E. coli can survive in drought and flooding conditions and can tolerate large changes in osmolality. However, the cell processes that limit bacterial growth under high osmotic stress conditions are not known. In this study, the dose of four different enzymes in E. coli was decreased by using deletion strains complemented by the gene carried in a tunable plasmid. Under conditions of limiting enzyme concentration (lower than that achieved by chromosomal gene expression), cell growth can be blocked by osmotic stress conditions that are normally tolerated. These observations indicate that E. coli has evolved to deal with variations in its osmotic environment and that normal protein levels are sufficient to buffer the cell from environmental changes. Additional factors involved in the osmotic pressure response may include altered protein concentration/activity levels, weak solute interactions with ligands which can make it more difficult for proteins to bind their substrates/inhibitors/cofactors in vivo, and/or viscosity effects.
KEYWORDS: R67 dihydrofolate reductase, methylenetetrahydrofolate reductase, serine hydroxymethyl transferase, chorismate mutase, macromolecular crowding, folate, enzyme catalysis, trehalose, osmotic stress
ABSTRACT
How enzymes behave in cells is likely different from how they behave in the test tube. Previous in vitro studies find that osmolytes interact weakly with folate. Removal of the osmolyte from the solvation shell of folate is more difficult than removal of water, which weakens binding of folate to its enzyme partners. To examine if this phenomenon occurs in vivo, osmotic stress titrations were performed with Escherichia coli. Two strategies were employed: resistance to an antibacterial drug and complementation of a knockout strain by the appropriate gene cloned into a plasmid that allows tight control of expression levels as well as labeling by a degradation tag. The abilities of the knockout and complemented strains to grow under osmotic stress were compared. Typically, the knockout strain could grow to high osmolalities on supplemented medium, while the complemented strain stopped growing at lower osmolalities on minimal medium. This pattern was observed for an R67 dihydrofolate reductase clone rescuing a ΔfolA strain, for a methylenetetrahydrofolate reductase clone rescuing a ΔmetF strain, and for a serine hydroxymethyltransferase clone rescuing a ΔglyA strain. Additionally, an R67 dihydrofolate reductase clone allowed E. coli DH5α to grow in the presence of trimethoprim until an osmolality of ∼0.81 is reached, while cells in a control titration lacking antibiotic could grow to 1.90 osmol.
IMPORTANCE E. coli can survive in drought and flooding conditions and can tolerate large changes in osmolality. However, the cell processes that limit bacterial growth under high osmotic stress conditions are not known. In this study, the dose of four different enzymes in E. coli was decreased by using deletion strains complemented by the gene carried in a tunable plasmid. Under conditions of limiting enzyme concentration (lower than that achieved by chromosomal gene expression), cell growth can be blocked by osmotic stress conditions that are normally tolerated. These observations indicate that E. coli has evolved to deal with variations in its osmotic environment and that normal protein levels are sufficient to buffer the cell from environmental changes. Additional factors involved in the osmotic pressure response may include altered protein concentration/activity levels, weak solute interactions with ligands which can make it more difficult for proteins to bind their substrates/inhibitors/cofactors in vivo, and/or viscosity effects.
INTRODUCTION
Osmolytes are small molecules produced by cells in response to harsh conditions, such as heat, dehydration, and high salt concentrations (1). Three classes of osmolytes are amino acids (e.g., proline, taurine, and glutamate), polyols (glycerol, sucrose, and trehalose), and methylamines (trimethylamine oxide and glycine betaine) (2). When Escherichia coli is perturbed by osmotic stress, it uses trehalose, K+, and glutamate as osmoprotectants (3–6). For cells grown in medium containing exogenous glycine betaine, it is the predominant osmoprotectant (7). As the osmolality increases, the cell becomes more crowded due to loss of water. An upper limit of osmolality occurs (∼1.9 osmol) where free water no longer exists and E. coli growth stops (7).
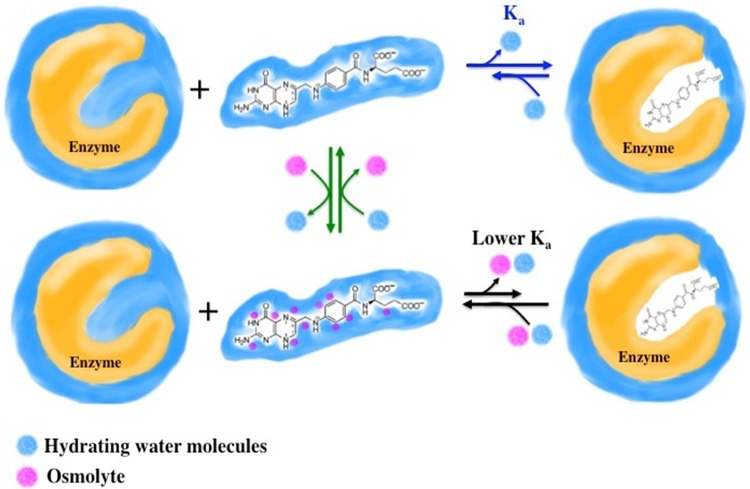
Previous studies have found that the in vitro addition of osmolytes to three different protein scaffolds that catalyze the dihydrofolate reductase (DHFR) reaction results in tighter binding of cofactor NADPH (8–10). Addition of osmolytes lowers the water activity of a solution; dehydration typically leads to tighter ligand binding, as less solvent needs to be removed to form a complex. In contrast, binding of DHF “breaks the rules,” as weaker binding upon osmolyte addition is observed in these three different proteins (8–10). To model this behavior, we proposed that osmolytes weakly associate with DHF. If the osmolyte-DHF interactions are more difficult to break than the water-DHF interactions, this shifts the equilibrium toward the unbound species and impedes binding to DHFR. This model, depicted in Fig. 1, is supported by small-molecule nuclear magnetic resonance (NMR) data (11) as well as high-hydrostatic-pressure experiments (12) and osmometry studies (13).
FIG 1.
Model of osmolyte interaction with DHF that results in weaker binding to DHFR. For enzyme assays in buffer, DHF binds tightly to DHFR (target enzyme) and water (blue) is released. Added osmolytes (magenta spheres) interact weakly with DHF. For DHF to bind to DHFR, both osmolytes and water need to be released. While these interactions are weak, if the osmolyte-DHF interaction is stronger than the water-DHF interaction, then binding to DHFR is weakened. (Note that osmolytes may additionally bind to sites on the enzyme [data not shown] and affect activity). We have used high hydrostatic pressure as an orthogonal technique to examine the top row of the model (blue equilibrium arrows) (12). We have also used NMR and vapor pressure osmometry to monitor interactions between folate and osmolytes (middle column, green equilibrium arrows) (11, 13). All data sets are consistent with this model.
Additional studies provide chemical guidance in understanding these weak interactions. For example, the Record group has also monitored the preferential interaction of betaine, glutamate, proline, and polyethylene glycols (PEGs) with numerous small molecules to determine which groups/atoms (e.g., aliphatic carbons, aromatic carbons, cationic nitrogens and amide nitrogens, phosphate oxygens, carboxylate oxygens, hydroxyls, and carbonyls) prefer to interact with water compared to several osmolytes (betaine, glutamate, and PEGs) (14–17). In a similar manner, Hong et al. have studied the preferential interaction of trehalose with many small molecules (18). In a recent investigation, we have studied the interaction of betaine with folate and other small aromatic compounds (13). We found that folate interacts almost equally well with betaine and water. Specifically, the glutamate tail prefers to interact with water, while the aromatic rings prefer betaine. As other redox states of folate, such as dihydrofolate, 5-methyltetrahydrofolate, and 5,10-methylenetetrahydrofolate, also contain these groups/atoms, these folates may also weakly associate with osmolytes and negatively impact protein function. Also, as the groups (hydroxyls, amides, cationic amines, etc.) found in osmolytes are also displayed on protein surfaces, DHF may also weakly associate with proteins.
In a recent review, Wood indicated that the cell processes that limit bacterial growth under osmotic stress conditions are not known (19). In this work we asked whether these “soft interactions” (20) or “quinary” behavior (21–24) between folate metabolites and osmolytes/crowders might occur in vivo and play a role in osmotic stress by helping to titrate enzyme activity. In other words, a high osmolyte concentration might sequester the substrate and lower the concentration of available, free substrate in the cell. This, in turn, could limit cell growth.
The enzyme concentrations/activities in E. coli are usually sufficient to overcome these weak folate-osmolyte interactions, even in the gel-like environment of the cell under conditions of high osmotic stress (25, 26). In this work, we asked what happens when the target enzyme concentration in the cell is less than that achieved by expression from the chromosomal gene. To achieve this condition, we cloned the gene of interest behind a tunable Ptet promoter in the pKTS plasmid (27). To achieve very low protein expression levels, the pKTS plasmid adds an SsrA tag to the C terminus of the protein, which targets the protein to be degraded by the ClpX protease. This plasmid is then transformed into a knockout strain of E. coli. The low protein dose lets us target the role of one specific protein in osmotic stress.
RESULTS
To determine which enzymes to use in our in vivo osmotic stress experiments, we considered whether the folate pathway enzyme is selectable by either antibiotic resistance or restoration of prototrophy to an auxotrophic strain. We also considered whether modifications to the C terminus would affect enzyme activity, which is the case for thymidylate synthase (28). Another factor considered was the oligomerization state of the enzyme, as additional SsrA degradation tags might help lower the protein concentration in the cell. Table 1 lists some pertinent parameters associated with the folate-mediated 1C metabolism enzymes we examined, and Fig. 2 shows where they occur in the pathway. The enzymes are serine hydroxymethyl transferase (SHMT; encoded by the glyA gene), methylene tetrahydrofolate reductase (MTHFR; metF), and the type II R67 DHFR. The last is unrelated to chromosomal DHFR (folA gene) and possesses a homotetrameric structure with a single active-site pore (29). A tandem array of 4 fused R67 DHFR genes produces a protein with four times the mass of the normal R67 DHFR monomer. This protein, named Quad4, was also used in our studies (30). We additionally studied osmotic stress effects on chorismate mutase (CM).
TABLE 1.
Enzyme parameters
| Enzyme | kcat (s−1) | Km (μM) | In vivo substrate concna | Rate-determining step | Oligomeric state |
|---|---|---|---|---|---|
| R67 dihydrofolate reductase | 1.3b | NADPH: 3.0 | NADPH: 120 μM | Hydride transferc | Tetramerd |
| DHF: 5.8b | Various polyglutamylated DHF species: ∼45 μM | ||||
| Quad4 dihydrofolate reductase | 1.8e | NADPH: 2.6 | NADPH: 120 μM | NDf | Dimer Kd of ∼1 μM; both monomer and dimer activee |
| DHF: 5.6e | Various polyglutamylated DHF species: ∼45 μM | ||||
| E. coli methylene tetrahydrofolate reductase (metF) | 10.4g | NADH: 20 | NADH: 83 μM | Reoxidation of the reduced flavin by CH2-THFg | Tetramer; active dimer at low protein concnh |
| 5,10-CH2-THF: 0.5g | Various polyglutamylated CH2-THF species: ∼12 μM | ||||
| E. coli serine hydroxymethyl transferase (glyA) | 10.6i | Serine: 800i | Serine: 68 μM | NDj | Dimerk |
| THF: 80 | Various polyglutamylated THF species: ∼6 μM | ||||
| Mutant hexameric chorismate mutase domain from E. coli bifunctional CM-prephenate dehydratasem (aroQ) | 0.16 (100-fold lower than E. coli CM domain)l | Chorismate: 600l | NAm | NA, but rate for the CM domain not limited by diffusionm | Hexamern |
From reference 89.
From reference 90.
From reference 36.
From reference 30.
ND, not determined, but presumably the same as R67 DHFR, as the enzyme rates and binding constants are similar.
From reference 91.
Main reaction catalyzed (94). Other reactions are listed in the text.
From reference 95.
From reference 96.
From reference 58. Also of interest, CM uses the diaxial form of its substrate; 44% glycerol and 33% sucrose shift the conformational equilibrium slightly (1.5 and 3.5%) toward the pseudo-diequatorial form (97). Additionally, no effect of 44% glycerol on either kcat or Km was observed, while an ∼2-fold-tighter Km for the substrate was monitored in 33% sucrose (97).
From reference 80.
NA, not available.
FIG 2.
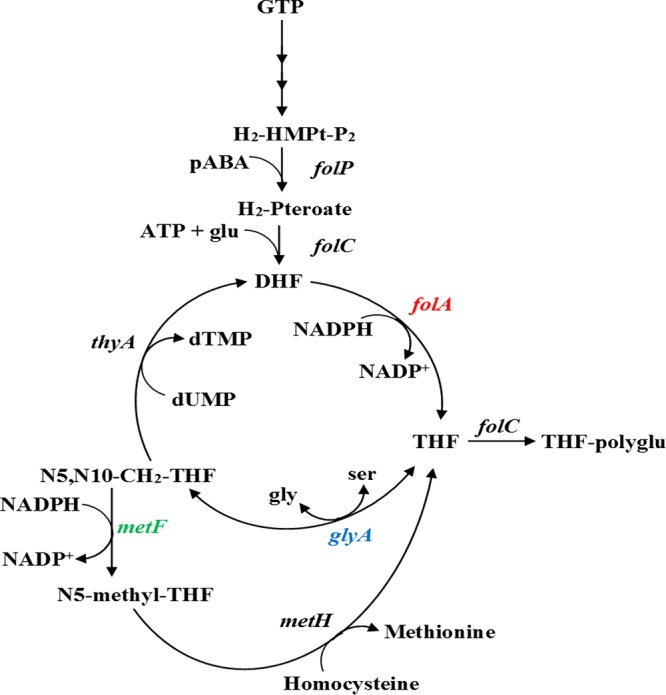
Selected folate cycle enzymes. The folP gene encodes dihydropteroate synthase (DHPS); this protein forms 7,8-dihydropteroate from 6-hydroxymethyl-dihydropterin diphosphate and p-amino benzoate. The folC gene encodes folylpolyglutamate synthase, which adds l-glutamate to dihydropteroate to form 7,8-dihydrofolate (DHF). The folA gene encodes dihydrofolate reductase (DHFR), which catalyzes the NADPH-dependent reduction of DHF to 5,6,7,8-tetrahydrofolate (THF). The glyA gene encodes serine hydroxymethyl transferase (SHMT), which interconverts l-serine and glycine using THF and 5,10-methylene-THF and pyridoxal phosphate as a cofactor. The metF gene encodes methylene-tetrahydrofolate reductase (MTHFR), which reduces 5,10-methylene-THF to 5-methyl-THF using NADPH as a cofactor. The thyA gene encodes thymidylate synthase (TS); this enzyme catalyzes the reductive methylation of 2′-deoxyuridine-5′-monophosphate (dUMP) using 5,10-methylene-THF to produce thymidine-5′-monophosphate (dTMP) and DHF. The metH gene encodes methionine synthase, which forms methionine and THF from 5-methy-THF and homocysteine. The in vivo activities of the colored enzymes were examined by our osmotic stress approach.
Osmotic stress titration of trimethoprim resistance associated with R67 DHFR.
The target of trimethoprim (TMP) is chromosomal E. coli DHFR (TMP Ki = 20 pM) (31); however, R67 DHFR provides resistance to this antibacterial drug, as its Ki is 7.5 million-fold higher (Ki = 0.15 mM) (32). We previously monitored the growth of E. coli DH5α expressing various R67 DHFR clones carried in pUC8 (9). Since wild-type (wt) R67 DHFR was overproduced and possessed sufficient activity, no selection by osmotic pressure was observed in the presence of TMP. The K32M R67 DHFR mutant clone in pUC8 had insufficient activity to allow growth in the presence of TMP, so again, no selection was observed. However, using osmotic pressure, we could titrate the ability of DH5α carrying a Y69L R67 DHFR clone to confer resistance to TMP. The Y69L mutant enzyme has levels of activity intermediate between those of the R67 DHFR and the K32M mutant (9, 33). This pattern of behavior indicates a window of enzyme activity that allows cell growth and selection by osmotic stress. Too low an activity does not enable growth, while too high an activity does not allow titration (34, 35). This situation is depicted in Fig. S1 in the supplemental material.
As mentioned above, R67 DHFR is a homotetramer with a single active-site pore (36). Another construct is Quad4, which has four R67 DHFR genes linked in frame (30). The Quad4 protein is almost fully active as a monomer. In addition, its oligomerization state depends on the protein concentration. Our in vitro ultracentrifugation studies found a Quad4 dimerization Kd (dissociation constant) of ∼1 μM (30). Since use of the pKTS vector leads to low expression levels, our in vivo studies likely describe monomeric Quad4.
To examine whether the wt R67 DHFR and Quad4 activities could be titrated by osmotic stress in vivo, their genes cloned into the pKTS plasmid (27) were transformed into DH5α and the tetracycline concentration that allowed confluent growth on M9 minimal medium was determined. Use of the Ptet promoter results in cell growth that is proportional to the amount of tetracycline added to the medium. Figure S2 shows that tetracycline concentrations of 50 to 200 ng/ml allowed DH5α carrying the R67 DHFR-pKTS clone to grow in the presence of 20 μg/ml TMP. DH5α carrying the Quad4-pKTS clone showed confluent growth in plates containing 10 to 200 ng/ml tetracycline.
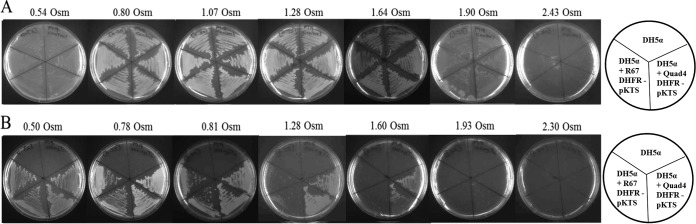
Our next step used osmotic stress to determine if growth of DH5α carrying an R67 DHFR-pKTS clone could be titrated using resistance to trimethoprim as our selection. As shown in Fig. 3, a control titration of DH5α on media lacking TMP showed confluent growth until 1.9 osmol was reached. The upper growth limit has been proposed by the Record lab to correspond to loss of free water in the cell (7). In contrast, the R67 DHFR-pKTS clone allowed confluent growth of host E. coli in medium containing 20 μg/ml TMP to 0.50 to 0.81 osmol. Sparse growth was observed until 1.28 osmol was reached. The Quad4-pKTS clone allowed growth until <1.93 osmol was reached. As R67 DHFR and Quad4 have the same active site, Quad4 serves as an internal control. Monomeric Quad4 possesses a single SsrA tag, in contrast to homotetrameric R67 DHFR, which displays 4 degradation tags. This difference in SsrA tag number likely leads to faster turnover of R67 DHFR than for Quad4. This proposed difference in enzyme concentration could provide higher DHFR activities for the Quad4 protein, which could more readily rescue the DH5α cells. Also, the fact that Quad4-pKTS continued to provide TMP resistance at high osmolalities (past where R67 DHFR-pKTS does) suggested that any osmotic pressure effects on the Tet repressor were not playing a large role in alteration of protein expression levels.
FIG 3.
Effect of osmolality on the ability of R67 and Quad4 DHFR to rescue E. coli DH5α from trimethoprim. Panel A shows the effect of sorbitol on cell growth in M9 minimal medium lacking TMP. Cells can grow up to 1.90 osmol, although growth is slow and sparse. Panel B shows the effect of sorbitol on growth in media containing 20 μg/ml TMP, 100 μg/ml ampicillin, and 100 ng/ml tetracycline. Cells carrying the R67 DHFR-pKTS clone can grow only until osmolalities between 0.81 and 1.28 are reached, while cells carrying the Quad4-pKTS clone can grow until osmolalities between 1.60 and 1.93 are reached. Table S1 lists the growth patterns as a function of osmolality. The drawings at the right show how various cultures were streaked on the plates. Two independent colonies were streaked per transformant.
Osmotic stress titration of R67 DHFR enzyme activity using a deletion strain.
While TMP is a competitive inhibitor of DHFR, antibiotics can also involve effects on uptake and efflux mechanisms as well as secondary targets (37). Thus, we also employed a more direct assay of DHFR activity using a deletion strain. The folA gene in E. coli strain NM522 was previously deleted (ΔfolA::kan) (38). These cells also carry an uncharacterized mutation in the thymidylate synthase (thyA) gene, and thymidine is required for cell growth. This strain, named LH18, grows on media supplemented with folate pathway end products (thymidine, adenine, pantothenate, glycine, and methionine) (38). Transformation of LH18 by R67 DHFR-pKTS or Quad4-pKTS rescues the cells from folate end product auxotrophy and allows growth of cells in Bonner-Vogel (BV) minimal medium (39) plus thymidine.
Next the tetracycline concentration was varied and the cell growth pattern monitored on minimal media (see Fig. S3). For LH18 carrying the R67 DHFR-pKTS plasmid (LH18 + R67 DHFR-pKTS), no growth was observed for the first 2 days at low tetracycline levels (0 to 25 ng/ml). This result indicates that the pKTS plasmid can restrict protein expression so that DHFR activity is lower than that encoded by the chromosome. This type of result has previously been reported for chorismate mutase (27). Confluent growth of the rescued cells was achieved at 100 ng/ml tetracycline. Growth for 5 days resulted in isolated colonies at low tetracycline concentrations, consistent with the occurrence of suppressing mutations. In contrast, LH18 carrying the Quad4-pKTS plasmid grew in the absence of tetracycline, consistent with some leakiness of the promoter as well as Quad4 possessing fewer degradation tags and sufficient activity to rescue the cell.
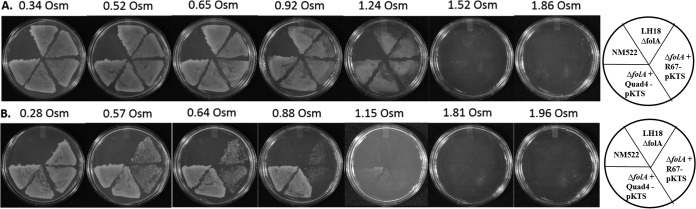
To monitor osmotic stress effects, the deletion and complemented strains were plated on BV minimal and supplemented plates containing increasing concentrations of sorbitol (Fig. 4). On the plates supplemented with thymidine, adenine, pantothenate, glycine, and methionine, all strains grew to osmolalities between 1.24 and 1.52 (see Table S1 for a list of osmolality growth limits). In minimal plates containing 200 ng/ml tetracycline, complemented strains of LH18 + R67 DHFR-pKTS grew until 0.57 osmol was reached. At this concentration and above, isolated colonies were observed after several days. These isolated colonies persisted until osmolalities between 0.88 and 1.15 were reached; however, no growth was observed at higher osmolalities.
FIG 4.
The effect of osmolality on R67 and Quad4 DHFR function in restoring the E. coli ΔfolA strain to prototrophy. Panel A shows the effect of sorbitol on cell growth in BV media supplemented with thymidine, adenine, pantothenate, glycine, and methionine and containing kanamycin. Cells can grow up to osmolalities between 1.24 and 1.52. Panel B shows the effect of sorbitol on growth in minimal BV media containing kanamycin, ampicillin, and 200 ng/ml tetracycline. Cell growth is reliant on enzyme activity encoded by the DHFR genes cloned into the pKTS plasmid. Cells carrying the R67 DHFR-pKTS plasmid can grow only until osmolalities between 0.28 and 0.57 are reached, with isolated colonies appearing at higher osmolalities (0.88 to 1.15). In contrast, cells carrying the Quad4-pKTS plasmid can grow confluently until 1.81 osmol is reached. Table S1 lists the osmolalities at which cells stop growing. The drawings indicate the positioning of the various streaks on the plates. Two independent colonies were streaked per transformant. Figure S4 in the supplemental material shows the titrations for NM522 and NM522 thyA.
In contrast to the R67 DHFR case, Quad4-pKTS readily rescues LH18, as cells grew in BV minimal medium until osmolalities between 1.15 and 1.81were reached. To determine if the activity of Quad4 DHFR could be titrated by osmotic stress, we redid the sorbitol titrations using 0 and 50 ng/ml tetracycline (data not shown). Under both these conditions, Quad4-pKTS still allowed LH18 to grow until ∼1.6 osmol was reached, which was very close to the osmolality limit of the deletion strain grown on supplemented media. Thus, the activity of Quad4 DHFR could not be titrated by osmotic stress. In contrast, LH18 complemented by R67 DHFR-pKTS did not grow on plates without tetracycline and grew only as isolated colonies when 50 ng/ml tetracycline was added. These observations are consistent with moving up and down the titration axis when considering Fig. S1. Conditions that favor low protein expression (e.g., R67 DHFR-pKTS with a low tetracycline concentration) and/or high degradation rates (higher number of SsrA tags) are not sufficient to rescue the cell from auxotrophy, while conditions that provide high enzyme activity do not allow titration.
Liquid cultures show effects similar to those with agar plates.
The growth of LH18 as well as its parent strains and complemented strains were additionally monitored in liquid media by plots of turbidity versus time. Figure 5 compares the growth rates in a bar graph, while Table S2 lists the doubling times. LH18 and LH18 carrying Quad4-pKTS showed comparable growth even though LH18 was grown in BV media supplemented with thymidine, adenine, pantothenate, glycine, and methionine, while the complemented strain was grown in BV minimal medium plus 100 ng/ml tetracycline. The deletion strain carrying R67 DHFR-pKTS grew more slowly in minimal medium with 100 ng/ml tetracycline.
FIG 5.
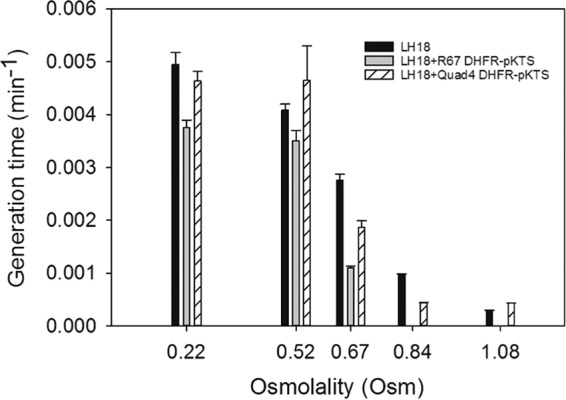
Growth rates of LH18 (ΔfolA) cells alone (black bars) or carrying the R67 DHFR-pKTS (gray bars) or Quad4-pKTS plasmid (bars with diagonal lines) were monitored in liquid culture. The growth rate is plotted as a function of medium osmolality. The growth media and the doubling times are listed in Table S2.
As the osmolality of the medium was increased, all cells grew more slowly. As shown in Fig. 5, the growth rate of LH18 cells carrying the R67 DHFR-pKTS clone was most impacted, with cell growth ceasing at osmolalities between 0.67 and 0.84. In contrast, the LH18 cells with the deletion and the complemented LH18 + Quad4-pKTS cells grew until osmolalities between 1.08 and 1.16 were reached. These trends echo those seen in the agar plate titrations, which are consistent with previous observations that immobilization of bacteria on agar plates or biofilms constrained growth compared to that under planktonic conditions and higher osmolalities could be required to inhibit growth (40, 41).
Lag times, in general, increased with increasing osmolality (42). This is likely due to initial plasmolysis (43); however, other factors, such as the number of CFU added to initiate growth, can play a role. The maximal turbidity of the cultures was also impacted, consistent with hyperosmolality leading to cells devoting more resources to osmoprotection than metabolism (44).
The above-described growth experiments were repeated using NaCl as the osmotic stressor. Doubling times are listed in Table S2. The LH18 cells with the deletion in media supplemented with thymidine, adenine, pantothenate, glycine, and methionine and the complemented Quad4-pKTS cells in minimal medium could grow until osmolalities between 1.12 and 1.4 were reached. However, the LH18 strain complemented by the R67 DHFR-pKTS clone was affected more severely by NaCl stress and grew only until osmolalities between 0.55 and 0.60 in minimal medium were reached. The trend is similar to that observed with sorbitol; however, NaCl is a slightly better stressor. This difference may arise from sorbitol being used as an alternate carbon source (45, 46). Alternatively, NaCl can have different effects than sorbitol; for example, transcriptional responses (47), induction of different proteins (48, 49), and ionic effects (50) could alter the magnitude of the osmotic stress effect.
Growth under osmotic stress is dependent on the enzyme concentration in the cell.
The DHFR concentration can be increased by addition of higher tetracycline concentrations, which drives expression of the Ptet promoter. To monitor growth rate as a function of varying the amount of sorbitol under different tetracycline concentrations, three-dimensional (3D) plots were constructed with the culture turbidity at 600 nm and the concentration of sorbitol as the axes. Figure S5A depicts the growth of R67 DHFR-pKTS-complemented LH18 cells in BV minimal media containing 0, 50, 100, and 200 ng/ml tetracycline. Without tetracycline, there was no growth. Increased growth occurred with increasing tetracycline concentrations. Figure S5B also shows the growth curves of the parent strains NM522 and NM522 thyA. Growth of the parent strains was faster than that of the pKTS-complemented strains, consistent with the hypothesis that use of the pKTS plasmid allows lower levels of protein concentration/activity than provided by the chromosome (27).
Addition of betaine to osmotic stress titrations enables growth to higher osmolalities.
When glycine betaine is added to the growth medium, it becomes the predominant osmolyte in the cell (7). Betaine is an osmoprotectant that functions by preferential exclusion from protein surfaces (51), which results in it often being used to fold proteins (52–54).
To study the effect of added betaine, we redid our sorbitol titrations of LH18 carrying the R67 DHFR-pKTS plasmid with 1 mM betaine in the agar plates. The cells with the deletion and complemented cells grew until osmolalities between 1.81 and 2.22 were reached in media supplemented with thymidine, adenine, pantothenate, glycine, and methionine (Fig. S6). Addition of betaine increased the osmolality to which cells were able to grow (7). When the complemented cells were plated on minimal medium, they also tolerated much higher osmolalities when grown in the presence of 1 mM betaine. The R67 DHFR-pKTS-complemented LH18 strain grew confluently at 0.71 osmol; however, many overgrowing colonies were observed at osmolalities between 0.98 and 1.67. The Quad4-pKTS-complemented LH18 cells grew to 1.97 osmol; however, no growth was observed at 2.15 osmol. These results were similar to growth of the LH18 strain with the deletion in supplemented media. Liquid titrations of deletion and complemented strains were done using sorbitol as the osmotic stressor. Trends similar to those seen for the agar plates were observed (Fig. S6C). The ability of betaine to extend growth to higher-osmolality conditions supports the hypothesis that water activity played a role in our experiments.
Can osmotic stress titrations apply to other folate pathway enzymes?
To test whether the activity of other folate-utilizing enzymes could be titrated by in vivo osmotic stress conditions, we used methylene tetrahydrofolate reductase (MTHFR; encoded by the metF gene) and serine hydroxymethyl transferase (SHMT; encoded by the glyA gene). Summaries of these 2 enzymes are provided in Table 1.
The metF and glyA genes from E. coli were cloned into the pKTS plasmid. Tetracycline titrations showed that 100 ng/ml tetracycline allowed the ΔmetF cells carrying the MTHFR-pKTS plasmid to grow in minimal medium, while 75 ng/ml tetracycline allowed the ΔglyA cells carrying the SHMT-pKTS cells to grow confluently in minimal medium (data not shown).
In vivo titrations of methylene tetrahydrofolate reductase activity.
MTHFR is functional as a homotetramer and catalyzes the reduction of 5,10-methylene-THF to 5-methyl-THF using NADH and FAD as cofactors. The 5-methyl-THF product is used by methionine synthase to produce methionine from homocysteine.
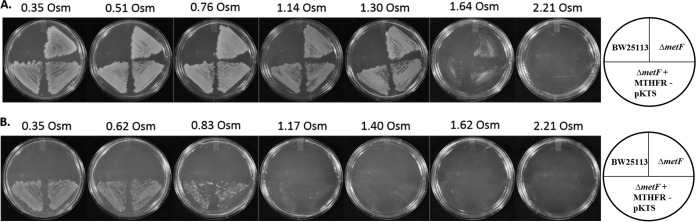
Addition of 100 ng/ml tetracycline to the MTHFR-pKTS-complemented ΔmetF strain resulted in cell growth in minimal medium lacking methionine. Figure 6 shows our in vivo sorbitol titrations. All strains grew confluently on media supplemented with methionine until 1.30 osmol was reached. Sparse growth was observed at 1.64 osmol. In contrast, two different colonies of the complemented strain (ΔmetF + MTHFR-pKTS) showed confluent growth on minimal medium until ∼0.62 osmol was reached, after which growth was dominated by isolated, overgrowing colonies. No growth was observed at osmolalities of ≥1.17. Since the complemented strains did not grow under high-osmolality conditions (1.17 osmol, minimal medium), while the parent strain grew well at 1.30 osmol (medium supplemented with methionine), we conclude that the MTHFR activity can be titrated by osmotic stress in vivo.
FIG 6.
Osmotic stress severely impairs the growth of the metF-complemented strains in minimal medium. Panel A shows the growth of the metF deletion and metF-complemented strains in minimal medium supplemented with methionine and kanamycin. BW25113 does not grow due to the presence of kanamycin. Osmotic stress was induced by the addition of sorbitol to the media. Panel B shows growth of the deletion strain carrying the MTHFR-pKTS plasmid where protein production was induced by addition of 100 ng/ml tetracycline. Cells in panel A grew until osmolalities between 1.30 and 1.64 were reached, while cells in panel B grew to 0.62 osmol (confluent growth). Table S1 lists the osmolalities at which cells stop growing. The drawings show how the cultures are streaked. Two independent colonies were streaked per transformant. Figure S4 shows how BW25113 is affected by increasing sorbitol concentrations.
Figure S7 shows titration of the ΔmetF strains using NaCl. Results similar to those with sorbitol were obtained, as all cells grew until 1.35 osmol was reached on methionine-supplemented medium. However, the complemented cells showed confluent growth on minimal medium until 0.22 osmol, with suppressor colonies until 0.48 osmol was reached and no growth after 0.71 osmol.
Doubling times of these strains in liquid media were also obtained, and the values are listed in Table S2. With increasing sorbitol stress, the growth rate of all strains decreased (see Fig. S8A). As with the agar plates, the deletion cells could survive in higher osmolalities in media supplemented with methionine than the complemented cells in minimal medium. A similar pattern was observed when NaCl was added to the media (Fig. S8B).
In vivo titrations of serine hydroxymethyl transferase activity.
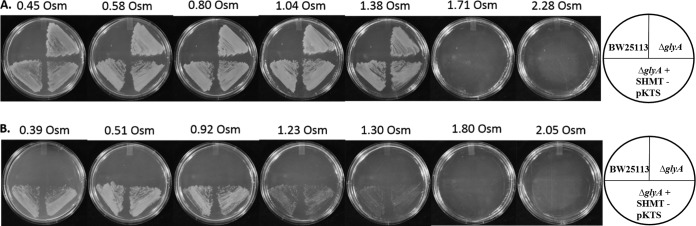
SHMT is a homodimer that interconverts glycine and serine using 5,10-methylene-THF, THF, and pyridoxal phosphate. It can also catalyze the conversion of 5,10-methenyl-THF to 5-formyl-THF (55). While this enzyme can also catalyze retroaldol cleavage, racemase, aminotransferase, and decarboxylase reactions (56), Contestabile et al. (57) suggest that the SHMT reaction is its primary in vivo function. Both serine and glycine are required for growth of ΔglyA cells. Transformation of the ΔglyA cells by SHMT-pKTS restored prototrophy to the strain in the presence of ≥75 ng/ml tetracycline. Sorbitol titrations of cell growth are shown in Fig. 7. Control titrations showed that all cells grew in BV media supplemented with glycine and serine until 1.38 osmol was reached. However, in minimal medium, the cells with the deletion complemented by the SHMT-pKTS plasmid grew confluently only until 0.92 osmol was reached; single colonies predominated from 1.23 to 1.3 osmol, while cell growth stopped at higher osmolalities.
FIG 7.
Addition of sorbitol to the growth media impacts the growth of the glyA-complemented strains. Panel A shows the growth patterns in BV supplemented medium containing serine and glycine. Panel B shows how the cells grew on minimal medium plus 75 ng/ml tetracycline. Cells in panel A grew until osmolalities between 1.38 and 1.71 were reached, while cells in minimal medium (B) grew until 0.92 osmol was reached (mat growth). Table S1 lists the osmolalities at which cells stop growing. The drawings at the right show how cultures were streaked on the plates. Two independent colonies were streaked per transformant. Figure S4 shows the titrations for BW25113.
The osmotic stress titrations were repeated using NaCl (see Fig. S9). Trends similar to those with sorbitol were observed, although NaCl had a more pronounced effect.
Osmotic stress titrations were also performed in liquid media. Table S2 provides the doubling times, while Fig. S10 shows the growth rates. The behavior was similar to that observed in the agar plates, with the complemented cells not being able to survive high-osmolality conditions on minimal media, while the cells with the deletion could grow on media supplemented with glycine and serine.
Can the activity of chorismate mutase be titrated with osmotic stress?
To test whether another enzyme can undergo in vivo osmotic stress titrations of catalytic activity, we used a chorismate mutase gene (aroQ) that was initially cloned into the pKTS plasmid (27). Chorismate sits at a metabolic branch point, and this enzyme commits chorismate to phenylalanine and tyrosine production. A gene encoding a mutant chorismate mutase was used where insertion of 5 amino acids in the middle of a helix in the protein resulted in a hexamer with an approximately 600-fold decrease in activity (58). This truncated mutant gene cloned in pKTS was then transformed into a CM-deficient strain (KA12/pKIMP-UAUC) to create a complemented strain (59, 60).
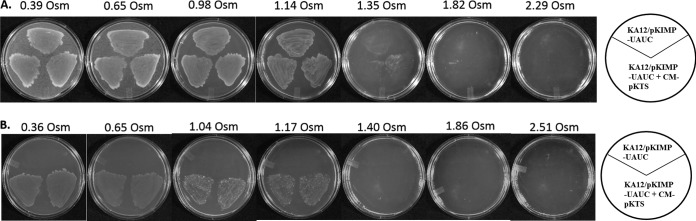
The chorismate mutase-deficient strain grew in media supplemented with tyrosine and phenylalanine, whereas prototrophy was restored in the CM-pKTS-complemented cells by the addition of ≥200 ng/ml tetracycline. As shown in Fig. 8, osmotic stress experiments with increasing sorbitol concentrations allowed the CM-deficient cells grown on media supplemented with tyrosine and phenylalanine to survive until 1.14 osmol was reached, with sparse growth seen at 1.35 osmol. The complemented cells showed confluent growth until 0.65 osmol was reached on minimal medium. An increasing number of overgrowing colonies dominated the growth from 1.04 to 1.17 osmol, with no growth at 1.40 osmol. From these titrations, it appears that CM activity can be titrated by osmotic stress.
FIG 8.
Effects of sorbitol on chorismate mutase function in vivo. Panel A shows the growth of the deletion and complemented cells in medium supplemented with tyrosine and phenylalanine. Panel B depicts the growth pattern in minimal medium plus 200 ng/ml tetracycline. Cells in supplemented medium (A) can grow until osmolalities between 1.35 and 1.82 are reached, while cells in minimal medium (B) can grow confluently until osmolalities between 0.65 and 1.04 are reached. Table S1 lists the osmolalities at which cells stop growing.
Suppressors in the SsrA tag.
Streaks on agar plates allow us to discern where confluent growth diverges from single-colony growth. We monitored agar plate growth daily and initially found confluent growth. After about 2 days, overgrowing colonies started to appear at lower tetracycline or higher sorbitol concentrations. Figure S11 shows a sample time course of colony growth as a function of time for the MTHFR-pKTS-rescued ΔmetF cells.
Twenty overgrowing colonies from each of the ΔfolA, ΔglyA, and ΔmetF knockout strains carrying their respective genes in the pKTS plasmid were subcultured in medium containing 1 M sorbitol. Plasmid DNA was extracted and retransformed into the appropriate parent strain. Typically <10 colonies continued to grow on 1 M sorbitol medium, suggesting that many original colonies had chromosomal mutations. DNA sequencing of the plasmids showed mutations in the SsrA tag or introduction of a stop codon near the end of the gene or in the SsrA tag sequence (see Table S3).
That all these mutations alter or delete the SsrA tag suggests a common mechanism of suppression, e.g., an increase in enzyme activity due to enhanced protein production. This points to the enzyme activity being limiting in the cell. This behavior has previously been observed by the Hilvert group in suppressors of chorismate mutase (27).
Are the observed effects bacteriostatic or bactericidal?
CFU were measured for the R67 DHFR-pKTS-rescued LH18 strain as well as the MTHFR-pKTS-rescued ΔmetF strain and the SHMT-pKTS-rescued ΔglyA strain. Cells were incubated in several sorbitol concentrations, ranging from 0 to 1.75 M, for up to 48 h. Complemented strains inoculated into BV minimal medium plus the appropriate tetracycline concentration did not grow at higher osmolalities, and dilutions of these cultures at 0, 24, and 48 h showed similar CFU (data not shown), indicating bacteriostatic effects (41, 61–63).
DISCUSSION
The complexity and heterogeneity of the cell provide many possible scenarios for how and why our osmotic stress titrations of enzyme activity may occur. Table 2 lists a few of the important variables, which include protein concentration, the activity of the enzyme under cellular conditions, how the various solutes interact, and the identity of the osmolytes in E. coli.
TABLE 2.
Potential contributing factors associated with in vivo titrations
| Variable | Mode of modulation |
|---|---|
| Protein concn | Removed in knockout E. coli strains; readded at tunable levels using the Ptet promoter and tetracycline in pKTSa |
| Minimized via use of SsrA degradation tag in pKTSa | |
| Proteins can be stabilized or destabilized by interaction with osmolytesb and/or crowders.c | |
| Enzyme activity can be modulated by interactions with neighboring macromolecules.d | |
| Altered protein expression levels possible due to osmotic stresse | |
| May be affected by folding mechanism associated with oligomerization; e.g., if monomer can fold coming off the ribosome, it may be more stable than if an unfolded monomer needs another monomer to be synthesized to form a dimer | |
| Enzyme activity | kon and koff rates as well as kcat/Km can be altered by interaction with crowders as well as increased viscosity upon addition of osmolytes and/or crowders. At high substrate concn, if chemistry is rate determining, then viscosity will likely have minimal effects.f |
| Substrate capture (e.g., kcat/Km) can be rate determining at low substrate concn.g | |
| If enzyme activity is inhibited by osmotic stress, the substrate concn will rise. | |
| Substrate concn in cell | Weak interaction of ligands with osmolytes, for example, folate interaction with trehalose and betaine, can lower the concn of free ligand.h |
| Cofactor concn in cell | Weak interaction of ligands with macromolecular surfaces; for example, folate interaction with lysozyme, BSA, and others can lower the concn of free ligand.i |
| Gene knockout results in buildup of the substrate concn for the encoded enzyme. | |
| No metabolomics information on folate redox state concn or NADPH in E. coli under osmotic stressj | |
| Exclusion of osmolytes from NADPH can tighten binding.k | |
| Domino effects | Buildup in substrate concn of inhibited enzymes can inhibit other enzymes in pathway. For example, inhibition of DHFR by trimethoprim results in buildup of DHF concn, which, in turn, inhibits folyl-polyglutamate synthase.l |
| Inhibition of one enzyme will deplete the concn of its product, which, as the substrate of another enzyme, will reduce this rate as well. For example, a mathematical model of folate liver metabolism predicts that >90% inhibition of DHFR is necessary to affect thymidylate synthase activity.m | |
| Identity of osmolytes in E. coli | Normally trehalose is produced as an osmoprotectant against low-water conditions.n Addition of betaine to the media results in its uptake and higher growth rates due to the preferential exclusion mechanism. Cells cannot grow when no free water remains (∼1.9 osmol).o |
| Addition of betaine switches E. coli to fermentative pathways at high NaCl concn.p |
A prime consideration is the protein concentration, which needs to be tightly controlled. Since E. coli is normally able to grow until ∼1.9 osmol is reached (7), the protein dose needs to be lower than that offered by chromosomal expression. The tunable plasmid pKTS provides a path to achieve low expression levels via use of the Ptet promoter and the SsrA degradation tag. Additionally, protein stability (and thus turnover) can be affected by the presence of osmolytes (64, 65), crowders (21, 66, 67), and/or volume exclusion (68, 69). Both stabilization (64, 66) and destabilization (66, 67) have been observed.
A second important parameter is the available ligand concentration. For example, the glutamate tail of folate prefers to interact with water, while the pterin and benzoyl groups prefer to interact with betaine (13). If the weak folate-osmolyte or folate-crowder interactions are more difficult to break than the folate-water interaction, then binding to an enzyme partner is weakened (or, restated, the binding equilibrium is shifted toward the free, unbound enzyme). This type of situation occurs with DHFR function in vitro (8–10). It may also apply to ATP, as an all-atom molecular dynamics model of the Mycoplasma genitalium cytoplasm found ATP often associated with protein surfaces (66). Most recently, ATP has been suggested to act as a biological hydrotrope, being able to decrease protein aggregation and liquid-liquid phase transitions (70). In these examples, ligands weakly interact with macromolecular surfaces. If dissociation from these surfaces is slower than desolvation, then the concentration of free ligand available for binding to its specific enzyme partner is decreased. Additionally, Zotter et al. have recently studied TEM1 β-lactamase activity in HeLa cells (71). As the logP for the microinjected substrate is 4.5, they found slower diffusion rates than for a fused mCherry-lactamase construct. They modeled the reaction as having a soft interaction with macromolecule(s) X and proposed release of substrate from X as the slow step in the in vivo reaction.
Ligand concentration can also be altered by use of a deletion strain or an inhibitor or decreased enzyme expression levels. With low to no enzyme activity, the concentration of its substrate builds up. This can provide a domino effect if the increased substrate concentration inhibits another enzyme in the pathway. An example of this is inhibition of chromosomal E. coli DHFR by trimethoprim, which results in an increased DHF concentration, which, in turn, inhibits folyl-polyglutamate synthase (72). A related factor upon enzyme inhibition is loss of its product, which can limit the activity of downstream enzymes (73).
A third issue is how enzyme activity can be modulated by osmolytes and/or crowders. Enzyme activity can be either enhanced or decreased in vitro, depending on the identity of the enzyme, ligand, and osmolyte or crowder added. Activities that are enhanced likely follow a preferential exclusion model in which osmolytes are excluded from protein surfaces (74), while activities that are decreased likely follow a preferential interaction model in which osmolytes act as a cosolvent and interact with the protein or ligand surface, albeit weakly. Acosta et al. summarized crowding effects for 32 different studies and concluded that “a one-size-fits-all theory cannot fully reduce or explain the effects of crowding on enzyme kinetics” (75). A related issue is altered viscosity, which can diminish enzyme activity by affecting either substrate binding or product release or altering any conformational changes in the enzyme (76–81). Most enzymes listed in Table 1 have chemistry as their rate-determining steps, suggesting that viscosity effects might be minimal under saturating substrate conditions. Also, as shown in Fig. 4, Quad4-pKTS allowed growth of the ΔfolA cells at 1.06 osmol, while R67 DHFR-pKTS allowed growth to ≤0.87 osmol (a larger difference would exist if confluent lawns are considered versus single-colony growth). Quad4 may serve as an internal control for R67 DHFR, as presumably the internal viscosity of the E. coli cells would be higher under the higher-osmolality conditions. These various considerations suggest that viscosity effects could play a role but likely do not dominate.
Another factor is the identity of the osmoprotectants produced by the organism in response to osmotic stress. E. coli produces trehalose, K+, and glutamate in response to osmotic pressure (3–6). E. coli takes up betaine when it is added to media; this typically allows cells to grow to higher osmolalities, as betaine has been proposed to be the most excluded osmolyte from protein surfaces (7, 51). The preferential interaction coefficient (μ23/RT) measures whether a molecule prefers to be solvated by water or to interact with osmolytes present. Table S4 provides predicted μ23/RT values for the substrates of the enzymes involved in this study with respect to trehalose and betaine. A positive value indicates a preference for solvation by water, while a negative value supports interaction with the osmolyte. A value of zero indicates that osmolyte and water interact equally well. All the folates show negative to zero values, suggesting their likely interaction with trehalose, betaine, and/or protein surfaces displaying these functional groups.
Even though the above discussion indicates that many factors may be involved, the prime effect in our experiments appears to be a decreased enzyme activity, modulated by the protein concentration and/or the availability of substrate. Another possibility is enzyme-osmolyte interactions that decrease kcat or kcat/Km. This statement arises from the above discussion as well as the pKTS plasmid, which lets us target the effect of a low protein dose for one specific protein in osmotic stress. Here we consider that all the strains are related and have sufficient protein doses to grow at high osmolalities except for the strain that underexpresses one protein. While the osmotic stress conditions could affect other proteins, their dose is sufficiently high to be able to tolerate these environmental changes. Rather it is the low-dose enzyme that is sensitive to the altered milieu.
Which titrations worked and why?
The activities of wt R67 DHFR, SHMT, MTHFR, and CM are titratable in vivo by osmotic stress. One parameter associated with these enzymes is that they all have relatively low kcat values (Table 1). This likely enables titration of enzyme activity.
Another observation is that all these enzymes are oligomers, which allows presentation of multiple SsrA tags. Avidity effects due to multiple SsrA tags may aid proteolysis. Also, folding pathways that involve unfolded monomeric intermediates could play a role.
While the folate pathway enzymes were predicted to be titratable based on their μ23/RT values (see Table S4), our results with chorismate mutase were a surprise. Why might the activity of CM be sensitive to osmotic stress in vivo? Possibly the hexameric mutant CM protein is unstable at low concentrations. Additionally, the presence of six SsrA tags should lead to rapid degradation. Another possibility is effects of the osmolytes on the enzyme.
Conclusion.
Mutations that provide some resistance to osmotic stress in bacteria identify differential expression of RNA polymerases, overproduction of osmolytes, increased transport of osmolytes, defective N-acetylglucosamine catabolism, and mutations in the cell shape-regulating protein MreB (82–85). In contrast, we took an underexpression approach and found that in the absence of external osmolyte, four different complemented strains can grow in minimal medium at a low protein dosage. However, placing these cells under high-osmolality conditions blocks growth. What is it about this combination of high osmolality and low enzyme activity that inhibits cell growth? Our in vitro studies suggest that water activity is critically important to catalytic efficiency and that weak folate-osmolyte interactions likely play a role, making it more difficult for proteins to find and bind their substrates/inhibitors/cofactors. While other factors may also be important (Table 2), our study introduces the new possibility of osmolyte/crowder interactions with ligands influencing catalytic efficiency in vivo. This scenario could be an example of negative design, as these difficulties are not normally seen: protein expression levels are sufficient to allow cell growth at high osmolalities, indicating that E. coli has evolved to deal with variations in the osmotic environment. Further study will let us know whether these titrations are restricted to folate metabolism or whether the activities of other enzymes can be titrated.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this work are listed in Table 3. The thyA ΔfolA::kan strain (named LH18) was constructed in 1988 (38). The Keio deletion strains (86) for metF and glyA were procured from the Coli Genetic Stock Center (CGSC; http://cgsc2.biology.yale.edu/). The deletion strains were unable to grow on Bonner-Vogel (BV) minimal media containing 40 μg/ml guanine, 50 μg/ml tyrosine, histidine, and tryptophan, and 1 mM thiamine (39). The ΔfolA::kan strain required addition of 200 μg/ml thymidine, 30 μg/ml adenine, 10 μg/ml pantothenate, and 50 μg/ml glycine and methionine for growth (87). The ΔmetF::kan strain required addition of methionine (50 μg/ml), while the ΔglyA::kan strain needed glycine and serine (50 μg/ml each) for growth. Kanamycin (50 μg/ml) was added to select for the deletion strain. Bonner-Vogel media for the chorismate mutase aroQ deletion strain included 20 μg/ml phenylalanine, 20 μg/ml tyrosine, and 20 μg/ml chloramphenicol (60).
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Source | Genotype or description |
|---|---|---|
| Strains | ||
| DH5α | Invitrogen | F− ϕ80lacZΔM15(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 |
| NM522 | Howella | supE thi Δ(lac-proAB) Δ(mcrB-hsdS) (r– m–) [F′ proAB lacIqZΔM15] |
| NM522 thyA | Howella | supE thi Δ(lac-proAB) hsdS (r− m−) [F′ proAB lacIqZΔM15] thyA |
| LH18 | Howella | NM522 thyA ΔfolA::kan |
| BW25113 | CGSC | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 |
| JW2535-1 | CGSC | BW25113 ΔglyA725::kan |
| JW3913-1 | CGSC | BW25113 ΔmetF728::kan |
| KA12/pKIMP-UAUC | Colquhounb | Δ(srlR-recA)306::Tn10 Δ(pheA-tyrA-aroF) thi-1 endA-1 hsdRl7 Δ(argF-lac) U169 supE44. The strain is complemented by the pKIMP plasmid (=pACYC184, which confers chloramphenicol resistance and carries the pheC and tyrA genes), which restores Phe and Tyr biosynthetic pathways. |
| Plasmids | ||
| CM-pKTS | Kast and Hilvertc | Ampicillin resistance, Ptet promoter control of CM expression, SsrA tag added |
| R67 DHFR-pKTS | This studyd | Ampicillin resistance, Ptet promoter control of R67 DHFR expression, SsrA tag added, trimethoprim resistance when protein expressed |
| Quad4-pKTS | This studyd | Ampicillin resistance, Ptet promoter control of Quad4 expression, SsrA tag added, trimethoprim resistance when protein expressed |
| MTHFR-pKTS | This studyd | Ampicillin resistance, Ptet promoter control of MTHFR expression, SsrA tag added |
| SHMT-pKTS | This studyd | Ampicillin resistance, Ptet promoter control of SHMT expression, SsrA tag added |
Plasmids.
Table 3 also lists the plasmids used. The pKTS plasmid was obtained from Donald Hilvert and Peter Kast (ETH Zurich) (27). This plasmid enables control of gene expression via the Ptet promoter. Addition of a C-terminal SsrA degradation tag targets the expressed protein to the ClpX protease, decreasing the gene product concentration in vivo. The protein expression level can be less than that associated with expression of the gene from the E. coli chromosome. The plasmid also confers ampicillin resistance.
A supplemental data section and Table 4 describe mutagenesis to add NdeI and XhoI restriction enzyme sites to the genes encoding methylene tetrahydrofolate reductase (MTHFR; metF) and serine hydroxymethyl transferase (SHMT; glyA). These constructs were then cloned into the pKTS plasmid. The R67 DHFR and Quad4 genes were synthesized by GenScript with NdeI and XhoI sites. All gene sequences were confirmed at the Molecular Biology Resource facility (University of Tennessee, Knoxville, TN).
TABLE 4.
List of PCR primer sequences used to introduce NdeI and XhoI restriction enzyme sitesa
| Enzyme (plasmid) | Primer sequence |
|---|---|
| MTHFR (pCAS30) | 5′ TATTTACATATGAGCTTTTTTCACGCCAGC 3′ (NdeI) |
| 5′ AAGGGGTTATGCTAGTTATTGCTCA 3′ (reverse primer) | |
| SHMT (pBSGlyA) | 5′ GGGAGGAGGCATATGTTAAAGCGTGAAATGAACATTGCCGATTATGATGCC 3′ (NdeI) |
| 5′ GAGAGAGAGCTCGAGTGCGTAAACCGGGTAACGTGC 3′ (XhoI) (reverse primer) |
Introduced restriction enzyme sequences are underlined.
Complementation.
Each deletion strain was transformed with several pKTS constructs. The deletion strain was complemented with the pKTS vector containing the corresponding gene; these are the complemented or rescued strains. The complemented strains could grow on minimal medium containing tetracycline, indicating that the plasmid restored prototrophy to the bacteria. For example, the ΔmetF strain was complemented by transformation with the MTHFR-pKTS clone. Vector controls consisted of deletion strains transformed with the pKTS vector containing genes encoding other folate pathway enzymes. For example, transformation of the ΔmetF strain by the SHMT-pKTS or R67 DHFR-pKTS plasmid did not rescue cells from methionine auxotrophy (data not shown).
Tetracycline titrations.
The complemented strains were grown overnight in Luria-Bertani broth with 50 μg/ml kanamycin and 100 μg/ml ampicillin. The cells were centrifuged and the pellet washed with 1× BV salts (39). These steps were repeated and the cell pellet was resuspended in 1× BV salts to a turbidity of 1.0 at 600 nm; 10 μl of solution was then streaked on solid media. The plates contained 50 μg/ml kanamycin, 100 μg/ml ampicillin, and various tetracycline concentrations (0, 10, 25, 50, 75, 100, 200, and 500 ng/ml). The plates were incubated at 37°C for up to 5 days. Good growth was defined as confluent growth without an overlay of single (suppressor) colonies. Table 5 lists the tetracycline concentrations used for each construct.
TABLE 5.
Antibiotic concentrations and supplements used for each strain
| Strain | Antibiotic |
Supplement(s) for growth in minimal media | ||
|---|---|---|---|---|
| Kanamycin | Ampicillin | Tetracycline | ||
| DH5α | NAa | NA | NA | NA |
| DH5α + R67 DHFR-pKTS | NA (20 μg trimethoprim/ml) | 100 μg/ml | 100 ng/ml | NA |
| DH5α + Quad4-pKTS | NA (20 μg trimethoprim/ml) | 100 μg/ml | 100 ng/ml | NA |
| NM522 | NA | NA | NA | NA |
| NM522 thyA8 | NA | NA | NA | 200 μg/ml thymidine |
| LH18 (ΔfolA) | 50 μg/ml | NA | NA | 200 μg/ml thymidine, 50 μg/ml methionine and glycine, 30 μg/ml adenine, 10 μg/ml pantothenate |
| LH18 (ΔfolA) + R67 DHFR-pKTS | 50 μg/ml | 100 μg/ml | 200 ng/ml for agar plates (100 for liquid media)b | NA |
| LH18 (ΔfolA) + Quad4-pKTS | 50 μg/ml | 100 μg/ml | 200 ng/ml for agar plates (100 ng/ml for liquid media) | NA |
| BW25113 | NA | NA | NA | NA |
| JW3913-1 (ΔmetF) | 50 μg/ml | NA | NA | 50 μg/ml methionine |
| JW3913-1 (ΔmetF) + MTHFR-pKTS | 50 μg/ml | 100 μg/ml | 100 ng/ml for agar plates (75 ng/ml for liquid media) | NA |
| JW2535-1 (ΔglyA) | 50 μg/ml | NA | NA | 50 μg/ml glycine and serine |
| JW2535-1 (ΔglyA) + SHMT-pKTS | 50 μg/ml | 100 μg/ml | 75 ng/ml for agar plates (50 ng/ml for liquid media) | NA |
| KA12/pKIMP-UAUC | NA (50 μg/ml chloramphenicol) | NA | NA | 50 μg/ml phenylalanine and tyrosine |
| KA12/pKIMP-UAUC + CM-pKTS | NA (50 μg/ml chloramphenicol) | 100 μg/ml | 200 ng/ml | NA |
NA, not applicable.
We used 200 ng/ml of tetracycline for the LH18::ΔfolA + R67 DHFR-pKTS titration, compared to 100 ng/ml for DH5α+ R67 DHFR-pKTS, to avoid overgrowing colonies at lower osmotic stress.
Osmotic stress titrations.
Once the appropriate tetracycline concentration was determined, osmotic stress titrations were performed. As described above, cells were streaked on BV minimal media containing different concentrations of sorbitol or NaCl and growth was monitored for up to 5 days at 37°C. The plates were incubated in the dark. To determine if the results of our osmotic stress titrations were affected by the identity of the major osmolyte present in the cell, BV medium containing 1 mM betaine was used.
The water activity of the solid medium was measured at room temperature using an AquaLab dew point water activity meter 4TE (Decagon Devices, Inc., Pullman, WA). Then, the osmolality was calculated using equation 1:
| (1) |
where AH2O is the water activity.
A positive control is the growth of the cells on BV-supplemented media containing increasing concentrations of sorbitol or NaCl. This allows discrimination between the lack of cell growth due to loss of free water in the cell (7) from the effect of osmolytes on enzyme activity. Each experiment was done at least in duplicate.
Osmotic stress titrations of trimethoprim resistance.
The activity of chromosomal E. coli DHFR is inhibited by trimethoprim, but R67 DHFR provides resistance to this drug. E. coli strain DH5α was transformed with R67 DHFR-pKTS or Quad4-pKTS vectors. The ability of these clones to confer TMP resistance was assessed by the ability of the transformed cells to grow on BV minimal media containing 20 μg/ml TMP, 100 μg/ml ampicillin, 100 ng/ml tetracycline, and various concentrations of sorbitol (0 to 1.50 M). The plates were incubated at 37°C and cell growth was observed for 5 days. As a control to evaluate the concentration of sorbitol that blocked DH5α growth, cells were grown on BV minimal media with increasing sorbitol concentrations but without antibiotics.
Isolation and characterization of suppressors.
Overgrowing colonies were frequently observed on plates with low tetracycline concentrations and/or high osmotic stress conditions. Plasmids were isolated from these colonies and retransformed into the appropriate deletion cells. Plasmid DNA from those transformants that were still able to grow on media containing 1 M sorbitol were sequenced to identify the mutation(s).
Measurement of growth in liquid media and doubling time calculations.
Cell growth was monitored on an automated 96-well plate reader (BioTek Cytation 5) in liquid BV minimal medium with shaking at 37°C. Turbidities at 600 nm of triplicate samples were observed every 20 min for 36 h. A gas-permeable sealing membrane (Breathe Easy; Diversified Biotech) for microtiter plates (Costar; 3370) reduced evaporation. Osmotic stress was applied using either sorbitol or NaCl. The osmolality of liquid medium was measured with a vapor pressure osmometer (VPO; Wescor Vapro; 5520). Natural log values of absorbance were plotted against time to obtain the slope, which was used to calculate the doubling time (DT) as per equation 2:
| (2) |
The solution osmolality increased ∼5% over 24 h. At longer times (36 h), the osmolality increased by ∼11%. The water activity meter is typically used to measure dry, solid foods while the VPO measures solutions. This difference in sample type led us to standardize and compare the Aqualab dew point water activity meter with the Vapro pressure instrument. The osmolalities of standard solutions with increasing concentrations of sorbitol (0 to 1.5 M) or NaCl (0 to 0.7 M) were determined using both instruments. A graph of the osmolalities from these two different instruments was used to correct the AquaLab values.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant GM 110669 (to E.E.H.).
We have no conflicts of interest with regard to the contents of this article.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Peter Kast and Don Hilvert for their gift of the pKTS vector, Elizabeth Trimmer (Grinnell College) for the pCAS30 plasmid carrying the E. coli MTHFR gene, and Roberto Contestabile (Sapienza University of Rome) for graciously providing the pBSGlyA plasmid carrying the E. coli SHMT gene. We gratefully acknowledge Thomas Colquhoun (University of Florida) as the source of the KA12/pKIMP-UAUC E. coli strain. We thank Faith Critzer (Food Science Department at UTK) for the use of her AquaLab water activity meter to measure agar osmolalities. We additionally thank Gladys Alexandre and Liz Fozo (UTK) for their helpful comments and discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01139-18.
REFERENCES
- 1.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. 1982. Living with water stress: evolution of osmolyte systems. Science 217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 2.Bolen DW. 2001. Protein stabilization by naturally occurring osmolytes. Methods Mol Biol 168:17–36. [DOI] [PubMed] [Google Scholar]
- 3.Larsen PI, Sydnes LK, Landfald B, Strom AR. 1987. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: betaines, glutamic acid, and trehalose. Arch Microbiol 147:1–7. doi: 10.1007/BF00492896. [DOI] [PubMed] [Google Scholar]
- 4.Wood JM. 2011. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu Rev Microbiol 65:215–238. doi: 10.1146/annurev-micro-090110-102815. [DOI] [PubMed] [Google Scholar]
- 5.Record MT Jr, Courtenay ES, Cayley DS, Guttman HJ. 1998. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem Sci 23:143–148. doi: 10.1016/S0968-0004(98)01196-7. [DOI] [PubMed] [Google Scholar]
- 6.Record MT Jr, Courtenay ES, Cayley S, Guttman HJ. 1998. Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments. Trends Biochem Sci 23:190–194. doi: 10.1016/S0968-0004(98)01207-9. [DOI] [PubMed] [Google Scholar]
- 7.Cayley S, Record MT Jr. 2003. Roles of cytoplasmic osmolytes, water, and crowding in the response of Escherichia coli to osmotic stress: biophysical basis of osmoprotection by glycine betaine. Biochemistry 42:12596–12609. doi: 10.1021/bi0347297. [DOI] [PubMed] [Google Scholar]
- 8.Bhojane P, Duff MR Jr, Patel HC, Vogt ME, Howell EE. 2014. Investigation of osmolyte effects on FolM: comparison with other dihydrofolate reductases. Biochemistry 53:1330–1341. doi: 10.1021/bi4014165. [DOI] [PubMed] [Google Scholar]
- 9.Chopra S, Dooling R, Horner CG, Howell EE. 2008. A balancing act: net uptake of water during dihydrofolate binding and net release of water upon NADPH binding in R67 dihydrofolate reductase. J Biol Chem 283:4690–4698. doi: 10.1074/jbc.M709443200. [DOI] [PubMed] [Google Scholar]
- 10.Grubbs J, Rahmanian S, Deluca A, Padmashali C, Jackson M, Duff MR, Howell EE. 2011. Thermodynamics and solvent effects on substrate and cofactor binding in Escherichia coli chromosomal dihydrofolate reductase. Biochemistry 50:3673–3685. doi: 10.1021/bi2002373. [DOI] [PubMed] [Google Scholar]
- 11.Duff MR Jr, Grubbs J, Serpersu EH, Howell EE. 2012. Weak interactions between folate and osmolytes in solution. Biochemistry 51:2309–2318. doi: 10.1021/bi3000947. [DOI] [PubMed] [Google Scholar]
- 12.Timson MJ, Duff MR Jr, Dickey G, Saxton AM, Reyes-De-Corcuera J, Howell EE. 2013. Further studies on the role of water in R67 dihydrofolate reductase. Biochemistry 52:2118–2127. doi: 10.1021/bi301544k. [DOI] [PubMed] [Google Scholar]
- 13.Bhojane PP, Duff MR Jr, Bafna K, Rimmer GP, Agarwal PK, Howell EE. 2016. Aspects of weak interactions between folate and glycine betaine. Biochemistry 55:6282–6294. doi: 10.1021/acs.biochem.6b00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capp MW, Pegram LM, Saecker RM, Kratz M, Riccardi D, Wendorff T, Cannon JG, Record MT Jr. 2009. Interactions of the osmolyte glycine betaine with molecular surfaces in water: thermodynamics, structural interpretation, and prediction of m-values. Biochemistry 48:10372–10379. doi: 10.1021/bi901273r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guinn EJ, Kontur WS, Tsodikov OV, Shkel I, Record MT Jr. 2013. Probing the protein-folding mechanism using denaturant and temperature effects on rate constants. Proc Natl Acad Sci U S A 110:16784–16789. doi: 10.1073/pnas.1311948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles DB, Shkel IA, Phan NM, Sternke M, Lingeman E, Cheng X, Cheng L, O'Connor K, Record MT. 2015. Chemical interactions of polyethylene glycols (PEGs) and glycerol with protein functional groups: applications to effects of PEG and glycerol on protein processes. Biochemistry 54:3528–3542. doi: 10.1021/acs.biochem.5b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng X, Guinn EJ, Buechel E, Wong R, Sengupta R, Shkel IA, Record MT Jr. 2016. Basis of protein stabilization by K glutamate: unfavorable interactions with carbon, oxygen groups. Biophys J 111:1854–1865. doi: 10.1016/j.bpj.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong J, Gierasch LM, Liu Z. 2015. Its preferential interactions with biopolymers account for diverse observed effects of trehalose. Biophys J 109:144–153. doi: 10.1016/j.bpj.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood JM. 2015. Bacterial responses to osmotic challenges. J Gen Physiol 145:381–388. doi: 10.1085/jgp.201411296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miklos AC, Li C, Sharaf NG, Pielak GJ. 2010. Volume exclusion and soft interaction effects on protein stability under crowded conditions. Biochemistry 49:6984–6991. doi: 10.1021/bi100727y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen RD, Pielak GJ. 2017. A cell is more than the sum of its (dilute) parts: a brief history of quinary structure. Protein Sci 26:403–413. doi: 10.1002/pro.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien P, Gierasch LM. 2014. Challenges and dreams: physics of weak interactions essential to life. Mol Biol Cell 25:3474–3477. doi: 10.1091/mbc.e14-06-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Zhuravleva A, Gierasch LM. 2011. Exploring weak, transient protein-protein interactions in crowded in vivo environments by in-cell nuclear magnetic resonance spectroscopy. Biochemistry 50:9225–9236. doi: 10.1021/bi201287e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith AE, Zhou LZ, Gorensek AH, Senske M, Pielak GJ. 2016. In-cell thermodynamics and a new role for protein surfaces. Proc Natl Acad Sci U S A 113:1725–1730. doi: 10.1073/pnas.1518620113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spitzer J. 2011. From water and ions to crowded biomacromolecules: in vivo structuring of a prokaryotic cell. Microbiol Mol Biol Rev 75:491–506. doi: 10.1128/MMBR.00010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spitzer J, Poolman B. 2013. How crowded is the prokaryotic cytoplasm? FEBS Lett 587:2094–2098. doi: 10.1016/j.febslet.2013.05.051. [DOI] [PubMed] [Google Scholar]
- 27.Neuenschwander M, Butz M, Heintz C, Kast P, Hilvert D. 2007. A simple selection strategy for evolving highly efficient enzymes. Nat Biotechnol 25:1145–1147. doi: 10.1038/nbt1341. [DOI] [PubMed] [Google Scholar]
- 28.Perry KM, Carreras CW, Chang LC, Santi DV, Stroud RM. 1993. Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2′-deoxyuridine 5′-monophosphate and 5,10-methylenetetrahydrofolate. Biochemistry 32:7116–7125. doi: 10.1021/bi00079a007. [DOI] [PubMed] [Google Scholar]
- 29.Howell EE. 2005. Searching sequence space: two different approaches to dihydrofolate reductase catalysis. Chembiochem 6:590–600. doi: 10.1002/cbic.200400237. [DOI] [PubMed] [Google Scholar]
- 30.Feng J, Grubbs J, Dave A, Goswami S, Horner CG, Howell EE. 2010. Radical redesign of a tandem array of four R67 dihydrofolate reductase genes yields a functional, folded protein possessing 45 substitutions. Biochemistry 49:7384–7392. doi: 10.1021/bi1005943. [DOI] [PubMed] [Google Scholar]
- 31.Stone SR, Morrison JF. 1986. Mechanism of inhibition of dihydrofolate reductases from bacterial and vertebrate sources by various classes of folate analogues. Biochim Biophys Acta 869:275–285. doi: 10.1016/0167-4838(86)90067-1. [DOI] [PubMed] [Google Scholar]
- 32.Amyes SG, Smith JT. 1976. The purification and properties of the trimethoprim-resistant dihydrofolate reductase mediated by the R-factor, R388. Eur J Biochem 61:597–603. doi: 10.1111/j.1432-1033.1976.tb10055.x. [DOI] [PubMed] [Google Scholar]
- 33.Stinnett LG, Smiley RD, Hicks SN, Howell EE. 2004. “Catch 222,” the effects of symmetry on ligand binding and catalysis in R67 dihydrofolate reductase as determined by mutations at Tyr-69. J Biol Chem 279:47003–47009. doi: 10.1074/jbc.M404485200. [DOI] [PubMed] [Google Scholar]
- 34.Newton MS, Guo X, Soderholm A, Nasvall J, Lundstrom P, Andersson DI, Selmer M, Patrick WM. 2017. Structural and functional innovations in the real-time evolution of new (βα)8 barrel enzymes. Proc Natl Acad Sci U S A 114:4727–4732. doi: 10.1073/pnas.1618552114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butz M, Neuenschwander M, Kast P, Hilvert D. 2011. An N-terminal protein degradation tag enables robust selection of highly active enzymes. Biochemistry 50:8594–8602. doi: 10.1021/bi2011338. [DOI] [PubMed] [Google Scholar]
- 36.Narayana N, Matthews DA, Howell EE, Xuong N. 1995. A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site. Nat Struct Biol 2:1018–1025. doi: 10.1038/nsb1195-1018. [DOI] [PubMed] [Google Scholar]
- 37.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howell EE, Foster PG, Foster LM. 1988. Construction of a dihydrofolate reductase-deficient mutant of Escherichia coli by gene replacement. J Bacteriol 170:3040–3045. doi: 10.1128/jb.170.7.3040-3045.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 40.Jeanson S, Floury J, Gagnaire V, Lortal S, Thierry A. 2015. Bacterial colonies in solid media and foods: a review on their growth and interactions with the micro-environment. Front Microbiol 6:1284. doi: 10.3389/fmicb.2015.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hachicho N, Birnbaum A, Heipieper HJ. 2017. Osmotic stress in colony and planktonic cells of Pseudomonas putida mt-2 revealed significant differences in adaptive response mechanisms. AMB Express 7:62. doi: 10.1186/s13568-017-0371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Qiu Y, Shi H, Yin H. 2016. The importance of lag time extension in determining bacterial resistance to antibiotics. Analyst 141:3059–3067. doi: 10.1039/C5AN02649K. [DOI] [PubMed] [Google Scholar]
- 43.Konopka MC, Shkel IA, Cayley S, Record MT, Weisshaar JC. 2006. Crowding and confinement effects on protein diffusion in vivo. J Bacteriol 188:6115–6123. doi: 10.1128/JB.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sévin DC, Stahlin JN, Pollak GR, Kuehne A, Sauer U. 2016. Global metabolic responses to salt stress in fifteen species. PLoS One 11:e0148888. doi: 10.1371/journal.pone.0148888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bren A, Park JO, Towbin BD, Dekel E, Rabinowitz JD, Alon U. 2016. Glucose becomes one of the worst carbon sources for E. coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci Rep 6:24834. doi: 10.1038/srep24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aidelberg G, Towbin BD, Rothschild D, Dekel E, Bren A, Alon U. 2014. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst Biol 8:133. doi: 10.1186/s12918-014-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirasawa T, Ashitani K, Yoshikawa K, Nagahisa K, Furusawa C, Katakura Y, Shimizu H, Shioya S. 2006. Comparison of transcriptional responses to osmotic stresses induced by NaCl and sorbitol additions in Saccharomyces cerevisiae using DNA microarray. J Biosci Bioeng 102:568–571. doi: 10.1263/jbb.102.568. [DOI] [PubMed] [Google Scholar]
- 48.Weber A, Kogl SA, Jung K. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J Bacteriol 188:7165–7175. doi: 10.1128/JB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ndimba BK, Chivasa S, Simon WJ, Slabas AR. 2005. Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5:4185–4196. doi: 10.1002/pmic.200401282. [DOI] [PubMed] [Google Scholar]
- 50.Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N. 2000. Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123:1047–1056. doi: 10.1104/pp.123.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Courtenay ES, Capp MW, Anderson CF, Record MT Jr. 2000. Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry 39:4455–4471. doi: 10.1021/bi992887l. [DOI] [PubMed] [Google Scholar]
- 52.Venkatesu P, Lee MJ, Lin HM. 2009. Osmolyte counteracts urea-induced denaturation of alpha-chymotrypsin. J Phys Chem B 113:5327–5338. doi: 10.1021/jp8113013. [DOI] [PubMed] [Google Scholar]
- 53.Belluzo S, Boeris V, Farruggia B, Pico G. 2011. Influence of stabilizers cosolutes on catalase conformation. Int J Biol Macromol 49:936–941. doi: 10.1016/j.ijbiomac.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 54.Bourot S, Sire O, Trautwetter A, Touze T, Wu LF, Blanco C, Bernard T. 2000. Glycine betaine-assisted protein folding in a lysA mutant of Escherichia coli. J Biol Chem 275:1050–1056. doi: 10.1074/jbc.275.2.1050. [DOI] [PubMed] [Google Scholar]
- 55.Stover P, Schirch V. 1990. Serine hydroxymethyltransferase catalyzes the hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate. J Biol Chem 265:14227–14233. [PubMed] [Google Scholar]
- 56.Matthews RG, Drummond JT. 1990. Providing one-carbon units for biological methylations: mechanistic studies on serine hydroxymethyltransferase, methylenetetrahydrofolate reductase, and methyltetrahydrofolate-homocysteine methyltransferase. Chem Rev 90:1275–1290. doi: 10.1021/cr00105a010. [DOI] [Google Scholar]
- 57.Contestabile R, Paiardini A, Pascarella S, di Salvo ML, D'Aguanno S, Bossa F. 2001. l-Threonine aldolase, serine hydroxymethyltransferase and fungal alanine racemase. A subgroup of strictly related enzymes specialized for different functions. Eur J Biochem 268:6508–6525. [DOI] [PubMed] [Google Scholar]
- 58.MacBeath G, Kast P, Hilvert D. 1998. Probing enzyme quaternary structure by combinatorial mutagenesis and selection. Protein Sci 7:1757–1767. doi: 10.1002/pro.5560070810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colquhoun TA, Schimmel BCJ, Kim JY, Reinhardt D, Cline K, Clark DG. 2010. A petunia chorismate mutase specialized for the production of floral volatiles. Plant J 61:145–155. doi: 10.1111/j.1365-313X.2009.04042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kast P, Asif-Ullah M, Jiang N, Hilvert D. 1996. Exploring the active site of chorismate mutase by combinatorial mutagenesis and selection: the importance of electrostatic catalysis. Proc Natl Acad Sci U S A 93:5043–5048. doi: 10.1073/pnas.93.10.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amyes SG, Smith JT. 1974. Trimethoprim action and its analogy with thymine starvation. Antimicrob Agents Chemother 5:169–178. doi: 10.1128/AAC.5.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlivan EP, McPartlin J, Weir DG, Scott J. 2000. Mechanism of the antimicrobial drug trimethoprim revisited. FASEB J 14:2519–2524. doi: 10.1096/fj.99-1037com. [DOI] [PubMed] [Google Scholar]
- 63.McMahon MA, Xu J, Moore JE, Blair IS, McDowell DA. 2007. Environmental stress and antibiotic resistance in food-related pathogens. Appl Environ Microbiol 73:211–217. doi: 10.1128/AEM.00578-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baskakov I, Bolen DW. 1998. Forcing thermodynamically unfolded proteins to fold. J Biol Chem 273:4831–4834. doi: 10.1074/jbc.273.9.4831. [DOI] [PubMed] [Google Scholar]
- 65.Parsegian VA, Rand RP, Rau DC. 2000. Osmotic stress, crowding, preferential hydration, and binding: a comparison of perspectives. Proc Natl Acad Sci U S A 97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu I, Mori T, Ando T, Harada R, Jung J, Sugita Y, Feig M. 2016. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. Elife 5:e19274. doi: 10.7554/eLife.19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stadmiller SS, Gorensek-Benitez AH, Guseman AJ, Pielak GJ. 2017. Osmotic shock induced protein destabilization in living cells and its reversal by glycine betaine. J Mol Biol 429:1155–1161. doi: 10.1016/j.jmb.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuznetsova IM, Turoverov KK, Uversky VN. 2014. What macromolecular crowding can do to a protein. Int J Mol Sci 15:23090–23140. doi: 10.3390/ijms151223090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van den Berg J, Boersma AJ, Poolman B. 2017. Microorganisms maintain crowding homeostasis. Nat Rev Microbiol 15:309–318. doi: 10.1038/nrmicro.2017.17. [DOI] [PubMed] [Google Scholar]
- 70.Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA. 2017. ATP as a biological hydrotrope. Science 356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 71.Zotter A, Bauerle F, Dey D, Kiss V, Schreiber G. 2017. Quantifying enzyme activity in living cells. J Biol Chem 292:15838–15848. doi: 10.1074/jbc.M117.792119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwon YK, Lu W, Melamud E, Khanam N, Bognar A, Rabinowitz JD. 2008. A domino effect in antifolate drug action in Escherichia coli. Nat Chem Biol 4:602–608. doi: 10.1038/nchembio.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwon YK, Higgins MB, Rabinowitz JD. 2010. Antifolate-induced depletion of intracellular glycine and purines inhibits thymineless death in E. coli. ACS Chem Biol 5:787–795. doi: 10.1021/cb100096f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parsegian VA, Rand RP, Rau DC. 1995. Macromolecules and water: probing with osmotic stress. Methods Enzymol 259:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- 75.Acosta LC, Perez Goncalves GM, Pielak GJ, Gorensek-Benitez AH. 2017. Large cosolutes, small cosolutes and dihydrofolate reductase activity. Protein Sci 26:2417–2425. doi: 10.1002/pro.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Snider MJ, Gaunitz S, Ridgway C, Short SA, Wolfenden R. 2000. Temperature effects on the catalytic efficiency, rate enhancement, and transition state affinity of cytidine deaminase, and the thermodynamic consequences for catalysis of removing a substrate “anchor.” Biochemistry 39:9746–9753. [DOI] [PubMed] [Google Scholar]
- 77.Gulotta M, Qiu L, Desamero R, Rosgen J, Bolen DW, Callender R. 2007. Effects of cell volume regulating osmolytes on glycerol 3-phosphate binding to triosephosphate isomerase. Biochemistry 46:10055–10062. doi: 10.1021/bi700990d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhadin N, Callender R. 2011. Effect of osmolytes on protein dynamics in the lactate dehydrogenase-catalyzed reaction. Biochemistry 50:1582–1589. doi: 10.1021/bi1018545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phillip Y, Harel M, Khait R, Qin S, Zhou HX, Schreiber G. 2012. Contrasting factors on the kinetic path to protein complex formation diminish the effects of crowding agents. Biophys J 103:1011–1019. doi: 10.1016/j.bpj.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mattei P, Kast P, Hilvert D. 1999. Bacillus subtilis chorismate mutase is partially diffusion-controlled. Eur J Biochem 261:25–32. doi: 10.1046/j.1432-1327.1999.00169.x. [DOI] [PubMed] [Google Scholar]
- 81.Gadda G, Sobrado P. 2018. Kinetic solvent viscosity effects as probes for studying the mechanisms of enzyme action. Biochemistry 57:3445–3453. doi: 10.1021/acs.biochem.8b00232. [DOI] [PubMed] [Google Scholar]
- 82.Jensen SI, Lennen RM, Herrgard MJ, Nielsen AT. 2015. Seven gene deletions in seven days: fast generation of Escherichia coli strains tolerant to acetate and osmotic stress. Sci Rep 5:17874. doi: 10.1038/srep17874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winkler JD, Garcia C, Olson M, Callaway E, Kao KC. 2014. Evolved osmotolerant Escherichia coli mutants frequently exhibit defective N-acetylglucosamine catabolism and point mutations in cell shape-regulating protein MreB. Appl Environ Microbiol 80:3729–3740. doi: 10.1128/AEM.00499-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guo Y, Winkler J, Kao KC. 2017. Insights on osmotic tolerance mechanisms in Escherichia coli gained from an rpoC mutation. Bioengineering (Basel) 4(3):61. doi: 10.3390/bioengineering4030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Csonka LN. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev 53:121–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Singer S, Ferone R, Walton L, Elwell L. 1985. Isolation of a dihydrofolate reductase-deficient mutant of Escherichia coli. J Bacteriol 164:470–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reece LJ, Nichols R, Ogden RC, Howell EE. 1991. Construction of a synthetic gene for an R-plasmid-encoded dihydrofolate reductase and studies on the role of the N-terminus in the protein. Biochemistry 30:10895–10904. doi: 10.1021/bi00109a013. [DOI] [PubMed] [Google Scholar]
- 90.Park H, Zhuang P, Nichols R, Howell EE. 1997. Mechanistic studies of R67 dihydrofolate reductase. Effects of pH and an H62C mutation. J Biol Chem 272:2252–2258. [DOI] [PubMed] [Google Scholar]
- 91.Trimmer EE, Ballou DP, Matthews RG. 2001. Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by steady-state and rapid-reaction studies. Biochemistry 40:6205–6215. doi: 10.1021/bi002789w. [DOI] [PubMed] [Google Scholar]
- 92.Guenther BD, Sheppard CA, Tran P, Rozen R, Matthews RG, Ludwig ML. 1999. The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia. Nat Struct Biol 6:359–365. doi: 10.1038/7594. [DOI] [PubMed] [Google Scholar]
- 93.Misra SK, Bhakuni V. 2003. Unique holoenzyme dimers of the tetrameric enzyme Escherichia coli methylenetetrahydrofolate reductase: characterization of structural features associated with modulation of the enzyme's function. Biochemistry 42:3921–3928. doi: 10.1021/bi0340200. [DOI] [PubMed] [Google Scholar]
- 94.Schirch V, Hopkins S, Villar E, Angelaccio S. 1985. Serine hydroxymethyltransferase from Escherichia coli: purification and properties. J Bacteriol 163:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schirch D, Delle Fratte S, Iurescia S, Angelaccio S, Contestabile R, Bossa F, Schirch V. 1993. Function of the active-site lysine in Escherichia coli serine hydroxymethyltransferase. J Biol Chem 268:23132–23138. [PubMed] [Google Scholar]
- 96.Florio R, Chiaraluce R, Consalvi V, Paiardini A, Catacchio B, Bossa F, Contestabile R. 2009. The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia coli serine hydroxymethyltransferase. FEBS J 276:132–143. doi: 10.1111/j.1742-4658.2008.06761.x. [DOI] [PubMed] [Google Scholar]
- 97.Guilford WJ, Copley SD, Knowles JR. 1987. On the mechanism of the chorismate mutase reaction. J Am Chem Soc 109:5013–5019. doi: 10.1021/ja00250a041. [DOI] [Google Scholar]
- 98.Wu F, Minteer S. 2015. Krebs cycle metabolon: structural evidence of substrate channeling revealed by cross-linking and mass spectrometry. Angew Chem 54:1851–1854. doi: 10.1002/anie.201409336. [DOI] [PubMed] [Google Scholar]
- 99.An SG, Kumar R, Sheets ED, Benkovic SJ. 2008. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320:103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 100.Metris A, George SM, Mulholland F, Carter AT, Baranyi J. 2014. Metabolic shift of Escherichia coli under salt stress in the presence of glycine betaine. Appl Environ Microbiol 80:4745–4756. doi: 10.1128/AEM.00599-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang EY, Mohler AM, Rohlman CE. 1997. Protein expression in response to folate stress in Escherichia coli. J Bacteriol 179:5648–5653. doi: 10.1128/jb.179.17.5648-5653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gunasekera TS, Csonka LN, Paliy O. 2008. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol 190:3712–3720. doi: 10.1128/JB.01990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmidt A, Kochanowski K, Vedelaar S, Ahrne E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M. 2016. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol 34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kocharunchitt C, King T, Gobius K, Bowman JP, Ross T. 2012. Integrated transcriptomic and proteomic analysis of the physiological response of Escherichia coli O157:H7 Sakai to steady-state conditions of cold and water activity stress. Mol Cell Proteomics 11:M111.009019. doi: 10.1074/mcp.M111.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fierke CA, Johnson KA, Benkovic SJ. 1987. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry 26:4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 106.Walsh CT. 1979. Enzymatic reaction mechanisms. WH Freeman & Co, New York, NY. [Google Scholar]
- 107.Segel IH. 1975. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. John Wiley and Sons, New York, NY. [Google Scholar]
- 108.Sévin DC, Sauer U. 2014. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat Chem Biol 10:266–272. doi: 10.1038/nchembio.1437. [DOI] [PubMed] [Google Scholar]
- 109.Nijhout H, Reed M, Budu P, Ulrich C. 2004. A mathematical model of the folate cycle: new insights into folate homeostasis. J Biol Chem 279:55008–55016. doi: 10.1074/jbc.M410818200. [DOI] [PubMed] [Google Scholar]
- 110.Giaever HM, Styrvold OB, Kaasen I, Strom AR. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J Bacteriol 170:2841–2849. doi: 10.1128/jb.170.6.2841-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.