Integration of viral genome sequencing into influenza surveillance for wild birds and domestic poultry can elucidate evolutionary pathways of economically costly poultry pathogens. Evolutionary analyses of H5 LPAIVs detected in domestic poultry in the United States and Canada during 2001 to 2017 suggest that these viruses originated from repeated introductions of IAVs from wild birds, followed by various degrees of reassortment. Reassortment was observed where biosecurity was low and where opportunities for more than one virus to circulate existed (e.g., congregations of birds from different premises, such as live-bird markets). None of the H5 lineages identified were maintained for the long term in domestic poultry, suggesting that management strategies have been effective in minimizing the impacts of virus introductions on U.S. poultry production.
KEYWORDS: low-pathogenic avian influenza, subtype H5, wild birds, domestic poultry, backyard poultry, live-bird market, dabbling duck, goose, swan, evolutionary network, reassortment, phylogenetic, United States, goose/swan
ABSTRACT
Wild-bird origin influenza A viruses (IAVs or avian influenza) have led to sporadic outbreaks among domestic poultry in the United States and Canada, resulting in economic losses through the implementation of costly containment practices and destruction of birds. We used evolutionary analyses of virus sequence data to determine that 78 H5 low-pathogenic avian influenza viruses (LPAIVs) isolated from domestic poultry in the United States and Canada during 2001 to 2017 resulted from 18 independent virus introductions from wild birds. Within the wild-bird reservoir, the hemagglutinin gene segments of H5 LPAIVs exist primarily as two cocirculating genetic sublineages, and our findings suggest that the H5 gene segments flow within each migratory bird flyway and among adjacent flyways, with limited exchange between the nonadjacent Atlantic and Pacific Flyways. Phylogeographic analyses provided evidence that IAVs from dabbling ducks and swans/geese contributed to the emergence of viruses among domestic poultry. H5 LPAIVs isolated from commercial farm poultry (i.e., turkey) that were descended from a single introduction typically remained a single genotype, whereas those from live-bird markets sometimes led to multiple genotypes, reflecting the potential for reassortment with other IAVs circulating within live-bird markets. H5 LPAIVs introduced from wild birds to domestic poultry represent economic threats to the U.S. poultry industry, and our data suggest that such introductions have been sporadic, controlled effectively through production monitoring and a stamping-out policy, and are, therefore, unlikely to result in sustained detections in commercial poultry operations.
IMPORTANCE Integration of viral genome sequencing into influenza surveillance for wild birds and domestic poultry can elucidate evolutionary pathways of economically costly poultry pathogens. Evolutionary analyses of H5 LPAIVs detected in domestic poultry in the United States and Canada during 2001 to 2017 suggest that these viruses originated from repeated introductions of IAVs from wild birds, followed by various degrees of reassortment. Reassortment was observed where biosecurity was low and where opportunities for more than one virus to circulate existed (e.g., congregations of birds from different premises, such as live-bird markets). None of the H5 lineages identified were maintained for the long term in domestic poultry, suggesting that management strategies have been effective in minimizing the impacts of virus introductions on U.S. poultry production.
INTRODUCTION
Influenza A viruses (IAVs) are single-stranded, negative-sense RNA viruses with eight genomic segments. Wild waterbirds, especially migratory waterfowl, such as geese and ducks, along with gulls and shorebirds, are purported to be the natural reservoir for IAVs of the hemagglutinin (HA) subtypes H1 to H16 and neuraminidase (NA) subtypes N1 to N9. IAVs are maintained in wild waterbirds, and those of low pathogenicity result in enteric infections with rare evidence of clinical illness. In contrast, IAVs typically result in respiratory disease in gallinaceous birds, such as chickens and turkeys; clinical signs and disease severity vary and are strain dependent. Subtypes H5 and H7 low-pathogenic avian influenza A viruses (LPAIVs) demonstrate the potential to evolve from low to highly pathogenic avian influenza A viruses (HPAIVs) through increased HA cleavability by acquiring multiple basic amino acids (1–3) or insertions (4–8) at the cleavage site during replication (8, 9). H5 and H7 HPAI causes high mortality among domestic poultry, leading to large economic losses.
Reassortment is another mechanism that can contribute to the generation of novel and economically costly IAVs in domestic birds. For example, H7N9 and H10N8 IAVs recently identified in China appear to have wild-bird origin HA and NA genes and internal genes from IAVs circulating among domestic poultry (10, 11). Both of these reassortant viruses have been associated with human disease that may have resulted from contact with infected birds at live-bird markets (LBMs). An H7N9 IAV that most likely resulted from reassortment of H7, N9, and H9N2 avian IAVs has caused >1,566 human infections and at least 613 deaths in China (11, 12); an H10N8 avian-origin IAV also originated from reassortment of H10N8 and H9N2 IAVs in LBMs and caused human infections (10, 13).
In the United States and Canada (collectively referred to as North America for the purposes of this study), poultry production systems include large commercial poultry farms (CPFs), backyard poultry operations (BYPs), game bird poultry farms (GBPs), and LBMs. CPFs are defined as large-scale commercial poultry farms with >1,000 domestic birds per year. BYPs are defined as residential farms raising small flocks of domestic birds; these typically produce ≤1,000 birds per year. GBPs are poultry operations raising small flocks of game birds, such as pheasants and quail, often released for sport harvest. LBMs are operations that typically supply live birds for on-site slaughter to consumers. Some, such as botanicas, may sell live birds as well. LBMs may acquire birds from both non-CPF and CPF sources. In the United States, CPFs are located primarily in the southern and midwestern regions, while many LBMs are located in urban areas of the western and northeastern regions, and BYPs and GBPs are present in all regions (14–16).
Detections of IAVs in domestic poultry are not uncommon (17); wild-bird origin IAVs are usually identified as the source of virus across a variety of North American poultry production systems (3, 9, 17–21). In rare cases, LPAIVs circulating in LBMs have been associated with outbreaks at commercial farms (22). However, there is limited information regarding the evolutionary patterns of IAVs detected in North American poultry. Such information is necessary to improve our collective understanding of the natural reservoirs (location and wild-bird taxa) in which viruses that ultimately lead to poultry outbreaks are maintained, how evolutionary mechanisms for IAVs might vary between poultry production systems, and the frequency with which IAVs are shared among CPFs, BYBs, GBPs, and LBMs.
In this study, we genetically characterized and compared the inferred evolutionary pathways of 78 H5 LPAIVs from the United States and Canada during 2001 to 2017. This study seeks to explore the evolutionary pathways of IAVs that move from wild birds into domestic poultry. A better understanding of viruses at this interface will help improve influenza surveillance and management strategies, reduce economic and animal losses, and decrease opportunities for the generation of novel pathogens, including those that can infect humans and cause pandemic influenza threats.
RESULTS
H5 LPAIVs were sporadically detected in CPFs and LBMs.
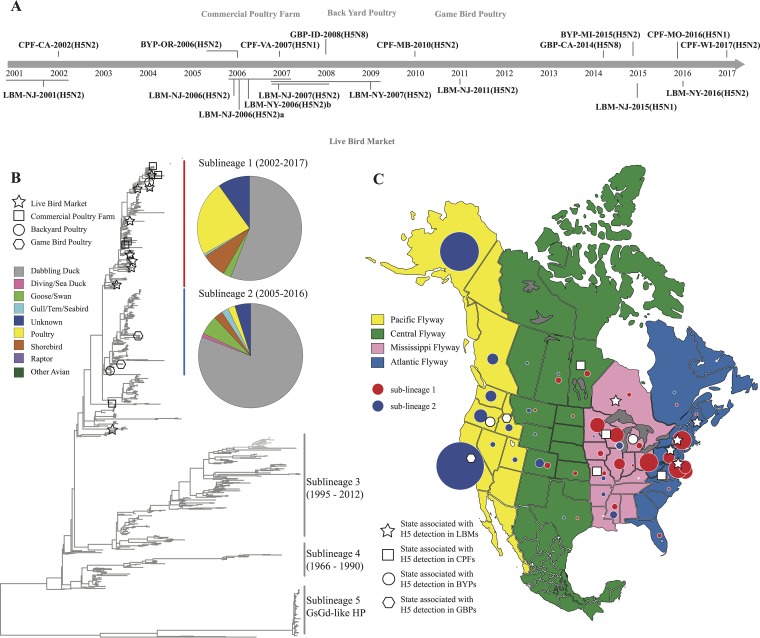
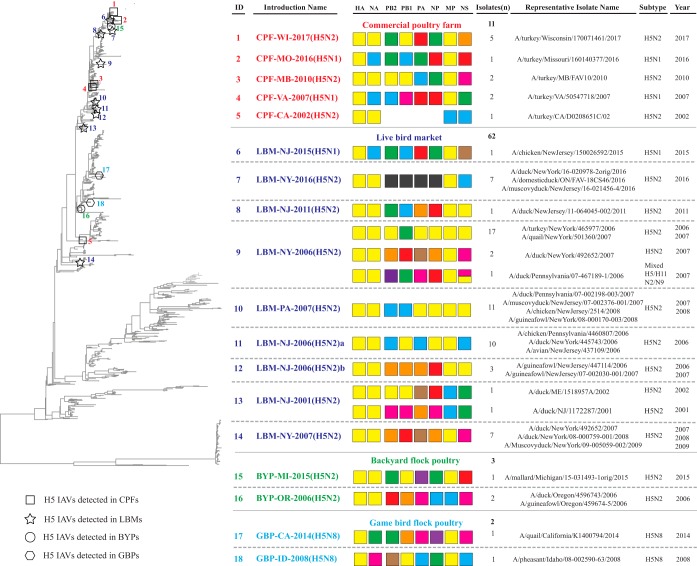
In this study, an isolate is defined as an IAV recovered from wild birds or domestic poultry, and an introduction is defined as a case of IAV infection in domestic poultry; one or more isolates may be recovered from the same introduction. A total of 78 H5 LPAI isolates from domestic poultry in North America during 2001 to 2017 were included in the study. Isolates were recovered from CPFs (n = 11), BYPs (n = 3), GBPs (n = 2), and LBMs (n = 62) (Table 1). H5 isolates were identified on CPFs in turkey (n = 11, commercial turkey growers are indoor operations with curtain-sides); from BYPs in a mallard duck, a domestic duck, and a guinea fowl; from GBPs in a pheasant and a quail; and in LBM samples collected from domestic ducks (n = 42), chickens (n = 7), guinea fowl (n = 6), turkey (n = 1), quails (n = 2), pheasant (n = 1), and unknown species (n = 3) (Table 1). Based upon the inferred phylogenetic relationships and nucleotide identities for the HA gene segment, these 78 H5 poultry isolations result from 18 independent introductions (see details in Materials and Methods) (Fig. 1A; see also Fig. S1 in the supplemental material). Among the 18 introductions, 5 were detected in CPFs, 9 in LBMs, 2 from a BYP, and 2 from a GBP (Fig. 1A). Each introduction event was identified by the operation type, state, year, and HA/NA subtype from the first isolate; for example, CPF-WI-2017(H5N2) denotes an H5N2 event in commercial turkeys from Wisconsin during 2017. When more than one event occurred in a state within 1 year, the events are distinguished by a letter; for example, LBM-NJ-2006(H5N2)a and LBM-NJ-2006(H5N2)b represent two independent H5N2 events in New Jersey LBMs during 2006.
TABLE 1.
H5 LPAI viruses detected in domestic poultry in the United States and Canada (2001 to 2017)a
| Introduction | Host(s) | State(s) or province | Yr | Isolate name | Sample date (yr-mo-day) | Subtype |
|---|---|---|---|---|---|---|
| BYP-MI-2015(H5N2) | Mallard | Michigan | 2015 | A/mallard/Michigan/15-031493-1orig/2015 | 2015-10-01 | H5N2 |
| BYP-OR-2006(H5N2) | Duck, guinea fowl | Oregon | 2006 | A/duck/Oregon/459674-3/2006 | 2006-09-29 | H5N2 |
| A/guineafowl/Oregon/459674-5/2006 | 2006-09-29 | H5N2 | ||||
| GBP-CA-2014(H5N8) | Quail | California | 2014 | A/quail/California/K1400794/2014 | 2014-04 | H5N8 |
| GBP-ID-2008(H5N8) | Pheasant | Idaho | 2008 | A/pheasant/Idaho/08-002590-63/2008 | 2008 | H5N8 |
| CPF-WI-2017(H5N2) | Turkey | Wisconsin | 2017 | A/turkey/Wisconsin/17-007146-1/2017 | 2017-03-02 | H5N2 |
| A/turkey/Wisconsin/17-007146-2/2017 | 2017-03-02 | H5N2 | ||||
| A/turkey/Wisconsin/17-007146-3/2017 | 2017-03-02 | H5N2 | ||||
| A/turkey/Wisconsin/17-007319-3/2017 | 2017-03-03 | H5N2 | ||||
| A/turkey/Wisconsin/17-007981-6/2017 | 2017-03-09 | H5N2 | ||||
| CPF-MO-2016(H5N1) | Turkey | Missouri | 2016 | A/turkey/Missouri/16-014037-7/2016 | 2016-04-29 | H5N1 |
| CPF-MB-2010(H5N2) | Turkey | Manitoba | 2010 | A/turkey/MB/FAV11/2010 | 2010-11-25 | H5N2 |
| A/turkey/MB/FAV10/2010 | 2010-11-26 | H5N2 | ||||
| CPF-VA-2007(H5N1) | Turkey | Virginia | 2007 | A/turkey/VA/505477-18/2007 | 2007-07-11 | H5N1 |
| A/turkey/Virginia/505477-17/2007 | 2007-07-11 | H5N1 | ||||
| CPF-CA-2002(H5N2) | Turkey | California | 2002 | A/turkey/CA/D0208651-C/02 | 2002 | H5N2 |
| LBM-NY-2016(H5N2) | Duck, muscovy duck | Ontario, New York, New Jersey | 2016 | A/domesticduck/ON/FAV-18CS46/2016 | 2016 | H5N2 |
| A/duck/NewYork/16-020978-2orig/2016 | 2016 | H5N2 | ||||
| A/duck/NewYork/16-021467-1orig/2016 | 2016 | H5N2 | ||||
| A/duck/NewYork/16-021916-1orig/2016 | 2016 | H5N2 | ||||
| A/duck/NewYork/16-021920-1orig/2016 | 2016 | H5N2 | ||||
| A/muscovyduck/NewJersey/16-021456-4/2016 | 2016 | H5N2 | ||||
| A/muscovyduck/NewJersey/16-021457-2/2016 | 2016 | H5N2 | ||||
| LBM-NJ-2015(H5N1) | Chicken | New Jersey | 2015 | A/chicken/New_Jersey/15_002659_2/2015 | 2015-01-20 | H5N1 |
| LBM-NJ-2011(H5N2) | Duck | New Jersey | 2011 | A/duck/NewJersey/11-064045-002/2011 | 2011-12-20 | H5N2 |
| LBM-NY-2007(H5N2) | Duck, muscovy duck | New York | 2007-2009 | A/duck/NewYork/07-002127-001/2007 | 2007-10-30 | H5N2 |
| A/muscovyduck/NewYork/08-000560-002/2008 | 2008-03-13 | H5N2 | ||||
| A/duck/NewYork/08-000759-001/2008 | 2008-04-22 | H5N2 | ||||
| A/duck/NewYork/08-000937-001/2008 | 2008-05-22 | H5N2 | ||||
| A/muscovyduck/NewYork/09-002670-002/2009 | 2009-03-04 | H5N2 | ||||
| A/duck/NewYork/09-005059-001/2009 | 2009-04-14 | H5N2 | ||||
| A/muscovyduck/NewYork/09-005059-002/2009 | 2009-04-14 | H5N2 | ||||
| LBM-PA-2007(H5N2) | Chicken, duck, guinea fowl, muscovy duck, pheasant | Pennsylvania, New York, New Jersey | 2007-2008 | A/duck/Pennsylvania/07-002198-003/2007 | 2007-11-09 | H5N2 |
| A/muscovyduck/NewJersey/07-002376-001/2007 | 2007-12-05 | H5N2 | ||||
| A/guineafowl/NewYork/08-000170-003/2008 | 2008-01-07 | H5N2 | ||||
| A/pheasant/NewYork/08-000170-002/2008 | 2008-01-07 | H5N2 | ||||
| A/guineafowl/NewYork/08-000238-001/2008 | 2008-01-28 | H5N2 | ||||
| A/chicken/NewJersey/251-4/2008 | 2008-02-01 | H5N2 | ||||
| A/chicken/NewJersey/577-6/2008 | 2008-03-27 | H5N2 | ||||
| A/chicken/NewJersey/08-000640-006/2008 | 2008-04-03 | H5N2 | ||||
| A/guineafowl/NewJersey/08-000640-008/2008 | 2008-04-03 | H5N2 | ||||
| A/guineafowl/NewJersey/08-000841-001/2008 | 2008-05-14 | H5N2 | ||||
| A/muscovyduck/NewJersey/08-000912-001/2008 | 2008-05-28 | H5N2 | ||||
| LBM-NY-2006(H5N2) | Duck, quail, turkey | New York, Pennsylvania | 2006-2007 | A/duck/NewYork/465571/2006 | 2006-10-23 | H5N2 |
| A/duck/NewYork/465976/2006 | 2006-10-24 | H5N2 | ||||
| A/duck/NewYork/466787/2006 | 2006-10-31 | H5N2 | ||||
| A/turkey/NewYork/465977/2006 | 2006-10-31 | H5N2 | ||||
| A/duck/NewYork/470179/2006 | 2006-11-07 | H5N2 | ||||
| A/avian/NewYork/466812/2006 | 2006-11-09 | H5N2 | ||||
| A/duck/Pennsylvania/07-467189-1/2006 | 2006-11-09 | Mixed | ||||
| A/duck/NewYork/469961/2006 | 2006-11-13 | H5N2 | ||||
| A/duck/NewYork/489761/2007 | 2007 | H5N2 | ||||
| A/duck/NewYork/481172/2007 | 2007-01-23 | H5N2 | ||||
| A/duck/NewYork/483239/2007 | 2007-02-02 | H5N2 | ||||
| A/duck/NewYork/484057/2007 | 2007-02-06 | H5N2 | ||||
| A/duck/NewYork/484680/2007 | 2007-02-12 | H5N2 | ||||
| A/duck/NewYork/490722/2007 | 2007-03-21 | H5N2 | ||||
| A/duck/NewYork/492652/2007 | 2007-04-05 | H5N2 | ||||
| A/duck/NewYork/494165/2007 | 2007-04-18 | H5N2 | ||||
| A/quail/NewYork/07-501360-1/2007 | 2007-06-02 | H5N2 | ||||
| A/quail/NewYork/501360/2007 | 2007-06-12 | H5N2 | ||||
| A/duck/NewYork/504371/2007 | 2007-06-22 | H5N2 | ||||
| A/duck/NewYork/504372/2007 | 2007-06-22 | H5N2 | ||||
| LBM-NJ-2006(H5N2)a | Chicken, duck, muscovy duck | New York, New Jersey, Pennsylvania | 2006 | A/avian/NewJersey/437109/2006 | 2006-05-09 | H5N2 |
| A/chicken/NewYork/439236/2006 | 2006-05-10 | H5N2 | ||||
| A/chicken/NewYork/439235/2006 | 2006-05-15 | H5N2 | ||||
| A/muscovyduck/NewYork/62095-1/2006 | 2006-05-15 | H5N2 | ||||
| A/duck/NewYork/440410/2006 | 2006-05-17 | H5N2 | ||||
| A/duck/NewYork/440409/2006 | 2006-05-23 | H5N2 | ||||
| A/duck/NewYork/445743/2006 | 2006-06-19 | H5N2 | ||||
| A/chicken/Pennsylvania/446080-7/2006 | 2006-07 | H5N2 | ||||
| A/duck/Pennsylvania/446080-6/2006 | 2006-07 | H5N2 | ||||
| A/duck/Pennsylvania/446080-7/2006 | 2006-07 | H5N2 | ||||
| LBM-NJ-2006(H5N2)b | Guinea fowl | New York, New Jersey | 2006-2007 | A/avian/NewYork/448534/2006 | 2006 | H5N2 |
| A/guineafowl/NewJersey/447114/2006 | 2006-07-19 | H5N2 | ||||
| A/guineafowl/NewJersey/07-002030-001/2007 | 2007-10-23 | H5N2 | ||||
| LBM-NJ-2001(H5N2) | Duck | Maine, New Jersey | 2001–2002 | A/duck/NJ/117228-7/2001 | 2001-07-16 | H5N2 |
| A/duck/ME/151895-7A/2002 | 2002-01-29 | H5N2 |
An isolate is defined as an avian influenza virus recovered from wild birds or domestic poultry. An introduction describes a case of avian influenza infection detected in domestic poultry, and one or multiple isolates can be recovered from the same introduction.
FIG 1.
Detections of low-pathogenic H5 avian influenza A viruses in domestic poultry in the United States and Canada (2001 to 2017). (A) Temporal distributions of introductions of H5 viruses detected on commercial poultry farms (CPFs), backyard poultry farms (BYPs), and game bird poultry farms (GBPs) and in live-bird markets (LBMs) during 2001 to 2017. An introduction was defined by a tree topology of the H5 gene with a bootstrap value of ≥70 (Fig. S1) and shared nucleotide sequence identity of ≥98%. (B) Simplified phylogenetic tree displaying the general topology for the North American sequence of influenza A viruses at the H5 hemagglutinin gene. The distribution of functional groups of wild-bird hosts for influenza A viruses in the tree are summarized by pie charts. (C) U.S. states and Canadian provinces of origin for H5 subtype influenza A virus isolates detected in North American domestic poultry during 2001 to 2017 and geographic distribution of the influenza A viruses of H5 sublineages 1 and 2 in wild birds based on the North American administrative definition of migratory bird flyways (https://www.fws.gov/birds/management/flyways.php). Circle sizes indicate the numbers of H5 influenza A virus isolates in the corresponding U.S. state/Canadian province. Based on ecologic attributes, the hosts of influenza viruses were categorized into 9 different groups: dabbling duck, diving/sea duck, goose/swan, gull/tern/seabird, poultry, raptor, shorebird, other avian, and unknown. Viruses from avian species that did not fit into any of these functional groups were categorized as “other avian,” and viruses for which the species sampled were unclear or for which the domestic status was ambiguous were categorized as “unknown.”
H5 LPAIVs from United States and Canadian domestic poultry are of North American lineage and share genetic ancestry with wild-bird origin IAVs.
Geographic lineage was assigned based upon phylogenetic analyses of the HA gene for the 78 H5 LPAIVs; we considered this lineage to be comprised primarily of four distinct sublineages (Fig. 1B). A total of 70 of the 78 domestic poultry isolates characterized in this study, as well as the majority of the wild-bird origin H5 LPAIVs identified in the United States and Canada from 2001 to 2017 clustered within sublineages 1 and 2. Viruses causing enzootic outbreaks of IAVs among domestic poultry in Mexico are clustered in sublineage 3, whereas IAVs circulating in both wild birds and domestic poultry in the United States and Canada during 1966 to 1990 were grouped in sublineage 4 (Fig. 1B). Eight additional H5 subtype IAVs identified in poultry from the United States and Canada, along with a relatively small number of wild-bird origin IAVs isolated from samples collected during 1987 to 2008, clustered in clades not assigned to these four sublineages (Fig. 1B).
We used the relationships inferred by phylogenetic and nucleotide sequence identities to further distinguish genetic groups within sublineages 1 and 2. The 78 H5 isolates from North American poultry clustered into 18 distinct genetic groups. Five genetic groups contained isolates detected in domestic birds sampled on CPFs, nine genetic groups contained isolates detected in birds sampled from LBMs, two genetic groups contained isolates detected in birds sampled from BYPs, and two genetic groups contained isolates detected in birds sampled from GBPs. For most poultry isolates (67%), nucleotide sequence identity at the HA gene segment of H5 LPAIVs was >99% similar to that for wild-bird origin viruses (Fig. 1B; see also Fig. S1 and Table S1 in the supplemental material); however, within a genetic group, the poultry isolates were most similar to each other, suggesting a common introduction event with subsequent spread (Fig. S1). Based upon this analysis, each of the 18 genetic groups was determined to represent discrete introduction events of IAVs from wild birds to domestic poultry. Additionally, only one production system was affected in each of the 18 inferred introduction events (e.g., CPFs, BYPs, GBPs, or LBMs) (Table 1).
Based upon phylogenetic analyses, the NA and internal gene segments of the H5 LPAI from poultry (including CPFs, LBMs, GBPs, and BYPs) were genetically diverse (Fig. 2). Phylogenetic analyses also identified potential precursor strains for 15 of the 18 purported introductions; three events were excluded: CPF-CA-2002(H5N2) had an incomplete genome, LBM-NY-2016(H5N2) lacked wild-bird progenitor genes, and LBM-NY-2007(H5N2) lacked wild-bird precursors (Table 2). For 13 of the inferred introductions, potential wild-bird progenitor strains shared similar phylogenetic positions in tree topologies with high nucleotide sequence similarity for 3 or 4 gene segments (Table 2). Potential wild-bird progenitor viruses across all eight gene segments were identified for two purported introductions: a Canada goose (Branta canadensis) virus for CPF-MO-2016(H5N1) and an American wigeon (Mareca americana) for BYP-OR-2006(H5N2) (Table 2).
FIG 2.
Summary of genotypic analysis of low-pathogenic H5 avian influenza A viruses detected in domestic poultry in the United States (2001 to 2017). Genotypes were assigned by unique combinations of sublineages for each gene, which were determined based on tree topology with a bootstrap value of ≥70 and a nucleotide sequence identity of ≥95%. To simplify the illustration, only the representative viruses were selected for each genotype, including those with unique combinations of location of detection (state/province) and year. Numbers on the tree indicate individual H5 low-pathogenic introductions and were linked to unique IDs of those introductions to the right.
TABLE 2.
Potential precursor viruses for predominate genotypes of low-pathogenic H5 avian influenza virus introductions in domestic poultry in the United States and Canada (2001 to 2017)
| Representative isolate | Introduction | Potential precursor virusa | No. of possible progenitor genes | Sequence identity (%)b |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HA | NA | PB2 | PB1 | PA | NP | MP | NS | ||||
| A/turkey/Wisconsin/17-007146-1/2017 | CPF-WI-2017(H5N2) | A/northernpintail/Ohio/15OS5861/2015(H5N2) | 4 | 99.14 | 98.81 | 98.26 | 98.96 | ||||
| A/turkey/Missouri/16-014037-7/2016 | CPF-MO-2016(H5N1) | A/canadagoose/DelawareBay/601/2016(H5N1) | 8 | 98.82 | 99.29 | 99.43 | 99.56 | 99.35 | 99.53 | 99.47 | 99.52 |
| A/turkey/MB/FAV10/2010 | CPF-MB-2010(H5N2) | A/americangreen-wingedteal/Illinois/2975/2009(Mixed) | 4 | 98.74 | 98.18 | 98.88 | 99.29 | ||||
| A/turkey/VA/50547718/2007 | CPF-VA-2007(H5N1) | A/mallard/PA/454069-9/2006(H5N1) | 4 | 98.74 | 99.39 | 98.31 | 99.40 | ||||
| A/turkey/CA/D0208651C/02c | CPF-CA-2002(H5N2) | ||||||||||
| A/duck/NewYork/16-020978-2orig/2016c | LBM-NY-2016(H5N2) | ||||||||||
| A/chicken/NewJersey/150026592/2015 | LBM-NJ-2015(H5N1) | A/americangreen-wingedteal/Wisconsin/11OS3580/2011 (H11N2) | 4 | 98.18 | 98.33 | 98.55 | 99.16 | ||||
| A/duck/NewJersey/11-064045-002/2011 | LBM-NJ-2011(H5N2) | A/mallard/Ohio/11OS1961/2011(H5N2) | 4 | 99.60 | 99.86 | 98.92 | 99.80 | ||||
| A/duck/NewYork/08-000759-001/2008c | LBM-NY-2007(H5N2) | ||||||||||
| A/chicken/NewJersey/2514/2008 | LBM-NJ-2007(H5N2) | A/mallard/Maryland/07OS2433/2007(H5N2) | 3 | 98.90 | 98.25 | 98.24 | |||||
| A/turkey/NewYork/465977/2006 | LBM-NY-2006(H5N2) | A/americanwigeon/Iowa/463993/2006(H5N2) | 4 | 99.08 | 98.54 | 98.53 | 98.29 | ||||
| A/chicken/Pennsylvania/4460807/2006 | LBM-NJ-2006(H5N2)a | A/mallard/Maryland/897/2004(H5N2) | 4 | 98.63 | 98.75 | 99.01 | 99.29 | ||||
| A/avian/NewYork/448534/2006 | LBM-NJ-2006(H5N2)b | A/mallard/Ohio/468158/2006(H5N2) | 4 | 98.34 | 98.63 | 98.14 | 98.30 | ||||
| A/duck/NJ/1172287/2001 | LBM-NJ-2001(H5N2) | A/mallard/Maryland/302/2001(H5N2) | 3 | 98.58 | 98.48 | 99.42 | |||||
| A/mallard/Michigan/15-031493-1orig/2015 | BYP-MI-2015(H5N2) | A/mallard/Ohio/11OS2156/2011(H5N2) | 4 | 98.68 | 98.96 | 98.57 | 99.30 | ||||
| A/duck/Oregon/4596743/2006 | BYP-OR-2006(H5N2) | A/americanwidgeon/Oregon/467919/2006(H5N2) | 8 | 99.60 | 100.00 | 99.83 | 99.78 | 99.86 | 99.81 | 99.73 | 99.88 |
| A/quail/California/K1400794/2014 | GBP-CA-2014(H5N8) | A/mallard/California/1479/2013(mixed) | 3 | 98.05 | 98.07 | 99.20 | |||||
| A/pheasant/Idaho/08-002590-63/2008 | GBP-ID-2008(H5N8) | A/ruddyturnstone/NewJersey/AI06-582/2006(H6N7) | 3 | 98.40 | 98.78 | 98.40 | |||||
A potential precursor virus was defined by the virus with at least three possible progenitor genes (see Materials and Methods for details).
Only sequence identity levels of >98% are shown.
No data for potential precursor virus, number of possible progenitor genes, or sequence identity because the complete genome was not available.
Geographic and temporal patterns of H5 LPAIV lineages in North America.
Fifteen of the 18 introductions were detected in only one flyway: seven within the Atlantic Flyway (one CPF and six LBM detections), three within the Mississippi Flyway (two CPF and one BYP detection), one in the Central Flyway (a CPF detection), and four within the Pacific Flyway (one CPF, one BYP, and two GBP detections). The remaining three introductions were LBM-associated detections in the Atlantic and Mississippi Flyways. Of the LBM events, eight were genetically related other IAVs detected in Canadian provinces or U.S. states (Table 1).
To further evaluate gene flow for the H5 HA gene segment, we performed phylogeographic analyses for viruses in sublineages 1 and 2. Viruses in sublineage 1, which included viruses associated with 13 introduction events, were predominantly from the Central (5.36%), Mississippi (31.07%), and Atlantic (63.57%) Flyways. Most (81.03%) of the viruses in sublineage 2, associated with three introduction events, were from wild birds sampled within the Pacific Flyway (Fig. 1C). Two events were not associated with either lineage 1 or 2, i.e., CPF-CA-2002(H5N2) and LBM-NY-2007(H5N2), the latter of which involved seven LPAIVs detected during 2007 to 2009. Bayesian analyses of viruses in sublineages 1 and 2 indicated unilateral or bilateral state transitions suggestive of H5 HA gene flow among the Atlantic, Mississippi, and Central Flyways and among the Mississippi, Central, and Pacific Flyways (Bayes factor > 3). However, no state transitions suggestive of H5 HA gene flow were supported between the Atlantic and Pacific Flyways (Bayes factor < 3; no significant transition was observed) (see Table S2 and Fig. S2 in the supplemental material). In summary, phylogeographic analyses suggested H5 HA gene flow across adjacent or nearby flyways but not between the Atlantic and Pacific Flyways.
H5 LPAIVs detected in domestic poultry are likely descendant from those in dabbling ducks or geese/swans.
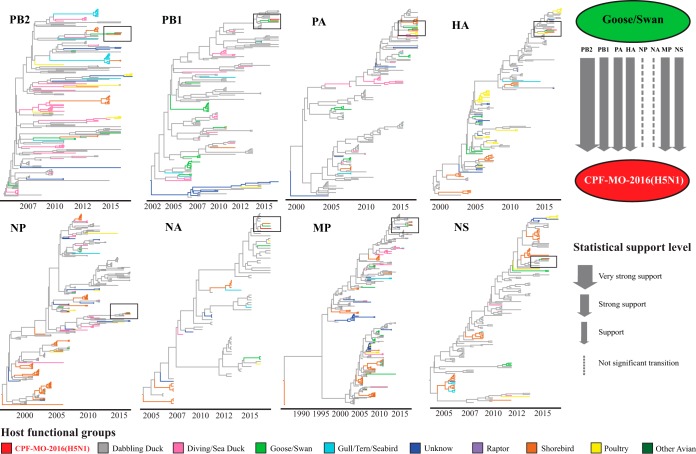
To understand whether specific hosts were associated with virus introductions detected in domestic poultry, we categorized bird host species into 9 functional groups based on taxonomic and ecologic attributes: dabbling duck, diving/sea duck, goose/swan, gull/tern/seabird, raptor, shorebird, other avian, unknown, and poultry (see Materials and Methods for details). Phylogeographic analyses were performed to estimate Bayes factors between the domestic poultry viruses and viruses from each functional group. A combination of a Bayes factor of ≥3 and a mean indicator of ≥0.5 was used as the threshold of statistical significance; a larger Bayes factor indicates a higher probability that a specific functional group is associated with an introduction event (3, 23).
Using phylogeographic analyses, we assessed the genetic origins for 16 of the 18 purported introduction events; two events were excluded: CPF-CA-2002(H5N2) had an incomplete genome, and LBM-NY-2016(H5N2) lacked related wild-bird samples. Our results suggest that IAVs in wild waterfowl (dabbling ducks, swans/geese, or diving/sea ducks) were the probable progenitor for at least one viral gene segment for 15 of 16 investigated introduction events; no probable wild-bird IAV progenitor was identified for LBM-NY-2007(H5N2) (see Tables S3 and S4 in the supplemental material). Neither gulls/terns/seabirds nor raptors were supported as probable sources of gene segments for H5 IAVs detected in domestic poultry, and shorebirds were supported as a probable progenitor for only one gene segment for a single introduction event [MP gene of CPF-VA-2007(H5N1)]. Dabbling ducks, in particular, were associated with numerous gene segments from North American poultry viruses, including all gene segments of viruses involved in the following outbreaks: CPF-WI-2017(H5N2) (Bayes factor, 6.59 to 35.12), CPF-MB-2010(H5N2) (Bayes factor, 3.24 to 189.74), LBM-NJ-2015(H5N1) (Bayes factor, 4.65 to 138.71), LBM-NY-2006(H5N2) (Bayes factor, 6.89 to 1,244.25), and LBM-NJ-2006a(H5N2) (Bayes factor, 12.21 to >10,000) (Table S4). For recent introductions (2015 to 2017), CPF-WI-2017(H5N2) was associated with dabbling duck origin viruses across all eight segments (Bayes factor, 6.59 to 35.12) (Table S3); CPF-MO-2016(H5N1) was associated with goose/swan origin viruses across six segments (PB2, PB1, PA, HA, MP, and NS) (Bayes factor, 15.69 to 149.91) (Fig. 3; Table S3); and LBM-NJ-2015(H5N1) was associated with dabbling duck viruses (HA and NA; Bayes factors, 53.98 and 8.12, respectively), diving/sea duck virus (NP; Bayes factor, 169.25), and goose/swan virus (NS; Bayes factor, 9.55) (Table S3). Functional groups associated with all eight gene segments were identified for only 4 of the 18 purported introductions into North American poultry during 2001 to 2017. Among them, all gene segments were associated with dabbling duck, goose/swan, or unknown functional groups (Tables S3 and S4).
FIG 3.
Summary of analyses to assess transition of influenza A viruses from a specific functional group of wild birds to the low-pathogenic H5 avian influenza A viruses detected in domestic poultry. In this figure, phylogeographical analyses showed one recent and representative H5 low-pathogenic introduction in North American poultry, CPF-MO-2016(H5N1), which is an introduction of virus in Missouri turkey as an example. The trees shown in the figure are constructed on the basis of the maximum clade credibility phylogenetic trees. Phylogeographical analyses were performed using all isolates for each inferred H5 introduction. Branches of the phylogenetic trees were colored according to the estimated ancestral state of the functional group of wild birds from discrete trait reconstruction. Arrow widths are based on the Bayes factor support levels. Statistical support is provided in greater detail in Table S3.
H5 LPAIVs in LBMs have potential opportunities for reassortment.
The opportunity for reassortment exists whenever two or more viruses circulate simultaneously. For CPFs, only a single genotype was associated with each introduction event, whereas multiple genotypes were identified among viruses associated with two of the LBM introductions [LBM-NY-2006(H5N2) and LBM-NJ-2001(H5N2)] (Fig. 2). Neither LBM virus was found to be related to other poultry introduction events in this study. Reassortment may have played a role in the evolutionary pathways of viruses associated with LBM-NY-2006(H5N2) and LBM-NJ-2001(H5N2). Phylogeographic analyses suggest that viruses from several functional groups may have contributed to the evolution of viruses detected in LBMs (Table S4). The viruses from the unknown group could include those from other reservoirs (e.g., wild bird) for which we lack data or uncontrolled viruses in the LBMs. Thus, these H5 viruses can be associated with a single introduction of the H5 gene, but NA or internal genes could be associated with viruses (all HA/NA subtypes rather than only the H5 subtype) that may circulate in LBMs or with introductions from other reservoirs (e.g., wild birds).
Compared with H5 viruses detected on CPFs, those detected in LBMs were more temporally and spatially diverse. For example, four LBM introductions events included detections over several years and/or multiple states [LBM-NJ-2007(H5N2), LBM-NY-2006(H5N2), LBM-NJ-2006(H5N2)b, and LBM-NJ-2001(H5N2)], whereas CPF introduction events did not occur across years.
Temporal gaps exist between emergence and detection of H5 LPAIVs in domestic poultry in North America.
The temporal gap between the time of H5 virus introduction into poultry and flock detection was estimated. Molecular clock analysis was used to determine the time of most-recent common ancestor (TMRCA) between inferred wild-bird progenitors and poultry isolates, and documented detection dates were obtained for 9 introduction events (Table 3). Estimated temporal gaps for three CPF introductions were 146, 80, and 142 days (average ± standard deviation, 123 ± 37 days) (Table 3). The temporal gaps for seven LBM introductions varied from 97 to 487 days (average ± standard deviation, 231± 137 days) (Table 3).
TABLE 3.
Time to most recent common ancestor estimation of HA gene for low-pathogenic H5 avian influenza A virus introductions from commercial farms (CPFs) and live-bird markets (LBMs) in the United States and Canada (2001 to 2017)a
| Source | Introduction | Mean TMRCA | 95% HPDb |
First detected datec | Differenced (no. of days) | Avg (days) | SD | |
|---|---|---|---|---|---|---|---|---|
| Low | High | |||||||
| Commercial farms | CPF-WI-2017(H5N2) | 2016-10-07 | 2016-01-01 | 2016-06-18 | 2017-03-02 | 146 | 123 | 37 |
| CPF-MB-2010(H5N2) | 2010-09-06 | 2010-11-16 | 2010-06-02 | 2010-11-25 | 80 | |||
| CPF-VA-2007(H5N1) | 2007-02-19 | 2007-06-10 | 2006-10-09 | 2007-07-11 | 142 | |||
| Live-bird markets | LBM-NY-2007(H5N2) | 2007-07-24 | 2007-10-18 | 2007-04-02 | 2007-10-30 | 97 | 231 | 137 |
| LBM-PA-2007(H5N2) | 2007-06-23 | 2007-09-23 | 2007-03-14 | 2007-11-09 | 138 | |||
| LBM-NY-2006(H5N2) | 2006-02-12 | 2006-06-17 | 2005-10-09 | 2006-10-23 | 253 | |||
| LBM-NJ-2006(H5N2)a | 2005-09-26 | 2006-01-18 | 2005-05-17 | 2006-05-09 | 225 | |||
| LBM-NJ-2006(H5N2)b | 2006-01-09 | 2006-06-22 | 2005-07-10 | 2006-07-19 | 191 | |||
| LBM-NJ-2001(H5N2) | 2000-03-16 | 2001-03-01 | 1999-01-23 | 2001-07-16 | 487 | |||
For each introduction, all isolates with an exact sampling date were included in the analysis. Dates are in the format year-month-day.
HPD, highest posterior density.
First detected date of an introduction was defined as the earliest sampling date among all isolates within this introduction.
Difference between date of first detection and mean time to most recent common ancestor.
DISCUSSION
We investigated evolutionary pathways for 78 H5 LPAIVs detected in North American domestic poultry across CPFs, BYPs, GBPs, and LBMs during 2001 to 2017; our data suggest that the events were the results of 18 discrete virus introductions from wild birds. The H5 LPAIVs from North American poultry in this study share ancestry with viruses circulating among wild waterfowl, a finding generally consistent with those from other investigations of outbreaks in U.S. poultry production systems (3, 9, 17–21). Our findings support that H5 LPAIVs maintained in certain wild bird species represent an ongoing threat to domestic poultry in North America.
A previous study investigating the ancestral origins of an H7 HPAIV reported among turkeys in Indiana, United States, suggested that IAVs from diving ducks were associated with evolution of the virus ultimately introduced into the CPFs, resulting in an outbreak (3). Our results show limited evidence for diving-duck-associated IAVs contributing to the ancestry of H5 IAVs detected in North American poultry; instead, we found proportionally more evidence for contributions from dabbling ducks and geese/swans (Fig. 3; Table S4). These findings suggest multiple potential evolutionary pathways for IAVs that are introduced to domestic poultry, and they highlight one of the challenges for avian influenza surveillance in wild birds: identifying the most pertinent wild bird species to target for sample collection to obtain meaningful reference information for better understanding the emergence of IAVs among poultry (24, 25).
We explored the time and location of H5 IAV introductions detected in various poultry production systems, including multiple introductions into CPFs and LBMs, but did not identify any clear patterns for introductions. Instead, H5 IAV introductions appeared to be sporadic throughout the year and occur in states/provinces in multiple regions of the United States and Canada. However, it is possible that the number of purported H5 introductions identified in this study was too small to detect patterns or that our data were of insufficient resolution to accurately identify the true times/locations of introduction events.
Our results suggest that sublineages of contemporary North American origin H5 gene segments have different geographic distributions: sublineage 1 was predominantly detected in the Atlantic Flyway but was also detected at a lower frequency in other flyways, whereas sublineage 2 was most frequently detected in the Pacific Flyway but was also identified at lower relative abundance in other flyways. Recent genomic evidence indicates that viruses initially have modest fidelity to migratory bird flyways; however, over multiple years, IAV lineages tend to disperse flyways (26, 27). Sublineages 1 and 2 have been cocirculating in wild birds in North America since 2002, so their restricted gene flow appears to be atypical. Post hoc phylogeographic analyses did not show similar geographic patterns for other HA gene segment sublineages or for those of other genes (data not shown). Additional research is needed to identify genetic barriers in North America for these two H5 sublineages.
Our results provide evidence that, compared with H5 IAV introductions in North American LBMs, those in each CPF introduction had a single genotype and a more recent estimated TMRCA with viruses circulating among wild birds. This may indicate that poultry management activities in North America have successfully identified and quickly stamped out low-pathogenic H5 IAVs in domestic birds raised on CPFs before the viruses have reassorted or become highly pathogenic. IAVs associated with introductions in LBMs sometimes had more than one genotype and, compared with CPF-associated IAVs, had longer mean TMRCAs with inferred wild bird predecessor viruses. Thus, viruses in North American LBMs may have a longer opportunity to cocirculate with other IAVs and subsequently form reassortants. Furthermore, the detection of genetically similar viruses associated with a single introduction event across multiple years and U.S. states/Canadian provinces suggests that management activities relative to the detection and stamping out of H5 IAVs may be less efficient in LBMs than on CPFs.
On the basis of our results, we propose a conceptual model (Fig. 4) describing generalized evolutionary pathways for low-pathogenic H5 avian IAVs detected in North American domestic poultry. Our results suggest that H5 viruses detected in domestic poultry in the United States and Canada are descended from IAVs circulating among wild birds, typically waterfowl, that are periodically introduced into poultry production systems. It appears that, on CPFs, H5 IAVs are detected relatively quickly through surveillance efforts and that the viruses are effectively eradicated through slaughter and additional preventative measures, as witnessed by apparent independent evolutionary pathways for viruses of a single genotype associated with each introduction event. In contrast, our study provides evidence that viruses in LBMs are not detected as rapidly as those on CPFs and, therefore, may have a greater potential for genetic reassortment with other wild-bird-associated IAVs. The high mobility of birds sold in LBMs also facilitates the geographic spread of viruses in the U.S. states and Canada.
FIG 4.
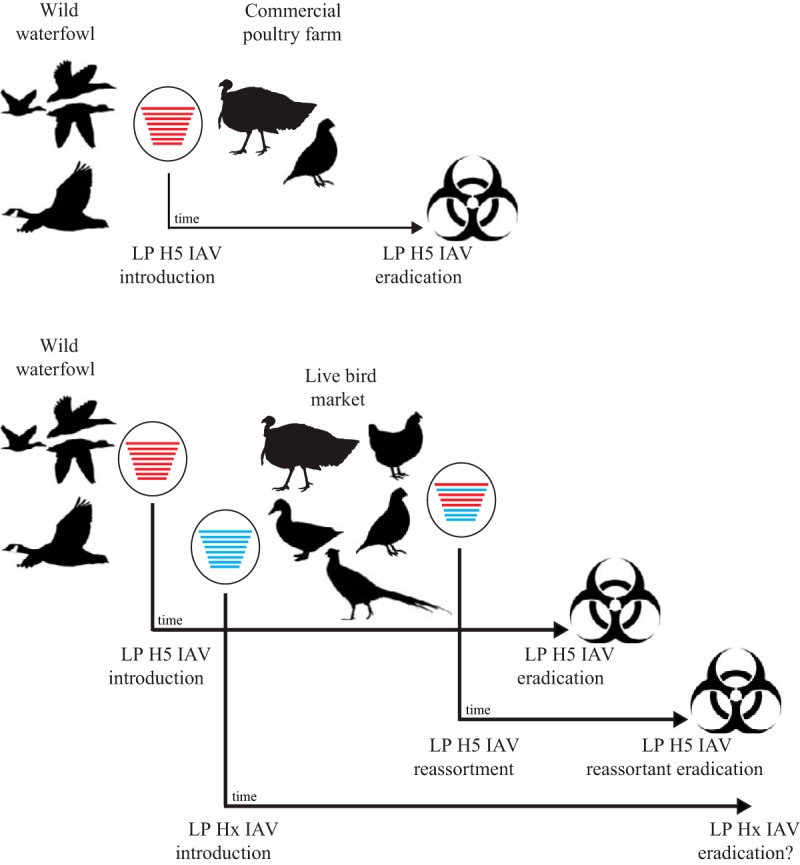
Conceptual model summarizing the generalized inferred evolutionary pathways for low-pathogenic (LP) H5 avian influenza A viruses (IAVs) detected on commercial poultry farms and in live-bird markets (LBMs) in the United States and Canada during 2001 to 2017. H5 viruses introduced by wild waterfowl were inferred to circulate for a longer time in LBMs than on commercial poultry farms. Furthermore, we found evidence suggesting reassortment between H5 viruses and other influenza A viruses in LBMs, resulting in multiple genotypes associated with a single introduction event. The blue and red lines denote genetically distinct gene segments; in each virus, the segments were vertically sorted in the order of PB2, PB1, PA, HA, NP, NA, MP, and NS.
In summary, the apparent repeated introduction of H5 IAVs from wild birds to domestic poultry in North America highlights the importance of avian IAV surveillance, particularly at the interface of wild birds and domestic poultry. Proactive and strategic surveillance covering multiple wild bird species and integrating genomic sequencing approaches is critical to understanding the evolutionary pathways of IAVs; such knowledge will be valuable in efforts to refine monitoring activities and optimize early-warning systems. In addition, our findings support the premise that avian H5 IAVs introduced from wild birds present an ongoing threat to domestic poultry and can be controlled effectively through management practices, particularly those implemented in North American CPFs.
MATERIALS AND METHODS
Data.
To understand the genesis of low-pathogenic avian H5 IAVs in North America, we sequenced 37 isolates recovered from domestic poultry samples collected during 2001 to 2017 (see Table S5 in the supplemental material); 27 of the isolates were H5N2 viruses (including mixed viruses) from LBMs, 5 were H5N2 viruses from CPFs, 1 was an H5N2 virus from a BYP, 1 was an H5N2 virus from a GBP, and 3 were H1N1 viruses from turkeys (n = 2) and a chicken (n = 1). Of note, if multiple isolates identified from the same case report had identical genomic sequences, only one of the isolates was selected for this study. We analyzed sequence data for these 34 poultry H5 isolates and 44 other North American poultry origin IAVs deposited in public databases (for a total of 78 H5 isolates) (Table 1). In addition, we sequenced 127 H5 IAV isolates originating from wild birds (mostly during 2015 and 2016, n = 116) in 30 U.S. states (Table S5); the isolates were from dabbling ducks (n = 94), shorebirds (n = 16), geese/swans (n = 10), gulls/terns/seabirds (n = 3), diving/sea ducks (n = 1), and unknown avian species (n = 3). Genomic sequencing and sequence assembly were performed as previously described (3); the GenBank accession numbers are listed in Table S5 and below. The geographic, temporal, and host distributions of all viruses that we sequenced are shown in Fig. S3 in the supplemental material. In this study, the IAVs from commercial flocks and LBMs were isolated during poultry premovement monitoring activities, suspected IAV outbreaks, or surveillance activities; the IAVs from wild birds were isolated during active influenza surveillance.
To perform systematic analyses, we retrieved the genomic sequences for all avian-origin IAVs from the following public databases in April 2017: the Influenza Virus Resources (28) (https://www.ncbi.nlm.nih.gov/genomes/FLU), the Influenza Research Database (29) (https://www.fludb.org), and GISAID (30) (https://www.gisaid.org). For those viruses for which we obtained redundant sequences, we only included the longest sequence contig per gene segment in analyses. In total, we used sequences from ∼20,000 influenza viruses worldwide, including >9,000 IAVs of North American origin (see Table S6 in the supplemental material). Because our preliminary phylogenetic analyses of these genomic sequences suggested that the low-pathogenic H5 IAV isolates in domestic poultry were genetically related to North American lineage IAVs (data not shown), we focused subsequent data analyses only on sequences for IAVs detected in North America. The temporal and host distributions of all viruses of North American origin included in this study are shown in Fig. S4 in the supplemental material.
Sequence alignment and phylogenetic analysis.
Multiple sequence alignments were generated using MAFFT v7.273 (31). Phylogenic analyses were performed using an approximate maximum-likelihood method with a generalized time-reversible substitution model and a “CAT” approximation rate model by using FastTree v2.1 (32). These preliminary trees provided initial inferences regarding tree topology among all sequences in the public databases, specifically to differentiate gene segment sequences of North American lineages from those of Eurasian lineages. A refined phylogenetic tree for each gene segment was then reconstructed using a maximum-likelihood method by running RAxML v 8.2.9 (33). A gamma model of rate heterogeneity and a generalized time-reversible substitution model were used for these phylogenetic analyses, and bootstrapping was conducted using the same rate and substitution models. Phylogenetic trees were visualized by ggtree v1.6.11 (34) and FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). Topologies of phylogenetic trees were validated using MrBayes 3.2.7 (35), PAUP* 4.0 (36), and PHYLIP 3.6 (37).
Genotype analyses and assignment of possible progenitor gene and potential precursor viruses for low-pathogenic H5 avian IAVs.
Genotypes for IAVs were assigned through analyses assessing genetic similarity among viral genome constellations. Gene segments were considered to be genotypically similar if they cooccurred in clades with other gene segment sequences (as determined using inferred tree topology) with a minimum bootstrap value of 70 and shared nucleotide sequence identities of ≥95% (18).
Possible progenitor gene segments and precursor viruses for H5 IAVs of North American poultry origin were also assessed using tree topology and sequence identities. Possible progenitor gene segments were inferred when the following criteria were met: (i) the candidate gene segment shared a phylogenetic clade with a minimum bootstrap value of 70 with a North American poultry gene segment; (ii) the candidate progenitor gene segment and North American poultry gene segment shared ≥98% nucleotide sequence identity; (iii) the candidate gene segment shared the highest nucleotide sequence identity with the poultry origin IAV gene segment in its genetic cluster; and (iv) the putative progenitor gene segment was detected prior to detection of the North American poultry IAV gene segment. A potential precursor virus was inferred when a virus had three or more possible progenitor gene segments from an H5 IAV identified in North American poultry.
Definition of purported H5 introduction into North American poultry.
Multiple H5 IAVs detected in North American domestic poultry were inferred to have resulted from the same introduction event if the following criteria were met: (i) the genetic sequence for the H5 HA gene segment of these two viruses shared a common clade with a minimum bootstrap value of 70; (ii) the H5 HA gene segment of these two viruses shared ≥98% nucleotide sequence identity; and (iii) the nucleotide sequence identity shared with H5 HA gene segments from IAVs originating from North American poultry was greater than that shared with gene segments from wild-bird origin IAVs.
Phylogeographic analyses to infer transition of viruses between wild birds and domestic poultry and between North American migratory bird flyways.
To enable inference of wild-bird hosts associated with H5 IAV introductions in North American domestic poultry, we performed phylogeographic analyses to assess support for associations between specific functional groups of wild-bird IAV hosts and H5 IAV gene segments detected in domestic poultry. Hosts of influenza viruses were categorized into 9 functional groups: dabbling duck, diving/sea duck, goose/swan, gull/tern/seabird, raptor, shorebird, other avian (i.e., American coot, double-crested cormorant, red-necked grebe, rock dove, and western grebe; n = 5), and unknown (i.e., hosts for whom the species was unclear or for which the domestic status was ambiguous, including ducks, geese, feces, fowl, and poultry; n = 22). To minimize sampling bias, we balanced the number of sequences in each host group by resampling: for each year, we randomly selected a maximum of 10 sequences from each functional group.
Phylogeographic analyses were performed as previously described (3, 23). An asymmetric substitution model with Bayesian stochastic search variable selection and a strict clock model were applied in the analyses. We used Markov chain Monte Carlo methods, setting the chain length to 100 million with sampling every 10,000 states. The convergence of each run was checked by Tracer v1.6 (http://beast.community/tracer) before continuing to the next step. All poorly configured states were removed according to a 10% burn-in rate. After that, maximum clade credibility phylogenetic trees were generated using TreeAnnotator v1.8.4 (38) (http://beast.community/treeannotator). The Bayes factor was calculated to indicate the statistical support level. Significant transition was indicated by a combination of a Bayes factor of ≥3 and a mean indicator of ≥0.5. Statistical support levels were interpreted from Bayes factors as follows: Bayes factor < 3, no support; 3 ≤ Bayes factor < 10, support; 10 ≤ Bayes factor < 100, strong support; 100 ≤ Bayes factor < 1,000, very strong support; and Bayes factor ≥ 1,000, decisive support. Maximum clade credibility phylogenetic trees were visualized by FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
In addition to the above-described analyses on the potential sources of wild bird hosts for the H5 IAV gene segments detected in domestic poultry, we also inferred the transition patterns of influenza A viruses across North American migratory bird flyways. We first designated each U.S. state and Canadian province into either the Atlantic, Mississippi, Central, or Pacific Flyway based on the administrative definition of North American migratory bird flyways (https://www.fws.gov/birds/management/flyways.php) (Fig. 1C). Phylogeographic analyses were then performed to assess support for transitions of IAVs between among migratory bird flyways. To minimize the biases for sample selection, we included all viruses in sublineages 1 and 2 (identified from the phylogenetic tree of the H5 gene) in the phylogeographic analyses. Phylogeographic analyses and data interpretation are the same as those described above.
TMRCA estimation.
The Bayesian Markov chain Monte Carlo method implemented in BEAST v1.8.4 (38) was used to estimate the substitution rates and TMRCA between inferred predecessor HA gene segments of IAVs identified in the wild-bird reservoir and H5 IAVs detected in poultry for which specific dates of detection were available. The SRD06 partitioned substitution model, uncorrelated lognormal relaxed clock model, and Bayesian skyline coalescent tree prior were implemented in the molecular clock analyses. Two independent runs with 100 million chain length (sampling frequency = 10,000) were combined by LogCombiner v1.8.4 (http://beast.community/logcombiner) and then analyzed by Tracer v1.6 using a 10% burn-in rate (http://tree.bio.ed.ac.uk/software/tracer/).
Accession number(s).
Sequences were deposited in GenBank under accession numbers CY235315 to CY235322, KU310460 to KU310475, KY131302 to KY131333, KY131357 to KY131364, KY550909 to KY550916, MF046190, MF046211, MF046222, MF046227, MF046237, MF046275, MF046276, MF046280, MF046298, MF046299, MF046306, MF046330, MF046351, MF046361, MF046366, MF046376, MF046389, MF046403, MF046406, MF046412, MF046415, MF046416, MF046434, MF046442, MF046448, MF046452, MF046468, MF046472, MF046481, MF046490, MF046496, MF046504, MF046507, MF046509, MF046514, MF046523, MF046531, MF046536, MF046544, MF046547, MF046556, MF046567, MF046570, MF359819 to MF359890, MF613674, MF613685, MF613713, MF613716, MF613724, MF613731, MF613745, MF613747, MF613753, MF613755, MF613762, MF613766, MF613772, MF613776, MF613777, MF613792, MF613809, MF613812, MF613817, MF613823, MF613869, MF613881, MF613903, MF613905, MF613918, MF613924, MF613928, MF613937, MF613940, MH341739 to MH341906, and MH546139 to MH547031.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Angela Danner and Karlie Woodard for their technical expertise, Kimberly Friedman for data management, and Canadian Wildlife Services, New Jersey Department of Environmental Protection, and Conserve Wildlife, LLC, for sample collection. We also thank numerous wildlife professionals at state and federal agencies who collected wild-bird samples and the National Animal Health Laboratory Network facilities that performed diagnostics on those samples.
This work was funded by the National Institutes of Health (NIH) (grant number R01AI116744), the U.S. Geological Survey through the Wildlife Program of the Ecosystem Mission Area, Centers of Excellence for Influenza Research and Surveillance, National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services, contract HHSN272201400006C, the American Lebanese Syrian Associated Charities; and the U.S. Department of Agriculture. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
A.S.B., T.J.D., S.K., M.K.T., A.M.R., D.E.S., R.J.W., and X.-F.W. initiated and designed this study; A.S.B., T.J.D., M.L.K., S.K., J.M.N., M.K.T., A.M.R., A.B.R., and D.E.S. collected data; L.L. and X.-F.W. performed experiments; L.L. and X.-F.W. wrote the first draft of the manuscript; and A.S.B., T.J.D., S.K., J.M.N., M.K.T., A.M.R., A.B.R., D.E.S., R.J.W., and X.-F.W. revised the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00913-18.
REFERENCES
- 1.Garcia M, Crawford JM, Latimer JW, Rivera-Cruz E, Perdue ML. 1996. Heterogeneity in the haemagglutinin gene and emergence of the highly pathogenic phenotype among recent H5N2 avian influenza viruses from Mexico. J Gen Virol 77:1493–1504. doi: 10.1099/0022-1317-77-7-1493. [DOI] [PubMed] [Google Scholar]
- 2.Killian ML, Kim-Torchetti M, Hines N, Yingst S, DeLiberto T, Lee D-H. 2016. Outbreak of H7N8 low pathogenic avian influenza in commercial turkeys with spontaneous mutation to highly pathogenic avian influenza. Genome Announc 4:e00457-16. doi: 10.1128/genomeA.00457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, Ramey AM, Bowman AS, DeLiberto TJ, Killian ML, Krauss S, Nolting JM, Torchetti MK, Reeves AB, Webby RJ, Stallknecht DE, Wan XF. 2017. Low-pathogenic influenza A viruses in North American diving ducks contribute to the emergence of a novel highly pathogenic influenza A(H7N8) virus. J Virol 91:e022080-16. doi: 10.1128/JVI.02208-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suarez DL, Senne DA, Banks J, Brown IH, Essen SC, Lee C-W, Manvell RJ, Mathieu-Benson C, Moreno V, Pedersen JC. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg Infect Dis 10:693. doi: 10.3201/eid1004.030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasick J, Handel K, Robinson J, Copps J, Ridd D, Hills K, Kehler H, Cottam-Birt C, Neufeld J, Berhane Y. 2005. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. J Gen Virol 86:727–731. doi: 10.1099/vir.0.80478-0. [DOI] [PubMed] [Google Scholar]
- 6.Khatchikian D, Orlich M, Rott R. 1989. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature 340:156–157. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- 7.Maurer-Stroh S, Lee RT, Gunalan V, Eisenhaber F. 2013. The highly pathogenic H7N3 avian influenza strain from July 2012 in Mexico acquired an extended cleavage site through recombination with host 28S rRNA. Virol J 10:139. doi: 10.1186/1743-422X-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee DH, Torchetti MK, Killian ML, Berhane Y, Swayne DE. 2017. Highly pathogenic avian influenza A(H7N9) virus, Tennessee, USA, March 2017. Emerg Infect Dis 23(11). doi: 10.3201/eid2311.171013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senne DA, Suarez DL, Stallnecht DE, Pedersen JC, Panigrahy B. 2006. Ecology and epidemiology of avian influenza in North and South America. Dev Biol (Basel) 124:37–44. [PubMed] [Google Scholar]
- 10.Xu Y, Cao H, Liu H, Sun H, Martin B, Zhao Y, Wang Q, Deng G, Xue J, Zong Y, Zhu J, Wen F, Long LP, Wong SS, Zhao N, Fu X, Liao M, Hu G, Webby R, Gao GF, Wan XF. 2015. Identification of the source of A (H10N8) virus causing human infection. Infect Genet Evol 30:159–163. doi: 10.1016/j.meegid.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. 25 January 2018. Influenza at the human-animal interface, summary and assessment, 8 December 2017 to 25 January 2018 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 13.Zhang T, Bi Y, Tian H, Li X, Liu D, Wu Y, Jin T, Wang Y, Chen Q, Chen Z. 2014. Human infection with influenza virus A (H10N8) from live poultry markets, China, 2014. Emerg Infect Dis 20:2076–2079. doi: 10.3201/eid2012.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardona C, Yee K, Carpenter T. 2009. Are live bird markets reservoirs of avian influenza? Poult Sci 88:856–859. doi: 10.3382/ps.2008-00338. [DOI] [PubMed] [Google Scholar]
- 15.Senne D, Suarez D, Pedersen J, Panigrahy B. 2003. Molecular and biological characteristics of H5 and H7 avian influenza viruses in live-bird markets of the northeastern United States, 1994-2001. Avian Dis 47:898–904. doi: 10.1637/0005-2086-47.s3.898. [DOI] [PubMed] [Google Scholar]
- 16.Suarez DL, Garcia M, Latimer J, Senne D, Perdue M. 1999. Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States. J Virol 73:3567–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halvorson DA, Frame DD, Friendshuh KAJ, Shaw DP. 1997. Outbreaks of low pathogenicity avian influenza in USA. Avian Dis 47:36–46. [Google Scholar]
- 18.Krauss S, Stucker KM, Schobel SA, Danner A, Friedman K, Knowles JP, Kayali G, Niles LJ, Dey AD, Raven G. 2015. Long-term surveillance of H7 influenza viruses in American wild aquatic birds: are the H7N3 influenza viruses in wild birds the precursors of highly pathogenic strains in domestic poultry? Emerg Microbes Infect 4:e35. doi: 10.1038/emi.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CW, Senne DA, Linares JA, Woolcock PR, Stallknecht DE, Spackman E, Swayne DE, Suarez DL. 2004. Characterization of recent H5 subtype avian influenza viruses from US poultry. Avian Pathol 33:288–297. doi: 10.1080/0307945042000203407. [DOI] [PubMed] [Google Scholar]
- 20.Lebarbenchon C, Pedersen JC, Sreevatsan S, Ramey AM, Dugan VG, Halpin RA, Ferro PJ, Lupiani B, Enomoto S, Poulson RL. 2015. H7N9 influenza A virus in turkeys in Minnesota. J Gen Virol 96:269–276. doi: 10.1099/vir.0.067504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramey AM, Torchetti MK, Poulson RL, Carter D, Reeves AB, Link P, Walther P, Lebarbenchon C, Stallknecht DE. 2016. Evidence for wild waterfowl origin of H7N3 influenza A virus detected in captive-reared New Jersey pheasants. Arch Virol 161:2519–2526. doi: 10.1007/s00705-016-2947-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yee KS, Novick CA, Halvorson DA, Dao N, Carpenter TE, Cardona CJ. 2011. Prevalence of low pathogenicity avian influenza virus during 2005 in two US live bird market systems. Avian Dis 55:236–242. doi: 10.1637/9427-061610-Reg.1. [DOI] [PubMed] [Google Scholar]
- 23.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown JD, Stallknecht DE. 2008. Wild bird surveillance for the avian influenza virus. Methods Mol Biol 436:85–97. doi: 10.1007/978-1-59745-279-3_11. [DOI] [PubMed] [Google Scholar]
- 25.Spackman E. 2009. The ecology of avian influenza virus in wild birds: what does this mean for poultry? Poult Sci 88:847–850. doi: 10.3382/ps.2008-00336. [DOI] [PubMed] [Google Scholar]
- 26.Lam TT, Ip HS, Ghedin E, Wentworth DE, Halpin RA, Stockwell TB, Spiro DJ, Dusek RJ, Bortner JB, Hoskins J, Bales BD, Yparraguirre DR, Holmes EC. 2012. Migratory flyway and geographical distance are barriers to the gene flow of influenza virus among North American birds. Ecol Lett 15:24–33. doi: 10.1111/j.1461-0248.2011.01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fries AC, Nolting JM, Bowman AS, Lin X, Halpin RA, Wester E, Fedorova N, Stockwell TB, Das SR, Dugan VG, Wentworth DE, Gibbs HL, Slemons RD. 2015. Spread and persistence of influenza A viruses in waterfowl hosts in the North American Mississippi migratory flyway. J Virol 89:5371–5381. doi: 10.1128/JVI.03249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J Virol 82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Squires RB, Noronha J, Hunt V, García-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN. 2012. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir Viruses 6:404–416. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bogner P, Capua I, Lipman DJ, Cox NJ. 2006. A global initiative on sharing avian flu data. Nature 442:981. doi: 10.1038/442981a. [DOI] [Google Scholar]
- 31.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 35.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 36.Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony, version 4.0 b10.
- 37.Felsenstein J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- 38.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.