When commercializing bacterial strains, like Bacillus spp., for feed applications or plant bioprotection, it is required that the strains are free of acquired antimicrobial resistance genes that could potentially spread to pathogenic bacteria, thereby adding to the pool of resistance genes that may cause treatment failures in humans or animals. Conversely, if antimicrobial resistance is intrinsic to a bacterial species, the risk of spreading horizontally to other bacteria is considered very low. Reliable susceptibility test methods and interpretation criteria at the species level are needed to accurately assess antimicrobial resistance levels. In the present study, tentative ECOFFs for five Bacillus species were determined, and the results showed that the variation in MICs followed the respective species. Moreover, putative resistance genes, which were detected by whole-genome sequencing and suggested to be intrinsic rather that acquired, could explain the resistance phenotypes in most cases.
KEYWORDS: antibiotic, breakpoint, intrinsic resistance, probiotic, antibiotic resistance
ABSTRACT
Bacillus megaterium (n = 29), Bacillus velezensis (n = 26), Bacillus amyloliquefaciens (n = 6), Bacillus paralicheniformis (n = 28), and Bacillus licheniformis (n = 35) strains from different sources, origins, and time periods were tested for the MICs for nine antimicrobial agents by the CLSI-recommended method (Mueller-Hinton broth, 35°C, for 18 to 20 h), as well as with a modified CLSI method (Iso-Sensitest [IST] broth, 37°C [35°C for B. megaterium], 24 h). This allows a proposal of species-specific epidemiological cutoff values (ECOFFs) for the interpretation of antimicrobial resistance in these species. MICs determined by the modified CLSI method were 2- to 16-fold higher than with the CLSI-recommended method for several antimicrobials. The MIC distributions differed between species for five of the nine antimicrobials. Consequently, use of the modified CLSI method and interpretation of resistance by use of species-specific ECOFFs is recommended. The genome sequences of all strains were determined and used for screening for resistance genes against the ResFinder database and for multilocus sequence typing. A putative chloramphenicol acetyltransferase (cat) gene was found in one B. megaterium strain with an elevated chloramphenicol MIC compared to the other B. megaterium strains. In B. velezensis and B. amyloliquefaciens, a putative tetracycline efflux gene, tet(L), was found in all strains (n = 27) with reduced tetracycline susceptibility but was absent in susceptible strains. All B. paralicheniformis and 23% of B. licheniformis strains had elevated MICs for erythromycin and harbored ermD. The presence of these resistance genes follows taxonomy suggesting they may be intrinsic rather than horizontally acquired. Reduced susceptibility to chloramphenicol, streptomycin, and clindamycin could not be explained in all species.
IMPORTANCE When commercializing bacterial strains, like Bacillus spp., for feed applications or plant bioprotection, it is required that the strains are free of acquired antimicrobial resistance genes that could potentially spread to pathogenic bacteria, thereby adding to the pool of resistance genes that may cause treatment failures in humans or animals. Conversely, if antimicrobial resistance is intrinsic to a bacterial species, the risk of spreading horizontally to other bacteria is considered very low. Reliable susceptibility test methods and interpretation criteria at the species level are needed to accurately assess antimicrobial resistance levels. In the present study, tentative ECOFFs for five Bacillus species were determined, and the results showed that the variation in MICs followed the respective species. Moreover, putative resistance genes, which were detected by whole-genome sequencing and suggested to be intrinsic rather that acquired, could explain the resistance phenotypes in most cases.
INTRODUCTION
The development of antimicrobial resistance has become a serious problem and is a threat to modern medicine (1). The extensive use of antimicrobials in both humans and animals and the direct selection of antimicrobial resistance are considered the main reasons for the development and spread of resistance (2, 3). It has been estimated that treatment failure due to antimicrobial resistance causes 25,000 deaths annually in the European Union and 23,000 deaths in the United States (4, 5).
Although antimicrobial resistance has dramatically increased in the postantibiotic era, the mechanisms responsible for resistance existed long before and are found in bacteria in all environments (2, 6). Consequently, nonpathogenic bacteria may contain antimicrobial resistance genes and pose a risk for disseminating these to pathogenic bacteria, especially if the resistance mechanism is encoded on a mobile genetic element (e.g., a plasmid or a conjugative transposon) (7). For this reason, legislation exists that requires viable microorganisms used as active agents in feed additives to be free of antimicrobial resistance genes that potentially could be transferred to other bacteria (8, 9).
Determination of antimicrobial resistance has traditionally been done by use of phenotypic methods, such as disk diffusion or microdilution assays. By use of predefined interpretation criteria, a given bacterium can be determined to be either susceptible or resistant to a given antimicrobial. Committees, such as the Clinical and Laboratory Standards Institute (CLSI) and European Committee of Antimicrobial Susceptibility Testing (EUCAST), define clinical breakpoints and standardized methodology for many pathogenic bacteria and antimicrobial compounds (10, 11, 12).
For the surveillance and determination of resistance in nonpathogenic bacteria, interpretation based on MIC distributions in the population and epidemiological cutoff values (ECOFFs) have been used to distinguish innocuous strains from strains with acquired resistance (12). Since the MIC distributions differ based on the antimicrobial compound, methodology, and bacterial species, the interpretation criteria (ECOFF) are often defined at the species level and in some cases even at the subspecies level (13, 14). If the ECOFF is not defined correctly, errors in interpretation may occur (12).
MIC distributions at the species level are also useful to reveal intrinsic resistance, especially when the absence of acquired resistance can be demonstrated. When a bacterial species is intrinsically resistant to a given antimicrobial, it is not relevant to define an ECOFF (8, 15).
Data on antimicrobial susceptibility are limited for several Bacillus species used as probiotic feed additives, and no species-specific ECOFFs exist (16, 17, 18). In the guidance documents provided by the European Food Safety Authority (EFSA), ECOFFs are defined at the genus level (8, 9). It is stated that the data used for the establishment of ECOFFs were derived from published research, monitoring programs, or from EUCAST. However, no data are available from EUCAST on Bacillus species, and only one scientific publication dealing with Bacillus spp. is listed as a reference (8, 9, 19). In a study by Noor Uddin et al. (20), resistance in Bacillus spp. used in aquaculture was interpreted based on breakpoints defined for other Gram-positive species (20). The use of interpretation criteria developed for other distantly related species may result in misleading interpretations of susceptibility.
The determination of species-specific MIC distributions requires correct species identification. Species identification has traditionally been based on phenotypic characterization, 16S rRNA sequencing, or sequence diversity of other target genes. More recently, methods based on genome sequencing and phylogenic analyses have been used (21). Several reclassifications within the genus Bacillus have been suggested based on comparative genome sequence analysis, with the division of Bacillus licheniformis into the species B. licheniformis and Bacillus paralicheniformis (22), as well as the classification of Bacillus methylotrophicus (KACC 18228) and Bacillus amyloliquefaciens subsp. plantarum (FZB42) to be later heterotypic synonyms of Bacillus velezensis (23).
The objective of this study was to determine the MICs toward five Bacillus species of nine antimicrobial agents and the MIC distribution in the population, as well as to identify genes conferring resistance and to propose species-specific ECOFFs for interpretations of resistance. The five Bacillus species were chosen because they are relevant species for use as probiotic feed additives or for plant protection.
RESULTS AND DISCUSSION
Species identification and multilocus sequence typing.
Overall, the strain collection displays a good representation of the species. Distinct species identification could be obtained for all strains by use of average nucleotide identity based on BLAST (ANIb), except for one strain which was closest and equally related to both B. licheniformis and B. paralicheniformis. Multilocus sequence typing (MLST) further confirmed the strains to be diverse (Table 1). In a comparison of sequence types (STs) registered in the MLST database for B. velezensis, B. amyloliquefaciens, B. paralicheniformis, and B. licheniformis, the collection of strains was considered representative of the respective species. Moreover, STs followed the ANIb clusters, and many of the STs were novel and not previously registered (Table 1).
TABLE 1.
Origin and sequence type of strains included in the studya
| Species | Source | Yr of isolation | Geographic area | MLSTb |
|---|---|---|---|---|
| B. megaterium (26) | Chicken feces (1) | 2005 | England | m12 |
| Soil (9) | 1958 (1), 1969 (1), 1980 (2), not given (5) | Germany (3), Washington, USA (1), Venezuela (1), not given (4) | m8, m9, m10, m14, m15, m17, m18, m19, m26 | |
| Rice (5) | 1996 (1), 2013 (4) | Philippines (4), California, USA (1) | m5, m20, m21, m22, m23 | |
| Vineyard rhizosphere (3) | 2013 | New York State, USA | m2, m4, m24 | |
| Wheat rhizosphere (1) | 2013 | New York State, USA | m25 | |
| Not given (10) | 1916 (1), not given (9) | USA (1), not given (9) | m1, m3, m5, m6, m7, m11, m13, m16, m17 (2) | |
| B. amyloliquefaciens (6) | Pig feces (2) | 2011 | Germany | 157*, 59 |
| Soil (3) | <1940 (1), 2005 (1), 2014 (1) | Not given (1), Japan (1), North Carolina, USA (1) | 117, 171*, 157* | |
| Not given (1) | Not given | Not given | 117 | |
| B. velezensis (26) | Pig feces (5) | 2011 (5) | Denmark (3), Germany (2) | 161* (3), 160*, 162* |
| Giant Panda feces (1) | <2012 | Japan | 173* | |
| Water (1) | 1999 | Spain | 168* | |
| Soil or soil related (6) | 2007 (1), 2012 (1), 2013 (3), 2014 (1) | Italy (1), New Jersey, USA (1), New York State, USA (1), North Carolina, USA (3) | 172*, 140, 163*, 166*, 42,174* | |
| Food (6) | <1978 (1), 2012 (1), 2014 (4) | Malaysia (1), North Carolina, USA (1), Ghana (4) | 167*, 42, 156*, 158*, 159* (2) | |
| Commercial probiotic for animals (5) | 2014 | Not given | 164*, 165*, 93, 42, 175* | |
| Not given (2) | <1946 (1), <1951 (1) | Not given (2) | 169*, 170* | |
| B. paralicheniformis (28) | Pig feces (3) | 2011 | Spain (2), Denmark (1) | 14 (2), 43* |
| Cattle abortion (1) | Not given | Not given | 17 | |
| Chinchilla feces (1) | 1965 | Not given | 4 | |
| Food products (6) | <1950 (1), ≤1953 (1), 2010 (1), 2014 (1), <2015 (1), not given (1) | Ghana (1), South Korea (1), Australia (1), not given (3) | 4, 14, 15, 21, 27, 33* | |
| Soil (7) | ≤1986 (1), 2011 (1), 2013 (1), not given (4) | Germany (1), United Kingdom (1), Denmark (1), New Jersey, USA (1), Sudan (2), not given (1) | 4 (2), 14, 18 (2), 35*, 36* | |
| Rice rhizosphere (1) | 2013 | California, USA | 4 | |
| Oil drilling (1) | Not given | Texas, USA | 37* | |
| Pharmaceutical filtration (1) | <1980 | Not given | 47* | |
| Wastewater treatment (1) | Not given | Not given | 21 | |
| Not given (6) | <1951 (3), <1954 (1), <1977 (1), Not given (1) | Not given (6) | 4, 14, 15 (3), 47* | |
| B. licheniformis (35)c | Bovine or bovine feed (5) | 1984 (1), 1993 (2), 1994 (1), ≤1996 (1) | Denmark (5) | 3 (3), 40*, 29* |
| Milk or milk products (5) | 1878 (1), <1950 (1), <1979 (1), 2010 (1), not given (1) | Egypt (1), Australia (1), not given (3) | 3, 5, 7, 25, 46* | |
| Pig feces (3) | 2011 (3) | Germany (2), Spain (1) | 1, 26 (2) | |
| Sheep feces (1) | <1980 | United Kingdom | 26 | |
| Human feces (1) | Not given | Vietnam | 3 | |
| Horse blood (1) | <1979 | Not given | 7 | |
| Insect (1) | 2012 | New Jersey, USA | 44 | |
| Food (6) | 1917 (1), 1921 (1), <1979 (1), <1996 (1), 2011 (2) | Sudan (2), Philippines (1), not given (3) | 3, 7, 9, 24, 44*, 34* | |
| Soil or soil related (4) | 1993 (1), 2011 (1), not given (2) | Denmark (1), Sweden (1), Japan (1), Arizona, USA (1) | 2, 13, 45*, 31* | |
| Sea animal or seawater (3) | 1944 (1), <1964 (1), not given (1) | Vietnam (1), not given (2) | 3, 39*, 30* | |
| Other sources (2) | Not given (2) | Norway (1), The Netherlands (1) | 3, 2 | |
| Not given (3) | <1944 (1), <1952 (1), <1963 (1) | Not given (3) | 1 (3) |
Numbers in parentheses represent the number of strains; ≤ in front of the year indicates that only the date of deposit is known.
*, novel ST identified in this study. STs for B. megaterium are indicated with an “m”, as no public database exists for those species.
One strain could not be identified to the species level as either B. licheniformis or B. paralicheniformis but is listed as B. licheniformis.
No MLST database for B. megaterium exists. ANIb analysis of this species revealed five clusters of related strains and a single strain. The single strain was more distantly related to the others (95.04% to 96.92%) and did not share any MLST alleles with other strains. Among the B. megaterium strains, 26 different STs were found, and the ST clustering followed the same clusters identified by ANIb, confirming that the MLST scheme reflects the phylogenetic relationship.
Comparison of MICs between species.
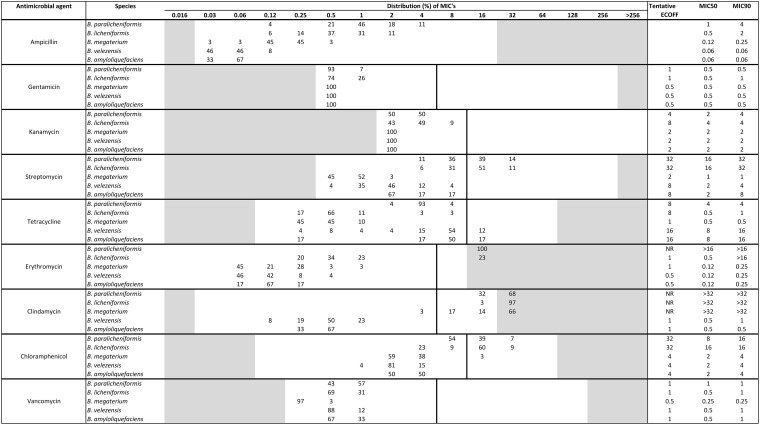
The nine antimicrobial agents tested are those recommended by the EFSA (8, 9). When using the modified CLSI method, all strains could grow under the test conditions, and growth was sufficient for clear interpretation of the outcome. The MIC distributions differed between the five species for seven of the antimicrobials (Table 2).
TABLE 2.
MIC distribution for nine antimicrobial agentsa
B. megaterium (n = 29), B. velezensis (n = 26), B. amyloliquefaciens (n = 6), B. paralicheniformis (n = 28), and B. licheniformis (n = 35) strains were tested by use of the modified CLSI method. The vertical solid lines indicate the EFSA ECOFF for the genus Bacillus. One strain could not be identified to the species level as either B. licheniformis or B. paralicheniformis but is listed as B. licheniformis. For ampicillin, the EFSA does not define an ECOFF and no other interpretation criteria exist.
For chloramphenicol, all strains of B. velezensis and B. amyloliquefaciens had MICs in the range of 1 to 4 mg/liter, which is severalfold under the EFSA ECOFF. The same is true for B. megaterium, except one resistant strain that had an MIC of 16 mg/liter. All B. paralicheniformis strains were resistant to chloramphenicol or had MICs just 2-fold under the EFSA ECOFF (MICs from 8 to 32 mg/liter). For B. licheniformis, most strains had MICs in the same range as B. paralicheniformis, except for 23% which had an MIC of 4 mg/liter. The same tendency was seen for streptomycin (Table 2).
For tetracycline, the B. megaterium isolates were all susceptible, with MICs ranging from 0.25 to 1 mg/liter. Most strains of B. velezensis and B. amyloliquefaciens had MICs from 4 to 16 mg/liter. The MIC distribution for B. paralicheniformis was below the EFSA ECOFF, ranging from 2 to 8 mg/liter. For B. licheniformis, most strains had MICs from 0.25 to 1 mg/liter, but three strains had MICs of 2 to 8 mg/liter.
All strains of B. megaterium, B. velezensis, and B. amyloliquefaciens were susceptible to erythromycin (MICs from 0.03 to 1 mg/liter) when using the current EFSA ECOFF (8, 9), whereas all B. paralicheniformis and 23% of the B. licheniformis strains were resistant (MIC, >8 mg/liter). For clindamycin, all strains of B. megaterium, except one, and all strains of B. paralicheniformis and B. licheniformis were resistant, whereas all strains of B. amyloliquefaciens and B. velezensis were susceptible (MIC, 0.12 to 1 mg/liter), being severalfold under the EFSA ECOFF.
MIC distributions differed between species for ampicillin. Because Bacillus spp., including species, such as B. licheniformis, can produce beta-lactamases, ampicillin susceptibility was not further evaluated, as testing is unreliable according to the CLSI (24, 25).
Even though the number of strains per species is limited, ranging from 6 to 35, the results clearly show that the MIC distributions follow the different species (Table 2). EUCAST recommends a minimum of 15 strains from the same laboratory, but data from at least five laboratories with a total of 100 strains are required for having an ECOFF formally defined by EUCAST (26). When data from fewer strains are used, tentative ECOFFs may be defined (26). Our results clearly support the recommendations by EUCAST that interpretation criteria for antimicrobial resistance should be evaluated and defined at the species level by use of MIC distributions (26). For Bacillus, the interpretation criteria set by the EFSA (8, 9) are defined at the genus level, and for several of the drug and species combinations, the EFSA ECOFFs are placed in the middle of the distributions and seem not to be applicable for many drug-species combinations. Therefore, we have proposed tentative ECOFFs values, shown in Table 2. These tentative ECOFFs may offer an objective basis to update or modify specific guidelines, such as those outlined by the EFSA (8, 9).
Comparison of the modified CLSI and CLSI-recommended methods.
While all strains could grow under both conditions used for susceptibility testing, differences in the MICs of some antimicrobials were observed (see Table S1 in the supplemental material). The largest differences were seen for tetracycline and erythromycin, where some isolates showed up to 16-fold higher MICs when tested with the modified CLSI method than the CLSI-recommended method. One strain which had at least a 4-fold higher MIC for erythromycin with the modified CLSI method was the B. paralicheniformis type strain KJ-16, which has an erythromycin resistance gene ermD (Table S1) (22).
It is well known that MICs depend on growth conditions, such as growth media, incubation temperature, and time (11). In a clinical setting, the most optimal test would distinguish between strains that can and cannot be treated by the antimicrobial in question (27). When dealing with nonpathogenic strains, such as commercial strains, the optimal method will clearly separate the susceptible population from the strains having acquired resistance (26). For the tested Bacillus species, the modified CLSI method seems most appropriate, as it offers a better correlation with the presence of resistance genes (see below) and gives the highest MIC, ensuring that a precautionary principle is followed.
Detection of known resistance genes and correlation with MICs obtained with the modified CLSI method. (i) B. megaterium.
The B. megaterium strains were predominantly susceptible to the antimicrobials tested. One strain had an MIC to chloramphenicol of 16 mg/liter, which was 4- to 8-fold higher than that of the B. megaterium population in general. This strain is more distantly related to the other B. megaterium strains and did not share any alleles for the genes included in the MLST scheme. It has a coding sequence (CDS) annotated by RAST as corresponding to chloramphenicol acetyltransferase (CAT; EC 2.3.1.28) (Table 3) (NCBI BioSample accession SAMN08399259). The gene is 648 bp in length and had the closest similarity (71% identity, 1% gaps, and 95% coverage), at the nucleotide level, to a cat gene found in Bacillus clausii (GenBank accession no. AY238971) (44). The predicted amino acid sequence is shorter than the Bacillus clausii CAT protein (215 versus 228 amino acids), and when aligning the sequences, 57% (130/228) amino acid identity was found. The putative cat gene was found on a 39-kb contig, with the cat gene located approximately 1 kb from the end of the contig. The flanking regions (1,440 bp upstream and 970 bp downstream of the cat gene) had high similarity and coverage to large B. megaterium plasmids; the downstream region had 74% coverage and 92% identity to pStB1A, and the upstream region had 98% coverage and 86% identity to pBMV. The upstream region also had high identity to (92%) and coverage (50%) to the B. megaterium chromosome. A comparison of the entire contig, however, revealed only 15% coverage and 85% identity to Bacillus chromosomes and plasmids. Based on the limited number of strains, it is impossible to conclude whether the putative cat gene is an intrinsic gene in a subclade of B. megaterium or an acquired resistance mechanism in a specific strain. The sequence homology to the B. clausii cat gene and reduced susceptibility to chloramphenicol strongly suggest that the gene is responsible for the reduced susceptibility to chloramphenicol, but this needs further study. To our knowledge, this specific cat gene has not been described previously.
TABLE 3.
Occurrence of putative antimicrobial resistance genesa
| Species (n) | % (no.) of strains with indicated antimicrobial resistance gene |
||||
|---|---|---|---|---|---|
| Chloramphenicol, putative cat | Clindamycin, putative lsa(B) | Streptomycin, putative aadK | Tetracycline, putative tet(L) | Erythromycin, ermD | |
| B. megaterium (29) | 3 (1) | 7 (2) | |||
| B. velezensis (26) | 58 (15) | 85 (22) | |||
| B. amyloliquefaciens (6) | 100 (6) | 86 (5) | |||
| B. paralicheniformis (28) | 100 (28) | ||||
| B. licheniformis (35) | 23 (8) | ||||
aadK, aminoglycoside 6-adenylyltransferase gene; tet(L), tetracycline efflux gene; lsa(B), ABC transporter gene; ermD, rRNA methylase gene; cat, chloramphenicol acetyltransferase gene.
All B. megaterium strains, except one with an MIC of 4 mg/liter, were resistant to clindamycin. However, no resistance genes were observed that could explain clindamycin resistance, except in two strains with unrelated STs and present in different ANIb clusters. The two strains contain a putative gene annotated by RAST as an ABC transporter (1,479 bp), with 80% identity at the nucleotide level to lsa(B). This gene is located on plasmid pSCFS1 from Staphylococcus sciuri (GenBank accession no. AJ579365) and encodes low-level clindamycin resistance (28). The gene also had 100% coverage and 97% identity to a gene annotated as an Lsa family ABC transporter in B. megaterium strain JX285 (GenBank accession no. CP018874). Although this gene may encode clindamycin resistance, its presence does not explain the clindamycin resistance observed in the majority of strains.
Another gene, annotated by RAST as macrolide-2 phosphotransferase, had the highest similarity to an mph(B) gene encoding macrolide resistance (GenBank accession no. D85892) (29). The gene was found in 14 strains, but it had only 33% identity over the entire gene and also had two 8-bp gaps leading to frameshifts. Moreover, since the strains were susceptible to erythromycin, it probably serves other functions in the cell.
A gene with low similarity and coverage (88% identity over 80 bp) to msrC (GenBank accession no. AY004350), encoding erythromycin resistance, was found in four strains, but no correlation with the erythromycin MIC was found. The gene was annotated by RAST as an ABC transporter and probably serves as such.
Four strains had a gene annotated by RAST as a trimethoprim resistance gene with similarity (69% identity over 470 bp and 1% gaps) to dfrD (GenBank accession no. Z50141). According to the guidance document recently adopted by the EFSA, when a suspected antimicrobial resistance gene is found, the strains should be tested for phenotypic resistance to the corresponding antimicrobial if the antimicrobial is on the WHO list of critically important antimicrobials (CIA) or highly important antimicrobials (HIA) (9). Trimethoprim is listed as an HIA (30), but the four strains were susceptible to trimethoprim (MIC, <0.12 to 0.5 mg/liter) (data not shown). Noor Uddin et al. (20) found genes with similarity to dfrD, dfrG, and dfrK (approximately 80% identity at the nucleotide level) in 91 strains of Bacillus nealsonii, but all strains were susceptible to trimethoprim (20), so these dfr-like genes are likely to serve other functions in these Bacillus strains.
(ii) B. velezensis and B. amyloliquefaciens.
MICs to streptomycin in both B. velezensis and B. amyloliquefaciens covered a larger range than for the other Bacillus species tested (Table 2). In 15 of 26 B. velezensis strains (NCBI BioSample accession no. SAMN08399260) and in all B. amyloliquefaciens strains (NCBI BioSample accession no. SAMN08399261), a putative aminoglycoside 6-adenylyltransferase gene, potentially providing streptomycin resistance, was found. The gene had the closest similarity to aadK from B. subtilis Marburg 168 (GenBank accession no. M26879), with identity of 68% (440/643) and coverage of 75% (643/855) (31). An identity of 39% (110/284) at the amino acid level was predicted, with an insert of six amino acids in position 280. Although none of the strains were resistant to streptomycin, based on the EFSA ECOFF of 8 mg/liter, the strains with the putative streptomycin resistance gene seemed to be reduced in susceptibility compared to the strains without the gene, as revealed by two overlapping MIC distributions represented in the population (Fig. 1). aadK from B. subtilis was originally found in susceptible derivatives of Marburg 168 strains but expressed resistance in Escherichia coli and B. subtilis when present in high copies (32). Whether this is the case with the putative aadK found in B. amyloliquefaciens and B. velezensis strains is unknown.
FIG 1.
Correlation between the occurrence of putative resistance genes and MICs in B. velezensis and B. amyloliquefaciens. (A) Putative tet(L) and the MIC toward tetracycline. (B) Putative aadK and the MIC toward streptomycin.
Most strains contain a putative tet(L) gene (1,377 bp; BioSample accession no. SAMN08399260), and the presence of this putative tet(L) gene correlates with reduced susceptibility to tetracycline (2 to 16 mg/liter), whereas strains with MICs from 0.25 to 1 mg/liter contain either a truncated version (639 bp; BioSample accession no. SAMN08399261) or no tet gene (Fig. 1). The putative tet(L) gene had 87% identity to tet(L) with GenBank accession no. (X08034) (33) at both the nucleotide level (1,202/1,377) and the amino acid level (398/458), with no gaps predicted. A BLAST search against the NCBI NR database showed the gene to have the highest similarity (87 to 99% identity over 100% coverage) to genes in the chromosomes of B. velezensis, B. amyloliquefaciens, and B. subtilis. Previously, tet(L) has been described in the chromosome of B. subtilis (33) but, to our knowledge, not in B. amyloliquefaciens or B. velezensis. The putative tet(L) gene may be intrinsic in the chromosome of these related Bacillus species, but further analysis of the flanking sequences is required to support this conclusion. Although classified as tet(L) according to the current nomenclature (>80% identity at the amino acid level) (34), they are more distantly related to the highly conserved tet(L) genes found on small plasmids in other Gram-positive bacteria (35).
A total of 19 out of 26 B. velezensis and all B. amyloliquefaciens strains contain a gene that shows similarity to the phenicol, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A resistance gene cfr (GenBank accession no. AM408573), with 72% identity over a coverage of 95%. All strains had an MIC from 1 to 4 mg/liter for chloramphenicol, and no correlation with the presence of the gene and reduced susceptibility was found (36). Since all strains were susceptible to clindamycin, this gene was not further assessed.
(iii) B. paralicheniformis and B. licheniformis.
All B. paralicheniformis strains were found to be resistant to erythromycin. The macrolide-lincosamide-streptogramin B (MLS) resistance gene ermD, with highest identity at the nucleotide level to ermD with GenBank accession no. M29832 (37) of 98.0 to 99.8%, was found in all strains. Similarly, all erythromycin-resistant strains of B. licheniformis possess ermD (93.8 to 98.1% identity to GenBank accession no. M29832). None of the B. licheniformis strains with an MIC of <16 mg/liter had any erythromycin resistance genes, except one strain with an MIC of 0.5 mg/liter; ermD in this strain had slightly lower similarity (93.6% identity) to ermD with GenBank accession no. M29832. Clindamycin belongs to the lincosamide class of antimicrobials, suggesting that the presence of ermD could confer resistance to clindamycin. However, all strains had an MIC to clindamycin of ≥16 mg/liter regardless of the presence or absence of ermD. Thus, resistance to clindamycin could not be explained by the presence of ermD.
Strains of B. licheniformis and B. paralicheniformis generally exhibited reduced susceptibilities to streptomycin and chloramphenicol compared to the other Bacillus species. Most strains had an MIC at or above the EFSA ECOFF, but no streptomycin or chloramphenicol resistance genes were detected by ResFinder in any of the strains. The ResFinder database was recently compared with three other databases and found to be, together with the Comprehensive Antibiotic Resistance Database (CARD) (38), the most reliable. The ResFinder database is considered suitable for the prediction of antibiotic resistance genes in Salmonella enterica serovar Typhimurium, E. coli, and enterococci (39). The database has, however, not been validated for Bacillus spp. or other bacterial species of more environmental origin. Moreover, the antimicrobial resistance genes currently known are mostly from bacteria relevant for human and animal infections, which could explain why no streptomycin or chloramphenicol resistance genes were found.
We are currently working on characterizing the genetic background for reduced susceptibility to streptomycin and chloramphenicol.
MIC values and induction of ermD variants found in B. licheniformis and B. paralicheniformis.
A subset of 10 B. paralicheniformis (including reference strains KJ-16 and ATCC 9945A) and five B. licheniformis strains (including reference strain DSM13) were selected and further tested for MIC toward erythromycin in the range of 8 to 256 mg/liter. The results showed strains with ermD identity to M29832 of 98.4 to 99.7% (one B. licheniformis and nine B. paralicheniformis strains) to have the highest MIC (≥256 mg/liter), whereas strains with identity of 94 to 97% (three B. licheniformis strains) had an MIC of 32 mg/liter. One strain with an ermD 94% identity had an MIC of 128 mg/liter; this was the strain that could not be identified as either B. licheniformis or B. paralicheniformis.
Three strains with nucleotide identity to ermD (M29832) of 99.7% and three strains with identity from 94 to 98.4% were tested for inducibility by erythromycin. Although induction varied between strains and biological replicates, ermD gene expression was always increased 7-fold or more.
Based on these results, inducible ermD in B. paralicheniformis probably provides high-level resistance (MIC, ≥256 mg/liter) to erythromycin. In B. licheniformis, susceptibility to erythromycin varies, but ermD can be induced, as previously described (37). Thus, we postulate that ermD is intrinsic in B. paralicheniformis and probably also in subpopulations of B. licheniformis. Additional studies are under way to support this conclusion.
Conclusion.
Use of the modified CLSI method and interpretation of resistance by use of species-specific tentative ECOFFs are recommended. The presence of several (putative) resistance genes follows related species, suggesting these genes to be intrinsic rather than horizontally acquired. However, this needs to be further investigated.
MATERIALS AND METHODS
Bacterial strains.
A total of 124 strains belonging to Bacillus megaterium (n = 29), B. velezensis (n = 26), Bacillus amyloliquefaciens (n = 6), B. paralicheniformis (n = 28), and B. licheniformis (n = 35) (including one strain which could not be clearly identified as either B. licheniformis or B. paralicheniformis) were included. The strains were bought from various culture collections, received from researchers, or isolated by Chr. Hansen A/S (CH). The strains originated from various sources and were isolated from different geographic areas over a time span of more than 100 years (Table 1). All strains were stored in the Chr. Hansen Culture Collection (CHCC) at −80°C.
DNA extraction, sequencing, quality control, and assembly of contigs.
For most strains, frozen cell pellets were shipped to BaseClear (Leiden, Netherlands), which performed DNA extraction and sequencing as a service. FASTQ output files were sent to CH.
Some strains were sequenced at CH; DNA was purified in a QIAcube using a Qiagen blood and tissue kit (Hilden, Germany), according the protocol provided. DNA libraries were made using the Kapa library preparation for Illumina kit (Wilmington, MA, USA), according to the manufacturer's instructions, and sequenced on an Illumina MiSeq platform using V2 flow cells and a 250-bp paired-end protocol.
FASTQ files were imported into CLC Genomics Workbench version 9.6 (Qiagen Bioinformatics, Aarhus, Denmark) and analyzed in a workflow comprising (i) PhiX removal by mapping against phiX, (ii) quality trimming with a quality limit of 0,005, and (iii) de novo assembly with mismatch cost of 2 and insertion and deletion cost of 3. After assembly, the sequences were decontaminated using a custom-built CLC tool to filter away contigs with coverage of <15× and/or <20% of the median coverage of the assembly. An average coverage of <40× or a genome size <80% of the median expected for the species resulted in an assembly being discarded. The remaining contigs were converted to FASTA files and used in the subsequent analysis.
Species identification ANIb and MLST.
Species identification was performed by whole-genome average nucleotide identity by BLAST (ANIb) calculation with the help of the JSpecies software, applying default parameters (21). An ANIb value of 95 to 96% was used as the species threshold. The following type strains were used: B. licheniformis DSM13, B. paralicheniformis KJ-16, B. sonorensis NBRC101234, B. aryabhattai B8W22, B. megaterium ATCC 14581, B. flexus NBRC15715, B. velezensis KCTC13012, B. amyloliquefaciens DSM7, and B. siamensis KCTC13613.
MLST of whole-genome sequences of B. amyloliquefaciens and B. velezensis was performed with the help of the microbial genomics module of the CLC software package, making use of the Bacillus subtilis multilocus sequence typing website (https://pubmlst.org/bsubtilis/) (40). For B. licheniformis and B. paralicheniformis, the Bacillus licheniformis multilocus sequencing typing website (https://pubmlst.org/blicheniformis/) (40) was used. For Bacillus megaterium, a new MLST scheme including the genes dnaK, fusA, groEL, gyrA, gyrB, ileS, lep, pheS, recA, rpoA, rpoB, and rpoC was developed and used.
Susceptibility testing.
The MICs of nine antimicrobial agents were determined by use of broth microdilution. All strains were tested by use of a modified CLSI method using Iso-Sensitest broth (Oxoid) with aerobic incubation at 37°C (35°C for B. megaterium) for 24 h. The inoculum was prepared and purity confirmed as recommended by the CLSI (11). Streptococcus thermophilus ATCC 15707 was included for quality control using the quality control ranges reported in the ISO 10932 standard (41).
In order to evaluate the modified CLSI method against the broth microdilution method recommended by the CLSI for Bacillus spp. other than the B. cereus group, a subset of strains was further tested by the CLSI-recommended method (24). In brief, the CLSI-recommended method used cation-adjusted Mueller-Hinton broth 2 (Sigma-Aldrich), with aerobic incubation at 35°C for 18 to 20 h using Staphylococcus aureus ATCC 29213 as a quality control (24).
All tests, regardless of method, were performed in duplicate on VetMIC panels Lact-1 and Lact-2 (SVA, Uppsala, Sweden) (see Table 2 for antimicrobials and ranges included). Retesting was performed if more than a 2-fold difference occurred for one or more antimicrobial agents. The results were accepted if they varied by three or fewer 2-fold concentrations, as previously described being within the technical variation for MIC broth dilution methods (42). If the MIC value in two tests differed 2-fold, the highest MIC is reported in Table 2, whereas if more than two tests were performed, the concentration closest to the average is reported.
Selected strains with an MIC to erythromycin of >8 mg/liter were further tested in the range of 8 to 256 mg/liter to obtain a more precise MIC value. Two-fold dilutions of erythromycin in IST broth were made manually in microtiter plates, and the plates were inoculated and incubated as described above for the modified CLSI method. The reference strains B. licheniformis DSM13, B. paralicheniformis ATCC 9945A, and B. paralicheniformis KJ-16 were included. As a quality control, S. thermophilus ATCC 15707 was tested both as described above for erythromycin at 8 to 256 mg/liter and on VetMIC panels Lact-1 and Lact-2.
Epidemiological MIC cutoff values for differentiation of susceptible and resistant populations.
MIC distributions were determined for each species-antimicrobial combination. The MIC distributions were used to define MIC50s, MIC90s, and a tentative ECOFF for differentiation between susceptible and resistant strains. MIC50 and MIC90 are defined as MICs inhibiting 50% and 90% of the strains, respectively. ECOFFs were determined from MIC distributions for each species-drug combination, as recommended by EUCAST (12, 26), with susceptible being the population with MIC at or below the ECOFF and resistant being the population with an MIC of > ECOFF. Moreover, according to EUCAST, the susceptible population, referred to as wild type by EUCAST, is also characterized by the absence of acquired resistance mechanisms and/or mutations leading to resistance (26). Our data were also evaluated with the interpretation criteria defined by the EFSA for Bacillus spp. (8).
Detection of known antimicrobial resistance genes and comparison with phenotypes.
The genome sequences of all strains were screened for the presence of genes with identity to known resistance genes contained in the curated ResFinder database (43). The database was downloaded and imported into CLC Main Workbench 7 on 10 October 2017. The assembled contigs of each strain were joined using the join function in CLC. The joined contigs were screened for resistance genes (using BLASTN) against the ResFinder database, with a minimum word size of 11 and a maximum E value of 1.0E−8 to ensure a broad screening. If an E value of <1.0E−11 was observed, a manual assessment was performed by RAST annotating selected genome sequences and assessing putative genes at both the amino acid and nucleotide levels by screening the putative genes and flanking sequences against the NCBI NR database. Genome sequences representing strains with specific putative resistance genes were uploaded to NCBI.
Induction of erythromycin resistance.
Six ermD-positive B. licheniformis and B. paralicheniformis strains were selected based on nucleotide identity to ermD (GenBank accession no. M29832), three strains with 99.77% identity, and three strains with identity in the range of 94.52 to 98.38%.
The strains were inoculated from an overnight culture into 2 × 100-ml brain heart infusion (BHI) to an optical density at 600 nm (OD600) of 0.1 and incubated aerobically in 500-ml flasks, with shaking at 175 rpm at 37°C. In exponential phase (OD600, 1), the cultures were split in two, one with addition of erythromycin (0.1 mg/liter) and one without, and incubated under the same conditions for 60 min. Prior to cell harvest for RNA extraction, the OD600 was adjusted to 1.0 with fresh medium. Total RNA was purified with the RNeasy kit (Qiagen GmbH). cDNA was prepared from total RNA with the high-capacity RNA-to-cDNA kit (Applied Biosystems).
Primers for the reverse transcription-PCRs (RT-PCRs) were designed online by the Universal ProbeLibrary assay design (Life Science) with 60°C as the annealing temperature in all cases (Roche Diagnostics GmbH). Unique DNA sequences from the Bacillus strains in this study were used. The dnaA gene was chosen as a reference. The comparative RT-PCR experiments were performed from two biological replicates with nine technical replicates each with the Fast SYBR green master mix (Applied Biosystems) on a QuantStudio PCR machine (Applied Biosystems). The RT-PCR program was 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s and 60°C for 1 min. The RT-PCR data were analyzed with the QuantStudio real-time PCR software (Applied Biosystems). Melt curves were generated at the end of the RT-PCR program.
The strain-specific primers designed and used were dnaA-forward, 5′-GTCGGCCAGCAGTTCAATA-3′; dnaA-reverse, 5′-CGATTTTGTCCGTTTCTTCG-3′; ermD-forward, 5′-AAGTCAAAAAGCCGGTAAGGT-3′; ermD-reverse1, 5′-TGTGCCGTTTTACGTGTGAG-3′; and ermD-reverse2, 5′-TGTGCTGTTTTACGTGTGAG-3′ (the underlined bases differ for ermD-reverse1 and ermD-reverse2).
Accession number(s).
Genome sequences were deposited as follows: B. megaterium with a chloramphenicol acetyltransferase (CAT) (EC2.3.1.28), NCBI BioSample accession no. SAMN08399259; B. velezensis with a putative aminoglycoside 6-adenylyl transferase (streptomycin resistance) and a putative tet(L) gene (tetracycline resistance), NCBI BioSample accession number SAMN08399260; and B. amyloliquefaciens with a putative aminoglycoside 6-adenylyltransferase (streptomycin resistance) and a truncated tet(L), NCBI BioSample accession number SAMN08399261.
Supplementary Material
ACKNOWLEDGMENTS
We thank Abdallah Albayasli, Anna Minor Nielsen, and Peter Breum for excellent technical assistance with the susceptibility testing. Jacob Bælum, Martin Abel-Kistrup, Karin Elisabeth Schlichter, and Helle Schack Andersen are thanked for assistance with the DNA sequencing. Roberto La Ragione, Animal Health and Veterinary Laboratories Agency, Surrey, United Kingdom; Lene Jespersen, Department of Food Science, University of Copenhagen, Denmark; Mark Turner, School of Agriculture and Food Sciences, University of Queensland, Australia; and Per Einar Granum, Department for Food Safety and Infection Biology, Norwegian University of Life Sciences, Norway, are thanked for sending strains.
This research was supported exclusively by internal funds.
This work was performed by employees of Chr. Hansen A/S, a company that produces strains for plant protection, animal and human health, as well as for the food and dairy industry. Some of the authors are shareholders in Chr. Hansen A/S.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01108-18.
REFERENCES
- 1.World Health Organization. 2015. Global action plan on antimicrobial resistance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10:122–129. doi: 10.1038/nm0204-122. [DOI] [PubMed] [Google Scholar]
- 4.WHO Regional Office for Europe. 2011. Tackling antibiotic resistance from a food safety perspective in Europe. World Health Organization Regional Office for Europe, Copenhagen, Denmark: http://www.euro.who.int/__data/assets/pdf_file/0005/136454/e94889.pdf?ua=1. [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 6.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 7.Martínez JL, Coque TM, Lanza VF, de la Cruz F, Baquero F. 2017. Genomic and metagenomic technologies to explore the antibiotic resistance mobilome. Ann N Y Acad Sci 1388:26–41. doi: 10.1111/nyas.13282. [DOI] [PubMed] [Google Scholar]
- 8.EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). 2012. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J 10:2740. doi: 10.2903/j.efsa.2012.2740. [DOI] [Google Scholar]
- 9.EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). 2018. Guidance on the charaterisation of microorganisms used as feed additives or as production organisms. EFSA J 16:5206. doi: 10.2903/j.efsa.2018.5206. [DOI] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (CLSI). 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Clinical and Laboratory Standards Institute (CLSI). 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grows aerobically; approved standard, 10th ed CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 13.Agersø Y, Torpdahl M, Zachariasen C, Seyfarth A, Hammerum A, Nielsen EM. 2012. Tentative colistin epidemiological cut-off value for Salmonella spp. Foodborne Pathog Dis 9:367–369. doi: 10.1089/fpd.2011.1015. [DOI] [PubMed] [Google Scholar]
- 14.Klare I, Konstabel C, Werner G, Huys G, Vankerckhoven V, Kahlmeter G, Hildebrandt B, Müller-Bertling S, Witte W, Goossens H. 2007. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J Antimicrob Chemother 59:900–912. doi: 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- 15.Leclercq R, Cantón R, Brown DFJ, Giske CG, Heisig P, Macgowan AP, Mouton JW, Nordmann P, Rodloff AC, Rossolini GM, Soussy CJ, Steinbakk M, Winstanley TG, Kahlmeter G. 2013. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect 19:141–160. doi: 10.1111/j.1469-0691.2011.03703.x. [DOI] [PubMed] [Google Scholar]
- 16.Larsen N, Thorsen L, Kpikpi EN, Stuer-Lauridsen B, Cantor MD, Nielsen B, Brockmann E, Derkx PMF, Jespersen L. 2014. Characterization of Bacillus spp. strains for use as probiotic additives in pig feed. Appl Microbiol Biotechnol 98:1105–1118. doi: 10.1007/s00253-013-5343-6. [DOI] [PubMed] [Google Scholar]
- 17.Mohammed MJ, Marston CK, Popovic T, Weyant RS, Tenover FC. 2002. Antimicrobial susceptibility testing of Bacillus anthracis: comparison of results obtained by using the National Committee for Clinical Laboratory Standards broth microdilution reference and Etest agar gradient diffusion methods. J Clin Microbiol 40:1902–1907. doi: 10.1128/JCM.40.6.1902-1907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luna VA, King DS, Gulledge J, Cannons AC, Amuso PT, Cattani J. 2007. Susceptibility of Bacillus anthracis, Bacillus cereus, Bacillus mycoides, Bacillus pseudomycoides and Bacillus thuringiensis to 24 antimicrobials using Sensititre automated microbroth dilution and Etest agar gradient diffusion methods. J Antimicrob Chemother 60:555–567. doi: 10.1093/jac/dkm213. [DOI] [PubMed] [Google Scholar]
- 19.Adimpong DB, Sørensen KI, Thorsen L, Stuer-Lauridsen B, Abdelgadir WS, Nielsen DS, Derkx PMF, Jespersen L. 2012. African traditional bread production and characterization of the bacitracin operon and bacitracin biosynthesis. Appl Environ Microbiol 78:7903–7914. doi: 10.1128/AEM.00730-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noor Uddin GM, Larsen MH, Christensen H, Aarestrup FM, Phu TM, Dalsgaard A. 2015. Identification and antimicrobial resistance of bacteria isolated from probiotic products used in shrimp culture. PLoS One 10:e0132338. doi: 10.1371/journal.pone.0132338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunlap CA, Kwon SW, Rooney AP, Kim SJ. 2015. Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste. Int J Syst Evol Microbiol 65:3487–3492. doi: 10.1099/ijsem.0.000441. [DOI] [PubMed] [Google Scholar]
- 23.Dunlap CA, Kim SJ, Kwon SW, Rooney AP. 2016. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and “Bacillus oryzicola” are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int J Syst Evol Microbiol 66:1212–1217. doi: 10.1099/ijsem.0.000858. [DOI] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute (CLSI). 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline, 2nd ed CLSI document M45-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Kelly J, Dideberg O, Charlier P, Wery J, Libert M, Moews P, Knox JR, Duez C, Fraipont C, Joris B, Dusart J, Frère JM, Ghuyson JN. 1986. On the origin of bacterial resistance to penicillin: comparison of a beta-lactamase and a penicillin target. Science 231:1429–1431. doi: 10.1126/science.3082007. [DOI] [PubMed] [Google Scholar]
- 26.European Committee on Antimicrobial Susceptibility Testing. 2017. MIC distributions and epidemiological cut-off values (ECOFF) setting, EUCAST SOP 10.0 European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/EUCAST_SOP_10.0_MIC_distributions_and_epidemiological_cut-off_value__ECOFF__setting_20171117.pdf. [Google Scholar]
- 27.Mouton JW, Brown DFJ, Apfalter P, Cantón R, Giske CG, Ivanova M, MacGowan AP, Rodloff A, Soussy CJ, Steinbakk M, Kahlmeter G. 2012. The role of pharmacokinetics/pharmacodynamics in setting clinical MIC breakpoints: the EUCAST approach. Clin Microbiol Infect 18:E37–E45. doi: 10.1111/j.1469-0691.2011.03752.x. [DOI] [PubMed] [Google Scholar]
- 28.Kehrenberg C, Ojo KK, Schwarz S. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J Antimicrob Chemother 54:936–939. doi: 10.1093/jac/dkh457. [DOI] [PubMed] [Google Scholar]
- 29.Noguchi N, Katayama J, O'Hara K. 1996. Cloning and nucleotide sequence of the mphB gene for macrolide 2′-phosphotransferase II in Escherichia coli. FEMS Microbiol Lett 144:197–202. doi: 10.1111/j.1574-6968.1996.tb08530.x. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. 2017. Critically important antimicrobials for human medicine, 5th rev World Health Organization, Geneva, Switzerland. [Google Scholar]
- 31.Ohmiya K, Tanaka T, Noguchi N, O'Hara K, Kono M. 1989. Nucleotide sequence of the chromosomal gene coding for the aminoglycoside 6-adenylyltransferase from Bacillus subtilis Marburg 168. Gene 78:377–378. doi: 10.1016/0378-1119(89)90241-2. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi Y, Ohshima H, Tsuge K, Itaya M. 2003. Far different levels of gene expression provided by an oriented cloning system in Bacillus subtilis and Escherichia coli. FEMS Microbiol Lett 221:125–130. doi: 10.1016/S0378-1097(03)00171-X. [DOI] [PubMed] [Google Scholar]
- 33.Sakaguchi R, Amano H, Shishido K. 1988. Nucleotide sequence homology of the tetracycline-resistance determinant naturally maintained in Bacillus subtilis Marburg 168 chromosome and the tetracycline-resistance gene of B. subtilis plasmid pNS1981. Biochim Biophys Acta 950:441–444. doi: 10.1016/0167-4781(88)90142-X. [DOI] [PubMed] [Google Scholar]
- 34.Levy SB, McMurry LM, Barbosa TM, Burdett V, Courvalin P, Hillen W, Roberts C, Rood JI, Taylor DE. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother 43:1523–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz S, Cardoso M, Wegener HC. 1992. Nucleotide sequence and phylogeny of the tet(L) tetracycline resistance determinant encoded by plasmid pSTE1 from Staphylococcus hyicus. Antimicrob Agents Chemother 36:580–588. doi: 10.1128/AAC.36.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob Agents Chemother 51:483–487. doi: 10.1128/AAC.01340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hue KK, Bechhofer DH. 1992. Regulation of the macrolide-lincosamide-streptogramin B resistance gene ermD. J Bacteriol 174:5860–5868. doi: 10.1128/jb.174.18.5860-5868.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xavier BB, Das AJ, Cochrane G, De Ganck S, Kumar-Singh S, Aarestrup FM, Goossens H, Malhotra-Kumar S. 2016. Consolidating and exploring antibiotic resistance gene data resources. J Clin Microbiol 54:851–859. doi: 10.1128/JCM.02717-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 40.Jolley KA, Maiden MCJ. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.International Organization for Standardization. 2010. Milk and milk products-determination of minimal inhibitory concentration (MIC) of antibiotic applicable to bifidobacteria and non-enterococcal lactic acid bacteria (LAB). ISO 10932:2010 (IDF 223:2010) International Organization for Standardization, Geneva, Switzerland: https://www.iso.org/standard/46434.html. [Google Scholar]
- 42.National Committee for Clinical Laboratory Standards (NCCLS). 2001. Development of in vitro susceptibility testing criteria and quality control parameters; approved guideline, 2nd ed NCCLS document M23-A2. National Committee for Clinical Laboratory Standards, Wayne, PA. [Google Scholar]
- 43.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.