Eukaryotic ribosomes contain the high-affinity protein kinase C βII (PKCβII) scaffold, receptor for activated C kinase (RACK1), but its role in protein synthesis control remains unclear. We found that RACK1:PKCβII phosphorylates eukaryotic initiation factor 4G1 (eIF4G1) at S1093 and eIF3a at S1364.
KEYWORDS: translation, eIF4G, RACK1, ribosome, PKCβII, eIF3, eIF4E
ABSTRACT
Eukaryotic ribosomes contain the high-affinity protein kinase C βII (PKCβII) scaffold, receptor for activated C kinase (RACK1), but its role in protein synthesis control remains unclear. We found that RACK1:PKCβII phosphorylates eukaryotic initiation factor 4G1 (eIF4G1) at S1093 and eIF3a at S1364. We showed that reversible eIF4G(S1093) phosphorylation is involved in a global protein synthesis surge upon PKC–Raf–extracellular signal-regulated kinase 1/2 (ERK1/2) activation and in induction of phorbol ester-responsive transcripts, such as cyclooxygenase 2 (Cox-2) and cyclin-dependent kinase inhibitor (p21Cip1), or in 5′ 7-methylguanosine (m7G) cap-independent enterovirus translation. Comparison of mRNA and protein levels revealed that eIF4G1 or RACK1 depletion blocked phorbol ester-induced Cox-2 or p21Cip1 expression mostly at the translational level, whereas PKCβ inhibition reduced them both at the translational and transcript levels. Our findings reveal a physiological role for ribosomal RACK1 in providing the molecular scaffold for PKCβII and its role in coordinating the translational response to PKC-Raf-ERK1/2 activation.
INTRODUCTION
In metazoans, a core set of 11 eukaryotic initiation factors (eIFs) facilitates translation initiation and thus determines the protein synthesis rate (1). Adaptive regulation of this machinery relies on signals to two multiprotein ensembles: the eIF4G scaffold (controlling ribosome recruitment to mRNAs) and the eIF2 module (controlling Met-tRNAi loading onto 40S ribosomal subunits). Both ensembles ensure unwinding of the 5′ untranslated region (5′UTR) while scanning for an initiation codon(s) (2). The best-known translation stimuli occur through AKT/mTOR; mTORC1 is involved in regulating the eIF4E-binding proteins (competitive inhibitors of eIF4E:4G binding [3]) and ribosomal protein S6 kinase (4). However, mTORC1 inhibition with rapamycin decreased a serum-induced translation surge in quiescent fibroblasts by only ∼15% (5).
A crucial role in adaptive protein synthesis control is assumed by the C-terminal portion of eIF4G, comprising 3 HEAT (Huntingtin/EF3/PP2A/Tor1) domains and the interdomain linker (IDL) separating HEAT domains 1 and 2 (Fig. 1A). The eIF4G C terminus coordinates pivotal roles in protein synthesis initiation, namely, 5′UTR unwinding/scanning by the eIF4G:4A:4B translation initiation helicase (6, 7) and 40S ribosomal subunit recruitment via eIF3c-e (8). Of particular interest is the IDL (Fig. 1A), since it harbors a plethora of important posttranslational modifications and its flexible structure allows for rapid adaptation of eIF4G:protein assemblies on template mRNA. For example, phosphorylation of S1232 by extracellular signal-regulated kinase 1/2 (ERK1/2) (upon PKC-Raf activation [6]) or by cyclin-dependent kinase 1 (CDK1):cyclin B1 (during mitosis [9]) coordinates anticooperative interactions of HEAT2 with eIF4A that control 5′UTR unwinding/scanning. A comprehensive understanding of eIF4G's role in adaptive protein synthesis regulation is hindered by the multitude of its binding partners (for eIF4G1's C terminus, eIF4A, eIF4B, eIF3c-e, and MNK), its complex association with template mRNA, clusters of posttranscriptional modifications and structural flexibility in the IDL, and the dynamics of the initiation complex in the scanning/initiation process.
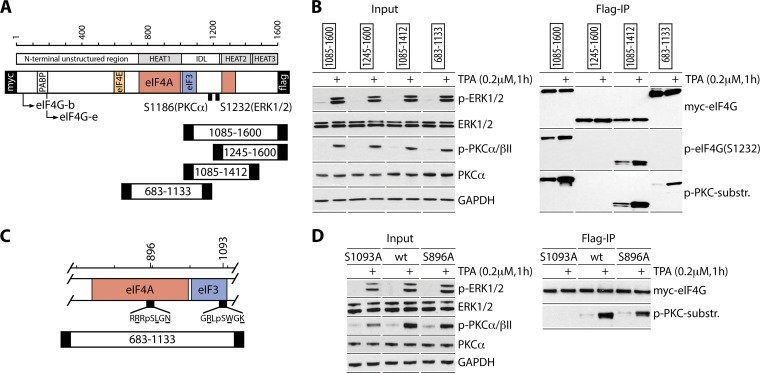
FIG 1.
Mapping of PKC-dependent phosphosites in the C-terminal portion of eIF4G. (A) eIF4G structural domains, translation factor interactions, isoforms and truncation fragments, and two previously investigated phosphosites (6, 10). (B) TPA-dependent phosphorylation of eIF4G. HEK293 cells were transfected (16 h) with expression vectors for the Myc-eIF4G-Flag fragments indicated, serum starved (24 h), and treated with DMSO or TPA. Input cell lysates and Flag IPs were tested by immunoblotting with the indicated antibodies. The assay was repeated 3 times with consistent results; a representative series is shown. (C) Sequence context and localization of two putative PKC-dependent phosphosites, predicted based on the sequence specificity of p(S)-PKC substrate antibodies. (D) TPA-dependent phosphorylation of eIF4G(S1093) in HEK293 cells transiently transfected for expression of the indicated fragments as described for panel B. The assay was conducted 3 times with similar outcomes; results from a representative test are depicted.
In this study, we investigated phosphorylation of eIF4G1 by members of the protein kinase C (PKC) superfamily. PKC, strictly defined by a diacylglycerol/Ca2+/phorbol ester activation spectrum, encompasses four classical/conventional isoforms (α, βI/βII, and γ) and 7 novel/atypical isoforms. We previously identified PKCα as the kinase for eIF4G(S1186) phosphorylation (10). Here, we identified S1093 in the eIF4G1 IDL as a substrate of PKCβII. An involvement for PKCβII in translation initiation control is plausible, since its high-affinity scaffold RACK1 is a stoichiometric component of the 40S ribosomal subunit (11). As a scaffold, RACK1 was shown to interact with more than 100 proteins, either directly or as a part of a complex. Not surprisingly, RACK1 has proposed roles in protein synthesis (14); the mechanisms of its involvement, however, remain unresolved. Our investigations implicate RACK1:PKCβII in phosphorylation of eIF4G1(S1093) and eIF3a(S1364) and revealed a defining role for RACK1:PKCβII in PKC-Raf-ERK1/2-mediated template-specific and global protein synthesis control. We report the effects of RACK1:PKCβII-mediated eIF4G1(S1093) phosphorylation on eIF4G function during translation initiation in a companion manuscript (15).
RESULTS
eIF4G1(S1093) is phosphorylated by PKC.
There are 3 eIF4G isoforms in mammals, eIF4G1, eIF4G2, and death-associated protein 5 (DAP5). Compared to eIF4G1, DAP5 lacks the N-terminal ∼700 amino acids (aa) (it does not bind eIF4E), HEAT1:eIF4A binding is relatively weak (16), HEAT2:eIF4A binding is absent (17), and its IDL is poorly conserved. DAP5 may perform critical translation initiation roles at specific templates, distinct from those for eIF4G1/2, that hinge on inducible DAP5:eIF2β binding (18). eIF4G2 is homologous to eIF4G1 with identical binding partners; however, it is expressed at an ∼1:10 ratio (19) and lacks key posttranslational IDL modifications, e.g., a PKCα site at S1186 or the adjacent S1188 (10) and an ERK1/2/CDK1 site at S1232 (6, 9). Accordingly, eIF4G1 (referred to as eIF4G from here on) dominates the protein synthesis response to PKC-Raf-ERK1/2 activation (see below) and is the focus of our studies.
To map PKC-dependent phosphosites in eIF4G's IDL, we carried out Flag immunoprecipitations (Flag IPs) of lysates from HEK293 cells expressing Myc/Flag-tagged truncated eIF4G fragments (Fig. 1A). 12-O-Tetradecanoyl-phorbol-13-acetate (TPA) stimulation yielded p-PKCα/βII-, p-ERK1/2-, and ERK1/2-mediated p-eIF4G(S1232) in the 1085-1600/1412 fragments (Fig. 1B). These fragments contained a TPA-dependent phosphosite detected by p-(S)-PKC substrate-specific antibodies, likely the known PKCα site at eIF4G(S1186) (Fig. 1B) (10). However, we observed a new TPA-stimulated phosphosite in the 683-1133 fragment, which does not include S1186 (Fig. 1B). Specificity of the p-(S)-PKC substrate antibody, p-(S) surrounded by R/K residues at −2/+2 and a hydrophobic/aromatic residue at the +1 position, indicated substrates at S896 or S1093 (Fig. 1C). To locate this novel phosphosite, we tested 683-1133 fragments with S896A or S1093A substitutions. Flag IP of lysates from HEK293 cells expressing wild-type (wt) or mutant fragments revealed lost reactivity with p-(S)-PKC substrate antibody for eIF4G(S1093A) (Fig. 1D). Thus, a newly recognized TPA-responsive phosphosite is located at eIF4G(S1093), in the eIF3e binding motif (aa 1052 to 1104 [8]) (Fig. 1C).
TPA-activated PKCβII phosphorylates eIF4G(S1093).
The eIF4G(S1093) site is in consensus context for AGC kinases ROCK1/2, DMPK1/2, MRCKα/β, and PKCα/β/γ/δ (Fig. 2A). To identify the kinase(s) for p-eIF4G(S1093), we first applied broad-spectrum inhibitors Bim1 (classic [α, βI, βII, and γ] and some novel PKCs [20]) and Y27632 (ROCK1/2 and MRCKα/β [21]). HEK293 cells expressing tagged 683-1133 fragment were treated with the indicated inhibitor (2 h) followed by TPA (1 h). TPA stimulation in the absence of inhibitors activated PKCα/βII and ERK1/2 (Fig. 2B, top panel, lane 2). Bim1 did not affect PKCα/βII phosphorylation but blocked downstream p-ERK1/2 and p-eIF4G(S1232) (Fig. 2B, lane 4). Y27632 inhibited neither PKCα/βII nor ERK1/2 phosphorylation (Fig. 2B, top panel, lane 6). Flag IP indicated an involvement of a PKC isoform in eIF4G(S1093) phosphorylation, because basal and TPA-induced phosphorylation of this site, detected by p-(S)-PKC substrate antibodies, was prevented only by Bim1 (Fig. 2B, bottom panel, lanes 3 and 4).
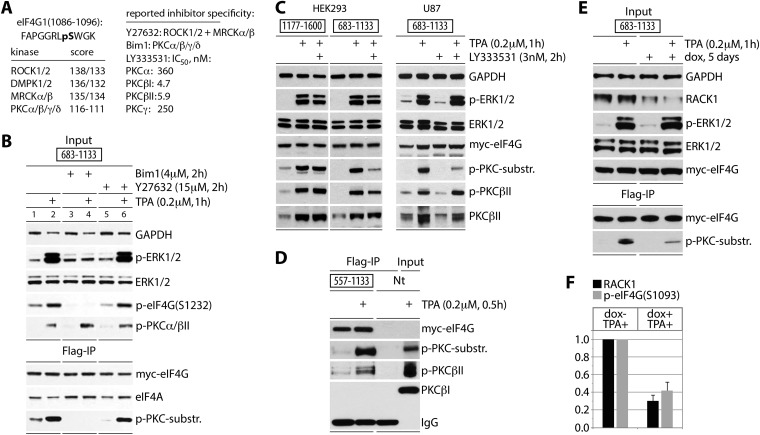
FIG 2.
eIF4G(S1093) is phosphorylated by RACK1:PKCβII. (A) Sequence context of eIF4G(S1093), quantitative computational prediction of kinases (http://www.phosphonet.ca), and specificity of inhibitors used in this study (20, 21). (B) Bim-1 inhibits TPA-induced eIF4G(S1093) phosphorylation. HEK293 cells were transfected for expression of the indicated tagged fragments (16 h), serum starved (24 h), pretreated with or without the indicated inhibitor (2 h), and treated with TPA. Cell lysates were subjected to immunoblotting (top panel) or anti-Flag IP/immunoblotting (bottom panel) with the indicated antibodies. The experiment was repeated three times with consistent results; results of a representative test are shown. (C) The PKCβ-specific inhibitor LY33353 (20) blocks TPA-induced eIF4G(S1093) phosphorylation. HEK293 and U87 cells were treated as described for panel B. Cell lysates were analyzed by immunoblotting with the indicated antibodies. Three test runs yielded similar results; results of a representative assay are shown. (D) TPA-dependent co-IP of active PKCβII with eIF4G. Nontransfected cells (Nt) were used as a negative IP control. (E) Dox-inducible RACK1 depletion prevents eIF4G(S1093) phosphorylation. HeLa cells were Dox induced (5 days), transfected with eIF4G(683-1133) as for panel B, and TPA stimulated. Cell lysates were analyzed by immunoblotting (input) and Flag IP followed by immunoblotting as shown. (F) Quantification of RACK1 depletion and reduction of p-eIF4G(S1093) was normalized by setting the value of +TPA/−Dox to 1; error bars represent SEM. For panels D and E, three repeat assays yielded similar outcomes, and results of representative tests are depicted.
Previously, we showed that PKCα does not phosphorylate S1093 in vitro (10). To test the involvement of PKCβ isoforms in eIF4G(S1093) phosphorylation, we used the PKCβ-specific inhibitor LY333531 at a 3 nM concentration, which is below the 50% inhibitory concentrations (IC50s) for PKCβI and II and ∼100-fold below the IC50s for PKCα/γ (22) (Fig. 2A). HEK293 and glioma (U87) cells were transfected for expression of Myc-eIF4G-Flag fragment 1177-1600 (containing only the PKCα site at S1186) or 683-1133 (bearing only the PKC-dependent site at S1093). Transfected cells were treated with LY333531 (3 nM, 2 h) and TPA stimulated (1 h). TPA induced phosphorylation of PKCβII and ERK1/2, which was not blocked by 3 nM LY333531 (Fig. 2C). Expression of the tagged eIF4G fragments was sufficiently high for detection with p-(S)-PKC substrate antibodies in lysates. Phosphorylation of the 683-1133 fragment was stimulated by TPA and inhibited by LY333531 pretreatment in both cell lines; in contrast, TPA-dependent phosphorylation of 1177-1600 (at the PKCα-dependent S1186) did not respond to 3 nM LY333531 (Fig. 2C). These results indicate an involvement of PKCβ in eIF4G(S1093) phosphorylation.
Inhibitors cannot distinguish between PKCβI and II isoforms. However, the presence of the high-affinity PKCβII scaffold RACK1 in the 40S ribosomal subunit (23), in direct proximity to eIF3 and the eIF4G C terminus (24), favors PKCβII involvement with the translation initiation apparatus. To test eIF4G:PKCβII interactions in response to TPA, we analyzed anti-Flag IPs from cells expressing eIF4G(557-1133) with p-PKCβII(S660) antibodies. This confirmed TPA-responsive co-IP of activated PKCβII but not PKCβI with eIF4G (Fig. 2D). Lastly, to directly implicate RACK1:PKCβII in the observed effects, we constructed an HEK293 cell line with doxycycline (Dox)-inducible RACK1 depletion (Fig. 2E). Dox treatment for 5 days reduced RACK1 abundance to ∼30% of endogenous levels (Fig. 2F). TPA treatment of RACK1-depleted cells reduced detection of the 683-1133 fragment with p-(S)-PKC substrate antibody to ∼40% of that in mock-induced cells (Fig. 2F). In aggregate, our findings suggest that TPA stimulation of cells leads to S1093 phosphorylation in the eIF4G IDL, catalyzed by RACK1:PKCβII on 40S ribosomal subunits.
PKCβII phosphorylates eIF4G(S1093) and eIF3a(S1364) and controls eIF4G:eIF3 assembly.
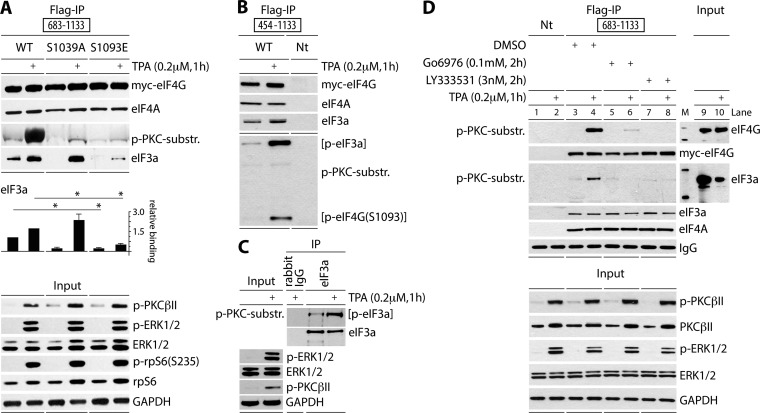
To begin investigating the effects of eIF4G(S1093) phosphorylation on translation initiation, we created Myc-eIF4G-Flag fragments carrying S1093A or -E substitutions (Fig. 3A). The 683-1133 fragment has the proximal eIF4A binding motif in HEAT1 and the eIF3 binding site in the IDL (Fig. 1C), but it lacks all other canonical eIF4G interactions. HEK293 cells were transfected with wt and mutant 683-1133 fragments, serum starved, and TPA stimulated as indicated (Fig. 3A). Anti-Flag IP showed equal, TPA-unresponsive binding of all fragments with eIF4A (Fig. 3A). Only the wt fragment reacted with p-(S)-PKC substrate-specific antibodies after TPA stimulation (Fig. 3A). TPA-induced co-IP of eIF3a with eIF4G(683-1133) changed substantially upon S1093 mutation. S1093A substitution reduced basal binding but enhanced TPA-inducible binding, and S1093E substitution almost abolished TPA-induced eIF3 binding with eIF4G(683-1133) (Fig. 3A). This showed that reversible S1093 phosphorylation is involved in controlling eIF4G:eIF3/40S ribosomal subunit assembly.
FIG 3.
PKCβII phosphorylates eIF4G(S1093) and eIF3a(S1364) and controls eIF4G:eIF3 assembly. (A) HEK293 cells were transfected (16 h) for expression of tagged 683-1133 fragments, serum starved (24 h), and treated with TPA (+). Cell lysates were subjected to immunoblotting (bottom panel) or Flag IP/immunoblotting (top panel) as shown. Relative binding of eIF3a was quantified and averaged between 3 assays. Quantification between experiments was normalized by setting the value of mock stimulation with wt 683-1133 fragment to 1. Error bars represent SEM; asterisks represent Student t test results (P < 0.05). (B) HEK293 cells were transfected (16 h) for expression of Myc/Flag-tagged 454-1133 fragment, serum starved (24 h), and treated with TPA (+). Cell lysates were subjected to Flag IP/immunoblotting with the indicated antibodies. (C) HEK293 cells were serum starved (24 h) and treated with DMSO or TPA (+). Cell lysates were subjected to immunoblotting, rabbit IgG IP/immunoblotting, or eIF3a IP/immunoblotting with the indicated antibodies. (D) PKCβ inhibitors prevent eIF4G(S1093) and eIF3(S1364) phosphorylation. HEK293 cells were transfected (16 h) for expression of Myc/Flag-tagged 683-1133 fragment, serum starved (24 h), pretreated with Go6976 or LY333531 (2 h), and treated with TPA. Cell lysates were subjected to immunoblotting (bottom panel) or Flag IP/immunoblotting (top panel) with the indicated antibodies. All experiments were repeated at least three times; results from representative assays are shown.
p-(S)-PKC substrate-specific antibody immunoblots after co-IP with eIF4G fragments, e.g., the 454-1133 fragment, revealed an additional, TPA-responsive band (Fig. 3B). This suggests that at least one more PKC-responsive site may be present in a protein bound to eIF4G; its size (∼175 kDa) suggested this protein to be eIF3a (Fig. 3B). eIF3a IP from TPA-stimulated HEK293 cells revealed bands of identical size that specifically reacted with eIF3a and p-(S)-PKC substrate-specific antibodies and yielded enhanced signal with the latter in TPA-stimulated cells (Fig. 3C). The eIF3a phosphosite in question may be S1364, which is in ideal PKCβII consensus and was identified using the same p-(S)-PKC substrate-specific antibodies before (https://www.phosphosite.org). To determine if PKCβII catalyzes the TPA-responsive site in eIF3a, we employed the broad (classical isoform) PKC inhibitor Go6967 and the PKCβ-specific inhibitor LY333531 (Fig. 2A) in co-IP assays with the eIF4G(683-1133) fragment (Fig. 3D). Pretreatment of transfected HEK293 cells with both inhibitors had no effect on PKCβII and ERK1/2 phosphorylation but blocked phosphorylation of both TPA-responsive sites in eIF4G and eIF3a as detected by p-(S)-PKC substrate-specific antibodies (Fig. 3D, lanes 6 and 8). For reasons pointed out above, abolition of TPA-responsive eIF3a phosphorylation by LY333531 (at 3 nM) strongly implicates PKCβ. Juxtaposing the p-PKC substrate-specific antibody-responsive band in the co-IP with eIF3a in the input lysate (Fig. 3D, input panels) provided additional evidence that the phosphorylated protein was eIF3a.
In summary, our investigations up to this point implicated RACK1:PKCβII in phosphorylation of eIF4G(S1093) and eIF3a(S1364). These results are in agreement with recent ultrastructural studies of eIF3 and the 40S ribosomal subunit, which place both substrates proposed here in close physical proximity to the RACK1 scaffold (24) (see Discussion). Our further studies diverged into two directions. We report on the influence of RACK1:PKCβII/eIF4G(S1093) and eIF3a(S1364) phosphorylation on global and template-specific translation upon PKC-Raf-ERK1/2 activation below. Our investigations of RACK1:PKCβII's role in shaping eIF4G's intramolecular arrangement and the assembly of translation initiation complexes are reported in a separate study (15).
eIF4G controls TPA-induced global and template-specific translation.
To investigate the role of RACK1:PKCβII-mediated eIF4G(S1093) phosphorylation in adaptive translation control, we performed puromycylation assays of global protein synthesis and tested a select group of eminent, high-impact TPA-inducible biological response modifiers. These include cyclooxygenase 2 (Cox-2) (the rate-limiting factor in prostaglandin E2 biosynthesis [25]), urokinase-type plasminogen activator receptor (uPAR) (a coordinator of cell signaling and extracellular matrix proteolysis [26]), hypoxia-inducible factor 1-α (HIF1-α) (a key node of the hypoxia response [27]), dual-specificity mitogen-activated protein kinase (MAPK) phosphatase (MKP-2) (a negative feedback regulator of ERK1/2, p38, and JNK [28]), and p21Cip1 (an inhibitor of cyclin-dependent kinases/cell cycle progression [29]). Biologically, these proteins share low constitutive expression with very strong stress/mitogen inducibility that prominently involves posttranscriptional regulation (30–33). This group of proteins was compared to (constitutively expressed) Cox-1, which lacks the inducible posttranscriptional control of Cox-2 (34).
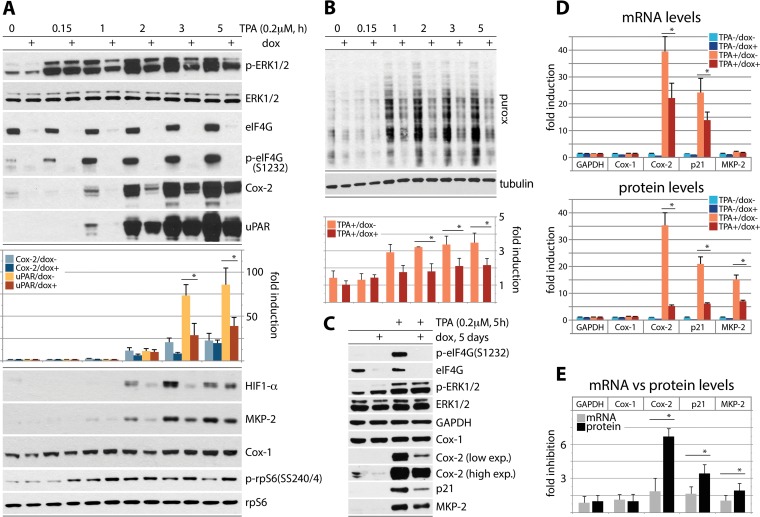
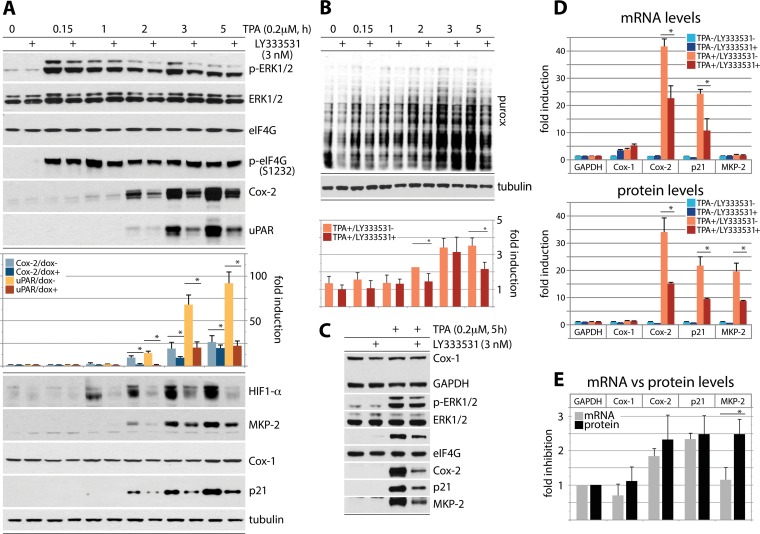
First, to test eIF4G's role in coordinating TPA-stimulated translation, we used HeLa cells with Dox-inducible eIF4G depletion (35). Cells were Dox induced (72 h), serum starved (24 h), and TPA stimulated for up to 5 h (Fig. 4A and B). For the last 15 min of the incubation interval, the cells were treated with 5 μM puromycin (Fig. 4B). As previously shown (9), these assay conditions do not interfere with signal transduction pathways (p-ERK1/2 occurred in all TPA-stimulated samples [Fig. 4A]) and do not lead to mTOR activation (data not shown). Dox induction decreased eIF4G and (ERK1/2-dependent) p-eIF4G(S1232) levels by ∼90% (35) (Fig. 4A). TPA-mediated induction of Cox-2 and uPAR was first evident at 1 h after TPA stimulation and reached ∼25-fold- and ∼80-fold-enhanced expression at 5 h after TPA stimulation (normalized to rpS6 [Fig. 4A]), respectively. eIF4G depletion reduced this effect up to ∼2.5-fold (Fig. 4A). Residual inducibility may be due to remnant eIF4G1 and expression of eIF4G2/DAP5 at endogenous levels. A similar induction pattern was observed for HIF1-α and MKP-2 (Fig. 4A). TPA induction of Cox-1 was too delicate to reliably quantify and did not respond to eIF4G depletion (Fig. 4A).
FIG 4.
Regulation of adaptive global and template-specific translation by eIF4G. (A and B) eIF4G depletion inhibits the TPA-induced surge of Cox-2, uPAR, HIF-1α, and MKP-2 translation (A) and of global protein synthesis (B). HeLa cells with Dox-inducible eIF4G depletion (35) were Dox induced (72 h), serum starved (24 h), and TPA stimulated (up to 5 h). For the last 15 min of the stimulation interval, the cells were treated with 5 μM puromycin. Cell lysates were subjected to immunoblotting with the indicated antibodies (A) or with an antipuromycin antibody (B). (C to E) HeLa cells with Dox-inducible eIF4G depletion were Dox induced (72 h), serum starved (24 h), and TPA stimulated (5 h); tandem samples were used either for immunoblotting with the indicated antibodies (C and D, bottom panel) or for RNA isolation and RT-qPCR (D, top panel). Relative inhibition upon eIF4G depletion of TPA-induced GAPDH, Cox-1, Cox-2, p21Cip1, and MKP-2 mRNA (D, top panel) versus the corresponding proteins (D, bottom panel) is also shown. (E) Dox−/Dox+ ratio of GAPDH, Cox-1, Cox-2, p21Cip1, and MKP-2 mRNA versus protein levels in TPA-stimulated cells, based on data shown in panel D. In panels B and D, quantifications for protein/transcript levels represent the average values from 3 independent series (normalized to mock-treated cells) and two independent RT-qPCR experiments, each analyzed in triplicate, for mRNA. The error bars represent SEM, and asterisks indicate significant (P < 0.05) Student t test (A, B, D, and E) results.
To assess global protein synthesis in our assay, we probed puromycylated polypeptides (puro:x) with an antipuromycin antibody by immunoblotting. The intensity of puromycin labeling was quantified and normalized to tubulin levels (Fig. 4B). In serum-starved HeLa cells, TPA stimulation steadily increased global translation up to ∼2.5-fold at 5 h. eIF4G depletion consistently suppressed TPA-induced global protein synthesis by ∼40% at 1 to 5 h after TPA stimulation (Fig. 4B).
Our investigations targeted genes with known, intricate posttranscriptional regulation of mRNA stability. Therefore, to decipher the effects of TPA on template abundance versus translation, we evaluated the effect of eIF4G depletion on Cox-1, Cox-2, p21Cip1, and MKP-2 mRNA and protein levels upon TPA stimulation in parallel (Fig. 4C to E). TPA did not significantly alter GAPDH (glyceraldehyde-3-phosphate dehydrogenase), Cox-1, or MKP-2 mRNA levels but potently induced Cox-2 and p21Cip1 template abundance (Fig. 4D). This is consistent with compelling prior evidence for regulation of Cox-2 and p21Cip1 mRNA stability via RNA-binding proteins interacting with AU-rich elements (AREs) in their 3′UTRs (36, 37). A TPA-induced surge in p21Cip1 template abundance was due to mRNA stabilization rather than enhanced transcription (38). eIF4G depletion had a minor, <2-fold suppressive effect on the TPA-induced increase of Cox-2 and p21Cip1 mRNA abundance (Fig. 4D), yet it reduced protein synthesis ∼7-fold and ∼4-fold, respectively (Fig. 4D). TPA-induced MKP-2 translation occurred without an accompanying effect on its mRNA (Fig. 4D).
Our data indicate combinatorial posttranscriptional regulation mediated by TPA-mediated PKC activation. This includes template stabilization, e.g., via PKC signals to RNA-binding proteins interacting with the AREs in Cox-2 (39, 40) and p21Cip1 (41) 3′UTRs and to translation initiation machinery, e.g., via RACK1:PKCβII to eIF4G. Since it occurred in the absence of mRNA increases (MKP-2 [Fig. 4D]), TPA inducibility is not merely a function of mRNA levels but involves active translation stimulation. This is also borne out by the finding that eIF4G depletion had a far greater effect on Cox-2/p21Cip1 translation than on the abundance of their templates (Fig. 4E).
PKCβII coordinates TPA-induced global and template-specific translation.
Our studies implicated eIF4G-dependent translation induction in TPA-stimulated protein synthesis. We next tested the role of PKCβ activity in this phenomenon, using the PKCβ-specific inhibitor LY333531 (Fig. 5). Serum-starved HeLa cells were TPA stimulated in the presence of dimethyl sulfoxide (DMSO) or LY33353 and analyzed as reported for Fig. 4. LY333531 did not affect levels of eIF4G or p-eIF4G(S1232), but, similar to the case for eIF4G depletion, it suppressed TPA-induced Cox-2, uPAR, HIF1-α, MKP-2, and p21Cip1 expression (Fig. 5A). Meanwhile, Cox-1 expression was virtually unchanged upon TPA stimulation/PKCβ inhibition (Fig. 5A). Puromycylation assays showed an ∼2.5-fold increase of global translation at 5 h after TPA stimulation. LY333531 decreased this effect by ∼25 to 30% (Fig. 5B), less efficiently than eIF4G depletion (Fig. 4B). Our observations indicate major roles for PKCβ activity in TPA-responsive template-specific translation and some influence over global protein synthesis (Fig. 5A and B). We next performed tandem tests of TPA-induced GAPDH, Cox-1, Cox-2, p21Cip1, and MKP-2 mRNA/protein upon LY333531 treatment, as reported for eIF4G depletion in Fig. 4C to E (Fig. 5C to E). In contrast to eIF4G depletion, LY333531 caused similar decreases in TPA-induced Cox-2/p21Cip1 mRNA and protein abundance (Fig. 5D and E). LY333531 had similar effects on MKP-2 mRNA/protein abundance as eIF4G depletion, likely reflecting the lack of stability-determining elements in the MKP-2 mRNA (Fig. 5D).
FIG 5.
Regulation of global and template-specific translation by PKCβ activity. (A and B) Specific inhibition of PKCβ with LY333531 reduces TPA-inducible translation of Cox-2, uPAR, HIF1-α, MKP-2, and p21Cip1 mRNAs (A) and global protein synthesis (B). HeLa cells were Dox induced (72 h), serum starved (24 h), pretreated with 3 nM LY333531 (2 h), TPA stimulated (up to 5 h), and analyzed as shown in Fig. 4. (C to E) HeLa cells were serum starved (24 h), pretreated with 3 nM LY333531 (2 h), and TPA stimulated (5 h) (C). Tandem samples were used for RNA isolation, RT-qPCR, and immunoblotting and analyzed as described for Fig. 4D and E (D and E).
RACK1 regulates the expression of TPA-inducible genes.
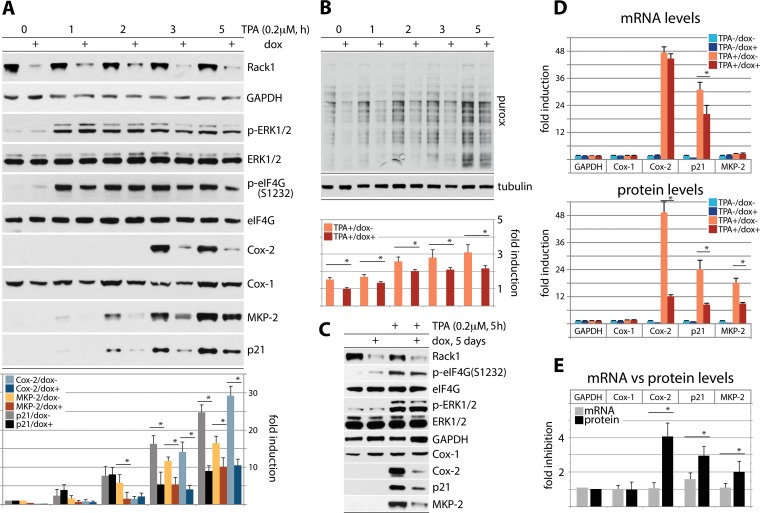
To empirically connect the effects of eIF4G depletion/PKCβ inhibition on template-specific or global translation, we tested TPA stimulation of HeLa cells with Dox-inducible depletion of RACK1 (Fig. 6). As for Fig. 4A and B, cells were Dox induced (72 h), serum starved (24 h), TPA stimulated (up to 5 h), and treated with 5 μM puromycin (Fig. 6A and B). Dox induction decreased RACK1 levels by ∼80% but did not alter total eIF4G levels or eIF4G(S1232) phosphorylation (Fig. 6A); however, TPA-induced p21Cip1, MKP-2, and Cox-2 accumulation was diminished ∼2- to 3-fold upon RACK1 depletion (Fig. 6A). Puromycylation assays showed that native and TPA-stimulated global protein expression was decreased by ∼25 to 30% with RACK1 depletion (Fig. 6B). Tandem analyses of GAPDH, Cox-1, Cox-2, p21Cip1, and MKP-2 mRNA versus protein expression upon RACK1 depletion were performed as shown in Fig. 4C to E and 5C to E (Fig. 6C to E). In contrast to the case for eIF4G depletion or PKCβ inhibition, Cox-2 and p21Cip1 mRNA levels were virtually unchanged or merely ∼1.6-fold reduced, respectively, upon RACK1 depletion (Fig. 6C and D). This could reflect the absence of obvious links of RACK1 to mechanisms controlling mRNA template stability. As in prior analyses, GAPDH, Cox-1, and MKP-2 mRNAs were unresponsive to TPA (Fig. 4D and 5D) or RACK1 depletion (Fig. 6D).
FIG 6.
Regulation of global and template-specific translation by RACK1. (A and B) HeLa cells with Dox-inducible RACK1 depletion were Dox induced (72 h), serum starved (24 h), TPA stimulated (up to 5 h), and analyzed for template-specific (A) and global (B) protein synthesis induction as shown in Fig. 4A and B and 5A and B. (C to E) HeLa cells with Dox-inducible RACK1 depletion were Dox induced (72 h), serum starved (24 h), and TPA stimulated (5 h) (C). Tandem samples were used for RNA isolation, RT-qPCR, and immunoblotting and analyzed as described for Fig. 4D and E (D and E).
TPA-induced Cox-2, p21Cip1, and MKP-2 translation was strongly opposed by RACK1 depletion (Fig. 6D), in accordance with RACK1's function as a ribosomal protein and a role for RACK1:PKCβII in coordinating translation initiation through phosphorylation of eIF4G(S1093). Our investigations suggest that template stabilization may be a prerequisite for proper translation induction of immediate-response genes such as that for Cox-2 or p21Cip1, but this effect requires activation of translation, e.g., through combinatorial signals to translation machinery.
Effect of eIF4G(S1093A/E) substitution on template-specific translation.
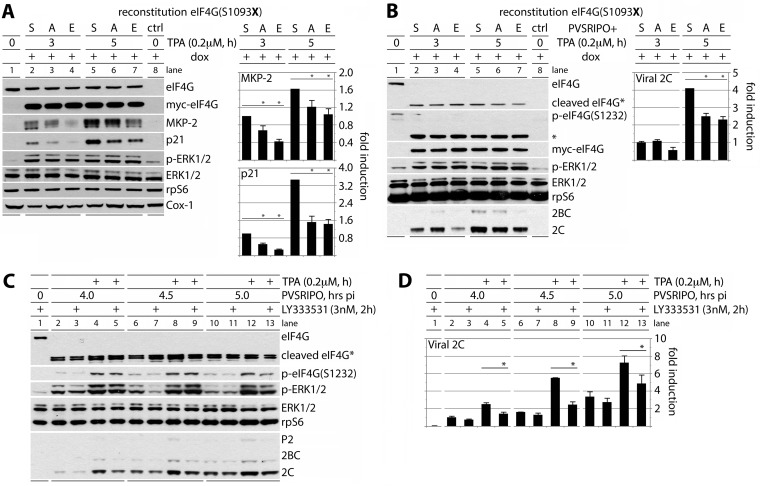
To study the involvement of reversible eIF4G(S1093) phosphorylation in TPA-mediated translation stimulation, we created HeLa cell lines with Dox-inducible eIF4G knockdown/knock-in of Myc-/Flag-tagged exogenous eIF4G. Upon treatment of such cells with Dox, endogenous eIF4G depletion was mock reconstituted with pcDNA5 (ctrl) (Fig. 7A and B, lanes 8) or reconstituted with wt eIF4G, eIF4G(S1093A), or eIF4G(S1093E) (Fig. 7A and B, lanes 2 to 7). Dox induction reduced eIF4G in mock-reconstituted cells by >90% (Fig. 7A and B, compare lane 8 to lane 1). Exogenous eIF4G reconstitution restored eIF4G to native levels in mock-induced cells (Fig. 7A, compare lane 1 to lanes 2 to 7). Cox-1 expression was unchanged upon Dox induction, TPA stimulation, or S1093A/E substitution. TPA stimulated MKP-2 and p21Cip1 translation, and this effect was modulated by S1093 substitution. Both S→A and S→E substitutions reduced translation; the latter consistently exerted greater effects (Fig. 7A). TPA-induced co-IP of eIF3a with eIF4G was slightly enhanced with the S1093A substitution and much reduced with S1093E (Fig. 3A). Thus, the loss of inducibility associated with the S1093 substitution variants in vivo is not due to reduced eIF3 binding introduced by modification of the eIF3-binding motif in the IDL. Rather, our data suggest that it is mediated by a loss of reversible phosphorylation associated with the S1093 nonphosphorylatable/phosphomimic substitutions.
FIG 7.
Modulation of template-specific translation by reversible RACK1:PKCβII-mediated eIF4G(S1093) phosphorylation. (A and B) Combined Dox-induced endogenous eIF4G depletion and eIF4G(S1093A/E) reconstitution inhibit TPA-induced translation of MKP-2 or p21Cip1 (A) or of recombinant poliovirus (PVSRIPO) (B). HeLa cells were Dox induced (96 h), serum starved (24 h), and treated with TPA as indicated. PVSRIPO infection was initiated at the time of TPA addition (B); PVSRIPO infection produces eIF4G cleavage (B, lanes 2 to 7 [*]). Cell lysates were analyzed by immunoblotting with the indicated antibodies (left panels), and MKP-2 and p21Cip1 (A) or viral 2C (B) levels were quantified (right panels). (C and D) PKCβ activation enhances IRES-mediated translation of PVSRIPO. HeLa cells were serum starved (24 h), pretreated with 3 nM LY333531 (2 h), stimulated by TPA (+), and infected with PVSRIPO (multiplicity of infection = 2). After the indicated intervals postinfection (pi), cell lysates were subjected to immunoblotting with the indicated antibodies. PVSRIPO infection produces eIF4G cleavage (panel C, lanes 2 to 13 [*]). Expression of viral 2C was quantified (D). Quantitations represent the averages from 3 tests normalized to wt eIF4G data (A and B) or samples without TPA or LY333531 (C and D). Error bars represent SEM; asterisks indicate significant (P < 0.05) Student t test (D) or ANOVA-protected t test (A and B) results.
We also tested the effect of eIF4G(S1093) substitution on enterovirus (type 1) internal ribosomal entry site (IRES)-mediated translation. Type 1 IRESs recruit ribosomes in an eIF4E/m7G(cap)-independent manner via eIF4G (42, 43), through a mechanism that responds to Raf-ERK1/2-MNK1/2 activation (35, 44, 45); RACK1 has been implicated in viral IRES-mediated translation (46). Our assays were carried out in HeLa cell lines with Dox-inducible eIF4G knockdown/knock-in of Myc-/Flag-tagged exogenous eIF4G carrying S1093 substitutions (Fig. 7B). These cells were infected with PVSRIPO, the attenuated poliovirus chimera containing a foreign IRES of human rhinovirus type 2 (47); Myc-eIF4G-Flag was cleaved in PVSRIPO-infected cells (Fig. 7B, lanes 2 to 7). As with host transcripts MKP-2 and p21Cip1, irreversible eIF4G(S1093A/E) substitutions decreased TPA-induced type 1 IRES-mediated viral translation in a similar pattern (Fig. 7B). We also analyzed the role of PKCβ activity in IRES-mediated viral translation (Fig. 7C and D). HeLa cells were pretreated with LY333531, stimulated with TPA, and infected with PVSRIPO (Fig. 7C). TPA stimulation enhanced viral translation 2- to 3-fold at 4 to 5 h postinfection (Fig. 7D, compare bars 2, 6, and 10 with bars 4, 8, and 12). The TPA stimulatory effect on IRES-mediated viral translation was blocked by PKCβ inhibition (Fig. 7D, compare bars 4, 8, and 12 with bars 5, 9, and 13).
Our investigations suggest key roles for RACK1:PKCβII-mediated eIF4G(S1093) phosphorylation in adaptive protein synthesis control. In a companion study, we unraveled mechanisms of how this event controls eIF4G's dynamic intramolecular configurations and assembly with its binding partners eIF4E and eIF3/40S ribosomal subunit (15).
DISCUSSION
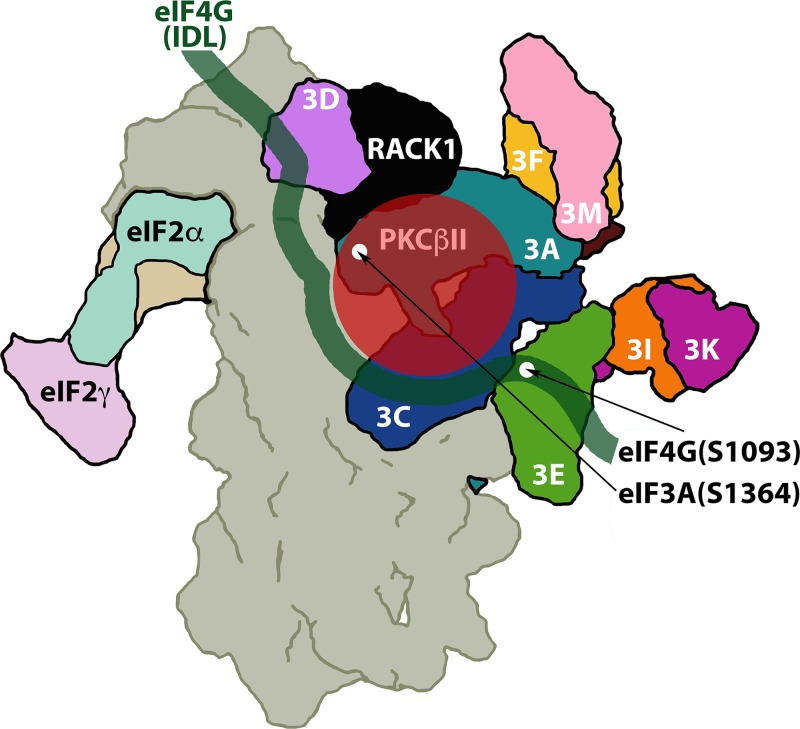
Adaptive regulation of protein synthesis in response to external signals is critical for cell survival. In this investigation, we discovered that ribosomal RACK1:PKCβII activation leads to phosphorylation of eIF4G(S1093) and eIF3a(S1364), stimulation of global protein synthesis, and drastic template-specific induction of key biological response modifiers. Only one activated PKC isoform (PKCβII) binds RACK1 with nanomolar affinity (48). Structural studies of the 40S ribosomal subunit in complex with eIF3 suggest that RACK1 is in direct proximity to the newly discovered PKCβII substrates, eIF4G(S1093) and eIF3a(S1364) (24) (Fig. 8).
FIG 8.
Hypothetical model for RACK1:PKCβII in relation to its substrates eIF4G(S1093) and eIF3a(S1364) (the structures of the 40S ribosomal subunit [gray], eIF3 [bright colors], RACK1 [black], and eIF2 [pastel colors], seen from the back side of the ribosome, are adapted from reference 24). RACK1:PKCβII is in close proximity to eIF4G(S1093) and eIF3a(S1364); a hypothetical arrangement of the flexible eIF4G IDL (spanning aa 1011 to 1104) with binding to the eIF3c, -d, and -e subunits (8) is superimposed.
Cox-2, HIF-1α, MKP-2, p21Cip1, HIF-1α, and uPAR were potently induced upon TPA stimulation of HeLa cells. eIF4G depletion, the PKCβ-specific inhibitor LY333531, or RACK1 depletion countered this effect, implicating RACK1:PKCβII-mediated eIF4G(S1093) phosphorylation. Similar effects were observed with TPA-induced global protein synthesis. eIF4G and RACK1 depletion mainly affected Cox-2 and p21Cip1 translation, whereas PKCβ inhibition had combinatorial effects on protein synthesis and Cox-2 and p21Cip1 mRNA abundance. Cox-2 and p21Cip1 mRNAs feature AU-rich elements (AREs) in their 3′UTRs. Rapid, posttranscriptional induction of such ARE-containing mRNAs is directed through trans-acting RNA-binding proteins with 3′UTR specificity. For example, ELAV1-like protein/human antigen R (HuR) was shown to enhance stability of the Cox-2 or p21Cip1 transcripts, increasing translation of these mRNAs (49, 50). HuR cytoplasmic translocation and its active role in posttranscriptional induction at ARE-containing templates were linked to phosphorylation by PKCα (40) and PKCδ (51). We therefore stipulate that TPA stimuli induce combinatorial effects on template stability and translation enhancement, e.g., via PKC-induced formation of 3′UTR ribonucleoprotein complexes combined with RACK1:PKCβII-mediated eIF4G(S1093)/eIF3a(S1364) phosphorylation. We did not distinguish transcriptional induction versus posttranscriptional stabilization of Cox-2 or p21Cip1 mRNAs in our assay; thus, it is possible that PKC-Raf-ERK1/2-induced transcriptional responses contributed to the observed effects.
Our findings reveal a physiological role for RACK1 in protein synthesis control via PKC-Raf-ERK1/2 signaling. In accordance with RACK1's role in scaffolding PKCβII—ostensibly to direct it to its substrates—the RACK1:PKCβII substrates eIF4G(S1093) and eIF3(S1364) are located in close proximity to RACK1 on the 40S ribosomal subunit (Fig. 8). The unstructured eIF4G IDL, connecting HEAT1 and -2 domains, interacts with eIF3c-e (8); eIF4G(S1093) maps to the eIF3e-binding motif (7) (Fig. 8). We report in a companion article that reversible eIF4G(S1093) phosphorylation controls dynamic long-range intramolecular contacts between eIF4G's flexible unstructured domains that govern assembly with its binding partners eIF4E and eIF3 (15).
MATERIALS AND METHODS
Cell lines, DNA transfections, and eIF4G expression plasmids.
HEK293, HeLa, and U87 cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and nonessential amino acids and were transfected with 16 μg of plasmid DNA using 40 μl of Lipofectamine 2000 (Invitrogen) per 15-cm petri dish. At 16 h posttransfection, the cells were serum starved (24 h) and treated with kinase inhibitors and/or activators. U87 cells were transfected using Continuum transfection reagent (Gemini Bio-Products). Construction of Myc-/Flag-tagged eIF4G expression plasmids has been described previously (10). The eIF4G fragments used in this study were generated by PCR with the corresponding primers (Table 1), and mutations were introduced by overlapping PCR as described earlier (10).
TABLE 1.
Oligonucleotide primers used in this study
| Primer no. | Primer name | Sequence (5′→3′)a |
|---|---|---|
| 1 | 1085(+) | GTAAGCTTTTTGCACCTGGAGGGCGACTG |
| 2 | 1245(+) | GTAAGCTTAAATCCAAGGCTATCATTGAGG |
| 3 | 1412(−) | TTCTCGAGCTGGCCTTCAGGTAGAAATTCC |
| 4 | 1600(−) | TTCTCGAGGTTGTGGTCAGACTCCTCCTC |
| 5 | 683(+) | TTAAGCTTGGGCCCCCAAGGGGTGG |
| 6 | 1177(+) | GTAAGCTTCGTGCGCGGACACCTGCTAC |
| 7 | 454(+) | ATAAGCTTGAGGAGGAAATGGAAGAAGAAGE |
| 8 | 557(+) | TTAAGCTTGAGTCTGAGGGCAGTGGTGTGC |
| 9 | 1133(−) | TTCTCGAGAGGTACCGCTTGTTGAAGG |
| 10 | S898A(+) | CGGCGGCGCGCTTTAGGGAATATC |
| 11 | S898A(−) | GATATTCCCTAAAGCGCGCCGCCG |
| 12 | S1093A(+) | GGCGACTGGGCTGGGGCAAGGGCAGC |
| 13 | S1093A(−) | GCCCCAGCCCAGTCGCCCTCCAGGTGC |
| 14 | S1093E(+) | GGCGACTGGAGTGGGGCAAGGGCAGC |
| 15 | S1093E(−) | GCCCCACTCCAGTCGCCCTCCAGGTGC |
| 16 | Rack-miR-145(+)b | GCCTCGAGATCTGCGACACAACGGCAGGGTAACCCAGAGTGAAGCCACAGATG |
| 17 | Rack-miR-145(−) | GCCTCGAGGATCCGCACCACAACGGCTGGGTAACCCAGACATCTGTGGCTTCAC |
| 18 | Rack-miR-307(+)c | GCCTCGAGATCTGCGGGATGTGGTTCTCTCCTCAGATGGTGAAGCCACAGATG |
| 19 | Rack-miR-307(−) | GCCTCGAGGATCCGCATGATGTGGTTATCTCCTCAGATGCATCTGTGGCTTCAC |
| 20 | Rack-miR-505(+)c | GCCTCGAGATCTGCGGACCCTGGGTCTGTGCAAATACAGTGAAGCCACAGATG |
| 21 | Rack-miR-505(−) | GCCTCGAGGATCCGCATACCCTGGGTGTGTGCAAATACACATCTGTGGCTTCAC |
Restriction sites used for cloning are underlined, mutated nucleotides are in bold italic, and RACK1 target sequences are in bold.
Based on previously validated siRNA (52).
Based on validated Mission shRNA clones (Sigma Aldrich).
Stable cell lines, puromycylation assays, and viral infections.
HeLa stable cell lines with Dox-inducible eIF4G knockdown and mock (pcDNA5) or wt eIF4G-b reconstitution were described earlier (9). Two additional such cell lines with eIF4G(S1093A) and (S1093E) mutant reconstitution were established. These cell lines were maintained in DMEM supplemented with 10% FBS, nonessential amino acids, 800 μg/ml G418, 5 μg/ml blasticidin, and 100 μg/ml zeocin (all from Invitrogen); hygromycin B (100 μg/ml; Corning) was used for reconstitution cell lines instead of zeocin. Stable HeLa and HEK293 cell lines with Dox-inducible RACK1 depletion were established using procedures employed for generating eIF4G knockdown lines (9). Briefly, the miR-4G sequence in pcDNA3.1/TO was replaced with RACK1-specific miRNAs designed according to validated shRNA clones (52) (Sigma-Aldrich) (Table 1). For endogenous eIF4G knockdown/exogenous variant reconstitution, cells were grown with 1 μg/ml Dox (Sigma) for at least 72 h prior to inhibitor treatment or virus infection. For puromycylation assays, 5 μM puromycin was added to the cells 15 min before lysis. PVSRIPO infections were carried out at a multiplicity of infection of 2 as described previously (35); where indicated, cells were treated with inhibitors at 2 h prior to infection, and TPA was added at the 0-h interval.
Kinase and translation inhibitors and activators.
Inhibitors of ROCK1/2, MRCK1/2 (Y27632), PKC (Bim1 and Go6967 [Tocris] and LY333531 [Calbiochem]), and the PKC activator 12-O-tetradecanoyl-phorbol-13-acetate (TPA) (Sigma) were dissolved in DMSO and used at the concentrations indicated. Puromycin 2HCl (Tocris) was dissolved in distilled H2O.
IP, immunoblotting, and antibodies.
Cell lysate preparation, IP with anti-Flag M2–agarose beads (Sigma), and immunoblotting were performed as described previously (10). For IPs, ∼70% confluent cultures grown in 150-mm dishes were lysed with polysome lysis buffer (10 mM HEPES [pH 7.5], 100 mM KCl, 5 mM MgCl2, 0.5% NP-40, and 1 mM dithiothreitol [DTT] [all from Sigma-Aldrich], and EDTA-free proteinase and phosphatase inhibitor [Thermo Scientific]). After overnight incubation for Flag IP (10), beads were washed 5 times in NT-2 buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40 and [all from Sigma-Aldrich], and EDTA-free proteinase and phosphatase inhibitor). eIF3a IP was performed using protein A-agarose beads (Thermo Scientific) and anti-eIF3a antibody (Cell Signaling). Nonspecific rabbit IgG (Cell Signaling) was used as a negative IP control. Prior to IP, all cell lysates were analyzed by immunoblotting to ensure the proper signaling pathway activation/inhibition, and equal loading was ascertained by immunoblotting of “housekeeping” proteins. Antibodies against tubulin and c-Myc tag (Sigma), puromycin (Millipore), PKCβI (LifeSpan BioSciences), PKCβII (Novus Biologicals), poliovirus 2C (35, 44), GAPDH, ERK1/2, PKCα, rpS6, eIF4A, eIF3a, RACK1, eIF4G1, Cox-1, Cox-2, p21Cip1, uPAR, HIF-1α, MKP-2, mTOR, and AKT (Cell Signaling), phospho-specific p-rpS6(S235/6), p-rpS6(S240/4), p-PKCα/βII(T638/641), p-PKCβII(S660), p-ERK1/2(T202/Y204), and p-(S)-PKC substrate (Cell Signaling), and p-eIF4G(S1232) (Novus Biologicals) were used in this study. Immunoblots were developed with SuperSignal West Pico (Thermo Scientific) or Western Bright (BioExpress) enhanced chemiluminescence (ECL) kits.
Reverse transcription–real-time quantitative PCR (RT-qPCR).
Total RNA from HeLa cells was isolated with TRIzol reagent (Life Technologies), followed by the RNeasy cleanup protocol (Qiagen). Target RNA amplification and quantification were performed using the ABI 7900HT real-time PCR system; reactions were set up with the TaqMan RNA-to-CT 1-step kit and gene-specific TaqMan gene expression assays (all from Applied Biosystems) according to the manufacturer's instructions. RNA quantifications were performed using the 2−ΔΔCT method.
Statistical analysis.
Quantification of immunoblot signals was performed using the Li-COR Odyssey FC2 imaging system and Image Studio software. All experiments were repeated at least 3 times. Quantified immunoblot data were normalized between experiments as described in the figure legends and were represented as averages and standard errors of the means (SEM). The paired Student t test was used to compare only two groups, and the analysis of variance (ANOVA)-protected t test was applied for multiple comparisons within data groups. Significance was defined as a P value of <0.05, and the tests used for each data group are described in the figure legends.
ACKNOWLEDGMENTS
This work is supported by PHS grants P50 CA190991 and R01 NS108773 (M.G.) and by the Lefkofsky Family Foundation.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/MCB.00306-18.
REFERENCES
- 1.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinnebusch AG. 2000. Mechanism and regulation of methionyl-tRNA binding to ribosomes, p 185–243. In Sonenberg N, Hershey JWB, Mathews MB (ed), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 3.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC Jr, Sonenberg N. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 4.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A 95:1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jefferies HB, Reinhard C, Kozma SC, Thomas G. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci U S A 91:4441–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrikov MI, Dobrikova EY, Gromeier M. 2013. Dynamic regulation of the translation initiation helicase complex by mitogenic signal transduction to eukaryotic translation initiation factor 4G. Mol Cell Biol 33:937–946. doi: 10.1128/MCB.01441-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villa N, Do A, Hershey JW, Fraser CS. 2013. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 288:32932–32940. doi: 10.1074/jbc.M113.517011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrikov MI, Shveygert M, Brown MC, Gromeier M. 2014. Mitotic phosphorylation of eukaryotic initiation factor 4G1 (eIF4G1) at Ser1232 by Cdk1:cyclin B inhibits eIF4A helicase complex binding with RNA. Mol Cell Biol 34:439–451. doi: 10.1128/MCB.01046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrikov M, Dobrikova E, Shveygert M, Gromeier M. 2011. Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase Cα regulates eIF4G1 binding to Mnk1. Mol Cell Biol 31:2947–2959. doi: 10.1128/MCB.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyle SM, Gilbert WV, Doudna JA. 2009. Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol 29:1626–1634. doi: 10.1128/MCB.01718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Reference deleted.
- 14.Gandin V, Senft D, Topisirovic I, Ronai ZA. 2013. RACK1 function in cell motility and protein synthesis. Genes Cancer 4:369–377. doi: 10.1177/1947601913486348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrikov MI, Dobrikova EY, Gromeier M. 2018. Ribosomal RACK1:protein kinase C βII modulates intramolecular interactions between unstructured regions of eukaryotic initiation factor 4G (eIF4G) that control eIF4E and eIF3 binding. Mol Cell Biol 38:e00306-18. doi: 10.1128/MCB.00306-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Virgili G, Frank F, Feoktistova K, Sawicki M, Sonenberg N, Fraser CS, Nagar B. 2013. Structural analysis of the DAP5 MIF4G domain and its interaction with eIF4A. Structure 21:517–527. doi: 10.1016/j.str.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imataka H, Sonenberg N. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol 17:6940–6947. doi: 10.1128/MCB.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant JD, Brown MC, Dobrikov MI, Dobrikova EY, Gemberling SL, Zhang Q, Gromeier M. 2018. Regulation of HIF-1α during hypoxia by DAP5-induced translation of PHD2. Mol Cell Biol 38:e00647-17. doi: 10.1128/MCB.00647-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. 2011. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol 7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Way KJ, Chou E, King GL. 2000. Identification of PKC-isoform-specific biological actions using pharmacological approaches. Trends Pharmacol Sci 21:181–187. doi: 10.1016/S0165-6147(00)01468-1. [DOI] [PubMed] [Google Scholar]
- 21.Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, Tamakawa H, Yamagami K, Inui J, Maekawa M, Narumiya S. 1997. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 22.Ishii H, Jirousek MR, Koya D, Takagi C, Xia P, Clermont A, Bursell SE, Kern TS, Ballas LM, Heath WF, Stramm LE, Feener EP, King GL. 1996. Amelioration of vascular dysfunctions in diabetic rats by an oral PKC beta inhibitor. Science 272:728–731. doi: 10.1126/science.272.5262.728. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J. 2004. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- 24.des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. 2015. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525:491–495. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hla T, Neilson K. 1992. Human cyclooxygenase-2 cDNA. Proc Natl Acad Sci U S A 89:7384–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith HW, Marshall CJ. 2010. Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol 11:23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 27.Wang GL, Jiang BH, Rue EA, Semenza GL. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92:5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan KL, Butch E. 1995. Isolation and characterization of a novel dual specific phosphatase, HVH2, which selectively dephosphorylates the mitogen-activated protein kinase. J Biol Chem 270:7197–7203. doi: 10.1074/jbc.270.13.7197. [DOI] [PubMed] [Google Scholar]
- 29.Besson A, Yong VW. 2000. Involvement of p21(Waf1/Cip1) in protein kinase C alpha-induced cell cycle progression. Mol Cell Biol 20:4580–4590. doi: 10.1128/MCB.20.13.4580-4590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cok SJ, Acton SJ, Morrison AR. 2003. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J Biol Chem 278:36157–36162. doi: 10.1074/jbc.M302547200. [DOI] [PubMed] [Google Scholar]
- 31.Galban S, Kuwano Y, Pullmann R Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, Lewis SM, Holcik M, Gorospe M. 2008. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol 28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R Jr, Martindale JL, Yang X, Gorospe M. 2008. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol 28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran H, Maurer F, Nagamine Y. 2003. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol Cell Biol 23:7177–7188. doi: 10.1128/MCB.23.20.7177-7188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore AE, Young LE, Dixon DA. 2011. MicroRNA and AU-rich element regulation of prostaglandin synthesis. Cancer Metastasis Rev 30:419–435. doi: 10.1007/s10555-011-9300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown MC, Bryant JD, Dobrikova EY, Shveygert M, Bradrick SS, Chandramohan V, Bigner DD, Gromeier M. 2014. Induction of viral, 7-methyl-guanosine cap-independent translation and oncolysis by mitogen-activated protein kinase-interacting kinase-mediated effects on the serine/arginine-rich protein kinase. J Virol 88:13135–13148. doi: 10.1128/JVI.01883-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon DA, Kaplan CD, McIntyre TM, Zimmerman GA, Prescott SM. 2000. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3′-untranslated region. J Biol Chem 275:11750–11757. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Wang W, Fan J, Lal A, Yang D, Cheng H, Gorospe M. 2004. Prostaglandin A2-mediated stabilization of p21 mRNA through an ERK-dependent pathway requiring the RNA-binding protein HuR. J Biol Chem 279:49298–49306. doi: 10.1074/jbc.M407535200. [DOI] [PubMed] [Google Scholar]
- 38.Schwaller J, Koeffler HP, Niklaus G, Loetscher P, Nagel S, Fey MF, Tobler A. 1995. Posttranscriptional stabilization underlies p53-independent induction of p21WAF1/CIP1/SDI1 in differentiating human leukemic cells. J Clin Invest 95:973–979. doi: 10.1172/JCI117806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doller A, Akool el S, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. 2008. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol Cell Biol 28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doller A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. 2007. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol Biol Cell 18:2137–2148. doi: 10.1091/mbc.e06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lafarga V, Cuadrado A, Lopez de Silanes I, Bengoechea R, Fernandez-Capetillo O, Nebreda AR. 2009. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol Cell Biol 29:4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. 2009. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci U S A 106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. 2014. The mechanism of translation initiation on type 1 picornavirus IRESs. EMBO J 33:76–92. doi: 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown MC, Dobrikov MI, Gromeier M. 2014. Mitogen-activated protein kinase-interacting kinase regulates mTOR/AKT signaling and controls the serine/arginine-rich protein kinase-responsive type 1 internal ribosome entry site-mediated translation and viral oncolysis. J Virol 88:13149–13160. doi: 10.1128/JVI.01884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown MC, Gromeier M. 2017. MNK controls mTORC1:substrate association through regulation of TELO2 binding with mTORC1. Cell Rep 18:1444–1457. doi: 10.1016/j.celrep.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, Baumert TF, Schuster C, Imler JL. 2014. RACK1 controls IRES-mediated translation of viruses. Cell 159:1086–1095. doi: 10.1016/j.cell.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown MC, Dobrikova EY, Dobrikov MI, Walton RW, Gemberling SL, Nair SK, Desjardins A, Sampson JH, Friedman HS, Friedman AH, Tyler DS, Bigner DD, Gromeier M. 2014. Oncolytic polio virotherapy of cancer. Cancer 120:3277–3286. doi: 10.1002/cncr.28862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stebbins EG, Mochly-Rosen D. 2001. Binding specificity for RACK1 resides in the V5 region of beta II protein kinase C. J Biol Chem 276:29644–29650. doi: 10.1074/jbc.M101044200. [DOI] [PubMed] [Google Scholar]
- 49.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. 2001. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest 108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. 2003. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol 23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doller A, Schlepckow K, Schwalbe H, Pfeilschifter J, Eberhardt W. 2010. Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol Cell Biol 30:1397–1410. doi: 10.1128/MCB.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mamidipudi V, Zhang J, Lee KC, Cartwright CA. 2004. RACK1 regulates G1/S progression by suppressing Src kinase activity. Mol Cell Biol 24:6788–6798. doi: 10.1128/MCB.24.15.6788-6798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]