Most microbial species inhabiting natural environments have not yet been isolated. One of the serious issues preventing their isolation is intrinsically slow and/or poor growth. Moreover, these slow and/or poor growers are likely to be highly sensitive to environmental stresses, especially oxidative stress. We reported previously that interaction between agar and phosphate during autoclave sterilization generates hydrogen peroxide, which adversely affects the culturability of environmental microorganisms, in particular, slow-growing organisms vulnerable to oxidative stress. In this study, we successfully isolated many slow-growing bacterial strains with phylogenetic novelty by simply modifying their cultivation on agar plates, i.e., autoclaving the phosphate and agar separately. The current limited repertoire of culture techniques still has room for improvement in the isolation of microorganisms previously considered unculturable.
KEYWORDS: isolation, agar plate, bacteria, poor growers, slow growers, uncultured
ABSTRACT
Most microorganisms living in the environment have yet to be cultured, owing at least in part to their slow and poor propagation properties and susceptibility to oxidative stress. Our previous studies demonstrated that a simple modification in the preparation of agar media, i.e., autoclaving the phosphate and agar separately (termed “PS” medium), can greatly improve the culturability of microorganisms by mitigating oxidative stress compared with the use of “PT” medium (autoclaving the phosphate and agar together). Here, we attempted to isolate phylogenetically novel bacteria by combining PS medium with prolonged cultivation. After inoculation with forest soil or pond sediment samples, significantly more colonies appeared on PS medium than on PT medium. A total of 98 and 74 colonies that emerged after more than 7 days of cultivation were isolated as slow growers from PS and PT media, respectively. Sequencing analysis of their 16S rRNA genes revealed that the slow growers recovered from PS medium included more phylogenetically novel bacteria than those from PT medium, including a strain that could be classified into a novel order in the class Alphaproteobacteria. Further physiological analysis of representative strains showed that they were actually slow and poor growers and formed small but visible colonies only on PS medium. This study demonstrates that the culturability of previously uncultured bacteria can be improved by using an isolation strategy that combines a simple modification in medium preparation with an extended incubation time.
IMPORTANCE Most microbial species inhabiting natural environments have not yet been isolated. One of the serious issues preventing their isolation is intrinsically slow and/or poor growth. Moreover, these slow and/or poor growers are likely to be highly sensitive to environmental stresses, especially oxidative stress. We reported previously that interaction between agar and phosphate during autoclave sterilization generates hydrogen peroxide, which adversely affects the culturability of environmental microorganisms, in particular, slow-growing organisms vulnerable to oxidative stress. In this study, we successfully isolated many slow-growing bacterial strains with phylogenetic novelty by simply modifying their cultivation on agar plates, i.e., autoclaving the phosphate and agar separately. The current limited repertoire of culture techniques still has room for improvement in the isolation of microorganisms previously considered unculturable.
INTRODUCTION
It is widely recognized that the number of viable microorganisms in the environment is several orders of magnitude greater than the number detected by cultivation on nutrient-rich laboratory media; this phenomenon is termed the “great plate count anomaly” (1, 2). The recent development of sequencing technologies, including metagenomic analysis, has enabled researchers to estimate the occurrence of large numbers of uncultured microorganisms and their potential functions in the environment without cultivation (3). However, there is no doubt that it is essential for microbiologists to isolate such as-yet-uncultured and functionally important microorganisms. Without their isolation, we will never know their biology and the functions that have been hypothesized as a result of culture-independent omics studies (4–7).
Many attempts have been made to develop efficient methods of isolating uncultured microorganisms (8–12). These include new cultivation platforms mimicking natural environments (13, 14), alteration of gelling agents (15), physicochemical separation of cells to decrease the negative effects of competitors and inhibitors (16, 17), addition of antioxidants (18), and supplementation with signal compounds (19). In addition, we recently revealed that a hidden pitfall in the preparation of agar media greatly lowers the culturability of microorganisms in various environments (20, 21). We demonstrated that reactive oxygen species (ROS) are generated and inhibit colony formation by some microorganisms when phosphate is autoclaved together with agar (termed “PT” medium, where “P” is phosphate and “T” represents “together”). In contrast, the generation of ROS and unfavorable compounds is minimized and the culturability of uncultured microorganisms improved by separate sterilization of phosphate and agar (termed “PS” medium, where “S” represents “separately”). Subsequent studies have further demonstrated that this technique is very effective for culturing recalcitrant microorganisms from various environments (22–24). However, the application of PS medium for isolation is still limited. Especially, the effect of PS medium on the isolation of slowly and/or poorly growing microorganisms has not yet been confirmed.
It is well accepted that many fastidious microorganisms, particularly those living in oligotrophic environments, are slow and/or poor growers. For example, long-term incubation for up to 24 weeks was required for the isolation of SAR11 clade bacteria, which are the most abundant and ubiquitous bacteria in the ocean (25, 26). Strategies used to improve the cultivation efficiency of slow and/or poor growers are (i) reducing the inoculum size to decrease the chance of encountering fast-growing competitors and (ii) increasing incubation time to allow sufficient growth (27). Indeed, there have been many reports that these strategies can substantially improve the culturability of microorganisms from a variety of environmental samples and the possibility of obtaining phylogenetically novel microorganisms (27–34). In contrast, Buerger et al. have reported that the probability of finding phylogenetically novel microorganisms is not improved simply by picking up slow-growing colonies (35, 36). They showed that most of the bacterial strains obtainable through simple long-term cultivation are not a pool of slow growers but instead are fast growers that take time to form colonies only for the first time upon “wake-up” from their in situ environments (12, 35, 36).
From these reports, we hypothesized that it would be possible to effectively acquire novel microorganisms by adding another strategy to a simple long-term cultivation method. We therefore prepared culture medium by separate sterilization of phosphate and agar (i.e., PS medium) to isolate slow-growing bacteria from natural environments. This is because PS medium has been demonstrated to improve the formation of colonies by some slow- and/or poorly growing microorganisms, such as Gemmatimonas aurantiaca (20). We also validated the effectiveness of this strategy by phylogenetic and physiological analyses of several slow-growing isolates.
RESULTS AND DISCUSSION
Validation of effectiveness of PS medium for improving culturability.
Throughout the experiments, forest soil and pond sediment were used as isolation sources, and the PYG medium (containing peptone, yeast extract, and glucose as the substrates) was used as a basal medium (20). To evaluate the effects of the method of preparation of agar plates (i.e., PT or PS) on the culturability of environmental microorganisms, numbers of CFU were monitored over 3 weeks of incubation (Fig. 1). In all of the cultivation experiments, the number of CFU increased linearly in the initial stage of incubation (days 1 to 5). Although the rate of increase in CFU became moderate thereafter, we confirmed that new colonies continued to emerge, even after a week of cultivation. For both types of environmental samples, the numbers of CFU obtained on PS medium were significantly greater than those obtained on PT medium (P < 0.05). This observed improvement in culturability by separate sterilization of phosphate and agar in solid medium preparation was consistent with our previous findings (20, 21).
FIG 1.
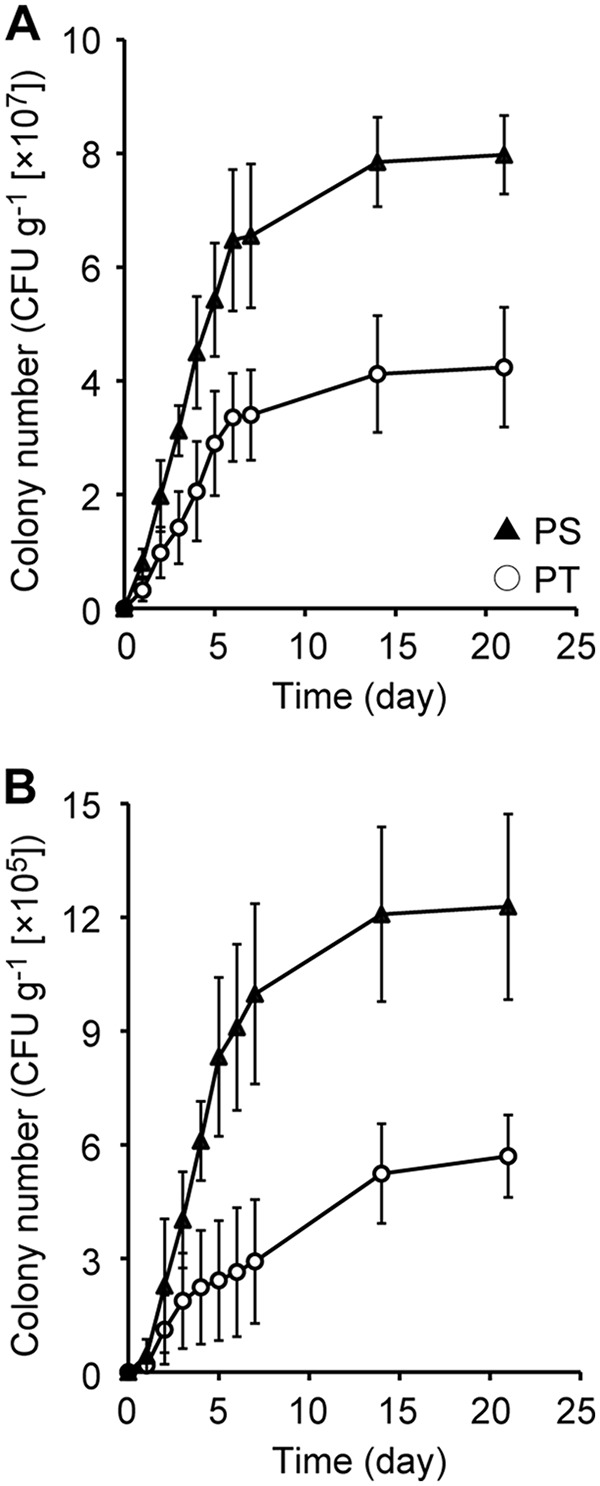
Total colony numbers from soil (A) and sediment (B) samples grown on PT (phosphates and agar autoclaved together) and PS (phosphates and agar autoclaved separately) agar media. CFU counts are averages from five replicate agar plates. Error bars represent standard deviations.
Isolation of slow growers and determination of their phylogenetic properties.
To isolate slow-growing microbes, the incubation period was extended to 3 weeks in this study, whereas cultivation was performed only for 7 days in our previous study (20). New colonies that appeared after more than 7 days of incubation were defined as slow growers and were picked up and purified on fresh agar medium. A total of 172 strains of slow growers were isolated, and their partial 16S rRNA gene sequences were determined. The results of phylogenetic analysis of the slow-growing isolates are summarized in Table 1. Table 1 also contains the results obtained for fast growers (colonies that emerged within 7 days of cultivation) reported in our previous article (20) for comparison.
TABLE 1.
Comparison of diversity and novelty of microorganisms isolated from soil and sediment samples by using different isolation procedures
| Microorganism | No. of isolates | No. of OTUsa | Shannon diversity index | No. of novel OTUsb | Novelty indexc |
|---|---|---|---|---|---|
| Slow-growing isolates | |||||
| Soil PT | 29 | 16 | 2.49 | 1 | 0.034 |
| Soil PS | 28 | 18 | 2.77 | 8 | 0.286 |
| Sediment PT | 45 | 13 | 1.75 | 2 | 0.044 |
| Sediment PS | 70 | 34 | 3.02 | 16 | 0.229 |
| Fast-growing isolatesd | |||||
| Soil PT | 169 | 27 | 2.48 | 2 | 0.012 |
| Soil PS | 355 | 52 | 2.97 | 6 | 0.017 |
| Sediment PT | 303 | 44 | 2.79 | 6 | 0.020 |
| Sediment PS | 503 | 69 | 3.30 | 22 | 0.044 |
Isolates were assigned to OTUs with a cutoff value of 97% identity on the basis of their partial 16S rRNA gene sequences.
OTUs with less than 80% classification reliability at the genus level in RDP Classifier analysis were defined as novel OTUs.
Novelty index was calculated by the number of novel OTUs/total number of isolates.
Data on fast-growing isolates were from our previous work (20).
Phylogenetic analysis of the slow-growing isolates was conducted by using Ribosomal Database Project (RDP) Classifier. The overall trends in phylum or class level are shown in Fig. 2. The slow growers recovered from the soil sample were dominated by Alphaproteobacteria and Actinobacteria and contained some Bacteroidetes and Firmicutes strains, regardless of the medium preparation protocol used. Notably, strains classified into Gammaproteobacteria and Acidobacteria were isolated only from PS medium. The overall phylogenetic trends in the slow growers from the sediment samples on PT and PS media were also similar, in that Alphaproteobacteria and Actinobacteria were the major bacterial taxa, and Betaproteobacteria and Bacteroidetes were the minor ones. These observations suggested that the effects of the medium preparation protocol on the phylogenetic trends of slow-growing isolates were not remarkable at least at the phylum or class level.
FIG 2.
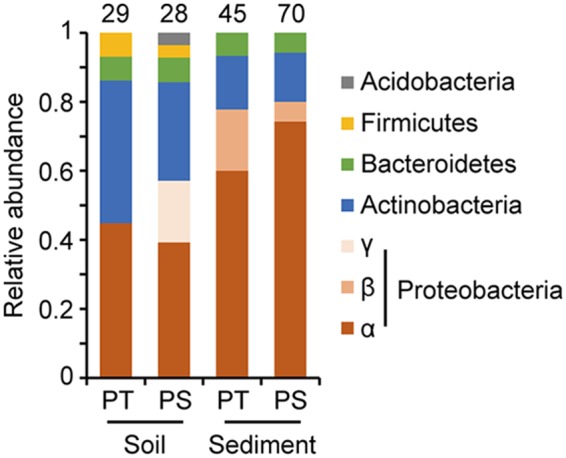
Phylogenetic distribution of strains isolated from soil and sediment by using the PT and PS media. Isolates were classified at the phylum/class level by RDP Classifier on the basis of their partial 16S rRNA gene sequences. The number above each bar indicates the number of isolates obtained with each isolation method.
Effects of isolation method on diversity of strains isolated.
On the basis of their partial 16S rRNA gene sequences, the slow-growing isolates were assigned to operational taxonomic units (OTUs) by using a cutoff value of 97% identity. Detailed phylogenetic information on the total of 67 OTUs is given in Table S1 in the supplemental material. To determine whether the modification of the culture method allowed us to isolate a greater variety of microbial species, the diversity of isolates obtained under each condition was evaluated by calculating the Shannon diversity index on the basis of the number of OTUs and the number of strains assigned to each OTU (Table 1). As reported in our previous study (20), the diversity indexes of fast-growing isolates from PS medium were higher than those from PT medium (soil, 2.97 versus 2.48; sediment, 3.30 versus 2.79, respectively). The same trend was also observed for slow-growing isolates; the diversity indexes were higher in PS culture than in PT culture (soil, 2.77 versus 2.49; sediment; 3.02 versus 1.75, respectively). These results indicated that modification of the preparation of solid media (i.e., use of the PS protocol) could increase the possibility of isolating a greater diversity of microbial species, regardless of their growth rate.
Effects of isolation method on novelty of strains isolated.
The frequency of isolation of phylogenetically novel microorganisms was compared among the respective isolation protocols. Representative sequences from each OTU were subjected to RDP Classifier analysis, and OTUs with less than 80% classification reliability at the genus level were defined as novel. The novelty index for each isolation protocol was calculated using the equation number of novel OTUs/total number of isolates (Table 1). As reported in our previous study (20), the novelty index of fast-growing isolates from PS medium was higher than that from PT medium (soil, 0.017 versus 0.012; sediment, 0.044 versus 0.020, respectively). Our present study demonstrated the same trend in slow growers; the isolates from PS medium contained phylogenetically novel bacteria in a greater proportion than those from PT medium (soil, 0.286 versus 0.034; sediment, 0.229 versus 0.044, respectively). Furthermore, the increase in novelty index by applying the PS protocol was more remarkable when targeting slow growers. These results clearly demonstrated that a combination of the two isolation strategies, i.e., the use of PS medium and a longer incubation time, greatly increased the possibility of capturing phylogenetically novel microorganisms.
Growth properties of phylogenetically novel isolates.
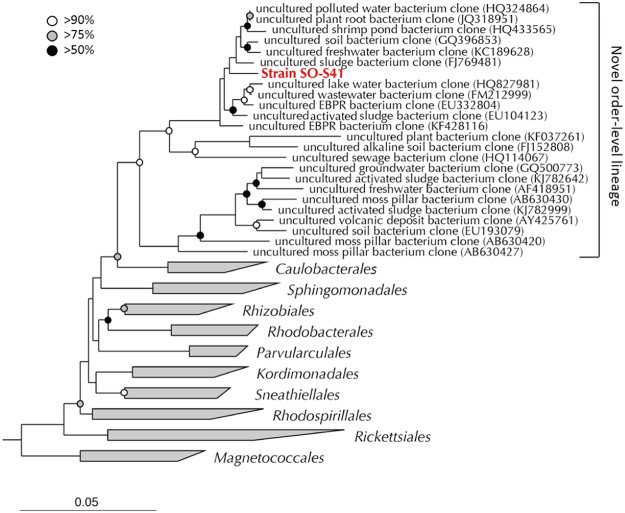
We conducted further phylogenetic and physiological analyses on three representative strains among the phylogenetically novel slow growers isolated from PS medium. Strain SO-S41 (OTU YG14) was isolated from soil, classified into the class Alphaproteobacteria, and was found to be closely related to Nitrospirillum amazonense (89.6% identity). To precisely evaluate its phylogenetic novelty, almost the full length of the 16S rRNA gene sequence was determined. The close relatives of strain SO-S41 were Pleomorphomonas oryzae (89.0% identity) and N. amazonense (88.3% identity). To obtain further phylogenetic information, we constructed a phylogenetic tree by using the alphaproteobacterial sequences collected from isolated strains and environmental samples (Fig. 3). Strain SO-S41 was not included in any known orders in the Alphaproteobacteria and formed a unique order-level lineage with the environmental sequences. Thus, strain SO-S41 might be a phylogenetically novel bacterium at the order level.
FIG 3.
Phylogenetic positions of the novel strain SO-S41 and its close relatives within the class Alphaproteobacteria. The tree was constructed by using the neighbor-joining method. Scale bar indicates 0.05 substitutions per nucleotide position. Nodes with bootstrap values of >50%, >75%, and >90% are shown as black circles, gray circles, and open circles, respectively. Three different sequences (Vibrio chagasii [accession no. AJ316199], Vibrio harveyi [accession no. EU130475], and Escherichia coli [accession no. EU014689]) of valid species within the Gammaproteobacteria were used as the outgroup. EBPR, enhanced biological phosphorus removal. Accession numbers are shown in parentheses next to the organism descriptions in the novel order-level lineage group.
After more than 1 week of incubation, strain SO-S41 formed tiny but visible colonies on the agar plates prepared by using the PS protocol, but no colonies were found on the same agar plates prepared by using the PT protocol, even after prolonged incubation of more than 6 weeks (Fig. 4A and B). This strain also grew slowly and poorly in the PYG liquid medium. It required about 7 days for full growth, and the final optical density at 600 nm (OD600) value was only about 0.11 (data not shown). Although the growth of strain SO-S41 was also tested in other conventional liquid media (e.g., diluted R2A or tryptic soy broth [TSB]) and at different pH values and ionic strengths, growth was not improved by any of these other conditions tested (data not shown).
FIG 4.
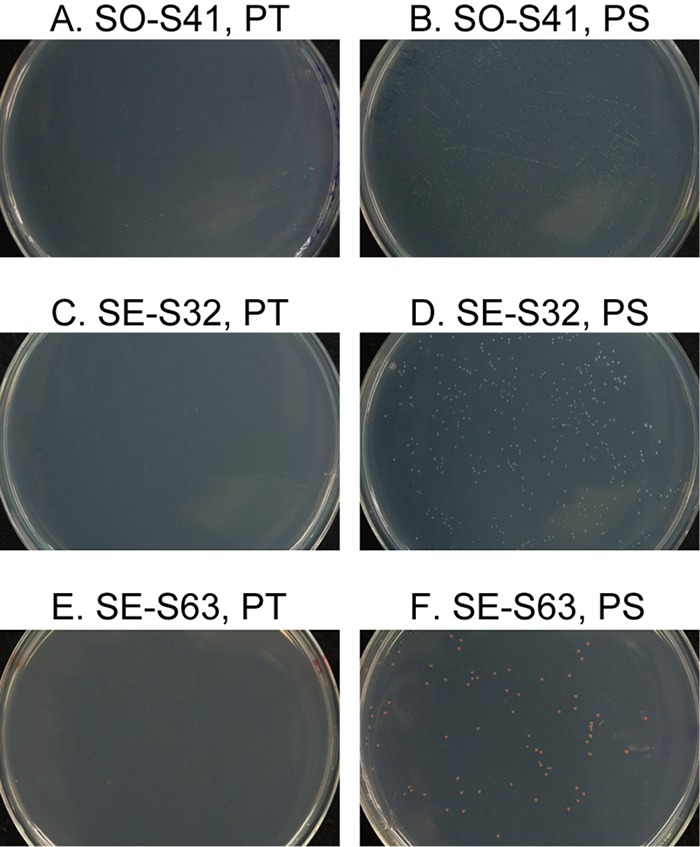
Colony formation by the three representative isolates on PT and PS agar plates. The photographs were taken after 2 weeks of incubation.
Strains SE-S32 (OTU YG27) and SE-S63 (OTU YG29) were isolated from sediment and are also alphaproteobacterial strains. The closest relative of strain SE-S32 on the basis of its nearly full-length 16S rRNA gene was Prosthecomicrobium hirschii (95.1% identity). The results of the phylogenetic tree analysis suggested that strain SE-S32 could be classified into a novel genus in the family Hyphomicrobiaceae (see Fig. S1 in the supplemental material). The closest relative of strain SE-S63 was Methylocella silvestris (92.3% identity). Strain SE-S63 was expected to be a novel genus in the family Methylobacteriaceae, as determined from the results of the phylogenetic tree analysis (Fig. S2). These two strains formed tiny but visible colonies after more than 1 week of cultivation on PS agar medium but not on PT agar medium even after more than 6 weeks of cultivation (Fig. 4C to F). Also, these strains grew slowly and poorly in the PYG liquid medium, as did strain SO-S41 (data not shown). Collectively, these results indicated that at least some of our novel isolates were actually slow and poor growers; in other words, they were recalcitrant microorganisms that could not be isolated without employing our cultivation strategies.
Conclusion.
Our results clearly demonstrated that the culturability of previously uncultured bacteria with phylogenetic novelty could be improved by using an isolation strategy that combined prolonged cultivation with a simple modification of the preparation of agar media. Although only aerobic chemoheterotrophs were targeted for isolation in this study, this strategy can be applicable to other microorganisms, such as anaerobes, lithotrophs, and autotrophs. We expect that application of this strategy to diverse environmental samples, especially in oligotrophic natural environments, such as freshwater and seawater, will enable the isolation of a number of phylogenetically and functionally novel microorganisms.
MATERIALS AND METHODS
Preparation of agar media.
The PYG agar medium used for isolation of microorganisms from environmental samples was prepared as described previously (20, 21). The medium constituents were grouped into three solutions (solutions A, B, and C). Solution A contained basal salts and Bacto agar (final concentration 15 g · liter−1), solution B was phosphate buffer (final concentration 20 mM phosphate [pH 7]), and solution C contained Bacto peptone, Bacto yeast extract, and glucose, with final concentrations of 0.1 g · liter−1 each. Using these solutions, agar media were prepared by using two different procedures, namely, the PT and PS protocols. For PT medium, solutions A and B were mixed together before autoclaving, and then the separately autoclaved solution C was added before pouring of the medium. For PS medium, solutions A, B, and C were separately autoclaved and subsequently mixed.
Cultivation of environmental samples on agar media.
Forest soil and pond sediment samples were collected at Hokkaido University, Sapporo, Hokkaido, Japan, as described previously (20). Soil samples from a depth of 5 to 10 cm below the surface were collected from a small deciduous forest. Sediment samples were collected from 0 to 10 cm below the bottom of a shallow pond. Each environmental sample was suspended in sterilized saline (0.9% NaCl) and diluted in a 10-fold series. Aliquots (100 μl) from each dilution were inoculated onto PT and PS agar media with five replicates and incubated at 25°C in the dark. The number of CFU on each agar plate was counted during 3 weeks of incubation. Only plates with 30 to 300 CFU were included in the cultivation results reported. New colonies that appeared more than 7 days after the start of incubation were picked up on days 14 and 21 and transferred to fresh agar plates for further purification. The data were analyzed for statistical significance using Student's t test. A total of 172 purified colonies were subjected to phylogenetic analysis based on their 16S rRNA gene sequences.
Phylogenetic analysis of isolates.
Sequencing analysis of the 16S rRNA gene was conducted as described previously (20). Purified colonies were transferred to PCR tubes containing lysis solution, and chromosomal DNA was extracted by heating the tubes in a microwave (50 s). The 16S rRNA gene was amplified by PCR with universal primers 27F (5′-AGA GTT TGA TCM TGG CTCAG-3′) and 1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) (37). The PCR products were purified by using a NucleoSpin Gel and PCR cleanup kit (TaKaRa), in accordance with the manufacturer's instructions. The sequences of the PCR products were determined by the TaKaRa Bio Company using the primer 357F (5′-CTC CTA CGG GAG GCA GCA G-3′). The 16S rRNA gene sequences from all isolates were checked for chimeras by using Decipher (38), and sequence quality was checked by using an ABI 3730xl base caller (Applied Biosystems). The sequences obtained were assigned to OTUs by using the BLASTClust program (39), with a cutoff value of 97% sequence identity. Phylogenetic classification of each OTU was performed using the RDP Classifier (40), and OTUs with less than 80% classification reliability at the genus level were defined as novel. The closest relatives of each OTU were inferred by using the BLAST program (39). Almost-full-length 16S rRNA gene sequences were determined by direct sequencing of the DNA fragment amplified with the primer pair of 27F and 1492R, as described previously (41). Phylogenetic analysis was performed with the ARB software package (http://www.arb-home.de/) (42), as described previously (15). In brief, after automatic and manual sequence alignments, phylogenetic trees were constructed by using the neighbor-joining method (43). Bootstrap values were determined from 1,000 resamplings using the neighbor-joining and maximum likelihood methods.
Accession number(s).
The nucleotide sequence data for the partial 16S rRNA genes of isolated OTUs (accession numbers LC341972 to LC342038) and the almost-full-length 16S rRNA genes of strains SE-S32, SO-S41, and SE-S63 (accession numbers LC342039, LC342040, and LC342041, respectively) have been submitted to GenBank.
Supplementary Material
ACKNOWLEDGMENTS
We declare no conflicts of interest.
This research was supported by the Institute for Fermentation, Osaka, Japan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00807-18.
REFERENCES
- 1.Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 2.Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Jia H. 2016. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol 14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 4.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayumi D, Mochimaru H, Tamaki H, Yamamoto K, Yoshioka H, Suzuki Y, Kamagata Y, Sakata S. 2016. Methane production from coal by a single methanogen. Science 354:222–225. doi: 10.1126/science.aaf8821. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K, Miyamoto K, Kimura Y, Oda K. 2016. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351:1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- 8.Alain K, Querellou J. 2009. Cultivating the uncultured: limits, advances and future challenges. Extremophiles 13:583–594. doi: 10.1007/s00792-009-0261-3. [DOI] [PubMed] [Google Scholar]
- 9.Vartoukian SR, Palmer RM, Wade WG. 2010. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett 309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 10.Pham VH, Kim J. 2012. Cultivation of unculturable soil bacteria. Trends Biotechnol 30:475–484. doi: 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Puspita ID, Kamagata Y, Tanaka M, Asano K, Nakatsu CH. 2012. Are uncultivated bacteria really uncultivable? Microbes Environ 27:356–366. doi: 10.1264/jsme2.ME12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein SS. 2013. The phenomenon of microbial uncultivability. Curr Opin Microbiol 16:636–642. doi: 10.1016/j.mib.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Kaeberlein T, Lewis K, Epstein SS. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari BC, Winsley T, Gillings M, Binnerup S. 2008. Cultivating previously uncultured soil bacteria using a soil substrate membrane system. Nat Protoc 3:1261–1269. doi: 10.1038/nprot.2008.102. [DOI] [PubMed] [Google Scholar]
- 15.Tamaki H, Hanada S, Sekiguchi Y, Tanaka Y, Kamagata Y. 2009. Effect of gelling agent on colony formation in solid cultivation of microbial community in lake sediment. Environ Microbiol 11:1827–1834. doi: 10.1111/j.1462-2920.2009.01907.x. [DOI] [PubMed] [Google Scholar]
- 16.Connon SA, Giovannoni SJ. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68:3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zengler K, Toledo G, Rappé M, Elkins J, Mathur EJ, Short JM, Keller M. 2002. Cultivating the uncultured. Proc Natl Acad Sci U S A 99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin SE, Flowers RS, Ordal ZJ. 1976. Catalase: its effect on microbial enumeration. Appl Environ Microbiol 32:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruns A, Cypionka H, Overmann J. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl Environ Microbiol 68:3978–3987. doi: 10.1128/AEM.68.8.3978-3987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka T, Kawasaki K, Daimon S, Kitagawa W, Yamamoto K, Tamaki H, Tanaka M, Nakatsu CH, Kamagata Y. 2014. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl Environ Microbiol 80:7659–7666. doi: 10.1128/AEM.02741-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki K, Kamagata Y. 2017. Phosphate-catalyzed hydrogen peroxide formation from agar, gellan, and κ-carrageenan and recovery of microbial cultivability via catalase and pyruvate. Appl Environ Microbiol 83:e01366-17. doi: 10.1128/AEM.01366-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decleyre H, Heylen K, Van Colen C, Willems A. 2015. Dissimilatory nitrogen reduction in intertidal sediments of a temperate estuary: small scale heterogeneity and novel nitrate-to-ammonium reducers. Front Microbiol 6:1124. doi: 10.3389/fmicb.2015.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dione N, Khelaifia S, La Scola B, Lagier JC, Raoult D. 2016. A quasi-universal medium to break the aerobic/anaerobic bacterial culture dichotomy in clinical microbiology. Clin Microbiol Infect 22:53–58. doi: 10.1016/j.cmi.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 24.Nishioka T, Mohamed Elsharkawy M, Suga H, Kageyama K, Hyakumachi M, Shimizu M. 2016. Development of culture medium for the isolation of Flavobacterium and Chryseobacterium from rhizosphere soil. Microbes Environ 31:104–110. doi: 10.1264/jsme2.ME15144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- 26.Song J, Oh HM, Cho JC. 2009. Improved culturability of SAR11 strains in dilution-to-extinction culturing from the East Sea, West Pacific Ocean. FEMS Microbiol Lett 295:141–147. doi: 10.1111/j.1574-6968.2009.01623.x. [DOI] [PubMed] [Google Scholar]
- 27.Button DK, Schut F, Quang P, Martin R, Robertson BR. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol 59:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis KE, Sangwan P, Janssen PH. 2011. Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ Microbiol 13:798–805. doi: 10.1111/j.1462-2920.2010.02384.x. [DOI] [PubMed] [Google Scholar]
- 29.Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sait M, Hugenholtz P, Janssen PH. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ Microbiol 4:654–666. doi: 10.1046/j.1462-2920.2002.00352.x. [DOI] [PubMed] [Google Scholar]
- 31.Joseph SJ, Hugenholtz P, Sangwan P, Osborne CA, Janssen PH. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl Environ Microbiol 69:7210–7215. doi: 10.1128/AEM.69.12.7210-7215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol 70:4748–4755. doi: 10.1128/AEM.70.8.4748-4755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis KE, Joseph SJ, Janssen PH. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol 71:826–834. doi: 10.1128/AEM.71.2.826-834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sangwan P, Kovac S, Davis KE, Sait M, Janssen PH. 2005. Detection and cultivation of soil verrucomicrobia. Appl Environ Microbiol 71:8402–8410. doi: 10.1128/AEM.71.12.8402-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. 2012. Microbial scout hypothesis and microbial discovery. Appl Environ Microbiol 78:3229–3233. doi: 10.1128/AEM.07308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. 2012. Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl Environ Microbiol 78:3221–3228. doi: 10.1128/AEM.07307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heuer H, Hartung K, Wieland G, Kramer I, Smalla K. 1999. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl Environ Microbiol 65:1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright ES, Yilmaz LS, Noguera DR. 2012. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78:717–725. doi: 10.1128/AEM.06516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato S, Goya E, Tanaka M, Kitagawa W, Kikuchi Y, Asano K, Kamagata Y. 2016. Enrichment and isolation of Flavobacterium strains with tolerance to high concentrations of cesium ion. Sci Rep 6:20041. doi: 10.1038/srep20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.