The receptor for activated C kinase (RACK1), a conserved constituent of eukaryotic ribosomes, mediates phosphorylation of eukaryotic initiation factor 4G1(S1093) [eIF4G1(S1093)] and eIF3a(S1364) by protein kinase C βII (PKCβII) (M. I.
KEYWORDS: translation, eIF4G, RACK1, ribosome, PKCβII, eIF3, eIF4E
ABSTRACT
The receptor for activated C kinase (RACK1), a conserved constituent of eukaryotic ribosomes, mediates phosphorylation of eukaryotic initiation factor 4G1(S1093) [eIF4G1(S1093)] and eIF3a(S1364) by protein kinase C βII (PKCβII) (M. I. Dobrikov, E. Y. Dobrikova, and M. Gromeier, Mol Cell Biol 38:e00304-18, 2018, https://doi.org/10.1128/MCB.00304-18). RACK1:PKCβII activation drives a phorbol ester-induced surge of global protein synthesis and template-specific translation induction of PKC–Raf–extracellular signal-regulated kinase 1/2 (ERK1/2)-responsive genes. For unraveling mechanisms of RACK1:PKCβII-mediated translation stimulation, we used sequentially truncated eIF4G1 in coimmunoprecipitation analyses to delineate a set of autoinhibitory elements in the N-terminal unstructured region (surrounding the eIF4E-binding motif) and the interdomain linker (within the eIF3-binding site) of eIF4G1. Computer-based predictions of secondary structure, mutational analyses, and fluorescent titration with the β-sheet dye thioflavin T suggest that eIF4G1(S1093) modulates a 4-stranded β-sheet composed of antiparallel β-hairpins formed by the autoinhibitory elements in eIF4G1's unstructured regions. The intact β-sheet “locks” the eIF4G configuration, preventing assembly with eIF3/40S ribosomal subunits. Upon PKC stimulation, activated RACK1:PKCβII phosphorylates eIF4G(S1093) in the tight 48S initiation complex, possibly facilitating dissociation/recycling of eIF4F.
INTRODUCTION
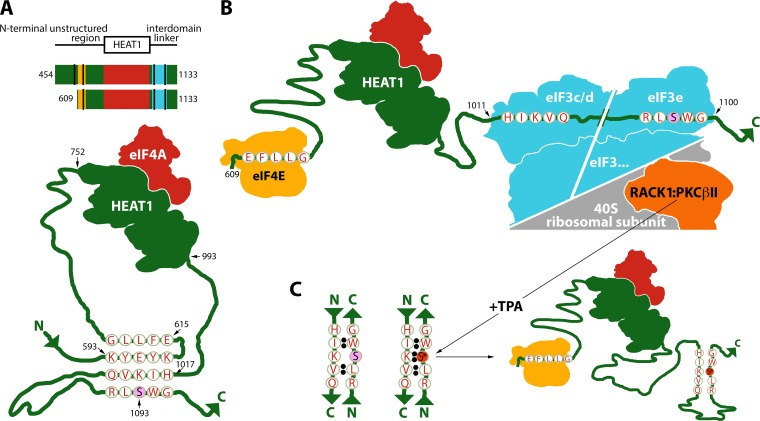
Phosphorylation is an important mechanism for modulating intramolecular interactions between intrinsically disordered protein regions. For example, disorder-to-order transitions occur upon phosphorylation of smooth-muscle myosin (1, 2) or phosphorylation of S65 in eIF4E-binding protein 1, which controls interactions with eIF4E (3). eIF4G contains two large disordered segments, the N-terminal unstructured region and the interdomain linker (IDL) separating HEAT (Huntingtin/EF3/PP2A/Tor1) domains 1 and 2 (4) (Fig. 1A). Both disordered regions harbor fundamental, essential functions of the eIF4G translation initiation scaffold: binding to the m7G cap-binding protein eIF4E (in the N-terminal unstructured region) and eIF3/40S ribosomal subunit (in the IDL). Adaptive regulation of the flexible, disordered regions of eIF4G is poorly understood. A presence of autoinhibitory conformations involving these regions, which are modulated by interactions with eIF4G binding partners, has been stipulated before (5).
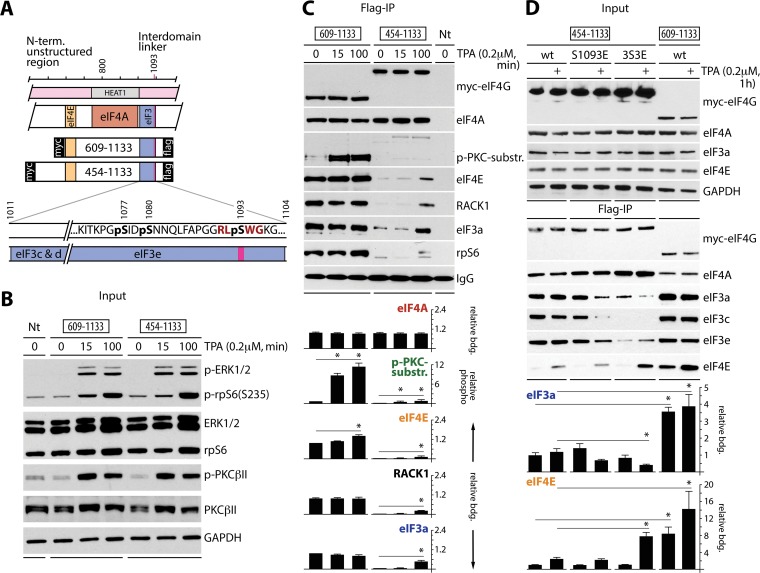
FIG 1.
Time course of eIF4G(S1093) phosphorylation. (A) Context of the N-terminal and IDL unstructured regions of eIF4G (pink boxes), the 609- and 454-1133 fragments with binding sites for eIF4E and eIF3e, the position of S1093 (with the PKCβII consensus context in red), and a cluster of adjacent phosphosites (bold black). (B) HEK293 cells were transfected (16 h) for expression of the indicated tagged eIF4G fragments, serum starved (24 h), and treated with DMSO (0) or 0.2 μM TPA for the indicated periods. Nt, nontransfected. Cell lysates were tested by immunoblotting with the indicated antibodies. The assay was repeated three times with consistent results; results of a representative test are shown. (C) Regulation of eIF4G(S1093) phosphorylation and of eIF4E, eIF3, eIF4A, and RACK1 interactions with eIF4G in response to TPA stimulation in HEK293 cells treated as described for panel B. Cell lysates were subjected to anti-Flag IP/immunoblotting with the indicated antibodies (top panel). Relative eIF4G binding and eIF4G(S1093) phosphorylation were quantified and averaged between 3 tests (bottom panel). Quantifications were normalized by setting the value of 0-min TPA (609-1133 fragment) to 1; error bars represent standard errors of the means (SEM), and the asterisks represent Student t test results (P < 0.05). (D) Mutational analysis of the phosphoserine cluster in the eIF3e-binding motif. 3S3E is the triple phosphomimic S1077E/S1080E/S1093E. HEK293 cells were transfected (16 h) for expression of tagged wt or mutant 454-1133 fragments, serum starved (24 h), and treated with TPA (+). Cell lysates were tested by immunoblotting with the indicated antibodies (top panel) or subjected to anti-Flag IP/immunoblotting with the indicated antibodies (middle panel). Relative binding of eIF3a and eIF4E was quantified and averaged between 3 assays (bottom panel). Error bars represent SEM; asterisks represent Student t test results (P < 0.05).
In a companion paper to the present report, we showed that RACK1:PKCβII phosphorylates eIF4G1 (referred to as eIF4G from here on) IDL at S1093 (within the eIF3e-binding motif) and that phorbol ester-induced PKC–Raf–extracellular signal-regulated kinase 1/2 (ERK1/2) signaling stimulates global and 12-O-tetradecanoyl-phorbol-13-acetate (TPA)-responsive template-specific translation in an eIF4G-, PKCβ-, and RACK1-dependent manner (4). We showed anomalous TPA-inducible eIF3 binding to nonphospho (S1093A) or phosphomimic (S1093E) variants of eIF4G, indicating that reversible S1093 phosphorylation regulates eIF4G's assembly with eIF3 and 40S ribosomal subunits (4).
In this companion study, we deciphered the molecular mechanisms of RACK1:PKCβII-induced translation adaptation. Coimmunoprecipitation (co-IP) studies with serial eIF4G truncation fragments suggested the presence of autoinhibitory features in the N-terminal unstructured region and the IDL of eIF4G, which control interactions with eIF4E and eIF3/RACK1/40S ribosomal subunits. Computational analyses of primary eIF4G sequences, coimmunoprecipitation studies with mutant eIF4G fragments, and fluorescence titration with thioflavin T (ThT) (6–9) suggest that the unstructured regions of eIF4G participate in a putative 4-stranded β-sheet structure. This structure is formed by two antiparallel β-hairpins located in the N-terminal unstructured region and the IDL, respectively. The IDL β-hairpin contains S1093. Upon S1093 phosphorylation by RACK1:PKCβII, altered electrostatic charges stabilize the IDL β-hairpin, which impedes eIF3 binding.
Our investigations revealed complex intramolecular autoinhibitory features in eIF4G, involving flexible disordered regions that are controlled by their posttranslational modifications and dynamic interactions with translation initiation factors. RACK1:PKCβII can phosphorylate eIF4G(S1093) only when the 40S ribosomal subunit is assembled with eIF4G. We hypothesize that ribosomal RACK1:PKCβII-mediated eIF4G(S1093) phosphorylation drives eIF4G dissociation from the 48S initiation complex after completion of scanning/recognition of the initiation codon.
RESULTS
Time course of eIF4G(S1093) phosphorylation.
The newly identified RACK1:PKCβII substrate eIF4G(S1093) (4) is located in the eIF3e-binding domain (10) as a part of the S1077/S1080/S1093 cluster (Fig. 1A), which is phosphorylated in mitosis (11). We analyzed the time course of TPA-dependent eIF4G(S1093) phosphorylation, eIF4E binding, and ribosome recruitment of two Myc-eIF4G-Flag-tagged fragments (609-1133 and 454-1133) (Fig. 1A), with distinct truncations of the N-terminal unstructured region (Fig. 1A). The fragments were expressed in serum-starved and TPA-stimulated HEK293 cells. TPA induced PKCβII (Fig. 1B) and eIF4G(S1093) (Fig. 1C) phosphorylation within 15 min. Flag IP revealed equal binding of both fragments with eIF4A but sharply diverging associations with eIF4E and eIF3/RACK1/rpS6 (Fig. 1C). The 609-1133 fragment showed near-constitutive, TPA-independent eIF4E binding and eIF3/RACK1/rpS6 recruitment (Fig. 1C). Interactions of 454-1133 with eIF4E/eIF3a/rpS6/RACK1, however, were much weaker and TPA responsive; S1093 phosphorylation in this fragment was less efficient, suggesting that eIF4G assembly with the 40S ribosomal subunit is a requisite for eIF4G(S1093) phosphorylation (Fig. 1C).
Mutational analysis of the eIF4G(S1093) context.
To investigate the role of cluster phosphorylation in eIF4G interactions with its binding partners, we created single S1093E and triple S1077E/S1080E/S1093E (3S3E) phosphomimic substitutions in the tagged 454-1133 eIF4G fragment (Fig. 1D). HEK293 cells were transfected with wild-type (wt) 609-1133 or wt/mutant 454-1133 fragments, serum starved, and TPA stimulated (Fig. 1D). Flag IP showed that S1093E substitution inhibited interactions with eIF3 but had no substantive effect on eIF4E binding (Fig. 1D). The 3S3E substitution exacerbated the effects on eIF3 binding and enhanced TPA-inducible eIF4E association (Fig. 1D). As observed before (Fig. 1C), the wt 609-1133 fragment exhibited strong, near-constitutive eIF3 and eIF4E binding (Fig. 1D).
Our data suggest that the N-terminal unstructured region in the range of amino acids (aa) 454 to 609 inhibits eIF4G's interactions with eIF4E and eIF3/40S ribosomal subunit. This is in agreement with prior work that showed eIF4G's unstructured N-terminal region (adjacent to the eIF4E-binding site) to harbor an autoinhibitory domain for eIF4A helicase activity (5). eIF4E:eIF4G binding overcame the inhibition and stimulated eIF4A helicase activity. Since eIF4A:HEAT1 binding itself was unaffected (5), the N-terminal autoinhibitory domain may influence eIF4A activity via interactions with the unstructured IDL, e.g., in the region of the newly identified S1093 phosphosite. To investigate this, we created a panel of Myc/Flag-tagged eIF4G fragments with sequentially truncated N termini or IDLs (Fig. 2A).
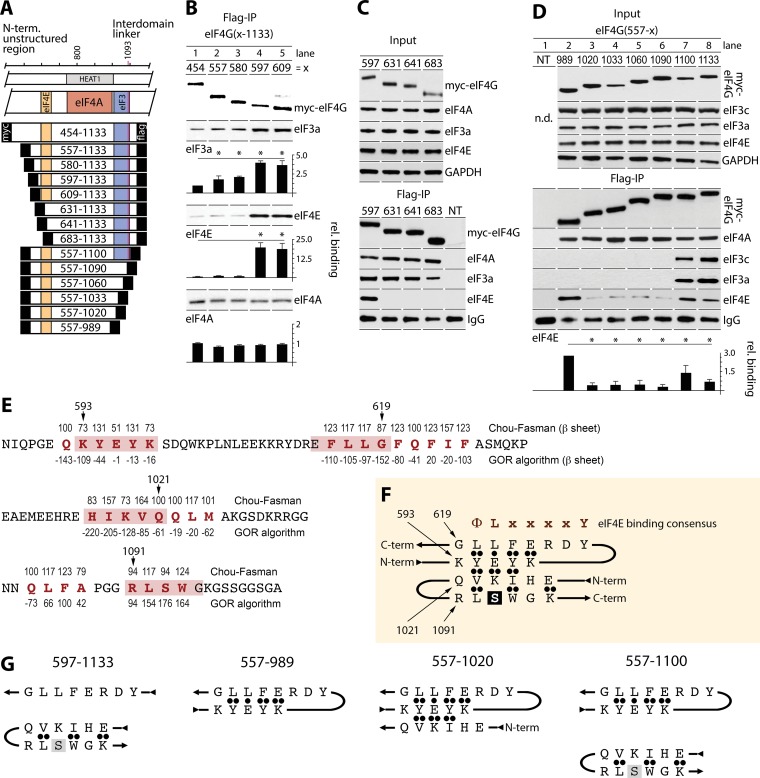
FIG 2.
Autoinhibitory regions within eIF4G's N-terminal unstructured region and the IDL regulate interactions with eIF4E/eIF3, and a putative β-sheet is formed by discontinuous nonstructured domains of eIF4G. (A) eIF4G(454-1133) and a panel of Myc/Flag-tagged fragments that were investigated. (B to D) HEK293 cells were transfected for expression of the indicated N-terminal (B and C) or C-terminal (D) truncation fragments (16 h) and synchronized by Th block (24 h). Cell lysates were subjected to immunoblotting (C and D, top panels) or anti-Flag IP/immunoblotting with the indicated antibodies (C and D, bottom and middle panels, respectively). (B and D) Relative binding of eIF3a, eIF4A, and eIF4E was quantified and averaged between 3 assays. Error bars represent SEM; asterisks represent Student t test results (P < 0.05). (E) Computational approaches for β-structure predictions in eIF4G. Sequences of N-terminal (aa 586 to 624) and C-terminal (aa 1008 to 1034 and 1082 and 1104) portions of eIF4G harboring elements putatively engaged in β-structures are shown. Numbers above the amino acid sequence represent Chou-Fasman conditional probability for β-structure formation (values should be >100). Numbers below the amino acid sequence represent GOR probability for β-structure formation (values should be negative). Elements predicted to participate in β-structures are shown in red letters, and pink boxes represent selected pentapeptides involved in putative β-structures. (F) A proposed 4-stranded β-sheet in eIF4G. ••, strongly favorable hydrophobic and electrostatic interactions between β-strands; •, favorable interactions for β-sheet formation. (G) Depiction of putative β-structures for eIF4G truncation fragments analyzed for eIF4G:eIF4E/eIF3 assembly in co-IP studies shown in panels B and D; see the text for a detailed explanation.
Long-range intramolecular contacts involving eIF4G's unstructured regions.
To define the autoinhibitory domain in eIF4G's N-terminal unstructured region, we used the 454-1133 fragment and compared it to successive N-terminal truncations at aa 557, 580, 597, and 609 (Fig. 2A and B); all of these fragments contained the intact eIF4E-binding motif. For consistency, HEK293 cells expressing these fragments were synchronized by thymidine (Th) block (this was employed in all assays without serum starvation). Flag IP of lysates revealed similar expression of fragments with even binding to eIF4A (Fig. 2B). Interactions with eIF3a and eIF4E, however, covaried with the length of the N terminus (Fig. 2A). Binding of eIF3a and eIF4E was increased with stepwise N-terminal truncation to reach a maximum of ∼4-fold (eIF3) or ∼20-fold (eIF4E) with the 597-1133 fragment compared to the 454-1133 fragment. The steepest binding differential occurred in the 580-to-597 range, particularly for eIF4E (Fig. 2B). This suggests an autoinhibitory feature in eIF4G (aa 580 to 597), which did not affect eIF4A:HEAT1 interactions, suppressed nearby binding of eIF4E, and impeded eIF3a binding at a distant site in the IDL. N-terminal truncations beyond 609-1133 (631, 641, and 683) (Fig. 2A) deleted the eIF4E binding motif. The 631, 641, and 683 fragments lost interaction with eIF4E, but binding with eIF3 or eIF4A did not change compared to that for the 597-1133 fragment (Fig. 2C).
Next, to evaluate the IDL's role in controlling eIF3/eIF4E binding to eIF4G, we compared Flag IP of eIF4G fragments containing the eIF4E-binding site, HEAT1, and variable IDL portions (Fig. 2A and D). The shortest fragment (557-989, lacking the IDL entirely) efficiently bound eIF4E (Fig. 2D). Extending the IDL up to aa 1090 inhibited eIF4E:eIF4G interactions ∼7-fold (Fig. 2D). Adding a further 10 aa (including the PKCβII site, S1093) partially restored binding with eIF4E (Fig. 2D). This implies (i) eIF4E-binding inhibitory sequences in the eIF4G IDL(989-1020) and (ii) a stimulatory element in the 1090-1100 range (where S1093 is located). eIF4G/IDL interacts with eIF3c/d (aa 1011 to 1051) and eIF3e (aa 1052 to 1104) (10). Accordingly, our data showed co-IP of eIF3a/c subunits with the longest eIF4G fragments (557-1100 and 557-1133); shorter fragments lacked eIF3 binding (Fig. 2D). Thus, eIF4G(1090-1100) is critical for stable eIF4G:eIF3 interactions (Fig. 2D).
A putative β-sheet structure formed by elements within eIF4G's nonstructured regions.
Our findings suggest autoregulatory features in eIF4G(580-597) (Fig. 2B), eIF4G(989-1020), and eIF4G(1090-1100) (Fig. 2D). We applied two algorithms for predicting protein secondary structures based on primary sequences. The Chou-Fasman homology approach (12–14) examines the statistical distribution of amino acids in β-sheets based on known structures. The Garnier-Osguthorpe-Robson (GOR) algorithm analyzes both conformational parameters for each amino acid and effects within a window of 8 aa before and after the position of interest (15). These methods yielded 4 elements predicted by both algorithms to be putatively involved in β-sheet formation: aa 592 to 597, 616 to 624, 1017 to 1024, and 1084 to 1087 (Fig. 2E, red letters). A fifth element containing S1093 (aa 1091 to 1094) was identified by the Chou-Fasman algorithm only (Fig. 2E, red letters).
Based on our analyses, amino acid conformational preferences, and side chain:side chain interactions in β-hairpins (16, 17), we selected 4 pentapeptides within the elements possibly involved in β-sheet formation: upstream of (593-KYEYK-597) and within (615-EFLLG-619) the eIF4E-binding motif and in the eIF3c/d (1017-HIKVQ-1021)- or eIF3e (1091-RLSWG-1095)-binding domains (Fig. 2E, boxed in pink). For higher β-sheet stability, alternating charged/hydrophobic amino acid sequences are preferred (16); this requirement is matched by 593-KYEYK-597. The 593-KYEYK-597 pentapeptide could form an antiparallel β-hairpin with 615-EFLLG-619 (Fig. 2F). This would account for eIF4E:eIF4G binding inhibition in eIF4G fragments containing aa 593 to 597 (Fig. 2B, lanes 1 to 3) and ∼20-fold increased eIF4E:eIF4G binding upon truncation of the 593-KYEYK-597 pentapeptide (Fig. 2B, lanes 4 and 5).
The 593-KYEYK-597 sequence may be involved in similar interactions with an antiparallel β-hairpin formed by 1017-HIKVQ-1021 and 1091-RLSWG-1095 (Fig. 2F), keeping eIF4G in a “locked” configuration and inhibiting eIF3:eIF4G interactions. As such, the presence of distinct β-structures in truncated eIF4G fragments may dictate interactions with eIF4E/eIF3. Thus, 580-1133 (and extended) eIF4G fragments may contain a 4-stranded β-sheet (Fig. 2F). Indeed, these fragments showed the lowest binding affinity for eIF4E and eIF3a (Fig. 2B, lanes 1 to 3). Shorter 597-1133 and 609-1133 fragments, lacking the β-hairpin in the eIF4E-binding site (Fig. 2G, left panel), efficiently bound eIF4E and eIF3 (Fig. 2B, lanes 4 and 5). The absence of 593-KYEYK-597 in these fragments should disrupt the 4-stranded β-sheet (Fig. 2G, left panel), destabilizing autoinhibitory secondary structures that suppress eIF4E and eIF3 binding (Fig. 2B, lanes 4 and 5).
eIF4E binding to the 557-989 fragment, which features only the 593-KYEYK-597/615-EFLLG-619 β-hairpin (Fig. 2G, center left panel), is inhibited by extending the IDL to aa 1020 (Fig. 2D, lane 3). This is possibly due to formation of a 3-stranded β-sheet involving the N-terminal β-hairpin and the 1017-HIKVQ-1021 pentapeptide (Fig. 2G, center right panel). Upon extension of the IDL in the 557-1100 fragment (Fig. 2G, right panel), the formation of a 4-stranded β-sheet and partial restoration of eIF4E binding (Fig. 2D, lane 7) suggest that the 4-stranded β-sheet may be in equilibrium with an arrangement of two separate β-hairpins (Fig. 2G, right panel).
Functional analysis of potentially β-structured segments in eIF4G.
Putative β-structures formed by N-terminal unstructured regions and the IDL in eIF4G involve eIF4E/eIF3 binding motifs. To investigate an involvement of such structures in regulating the association of eIF4G with initiation factors, we used mutational analyses (Fig. 3). Based on the eIF4G(580-1133) fragment containing all autoinhibitory features identified in this study (see Fig. 2B, lane 3, for interactions with eIF4E and eIF3), we created seven distinct Myc/Flag-tagged fragments with targeted disruption of all elements predicted by both the Chou-Fasman and Garnier-Osguthorpe-Robson algorithms to be putatively involved in β-sheet formation (Fig. 2E). The 593-KGGGK, 615-EGGGG, 621-QGGGA, 1017-HGGGQ, 1021-QGGGA, 1083-NGGGA, and 1091-RGGGG pentapeptides were introduced with central triplets of aromatic/hydrophobic amino acids replaced by glycines to prevent secondary structure formation (Fig. 3A). These mutant fragments were tested with Flag IP for binding with eIF3/eIF4E (Fig. 3B and C).
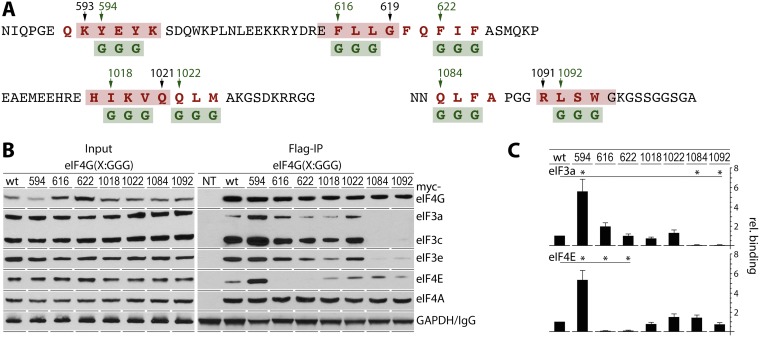
FIG 3.
Functional analysis of the sequences involved in β-sheet formation. (A) Substitutions in 7 tagged eIF4G(580-1133) fragments. Triplets of aromatic/hydrophobic/charged amino acids (red letters) involved in putative β-sheet structures were replaced with glycines (green letters). Amino acid numbers in green represent the first glycine substitution in each mutated fragment. (B) HEK293 cells were transfected for expression of the indicated fragments (16 h) and synchronized by Th block (24 h). Cell lysates were subjected to immunoblotting or anti-Flag IP/immunoblotting with the indicated antibodies. (C) Binding of eIF3a and eIF4E with eIF4G fragments was quantified and averaged between 3 tests. Error bars represent SEM; asterisks represent Student t test results (P < 0.05).
Replacing 593-KYEYK-597 with KGGGK should disrupt the core of the proposed 4-stranded β-sheet (Fig. 2F) and abolish inhibitory features relative to binding with eIF3/eIF4E. This was indeed the case, as eIF4G(580-1133) containing 593-KGGGK-597 displayed ∼6-fold-increased binding with eIF3 and eIF4E compared to the wild type (Fig. 3B and C). This agrees with unencumbered eIF4E/eIF3 binding of the 597-1133 and 609-1133 truncation fragments (Fig. 2B, lanes 4 and 5). Substitutions within the eIF4E-binding motif (615-EGGGG-619 and 621-QGGGA-625) (Fig. 3A), as expected, abolished eIF4E binding and retained approximately wild-type levels of eIF3 binding (Fig. 3B and C). Substitutions 1017-HGGGQ-1021 and 1021-QGGGA-1025 did not significantly alter eIF3/eIF4E interactions, indicating minor contributions to the formation of a 4-stranded β-sheet (Fig. 3B and C). Not surprisingly, the 1083-NGGGA-1087 and 1091-RGGGG-1095 substitutions (within the eIF3e-binding motif in the IDL) abolished co-IP of eIF3 subunits (Fig. 3B and C). Thus, our mutational analyses of a putative 4-stranded β-sheet confirmed a major role for 593-KYEYK-597 in long-range autoinhibitory configurations of the N-terminal structural region and IDL of eIF4G.
Design of the 4-stranded β-sheet in eIF4G with high-affinity binding to ThT.
As shown in Fig. 1D, the single phosphomimic S1093E and, especially, the triple phosphomimic 3S3E substitution in the IDL reduced association with eIF3 compared to that for the wild-type eIF4G(454-1133) fragment. We propose that S1093E and 3S3E increase β-hairpin stability with the antiparallel 1017-HIKVQ-1021 pentapeptide (Fig. 2F), due to strong electrostatic interactions between the positively charged K1019 and negatively charged E1093, e.g., within the putative 4-stranded β-sheet formation (Fig. 4B, right panel).
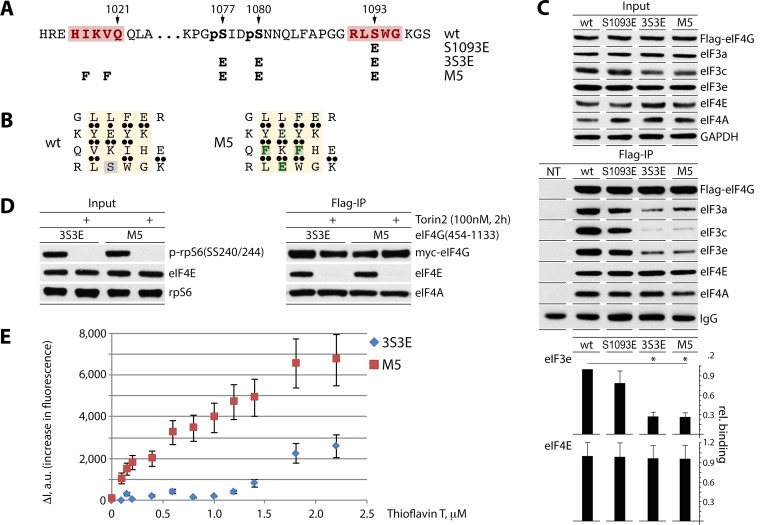
FIG 4.
Design of the 4-stranded β-sheet with a high-affinity ThT binding site and fluorescent titration of eIF4G:eIF4A complexes by ThT. (A and B) Mutations S1093E, 3S3E and M5 in eIF4G(454-1133) (A). M5 carries a high-affinity ThT site in the putative 4-stranded β-sheet (see the text). Substituted amino acids are colored green (B). (C) HEK293 cells were transfected for expression of the indicated fragments (16 h) and synchronized by Th block (24 h). Cell lysates were subjected to immunoblotting (top panel) or anti-Flag IP/immunoblotting (middle panel) with the indicated antibodies. Results from Flag co-IP of eIF3e/eIF4E were quantified and averaged between 3 tests (bottom panel); error bars represent SEM, and asterisks represent Student t test results (P < 0.05). (D and E) Fluorescent titration of eIF4G:eIF4A complexes by ThT. HEK293 cells were transfected for expression of 3S3E and M5 fragments (16 h) and synchronized by Th block (24 h), and Torin2 was added 2 h prior to lysis to block eIF4E:eIF4G binding (D). For isolation of eIF4G:eIF4A complexes, cell lysates were subjected to sequential anti-Flag IP/Flag-peptide elution and anti-Myc IP/Myc-peptide elution. ThT:eIF4G(454-1133) binding isotherms were obtained by measuring the increase in dye fluorescence intensity (ΔI) (480 nm) relative to that in buffer (E). Fluorescence titrations were performed in triplicate; error bars represent SEM.
Empirically documenting the 4-stranded β-sheet in eIF4G is not trivial, since this requires an approach distinguishing this specific formation from any secondary structural arrangement that may occur. Thus, for empirically demonstrating the 4-stranded β-sheet in eIF4G, we decided to use the benzathiole dye thioflavin T (ThT) as a fluorescent marker of β-sheets (6–9). The minimal binding site for ThT has been determined to be 4-stranded β-sheets containing 4 aromatic cross-strand residues (6). Binding of ThT to the aromatic or hydrophobic grooves in β-sheets, e.g., in the amyloid fibril, results in fluorescence enhancement (18, 19). In order to create a high-affinity ThT binding site in eIF4G, we replaced two hydrophobic residues in the 1018-IKV-1020 triplet with aromatic residues to give 1018-FKF-1020 in the 3S3E eIF4G(580-1133) fragment (mutant 5 [M5]) (Fig. 4A and B). As a result, M5 contains four aromatic cross-strand residues: F(616):Y(596):F(1018):W(1094) (Fig. 4B). Anti-Flag IP revealed, as expected, decreased binding of S1093E and especially the 3S3E mutant fragments with eIF3 subunits (Fig. 4C), as observed before (Fig. 1D). The aromatic substitution 1018-FKF-1020 did not alter affinity for eIF3, eIF4E, or eIF4A compared to 3S3E (Fig. 4C).
Fluorescent titration of an eIF4G:eIF4A complex by ThT.
For fluorescent titration of an eIF4G:eIF4A complex with ThT, we isolated tagged eIF4G(M5) and eIF4G(3S3E) fragments by consecutive Flag/Myc IP from transfected HEK293 cells. eIF3 and eIF4E binding to eIF4G likely discourages the formation of the 4-stranded β-sheet. eIF3 binding to the 3S3E and M5 fragments is inherently low (Fig. 1D and 4C). To prevent eIF4E:eIF4G binding, the cells were treated with the mTOR inhibitor Torin2. This blocked rpS6(SS240/244) phosphorylation and eIF4E co-IP with 3S3E/M5 (Fig. 4D). For purification of fragments, cell lysates were subjected to sequential anti-Flag IP/Flag-peptide elution followed by anti-Myc IP/Myc-peptide elution. Equal concentrations of eIF4A:eIF4G(3S3E) and eIF4A:eIF4G(M5) were titrated with increasing concentrations of ThT. In contrast to the case for eIF4G(3S3E), binding of ThT with the M5 mutant induced a strong increase in fluorescence intensity (ΔI), in the nanomolar range at 480 nm (Fig. 4E), suggesting that this eIF4G fragment had a high-affinity binding site for ThT.
These observations, combined with comprehensive analyses of eIF4G interactions with its canonical binding partners eIF4A, eIF4E, and eIF3, support the notion of autoinhibitory secondary structures formed by long-range contacts of eIF4G's N-terminal and IDL flexible unstructured segments. RACK1:PKCβII may play an important role in adaptive regulation of these structures, due to its phosphorylation site in eIF4G(S1093).
DISCUSSION
We identified eIF4G(S1093) and eIF3a(S1364) as phosphorylation sites for RACK1:PKCβII, revealing physiological significance of ribosomal RACK1 in the control of translation initiation (4). In depletion/inhibitor studies, eIF4G, PKCβII, and RACK1 accounted for major contributions to a surge of global and template-specific protein synthesis upon PKC-Raf-ERK1/2 activation (4). In the present study, we deciphered RACK1:PKCβII's influence over translation mechanistically, by delineating their role in modulating inhibitory intramolecular arrangements within eIF4G that control interactions with eIF4E and eIF3.
We found that a short segment in eIF4G's N-terminal unstructured region, immediately upstream of the eIF4E-binding motif, strongly inhibits eIF4G:eIF4E and eIF3 association. Truncation or mutation of the 593-KYEYK-597 pentapeptide yielded constitutively high, almost TPA-unresponsive assembly of eIF4E/eIF3 with eIF4G. Computational and mutational analyses suggest that 593-KYEYK-597 inhibits eIF4E binding by participating in a β-hairpin involving the eIF4E-binding motif in eIF4G's N-terminal unstructured domain (Fig. 5A). The influence of 593-KYEYK-597 on eIF3 binding in the distant IDL may be explained by its contribution to a 4-stranded β-sheet, composed of two antiparallel β-hairpins, in the N-terminal unstructured domain and in the IDL spanning the eIF3-binding region (Fig. 5A). We employed fluorescence titration with ThT to experimentally demonstrate the formation of this 4-stranded β-sheet. Deleting 593-KYEYK-597 disrupts eIF4G's “locked” conformation and the 4-stranded β-sheet and permits constitutive, unimpeded eIF4E and eIF3 assembly with eIF4G (Fig. 5B).
FIG 5.
Model for RACK1:PKCβII phosphorylation of eIF4G(S1093) and its role in controlling dynamic eIF4G:eIF3 assembly. (A) Top panel, eIF4G fragments bearing eIF4E (gold), eIF4A (red), and eIF3 (blue) binding sites; sequences constituting a putative 4-stranded β-sheet are indicated by black stripes. Bottom, fragment 454-1133 contains the intact 4-stranded β-sheet; in unstimulated cells 593-KYEYK-597 anchors an antiparallel β-hairpin with the eIF4E-binding domain. This β-hairpin in the N-terminal unstructured region stabilizes a more complex β-sheet structure including an antiparallel β-hairpin forming in the IDL. The 4-stranded β-sheet may lock the eIF4G configuration and prevent binding to the 43S preinitiation complex. (B) The 609-1133 eIF4G fragment lacks the core inhibitory 593-KYEYK-597 pentapeptide. The 4-stranded β-sheet cannot form, allowing eIF4E/eIF3 assembly, mimicking TPA-stimulated conditions. In this tight complex, during TPA stimulation, activated PKCβII phosphorylates eIF4G(S1093). (C) Phosphorylated 1091-RLpSWG-1095 favors a stable antiparallel β-hairpin with 1017-HIKVQ-1021 in the eIF3c/d-binding site, thus facilitating dissociation of eIF4G from the initiation complex.
eIF4G(S1093) is located in the center of the putative β-hairpin in the IDL, encompassing the eIF3-binding region (Fig. 5B and C). Thus, phosphorylation of eIF4G(S1093), by stabilizing the β-hairpin, antagonizes eIF3 binding (Fig. 5C). This explains low eIF3 binding with the eIF4G(S1093E) phosphomimic and high binding with the nonphosphorylatable eIF4G(S1093A) mutant (see Fig. 3A in reference 4).
During early translation initiation, prior to 40S ribosomal subunit recruitment by eIF4F, eIF4E and eIF3 binding to eIF4G would prevent the formation of the 4-stranded β-sheet, akin to the effect of 593-KYEYK-597 deletion (Fig. 5B). Phosphorylation of eIF4G(S0193) can occur only later in the initiation process, with eIF4G bound to either the 43S preinitiation complex during scanning or the 48S initiation complex at the initiation codon, because it requires proximity of eIF4G to ribosomal RACK1. We hypothesize that TPA-induced eIF4G(S1093) phosphorylation stimulates translation by spurring eIF4G:4A:4B dissociation from 48S initiation complexes (Fig. 5C), thus recycling them for new initiation rounds.
MATERIALS AND METHODS
Cell lines, DNA transfections, and eIF4G expression plasmids.
HEK293 cells were grown, transfected, synchronized, and treated with kinase inhibitors and/or activators as described earlier (4). Construction of Myc/Flag-tagged eIF4G expression plasmids has been described previously (4); the eIF4G fragments used in this study were generated by PCR with the corresponding primers (Table 1), and mutations were introduced by overlapping PCR as described earlier (4).
TABLE 1.
Oligonucleotide primers used in this study
| Primer no. | Primer name | Sequence (5′→3′)a |
|---|---|---|
| 1 | 454(+) | ATAAGCTTGAGGAGGAAATGGAAGAAGAAGE |
| 2 | 557(+) | TTAAGCTTGAGTCTGAGGGCAGTGGTGTGC |
| 3 | 580(+) | CCAAGCTTAAAATTCACAATGCTGAGAACATCC |
| 4 | 597(+) | CCAAGCTTAAGTCAGATCAGTGGAAGCCTC |
| 5 | 609(+) | CCAAGCTTAAAAAACGTTACGACCGTGAGTTCC |
| 6 | 631(+) | ACAAGCTTGAGGGATTGCCACATATCAG |
| 7 | 641(+) | TCAAGCTTCTGGACAAGGCCAATAAAACACC |
| 8 | 683(+) | TTAAGCTTGGGCCCCCAAGGGGTGG |
| 9 | 989(−) | CCCTCGAGATTGCTCCCTCGCAGATCC |
| 10 | 1020(−) | TTCTCGAGCACTTTGATGTGCTCTCGATG |
| 11 | 1033(−) | TTCTCGAGGCCCCGACGCTTGTCACTG |
| 12 | 1060(−) | GTCTCGAGACCTTTGCTGATGGGAACTGTG |
| 13 | 1090(−) | TTCTCGAGCCCTCCAGGTGCAAAGAGC |
| 14 | 1100(−) | TTCTCGAGGCCTCCGCTGCTGCCCTTGC |
| 15 | 1133(−) | TTCTCGAGAGGTACCGCTTGTTGAAGG |
| 16 | 594GGG(+) | GAAGGGTGGAGGTAAGTCAGATCAGTGGAAGCC |
| 17 | 594GGG(−) | CTTACCTCCACCCTTCTGTTCCCCGGGC |
| 18 | 616GGG(+) | CGTGAGGGCGGCGGTGGTTTTCAGTTCATCTTTGCC |
| 19 | 616GGG(−) | ACCACCGCCGCCCTCACGGTCGTAACGTTT |
| 20 | 622GGG(+) | TTTCAGGGCGGCGGTGCCAGTATGCAGAAGCC |
| 21 | 622GGG(−) | GGCACCGCCGCCCTGAAAACCAAGCAGGAAC |
| 22 | 1018GGG(+) | GCACGGCGGAGGGCAGCAGCTCATGGCCA |
| 23 | 1018GGG(−) | CTGCCCTCCGCCGTGCTCTCGATGTTCTTCC |
| 24 | 1022GGG(+) | GCAGGGCGGCGGTGCCAAGGGCAGTGACAAGCG |
| 25 | 1022GGG(−) | TGGCACCGCCGCCCTGCACTTTGATGTGCTCTCG |
| 26 | 1083GGG(+) | CAACGGCGGCGGTGCACCTGGAGGGCGACTG |
| 27 | 1083GGG(−) | GGTGCACCGCCGCCGTTGTTAGAATCGATGGAGC |
| 28 | 1092GGG(+) | GGGCGAGGCGGCGGTGGCAAGGGCAGCAGCGG |
| 29 | 1092GGG(−) | GCCACCGCCGCCTCGCCCTCCAGGTGCAAAGAG |
| 30 | 1018/20FF(+) | CACTTCAAATTCCAGCAGCTCATGGCC |
| 31 | 1018/20FF(−) | CTGGAATTTGAAGTGCTCTCGATGTTCTTCC |
| 32 | S1093E(+) | GGCGACTGGAGTGGGGCAAGGGCAGC |
| 33 | S1093E(−) | GCCCCACTCCAGTCGCCCTCCAGGTGC |
| 34 | SS1077/80EE(+) | GAGATCGATGAGAACAACCAGCTCTTTGC |
| 35 | SS1077/80EE(−) | GGTTGTTCTCATCGATCTCGCCAGGC |
Restriction sites used for cloning are underlined, and mutated nucleotides are in bold italic.
Kinase and translation inhibitors and activators.
12-O-Tetradecanoyl-phorbol-13-acetate (TPA) (Sigma-Aldrich), Torin2 (Tocris) (dissolved in dimethyl sulfoxide [DMSO]), and thioflavin T (Sigma-Aldrich) (dissolved in water) were used at the concentrations indicated.
IP, immunoblotting, and antibodies.
Cell lysate preparation, immunoprecipitation, and immunoblotting methods are described elsewhere (4). For isolation of eIF4G:eIF4A complexes, HEK293 cells were transfected with 16 μg of Myc-eIF4G-Flag expression plasmid DNA using 40 μl of Lipofectamine 2000 (Invitrogen) per 15-cm petri dish. At 16 h posttransfection, the cells were synchronized by thymidine (Th) block (24 h); 150 nM Torin2 was added 2 h prior to lysis. Cell lysates were subjected to sequential IP; the first round was with anti-Flag–agarose beads followed by Flag-peptide elution, and the second round was with anti-Myc–agarose beads followed by Myc-peptide elution (all from Sigma). The antibodies used were against c-Myc tag (Sigma), eIF3e and PKCβII (Novus Biologicals), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), rpS6, eIF3c, eIF3a eIF4A, eIF4E, RACK1, and ERK1/2 (Cell Signaling), and phospho-specific p-rpS6(S240/4), p-PKCβII(T638/41), p-ERK1/2(T202/Y204), and p-(S)-PKC-substrate (Cell Signaling). Immunoblots were developed with SuperSignal West Pico (Thermo Scientific) or Western Bright (BioExpress) enhanced chemiluminescence (ECL) kits. Immunoblot signals were quantified using the Li-COR Odyssey FC2 imaging system and Image Studio software.
Fluorescent titration of the 4-stranded β-sheet in the eIF4G:4A complex.
Fluorescence of ThT was measured using a SpectraMax M5 microplate reader (Molecular Devices) in a 96-well clear-bottomed, black-walled microplate (Greiner Bio-One) with excitation at 380 nm and emission at 480 nm.
ACKNOWLEDGMENTS
This work is supported by PHS grants P50 CA190991 and R01 NS108773 (M.G.) and by the Lefkofsky Family Foundation.
We thank Timothy Smith and Pei Zhou for helpful discussions and technical assistance with the fluorescence titration assay.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/MCB.00304-18.
REFERENCES
- 1.Espinoza-Fonseca LM, Kast D, Thomas DD. 2007. Molecular dynamics simulations reveal a disorder-to-order transition on phosphorylation of smooth muscle myosin. Biophys J 93:2083–2090. doi: 10.1529/biophysj.106.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kast D, Espinoza-Fonseca LM, Yi C, Thomas DD. 2010. Phosphorylation-induced structural changes in smooth muscle myosin regulatory light chain. Proc Natl Acad Sci U S A 107:8207–8212. doi: 10.1073/pnas.1001941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tait S, Dutta K, Cowburn D, Warwicker J, Doig AJ, McCarthy JE. 2010. Local control of a disorder-order transition in 4E-BP1 underpins regulation of translation via eIF4E. Proc Natl Acad Sci U S A 107:17627–17632. doi: 10.1073/pnas.1008242107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobrikov MI, Dobrikova EY, Gromeier M. 2018. Ribosomal RACK1:protein kinase C βII phosphorylates eukaryotic initiation factor 4G1 at S1093 to modulate cap-dependent and -independent translation initiation. Mol Cell Biol 38:e00304-18. doi: 10.1128/MCB.00304-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. 2013. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A 110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biancalana M, Koide S. 2010. Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta 1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biancalana M, Makabe K, Koide A, Koide S. 2009. Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies. J Mol Biol 385:1052–1063. doi: 10.1016/j.jmb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girych M, Gorbenko G, Trusova V, Adachi E, Mizuguchi C, Nagao K, Kawashima H, Akaji K, Lund-Katz S, Phillips MC, Saito H. 2014. Interaction of thioflavin T with amyloid fibrils of apolipoprotein A-I N-terminal fragment: resonance energy transfer study. J Struct Biol 185:116–124. doi: 10.1016/j.jsb.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Wu C, Biancalana M, Koide S, Shea JE. 2009. Binding modes of thioflavin-T to the single-layer beta-sheet of the peptide self-assembly mimics. J Mol Biol 394:627–633. doi: 10.1016/j.jmb.2009.09.056. [DOI] [PubMed] [Google Scholar]
- 10.Villa N, Do A, Hershey JW, Fraser CS. 2013. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 288:32932–32940. doi: 10.1074/jbc.M113.517011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobrikov MI, Shveygert M, Brown MC, Gromeier M. 2014. Mitotic phosphorylation of eukaryotic initiation factor 4G1 (eIF4G1) at Ser1232 by Cdk1:cyclin B inhibits eIF4A helicase complex binding with RNA. Mol Cell Biol 34:439–451. doi: 10.1128/MCB.01046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou PY, Fasman GD. 1974. Prediction of protein conformation. Biochemistry 13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 13.Chou PY, Fasman GD. 1978. Empirical predictions of protein conformation. Annu Rev Biochem 47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 14.Chou PY, Fasman GD. 1978. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol 47:45–148. [DOI] [PubMed] [Google Scholar]
- 15.Garnier J, Gibrat JF, Robson B. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol 266:540–553. doi: 10.1016/S0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 16.Hutchinson EG, Sessions RB, Thornton JM, Woolfson DN. 1998. Determinants of strand register in antiparallel beta-sheets of proteins. Protein Sci 7:2287–2300. doi: 10.1002/pro.5560071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wouters MA, Curmi PM. 1995. An analysis of side chain interactions and pair correlations within antiparallel beta-sheets: the differences between backbone hydrogen-bonded and non-hydrogen-bonded residue pairs. Proteins 22:119–131. doi: 10.1002/prot.340220205. [DOI] [PubMed] [Google Scholar]
- 18.Wu C, Bowers MT, Shea JE. 2011. On the origin of the stronger binding of PIB over thioflavin T to protofibrils of the Alzheimer amyloid-beta peptide: a molecular dynamics study. Biophys J 100:1316–1324. doi: 10.1016/j.bpj.2011.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Younan ND, Viles JH. 2015. A comparison of three fluorophores for the detection of amyloid fibers and prefibrillar oligomeric assemblies. ThT (thioflavin T); ANS (1-anilinonaphthalene-8-sulfonic acid); and bisANS (4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid). Biochemistry 54:4297–4306. doi: 10.1021/acs.biochem.5b00309. [DOI] [PubMed] [Google Scholar]