The presence of ergot alkaloid synthesis genes in the genome of Penicillium camemberti is significant, because the fungus is widely consumed in Brie and Camembert cheeses. Our results show that, although the fungus has several functional genes from the ergot alkaloid pathway, it produces only an early pathway intermediate in culture and does not produce ergot alkaloids in cheese. Penicillium biforme, a close relative of P. camemberti, contains a similar but fully functional set of ergot alkaloid synthesis genes and produces ergot alkaloids chanoclavine-I, chanoclavine-I aldehyde, and rugulovasine A and B. Our reconstruction of the P. camemberti pathway in the model fungus Neosartorya fumigata indicated that P. camemberti formerly had the capacity to produce these same ergot alkaloids. Neither P. camemberti nor P. biforme produced ergot alkaloids in cheese, indicating that nutritionally driven gene regulation prevents these fungi from producing ergot alkaloids in a dairy environment.
KEYWORDS: specialized metabolism, aldehyde dehydrogenase, Camembert cheese, rugulovasines
ABSTRACT
Ergot alkaloids are specialized fungal metabolites with potent biological activities. They are encoded by well-characterized gene clusters in the genomes of producing fungi. Penicillium camemberti plays a major role in the ripening of Brie and Camembert cheeses. The P. camemberti genome contains a cluster of five genes shown in other fungi to be required for synthesis of the important ergot alkaloid intermediate chanoclavine-I aldehyde and two additional genes (easH and easQ) that may control modification of chanoclavine-I aldehyde into other ergot alkaloids. We analyzed samples of Brie and Camembert cheeses, as well as cultures of P. camemberti, and did not detect chanoclavine-I aldehyde or its derivatives. To create a functioning facsimile of the P. camemberti eas cluster, we expressed P. camemberti easH and easQ in a chanoclavine-I aldehyde-accumulating easA knockout mutant of Neosartorya fumigata. The easH-easQ-engineered N. fumigata strain accumulated a pair of compounds of m/z 269.1288 in positive-mode liquid chromatography-mass spectrometry (LC-MS). The analytes fragmented in a manner typical of the stereoisomeric ergot alkaloids rugulovasine A and B, and the related rugulovasine producer Penicillium biforme accumulated the same isomeric pair of analytes. The P. camemberti eas genes were transcribed in culture, but comparison of the P. camemberti eas cluster with the functional cluster from P. biforme indicated 11 polymorphisms. Whereas other P. camemberti eas genes functioned when expressed in N. fumigata, P. camemberti easC did not restore ergot alkaloids when expressed in an easC mutant. The data indicate that P. camemberti formerly had the capacity to produce the ergot alkaloids rugulovasine A and B.
IMPORTANCE The presence of ergot alkaloid synthesis genes in the genome of Penicillium camemberti is significant, because the fungus is widely consumed in Brie and Camembert cheeses. Our results show that, although the fungus has several functional genes from the ergot alkaloid pathway, it produces only an early pathway intermediate in culture and does not produce ergot alkaloids in cheese. Penicillium biforme, a close relative of P. camemberti, contains a similar but fully functional set of ergot alkaloid synthesis genes and produces ergot alkaloids chanoclavine-I, chanoclavine-I aldehyde, and rugulovasine A and B. Our reconstruction of the P. camemberti pathway in the model fungus Neosartorya fumigata indicated that P. camemberti formerly had the capacity to produce these same ergot alkaloids. Neither P. camemberti nor P. biforme produced ergot alkaloids in cheese, indicating that nutritionally driven gene regulation prevents these fungi from producing ergot alkaloids in a dairy environment.
INTRODUCTION
Ergot alkaloids are a diverse family of specialized metabolites produced by at least two lineages of fungi. Lysergic acid-derived ergot alkaloids produced by certain members of the Clavicipitaceae have poisoned humans and animals when produced in agricultural crops (1, 2) and been exploited by humans for clinical applications and as recreational drugs (3–6). Clavine-derived ergot alkaloids produced by certain members of the Eurotiales have not been characterized as thoroughly from an activity perspective, although recent evidence indicates that the clavine alkaloids of the animal pathogen Neosartorya fumigata (Aspergillus fumigatus) contribute to virulence (7).
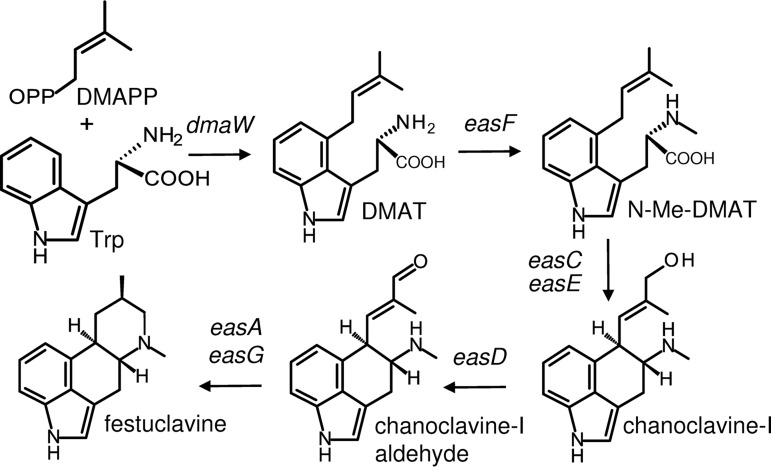
All ergot alkaloids are derived from 4-prenylated tryptophan via a pathway that starts with several steps carried out by homologous enzymes (Fig. 1). The ergot alkaloid pathway has been reviewed in detail recently (e.g., see references 1, 2, 8, 9, and 10), and the early steps will be summarized briefly here. After the addition of a single prenyl group at position 4 of tryptophan by the enzyme dimethylallyltryptophan synthase (encoded by dmaW) (11), the amino group of the prenylated tryptophan is methylated (12) before closure of the third ring to form chanoclavine-I by a decarboxylative mechanism involving the products of easC and easE (13, 14). The primary alcohol of chanoclavine-I may then be oxidized to the aldehyde form by the product of easD (the allele for which is called fgaDH in A. fumigatus [15]). The resulting product, chanoclavine-I aldehyde, serves as a branch point intermediate that may be acted on by the products of different alleles of ergot alkaloid synthesis genes in different lineages to yield lysergic acid and its derivatives in the Clavicipitaceae or various clavine-type ergot alkaloids in the Eurotiales (including Neosartorya and Penicillium species).
FIG 1.
Pathway steps to festuclavine in N. fumigata. Genes controlling relevant steps are indicated. DMAPP, dimethylallylpyrosphosphate; DMAT, dimethylallyltryptophan.
The ergot alkaloid synthesis (eas) genes are found clustered in the genomes of ergot alkaloid-producing fungi (Fig. 2) (e.g., see references 16, 17, 18, and 19). The five genes required to synthesize chanoclavine-I aldehyde are conserved among ergot alkaloid-producing fungi (whose pathways extend to that point) and have predictable activities. Genes for enzymes downstream from chanoclavine-I aldehyde are in some cases unique and lineage specific, such as the genes for the lysergyl peptide synthetase complex of the Clavicipitaceae (20) or those for fumigaclavine production in N. fumigata (21). In other cases, downstream genes are represented by alleles of homologous genes encoding enzymes with differing activities; examples include the reductase versus isomerase alleles of easA (22–25) and alleles of easH encoding enzymes that act at different steps in pathways in the Clavicipitaceae (26) versus those in the Eurotiales (27).
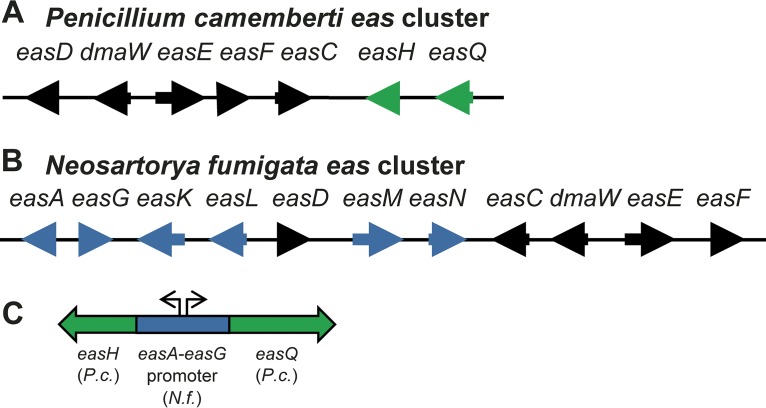
FIG 2.
Ergot alkaloid synthesis (eas) gene clusters of P. camemberti (A) and N. fumigata (B). Genes drawn in black are shared between the two clusters. Genes shown in green are unique to the P. camemberti cluster, whereas those in blue are unique to the N. fumigata cluster. The related fungus Penicillium biforme has an eas cluster identical to that of P. camemberti with respect to gene presence, order, and orientation. The N. fumigata cluster is redrawn from Coyle and Panaccione (17) and Unsöld and Li (18). (C) Construct for coexpressing P. camemberti (P.c.) easH and easQ under the control of the bidirectional easA-easG promoter of N. fumigata (N.f.).
Penicillium camemberti is a domesticated fungus that contributes to the unique flavor and texture of Brie and Camembert cheeses and is not known outside cheese-making environments (28, 29). Publicly available DNA sequence data revealed that the P. camemberti genome (30, 31) contains a cluster of genes containing homologs to several eas genes. The presence of these genes was recently noted in a review article by Martín et al. (32). Moreover, Gerhards and Li (33) showed that the product of the P. camemberti homolog of easD was functional when expressed in vitro. We thus hypothesize that P. camemberti has or had (at one time in its evolutionary past) the ability to produce ergot alkaloids. Considering the role of P. camemberti in cheese ripening and its direct consumption by people in Brie and Camembert cheese rind, we used a heterologous expression strategy to investigate the capacity of P. camemberti to produce ergot alkaloids and the theoretical product(s) of its ergot alkaloid synthesis gene cluster.
RESULTS AND DISCUSSION
Structural and functional analysis of the Penicillium camemberti eas gene cluster.
Since the ergot alkaloid synthesis (eas) cluster of P. camemberti contained homologs of five genes previously established to yield chanoclavine-I aldehyde in N. fumigata (dmaW, easF, easC, easE, and easD) (Fig. 1 and 2), we investigated the possibility that P. camemberti had the capacity to produce chanoclavine-I aldehyde or a derivative thereof. Four samples of commercial cheese and cultures of P. camemberti strains NRRL 874 and NRRL 875 grown on malt extract agar failed to yield chanoclavine-I aldehyde or known derivatives upon analysis by high-performance liquid chromatography (HPLC) with fluorescence detection and liquid chromatography-mass spectrometry (LC-MS).
The P. camemberti cluster also contains an allele of easH, homologs of which encode products that catalyze different steps in the ergot alkaloid pathways of divergent fungi. The version of EasH from Aspergillus japonicus catalyzes an oxidation step required for formation of a cyclopropane ring in cycloclavine (27), whereas the version of EasH found in Claviceps purpurea catalyzes an oxidation step required for cyclolization of the amino acid side chain of ergopeptines (26). The allele of easH from P. camemberti encoded a product that was more similar to EasH of A. japonicus (52% amino acid sequence identity) than it was to the version of EasH from C. purpurea (35% amino acid sequence identity).
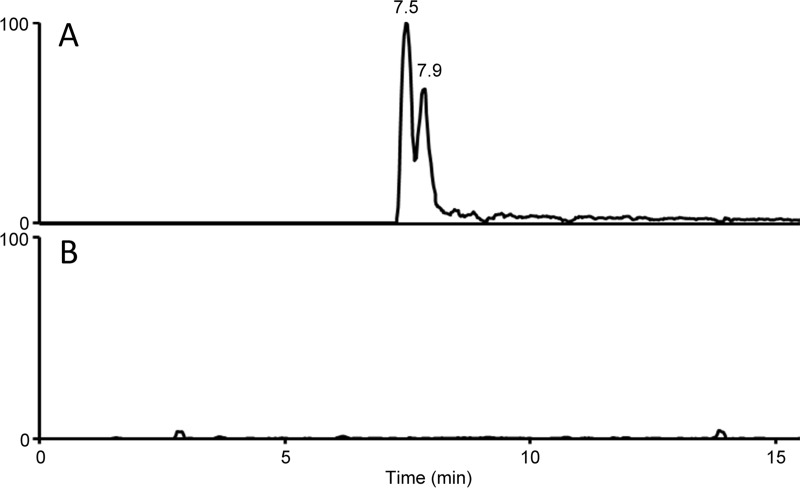
Adjacent to the previously annotated eas genes in the P. camemberti cluster was an aldehyde dehydrogenase-encoding gene which we named easQ (Fig. 2). Homologs of this gene had not been associated with eas clusters previously. Considering the location of easQ and its predicted activity, we expressed easQ along with easH of P. camemberti in the N. fumigata easA knockout (22). This strain was chosen as an expression host because it has a functional eas genetic background that simulates the remainder of the P. camemberti eas gene cluster (dmaW, easF, easC, easE, and easD) and terminates its pathway at chanoclavine-I aldehyde (22). Eight colonies were obtained from cotransformation of the easH-easQ expression plasmid and the separate, phleomycin resistance-conferring plasmid. One colony was found to express both easH and easQ of P. camemberti, as evidenced by accumulation of mRNA from the introduced genes (see Fig. S1 in the supplemental material). Chemical analyses revealed that the easH-easQ transformant cultured on malt extract agar accumulated a pair of analytes of m/z 269.1 in positive-mode LC-MS (Fig. 3). High-resolution mass spectrometry of these analytes yielded an m/z of 269.1288, indicating a molecular formula of C16H16N2O2 (calculated m/z of 269.1285). There was no evidence of these analytes in the nontransformed recipient N. fumigata easA knockout strain (Fig. 3).
FIG 3.
Extracted ion chromatogram (m/z 269.1) showing accumulation of a pair of novel compounds in easH- and easQ-transformed N. fumigata easA knockout (A) compared to the results for the nonmodified recipient strain (B). Retention times of m/z 269.1 analytes are indicated above peaks. Data were collected on a Thermo LCQ Deca XP mass spectrometer operated in positive mode. Retention times of major analytes are indicated.
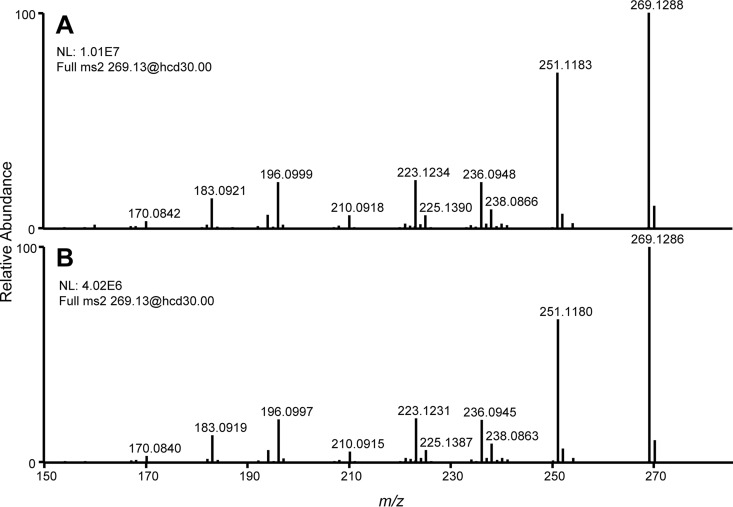
Two previously characterized classes of ergot alkaloids share the indicated molecular formula: lysergic acid (and its diastereoisomer isolysergic acid) and the diastereoisomeric alkaloids rugulovasine A and B. The easH-easQ-engineered strain of N. fumigata lacked two genes (easA and cloA) previously demonstrated to be required for lysergic acid synthesis (34, 35), eliminating lysergic acid/isolysergic acid as candidates for the analytes. Because rugulovasine A and B are produced by the closely related fungus Penicillium biforme (36), P. biforme strain NRRL 885 was cultured on malt extract agar and found to produce a pair of analytes with the same m/z value and retention times as the analytes produced in the easH-easQ-engineered N. fumigata strain. The m/z 269.1 analytes of the easH- and easQ-transformed N. fumigata easA knockout strain and P. biforme NRRL 885 fragmented similarly (Fig. 4) and in a manner consistent with the high-resolution-fragment ions obtained from rugulovasine A and B in previous work (37). Whereas rugulovasines were easily detected by LC-MS, they were not detected by HPLC with fluorescence detection (monitoring at excitation/emission wavelengths of 272 nm/372 nm or 310 nm/410 nm, respectively), which contrasts with other ergot alkaloids. The fluorophore in rugulovasines may require different excitation and/or emission wavelengths, or the conjugated double-bond structure outside the indole ring of rugulovasines may quench fluorescence.
FIG 4.
High-resolution MS spectra of ions resulting from fragmentation of the parent ion of m/z 269.13 in malt extract medium cultures of easH- and easQ-transformed N. fumigata easA knockout (A) and P. biforme (B). Data were collected in positive mode. No precursor ion for this compound was isolated from extracts of nontransformed N. fumigata easA knockout (Fig. 3), and thus, no spectrum is provided for the nontransformed recipient strain.
Separate functional analyses of easH and easQ.
The expression of P. camemberti easH alone in the N. fumigata easA knockout did not result in the formation of any metabolites downstream from chanoclavine-I aldehyde, indicating that chanoclavine-I aldehyde is not a substrate for the product of this gene. PCR analyses of genomic DNA and cDNA templates demonstrated that the easH gene was present and transcribed in transformants (Fig. S2).
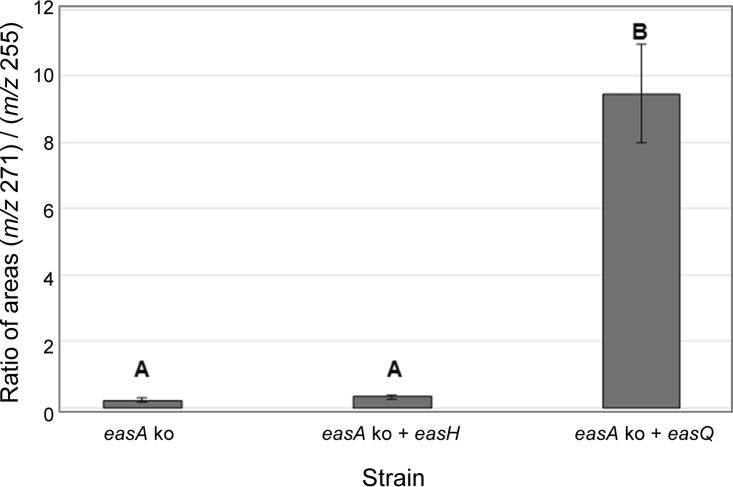
The expression of easQ alone in the N. fumigata easA knockout strain resulted in the accumulation of an ion at m/z 271.2 in positive-mode LC-MS analyses but not in the accumulation of rugulovasine A and B. In addition to the accumulation of the m/z 271.2 ion, the relative concentration of chanoclavine-I aldehyde decreased in the easQ transformants. These changes were evident in the ratio of relative peak areas of the m/z 271.2 ion (presumed oxidized chanoclavine-I aldehyde) compared to that of chanoclavine-I aldehyde (m/z 255.2). The ratio was significantly increased in the easQ transformants compared to that of the nontransformed N. fumigata easA knockout (P = 0.002) or the easH-transformed N. fumigata easA knockout (P = 0.007) (Fig. 5). The change in mass and shifting proportions of these two analytes indicate that the product of easQ adds an oxygen to chanoclavine-I aldehyde. Considering the structural similarity of EasQ to aldehyde dehydrogenases, a likely activity for the enzyme is the oxidation of chanoclavine-I aldehyde to the corresponding carboxylic acid. Considerably more research will be required to demonstrate this activity.
FIG 5.
Ratios of areas corresponding to presumed oxidized chanoclavine-I aldehyde (m/z 271) and chanoclavine-I aldehyde (m/z 255) based on relative peak areas observed in the LC-MS data of N. fumigata easA knockout (easA ko; n = 3), N. fumigata easA knockout transformed with easH (easA ko + easH; n = 4), and N. fumigata easA knockout transformed with easQ (easA ko + easQ; n = 4). Mean values labeled with different capital letters differ significantly (P < 0.05) in a Tukey-Kramer honestly significant difference test. Error bars represent standard errors.
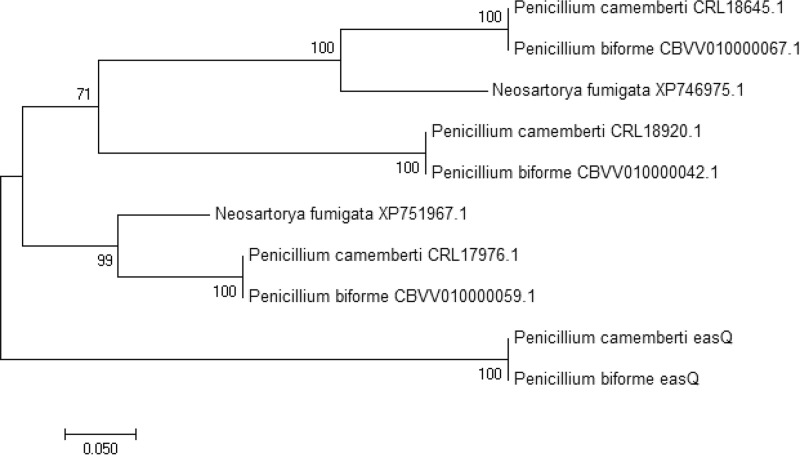
Phylogenetic analysis of easQ indicated that the alleles in the P. camemberti and P. biforme eas clusters diverged from a gene family shared among the two Penicillium species and N. fumigata (Fig. 6). In total, P. camemberti and P. biforme have four easQ-like aldehyde dehydrogenase-encoding homologs, whereas N. fumigata has alleles of two of the four aldehyde dehydrogenase-encoding genes (but no allele of easQ) (Fig. 6). In addition to close sequence identities, the homology of the four aldehyde dehydrogenase-encoding genes of P. camemberti/P. biforme and the two aldehyde dehydrogenase-encoding genes of N. fumigata is supported by the presence of introns in similar positions (43rd codon and 110th codon) in each of the genes. Limited activity from the products of the N. fumigata aldehyde dehydrogenase paralogs may account for low levels of m/z 271.2 ion detected in the N. fumigata easA knockout (Fig. 5). Additional aldehyde dehydrogenase-encoding homologs with lesser amino acid sequence identities were detected in the genomes of P. camemberti, P. biforme, and N. fumigata, but the genes encoding these additional enzymes did not have the same exon/intron structure as easQ and its paralogs.
FIG 6.
Maximum-likelihood tree indicating relatedness of easQ-like aldehyde dehydrogenase genes from the indicated fungi. Sequences were aligned by MUSCLE, and tree was drawn with the maximum-likelihood method in MEGA7 (52) based on the LG model (53). Bootstrap values (percentages, obtained from 1,000 pseudoreplicates) are listed at nodes. Tree is unrooted. Scale bar indicates number of substitutions per site. GenBank or Swiss-Prot accession numbers are shown for paralogs. The accession number for EasQ of P. camemberti is CRL19771.1; EasQ of P. biforme is identical to that of P. camemberti and derived from the sequence with GenBank accession number CBXO010000115.1.
Hypothesized roles for EasH and EasQ in synthesis of rugulovasines.
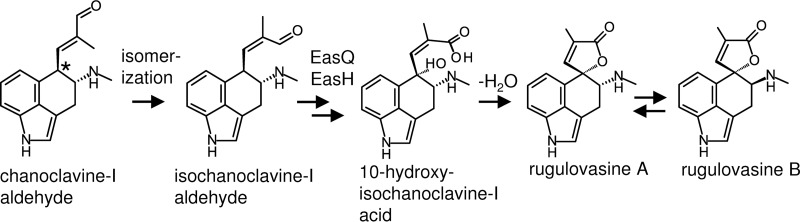
The analyses of the ergot alkaloid gene clusters of P. camemberti and P. biforme provide the first genetic explanation for the biosynthesis of rugulovasine A and B. The mass spectral data indicate that EasQ hydroxylates chanoclavine-I aldehyde, and based on the DNA sequence data, we hypothesize that oxidation occurs at the aldehyde carbon. Our data demonstrate that the product of easH, a predicted dioxygenase, is also required to convert chanoclavine-I aldehyde to rugulovasine A and B. We hypothesize that the product of easH oxidizes the aromatic carbon customarily labeled carbon 10 in chanoclavine-I aldehyde or, perhaps, that same carbon in the corresponding carboxylic acid, isochanoclavine-I acid (Fig. 7). Dehydration of the 10-hydroxylated isochanoclavine-I acid would result in the formation of rugulovasine A. This hypothesis is supported by a previously proposed mechanism for the homolog of EasH from A. japonicus (27). Jakubczyk et al. (27) proposed three mechanisms by which EasH might oxidize the very same carbon in a different ergot alkaloid intermediate to result in the formation of cycloclavine. One of the proposed mechanisms involved hydroxylation of carbon 10. Such an activity on isochanoclavine-I acid would set the stage for ring closure to rugulovasine A via dehydration (Fig. 7). Once rugulovasine A is formed, it readily interconverts with rugulovasine B in polar solvents (38, 39). The lack of any observed product when easH was introduced alone into the chanoclavine-I aldehyde-producing N. fumigata easA knockout suggests that EasH does not act directly on chanoclavine-I aldehyde but, rather, on the product resulting from oxidation of chanoclavine-I aldehyde by EasQ.
FIG 7.
Hypothesized biosynthesis of rugulovasines from chanoclavine-l aldehyde. EasQ, predicted to be an aldehyde dehydrogenase, is proposed to oxidize the aldehyde to a carboxylic acid. EasH, a predicted dioxygenase, is predicted to oxidize the carbon customarily labeled carbon 10 (indicated by the asterisk). Rugulovasine A and B spontaneously isomerize by a previously documented mechanism (38, 39).
In order for the hypothesized oxidation and dehydration steps to yield rugulovasine A and B, chanoclavine-I aldehyde must first isomerize around the carbon adjacent to the aldehyde group (Fig. 7). An isomerization appears necessary to bring the oxidized side chain into close proximity to carbon 10. It is likely that isomerization occurs prior to oxidation of the aldehyde to a carboxylic acid. Isomerization may occur spontaneously via keto-enol tautomerization (22). Additionally, nonenzymatic isomerization of chanoclavine-I aldehyde via a glutathione-promoted, temporary bond reduction has also been described (40). Alternatively, if the isomerization is enzyme catalyzed, it would have to rely on the product of easH, easQ, or an enzyme already encoded in the N. fumigata background, because it occurred readily in the easH- and easQ-transformed N. fumigata easA knockout strain.
Genetic basis for lack of accumulation of rugulovasines in P. camemberti.
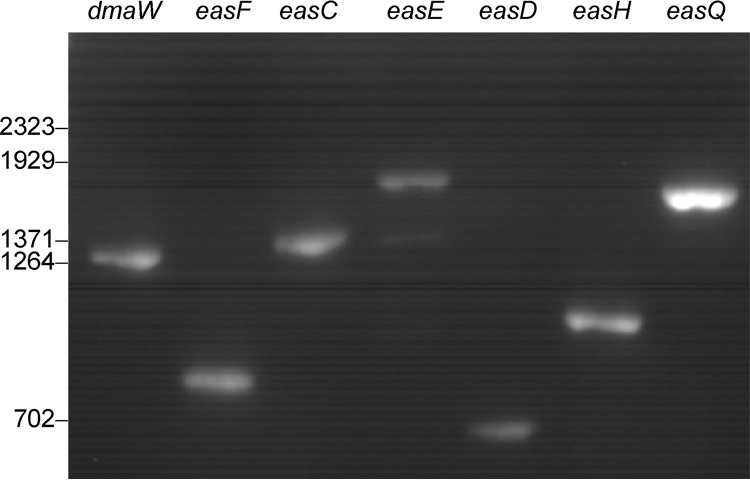
Possible reasons for the lack of accumulation of rugulovasines in P. camemberti include lack of expression of eas genes, misprocessing of eas gene transcripts, or null mutations in individual eas genes. Whether the eas genes of P. camemberti were transcriptionally active in vitro was investigated in qualitative reverse transcriptase PCR analyses. mRNAs from dmaW, easF, easC, easE, easD, easH, and easQ were detected in P. camemberti strain NRRL 874 grown in malt extract broth (Fig. 8). Sequencing of the cDNA products indicated that the mRNAs were processed properly, as the cDNAs contained open reading frames corresponding to those for functional eas genes from N. fumigata.
FIG 8.
PCR products amplified from cDNA prepared from P. camemberti NRRL 874 grown in malt extract broth for 3 days. Lanes labeled dmaW, easF, easC, easE, easD, easH, and easQ contain products amplified with gene-specific primer combinations 13 to 19 (Table 1), respectively. Relative migrations of BstEII-digested bacteriophage lambda fragments (sizes listed in bp) are indicated at left of gel.
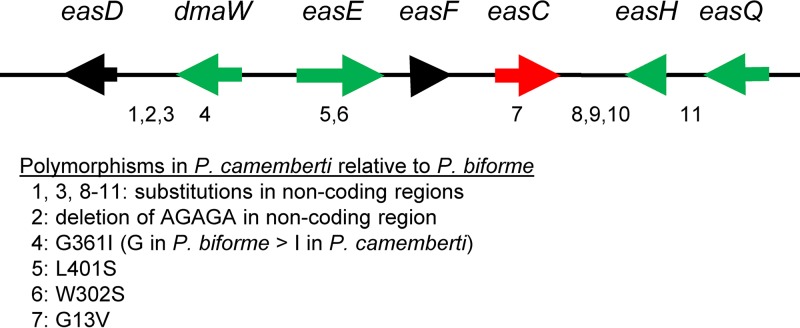
The possibility that individual eas genes of P. camemberti were mutated was investigated by comparing the P. camemberti eas gene cluster to the functional eas gene cluster of P. biforme. The two eas clusters were nearly identical, with the exception of four polymorphisms in the coding regions of eas genes and seven intergenic polymorphisms (Fig. 9). Each of the four polymorphisms that occurred within coding sequences resulted in changes in amino acids in the respective enzymes (Fig. 9). One amino acid substitution was encoded in dmaW, two in easE, and one in easC. Since the alleles of easD, easF, easH, and easQ from P. camemberti were identical to those of P. biforme, which produced rugulovasine in culture, they were presumed to be functional. Moreover, the heterologous expression studies with N. fumigata showed that the P. camemberti alleles of easH and easQ were functional. The functionality of the three polymorphic alleles of P. camemberti (dmaW, easE, and easC) was tested by transforming them into previously constructed N. fumigata knockout strains (14, 17) in complementation studies. Transformed strains that contained the dmaW or easE constructs each accumulated fumigaclavine C (Table S1), the pathway end product and typically most abundant ergot alkaloid in wild-type N. fumigata (41, 42), indicating that the P. camemberti alleles restored the N. fumigata ergot alkaloid pathway. Attempts to complement an easC knockout mutant of N. fumigata (14) with the P. camemberti allele of easC did not result in the accumulation of fumigaclavine C (Table S1). Analysis of a P. camemberti easC-transformed N. fumigata easC knockout by reverse transcriptase PCR and DNA sequencing of the easC cDNA product indicated that the P. camemberti gene was present, transcribed, and properly processed (Fig. S3).
FIG 9.
Positions and consequences of polymorphisms in the P. camemberti eas cluster relative to the sequences of the functional P. biforme eas cluster. Genes drawn in green were shown to function when heterologously expressed in N. fumigata strains. Genes drawn in black were not functionally tested but have sequences identical to those of functional alleles in P. biforme. The allele of easC, drawn in red, failed to complement the easC mutant of N. fumigata. Numerals 1 through 11 indicate the positions of polymorphisms in P. camemberti compared to the sequences observed in P. biforme. The table describes the polymorphisms based on the assumption that the P. biforme sequence is ancestral and the P. camemberti sequence is derived.
Because blocking the N. fumigata pathway by knocking out easC in previous studies (14) resulted in the accumulation of the early pathway intermediate N-methyl-dimethylallyltryptophan (N-Me-DMAT) (Fig. 1), we tested whether N-Me-DMAT would accumulate in cultures of P. camemberti NRRL 874 cultured in malt extract medium, which was conducive to rugulovasine accumulation in P. biforme. An analyte with an m/z value of 287.1758, which is consistent with the m/z value of 287.1754 calculated for [N-Me-DMAT + H]+, was observed in positive-mode LC-MS (Fig. S4). The analyte of m/z 287.1758 fragmented similarly to an N-Me-DMAT standard (Fig. S5). The same analyte was produced when P. camemberti NRRL 874 was cultured in a variant of Czapek's medium in which succinic acid was the only carbon source. These data indicate that under conditions conducive to ergot alkaloid production, P. camemberti expresses the limited part of the pathway it possesses. Cheese samples also were checked for N-Me-DMAT, and no evidence of accumulation of this compound was detected.
Lack of ergot alkaloid accumulation in cheese.
To test for accumulation of ergot alkaloids in cheese ripened with the same isolates used in this present study, four different wheels of Camembert cheese were cultured and ripened for 8 weeks with P. biforme NRRL 885 and P. camemberti NRRL 874. Samples were analyzed by LC-MS, and no ergot alkaloids (including rugulovasine A or B, chanoclavine-I, chanoclavine-I aldehyde, or N-Me-DMAT) were detected in any cheese sample. Four samples of commercial cheese rind containing P. camemberti also were investigated and found to be free of ergot alkaloids. Since P. biforme accumulated ergot alkaloids in vitro (and P. camemberti produced the precursor N-Me-DMAT in vitro), these observations indicate that nutritionally driven gene regulation prevents the accumulation of ergot alkaloids in Camembert cheese. This hypothesis was supported by our lack of detection of transcripts for any of the seven P. camemberti eas cluster genes in the cheese rind metatranscriptome data of Lessard et al. (43), which includes seven samples of Camembert cheese rind collected between 7 and 77 days postinoculation. The potential presence of transcripts of P. biforme eas genes in cheese rind was not investigated. The lack of accumulation of ergot alkaloids in P. biforme-ripened cheese (in spite of a functioning eas pathway in vitro) and the lack of transcripts of P. camemberti eas genes in cheese rind (43) indicate that the lack of ergot alkaloids in P. biforme cheese rind is due to gene regulation.
Conclusions and implications.
Our data indicate that P. camemberti previously had the genetic capacity to produce ergot alkaloids, including rugulovasine A and B and precursors to these compounds, such as chanoclavine-I and chanoclavine-I aldehyde. While the health effects of the rugulovasines are largely understudied, the available data indicate that they induce negative health consequences. Ingestion of rugulovasines by day-old poultry caused acute toxicity leading to death (36); in fact, chick death was used as a bioassay to purify the rugulovasines. Rugulovasines also decreased blood pressure when administered to cats (44). Chlorinated and brominated forms of rugulovasines have been reported in some rugulovasine producers (37); however, chlorinated and brominated forms were not detected in P. biforme extracts, based on the absence of analytes of the calculated m/z values.
Through our heterologous expression studies, we have identified a new ergot alkaloid biosynthesis gene, easQ, which appears to encode an aldehyde dehydrogenase involved in synthesis of rugulovasines. We also provided indirect evidence for the function of the product of an allele of the gene easH and have proposed a biosynthetic pathway for the rugulovasine class of ergot alkaloids. We have used similar heterologous expression and analytical approaches to study other genes in the ergot alkaloid pathway; for example, easA from Claviceps purpurea (22) and cloA from Epichloë festucae × Epichloë typhina (35).
Finally, our data indicate that the lack of accumulation of rugulovasines in P. camemberti in vitro results from a mutation in easC and that the lack of production of precursors in cheese is due to substrate-specific control of the pathway. Phylogenetic analyses indicate that P. biforme is ancestral to P. camemberti and that Penicillium commune may be ancestral to both (31, 45). The close phylogenetic relationship between P. biforme, P. fuscoglaucum, and P. camemberti was demonstrated by data from 17 different microsatellite markers (28). There are two chemotypes of P. commune, with chemotype I being a producer of cyclopenoxoic acid and rugulovasines and chemotype II producing fumigaclavines (46). Penicillium camemberti is hypothesized to have descended from P. commune chemotype I. These phylogenetic and mycotoxin data suggest that the ancestor to P. camemberti had the genetic capacity to produce rugulovasines, but at some point in the evolution of domesticated P. camemberti, nonproducing strains evolved. It is possible that P. biforme may conditionally produce rugulovasines as a means of defense of resources. Since domesticated P. camemberti occurs only in hygienic environments with defined microflora, pressure to maintain this pathway may have been lost.
MATERIALS AND METHODS
Characterization of P. camemberti and P. biforme ergot alkaloid gene clusters.
The ergot alkaloid gene clusters of P. camemberti and P. biforme were identified by comparing individual eas genes from N. fumigata (17) to genomes of P. camemberti strain FM 013 (GenBank accession number CBVV000000000.1) and P. biforme strain FM 169 (GenBank accession number CBXO000000000.1). Contigs containing eas gene clusters of P. camemberti (GenBank accession number CBVV010000119.1) and P. biforme (GenBank accession number CBXO010000115.1) were downloaded, and additional genes were annotated by blastx, searching individual, contiguous 5-kb fragments. Polymorphisms between P. camemberti and P. biforme eas gene clusters were identified by blastn analysis of the aligned sequences of the two fungi. The potential presence of transcripts originating from P. camemberti eas cluster genes in the metatranscriptome of Camembert cheese rind (43) was assessed by megablast searching the data available under GenBank Sequence Read Archive accession number SRX360429.
Sample preparation for alkaloid analysis.
Cultures of P. camemberti NRRL 874, P. camemberti NRRL 875, P. biforme NRRL 885, and N. fumigata strains were grown on malt extract agar (6 g malt extract, 6 g dextrose, 1.8 g maltose, 1.2 g yeast extract, 15 g agar per liter) at 22°C for 7 days prior to alkaloid analysis. These same isolates were also cultured in a variant of Czapek's medium in which succinic acid was the sole carbon source (1.2 g succinic acid, 2 g sodium nitrate, 1 g dipotassium phosphate, 0.5 g magnesium sulfate, 0.5 g potassium chloride, 0.01 g ferrous sulfate per liter). For HPLC with fluorescence detection, samples of 50-mm2 surface area malt extract agar or 400 μl modified Czapek's medium were extracted with 400 μl HPLC-grade methanol for 1 h and then clarified by centrifugation before injecting 20 μl. For LC-MS analyses, an entire petri dish culture was extracted by repeated washing with 4 ml of methanol, and then 2 ml of the extract was concentrated to 100 μl in a vacuum concentrator prior to injecting 10 μl. Samples of Brie and Camembert cheese bought commercially and those made in the laboratory were also analyzed by HPLC and LC-MS by extracting 200-mg portions of cheese rind in 1 ml of HPLC-grade methanol. Laboratory-generated cheese wheels (approximately 300 ml) were made with mesophilic cheese culture and animal rennet (both from New England Cheesemaking Supply Co., South Deerfield, MA) and approximately 1 × 107 conidia of either P. camemberti NRRL 874 or P. biforme NRRL 885 as the ripening fungi. Curds were allowed to mature for 8 weeks at 4°C prior to sampling.
HPLC and LC-MS analyses.
Fluorescence HPLC was performed essentially as described previously (47). Briefly, analytes were separated by reverse-phase chromatography on a C18 column (Prodigy 5-μm ODS3, 150 mm by 4.6 mm; Phenomenex, Torrance, CA) with a multilinear, binary gradient from 5% acetonitrile plus 95% 50 mM ammonium acetate to 75% acetonitrile plus 25% 50 mM ammonium acetate. Ergot alkaloids were detected by two fluorescence detectors connected in series. One was set with excitation at 272 nm and emission at 372 nm, whereas the other had excitation and emission wavelengths of 310 nm and 410 nm, respectively.
Most LC-MS analyses were conducted on a Thermo LCQ Deca XP plus mass spectrometer connected to a Thermo Surveyor HPLC system (Thermo Scientific, Waltham, MA). Analyses were essentially as described by Ryan et al. (48, 49), except that the linear, binary gradient was as follows: mobile phases A (5% acetonitrile, 0.1% formic acid) and B (75% acetonitrile, 0.1% formic acid) were initially combined at 86% A plus 14% B and linearly ramped up to 100% B over 20 min. The column was a Phenomenex 4-μm polar-RP (150 mm by 2 mm) maintained at 30°C, and the flow rate 200 μl/min.
High-resolution mass spectra were collected on a Thermo Scientific Q Exactive mass spectrometer coupled to a Thermo Accela 1250 ultra-high-performance liquid chromatography (UHPLC) system. The mass spectrometer was operated in positive ion mode and programmed with data-dependent acquisition settings. Precursor scans were acquired at 70,000 resolution (at m/z = 200) over an m/z range of 80 to 1,200. The 10 most abundant ions from each precursor scan were selected for higher-energy collisional-dissociation (HCD) fragmentation (normalized collisional energy [NCE] = 30) and were analyzed at 35,000 resolution. Separations were performed on a 2.1-mm by 100-mm Zorbax Eclipse XDB-C18 column (Agilent, Santa Clara, CA) subjected to a gradient prepared by combining mobile phase A (0.1% formic acid) and mobile phase B (acetonitrile, 0.1% formic acid). The sample was loaded at 95% A plus 5% B and held for 1 min before ramping linearly to 40% A plus 60% B at 20 min at a flow rate of 300 μl/min.
Preparation of gene constructs for fungal transformation.
Constructs for heterologous expression were assembled by a combination of PCR, fusion PCR, and cloning into plasmids by restriction enzyme-mediated strategies. A typical PCR mixture was comprised of 11 μl distilled/deionized water, 5 μl 5× Phusion high-fidelity (HF) buffer (100 mM KCl, 20 mM Tris-HCl, pH 7.4, 1.5 mM MgCl2; Thermo Scientific, Waltham, MA), 4 μl 1.25 mM deoxynucleoside triphosphates [dNTPs], 1.25 μl 20 μM forward primer, 1.25 μl 20 μM reverse primer, 2 μl template DNA, and 0.5 μl Phusion HF polymerase (Thermo Scientific). The various reaction mixtures underwent the same general temperature-cycling regimen, with an initial denaturation at 98°C for 30 s, followed by 35 cycles of 98°C (15 s), annealing temperature (as defined by primer pair in Table 1) (15 s), and extension at 72°C for the extension time defined in Table 1, followed by a final extension at 72°C for 60 s. The primers, annealing temperatures, and extension times are listed in Table 1. PCR products were cleaned with the Zymogen DNA Clean & Concentrator kit (Zymo Research Corp., Irvine, CA) prior to a subsequent PCR, restriction digestion, or ligation.
TABLE 1.
Primers and PCR protocol informationa
| Primer pair | Primer sequence (5′ to 3′) | Product(s) (length in bp)b | Annealing temp (°C), extension time (s) |
|---|---|---|---|
| 1 | GAGTAGGCACTCCGCACCATGACGATTTCCAATTGCGCCAAC + | easH with promoter extension (1,428) | 64, 45 |
| CCACCGCGGTGGCGGCCGCTATTTCGTCACTTTGGCTTGCATG | |||
| 2 | GTACTTGGTGGATTAGAAGCAATGTGTGAGACATCAATTGATCTGAC + | easQ with promoter extension (1,927) | 64, 60 |
| GGGGATCCGGTCTATGTGAAGCATGCGGGAATG | |||
| 3 | GTTGGCGCAATTGGAAATCGTCATGGTGCGGAGTGCCTACTC + | N. fumigata promoter with easH and easQ extensions (837) | 64, 45 |
| GTCAGATCAATTGATGTCTCACACATTGCTTCTAATCCACCAAGTAC | |||
| 4 | CCACCGCGGTGGCGGCCGCTATTTCGTCACTTTGGCTTGCATG + | easH-N. fumigata promoter-easQ fusion (4,111) | 64, 120 |
| GGGGATCCGGTCTATGTGAAGCATGCGGGAATG | |||
| 5 | GTATACTAGTGCAATCCGCAATGAATCTGCAGG + | easQ with native promoter (2,967) | 62, 90 |
| CAGAGCGGCCGCTCTATGTGAAGCATGCGGGAATG | |||
| 6 | ACCGGATCCGTGCCGTAGTCCTATACTAAG + | easH with native promoter (2,213) | 62, 90 |
| CCACCGCGGGTGGCGGCCGCTATTTCGTCACTTTGGCTTGCATG | |||
| 7 | CATCGGATCCGGCCAACATGACTCCCACGGC + | dmaW with native promoter (2,686) | 67, 90 |
| CTACGCGGCCGCGTGTAACTATGGAGGATGACAAGC | |||
| 8 | CTCAGAATTCGAGCTGGTCCGCGTTCAGG + | easC with native promoter (3,070) | 67, 90 |
| CTACGGATCCGTCGCTTGATGGCTGTGATGTAACG | |||
| 9 | CAACGCGGCCGCAAGAGGATAGAGCTTTCAGCTGG + | easE with native promoter (3,642) | 67, 90 |
| CAGAACTAGTGGTGATTCAACGGAGCCCATGG | |||
| 10 | GAGTAGGCACTCCGCACCATGATCTCAATAGATCATATTCC + | easC with promoter extension (2,098) | 59, 40 |
| GTGGCCTTGAGTTAATCTGAAGC | |||
| 11 | GGAATATGATCTATTGAGATCATGGTGCGGAGTGCCTACTC + | N. fumigata promoter with easC extension (809) | 59, 40 |
| GCTTCTAATCCACCAAGTACTTGG | |||
| 12 | CTCGGAATTCGCTTCTAATCCACCAAGTACTTGG + | N. fumigata promoter-easC fusion (2,886) | 59, 90 |
| GAGTACTAGTGTGGCCTTGAGTTAATCTGAAGC | |||
| 13 | GGACTTCCCCAACCATGATCAG + | dmaW cDNA (1,282) | 62, 45 |
| CGAAGGAGGTAGGATGGCCAC | |||
| 14 | GGAATCCGGCACTGGATTGAAC + | easF cDNA (853) | 62, 45 |
| CTCTCAATCCCGCCGACCC | |||
| 15 | GCCTGGAGCAAGTCAAGTTCTCA + | easC cDNA (1,323), easC genomic DNA (1,391) | 62, 45 |
| TGATTCGACGATTCTCTCCACC | |||
| 16 | GGAGTCATGTTGGCCATCTCGC + | easE cDNA (1,570) | 62, 45 |
| GTCCCACTGCCGCTTCAGC | |||
| 17 | CGCATCGGGTATTGGTGCCG + | easD cDNA (670) | 62, 45 |
| CTAGCCACATCCTCGGGCTC | |||
| 18 | GTACTTGGTGGATTAGAAGCAATGTGTGAGACATCAATTGATCTGAC + | easQ cDNA (1,659) | 62, 45 |
| GACTAGGTCAAGCATGACTGGC | |||
| 19 | GAGTAGGCACTCCGCACCATGACGATTTCCAATTGCGCCAAC + | easH cDNA (951) | 62, 45 |
| CCTTGTAGGGATACACAGTTGG |
Underlining indicates unique restriction sites for cloning PCR products: ACTAGT, SpeI; GCGGCCGC, NotI; GGATCC, BamRI; and, GAATTC, EcoRI.
“Extension” refers to incorporation of an additional 17 to 23 nt at the 5′ end of a primer to add sequences that will facilitate a later fusion PCR.
Constructs were prepared in general by cloning PCR products containing genes of interest into pBCphleo (50), which was obtained from the Fungal Genetics Stock Center (Kansas State University, Manhattan, KS). Most cloning strategies were based on restriction sites engineered into PCR primers and also contained within pBCphleo. In some cases (described individually below), genes were connected to the N. fumigata bidirectional easA-easG promoter (35) by fusion PCR. In other cases (particularly after reverse transcriptase PCR analyses indicated that P. camemberti eas genes were transcribed under routine culture conditions), P. camemberti genes were expressed from their native promoters. Ligations were conducted with T4 DNA ligase (New England BioLabs, Ipswich, MA) and transformed into Escherichia coli. Plasmids were purified with the Zyppy plasmid miniprep kit (Zymo Research Corp., Irvine, CA).
For coexpression of P. camemberti easH and easQ genes, the individual genes were PCR amplified with primer pairs 1 and 2 (Table 1). The cleaned PCR products were then attached to an N. fumigata easA-easG bidirectional promoter, which was amplified with primer pair 3 (Table 1). A fusion PCR with primer pair 4 (Table 1) assembled the final construct (Fig. 2C), which was cotransformed along with pBCphleo in an equal molar concentration into a previously created chanoclavine-l aldehyde-accumulating easA knockout strain of N. fumigata (22) by the transformation protocol described below.
To assess the individual functions of P. camemberti easQ and easH, gene fragments under the control of their native promoters were PCR amplified using primer pairs 5 and 6, respectively (Table 1), digested with their corresponding restriction enzymes (sites included in PCR primers), cleaned, and then ligated with similarly digested pBCphleo. The resulting plasmids were transformed into the N. fumigata easA knockout strain (22).
The functionality of P. camemberti dmaW, easC, and easE was tested by using those genes in attempted complementation of previously characterized N. fumigata knockout mutants. The P. camemberti gene fragments were PCR amplified from primer pairs 7, 8, and 9 (Table 1), restriction digested, and cloned into pBCphleo. The resulting plasmids were transformed into dmaW, easC, and easE knockout strains of N. fumigata (14, 17). To further test easC, the coding sequences of P. camemberti easC were amplified with primer combination 10 (Table 1) and joined by fusion PCR to the easA-easG promoter of N. fumigata (which had been amplified with primer pair 11 [Table 1]) in a fusion PCR primed with combination 12 (Table 1).
Fungal transformation.
Transformation of N. fumigata strains was as described previously (35, 51). Each of the recipient strains was a previously prepared mutant with knockout of an individual eas gene and already resistant to hygromycin. For this reason, phleomycin resistance as conferred by pBCphleo was the selectable marker employed. The easA knockout of N. fumigata (22), which stops its pathway after easD (Fig. 1) and, thus, mimics the core eas gene cluster of P. camemberti (Fig. 2), was the recipient for transformations involving the easH-promoter-easQ construct, as well as for individual easH and easQ constructs. Strains with knockouts of dmaW (17), easC (14), and easE (14) served as recipients of constructs containing individual alleles of homologs from P. camemberti in genetic complementation tests. Transformants were checked for the presence of the introduced construct by PCR. Colonies from transformation experiments that did not yield a change in phenotype were checked by qualitative reverse transcriptase PCR for accumulation of transcript from the introduced construct (as described below). Fragments amplified from cDNA were sequenced by Sanger technology (Eurofins Genomics, Louisville, KY) to demonstrate the lack of introns, confirming the absence of genomic DNA in the template.
Extraction of mRNA and preparation of cDNA.
Cultures of P. camemberti NRRL 874 or N. fumigata transformants were grown as surface cultures on malt extract broth for 3 days at room temperature. A 100-mg sample of the fungal mat was then frozen with liquid nitrogen and ground into a powder with a mortar and pestle. RNA was extracted by following the instructions of the Qiagen RNeasy plant kit (Qiagen, Germantown, MD) with the on-column DNase I treatment. cDNA was obtained by reverse transcribing the mRNA from an oligo(dT) primer with SuperScript IV reverse transcriptase (Thermo Scientific, Waltham, MA). The cDNA was then used as the template in individual reaction mixtures with primer pairs 13 through 19 (Table 1). PCR products were sequenced by Sanger technology at Eurofins Genomics (Louisville, KY).
Statistical analysis.
To compare relative proportions of m/z 271 ion (presumed oxidized chanoclavine-I aldehyde) and m/z 255 ion (chanoclavine-I aldehyde), the mean ratio of peak areas was calculated from analyses of three independent cultures of nontransformed N. fumigata easA knockout and four independent cultures each of N. fumigata easA knockout transformed with P. camemberti easH or P. camemberti easQ. Data were log10 transformed so that variances approximated equality in a Brown-Forsythe test (P = 0.80). A single-factor analysis of variance (ANOVA) showed that fungal strain had a significant effect on the m/z 271-to-m/z 255 ratio (P = 0.002). Individual-treatment mean values were then compared in a Tukey-Kramer honestly significant difference test. Analyses were performed with JMP (SAS, Cary, NC).
Supplementary Material
ACKNOWLEDGMENTS
High-resolution mass spectrometry analyses were conducted at the West Virginia University Shared Research Facilities.
We thank Sarah O'Connor (John Innes Institute) for the N-Me-DMAT standard.
This work was supported by grant number R15GM114774 from the National Institutes of Health, National Institute of General Medical Sciences, and by Hatch project WVA00669 from the United States Department of Agriculture, National Institute of Food and Agriculture.
Footnotes
This article was published with permission of the West Virginia Agriculture and Forestry Experiment Station as scientific article number 3343.
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01583-18.
REFERENCES
- 1.Florea S, Panaccione DG, Schardl CL. 2017. Ergot alkaloids of the family Clavicipitaceae. Phytopathology 107:504–518. doi: 10.1094/PHYTO-12-16-0435-RVW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haarmann T, Rolke Y, Giesbert S, Tudzynski P. 2009. Ergot: from witchcraft to biotechnology. Mol Plant Pathol 10:563–577. doi: 10.1111/j.1364-3703.2009.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferriere A, Cortet C, Chanson P, Delemer B, Caron P, Chabre O, Reznik Y, Bertherat J, Rohmer V, Briet C, Raingeard I. 2017. Cabergoline for Cushing's disease: a large retrospective multicenter study. Eur J Endocrinol 176:305–314. doi: 10.1530/EJE-16-0662. [DOI] [PubMed] [Google Scholar]
- 4.Lei X, Yu J, Niu Q, Liu J, Freering PC, Wu F. 2015. The FDA-approved natural product dihydroergocristine reduces the production of the Alzheimer's disease amyloid-β peptides. Sci Rep 5:16541. doi: 10.1038/srep16541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zajdel P, Bednarski M, Sapa J, Nowak G. 2015. Ergotamine and nicergoline—facts and myths. Pharmacol Rep 67:360–363. doi: 10.1016/j.pharep.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann A. 1980. LSD—my problem child. McGraw-Hill, New York, NY. [Google Scholar]
- 7.Panaccione DG, Arnold SL. 2017. Ergot alkaloids contribute to virulence in an insect model of invasive aspergillosis. Sci Rep 7:8930. doi: 10.1038/s41598-017-09107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerhards N, Neubauer L, Tudzynski P, Li SM. 2014. Biosynthetic pathways of ergot alkaloids. Toxins (Basel) 6:3281–3295. doi: 10.3390/toxins6123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young CA, Schardl CL, Panaccione DG, Florea S, Takach JE, Charlton ND, Moore N, Webb JS, Jaromczyk J. 2015. Genetics, genomics and evolution of ergot alkaloid diversity. Toxins (Basel) 7:1273–1302. doi: 10.3390/toxins7041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson SL, Panaccione DG. 2015. Diversification of ergot alkaloids in natural and modified fungi. Toxins (Basel) 7:201–218. doi: 10.3390/toxins7010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai HF, Wang H, Gebler JC, Poulter CD, Schardl CL. 1995. The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem Biophys Res Commun 216:119–125. doi: 10.1006/bbrc.1995.2599. [DOI] [PubMed] [Google Scholar]
- 12.Rigbers O, Li SM. 2008. Ergot alkaloid biosynthesis in Aspergillus fumigatus overproduction and biochemical characterization of a 4-dimethylallyltryptophan N-methyltransferase. J Biol Chem 283:26859–26868. doi: 10.1074/jbc.M804979200. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz N, Olšovská J, šSulc M, Tudzynski P. 2010. Alkaloid cluster gene ccsA of the ergot fungus Claviceps purpurea encodes chanoclavine I synthase, a flavin adenine dinucleotide-containing oxidoreductase mediating the transformation of N-methyl-dimethylallyltryptophan to chanoclavine I. Appl Environ Microbiol 76:1822–1830. doi: 10.1128/AEM.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goetz KE, Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG. 2011. Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus. Curr Genet 57:201–211. doi: 10.1007/s00294-011-0336-4. [DOI] [PubMed] [Google Scholar]
- 15.Wallwey C, Matuschek M, Li SM. 2010. Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I to chanoclavine-I aldehyde catalyzed by a short-chain alcohol dehydrogenase FgaDH. Arch Microbiol 192:127–134. doi: 10.1007/s00203-009-0536-1. [DOI] [PubMed] [Google Scholar]
- 16.Tudzynski P, Hölter K, Correia T, Arntz C, Grammel N, Keller U. 1999. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol Gen Genet 261:133–141. doi: 10.1007/s004380050950. [DOI] [PubMed] [Google Scholar]
- 17.Coyle CM, Panaccione DG. 2005. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl Environ Microbiol 71:3112–3118. doi: 10.1128/AEM.71.6.3112-3118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unsöld IA, Li SM. 2005. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- 19.Schardl CL, Young CA, Hesse U, Amyotte SG, Andreeva K, Calie PJ, Fleetwood DJ, Haws DC, Moore N, Oeser B, Panaccione DG, Schweri KK, Voisey CR, Farman ML, Jaromczyk JW, Roe BA, O'Sullivan DM, Scott B, Tudzynski P, An Z, Arnaoudova EG, Bullock CT, Charlton ND, Chen L, Cox M, Dinkins RD, Florea S, Glenn AE, Gordon A, Güldener U, Harris DR, Hollin W, Jaromczyk J, Johnson RD, Khan AK, Leistner E, Leuchtmann A, Li C, Liu JG, Liu J, Liu M, Mace W, Machado C, Nagabhyru P, Pan J, Schmid J, Sugawara K, Steiner U, Takach JE, Tanaka E, Webb JS, Wilson EV, Wiseman JL, Yoshida R, Zeng Z. 2013. Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet 9:e1003323. doi: 10.1371/journal.pgen.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haarmann T, Machado C, Lübbe Y, Correia T, Schardl CL, Panaccione DG, Tudzynski P. 2005. The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution. Phytochemistry 66:1312–1320. doi: 10.1016/j.phytochem.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Unsöld IA, Li SM. 2006. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. Chembiochem 7:158–164. doi: 10.1002/cbic.200500318. [DOI] [PubMed] [Google Scholar]
- 22.Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG. 2010. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl Environ Microbiol 76:3898–3903. doi: 10.1128/AEM.02914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng JZ, Coyle CM, Panaccione DG, O'Connor SE. 2010. A role for old yellow enzyme in ergot alkaloid biosynthesis. J Am Chem Soc 132:1776–1777. doi: 10.1021/ja910193p. [DOI] [PubMed] [Google Scholar]
- 24.Cheng JZ, Coyle CM, Panaccione DG, O'Connor SE. 2010. Controlling a structural branch point in ergot alkaloid biosynthesis. J Am Chem Soc 132:12835–12837. doi: 10.1021/ja105785p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallwey C, Matuschek M, Xie XL, Li SM. 2010. Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I aldehyde to festuclavine by the festuclavine synthase FgaFS in the presence of the old yellow enzyme FgaOx3. Org Biomol Chem 8:3500–3508. doi: 10.1039/c003823g. [DOI] [PubMed] [Google Scholar]
- 26.Havemann J, Vogel D, Loll B, Keller U. 2014. Cyclolization of D-lysergic acid alkaloid peptides. Chem Biol 21:146–155. doi: 10.1016/j.chembiol.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Jakubczyk D, Caputi L, Hatsch A, Nielsen CA, Diefenbacher M, Klein J, Molt A, Schröder H, Cheng JZ, Naesby M, O'Connor SE. 2015. Discovery and reconstitution of the cycloclavine biosynthetic pathway—enzymatic formation of a cyclopropyl group. Angew Chem Int Ed Engl 54:5117–5121. doi: 10.1002/anie.201410002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giraud F, Giraud T, Aguileta G, Fournier E, Samson R, Cruaud C, Lacoste S, Ropars J, Tellier A, Dupont J. 2010. Microsatellite loci to recognize species for the cheese starter and contaminating strains associated with cheese manufacturing. Int J Food Microbiol 137:204–213. doi: 10.1016/j.ijfoodmicro.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Ropars J, Cruaud C, Lacoste S, Dupont J. 2012. A taxonomic and ecological overview of cheese fungi. Int J Food Microbiol 155:199–210. doi: 10.1016/j.ijfoodmicro.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Cheeseman K, Ropars J, Renault P, Dupont J, Gouzy J, Branca A, Malagnac F. 2014. Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nature Comm 5:2876. doi: 10.1038/ncomms3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ropars J, De la Vega RCR, López-Villavicencio M, Gouzy J, Sallet E, Dumas É, Giraud T. 2015. Adaptive horizontal gene transfers between multiple cheese-associated fungi. Curr Biol 25:2562–2569. doi: 10.1016/j.cub.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martín JF, Álvarez-Álvarez R, Liras P. 2017. Clavine alkaloids gene clusters of Penicillium and related fungi: evolutionary combination of prenyltransferases, monooxygenases and dioxygenases. Genes (Basel) 8:E342. doi: 10.3390/genes8120342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerhards N, Li SM. 2017. A bifunctional old yellow enzyme from Penicillium roqueforti is involved in ergot alkaloid biosynthesis. Org Biomol Chem 15:8059–8071. doi: 10.1039/C7OB02095C. [DOI] [PubMed] [Google Scholar]
- 34.Haarmann T, Ortel I, Tudzynski P, Keller U. 2006. Identification of the cytochrome P450 monooxygenase that bridges the clavine and ergoline alkaloid pathways. Chembiochem 7:645–652. doi: 10.1002/cbic.200500487. [DOI] [PubMed] [Google Scholar]
- 35.Robinson SL, Panaccione DG. 2014. Heterologous expression of lysergic acid and novel ergot alkaloids in Aspergillus fumigatus. Appl Environ Microbiol 80:6465–6472. doi: 10.1128/AEM.02137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorner JW, Cole RJ, Hill R, Wicklow D, Cox RH. 1980. Penicillium rubrum and Penicillium biforme, new sources of rugulovasines A and B. Appl Environ Microbiol 40:685–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Medeiros LS, da Silva JV, Abreu LM, Pfenning LH, Silva CL, Thomasi SS, Venâncio T, van Pée KH, Nielsen KF, Rodrigues-Filho E. 2015. Dichlorinated and brominated rugulovasines, ergot alkaloids produced by Talaromyces wortmannii. Molecules 20:17627–17644. doi: 10.3390/molecules200917627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole RJ, Kirksey JW, Clardy J, Eickman N, Weinreb SM, Singh P, Kim D. 1976. Structures of rugulovasine-A and-B and 8-chlororugulovasine-A and -B. Tetrahedron Lett 17:3849–3852. doi: 10.1016/0040-4039(76)80164-5. [DOI] [Google Scholar]
- 39.Rebek J, Shue YK, Tai DF. 1984. The rugulovasines: synthesis, structure and interconversions. J Org Chem 49:3540–3545. doi: 10.1021/jo00193a018. [DOI] [Google Scholar]
- 40.Matuschek M, Wallwey C, Xie X, Li SM. 2011. New insights into ergot alkaloid biosynthesis in Claviceps purpurea: an agroclavine synthase EasG catalyses, via a non-enzymatic adduct with reduced glutathione, the conversion of chanoclavine-I aldehyde to agroclavine. Org Biomol Chem 9:4328–3435. doi: 10.1039/c0ob01215g. [DOI] [PubMed] [Google Scholar]
- 41.Panaccione DG, Coyle CM. 2005. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl Environ Microbiol 71:3106–3111. doi: 10.1128/AEM.71.6.3106-3111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson SL, Panaccione DG. 2012. Chemotypic and genotypic diversity in the ergot alkaloid pathway of Aspergillus fumigatus. Mycologia 104:804–812. doi: 10.3852/11-310. [DOI] [PubMed] [Google Scholar]
- 43.Lessard M-H, Viel C, Boyle B, St Gelais D, Labrie S. 2014. Metatranscriptome analysis of fungal strains Penicillium camemberti and Geotrichum candidum reveal cheese matrix breakdown and potential development of sensory properties of ripened Camembert-type cheese. BMC Genomics 15:235. doi: 10.1186/1471-2164-15-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meurant G. 1981. Roquefortines, p 528–544. In Cole RJ, Cox RH (ed), Handbook of toxic fungal metabolites. Academic Press, New York, NY. [Google Scholar]
- 45.Polonelli L, Morace G, Rosa R, Castagnola MA, Frisvad JC. 1987. Antigenic characterization of Penicillium camemberti and related common cheese contaminants. Appl Environ Microbiol 53:872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frisvad JC, Filtenborg O. 1989. Terverticillate penicillia: chemotaxonomy and mycotoxin production. Mycologia 1:837–861. doi: 10.2307/3760103. [DOI] [Google Scholar]
- 47.Panaccione DG, Ryan KL, Schardl CL, Florea S. 2012. Analysis and modification of ergot alkaloid profiles in fungi. Methods Enzymol 515:267–290. doi: 10.1016/B978-0-12-394290-6.00012-4. [DOI] [PubMed] [Google Scholar]
- 48.Ryan KL, Moore CT, Panaccione DG. 2013. Partial reconstruction of the ergot alkaloid pathway by heterologous gene expression in Aspergillus nidulans. Toxins (Basel) 5:445–455. doi: 10.3390/toxins5020445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan KL, Akhmedov NG, Panaccione DG. 2015. Identification and structural elucidation of ergotryptamine, a new ergot alkaloid produced by genetically modified Aspergillus nidulans and natural isolates of Epichloë species. J Agric Food Chem 63:61–67. doi: 10.1021/jf505718x. [DOI] [PubMed] [Google Scholar]
- 50.Silar P. 1995. Two new easy to use vectors for transformations. Fungal Genet Rep 42:73. doi: 10.4148/1941-4765.1353. [DOI] [Google Scholar]
- 51.Bilovol Y, Panaccione DG. 2016. Functional analysis of the gene controlling hydroxylation of festuclavine in the ergot alkaloid pathway of Neosartorya fumigata. Curr Genet 62:853–860. doi: 10.1007/s00294-016-0591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.