Microbial secondary metabolites are an important source of biologically active and therapeutically relevant small molecules. However, much of this active molecular diversity is challenging to access due to low production levels or difficulty in discerning secondary metabolites within complex microbial extracts prior to isolation. Here, we demonstrate that ecological stimuli increase secondary metabolite production in phylogenetically diverse actinobacteria isolated from understudied hypogean environments. Additionally, we show that comparative metabolomics linking stimuli to metabolite response data can effectively reveal secondary metabolites within complex biological extracts. This approach highlighted secondary metabolites in almost all observed natural product classes, including low-abundance analogs of biologically relevant metabolites, as well as a new linear aminopolyol polyketide, funisamine. This study demonstrates the generality of activating stimuli to potentiate secondary metabolite production across diverse actinobacterial genera.
KEYWORDS: natural products, metabolomics, antibiotics, mixed culture, secondary metabolites, actinomycetes
ABSTRACT
Microorganisms within microbial communities respond to environmental challenges by producing biologically active secondary metabolites, yet the majority of these small molecules remain unidentified. We have previously demonstrated that secondary metabolite biosynthesis in actinomycetes can be activated by model environmental chemical and biological stimuli, and metabolites can be identified by comparative metabolomics analyses under different stimulus conditions. Here, we surveyed the secondary metabolite productivity of a group of 20 phylogenetically diverse actinobacteria isolated from hypogean (cave) environments by applying a battery of stimuli consisting of exposure to antibiotics, metals, and mixed microbial culture. Comparative metabolomics was used to reveal secondary metabolite responses from stimuli. These analyses revealed substantial changes in global metabolomic dynamics, with over 30% of metabolomic features increasing more than 10-fold under at least one stimulus condition. Selected features were isolated and identified via nuclear magnetic resonance (NMR), revealing several known secondary metabolite families, including the tetarimycins, aloesaponarins, hypogeamicins, actinomycins, and propeptins. One prioritized metabolite was identified to be a previously unreported aminopolyol polyketide, funisamine, produced by a cave isolate of Streptosporangium when exposed to mixed culture. The production of funisamine was most significantly increased in mixed culture with Bacillus species. The biosynthetic gene cluster responsible for the production of funisamine was identified via genomic sequencing of the producing strain, Streptosporangium sp. strain KDCAGE35, which facilitated a deduction of its biosynthesis. Together, these data demonstrate that comparative metabolomics can reveal the stimulus-induced production of natural products from diverse microbial phylogenies.
IMPORTANCE Microbial secondary metabolites are an important source of biologically active and therapeutically relevant small molecules. However, much of this active molecular diversity is challenging to access due to low production levels or difficulty in discerning secondary metabolites within complex microbial extracts prior to isolation. Here, we demonstrate that ecological stimuli increase secondary metabolite production in phylogenetically diverse actinobacteria isolated from understudied hypogean environments. Additionally, we show that comparative metabolomics linking stimuli to metabolite response data can effectively reveal secondary metabolites within complex biological extracts. This approach highlighted secondary metabolites in almost all observed natural product classes, including low-abundance analogs of biologically relevant metabolites, as well as a new linear aminopolyol polyketide, funisamine. This study demonstrates the generality of activating stimuli to potentiate secondary metabolite production across diverse actinobacterial genera.
INTRODUCTION
Secondary metabolites are used by microorganisms to communicate and compete within ecological systems. As selective and potent mediators of clinically relevant biological processes, they often become lead compounds for drug development and have been used to identify druggable biological targets (1, 2). The biosynthetic gene clusters responsible for the production of many classes of secondary metabolites can be readily identified within genomic sequence data using automated bioinformatics analyses (3–6). However, an inspection of microbial genomes reveals that the products of a large portion of the genetically encoded secondary metabolites remain unidentified in culture extracts (7). This discrepancy between potential and isolable secondary metabolite production may stem from low expression and/or translation of secondary metabolite biosynthetic genes and limitations in our methods of detection via abundance or biological activity (8). So called “brothological” methods (one strain many compounds [OSMAC]) (9) that vary in cultivation parameters, such as medium composition, pH, aeration, and temperature, can effectively increase the observable secondary metabolite output from microbial producers. These general approaches may increase secondary metabolite production by altering precursor levels or biosynthetic enzyme activity/stability, or through changes in gene regulation, microbial metabolism, and growth phase timing. An alternative strategy employs, in a single growth medium, defined chemical or biological agents that model ecological and environmental stimuli. Discrete stimuli found to elicit natural product production include histone deacetylase inhibitors (10, 11), exposure to subinhibitory levels of antibiotics (12–14), selection for spontaneous antibiotic resistance (15, 16), low-level metal exposure (17), and mixed microbial culture (18, 19). The finding that variations in discrete chemical and biological stimuli can alter secondary metabolite production is consistent with the hypothesis that the role of most natural products is to respond to environmental interactions.
Regardless of the means of stimulating secondary metabolite production, the identification of the often low-abundance secondary metabolites of interest within the observable metabolome remains a substantial challenge. High-performance liquid chromatography-mass spectrometry (HPLC-MS) is frequently used to detect secondary metabolites from biological extracts, but these analyses often result in the observation of thousands of small-molecule metabolites and by-products of similar abundance to potential secondary metabolites within the extract. The application of comparative metabolomics methods, such as multivariate statistical methods (MVSA) and/or molecular networking (16, 20, 21), has superseded the “stare and compare” approach, providing unbiased assessments of metabolite covariance across samples. Combining comparative metabolomics with the application of discrete stimuli can effectively prioritize secondary metabolites for isolation (22). This facilitates secondary metabolite identification within extracts, as they should thereby be identifiable by their responses to multiply applied stimuli. Unbiased comparative metabolomics workflows are more difficult to interpret productively in brothological approaches because the variations in medium composition often eclipse changes in metabolite inventories resulting from altered secondary metabolite production.
While the combination of chemical/biological stimuli and comparative metabolomics has led to the activation, production, and identification of natural products from actinomycetes of the genus Streptomyces (13, 23–25) and, less commonly, other actinomycetes (16, 19, 26, 27), these strategies have not been thoroughly evaluated across other actinobacterial genera. Here, we applied subinhibitory antibiotic exposure, rare earth metal exposure, and microbial competition activation strategies to a selection of 20 phylogenetically diverse cave-derived actinomycetes and assessed the resulting global secondary metabolite responses through comparative metabolomics. Marked changes in the observed metabolomes and increased production of secondary metabolites were evident in response to environmental stimuli. We isolated a subset of metabolites prioritized through comparative metabolomics analyses and identified members of natural product families, including the previously described actinomycin, hypogeamicin, tetarimycin, aloesaponarin, and propeptin classes, as well as a novel linear polyketide (funisamine) produced by a Streptosporangium strain under mixed-culture stimulus conditions.
RESULTS
Phylogenetically diverse actinomycetes isolated from hypogean environments.
Phylogenetically diverse microorganisms from unique unexplored environments have a strong track record of revealing useful chemical diversity (28, 29). Hypogean sediments are unique environments that have been shown to be replete with bacterial diversity (30–32) but not extensively explored for secondary metabolite-producing organisms. Caves maintain microbial habitats distinct from epigean (surface) ecosystems. The interior regions of limestone caves in the central United States are generally isothermal year round (∼13°C), highly oligotrophic (33), and completely aphotic. Formed millions of years ago by chemical and biological (34) dissolution of limestone, they are often highly humid due to hydrological activity and therefore present vast wet highly oxygenated surface areas for maintaining microbial biofilms competing for allochthonous dissolved organic matter. Based on these unique environmental parameters, we selected actinomycetes sourced from cave environs for stimulus response mapping.
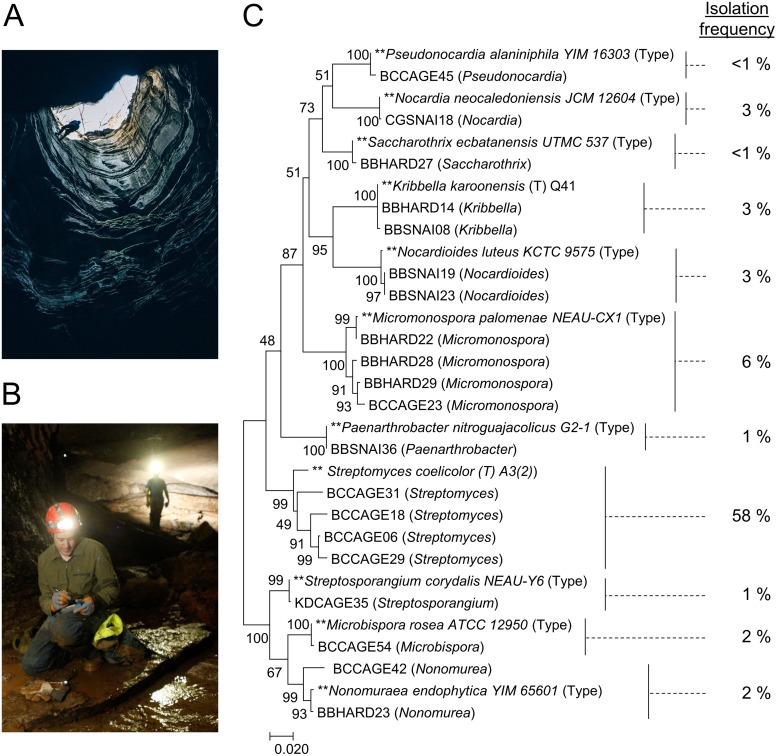
To investigate the secondary metabolic responses of cave-derived actinomycetes to environmental stimuli, environmental samples were acquired from Hardin's (Ashland City), Snail Shell (Rockvale), and Cagle's Chasm (South Pittsburg) caves in Tennessee by aseptically collecting cave sediments and swabbing cave formations. These formations (speleothems) included flowstone, stalactites, stalagmites, wall coralloids, and columns. To provide a basis for comparison of cultivatable diversity to studies of actinomycetes from surface environments, we used established actinobacterial isolation procedures (35). After collection, cave sediment samples were dried, and we used 100-fold diluted International Streptomyces Protocol 2 (ISP2) agar for dilution plating, while swabs were suspended and directly applied to agar plates without prior desiccation. Hundreds of individual bacterial colonies were isolated using this approach, and 16S rRNA gene sequences were acquired for 155 strains. The majority of these (58%) were most closely related to actinomycetes of the genus Streptomyces; however, a number of rare actinomycetes were isolated (see Table S1 in the supplemental material), which are important sources of novel secondary metabolites (36). A group of 20 cave strains from this sequenced collection were selected for this study, consisting of four Streptomyces species and 16 less commonly described actinomycetes with high 16S rRNA gene sequence similarity (GenBank accession numbers available in Table S2) to those of members of the genera Microbispora, Micromonospora, Streptosporangium, Saccharothrix, Nonomuraea, Pseudonocardia, Nocardioides, Nocardia, Kribbella, and Paenarthrobacter (Fig. 1).
FIG 1.
(A and B) Images from Cagle's Chasm (A) and Snail Shell (B) caves. (C) Phylogenetic tree constructed using MEGA7 with maximum likelihood analysis from 16S sequence alignment of the 20 strains selected for stimulus exposure studies and 11 type strains for reference. Bootstrap values based on 1,000 pseudoreplicates are shown next to the branch points. The initial tree for the heuristic search was obtained automatically by applying neighbor-joining and BIONJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Isolation frequency of genera in our study is calculated as the number of strains of a given genus identified divided by the total number (155) of sequenced isolates.
Stimuli increase natural product biosynthesis across actinomycete genera.
Selected strains were exposed to the following six different stimuli previously reported to activate secondary metabolite production in actinomycetes: subinhibitory antibiotic concentrations (1/10 the MIC) of either rifampin or streptomycin (37), rare earth metal exposure with lanthanum or scandium (38), and microbial competition with mycolic acid containing bacteria Tsukamurella pulmonis or Rhodococcus sp. strain BBSNAI13 (25). All organisms were cultured in ISP2 medium, a benchmark-defined medium, and metabolites were extracted after 7 days. These total-cell extracts were then analyzed through HPLC-electrospray ionization-MS (HPLC-ESI-MS) in both positive- and negative-ionization modes. In this study, the combined positive and negative ESI data for all extracts were prepared for initial comparative analysis. The metabolomic platform XCMS (39) was used to process data sets, providing peak detection, retention time correction, and chromatogram alignment to generate a combined list of ∼19,000 features, ions with distinct m/z and retention times. Feature ion abundances from technical replicates were averaged, and pairwise comparisons were performed between each stimulus condition and the respective controls to determine the number of features with increased production through stimulus exposure.
The overall responses to stimuli were measured by the number of features with ≥10-fold increased ion abundance compared to the control conditions. Responses varied between genera (see Fig. S1 in the supplemental material), and even phylogenetically similar isolates displayed markedly differing metabolomic response patterns. For instance, of the two Kribbella isolates, one (BBHARD14) responded most strongly to metal exposure, while the other (BBSNAI08) responded most strongly to mixed culture. Of the four Micromonospora isolates, two responded most strongly to mixed culture (BCCAGE23 and BBHARD28), while two responded most strongly to metal exposure (BBHARD22 and BBHARD29). Interestingly, across strains, there was relatively little overlap of overproduced features between stimuli of the same type. For example, within the antibiotic category, rifampin and streptomycin treatment generally led to the accumulation of different metabolites (Fig. S2). Similar results were observed for metal and mixed-culture exposure (Fig. S3 and S4), with the exception of two Nocardioides isolates (BBSNAI19 and BBSNAI23) which exhibited a high degree of overlap between responses under lanthanum and scandium exposure conditions. The four Streptomyces isolates demonstrated relatively fewer responses, suggesting that rare actinomycetes are particularly sensitive to applications of stimuli.
At least eight families of natural products were identified within the extracts (see below), and increased production was observed for seven of these under at least one stimulus condition. Two of these natural product families, an unidentified polyene natural product and a novel aminopolyol, funisamine, were detectable only under stimulus conditions, and others, like tetarimycin, were barely detectable under control conditions and would have been overlooked if not for enhanced production under stimulus conditions (Tables S3 and S4). The unidentified polyene was activated in a cave isolate, Nonomuraea sp. BCCAGE42, under antibiotic exposure conditions, and funisamine was produced by Streptosporangium sp. strain KDCAGE35 under mixed-culture conditions. Interestingly, while exposure to either rifampin or streptomycin elicited comparatively few bulk metabolomic responses across strains relative to mixed culture and metal stimuli, antibiotic exposure demonstrated a substantial effect on the accumulation of individual natural product features, which were conspicuous in pairwise analysis. Of 23 putative natural product features with abundances of ≥10-fold under stimulus conditions versus the controls, 17 were observed in greater abundance under subinhibitory antibiotic exposure conditions (Fig. S5).
Comparative metabolomics prioritizes secondary metabolites from stimulus exposure.
Two multivariate statistical analysis methods were used to prioritize metabolomic features for isolation and dereplication, volcano plots and S-plots (Fig. 2). Volcano plots organize features on the y axis by the probability of observing such a fold change under the null hypothesis of no-change (P value) and by relative ion abundance fold change on the x axis but not by feature ion abundance. In comparison, S-plots organize features on the y axis by their Pearson correlation coefficient, a measure of their linear correlation, and on the x axis by their component coefficients, a measure of a feature's contribution to the total variance between conditions, which is influenced by feature ion abundance. It is also important to note that the relative intensities of ions from a mixture of small molecules as detected by MS do not necessarily correlate with the relative concentrations of those compounds, due to various ionization efficiencies. While these analyses aid in prioritizing a number of features, the practical isolability of compounds from an extract is limited by many factors, including abundance, chromatographic properties, and chemical stability. Both MS and UV-Vis absorption spectroscopy were utilized to characterize culture extracts, and the absorption spectra aided in determining the abundance and isolability of prioritized features.
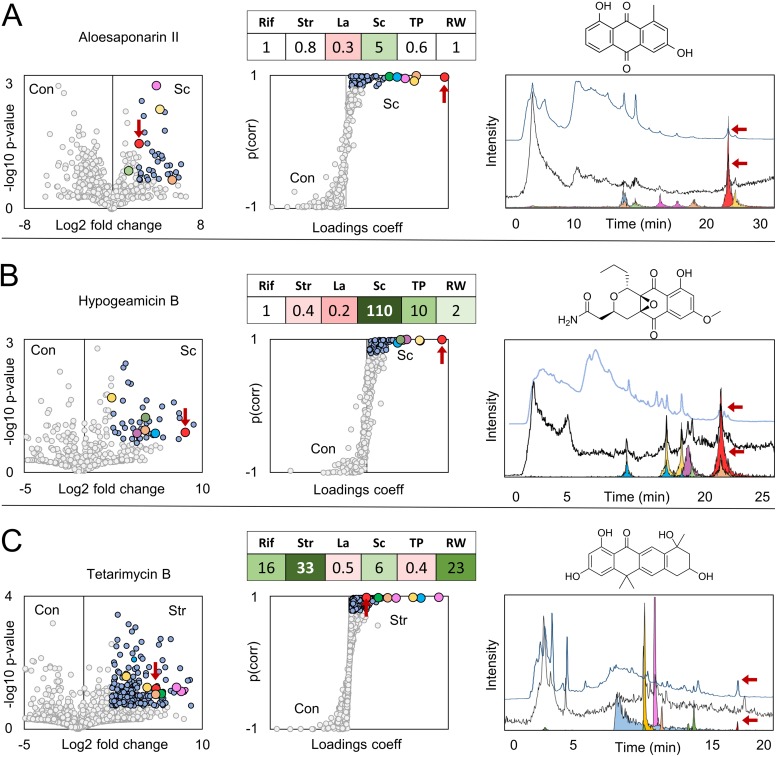
FIG 2.
Prioritization of natural products from comparative metabolomics. (A to C) Volcano and S-plot prioritization of aloesaponarins (ESI−) from Micromonospora sp. BBHARD22 exposed to scandium (Sc) (A), hypogeamicins (ESI+) from Nonomuraea sp. BBHARD23 exposed to scandium (Sc) (B), and tetarimycins (ESI+) from Microbispora sp. BCCAGE54 exposed to streptomycin (Str) (C). Circles are colored for features outside (gray) and within (blue) 0.8 Pearson correlation coefficient (coeff) thresholds for S-plots, and 0.8 probability value and 5-fold change thresholds for volcano plots. Features corresponding to aloesaponarin (A), hypogeamicin (B), and tetarimycin (C) are colored red in volcano and S-plots and are additionally distinguished by red arrows in volcano and S-plots as well as the UV-Vis and MS chromatograms. Overlays of TIC chromatograms, total absorption spectrums, and extracted ion chromatograms for abundant features were prioritized through S-plot analyses. Within each row, the color of the peak corresponds to the color of the circle in the volcano and S-plots. The fold change of the highlighted natural product under stimulus conditions relative to the controls is shown in tables over the S-plots for rifampin (Rif), streptomycin (Str), lanthanum (La), scandium (Sc), Tsukamurella pulmonis (TP), and Rhodococcus sp. BBSNAI13 (RW) conditions. Con, control.
Comparative metabolomics analyses of extracts from the cave isolate Micromonospora sp. BBHARD22 revealed a number of abundant features increased under scandium-supplemented conditions (Fig. 2A and Tables S3 and S4). Overlaying extracted ion chromatograms for S-plot prioritized features onto the total ion and UV-Vis chromatograms indicated that one feature was readily isolable based on abundance in both MS and UV-Vis detection, as well as elution in a noncomplex region of the chromatogram. The production of this feature was increased 6-fold under scandium metal exposure conditions and was ultimately identified by nuclear magnetic resonance (NMR) spectroscopy to be the known anthraquinone natural product aloesaponarin II, originally isolated from the plant Aloe saponaria (40) but also previously observed in extracts of both marine (41) and terrestrial (42) Streptomyces species. Another abundant feature prioritized from scandium exposure culture extracts was additionally identified as the anthraquinone okicenone (43). In addition to these confirmed anthraquinones, seven additional features were identified across multiply applied stimuli that were assigned as likely secondary metabolites based on their MS, tandem MS (MS/MS), and UV (Fig. S12 and S13) spectra in comparison to compounds isolated and characterized from this strain. Notably, these putative secondary metabolites, which are likely congeners of aloesaponarin and okicenone, vary in concentration across stimuli, with discrete stimuli facilitating increased production of individual congeners.
Analysis of the cave isolate Nonomuraea sp. BBHARD23 under stimulus conditions prioritized several features with enhanced production under scandium metal exposure conditions (Fig. 2B and Tables S3 and S4). Under these conditions, production of the pyronapthoquinone hypogeamicins A, B, and C, was increased, and these secondary metabolites were prioritized through comparative metabolomics; the most abundant of these was identified as hypogeamicin B (44). Additionally, several unidentified features were prioritized, with m/z and UV absorbance spectra similar to those of the identified hypogeamicins but with distinct chromatographic retention times. These putative hypogeamicin analogs were increased under mixed-culture conditions, with production increased almost 600-fold under exposure to competition with Rhodococcus species. In both of these examples, the highlighted secondary metabolite classes had very high ion abundances relative to other prioritized metabolites. Again, individual characterized and putative hypogeamicin congeners vary across stimuli (Tables S3 and S4). For example, the production of hypogeamicin C and four other putative analogs could be elicited by the administration of subinhibitory streptomycin, whereas hypogeamicin A could not. Scandium exposure led to increases of ∼2-, 110-, and 6-fold in hypogeamicin A, B, and C, respectively, but resulted in a decrease in the abundances of some putative hypogeamicin analogs.
Comparative analysis of resulting stimulus extracts from Microbispora sp. strain BCCAGE54 revealed increased production of a large number of metabolites. Volcano and S-plot analyses of subinhibitory streptomycin exposure prioritized 295 features, but the features with the highest ion intensity coeluted in a crowded region of the chromatogram and eluded isolation attempts (Fig. 2C). In comparison, one prioritized feature with relatively low ion abundance, yet a 33-fold increased production under antibiotic exposure conditions, eluted in a chromatographically tractable region and was correlated with a relatively large UV-Vis absorption peak. This induced feature was identified as tetarimycin B, a polyketide secondary metabolite recently discovered via the heterologous expression of an environmentally derived type II polyketide synthase (PKS) gene cluster (45). Sequence homology searches have indicated that tetarimycin-encoding gene clusters may be widespread in the environment, and the lack of identified tetarimycins from culture-based screens indicates the difficult-to-detect nature of this secondary metabolite family (45). Indeed, while tetarimycin B was produced in detectable levels within our unstimulated Microbispora cultures, the low-abundance ions detected under these conditions would likely have been overlooked entirely without prioritization through comparative metabolomics. As in previous examples, the production of several putative new tetarimycin analogs was elicited in multiple stimuli, along with several features displaying properties of secondary metabolites with high molecular weight, extended UV-Vis chromophores, and responses profiles matching tetarimycins (Fig. S21). During the isolation of tetarimycin, an additional pair of metabolites was isolated from large-scale culture extracts and identified as propeptin 1 and 2. The relative abundances of these two congeners varied subtly but consistently across stimulus classes with mixed-culture and antibiotic exposure conditions, resulting in higher ratios of propeptin 1, and metal exposure conditions resulting in higher ratios of propeptin 2 (Table S3).
As noted, volcano and S-plots emphasize different m/z features within the data. Features with significant fold changes are prioritized by volcano plots, whereas those with significant differences in abundance are prioritized by S-plots. The chromatogram overlays in Fig. 2 show features emphasized through S-plot analyses, which were found to be the most reliable method for prioritizing features for isolation. Similar overlays for features prioritized by volcano plot thresholds are available in the supplemental material (Fig. S6). Tables of observed secondary metabolite fold change across stimulus conditions (Tables S3 and S4) along with extracted ion (Fig. S7 to S11) and UV (Fig. S13 to S22) chromatograms for prioritized metabolites and NMR spectra of identified metabolites (Fig. S23 to S28) are also provided in the supplemental material.
Novel polyketide produced from interactions between rare Streptosporangium sp. and microbial coculture.
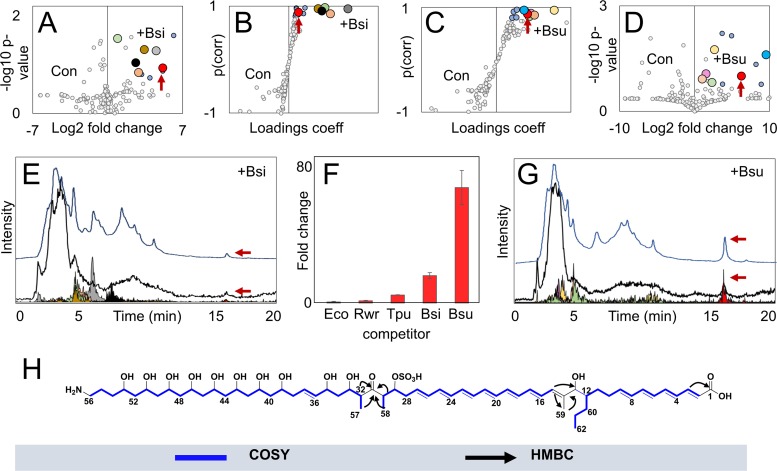
A cave isolate designated Streptosporangium sp. KDCAGE35 with close 16S rRNA gene sequence similarity to type strain Streptosporangium corydalis (99.6% identity) was shown to be highly responsive to mixed-culture stimulus, with ∼16% of the total detected features displaying increased ion intensity at 10-fold or higher levels under these conditions. While the production of many features under mixed-culture conditions was increased relative to that in unstimulated cultures, most remained insufficiently abundant for isolation. Further mixed-culture screens were performed with Streptosporangium sp. KDCAGE35 using Escherichia coli, Bacillus sp. strain KDCAGE13 (99.9% 16S rRNA gene similarity to Bacillus simplex), and Bacillus subtilis in addition to the previously screened Rhodococcus sp. BBSNAI13 and T. pulmonis. Volcano and S-plot analyses comparing mixed cultures with Bacillus strains and Streptosporangium sp. KDCAGE35 monocultures identified a metabolite with a mass of 1,176.6 Da that eluted in a tractable region of the chromatogram. This mass was correlated with a UV spectrum (λ maxima, 370, 389, and 413 nm) characteristic of conjugated polyene systems (e.g., heptaenes). The production of this feature was also induced in screens with T. pulmonis, but the significantly enhanced production elicited from cocultivations with B. subtilis (∼70-fold increase) enabled isolation and identification. Comparisons of production yields of this prioritized feature from cocultivation of different species with Streptosporangium sp. KDCAGE35 are shown in Fig. 3F. The target compound was isolated as a bright-yellow solid from several small-scale mixed cultures (totaling 2 liters) of the Streptosporangium sp. KDCAGE35 and B. subtilis mixed culture using a combination of adsorbent resin, size-exclusion chromatography, and reverse-phase chromatography. Electrospray Fourier-transform ion cyclotron resonance MS of the isolated compound yielded an accurate mass of 1,176.6507 [M-H]− (calculated for C62H98NO18S−, 1,176.6510, 0.25 ppm), which in combination with formula constraints from NMR data and isotope analyses suggested a molecular formula of C62H99NO18S. The full structure was derived from correlated spectroscopy (COSY), total correlated spectroscopy (TOCSY), heteronuclear single quantum coherence (HSQC), heteronuclear multiple bond correlation (HMBC), and nuclear Overhauser effect spectroscopy (NOESY) NMR (Fig. S29 to S41 and Table S5), which identified two conjugated systems assignable to four and seven double bonds, respectively (Fig. 3H). The structure was separated into three spin systems by two quaternary carbons at C-14 and C-31. Figure 3H shows important HMBCs which facilitated connections across these spin systems. HMBCs show interactions from the proton at position CH-13 and methyl protons CH3-59 to the C-14 quaternary carbon and to CH-15 of the next spin system. Methyl protons at CH3-59 also showed HMBCs with CH-13 and long-range coupling in the COSY spectra with the proton at CH-15. The second quaternary carbon, a ketone at C-31, was bridged by HMBCs from protons at CH-30, CH-32, CH3-57, and CH3-58 to the C-31 carbon. NOESY correlations between methine protons CH-29, CH-30, CH-32, and CH-33 to methyl protons CH3-57 and CH3-58 confirmed connectivity in these spin systems. A shift of 4.61 ppm for the proton at CH-29 was consistent with a sulfate group at C-29. This is additionally supported by the identification of a fragment consistent with the loss of a sulfate group by MS (Fig. S42 to S44 and Table S6). We named this compound funisamine (from the Latin noun for rope, funis).
FIG 3.
Activation of funisamine biosynthesis through mixed culture. (A to D) Volcano and S-plot comparisons between Streptosporangium sp. KDCAGE35 and Bacillus sp. KDCAGE13 mixed (+Bsi) and mono- (Con) cultures and Streptosporangium sp. KDCAGE35 and B. subtilis mixed (+Bsu) and mono- (Con) cultures (ESI−). Circles are colored for features outside (gray) and within (blue) 0.8 Pearson correlation coefficient thresholds for S-plots, and 0.8 probability value and 5-fold change thresholds for volcano plots. The feature corresponding to funisamine is enlarged and colored red. (E and G) Overlays of total ion chromatograms, total absorption spectrums, and extracted ion chromatograms for funisamine (m/z 1,176.6) and other features prioritized from Streptosporangium sp. KDCAGE35 mixed cultures with B. subtilis and Bacillus sp. KDCAGE13. (F) Fold change of funisamine production in mixed cultures with Rhodococcus sp. BBSNAI13 (Rwr), E. coli (Eco), T. pulmonis (Tpu), B. sp. KDCAGE13 (Bsi), and B. subtilis (Bsu) versus control quantified by UV absorption at 413 nm. (H) Proposed structure of funisamine generated from HSQC, COSY, TOCSY, HMBC, and NOESY spectra. Major COSY correlations and HMBCs are shown. Stereochemical configurations of substituent groups and double bonds were inferred from bioinformatic analysis.
Cocultivated elicitor strains E. coli and B. subtilis had similar growth rates and doubling times under the culture conditions (Fig. S46) yet had differing impacts on funisamine compound production (Fig. 3F). The mechanism and factors governing the apparent Bacillus species selectivity of this activation remain unknown, though there appears to be no correlation to the elicitor strain growth rate. While some related aminopolyols do have reported antimicrobial properties against Bacillus strains (46), funisamine exhibited antimicrobial activity only at very high concentrations, at ∼25 times the levels produced under mixed-culture conditions. Microtiter plate antibacterial assays with Staphylococcus aureus, E. coli, and Candida albicans yielded MIC values of ∼1 mM. Funisamine (and perhaps other long-chain aminopolyols) may also serve a role other than as an antibiotic in the producer's natural environment.
An issue with mixed-culture stimulus approaches is the uncertainty of which organism(s) within the culture are ultimately responsible for producing compounds observed within an extract. Funisamine was produced under mixed-culture conditions with the Streptosporangium sp. KDCAGE35 and three separate cocultivated species, strongly suggesting that Streptosporangium sp. KDCAGE35 was likely responsible for the production of funisamine. Another prioritized and isolable metabolite was observed to be entirely unique to B. subtilis and Streptosporangium sp. KDCAGE35 mixed cultures (Fig. S45). This feature was isolated and identified through NMR spectroscopy to be the antibiotic amicoumacin B, which is known to be produced by Bacillus strains and appears to be produced in response to Streptosporangium sp. KDCAGE35. To test if amicoumacin induced funisamine production, we added purified amicoumacin to monocultures of Streptosporangium sp. KDCAGE35 and did not detect funisamine production (data not shown).
Biosynthesis of funisamine.
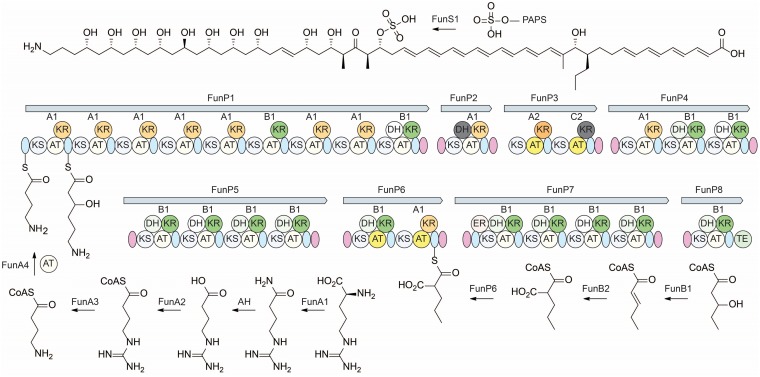
To understand the biosynthesis of funisamine, we endeavored to identify the cognate biosynthetic gene cluster within Streptosporangium sp. KDCAGE35. The genome of Streptosporangium sp. KDCAGE35 was determined using a combination of Pacific Biosciences RSII and Illumina sequencing to reveal a chromosome of 11,340,955 bp. Potential secondary metabolite gene clusters were then identified using the antibiotics and secondary metabolite analysis shell algorithm (antiSMASH version 4) (47). This analysis identified 22 putative secondary metabolite-producing gene clusters (Table S7), including 6 terpenes, 5 nonribosomal peptide synthetases, 3 lantipeptides, 2 bacteriocins, 1 lasso peptide, and 3 PKS gene clusters. One of these PKS gene clusters, a type I PKS consisting of 26 modules (GenBank accession number MH203088), contained a modular organization of catalytic domains consistent with the determined structure of funisamine. The modular PKS diagram of this cluster is shown in Fig. 4, and gene descriptions and a genetic organization are provided in the supplemental material (Table S8). While the stereochemistry of substituent hydroxyl and methyl groups has not been experimentally validated, analysis of the ketoreductase (KR) domains within each module (Fig. S47) facilitated the estimation of the stereochemistry of the hydroxyl and methyl substituents in the structure based on the amino acids present in six distinguishing positions along the loop and catalytic regions of the KR, as described by Keatinge-Clay (48). Predicted stereochemical configurations of products for each polyketide synthase module in funisamine biosynthesis are shown in Fig. S48. Methyl groups at positions C-14, C-30, and C-32 were consistent with antiSMASH methylmalonyl extender unit predictions based on acyltransferase (AT) sequences for modules at those positions. The AT for the module predicted to activate the precursor at position C-12 was predicted to use a modified malonyl (ethylmalonyl) coenzyme A (CoA) unit for polyketide extension, consistent with the propyl group at this position.
FIG 4.
Proposed biosynthesis of funisamine. Biosynthesis of the 4-aminobutanoyl-CoA starter unit from arginine and loading of the polyketide synthase is shown on the bottom left. Propylmalonyl-CoA biosynthesis via beta-oxidation of odd-chain fatty acid pentanoate is shown on the bottom right. Stereochemical assignment of funisamine structure shown at the top was inferred based on ketoreductase type and geometry indicated as A1, A2, B1, B2, C1, or C2, as previously described (48). Sulfation by putative sulfotransferase FunS1 and 3′-phosphoadenosine 5′-phosphosulfate (PAPS) cofactor shown at the top. The modular organization of funisamine polyketide synthase genes (FunP1 to FunP8) in Streptosporangium sp. KDCAGE35 is shown in the center. The layout of catalytic domains of ketosynthase (KS), acyltransferase (AT), ketoreductase (KR), dehydratase (DH), enoylreductase (ER), and thioesterase (TE) present within the polyketide synthases is shown with inactive domains colored gray. A-type and B-type KR domains are colored orange and green, respectively. Acyltransferase domains predicted to use methyl or propylmalonyl extender units are colored yellow. Acyl-carrier-proteins are colored light blue, and docking domains are purple. Stereochemical configurations of substituent groups and double bonds were inferred from bioinformatic analysis.
The proposed 4-aminobutyryl-CoA starter precursor unit is likely derived from arginine through the unusual pathway described by Leadlay and coworkers (49) (Fig. 4a). Genes encoding the essential arginine monooxygenase (funA1), 4-guanidinylbutanoate-CoA ligase (funA2), agmatinase (funA3), and 4-aminobutyryl-CoA-acyl-carrier-protein (4-aminobutyryl-CoA-ACP) acyltransferase (funA4) were all evident within the gene cluster. A putative sulfotransferase, encoded by funS1, likely catalyzes the sulfation at C-29. FunS1 is similar yet distinct in sequence to other observed sulfotransferase enzymes involved in the biosynthesis of aminopolyol polyketides, such as MedB (NCBI accession no. BAW35627.1, 47% identical, 60% similar) and SMALA2697 (NCBI accession no. WP_099013767.1, 44% identical, 59% similar), which facilitate the sulfation of mediomycin and clethramycin. Production of the evident propyl-malonyl-CoA unit added at this position most likely derives from a 3-hydroxypentyl-CoA by dehydrogenase and carboxylase enzymes encoded by funB1 and funB2 observed within the cluster. Expanded versions of Fig. 4 showing all thioester intermediates are included in the supplemental material (Fig. S49 to S51).
DISCUSSION
Despite the oligotrophic, relatively cold, and aphotic nature of hypogean ecosystems, caves have been demonstrated to harbor rich, stable, and diverse communities of microorganisms. Metagenomic (50, 51) and metatranscriptomic (52) studies indicate that hypogean bacteria are primarily heterotrophic but include some autotrophic bacteria, which may be more represented in regions that are more remote from allochthonous energy inputs. Actinobacteria have been noted as one of the most abundant phyla associated with hypogean ecosystems (53), have been found to be associated with speleothems (54), and constitute some of the first studied hypogean microbial families due to their association with degradation of prehistoric cave paintings (55). Here, we confirm that actinomycetes are abundant in the three oligotrophic cave locations we sampled using standard actinomycete protocols, and that the actinobacterial generic distribution we observed mirrors what is found on epigean ecosystems.
We selected 20 diverse hypogean actinobacteria for analysis of their secondary metabolism using a stimulus-response paradigm in a single growth medium. The organism set was biased for less commonly described actinobacterial genera, including several from which few or no secondary metabolites have been previously reported. Overall response data from stimulated culture extracts indicate that stimulus exposure, even for stimuli within the same class, leads to distinct changes in the metabolic inventories of exposed strains. This global trend is also evident within the response of identified and putative natural product features, which largely demonstrate responsiveness to a specific stimulus rather than to a class of stimuli. Previous studies have indicated that the production of some classes of secondary metabolites may be induced by general bacterial stressors, such as oxidative stress (56) and guanosine tetraphosphate (ppGpp) (17, 57), or by master regulators, such as ScmR in Burkholderia spp. (58). However, the trends observed within our data set, particularly the variations in congener abundance across stimuli, suggest that the increased abundance of natural products under stimulated conditions stems largely from the results of specific interactions with a given stimulus rather than a general response to stress.
Analysis of abundant and covarying metabolites along with manual inspections of HPLC-MS chromatograms identified ∼58 putative secondary metabolites from an estimated 13 classes based upon MS and UV analyses, spectroscopic similarity to validated congeners isolated in producing strains, and the matching of productive response of putative metabolites to multiplexed stimulus conditions. The isolation of a subset of secondary metabolites from these candidates for structural identification was biased toward abundant compounds found in more isolable chromatographic regions with regard to reverse-phase HPLC techniques. In all, the subset of isolated compounds revealed 12 identified secondary metabolites within eight classes of compounds, seven of which have been previously isolated from epigean ecosystems. One, funisamine, represents a new secondary metabolite based on its carbon framework, though it bears similarity to a class of giant linear polyenes isolated from other actinobacteria (59–61). This subset of isolated structurally validated compounds revealed a moderate natural product rediscovery rate, with one of eight compound families representing a new carbon scaffold. However, we note that focusing on abundant chromatographically tractable compounds may have biased our campaign toward rediscovery, and that the products of a significant portion of putative secondary metabolites identified using this workflow remain to be isolated and characterized.
The conservation of core structural elements within the aminopolyol class of secondary metabolites suggests that they serve important functions for producing strains within their natural environment, and the evolution (62) and ecological role (63) of aminopolyol natural products have recently been investigated. This class of natural products commonly exhibits antibiotic and/or antifungal activity. The linearmycins were found to inhibit the growth of several Gram-positive bacteria and lyse Bacillus species by directly targeting the cytoplasmic membrane (64). A role in vesicle biogenesis within producing Streptomyces spp. has also been suggested for linearmycins (63). To our knowledge, funisamine is the only known long-chain aminopolyol produced by a Streptosporangium sp., and phylogenetic analyses reveal that many of the ketosynthase and acyltransferase sequences within the funisamine PKS are divergent from domains observed in Streptomyces-derived aminopolyol gene clusters. Thus, while funisamine is structurally similar to other known aminopolyols, the low sequence similarity of the funisamine gene cluster from Streptosporangium sp. KDCAGE35 to Streptomyces gene clusters suggests a substantial amount of evolutionary distance. A number of likely homologous sequences with high identity to the genes involved in funisamine biosynthesis were also observed in the genome of a close relative, Streptosporangium subroseum CGMCC 4.2132, including the arginine monooxygenase (funA1), 4-guanidinylbutanoate-CoA ligase (funA2), agmatinase (funA3), 4-aminobutyryl-CoA-ACP acyltransferase (funA4), sulfotransferase (funS1), and 2-pentenyl-CoA carboxylase (funB2), all with >97% identity. The antiSMASH analysis of publicly available genome scaffolds for this strain (NCBI RefSeq accession number NZ_FZOD00000000.1) revealed a fragmented biosynthetic gene cluster (BGC) with sequence similarity to the type I PKS involved in the aminopolyol ECO-02301 biosynthesis. This indicates that funisamine or related congeners may be produced in other Streptosporangium species.
Genome mining methods have empowered the development of a variety of complementary methods to expedite the labor-intensive process of natural product discovery. For example, native and heterologous expression technologies, using biosynthetic gene clusters obtained from genetically “interesting” producers or environmental DNA, identify secondary metabolites via comparative analysis between an expressing strain and the nonexpressing host strain. Herein, we demonstrate that comparisons of metabolite responses to multiple stimulus conditions can highlight low-abundance secondary metabolites, in this case across a wide variety of actinobacterial genera. In comparison to bioactivity-based approaches, the advantage of comparative metabolomics approaches is that they are not constrained by specificity of a given bioassay and permit the discovery of compounds active against a broader array of targets. Indeed, nearly all of the compounds of this study identified solely via comparative metabolomics are reported to possess a variety of useful biological activities.
However, one disadvantage of prioritization of metabolites by comparative metabolomics is that it is a “shoot first, ask questions later” proposition. There is no guarantee that the compounds identified possess therapeutically relevant biological activity. This study highlights a critical need for prioritization in discovery. In addition to the discovery of a new scaffold, within the known compound classes we identified here, we observed stimulus-dependent amplification of multiple individual congeners of compounds that were previously not described. These were not prioritized for isolation based on comparative metabolomics covariance ranking, as they represented analogs of known compounds, and we had no knowledge of beneficial activity. Thus, a potential future avenue for these techniques is to combine bioactivity measurements with comparative metabolomics data sets. Examples of this combined approach include overlaying bioactivity to molecular networking (65) and our own multiplexed activity metabolomics (MAM) approach (66). In both cases, these methods enable an estimation of the activity of unknown compounds and structure-activity relationships within compound families prior to compound isolation using activity metabolomics techniques (65, 66).
MATERIALS AND METHODS
Reagents and strains.
All reagents were obtained from the Sigma-Aldrich chemical company, unless otherwise specified. Mixed-culture strains Tsukamurella pulmonis, Bacillus subtilis, and Escherichia coli were obtained from the American Type Culture Collection (ATCC 700081, ATCC 23857, and ATCC 10536), and Rhodococcus sp. BBSNAI13 and Bacillus sp. KDCAGE13 were obtained via dilution plating from hypogean sediments (as described below) and taxonomically assigned by 16S rRNA gene analysis.
Cave strain isolation and identification.
Desiccated cave sediments from Hardin's (Ashland City), Snail Shell (Rockvale), and Cagle's Chasm (South Pittsburg) caves in Tennessee collected between 2010 and 2015 were vortexed in sterile water (100 mg/ml), and supernatants were serially diluted (10-fold, 100-fold, and 1,000-fold) and plated on minimal medium agar plates (ISP2 medium/100-fold diluted, 1.5% agar). Plates were then incubated at 30°C, and colonies were picked over a 3-week period. DNA isolations for purified colonies were performed with a commercial kit (Wizard DNA isolation kit; Promega, Inc.). The 16S rRNA genes for these were then amplified with universal primers 27F (AGA GTT TGA TCC TGG CTC AG) and 1525R (AAG GAG GTG ATC CAG CCG CA) and the use of high-fidelity DNA polymerase (Phusion; Thermo, Inc.). The PCR thermocycler conditions were initial incubation at 98°C for 1 min and 30 cycles of (i) denaturation at 98°C for 0.5 min, (ii) annealing at 59°C for 1 min, and (iii) extension at 72°C for 1.5 min. The final extension step was conducted at 72°C for 10 min. Target 16S rRNA amplicons were purified using a gel extraction kit (QIAquick gel extraction kit). Purified PCR products were directly sequenced from both the 5′ and 3′ ends, and these sequences were combined to give the near-full-length 16S rRNA gene sequences. Preliminary genus identifications were determined by a comparison of 16S rRNA gene sequences to type strains in the EzBioCloud database (30).
Microbial stimulation and culture conditions.
For all conditions, a spore suspension of the selected strain was inoculated on ISP2 agar plates from frozen stocks and cultured for 7 days at 30°C. These plates were then used to inoculate 25-ml (in 250-ml Erlenmeyer flasks) ISP2 liquid seed cultures, which were also cultured for 7 days at 30°C on a rotary shaker at 180 rpm. For subinhibitory antibiotic exposure fermentations, 1.25 ml of the seed culture was transferred into 25-ml (5% inoculum) ISP2 medium experimental cultures containing either rifampin (120 nM) or streptomycin (170 nM). For rare earth metal fermentations, 1.25 ml of the seed culture was transferred into 25-ml (5% inoculum) ISP2 medium experimental cultures containing either scandium chloride (200 μM) or lanthanum chloride (1,500 μM). For mixed-culture fermentations, 1.25 ml of the seed culture was transferred into 25-ml (5% inoculum) ISP2 medium experimental cultures. After 24 h, 250 μl (1%) of either T. pulmonis or Rhodococcus sp. BBSNAI13 overnight cultures was added to the actinomycete culture. The controls were prepared by inoculating 1.25 ml (5%) of seed cultures into 25 ml of ISP2 without additives. All cultures were allowed to ferment at 30°C and 180 rpm for 7 days before extraction.
Extraction of liquid fermentations.
Whole-culture metabolite extracts were generated by adding an equal volume of methanol (25 ml) to each culture. These were then shaken on a rotary shaker for 1 h before the extractions were centrifuged at 3,750 rpm. Supernatants were dried in vacuo (Genevac) to yield crude extracts.
LC-MS data acquisition and processing.
Extract samples were resuspended in 50:50 methanol-water at a concentration of 200 mg/ml and centrifuged at 13,000 rpm, and the supernatant was transferred to a fresh vial to remove insoluble extract components. LC-MS data acquisition was performed with a 30-min gradient. Mobile phase A consisted of 95% water and 5% acetonitrile with 10 mM ammonium acetate, and mobile phase B consisted of 95% acetonitrile and 5% water with 10 mM ammonium acetate. A Luna 4.6- by 250-mm 5-μm particle C18 100 Å column (Phenomenex) was used for chromatographic separations, with a flow rate of 1 ml/min and a column temperature of 25°C. An autosampler with a loop size of 20 μl was used for sample injection. The solvent composition began at 100% A, which was held for 1 min and ramped linearly to 100% B over the next 29 min, held at 100% B for 15 min, and returned to 100% A over a 1-min period. The gradient was held at 100% A for the next 10 min for equilibration. The flow was split 3:1 using a flow splitter, with 750 μl/min directed through a Surveyor PDA Plus detector and 250 μl/min diverted to the mass spectrometer. Mass spectra were acquired at a rate of 1 Hz from 150 to 2,000 Da in both positive- and negative-ion modes for the duration of each sample analysis on a TSQ H-ESI mass spectrometer (Thermo Scientific). The source capillary was held at 350°C and 3.0 kV, with a desolvation gas flow of 35 liters/h and a vaporizer temperature of 300°C. Data were converted to mzXML format using the msconvert tool from the ProteoWizard package (31). Peak picking and alignment were performed using XCMS in R (21). See the supplemental material for details and package locations. The resulting data matrices were formatted in Microsoft Excel. Data were filtered by abundance to remove features with a maximal intensity of less than 1 × 105, and features eluting after the 30-min gradient (i.e., wash) were removed. The total ion current (TIC) for each injection was normalized to a total value of 10,000, such that feature abundance values of 100 represent 1% of the total abundance for that injection. Principal component and S-plot analyses were performed using Umetrics (Waters Corp.), and formatting for Umetrics was performed by transposing the XCMS-generated feature intensity matrix into the Umetrics software. Volcano plots were generated in Excel using P values determined by the pairwise XCMS analysis of LC-MS data of selected stimulus conditions versus runs with ISP2 medium without stimulus. Fold change calculations for each feature were performed in Excel after data normalization.
Funisamine isolation.
A spore suspension of Streptosporangium sp. KDCAGE35 was inoculated on ISP2 agar plates from frozen stocks and cultured for 7 days at 30°C. The plate cultures were then used to inoculate 25-ml ISP2 liquid seed cultures, which were also cultured for 7 days (30°C, 180 rpm). These were then used to inoculate 50-ml (in 250-ml Erlenmeyer flasks) production cultures (5% inoculum) in ISP2 medium, totaling 2 liters. After 24 h, 500 μl (1% inoculum) of overnight B. subtilis culture was added to the production culture. After 7 days, the cultures were combined and extracted with Diaion HP-20 resin (50 g/liter) at 30°C for 3 h to allow for compound partitioning within the resin. Resin and mycelia were then isolated via centrifugation and extracted with methanol for 1 h at room temperature. The methanol extract was separated from resin and mycelia via centrifugation and concentrated in vacuo. Funisamine was then purified from this concentrated extract by following its m/z and UV elution through (i) size-exclusion chromatography on a Sephadex LH-20 column (2 cm diameter by 100 cm length) in methanol, (ii) preparative C18 HPLC with a linear gradient of water-acetonitrile buffered with 10 mM ammonium acetate, and (iii) a Sephadex LH-20 column (2 cm diameter, 50 cm length) in methanol to yield 3 mg of pure funisamine.
High-resolution mass spectrometry.
Mass accuracy and fragmentation measurements were performed using an electrospray 15T solariX Fourier transform ion cyclotron resonance (ICR) mass spectrometer (Bruker Daltonics, Billerica, MA, USA). External mass calibration was performed prior to analysis (ESI-L tuning mix; Agilent Technologies, Santa Clara, CA). Funisamine was detected in negative mode as the doubly charged ion ([M-2H]2−, m/z 587.8220; resolving power, 497,566) and singly charged ion ([M-H]−, m/z 1,176.6507; resolving power, 250,639) by electrospray ionization. The calculated m/z for C62H98NO18S− was 1,176.6510, yielding an error of 0.25 ppm. For fragmentation experiments, the singly charged ion was isolated in the source region of the instrument (quadrupole isolation window, 5.0 Da), accumulated in the collisional hexapole (20 s), and fragmented by sustained off-resonance irradiation collision-induced dissociation (SORI-CID) in the ICR cell using pulsed argon (pulse length, 0.25 s; frequency offset, 500 Hz; SORI power, 2.5%).
Genome sequencing of Streptosporangium sp. KDCAGE35.
Genomic DNA was isolated from Streptosporangium sp. KDCAGE35 using cetyltrimethylammonium bromide (67). Whole-genome sequencing was performed by Novogene, Inc., using a PacBio RSII platform with >40× coverage, followed by short-read Illumina sequencing performed by the Vanderbilt Vantage sequencing core facility. Illumina reads were trimmed using FaQCs to remove Illumina adapters and trim at minimum quality (68). PacBio sequences and trimmed Illumina reads were then assembled using the SPAdes genome assembler version 3.11.1 (69), using default settings and the careful flag. The resulting assembled scaffolds were polished by using the same trimmed Illumina reads using Pilon version 1.22 (70), with default settings. The Bam file required for Pilon was generated using BWA-mem (71).
Accession number(s).
The 16S rRNA gene sequences for cave isolates used in the stimulus exposure study were deposited in GenBank with the accession numbers MH182596, MH182597, MH182598, MH182599, MH182600, MH182601, MH182602, MH182603, MH182604, MH182605, MH182606, MH182607, MH182608, MH182609, MH182610, MH182611, MH182612, MH182613, MH182614, and MH277690 (Table S2). The sequence for the described putative funisamine BGC was deposited in GenBank under the accession number MH203088.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grants RO1 GM092218, T32 GM 0650086, and 065086-13), the National Science Foundation (NSF-GRFP grans 1445197), the Vanderbilt Department of Chemistry, and the Vanderbilt Institute of Chemical Biology. J.M.S. is supported by the National Institutes of Health (grant P41 GM103391-07; principal investigator [PI], R. M. Caprioli) and the Defense Advanced Research Projects Agency (grant W911NF-14-2-0022; PI, R. M. Caprioli). The 15T MALDI FT-ICR mass spectrometer was acquired through the National Institutes of Health Shared Instrumentation Grant Program (grant 1S10OD012359-01). The Vanderbilt VANTAGE Core provided technical assistance for this work. VANTAGE is supported in part by CTSA Grant (5UL1 RR024975-03), the Vanderbilt Ingram Cancer Center (grant P30 CA68485), the Vanderbilt Vision Center (grant P30 EY08126), and NIH/NCRR (grant G20 RR030956).
B.C.C. and Z.B.H. performed culture experiments. B.C.C. processed the metabolomics data and discovered, isolated, and solved the NMR structure for funisamine. J.M.S. performed high-resolution mass spectrometry experiments. B.C.C., A.E.Y.-G., and B.O.B. performed analysis of the funisamine gene cluster. B.C.C., J.M.S., and B.O.B. designed the experiments and wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01125-18.
REFERENCES
- 1.Genilloud O. 2017. Actinomycetes: still a source of novel antibiotics. Nat Prod Rep 34:1203–1232. doi: 10.1039/C7NP00026J. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. 2016. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 3.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh CT, Fischbach MA. 2010. Natural products version 2.0: connecting genes to molecules. J Am Chem Soc 132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster ALH, Wyatt MA, Magarvey NA. 2015. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res 43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston CW, Skinnider MA, Wyatt MA, Li X, Ranieri MRM, Yang L, Zechel DL, Ma B, Magarvey NA. 2015. An automated Genomes-to-Natural Products platform (GNP) for the discovery of modular natural products. Nat Commun 6:8421. doi: 10.1038/ncomms9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherlach K, Hertweck C. 2009. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem 7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 8.Nett M, Ikeda H, Moore BS. 2009. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bode HB, Bethe B, Höfs R, Zeeck A. 2002. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3:619–627. doi:. [DOI] [PubMed] [Google Scholar]
- 10.Henke MT, Soukup AA, Goering AW, McClure RA, Thomson RJ, Keller NP, Kelleher NL. 2016. New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans. ACS Chem Biol 11:2117–2123. doi: 10.1021/acschembio.6b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albright JC, Henke MT, Soukup AA, McClure RA, Thomson RJ, Keller NP, Kelleher NL. 2015. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem Biol 10:1535–1541. doi: 10.1021/acschembio.5b00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seyedsayamdost MR, Chandler JR, Blodgett JAV, Lima PS, Duerkop BA, Oinuma K, Greenberg EP, Clardy J. 2010. Quorum-sensing-regulated bactobolin production by Burkholderia thailandensis E264. Org Lett 12:716–719. doi: 10.1021/ol902751x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu F, Nazari B, Moon K, Bushin LB, Seyedsayamdost MR. 2017. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J Am Chem Soc 139:9203–9212. doi: 10.1021/jacs.7b02716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai Y, Sato S, Tanaka Y, Ochi K, Hosaka T. 2015. Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl Environ Microbiol 81:3869–3879. doi: 10.1128/AEM.04214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochi K, Okamoto S, Tozawa Y, Inaoka T, Hosaka T, Xu J, Kurosawa K. 2003. Ribosome engineering and secondary metabolite production. Adv Appl Microbiol 56:155–184. doi: 10.1016/S0065-2164(04)56005-7. [DOI] [PubMed] [Google Scholar]
- 16.Derewacz DK, Goodwin CR, McNees RC, McLean JA, Bachmann BO. 2013. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc Natl Acad Sci U S A 110:2336–41. doi: 10.1073/pnas.1218524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai K, Wang G, Okamoto S, Ochi K. 2007. The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol Lett 274:311–315. doi: 10.1111/j.1574-6968.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- 18.Angell S, Bench BJ, Williams H, Watanabe CMH. 2006. Pyocyanin isolated from a marine microbial population: synergistic production between two distinct bacterial species and mode of action. Chem Biol 13:1349–1359. doi: 10.1016/j.chembiol.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 19.Derewacz DK, Covington BC, McLean JA, Bachmann BO. 2015. Mapping microbial response metabolomes for induced natural product discovery. ACS Chem Biol 10:1998–2006. doi: 10.1021/acschembio.5b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chanana S, Thomas C, Braun D, Hou Y, Wyche T, Bugni T. 2017. Natural product discovery using Planes of Principal Component Analysis in R (PoPCAR). Metabolites 7:E34. doi: 10.3390/metabo7030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin CR, Covington BC, Derewacz DK, McNees CR, Wikswo JP, McLean JA, Bachmann BO. 2015. Structuring microbial metabolic responses to multiplexed stimuli via self-organizing metabolomics maps. Chem Biol 22:661–670. doi: 10.1016/j.chembiol.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Covington BC, McLean JA, Bachmann BO. 2016. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat Prod Rep 34:6–24. doi: 10.1039/C6NP00048G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K. 2009. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol 27:462–464. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- 24.Wu C, Du C, Gubbens J, Choi YH, van Wezel GP. 2015. Metabolomics-driven discovery of a prenylated isatin antibiotic produced by Streptomyces species MBT28. J Nat Prod 78:2355–2363. doi: 10.1021/acs.jnatprod.5b00276. [DOI] [PubMed] [Google Scholar]
- 25.Onaka H, Mori Y, Igarashi Y, Furumai T. 2011. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl Environ Microbiol 77:400–406. doi: 10.1128/AEM.01337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adnani N, Chevrette MG, Adibhatla SN, Zhang F, Yu Q, Braun DR, Nelson J, Simpkins SW, McDonald BR, Myers CL, Piotrowski JS, Thompson CJ, Currie CR, Li L, Rajski SR, Bugni TS. 2017. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chem Biol 12:3093–3102. doi: 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adnani N, Vazquez-Rivera E, Adibhatla SN, Ellis GA, Braun DR, Bugni TS. 2015. Investigation of interspecies interactions within marine Micromonosporaceae using an improved co-culture approach. Mar Drugs 13:6082–6098. doi: 10.3390/md13106082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamjam M, Sivalingam P, Deng Z, Hong K. 2017. Deep sea actinomycetes and their secondary metabolites. Front Microbiol 8:760. doi: 10.3389/fmicb.2017.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chávez R, Fierro F, García-Rico RO, Vaca I. 2015. Filamentous fungi from extreme environments as a promising source of novel bioactive secondary metabolites. Front Microbiol 6:903. doi: 10.3389/fmicb.2015.00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomczyk-Żak K, Zielenkiewicz U. 2016. Microbial diversity in caves. Geomicrobiol J 33:20–38. doi: 10.1080/01490451.2014.1003341. [DOI] [Google Scholar]
- 31.Adetutu E, Thorpe K, Shahsavari E, Bourne S, Cao X, Mazaheri R, Kirby G, Ball S. 2012. Bacterial community survey of sediments at Naracoorte Caves, Australia. Int J Speleol 41:137–147. doi: 10.5038/1827-806X.41.2.2. [DOI] [Google Scholar]
- 32.Wu Y, Tan L, Liu W, Wang B, Wang J, Cai Y, Lin X. 2015. Profiling bacterial diversity in a limestone cave of the western Loess Plateau of China. Front Microbiol 6:244. doi: 10.3389/fmicb.2015.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barton HA, Taylor MR, Pace NR. 2004. Molecular phylogenetic analysis of a bacterial community in an oligotrophic cave environment. Geomicrobiol J 21:11–20. doi: 10.1080/01490450490253428. [DOI] [Google Scholar]
- 34.Barton HA, Luiszer F. 2005. Microbial metabolic structure in a sulfidic cave hot spring: potential mechanisms of biospeleogenesis. J Cave Karst Stud 67:28–38. [Google Scholar]
- 35.Hamaki T, Suzuki M, Fudou R, Jojima Y, Kajiura T, Tabuchi A, Sen K, Shibai H. 2005. Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J Biosci Bioeng 99:485–492. doi: 10.1263/jbb.99.485. [DOI] [PubMed] [Google Scholar]
- 36.Tiwari K, Gupta RK. 2012. Rare actinomycetes: a potential storehouse for novel antibiotics. Crit Rev Biotechnol 32:108–132. doi: 10.3109/07388551.2011.562482. [DOI] [PubMed] [Google Scholar]
- 37.Goh E-B, Yim G, Tsui W, McClure J, Surette MG, Davies J. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A 99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka Y, Hosaka T, Ochi K. 2010. Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2). J Antibiot 63:477–481. doi: 10.1038/ja.2010.53. [DOI] [PubMed] [Google Scholar]
- 39.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 40.Yagi A, Makino K, Nishioka I. 1974. Studies on the constituents of Aloe sapnaria HAW. I. The structures of tetrahydroanthracene derivatives and the related anthraquinones. Chem Pharm Bull 22:1159–1166. doi: 10.1248/cpb.22.1159. [DOI] [PubMed] [Google Scholar]
- 41.Cui H, Shaaban KA, Qin S. 2006. Two anthraquinone compounds from a marine actinomycete isolate M097 isolated from Jiaozhou Bay. World J Microbiol Biotechnol 22:1377–1379. doi: 10.1007/s11274-006-9197-2. [DOI] [Google Scholar]
- 42.Abdalla MA, Win HY, Islam MT, von Tiedemann A, Schuffler A, Laatsch H. 2011. Khatmiamycin, a motility inhibitor and zoosporicide against the grapevine downy mildew pathogen Plasmopara viticola from Streptomyces sp. ANK313. J Antibiot (Tokyo) 64:655–659. doi: 10.1038/ja.2011.68. [DOI] [PubMed] [Google Scholar]
- 43.Funayama S, Ishibashi M, Komiyama K, Omura S. 1991. A new antibiotic, okicenone. J Antibiot (Tokyo) 44:819–823. doi: 10.7164/antibiotics.44.819. [DOI] [PubMed] [Google Scholar]
- 44.Derewacz DK, McNees CR, Scalmani G, Covington CL, Shanmugam G, Marnett LJ, Polavarapu PL, Bachmann BO. 2014. Structure and stereochemical determination of hypogeamicins from a cave-derived actinomycete. J Nat Prod 77:1759–1763. doi: 10.1021/np400742p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kallifidas D, Kang H, Brady SF. 2012. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J Am Chem Soc 134:19552–19555. doi: 10.1021/ja3093828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stubbendieck RM, Straight PD. 2017. Linearmycins activate a two-component signaling system involved in bacterial competition and biofilm morphology. J Bacteriol 199:e00186-17. doi: 10.1128/jb.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. 2017. antiSMASH 4.0–improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keatinge-Clay AT. 2007. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol 14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 49.Hong H, Fill T, Leadlay PF. 2013. A common origin for guanidinobutanoate starter units in antifungal natural products. Angew Chem Int Ed Engl 52:13096–13099. doi: 10.1002/anie.201308136. [DOI] [PubMed] [Google Scholar]
- 50.De Mandal S, Chatterjee R, Kumar NS. 2017. Dominant bacterial phyla in caves and their predicted functional roles in C and N cycle. BMC Microbiol 17:90. doi: 10.1186/s12866-017-1002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D'Auria G, Artacho A, Rojas RA, Bautista JS, Méndez R, Gamboa MT, Gamboa JR, Gómez-Cruz R. 2018. Metagenomics of bacterial diversity in Villa Luz caves with sulfur water springs. Genes 9:55. doi: 10.3390/genes9010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Portillo MC, Gonzalez JM, Saiz-Jimenez C. 2008. Metabolically active microbial communities of yellow and grey colonizations on the walls of Altamira Cave, Spain. J Appl Microbiol 104:681–691. doi: 10.1111/j.1365-2672.2007.03594.x. [DOI] [PubMed] [Google Scholar]
- 53.Oliveira C, Gunderman L, Coles CA, Lochmann J, Parks M, Ballard E, Glazko G, Rahmatallah Y, Tackett AJ, Thomas DJ. 2017. 16S rRNA gene-based metagenomic analysis of Ozark cave bacteria. Diversity 9:31. doi: 10.3390/d9030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adam D, Maciejewska M, Naômé A, Martinet L, Coppieters W, Karim L, Baurain D, Rigali S. 2018. Isolation, characterization, and antibacterial activity of hard-to-culture actinobacteria from cave moonmilk deposits. Antibiotics (Basel) 7:28. doi: 10.3390/antibiotics7020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gonzalez J, Portillo M, Saiz-Jimenez C. 2008. Microbes pose a risk to prehistoric cave paintings. Microbe 3:72–77. doi: 10.1128/microbe.3.72.1. [DOI] [Google Scholar]
- 56.Hong SY, Roze LV, Linz JE. 2013. Oxidative stress-related transcription factors in the regulation of secondary metabolism. Toxins (Basel) 5:683–702. doi: 10.3390/toxins5040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ochi K. 1987. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J Bacteriol 169:3608–3616. doi: 10.1128/jb.169.8.3608-3616.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao D, Bushin LB, Moon K, Wu Y, Seyedsayamdost MR. 2017. Discovery of scmR as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264. Proc Natl Acad Sci U S A 114:E2920–E2928. doi: 10.1073/pnas.1619529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakuda S, Guce-Bigol U, Itoh M, Nishimura T, Yamada Y. 1995. Linearmycin A, a novel linear polyene antibiotic. Tetrahedron Lett 36:2777–2780. doi: 10.1016/0040-4039(95)00392-P. [DOI] [Google Scholar]
- 60.Igarashi Y, Iwashita T, Fujita T, Naoki H, Yamakawa T, Yoshida R, Furuma T. 2003. Clethramycin, a new inhibitor of pollen tube growth with antifungal activity from Streptomyces hygroscopicus TP-A0623. II. Physico-chemical properties and structure determination. J Antibiot (Tokyo) 56:705–708. [DOI] [PubMed] [Google Scholar]
- 61.McAlpine JB, Bachmann BO, Piraee M, Tremblay S, Alarco AM, Zazopoulos E, Farnet CM. 2005. Microbial genomics as a guide to drug discovery and structural elucidation: ECO-02301, a novel antifungal agent, as an example. J Nat Prod 68:493–496. doi: 10.1021/np0401664. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Hashimoto T, Qin B, Hashimoto J, Kozone I, Kawahara T, Okada M, Awakawa T, Ito T, Asakawa Y, Ueki M, Takahashi S, Osada H, Wakimoto T, Ikeda H, Shin-ya K, Abe I. 2017. Characterization of giant modular PKSs provides insight into genetic mechanism for structural diversification of aminopolyol polyketides. Angew Chem Int Ed 56:1740–1745. doi: 10.1002/anie.201611371. [DOI] [PubMed] [Google Scholar]
- 63.Hoefler BC, Stubbendieck RM, Josyula NK, Moisan SM, Schulze EM, Straight PD. 2017. A link between linearmycin biosynthesis and extracellular vesicle genesis connects specialized metabolism and bacterial membrane physiology. Cell Chem Biol 24:1238.e7–1249.e7. doi: 10.1016/j.chembiol.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 64.Stubbendieck RM, Brock DJ, Pellois J-P, Gill JJ, Straight PD. 2018. Linearmycins are lytic membrane-targeting antibiotics. J Antibiot 71:372–381. doi: 10.1038/s41429-017-0005-z. [DOI] [PubMed] [Google Scholar]
- 65.Nothias L-F, Nothias-Esposito M, da Silva R, Wang M, Protsyuk I, Zhang Z, Sarvepalli A, Leyssen P, Touboul D, Costa J, Paolini J, Alexandrov T, Litaudon M, Dorrestein PC. 2018. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J Nat Prod 10.1021/acs.jnatprod.7b00737. [DOI] [PubMed] [Google Scholar]
- 66.Earl DC, Ferrell PB Jr, Leelatian N, Froese JT, Reisman BJ, Irish JM, Bachmann BO. 2018. Discovery of human cell selective effector molecules using single cell multiplexed activity metabolomics. Nat Commun 9:39. doi: 10.1038/s41467-017-02470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kieser T, Bibb MJ, Buttner M, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England. [Google Scholar]
- 68.Lo C-C, Chain PSG. 2014. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinformatics 15:366. doi: 10.1186/s12859-014-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.