidates a polyploid bipartite dsRNA genome (segments A and B; 3.2 and 2.8 kbp, respectively), organized as ribonucleoprotein complexes (RNP) (19, 20). The CP VP2 is synthesized as a precursor (pVP2) within a polyprotein that also includes the protease VP4 and the multifunctional protein VP3 (21, 22). In addition to its RNA-binding activity (20, 23, 24), VP3 interacts with itself (25), with the viral polymerase (4, 26, 27), and/or with pVP2 (17, 28). The proteolyzed pVP2 C terminus bears the α5-helix (17, 29), a conformational switch responsible for the inherent VP2 polymorphism. The VP3 C terminus interacts with the α5-helix to modulate the structural polymorphism of VP2; VP3 has thus been suggested to be a scaffold protein during capsid morphogenesis (17).
Whereas IBDV RNP and genome polyploidy are common among negative ssRNA viruses (30), the VP2 fold has structural similarities to other picornavirus-like CP (16), as it is notably similar to the CP of noda- and tetraviruses (31, 32). Here we found that IBDV capsid assembly has a structural framework like that of HK97-like lineage viruses. In genome-lacking virion particles, the VP3 forms a flexible inner shell that could act as a scaffold and functions as an RNA-binding protein in mature virions.
RESULTS
Cryo-ET of natural IBDV populations.
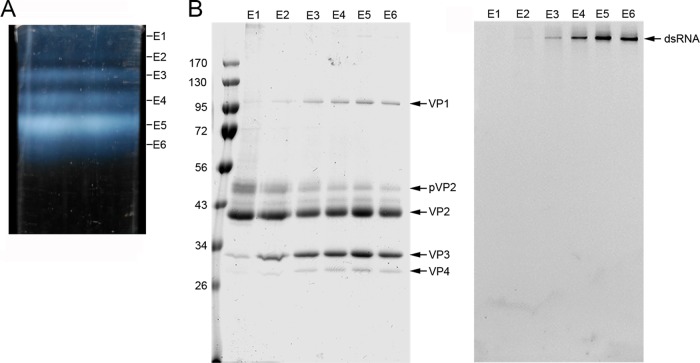
In IBDV-infected cells, up to six extracellular viral particles (E1 to E6) can be purified by ultracentrifugation in CsCl gradients (Fig. 1A), based on the copy number of packaged dsRNA segments. Virions that package four RNP (viral particles E5 and E6) are nonetheless the most abundant and make up 64% of total particles. E2 to E6 are T=13 capsids with a similar protein composition (Fig. 1B), with increasing RNP content from 1, 2, or 3 (for E2, E3, and E4, respectively) to 4 (for E5 and E6) dsRNA segments. E1 particles are empty and, in addition to T=13 capsids (∼70 nm in diameter), include T=7 (∼53 nm) and T=1 (∼26 nm) capsids, CP helical assemblies, and partially broken capsids (17). Due to this structural heterogeneity, the global stoichiometry of E1 particles differs from that of E2 to E6 particles (for example, E1 T=1 capsids are built only of VP2 trimers).
FIG 1.
Purification and biochemical analysis of IBDV natural populations. (A) A CsCl linear gradient for IBDV purification, illuminated from the bottom after centrifugation to equilibrium, containing at least six IBDV fractions (E1 to E6, from top to bottom). E1 to E6 bands contain 3, 5, 12, 16, 48, and 16% of the total virion particles, respectively. E6 has a slightly higher buoyant density, probably due to the presence of unspecific viral RNA fragments or even cellular RNA, as described in other viruses; E4 and E5 populations that contain more than 1 complete genome account for ∼80% of total particles. (B) Coomassie blue-stained (left) and ethidium bromide-stained (right) SDS-PAGE gel of E1 to E6 populations (on the same 11% gel). Molecular size markers (in 103 Da) are shown at the left; bands corresponding to proteins VP1, pVP2, VP2, VP3, and VP4 and dsRNA are indicated.
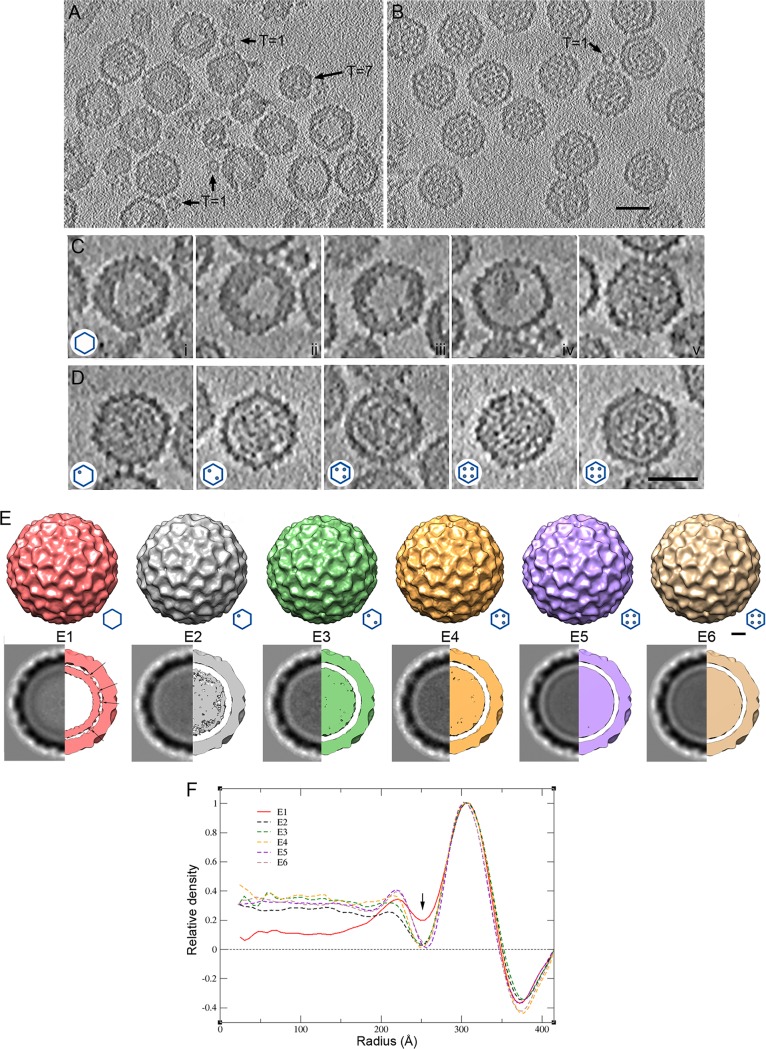
To analyze the organization of IBDV particle internal content, we used cryo-electron tomography (cryo-ET) to generate density maps of individual particles. A total of 46 tomograms were reconstructed (Fig. 2A and B; see Movies S1 and S2 in the supplemental material). Detailed inspection of central sections of individual particle tomograms (with no imposed symmetry) indicated that ∼50% of the E1 T=13 capsids showed an increased, irregular thickness of the capsid wall, with an internal density similar to the background (i.e., empty) (Fig. 2Ci to iii). Minor E1 particles were empty (14%; Fig. 2Civ) or had a homogeneous internal density (36%) that might reflect contaminating RNP-containing capsids (Fig. 2Cv). Although the capsid shell appeared to have considerable icosahedral symmetry (indicated by the hexagonal and pentagonal profiles), the underlying layer is variable and probably asymmetric (although it might be distorted during particle purification). E2 to E6 particles showed increased internal content, but no structural pattern was identified (Fig. 2D). A total of 1,517 particles were extracted, and for each IBDV population, icosahedral orientations were determined and maps were averaged (see Materials and Methods). The resolutions achieved based on a 0.5 Fourier shell correlation (FSC) threshold were ∼50 Å.
FIG 2.
Cryo-electron tomography of T=13 capsids from E1 to E6 IBDV populations. (A, B) Sections through a cryotomogram of a field of E1 (A) and E5 (B) IBDV populations. (A) At least two classes of E1 particles can be distinguished: E1 particles with an irregular thickened shell and empty central region and E1 particles with a uniform density in the capsid interior. T=1 and T=7 capsids are indicated. (B) E5 particles are uniformly full capsids. A contaminant T=1 capsid is indicated. Bar = 50 nm. (C) Gallery of sections from E1 capsid tomograms. Sections i to iii are empty E1 particles with a thickened capsid shell (50% of total); section iv is an empty E1 particle with collapsed internal material, and section v shows an E1 particle with a homogeneous internal density (∼36%). (D) Gallery of sections of E2, E3, E4, E5, and E6 IBDV capsids (from left to right, respectively). (Insets, bottom left) Hexagon diagrams show the amount of RNP (dots) packed within the particle. Bar = 50 nm. (E) (Top) Surface-shaded representations of the outer surface, viewed along an icosahedral 2-fold axis contoured at 1 σ above the mean density for the averaged E1 to E6 T=13 capsids. (Bottom) Transverse central sections of the 3DR (left) and 73-Å-thick slabs (right) contoured at 0.5 σ above the mean density to highlight internal content. Dark shading indicates a higher density. Arrows indicate contacts between the E1 T=13 capsid shell and the underlying shell. Bar, 100 Å. (F) Radial density profiles from averaged 3D maps of E1 to E6 particles. E2 to E6 profiles are practically superimposable along all radii. E1 and E2 to E6 profiles are superimposable only at the protein shell (radius, ∼260 to 350 Å). Note that the E1 profile has a less intense Fresnel ring (at a radius of ∼250 Å; arrow) due to numerous contacts between the capsid and the underlying layers (centered at a radius of ∼225 Å) and that the E1 internal volume is empty.
The outer surface of the E1 to E6 capsids showed the T=13l geometry (Fig. 2E, top and bottom). E1 to E6 T=13 particle diameters were identical, as determined from spherically averaged radial density plots of three-dimensional (3D) reconstructions (3DR) (Fig. 2F). After imposition of icosahedral symmetry, the E2 to E6 internal content was distributed homogeneously (Fig. 2F, dashed lines) and, as predicted, the density was greater than that in E1 particles (Fig. 2F, red line). The inner surface of the E2 to E6 protein shells was clearly separated from the underlying density ascribed to encapsidated RNP. The inner surface of the E1 protein shell nonetheless had extensive interactions with inner densities centered at a radius of ∼225 Å. This was reflected in the smaller Fresnel ring (∼250-Å radius) on the E1 capsid inner surface (Fig. 2F, arrow). Based on analysis of the radial density distribution of the E1 capsid, the density beneath the E1 capsid was delimited, and its average thickness was estimated to be ∼40 to 50 Å. Based on these findings, we carried out a detailed characterization of the internal density of the E1 T=13 capsid, which lacks dsRNA.
Biochemical analysis and cryo-EM of the empty E1 capsid: the IBDV procapsid-like assembly.
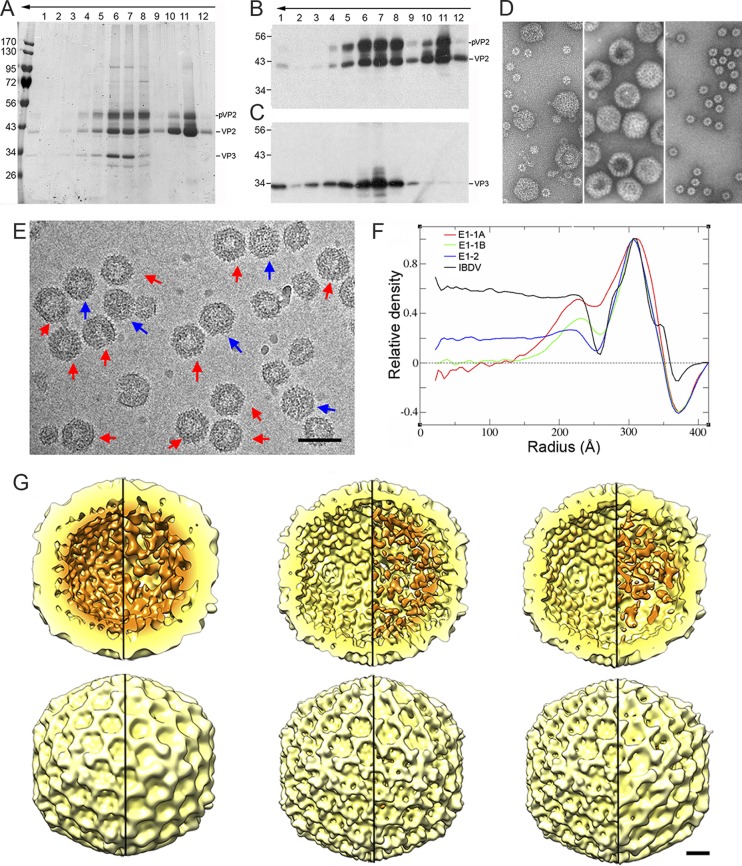
As E1 empty assemblies are heterogeneous, we optimized T=13 capsid purification by pooling several E1 fractions, which were ultracentrifuged in a linear sucrose gradient. Gradient fractions were characterized by SDS-PAGE and Western blotting (Fig. 3A to C) and by electron microscopy and negative staining (Fig. 3D). Whereas the middle fractions (fractions 6 to 8) were enriched in T=13 capsids (Fig. 3D, center), top fractions (10, 11) contained T=1 capsids (Fig. 3D, right). T=13 capsids comprised three major proteins, pVP2, VP2, and VP3, observed as ∼52-, ∼43-, and ∼34-kDa bands, respectively; T=1 capsids had mainly VP2 (Fig. 3A). Protein identity was confirmed by analysis of sucrose gradient fractions by SDS-PAGE and Western blotting using rabbit anti-VP2 (Fig. 3B) and anti-VP3 (Fig. 3C) sera. The results showed that, in the absence of any other viral protein, the inner density layer adjacent to the inner capsid shell is probably built of VP3, indicating its scaffold role in situ. We used stoichiometric analysis based on Coomassie blue-stained fractions 6 to 8 to calculate the molar ratio between the pVP2 and VP2 bands. Anti-VP2 serum recognized a minor, intermediate, processed pVP2 band between pVP2 and VP2 (33) (Fig. 3B). The E1 T=13 capsid had a 1:1 pVP2/VP2 ratio, indicating incomplete pVP2 processing. Based on these features, we suggest that E1 T=13 capsids are a by-product reminiscent of maturation and assembly intermediates or, in other words, the IBDV procapsid-like assemblies.
FIG 3.
Biochemical characterization and 3D cryo-EM reconstructions of IBDV E1 capsids. (A to C) The E1 pooled preparation was ultracentrifuged through a sucrose gradient, and 12 fractions were collected, concentrated, analyzed by SDS-PAGE, and developed by Coomassie blue staining (A) or by Western blotting using anti-VP2 (B) and anti-VP3 (C) antibodies. The direction of sedimentation was from right to left, with fraction 12 being at the gradient top. Molecular size markers (103 Da) are indicated at the left; the pVP2, VP2, and VP3 bands are indicated. (D) Electron microscopy of negatively stained E1 particles. (Left) Purified E1 particles after ultracentrifugation in a CsCl gradient; (center) T=13 capsid-enriched E1 fraction obtained after consecutive ultracentrifugation in a sucrose gradient, corresponding to central fractions (fractions 6 and 7); (right) T=1 capsid-enriched E1 fraction obtained after consecutive ultracentrifugation in a sucrose gradient, corresponding to upper fractions (fractions 10 and 11). (E) Cryo-electron micrograph of twice-purified E1 T=13 capsids from fractions 6 and 7 in panels A and D. Red arrows indicate E1-1 particles whose morphology is characterized by a thickened capsid wall with an empty central region; blue arrows indicate E1-2 particles that are homogeneously dark. Bar, 100 nm. (F) Radial density profiles from averaged 3D maps of E1-1A, E1-1B, and E1-2 particles. A radial profile calculated from a previous IBDV map is included as a control. Note that the VP3 layer (centered at an ∼225-Å radius), as well as the Fresnel ring at the inner capsid surface (at an ∼250-Å radius), decreases from E1-1A to E1-1B to E1-2. The E1-1A and E1-1B capsids are completely empty in the central region (to an ∼150-Å radius). (G) Radially color-coded inner (top) and outer (bottom) surfaces of E1-1A (left), E1-1B (center), and E1-2 capsids (right) viewed along an icosahedral 2-fold axis (yellow, T=13 shell; orange, VP3 density). Maps are calculated with (left) and without (right) icosahedral symmetry and are contoured at 1.25 σ above the mean density. Bar, 100 Å.
We analyzed E1 T=13 particles by cryo-electron microscopy (cryo-EM). In accordance with the findings of our previous cryo-ET analysis, visual inspection confirmed two classes of particles, E1-1 capsids (5,565 images), characterized by an empty central region and irregular capsid wall thickening (Fig. 3E, red arrows), and E1-2 capsids (6,565 images) with a uniform internal density (Fig. 3E, blue arrows). Particles were classified by two-dimensional (2D) and 3D analysis; based on the organization level of the VP3 inner layer, we defined two E1-1 subclasses, the E1-1A capsid (2,095 images) with many connections between the VP3 shell and the T=13 layer inner surface (Fig. 3F and G, left) and E1-1B (2,027 images) (Fig. 3F and G, middle). The E1-2 final map (2,377 images) had a VP3-related internal density distributed uniformly in its interior (Fig. 3F and G, right). Radial density profiles of the E1-1A map showed that the VP3 layer underlying the VP2 T=13 capsid is, on average, ∼4 nm thick (Fig. 3F). The minor density of the E1-1B and E1-2 maps might result from the low copy number of bound VP3 and/or from pVP2 unprocessed C-terminal ends. The three E1 capsid profiles, based on the VP3 internal organization, were reproduced computationally with the 3D maps (Fig. 4).
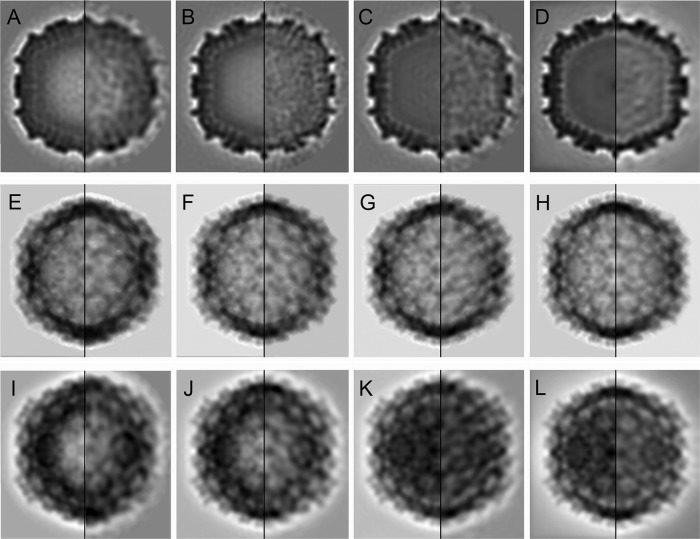
FIG 4.
Structural comparison of T=13 capsids from an E1 natural population. (A to D) Transverse central sections from the 3D reconstructions of E1-1A (A), E1-1B (B), E1-2 (C), and IBDV (D) particles, viewed along an icosahedral 2-fold axis and filtered at a 30-Å resolution (with icosahedral symmetry; left) or at a 40-Å resolution (without imposing icosahedral symmetry; right). (E to H) Two-dimensional reprojected views of the 3D reconstructions viewed along a 2-fold icosahedral axis of E1-1A (E), E1-1B (F), E1-2 (G), and IBDV (H) particles after computationally removing their internal content (with and without icosahedral symmetry; left and right, respectively). Reprojections are basically identical. (I to L) Two-dimensional reprojected views of the 3D reconstructions viewed along a 2-fold icosahedral axis of E1-1A (I), E1-1B (J), E1-2 (K), and IBDV (L) particles without removing the internal content (with and without icosahedral symmetry; left and right, respectively).
Location of (p)VP2 and VP3 in E1 and E5 T=13 capsids by immunoelectron microscopy.
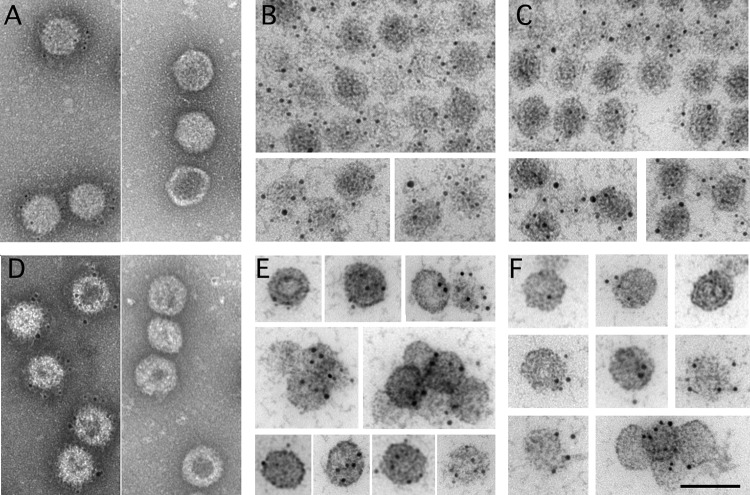
The location of pVP2/VP2 and VP3 on E1 T=13 particles, as well as E5 T=13 capsids, was also determined using immunogold labeling. Labeling anti-VP2 antiserum was clearly visible around the T=13 icosahedral structures (Fig. 5A and D, left), which blurred the peripheral edges of IBDV particles (compare Fig. 5A and D and Fig. 3D). This is due to the abundant immune complexes that surround these particles. Tests of preimmune antiserum (internal controls) showed no labeling. In contrast to VP2, when the same experiment was performed with anti-VP3 antiserum (Fig. 5A and D, right), no labeling was observed. This result suggests that VP3 is not present or is inaccessible in these particles. To test these hypotheses, the fractions were embedded in Lowicryl HM20 resin; in thin sections of this resin, capsid antigenicity is well preserved and internal inaccessible epitopes (inaccessible from the outside) would be exposed. Transverse sections of capsids were observed after labeling with polyclonal anti-VP2 or -VP3 antibody. Anti-VP2-labeled spherical/icosahedral particles (Fig. 5B and E) were not labeled by the preimmune serum. Anti-VP3 (Fig. 5C and F) showed positive VP3 labeling in the E1 and E5 T=13 capsids. These data strongly suggest that VP3 is not exposed on the E1 T=13 capsid surface and that it builds the innermost shell beneath the pVP2/VP2 layer, thus becoming accessible to antibodies only in sectioned particles.
FIG 5.
Immunogold labeling of pVP2/VP2 and VP3 on the outer and inner surfaces of E1 and E5 T=13 capsids. (A) E5 T=13 capsids were attached to coated grids and incubated with polyclonal rabbit anti-VP2 (left) or anti-VP3 (right) antiserum. Bound antibody was detected with goat anti-rabbit immunoglobulin conjugated to 5-nm colloidal gold particles. (B, C) Immunogold localization of pVP2/VP2 (B) and VP3 (C) of resin-embedded E5 T=13 capsids. (D) E1 T=13 capsids were attached to coated grids and incubated with polyclonal rabbit anti-VP2 (left) or anti-VP3 (right) antiserum. Bound antibody was detected as described in the legend to panel A. (E, F) Immunogold localization of pVP2/VP2 (E) and VP3 (F) of resin-embedded E1 T=13 capsids. The locations of pVP2/VP2 (B and E) and VP3 (C and F) with polyclonal anti-VP2 and -VP3 antisera and a 5-nm colloidal gold conjugate on LR-WHITE sections are shown. Cytochemical controls with the preimmune sera for VP2 and VP3 showed no label. Bar, 100 nm.
Mechanical properties of IBDV T=13 procapsid and T=1 subviral particle.
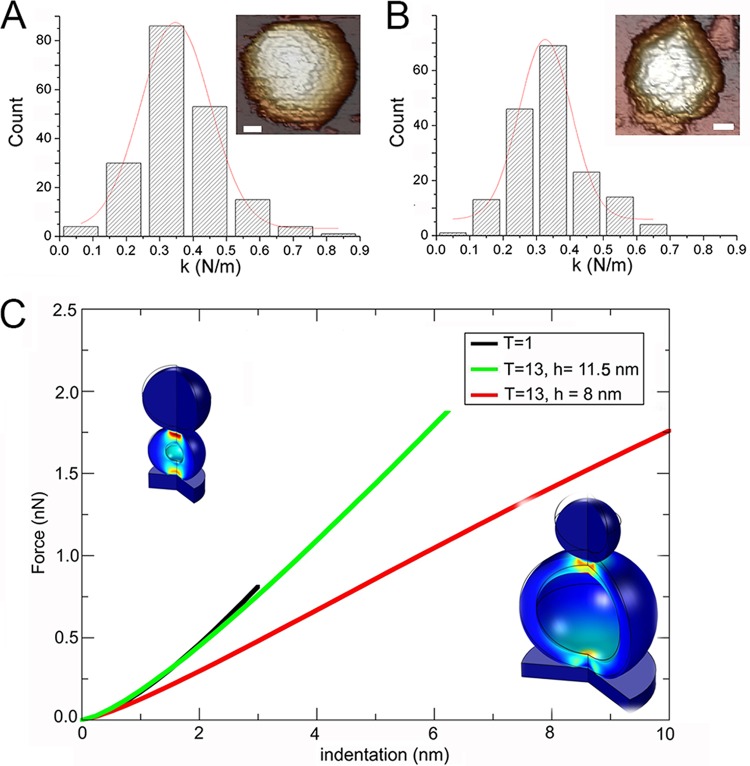
Additional evidence for the presence of VP3 as a genuine concentric layer beneath the T=13 capsid arose from atomic force microscopy (AFM) experiments. We used AFM nanoindentation to measure the rigidity of empty T=13 and T=1 capsids. T=1 capsids were analyzed twice, from the E1 fraction (T=1 and T=13 copurify in CsCl gradients and, based on their heights, are easily distinguished) and from VP2 (441 residues) expression in the absence of other IBDV components in a recombinant baculovirus system (18). The results were very similar in both cases, which ruled out any relevant structural/mechanical difference. We performed 193 indentations for 33 E1 T=13 particles and 170 indentations for 29 T=1 particles. The spring constants, k, of the E1 T=13 and T=1 capsids were calculated from their slope histograms (Fig. 6A and B). Although the capsid size was quite different, the rigidity was similar, as kE1-T=13 was equal to 0.347 ± 0.104 N/m and kT=1 was equal to 0.327 ± 0.085 N/m (for E1 T=1 capsids, kE1-T=1 was equal to 0.309 ± 0.127 N/m from 73 indentations for 13 T=1 capsids). The relatively broad distribution of rigidity slopes from indentation experiments with individual intact E1 T=13 capsids could account for their structural variability. This result would not be anticipated if both capsids were built exclusively of a similar VP2 protein layer since, according to continuum elasticity theory, the spring constant is inversely proportional to the capsid radius.
FIG 6.
Mechanical rigidity of E1 T=13 and T=1 capsids and finite element simulations that mimic AFM indentations. (A, B) Histograms of slopes of the indentation curves for E1 T=13 (A) and T=1 capsids (B), showing rigidity values (spring constant, k) for individual particles after nanoindentation. The k value for each population was calculated by Gaussian fits, with kE1-T=13 being equal to 0.347 ± 0.104 N/m (A) and kT=1 being equal to 0.327 ± 0.085 N/m (B). For E1 T=1 capsids, kE1-T=1 was equal to 0.309 ± 0.127 N/m. (Insets, top right) AFM images of individual T=13 (bar, 200 Å) and T=1 (bar, 100 Å) particles are shown. (C) Force-indentation curves obtained by FE simulation to mimic AFM experiments for IBDV T=13 and T=1 capsids. Values are shown for the T=1 capsid (black), the T=13 capsid using r equal to 35 nm and an 8-nm-thick layer (red), and the T=13 capsid with the thickness increased to 11.5 nm (green) to match the spring constant measured in the experiments.
To resolve this apparent paradox, we implemented finite element (FE) simulations of capsid deformation by mimicking AFM nanoindentation experiments (see Materials and Methods) (34, 35). In the FE simulations, the E1 T=13 and T=1 capsids were modeled as homogeneous spherical shells with an external radius, r, and thickness, h, derived from cryo-EM and X-ray data. The external radius and thickness of the spherical shell representing the empty T=1 capsid were 13 nm and 8 nm, respectively (Fig. 6C, left inset). A value of Young's modulus (E) of 120 MPa was selected to yield a spring constant in the simulations the same as that measured experimentally for the T=1 capsid. The E1 T=13 capsid (Fig. 6C, right inset) was modeled initially as a spherical shell with r equal to 35 nm and the same Young's modulus (E = 120 MPa) and thickness (h = 8 nm) as the T=1 capsid, to ascertain the spring constant of a T=13 particle built exclusively of a VP2 protein layer. The indentation curves for the 8-nm-thick T=1 and T=13 capsids are shown in Fig. 6C (T=1 capsid, black line; T=13 capsid, red line). At difference from the findings from the AFM experiments, the spring constant of a T=13 shell with the same thickness as the T=1 particle was notably smaller. These results suggested an additional internal protein layer that increases capsid rigidity. FE simulations for the E1 T=13 capsid were repeated using the same external radius with various thicknesses. A total thickness of 11.5 nm was needed to yield an E1 T=13 capsid with a spring constant that matched that of the empty T=1 capsid (Fig. 6C, green line). Assuming the same Young's modulus for T=1 and T=13 capsids, their similar experimental spring constants are thus due to an ∼3.5-nm-thick protein layer below the VP2 layer in the E1 T=13 capsid, which supports our inferences from cryo-ET and cryo-EM analyses of the E1 T=13 capsids.
DISCUSSION
The virus genetic program is directed to produce multiple copies of infectious particles, although the ratio of defective particles during the replication cycle varies greatly (36). For IBDV, we showed a direct correlation between the number of packaged RNP and particle infectivity; 96% of total infectivity is thus assigned to the E5 and E6 virus population (64% of total particles, with 4 RNP/particle) (19). The E1 to E4 populations are probably incorrectly assembled intermediates or abortive assemblies, a result of assembly pathway failure at longer postinfection times, when the host is severely impaired.
Our cryo-ET and cryo-EM analyses of IBDV populations demonstrated that E1 capsids bear a somewhat smaller, less ordered inner shell closely connected to the T=13 capsid inner surface. Our studies suggest that the empty E1 capsid is an assembly that resembles the IBDV procapsid. This is evidence that the RNA-binding VP3 protein can act as an SP by building a micellar-like layer in the IBDV procapsid-like E1 particles. In addition to recruiting CP during assembly and directing fidelity and the correct capsid size, the VP3 layer provides a defined set of mechanical properties to the capsid shell, as shown by our AFM indentations of empty T=13 and T=1 capsids. The additional internal VP3 layer increases procapsid-like rigidity; that is, it reduces excessive T=13 procapsid flexibility that might compromise productive virus capsid assembly.
At initial capsid assembly stages, VP3-pVP2 contacts are necessary, which could be due to the instability of the immature T=13 capsid. The relatively constant VP3 copy number (∼450 VP3 copies from the E2 to E6 populations [19]), which is independent of the number of packaged dsRNA segments, suggests that VP3 incorporation is determined by its role as an SP. In T=13 capsids purified from the E1 fraction (Fig. 3A, fractions 6 to 8), pVP2 and VP2 are present at a 1:1 ratio, which indicates that these capsids can be considered procapsids that require pVP2 maturation to become fully icosahedral capsids, such as those of infectious IBDV virions. Based on IBDV-related assemblies generated using various baculovirus-based expression systems, we showed that electrostatic interactions between pVP2 and VP3 are important in the assembly of T=13 virus-like particles; acid residues at the VP3 C terminus interact with the basic residues of the pVP2 amphipathic α5-helix (28).
The classification methods for the structural analysis of E1 particles resolved three particle classes probably related to assembly and/or maturation phases. The double-layered E1-1A particle shares many similarities with the herpesvirus procapsid, particularly with its B capsid (37), a dead-end side particle in which the SP layer remains at the T=16 capsid interior with several proteases (38). In addition to the mature VP2 B domain (built of N- and C-terminal α-helices) facing the capsid interior, the E1 procapsid-like inner surface is lined with the 71-residue pVP2 C-terminal domain that includes at least four α-helices and the disordered regions of the C-terminal-most residues (29, 39). These loops, together with the high α-helical content, might be a suitable interface for SP binding to initiate procapsid assembly. The labile E1-1A capsid matures to the E1-1B capsid and CP-CP connections become more robust, whereas CP-SP connections tend to disappear. In the E1-2 capsid, most VP3 is completely disengaged from the capsid and distributed homogeneously in the capsid interior; in vivo, VP3 would be bound to the viral genome.
These dynamic assembly states might coexist in the same particle. Once virions are nearly or recently assembled as provirions, VP3 probably stops its scaffold function when the pVP2 C-terminal region is proteolyzed during virus maturation by VP4, by the host protease puromycin-sensitive aminopeptidase (purSA) (33), or by VP2 itself (40). VP3 then becomes an RNA-binding protein. When VP3 is bound to the viral dsRNA genome in mature virions, it also mediates capsid stability by acquiring a conformation able to interact with the mature VP2 (without helix α5), which involves a new set of dynamic interactions between VP2 and VP3 (41). VP3 thus is able to interact with VP2 and with the dsRNA in the mature E5 particle. The battery of proteases that acts mainly on the pVP2 shell might trigger major conformational changes that transform the unstable capsid into a robust infectious particle (6).
As IBDV lacks an ATP-driven pump for genome translocation (and there is no candidate protein), the genome has to be incorporated early in the nascent capsid. Considering that VP3 might function as an adaptor molecule between the capsid shell and the viral genome, dsRNA genomic segments or their corresponding ssRNA molecules and their associated VP3 molecules can act together as a scaffold for the CP; i.e., capsid assembly and packaging of the genome are concomitant, as in many ssRNA viruses (42). VP3 might bind to the viral ssRNA of IBDV A and B segments, as in other dsRNA viruses (14, 15). Although E1 particles possibly represent unsuccessful viral assemblies (VP3 only as an SP without bound RNA), they allowed visualization of the VP3 layer as a scaffold bound to the capsid shell. We do not rule out the possibility that genome encapsidation could be mediated by VP3-VP1 interactions (43), as a VP1 fraction is covalently bound to dsRNA (20). The multiple VP3 binding activities during capsid assembly and genome packaging might then be either sequential or simultaneous.
In IBDV and birnaviruses in general, the CP is encoded as a precursor within the polyprotein. The genomic module for IBDV capsid assembly includes the CP, the VP4 protease (21, 22), and the VP3 SP (17, 28). This organization is similar to that of many HK97-like viruses (except that the portal complex is lacking), but VP2 shares the fold of the picornavirus-like CP rather than the HK97-like CP. The picornavirus-like positive ssRNA viruses do not require an SP, as their capsids are much simpler than the T=13 of IBDV (3). IBDV thus resembles a hybrid of picornavirus-like (CP fold) and HK97-like (SP and procapsid) viruses. HK97- and PRD1-like viruses have an empty procapsid and a packaging ATPase (7, 10, 44) and are more closely related to each other than to IBDV. Additional data are needed to discern whether SP-mediated assembly is indicative of an evolutionary link or is a consequence of mechanical and structural constraints.
Like the SP of many HK97-like viruses, VP3 has a pivotal function in the IBDV capsid assembly pathway. The herpesvirus- or HK97-like SP are nonetheless expelled intact or degraded from the procapsid during maturation (45, 46). Once its role in procapsid assembly has concluded and the pVP2 interaction is no longer needed, the multifunctional VP3 protein switches to an RNA-binding function, in which VP3 might interact with itself and/or activate the VP1 polymerase, thus extending its central role in the virus life cycle. In the mature virion, VP3 is bound to dsRNA, and RNP reinforces virus rigidity through a set of dynamic interactions that differ from those that take place during capsid assembly (41).
The multifunctionality of viral proteins is becoming apparent (47). The oligomeric state and the existence of intrinsically disordered regions of VP3 might be the key to determining the molecular basis of its ability to generate a variety of functions.
MATERIALS AND METHODS
Virion purification.
IBDV virions were purified from QM7 quail muscle cells (American Type Culture Collection [ATCC] CRL-1962) infected with the IBDV Soroa strain by ultracentrifugation on a 25% (wt/wt) sucrose cushion and a linear 25 to 50% sucrose gradient, as described previously (17). Alternatively, after pelleting through the sucrose cushion, particles were centrifuged through a CsCl equilibrium gradient. Six virus-containing bands (E1 to E6) were visible and were collected separately by side puncturing (19).
Purification of RNP and dsRNA from IBDV virions.
RNP complexes were purified from viral particles disrupted by dialysis against Tris-EDTA buffer (5 mM Tris-HCl, pH 8.0, 5 mM EDTA) (72 h, 20°C). The extract was concentrated in 0.5 ml 10 K Amicon ultracentrifuge tubes (Merck) and centrifuged twice on a glycerol step gradient, as reported previously (20). The middle fractions were dialyzed against Tris-EDTA buffer and analyzed by electron microscopy (EM) and immunoelectron microscopy using anti-VP3 antiserum. dsRNA segments A and B were purified as described previously (19).
SDS-PAGE and Western blotting.
Concentrated gradient fractions (2 to 5 μl) were added to Laemmli sample buffer to a 1× final concentration (62.5 mM Tris-HCl, 2% SDS, 5% glycerol, 0.012% bromophenol blue, 2 mM dithiothreitol, pH 6.8), heated (3 min, 100°C), and resolved in 6 to 11% or 11% polyacrylamide gels, developed by Coomassie blue staining, washed with distilled water, and visualized with ethidium bromide. Alternatively, electrophoresis was followed by Western blotting analyses using anti-VP2 or -VP3 antibodies.
Cryo-EM and image processing.
Purified E1 particles (5 μl) were applied to Quantifoil R 2/2 holey grids, blotted, and plunged into liquid ethane. Samples were observed in a Tecnai G2 electron microscope. Images were recorded under low-dose conditions with an FEI Eagle charge-coupled-device (CCD) camera at a detector magnification of ×69,444 (2.16-Å-per-pixel sampling rate). General image processing operations were performed using the Xmipp (http://xmipp.cnb.csic.es/ [48]) and RELION (49) software packages integrated into the SCIPION platform (50) and with Bsoft (https://lsbr.niams.nih.gov/bsoft/ [51]). Graphics were produced by UCSF Chimera (http://www.cgl.ucsf.edu/chimera/ [52]). The Xmipp automatic picking routine was used to select 6,909 and 5,241 individual particle images of the E1-1 and E1-2 capsids, respectively. E1-1 particles have a ring-like morphology with an empty central region, whereas E1-2 particles are homogeneously dark. A 0.7- to 5-μm defocus range was determined for each image with CTFfind4 (53). Images were down-sampled to a factor of 2, with a final sampling ratio of 4.32 Å/pixel. 2D classification with RELION, used to discard low-quality particles, was run for 25 iterations with 60 classes, an angular sampling of 5°, and a regularization parameter of T=1. 3D classification was run for 40 iterations with four classes for each image set of E1-1 and E1-2 capsids, starting with an angular sampling of 3.7° that was decreased sequentially by 0.2° and a regularization parameter of T=2. The published IBDV structure (31), low-pass filtered to 35 Å, was used as the initial reference for 3D classifications and refinements. Two classes were selected for E1-1 capsids, which contained 2,095 (E1-1A) and 2,027 (E1-1B) particles. The best two classes for E1-2 capsids were indistinguishable and were combined in a single map that contained 2,377 particles. Map resolutions were estimated from two independent hal data sets using the 0.3 criterion of the Fourier shell correlation (FSC), and the values for the E1-1A, E1-1B, and E1-2 maps were 28.2, 20.6, and 23.8 Å, respectively. Spherically averaged radial density profiles were calculated using Xmipp, normalized, and scaled to match the fit between the cryo-EM maps. The UCSF Chimera fitting routine was used to dock the IBDV T=13 asymmetric unit (PDB accession number 1WCE [16]) in the cryo-EM map, and icosahedral symmetry was applied. The mask generated was used to segment the E1-1A capsid.
Cryo-ET and image processing.
Samples of the E1 to E6 particles were mixed with 10-nm colloidal gold particles (Aurion) and vitrified as described above. Tomographic tilt series were recorded in a Tecnai-12 (FEI) electron microscope operating at 120 kV with a LaB6 source and equipped with an energy filter (GIF 2002; Gatan). Images were recorded on a 2,048- by 2,048-pixel CCD camera (Gatan) at a ×53,600 magnification (5.6 Å/pixel) with a ∼4-μm underfocus, using the serialEM package (54). Tilt series covering the range ±70° were collected in 2° increments with a total dose of ∼75 electrons/Å2 per series.
Data were preprocessed and aligned using IMOD software (55), with gold particles being used as fiducial markers. The final aligned tilt series was normalized and reconstructed using weighted back-projection algorithms implemented in IMOD; 11, 10, 3, 4, 10, and 8 tomograms of the E1, E2, E3, E4, E5, and E6 viral populations, respectively, were reconstructed. Individual virus particles were manually selected, extracted, and denoised by 100 iterations of anisotropic nonlinear diffusion (56) using Bsoft. A total of 247, 119, 95, 124, 475, and 457 particles were selected from the E1, E2, E3, E4, E5, and E6 populations, respectively. The extracted viruses were aligned to a scale and low-pass filtered to a 35-Å 3D map of IBDV (31) using maximum likelihood procedures for tomography. A new density map was calculated and iteratively refined. The resolution of the averaged E1 to E6 maps was in the 47- to 54-Å range. Spherically averaged radial density profiles of E1 to E6 capsids were calculated with Xmipp, normalized, and scaled to match the fit between the cryo-ET maps.
Immunogold labeling of E1 and E5 T=13 capsids.
(p)VP2 and VP3 were localized and identified by immunogold labeling on the outer or inner surfaces of E1 and E5 capsids directly from sucrose-purified (E1 capsids) or CsCl-purified (E5 capsids) material or from material embedded in Lowicryl HM20 resin. Fractions containing the E1 and E5 capsids were pooled and centrifuged (240,000 × g, 2 h, 4°C), resuspended in 2% low-melting-point agarose in PES buffer [25 mM piperazine-N,N-bis(2-ethanesulfonic acid) (pH 6.2), 150 mM NaCl, and 20 mM CaCl2], and subjected to mild fixation with a 4% paraformaldehyde solution containing 0.1% glutaraldehyde in phosphate-buffered saline (PBS) (30 min, 4°C). Fixed capsids were washed in buffered glycerol and quickly immersed in liquid ethane in the Leica EM CPC unit. The capsids were freeze substituted with 0.5% uranyl acetate in methanol (54 h, −90°C) and then embedded in Lowicryl HM20 resin (48 h at −40°C and 48 h at 20°C) in the Leica EM AFS2 unit. Thin sections (60 to 70 nm) of particles were obtained with a Leica EM UC6 ultramicrotome and stained with saturated uranyl acetate and lead citrate. Purified E1 and E5 capsids were applied to carbon-coated grids, which were then blocked with 1% bovine serum albumin (BSA)–PBS (30 min). Polyclonal rabbit anti-VP2 and -VP3 antisera, diluted 1/50 and 1/10 in 1% BSA–PBS, respectively, were incubated (60 min), followed by three washes with PBS, and incubated with 1% BSA–PBS (15 min). Goat anti-rabbit immunoglobulin antiserum conjugated with 5-nm gold particles (1/50; BBI Solutions) was added to the grid (45 min, room temperature). To remove nonspecific binding, the grids were washed three times with PBS followed by four washes with water. The grids were then negatively stained with 2% (wt/vol) aqueous uranyl acetate. Immunogold was detected on ultrathin sections of E1 and E5 capsids at room temperature using rabbit polyclonal anti-VP2 and -VP3 antisera essentially as described above.
Atomic force microscopy.
AFM experiments were as described previously (41). Surface-attached capsids were imaged under physiological conditions using the jumping mode. We used 0.05-N/m-force-constant Olympus silicon nitride cantilevers. Data were analyzed with WSxM software (57). The capsid position was determined in low-resolution images (128 by 128 pixels) at the maximal scanning force (<100 pN). The applied force was calibrated by single force-versus-Z piezo displacement curves (FZ) on a highly oriented pyrolytic graphite surface (HOPG) next to the capsid. After individual details about the capsids were resolved, indentation measurements were performed to quantitate the particle elastic response. A series of three to five successive FZ curves was obtained, and measurements were performed on ∼30 particles per population.
FE simulations of IBDV capsids.
FE simulations of AFM indentation were done using the COMSOL Multiphysics (version 4.3) program (Comsol, Stockholm, Sweden). In all cases, the capsid wall was considered a spherical shell made of a homogeneous material with a Young's modulus (E) equal to 120 MPa and a Poisson ratio (ν) equal to 0.3 (a standard value for protein-like materials). This model capsid was placed on a hard, flat substrate and indented by a hard spherical object with a radius (Rin) equal to 15 nm to mimic the AFM tip. The system was simulated using a 2D axisymmetric model meshed with over 3,000 to 6,000 triangular elements. Contacts were implemented between the shell and the tip as well as the supporting surface during indentation with a contact-penalty stiffness method based on the manufacturer's manual. A parametric, nonlinear solver was used to simulate the stepwise lowering of the tip onto the capsid. The spring constant was derived from a linear fit of force versus indentation for small indentations. For the T=1 capsid, the spring constant was measured as the slope of the force-indentation curve at 2 nm, and for the empty T=13 E1 capsid, the spring constant was measured as the slope of the force-indentation curve at 4 nm. These values were chosen to mimic the typical indentations used in the experiments.
Data availability.
The E1-1A, E1-1B, and E1-2 cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB; http://www.ebi.ac.uk/pdbe/emdb) with accession numbers emd-3507, emd-3509, and emd-3510, respectively. E1 to E6 cryo-ET averaged maps have been deposited in EMDB with accession numbers emd-3511 to emd-3516, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pedro J. de Pablo for technical support and stimulating discussions for AFM experiments and Santiago Casado for technical assistance with AFM. We also thank the CNB-CSIC Electron Microscopy Service for technical assistance with immunoelectron microscopy and Catherine Mark for editorial assistance.
C.P.M. was a Ph.D. fellow of the La Caixa Foundation International Fellowship Program (La Caixa/CNB). This work was supported by grants from the Spanish Ministry of Economy and Competitivity (BFU2013-43149-R to D.L., FIS2015-67837-P to D.R., MAT2015-66605-P to J.M., BFU2014-54181 to J.L.C., and BFU2017-88736-R to J.R.C.), the Generalitat de Catalunya (AGAUR 2014-SGR-922 to D.R.), the Comunidad Autónoma de Madrid (S2013/MIT-2850 to J.L.C. and S2013/MIT-2807 to J.R.C.), and the NIH Intramural Research Program of the Center for Information Technology (to B.L.T.) and of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (to A.C.S.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00968-18.
REFERENCES
- 1.Abrescia NG, Bamford DH, Grimes JM, Stuart DI. 2012. Structure unifies the viral universe. Annu Rev Biochem 81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 2.Bamford DH, Grimes JM, Stuart DI. 2005. What does structure tell us about virus evolution? Curr Opin Struct Biol 15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Rossmann M, Johnson J. 1989. Icosahedral RNA virus structure. Annu Rev Biochem 58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 4.Bahar MW, Graham SC, Chen RA, Cooray S, Smith GL, Stuart DI, Grimes JM. 2011. How vaccinia virus has evolved to subvert the host immune response. J Struct Biol 175:127–134. doi: 10.1016/j.jsb.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyun JK, Coulibaly F, Turner AP, Baker EN, Mercer AA, Mitra AK. 2007. The structure of a putative scaffolding protein of immature poxvirus particles as determined by electron microscopy suggests similarity with capsid proteins of large icosahedral DNA viruses. J Virol 81:11075–11083. doi: 10.1128/JVI.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol 15:227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feiss M, Rao VB. 2012. The bacteriophage DNA packaging machine. Adv Exp Med Biol 726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- 8.Krupovic M, Bamford DH. 2011. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol 1:118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Veesler D, Johnson JE. 2012. Virus maturation. Annu Rev Biophys 41:473–496. doi: 10.1146/annurev-biophys-042910-155407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong C, Oksanen HM, Liu X, Jakana J, Bamford DH, Chiu W. 2014. A structural model of the genome packaging process in a membrane-containing double stranded DNA virus. PLoS Biol 12:e1002024. doi: 10.1371/journal.pbio.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton JT. 2008. Segmented double-stranded RNA viruses: structure and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 12.Zhang X, Ding K, Yu X, Chang W, Sun J, Zhou ZH. 2015. In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature 527:531–534. doi: 10.1038/nature15767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Cheng L. 2015. Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science 349:1347–1350. doi: 10.1126/science.aaa4938. [DOI] [PubMed] [Google Scholar]
- 14.Sung PY, Roy P. 2014. Sequential packaging of RNA genomic segments during the assembly of bluetongue virus. Nucleic Acids Res 42:13824–13838. doi: 10.1093/nar/gku1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajardo T Jr, Sung PY, Roy P. 2015. Disruption of specific RNA-RNA interactions in a double-stranded RNA virus inhibits genome packaging and virus infectivity. PLoS Pathog 11:e1005321. doi: 10.1371/journal.ppat.1005321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Saugar I, Luque D, Ona A, Rodriguez JF, Carrascosa JL, Trus BL, Caston JR. 2005. Structural polymorphism of the major capsid protein of a double-stranded RNA virus: an amphipathic alpha helix as a molecular switch. Structure 13:1007–1017. doi: 10.1016/j.str.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Caston JR, Martinez-Torrecuadrada JL, Maraver A, Lombardo E, Rodriguez JF, Casal JI, Carrascosa JL. 2001. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J Virol 75:10815–10828. doi: 10.1128/JVI.75.22.10815-10828.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luque D, Rivas G, Alfonso C, Carrascosa JL, Rodriguez JF, Caston JR. 2009. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci U S A 106:2148–2152. doi: 10.1073/pnas.0808498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luque D, Saugar I, Rejas MT, Carrascosa JL, Rodriguez JF, Caston JR. 2009. Infectious bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J Mol Biol 386:891–901. doi: 10.1016/j.jmb.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birghan C, Mundt E, Gorbalenya AE. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the Ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J 19:114–123. doi: 10.1093/emboj/19.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldman AR, Lee J, Delmas B, Paetzel M. 2006. Crystal structure of a novel viral protease with a serine/lysine catalytic dyad mechanism. J Mol Biol 358:1378–1389. doi: 10.1016/j.jmb.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 23.Kochan G, Gonzalez D, Rodriguez JF. 2003. Characterization of the RNA-binding activity of VP3, a major structural protein of infectious bursal disease virus. Arch Virol 148:723–744. doi: 10.1007/s00705-002-0949-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Cleef KW, van Mierlo JT, Miesen P, Overheul GJ, Fros JJ, Schuster S, Marklewitz M, Pijlman GP, Junglen S, van Rij RP. 2014. Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res 42:8732–8744. doi: 10.1093/nar/gku528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casanas A, Navarro A, Ferrer-Orta C, Gonzalez D, Rodriguez JF, Verdaguer N. 2008. Structural insights into the multifunctional protein VP3 of birnaviruses. Structure 16:29–37. doi: 10.1016/j.str.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Garriga D, Navarro A, Querol-Audi J, Abaitua F, Rodriguez JF, Verdaguer N. 2007. Activation mechanism of a noncanonical RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 104:20540–20545. doi: 10.1073/pnas.0704447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrero D, Garriga D, Navarro A, Rodriguez JF, Verdaguer N. 2015. Infectious bursal disease virus VP3 upregulates VP1-mediated RNA-dependent RNA replication. J Virol 89:11165–11168. doi: 10.1128/JVI.00218-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saugar I, Irigoyen N, Luque D, Carrascosa JL, Rodriguez JF, Caston JR. 2010. Electrostatic interactions between capsid and scaffolding proteins mediate the structural polymorphism of a double-stranded RNA virus. J Biol Chem 285:3643–3650. doi: 10.1074/jbc.M109.075994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galloux M, Libersou S, Alves ID, Marquant R, Salgado GF, Rezaei H, Lepault J, Delmas B, Bouaziz S, Morellet N. 2010. NMR structure of a viral peptide inserted in artificial membranes: a view on the early steps of the birnavirus entry process. J Biol Chem 285:19409–19421. doi: 10.1074/jbc.M109.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortin J, Martin-Benito J. 2015. The RNA synthesis machinery of negative-stranded RNA viruses. Virology 479-480:532–544. doi: 10.1016/j.virol.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Luque D, Saugar I, Rodriguez JF, Verdaguer N, Garriga D, Martin CS, Velazquez-Muriel JA, Trus BL, Carrascosa JL, Caston JR. 2007. Infectious bursal disease virus capsid assembly and maturation by structural rearrangements of a transient molecular switch. J Virol 81:6869–6878. doi: 10.1128/JVI.00077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahlquist P. 2005. Virus evolution: fitting lifestyles to a T. Curr Biol 15:R465–R467. doi: 10.1016/j.cub.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 33.Irigoyen N, Caston JR, Rodriguez JF. 2012. Host proteolytic activity is necessary for infectious bursal disease virus capsid protein assembly. J Biol Chem 287:24473–24482. doi: 10.1074/jbc.M112.356113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibbons MM, Klug WS. 2007. Nonlinear finite-element analysis of nanoindentation of viral capsids. Phys Rev E Stat Nonlin Soft Matter Phys 75:031901. doi: 10.1103/PhysRevE.75.031901. [DOI] [PubMed] [Google Scholar]
- 35.de Pablo PJ, Mateu MG. 2013. Mechanical properties of viruses. Subcell Biochem 68:519–551. doi: 10.1007/978-94-007-6552-8_18. [DOI] [PubMed] [Google Scholar]
- 36.Alfson KJ, Avena LE, Beadles MW, Staples H, Nunneley JW, Ticer A, Dick EJ Jr, Owston MA, Reed C, Patterson JL, Carrion R Jr, Griffiths A. 2015. Particle-to-PFU ratio of Ebola virus influences disease course and survival in cynomolgus macaques. J Virol 89:6773–6781. doi: 10.1128/JVI.00649-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardone G, Newcomb WW, Cheng N, Wingfield PT, Trus BL, Brown JC, Steven AC. 2012. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol 86:4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steven AC, Baumeister W, Johnson LN, Perham RN. 2016. Molecular biology of assemblies and machines. Garland Science, New York, NY. [Google Scholar]
- 39.Garriga D, Querol-Audi J, Abaitua F, Saugar I, Pous J, Verdaguer N, Caston JR, Rodriguez JF. 2006. The 2.6-angstrom structure of infectious bursal disease virus-derived T=1 particles reveals new stabilizing elements of the virus capsid. J Virol 80:6895–6905. doi: 10.1128/JVI.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irigoyen N, Garriga D, Navarro A, Verdaguer N, Rodriguez JF, Caston JR. 2009. Autoproteolytic activity derived from the infectious bursal disease virus capsid protein. J Biol Chem 284:8064–8072. doi: 10.1074/jbc.M808942200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mertens J, Casado S, Mata CP, Hernando-Perez M, de Pablo PJ, Carrascosa JL, Caston JR. 2015. A protein with simultaneous capsid scaffolding and dsRNA-binding activities enhances the birnavirus capsid mechanical stability. Sci Rep 5:13486. doi: 10.1038/srep13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneemann A. 2006. The structural and functional role of RNA in icosahedral virus assembly. Annu Rev Microbiol 60:51–67. doi: 10.1146/annurev.micro.60.080805.142304. [DOI] [PubMed] [Google Scholar]
- 43.Lombardo E, Maraver A, Castón JR, Rivera J, Fernandez-Arias A, Serrano A, Carrascosa JL, Rodriguez JF. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J Virol 73:6973–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W. 2011. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A 108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prevelige PE, Fane BA. 2012. Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Adv Exp Med Biol 726:325–350. doi: 10.1007/978-1-4614-0980-9_14. [DOI] [PubMed] [Google Scholar]
- 46.Suhanovsky MM, Teschke CM. 2015. Nature's favorite building block: deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 479-480:487–497. doi: 10.1016/j.virol.2015.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jayaraman B, Smith AM, Fernandes JD, Frankel AD. 2016. Oligomeric viral proteins: small in size, large in presence. Crit Rev Biochem Mol Biol 51:379–394. doi: 10.1080/10409238.2016.1215406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de la Rosa-Trevin JM, Oton J, Marabini R, Zaldivar A, Vargas J, Carazo JM, Sorzano CO. 2013. Xmipp 3.0: an improved software suite for image processing in electron microscopy. J Struct Biol 184:321–328. doi: 10.1016/j.jsb.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 49.Scheres SH. 2012. A Bayesian view on cryo-EM structure determination. J Mol Biol 415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de la Rosa-Trevin JM, Quintana A, Del Cano L, Zaldivar A, Foche I, Gutierrez J, Gomez-Blanco J, Burguet-Castell J, Cuenca-Alba J, Abrishami V, Vargas J, Oton J, Sharov G, Vilas JL, Navas J, Conesa P, Kazemi M, Marabini R, Sorzano CO, Carazo JM. 2016. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J Struct Biol 195:93–99. doi: 10.1016/j.jsb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 51.Heymann JB, Belnap DM. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol 157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 56.Frangakis AS, Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol 135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 57.Horcas I, Fernandez R, Gomez-Rodriguez JM, Colchero J, Gomez-Herrero J, Baro AM. 2007. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev Sci Instrum 78:013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The E1-1A, E1-1B, and E1-2 cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB; http://www.ebi.ac.uk/pdbe/emdb) with accession numbers emd-3507, emd-3509, and emd-3510, respectively. E1 to E6 cryo-ET averaged maps have been deposited in EMDB with accession numbers emd-3511 to emd-3516, respectively.