IAVs emerge in humans from animal reservoirs, causing unpredictable pandemics. One of these pandemics was caused by an H1N1 virus in 2009, and this virus is still circulating seasonally. To analyze host-virus adaptations likely affecting influenza virus pathogenesis, protein amino acid sequences from viruses circulating at the beginning of the pandemic and those circulating currently were compared. Currently circulating viruses have incorporated amino acid changes in two viral proteins (NS1 and PA-X), affecting innate immune responses, and in the PA gene. These amino acid differences led to increased NS1-mediated and decreased PA-X-mediated inhibition of host gene expression. A recombinant pH1N1 virus containing PA, PA-X, and NS1 genes from recently circulating viruses is fitter in replication in tissue culture cells and in mice, and the virus is more pathogenic in vivo. Importantly, these results suggest that a balance in the abilities of NS1 and PA-X to induce host shutoff is beneficial for IAVs.
KEYWORDS: NS1, PA-X, gene expression inhibition, inflammatory responses, influenza virus, innate immunity, interferon responses
ABSTRACT
In 2009, a pandemic H1N1 influenza A virus (IAV) (pH1N1) emerged in the human population from swine causing a pandemic. Importantly, this virus is still circulating in humans seasonally. To analyze the evolution of pH1N1 in humans, we sequenced viral genes encoding proteins inhibiting general gene expression (nonstructural protein 1 [NS1] and PA-X) from circulating seasonal viruses and compared them to the viruses isolated at the origin of the pandemic. Recent pH1N1 viruses contain amino acid changes in the NS1 protein (E55K, L90I, I123V, E125D, K131E, and N205S), as previously described (A. M. Clark, A. Nogales, L. Martinez-Sobrido, D. J. Topham, and M. L. DeDiego, J Virol 91:e00721-17, 2017, https://doi.org/10.1128/JVI.00721-17), and amino acid changes in the PA-X protein (V100I, N204S, R221Q, and L229S). These amino acid differences between early and more recent pH1N1 isolates are responsible for increased NS1-mediated inhibition of host gene expression and decreased PA-X-mediated shutoff, including innate immune response genes. In addition, currently circulating pH1N1 viruses have acquired amino acid changes in the PA protein (V100I, P224S, N321K, I330V, and R362K). A recombinant pH1N1 virus containing PA, PA-X, and NS1 genes from currently circulating viruses is fitter in replication in cultured cells and in mice and is slightly more pathogenic than the original ancestor pH1N1 virus. These results demonstrate the need to monitor the evolution of pH1N1 in humans for mutations in the viral genome that could result in enhanced virulence. Importantly, these results further support our previous findings suggesting that inhibition of global gene expression mediated by NS1 and PA-X proteins is subject to a balance which can determine virus pathogenesis and fitness.
IMPORTANCE IAVs emerge in humans from animal reservoirs, causing unpredictable pandemics. One of these pandemics was caused by an H1N1 virus in 2009, and this virus is still circulating seasonally. To analyze host-virus adaptations likely affecting influenza virus pathogenesis, protein amino acid sequences from viruses circulating at the beginning of the pandemic and those circulating currently were compared. Currently circulating viruses have incorporated amino acid changes in two viral proteins (NS1 and PA-X), affecting innate immune responses, and in the PA gene. These amino acid differences led to increased NS1-mediated and decreased PA-X-mediated inhibition of host gene expression. A recombinant pH1N1 virus containing PA, PA-X, and NS1 genes from recently circulating viruses is fitter in replication in tissue culture cells and in mice, and the virus is more pathogenic in vivo. Importantly, these results suggest that a balance in the abilities of NS1 and PA-X to induce host shutoff is beneficial for IAVs.
INTRODUCTION
Influenza A viruses (IAVs) are members of the Orthomyxoviridae family of eight-segmented, single-stranded, negative-sense RNA viruses. IAVs are one of the main causes of respiratory infections in humans and are responsible for seasonal epidemics each year and occasional pandemics of great consequences. The first IAV pandemic in the 21st century started in 2009 with the emergence of a quadruple-reassortant swine-origin H1N1 IAV (pH1N1) (1, 2). Importantly, this virus is still circulating seasonally. Despite comprehensive vaccination programs, the WHO estimates that the global disease burden from seasonal influenza results in 1 billion infections, 3 million to 5 million cases of severe disease, and between 250,000 and 500,000 deaths annually (3).
The interferon (IFN) responses induced by the host after IAV infection limit virus replication (4). Therefore, to efficiently replicate within the host, IAVs encode at least 2 viral proteins (PA-X and nonstructural protein 1 [NS1]) displaying IFN antagonism activities (5).
IAV segment 3 encodes the PA and the PA-X proteins. PA is translated directly from the PA mRNA and required for virus replication and transcription (6). PA-X is translated as a +1 frameshift open reading frame (ORF) within the PA viral segment (7). PA-X shares the same first N-terminal 191 amino acids with the PA protein. However, PA-X contains a short C-terminal sequence (either 61 or 41 amino acids) produced by ribosomal frameshifting of the +1 reading frame of PA (7). PA-X shuts off host protein expression, contributing to blocking the cellular antiviral responses (7–12). This host shutoff activity of IAV PA-X is mediated by an endonucleolytic domain involved in degradation of host mRNAs, as mutations in the endonuclease active site render the protein inactive in inducing host shutoff (13). The host cellular shutoff activity of PA-X is stronger than that of PA or the N-terminal PA domain, indicating that the C-terminal region of PA-X contributes to the inhibition of host protein expression (13–15). Moreover, the PA-X protein has also been shown to be involved in modulating host inflammation, immune responses, apoptosis, and virus pathogenesis (7, 8, 16–19). Segment 3 of the pH1N1 IAV, encoding the PA and PA-X proteins, most likely originated from an avian virus (1).
IAV NS1 protein is encoded by segment 8 (or NS), a segment in human pH1N1 viruses likely derived from swine H1N1 viruses (1). IAV NS1 protein is the main protein responsible for counteracting innate immune responses induced by the host during infection (20). Accordingly, an IAV lacking NS1 (delNS1) or viruses containing deletions or mutations affecting NS1 functions have been shown to be attenuated and replication defective in IFN-competent systems, compared to wild-type (WT) viruses (21–27). Mechanisms involved in the ability of NS1 to counteract innate immune responses include inhibiting cellular transcription elongation, blocking posttranscriptional RNA processing and nuclear export, decreasing retinoic acid-inducible gene I (RIG-I) activation (28–31), interfering with IFN signaling, and directly inhibiting specific IFN-stimulated genes (ISGs) (20). In addition, NS1 proteins from some IAVs bind to cleavage- and polyadenylation-specific factor 30 (CPSF30), blocking pre-mRNA processing and the nuclear export of mRNAs (32–35), and to poly(A)-binding protein II (PABPII), inhibiting the ability of PABPII to stimulate the synthesis of long poly(A) tails (36).
Interestingly, the PA-X protein from pH1N1 viruses circulating at the beginning of the 2009 pandemic displayed host shutoff activity, inhibiting general gene expression (8), whereas the NS1 protein from the same viruses did not inhibit general host gene expression (37). In addition, using a live attenuated influenza virus backbone, we have shown that recombinant pH1N1 viruses encoding both NS1 and PA-X inhibiting or not general gene expression are impaired in viral growth in cultured cells and attenuated in vivo compared to recombinant pH1N1 viruses in which only one of the viral proteins (NS1 or PA-X) inhibited host gene expression (19), suggesting that inhibition of host protein expression by pH1N1 is subject to a strict balance which can determine the successful progression of viral infection (19). However, it is unknown whether the relationship between NS1 and PA-X host cell-inhibitory activities still exists in pH1N1 viruses currently circulating in humans, more notably since the gain of inhibition of host gene expression in NS1 of currently circulating pH1N1 viruses in recent viral human isolates (38).
We found that compared to the pH1N1 viruses infecting humans at the beginning of the 2009 pandemic, currently circulating pH1N1 viruses contain 4 amino acid changes in the PA-X protein (V100I, N204S, R221Q, and L229S) as well as 5 amino acid changes in the PA protein (V100I, P224S, N321K, I330V, and R362K). Interestingly, our data indicate that the PA-X protein from currently circulating pH1N1 viruses possesses decreased host shutoff activity. Moreover, we previously showed that currently circulating pH1N1 viruses contain 6 amino acid changes in NS1 (E55K, L90I, I123V, E125D, K131E, and N205S) that restored the ability of NS1 to inhibit host gene expression (38). In this study, we have analyzed the interplay between PA-X and NS1 of pH1N1 viruses from the beginning of the pandemic and those that are currently circulating in humans. To evaluate the effect of both the NS1 and PA genes on virus fitness and virulence, recombinant pH1N1 viruses encoding NS1 and PA genes from the original or currently circulating pH1N1 viruses were generated. We show that a recombinant pH1N1 virus containing both PA and NS1 genes from currently circulating viruses is fitter in replication in tissue culture cells and in vivo. In addition, this virus is slightly more pathogenic in mice than the pH1N1 virus at the start of the pandemic. The data presented here are consistent with our previous findings and suggest that optimal control of gene expression by NS1 and PA-X is important for pH1N1 pathogenesis. Moreover, our data highlight the fact that studying more than one gene at a time is a fruitful exercise that provides important insights into host-virus adaptations affecting viral pathogenesis and evolution.
RESULTS
Identification of amino acid changes in the PA gene from currently circulating pH1N1 viruses.
To analyze whether the PA segment from currently circulating human pH1N1 viruses has incorporated mutations since the virus emerged in 2009, we sequenced the PA segments of human clinical viruses circulating during the 2015–2016 season in the Rochester, NY, area (subjects ACU001, ACU004, ACU005, ACU007, ACU008, ACU009, ACU012, ACU017, and ACU022, named with the last three numbers). The amino acid sequences from the 9 human clinical isolates were identical and showed 4 amino acid changes (V100I, N204S, R221Q, and L229S) in the PA-X protein (Table 1) and 5 amino acid changes (V100I, P224S, N321K, I330V, and R362K) in the PA protein (Table 2), compared to the A/California/04/2009 pH1N1 strain, circulating at the origin of the pandemic.
TABLE 1.
Amino acid changes in PA-X proteins from influenza pH1N1 viruses circulating in the Rochester, NY, area during the 2015–2016 season
| Virus | Amino acid at position: |
|||
|---|---|---|---|---|
| 100 | 204 | 221 | 229 | |
| A/California/04/2009 | V | N | R | L |
| Isolate from patient: | ||||
| 001 | I | S | Q | S |
| 004 | I | S | Q | S |
| 005 | I | S | Q | S |
| 007 | I | S | Q | S |
| 008 | I | S | Q | S |
| 009 | I | S | Q | S |
| 012 | I | S | Q | S |
| 017 | I | S | Q | S |
| 022 | I | S | Q | S |
TABLE 2.
Amino acid changes in PA proteins from influenza pH1N1 viruses circulating in the Rochester, NY, area during the 2015–2016 season
| Virus | Amino acid at position: |
||||
|---|---|---|---|---|---|
| 100 | 224 | 321 | 330 | 362 | |
| A/California/04/2009 | V | S | N | I | R |
| Isolate from patient: | |||||
| 001 | I | P | K | V | K |
| 004 | I | P | K | V | K |
| 005 | I | P | K | V | K |
| 007 | I | P | K | V | K |
| 008 | I | P | K | V | K |
| 009 | I | P | K | V | K |
| 012 | I | P | K | V | K |
| 017 | I | P | K | V | K |
| 022 | I | P | K | V | K |
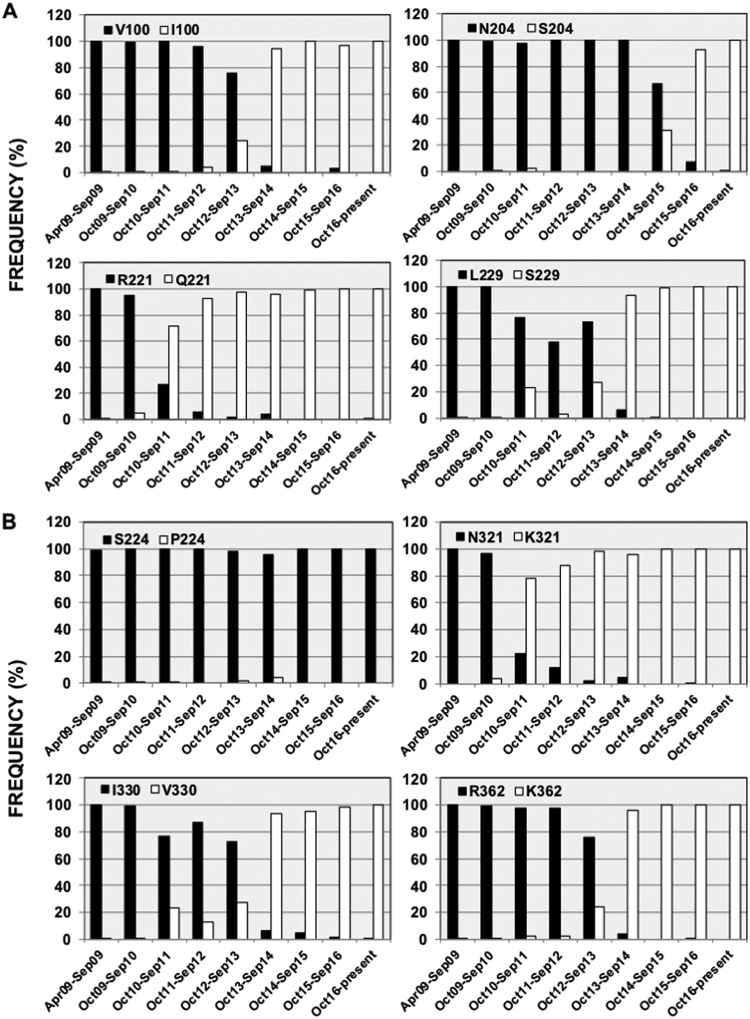
To analyze whether these amino acid changes in the PA and PA-X proteins of the 2015–2016 circulating pH1N1 virus are also found in viruses circulating globally, sequences available in the Influenza Research Database (https://www.fludb.org/) were analyzed. Notably, since October 2016, almost 100% of the pH1N1 viruses circulating worldwide encode these amino acid changes in both the PA (Fig. 1A) and PA-X (Fig. 1B) proteins. We plotted the prevalence of these mutations over time, showing the percentage of sequences encoding the different amino acids at each particular position in the PA-X (Fig. 1A) and PA (Fig. 1B) proteins. The first mutation selected in PA-X led to the R221Q amino acid change, and during the 2011–2012 season, more than 90% of the pH1N1 virus circulating contained this mutation (Fig. 1A). Mutations V100I and L229S were then selected, with more than 90% of the sequences encoding these amino acid changes during the 2013–2014 season (Fig. 1A). Finally, mutation N204S was selected, and during the 2013–2014 season, more than 90% of the circulating pH1N1 viruses contained this mutation (Fig. 1A). For the PA protein (Fig. 1B), mutation P224S appeared right after the pandemic started, with more than 98% of the sequences encoding this mutation in as early as April to September 2009 (Fig. 1B). At the origin of the pandemic, just taking into account sequences from people infected during April 2009, the percentage of sequences containing the P224S mutation increased to 92% (285 sequences [data not shown]), suggesting that this is an adaptive mutation that was selected very quickly. Next, the amino acid change N321K was selected, with more than 90% of the sequences encoding this amino acid change in PA during the 2011–2012 season (Fig. 1B). Finally, amino acid changes V100I (shared with the PA-X protein) (Fig. 1A), I330V, and R362K were selected, with more than 90% of the pH1N1 PA sequences containing these amino acid changes during the 2013–2014 season (Fig. 1B). Our data suggest that these changes in the pH1N1 PA and PA-X proteins are likely beneficial for the virus as they became fixed at the global level.
FIG 1.
Frequencies of identified mutations in currently circulating pH1N1 viruses over time. Publicly available sequences in the Influenza Research Database were downloaded, and the frequencies of PA-X (A) and PA (B) sequences containing the original amino acids present in A/California/04/2009 (black) and the mutant amino acids (white) are represented according to the month and season of virus isolation. Mutation V100I affecting both PA-X and PA amino acid sequences is represented only in panel A for simplicity. The numbers of sequences available in the Influenza Research Database (https://www.fludb.org/) for the periods April to September 2009, October 2009 to September 2010, October 2010 to September 2011, October 2011 to September 2012, October 2012 to September 2013, October 2013 to September 2014, October 2014 to September 2015, October 2015 to September 2016, and October 2016 to the present were 2,654, 1,744, 599, 44, 611, 238, 129, 1,766, and 960, respectively.
Increased NS1 and decreased PA-X shutoff activity in currently circulating pH1N1 viruses.
PA-X proteins from currently circulating pH1N1 viruses have an intact frameshift sequence but contain 4 amino acid changes (3 of them in the C-terminal end, after the frameshifting sequence, not affecting the PA protein amino acid sequence) compared to the 2009 pH1N1 strain A/California/04/2009 (Table 1 and Fig. 1A; see also Fig. 4). The NS1 protein from currently circulating pH1N1 viruses encodes 6 amino acid changes increasing the ability of the NS1 protein to block host gene expression, as we previously described (38). In addition, we have recently reported that pH1N1 viruses encoding PA-X and NS1 proteins that simultaneously inhibit or do not inhibit host gene expression are less fit in vitro and more attenuated in vivo than pH1N1 viruses encoding only one of the viral proteins (either NS1 or PA-X) able to inhibit host gene expression (19). In light of those previous data, we hypothesized that since the NS1 proteins of currently circulating human pH1N1 viruses have restored their ability to inhibit host gene expression, the mutations found in the PA-X proteins of recent pH1N1 isolates would result in decreased cellular host shutoff activity. To analyze whether this was the case, HEK293T cells were cotransfected with pCAGGS expression plasmids encoding green fluorescent protein (GFP) and Gaussia luciferase (Gluc) together with plasmids encoding hemagglutinin (HA) epitope-tagged WT (NS1WT and PA-XWT) or mutant (NS1MUT and PA-XMUT) viral proteins alone or in combination (Fig. 2), and the levels of Gluc (Fig. 2A) and GFP (Fig. 2B) expression were determined at 24 h posttransfection (hpt). In this work, WT refers to the sequence of the A/California/04/2009 H1N1 strain, circulating at the origin of the pandemic, whereas MUT refers to the sequence of currently circulating H1N1 viruses. As expected, pH1N1 NS1WT did not inhibit host gene expression as determined by levels of Gluc and GFP (Fig. 2A and B, respectively), whereas pH1N1 NS1MUT efficiently blocked host gene expression (37, 38). In contrast, pH1N1 PA-XWT inhibited host gene expression, while PA-XMUT was not as efficient in inhibiting host gene expression, as determined by Gluc (Fig. 2A) or GFP (Fig. 2B) expression. When expressed together, NS1MUT and PA-XWT exhibited the greatest inhibition of host gene expression, whereas NS1WT and PA-XMUT had the least efficacy in inhibiting reporter gene expression (Fig. 2A and B). Notably, when PA-X and NS1 protein expression levels were evaluated by Western blotting, NS1WT protein expression levels were higher than those of NS1MUT, while the levels of PA-XWT were lower than those of PA-XMUT (Fig. 2C). When simultaneously expressed, only pH1N1 NS1 proteins were detected by Western blotting. This is likely due to the lower levels of PA-X expression detected by Western blotting when expressed alone (Fig. 2C). Altogether, these data confirm that the NS1 protein of currently circulating human pH1N1 viruses acquired the ability to inhibit host gene expression (38) and that the 4 amino acid changes (V100I, N204S, R221Q, and L229S) present in the PA-X protein of recently circulating human pH1N1 viruses reduce PA-X's ability to inhibit host gene expression.
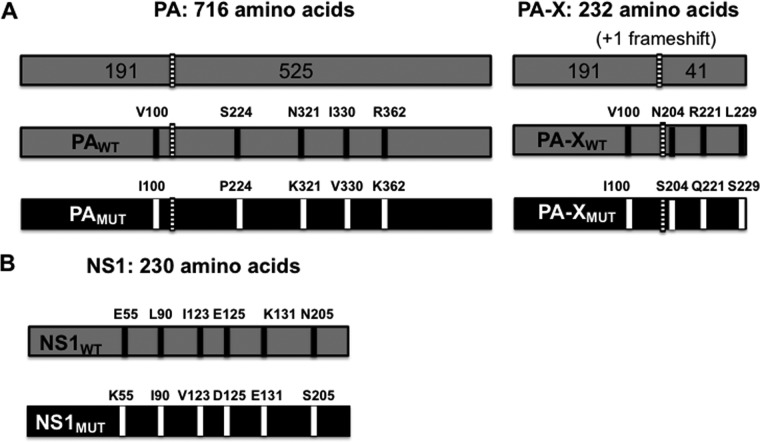
FIG 4.
Schematic representation of the recombinant pH1N1 viruses. (A) PA (left) and PA-X (right) WT (gray) and MUT (black) viral proteins. The 5 amino acid changes in pH1N1 PA (V100I, P224S, N321K, I330V, and R362K) and the 4 amino acid changes in pH1N1 PA-X (V100I, N204S, R221Q, and L229S) selected in currently circulating pH1N1 viruses are indicated. Numbers on the top indicate the amino acid lengths of the PA and PA-X proteins. The PA and PA-X proteins share the N-terminal 191 amino acids. The +1 frameshift is indicated with a striped bar. (B) pH1N1 NS1 WT (gray) (top) and MUT (black) (bottom) proteins containing 6 amino acid differences (E55K, L90I, I123V, E125D, K131E, and N205S) in currently circulating strains (38) are indicated. The number on the top indicates the amino acid length of WT and MUT NS1 proteins.
FIG 2.
Effect of pH1N1 NS1 and PA-X amino acid changes on inhibition of host gene expression. HEK293T cells were transiently cotransfected with pCAGGS plasmids expressing the indicated HA epitope-tagged NS1 and PA-X proteins, along with Gluc- and GFP-expressing plasmids. An empty pCAGGS plasmid was included as an internal control. NS1MUT encodes the amino acid changes E55K, L90I, I123V, E125D, K131E, and N205S. PA-XMUT encodes the amino acid changes V100I, N204S, R221Q, and L229S. At 24 hpt, Gluc (A), GFP (B), and NS1 (C) protein expression levels were analyzed. (A) Gluc expression was quantified in a Lumicount luminometer. Error bars represent the standard deviations for triplicates. *, P < 0.05 (NS1WT versus NS1MUT, PA-XWT versus PA-XMUT, NS1MUT/PA-XMUT versus NS1MUT/PA-XWT, and NS1WT/PA-XMUT versus NS1WT/PA-XWT) using Student's t test (n = 3 per time point). (B) GFP was visualized using a fluorescence microscope, and representative images obtained with a 20× objective are shown. Bars, 100 μm. (C) pH1N1 NS1 and PA-X and cellular actin protein expression levels were analyzed by Western blotting using cell extracts and antibodies specific to the HA epitope tag (to detect PA-X and NS1 proteins [top and bottom bands in the top blot, respectively]) and actin (loading control) (bottom blot). Western blots were quantified by densitometry using ImageJ software (v1.46), and the amounts of PA-X and NS1 proteins were normalized to the amounts of actin (top and bottom numbers, respectively, between the two blots). Protein expression levels in cells transfected with the pCAGGS plasmid expressing NS1WT and PA-XWT were considered 100%. ND, not detected. Molecular mass markers (in kilodaltons) are indicated on the left. The experiments were repeated three times, with similar results.
IAV NS1 and PA-X proteins have been shown to thwart the innate immune system (reviewed in references 20 and 39). To investigate how WT and MUT NS1 and PA-X proteins alone or in combination modulate innate immune responses, HEK293T cells were cotransfected with pCAGGS plasmids expressing WT or MUT NS1 and PA-X, alone or in combinations, together with reporter plasmids expressing firefly luciferase (Fluc) under the control of IFN-β (Fig. 3A) or IFN-stimulated response element (ISRE) (Fig. 3B) promoters. At 24 hpt, cells were mock infected (M) or infected (multiplicity of infection [MOI] of 3) with Sendai virus (SeV), and promoter activation was evaluated at 16 h postinfection (hpi) by assessing Fluc expression levels. As expected, SeV infection induced high levels of Fluc expression driven by the IFN-β (Fig. 3A) and ISRE (Fig. 3B) promoters in cells transfected with an empty (E) plasmid. However, activation of IFN-β and ISRE promoters in cells transfected with PA-X and/or NS1 proteins was significantly reduced (Fig. 3A and B, respectively), consistent with previous data showing that pH1N1 PA-X and NS1 proteins efficiently counteract IFN responses (7, 20, 38). As expected, inhibition of IFN-β and ISRE promoter activation by NS1MUT was more efficient than that by NS1WT (Fig. 3A and B, respectively) (38). Remarkably, PA-XMUT did not inhibit SeV-mediated activation of the IFN-β or ISRE promoters as efficiently as PA-XWT, when the viral protein was expressed either alone or together with WT or MUT pH1N1 NS1 (Fig. 3A and B, respectively). Importantly, the levels of NS1 and PA-X protein expression, as determined by Western blotting (Fig. 3C), were similar to those observed when we assessed their ability to inhibit host gene expression (Fig. 2C). We observed higher levels of NS1WT expression than of NS1MUT (Fig. 2C and 3C). In contrast, higher levels of PA-XMUT than of PA-XWT were observed when transfected alone (Fig. 2C and 3C). In contrast, we were not able to detect PA-X protein expression in the presence of either NS1WT or NS1MUT, likely due to lower levels of PA-X expression (Fig. 2C and 3C). These data indicate that the NS1 protein from currently circulating human pH1N1 viruses has evolved to increase its ability to counteract innate immune responses, whereas the PA-X protein has evolved to have a decreased ability to counteract IFN and IFN-stimulated responses.
FIG 3.
Effect of pH1N1 NS1 and PA-X amino acid changes on IFN responses induced by SeV infection. (A and B) HEK293T cells were transiently cotransfected, using calcium phosphate, with the indicated HA-tagged NS1- and PA-X-expressing pCAGGS plasmids, together with plasmids expressing Fluc under the control of an IFN-β (A) or an ISRE (B) promoter. NS1MUT encodes the amino acid changes E55K, L90I, I123V, E125D, K131E, and N205S. PA-XMUT encodes the amino acid changes V100I, N204S, R221Q, and L229S. An empty (E) pCAGGS plasmid was included as an internal control. At 24 hpt, cells were mock infected (M) or infected (MOI of 3) with the SeV Cantell strain (+SeV) to induce the activation of the IFN-β (A) or the ISRE (B) promoters. At 16 hpi, cell lysates were prepared for reporter gene expression. Fluc expression was measured by luminescence. Data show the means and standard deviations of the results determined for triplicate wells. Experiments were repeated 3 times in triplicate, with similar results. *, P < 0.05 (NS1WT versus NS1MUT, PA-XWT versus PA-XMUT, NS1MUT/PA-XMUT versus NS1MUT/PA-XWT, and NS1WT/PA-XMUT versus NS1WT/PA-XWT) using Student's t test (n = 3 per time point). (C) pH1N1 PA-X and NS1 and cellular actin protein expression levels were analyzed by Western blotting using cell extracts and antibodies specific to the HA tag (to detect PA-X and NS1 proteins [top and bottom bands in the top blot, respectively]) and actin (loading control) (bottom blot). Western blots were quantified by densitometry using ImageJ software (v1.46), and the amounts of PA-X and NS1 proteins were normalized to the amounts of actin (top and bottom numbers, respectively, between the two blots). Protein expression in cells transfected with the pCAGGS plasmid expressing NS1WT and PA-XWT was considered 100%. ND, not detected. Molecular mass markers (in kilodaltons) are indicated on the left. The experiments were repeated 3 times, with similar results.
Effect of NS1 and PA-X pH1N1 mutations on virus growth.
To analyze whether NS1 and PA proteins encoded by recently circulating human pH1N1 viruses exert an effect on virus growth and pathogenesis, recombinant viruses were generated (Fig. 4). To introduce amino acid changes only in the NS1 protein, and not in the overlapping nuclear export protein (NEP), we used a modified pH1N1 NS segment in which we split the ORFs of NS1 and NEP, as previously described (38, 40). All recombinant viruses encoded the HA protein from an egg-adapted pH1N1 strain (A/California/04/2009/E3), which is highly pathogenic in mice (38, 41). To analyze whether the identified PA and NS1 amino acid changes affect pH1N1 growth, Madin-Darby canine kidney (MDCK) cells were infected at a low MOI (0.001) and incubated at 33°C (Fig. 5A) or 37°C (Fig. 5B), and virus titers in cell culture supernatants were determined at different hours postinfection using an immunofocus assay. At earlier times after infection (24 and 48 hpi at 33°C and 12 and 24 hpi at 37°C), recombinant pH1N1 viruses encoding the currently circulating PA gene (PAMUT and PA-XMUT) grew with titers 5- to 10-fold higher than those of viruses encoding the PA gene of the original pH1N1 2009 virus (PAWT and PA-XWT). Correlating with the virus titers, the lysis plaques produced by infection with the recombinant viruses encoding PAMUT (NS1MUT/PAMUT and NS1WT/PAMUT) were larger at 33°C or 37°C than those observed for recombinant viruses encoding PAWT (NS1MUT/PAWT and NS1WT/PAWT) (Fig. 5C). To analyze whether the recombinant viruses carrying the currently circulating PA gene (PAMUT and PA-XMUT) grow slightly better in the more relevant human A549 lung cells, which have functional IFN induction and signal transduction pathways (42), A549 cells were infected (MOI of 0.1) and incubated at 33°C (Fig. 6A) and 37°C (Fig. 6B). As observed in MDCK cells, virus titers at 33°C were statistically significantly higher (5- to 10-fold) at 24 and 48 hpi for viruses carrying the PAMUT gene than for viruses carrying the PAWT gene (Fig. 6A). At 37°C, the viruses encoding PAMUT grew with slightly higher titers at 48 hpi (Fig. 6B). These results suggest that recombinant pH1N1 viruses encoding PAMUT (particularly NS1MUT/PAMUT, currently circulating in the human population) replicate to higher titers than viruses encoding PAWT in canine MDCK and human A549 cells. However, the presence of NS1WT or NS1MUT had a very minor effect on virus titers in cell cultures.
FIG 5.
Recombinant pH1N1 mutant virus growth kinetics in MDCK cells. (A and B) Canine MDCK cells were infected (MOI of 0.001) in triplicates with the recombinant viruses encoding different variants of NS1 and PA proteins (NS1MUT/PAMUT, NS1MUT/PAWT, NS1WT/PAMUT, and NS1WT/PAWT) and incubated at 33°C (A) or 37°C (B). Virus titers in infected cell TCS were determined at the indicated hours postinfection by an immunofocus assay. ns, not significant. (C) MDCK cells were infected with the indicated pH1N1 viruses and incubated at 33°C (top) or 37°C (bottom) for 3 days. Plaque phenotypes were assessed by immunostaining with the anti-NP monoclonal antibody HB-65.
FIG 6.
pH1N1 mutant virus growth kinetics in human A549 cells. (A and B) Human A549 cells were infected (MOI of 0.1) in triplicates with the recombinant viruses encoding different variants of NS1 and PA proteins (NS1MUT/PAMUT, NS1MUT/PAWT, NS1WT/PAMUT, and NS1WT/PAWT) and incubated at 33°C (A) or 37°C (B). Virus titers in infected cell TCS were determined at the indicated hours postinfection by an immunofocus assay. *, P < 0.05 (NS1MUT/PAMUT versus NS1MUT/PAWT, NS1MUT/PAMUT versus NS1MUT/PAWT, and NS1MUT/PAMUT versus NS1WT/PAWT) using Student's t test (n = 3 per time point); ns, not significant (P > 0.05). (C and D) Human A549 cells were infected (MOI of 0.1) in triplicates with viruses isolated from two patients (ACU009 and ACU022) infected during the 2015–2016 season (43) and with the A/California/04/2009 strain and incubated at 33°C (C) or 37°C (D). Virus titers in infected cell TCS were determined at the indicated hours postinfection by an immunofocus assay. *, P < 0.05 (virus isolates ACU009 and ACU022 versus A/California/04/2009).
To analyze whether the viruses circulating currently are more fit than the viruses circulating at the origin of the pandemic in 2009, the growth of two isolates from subjects ACU009 and ACU022, infected in the Rochester, NY, area during the 2015–2016 season (43) was compared to the growth of the A/California/04/2009 strain isolated at the beginning of the pandemic. To this end, human A549 lung cells were infected (MOI of 0.1), and virus titers in cell supernatants were analyzed at different times after infection (Fig. 6C and D). Currently circulating viruses grew with statistically significantly higher titers (around 10,000-fold higher) than the original A/California/04/2009 strain, at either 33°C (Fig. 6C) or 37°C (Fig. 6D). However, the recombinant viruses with mutations in the PA and NS1 genes grew to titers only 10-fold higher than those of the A/California/04/2009 strain (Fig. 5A and B and 6A and B), suggesting that mutations in other segments are also contributing to the higher fitness observed for currently circulating viruses in A549 cells. Accordingly, we have shown that the currently circulating H1N1 viruses have mutations in the HA and NA genes (43), likely contributing to virus fitness. Furthermore, mutations in other nonsequenced genes, such as the PB2, PB1, NP, M1, M2, and/or NEP genes, could also be contributing to the higher fitness observed for these virus isolates.
Effect of NS1 and PA-X mutations on pH1N1 virus pathogenesis in vivo.
To determine whether the amino acid changes selected in the NS1 and PA genes since the pH1N1 virus emerged in humans in 2009 provide the viruses a selective advantage in vivo, groups of mice (n = 5) were intranasally inoculated with 100 focus-forming units (FFU) (Fig. 7A), 1,000 FFU (Fig. 7B), and 10,000 FFU (Fig. 7C) of the recombinant pH1N1 viruses carrying the NS1 and PA genes in different combinations. Mice were then monitored for morbidity (weight loss [Fig. 7A to C, left]) and mortality (survival [Fig. 7A to C, right]) for 14 days. As expected, weight loss and survival correlated with the inoculated dose, with virulence being highest in animals infected with the highest viral doses (Fig. 7). The least virulent pH1N1 virus encoded NS1MUT/PAWT, as none of the mice inoculated with 100 or 1,000 FFU died, and only 20% died with the highest dose (10,000 FFU), with a maximum of 15% body weight loss by day 9 (Fig. 7). The next less virulent virus was pH1N1 NS1WT/PAWT. Mice inoculated with 100, 1,000, and 10,000 FFU lost averages of 13%, 13%, and 25% of their initial weight, respectively, and 100%, 80%, and 20% of the mice survived viral infection, respectively (Fig. 7). These results are in agreement with our previous data showing that recombinant pH1N1 viruses carrying an NS1MUT gene (and the PAWT gene) are slightly attenuated compared to a pH1N1 virus encoding NS1WT (38). Interestingly, and correlating with our in vitro observations, recombinant pH1N1 viruses encoding PAMUT showed increased virulence compared to recombinant pH1N1 viruses encoding WT PA (PAWT). Mice inoculated with 100 FFU of recombinant pH1N1 NS1MUT/PAMUT and NS1WT/PAMUT viruses had mortality rates of 20% and 40%, respectively (Fig. 7A). Furthermore, 60% and 100% of mice inoculated with 1,000 FFU of recombinant pH1N1 NS1MUT/PAMUT and NS1WT/PAMUT viruses succumbed to viral infection (Fig. 7B), and all the mice inoculated with 10,000 FFU of recombinant pH1N1 NS1MUT/PAMUT and NS1WT/PAMUT viruses rapidly lost weight and died between days 6 and 10 postinfection (Fig. 7C). From these experiments, the 50% mouse lethal doses (MLD50) for recombinant pH1N1 NS1MUT/PAMUT, NS1MUT/PAWT, NS1WT/PAMUT, and NS1WT/PAWT viruses were 480, >10,000, 147, and 3,162, respectively (Table 3). These data indicated that the most virulent virus was that containing NS1WT/PAMUT, followed by NS1MUT/PAMUT (∼3.3-fold-higher MLD50 than that of NS1WT/PAMUT), NS1WT/PAWT (MLD50 ∼21.5-fold higher than that of NS1MUT/PAMUT), and NS1MUT/PAWT, the most attenuated recombinant pH1N1 virus, with a MLD50 at least 68-fold higher than that of NS1MUT/PAMUT (Fig. 7 and Table 3).
FIG 7.
Virulence of recombinant pH1N1 viruses containing amino acid changes in NS1 and PA. Groups of 7- to 8-week-old C57BL/6 female mice (n = 5/group) were infected with 100 FFU/mouse (A), 1,000 FFU/mouse (B), or 10,000 FFU/mouse (C) of the recombinant viruses encoding different variants of NS1 and PA proteins (NS1MUT/PAMUT, NS1MUT/PAWT, NS1WT/PAMUT, and NS1WT/PAWT). Weight loss (left) and survival (right) were evaluated daily for 2 weeks.
TABLE 3.
MLD50 of pH1N1 viruses
a Mortality was determined over 2 weeks (n = 5).
To analyze whether the virulence observed correlates with viral replication in the lungs of infected animals, mice were infected with 1,000 FFU of each recombinant pH1N1 virus, and virus titers in lungs and nasal turbinates were evaluated at 2 and 4 days postinfection (dpi) (Fig. 8A and B, respectively). At day 2 after infection, recombinant pH1N1 viruses encoding PAMUT grew with titers ∼5- to 10-fold higher than those of the recombinant pH1N1 viruses carrying the PAWT gene in both the lungs (Fig. 8A) and the nasal turbinates (Fig. 8B). These results are in accordance with those obtained in vitro in MDCK cells (Fig. 5) and correlate with the higher virulence observed in mice (Fig. 7 and Table 3). Furthermore, the recombinant pH1N1 NS1MUT/PAMUT virus, similar to the pH1N1 virus currently circulating in humans, grew with slightly higher titers (∼2- to 3-fold) than the recombinant pH1N1 virus encoding NS1WT/PAMUT (Fig. 8A and B), likely explaining the selection of this virus in the human population.
FIG 8.
Growth and induction of innate immune responses in vivo of recombinant pH1N1 viruses encoding NS1 and/or PA variants. Groups of 7- to 8-week-old C57BL/6 female mice (n = 6/group) were infected with 1,000 FFU/mouse of NS1MUT/PAMUT, NS1MUT/PAWT, NS1WT/PAMUT, and NS1WT/PAWT recombinant pH1N1 viruses. (A and B) Mice were sacrificed at 2 dpi (n = 3) and 4 dpi (n = 3), and right lungs (A) and nasal turbinates (B) were harvested, homogenized, and used to quantify viral titers by an immunofocus assay (FFU per milliliter). (C) Left lungs were collected (n = 3), and total RNA was extracted to quantify levels of CCL2, TNF, IFN-β, and viral gene M1 mRNAs by RT-qPCR. *, P < 0.05 (NS1MUT/PAMUT versus NS1MUT/PAWT, NS1MUT/PAMUT versus NS1MUT/PAWT, and NS1MUT/PAMUT versus NS1WT/PAWT) using Student's t test (n = 3 per time point); ns, not significant (P > 0.05). r.u., relative units.
Host responses to infections are characterized by mechanisms mediated by the immune system in an attempt to stop and eliminate pathogens and to promote healing and recovery (44). However, an exacerbated inflammatory response mediated by IAV infection can be deleterious to the host (45–47). To analyze whether the different recombinant pH1N1 viruses induced differential innate immune responses, the lungs of infected mice in each group were subjected to reverse transcription-quantitative PCR (RT-qPCR) to evaluate the induction of IFN-β, tumor necrosis factor (TNF), and chemokine (C-C) motif ligand 2 (CCL2) genes at the mRNA level (Fig. 8C). The recombinant pH1N1 NS1MUT/PAMUT virus induced the strongest cytokine responses in vivo on day 2, likely because of the increased viral replication (Fig. 8A and B) and decreased host shutoff mediated by PAMUT (Fig. 2). Animals that were inoculated with PAWT-encoding recombinant pH1N1 viruses (NSWT/PAWT and NS1MUT/PAWT) showcased the lowest induction of inflammatory cytokines (such as CCL2 and TNF) and IFN-β (Fig. 8C). This was particularly true for mice infected with the recombinant pH1N1 virus encoding NS1MUT/PAWT, which showed lower levels of proinflammatory cytokines and IFN-β than mice infected with the recombinant pH1N1 NSWT/PAWT virus (Fig. 8C), as previously described (38). This is likely due to the increased inhibition of host gene expression mediated by NS1MUT compared to the NSWT protein (Fig. 2). By day 4, the levels of cytokines were very similar across all infected groups, but intriguingly, IFN-β levels appeared to be suppressed in mice infected with the two NS1MUT-encoding recombinant pH1N1 viruses (Fig. 8C), likely due to the increased inhibition of host gene expression mediated by NS1MUT compared to NS1WT (Fig. 2). Notably, mRNA expression levels of the viral M1 gene (Fig. 8C) correlated with lung viral titers (Fig. 8A).
DISCUSSION
IAV pathogenesis depends on the function of viral proteins and on host immune responses induced by viral infection (47). For IAV to replicate successfully in a host, the virus encodes proteins to either evade or block host responses (39). IAV NS1 and PA-X proteins are examples of such proteins able to block responses from the host, using diverse mechanisms. One of NS1's mechanisms to inhibit host gene expression involves binding to the 30-kDa subunit of CPSF30 (34, 48), effectively hindering cellular pre-mRNA processing and causing a global inhibition of host gene expression, including genes for the innate antiviral response (49). However, the ability of NS1 to bind to CPFS30 is not conserved among all IAVs, such as A/PuertoRico/8/34 (PR8) (33), pH1N1 (37), and H7N9 (50). The 2009 pH1N1 virus appears to compensate for this inability by encoding a PA-X protein that can block host protein expression (8). However, the impact of PA-X on viral pathogenicity is strain specific, and the loss of PA-X expression can increase (51) or decrease (52) viral replication and virulence.
Four amino acid changes in PA-X (V100I, N204S, R221Q, and L229S) (Fig. 1A and 4 and Table 1) and five amino acid changes in PA (V100I, P224S, N321K, I330V, and R362K) (Fig. 1B and 4 and Table 2) were observed between viruses circulating currently and pH1N1 viruses circulating at the origin of the pandemic. Interestingly, three out of the four mutations selected in PA-X are located in the short C-terminal end (41 amino acids) produced by ribosomal frameshifting of the PA gene (7) and do not affect the amino acid sequence of the PA protein. Host shutoff activity of pH1N1 PA-X is stronger than that of viral PA, indicating that the C-terminal region of PA-X contributes to the inhibition of host protein expression (13–15) and that these three amino acid changes could affect PA-X's endonuclease activity in currently circulating human pH1N1 viruses. Notably, PA and PA-X amino acid mutations appeared gradually in sequential order since 2009 (Fig. 1), and the selection of these mutations could be influenced by amino acids changes that occurred somewhere else in the viral genome, including NS1 (38). Nonetheless, these results indicate that the NS1 (38) and PA-X proteins of the 2009 pH1N1 virus have mutated to be better adapted in humans.
Viruses encoding NS1 and PA-X proteins that simultaneously inhibit or do not inhibit host gene expression were recently found to be less fit and highly attenuated in vivo using a live attenuated pH1N1 virus (19). In this case, mutations in the PA frameshift sequence, downregulating the expression of PA-X proteins, were introduced (19). Interestingly, we also found that the NS1 protein from currently circulating human pH1N1 viruses has regained the ability to inhibit general gene expression (38) (Fig. 2), whereas PA-X has decreased its ability to inhibit host gene expression (Fig. 2). These data suggest that a balance between the abilities of both pH1N1 viral proteins to inhibit host gene expression is needed during viral adaptation in the human population (19). We also evaluated the effect of pH1N1 NS1 and PA-X proteins on innate immune responses. As expected, amino acid changes in PA-X decreased its ability to inhibit innate immune responses (Fig. 3), whereas the amino acid changes increased the ability of pH1N1 NS1 to counteract host innate immune responses (Fig. 3). To analyze the effect of NS1 and PA-X variants on virus pathogenesis, recombinant pH1N1 viruses encoding the NS1 protein from the original 2009 virus (NS1WT) or currently circulating viruses (NS1MUT) and/or carrying PA genes (including PA-X) from the 2009 (PAWT) or present (PAMUT) pH1N1 viruses were generated. Recombinant pH1N1 viruses encoding PAMUT (particularly the virus encoding NS1MUT) grew to higher titers than recombinant pH1N1 viruses encoding PAWT (Fig. 5 and 8A). It was recently shown that mutation P224S in the PA gene of pH1N1 viruses (one of the mutations in currently circulating pH1N1 viruses in this study) increases virus replication and viral titers in the lungs of infected mice (53). These data suggest that the increased viral titers observed with our recombinant pH1N1 viruses encoding the PAMUT protein (Fig. 5 and 8A) might be due, at least in part, to this P224S mutation. It has also been shown that the absence or downregulation of PA-X expression in pH1N1 and H5N1 viruses actually increases viral replication and virulence in different animal model systems such as mice and avians (9, 51), providing another mechanism by which the recombinant pH1N1 viruses encoding the PAMUT (and PA-XMUT) proteins, defective in PA-X functions, grow better than recombinant pH1N1 viruses expressing WT PA (and PA-X) (Fig. 5 and 8A). In contrast to the PA gene, the presence or NS1WT or NS1MUT had a very minor effect on virus titers, as previously described (38). The recombinant pH1N1 NS1MUT/PAMUT virus reflects currently circulating pH1N1 viruses in humans, and the fact that this virus replicates and grows better likely explains its selection in the human population.
We found the NS1MUT/PAXMUT pH1N1 virus to be the most virulent, causing 100% mortality in mice (Fig. 7 and Table 3). Conversely, recombinant pH1N1 viruses that contained PAWT were more attenuated, with the most significant being NS1MUT/PAWT virus (Fig. 7 and Table 3). Finally, we examined the response of proinflammatory cytokines and chemokines upon viral infection with the recombinant pH1N1 NS1 and PA variants. The innate immune system has two distinct roles in the pathogenesis of IAV, where activation is protective against viral infection, but an uncontrolled response can cause severe damage (47). Remarkably, the recombinant pH1N1 NS1MUT/PAMUT virus induced the fastest and highest levels of cytokine responses (Fig. 8C), most likely because of the decreased shutoff properties of PAMUT and the highest titers observed for this virus in vitro and in vivo (Fig. 5 and 8A and B). These highest levels of proinflammatory cytokine induction (47) are likely responsible for the higher virulence of the recombinant pH1N1 NS1MUT/PAMUT virus than of recombinant viruses expressing the PAWT protein. According to these results, higher innate immune responses (including proinflammatory responses), correlating with higher virulence, were observed after infection with recombinant viruses expressing decreased levels of PA-X protein in vivo (7, 9, 18, 51). Similarly, it has been shown that increased levels of proinflammatory cytokine induction correlate with higher virulence with H5N1 IAVs (45, 46). Furthermore, mice and macaques infected with the 2009 pH1N1 virus induced higher levels of proinflammatory cytokines and chemokines than animals infected with a seasonal H1N1 A/Kawasaki/UTK-4/09 virus, resulting in higher pathogenicity (54). Two swine-origin pH1N1 viruses, one derived from a human patient and the other one derived from swine, were more virulent than a swine-origin 1918-like classical IAV in pigs (55). In addition, these viruses upregulated the expression of proinflammatory genes to a higher extent than the swine-origin 1918-like classical IAV, suggesting that both pH1N1 isolates are more virulent due in part to differences in the host transcriptional response during acute infection (55). This same argument is likely valid for the low virulence induced by our recombinant pH1N1 NS1MUT/PAWT virus. Compared to the recombinant pH1N1 NS1WT/PAWT virus, the NS1MUT/PAWT recombinant pH1N1 virus is slightly attenuated, likely due to the lower induction of proinflammatory cytokines, such as TNF and CCL2 (Fig. 8C) (38). Even though mice infected with the recombinant pH1N1 NS1WT/PAMUT virus showed a slight decrease in the induction of inflammatory cytokines compared to the NS1MUT/PAMUT recombinant pH1N1 virus, there was little initiation of IFN-β (Fig. 8C), possibly explaining the higher virulence of this virus than of the pH1N1 NS1WT/PAWT recombinant virus, suggesting that the balance between proinflammatory and IFN protective responses regulates virus pathogenesis.
In summary, we have shown that during the evolution of pH1N1 in humans, the virus has acquired mutations in both the PA-X and NS1 proteins. Mutations acquired in the PA-X protein resulted in decreased inhibition of host gene expression, while mutations in NS1 restored the ability of the viral protein to inhibit host gene expression (38). Our results support previous findings suggesting that inhibition of gene expression mediated by pH1N1 NS1 and PA-X proteins is subject to a strict balance which can determine viral pathogenesis, fitness, and host adaptation.
MATERIALS AND METHODS
Cell lines and viruses.
Madin-Darby canine kidney (MDCK) cells, human lung epithelial carcinoma A549 cells, and human embryonic kidney 293T (HEK293T) cells were grown in Dulbecco's modified Eagle's medium (DMEM; Mediatech, Inc.) that had been enriched with 10% fetal bovine serum (FBS) and 1% PSG (100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine) at 37°C with 5% CO2.
Recombinant pH1N1 viruses encoding a NS split segment where the coding regions of the NS1 and NEP genes were separated by the 2A autoproteolytic cleavage site of porcine teschovirus (38) were generated as previously described (56). Recombinant viruses encoded the HA protein from an egg-adapted pH1N1 strain (A/California/04/2009/E3), which is highly pathogenic in mice (38, 41). Recombinant pH1N1 viruses containing mutations in the NS1, PA, and PA-X proteins were generated using reverse genetics as previously described (57). The Cantell strain of Sendai virus (SeV) was used for the activation of IFN in the cell-based reporter assays (33). The influenza pH1N1 A/California/04/2009 strain was obtained from Bei Resources (catalog no. NR-13659). The currently circulating pH1N1 viruses ACU009 and ACU022 were isolated from nasal wash/swab specimens from patients named with the same IDs infected during the 2015–2016 season and amplified in MDCK cells as previously described (43).
PA and PA-X sequences.
RNA was obtained from 300 μl of patient nasal wash/swab specimens (subjects ACU001, ACU004, ACU005, ACU007, ACU008, ACU009, ACU012, ACU017, and ACU022, named with the last three numbers) using the QIAamp viral RNA extraction kit (Qiagen) according to the manufacturer's instructions. Reverse transcription (RT) reactions were performed for 2 h at 37°C by use of a high-capacity cDNA reverse transcription kit (Applied Biosystems) and the primers PA-NCR-5′-VS (5′-AGCGAAAGCAGGTACTGATCCAAAATGGAAGACTTTGTGCGACAATGC-3′), complementary to the 5′ noncoding region (NCR) and the first 24 nucleotides (nt) of the pandemic A/California/04/2009 H1N1 (pH1N1) influenza virus PA gene (GenBank accession no. FJ969514.1), and PA-3′-RS (5′-TAGTAGCATTGCCACAACTACTTCAGTGCATGTGTGAGGAAGG-3′), complementary to the last 26 nt of the pandemic A/California/04/2009 H1N1 (pH1N1) influenza virus PA gene and the 3′ NCR. The cDNAs were amplified by PCR using Platinum Pfx polymerase (Life Technologies) and the same VS and RS primers. Amplified PCR products were used for Sanger sequencing (Genewiz).
Human pH1N1 PA (and PA-X) sequences available at the Influenza Research Database (https://www.fludb.org/) were downloaded, introducing in the search the time periods shown in Fig. 1, and the sequences were aligned using the “analyze sequence variation (SNP)” function available in this database.
Plasmids.
pCAGGS protein expression plasmids encoding NS1 and PA-X variants N-terminally fused to an HA tag were obtained by PCR. Total RNAs extracted from MDCK cells infected with pH1N1 WT (A/California/04/2009) or with currently circulating pH1N1 viruses (referred to as MUT), isolated from nasal wash/swab specimens from infected subjects, were used for RT reactions. The RT reactions were performed at 37°C for 2 h by using a high-capacity cDNA reverse transcription kit (Applied Biosystems) and an oligo(dT) primer, to obtain the cDNAs. The cDNAs were amplified by PCR using Platinum Pfx polymerase (Life Technologies) and primers specific for NS1 and PA-X ORFs flanked by the restriction sites SphI and NheI. PCR products were then cloned into the pCAGGS expression plasmid digested with SphI and NheI.
To generate the pDZ-NSMUT and pDZ-NSWT plasmids, we previously cloned the coding regions of NS1WT and NS1MUT (containing the amino acid changes E55K, L90I, I123V, E125D, K131E, and N205S) into a pDZ plasmid carrying nonoverlapping NS1 and NEP genes (pDZ-NSs) separated by the porcine teschovirus 2A autoproteolytic site (38). The pDZ plasmid encoding the PAWT protein was kindly provided by Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai, New York, NY). The PAMUT plasmid (encoding the amino acid changes V100I, N204S, R221Q, and L229S in PA-X and the amino acid changes V100I, P224S, N321K, I330V, and R362K in PA) was constructed by obtaining the PAMUT gene by PCR. To this end, starting from the RNA extracted from infected cells, the cDNA was generated as described above for the generation of pCAGGS plasmids, and the cDNA was amplified using primers hybridizing at the 5′ and 3′ ends of the PA gene flanked by the restriction site SapI. PCR products were digested with the SapI restriction enzyme and cloned into the pDZ plasmid.
Virus rescues.
Cocultures (1:1) of HEK293T and MDCK cells (6-well plate format, 1 × 106 cells/well, in triplicates) were transiently cotransfected in suspension using DNA-In (MTI-GlobalStem) with 1 μg each of the six ambisense pDZ pH1N1 WT plasmids (pDZ-PB2, -PB1, -HA, -NP, -NA, and -M), kindly provided by A. García-Sastre at the Icahn School of Medicine at Mount Sinai, plus the pDZ-NSs plasmid encoding WT or MUT NS1 and/or the pDZ plasmid encoding WT and/or MUT PA (57). At 12 h posttransfection (hpt), transfection medium was replaced with DMEM containing 0.3% bovine serum albumin (BSA), antibiotics, and 1 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma). At 96 hpt, tissue culture supernatants (TCS) were collected and used to infect fresh monolayers of MDCK cells. At 3 days postinfection (dpi), recombinant viruses were plaque purified and scaled up using confluent monolayers of MDCK cells. Virus stocks were generated by infecting confluent 10-cm dishes of MDCK cells at a low multiplicity of infection (MOI of 0.001). Stocks were titrated by an immunofocus assay (focus-forming units [FFU] per milliliter) on MDCK cells (58). Briefly, MDCK cells were infected with 10-fold serial dilutions of the TCS. After 12 h at 33°C, cells were fixed and permeabilized with 1× phosphate-buffered saline (PBS) containing 4% formaldehyde and 0.5% Triton X-100 for 20 min at room temperature. Subsequently, cells were incubated in a blocking solution (1× PBS with 2.5% BSA) for 1 h at room temperature. Cells were then incubated with the anti-NP monoclonal antibody (MAb) HB-65 (ATCC HB-65) for 2 h at 37°C and thereafter washed with 1× PBS. Finally, cells were incubated at 37°C for 1 h with a fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG secondary antibody (Life Technologies). Viral titers (FFU per milliliter) were quantified by counting all the cells that were positive for NP expression (58). Microsoft Excel was used to calculate the means and standard deviations. The identities of the NS1 ORFs in the rescued viruses were confirmed by restriction analysis (data not shown) and sequencing (Genewiz).
Inhibition of host gene expression.
HEK293T cells (96-well plate format, 5 × 104 cells/well, in triplicates) were transiently cotransfected, using Lipofectamine 3000 (Thermo Fisher Scientific), with 200 ng/well of pCAGGS plasmids encoding either NS1 or PA-X proteins from the 2009 pH1N1 virus (NS1WT and PA-XWT) and/or from a currently circulating pH1N1 virus (NS1MUT and PA-XMUT) N-terminally fused to an HA tag, or an empty plasmid as an internal control, together with 50 ng/well of pCAGGS plasmids encoding green fluorescent protein (GFP) or Gaussia luciferase (Gluc). After 24 h, cells were evaluated for GFP expression under a fluorescence microscope, and Gluc activity was determined using a Biolux Gaussia luciferase reagent (New England BioLabs) and a Lumicount luminometer (Packard). Microsoft Excel was used to calculate the means and standard deviations. Statistical analysis was performed using a two-tailed Student t test.
Inhibition of IFN and ISRE promoters.
To evaluate the inhibition of IFN and IFN-stimulated response element (ISRE) promoter activation, HEK293T cells (96-well plate format, 5 × 104 cells/well, in triplicates) were transiently cotransfected, using a calcium phosphate mammalian transfection kit (Agilent Technologies), with 100 ng/well of pCAGGS plasmids encoding the indicated WT and/or MUT NS1 or PA-X proteins fused to an HA tag at the N terminus, or an empty plasmid as a control, together with 50 ng/well of plasmids expressing firefly luciferase (Fluc) under the control of the IFN-β or ISRE promoters (pIFN-β-Fluc and pISRE-Fluc, respectively) (33). After 20 hpt, cells were washed with 1× PBS and infected (MOI of 3) with the SeV Cantell strain for promoter activation. At 24 h postinfection (hpi), cells were harvested and lysed using passive lysis buffer (Promega). Luciferase expression in the cell lysates was determined using a dual-luciferase kit (Promega) according to the manufacturer's instructions. Measurements were recorded with a Lumicount luminometer (Packard), and the mean values and standard deviations were calculated with Microsoft Excel software. Statistical analysis was performed using a two-tailed Student t test.
SDS-PAGE and Western blot analysis.
Transfected cells were lysed in buffer containing 100 mM Tris-HCl (pH 6.8), 4% SDS, 20% glycerol, 0.2% bromophenol blue, and 20% β-mercaptoethanol and boiled for 10 min. Cell lysates were separated by SDS-PAGE and then transferred onto nitrocellulose membranes. Membranes were blocked for 1 h in 1× PBS containing 5% BSA and 0.1% Tween and then incubated with an anti-HA polyclonal antibody (Sigma) at 4°C overnight. An anti-β-actin monoclonal antibody (Sigma) was used as a loading control. Horseradish peroxidase (HRP) secondary antibodies (GE Healthcare) against either mouse or rabbit immunoglobulins (Ig) were used to detect bound primary antibodies. Protein expression was detected with a SuperSignal West Femto maximum-sensitivity chemiluminescent substrate kit (Thermo Scientific) in accordance with the manufacturer's instructions.
Virus growth kinetics.
Confluent monolayers of canine MDCK and human A549 cells (4 × 105 cells/well, 12-well plates, in triplicates) were infected at MOIs of 0.001 and 0.1, respectively, for 1 h at room temperature, using recombinant viruses; viruses isolated from subjects ACU009 and ACU022, infected during the 2015–2016 season (43); and the A/California/04/2009 strain (obtained from Bei Resources [catalog no. NR-13659]). Cells were then incubated in DMEM supplemented with 0.3% BSA, 1% ammonium persulfate, and 1 μg/ml TPCK-treated trypsin (Sigma) at 33°C or 37°C. At 12, 24, 48, and 72 hpi, tissue culture supernatants were collected and titrated on MDCK cells in 96-well plates (5 × 104 cells/well, in triplicates) by an immunofluorescence assay (FFU per milliliter) as described above. Microsoft Excel was used to calculate the means and standard deviations. Statistical analysis was performed using a two-tailed Student t test.
Plaque assay and immunostaining.
Confluent monolayers of MDCK cells (106 cells/well in a 6-well-plate format) were infected for 1 h at room temperature, and after virus adsorption, cells were overlaid with agar and incubated at 33°C or 37°C. At 3 dpi, cells were fixed with 4% paraformaldehyde for 15 min at room temperature. After the overlays were removed, cells were permeabilized (0.5% Triton X-100 in 1× PBS) for 15 min at room temperature, and immunostaining was performed as previously described (58, 59), using the NP MAb HB-65 (ATCC HB-65, clone HL16-L10-4R5) and vector kits (Vectastain ABC kit and DAB HRP substrate kit; Vector) according to the manufacturer's specifications.
In vivo experiments.
Seven- to eight-week-old wild-type (WT) female C57BL/6 mice were purchased from the Jackson Laboratory and maintained under pathogen-free conditions at the University of Rochester. All animal experiments were approved by the University Committee of Animal Resources and conformed to the recommendations contained in the Guide for the Care and Use of Laboratory Animals of the National Research Council (60). For viral infections, mice were first anesthetized intraperitoneally with 240 mg/kg of body weight of 2,2,2-tribromoethanol (Avertin) and then infected intranasally with 30 μl of the indicated virus preparations. Mice were examined each day for morbidity and mortality. Morbidity was determined by body weight loss and any symptoms of infection such as malaise or respiratory distress (data not shown). Percent body weight loss was determined relative to the starting weight. Mice that lost 25% of their initial body weight were considered to have reached the experimental endpoint and were humanely euthanized. The 50% mouse lethal dose (MLD50) for the recombinant pH1N1 viruses was determined using the method of Reed and Muench (61). At 2 and 4 dpi, lungs were surgically extracted and separated. The right lungs were placed into Eppendorf tubes and kept on dry ice for the determination of viral titers, while the left lungs were stored in RNAlater solution (Ambion). Nasal turbinates were also surgically extracted. Homogenization of the lungs and nasal turbinates was done via a Tenbroeck glass tissue grinder containing 1 ml of 1× PBS. Viral titers from homogenized right lungs and nasal turbinates were determined by an immunofluorescence assay (FFU per milliliter) as outlined above. Total RNAs were extracted with an RNeasy minikit (Qiagen) according to the manufacturer's recommendations. Reverse transcriptase reactions were conducted with a high-capacity cDNA transcription kit (Applied Biosystems) at 37°C for 2 h. Quantitative PCRs (qPCRs) were done by a TaqMan gene expression assay (Applied Biosystems) that is specific for the mRNA of the pH1N1 M1 gene, chemokine (C-C) motif ligand 2 (CCL2) (Mm00441242_m1), tumor necrosis factor (TNF) (Mm00443258_m1), and IFN-β (Mm00439552_s1). The 2−ΔΔCT method was used for quantification, and values are presented as fold induction (62). Statistical analysis for virus titers and qPCR results was performed using a two-tailed Student t test.
ACKNOWLEDGMENTS
We thank John J. J. Treanor and the Vaccine Research Unit at the University of Rochester Medical Center for providing the nasal wash/swab specimens from the “acute influenza” surveillance protocol (institutional review board [IRB] approval no. 09-0034). We thank Adolfo García-Sastre (Icahn School of Medicine at Mount Sinai, New York, NY) for providing the reverse genetics for influenza A/California/04/2009 H1N1.
This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Centers of Excellence in Influenza Research and Surveillance (CEIRS) contract no. HHSN272201400005C to D.J.T. and L.M.-S.; with funds from a University of Rochester research award to M.L.D. and A.N.; and with funds from the Comunidad de Madrid, Spain, to M.L.D. (reference no. 2017-T1/BMD-5155).
REFERENCES
- 1.Trifonov V, Khiabanian H, Rabadan R. 2009. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N Engl J Med 361:115–119. doi: 10.1056/NEJMp0904572. [DOI] [PubMed] [Google Scholar]
- 2.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, et al. . 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girard MP, Cherian T, Pervikov Y, Kieny MP. 2005. A review of vaccine research and development: human acute respiratory infections. Vaccine 23:5708–5724. doi: 10.1016/j.vaccine.2005.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klemm C, Boergeling Y, Ludwig S, Ehrhardt C. 17 January 2018. Immunomodulatory nonstructural proteins of influenza A viruses. Trends Microbiol doi: 10.1016/j.tim.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Resa-Infante P, Jorba N, Coloma R, Ortin J. 2011. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol 8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi T, MacDonald LA, Takimoto T. 2015. Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J Virol 89:6442–6452. doi: 10.1128/JVI.00319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, Mo Y, Wang X, Gu M, Hu Z, Zhong L, Wu Q, Hao X, Hu S, Liu W, Liu H, Liu X, Liu X. 2015. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J Virol 89:4126–4142. doi: 10.1128/JVI.02132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khaperskyy DA, McCormick C. 2015. Timing is everything: coordinated control of host shutoff by influenza A virus NS1 and PA-X proteins. J Virol 89:6528–6531. doi: 10.1128/JVI.00386-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khaperskyy DA, Schmaling S, Larkins-Ford J, McCormick C, Gaglia MM. 2016. Selective degradation of host RNA polymerase II transcripts by influenza A virus PA-X host shutoff protein. PLoS Pathog 12:e1005427. doi: 10.1371/journal.ppat.1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oishi K, Yamayoshi S, Kawaoka Y. 2015. Mapping of a region of the PA-X protein of influenza A virus that is important for its shutoff activity. J Virol 89:8661–8665. doi: 10.1128/JVI.01132-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi T, Chaimayo C, McGuinness J, Takimoto T. 2016. Critical role of the PA-X C-terminal domain of influenza A virus in its subcellular localization and shutoff activity. J Virol 90:7131–7141. doi: 10.1128/JVI.00954-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bavagnoli L, Cucuzza S, Campanini G, Rovida F, Paolucci S, Baldanti F, Maga G. 2015. The novel influenza A virus protein PA-X and its naturally deleted variant show different enzymatic properties in comparison to the viral endonuclease PA. Nucleic Acids Res 43:9405–9417. doi: 10.1093/nar/gkv926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desmet EA, Bussey KA, Stone R, Takimoto T. 2013. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol 87:3108–3118. doi: 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao HJ, Sun HL, Hu J, Wang JL, Xiong X, Wang Y, He QM, Lin Y, Kong WL, Seng LG, Pu J, Chang KC, Liu XF, Liu JH, Sun YP. 2015. Twenty amino acids at the C-terminus of PA-X are associated with increased influenza A virus replication and pathogenicity. J Gen Virol 96:2036–2049. doi: 10.1099/vir.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao HJ, Xu GL, Sun YP, Qi L, Wang JL, Kong WL, Sun HL, Pu J, Chang KC, Liu JH. 2015. PA-X is a virulence factor in avian H9N2 influenza virus. J Gen Virol 96:2587–2594. doi: 10.1099/jgv.0.000232. [DOI] [PubMed] [Google Scholar]
- 18.Hu J, Mo YQ, Gao Z, Wang XQ, Gu M, Liang YY, Cheng X, Hu SL, Liu WB, Liu HM, Chen SJ, Liu XW, Peng DX, Liu XF. 2016. PA-X-associated early alleviation of the acute lung injury contributes to the attenuation of a highly pathogenic H5N1 avian influenza virus in mice. Med Microbiol Immunol 205:381–395. doi: 10.1007/s00430-016-0461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogales A, Rodriguez L, DeDiego ML, Topham DJ, Martinez-Sobrido L. 2017. Interplay of PA-X and NS1 proteins in replication and pathogenesis of a temperature-sensitive 2009 pandemic H1N1 influenza A virus. J Virol 91:e00720-17. doi: 10.1128/JVI.00720-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 22.DeDiego ML, Nogales A, Lambert-Emo K, Martinez-Sobrido L, Topham DJ. 2016. NS1 protein mutation I64T affects interferon responses and virulence of circulating H3N2 human influenza A viruses. J Virol 90:9693–9711. doi: 10.1128/JVI.01039-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. 2017. NS1 protein amino acid changes D189N and V194I affect interferon responses, thermosensitivity, and virulence of circulating H3N2 human influenza A viruses. J Virol 91:e01930-16. doi: 10.1128/JVI.01930-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nogales A, Huang K, Chauche C, DeDiego ML, Murcia PR, Parrish CR, Martinez-Sobrido L. 2017. Canine influenza viruses with modified NS1 proteins for the development of live-attenuated vaccines. Virology 500:1–10. doi: 10.1016/j.virol.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol 79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pica N, Langlois RA, Krammer F, Margine I, Palese P. 2012. NS1-truncated live attenuated virus vaccine provides robust protection to aged mice from viral challenge. J Virol 86:10293–10301. doi: 10.1128/JVI.01131-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donelan NR, Basler CF, Garcia-Sastre A. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol 77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Z, Chen LM, Zeng H, Gomez JA, Plowden J, Fujita T, Katz JM, Donis RO, Sambhara S. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol 36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- 29.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M Jr, Garcia-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opitz B, Rejaibi A, Dauber B, Eckhard J, Vinzing M, Schmeck B, Hippenstiel S, Suttorp N, Wolff T. 2007. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol 9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 31.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, Krug RM, Montelione GT. 2008. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A 105:13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell 1:991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 35.Noah DL, Twu KY, Krug RM. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAs. Virology 307:386–395. doi: 10.1016/S0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Li Y, Krug RM. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J 18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hale BG, Steel J, Medina RA, Manicassamy B, Ye J, Hickman D, Hai R, Schmolke M, Lowen AC, Perez DR, Garcia-Sastre A. 2010. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J Virol 84:6909–6922. doi: 10.1128/JVI.00081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark AM, Nogales A, Martinez-Sobrido L, Topham DJ, DeDiego ML. 2017. Functional evolution of influenza virus NS1 protein in currently circulating human 2009 pandemic H1N1 viruses. J Virol 91:e00721-17. doi: 10.1128/JVI.00721-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasin AV, Temkina OA, Egorov VV, Klotchenko SA, Plotnikova MA, Kiselev OI. 2014. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res 185:53–63. doi: 10.1016/j.virusres.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Nogales A, DeDiego ML, Topham DJ, Martinez-Sobrido L. 2016. Rearrangement of influenza virus spliced segments for the development of live-attenuated vaccines. J Virol 90:6291–6302. doi: 10.1128/JVI.00410-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo H, Santiago F, Lambert K, Takimoto T, Topham DJ. 2011. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J Virol 85:448–455. doi: 10.1128/JVI.01812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J Virol 78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark AM, DeDiego ML, Anderson CS, Wang J, Yang H, Nogales A, Martinez-Sobrido L, Zand MS, Sangster MY, Topham DJ. 2017. Antigenicity of the 2015-2016 seasonal H1N1 human influenza virus HA and NA proteins. PLoS One 12:e0188267. doi: 10.1371/journal.pone.0188267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akira S. 2009. Pathogen recognition by innate immunity and its signaling. Proc Jpn Acad Ser B Phys Biol Sci 85:143–156. doi: 10.2183/pjab.85.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheung CY, Poon LL, Lau AS, Luk W, Lau YL, Shortridge KF, Gordon S, Guan Y, Peiris JS. 2002. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 360:1831–1837. doi: 10.1016/S0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 46.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJD, Chau TNB, Hoang DM, Chau NVV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha DQ, Guan Y, Peiris JSM, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavares LP, Teixeira MM, Garcia CC. 2017. The inflammatory response triggered by influenza virus: a two edged sword. Inflamm Res 66:283–302. doi: 10.1007/s00011-016-0996-0. [DOI] [PubMed] [Google Scholar]
- 48.Twu KY, Noah DL, Rao P, Kuo RL, Krug RM. 2006. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J Virol 80:3957–3965. doi: 10.1128/JVI.80.8.3957-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmolke M, Garcia-Sastre A. 2010. Evasion of innate and adaptive immune responses by influenza A virus. Cell Microbiol 12:873–880. doi: 10.1111/j.1462-5822.2010.01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ayllon J, Domingues P, Rajsbaum R, Miorin L, Schmolke M, Hale BG, Garcia-Sastre A. 2014. A single amino acid substitution in the novel H7N9 influenza A virus NS1 protein increases CPSF30 binding and virulence. J Virol 88:12146–12151. doi: 10.1128/JVI.01567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao H, Sun Y, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Sun H, Pu J, Chang KC, Liu X, Liu J. 2015. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep 5:8262. doi: 10.1038/srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng KH, Sun M, Iketani S, Holmes EC, Parrish CR. 2016. Comparing the functions of equine and canine influenza H3N8 virus PA-X proteins: suppression of reporter gene expression and modulation of global host gene expression. Virology 496:138–146. doi: 10.1016/j.virol.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, Xu Q, Shen Y, Liu L, Wei K, Sun H, Pu J, Chang KC, Liu J. 2014. Naturally occurring mutations in the PA gene are key contributors to increased virulence of pandemic H1N1/09 influenza virus in mice. J Virol 88:4600–4604. doi: 10.1128/JVI.03158-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li CJ, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Takahashi K, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, et al. . 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma WJ, Belisle SE, Mosier D, Li X, Stigger-Rosser E, Liu QF, Qiao CL, Elder J, Webby R, Katze MG, Richt JA. 2011. 2009 pandemic H1N1 influenza virus causes disease and upregulation of genes related to inflammatory and immune responses, cell death, and lipid metabolism in pigs. J Virol 85:11626–11637. doi: 10.1128/JVI.05705-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baker SF, Guo H, Albrecht RA, García-Sastre A, Topham DJ, Martínez-Sobrido L. 2013. Protection against lethal influenza with a viral mimic. J Virol 87:8591–8605. doi: 10.1128/JVI.01081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez-Sobrido L, Garcia-Sastre A. 2010. Generation of recombinant influenza virus from plasmid DNA. J Vis Exp 2010:2057. doi: 10.3791/2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nogales A, Baker SF, Martinez-Sobrido L. 2015. Replication-competent influenza A viruses expressing a red fluorescent protein. Virology 476:206–216. doi: 10.1016/j.virol.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nogales A, Rodriguez-Sanchez I, Monte K, Lenschow DJ, Perez DR, Martinez-Sobrido L. 2015. Replication-competent fluorescent-expressing influenza B virus. Virus Res 213:69–81. doi: 10.1016/j.virusres.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 61.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg (Lond) 27:493–497. [Google Scholar]
- 62.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]