Ethyl acetate is a common bulk chemical that is currently produced from petrochemical sources. Several Eat1-containing yeast strains naturally produce large amounts of ethyl acetate and are potential cell factories for the production of bio-based ethyl acetate. Rational design of the underlying metabolic pathways may result in improved production strains, but it requires fundamental knowledge on the function of Eat1. A key feature is the location of Eat1 in the yeast cell. The precursors for ethyl acetate synthesis can be produced in multiple cellular compartments through different metabolic pathways. The location of Eat1 determines the relevance of each pathway, which will provide future targets for the metabolic engineering of bulk ethyl acetate production in yeast.
KEYWORDS: AAT, Kluyveromyces, Wickerhamomyces, alcohol acetyltransferase, ethyl acetate, mitochondria, yeast
ABSTRACT
Eat1 is a recently discovered alcohol acetyltransferase responsible for bulk ethyl acetate production in yeasts such as Wickerhamomyces anomalus and Kluyveromyces lactis. These yeasts have the potential to become efficient bio-based ethyl acetate producers. However, some fundamental features of Eat1 are still not understood, which hampers the rational engineering of efficient production strains. The cellular location of Eat1 in yeast is one of these features. To reveal its location, Eat1 was fused with yeast-enhanced green fluorescent protein (yEGFP) to allow intracellular tracking. Despite the current assumption that bulk ethyl acetate production occurs in the yeast cytosol, most of Eat1 localized to the mitochondria of Kluyveromyces lactis CBS 2359 Δku80. We then compared five bulk ethyl acetate-producing yeasts in iron-limited chemostats with glucose as the carbon source. All yeasts produced ethyl acetate under these conditions. This strongly suggests that the mechanism and location of bulk ethyl acetate synthesis are similar in these yeast strains. Furthermore, an in silico analysis showed that Eat1 proteins from various yeasts were mostly predicted as mitochondrial. Altogether, it is concluded that Eat1-catalyzed ethyl acetate production occurs in yeast mitochondria. This study has added new insights into bulk ethyl acetate synthesis in yeast, which is relevant for developing efficient production strains.
IMPORTANCE Ethyl acetate is a common bulk chemical that is currently produced from petrochemical sources. Several Eat1-containing yeast strains naturally produce large amounts of ethyl acetate and are potential cell factories for the production of bio-based ethyl acetate. Rational design of the underlying metabolic pathways may result in improved production strains, but it requires fundamental knowledge on the function of Eat1. A key feature is the location of Eat1 in the yeast cell. The precursors for ethyl acetate synthesis can be produced in multiple cellular compartments through different metabolic pathways. The location of Eat1 determines the relevance of each pathway, which will provide future targets for the metabolic engineering of bulk ethyl acetate production in yeast.
INTRODUCTION
Ethyl acetate is a valuable bulk chemical and an important aroma compound in fermented foods (1). Industrially, ethyl acetate is produced from petrochemical resources, but biological production routes have been explored in recent years. Yeasts are prominent natural ethyl acetate producers. Ester production is well known in Saccharomyces cerevisiae, which typically produces between 8 and 32 mg/liter ethyl acetate in beer fermentations (2). Several non-Saccharomyces yeast species produce ethyl acetate from carbohydrates at a much higher yield than that of S. cerevisiae (3). Ethyl acetate yields up to 51.4% of the theoretical pathway maximum have been reported in Kluyveromyces marxianus (4). Other bulk ethyl acetate-producing yeasts include Wickerhamomyces anomalus (5, 6), Cyberlindnera fabianii (7), and Kluyveromyces lactis (8).
Alcohol acetyl transferases (AATs) are the main ethyl acetate-producing enzymes which use acetyl-coenzyme A (CoA) and ethanol as the substrate. Most research on ethyl acetate-producing AATs in yeast is based on Atf1 and Atf2 from S. cerevisiae (9, 10). An S. cerevisiae strain lacking atf1 and atf2 produced 50% less ethyl acetate compared to the parental strain (11). Homologs of Atf1 and Atf2 are present in bulk ethyl acetate-producing yeasts (12, 13).
The prevailing hypothesis on the physiological function of bulk ethyl acetate production suggests that it is produced as an overflow metabolite under conditions where the tricarboxylic acid (TCA) cycle does not function optimally (3, 14). Yeasts that naturally produce bulk amounts of ethyl acetate are Crabtree negative. They oxidize glucose and other carbohydrates to pyruvate in the cytosol. Under aerobic conditions, Crabtree-negative yeasts preferentially transport the pyruvate to the mitochondria. There, it is further oxidized via pyruvate dehydrogenase to acetyl-CoA (Fig. 1, reaction I) and subsequently oxidized in the TCA cycle (15). Ethyl acetate is formed under conditions where the efficiency of the TCA cycle is impaired by, e.g., iron or oxygen limitation (16, 17). As a consequence, acetyl-CoA cannot enter the TCA cycle and accumulates in the mitochondria. It is assumed that yeasts use an AAT-catalyzed reaction to relieve the acetyl-CoA accumulation and regenerate free CoA (3, 18). Ethyl acetate is formed in the process. This hypothesis would imply that mitochondrial acetyl-CoA accumulation causes ethyl acetate production (19).
FIG 1.
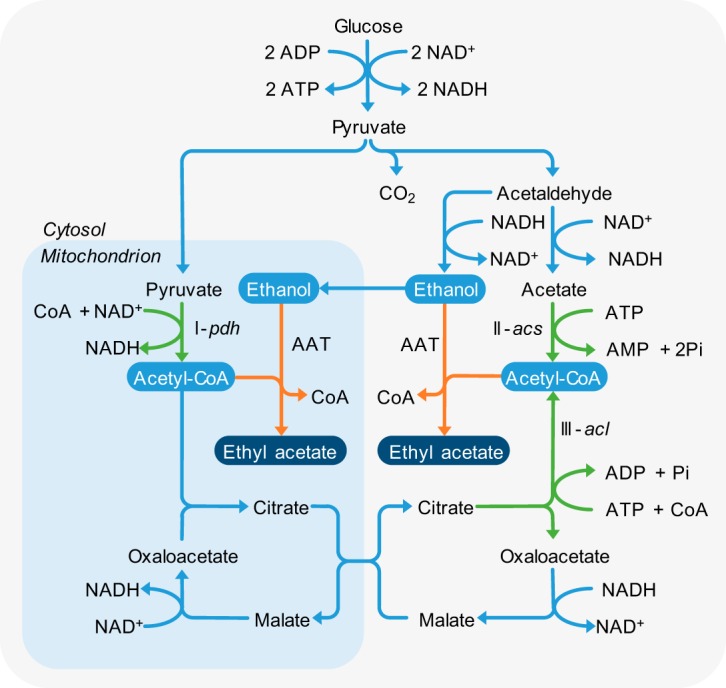
Potential pathways of ethyl acetate production via an AAT in yeast. The AAT-catalyzed reaction is indicated in orange. The three reactions forming acetyl-CoA during glucose catabolism are shown in green. Reaction I, pyruvate dehydrogenase (pdh); reaction II, acetyl-CoA synthetase (acs); and reaction III, ATP citrate lyase (acl).
Ethanol is the second substrate needed for ethyl acetate synthesis by AATs. Crabtree-negative yeasts typically do not form ethanol under aerobic conditions. However, unfavorable conditions, such as iron limitation, lead to ethanol formation in K. marxianus, even in the presence of oxygen (17, 20). Ethanol is produced from pyruvate via acetaldehyde in the cytosol (Fig. 1). The acetaldehyde may also be converted to acetate and further to cytosolic acetyl-CoA via acetyl-CoA synthetase (Fig. 1, reaction II). This reaction is essential in most yeasts, as it supplies acetyl-CoA for fatty acid synthesis (21). However, during aerobic growth on sugars, the acetyl-CoA flux in the cytosol is much lower than that in the mitochondria (22). It is therefore unlikely that it contributes significantly to bulk ethyl acetate synthesis. Moreover, bulk ethyl acetate synthesis in yeast does not occur in the absence of oxygen (19). Under anaerobic conditions, carbohydrate catabolism occurs in the cytosol, and mitochondrial acetyl-CoA cannot accumulate. These observations strongly suggest that acetyl-CoA used to synthesize ethyl acetate is derived from the mitochondria.
Atf1, Atf2, and their homologs appear to be cytosolic or located in the endoplasmic reticulum (23–25). A translocation step would therefore be required to transfer acetyl-CoA from the mitochondria to the cytosol. Some yeasts are able to translocate acetyl-CoA to the cytosol in the form of citrate. This shunt relies on the presence of ATP citrate lyase, which converts citrate to acetyl-CoA and oxaloacetate at the expense of one ATP (Fig. 1, reaction III). The reaction is typically present in oleaginous yeasts, such as Yarrowia lipolytica or Rhodosporidium toruloides (26, 27). It is not known if ATP citrate lyase is present in any of the yeasts that produce large amounts of ethyl acetate. Without this enzyme, transport of acetyl-CoA from the mitochondria to the cytosol is unlikely.
The hypothetical function of bulk ethyl acetate production is the release of excess mitochondrial acetyl-CoA. However, the previously assumed ethyl acetate-producing enzymes are located either in the cytosol or in the endoplasmic reticulum. These locations do not match with the mitochondrial function of ethyl acetate formation. Recently, a new family of AATs was discovered and designated Eat1. This family catalyzes ethyl acetate synthesis in S. cerevisiae, K. marxianus, W. anomalus, K. lactis, and other yeasts (28). It was shown that Eat1 is responsible for 80% and 50% of ethyl acetate production in K. lactis and S. cerevisiae, respectively. In this study, we show that the Eat1 of K. lactis is located in the mitochondria. In addition, we used in silico analyses and fermentations of bulk ethyl acetate-producing yeasts to support this view for the location of Eat1 in other yeasts as well.
RESULTS
Localization of Eat1 in yeast.
Huh et al. performed a global protein localization study in S. cerevisiae (29). This included the hypothetical protein YGR015C, which was later identified as the S. cerevisiae homolog of Eat1 (28). The S. cerevisiae Eat1 was tracked to the mitochondria (29), which suggests that Eat1 may be located in the mitochondria of bulk ethyl acetate-producing yeast as well. We initially tested the hypothesis by overexpressing the W. anomalus eat1 fused with mCherry at the C terminus from a multicopy plasmid in S. cerevisiae. The fusion protein was generally localized to the mitochondria of S. cerevisiae. However, we also observed a number of artifacts associated with heterologous overexpression of fluorescent protein fusions, such as variations of the fluorescence within the cell population or protein aggregation (unpublished result). To obtain more conclusive results, we fused Eat1 with yeast-enhanced green fluorescent protein (yEGFP) at the C terminus in K. lactis CBS 2359 Δku80. K. lactis CBS 2359 was chosen because it was previously demonstrated that Eat1 is the main enzyme responsible for bulk ethyl acetate production in this yeast (28).
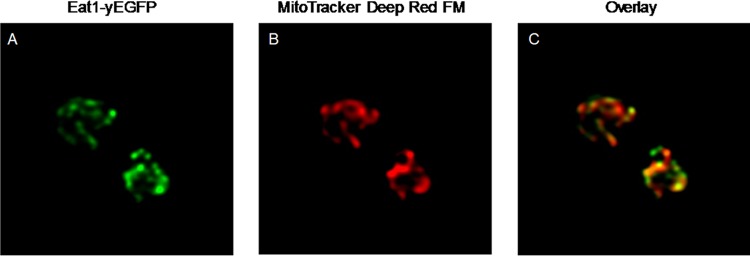
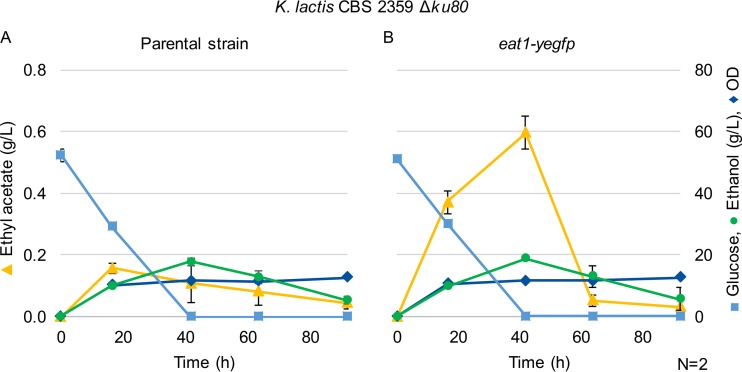
The location of Eat1-yEGFP in living cells of K. lactis CBS 2359 Δku80 eat1-yegfp was visualized using confocal microscopy. Eat1-yEGFP was clearly concentrated in structures within the cell (Fig. 2A). The mitochondria of these cells were stained with MitoTracker Deep Red FM (Fig. 2B). The overlay of the two images showed that the signals of yEGFP and the mitochondrial marker overlap almost completely (Fig. 2C). However, there are some areas where Eat1 fluorescence did not overlap the MitoTracker stain. This may be an artifact of the dye, but it could also indicate that Eat1 is located in multiple organelles. Nevertheless, the in vivo experiments showed that Eat1 is mostly located in the mitochondria of K. lactis CBS 2359 Δku80. To exclude the possibility that the Eat1-yEGFP fusion affected the function of Eat1, we compared the ethyl acetate production of K. lactis CBS 2359 Δku80 eat1-yegfp and its parental strain. The strains were cultivated in 50 ml of yeast minimal (YM) medium without iron supplementation. The strain producing the Eat1-yEGFP fusion was still able to synthesize ethyl acetate (Fig. 3). Surprisingly, the ethyl acetate titer achieved by K. lactis CBS 2359 Δku80 eat1-yegfp was 3.75-fold higher than that of K. lactis CBS 2359 Δku80. The reason for this increase is not clear, but it demonstrated that the Eat1-yEGFP fusion is still functional. These results show that ethyl acetate itself is primarily a mitochondrial product of K. lactis CBS 2359 Δku80.
FIG 2.
Visualization of Eat1 in the mitochondria of K. lactis CBS 2359 Δku80 eat1-yegfp. (A) Visualization of Eat1-yEGFP. (B) Mitochondria visualized by MitoTracker Deep Red FM. (C) Overlay of the two signals. The images shown are representative of the entire cell population.
FIG 3.
Fermentation profiles of (A) K. lactis CBS 2359 Δku80 and (B) K. lactis CBS 2359 Δku80 eat1-yegfp growing in shake flasks in 50 ml YM medium without iron supplementation. The numbers shown are the averages of biological duplicates. Error bars represent the standard deviation. Ethyl acetate evaporation was not measured.
Continuous fermentations indicate a common mechanism of ethyl acetate synthesis in yeast.
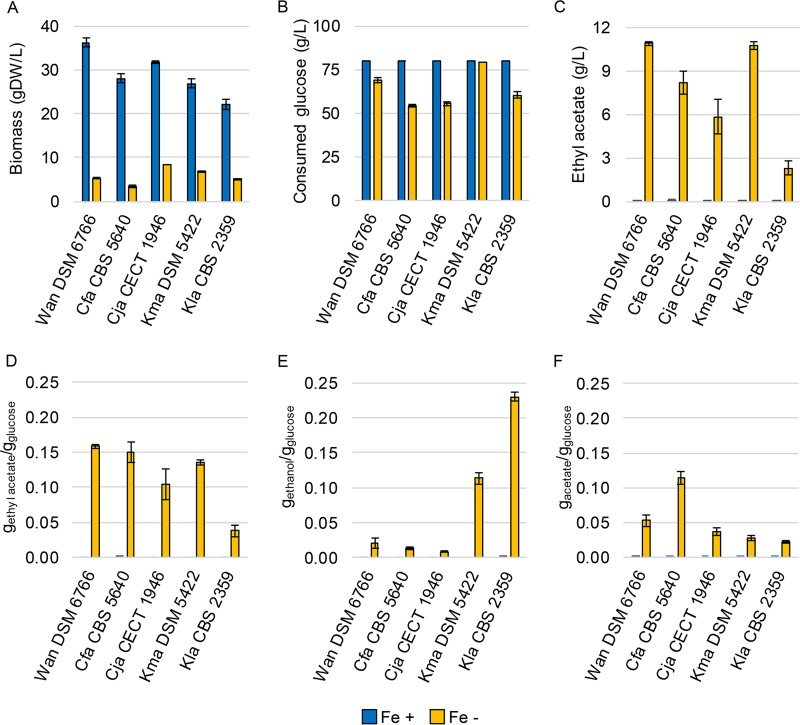
As Eat1 is located in the mitochondria of S. cerevisiae and K. lactis CBS 2359 Δku80, we were wondering whether it is located in the mitochondria of other bulk ethyl acetate-producing yeasts as well. The expression of green fluorescent protein (GFP)-fused proteins in these yeasts is cumbersome because of their poor genetic accessibility. We attempted to transform W. anomalus DSM 6766 and K. marxianus DSM 5422, but were not successful. To gain further insight on the location of ethyl acetate synthesis in other yeast strains, we compared the natural producers in vivo instead. We reasoned that if the conditions that trigger bulk ethyl acetate formation are similar, the underlying pathways are likely shared as well, including the cellular location of Eat1. However, there are no studies that accurately compare ethyl acetate production by multiple yeasts under the same conditions. Moreover, many studies on bulk ethyl acetate synthesis in yeast often did not control or measure parameters such as oxygen levels or ethyl acetate evaporation (3). This makes metabolic comparisons between different yeast species impossible. To resolve the issue, we examined bulk ethyl acetate production in five yeast species under the same controlled conditions. We used aerobic, iron-limited chemostats to induce ethyl acetate production in W. anomalus DSM 6766, C. fabianii CBS 5640, Cyberlindnera jadinii CECT 1946, K. marxianus DSM 5422, and K. lactis CBS 2359. When 1 mM FeSO4 was added to the medium, all five yeasts fully consumed the glucose, and virtually no ethyl acetate or other fermentation products were formed (Fig. 4). To induce ethyl acetate production, iron was omitted from the medium. Sufficient iron impurities were present to stably support between 3.4 ± 0.2 and 8.3 ± 0.0 g dry weight/liter biomass (Fig. 4A). Under iron-limited conditions, the yeast strains consumed between 54.5 ± 0.0 and 79.3 ± 0.0 g/liter glucose (Fig. 4B). Iron limitation induced ethyl acetate production in the five yeast species (Fig. 4C). The amount of ethyl acetate removed through gas stripping was added to the concentrations measured in the liquid. The headspace contained 25.9 ± 0.0% of the total ethyl acetate produced. The highest ethyl acetate titers were obtained with W. anomalus DSM 6766 and K. marxianus DSM 5422 (11.6 ± 0.2 and 10.7 ± 0.3 g/liter, respectively). However, K. marxianus DSM 5422 consumed more sugar, resulting in a lower ethyl acetate yield (Fig. 4D). W. anomalus DSM 6766 and C. fabianii CBS 5640 were the best ethyl acetate producers in terms of yield. They produced 0.17 ± 0.00 g ethyl acetate/g glucose and 0.16 ± 0.01 g ethyl acetate/g glucose, respectively. K. lactis CBS 2359 produced the least ethyl acetate per glucose (0.04 ± 0.01 g ethyl acetate/g glucose). The maximum theoretical pathway yield of ethyl acetate on glucose is 0.49 g ethyl acetate/g glucose.
FIG 4.
Cultivation parameters of five different yeast species that were grown in aerobic, pH-controlled chemostats in the presence or absence of 1 mM FeSO4. The numbers shown are averages of three steady states. Error bars indicate standard deviation. The amount of ethyl acetate that was removed by gas stripping was determined by headspace measurements and was added to the amount that was measured in the liquid phase. (A) Biomass concentration. (B) Glucose consumption. (C) Ethyl acetate titer. (D, E, and F) The yields of ethyl acetate, ethanol, and acetate, respectively, on glucose consumed. (See the Fig. 5 legend for a list of the species name abbreviations used in this figure.)
Besides ethyl acetate, the yeasts also formed significant amounts of ethanol or acetate as by-products (Fig. 4E and F, respectively). Crabtree-negative yeasts generally do not produce ethanol under aerobic conditions like the ones used in this study. However, iron limitation results in a metabolic deregulation, which leads to ethanol production (17, 20). W. anomalus DSM 6766, C. fabianii CBS 5640, and C. jadinii CECT 1946 produced between 0.02 ± 0.01 and 0.01 ± 0.00 g ethanol/g glucose. This was significantly lower compared to K. marxianus DSM 5422 and K. lactis CBS 2359, which produced 0.11 ± 0.01 and 0.23 ± 0.01 g ethanol/g glucose (Fig. 4E). The yeasts also produced between 0.02 ± 0.00 and 0.12 ± 0.01 g acetate/g glucose (Fig. 4F).
The five tested yeasts produced ethyl acetate under aerobic and iron-limited conditions. However, there were significant levels of ethanol and acetate produced. These products indicate that glucose is catabolized in the cytosol as well, despite the aerobic conditions. The effect was most pronounced in K. lactis CBS 2359, which produced 6.1-fold more ethanol than ethyl acetate. Most of this ethyl acetate was produced by the mitochondrial Eat1 (Fig. 2), despite the high carbon flux in the cytosol (Fig. 4E and F). The remaining four yeasts produced ethyl acetate under the same iron-limited conditions as K. lactis CBS 2359, which suggests that the ethyl acetate produced by these yeasts is of mitochondrial origin as well.
In silico indications for Eat1 localization in yeast.
Acetyl-CoA used for bulk ethyl acetate synthesis is produced in the mitochondria. We investigated whether the five yeast strains are able to transport acetyl-CoA from the mitochondria to the cytosol via citrate. The enzyme needed for the realization of this pathway is the cytosolic ATP citrate lyase (Fig. 1). We used BLASTP to search for homologs of three fungal ATP citrate lyase proteins in W. anomalus, C. jadinii, C. fabianii, K. marxianus, and K. lactis. The ATP citrate lyase homologs originated from Yarrowia lipolytica CLIB 122, Aspergillus nidulans FGSC A4, and Rhodosporidium toruloides IFO 0880. None of the bulk ethyl acetate-producing yeasts contained apparent ATP citrate lyase homologs. The absence of the ATP citrate lyase suggests that ethyl acetate-producing yeasts cannot transport acetyl-CoA to the cytosol (22, 26). Bulk ethyl acetate synthesis by Eat1 is therefore more likely to be located in the mitochondria.
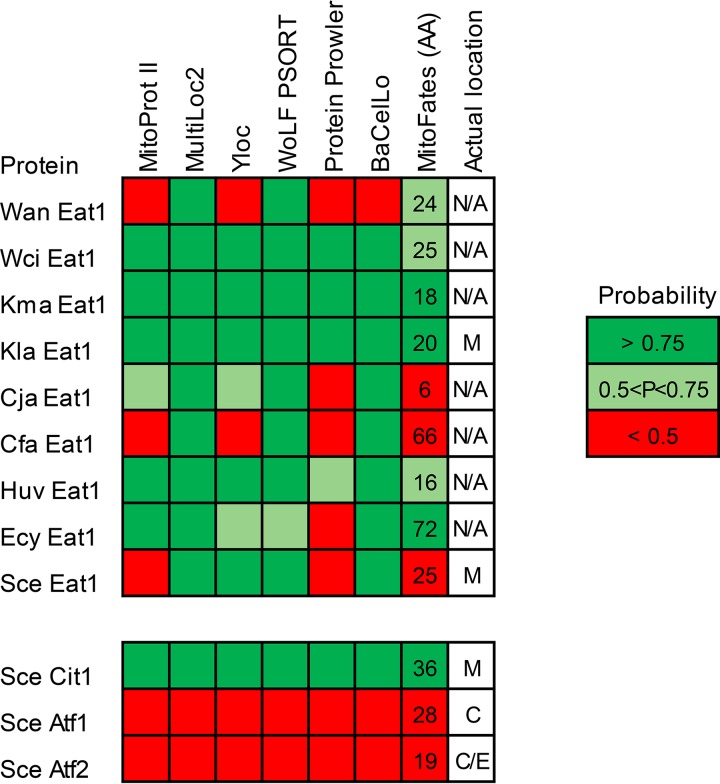
The subcellular location of proteins can also be predicted in silico based on their primary sequence. We predicted the location of nine Eat1 homologs originating from nine bulk ethyl acetate-producing yeast species (28). The mitochondrial citrate synthase (Cit1) and the cytosolic Atf1 and Atf2 from S. cerevisiae were included in the analysis as controls. Seven tools were used in the analysis, namely, MitoProt II (30), MultiLoc 2 (31), Yloc (32), WoLF PSORT (33), Protein Prowler (34), BacCelLo (35), and MitoFates (36). The Eat1 homologs were generally predicted as mitochondrial (Fig. 5). The Eat1 homologs of K. lactis, K. marxianus and Wickerhamomyces ciferrii were predicted as mitochondrial by all seven tools. The least-mitochondrial localization predictions were given to the W. anomalus and C. fabianii Eat1 homologs. Most of the nonmitochondrial predictions were assigned by Protein Prowler, Mito Prot II, and MitoFates. These tools also did not identify the S. cerevisiae Eat1 protein as mitochondrial. This indicates that the predictions made by these tools may not be reliable for the Eat1 homologs. On the other hand, MultiLoc2 and WoLF PSORT predicted all the proteins as mitochondrial, including the S. cerevisiae and K. lactis Eat1 homologs. These two tools also performed better than Protein Prowler (31, 37).
FIG 5.
Predicted probabilities of mitochondrial localization of the Eat1 proteins. Seven tools were used to predict the mitochondrial localization of nine Eat1 homologs from 9 yeast species. The numbers in the column represent the length of the presequences predicted by MitoFates. Wan, Wickerhamomyces anomalus; Wci, Wickerhamomyces ciferrii; Kma, Kluyveromyces marxianus; Kla, Kluyveromyces lactis; Cja, Cyberlindnera jadinii; Cfa, Cyberlindnera fabianii; Huv, Hanseniaspora uvarum; Ecy, Eremothecium cymbalarie; Sce, Saccharomyces cerevisiae; M, mitochondria; C, cytosol; E, endoplasmic reticulum; N/A, not available.
DISCUSSION
In this study, we included the cellular location of Eat1 in the hypothetical model of bulk ethyl acetate production in yeast. This AAT was previously linked to 80% and 50% of ethyl acetate synthesis in K. lactis and S. cerevisiae, respectively (28). Results presented here showed that Eat1 is a mitochondrial protein in both yeasts. However, the current model of bulk ethyl acetate production assumes that cytosolic AATs are responsible for ethyl acetate formation (19). The mitochondrial location of Eat1 in K. lactis CBS 2359 described in this study disagrees with this assumption. It is also likely that bulk ethyl acetate by other yeasts is of mitochondrial origin as well. Ideally, confocal microscopy could be used to confirm this. However, this is hampered by the lack of genetic tools needed to perform gene fusions in other bulk ethyl acetate-producing yeast. It is also not possible to discriminate between the mitochondrial and cytosolic acetyl-CoA flux using 13C tracking experiments, as the same carbon atoms are removed during the cleavage of pyruvate in both compartments. The acetyl-CoA produced in the mitochondria is therefore identical to the one produced in the cytosol. The in silico tools used in this study generally predicted that all known Eat1 homologs are mitochondrial. The apparent lack of ATP citrate lyase homologs also seems to indicate that acetyl-CoA cannot be transported to the cytosol. Mitochondrial acetyl-CoA may play a role in ethyl acetate synthesis in S. cerevisiae as well. This yeast contains a functional Eat1 homolog but produces only traces of ethyl acetate (2). This is likely caused by the Crabtree-positive nature of S. cerevisiae. When glucose is present in excess, the main carbon flux in S. cerevisiae bypasses the mitochondria in favor of cytosolic ethanol formation (15, 38). It is possible that the low ethyl acetate production in S. cerevisiae is caused by the low mitochondrial acetyl-CoA flux.
W. anomalus DSM 6766, C. fabianii CBS 5640, C. jadinii CECT 1946, K. marxianus DSM 5422, and K. lactis CBS 2359 are all Crabtree-negative yeasts. Under aerobic conditions, such yeasts convert glucose to cytosolic pyruvate and further to mitochondrial acetyl-CoA. Conditions such as iron limitation repress the synthesis of enzymes in the TCA cycle and respiratory chain (39, 40). As a consequence, ethanol is produced in the cytosol even under aerobic conditions (20, 41). In the mitochondria, iron limitation leads to the accumulation of acetyl-CoA. The accumulation may be resolved by Eat1 in the mitochondria by formation of ethyl acetate (14, 18, 41). The mitochondrial localization of Eat1 thus agrees with its proposed physiological function of preventing the accumulation of acetyl-CoA in yeast. However, more research is needed to confirm this hypothesis. It has been shown that Eat1 can function as a thioesterase in vitro (28). This activity could relieve acetyl-CoA accumulation by releasing acetate instead of ethyl acetate. However, it is possible that ethyl acetate formation provides ancillary benefits to the yeasts. It has been shown that ethyl acetate inhibits the growth of competitive microorganisms (42) and helps yeast disperse in the environment by attracting fruit flies (43).
The demonstration that Eat1 is a mitochondrial enzyme is critical for improving ethyl acetate production by microorganisms. Yeasts like K. marxianus and W. anomalus are naturally able to produce ethyl acetate at high yields. However, they also produce considerable amounts of ethanol and acetate as by-products. The acetyl-CoA and ethanol used for ethyl acetate synthesis are produced in the mitochondria and cytosol, respectively. Cytosolic pyruvate is the precursor of both substrates. Efficient ethyl acetate production would therefore require precise control over the pyruvate flux so that acetyl-CoA and ethanol production are stoichiometrically balanced. Alternatively, Eat1 can be used to produce ethyl acetate in heterologous hosts. In such cases, consideration should be given to identifying the localization presequence. In eukaryotes, N-terminal presequences are cleaved from the nascent protein during translocation to the mitochondria, giving rise to the mature protein (44). Unrelated hosts may not be able to perform this cleavage. The presence of the N-terminal localization sequence has resulted in lower protein activity and stability in some cases (45, 46). Proper N-terminal processing of the Eat1 proteins may therefore improve the activity of the protein in heterologous hosts.
Until now, it was assumed that yeasts such as K. lactis, K. marxianus, and W. anomalus produce ethyl acetate in the cytosol. The present study has established that the synthesis occurs in mitochondria instead. This finding agrees with the proposed biological function of bulk ethyl acetate synthesis. Our understanding of ester synthesis in yeast is hereby expanded, which will enable the design of more efficient processes for the production of bio-based ethyl acetate.
MATERIALS AND METHODS
Strain and plasmid construction.
Strains and plasmids that were used in this study are listed in Table 1. Wild-type yeast strains were obtained from culture collections. Kluyveromyces lactis CBS 2359 Δku80 (47) was a gift from Paul Hooykaas (Leiden University). Plasmid pCY 3040-01 (Addgene plasmid 36217) was a gift from Anne Robinson. Plasmid pUG75 (48) was obtained from Euroscarf (plasmid P30671). K. lactis CBS 2359 Δku80 eat1-yegfp was constructed by integrating the yegfp gene (49) in frame at the 3′ end of eat1. A (GGTGGTAGTGGT)2 linker was inserted between eat1 and yegfp. The native eat1 stop codon was removed. The linear integration cassette contained the linker, yegfp, and pAgTEF1-hphMX-tAgTEF1, flanked by 1,000-bp sequences upstream and downstream of the integration site. The flanking regions, yegfp, and pAgTEF1-hphMX-tAgTEF1 were amplified from the K. lactis CBS 2359 Δku80 genome, pCY-3040-01, and pUG75, respectively. The parts were assembled to yield the plasmid pYES2-KlaEat1-yEGFP-hphMX-1000 with the HiFi assembly kit (NEB), according to the manufacturer's instructions. pYES2 (Invitrogen) was used as the backbone. The linear integration cassette was PCR amplified from pYES2-KlaEat1-yEGFP-hphMX-1000. An aliquot of 1 μg of the cassette was transformed into K. lactis CBS 2359 Δku80 with the Li-acetate method (50). Transformants were selected on plates containing 100 μg/ml hygromycin B. Correct clones were confirmed by PCR and sequencing.
TABLE 1.
Strains and plasmids used and created during this study
| Strain or plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Cyberlindnera fabianii CBS 5640 | Wild-type strain | CBS |
| Cyberlindnera jadinii CECT 1946 | Wild-type strain | CECTa |
| Kluyveromyces marxianus DSM 5422 | Wild-type strain | DSMZ |
| Wickerhamomyces anomalus DSM 6766 | Wild-type strain | DSMZ |
| Kluyveromyces lactis CBS 2359 | Wild-type strain | CBS |
| Kluyveromyces lactis CBS 2359 Δku80 | Disruption of KU80 | 47 |
| Kluyveromyces lactis CBS 2359 Δku80 eat1-yegfp | Disruption of KU80, Eat1 labeled with a C-terminal yEGFP | This study |
| E. coli NEB5α | Cloning strain | New England Biolabs |
| Plasmids | ||
| pUG75 | HygR marker template | 48 |
| pCY-3040-01 | yEGFP template | 49 |
| pYES2 | Invitrogen | |
| pYES2-KlaEat1-yEGFP-hphMX-1000 | Plasmid carrying the yegfp integration cassette targeting the 3′ end of the K. lactis eat1 locus with 1,000-bp homologous regions | This study |
CECT, Colección Española de Cultivos Tipo (Spanish Type Culture Collection).
Cultivation conditions.
Yeast and Escherichia coli cultures were routinely cultivated in yeast extract-peptone-dextrose (YPD) (20 g/liter glucose, 10 g/liter yeast extract, and 20 g/liter peptone) and LB (10 g/liter tryptone, 5 g/liter yeast extract, and 10 g/liter NaCl) media, respectively. Bacteriological agar (15 g/liter) was added to make plates. Ampicillin (50 μg/ml) and hygromycin B (100 μg/ml) were added to the media when appropriate. Yeast and E. coli cultures were grown at 30°C and 37°C, respectively, unless stated otherwise. All strains were stored as glycerol stocks at −80°C. Yeast and E. coli strains were revived by streaking frozen cultures on agar plates and cultivating until colonies appeared. Single colonies were used to inoculate liquid precultures used in further experiments.
The ethyl acetate production of K. lactis CBS 2359 Δku80 and K. lactis CBS 2359 Δku80 eat1-yegfp was assessed as reported previously (28). The cells were grown in 250-ml Erlenmeyer flasks containing 50 ml yeast minimal (YM) medium adapted from Thomas and Dawson (18). YM medium contained glucose (50 g/liter), (NH4)2SO4 (2.5 g/liter), KH2PO4 (2.5 g/liter), 3-(N-morpholino) propanesulfonic acid (MOPS, 23.1 g/liter), MgSO4 (60 mg/liter), ZnSO4 · 7H2O (25.0 mg/liter), MnCl2 · 4H2O (4.0 mg/liter), CuSO4 · 5 H2O (2.5 mg/liter), CaCl2 · 2H2O (1.5 mg/liter), H3BO3 (1.5 mg/liter) Na2MoO4 · 2H2O (0.4 mg/liter), CoCl2 (0.2 mg/liter), and KI (0.3 mg/liter). The pH of the medium was set to 6.0 with 3 M NaOH. Iron was omitted from the medium. The medium was supplemented with 1 ml of 1,000× vitamin mix, according to Verduyn et al. (51). The vitamin mix contained biotin (0.05 mg/liter), Ca-pantothenate (1 mg/liter), nicotinic acid (1 mg/liter), inositol (25 mg/liter), thiamine-HCl (1 mg/liter), pyridoxine-HCl (1 mg/liter), and 4-amino benzoic acid (0.2 mg/liter). Erlenmeyer flasks (neck width, 34 mm) were inoculated with 0.5 ml preculture grown overnight in liquid YPD medium to an initial optical density at 600 nm (OD600) of 0.03. The flasks were closed with aluminum foil and shaken at 250 rpm. Experiments were performed as biological duplicates. Ethyl acetate evaporation was not measured in shake flasks.
Continuous fermentations.
The ethyl acetate production of W. anomalus DSM 6766, C. fabianii CBS 5640, C. jadinii CECT 1946, K. marxianus DSM 5422, and K. lactis CBS 2359 was studied in aerobic continuous fermentations. The culturing was performed in 1-L DasGip bioreactors (Eppendorf). The working volume was 0.5 liter. The pH was controlled at 5.0 (± 0.05) by automatic addition of 3 M KOH. The temperature was controlled at 30°C. Cultures were kept aerobic by controlling the dissolved oxygen (DO) at 40%. Sufficient oxygen transfer was achieved by stirring the fermenter at 1,200 rpm and automatically varying the oxygen fraction from 21% to 100%. Sparging was kept constant at 6 liters/h. The defined feed medium was designed to emulate the mineral composition of concentrated whey permeate augmented with ammonium sulfate (52) as closely as possible. Distilled water (dH2O) was used to prepare the medium. The medium contained (NH4)2SO4 (13.16 g/liter), Na2HPO4 · 2H2O (2.08 g/liter), NaCl (1.39 g/liter), KCl (1.89 g/liter), MgSO4 · 7H2O (0.81 g/liter), CaCl2 · H2O (0.139 g/liter), ZnSO4 · 7H2O (50 mg/liter), CuCl2 · 2H2O (6.6 mg/liter), Na2MoO4 · 2H2O (1 mg/liter), H3BO3 (2 mg/liter), MnSO4 · 1H2O (1.51 mg/liter), CoSO4 · 7H2O (2 mg/liter), NiSO4 · 6H2O (1 mg/liter), and 4 ml/liter of a 1,000× vitamin mix (51). Glucose (80 g/liter) was used as the carbon source. Twenty ml of 37% HCl was added to the 20-liter medium vessel to lower the pH of the medium. The dilution rate was 0.1 h−1. Ethyl acetate production was controlled by iron limitation. The producing (iron-limited) condition was achieved by omitting all sources of iron from the medium. The nonproducing (iron-rich/glucose-limited) condition was achieved by adding 1 mM FeSO4 to the medium vessel. Steady state was achieved after five culture volumes were exchanged, during which exchanges the physiological parameters remained stable. The reported numbers are an average of three sequential steady states achieved in one bioreactor. The amount of ethyl acetate in the headspace was quantified by extracting 250 μl of the bioreactor headspace through a septum with a syringe and immediately analyzing the ethyl acetate content by gas chromatography (GC). The concentration of ethyl acetate in the headspace was used to calculate the mass flow of ethyl acetate continuously discharged from the fermenters. The mass flow of ethyl acetate was related to the liquid flow of 0.05 liter/h and added to the ethyl acetate concentration in the liquid phase. Dry cell weight was determined in 50 ml of fermentation broth.
Confocal microscopy.
Confocal microscopy was carried out on a Leica TCS SP8 X system equipped with a 63×/1.20 numeric aperture water-immersion objective. Excitation of yeast-enhanced GFP (yEGFP) and MitoTracker Deep Red FM (Thermo Scientific) was performed using a white light laser, selecting the lasers lines at 488 nm and 633 nm, respectively. Confocal imaging was executed using internal filter-free spectral hybrid detectors. For yEGFP detection, a spectral window of 495 to 545 nm was selected, whereas MitoTracker Deep Red FM was detected using a window of 640 to 670 nm. Images with 1,024 × 1,024 pixels were acquired using the HyVolution software interface (Leica), operating in a sequential imaging configuration. The HyVolution software includes deconvolution of the confocal images by Huygens deconvolution software (53).
Bioinformatics.
The subcellular locations of proteins were predicted with six tools, as follows: MitoProt II (30), MultiLoc 2 (31), Yloc (32), WoLF PSORT (33), Protein Prowler (34), and BacCelLo (35). Where applicable, the prediction settings were set to fungal. MitoFates (36) was used to predict mitochondrial presequences under fungal prediction settings. BLASTP under default settings was used to look for homologs of ATP citrate lyase in C. fabianii CBS 5640, C. jadinii CECT 1946, K. marxianus DSM 5422, W. anomalus DSM 6766, and K. lactis CBS 2359. The ATP citrate lyase from Yarrowia lipolytica CLIB122 (NCBI reference sequence XP_503231), the ATP citrate lyase subunit 1 (NCBI reference sequence XP_660040) from Aspergillus nidulans FGSC A4, and the ATP citrate synthase from Rhodosporidium toruloides IFO 0880 (NCBI reference sequence PRQ71611) were used as query (54–56).
Analytical methods.
Glucose and organic acids were analyzed by high-performance liquid chromatography (HPLC) on a Thermo Scientific ICS5000 HPLC system equipped with a Dionex DP pump, a Dionex AS-AP autosampler, a Dionex variable wavelength detector (VWD) UV detector operated at 210 nm, and a Shodex refractive index (RI) detector operated at 35°C. An Aminex HPX-87H cation-exchange column (Bio-Rad) was used with a mobile phase of 0.008 M H2SO4 and was operated at 0.8 ml/min and 60°C. Dimethyl sulfoxide (10 mM) or propionic acid (125 mM) was used as the internal standard.
Volatile compounds were analyzed on two gas chromatography systems equipped with a flame ionization detector (GC-FID). In both cases, 0.5 μl of liquid or 250 μl of headspace sample was analyzed. For liquid samples, 2 mM 1-butanol was used as internal standard. The first system used a Shimadzu 2010 gas chromatograph equipped with a 20i-s autosampler. Samples were analyzed on a Stabilwax column (30 m × 0.53 mm, 0.5 μm coating; Restek). The column temperature was held at 60°C for 1 min and increased to 120°C at a rate of 20°C/minute. The split ratio was 20. The second system used an Agilent 7890B gas chromatograph equipped with an Agilent 7693 autosampler. The compounds were separated on a Nukol column (30 m × 0.53 mm, 1.0 μm coating; Supelco). The column temperature was maintained at 50°C for 2 min and increased to 200°C at a rate of 50°C/minute. The split ratio was 10.
Dry cell weight in continuous fermentations was measured by centrifuging 50 ml culture for 5 min at 4,000 × g. The pellet was then washed with 50 ml ultrapure MilliQ water (MQ), resuspended in 3 ml MQ, and dried overnight at 120°C in a preweighted aluminum tray before weighing.
ACKNOWLEDGMENTS
We acknowledge the BE-Basic Foundation and AkzoNobel Specialty Chemicals for funding the research.
REFERENCES
- 1.Park YC, Shaffer CEH, Bennett GN. 2009. Microbial formation of esters. Appl Microbiol Biotechnol 85:13–25. doi: 10.1007/s00253-009-2170-x. [DOI] [PubMed] [Google Scholar]
- 2.Saerens SMG, Delvaux FR, Verstrepen KJ, Thevelein JM. 2010. Production and biological function of volatile esters in Saccharomyces cerevisiae. Microb Biotechnol 3:165–177. doi: 10.1111/j.1751-7915.2009.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loser C, Urit T, Bley T. 2014. Perspectives for the biotechnological production of ethyl acetate by yeasts. Appl Microbiol Biotechnol 98:5397–5415. doi: 10.1007/s00253-014-5765-9. [DOI] [PubMed] [Google Scholar]
- 4.Löser C, Urit T, Stukert A, Bley T. 2013. Formation of ethyl acetate from whey by Kluyveromyces marxianus on a pilot scale. J Biotechnol 163:17–23. doi: 10.1016/j.jbiotec.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Passoth V, Olstorpe M, Schnurer J. 2011. Past, present and future research directions with Pichia anomala. Antonie Van Leeuwenhoek 99:121–125. doi: 10.1007/s10482-010-9508-3. [DOI] [PubMed] [Google Scholar]
- 6.Fredlund E, Beerlage C, Melin P, Schnürer J, Passoth V. 2006. Oxygen and carbon source-regulated expression of PDC and ADH genes in the respiratory yeast Pichia anomala. Yeast 23:1137–1149. doi: 10.1002/yea.1428. [DOI] [PubMed] [Google Scholar]
- 7.Meersman E, Steensels J, Struyf N, Paulus T, Saels V, Mathawan M, Allegaert L, Vrancken G, Verstrepen KJ. 2016. Tuning chocolate flavor through development of thermotolerant Saccharomyces cerevisiae starter cultures with increased acetate ester production. Appl Environ Microbiol 82:732–746. doi: 10.1128/AEM.02556-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Löser C, Urit T, Nehl F, Bley T. 2011. Screening of Kluyveromyces strains for the production of ethyl acetate: design and evaluation of a cultivation system. Eng Life Sci 11:369–381. doi: 10.1002/elsc.201000178. [DOI] [Google Scholar]
- 9.Minetoki T, Bogaki T, Iwamatsu A, Fujii T, Hamachi M. 1993. The purification, properties and internal peptide sequences of alcohol acetyltransferase isolated from Saccharomyces cerevisiae Kyokai no. 7. Biosci Biotechnol Biochem 57:2094–2098. doi: 10.1271/bbb.57.2094. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimoto H, Fujiwara D, Momma T, Tanaka K, Sone H, Nagasawa N, Tamai Y. 1999. Isolation and characterization of the ATF2 gene encoding alcohol acetyltransferase II in the bottom fermenting yeast Saccharomyces pastorianus. Yeast 15:409–417. doi:. [DOI] [PubMed] [Google Scholar]
- 11.Verstrepen KJ, Van Laere SDM, Vanderhaegen BMP, Derdelinckx G, Dufour J-PP, Pretorius IS, Winderickx J, Thevelein JM, Delvaux FR. 2003. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl Environ Microbiol 69:5228–5237. doi: 10.1128/AEM.69.9.5228-5237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider J, Rupp O, Trost E, Jaenicke S, Passoth V, Goesmann A, Tauch A, Brinkrolf K. 2012. Genome sequence of Wickerhamomyces anomalus DSM 6766 reveals genetic basis of biotechnologically important antimicrobial activities. FEMS Yeast Res 12:382–386. doi: 10.1111/j.1567-1364.2012.00791.x. [DOI] [PubMed] [Google Scholar]
- 13.Van Laere SDM, Saerens SMG, Verstrepen KJ, Van Dijck P, Thevelein JM, Delvaux FR. 2008. Flavour formation in fungi: characterisation of KlAtf, the Kluyveromyces lactis orthologue of the Saccharomyces cerevisiae alcohol acetyltransferases Atf1 and Atf2. Appl Microbiol Biotechnol 78:783–792. doi: 10.1007/s00253-008-1366-9. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong DW, Yamazaki H. 1984. Effect of iron and EDTA on ethyl acetate accumulation in Candida utilis. Biotechnol Lett 6:819–824. doi: 10.1007/BF00134726. [DOI] [Google Scholar]
- 15.Pfeiffer T, Morley A. 2014. An evolutionary perspective on the Crabtree effect. Front Mol Biosci 1:17. doi: 10.3389/fmolb.2014.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fredlund E, Blank LM, Schnürer J, Sauer U, Passoth V. 2004. Oxygen- and glucose-dependent regulation of central carbon metabolism in Pichia anomala. Appl Environ Microbiol 70:5905–5911. doi: 10.1128/AEM.70.10.5905-5911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urit T, Stukert A, Bley T, Löser C. 2012. Formation of ethyl acetate by Kluyveromyces marxianus on whey during aerobic batch cultivation at specific trace element limitation. Appl Microbiol Biotechnol 96:1313–1323. doi: 10.1007/s00253-012-4107-z. [DOI] [PubMed] [Google Scholar]
- 18.Thomas KC, Dawson PSS. 1978. Relationship between iron-limited growth and energy limitation during phased cultivation of Candida utilis. Can J Microbiol 24:440–447. doi: 10.1139/m78-073. [DOI] [PubMed] [Google Scholar]
- 19.Löser C, Urit T, Keil P, Bley T. 2015. Studies on the mechanism of synthesis of ethyl acetate in Kluyveromyces marxianus DSM. 5422. Appl Microbiol Biotechnol 99:1131–1144. doi: 10.1007/s00253-014-6098-4. [DOI] [PubMed] [Google Scholar]
- 20.van Dijken JP, Weusthuis RA, Pronk JT. 1993. Kinetics of growth and sugar consumption in yeasts. Antonie Van Leeuwenhoek 63:343–352. doi: 10.1007/BF00871229. [DOI] [PubMed] [Google Scholar]
- 21.Van den Berg MA, Steensma HY. 1995. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur J Biochem 231:704–713. [DOI] [PubMed] [Google Scholar]
- 22.van Rossum HM, Kozak BU, Pronk JT, van Maris AJA. 2016. Engineering cytosolic acetyl-coenzyme A supply in Saccharomyces cerevisiae: Pathway stoichiometry, free-energy conservation and redox-cofactor balancing. Metab Eng 36:99–115. doi: 10.1016/j.ymben.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Verstrepen KJ, Van Laere SDM, Vercammen J, Derdelinckx G, Dufour JP, Pretorius IS, Winderickx J, Thevelein JM, Delvaux FR. 2004. The Saccharomyces cerevisiae alcohol acetyl transferase Atf1p is localized in lipid particles. Yeast 21:367–376. doi: 10.1002/yea.1100. [DOI] [PubMed] [Google Scholar]
- 24.Lin JL, Wheeldon I. 2014. Dual N- and C-terminal helices are required for endoplasmic reticulum and lipid droplet association of alcohol acetyltransferases in Saccharomyces cerevisiae. PLoS One 9:e104141. doi: 10.1371/journal.pone.0104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J, Lin J-L, Palomec L, Wheeldon I. 2015. Microbial host selection affects intracellular localization and activity of alcohol-O-acetyltransferase. Microb Cell Fact 14:35. doi: 10.1186/s12934-015-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulton CA, Ratledge C. 1981. Correlation of lipid accumulation in yeasts with possession of ATP:citrate lyase. J Gen Microbiol 127:169–176. [Google Scholar]
- 27.Shashi K, Bachhawat AK, Joseph R. 1990. ATP:citrate lyase of Rhodotorula gracilis: purification and properties. Biochim Biophys Acta 1033:23–30. doi: 10.1016/0304-4165(90)90189-4. [DOI] [PubMed] [Google Scholar]
- 28.Kruis AJ, Levisson M, Mars AE, van der Ploeg M, Garcés Daza F, Ellena V, Kengen SWM, van der Oost J, Weusthuis RA. 2017. Ethyl acetate production by the elusive alcohol acetyltransferase from yeast. Metab Eng 41:92–101. doi: 10.1016/j.ymben.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 30.Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 31.Blum T, Briesemeister S, Kohlbacher O. 2009. MultiLoc2: integrating phylogeny and gene ontology terms improves subcellular protein localization prediction. BMC Bioinformatics 10:274. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Briesemeister S, Rahnenführer J, Kohlbacher O. 2010. YLoc-an interpretable web server for predicting subcellular localization. Nucleic Acids Res 38:W497–W502. doi: 10.1093/nar/gkq477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boden M, Hawkins J. 2005. Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics 21:2279–2286. doi: 10.1093/bioinformatics/bti372. [DOI] [PubMed] [Google Scholar]
- 35.Pierleoni A, Martelli PL, Fariselli P, Casadio R. 2006. BaCelLo: a balanced subcellular localization predictor. Bioinformatics 22:e408–e416. doi: 10.1093/bioinformatics/btl222. [DOI] [PubMed] [Google Scholar]
- 36.Fukasawa Y, Tsuji J, Fu S-C, Tomii K, Horton P, Imai K. 2015. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics 14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casadio R, Martelli PL, Pierleoni A. 2008. The prediction of protein subcellular localization from sequence: a shortcut to functional genome annotation. Brief Funct Genomic Proteomic 7:63–73. doi: 10.1093/bfgp/eln003. [DOI] [PubMed] [Google Scholar]
- 38.De Deken RH. 1966. The Crabtree effect: a regulatory system in yeast. J Gen Microbiol 44:149–156. doi: 10.1099/00221287-44-2-149. [DOI] [PubMed] [Google Scholar]
- 39.Puig S, Askeland E, Thiele DJ. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 40.Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, Rolfes R, Brown PO, Botstein D, Philpott CC. 2004. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol Biol Cell 15:1233–1243. doi: 10.1091/mbc.e03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Löser C, Urit T, Forster S, Stukert A, Bley T. 2012. Formation of ethyl acetate by Kluyveromyces marxianus on whey during aerobic batch and chemostat cultivation at iron limitation. Appl Microbiol Biotechnol 96:685–696. doi: 10.1007/s00253-012-4205-y. [DOI] [PubMed] [Google Scholar]
- 42.Fredlund E, Druvefors UA, Olstorpe MN, Passoth V, Schnurer J. 2004. Influence of ethyl acetate production and ploidy on the anti-mould activity of Pichia anomala. FEMS Microbiol Lett 238:133–137. [DOI] [PubMed] [Google Scholar]
- 43.Christiaens JFF, Franco LMM, Cools TLL, De Meester L, Michiels J, Wenseleers T, Hassan BAA, Yaksi E, Verstrepen KJJ, De Meester L, Michiels J, Wenseleers T, Hassan BAA, Yaksi E, Verstrepen KJJ. 2014. The fungal aroma gene ATF1 promotes dispersal of yeast cells through insect vectors. Cell Rep 9:425–432. doi: 10.1016/j.celrep.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Wiedemann N, Pfanner N, Ryan MT. 2001. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J 20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veling MT, Reidenbach AG, Freiberger EC, Kwiecien NW, Hutchins PD, Drahnak MJ, Jochem A, Ulbrich A, Rush MJP, Russell JD, Coon JJ, Pagliarini DJ. 2017. Multi-omic mitoprotease profiling defines a role for Oct1p in coenzyme Q production. Mol Cell 68:970–977. doi: 10.1016/j.molcel.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vögtle F-N, Prinz C, Kellermann J, Lottspeich F, Pfanner N, Meisinger C. 2011. Mitochondrial protein turnover: role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol Biol Cell 22:2135–2143. doi: 10.1091/mbc.e11-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kooistra R, Hooykaas PJJ, Steensma HY. 2004. Efficient gene targeting in Kluyveromyces lactis. Yeast 21:781–792. doi: 10.1002/yea.1131. [DOI] [PubMed] [Google Scholar]
- 48.Hegemann JH, Heick SB. 2011. Delete and repeat: a comprehensive toolkit for sequential gene knockout in the budding yeast Saccharomyces cerevisiae. Methods Mol Biol 765:189–206. doi: 10.1007/978-1-61779-197-0_12. [DOI] [PubMed] [Google Scholar]
- 49.Young CL, Raden DL, Caplan JL, Czymmek KJ, Robinson AS. 2012. Cassette series designed for live-cell imaging of proteins and high-resolution techniques in yeast. Yeast 29:119–136. doi: 10.1002/yea.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gietz DR, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 51.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 52.Urit T, Löser C, Wunderlich M, Bley T. 2011. Formation of ethyl acetate by Kluyveromyces marxianus on whey: studies of the ester stripping. Bioprocess Biosyst Eng 34:547–559. doi: 10.1007/s00449-010-0504-9. [DOI] [PubMed] [Google Scholar]
- 53.Borlinghaus RT, Kappel C. 2016. HyVolution—the smart path to confocal super-resolution. Nat Methods 13:i–iii. [Google Scholar]
- 54.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MÁ Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 55.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, de Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich J-M, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, de Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud J-M, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard G-F, Straub M-L, Suleau A, Swennen D, Tekaia F, Wésolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet J-L. 2004. Genome evolution in yeasts. Nature 430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 56.Coradetti ST, Pinel D, Geiselman G, Ito M, Mondo S, Reilly MC, Cheng Y-F, Bauer S, Grigoriev I, Gladden JM, Simmons BA, Brem R, Arkin AP, Skerker JM. 2018. Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides. Elife 7:e32110. doi: 10.7554/eLife.32110. [DOI] [PMC free article] [PubMed] [Google Scholar]