Abstract
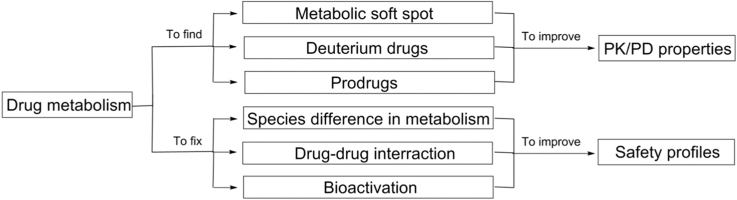
Drug metabolism as a discipline plays an important role in drug discovery and development and the effects of drug metabolism on pharmacokinetics (PK), pharmacodynamics (PD), and safety should be carefully considered. This communication provides an overview of common strategies in the area of drug metabolism for improving PK/PD and safety profiles of drug candidates; these include, but are not limited to, collaboration with medicinal chemists on structure–activity relationships (SAR) to overcome high clearance, using deuterium replacement to further optimize a lead, prodrug approaches to circumvent formulation and delivery difficulties, and addressing issues such as species differences in metabolism, drug–drug interactions (DDI) and formation of reactive metabolites.
KEY WORDS: Bioactivation, Drug discovery and development, Drug metabolism, Metabolite, Pharmacokinetics, Pharmacodynamics, Safety, Toxicity
Graphical abstract
Drug metabolism plays important roles in optimizing pharmacokinetics (PK), pharmacodynamics (PD), and safety profiles of drug candidates in drug discovery and development.
1. Introduction
Drug discovery and development is a time-consuming and costly process. Based on the study by the Tufts Center for the Study of Drug Development (2016)1, on average, it takes more than 10 years and over $2.6 billion to develop a new drug. One of the biggest challenges for the pharmaceutical research community is therefore to execute an optimal process for drug discovery and development. Rational drug design represents an approach to expedite such a process with efficiency as one of the primary objectives, combining the latest science and technology to advance medicines rapidly from laboratory bench side to hospital bed side.
The disposition of a drug in the body involves absorption, distribution, metabolism, and excretion (ADME). ADME is an important component in the drug design process, which studies the fate of a drug molecule after administration. It is a complex process involving transporters and metabolizing enzymes with physiological consequences on pharmacological and toxicological effects, and can play a major role in drug design for identifying better drug molecules in a more efficient way. Metabolism of drugs in the body is a complex biotransformation process where drugs are structurally modified to different molecules (metabolites) by various metabolizing enzymes. Studies on drug metabolism are key processes to optimize lead compounds for optimal PK/PD properties, to identify new chemical entities based on the finding of active metabolites, to minimize potential safety liabilities due to formation of reactive or toxic metabolites, and to compare preclinical metabolism in animals with humans for ensuring potential adequate coverage of human metabolites in animals and for supporting human dose prediction, etc.2 This review focuses on the study of drug metabolism as a discipline for its roles in optimizing pharmacokinetics (PK), pharmacodynamics (PD), and safety profiles of drug candidates in drug discovery and development. The impact of protein binding and transporter on PK and PD properties of drug candidates are beyond the scope of this review.
2. Improving PK and PD properties
2.1. Metabolic soft spot
Drugs need to reach the site(s) of action to elicit their pharmacological effects after administration into the body. However, if drugs show inferior PK properties, e.g., high clearance, short half-life (t1/2) and low bioavailability following oral dosing, it is likely that their PD effects will be sub-optimal. In vitro metabolism studies in human and animal tissue (e.g., liver) preparations and/or in vivo metabolism studies in animals are useful approaches to identify major metabolism pathways (“soft spots”) of drugs3. It is known that the benzylic C–H bond, the allylic methyl and the O-, N-, S-methyl groups are the mostly preferred metabolic soft spots when these groups are not sterically hindered, subjecting to P450 mediated metabolism4. Because of the nature of the enzyme-catalyzed hydroxylation reactions, the chemo- and regiospecicity of substrate oxidation as well as the rate of metabolism is largely determined by the intrinsic reactivity of the substrate sites that are accessible to the ferryl oxidizing species in the P450-substrate complex5, 6. Thus, the tendency to be metabolic soft spots will be depending on the intrinsic reactivity of the functional groups and the substrate specificity of the particular molecule bearing this particular functional group (soft spot) in metabolizing enzyme systems. One of common approaches to address the metabolic soft spot issue is to use bioisosteres to replace those identified soft spots. Bioisosteres are substituents or groups that have chemical or physical similarities and related molecular shapes and may produce roughly similar biological properties7. For example, in some cases, when a benzylic methyl group is identified as a metabolic soft spot, a fluorine or a chlorine atom, or a -CF3 group, could be used to replace the benzylic methyl group.
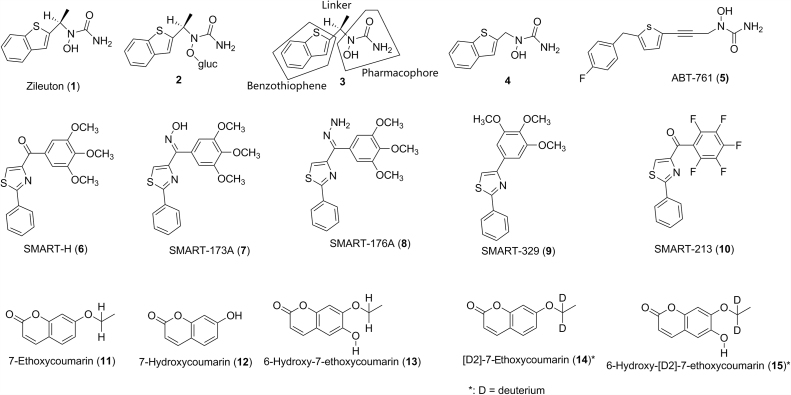
In cases of structure–metabolism relationship studies, blocking a metabolic soft spot is one of the approaches to lower intrinsic clearance which may lead to lower total clearance and, assuming no change in volume of distribution, longer half-life for modified molecules. For example, zileuton (1, Fig. 1) is a 5-lipoxygenase (5-LO) inhibitor used for the maintenance treatment of asthma in human. Zileuton showed half-lives of 0.4 h in cynomolgus monkey, and 2.4 h in human in vivo8. The major metabolism pathway for zileuton in cynomolgus monkey and human is glucuronidation at the N-hydroxyurea moiety to afford metabolite 2 (Fig. 1) with subsequent urinary excretion9. Structure–activity relationship (SAR) studies indicated that the N-hydroxyurea portion is a pharmacophore required for activity. Thus, structural modification on zileuton to minimize the glucuronidation could only be focused on the linker and the benzothiophene portions of zileuton (3, Fig. 1). Compound 4, which resulted from the removal of the methyl group from the linkage group of zileuton showed a 6-fold increase in the intrinsic clearance (Clint) in cynomolgus monkey liver microsomes as well as a 2-fold decrease in the half-life and 4-fold increase in plasma clearance in cynomolgus monkey in vivo8. This suggests that the steric hindrance of the neighboring methyl group from the linker of zileuton indeed diminished the glucuronidation at the N-hydroxyurea portion. SAR studies on the linker portion indicated that compounds with the acetylene linkage generally had lower rates of glucuronidation in cynomolgus monkey liver microsomes as well as longer half-lives and lower plasma clearances in cynomolgus monkey in vivo8. SAR studies on the benzothiophene portion indicated that the use of a simple thiophene ring in place of benzothiophene portion could significantly reduce the glucuronidation rate8. Finally, ABT-761 (5, Fig. 1) with a single furan ring and an acetylene linker was identified as a second generation 5-LO inhibitor which showed >12-fold and >29-fold lower rate of glucuronidation in cynomolgus monkey and human liver microsomes than that of zileuton, respectively. Accordingly, ABT-761 showed >40-fold and >29-fold longer half lives in cynomolgus monkey and human in vivo than that of zileuton, respectively8. Consequently, ABT-761 showed a significant increase in efficacy relative to zileuton at the same dose10. Thanks to the increased potency and improved PK properties, ABT-761 requires only once-daily dosing in human, compared to the multiple daily dosing regimen for zileuton10. This study demonstrated that blocking a metabolism soft spot indeed could improve PK properties of a new chemical entity (NCE) while maintaining the same or producing better pharmacological activity.
Figure 1.
Structures of zileuton (1), SMART-H (6), 7-ethoxycoumarin (11) and their analogs or metabolites.
In studies of 4-substituted methoxybenzoyl-aryl-thiozoles as novel anti-cancer agents, 4-(3,4,5-trimethoxybenzoyl)-2-phenylthiazole SMART-H (6, Fig. 1) was identified as a lead with potent inhibition activity against tubulin polymerization and cancer cell growth11. However, SMART-H showed high metabolic instability in human, dog, rat and mouse liver microsomes with in vitro half-lives ranging from <5 to 30 min11. Metabolite profiling of SMART-H in human liver microsomes indicated that the main metabolism pathway was the reduction of the ketone functional group. The O-demethylation of the methoxy groups of SMART-H was the second most prevalent metabolism pathway. Thus, the ketone and the methoxy groups of SMART-H were considered as metabolic soft spots. When the ketone group was replaced with an oxime group for SMART-173A (7) and a hydrazide group for SMART-176A (8, Fig. 1), SMART-173A and SMART-176A demonstrated an increase in metabolic stability in human liver microsomes by 3-fold and 2-fold, respectively. SMART-173A partially retained the anticancer activity with the average IC50 value of 143 nmol/L against four prostate tumor cell lines (compared to IC50 of 44 nmol/L of SMART-H), while SMART-176A significantly lost the anticancer activity with the average IC50 value of 1250 nmol/L11. With the removal of the most labile ketone functional group of SMART-H, SMART-329 (9, Fig. 1) increased the metabolic stability by 2-fold11. However, SMART-329 completely lost anticancer activity, suggesting that the ketone functional group is critical for anticancer activity. When the second soft spot (three methoxy groups) of SMART-H was replaced with fluorine atoms, the resulting SMART-213 (10, Fig. 1) showed little change in metabolic stability and no anticancer activity11. This suggests that the methoxy groups might also contribute to anticancer activity. Thus, it is possible that structural modification on or around the metabolic soft spots of a lead compound could negatively affect the biological activity.
2.2. Metabolic switching and deuterium replacement
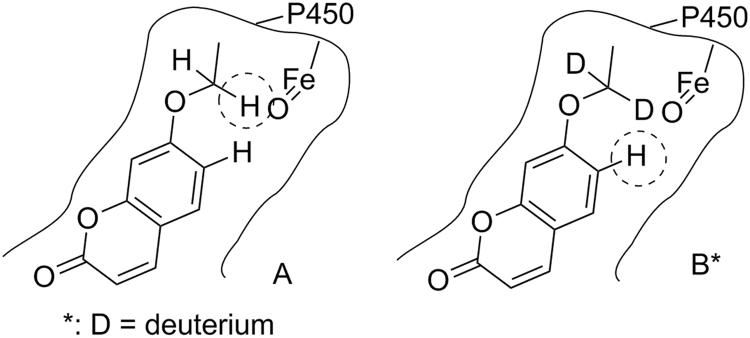
Many small molecule drugs are metabolized by cytochrome P450 (CYP) enzymes in the body. CYPs, predominantly residing in the endoplasmic reticulum of hepatocytes, are a class of enzymes which catalyze a range of oxidative and reductive biotransformation, including carbon hydroxylation, heteroatom oxidation, bond oxidation, hydrocarbon desaturation, and halocarbon dehalogenation, etc.4 CYP-catalyzed biotransformation of drugs to metabolites generally involves an initial binding of a drug to the active site of the enzyme, followed by catalytic turnover of a drug to a metabolite through a so called P450 catalytic cycle12. Whereas the structure–metabolism relationship of drugs involving CYPs is complex and not completely understood, it is generally accepted that major drug-metabolizing CYP enzymes, e.g., CYP3A, have a large active site and prefer lipophilic substrates. The binding and orientation of a substrate are often determined by hydrophobic and steric interactions of the substrate with the specific amino acids at the active site of a CYP enzyme, although infrequently substrates interact with CYP via hydrogen bonding or ionic interactions5, 12, 13, 14, 15, 16. These hydrophobic and steric interactions mainly depend on the lipophilicity (e.g, logP) and steric structures of the substrates17, 18. Subsequent catalytic turnover will then oxidize a C-H bond of the substrate to a C-OH bond to give a specific metabolite. One of the common approaches to slow down or block the metabolism on a specific site of a molecule is to use a substituent, (e.g., a bioisostere, a bulky group, a halogen or a deuterium atom) to replace the H-atom of the C-H bond of the molecule or to place a bulky group in a neighboring position to reduce or block the accessibility of the iron-bound oxidizing species of the P450 enzyme. Once the molecule is bound within the active site, the specific site of metabolism in the substrate where the structural modification occurs, is then largely determined by the intrinsic reactivity of the sites on the molecule that are accessible to the iron-bound oxidizing species in the active site of the CYP. Due to the nature of the active site(s) of CYP enzymes (e.g., CYP 3A4) with a large space binding pocket, blockage of the metabolically labile site (or soft spot) of a lead molecule using one of the above mentioned substituents may result in a change of the binding and orientation of the newly synthesized molecule, switching the original metabolism site to a different C-H bond and resulting in the formation of other metabolite(s) to a greater extent, as compared to that of the lead compound. Because CYP enzymes may have multiple binding sites, it is also possible that the blockage of a soft spot of a lead molecule via structural modification may cause the binding of the new molecule to the alternative binding site of the metabolizing enzyme, resulting in metabolism on a different site of new molecule. This metabolic switching phenomenon is not uncommon. For example, 7-ethoxycoumarin (11, Fig. 1) was extensively metabolized to 7-hydroxycoumarin (12) with a minor metabolite 6-hydroxy-7-ethoxycoumarin (13) through a CYP1A1-mediated oxidations in rat19. Mechanistically, the C-H bond at the α-carbon of the ethoxy group of 7-ethoxycoumarin 11 is initially oxidized to give an unstable hemiacetal intermediate, which subsequently loses an acetaldehyde to give the metabolite 7-hydroxycoumarin (12). [D2]-7-Ethoxycoumarin (14), resulting from the replacement of two hydrogen atoms with deuterium atoms at the α-carbon of the ethoxy group of 7-ethoxycoumarin 11, showed a 2-fold reduction of the formation of 7-hydroxycoumarin (12) in rat liver microsomes. However, [D2]-7-ethoxycoumarin (14) showed a 5-fold increase in the formation of 6-hydroxy-[D2]-7-ethoxycoumarinin (15) in rat liver microsomes, resulting in almost no change of total metabolic stability, compared to 7-ethoxycoumarin (11)19. This result suggests that the oxidation of the C-H bond at the α-carbon of the ethoxy group of 7-ethoxycoumarin 11 (Fig. 2A) was switched to the oxidation of the C-H bond at 6 position of [D2]-7-ethoxycoumarin 14 (Fig. 2B). Thus, addressing the metabolically labile sites (or soft spots) of a lead molecule is a systematic SAR process, where multiple factors, including lipophilicity, stereochemistry, and intrinsic reactivity should be considered in the drug design process.
Figure 2.
Schematic presentation for oxidation of the C-H bond at the α-carbon of the ethoxy group of 7-ethoxycoumarin (A) and for oxidation of the C-H bond at the 6-position of [D2]-7-ethoxycoumarin (B) at the active site of a cytochrome P450 enzyme.
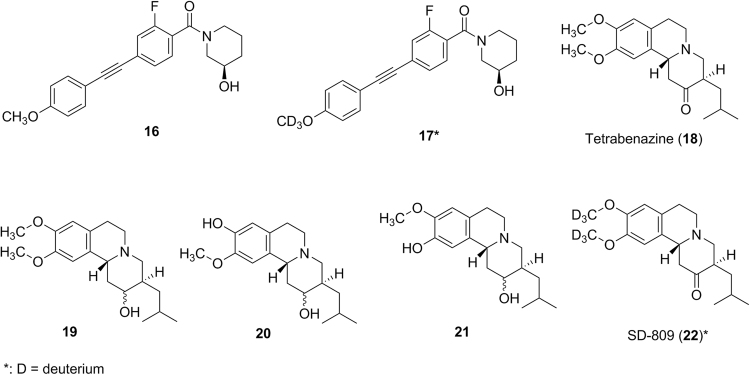
Replacement of hydrogen atoms with deuterium atoms to block a metabolic soft spot or to alter the route of metabolism is an approach to utilize the so-called “isotope effect” when designing new bioactive molecules. Because the carbon-deuterium bond is more difficult to break than the carbon-hydrogen bond, the deuterated molecule may have reduced metabolism on the carbon atom where the deuterium atom is attached, potentially lowering the in vitro and in vivo clearance or altering the metabolism. This was exemplified by the effort to identify new mGlu3-selective and CNS-penetrant negative allosteric modulators20. In SAR studies, compound 16 (Fig. 3) was identified as a lead with good biological activity. However, compound 16 was metabolically unstable in human and rat liver microsomes with a calculated hepatic clearance of 18.9 mL/min/kg in human and 54.1 mL/min/kg in rats. The major metabolic pathway (soft spot) for 16 was the CYP-mediated O-demethylation. This soft spot issue could not be fixed through traditional electronic or steric perturbations due to extremely shallow allosteric ligand SAR20. However, by replacing the hydrogen atoms of the -OCH3 group of 16 with the deuterium atoms (17, Fig. 3), the calculated hepatic clearance in rats decreased from 54.1 mL/min/kg for compound 16 to 35.9 mL/min/kg for 17. Correspondingly, the plasma clearance in rats after IV dosing decreased from 5.2 mL/min/kg for 16 to 2.9 mL/min/kg for 1720. The observed reduced deuterium effect might be due to the possible metabolic switching for 17. This deuterium replacement approach resulted in the discovery of a new compound which warranted further in vitro and in vivo biological testing20.
Figure 3.
Structures of compound 16, tetrabenazine (18) and their analogs or metabolites.
Another example involves tetrabenazine (18, Fig. 3), where its deuterated version represents a new drug form with much improved safety profiles. Tetrabenazine is a marketed drug for treatment of chorea associated with Huntington's disease and is extensively metabolized to form an active metabolite 19 (Fig. 3), which then is further metabolized to active metabolites 20 and 21 (Fig. 3) by the polymorphic enzyme CYP2D621. However, the observed clinical adverse effects, e.g., sedation, somnolence, fatigue and insomnia might be associated with not only the high Cmax of the drug itself but also with the variable levels of active metabolites in patients due to CYP2D6 polymorphism21. Thus, it is required to individualize and slowly titrate dosage over several weeks to identify a dose that reduces chorea while still being well tolerated. SD-809 (22, Fig. 3) from Auspex Pharmaceuticals is a deuterated version of tetrabenazine with an attempt to alter the O-demethylation reaction22. SD-809 showed a longer half-life and higher exposure with a slight Cmax increase relative to tetrabenazine in humans22. This may allow patients to take a much lower dose of SD-809 to achieve a similar exposure with a lower Cmax than tetrabenazine. Thus, if the observed adverse effects in humans associated with administration of tetrabenazine are Cmax-driven, SD-809 may provide an improved safety profile. Auspex Pharmaceuticals announced that SD-809 showed strong efficacy and safety results in its phase 3 clinical registration trial studies in its news release on December 16, 201423. FDA approved SD-809 (Austedo) for the treatment of chorea associated with Huntington's disease on April 3, 2017. SD-809 (Austedo) is indeed the first deuterated product approved by the FDA for human use.
2.3. Prodrugs or active metabolites as new drug candidates
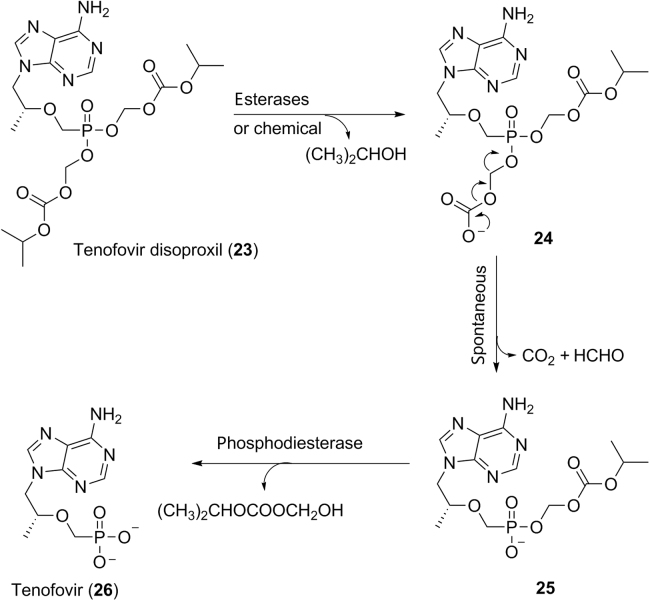
Prodrugs are a class of drugs administered in a pharmacologically inactive form which is enzymatically or chemically transformed to a pharmacologically active form in vivo. This is a traditional strategy to chemically modify pharmacologically active molecules to overcome problems associated with absorption barrier, route of administration, metabolism, excretion, toxicology, as well as to have a site-selective delivery. Prodrugs commonly have functional groups of esters, amides, phosphates, carbonates, or carbamates which are easily cleaved enzymatically or chemically in the body. One of the common usages of prodrugs is to mask the polar or ionizable functional groups of active molecules to improve oral bioavailability. For example, tenofovir (26, Scheme 1) is a nucleotide inhibitor of reverse transcriptase, a crucial viral enzyme in human immunodeficiency virus 1 (HIV-1) infections. Because of the high hydrophilicity of the phosphonic acid group, tenofovir exhibited very low oral bioavailability in human (<5%), which limited its use as a drug24. SAR studies on bis-carbonate esters of tenofovir led to the discovery of the prodrug tenofovir disoproxil (23, Scheme 1) with oral bioavailability of 39% in human24, 25. Mechanistically, tenofovir disoproxil is initially hydrolyzed to the intermediate 24 enzymatically (carboxyesterases) or chemically, followed by spontaneous loss of CO2 and formaldehyde to afford the monoester intermediate 25 (Scheme 1)26. Phosphodiesterases are probably the enzymes responsible for the final hydrolysis of the monoester intermediate 25 to the active drug tenofovir 26 (Scheme 1)26.
Scheme 1.
Proposed mechanism for the formation of active drug tenofovir (26) from the prodrug tenoforvir disoproxil (23).
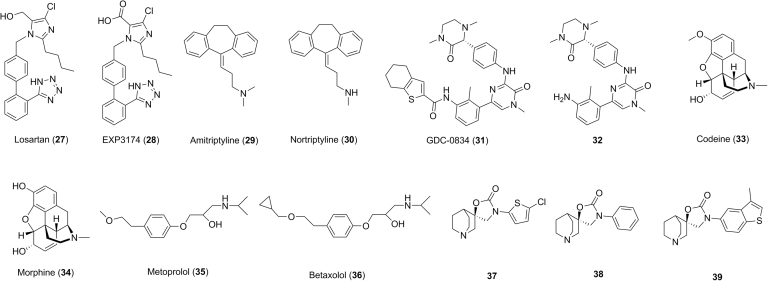
After administration to the body, drugs will be cleared through metabolism and/or excretion in intact form. The latter process sometimes involves active transport, which is beyond the scope of this manuscript. In a majority of cases, sites of metabolism are unpredictable and metabolites could have no pharmacological activity or have activity less than, equivalent to, or more than that of the parent molecules. Metabolites with similar or better pharmacological activity are commonly considered as active metabolites. The conversion of drugs to active metabolites is distinct from the conversion of prodrugs to active drugs in the following aspects. In the case of the conversion of drugs to active metabolites, drugs and active metabolites are pharmacologically active. Metabolism (biotransformation) of drugs is enzymatic and the sites of metabolism are not predictable. However, the conversion of the pharmacologically inactive prodrugs to active drugs can be either an enzymatic or chemical process, and is designed with intended purposes. The unpredictability of the formation of metabolites through metabolism of a drug in the body offers an opportunity for identifying active metabolites as NCEs (new drugs) or new structural templates for further optimization in drug discovery. A hint of the presence of active metabolites may come from a lack of PK–PD correlation or a lack of in vivo efficacy–in vitro potency correlation of a drug molecule. For example, losartan (27, Fig. 4) is used as an angiotensin II receptor antagonist for treatment of hypertension in human. The alcohol functional group of losartan is oxidized to the carboxylic acid group to afford the metabolite EXP3174 (28, Fig. 4) in the body27. EXP3174 is one of the major circulating metabolites in human. In vitro studies showed that EXP3174 has an IC50 of 0.2 nmol/L against the angiotensin II receptor, compared to 4 nmol/L for losartan. By considering the in vitro potency, plasma exposure and the free fraction in plasma, it was estimated that the active metabolite EXP3174 may have contributed approximately 14-times the activity in vitro–in vivo losartan itself27, 28, suggesting that further in vitro and in vivo studies on EXP3174 is warranted.
Figure 4.
Structures of losartan (27), amitriptyline (29), GDC-0834 (31), codeine (33), metoprolol (35), compound 37 and their analogs or metabolites.
In some cases, active metabolites have been developed as new drugs. For example, amitriptyline (29, Fig. 4) is a widely used drug for treatment of mental disorders, including depression and anxiety. Amitriptyline is metabolized by CYP2D6, 3A4 and 2C19 to a demethylated metabolite 30 (Fig. 4)29, 30. This metabolite 30 is a more potent and selective norepinephrine reuptake inhibitor with Ki of 4.4 nmol/L, compared to 22.4 nmol/L for amitriptyline, against norepinephrine transporter. Consequently, metabolite 30 underwent development and eventually became a marketed drug, nortriptyline (Fig. 4).
3. Improving safety profile of drug candidates
3.1. Species difference in metabolism of drugs
Drugs are converted to various metabolites by metabolizing enzymes in the body, and some metabolites may lead to toxicological consequences. In vitro metabolism studies of drug candidates should be initially conducted to compare the similarity of metabolism fate of drug candidates between humans and animal species, and these in vitro results need to be further confirmed in animals in vivo. Animals with similar metabolism fate to human would be selected as safety species in the hope that any major metabolite(s) formed in human will be present in animals to a similar extent in preclinical safety assessment studies31. In addition, the similarity of in vitro and in vivo metabolism of drug candidates in animals would provide a supporting evidence for us to use the in vitro–in vivo extrapolation (IVIVE) to predict human PK properties. For example, (R)-N-(3-(6-(4-(1,4-dimethyl-3-oxopiperazin-2-yl)phenylamino)-4-methyl-5-oxo-4,5-dihydropyrazin-2-yl)-2-methylphenyl)-4,5,6,7-tetrahydrobenzo[b]thiophene-2-carboxamide GDC-0834 (31, Fig. 4) is a potent and selective inhibitor of Bruton's tyrosine kinase (BTK) for potential treatment of rheumatoid arthritis32. In vitro metabolism studies in hepatocytes indicated that GDC-0834 was extensively metabolized in human (80% turnover), moderately metabolized in mouse (56% turnover) and cynomolgus monkey (53% turnover) and relatively stable in rat (20% turnover) and dog (17% turnover) after 3-h incubations. The major hydrolyzed aniline-like metabolite 32 (Fig. 4) detected in hepatocyte incubations accounted for about 62% (human), 9% (mouse), 4% (cynomolgus monkey), 9% (rat) and 12% (dog) of GDC-083432. The amount of metabolite 32 was 2-fold more than that of GDC-0834 in human hepatocytes at 3 h, while the amount of GDC-0834 was the major drug-related component in hepatocytes of mouse, cynomolgus monkey, rat and dog. In vivo studies indicated that metabolite 32 was minor (in rat and cynomolgus monkey) to moderate (in dog), and GDC-0834 was the major drug-related component in plasma after oral administration of GDC-0834 to cynomolgus monkey, rat and dog32. If this in vitro–in vivo correlation for 32 in animals was to hold true for human, then metabolite 32 might be a circulating metabolite whose exposure might be much higher than that of GDC-0834. Additional studies indicated that GDC-0834 was stable in intestinal S9 fractions, urine, blood and simulated gastric fluid. These data suggest that the conversion of GDC-0834 to metabolite 32 occurs mainly in the liver. Subsequent human PK prediction using IVIVE and allometric scaling methods provided a wide range of human blood clearance (5.4–19 mL/min/kg) with low confidence32. This significant difference in the major route of metabolism in human and animals, and low confidence of human PK prediction, prompted the investigators to conduct a single-dose study in healthy volunteers to assess quickly the human PK of GDC-0834. At the doses of 35 and 105 mg, concentrations of GDC-0834 were below the limit of quantitation (<1 ng/mL)32. However, substantial amount of metabolite 32 was detected with Cmax and AUC of 142 ng/mL (238 nmol/L) and 837 h·ng/mL (1404 nmol/L·h) at a dose of 35 mg, and 390 ng/mL (654 nmol/L) and 2448 h·ng/mL (4105 nmol/L·h) at a dose of 105 mg, respectively. Anilines or aromatic amines are a known class of compounds which have a potential to cause methaemoglobinemia, agranulocytosis, aplastic anaemia, hepatotoxicity, skin hypersensitivity and increased risk of mutagenicity in animals and humans33. Mechanistically, the aniline nitrogen could be oxidized to hydroxylamine, nitroso, nitro and related species. These metabolites or intermediates are either chemically reactive or could be further metabolized to reactive metabolites toward proteins and DNAs. Redox cycling between oxidized aniline species (e.g., nitroso and nitro) leads to formation of reactive oxygen species (e.g., hydrogen peroxide, superoxide, etc.) which may cause oxidative stress to cells33. Covalent binding to proteins or DNAs as well as oxidative stress in living cells might be one of the possible causes for the aniline-associated side effects or toxicities. Due to the high exposure of the aniline-like metabolite 32 in human with a possible safety liability for the aniline structural alert, the clinical development of GDC-0834 was terminated32. This case study suggests that it is critical to consider potential impacts of species differences in metabolism of drugs and the potential coverage of major human metabolites in animals should be carefully assessed in drug design. From a regulatory perspective, it is strongly encouraged to identify circulating metabolite(s) as early as possible in clinical trials. This will ensure that the exposure of human metabolite(s) is covered in animals in preclinical safety assessment studies.
3.2. Minimizing polymorphic drug metabolizing enzyme-related risk and drug–drug interaction potential
Most drugs are mainly cleared from the body through metabolizing enzyme-mediated processes34. Some metabolizing enzymes (e.g., CYP2D6, 2C9 and 2C19, etc.) are polymorphic in nature and exist in two or more variant forms with different enzymatic activity in different individuals. Poor metabolizers (individuals with low or no enzymatic activity) could have much higher drug exposure than extensive metabolizers (individuals with high enzymatic activity) when a given dose of a drug is administered. In the case of prodrugs where metabolizing enzymes are required to transform prodrugs to active drugs, the concentrations of active drugs could be lower in poor metabolizers than that in normal patients, resulting in less optimal therapeutic effects. Thus, drug candidates which are mainly metabolized by a polymorphic enzyme could have different pharmacological and toxicological effects among poor metabolizers and extensive metabolizers. The opioid analgesic drug codeine 33 (Fig. 4) is a good example of this. Codeine is a prodrug that only weakly binds to the opioid receptors for analgesic effect. Polymorphic enzyme CYP2D6 catalyzes the O-demethylation of codeine to the active drug morphine 34 (Fig. 4) which has 200-fold greater affinity than codeine. Poor metabolizer patients with low CYP2D6 activities have lower levels of morphine, resulting in less optimal analgesic effect. However, ultrarapid metabolizer patients with higher CYP2D6 activities can have much higher levels of morphine. It was reported that respiratory depression and death occurred in children who received codeine following tonsillectomy and/or adenoidectomy and had evidence of being ultra-rapid metabolizers of codeine due to a CYP2D6 polymorphism35. To avoid treatment complications in patients who are either ultrarapid or poor metabolizers, opioids that are not metabolized by CYP2D6 may be used (e.g., morphine, oxymorphone, buprenorphine, fentanyl, methadone, hydromorphone), alongside non-opioids, depending upon the type of pain being treated36.
In SAR studies, structural modification of a lead compound which is mainly metabolized by a polymorphic enzyme could lead to the discovery of a new drug candidate which is mainly metabolized by multiple enzymes. For example, metoprolol (35, Fig. 4), a selective β1 receptor blocker for treatment of high blood pressure, is mainly metabolized by CYP2D6 in human liver microsomes37. In humans, metroprolol demonstrated low oral bioavailability and a relatively short duration of action, due to fast hepatic metabolism to an O-demethylated metabolite, accounting for 65% of the total metabolism of metoprolol, with α-hydroxylation and N-demethylation accounting for 10% each2. To reduce the major O-demethylation pathway, a bulky cyclopropyl group was used to replace the methyl group of metoprolol to give betaxolol (36, Fig. 4), which showed much slower metabolism. Betaxolol is metabolized by both CYP2D6 and CYP1A2, while CYP2D6 accounted for only 40% of metabolism in human2. Indeed, a clinical study examining metoprolol polymorphic metabolism by CYP2D6 showed a 100-fold variation for both plasma concentrations of metoprolol and its metabolite α-hydroxy metoprolol in patients with cardiovascular diseases2. It is expected that betaxolol (36, Fig. 4) should possess much lower CYP2D6-dependent polymorphism-related risk in human. This example illustrates that reaction phenotyping followed by structural optimization can result in a molecule with a minimized polymorphic metabolizing enzyme-related risk.
Some drugs could be inhibitors or inducers of metabolizing enzymes. If a drug is an inhibitor of a metabolizing enzyme for another drug, when these two drugs are co-administered to the body, the exposure of the other drug could be higher than expected, resulting in a potential safety issue if the safety margin of that drug is limited. If a drug is an inducer of a metabolizing enzyme for another drug, when the two drugs are co-administered, the exposure of another drug could be lower than expected, resulting in potential suboptimal pharmacological effects on the body. This phenomenon where the systemic exposure of a drug is affected by co-administration of another drug is commonly referred to as a drug–drug interaction (DDI). Thus, it is desirable to minimize DDI potential in the drug design process. In cases where a drug candidate has been found to have a high DDI potential with other drugs, development is generally terminated and new drug candidates with lower DDI potential may need to be identified. For example, in an effort to develop potent α7 nicotinic acetylcholine receptor (α7 nAChR) partial agonists for potential treatment of several neurological and psychiatric disorders including cognitive dysfunction, (R)-3′-(5-chlorothiophen-2-yl)spiro[1-azabicyclo[2.2.2]octane-3,5′-oxazolidin]-2′-one (37, Fig. 4) was identified as a lead with a high affinity (Ki=9 nmol/L) toward the α7 nAChR38. However, compound 37 also exhibited potent inhibition toward CYP2D6 with an IC50 of 2.0 mol/L. By contrast, compound 38 (Fig. 4), which was derived from the replacement of the 5-chlorothiophene ring with a benzene ring, showed minimal inhibition toward CYP2D6 (IC50>30 mol/L), suggesting that the CYP2D6 inhibition issue might be addressed by structural modification around the 5-chlorothiophene ring of 3738. After an intensive SAR investigation, a novel and potent α7 nAChR partial agonist (R)-3′-(3-methylbenzo[b]thiophen-5-yl)spiro-[1-azabicyclo[2.2.2]-octane-3,5′-oxazolidin]-2′-one (39, Fig. 4) with a high affinity (Ki=3 nmol/L) toward the α7 nAChR was discovered, which possessed minimal inhibition toward CYP2D6 (IC50 >30 μmol/L)38.
3.3. Minimizing toxicity potential associated with bioactivation
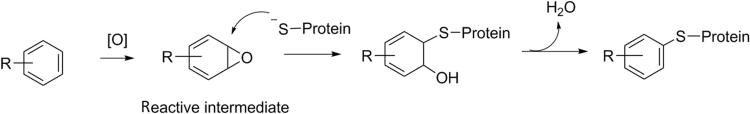
Having an acceptable safety profile is one of the most important requirements for an NCE to become a successful drug. However, in some cases, development of drugs is terminated due to preclinical or clinical observations of toxicity. Drug-induced liver injury (DILI) and genotoxicity are among the most commonly observed toxicities. There are many possible causes leading to such preclinical and clinical toxicity. One of the causes is thought to be metabolism-related bioactivation39. In some cases, metabolism may convert drugs to chemically reactive metabolites/intermediates. Due to the nature of their high electrophilicity, those reactive metabolites may react with components of cellular proteins, DNA, or even the metabolizing enzymes (which catalyze the formation of reactive metabolites) to form corresponding drug-protein adducts, drug-DNA adducts, etc. (Scheme 2). Even though there is no clear correlation between formation of drug-protein adducts/drug-DNA adducts and DILI/genetoxicity, it is believed that bioactivation processes may disrupt normal cellular functions or trigger sequential immune responses and may represent one of the possible causes leading to the observed DILI or genetoxicity. In drug design, it is prudent to design/select compounds which possess a low propensity to form reactive metabolites for further development.
Scheme 2.
Formation of a drug-protein adduct through a bioactivation process where a drug is metabolized to a reactive intermediate which can subsequently bind to a protein.
Electrophilic reactive metabolites formed from bioactivation of drugs could be roughly grouped into two categories: soft electrophiles and hard electrophiles. Based on the hard and soft (Lewis) acids and bases theory (HSAB), hard electrophiles have either a high positive charge density or a formal positive charge at the electrophilic center40. Conversely, soft electrophiles have a lower positive charge density. Hard nucleophiles have high electronegativity and low polarization of valence electrons, whereas soft nucleophiles have low electronegativity and are more polarizable. The reaction rates and selectivity of electrophiles and nucleophiles are mainly dependent upon comparable states of “hardness”40. For example, a soft electrophile such as the α,β-unsaturated ketone can react predominantly with a soft nucleophile such as the thiol group of glutathione (GSH). Similarly, a hard electrophile such as the methyl carbonium ion formed from dimethyl nitrosamine will react with hard nucleophiles such as the nitrogen atoms of purine/pyrimidine bases in DNA. It is worthwhile mentioning that some electrophiles may react with both soft and hard nucleophiles. For example, styrene oxide is generally considered as a soft electrophile. It can react with either GSH (a soft nucleophile) to form GSH adducts41 or react with one of the endocyclic nitrogen atoms of guanine in DNA (a hard nucleophile) to form 7-alkylguanine adducts42.
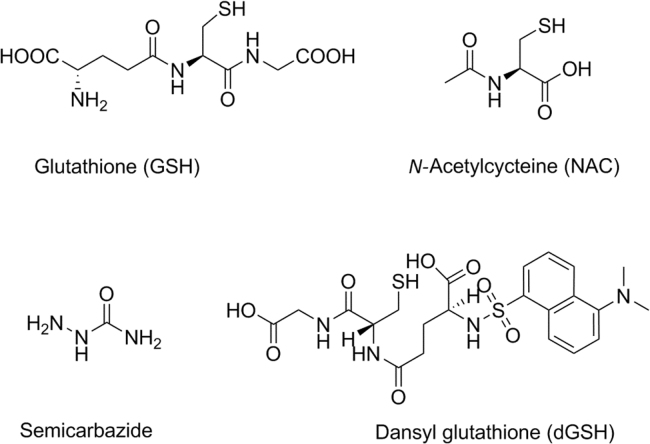
Electrophilic reactive metabolites, in general, are highly unstable, and readily react with nucleophilic macromolecules (proteins, DNA etc.) in biological systems. Due to their highly reactive nature, these metabolites are often short-lived and rarely detectable per se even using the state-of-art modern instrumentation. One approach to deduce the structures of reactive metabolites is via in vitro trapping studies43, 44. In such a practice, one of the nucleophilic trapping agents of GSH, N-acetylcysteine (NAC), potassium cyanide or semicarbazide (Fig. 5) is added in incubations of drugs with liver microsomes or recombinant metabolizing enzymes. If drugs are incubated in hepatocytes, no additional GSH is needed due to the fact that concentration of the endogenous GSH in hepatocytes is high enough for trapping purposes. Structures of corresponding adducts can be detected and characterized by LC–MS and/or NMR. Based on the structures of those adducts, we can postulate the structures of unstable reactive metabolites. Interested readers may refer to additional reviews43, 44.
Figure 5.
Structures of commonly used trapping agents in vitro.
It is believed that covalent protein binding of reactive metabolites formed through a bioactivation process is one of the possible causes leading to DILI signals in animals and human. To quantify covalent protein binding of drugs in biological systems, 3H- or 14C- labeled drugs are required. This is a very resource consuming process and has been a subject of other review articles39, 43, 44. In cases where the 3H- or 14C-labeled drugs are not available, several approaches were attempted to provide semi-quantitative measurement of bioactivation potential of drugs in biological system. One of the approaches is to use commercially available radiolabeled trapping agents (e.g., [35S]cysteine or [14C]sodium cyanide, etc.) in trapping studies with unlabeled test compounds in biological systems. Then, the relative amount of the radioactive peaks of the drug adducts could be used to rank compounds with similar structures45, 46. Dansyl glutathione (dGSH, Fig. 5), a fluorescent-tagged GSH derivative, is one of the other trapping agents used for semi quantification of bioactivation potential of test compounds47. The drug-dGSH adducts could be detected using both the fluorescence spectroscopy detection and LC–MS. Structural information of the drug-dGSH adducts obtained in LC–MS is used to postulate structures of reactive metabolites formed in the biological systems. At the same time, the relative amount of the dGSH adducts could be quantified by fluorescent detection. This approach also could be used to rank compounds with similar structures.
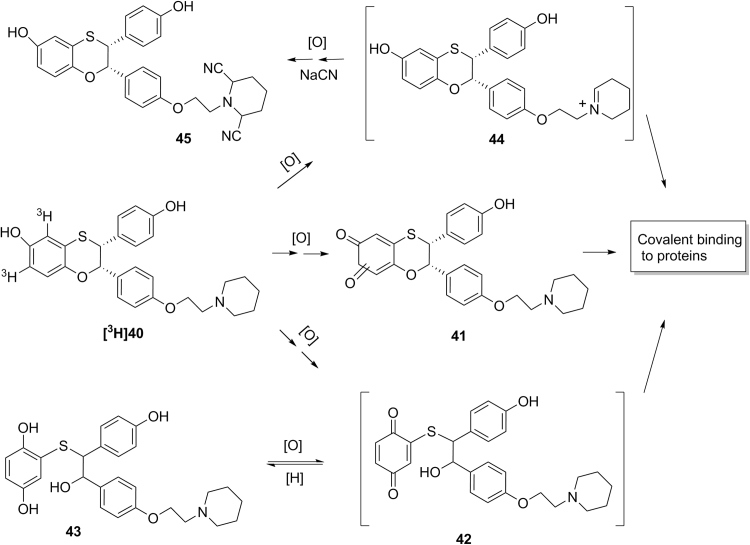
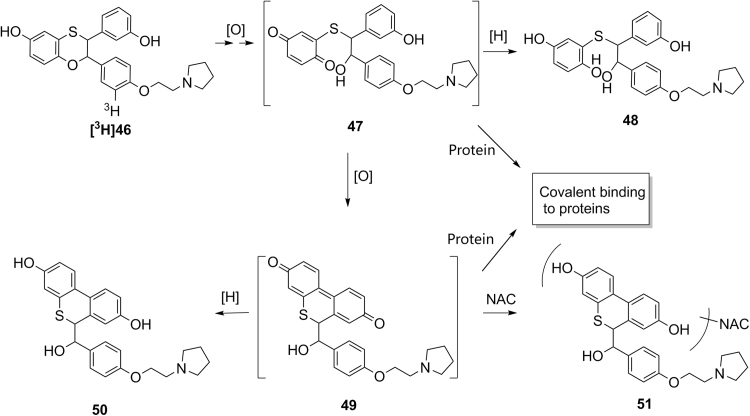
In our previous effort to develop a selective estrogen receptor modulator (SERM) with an estrogen receptor-α (ER-α) subtype selectivity for potential treatment of osteoporosis, (2S,3R)-3-(4-hydroxyphenyl)-2-[4-(2-piperidin-1-ylethoxy)phenyl]-2,3-dihydro-1,4-benzoxathiin-6-ol 40 (Scheme 3) was identified as a lead with potent and selective ER-α antagonist activities in vitro and in vivo in animals48. In vitro covalent protein binding values of [3H]40 were 1106 pmol-equiv/mg protein in human liver microsomes, and 170 pmol-equiv/mg protein in human hepatocytes, when [3H]40 (10 µmol/L) was incubated in human liver microsomes (1 mg protein/mL) for 45 min or in human hepatocytes (1×106 cells/mL) for 60 min48. In vitro metabolism studies identified a quinone metabolite 41 and a hydroquinone metabolite 43 which might be formed from reduction of the reactive para-quinone metabolite 42. Subsequent in vitro trapping studies identified a bis-cyano adduct 45, indicating the formation of reactive iminium ion species 4448. These results suggest that compound 40 is subject to bioactivation through pathways involving at least reactive quinone species 41 and 42 and iminium species 44 (Scheme 3). Subsequently, (2S,3R)-(+)-3-(3-hydroxyphenyl)-2-[4-(2-pyrrolidin-1-ylethoxy)phenyl]-2,3-dihydro-1,4-benzoxathiin-6-ol (46, Scheme 4) was synthesized in the effort to minimize formation of reactive metabolites48. In vitro covalent protein binding values of [3H]46 (10 µmol/L) showed covalent protein values of 461 pmol-equiv/mg protein in human liver microsomes and 48 pmol-equiv/mg protein in human hepatocytes, when [3H]46 (10 µmol/L) was incubated in human liver microsomes (1 mg proteins/mL) for 45 min or in human hepatocytes (1×106 cells/mL) for 60 min48. These in vitro covalent protein binding values of [3H]46 were much lower than that of [3H]4047. In vitro metabolism studies of [3H]46 identified a hydroquine metabolite 46 and a biphenyl hydroquinone metabolite 50 (Scheme 4)48. Metabolites 48 and 50 could be formed from reduction of corresponding reactive quinone intermediates 47 and 49, respectively. Additional in vitro trapping studied identified a NAC adduct 51 with the structure confirmed by LC–MS/MS and NMR48. However, no cyano adduct was detected in trapping studies in the presence of potassium cyanide. These results suggest that the replacement of the piperidine group of 40 with a pyrrolidine group did block the pathway for the formation of the reactive iminium ion 4448. However, the pathways leading to the formation of the reactive quinone metabolites 47 and 49 still existed, which might be responsible for the observed covalent protein binding in human liver microsomes and hepatocytes48. These studies indicate that understanding of bioactivation mechanism could help chemists to better design molecules with low propensity toward bioactivation and eventually to find drug candidates with lower risk of drug metabolism-induced toxicity.
Scheme 3.
Proposed mechanism for the bioactivation of [3H]40.
Scheme 4.
Proposed mechanism for the bioactivation of [3H]46.
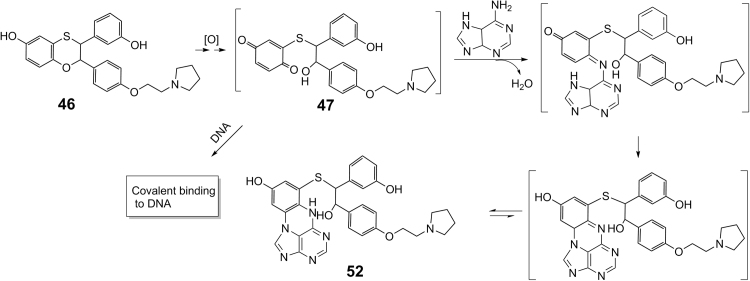
In some cases where bioactivation is believed to likely be one of the causes for observed genetoxicity, trapping studies of drugs with DNA or DNA bases might be performed to elucidate the structures of reactive metabolites formed in biological systems. For example, the above mentioned new lead compound 46 showed genetoxicity in chromosomal aberration assay in Chinese hamster ovary (CHO) cells in vitro and micronucleus induction assay in mouse bone marrow in vivo. Subsequent in vitro trapping studies using DNA bases indicated that up to five adenine adducts were detected in incubations of 46 with human and monkey liver microsomes or recombinant human CPY3A449. Based on the LC–MS/MS and NMR data, the major adenine adduct 50 has a cyclized structure (Scheme 5). This further confirmed the existence of the reactive ring-opened para-quinone intermediate 47 as discussed above. A single cell gel electrophoresis assay (Comet assay) in human hepatocytes further indicated that 46 caused DNA damage in a dose-dependent manner49. It is possible that bioactivation of 46 may be related to the observed genetoxicity.
Scheme 5.
Proposed mechanism for the formation of the adenine adduct 52 through bioactivation processes of 46 in the presence of a trapping agent adenine.
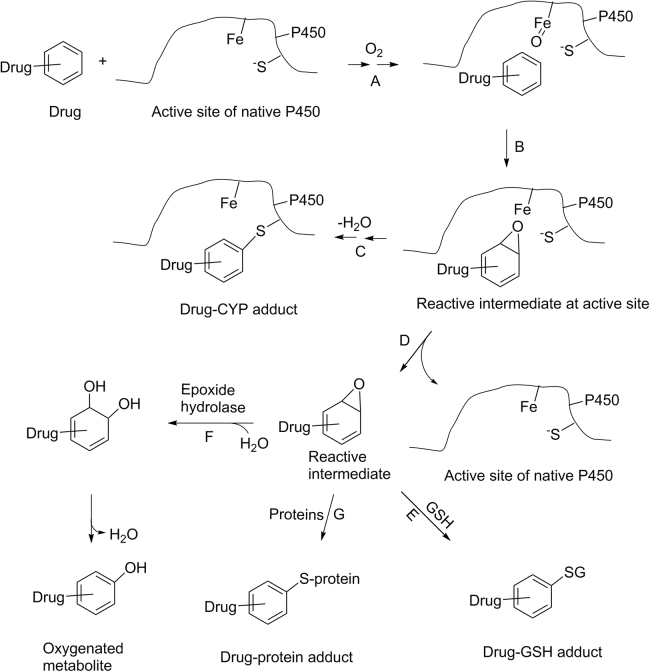
It is worth mentioning that there might be several pathways affecting the fate of the reactive intermediates formed in biological systems. Scheme 6 is a schematic presentation for multiple possible pathways involving a CYP-catalyzed formation of a reactive epoxide intermediate. Binding of a drug to the active site of a CYP enzyme, followed by the assistance of cytochrome P450 reductase and cytochrome b5, triggers the formation of an oxidative oxygen species associated with a CYP enzyme (Scheme 6, pathway A). The oxygen species oxidizes the drug molecule in its proximity to form a reactive epoxide intermediate at the active site (Scheme 6, pathway B). This reactive epoxide intermediate may react with amino acid residues of a CYP enzyme to form drug-CYP (drug-protein) adducts (Scheme 6, pathway C). The formation of the drug-CYP adducts may or may not demonstrate a time-dependent inactivation of the catalytic activity of this CYP enzyme, depending on the covalent binding of the reactive intermediates to the amino acid residues in the specific regions of the CYP enzyme. In a hypothetical extreme case where the intermediate is highly reactive, a majority of the reactive intermediate formed at the active site may react with amino acid residues in situ to form drug-CYP adducts. Thus, only a small amount of reactive intermediate escapes from the active site to the biological system (Scheme 6, pathway D) to be trapped by a trapping agent (e.g., GSH) to form drug-GSH adducts (Scheme 6, pathway E). Unfortunately, due to the complexity of the drug-CYP adducts, the extent of the formation of these drug-protein adducts cannot be easily assessed in bioanalysis of samples from trapping studies. In this case, the amount of the drug-adducts with the trapping agents detected from semi-quantitative approaches is inversely proportional to the actual amount of drug-protein adducts. In another case where the reactive epoxide intermediate is stable enough, a portion of this reactive intermediate can escape from the CYP active site (Scheme 6, pathway D). This epoxide intermediate can either be hydrated by epoxide hydrolase, followed by dehydration to give a mono-oxygenated metabolite (Scheme 6, pathway F) or react with surrounding proteins, including CYP enzymes, to form drug-protein adducts (Scheme 6, pathway G). In the presence of a trapping agent (e.g., GSH), this reactive epoxide intermediate can also react with the trapping agent to form the drug-GSH adducts (Scheme 6, pathway E). In this case, pathways E, F and G are competitive in nature, and the amount of the detected drug-GSH adduct depends on the partitioning among these pathways. The nature of the partitioning among these pathways will be compound-dependent and cannot be easily predicted. In addition, the mono-oxygenated metabolite, formed through the epoxide hydrolase Pathway F, may also be formed through a CYP-mediated direct hydrogen abstraction, followed by oxygen rebounding mechanism. This makes it impossible to use the formation of this metabolite formed from the epoxide intermediate as an additional measure for assessing the bioactivation potential of a compound. It is possible that one compound with less amount of GSH adducts may have higher bioactivation potential to form more drug-protein adducts than the other compound with more GSH adducts. Thus, caution needs to be taken when a comparison of bioactivation potential is used to rank compounds based on semi-quantification or the mass spectrometry responses of drug adducts with trapping agents as a relative percentage of total drug-related components.
Scheme 6.
Schematic presentation of the processes for bioactivation of a drug catalyzed by a cytochrome P450 enzyme. A: binding of a drug to P450 active site; B: formation of reactive intermediate at active site; C: binding of reactive intermediate to P450; D: release of reactive intermediate from active site; E: formation of a drug-GSH adduct; F: hydration of reactive intermediate; G: binding of reactive intermediate to proteins in biological system.
Even though the bioactivation-mediated covalent binding of a reactive metabolite to proteins of human and animals may have a potential to cause toxicity, a specific group of drugs, called covalent drugs, indeed efficiently utilize the covalent mechanism toward its biological targets for action50. Clopidogrel, lansoprazole and esomeprazole are among the marketed covalent drugs50. In developing covalent drugs, the balance of the non-covalent binding affinity and the reactivity of the electrophilic warhead(s) towards the biological targets should be carefully considered and safety profiles of these covalent drugs should be closely monitored50.
4. Closing remarks
Drug discovery is an extremely complicated process involving scientists from various areas. In preclinical research, it involves medicinal chemistry, biology and pharmacology, drug metabolism and pharmacokinetics, formulation, and toxicology. Drug metabolism plays an important role in determining pharmacological and toxicological effects of a drug in human. From a drug metabolism perspective, the characteristics of an ideal drug molecule should include high oral bioavailability (for PO dosing) or a good aqueous solubility (for intravenous dosing), adequate elimination t1/2 for intended dosing intervals, balanced clearance among hepatic metabolism, biliary and renal excretion, metabolism by multiple enzymes, low potential to inhibit or induce drug metabolizing enzymes and transporters, low propensity for bioactivation, and comparable in vitro and plasma metabolite profiles in humans and toxicological species22. All contributions from drug metabolism should be considered in designing drugs for a targeted patient population with respect to safety and efficacy profiles. The common approaches in drug metabolism to improve PK/PD and safety of drug candidates include blocking a metabolic soft spot to lower total clearance, deuterium replacement to alter metabolism of a lead, prodrug for improved absorption and distribution, minimizing DDI and bioactivation potential, and selection of preclinical species for safety assessment. Taken together, drug metabolism as a discipline is a critical component in drug discovery and development, contributing significantly to the process of identifying a new drug molecule and bringing it to patients for meeting unmet medical needs.
Acknowledgements
The authors would like to thank Charles Thompson, Mark Cancilla and Christine Fandozzi for critical reading of the manuscript and invaluable suggestions.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.DiMasi J.A., Grabowski H.G., Hansen R.W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z., Zhu M., Tang W. Metabolite identification and profiling in drug design: current practice and future directions. Curr Pharm Des. 2009;15:2220–2235. doi: 10.2174/138161209788682460. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D., Luo D., Ding D., Lu C. Preclinical experimental models of drug metabolism and disposition in drug discovery and development. Acta Pharm Sin B. 2012;2:549–561. [Google Scholar]
- 4.Trunzer M., Faller B., Zimmerlin A. Metabolic soft spot identification and compound optimization in early discovery phases using MetaSite and LC–MS/MS validation. J Med Chem. 2009;52:329–335. doi: 10.1021/jm8008663. [DOI] [PubMed] [Google Scholar]
- 5.De Voss J.J., Sibbesen O., Zhang Z., De Montellano P.R. Substrate docking algorithms and prediction of the substrate specificity of cytochrome P450cam and its L244A mutant. J Am Chem Soc. 1997;24:5489–5498. [Google Scholar]
- 6.Zhang Z., Sibbesen O., Johnson R.A., De Montellano P.R. The substrate specificity of cytochrome P450cam. Bioorg Med Chem. 1998;6:1501–1508. doi: 10.1016/s0968-0896(98)00091-1. [DOI] [PubMed] [Google Scholar]
- 7.Silverman R.B., Holladay M.W. Lead discovery and lead modification. In: Silverman R.B., Holladay M.W., editors. The Organic Chemistry of Drug Design and Drug Action. 3rd ed. Academic Press; San Diego, CA: 2014. pp. 19–122. [Google Scholar]
- 8.Bouska J.J., Bell R.L., Goodfellow C.L., Stewart A.O., Brooks C.D., Carter G.W. Improving the in vivo duration of 5-lipoxygenase inhibitors: application of an in vitro glucuronosyltransferase assay. Drug Metab Dispos. 1997;25:1032–1038. [PubMed] [Google Scholar]
- 9.Braeckman R.A., Granneman G.R., Rubin P.R., Kesterson J.W. Pharmacokinetics and metabolism of the new 5-lipoxygenase inhibitor A-64077 after single oral administration in man. J Clin Pharmacol. 1989;29:A22. [Google Scholar]
- 10.Reid J.J. ABT-761 (Abbott) Curr Opin Investig Drugs. 2001;2:68–71. [PubMed] [Google Scholar]
- 11.Li C.M., Lu Y., Narayanan R., Miller D.D., Dalton J.T. Drug metabolism and pharmacokinetics of 4-substituted methoxybenzoyl-aryl-thiazoles. Drug Metab Dispos. 2010;38:2032–2039. doi: 10.1124/dmd.110.034348. [DOI] [PubMed] [Google Scholar]
- 12.Dierks E.A., Zhang Z., Johnson E., De Montellano P.R. The catalytic site of cytochrome P4504A11 (CYP4A11) and its L131F mutant. J Biol Chem. 1998;273:23055–23061. doi: 10.1074/jbc.273.36.23055. [DOI] [PubMed] [Google Scholar]
- 13.White R.E., McCarthy M.B. Active site mechanics of liver microsomal cytochrome P-450. Arch Biochem Biophys. 1986;246:19–32. doi: 10.1016/0003-9861(86)90445-5. [DOI] [PubMed] [Google Scholar]
- 14.Duquette P.H., Erickson R.R., Holtzman J.L. Role of substrate lipophilicity on the N-demethylation and type I binding of 3-O-alkylmorphine analogs. J Med Chem. 1983;26:1343–1348. doi: 10.1021/jm00364a002. [DOI] [PubMed] [Google Scholar]
- 15.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 16.Cupp-Vickery J.R., Poulos T.L. Structure of cytochrome P450eryF involved in erythromycin biosynthesis. Nat Struct Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 17.Lewis D.F., Dickins M. Baseline lipophilicity relationships in human cytochromes P450 associated with drug metabolism. Drug Metab Rev. 2003;35:1–18. doi: 10.1081/dmr-120018245. [DOI] [PubMed] [Google Scholar]
- 18.Lewis D.F., Jacobs M.N., Dickins M. Compound lipophilicity for substrate binding to human P450s in drug metabolism. Drug Discov Today. 2004;9:530–537. doi: 10.1016/S1359-6446(04)03115-0. [DOI] [PubMed] [Google Scholar]
- 19.Harada N., Miwa G.T., Walsh J.S., Lu A.Y. Kinetic isotope effects on cytochrome P-450-catalyzed oxidation reactions. Evidence for the irreversible formation of an activated oxygen intermediate of cytochrome P-448. J Biol Chem. 1984;259:3005–3010. [PubMed] [Google Scholar]
- 20.Wenthur C.J., Morrison R., Felts A.S. Discovery of (R)-(2–fluoro–4-((-4-methoxyphenyl)ethynyl)phenyl) (3–hydroxypiperidin-1-yl)methanone (ML337), an mGlu3 selective and CNS penetrant negative allosteric modulator (NAM) J Med Chem. 2013;56:3713–3718. doi: 10.1021/jm400439t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huntington Study Group Tetrabenazine as antichorea therapy in Huntington disease: a randomized controlled trial. Neurology. 2006;66:366–372. doi: 10.1212/01.wnl.0000198586.85250.13. [DOI] [PubMed] [Google Scholar]
- 22.FDA Clinical Pharmacology and Biopharmaceutics Review(s) [Cited 2018 Mar 1]. Available from: 〈https://www.accessdata.fda.gov/drugsatfda_docs/nda/2017/208082Orig1s000ClinPharmR.pdf〉.
- 23.Auspex Pharmaceuticals, Inc., Auspex Announces Positive Topline Results from Registration Trial of SD-809 for Chorea Associated with Huntington's Disease. [Cited 2018 Mar 1]. Available from: 〈https://globenewswire.com/news-release/2014/12/16/691862/10112630/en/Auspex-Announces-Positive-Topline-Results-From-Registration-Trial-of-SD-809-for-Chorea-Associated-With-Huntington-s-Disease.html〉.
- 24.Gallant J.E., Deresinski S. Tenofovir disoproxil fumarate. Clin Infect Dis. 2003;37:944–950. doi: 10.1086/378068. [DOI] [PubMed] [Google Scholar]
- 25.Chapman T.M., McGavin J.K., Noble S. Tenofovir disoproxil fumarate. Drugs. 2003;63:1597–1608. doi: 10.2165/00003495-200363150-00006. [DOI] [PubMed] [Google Scholar]
- 26.Van Gelder J., Deferme S., Naesens L. Intestinal absorption enhancement of the ester prodrug tenofovir disoproxil fumarate through modulation of the biochemical barrier by defined ester mixtures. Drug Metab Dispos. 2002;30:924–930. doi: 10.1124/dmd.30.8.924. [DOI] [PubMed] [Google Scholar]
- 27.Lo M.W., Goldberg M.R., McCrea J.B., Lu H., Furtek C.I., Bjornsson T.D. Pharmacokinetics of losartan, an angiotensin II receptor antagonist, and its active metabolite EXP3174 in humans. Clin Pharmacol Ther. 1995;58:641–649. doi: 10.1016/0009-9236(95)90020-9. [DOI] [PubMed] [Google Scholar]
- 28.Le M.T., Vanderheyden P.M., Szaszák M., Hunyady L., Kersemans V., Vauquelin G. Peptide and nonpeptide antagonist interaction with constitutively active human AT1 receptors. Biochem Pharmacol. 2003;65:1329–1338. doi: 10.1016/s0006-2952(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 29.Venkatakrishnan K., Greenblatt D.J., Von Moltke Ll, Schmider J., Harmatz J.S., Shader R.I. Five distinct human cytochromes mediate amitriptyline N-demethylation in vitro: dominance of CYP 2C19 and 3A4. J Clin Pharmacol. 1998;38:112–121. doi: 10.1002/j.1552-4604.1998.tb04399.x. [DOI] [PubMed] [Google Scholar]
- 30.Ulrich S., Läuter J. Comprehensive survey of the relationship between serum concentration and therapeutic effect of amitriptyline in depression. Clin Pharmacokinet. 2002;41:853–876. doi: 10.2165/00003088-200241110-00004. [DOI] [PubMed] [Google Scholar]
- 31.FDA Safety Testing of Drug Metabolites Guidance for Industry [Cited 2017 Jul 12]. Available from: 〈https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugs-gen/documents/document/ucm079266.pdf〉.
- 32.Liu L., Halladay J.S., Shin Y. Significant species difference in amide hydrolysis of GDC-0834, a novel potent and selective Bruton's tyrosine kinase inhibitor. Drug Metab Dispos. 2011;39:1840–1849. doi: 10.1124/dmd.111.040840. [DOI] [PubMed] [Google Scholar]
- 33.Smith G.F. Designing drugs to avoid toxicity. Prog Med Chem. 2011;50:1–47. doi: 10.1016/B978-0-12-381290-2.00001-X. [DOI] [PubMed] [Google Scholar]
- 34.Meyer U.A. Overview of enzymes of drug metabolism. J Pharmacokinet Biopharm. 1996;24:449–459. doi: 10.1007/BF02353473. [DOI] [PubMed] [Google Scholar]
- 35.Racoosin J.A., Roberson D.W., Pacanowski M.A., Nielsen D.R. New evidence about an old drug—risk with codeine after adenotonsillectomy. N Engl J Med. 2013;368:2155–2157. doi: 10.1056/NEJMp1302454. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson W.T., Formea C.M. Clinical perspective on the clinical pharmacogenetics implementation consortium updated 2014 guidelines for CYP2D6 and codeine. Clin Chem. 2015;61:319–321. doi: 10.1373/clinchem.2014.226795. [DOI] [PubMed] [Google Scholar]
- 37.Otton S.V., Crewe H.K., Lennard M.S., Tucker G.T., Woods H.F. Use of quinidine inhibition to define the role of the sparteine/debrisoquine cytochrome P450 in metoprolol oxidation by human liver microsomes. J Pharmacol Exp Ther. 1988;247:242–247. [PubMed] [Google Scholar]
- 38.Tatsumi R., Fujio M., Satoh H. Discovery of the α7 nicotinic acetylcholine receptor agonists. (R)-3′-(5-Chlorothiophen-2-yl)spiro-1-azabicyclo[2.2.2]octane-3,5′-[1′,3′]oxazolidin-2′-one as a novel, potent, selective, and orally bioavailable ligand. J Med Chem. 2005;48:2678–2686. doi: 10.1021/jm049188d. [DOI] [PubMed] [Google Scholar]
- 39.Evans D.C., Watt A.P., Nicoll-Griffith D.A., Baillie T.A. Drug–protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem Res Toxicol. 2004;17:3–16. doi: 10.1021/tx034170b. [DOI] [PubMed] [Google Scholar]
- 40.LoPachin R.M., DeCaprio A.P. Protein adduct formation as a molecular mechanism in neurotoxicity. Toxicol Sci. 2005;86:214–225. doi: 10.1093/toxsci/kfi197. [DOI] [PubMed] [Google Scholar]
- 41.Yagen B., Foureman G.L., Ben-Zvi Z. The metabolism and excretion of 14C-styrene oxide-glutathione adducts administered to the winter flounder, Pseudopleuronectes americanus, a marine teleost. Identification of the corresponding S-cysteine derivatives as major urinary metabolites. Drug Metab Dispos. 1984;12:389–395. [PubMed] [Google Scholar]
- 42.Vodicka P., Stetina R., Kumar R., Plna K., Hemminki K. 7-Alkylguanine adducts of styrene oxide determined by 32P-postlabelling in DNA and human embryonal lung fibroblasts (HEL) Carcinogenesis. 1996;17:801–808. doi: 10.1093/carcin/17.4.801. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z., Gan J. Protocols for assessment of in vitro and in vivo bioactivation potential of drug candidates. In: Zhang D., Zhu M., Humphreys W.G., editors. Drug Metabolism in Drug Design and Development: Basic Concepts And Practice. John Wiley & Sons; New Jersey: 2008. pp. 447–476. [Google Scholar]
- 44.Tang W., Zhang Z. Bioactivation and reactive metabolite assays. In: Lyubimov A.V., editor. Encyclopedia of Drug Metabolism and Interactions. John Wiley & Sons; New Jersey: 2012. pp. 627–656. [Google Scholar]
- 45.Inoue K., Shibata Y., Takahashi H., Ohe T., Chiba M., Ishii Y. A trapping method for semi-quantitative assessment of reactive metabolite formation using [35S]cysteine and [14C]cyanide. Drug Metab Pharmacokinet. 2009;24:245–254. doi: 10.2133/dmpk.24.245. [DOI] [PubMed] [Google Scholar]
- 46.Meneses-Lorente G., Sakatis M.Z., Schulz-Utermoehl T., De Nardi C., Watt A.P. A quantitative high-throughput trapping assay as a measurement of potential for bioactivation. Anal Biochem. 2006;351:266–272. doi: 10.1016/j.ab.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 47.Gan J., Harper T.W., Hsueh M.M., Qu Q., Humphreys W.G. Dansyl glutathione as a trapping agent for the quantitative estimation and identification of reactive metabolites. Chem Res Toxicol. 2005;18:896–903. doi: 10.1021/tx0496791. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z., Chen Q., Li Y. In vitro bioactivation of dihydrobenzoxathiin selective estrogen receptor modulators by cytochrome P450 3A4 in human liver microsomes: formation of reactive iminium and quinone type metabolites. Chem Res Toxicol. 2005;18:675–685. doi: 10.1021/tx0496789. [DOI] [PubMed] [Google Scholar]
- 49.Li Y., Doss G.A., Li Y., Chen Q., Tang W., Zhang Z. In vitro bioactivation of a selective estrogen receptor modulator (2S,3R)-(+)-3-(3-hydroxyphenyl)-2-[4-(2-pyrrolidin-1-ylethoxy)phenyl]-2,3-dihydro-1,4-benzoxathiin-6-ol (I) in liver microsomes: formation of adenine adducts. Chem Res Toxicol. 2012;25:2368–2377. doi: 10.1021/tx3002466. [DOI] [PubMed] [Google Scholar]
- 50.Singh J., Petter R.C., Baillie T.A., Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]