This study revealed the rumen bacteriome from a large dairy cattle cohort (n = 334) raised under the same management and showed the linkages among the rumen core and pan bacteriomes, rumen short-chain fatty acids, and milk production phenotypes. The findings from this study suggest that the pan rumen bacteriome, together with the core bacteriome, potentially contributes to variations in host milk production traits. Fundamental knowledge on the rumen core and pan microbiomes and their roles in contributing to lactation performance provides novel insights into future strategies for manipulating rumen microbiota to enhance milk production in dairy cattle.
KEYWORDS: core rumen bacteriome, pan rumen bacteriome, interindividual variation, milk production phenotypes, dairy cattle
ABSTRACT
Currently, knowledge on the extent to which rumen microbiota differ in a large population of cattle fed the same diet and whether such differences are associated with animal performance is limited. This study was conducted to characterize the rumen microbiota of a large cohort of lactating Holstein dairy cows (n = 334) that were fed the same diet and raised under the same environment, aiming to uncover linkages between core and pan rumen microbiomes and host phenotypes. Amplicon sequencing of the partial 16S rRNA gene identified 391 bacterial genera in the pan bacteriome and 33 genera in the core bacteriome. Interanimal variation existed in the pan and core bacteriomes, with the effect of lactation stage being more prominent than that of parity (the number of pregnancies, ranging from 2 to 7) and sire. Spearman's correlation network analysis revealed significant correlations among bacteria, rumen short-chain fatty acids, and lactation performance, with the core and noncore genera accounting for 53.9 and 46.2% of the network, respectively. These results suggest that the pan rumen bacteriome together with the core bacteriome potentially contributes to variations in milk production traits. Our findings provide an understanding of the potential functions of noncore rumen microbes, suggesting the possibility of enhancing bacterial fermentation using strategies to manipulate the core and noncore bacteriomes for improved cattle performance.
IMPORTANCE This study revealed the rumen bacteriome from a large dairy cattle cohort (n = 334) raised under the same management and showed the linkages among the rumen core and pan bacteriomes, rumen short-chain fatty acids, and milk production phenotypes. The findings from this study suggest that the pan rumen bacteriome, together with the core bacteriome, potentially contributes to variations in host milk production traits. Fundamental knowledge on the rumen core and pan microbiomes and their roles in contributing to lactation performance provides novel insights into future strategies for manipulating rumen microbiota to enhance milk production in dairy cattle.
INTRODUCTION
Ruminants convert a wide variety of indigestible feed plant mass into human-edible products, such as milk (a low-cost product with rich nutrients and high proteins), and this process is mediated mainly by microbial degradation and fermentation in the rumen. The rumen harbors a diverse microbial population, including bacteria, archaea, protozoa, and fungi (1). Recent studies have revealed that the rumen microbiome may directly or indirectly contribute to animal performance. For instance, differences in rumen microbial compositions have been linked to feed efficiency in both beef (2, 3) and dairy (4) cattle. Recent studies have also revealed that genetic and functional aspects of the rumen microbiome are associated with feed efficiency of dairy (5) and beef (6) cattle, highlighting that manipulation of both compositional and functional outcomes of the rumen microbiome is vital for managing cattle and improving feed efficiency. The rumen microbial metagenome is also associated with methane production in sheep (7) and cattle (8). Therefore, improving rumen microbial functionality is vital for achieving better production and having fewer negative environmental impacts.
Recent molecularly based microbial identifications have advanced our understanding of the composition and function of rumen microbiota. Many efforts have been made to survey rumen microbiota, revealing that numerous factors can affect the rumen microbial composition, with diet being the main driving force for the microbial compositional shift (9). In addition, multiple studies have reported the “core rumen microbiota” in beef cattle (6, 10) and dairy cows (11, 12), as well as that among all ruminants (13). However, most of these previous studies involved only a small number of animals (6, 10–12) or lacked standardized variables, such as age, gender, and diet (13). A few studies have investigated associations between rumen microbiota and milk composition (4), how the rumen microbiota shifts during lactation, and how cattle breeds affect such relationships (4, 14, 15), indicating that the rumen microbiota can be prominently affected by the lactation stage. However, these studies used only a few animals (sample sizes of 15, 20, and 23, respectively), and the identified linkage between rumen microbiota and milk production-related traits needs to be verified in a larger population.
To date, how and to what extent these factors affect the rumen microbiota, especially in dairy cows, remain unknown. In this study, the composition of the rumen bacteriome was assessed in a large cohort (n = 334) of dairy cows with the aim of identifying (i) to what extents rumen microbiota differ when the cows are fed the same diet under the same management regime and (ii) whether the core and/or pan bacteriome contributes to the variation in rumen fermentation measurements and subsequent cow phenotypes. We focused on the bacteriome because among the complex rumen microbes, bacteria are the major microbial group contributing to the production of volatile fatty acids (VFAs) and microbial proteins that are utilized by dairy cows for milk production, and the shifts in ruminal VFAs and microbial proteins can directly affect milk yield and milk protein content (16). Fundamental knowledge of the rumen core and pan bacteriomes and their relationships with lactation performance will provide novel insights into the manipulation of rumen microbiota for enhancing milk production.
RESULTS
Identification of pan and core rumen microbiota.
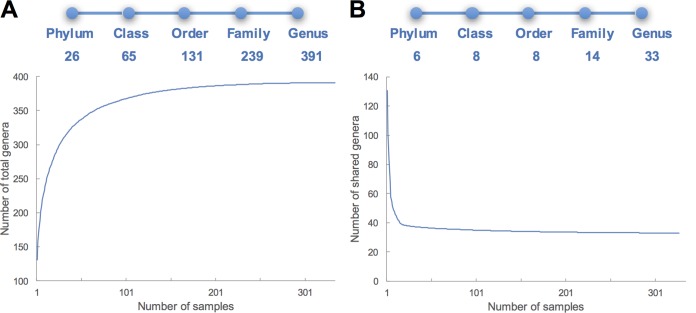
Amplicon sequencing generated a total of 18,613,732 high-quality sequences, which were assigned to 55,730 operational taxonomic units (OTUs) from 334 individuals. The distributions of alpha diversity indices showed that Shannon indices ranged from 4.91 to 6.52 (coefficient of variation [CV] = 6.2%), and Chao1 indices ranged from 2,127 to 2,931 (CV = 16.2%) (see Fig. S1 and Table S1 in the supplemental material). Totals of 26 phyla, 65 classes, 131 orders, 239 families, and 391 genera were identified and defined as the pan rumen bacteriome (Fig. 1A).
FIG 1.
Pan and core rumen bacteriomes identified across the cohort of Holstein dairy cows (n = 334). (A) Total number of genera comprising the rumen pan bacteria increasing with numbers of individuals of the cohort. (B) Number of core genera (defined as present in all individuals) decreasing with numbers of individuals of the cohort.
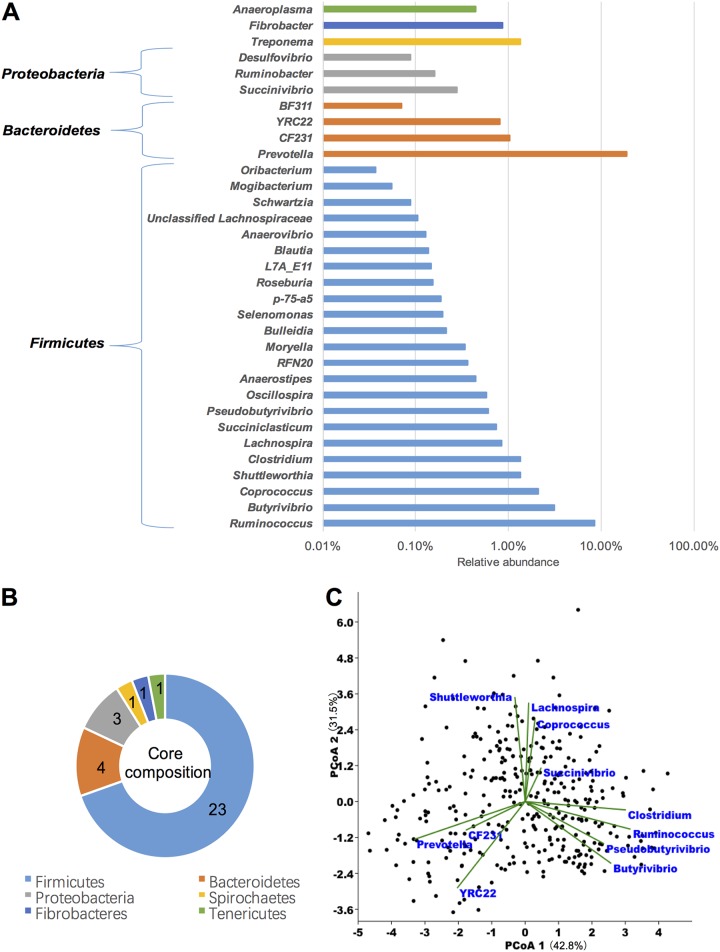
Certain bacterial taxa were detected in the rumen of all 334 cows, with 33 genera belonging to 6 phyla identified and thus defined as the core bacteriome (Fig. 1B and 2A). These phyla accounted for 45.53% ± 6.42% (mean ± standard error of the mean [SEM]) of all bacterial sequences, which included Firmicutes (21.67% ± 0.18%, 23 genera), Bacteroidetes (20.68% ± 0.49%, 4 genera), Proteobacteria (0.52% ± 0.01%, 3 genera), Spirochaetes (1.35% ± 0.04%, 1 genus), Fibrobacteres (0.86% ± 0.02%, 1 genus), and Tenericutes (0.44% ± 0.01%, 1 genus) (Fig. 2B; see also Table S2 in the supplemental material). From the identified core genera, eight of them were considered abundant core genera (relative abundance, >1%), including Prevotella (18.79% ± 0.46%), Ruminococcus (8.42% ± 0.10%), Butyrivibrio (3.14% ± 0.05%), Coprococcus (2.10% ± 0.03%), Treponema (1.35% ± 0.04%), Shuttleworthia (1.33% ± 0.05%), Clostridium (1.33% ± 0.02%), and CF231 (1.02% ± 0.03%). Notably, large interanimal variations in the abundances of these core taxa were observed, with the CVs ranging from 14.1 to 64.8% (Table S2). In addition, principal-coordinate analysis (PCoA) based on genus-level Bray-Curtis dissimilarity identified the top genera contributing to rumen bacteriome variation, including the above eight core abundant genera together with Succinivibrio, Lachnospira, and YRC22 (Fig. 2C). Among them, the abundant core genera contributed to 73% of the total sequences.
FIG 2.
Core rumen bacteriome. (A) Distribution of core bacterial genera. (B) Composition of core bacterial phyla. (C) Rumen bacterial community variation in the cohort, represented by principal-coordinate analysis (PCoA) based on genus-level Bray-Curtis dissimilarity. The top 11 contributors to community variation were presented.
Nondietary effects on rumen microbiota.
As all the cows were from the same breed, fed the same diet, and raised under the same environment and management regime, we further explored the nondietary effects, including parity, lactation stage, and sire, on the rumen microbiota.
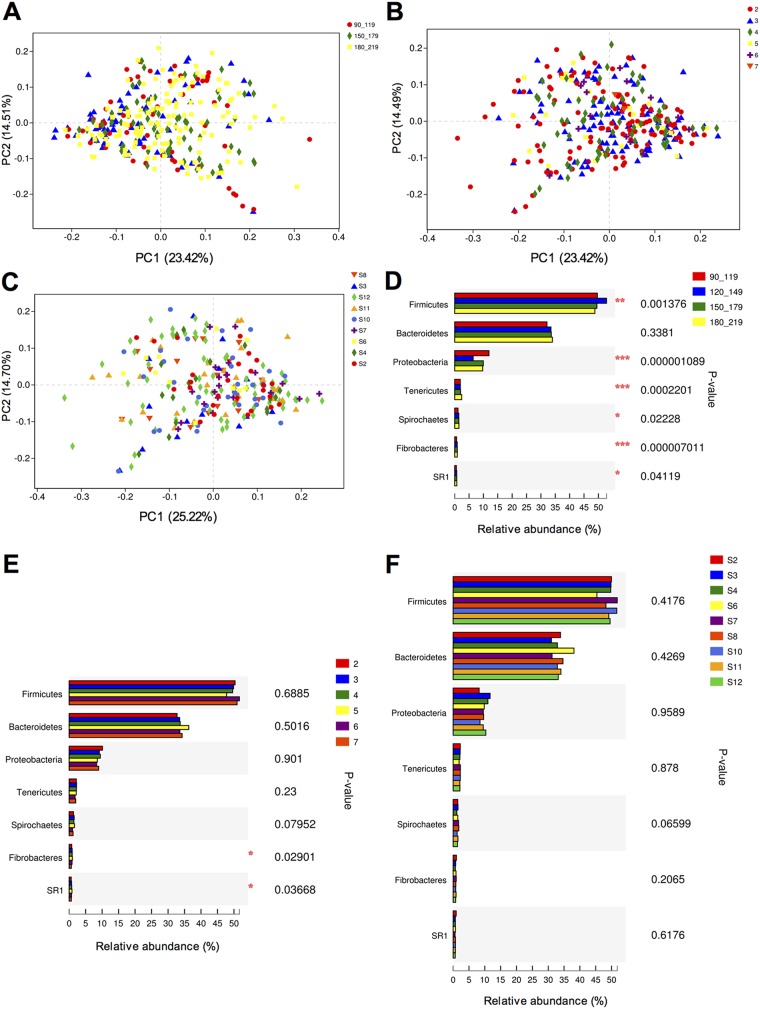
The lactation period of the entire cohort ranged from 90 to 219 days in milk (DIM). To define the lactation stage that affected the rumen microbiota throughout the whole period, we first analyzed the relationship between rumen VFAs and DIM (see Fig. S2 in the supplemental material). We speculated that differences in rumen fermentation measurements were the result of microbial compositional shifts. When the total VFAs were compared, significant differences (P < 0.01) were detected among the following four DIM groups: 90 to 19 DIM (106.97 ± 1.75 mmol/liter), 120 to 149 DIM (97.62 ± 2.18 mmol/liter), 150 to 179 DIM (97.75 ± 1.81 mmol/liter), and 180 to 219 DIM (106.15 ± 1.78 mmol/liter) (Fig. S2 and Tables S3 and S4 in the supplemental material). Therefore, these four DIM groups were used to identify the lactation stage effects on rumen microbiota. PCoA plots (based on the Bray-Curtis distance) revealed no separation in bacterial composition at the genus level according to either parity or DIM (Fig. 3A and B). Further Kruskal-Wallis H analysis of the relative abundances of abundant taxa (pan bacteria with a relative abundance of >0.10% in at least 60% of the cows) at the phylum (Fig. 3D and E) and genus levels (see Fig. S3 in the supplemental material) revealed that the relative abundances of 3 phyla and 17 genera were significantly affected by the DIM (P < 0.05), while those of 3 genera were affected by the parity (P < 0.05). No sire effect was detected by either PCoA analysis of bacterial profiles (Fig. 3C) or comparisons of relative abundances among the nine sire groups (Fig. 3F; see also Fig. S4 in the supplemental material).
FIG 3.
Nondietary effects on rumen microbiota. (A) Principal-coordinate analysis (PCoA) based on genus level Bray-Curtis dissimilarity in relation to days in milk (DIM); different colors represent different lactation period groups. (B) PCoA based on genus level Bray-Curtis dissimilarity in relation to parity; different colors represent different parity groups. (C) PCoA based on genus level Bray-Curtis dissimilarity in relation to sire; different colors represent sires of cows. (D to F) Relative abundances of seven predominant phyla between different DIM (D), parity (E), and sire (F) groups. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Kruskal-Wallis H test).
Relationship between ruminal VFAs and lactation traits and the rumen bacteriome.
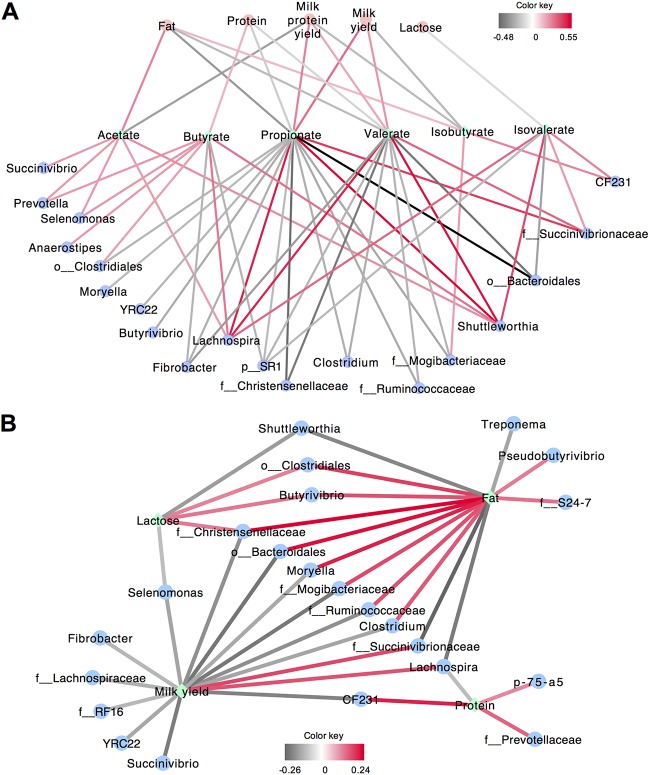
Several lactation performance parameters were measured (Table 1), including milk yield (31.74 ± 0.43 kg/day, CV = 24.9%), milk protein content (3.12% ± 0.02%, CV = 9.4%), milk fat content (3.20% ± 0.05%, CV = 28.5%), and lactose content (4.97% ± 0.01%, CV = 4.3%) (see Table S5 in the supplemental material). The distributions of lactation performance parameters also showed variations in lactation performance among the cohort (CVs ranging from 4.34 to 24.94%; see also Fig. S5 in the supplemental material). Additionally, the VFA concentrations also had high variability, reflective of the high CV values for each individual VFA, including acetate (70.33 ± 0.65 mmol/liter, CV = 17.0%), propionate (17.53 ± 0.27 mmol/liter, CV = 28.1%), butyrate (10.96 ± 0.14 mmol/liter, CV = 23.5%), isobutyrate (0.98 ± 0.02 mmol/liter, CV = 28.7%), valerate (1.16 ± 0.01 mmol/liter, CV = 22.0%), and isovalerate (1.47 ± 0.02 mmol/liter, CV = 21.1%) (see Fig. S6 and Table S5 in the supplemental material). Analysis of Spearman correlations between lactation traits and rumen fermentation measurements showed that several VFAs were significantly associated in a moderate way with milk yield and composition (0.1 < ∣R∣ < 0.4, P < 0.05), and these VFAs were also correlated with the relative abundances of specific bacterial genera (0.2 < ∣R∣ < 0.6, P < 0.05) (Fig. 4A; see also Tables S6 and S7 in the supplemental material). Specifically, positive correlations were found between milk yield and proportions of propionate (R = 0.31, P < 0.05) and valerate (R = 0.23, P < 0.05). These two VFAs were positively correlated with relative abundances of the genera Lachnospira and Shuttleworthia as well as with an unclassified genus from the family Succinivibrionaceae (0.33 < R < 0.55, P < 0.05). Milk protein content was positively correlated with butyrate proportion (R = 0.16, P < 0.05), and butyrate was positively correlated with the relative abundances of the genera Prevotella, Selenomonas, Anaerostipes, Lachnospira, and Shuttleworthia as well as with an unclassified genus that belongs to the order Clostridiales (0.20 < R < 0.30, P < 0.05). Positive correlations were found between milk fat content and acetate proportion (R = 0.26, P < 0.05), while acetate concentration was positively correlated with the genera Succinivibrio, Prevotella, Selenomonas, Lachnospira, Shuttleworthia and an unclassified genus from the family Succinivibrionaceae (0.21 < R < 0.43, P < 0.05). Positive correlations were also detected between milk fat content and isobutyrate proportion (R = 0.15, P < 0.05), and isobutyrate was positively correlated with relative abundances of the genus CF231 and an unclassified genus from the family Mogibacteriaceae (0.20 < R < 0.26, P < 0.05).
TABLE 1.
Lactation performance and rumen fermentation measurements
| Measurement | Mean | SEM |
|---|---|---|
| Lactation performance measures | ||
| Milk yield (kg/day) | 31.74 | 0.43 |
| Milk protein (%) | 3.12 | 0.02 |
| Milk fat (%) | 3.20 | 0.05 |
| Milk lactose (%) | 4.97 | 0.01 |
| Rumen fermentation products (mmol/liter) | ||
| Acetate | 70.33 | 0.65 |
| Propionate | 17.53 | 0.27 |
| Isobutyrate | 0.98 | 0.02 |
| Butyrate | 10.96 | 0.14 |
| Isovalerate | 1.47 | 0.02 |
| Valerate | 1.16 | 0.01 |
| Total VFAs | 102.44 | 1.01 |
FIG 4.
Correlation networks showing associations between lactation performance, rumen fermentation products, and bacterial genera (with relative abundance of >0.1% in at least 60% of all the cows). (A) The correlation network of lactation performance parameters, rumen fermentation parameters, and rumen bacterial genera. Only significant (P < 0.05) correlations were chosen to be displayed in the network. (B) The correlation network of lactation performance and rumen bacterial genera. Only strong (correlation coefficient R > 0.2 or <−0.2) and significant (P < 0.05) correlations were chosen to be displayed in the network. The edge width and color (red, positive; black, negative) are proportional to the correlation strength.
Relationship between the rumen bacteriome and lactation performance.
Further Spearman correlation analysis was performed to detect whether and how the rumen bacteriome could be attributed to milk yield and composition. Significant associations (P < 0.05) were found between lactation performance and alpha diversity indices (Shannon index and Chao1 index) (see Fig. S7 in the supplemental material). The Spearman correlation coefficients revealed several significant linkages (P < 0.05), including a negative association between milk yield and the Shannon index (R = −0.24, P < 0.05); positive associations between lactose (R = 0.15, P < 0.05), milk fat (R = 0.26, P < 0.05), and milk protein (R = 0.19, P < 0.05) and the Shannon index; a negative association between milk yield and the Chao1 index (R = −0.17, P < 0.05); and a positive association between milk fat and the Chao1 index (R = 0.13, P < 0.05) (Fig. S7).
Further Spearman correlation analysis based on abundant bacterial genera revealed the networks among bacterial taxa and milk-related traits. The correlation networks contain 26 genera in total, with the core and noncore genera accounting for 53.9 and 46.2% (14 core genera and 12 noncore genera), respectively (Fig. 4B; see also Table S8 in the supplemental material). Milk yield possessed the most complex relationships with bacterial genera, including two positive relationships (R > 0.16, P < 0.05) and 13 negative relationships (R < −0.12, P < 0.05) (Fig. 4B). Among them, the two positive correlations between milk yield and the relative abundance of the genus Lachnospira (R = 0.16, P < 0.05) and that of an unclassified genus belonging to the Succinivibrionaceae family (R = 0.17, P < 0.05) (Fig. 4B) were consistent with the positive relationships between the relative abundances of these bacterial taxa and proportions of propionate and valerate (0.33 < R < 0.55, P < 0.05) (Fig. 4A). Milk fat was positively correlated with relative abundances of nine genera (R > 0.12, P < 0.05), including Butyrivibrio, Pseudobutyrivibrio, Clostridium, and Moryella and unclassified genera belonging to the families Christensenellaceae, Mogibacteriaceae, Ruminococcaceae, Clostridiales, and S24-7, and the order Bacteroidales. Milk fat was negatively correlated with the relative abundances of four genera (R < −0.15, P < 0.05), including Lachnospira, Shuttleworthia, and Treponema and an unclassified genus belonging to the Succinivibrionaceae (Fig. 4B). The positive relationship between milk fat and the relative abundance of an unclassified genus belonging to Mogibacteriaceae (R = 0.17, P < 0.05) was consistent with the positive relationship between this taxon and isobutyrate (R = 0.21, P < 0.05) (Fig. 4A). Milk protein was positively correlated with the relative abundances of three genera (R > 0.11, P < 0.05), including CF231 and p-75-a5 and an unclassified genus belonging to the family Prevotellaceae and was negatively correlated with the relative abundance of Lachnospira (R = −0.11, P < 0.05) (Fig. 4B). Among the genera in the correlation network, as shown in Fig. 4B, Pseudobutyrivibrio, Treponema, Shuttleworthia, Butyrivibrio, Moryella, Clostridium, Lachnospira, Selenomonas, Fibrobacter, Succinivibrio, p-75-a5, CF231, and YRC22 were core bacterial taxa.
DISCUSSION
Although recent studies have identified the core rumen microbiome of cattle (6, 11, 13, 17), our study has provided a more comprehensive understanding of the core rumen bacteriome using a large number of dairy cows under the same dietary conditions and management. Some of the core bacterial taxa identified in our study are consistent with those identified in previous studies (6, 11, 13, 17). For example, the genus Prevotella, which utilizes starch and proteins to mainly produce succinate and acetate (18), was the most abundant (18.8%) in the rumen of lactating dairy cows. The genus Ruminococcus, which can break down fibrous plant materials to produce acetate, formate, succinate, and others (13), was identified as the second most predominant (8.42%) core taxon in this study. The genus Butyrivibrio, which has functions including fiber degradation, protein breakdown, and butyrate production, was the third most abundant genus (3.14%) in our core bacteriome (19). The consistent presence of these genera as core taxa in the rumen suggests their vital functions in the rumen ecological niches of dairy cows. However, our results indicate that the abundances of these core bacterial genera varied across individuals and the bacteriome beta diversity variation was driven mainly by differences in the relative abundances of the abundant core taxa, suggesting that the abundant core members significantly contribute to the interindividual variations of bacterial composition in the rumen.
To date, only a few studies have focused on identifying the core bacteriome in dairy cows. Notably, some taxa were detected only as members of the core rumen bacteriome of dairy cows in our study. For example, compared with the 32 core bacterial genera identified from 16 dairy cows by Jami and Mizrahi (11), only 16 of them were part of core bacterial taxa in our study (see Fig. S8 in the supplemental material). Among these 16 genera, Fibrobacter, Mogibacterium, Treponema, CF231, YRC22, and BF311 were identified as core bacterial taxa in the rumen of various species (e.g., cattle, sheep, goats, cervids, and camelids) from different geographic locations (13), suggesting that colonization of these taxa is likely driven by ruminants. The interstudy variations in the identified core rumen bacteriome of dairy cows may be attributed to the differences in dietary conditions (forage-to-concentrate ratio), geographical locations, management regimes, and/or lactation stages among different studies. For instance, the phylum Fibrobacteres, was identified as a core rumen bacterial taxon of dairy cows only in this study. This may be because the forage-to-concentrate ratio in our study (45:55) was higher than that in the previous study (30:70) (11). Organisms belonging to this phylum are involved in cellulose degradation and have been reported to be influenced by diet (20–22). The identification of Fibrobacteres as part of the core bacterial taxa suggests its essential role in the fibrolytic community of rumen ecosystems in dairy cows, especially in cows under a corn-based high-grain diet. The core bacteriome identified in this study indicates that some taxa occupy certain specific ecosystems in the rumen and play important roles in rumen ecological niches under certain dietary conditions and management regimes.
Since all the animals were fed the same diet and raised under the same feeding and management regimes, the interanimal variations in the rumen bacteriome in this study may be explained by animal genetics (sire), age (parity), and lactation stage. Our results revealed that among these factors, lactation stage is a more effective driver than parity and sire. The prominent effect of lactation stage on rumen bacterial communities has been previously reported by Bainbridge et al. (14) who showed that the three most predominant phyla (Bacteroidetes, Firmicutes, and Proteobacteria) were significantly affected by lactation stage. Although the Bainbridge et al. study used primiparous (parity = 1) cows from various breeds across three lactation stages (early [3 DIM], middle [93 and 183 DIM], and late [273 DIM]), the effect of lactation stage on rumen bacterial communities was consistent between the two studies. Due to host physiological (such as energy demand, feed intake, hormone, and metabolism) changes across the lactation period, it is not surprising to detect different lactation stages (early, middle, and late) affecting the rumen microbiota, as reported by Bainbridge et al. (14). However, the further observation of a lactation effect during the midlactation stage in our study suggests that the shifts in microbiota also exist in the period when host physiology is relatively stable. Age is also a determinant of the rumen bacterial composition, and most previous studies focused on the establishment of the bacterial community in the rumen of preweaned calves (23, 24). Knowledge of the age effect on the adult rumen microbiota is limited. In our study, age (commonly indicated by parity of the dairy cows) did not affect the rumen bacterial composition, suggesting that there is a “stable” adult microbiota after establishment of the microbiome during early life. Furthermore, host genetics (such as breed and sire) has been reported to influence the rumen microbial composition in beef cattle (25, 26). However, to date, information on the genetic effects on the rumen microbiome in dairy cows is limited. Similar to our findings of no sire effect on rumen microbiota, Bainbridge et al. (14) also did not detect a breed effect when the rumen microbiota of three breeds (Holstein, Jersey, and Holstein × Jersey dairy cows) were compared. The discrepancy of results among various studies suggests that the mechanisms underlying the host genetic effects in previous studies may have been driven by multiple factors and thus need to be further investigated.
In this study, we further identified significant linkages between VFAs and lactation performance as well as with the abundance of rumen bacteria. For instance, a positive correlation was found between propionate proportions and milk yield. Propionate is the major precursor for gluconeogenesis in the liver of lactating cows, which contributes to 45 to 60% of the total glucose (27). It has been reported that increasing rumen propionate production through altering rumen microbiota, for example by introducing antibiotics such as monensin, ultimately improves feed efficiency, milk yield, and milk composition (28, 29). The positive correlation between propionate and milk yield in this study further confirms the important role of this VFA in milk production. In addition, positive relationships were also found between propionate proportion and the relative abundance of bacterial genera Lachnospira and Shuttleworthia and an unclassified genus belonging to the family Succinivibrionaceae in our study. As a major pectinolytic bacterial taxon inhabiting the rumen, Lachnospira has been reported to produce butyrate (30). The relationship between Lachnospira and propionate in our study suggests that some species belonging to this genus may positively interact with propionate-producing taxa in the rumen, which needs further validation in the future using the isolates from this genus. Taxa belonging to Succinivibrionaceae family have been reported to produce succinate (a precursor of propionate); thus, the detected positive relationship between an unclassified genus belonging to the family Succinivibrionaceae and propionate further indicates the involvement of this microbial group in propionate production. Our findings underline some potential contributions of rumen bacterial taxa to rumen VFAs, which influence lactation performance. In addition, the relationships between the rumen bacteriome and lactation performance, mainly milk yield and milk fat content, were identified through correlation network analysis. Recent studies have reported that milk fat possesses multiple and strong relationships with rumen microbes and microbial pathways in Holstein dairy cattle (4, 14) and buffalo (15). Compared with previous studies (4, 14, 15), a lower number of strong correlations in our study were observed, which could be due to the interindividual variations in both bacteriomes (CVs of core taxa abundances ranging from 14.1% to 64.8%) and phenotypes (CVs ranging from 4.3% to 28.5%). Nevertheless, compared with the small sample sizes in previous studies (ranging from 15 to 22), the linkage identified between rumen microbiota and milk traits in this study is more biologically sound based on the high statistical power, with data collected from hundreds (n = 334) of dairy cows, with fewer external variables. In addition, the abundance of some bacterial genera (e.g., Butyrivibrio) showed positive relationships with VFA concentrations in relation to milk production and composition (Fig. 4A) but showed contradictory relationships with milk production and composition (Fig. 4B). Such contradictory relationships could be due to the higher taxonomic level (genus) used for such analysis. It is known that different species within the same genus can have varied functions (e.g., Butyrivibrio spp.) (31); therefore, a further understanding based on deeper taxonomic levels (species and/or stain levels) and a functional analysis of identified bacterial taxa using bacterial cultures are needed to identify the causal relationships behind the identified correlations.
Moreover, our findings revealed that the core and pan bacterial genera accounted for 53.9 and 46.2% of the correlation network associated with milk-production traits, suggesting that core and pan bacteriomes may interact with each other to influence host performance. For example, the genus Sharpea was identified as a noncore genus in this study, accounting for 0.16% of the relative abundance, with large variation (CV = 196.6%). This taxon has been reported to produce lactate and has a high abundance in the rumen of sheep with low methane yield (27). In addition, two bacterial taxa, Bifidobacterium (0.02% on average, CV = 191.1%) and Lactobacillus (0.01% on average, CV = 110.0%), were also determined to be pan bacterial taxa in this study. These organisms produce lactate, which could lead to the lower pH in the rumen and metabolic dysfunction (such as ruminal acidosis) (27). The varied abundance of these taxa suggests that the health of rumen may also differ among the cows. Future studies that evaluate the methane emission and ruminal pH would provide more knowledge on how the pan bacteriome contributes to rumen function in terms of rumen metabolic health and fermentation efficiency.
In conclusion, this study identified variations in the rumen bacteriome, VFAs, and milk production traits among a large cohort of dairy cows. Interanimal variation existed in both the pan and core bacteriomes, with the effect of lactation stage being more prominent than those of parity and sire. In addition, candidate bacterial taxa from both the core bacteriome and the pan bacteriome were associated with the lactation performance of the host, which has provided a further understanding of the pan bacteriome that may contribute to various rumen functions, in addition to the core members in rumen, which are known to contribute to basic rumen functions. However, future studies on functional analysis through metagenomics and/or culture-based microbial genomics as well as quantitative validation using quantitative PCR are needed, which can provide more a comprehensive understanding of the causal relationships behind the identified correlations. Due to the complexity of the rumen microbiota, future studies on other microbial groups in the rumen, including archaea, protozoa, and fungi, are needed to provide a more comprehensive understanding of the core and pan rumen microbiomes and their roles in milk production of dairy cows.
MATERIALS AND METHODS
Animals and sampling.
The experimental procedures used in this study were approved by the Animal Care Committee of Zhejiang University (Hangzhou, China) and were in accordance with the university's guidelines for animal research.
A total of 334 Holstein dairy cows (parity, 3.12 ± 1.16; DIM, 159 ± 34 [mean ± SD]) housed at a commercial dairy farm were selected for the experiment. Cows with histories of disease were not included in the experiment. The animals were fed three times daily at 0630, 1400, and 2000 h at a forage-to-concentrate ratio of 45:55 and had free access to water. The cohort was grouped according to DIM (classified at four levels, ranging from 90 to 119, 120 to 149, 150 to 179, and 180 to 219 DIM), parity (classified into six groups, ranging from 2 to 7), and sire (cows with the same father, classified into nine groups) to detect the impact of nondietary factors on rumen bacterial communities in the cohort (see Table S4 in the supplemental material). Milk yield was recorded for three consecutive days. Milk samples were collected on the third day (sampling day) for the measurement of milk components (milk protein, milk fat, and lactose) by infrared analysis (32) with a spectrophotometer (Foss-4000; Foss Electric A/S, Hillerod, Denmark). Rumen fluid was collected using oral stomach tubes before the morning feeding on the sampling day, according to a previously reported procedure (33). Rumen digesta samples were stored at −80°C until further analysis.
VFA measurement.
To measure the concentrations of VFAs, including acetate, propionate, butyrate, valerate, isobutyrate, and isovalerate, 2 ml of rumen sample was mixed with 20 μl of 25% orthophosphate acid, and was then centrifuged at 10,000 rpm and 4°C for 10 min. The supernatant was then subjected to VFA measurement using a gas chromatograph (GC-2010, Shimadzu, Kyoto, Japan). The temperatures of the injector/detector and the column were 260 and 220°C, respectively. Nitrogen was used as a carrier. The total VFA concentration was calculated as sum of the six VFA concentrations, and the proportion of each VFA was calculated for further analysis (34).
DNA extraction and sequencing.
Total DNA was extracted from each rumen fluid sample using the bead-beating method (35). The qualities and quantities of the DNA samples were measured using the NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). DNA was amplified by using the 341F/806R primer set (341F, 5′-CCTAYGGGRBGCASCAG-3′; 806R, 5′-GGACTACNNGGGTATCTAAT-3′), which targets the V3 to V4 region of the bacterial 16S rRNA gene, with the reverse primer containing a 6-bp error-correcting barcode unique to each sample. The PCR solution contained 0.5 U of Taq polymerase (TransGen Biotech, Beijing, China) in 25 μl of 10× PCR buffer, 200 μM each dNTP, 0.2 μM each primer, and 2 μl of DNA (50 ng/μl). PCRs were performed using Phusion high-fidelity PCR mastermix (New England BioLabs Ltd., Beijing, China) with the following program: 94°C for 3 min; 35 cycles of 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s; followed by 72°C for 10 min. PCR products were visualized on 2% agarose gels and purified using the QIAquick gel extraction kit (Qiagen, Dusseldorf, Germany). Amplicon sequencing was conducted on an Illumina HiSeq platform using the paired-end 2 × 250-bp protocol (36).
Sequencing data analysis.
Paired-end reads were merged using FLASH v1.2.7 (see http://ccb.jhu.edu/software/FLASH/) (37). Sequences were demultiplexed and quality filtered using QIIME v1.7.0 (see http://qiime.org/index.html), and bases with quality scores higher than 20 were retained for further analysis (38). Chimeric sequences were identified and removed (see Table S9 in the supplemental material) using the UCHIME algorithm (39, 41). OTUs were clustered with a 97% similarity threshold using UPARSE v7.0.1001 (40), and taxonomy was assigned using the latest Greengenes database (May 2013 release). The alpha diversities of the bacterial communities were determined using various diversity indices (Chao1 and Shannon indexes) and calculated using the procedures within QIIME v1.7.0. Jackknifed beta diversity was calculated based on Bray-Curtis dissimilarity calculated by QIIME v1.7.0 and visualized by PCoA using PAST v3.18 (see http://folk.uio.no/ohammer/past/).
The core rumen bacterial microbiota was defined as taxa observed in 100% of the samples, and pan microbiota was defined as the total observed richness in all the samples. The contributions of bacterial genera to microbiome variation were derived from canonical correspondence analysis (CCA) using PAST v3.18.
Network analysis.
Correlation networks were generated to explore the relationships among rumen microbiota, rumen fermentation, and lactation performance. Spearman's correlation analysis between rumen bacterial taxa (existing in at least 60% of all the cows), VFAs, and lactation traits was performed using SPSS, with significant (P < 0.05) correlations chosen to be displayed in the network. Spearman's correlation analysis was performed directly between phenotypes and bacterial genera (with relative abundances of >0.10% in at least 60% of the cows), and significant correlation coefficients with R > 0.2 or <−0.2 (P < 0.05) were determined for generating the correlation network. The correlation networks were generated by Spearman's rank correlation coefficients and visualized by Cytoscape v3.2.1.
Statistical analysis.
Lactation performance (milk yield, milk fat content, milk protein content, lactose content, and milk protein yield) and rumen VFA (acetate, propionate, butyrate, isobutyrate, valerate, isovalerate, and total VFA) concentrations were analyzed using one-way ANOVA by SPSS (v22), with statistical significances declared at a P value of <0.05. The effects of parity, lactation stage, and sire on the abundances of bacteria phyla and genera (pan microbiota) were determined using the Kruskal-Wallis H test (PAST), with statistical significances declared at a P value of <0.05.
Accession number(s).
The identified sequences were deposited in the NCBI Sequence Read Archive (SRA) under the accession no. SRP149811.
Supplementary Material
ACKNOWLEDGMENTS
The research described herein was supported by grants from the National Natural Science Foundation of China (31472121 to J.L. and 31729004 to L.L.G. and J.L.), the University of Albert China Opportunity Fund and NSERC Discovery (to L.L.G.), and the China Agriculture (Dairy) Research System (CARS-36 to J.L.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00970-18.
REFERENCES
- 1.Deng W, Xi D, Mao H, Wanapat M. 2008. The use of molecular techniques based on ribosomal RNA and DNA for rumen microbial ecosystem studies: a review. Mol Biol Rep 35:265–274. doi: 10.1007/s11033-007-9079-1. [DOI] [PubMed] [Google Scholar]
- 2.Carberry CA, Kenny DA, Han S, Mccabe MS, Waters SM. 2012. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl Environ Microbiol 78:4949–4958. doi: 10.1128/AEM.07759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernandez-Sanabria E, Goonewardene LA, Wang Z, Durunna ON, Moore SS, Guan Luo L. 2012. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl Environ Microbiol 78:1203–1214. doi: 10.1128/AEM.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jami E, White BA, Mizrahi I. 2014. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One 9:e85423. doi: 10.1371/journal.pone.0085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shabat SKB, Sasson G, Doronfaigenboim A, Durman T, Yaacoby S, Miller MEB, White BA, Shterzer N, Mizrahi I. 2016. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J 10:2958–2972. doi: 10.1038/ismej.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, Guan LL. 2017. Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. Appl Environ Microbiol 83:e00061-17. doi 10.1128/AEM.00061-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi W, Moon CD, Leahy SC, Kang D, Froula J, Kittelmann S, Fan C, Deutsch S, Gagic D, Seedorf H, Kelly WJ, Atua R, Sang C, Soni P, Li D, Pinares-Patino CS, McEwan JC, Janssen PH, Chen F, Visel A, Wang Z, Attwood GT, Rubin EM. 2014. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res 24:1517–1525. doi: 10.1101/gr.168245.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace RJ, Rooke JA, McKain N, Duthie CA, Hyslop JJ, Ross DW, Waterhouse A, Watson M, Roehe R. 2015. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:1–14. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghaffari MH, Tahmasbi AM, Khorvash M, Naserian AA, Ghaffari AH, Valizadeh H. 2014. Effects of pistachio by-products in replacement of alfalfa hay on populations of rumen bacteria involved in biohydrogenation and fermentative parameters in the rumen of sheep. J Anim Physiol Anim Nutr (Berl) 98:578–586. doi: 10.1111/jpn.12120. [DOI] [PubMed] [Google Scholar]
- 10.Petri RM, Schwaiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, McAllister TA. 2013. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS One 8:e83424. doi: 10.1371/journal.pone.0083424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jami E, Mizrahi I. 2012. Composition and similarity of bovine rumen microbiota across individual animals. PLoS One 7:e33306. doi: 10.1371/journal.pone.0033306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lettat A, Benchaar C. 2013. Diet-induced alterations in total and metabolically active microbes within the rumen of dairy cows. PLoS One 8:e60978. doi: 10.1371/journal.pone.0060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson G, Cox F, Ganesh S, Jonker A, Young W, Janssen PH. 2015. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep 5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bainbridge ML, Cersosimo LM, Wright AD, Kraft J. 2016. Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein × Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol Ecol 92:fiw059. doi: 10.1093/femsec/fiw059. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Xu C, Huo D, Hu Q, Peng Q. 2017. Comparative study of the gut microbiome potentially related to milk protein in Murrah buffaloes (Bubalus bubalis) and Chinese Holstein cattle. Sci Rep 7:42189. doi: 10.1038/srep42189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berthiaume R, Benchaar C, Chaves AV, Tremblay GF, Castonguay Y, Bertrand A, Belanger G, Michaud R, Lafreniere C, McAllister TA, Brito AF. 2010. Effects of nonstructural carbohydrate concentration in alfalfa on fermentation and microbial protein synthesis in continuous culture. J Dairy Sci 93:693–700. doi: 10.3168/jds.2009-2399. [DOI] [PubMed] [Google Scholar]
- 17.Kim M, Yu Z. 2014. Variations in 16S rRNA-based microbiome profiling between pyrosequencing runs and between pyrosequencing facilities. J Microbiol 52:355–365. doi: 10.1007/s12275-014-3443-3. [DOI] [PubMed] [Google Scholar]
- 18.Stevenson DM, Weimer PJ. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 75:165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- 19.Forster R, Gong J, Teather R. 1997. Group-specific 16S rRNA hybridization probes for determinative and community structure studies of Butyrivibrio fibrisolvens in the rumen. Appl Environ Microbiol 63:1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tajima K, Aminov RI, Nagamine T, Matsui H, Nakamura M, Benno Y. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl Environ Microbiol 67:2766–2774. doi: 10.1128/AEM.67.6.2766-2774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brulc J, Antonopoulos D, Miller M, Wilson M, Yannarell A, Dinsdale E, Edwards R, Frank E, Emerson J, Wacklin P, Coutinho P, Henrissat B, Nelson K, White B. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci U S A 106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernando SC, Purvis HT II, Najar FZ, Sukharnikov LO, Krehbiel CR, Nagaraja TG, Roe BA, Desilva U. 2010. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl Environ Microbiol 76:7482–7490. doi: 10.1128/AEM.00388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li RW, Connor EE, Li C, Baldwin RL VI, Sparks ME. 2012. Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environ Microbiol 14:129–139. doi: 10.1111/j.1462-2920.2011.02543.x. [DOI] [PubMed] [Google Scholar]
- 24.Jami E, Israel A, Kotser A, Mizrahi I. 2013. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J 7:1069–1079. doi: 10.1038/ismej.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez-Sanabria E, Goonewardene LA, Wang Z, Zhou M, Moore SS, Guan LL. 2013. Influence of sire breed on the interplay among rumen microbial populations inhabiting the rumen liquid of the progeny in beef cattle. PLoS One 8:e58461. doi: 10.1371/journal.pone.0058461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roehe R, Dewhurst RJ, Duthie CA, Rooke JA, Mckain N, Ross DW, Hyslop JJ, Waterhouse A, Freeman TC, Watson M. 2016. Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet 12:e1005846. doi: 10.1371/journal.pgen.1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagaraja TG, Newbold CJ, Nevel CJV, Demeyer DI. 1997. Manipulation of ruminal fermentation, p 523–632. In Hobson PN, Stewart CS (ed), The rumen microbial ecosystem. Springer, Rotterdam, The Netherlands. [Google Scholar]
- 28.Wiltrout DW, Satter LD. 1972. Contribution of propionate to glucose synthesis in the lactating and nonlactating cow. J Dairy Sci 55:307–317. doi: 10.3168/jds.S0022-0302(72)85487-0. [DOI] [PubMed] [Google Scholar]
- 29.Thornton JH, Owens FN. 1981. Monensin supplementation and in vivo methane production by steers. J Anim Sci 52:628–634. doi: 10.2527/jas1981.523628x. [DOI] [PubMed] [Google Scholar]
- 30.Duffield TF, Merrill JK, Bagg RN. 2012. Meta-analysis of the effects of monensin in beef cattle on feed efficiency, body weight gain, and dry matter intake. J Anim Sci 90:4583–4592. doi: 10.2527/jas.2011-5018. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Hatem A, Catalyurek UV, Morrison M, Yu Z. 2013. Metagenomic insights into the carbohydrate-active enzymes carried by the microorganisms adhering to solid digesta in the rumen of cows. PLoS One 8:e78507. doi: 10.1371/journal.pone.0078507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laporte MF, Paquin P. 1999. Near-infrared analysis of fat, protein, and casein in cow's milk. J Agri Food Chem 47:2600–2605. doi: 10.1021/jf980929r. [DOI] [PubMed] [Google Scholar]
- 33.Imhasly S, Naegeli H, Baumann S, Von BM, Luch A, Jungnickel H, Potratz S, Gerspach C. 2014. Metabolomic biomarkers correlating with hepatic lipidosis in dairy cows. BMC Vet Res 10:1–9. doi: 10.1186/1746-6148-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang JK, He B, Du W, Luo Y, Yu Z, Liu JX. 2015. Yeast with surface displayed xylanase as a new dual purpose delivery vehicle of xylanase and yeast. Animal Feed Sci Technol 208:44–52. doi: 10.1016/j.anifeedsci.2015.07.002. [DOI] [Google Scholar]
- 35.Li M, Penner GB, Hernandez-Sanabria E, Oba M, Guan LL. 2009. Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. J Appl Microbiol 107:1924–1934. doi: 10.1111/j.1365-2672.2009.04376.x. [DOI] [PubMed] [Google Scholar]
- 36.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso J, Kuczynski J, Stombaugh J, Bittinger K, Bushman F. 2010. QIIME allows integration and analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 41.Edgar R. 2016. UCHIME2: improved chimera prediction for amplicon sequencing. bioRxiv doi: 10.1101/074252. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.