Abstract
Optical coherence tomography (OCT) has revolutionized diagnosis and prognosis of ophthalmic diseases by visualization and measurement of retinal layers. To speed up quantitative analysis of disease biomarkers, an increasing number of automatic segmentation algorithms have been proposed to estimate the boundary locations of retinal layers. While the performance of these algorithms has significantly improved in recent years, a critical question to ask is how far we are from a theoretical limit to OCT segmentation performance. In this paper, we present the Cramèr-Rao lower bounds (CRLBs) for the problem of OCT layer segmentation. In deriving the CRLBs, we address the important problem of defining statistical models that best represent the intensity distribution in each layer of the retina. Additionally, we calculate the bounds under an optimal affine bias, reflecting the use of prior knowledge in many segmentation algorithms. Experiments using in vivo images of human retina from a commercial spectral domain OCT system are presented, showing potential for improvement of automated segmentation accuracy. Our general mathematical model can be easily adapted for virtually any OCT system. Further, the statistical models of signal and noise developed in this paper can be utilized for future improvements to OCT image denoising, reconstruction, and many other applications.
I. Introduction
Optical coherence tomography (OCT) is a photonic imaging technology developed in the early 1990s for 3-dimensional imaging of reflectance [1]. OCT can be viewed as an optical analogue to ultrasound, in that it rejects multiple-scattered photons based on their arrival time. This is accomplished using an interferometer and either a broadband or wavelength-swept light source. OCT acquires a depth profile of reflectance, or A-scan, at a single location and then laterally samples the region of interest to produce a cross-sectional or volumetric image. A-scans in a single plane are frequently grouped together into 2-D images known as B-scans. Modern clinical OCT systems acquire the entire A-scan simultaneously using a technique called Fourier Domain OCT (FD-OCT). FD-OCT can be performed either in the spectral domain (SD) using a broadband source and a spectrometer, or using a spectrally swept source (SS). OCT has been adapted for a variety of applications including diagnosis and prognosis of cancer [2], [3], cardiovascular diseases [4], [5], and neurodegenerative diseases [6], [7].
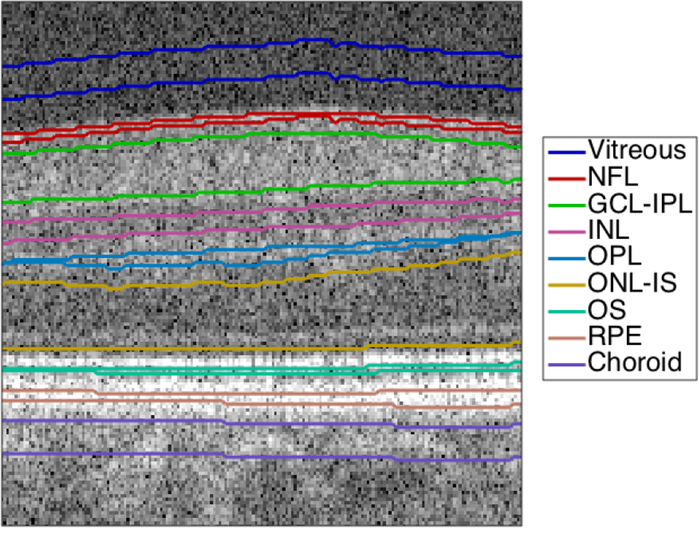
The highest-impact application of OCT is the diagnosis of ophthalmic diseases, including age-related macular degeneration (AMD) [8], [9], diabetes [10]–[12] and glaucoma [13], [14]. A critical task for retinal diagnostics is segmentation of the retina into its anatomical layers, which correspond to different functional regions. An OCT image of the retina with expert manual segmentation is shown in Figure 1.
Fig. 1.
Manually segmented 2-D OCT image (B-scan) of the retina, centered on the fovea. The layers shown are the vitreous, the nerve fiber layer (NFL), the ganglion cell layer (GCL) and inner plexiform layer (IPL) complex, the inner nuclear layer (INL), the outer plexiform layer (OPL), the outer nuclear layer (ONL) and photoreceptor inner segment (IS), the photoreceptor outer segment (OS), the retinal pigment epithelium (RPE), and the choroid.
Manual segmentation of large OCT datasets requires significant time and attention from expert graders. Therefore, several techniques have been developed to automatically segment the layers of retinal OCT images. These include boundary tracking/dynamic programming (Djikstra’s algorithm) techniques [15]–[17], pixel classification [18], active contours [19], [20], graph search [8], [21], kernel regression [10], and deep learning [22], [23]. These segmentation algorithms are benchmarked against expert manual graders, the current gold standard. While the performance of these algorithms has been significantly improved in the recent years, a critical question has not yet been addressed: “how far is OCT segmentation performance from its theoretical limit?” The answer to this question justifies further investment of time and financial resources to gain further segmentation accuracy.
A popular tool for determining the theoretically achievable accuracy of an estimator is the Cramèr-Rao Lower Bound (CRLB). The CRLB has been extensively used to quantify the performance limits of image processing tools such as image denoising [24], registration [25], particle displacement [26], frequency estimation [27], digital super-resolution [28], signal-to-noise ratio (SNR) estimation [29], stellar photometry [30], ballistic photon based object detection in scattering media [31], and spectral peak estimation [32]. The CRLB has also been used previously to evaluate the performance of several segmentation methods [33]–[36]. Two of these methods are of particular interest for the segmentation of retinal OCT images: determining the location of changes in a steplike signal [35] and the more general problem of 2D image segmentation [36].
The work of [35] provides a useful point of reference, because the layer boundaries in an OCT A-scan can be modeled as steplike changes. However, this method does not perfectly match the problem at hand, because the shape of a steplike change in OCT comes from the convolution of a step function with a system-specific point spread function (PSF), usually modeled as a Gaussian [37]. This convolution yields an error function-shaped curve, whereas [35] uses a logistic curve. Moreover, [35] assumes an un ed estimator, which does not reflect a priori models utilized in many modern image segmentation algorithms. The work of [36] provides an excellent general framework for fuzzy segmentation of 2D images and introduces an optimal affine bias model to determine the CRLB for biased operators. However, as we discuss below, the layered structure of the retina lends itself better to non-fuzzy segmentation.
An additional critical shortcoming that precludes direct application of the studies in [35] and [36] to OCT images is their simplistic modeling of noise as uniform additive white Gaussian (UAWG). While UAWG is a reasonable model for some consumer electronic imaging applications, this is not an accurate model of noise in OCT images. This is because at least two processes, detection noise and speckle, affect OCT images.
The detection noise results from many sources and has a lower limit determined by shot noise (i.e. bandlimited quantum noise). In FD-OCT, the detection noise can be approximated as an additive circular complex Gaussian produced by the detection process [37]. Speckle is an artifact of coherent imaging processes; it is deterministic based on the interaction of light with subvoxel features and is best modeled as multiplicative noise [38]. Henceforth in this paper, we will refer to the value of the observed signal (including noise) as intensity, in keeping with standard image processing terminology. This intensity is linearly proportional, but not equivalent, to the optical intensity of the backscattered light.
Various probability density functions (PDFs) of the intensity have been proposed to model speckle in OCT. The negative exponential distribution [39]–[42] and the gamma distribution [43]–[45] are among the more popular models. More recently, the negative exponential distribution and the K distribution have been compared for modeling OCT intensity in microsphere phantoms and skin [46], [47]. However, there has not been a full, comparative accounting of all the suggested distributions on a commercially available OCT system, especially for in vivo human retinal OCT images.
Multiple manuscripts in the literature have now specifically used noise distributions for OCT denoising [41], [42]. Both Ralston et al. [41] and Yin et al. [42] derive OCT denoising methods by modeling the noise as Gaussian in the log domain. However, the Gaussian distribution has not been empirically and statistically validated in the retina. Indeed, we will show below that it is possible to reject the Gaussian distribution with statistical significance. Therefore, we hope to provide superior and validated noise distributions both to calculate the CRLB and so that other techniques may be improved.
Notably, none of these studies take into account the effect of detection noise. Moreover, as the cellular composition varies between different retinal layers, a single distribution might not efficiently model the speckle pattern in each layer.
We note that a recent paper has attempted to statistically model noise in retinal OCT images with Normal-Laplace PDF, which is shown to have a lower chi-square error than Gaussian [48]. However, neither distribution is a good fit to their experimental data, as is evident in Fig. 4 of [48]. The denoising technique described in [48] is an example of the applications which we hope will benefit from using empirically validated noise distributions.
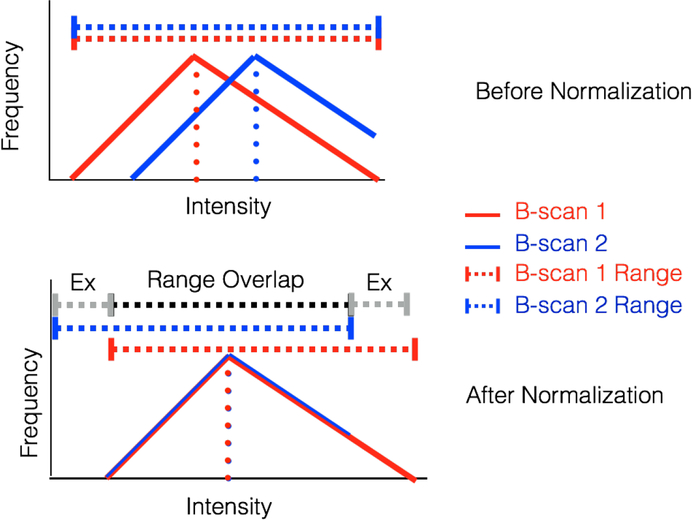
Fig. 4.
Schematic diagram of B-scan intensity normalization and range exclusion. The diagram shows the histograms of two layers before and after normalization (top and bottom, respectively). Before normalization, the histograms have the same shape and range but are separated; after normalization, the histograms overlap but the ranges have shifted. The range used in analysis is the intersection of all ranges, denoted in black; the ranges excluded from analysis are denoted in grey.
The novelty of our paper is as follows: 1) we developed physically derived and empirically validated layer-specific statistical models of the intensity in retinal OCT images; 2) using these models, we calculated the unbiased and biased CRLB for estimating the layer boundary locations in retinal OCT images. The impact of our paper goes beyond ophthalmic OCT applications as our proposed approach is general and can be adopted for modeling many other speckle dominated imaging scenarios.
The rest of the paper is organized as follows: Section II describes our calculation of the unbiased CRLB for OCT A-Scans and the biased CRLB for OCT B-scans. Section III describes our use of clinical OCT images to construct empirical statistical models of intensity. The results of both the modeling and the CRLB calculations are described in Section IV. Finally, in Section V, we discuss the results in context of the previous literature.
II. Boundary Segmentation CRLB for Layered Structures
Given a noisy signal g[k; h] dependent on a parameter vector h and position k, the CRLB states that the covariance of an unbiased estimator of h is bounded by:
| (1) |
where J is the Fisher information matrix. The elements of J are given by
| (2) |
where E[] is the expected value operator, p(g[k; h], k) is the probability of observing the intensity g[k; h] at position k and hi and hj are the ith and jth elements of h [49].
A. Calculation of Unbiased CRLB at Layer Boundaries in Single A-scan
In this section, we define our signal model for OCT A-scans. Then, based on that signal model, we derive a expression for the unbiased CRLB of a single-layer boundary position estimate. Taking into account both additive detection noise wadd[k; h] and speckle wsp[k; h], the observed intensity of an OCT A-scan can be modeled as
| (3) |
where s[k; h] is the noiseless intensity at depth pixel number k, fs is the axial spatial sampling frequency such that the optical depth is k/fs, s0 is a constant factor related to the system configuration, PSF(k/fs) is the OCT axial PSF, R(k/fs; h) is the depth reflectivity profile of the sample, and ⊗ is the convolution operator [37].
Each g[k; h] is a random variable with PDF p(g[k; h]) determined by s[k; h] and the PDFs of the additive and multiplicative noise processes. We rewrite this noise as purely additive to enhance mathematical tractability. This is accomplished by expressing g[k; h] as a fixed mean intensity plus a random noise variable w[k; h] with a zero expected value (ZEV) PDF .
To apply the CRLB to OCT segmentation, we first consider a model dependent only on a single parameter, the position of the ith layer boundary zi. We model the noiseless signal in a single A-scan near the ith boundary as a Heaviside step function H(x) at position zi convolved with a Gaussian OCT PSF [37]:
| (4) |
| (5) |
where A is a system-dependent scaling factor, Ii and Ii+1 are the average intensities of the layers above and below the boundary, σPSF is the standard deviation of the axial PSF, erf() is the error function, and erfc() is the complementary error function. Additionally, because most OCT images are viewed in the log domain to enhance the visibility of dimmer layers [50], we carry out these calculations in the log domain.
We make two physically and anatomically rational assumptions about noise in retinal layers. First, we note that the cellular and sub-cellular structures in each layer differ in shape and size. Thus, each layer has unique speckle characteristics and the noise is not stationary for the whole image. Moreover, because of optical blurring, the noise smoothly transitions from one layer’s noise PDF to the other. Therefore, we treat the noise PDF near the ith layer boundary wi[k; zi] as
| (6) |
where ni and ni+1 are the noise in layer i and i + 1, respectively, and each ni is drawn from a ZEV PDF qi(ni).
The observed signal near the ith layer boundary gi[k; zi] is therefore:
| (7) |
The PDF of gi[k; zi] is then:
| (8) |
To calculate the CRLB, we use the scalar version of Eq. (2) for a single variable h [49]:
| (9) |
We then calculate the Fisher Information for each layer boundary:
| (10) |
where . For many non-Gaussian noise distributions, the above integral cannot be evaluated analytically. In such cases, we numerically calculated the integral in Mathematica.
B. Biased CRLBs for Segmentation of B-scans
Most OCT layer segmentation algorithms achieve improved performance by utilizing a priori information and thus are categorized as biased estimators. These include the popular assumption that the layer boundaries are smooth, which can be exploited by analyzing a whole B-scan rather than an individual A-scan [15], [16], [51]. Biased estimators can lead to improved performance, defined as reduced mean square error (MSE) [52]. Thus, to further broaden the applicability of our bounds, we extend our CRLB findings to the case of biased estimators.
Our approach follows Peng and Varshney’s [36] use of an optimal affine bias model to determine the CRLB for a biased fuzzy segmentation estimator. The optimal affine bias CRLB puts a bound on the covariance of a biased estimate ĥ of a parameter vector h. If the bias ψ = E[ĥ] − h is affine, i.e. ψ = Kh + u, then it can be shown that the covariance of ĥ is bounded by [36]
| (11) |
We justify the use of an affine bias model for a whole B-scan in Appendix A. When segmenting a whole B-scan, the parameter vector h contains all eight layer boundary locations in each constitutent A-scan. Consequently, the diagonal elements of J−1 are the unbiased CRLBs for each layer boundary, repeated for every A-scan. The diagonal of CRLBBiased gives the lower bound on the variance of the layer boundary positions across the B-scan. We average the bound across the B-scan for each boundary to obtain an average bound for a biased estimator, which is a useful comparison to the position-independent bound for an unbiased estimator.
We estimate Cov [h] using the same set of expert segmented B-scans used for generating the empirical distributions (see Section III-A). We use a statistical bootstrapping technique as per Peng and Varshney [36] to generate more accurate estimates of the covariance. Starting with 100 B-scans as sample data, we picked 100 B-scans with replacement and calculated the covariance of the boundary locations between the B-scans. We repeated this procedure 100 times and averaged the covariance matrices from each set of generated data to give an accurate estimate of the true covariance matrix of boundary positions in B-scans.
III. Empirical Model of OCT Intensity
In this section, we describe our method to construct an empirical, layer-specific model of the signal and noise in retinal OCT images. First, we discuss the acquisition and segmentation of the data. Then, we discuss the data processing performed to generate accurate representations of intensity across the entire population. Next, we discuss the different distributions and parameter selection process for modeling the retinal layers. Finally, we discuss how to choose the best model and estimate the mean intensity and ZEV noise PDF for each layer. The steps of this process are shown in Figure 2.
Fig. 2.
Schematic indicating the process by which we construct empirical, layer-specific intensity models.
A. Collection and Processing of OCT Data
While our technique to obtain layer-specific intensity models is general, for this study we utilized an SD-OCT system from Bioptigen Inc. (Research Triangle Park, NC). We chose this system because it is commercially available, performs minimal image processing, and offers ready access to raw data.
We acquired volumetric scans (6.7 × 6.7 mm) from 10 normal adult subjects under an IRB approved protocol. The Bioptigen SD-OCT system had a Gaussian axial PSF with a full-width at half-maximum (FWHM) resolution of 4.6 μm (in tissue), an axial pixel sample spacing of 3.23 μm, and a total axial depth range of 3.3 mm. The illumination from the SD-OCT had a central wavelength of 830 nm with a FWHM bandwidth of 50 nm. The volumetric scans had lateral and azimuthal pixel sampling spacings of 6.7 μm and 67 μm (1000 A-scans scans per B-scan, 100 B-scans per volume), respectively [15]. Each B-scan was cropped laterally to the central 800 A-scans, for a final image size of 800 × 1024. The intensity of each voxel in the volume was represented by a 16-bit integer normalized from 0 to 255.
A grader semi-automatically segmented 10 evenly spaced B-scans from each subject by using DOCTRAP software [9]. In some layers, blood vessels and specular reflections led to intensity distortions above and below the vessel. Therefore, a separate grader manually removed A-scans containing large vessels or specular reflections. We excluded these A-scans from the subsequent analysis. An example of an excluded vessel is shown in the green box of Figure 3.
Fig. 3.
Raw OCT B-scan showing a retinal ghost/autocorrelation artifact (red box) and intensity distortions from blood vessels (green box).
Additionally, particularly bright images can yield autocorrelation artifacts in OCT images [37]. The result of this is a ghost image of the retina with a prominent peak corresponding to the autocorrelation between the NFL and the RPE, at a depth determined by the difference between the RPE and NFL depths. Manual examination of the OCT images indicated that the autocorrelation artifact only occupies the top 0.55 mm of the 3.3 mm-deep scan. In most OCT images, the OCT photographer attempts to ensure that the NFL does not overlap with the autocorrelation artifact. Therefore, to represent the typical OCT image, we ignored any pixels in the top 0.55 mm of the B-scans. An example of an autocorrelation artifact is shown in the red box of Figure 3.
B. Normalization of B-scans
Differences in beam placement, ocular or corneal clarity, or other factors may change the recorded intensity from the layers of the retina by a constant multiplicative factor between B-scans from a single subject, as well as between subjects. Additionally, different amounts of absorption or scattering in OCT images can lead to a secondary, layer-specific multiplicative factor. In the log domain, this appears as a constant increase or decrease of all pixel intensities in a particular B-scan or layer, as illustrated schematically in Figure 4.
To compensate for this imaging artifact, we calculated the average intensity of each layer across all B-scans. We then added a constant value to the same layer region of each B-scan so that its average intensity value is equal to the global average intensity for that layer. As shown in Figure 4, this procedure results in a different gray-scale range for each post-normalization layer. Therefore, when considering the combined intensity histograms of multiple layers (see Section III-C), we ignored post-normalization gray-scale values that were not within the range of all other layers.
C. Generation of OCT Intensity Histograms
For each B-scan, we utilized the segmented retinal layer boundaries to isolate individual layers. First, we isolated each layer, defined as all pixels at or between the segmented boundaries. Then, to minimize the effect of possible segmentation errors, we excluded pixels in a 3 pixel-wide region next to the borders, as shown in Figure 5. Noting that the choroid lower boundary is not visible in all B-scans, we set it at 13 pixels below the RPE/choroid segmentation line. Since the vitreous has no upper boundary, we represented it as the region 10 to 20 pixels above the vitreous/NFL segmentation line.
Fig. 5.
Diagram of the boundaries used for each layer. The data for each layer is taken from between the lines with colors corresponding to that layer in the legend. These lines are calculated from segmentation lines with an offset to account for possible segmentation inaccuracies.
We then created a histogram from the intensity values for each layer in each B-scan with one gray-scale value wide bins. Finally, we added the histograms for each layer from all B-scans across all subjects to produce global layer-specific histograms.
D. Mathematical Modeling of Intensity Histograms
We considered three categories of mathematical models to represent the probability distribution of the intensity histograms: distributions based on theoretical models of speckle and additive noise in OCT images, mixture models based on the theoretical distributions, and Gaussian related distributions.
1). Theoretical Models:
In this section, based on first principles, we derive progressively more complex models of noise. In Section IV, we will experimentally evaluate the practical suitability of these complex models versus their more simplistic counter parts for representing OCT signal intensity.
First, we consider the speckle in OCT images. Although in principle speckle is a deterministic process, it is best modeled using stochastic tools because it is the result of the coherent addition of myriad sub-voxel scatterers. The intensity of a single, fully-developed speckle pattern Isp has a negative exponential distribution [38]:
| (12) |
where σS is the expected value of the distribution. If the layer varies such that σS is distributed according to a Gamma distribution with shape parameter α and expected value μ, the intensity for the whole layer will have a special form of the K distribution [47] with PDF:
| (13) |
where Kα (x) is the modified Bessel function of the second kind with order α. The presence of different polarizations, frequencies, and scatter angles can lead to multiple uncorrelated speckle realizations being added incoherently in the OCT images. We note that if m uncorrelated speckle patterns are present, the PDF is a full K distribution with PDF:
| (14) |
Second, we account for the additive noise. The additive noise takes the form of a complex Gaussian added to a deterministic phasor with intensity Isp [37]. The PDF of the signal’s intensity is therefore a modified Rice distribution:
| (15) |
where σR is a scale parameter and I0(x) is the modified Bessel function of the first kind with order zero [37]. Because we model Isp as a random variable, the PDF of the total intensity I will be a compound distribution between the modified Rice distribution pModRice(I|σR, Isp) and the PDF of the multiplicative speckle noise psp(Isp):
| (16) |
If psp(Isp) is a negative exponential distribution, p(I) is also a negative exponential distribution with scale parameter σC = σS + σR. If psp(Isp) is a K distribution (either the special case or full K distribution), the compound distribution integral cannot be evaluated analytically and must be examined with numerical methods as described in Section III-E. Due to computational complexity the only compound method that we could feasibly evaluate was the special K-Rice compound.
2). Mixture Models:
In addition to the purely theoretical distributions, we considered the biologically plausible case that there are two well-defined populations of voxel intensities within a layer (e.g. two dominant types of cellular substructures). To account for these cases, we used a mixture of two negative exponential distributions with PDF:
| (17) |
where B is the proportionality constant. Similarly, the PDF for a mixture of two special-case K distributions is:
| (18) |
and finally, the PDF for a mixture of two full K distributions is:
| (19) |
3). Gaussian-Related Models:
We also considered two models related to the Gaussian noise because of its explicit or implicit prevalence in the OCT literature. First of these models is the lognormal distribution, used in papers that assume the log domain OCT images are corrupted by Gaussian noise [53], [54]. The PDF of the lognormal distribution is:
| (20) |
where μ is the location parameter and σ is the scale parameter.
Second, we considered multiplicative Gaussian noise, which assumes a truncated Gaussian distribution in the linear, rather than log, domain. Note that the multiplicative noise cannot take negative values and therefore the Gaussian distribution must be truncated at 0. The resulting normalized PDF is:
| (21) |
where μ is the location parameter, σ is the scale parameter, and Ф(z) is the cumulative distribution function (CDF) of the standard normal distribution.
E. Signal Modeling and Parameter Estimation
Because OCT images are commonly viewed in the log domain to enhance visibility of the dimmer layers [50], we transform each of the above distributions to the log domain. The log domain intensity of a pixel with intensity I is IL = A log10(I), where A is a deterministic scaling factor determined by the OCT system’s software. Therefore, the log domain PDF, given linear domain PDF P(I), is given by the chain rule:
| (22) |
For all distributions except the Rice-K compound, we determined the best parameter values by fitting the log domain distributions to the data based on a weighted nonlinear least squares criterion. We used a two-step procedure for parameter estimation. In the first step, we gave the data at bin k with count number M[k] weight 1/M[k], as per [55]. The fit of the jth distribution to the ith layer’s data with these weights was aij[k], our initial estimate. This estimate might be affected by the over-emphasis of the bins with fewer counts [56]. Therefore, following [56], we refit the distribution giving the data at bin k weight 1/aij[k]. The fit of the jth distribution to the ith layer’s data with these new weights was bij[k], our second and final estimate.
We calculated the goodness of fit for each model using Pearson’s χ2 test, which provides a quantitative p-value. First, we calculated the test statistic:
| (23) |
We then calculated a p-value from the value:
| (24) |
where χ2(x, v) is the PDF of the χ2 distribution with v degrees of freedom (DOF) at value x, Nbins is the number of bins where bij[k] ≥ 1 and the gray-scale ranges of all B-scans overlap, and dj is the number of parameters in the jth distribution.
The Rice-special K compound distribution cannot be evaluated analytically and thus we numerically calculated the PDF at every point in a 200 × 200 × 200 grid in parameter space, for parameters α, μ, and σR. For every Rice-special K PDF/layer combination, we then calculated a χ2 test statistic as above. For each layer, we examined the 100 PDFs with the lowest χ2s and calculated the mean τ and range ρ of the corresponding parameters. We then performed a refined search for each layer on a 100 × 100 × 100 point grid, where each parameter spanned the range τ ± max(ρ, τ × 0.02)/2. Again, we calculated the χ2 test statistics. We considered the distribution with minimum χ2 to be the best Rice-special K compound distribution for a given layer.
To determine the best distribution to use for each layer, we calculated the Aikake Information Criterion (AIC) value for each distribution/layer combination. The AIC value is given by
| (25) |
where L is the log-likelihood value of the data given the distribution and d is the number of parameters of the distribution [57]. In this case, we approximated each the counts in each bin as an independent Poisson process and calculated the log-likelihood L of the model f[k] on the data M[k] as:
| (26) |
For each layer, we chose the model with the lowest AIC value. This is the most parsimonious model, i.e. the model that describes the data best without overfitting [57]. We then used the chosen intensity distribution p̂i(I) to calculate an expected value,
| (27) |
and a ZEV noise distribution,
| (28) |
IV. Results
A. Empirical Model
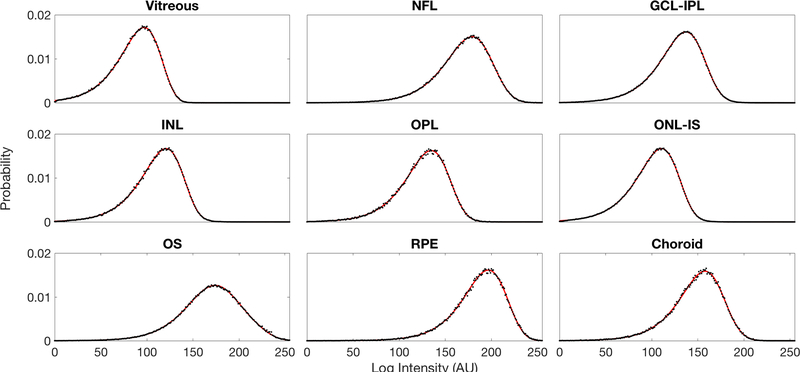
We used the AIC model selection process as discussed in Section III-E to attain the most parsimonious distribution representing each retinal layer intensity. The AIC values are shown in Table I. The summary of resulting AIC-chosen intensity distributions and parameters for each layer are shown in Table II. Among the nine models considered for representing retinal layers, three K-family models were found to be most suitable for our data: the full K mixture model for the vitreous, GCL-IPL, INL, OPL, ONL-IS, and OS; the special case K mixture model for the NFL, and Choroid; and the special case K model for RPE. Each of the chosen models has a χ2 p-value of at least 0.34, indicating that these models should not be rejected on the basis of goodness of fit. The intensity distributions of each layer from our experimental data are plotted along with the best theoretical models in Figure 6. For each layer based on the corresponding best model, we calculated the average intensity Ii and ZEV noise distributions qi(n) shown in Figure 7.
Table I.
Table indicating AIC values of each distribution/layer combination. A smaller AIC Value indicates a more parsimonious model.
| Layer | Neg. Exp. | Neg. Exp. Mix. | Spec. K | Spec. K Mix. | K | KMix | Gaussian | Lognormal | Rice-K |
|---|---|---|---|---|---|---|---|---|---|
| Equation Reference | 12 | 17 | 13 | 18 | 14 | 19 | 20 | 21 | 16 |
| Vitreous | 2152 | 1669 | 1807 | 1579 | 1798 | 1578 | 68423 | 5148 | 1875 |
| NFL | 16315 | 2061 | 1856 | 1844 | 1858 | 1848 | 45295 | 28505 | 11420 |
| GCL-IPL | 31621 | 3230 | 3900 | 1972 | 3484 | 1964 | 91303 | 39925 | 9109 |
| INL | 5894 | 1805 | 2246 | 1634 | 2148 | 1634 | 43465 | 9808 | 2966 |
| OPL | 4164 | 1662 | 1663 | 1532 | 1625 | 1531 | 17457 | 5857 | 1885 |
| ONL-IS | 21416 | 2726 | 4871 | 1908 | 4405 | 1889 | 82173 | 43559 | 9470 |
| OS | 62798 | 2749 | 3790 | 1692 | 3429 | 1685 | 7149 | 24294 | 5508 |
| RPE | 1801 | 1453 | 1453 | 1458 | 1452 | 1459 | 9164 | 3980 | 1675 |
| Choroid | 5898 | 1679 | 1683 | 1603 | 1653 | 1607 | 18631 | 8322 | 3338 |
Table II.
Per-layer distributions and parameters for the empirical model. Each distribution was chosen by an AIC selection process. The high χ2 p-values indicate that the fits should not be rejected.
| Layer | Distribution Name | B | μ1 | μ2 | α1 | α2 | m1 | m2 | X2 | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Vitreous | Spec. K Mix. | 0.999215 | 96.58 | 509.9 | 50 | 31.66 | – | – | 0.9945 | |
| NFL | Spec. K Mix. | 0.86722 | 5974.7 | 4004.1 | 3.149 | 49.998 | – | – | 0.68358 | |
| GCL-IPL | Full K Mix. | 0.062305 | 1676.5 | 677.32 | 1.8277 | 8.3798 | 1.8313 | 1.0009 | 0.6193 | |
| INL | Spec. K Mix. | 0.9296 | 300 | 564.85 | 22.254 | 2.7856 | – | – | 0.97519 | |
| OPL | Full K Mix | 0.12893 | 809.09 | 615.85 | 1.3218 | 9.3597 | 1.3283 | 1.0317 | 0.67172 | |
| ONL-IS | Full K Mix. | 0.87569 | 193.9 | 228.85 | 14.084 | 1.0442 | 1.0855 | 1.0425 | 0.81396 | |
| OS | Full K Mix. | 0.66416 | 9887.9 | 3328.1 | 1.0645 | 1.9204 | 1.0704 | 1.7793 | 0.6062 | |
| RPE | Full K | – | 11747 | – | 6.7792 | – | 1.014 | – | 0.77231 | |
| Choroid | Spec. K Mix. | 0.70409 | 1613.8 | 2854.7 | 9.6091 | 5.799 | – | – | 0.68537 |
Fig. 6.
Experimental PDFs (black dots) compared to the proposed best theoretical model PDFs (red line) for each layer. The data for the experimental PDFs comes from intensity-normalized data.
Fig. 7.
Expected value (left) and ZEV additive noise PDFs (right) derived from the chosen intensity distributions for each of the nine retinal layers.
B. CRLBs for Layer Boundary Locations
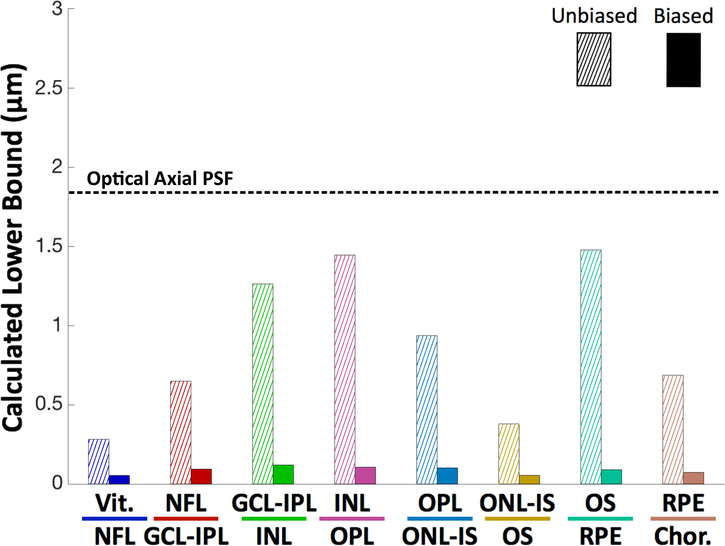
Following the methodology of Section II, we calculated unbiased and biased CRLBs for each of the eight retinal boundary positions. The unbiased estimator for the boundary location in a single A-scan is shown by the hatched bars of Figure 8.
Fig. 8.
CRLBs for the locations of layer boundaries in the unbiased single A-scan case (hatched) and laterally averaged biased single B-scan case (solid). For the sake of comparison, the dashed horizontal line indicates the accuracy of automatic retinal layer segmentation algorithms for normal subjects. The optical resolution of the OCT system is indicated by the dotted line.
We compared the unbiased to the optical axial resolution of the Bioptigen OCT system, defined as the standard deviation of the axial PSF. We also tested the performance of a state-of-the-art deep learning based OCT layer segmentation algorithm [23] and the publicly available OCTExplorer software (downloaded at: https://www.iibi.uiowa.edu/content/iowa-reference-algorithms-human-and-murine-oct-retinal-layer-analysis-and-display [58]–[60], on this dataset and compared their results with the manually corrected segmentations. The layer-specific RMS error values for the Deep Learning and OCTExplorer, respectively, are as follows: Vitreous/NFL: 2.41 μm and 3.39 μm; NFL/GCL-IPL: 7.16 μm and 6.47 μm; GCL-IPL/INL: 5.21 μm and 5.38 μm; INL/OPL: 5.05 μm and 5.29 μm; OPL/ONL-IS: 4.36 μm and 7.86 μm; ONL-IS/OS: 2.46 μm and 2.93 μm; OS/RPE: 3.23 μm and 4.92 μm; RPE/Choroid: 4.96 μm and 4.45 μm.
The biased minimum covariance matrix for the 8 boundary positions at 800 A-scan positions in each B-scan is shown in Figure 9, and the position-dependent are shown in Figure 10. For the sake of comparison, we also show the biased averaged across the 800 A-scans in each B-scan as the solid bars Figure 8.
Fig. 9.
Position-dependent lower bounds on the covariance of each layer’s location. The axes are arranged so that the first 800 pixels correspond to the Vitreous/NFL boundary at positions 1–800, the second 800 pixels correspond to the NFL/GCL-IPL boundary at positions 1–800, and so on.
Fig. 10.
Position-dependent for a biased estimator of each layer boundary’s location at each A-scan of a B-scan.
V. Discussion
The empirical K distribution family based models of the intensity in each layer of the retina derived in this work are logical extensions to existing knowledge about the structure of the eye and the nature of speckle and detection noise. Note that the the special K distribution used was previously suggested for modeling OCT intensity [47]. However, we showed that the special K distribution used by [47] is only the best model for one out of the nine layers studied in this work. The other layers all conformed better to two other K distribution mixtures, indicating that the complex nature of retinal tissue cannot be fully described with a single distribution. Additionally, six of the layers conformed best to the non-special K distribution mixture, which has two more parameters than the special K distribution mixture. This may indicate additional complexity in the layer structure or inter-subject variance.
Although R2 is popular for evaluating goodness of fit [46], [47], it is not quantitatively meaningful for nonlinear models such as these [61]. Therefore, we used the χ2 goodness of fit metric and its corresponding p-value, which gives a true quantitative metric for goodness of fit.
The results of the unbiased CRLB are intuitive . The bounds are on the order of microns; the higher-contrast layers (for example, Vitreous/NFL) have bounds lower than the resolution of the system (as defined by the FWHM of the PSF), and the lower-contrast layers have bounds of less than two times the resolution.
The biased follow the same trend but were markedly smaller. The smallest averaged biased bound, at 0.06 μm, was far below subcellular resolution. The largest biased averaged bound, at 0.14 μm, was still quite small and over an order of magnitude below the resolution of the system. This can be attributed to the smoothness of the retinal surfaces; the retina’s shape is so highly correlated that many A-scans effectively act as independent measurements for adjacent A-scans. The averaged biased bounds are also more than order of magnitude smaller than the the accuracy of the published segmentation techniques as per Section IV-B. This indicates that there is potential to improve the performance of retinal layer segmentation algorithms.
An exciting novel development in retinal layer segmentation is the utilization of deep learning based algorithms [22], [23]. While it is hard to model the prior learned in these black-box algorithms, smoothness discussed in this paper is expected to be their major component. Note that no deep learning paper to date has provided a significantly more accurate segmentation of normal tissue than classic segmentation techniques. The utility of deep learning methods has been mainly on improving the performance of diseased eyes in segmenting hard to segment features, at an accuracy similar to normal tissue. Of course, the method for CRLB estimation presented in this paper is general and the prior utilized in this work can be easily replaced by alternate priors, including those learned via deep learning. Future studies on this topic are warranted to better characterize the limits on segmentation accuracy of deep learning techniques.
The position-dependent biased bounds are space variant. The bounds are larger at the edges of the image due to two sources. First, OCT images are blurrier at the edges, leading to increased variance in manual segmentation [62]. Second, fewer A-scans surround each individual A-scan in the image boundaries and thus negatively affecting the utility of smoothness prior.
We constructed the covariance data for our biased CRLBs from the manual segmentation of expert graders. Although manual segmentation is the gold standard, it is not perfect due to the noise and limited resolution of OCT images. The results of the biased CRLB must therefore be regarded as the lower bound of algorithmic segmentation against this particular set of human graders, not against the true anatomical structure of the retina. Indeed, it is practically impossible to discuss the theoretical bounds on all different OCT machines, retinal diseases, or alternative manual segmentation results in one paper. However, since the proposed methodology is general, one can attain bounds for different imaging scenarios by simply replacing the test dataset.
Noting that in diseased eyes layer boundary delineation is often less clear than the normal tissue, we expect that the lower bounds derived here for normal tissue to be valid for some (but not all) diseased tissues. To achieve tighter (and less optimistic) bounds, we are also pursuing the modeling and segmentation bounds of non-healthy retinas. This requires new datasets and segmentations, but with those segmentations completed, diseased tissue of the retina can be effectively modeled with the approach outlined in this manuscript. To ease adaptation of the proposed method to diseased eyes, we have released the source code of our paper so that the readers can incorporate our findings with alternative datasets. We have made the open-source code for our paper freely available online at http://people.duke.edu/~sf59/Dubose TMI 2018.htm to allow other researchers to test and modify the algorithm for their specific applications and dataset.
In conclusion, we present retina layer-specific statistical intensity models of the OCT images and biased and unbiased CRLBs for segmentation of those layers. The intensity in the OCT images was best modeled by the K distribution family. For our dataset, the unbiased were on the order of microns, and the biased were on the order of hundreds of microns. These results suggest that there may be room to improve the accuracy of OCT segmentation techniques. In our future work, we will extend our work to include diseased retina, other OCT imaging systems and 3-D segmentation algorithms.
Acknowledgement
The authors would like to thank Dr. Cynthia Toth, Du Tran-Viet, and the Duke DARSI Lab for their assistance in obtaining the images used in this manuscript. Dr. Farsiu acknowledges support from NIH R01-EY022691 and NIH P30-EY5722. Mr. Cunefares effort is supported in part by NIH T32-EB001040. Dr. Izatt acknowledges support from NIH R01-EY23039.
Appendix A Justification of the Affine Bias Model
As stated in Section II-B, we used an affine model to approximate the bias introduced by segmentation algorithms.
In this appendix, we justify the optimum affine bias model for layer segmentation and demonstrate empirically how it accurately reflects the bias of most layer segmentation algorithms towards a smooth layer boundary.
Each B-scan is composed of A-scans of the form g[k] = s[k] + w[k], where s[k] is the true signal value and w[k] is a noise value. If we consider each boundary separately as in [15], a signal model for a single boundary in a single A-scan is a Heaviside step function at true boundary locations z convolved with a Gaussian as per Section II. The derivative of this signal is a Gaussian centered at the boundary location. A reasonable unbiased estimator for the boundary locations ẑ in the A-scan can thus be expressed as the centroid of the derivative of the A-scan:
| (29) |
where C is a centroiding matrix and D is the derivative matrix. Given several adjacent A-scans, we can produce a biased estimate ẑ’ by a weighted averaging with the estimates of N adjacent A-scans
| (30) |
where W is a weighting matrix that might take into account, for example, distance and radiometric similarity of the noisy A-scans. The expected value of the estimate ẑ’ is
| (31) |
The bias is then
| (32) |
which conforms to the bias model ψ = Kh + u.
Contributor Information
Theodore B. DuBose, IEEE.
David Cunefare, IEEE.
Elijah Cole, IEEE.
Peyman Milanfar, IEEE.
Joseph A. Izatt, IEEE.
Sina Farsiu, IEEE.
References
- [1].Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, and Fujimoto JG, “Optical Coherence Tomography,” Science, vol. 254, no. 5035, pp. 1178–1181, November 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tearney GJ, Brezinski ME, Bouma BE, Boppart SA, Pitris C, Southern JF, and Fujimoto JG, “In vivo endoscopic optical biopsy with optical coherence tomography,” Science, vol. 276, no. 5321, pp. 2037–2039, June 1997. [DOI] [PubMed] [Google Scholar]
- [3].Sivak MV, Kobayashi K, Izatt JA, Rollins AM, Ung-Runyawee R, Chak A, Wong RC, Isenberg GA, and Willis J, “High-resolution endoscopic imaging of the GI tract using optical coherence tomography,” Gastrointestinal Endoscopy, vol. 51, no. 4, pp. 474–479, April 2000. [DOI] [PubMed] [Google Scholar]
- [4].Yabushita H, Bouma BE, Houser SL, Aretz HT, Jang I-K, Schlendorf KH, Kauffman CR, Shishkov M, Kang D-H, Halpern EF, and Tearney GJ, “Characterization of human atherosclerosis by optical coherence tomography,” Circulation, vol. 106, no. 13, pp. 1640–1645, September 2002. [DOI] [PubMed] [Google Scholar]
- [5].Jang I-K, Bouma BE, Kang D-H, Park S-J, Park S-W, Seung K-B, Choi K-B, Shishkov M, Schlendorf K, Pomerantsev E, Houser SL, Aretz H, and Tearney GJ, “Visualization of coronary atherosclerotic plaques in patients using optical coherence tomography: comparison with intravascular ultrasound,” J. Am. Coll. Card, vol. 39, no. 4, pp. 604–609, February 2002. [DOI] [PubMed] [Google Scholar]
- [6].Frohman EM, Fujimoto JG, Frohman TC, Calabresi PA, Cutter G, and Balcer LJ, “Optical coherence tomography: a window into the mechanisms of multiple sclerosis,” Nature Clin. Pract. Neuro, vol. 4, no. 12, pp. 664–675, December 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zaveri M, Conger A, Salter A, Frohman T, Galetta S, Markowitz C, Jacobs D, Cutter G, Ying G, Maguire M, Calabresi P, Balcer L, and EM F, “Retinal imaging by laser polarimetry and optical coherence tomography evidence of axonal degeneration in multiple sclerosis,” Arch. Neuro, vol. 65, no. 7, pp. 924–928, July 2008. [DOI] [PubMed] [Google Scholar]
- [8].Garvin MK, Abràmoff MD, Kardon R, Russell SR, Wu X, and Sonka M, “Intraretinal layer segmentation of macular optical coherence tomography images using optimal 3-d graph search,” IEEE Trans. Med. Imag, vol. 27, no. 10, pp. 1495–1505, April 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Farsiu S, Chiu SJ, O’Connell RV, Folgar FA, Yuan E, Izatt JA, and Toth CA, “Quantitative classification of eyes with and without intermediate age-related macular degeneration using optical coherence tomography,” Ophthalmology, vol. 121, no. 1, pp. 162–172, January 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chiu SJ, Allingham MJ, Mettu PS, Cousins SW, Izatt JA, and Farsiu S, “Kernel regression based segmentation of optical coherence tomography images with diabetic macular edema,” Biomed. Opt. Expr, vol. 6, no. 4, pp. 1172–1194, April 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hee MR, Puliafito CA, Wong C, Duker JS, Reichel E, Rutledge B, Schuman JS, Swanson EA, and Fujimoto JG, “Quantitative assessment of macular edema with optical coherence tomography,” Arch. Ophth, vol. 113, no. 8, pp. 1019–1029, August 1995. [DOI] [PubMed] [Google Scholar]
- [12].DeBuc DC and Somfai GM, “Early detection of retinal thickness changes in diabetes using optical coherence tomography,” Med. Sci. Monit, vol. 16, no. 3, pp. 15–21, February 2010. [PubMed] [Google Scholar]
- [13].Schuman JS, Hee MR, Puliafito CA, Wong C, Pedut-Kloizman T, Lin CP, Hertzmark E, Izatt JA, Swanson EA, and Fujimoto JG, “Quantification of nerve fiber layer thickness in normal and glaucomatous eyes using optical coherence tomography: a pilot study,” Arch. Ophth, vol. 113, no. 5, pp. 586–596, May 1995. [DOI] [PubMed] [Google Scholar]
- [14].Hood DC, Raza AS, de Moraes CGV, Johnson CA, Liebmann JM, and Ritch R, “The nature of macular damage in glaucoma as revealed by averaging optical coherence tomography data,” Transl. Vis. Sci & Tech, vol. 1, no. 1, pp. 3–3, May 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chiu SJ, Li XT, Nicholas P, Toth CA, Izatt JA, and Farsiu S, “Automatic segmentation of seven retinal layers in sdoct images congruent with expert manual segmentation,” Opt. Expr, vol. 18, no. 18, pp. 19 413–19 428, August 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mishra A, Wong A, Bizheva K, and Clausi DA, “Intra-retinal layer segmentation in optical coherence tomography images,” Opt. Expr, vol. 17, no. 26, pp. 23 719–23 728, December 2009. [DOI] [PubMed] [Google Scholar]
- [17].Tian J, Varga B, Tatrai E, Fanni P, Somfai GM, Smiddy WE, and DeBuc DC, “Performance evaluation of automated segmentation software on optical coherence tomography volume data,” J. Biophotonics, vol. 9, no. 5, pp. 478–489, May 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vermeer K, Van der Schoot J, Lemij H, and De Boer J, “Automated segmentation by pixel classification of retinal layers in ophthalmic OCT images,” Biomed. Opt. Expr, vol. 2, no. 6, pp. 1743–1756, June 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fernandez DC, “Delineating fluid-filled region boundaries in optical coherence tomography images of the retina,” IEEE Trans. Med. Imag, vol. 24, no. 8, pp. 929–945, August 2005. [DOI] [PubMed] [Google Scholar]
- [20].Farsiu S, Chiu SJ, Izatt JA, and Toth CA, “Fast detection and segmentation of drusen in retinal optical coherence tomography images,” in Proceedings of Biomed. Opt. (BiOS) 2008, Manns F, Söderberg PG, Ho A, Stuck BE, and Belkin M, Eds. International Society for Optics and Photonics, January 2008, p. 68440D. [Google Scholar]
- [21].Keller B, Cunefare D, Grewal DS, Mahmoud TH, Izatt JA, and Farsiu S, “Length-adaptive graph search for automatic segmentation of pathological features in optical coherence tomography images,” J. Biomed. Opt, vol. 21, no. 7, p. 076015, June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sui X, Zheng Y, Wei B, Bi H, Wu J, Pan X, Yin Y, and Zhang S, “Choroid segmentation from optical coherence tomography with graph-edge weights learned from deep convolutional neural networks,” Neurocomputing, vol. 237, pp. 332–341, May 2017. [Google Scholar]
- [23].Fang L, Cunefare D, Wang C, Guymer RH, Li S, and Farsiu S, “Automatic segmentation of nine retinal layer boundaries in oct images of non-exudative amd patients using deep learning and graph search,” Biomed. Opt. Express, vol. 8, no. 5, pp. 2732–2744, May 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chatterjee P and Milanfar P, “Is denoising dead?” IEEE Trans. Image Process, vol. 19, no. 4, pp. 895–911, April 2010. [DOI] [PubMed] [Google Scholar]
- [25].Robinson D and Milanfar P, “Fundamental performance limits in image registration,” IEEE Trans. Image Process, vol. 13, no. 9, pp. 1185–1199, August 2004. [DOI] [PubMed] [Google Scholar]
- [26].Wernet MP and Pline A, “Particle displacement tracking technique and Cramèr–Rao lower bound error in centroid estimates from CCD imagery,” Experiments in Fluids, vol. 15, no. 4, pp. 295–307, September 1993. [Google Scholar]
- [27].Yazdanfar S, Yang C, Sarunic MV, and Izatt JA, “Frequency estimation precision in doppler optical coherence tomography using the Cramèr–Rao lower bound,” Opt. Expr, vol. 13, no. 2, pp. 410–416, January 2005. [DOI] [PubMed] [Google Scholar]
- [28].Robinson D and Milanfar P, “Statistical performance analysis of super-resolution,” IEEE Trans. Image Process, vol. 15, no. 6, pp. 1413–1428, May 2006. [DOI] [PubMed] [Google Scholar]
- [29].Alagha NS, “Cramèr–Rao bounds of SNR estimates for BPSK and QPSK modulated signals,” IEEE Commun. Lett, vol. 5, no. 1, pp. 10–12, August 2001. [Google Scholar]
- [30].Jakobsen P, Greenfleld P, and Jedrzejewski R, “The Cramèr–Rao lower bound and stellar photometry with aberrated HST images,” Astronomy and Astrophysics, vol. 253, pp. 329–332, January 1992. [Google Scholar]
- [31].Farsiu S, Christofferson J, Eriksson B, Milanfar P, Friedlander B, Shakouri A, and Nowak R, “Statistical detection and imaging of objects hidden in turbid media using ballistic photons,” Appl. Opt, vol. 46, no. 23, pp. 5805–5822, August 2007. [DOI] [PubMed] [Google Scholar]
- [32].Rye BJ and Hardesty R, “Discrete spectral peak estimation in incoherent backscatter heterodyne lidar. I. spectral accumulation and the Cramèr–Rao lower bound,” IEEE Trans. Geosci. Remote Sens, vol. 31, no. 1, pp. 16–27, August 1993. [Google Scholar]
- [33].Lei T and Udupa JK, “Performance evaluation of finite normal mixture model-based image segmentation techniques,” IEEE Trans. Image Process, vol. 12, no. 10, pp. 1153–1169, October 2003. [DOI] [PubMed] [Google Scholar]
- [34].Wörz S and Rohr K, “Cramèr–Rao bounds for estimating the position and width of 3d tubular structures and analysis of thin structures with application to vascular images,” J. Math. Imag. and Vis, vol. 30, no. 2, pp. 167–180, November 2008. [Google Scholar]
- [35].Reza AM and Doroodchi M, “Cramèr–Rao lower bound on locations of sudden changes in a steplike signal,” IEEE Trans. Signal Process, vol. 44, no. 10, pp. 2551–2556, August 1996. [Google Scholar]
- [36].Peng R and Varshney PK, “On performance limits of image segmentation algorithms,” Comp. Vis. and Image Und, vol. 132, pp. 24–38, March 2015. [Google Scholar]
- [37].Izatt JA and Choma MA, Optical Coherence Tomography: Technology and Applications. Springer Science & Business Media, 2008, ch. 2: Theory of Optical Coherence Tomography. [Google Scholar]
- [38].Goodman JW, Speckle Phenomena in Optics: Theory and Applications. Roberts and Company Publishers, 2007. [Google Scholar]
- [39].Pircher M, Götzinger E, Leitgeb R, Fercher AF, and Hitzenberger CK, “Speckle reduction in optical coherence tomography by frequency compounding,” J. Biomed. Opt, vol. 8, no. 3, pp. 565–569, July 2003. [DOI] [PubMed] [Google Scholar]
- [40].Karamata B, Hassler K, Laubscher M, and Lasser T, “Speckle statistics in optical coherence tomography,” J. Opt. Soc. Am. A, vol. 22, no. 4, pp. 593–596, April 2005. [DOI] [PubMed] [Google Scholar]
- [41].Ralston TS, Atkinson I, Kamalabadi F, and Boppart SA, “Multidimensional denoising of real-time OCT imaging data,” in 2006 IEEE International Conference on Acoustics Speech and Signal Processing Proceedings, vol. 2 IEEE, May 2006, pp. 1148–1151. [Google Scholar]
- [42].Yin D, Gu Y, and Xue P, “Speckle-constrained variational methods for image restoration in optical coherence tomography,” J. Opt. Soc. Am. A, vol. 30, no. 5, pp. 878–885, May 2013. [DOI] [PubMed] [Google Scholar]
- [43].Schmitt JM, Xiang S, and Yung KM, “Speckle in optical coherence tomography,” J. Biomed. Opt, vol. 4, no. 1, pp. 95–105, January 1999. [DOI] [PubMed] [Google Scholar]
- [44].Lindenmaier AA, Conroy L, Farhat G, DaCosta RS, Flueraru C, and Vitkin IA, “Texture analysis of optical coherence tomography speckle for characterizing biological tissues in vivo,” Opt. Lett, vol. 38, no. 8, pp. 1280–1282, April 2013. [DOI] [PubMed] [Google Scholar]
- [45].Kirillin MY, Farhat G, Sergeeva EA, Kolios MC, and Vitkin A, “Speckle statistics in OCT images: Monte Carlo simulations and experimental studies,” Opt. Lett, vol. 39, no. 12, pp. 3472–3475, June 2014. [DOI] [PubMed] [Google Scholar]
- [46].Weatherbee A, Sugita M, Bizheva K, Popov I, and Vitkin A, “Probability density function formalism for optical coherence tomography signal analysis: a controlled phantom study,” Opt. Lett, vol. 41, no. 12, pp. 2727–2730, June 2016. [DOI] [PubMed] [Google Scholar]
- [47].Sugita M, Weatherbee A, Bizheva K, Popov I, and Vitkin A, “Analysis of scattering statistics and governing distribution functions in optical coherence tomography,” Biomed. Opt. Expr, vol. 7, no. 7, pp. 2551–2564, July 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Amini Z and Rabbani H, “Statistical modeling of retinal optical coherence tomography,” IEEE Trans. Med. Imag, vol. 35, no. 6, pp. 1544–1554, January 2016. [DOI] [PubMed] [Google Scholar]
- [49].Kay SM, Fundamentals of Statistical Signal Processing, Volume I: Estimation Theory. Prentice-Hall PTR, 1993. [Google Scholar]
- [50].Fercher AF, Optical Coherence Tomography: Technology and Applications. Springer Science & Business Media, 2008, ch. 4: Inverse Scattering, Dispersion, and Speckle in Optical Coherence Tomography. [Google Scholar]
- [51].Srinivasan PP, Heflin SJ, Izatt JA, Arshavsky VY, and Farsiu S, “Automatic segmentation of up to ten layer boundaries in sd-oct images of the mouse retina with and without missing layers due to pathology,” Biomed. Opt. Expr, vol. 5, no. 2, pp. 348–365, February 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Eldar YC, “Rethinking biased estimation: Improving maximum likelihood and the Cramér–Rao bound,” Found. and Trends ® in Sig. Proc, vol. 1, no. 4, pp. 305–449, July 2008. [Google Scholar]
- [53].Boyer KL, Herzog A, and Roberts C, “Automatic recovery of the optic nervehead geometry in optical coherence tomography,” IEEE Trans. Med. Imag, vol. 25, no. 5, pp. 553–570, May 2006. [DOI] [PubMed] [Google Scholar]
- [54].Salinas HM and Fernández DC, “Comparison of PDE-based nonlinear diffusion approaches for image enhancement and denoising in optical coherence tomography,” IEEE Trans. Med. Imag, vol. 26, no. 6, pp. 761–771, May 2007. [DOI] [PubMed] [Google Scholar]
- [55].Berkson J, “Minimum chi-square, not maximum likelihood!” The Annals of Statistics, pp. 457–487, May 1980. [Google Scholar]
- [56].Turton DA, Reid GD, and Beddard GS, “Accurate analysis of fluorescence decays from single molecules in photon counting experiments,” Analytical Chemistry, vol. 75, no. 16, pp. 4182–4187, June 2003. [DOI] [PubMed] [Google Scholar]
- [57].Akaike H, “A new look at the statistical model identification,” IEEE Trans. Autom. Control, vol. 19, no. 6, pp. 716–723, December 1974. [Google Scholar]
- [58].Antony B, Abràmoff MD, Tang L, Ramdas WD, Vingerling JR, Jansonius NM, Lee K, Kwon YH, Sonka M, and Garvin MK, “Automated 3-d method for the correction of axial artifacts in spectral-domain optical coherence tomography images,” Biomed. Opt. Express, vol. 2, no. 8, pp. 2403–2416, August 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen X, Niemeijer M, Zhang L, Lee K, Abràmoff MD, and Sonka M, “Three-dimensional segmentation of fluid-associated abnormalities in retinal oct: Probability constrained graph-search-graph-cut,” IEEE Trans. Med. Imag, vol. 31, no. 8, pp. 1521–1531, August 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Abràmoff MD, Garvin MK, and Sonka M, “Retinal imaging and image analysis,” IEEE Rev. Biomed. Eng, vol. 3, pp. 169–208, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Spiess A-N and Neumeyer N, “An evaluation of r2 as an inadequate measure for nonlinear models in pharmacological and biochemical research: a Monte Carlo approach,” BMC Pharmacology, vol. 10, no. 1, pp. 1–11, June 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Polans J, Keller B, Carrasco-Zevallos OM, LaRocca F, Cole E, Whitson HE, Lad EM, Farsiu S, and Izatt JA, “Wide-field retinal optical coherence tomography with wavefront sensorless adaptive optics for enhanced imaging of targeted regions,” Biomed. Opt. Expr, vol. 8, no. 1, pp. 16–37, January 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]