In spite of the limited effectiveness of the considered inoculated starter strains, this study provides new information on the microbial development of box and heap cocoa fermentations, under inoculated and noninoculated conditions, as we coupled yeast/bacterial amplicon-based sequencing data with microbial metabolite detection. The information so far available suggests that microbial communities have played an important role in the evolution of aroma compounds. Understanding the pathways that microorganisms follow during the formation of aromas could be used to improve the fermentation processes and to enhance chocolate quality.
KEYWORDS: cocoa beans, HTS, bacteria, fermentation, nonvolatile organic compounds, volatile organic compounds, yeast
ABSTRACT
Forastero hybrid cocoa bean fermentations have been carried out in a box (B) and in a heap (H), with or without the inoculation of Saccharomyces cerevisiae and Torulaspora delbrueckii as starter cultures. The bacteria, yeasts, and microbial metabolites (volatile and nonvolatile organic compounds) were monitored during fermentation to assess the connection between microbiota and the release of metabolites during this process. The presence of starter cultures was detected, by means of culture-dependent analysis, during the first 2 days of both fermentations. However, no statistical difference was observed in any of the physicochemical or microbiological analyses. Plate counts revealed the dominance of yeasts at the beginning of both fermentations, and these were followed by acetic acid bacteria (AAB) and lactic acid bacteria (LAB). Hanseniaspora opuntiae, S. cerevisiae, Pichia pijperi, Acetobacter pasteurianus, and Lactobacillus fermentum were the most abundant operational taxonomic units (OTUs) during both fermentation processes (B and H), although different relative abundances were observed. Only the diversity of the fungal species indicated a higher level of complexity in the B fermentations than in the H fermentations (P < 0.05), as well as a statistically significant difference between the initially inoculated starter cultures (P < 0.01). However, the microbial metabolite analysis indicated different distributions of the volatile and nonvolatile compounds between the two procedures, that is, B and H (P < 0.05), rather than between the inoculated and noninoculated fermentations. The box fermentations showed faster carbohydrate metabolism and greater production of organic acid compounds, which boosted the formation of alcohols and esters, than did the heap fermentations. Overall, the microbial dynamics and associations between the bacteria, yeasts, and metabolites were found to depend on the type of fermentation.
IMPORTANCE In spite of the limited effectiveness of the considered inoculated starter strains, this study provides new information on the microbial development of box and heap cocoa fermentations, under inoculated and noninoculated conditions, as we coupled yeast/bacterial amplicon-based sequencing data with microbial metabolite detection. The information so far available suggests that microbial communities have played an important role in the evolution of aroma compounds. Understanding the pathways that microorganisms follow during the formation of aromas could be used to improve the fermentation processes and to enhance chocolate quality.
INTRODUCTION
Cocoa (Theobroma cacao L.) is an important plant crop throughout the world, and its production serves as a main source of income in several developing countries (1). According to the Food and Agriculture Organization (FAO), the world cocoa bean production was 4,466,574 tonnes in 2016 (http://www.fao.org/faostat). In terms of overall amount of beans per country, the main cocoa-producing countries in 2016 were the Ivory Coast, followed by Ghana, Indonesia, and Cameroon (http://www.fao.org/faostat). Chocolate production begins with the harvesting of the cocoa fruit, where cocoa beans and the surrounding mucilaginous pulp inside the pods are removed. At this point, the product has an astringent characteristic and needs to be fermented, dried, and roasted in order to acquire the optimal features of cocoa flavor and taste (3). Spontaneous fermentation normally lasts from 3 to 10 days in heaps, boxes, baskets, or trays.
According to Schwan and Fleet (4), the microbiota present during cocoa fermentation is composed of yeasts, lactic acid bacteria (LAB), and acetic acid bacteria (AAB). Two important stages are present during cocoa fermentation; in the first stage, yeasts proliferate in the reducing sugars and citric acid from the pulp and produce ethanol and carbon dioxide. At the same time, the temperatures and pH increase, due to aerobic and oxidative reactions, thus allowing LAB and AAB to grow (4). LAB mainly transforms sugars and organic acids into lactic acid, and, under aerobic conditions, AAB converts ethanol to acetic acid (3). The second stage involves the death of the seed embryo, due to the high concentrations of ethanol and acetic acid, and an increase in temperatures (3). The quality of the end product chocolate depends on the three previously cited groups of microorganisms, since they are able to produce metabolites and flavor precursors (4).
Although the importance of yeasts during cocoa fermentation has emerged in recent studies (5, 6), fungal biodiversity in fermented food has been studied far less than bacteria. In spite of the use of high-throughput sequencing (HTS) this decade, this new technology has mainly been used to obtain new insights into the domain of fermented foods, as it enables the genetic variants of a complex ecosystem to be discovered, validated, and screened (7). The importance of identifying the microbial composition of food ecosystems involves finding appropriate starter cultures that enhance a particular aspect of the product. Saccharomyces cerevisiae and Torulaspora delbrueckii have recently been detected and used as starter cultures in cocoa fermentation, and they have shown a positive impact on the aroma profile of the end product (8, 9). However, there has been much controversy concerning the choice of the starter cultures used in cocoa fermentation to improve the quality of the end product. This paper explores the impact of two cocoa fermentation starter cultures introduced to guarantee the production of cocoa beans with specific and reproducible features exposed to different fermentation methods. We point out the challenges related to the reproducibility of the effect of the starter cultures on cocoa bean fermentations and their correlations with the initial microbial populations and importance for aroma development.
The present study was aimed at determining the dynamics and biodiversity of both bacteria and yeasts by means of amplicon-based sequencing of the 16S rRNA genes and the internal transcribed spacer 2 (ITS2) gene, respectively, during cocoa bean fermentation carried out both spontaneously and in the presence of yeast starter cultures, in both boxes and heaps, in order to acquire more detailed knowledge about the relationship between microorganisms and their surroundings. The nonvolatile and volatile organic compounds were also assessed, with the aim of investigating how the use of cultures can affect the volatilome profile of fermented cocoa from the two different fermentation processes. In this study, we have also proposed the measurements of associations between microbial communities and the development of microbial volatile and nonvolatile compounds. A better understanding of the microbial communities and physicochemical dynamics during box and heap fermentations will undoubtedly help the development of new management procedures for the production of high-quality cocoa.
RESULTS
Physical and microbiological changes that take place in box and heap fermentations.
Temperature and pH were measured during the box and heap (B and H, respectively) fermentations at time zero and after 48, 96, and 120 h, as shown in Table 1. No significant difference (P > 0.05) between the considered conditions (inoculated and noninoculated) was observed from the physical or microbiological analysis, while the temperature observed during the B and H fermentations increased significantly from the initial values of 27°C to 43°C and 40°C, respectively, at the end of the fermentation (P < 0.05). The pH of the cocoa bean pulp was 3.5 at the beginning of the trial, and it increased to 4.2 and 4.7 at the end of the fermentation for the B and H fermentations, respectively (P < 0.05).
TABLE 1.
Average changes in the physical and microbiological parameters during the inoculated and noninoculated box and heap fermentations of cocoa bean pulp turned after 48 and 96 h
| Parameter by fermentation | Inoculationa | Data by fermentation time (h)b |
|||
|---|---|---|---|---|---|
| 0 | 48 | 96 | 120 | ||
| Box | |||||
| °C | S | 26.73 ± 0.60 | 36.20 ± 1.53 | 41.88 ± 1.78 | 43.70 ± 2.94 |
| ST | 26.48 ± 0.34 | 35.10 ± 2.46 | 41.73 ± 2.06 | 42.78 ± 3.68 | |
| C | 26.73 ± 0.62 | 36.20 ± 1.53 | 41.88 ± 1.78 | 43.70 ± 2.94 | |
| Avg | 26.58 ± 0.08 C | 35.80 ± 1.22 B | 42.09 ± 0.50 A | 43.33 ± 0.70 A | |
| pH | S | 3.55 ± 0.03 | 3.88 ± 0.16 | 4.15 ± 0.11 | 3.96 ± 0.18 |
| ST | 3.54 ± 0.01 | 4.00 ± 0.16 | 4.20 ± 0.15 | 4.18 ± 0.26 | |
| C | 3.55 ± 0.03 | 3.88 ± 0.17 | 4.27 ± 0.11 | 3.96 ± 0.18 | |
| Avg | 3.57 ± 0.03 C | 4.08 ± 0.11 B | 4.31 ± 0.09 A | 4.15 ± 0.17 AB | |
| Log CFU | |||||
| Yeast | S | 7.08 ± 0.05 | 7.19 ± 0.15 | NC | NC |
| ST | 7.14 ± 0.01 | 7.05 ± 0.06 | NC | NC | |
| C | 7.19 ± 0.17 | 7.55 ± 0.27 | NC | NC | |
| Avg | 7.14 ± 0.11 A | 7.26 ± 0.29 A | NC | NC | |
| LAB | S | 5.38 ± 0.30 | 7.18 ± 0.02 | 5.11 ± 1.28 | 6.75 ± 0.17 |
| ST | 6.13 ± 0.22 | 6.85 ± 0.27 | 4.33 ± 0.38 | 7.35 ± 0.25 | |
| C | 6.21 ± 0.74 | 6.88 ± 0.40 | 4.00 ± 0.00 | 5.49 ± 0.59 | |
| Avg | 5.91 ± 0.61 B | 6.97 ± 0.31 A | 5.44 ± 2.85 C | 6.55 ± 0.94 A | |
| AAB | S | 6.41 ± 0.11 | 7.01 ± 0.12 | 5.54 ± 1.78 | 5.63 ± 1.88 |
| ST | 6.28 ± 0.21 | 6.96 ± 0.06 | 5.60 ± 1.84 | 7.34 ± 0.09 | |
| C | 6.28 ± 0.25 | 7.31 ± 0.26 | 4.00 ± 0.00 | 5.76 ± 0.49 | |
| Avg | 6.32 ± 0.20 A | 7.09 ± 0.23 A | 5.05 ± 2.01 B | 6.69 ± 2.85 A | |
| Heap | |||||
| °C | S | 28.20 ± 1.15 | 38.17 ± 0.75 | 36.57 ± 1.80 | 40.07 ± 0.12 |
| ST | 27.37 ± 0.32 | 39.00 ± 2.21 | 36.57 ± 0.90 | 39.38 ± 0.32 | |
| C | 26.27 ± 0.06 | 38.97 ± 0.32 | 39.37 ± 2.57 | 40.30 ± 0.56 | |
| Avg | 27.28 ± 0.97 C | 38.71 ± 0.47 B | 38.40 ± 1.59 B | 40.07 ± 0.23 A | |
| pH | S | 3.55 ± 0.01 | 4.24 ± 0.17 | 4.48 ± 0.79 | 4.90 ± 0.97 |
| ST | 3.53 ± 0.01 | 4.32 ± 0.23 | 4.05 ± 0.40 | 4.52 ± 0.74 | |
| C | 3.50 ± 0.05 | 3.87 ± 0.08 | 4.24 ± 0.29 | 3.99 ± 0.33 | |
| Avg | 3.54 ± 0.02 B | 4.28 ± 0.24 A | 4.26 ± 0.21 A | 4.71 ± 0.90 A | |
| Log CFU | |||||
| Yeast | S | 7.16 ± 0.92 | 7.80 ± 0.15 | 7.13 ± 0.16 | 8.03 ± 0.29 |
| ST | 6.76 ± 0.85 | 7.72 ± 0.15 | 7.24 ± 1.41 | 7.43 ± 0.07 | |
| C | 7.02 ± 0.71 | 6.62 ± 0.02 | 6.34 ± 0.04 | 7.24 ± 0.28 | |
| Avg | 6.98 ± 0.20 C | 7.38 ± 0.66 B | 6.90 ± 0.48 D | 7.57 ± 0.41 A | |
| LAB | S | 5.67 ± 0.25 | 7.28 ± 0.19 | 7.36 ± 0.04 | 7.69 ± 0.28 |
| ST | 5.95 ± 0.29 | 7.07 ± 0.09 | 7.00 ± 0.01 | 7.50 ± 0.04 | |
| C | 5.72 ± 0.03 | 6.17 ± 0.21 | 7.40 ± 0.04 | 8.10 ± 0.13 | |
| Avg | 5.78 ± 0.15 D | 6.84 ± 0.59 C | 7.25 ± 0.22 B | 7.76 ± 0.30 A | |
| AAB | S | 6.20 ± 0.23 | 6.81 ± 0.14 | 8.33 ± 0.02 | 7.80 ± 0.18 |
| ST | 5.70 ± 0.04 | 7.15 ± 0.28 | 8.08 ± 0.11 | 7.66 ± 0.01 | |
| C | 6.60 ± 0.04 | 5.65 ± 0.15 | 8.42 ± 0.02 | 8.56 ± 0.04 | |
| Avg | 6.17 ± 0.45 D | 6.54 ± 0.79 C | 8.28 ± 0.18 A | 8.00 ± 0.49 B | |
S, S. cerevisiae; ST, S. cerevisiae and T. delbrueckii; C, noninoculated.
Values are expressed as the mean ± SD from triplicate determinations. Different letters indicate statistical differences related to the fermentation period using the least significant difference test (P < 0.05). P values were adjusted using Bonferroni's method. NC, below the detection limit.
The yeast, LAB, and AAB population dynamics are reported in Table 1. The yeasts constituted the dominant population for the first 48 h in both processes (B and H), and they were already detected at high loads in the cocoa beans before the introduction of the starter strain inoculum, with an average value of 6.98 log CFU · g−1 in the H fermentation and 7.14 log CFU · g−1 in the B fermentation. On the other hand, the yeast population in the H fermentation remained at around 7 log CFU · g−1, even after 48 h, with the highest count recorded at the end of the process (7.57 log CFU · g−1). A significant difference between B and H fermentations was also observed in the LAB dynamics during the fermentation time, with a marked increase in the counts after 48 h in both fermentation processes (B, 5.91 to 6.55 log CFU · g−1; H, 5.78 to 7.76 log CFU · g−1), as shown in Table 1 (P < 0.01). High counts of AAB were observed at the beginning of the B and H fermentations (6.32 and 6.17 log CFU · g−1, respectively). However, this population showed a fluctuating trend during the B fermentation, whereas it increased over time during the H fermentation to a final count of 8.00 log CFU · g−1 (P < 0.01). It should be noted that after 96 h, the AAB dominated the LAB and yeasts in both fermentation processes. Overall, higher counts were observed for the three considered microbial groups (yeasts, LAB, and AAB) in the H fermentations than in the B ones at 96 h, as shown in Table 1 (P < 0.05).
Identification of isolated yeast colonies and assessment of the dominance of the starter strains.
In order to establish the yeast dynamics in the B and H fermentations, 104 yeast colonies were isolated from WL nutrient agar plates. The ITS-restriction fragment length polymorphism (ITS-RFLP) fingerprints identified S. cerevisiae and T. delbrueckii in 70% of the isolated colonies. Furthermore, repetitive extragenic palindromic-PCR (REP-PCR) fingerprints and a comparison with the starter profiles highlighted the presence of S. cerevisiae ID76 and T. delbrueckii ID103 in the cultivable mycobiota during the first 48 h of both the B and H fermentations. S. cerevisiae ID76 represented 68% of the isolates from the fermentations inoculated with S. cerevisiae (S) and 51% of the colonies isolated from the fermentations inoculated with S. cerevisiae and T. delbrueckii (ST). Finally, 38% of the colonies isolated from the ST fermentations were ascribed to a T. delbrueckii ID103 profile. Apart from the identification of the starter strains, Hanseniaspora opuntiae represented the most abundant autochthonous species, representing 31% of the colonies isolated from the noninoculated fermentations (data not shown).
Dynamics of the nonvolatile organic compounds during cocoa bean fermentation.
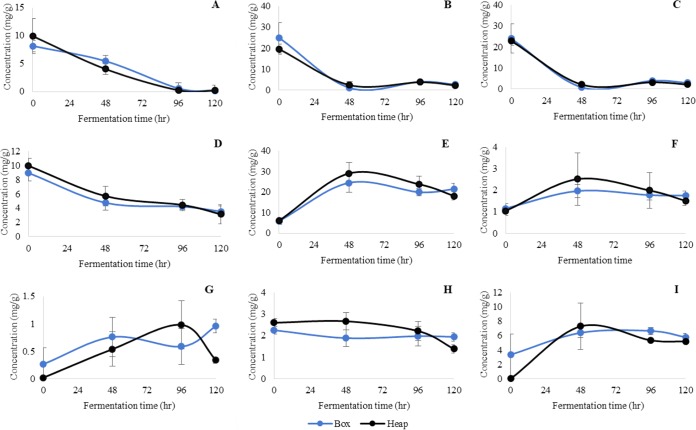
The evolution of nonvolatile compounds was determined during the B and H fermentations of the cocoa beans by means of high-performance liquid chromatography (HPLC), as shown in Fig. 1. No significant differences were observed between the inoculated and noninoculated fermentations through the analysis of the nonvolatile compounds. At the beginning of the process, the B fermentations showed higher concentrations of glucose, fructose, and sucrose (24, 25, and 8 mg/g, respectively) than did the H fermentations (20, 23, and 10 mg/g, respectively), and significantly decreased levels of glucose, fructose, and sucrose during both fermentation processes (B and H) were observed over the fermentation period (P < 0.05).
FIG 1.
Dynamics of the sugar and organic acid compounds in the cocoa bean pulp inoculated and noninoculated during box and heap fermentations, expressed as milligrams per gram. Data are expressed as mean ± standard deviation (SD) values from triplicate determinations. (A) Sucrose; (B) glucose; (C) fructose; (D) citric acid; (E) succinic acid; (F) malic acid; (G) acetic acid; (H) gluconic acid; and (I) lactic acid.
As far as the overall content of organic acids is concerned, the highest concentration in the cocoa bean pulp before the start of the fermentation was that of citric acid, and this was followed by succinic and gluconic acid in both fermentation processes (Fig. 1). It should be pointed out that greater amounts of lactic and succinic acid were detected at 48 h, whereas the maximum production of acetic acid was observed at 96 h. Succinic acid was found to be the most abundant organic acid from 48 h to the end of both fermentation processes (B and H), with concentrations of up to 21 and 18 mg/g, respectively. The dynamics over time observed for the organic acids during both fermentation processes (B and H) were similar. A statistically significant decrease in the citric and gluconic acid concentrations was observed during the B and H fermentations, and the lowest values were reached at the end (P < 0.01). On the other hand, an increase in the malic, succinic, lactic, and acetic acid concentrations was found during the fermentation period (P < 0.01) for both processes (B and H). No significant changes were observed for the oxalic, pyruvic, tartaric, or fumaric acids during B or H over the fermentation period (see Table S1 in the supplemental material).
Volatilome during cocoa bean pulp fermentation.
A total of 72 volatile organic compounds (VOCs) were identified by means of headspace solid-phase microextraction gas chromatography–quantitative mass spectrometry (HS-SPME/GC-qMS) on fermented cocoa bean pulp (Table S1). No significant differences were observed between the inoculated and noninoculated fermentations from the VOC analysis. At the beginning of the B and H fermentation processes, 2-pentanol, ethyl acetate, limonene, and 1,2-propanediol diacetate were found to be the most abundant volatile compounds, whereas acetic acid, limonene, 2-heptanol, phenylethyl alcohol, isopentyl alcohol, isovaleric acid, and benzeneacetaldehyde represented the most retrieved VOCs in the headspace at the end of both fermentations (Table S1). It should be pointed out that the total peak area of the VOCs at the end of the B fermentation was about twice as high as that of the H fermentation (Table S1, P < 0.01).
Mycobiota of cocoa beans during fermentation.
A total of 1,304,936 raw reads (2 × 250 bp) were obtained, and 1,217,061 reads passed the filters applied through QIIME, with an average value of 31,975 ± 22,635 reads/sample and a mean sequence length of 411 bp. The rarefaction analysis and the estimated sample coverage (ESC) were satisfactory for all of the samples, with an ESC average of 97% (Table S2), while the alpha diversity indicated a higher level of complexity in the B fermentations than in the H fermentations (P < 0.05). Overall, 18 fungal OTUs were identified during the fermentations, as shown in Table 2. A statistically significant difference between conditions was found, with a higher relative abundance of Hanseniaspora opuntiae in the noninoculated fermentation (46.23%) than in those inoculated with S. cerevisiae and T. delbrueckii (ST; 25.60%) (P < 0.05). In addition, a significantly higher presence of T. delbrueckii was observed in the fermentations inoculated with the mixed yeast culture (ST, 22.23%) than in the fermentations inoculated only with S. cerevisiae (S) and than in the noninoculated ones (0.03 and 0.11%, respectively) (P < 0.01, Table 2).
TABLE 2.
Incidence of the fungal taxonomic groups, achieved by means of amplicon sequencing, expressed as relative abundancesa
| Taxonomic group | Relative abundance |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. cerevisiae |
S. cerevisiae + T. delbrueckii |
Noninoculated |
|||||||||||||||||||||
| Box |
Heap |
Box |
Heap |
Box |
Heap |
||||||||||||||||||
| T0 | T48 | T96 | T120 | T0 | T48 | T96 | T120 | T0 | T48 | T96 | T120 | T0 | T48 | T120 | T0 | T48 | T96 | T120 | T0 | T48 | T96 | T120 | |
| Botryosphaeria | 1.33 | 0.43 | 0.46 | 0.21 | 0.59 | 1.55 | 0.13 | 0.13 | 0.53 | 0.44 | 0.91 | 3.51 | 0.56 | 0.32 | 0.59 | 0.66 | 0.20 | 1.08 | 0.16 | 11.07 | 0.00 | 0.47 | 0.12 |
| Candida | 3.33 | 1.35 | 1.21 | 1.27 | 1.03 | 0.46 | 0.31 | 0.74 | 1.89 | 0.97 | 0.77 | 5.56 | 0.53 | 0.34 | 0.72 | 2.83 | 1.48 | 2.59 | 0.49 | 0.89 | 0.24 | 1.39 | 1.73 |
| Candida butyri | 2.23 | 0.27 | 0.42 | 0.50 | 0.16 | 0.03 | 0.07 | 0.09 | 0.77 | 0.69 | 0.34 | 0.40 | 0.15 | 0.04 | 0.07 | 0.70 | 0.21 | 1.16 | 0.09 | 0.07 | 0.00 | 0.07 | 0.03 |
| Candida diversa | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 5.50 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.96 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Candida inconspicua | 0.00 | 0.58 | 2.63 | 7.82 | 0.06 | 0.03 | 0.25 | 0.32 | 0.00 | 0.74 | 1.51 | 12.97 | 0.00 | 0.04 | 0.94 | 0.03 | 2.61 | 0.96 | 13.47 | 0.00 | 0.87 | 1.62 | 1.43 |
| Candida jaroonii | 13.07 | 4.40 | 3.63 | 1.24 | 4.01 | 1.65 | 0.59 | 1.00 | 8.06 | 3.80 | 2.29 | 1.24 | 3.41 | 1.55 | 1.96 | 4.34 | 2.18 | 3.04 | 0.86 | 1.89 | 0.24 | 1.27 | 0.80 |
| Candida quercitrusa | 2.27 | 1.03 | 0.78 | 0.32 | 1.31 | 0.94 | 0.32 | 0.81 | 1.40 | 0.72 | 1.17 | 0.49 | 1.27 | 0.99 | 1.34 | 2.43 | 1.08 | 1.04 | 0.16 | 0.47 | 0.09 | 1.46 | 0.89 |
| Ceratocystis | 3.08 | 5.98 | 2.93 | 1.42 | 1.43 | 0.99 | 0.50 | 0.50 | 2.46 | 2.91 | 2.91 | 3.41 | 1.27 | 1.56 | 2.32 | 4.19 | 2.45 | 9.32 | 4.81 | 2.07 | 0.87 | 1.53 | 0.80 |
| Hanseniaspora opuntiae | 38.33 | 40.98 | 39.72 | 28.21 | 49.76 | 39.10 | 48.67 | 64.37 | 36.78 | 37.25 | 15.62 | 0.38 | 43.73 | 29.51 | 11.70 | 54.21 | 48.11 | 29.13 | 21.78 | 69.22 | 72.35 | 35.05 | 44.81 |
| Issatchenkia | 0.09 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.27 | 0.25 | 0.00 | 0.37 | 0.00 | 0.41 | 0.37 | 0.00 | 0.00 | 0.00 | 2.46 | 1.18 |
| Kluyveromyces marxianus | 0.00 | 0.28 | 0.18 | 8.91 | 0.06 | 0.13 | 11.18 | 1.79 | 0.00 | 0.42 | 0.38 | 15.76 | 0.00 | 0.13 | 2.80 | 0.01 | 4.61 | 1.03 | 43.54 | 0.09 | 7.76 | 7.23 | 1.98 |
| Lasiodiplodia theobromae | 0.83 | 0.61 | 2.07 | 1.33 | 0.49 | 0.49 | 0.09 | 0.07 | 3.30 | 0.91 | 1.22 | 0.90 | 0.80 | 0.09 | 0.25 | 4.94 | 0.61 | 0.92 | 0.27 | 0.10 | 0.16 | 0.07 | 0.16 |
| Penicillium | 0.62 | 0.39 | 1.09 | 0.16 | 0.19 | 0.13 | 0.00 | 0.06 | 0.12 | 0.16 | 0.31 | 0.25 | 0.10 | 0.19 | 0.27 | 0.55 | 0.12 | 0.24 | 0.10 | 0.15 | 0.02 | 0.52 | 0.21 |
| Pichia | 0.56 | 0.09 | 0.05 | 0.16 | 0.12 | 0.12 | 0.01 | 0.78 | 0.06 | 0.09 | 0.15 | 0.60 | 0.07 | 0.01 | 0.49 | 0.37 | 0.41 | 0.55 | 0.19 | 0.19 | 0.00 | 1.21 | 1.28 |
| Pichia pijperi | 10.90 | 10.23 | 12.75 | 15.36 | 10.21 | 14.55 | 6.79 | 8.42 | 7.67 | 9.57 | 9.05 | 18.01 | 13.62 | 18.35 | 17.13 | 11.32 | 10.91 | 14.24 | 8.38 | 10.37 | 6.29 | 29.42 | 24.14 |
| Saccharomyces cerevisiae | 12.95 | 28.62 | 28.30 | 30.22 | 26.08 | 37.67 | 30.29 | 19.11 | 7.73 | 21.50 | 31.97 | 15.98 | 7.67 | 21.57 | 33.96 | 0.28 | 19.95 | 27.12 | 3.35 | 0.37 | 10.28 | 11.92 | 17.96 |
| Saccharomycopsis | 0.72 | 0.35 | 0.32 | 0.15 | 0.13 | 0.15 | 0.07 | 0.13 | 1.12 | 0.30 | 0.32 | 0.03 | 0.27 | 0.10 | 0.32 | 0.82 | 0.49 | 1.21 | 0.22 | 0.52 | 0.13 | 0.32 | 0.34 |
| Torulaspora delbrueckii | 0.06 | 0.01 | 0.04 | 0.01 | 0.03 | 0.00 | 0.01 | 0.04 | 25.35 | 16.32 | 28.18 | 8.48 | 23.02 | 20.45 | 22.50 | 0.02 | 0.07 | 0.27 | 0.03 | 0.12 | 0.09 | 0.13 | 0.09 |
Only OTUs with an incidence above 1% in at least 2 samples are shown. Values are expressed as the mean of duplicate determinations. The abundances of OTUs in the 2 biological replicates of each sampling time were averaged. Samples are labeled according to the fermentation period (0 h [T0], 48 h [T48], 96 h [T96], and 120 h [T120]), fermentation method (box and heap), and condition (inoculated with S, inoculated with ST, and noninoculated).
The inoculated cocoa beans (S and ST) in both fermentation processes (B and H) showed a dominance of H. opuntiae, Candida jaroonii, S. cerevisiae, T. delbrueckii, and Pichia pijperi at time zero (Table 2). In addition, H. opuntiae, P. pijperi, and C. jaroonii were the most predominant in the noninoculated B fermentations at the beginning of the process, while H. opuntiae, P. pijperi, and Botryosphaeria spp. reached the highest incidence in the noninoculated H fermentations. However, H. opuntiae, S. cerevisiae, P. pijperi, and Kluyveromyces marxianus were the most abundant at the end of both fermentations (B and H). The mycobiota dynamics were similar over time for the inoculated and noninoculated B and H fermentations. S. cerevisiae significantly increased over time in both processes, while H. opuntiae significantly decreased, as shown in Table 2 (P < 0.01).
Bacterial community of the fermented cocoa beans.
The total number of paired sequences obtained from the fermented cocoa beans reached 4,159,213 raw reads. After merging, a total of 2,655,230 reads passed the filters applied through QIIME, with an average value of 63,220 ± 45,781 reads/sample and a mean sequence length of 445 bp. The rarefaction analysis and Good's coverage, expressed as a percentage (91%), also indicated satisfactory coverage of all the samples (Table S3). Alpha diversity only indicated a higher level of complexity over the fermentation period (Table S3, P < 0.05). No significant difference was observed when the different conditions (inoculated with S or ST and noninoculated) were compared or between processes (B and H). The taxonomic classification of bacterial communities includes the family, genus, and species levels.
Overall, the most abundant OTUs detected at 48 h in both the inoculated and noninoculated B fermentations were Acetobacter pasteurianus, Lactobacillus fermentum, and Lactobacillus plantarum (Table 3). It should be noted that A. pasteurianus and L. fermentum remained the two most abundant OTUs at the end of the box fermentation under both conditions (inoculated or noninoculated), and these were followed by Bacillus species. As far as the inoculated H fermentations are concerned, A. pasteurianus, L. fermentum, and Acetobacteraceae were the most abundant OTUs detected at 48 h, and A. pasteurianus and L. fermentum remained the dominant OTUs over the entire fermentation period (Table 3). Instead, the noninoculated H fermentations were characterized by high relative abundances of L. fermentum, A. pasteurianus, and L. plantarum at 48 h, while L. fermentum, Bacillus spp., and Klebsiella spp. took over and dominated at the end of the process. As far as the dynamics are concerned, we observed an increase in the relative abundances under different conditions for L. fermentum, L. plantarum, A. pasteurianus, Bacillus spp., Acetobacteraceae, and Lactobacillaceae over the fermentation period, while Erwinia spp., Gluconobacter spp., Trabulsiella spp., and Enterobacteriaceae decreased over time (P < 0.01), as shown in Table 3.
TABLE 3.
Classification at the family/genus level of the occurrence of the bacterial taxonomic groups, achieved by means of amplicon sequencing, expressed as relative abundancesa
| Taxonomic group | Relative abundance |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. cerevisiae |
S. cerevisiae + T. delbrueckii |
Noninoculated |
||||||||||||||||||||||
| Box |
Heap |
Box |
Heap |
Box |
Heap |
|||||||||||||||||||
| T0 | T48 | T96 | T120 | T0 | T48 | T96 | T120 | T0 | T48 | T96 | T120 | T0 | T48 | T96 | T120 | T0 | T48 | T96 | T120 | T0 | T48 | T96 | T120 | |
| Acetobacter pasteurianus | 3.16 | 13.81 | 62.90 | 52.83 | 1.05 | 40.27 | 76.99 | 69.22 | 3.02 | 42.67 | 63.42 | 86.96 | 0.96 | 57.24 | 79.19 | 24.74 | 2.40 | 18.22 | 42.67 | 64.24 | 4.70 | 28.52 | 36.67 | 45.93 |
| Acetobacteraceae | 0.19 | 1.73 | 1.01 | 1.29 | 0.00 | 16.87 | 6.23 | 5.08 | 0.29 | 4.70 | 0.86 | 1.05 | 0.10 | 11.79 | 4.41 | 0.86 | 0.10 | 0.58 | 3.45 | 2.40 | 0.38 | 5.42 | 1.44 | 0.10 |
| Acinetobacter | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.29 | 0.00 | 0.00 | 0.05 | 0.00 | 0.00 | 0.00 | 0.00 | 4.99 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.05 | 0.00 |
| Acinetobacter guillouiae | 0.24 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.86 | 0.10 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 1.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.05 | 0.00 | 0.00 | 0.14 |
| Acinetobacter rhizosphaerae | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.19 | 1.44 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.25 | 0.00 | 0.00 | 0.00 | 0.00 | 0.05 | 0.00 | 0.19 | 0.24 |
| Bacillus | 0.00 | 0.00 | 0.00 | 16.35 | 0.00 | 0.00 | 0.00 | 0.77 | 0.00 | 0.00 | 11.94 | 0.29 | 0.00 | 0.00 | 0.00 | 3.36 | 0.00 | 0.10 | 0.00 | 4.41 | 0.05 | 0.00 | 0.96 | 8.96 |
| Dyella | 1.68 | 0.58 | 0.10 | 0.14 | 0.77 | 0.10 | 0.00 | 0.00 | 0.43 | 0.38 | 0.10 | 0.00 | 0.19 | 0.19 | 0.00 | 0.00 | 1.25 | 0.29 | 0.58 | 0.19 | 0.96 | 1.10 | 0.53 | 0.14 |
| Enterobacteriaceae | 3.26 | 1.34 | 1.20 | 0.29 | 0.00 | 0.00 | 0.00 | 0.10 | 0.96 | 0.58 | 0.10 | 0.00 | 0.19 | 0.00 | 0.00 | 0.10 | 1.82 | 1.53 | 0.58 | 0.19 | 2.21 | 1.53 | 0.62 | 0.34 |
| Erwinia | 4.94 | 1.68 | 1.10 | 0.53 | 0.19 | 0.19 | 0.00 | 0.00 | 1.63 | 0.72 | 0.19 | 0.00 | 0.29 | 0.10 | 0.00 | 0.00 | 3.45 | 1.53 | 0.77 | 0.58 | 2.30 | 1.82 | 1.05 | 0.34 |
| Gluconobacter | 3.36 | 1.25 | 0.38 | 0.05 | 1.63 | 1.34 | 0.38 | 0.19 | 4.07 | 1.15 | 0.05 | 0.10 | 2.97 | 1.25 | 0.38 | 0.10 | 2.40 | 0.67 | 0.58 | 0.00 | 2.88 | 1.34 | 0.10 | 0.10 |
| Klebsiella | 0.34 | 0.43 | 0.19 | 0.24 | 0.00 | 0.00 | 0.19 | 0.19 | 1.10 | 0.19 | 0.10 | 0.00 | 0.00 | 0.00 | 0.10 | 0.48 | 1.25 | 1.15 | 0.48 | 0.10 | 0.38 | 0.34 | 0.34 | 3.93 |
| Lactobacillaceae | 0.05 | 2.06 | 3.88 | 1.20 | 0.00 | 0.38 | 0.00 | 0.10 | 0.05 | 1.68 | 0.96 | 0.00 | 0.00 | 0.10 | 0.19 | 0.10 | 0.10 | 5.37 | 2.21 | 1.25 | 0.19 | 0.62 | 1.10 | 0.34 |
| Lactobacillus plantarum group | 1.44 | 15.58 | 12.27 | 4.31 | 0.00 | 2.11 | 0.38 | 0.19 | 0.91 | 10.74 | 1.92 | 0.67 | 0.00 | 1.15 | 0.19 | 1.63 | 0.58 | 28.57 | 14.09 | 5.47 | 0.29 | 3.74 | 6.62 | 1.49 |
| Lactobacillus fermentum | 0.19 | 31.59 | 3.40 | 8.63 | 0.00 | 10.07 | 7.19 | 5.75 | 0.14 | 6.14 | 10.12 | 1.44 | 0.00 | 9.20 | 6.90 | 12.18 | 0.10 | 3.55 | 0.58 | 7.00 | 0.05 | 30.87 | 13.71 | 16.73 |
| Lysinibacillus | 0.00 | 0.00 | 0.00 | 0.58 | 0.00 | 0.00 | 0.00 | 0.67 | 0.00 | 0.00 | 1.63 | 0.00 | 0.00 | 0.00 | 0.00 | 0.96 | 0.00 | 0.00 | 0.00 | 0.67 | 0.00 | 0.00 | 0.19 | 0.05 |
| Trabulsiella | 5.13 | 3.12 | 2.49 | 1.53 | 0.38 | 0.29 | 0.00 | 0.00 | 2.64 | 0.81 | 0.53 | 0.19 | 0.58 | 0.10 | 0.00 | 0.10 | 5.75 | 1.73 | 1.53 | 0.86 | 2.97 | 4.12 | 0.96 | 0.38 |
Only OTUs with an incidence above 1% in at least 2 samples are shown. Values are expressed as the mean of duplicate determinations. The abundances of OTUs in the 2 biological replicates of each sampling time were averaged. Samples are labeled according to the fermentation period (0 h [T0], 48 h [T48], 96 h [T96], and 120 h [T120]), fermentation method (box and heap), and condition (inoculated with S, inoculated with ST, and noninoculated).
OTU cooccurrence and/or coexclusion during cocoa bean fermentation.
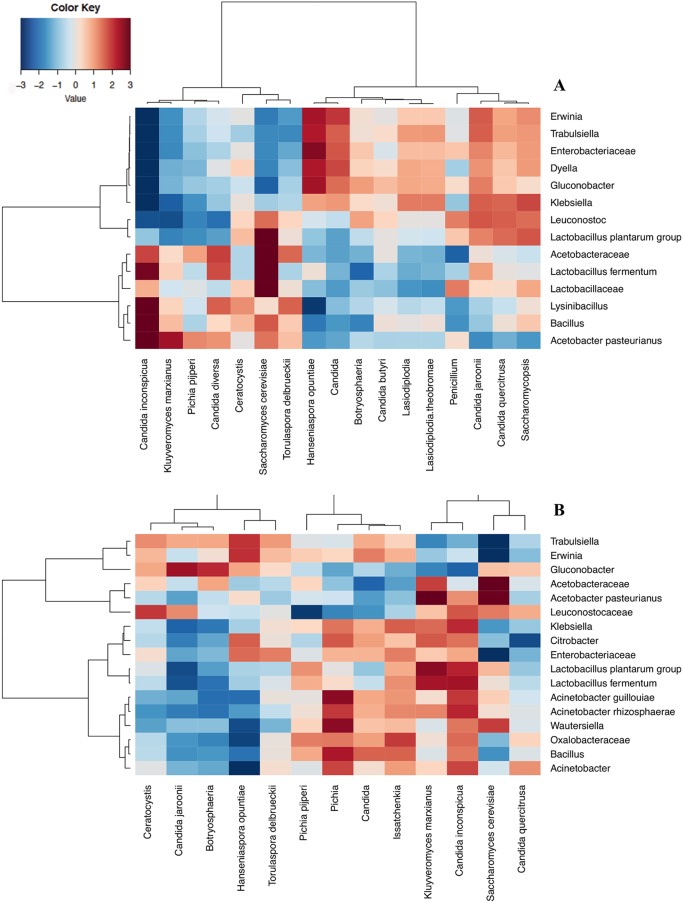
When the relative abundance of the bacterial and yeast populations was plotted, considering the OTUs of all the conditions (inoculated with S, inoculated with ST, and noninoculated) of each fermentation method (B and H) together, it was possible to observe microbial cooccurrence or coexclusion dynamics between the two different communities, as shown in Fig. 2.
FIG 2.
Spearman's correlation between the microbial OTUs observed with an incidence >1% in at least 2 samples. The samples are labeled according to the box (A) and heap (B) fermentation methods. Rows and columns are clustered by means of Ward's linkage hierarchical clustering. The intensity of the colors represents the degree of correlation between the fungal and bacterial OTUs, as measured by Spearman's correlation. The intensity of the colors represents the degree of correlation between the yeast and bacteria, where blue represents a negative degree of correlation and red a positive degree of correlation.
Overall, L. plantarum, A. pasteurianus, and Enterobacteriaceae were negatively associated with the main yeast OTUs (S. cerevisiae, K. marxianus, Candida inconspicua, and P. pijperi) in the B fermentations. In short, S. cerevisiae was positively correlated with Acetobacteraceae and Lactobacillaceae, whereas A. pasteurianus was positively correlated with K. marxianus and C. inconspicua and negatively correlated with C. jaroonii and H. opuntiae (P < 0.05). However, H. opuntiae was positively associated with the presence of the Enterobacteriaceae family, as well as with Gluconobacter spp. (P < 0.05). It is worth noting that H. opuntiae and C. jaroonii were found to be positively associated with the minor OTUs Citrobacter and Erwinia (P < 0.05, Fig. 2A).
The L. fermentum in the H fermentations showed a positive correlation with K. marxianus and C. inconspicua and a negative correlation with C. jaroonii (P < 0.05), as can be observed in Fig. 2B. S. cerevisiae was positively correlated with Acetobacteraceae and with A. pasteurianus (Fig. 2B).
Correlation between sugar and the organic acid compounds and microbiota populations detected by means of HPLC.
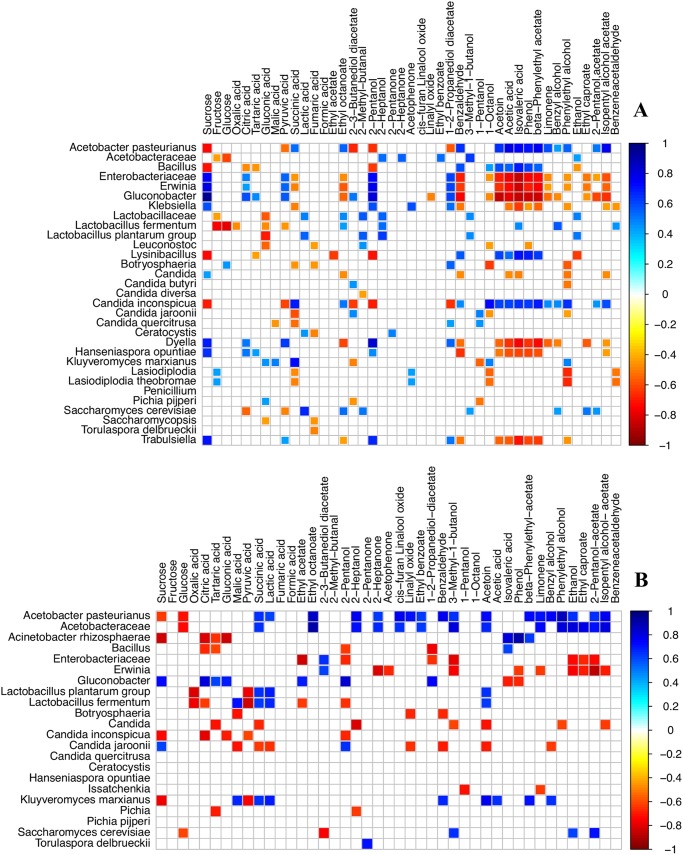
Significantly different correlations were observed between the changes found in the concentration of sugars, organic acids, and microbes in the B and H fermentations, as shown in Fig. 3 (P < 0.05). Overall, the most abundant microbial species in the fermented cocoa beans in the B fermentations, that is, H. opuntiae, A. pasteurianus, K. marxianus, L. plantarum, and S. cerevisiae, were statistically positively associated with intermediate metabolites, such as the citric, lactic, and succinic acids (P < 0.05), while Bacillus spp., L. plantarum, A. pasteurianus, and L. fermentum were statistically negatively correlated with the energy/carbon substrates (P < 0.05). In addition, sucrose was positively correlated with the presence of H. opuntiae (P < 0.05) and negatively correlated with A. pasteurianus and Bacillus spp. (P < 0.05). Citric acid was negatively correlated with Bacillus spp. and S. cerevisiae but positively correlated with H. opuntiae, Gluconobacter spp., and Erwinia spp. (P < 0.05). L. fermentum was negatively correlated with fructose, glucose, gluconic acid, and pyruvic acid (P < 0.05). Finally, succinic acid was positively associated with A. pasteurianus, C. inconspicua, and K. marxianus, and lactic acid was positively related to L. plantarum and S. cerevisiae, as shown in Fig. 3A (P < 0.05).
FIG 3.
Correlation plot showing Spearman's correlation between the microbial OTUs and metabolites observed with an incidence >1% in at least 2 samples. The samples are labeled according to the box (A) and heap (B) fermentation methods. Only significant associations between the OTUs and metabolites are shown (P < 0.05). The intensity of the colors represents the degree of correlation between the fungal and bacterial OTUs, as measured by Spearman's correlation, where the blue color represents a positive degree of correlation and red a negative correlation between the sugars, organic acids, and OTUs.
However, few statistically significant correlations were found in the H fermentations (Fig. 3B); A. pasteurianus was found to be negatively associated with sucrose, while Gluconobacter spp. were positively related with sucrose (P < 0.05). In addition, A. pasteurianus, K. marxianus, L. plantarum, and L. fermentum were positively associated with succinic and lactic acid, as shown in Fig. 3B (P < 0.05).
Correlation between the microbiota and volatilome profile.
Significantly different associations were observed between the secondary metabolites and the main OTUs in the B and H fermentations, as shown in Fig. 3 (P < 0.05). The main bacterial and fungal taxa in the B fermentations (Fig. 3A), that is, S. cerevisiae, H. opuntiae, L. plantarum, A. pasteurianus, K. marxianus, C. inconspicua, and L. fermentum, were statistically correlated with the key aroma and fermentative markers, while the minor OTU bacteria (Enterobacteriaceae, Trabulsiella, Erwinia, and Gluconobacter) and H. opuntiae were statistically negatively correlated with the acids and phenols. In short, positive correlations were found between S. cerevisiae and ethyl octanoate, 2-methyl-butanal, and 3-methyl-butanol, between H. opuntiae and 2-pentanol (P < 0.05), between L. plantarum and 2-heptanol, 2-methyl-butanal, 3-methyl-1-butanol, and ethanol, and between L. fermentum and ethyl octanoate, 2-heptanol, benzyl alcohol, and isovaleric acid (P < 0.05). In addition, A. pasteurianus, C. inconspicua, and Bacillus spp. were also positively correlated with acetoin, acetic acid, isovaleric acid, phenol, limonene, benzyl alcohol, and phenylethyl alcohol (P < 0.05), while these compounds were positively correlated with H. opuntiae and the minor bacterial OTUs, as shown in Fig. 3A (P < 0.05).
Fewer correlations were observed in the H fermentations than in the B fermentations (Fig. 3B). In general, some of the most abundant microbes (A. pasteurianus, T. delbrueckii, S. cerevisiae, and K. marxianus) and Acetobacteraceae showed several significantly positive correlations with VOCs. A. pasteurianus was positively correlated with ethyl octanoate, 2-heptanol, 2-hepanone, cis-furan-linalool oxide, benzaldehyde, acetoin, β-phenylethylacetate, 3-methyl-1-butanol, limonene, 2-pentanol acetate, phenylethyl alcohol, ethanol, and isopentyl alcohol (P < 0.05). T. delbrueckii was positively associated with 2-heptanone (P < 0.05). S. cerevisiae was positively correlated with 3-methyl-1-butanol and ethanol (P < 0.05). Finally, Acetobacteraceae was positively correlated with ethyl octanoate, 2-heptanol, 2-heptanone, cis-furan-linalool oxide, 3-methyl-1-butanol, acetoin, limonene, phenylethyl alcohol, ethanol, and isopentyl alcohol, while K. marxianus was positively correlated with benzaldehyde, acetoin, acetic acid, benzyl alcohol, and β-phenylethylacetate (P < 0.05).
DISCUSSION
In this study, the changes that have taken place in the physicochemical composition, microbial counts, and microbiota diversity of cocoa beans in two different fermentation processes, that is, in boxes (B) and heaps (H), inoculated or not inoculated with yeasts as a starter culture, have been investigated. The ability of the survival and growth of selected starter strains, in this case, S. cerevisiae ID67 and T. delbrueckii ID103, during cocoa fermentation is one of the most important features to ensure their effect during this process. These starter strains have shown the ability to coexist with autochthonous microbial communities in fermented cocoa beans. However, the yeast cultures used in the present study did not significantly modify the microbiological dynamics, physicochemical parameters, or metabolites produced during fermentation, whereas the same starter strains influenced the fermentative process and the quality of the end products in at least one cocoa hybrid variety (9). It is important to note that the initial yeast load in a previous study was lower than those observed in our study, and this might explain the discrepancies on the impact of the same yeast culture during cocoa fermentation. The different fermentation practices, the cocoa variety, and the use of different starter cultures on site during cocoa bean fermentation play important roles in the success of the starter culture used during fermentation and might also explain the discrepancies found between studies (5, 6, 8–14). Our results confirmed that the performance of starter cultures on cocoa fermentation might change from the geographic origin. Moreover, the effectiveness of the cultures depends on the complexity of the microbial consortia. This in turn is influenced directly by the used fermentation method, each of which is characterized by its own microenvironment and is affected by oxygen availability, local agriculture practices, temperature, amount of cocoa mass used, etc.
During fermentation, cocoa beans constitute an ecological niche for a wide range of microbes. The advances made in studying the dynamics of cocoa microbial communities have shown that the composition of these communities follows predictable patterns that report a rapid decline in yeast counts after 48 h, when the sugars are depleted, a rise in temperature, and an increase in LAB and AAB (15–19). The great impact on the microbial dynamics and succession during cocoa fermentation have been explained by considering the use of different cultivar varieties, fermentation methods, environmental conditions, and harvesting and postharvesting methods, as well as external factors, such as cross-contamination (equipment, operators, insect interactions, and microbial populations from previous fermentations) (14, 16).
The use of molecular biology tools and the improvement of culturing techniques have facilitated the detection of new yeast, LAB, and AAB species. A restricted microbial population that includes H. opuntiae, A. pasteurianus, and L. fermentum has already been reported for fermented cocoa beans and has also been detected in our study (8, 13, 20). However, some discrepancies can be observed among the most abundant microbial species in fermented cocoa beans, which may vary considerably from country to country. Through the application of amplicon-based sequencing in our study, we have been able to detect unusual yeasts, such as C. jaroonii, Lasiodiplodia theobromae, and Botryosphaeria, during cocoa fermentation, none of which had previously been detected. Noteworthy, there is a lack of information available regarding the incidence of minor microbial groups (21). In spite of the great advances made in microbial ecology, through next-generation sequencing, microbial species-level identification and strain-level differentiation still represent a challenge which needs to be addressed in the future to achieve an accurate identification.
This study, in an attempt to gain more knowledge about the range of potential interactions between microbial communities, describes a possible cooccurrence and coexclusion. Our results showed a modulation of the LAB due to the presence of the yeast culture, in agreement with previous observations (22). It should be noted that in our study, these associations depended on the type of fermentation process, and the correlation data set was used to explore the possible microbial dynamics, interactions, and metabolism. This information can offer information about the kinetics of substrate consumption and aroma production by the microbiota present in fermented cocoa beans. However, it has been found that the correlations depend on the number of samples in which a type II error reflects the failure to reject a null hypothesis that is not true.
The dynamics of the nonvolatile compounds have shown successful competition for nutrients by the microbial populations within the cocoa fermentations. The ability of the fungal and bacterial communities to reduce sugars that has been observed in our results has been studied in detail and is supported by previous studies (23, 24). As far as the organic acid dynamics are concerned, citric acid showed the highest concentration at the beginning of both fermentations and then decreased over time. This utilization of citrate has been attributed to bacteria, which metabolize it into acetic acid, carbon dioxide, and lactic acid (25). However, not only bacteria can utilize citrate as an energy source: some isolates within Candida krusei have also been reported to assimilate citrate during cocoa fermentation (26). However, C. krusei was not detected in our study during cocoa fermentation; however, the most abundant yeasts found in this study, H. opuntiae and S. cerevisiae, have never shown the capability to assimilate citrate in vitro (27). Therefore, it has been hypothesized that the observed citrate assimilation was due to LAB, such as the highly abundant L. fermentum, as also supported in a previous study (28). The high concentrations of succinic acid from 48 h to the end of the fermentation are likely related to the metabolic activity of the LAB, since these bacteria have shown the capability to produce succinic acid from citrate fermentation or convert fumaric and malic acids to succinic acid (29, 30). The reduction in pH in the pulp caused by LAB producing lactic acid favors the growth of AAB species, such as A. pasteurianus, which are capable of producing acetic and malic acids (29, 31).
Biochemical reactions play key roles in the formation of VOCs in fermented cocoa beans (22, 32). In our study, we observed that the dynamics of VOCs during fermentation changed in the concentration, as did their composition. According to Koné et al. (33), P. kudriavzevii and S. cerevisiae are the most important producers and contributors of cocoa aroma compounds, and these are followed by Wickerhamomyces anomalus, Geotrichum spp., and Pichia galeiformis. In our study, desirable cocoa aroma compounds, such as 2-heptanol, ethyl acetate, and 2-phenylethanol, were found in both fermentation processes, as previously identified by Ramos et al. (6). The principal producers of alcohol, ester, and acid compounds have been linked to yeasts, such as S. cerevisiae, Candida spp., and to other yeast species that have not been identified in this study on fermented cocoa beans (33–35). Apart from the production of VOCs by fungi, AAB are known to oxidize alcohols, such as ethanol, isoamyl alcohol, and 2-phenylethanol, to produce acids and acetaldehydes (36, 37).
We observed that the main bacterial group found in our study increased the concentration of succinic, acetic, and lactic acids, acetoin, alcohols, esters, and acetaldehydes. Overall, the biochemical contribution to food ecosystems might change according to the complexity of the microbial consortia (38). Therefore, further research is needed to understand the role of other compounds, such as free amino acids, oligopeptides, and polyphenols, in the development of microbes and aroma compounds (3, 22).
Conclusion.
Overall, the polyphasic approach applied in this study has allowed us to obtain new insights into the microbial development and aroma formation that take place during cocoa fermentation. Here, we observed that the starter culture modulated the microbiota composition of fermented cocoa beans and only marginally affected the metabolites, which were influenced more by the type of process that was carried out. Accordingly, the difference found between box and heap fermentations might be explained by considering the environmental and processing conditions, in which the microenvironment of each process plays an important role. The application of the omics approach has confirmed that fermented cocoa beans have complex microbial communities that are dominated by restricted bacterial and yeast populations. Future research is needed to assess how fermentation methods or the presence of the starter cultures can affect the final characteristics of chocolate.
MATERIALS AND METHODS
Cocoa bean fermentations.
The lyophilized S. cerevisiae ID67 and T. delbrueckii ID103 strains were provided by Lallemand (Montreal, Quebec, Canada) and were used as starter cultures in farmer-scale cocoa bean fermentations carried out in Ngoumou (Yaoundé, Cameroon) at the end of the mid-crop in 2016 (September to October 2016). The strains were chosen according to the study by Visintin et al. (9). Briefly, cocoa pods of the forastero hybrid were harvested by traditional methods and stored on the ground for 2 to 3 days before opening the pods. The cocoa pods were cut with nonsterile machetes, and the beans and the adhering pulp were removed by hand. Approximately 3 h after breaking the pods, the cocoa bean pulp was grouped into two independent lots (for the box and heap processes). Approximately 200 kg of fresh cocoa bean pulp was used for the B fermentation; it was placed in a wooden box (0.06 m3), covered with banana leaves, and closed with a wooden lid to protect it from the open air. The heap fermentations were set up with smaller amounts of beans than the box fermentations, due to the fact that an adult can manually turn no more than 100 kg of bean pulp. These beans were piled on top of banana leaves and covered with other banana leaves and jute rags. The field experiment involved inoculating the cocoa bean pulp with S. cerevisiae ID67 (S) or with S. cerevisiae ID67 in cocultures with T. delbrueckii ID103 (ST) in a 1:1 ratio (wt/vol) at the beginning of both fermentation processes (B and H). The lyophilized starter cultures were revitalized in a sterile saline solution for 30 min at room temperature and were progressively added and mixed with the cocoa pulp mass to final concentrations of 7.0 ± 0.2 log CFU · g−1. Moreover, noninoculated fermentations were carried out, without adding any starter culture to either fermentation process (B and H), and were used as a control. All trials were performed in duplicate (n = 12), according to the local agricultural practices; the cocoa bean pulp mass was turned manually at 48 and 96 h, and the fermentations were stopped after 120 h by spreading the beans on a drying platform. An aliquot of 1 to 1.5 kg of cocoa pulp was collected in sterile bags after 0, 48, 96, and 120 h for each of the six experimental trials. The pulp was taken randomly from at least five different zones of the fermentative mass in both the B and H fermentations. It should be noted that sampling was performed at 48 and 96 h before mixing the mass. Approximately 20 g of sample was collected, stored at −20°C, and transported on dry ice to the Department of Agriculture, Forestry and Food Sciences (University of Turin, Italy) for further metabolite analysis. Aliquots of 25 g of each sample were subjected to microbiological analyses at an experimental laboratory that had been set up on site. The pH values and temperatures were measured at the same sampling times during fermentation considering an average of five random zones of the cocoa bean pulp mass, and using a pH thermometer (Crison, Modena, Italy).
Culture-dependent microbial community dynamics.
A classical microbiological analysis was performed on samples recovered at 0, 48, 96, and 120 h. Twenty-five grams of cocoa beans and the adhering pulp were homogenized with 225 ml of Ringer's solution (Oxoid, Milan, Italy). Decimal dilutions were prepared in quarter-strength Ringer's solution. Aliquots of 0.1 ml of the appropriate dilutions were spread in triplicate on the following media: WL nutrient agar (WLN; Lab M, Heywood, Lancashire, UK) plus 1 μg/ml tetracycline (Sigma-Aldrich, Milan, Italy) to count the total yeasts incubated for 5 days at 30°C; de Man-Rogosa-Sharpe (MRS) agar (Oxoid) plus 2 μg/ml natamycin (Sigma-Aldrich) for the growing LAB, incubated at 30°C for 48 h; and acetic acid medium (1% glucose, 0.8% yeast extract, 0.5% bacteriological peptone, 15 g/liter agar, 0.5% ethanol, 0.3% acetic acid) plus 2 μg/ml of natamycin (Sigma-Aldrich) for the growing acetic acid bacteria (AAB) incubated at 30°C for 5 days. The results obtained from three independent determinations were expressed as the means of the log CFU per gram. Yeast colonies (5 to 8 for each sampling point) were randomly isolated from the plate with the highest WLN dilution. These colonies were further purified by streaking and were then stored in 20% (vol/vol) glycerol. A 1-ml aliquot of the first 10-fold serial dilution was collected at each sampling and centrifuged at the maximum speed for 30 s.
Assessment of the yeast ecology by means of culture-dependent analysis.
DNA extraction from single isolates was performed as described by Cocolin et al. (39) and normalized at 100 ng · liter−1. Isolates were grouped in relation to their restriction fragment length polymorphism (RFLP) profiles, which were obtained after enzymatic restriction of the amplified ITS-5.8S rDNA region, as previously described by Korabecná et al. (40). The ITS-5.8S rDNA region of at least three representative isolates of each RFLP group was used for sequencing (GATC Biotech, Cologne, Germany). An REP-PCR assay was performed on all the isolates previously identified as S. cerevisiae and T. delbrueckii, according to the procedure outlined in a previous study by Visintin et al. (9). A starter culture from the REP-PCR profiles was compared with those of S. cerevisiae ID67 and T. delbrueckii ID103.
Chemical analysis.
Fermented lyophilized bean pulp samples (0.20 g) were washed with 2 ml of pure hexane (Sigma-Aldrich, Milan, Italy) and vortexed for 5 min. The homogenate was centrifuged (6,000 × g, 4°C, 15 min), and the supernatant was removed. The washing process was repeated twice, and the precipitate was dried after the washings and resuspended with 10 ml of a 70:29.5:0.5 acetone–Milli-Q water–formic acid solution (Sigma-Aldrich). The solution was vortexed, centrifuged, and clarified by filtration through 0.45-μm syringe filters (LLG-Labware, CA, USA) and then evaporated. The extract was resuspended with 5 ml of Milli-Q water and passed through a C18 cartridge (Sep-Pack, USA). The column was washed with 5 ml of Milli-Q water to recover the samples.
The HPLC system (ThermoQuest Corporation, San Jose, CA, USA) was equipped with an isocratic pump (P1000), a multiple autosampler (AS3000) fitted with a 20-μl loop, a UV detector (UV100) set at 210 nm, and a refractive index detector (Spectra system RI-150; Thermo Electro Corporation). The analyses of the sugars (glucose, fructose, and sucrose) were performed isocratically, at 0.6 ml · min−1 and 80°C, with a 300 by 7.8 mm inner diameter (i.d.) cation exchange column (Aminex HPX-87P) equipped with a cation Carbo-P Micro-Guard cartridge (Bio-Rad Laboratories, Hercules, CA, USA). The analyses of the organic acids (acetic, lactic, malic, succinic, oxalic, gluconic, tartaric, pyruvic, fumaric, and citric acid) were performed isocratically, at 0.8 ml · min−1 and 60°C, with a 300 by 7.8 mm i.d. cation exchange column (Aminex HPX-87H) equipped with a Cation H+ Micro-Guard cartridge (Bio-Rad Laboratories). The data treatments were carried out using the ChromQuest chromatography data system (ThermoQuest, Inc.). Analytical-grade reagents were used as standards (Sigma-Aldrich, St. Louis, MO). All the samples of each biological replicate were analyzed in triplicate, and the identification of compounds was performed by comparing the retention time against the standard. The calibration curves of the standards were obtained by injecting serial dilutions of glucose, sucrose, fructose, and acetic, lactic, malic, succinic, oxalic, gluconic, tartaric, pyruvic, fumaric, and citric acid, under the same conditions as the sample analyses. The concentrations of the compounds were calculated by plotting a linear curve of the areas obtained in each sample.
Volatile metabolites produced by the microbiota consortia.
The dynamics of the volatile organic compounds (VOCs) of the fermented cocoa bean pulp were obtained under different previously lyophilized conditions using the headspace solid-phase microextraction (HS-SPME) technique, in which the fiber conditions and oven temperatures were set as previously described by Rodriguez-Campos et al., with some modifications (41). Samples of each biological replicate were analyzed in triplicate. The analysis was conducted in a 20-ml vial filled with 2 ml of 20% NaCl and 0.1 g of the sample, and 10 μl of 5-nonanol in ultrapure water was added to each sample at a 50 mg/liter concentration as an internal standard for the semiquantification. The fibers with VOCs were injected into the gas chromatograph-quantitative mass spectrometer (GC-qQP2010 Plus; Shimadzu, USA), which was equipped with an autosampler (AOC-5000, PAL system; CombiPAL, Switzerland) and a DB-WAXETR capillary column (30 m by 0.25 mm, 0.25-μm film thickness; J&W Scientific, Inc., Folsom, CA). The injection mode was established at 260°C (1 min), and helium was used, at a constant flow rate of 1 ml/min, as the carrier gas. The detection was carried out by means of the electron impact mass spectrometer in total ion current mode, using an ionization energy of 70 eV. The acquisition range was set at m/z 33 to 350 atomic mass units (amu). The peaks were identified by comparing the mass spectra of the peaks with the spectra of the MIST05 library and through a comparison of the retention indices (a matrix of a homologous series of C8 to C24 was used) with an injected pure standard under the same sample conditions described above. Semiquantitative data (micrograms per kilogram) were obtained by measuring the characteristic m/z peak area of each identified compound in relation to the added internal standard.
Statistical analyses.
Statistical analyses were carried out using generalized linear mixed-effect models for a nonnormally distributed data set. Mixed models were chosen because of their ability to capture both fixed (fermentation condition, inoculated with S, inoculated with ST, and noninoculated; fermentation time, 0 to 120 h) and random effects (fermentation types, B and H) (42). The P values were adjusted using Bonferroni's method and, when the linear mixed model revealed significant differences (P < 0.05), the Duncan honestly significant difference (HSD) test was applied. Mixed models were built and evaluated according to Crawley (43) using R version 3.3.2. The assessment of the mean difference between the box and heap fermentations over a specific fermentation period was subjected to a t test, in which each fermentation condition was compared between fermentation methods (B and H). In addition, Spearman's correlation test was used to assess the correlations between the OTUs and to establish any changes in concentration over the fermentation period.
DNA extraction, library preparation, and sequencing.
The total DNA was extracted from pellets of the cocoa matrices using a MasterPure complete DNA and RNA purification kit (Illumina, Inc., San Diego, CA), according to the manufacturer's instructions. Bacterial communities were studied by amplifying the V3 and V4 regions of 16S rRNA using the primers and under the conditions described by Klindworth et al. (44). The yeast communities were studied by amplifying the ITS2 region using ITS3tagmix (5′-CTAGACTCGTCACCGATGAAGAACGCAG) and ITS-4ngs (5′-TTCCTSCGGCTTATTGATATGC) (45). The PCR products were purified twice by means of an Agencourt AMPure kit (Beckman Coulter, Milan, Italy), and the resulting products were tagged using a Nextera XT index kit (Illumina), according to the manufacturer's instructions. After the second clean-up step with the Agencourt AMPure kit, a 4 nM pool was obtained in which the weight of the library and measured by means of Qubit fluorometric quantitation (Thermo Fisher Scientific), and the mean amplicon size was taken into account. A denatured 20 pM pool was obtained by mixing 5 μl of 0.2 N NaOH with 5 μl of the 4-nm pool. A final 10 pM library was combined with 10% PhiX. Sequencing was performed using a MiSeq instrument (Illumina) with V3 chemistry, according to the manufacturer's instructions, and 250-bp paired-end reads were generated.
Bioinformatics.
The obtained paired-end reads were first assembled with the FLASH software (46), with default parameters. The joint reads were further quality filtered (Phred < Q20) using the QIIME 1.9.0 software (47). Reads shorter than 250 bp were discarded using Prinseq. For the 16S data, the OTUs were picked at a 99% of similarity threshold, and centroid sequences of each cluster were used to assign the taxonomy by mapping against the Greengenes 16S rRNA gene database, version 2013, as recently described (48). The chloroplast and mitochondrial sequences were removed from the data set. For the ITS data set, 97% similarity was picked for the OTUs, by means of UCLUST clustering methods (49), and representative sequences of each cluster were used to assign the taxonomy using the UNITE rDNA ITS database, version 2012, by means of the RDP Classifier. Weighted and unweighted UniFrac distance matrices, as well as the OTU table, were used to find differences between the fermentation processes (B and H) and under different conditions (inoculated and noninoculated) in the Adonis and analysis of similarity (ANOSIM) statistical test in the R environment in order to avoid biases due to different sequencing depths. All of the samples of each data set were rarefied at the lowest number of reads after raw read quality filtering. QIIME was used to produce a filtered OTU table at 1% in at least 2 samples. The OTU table displays the highest taxonomy resolution reached when the taxonomy assignment was not able to reach the species level or when the genus or family name was displayed. The Kruskal-Wallis statistical package and Mann-Whitney tests were used to find significant differences (P < 0.05) in the microbial taxon abundance profiles and in the Shannon-Wiener diversity index (H′), according to the time, conditions, and methods. Spearman's rank correlation coefficient was obtained as a measure of the association between the microbial OTUs that occurred in at least 2 samples and the chemical variables through the psych function and plotted through the corrplot package of R. The OTUs that occurred in at least 2 samples of the microbial communities were conglomerated, by means of hierarchical clustering analysis, using Ward's method, which was acquired thorough the heatplot function plotted by the made4 package of R.
Accession number(s).
The 16S and ITS rRNA gene sequences are available at the NCBI Sequence Read Archive under accession numbers SRP126069 and SRP126081, respectively.
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01164-18.
REFERENCES
- 1.Beg MS, Ahmad S, Jan K, Bashir K. 2017. Status, supply chain and processing of cocoa. Trends Food Sci Technol 66:108–116. doi: 10.1016/j.tifs.2017.06.007. [DOI] [Google Scholar]
- 2.Reference deleted. [Google Scholar]
- 3.Thompson SS, Miller KB, Lopez AS, Camu N. 2001. Cocoa and coffee, p 721–733. In Doyle MP, Buchanan RL (ed), Food microbiology: fundamentals and frontiers. American Society for Microbiology, Washington, DC. [Google Scholar]
- 4.Schwan RF, Fleet GH. 2014. Cocoa and coffee fermentations. CRC Press, Boca Raton, FL. [Google Scholar]
- 5.Batista NN, Ramos CL, Ribeiro DD, Pinheiro ACM, Schwan RF. 2015. Dynamic behavior of Saccharomyces cerevisiae, Pichia kluyveri and Hanseniaspora uvarum during spontaneous and inoculated cocoa fermentations and their effect on sensory characteristics of chocolate. LWT-Food Sci Technol 63:221–227. doi: 10.1016/j.lwt.2015.03.051. [DOI] [Google Scholar]
- 6.Ramos CL, Dias DR, Miguel MGDCP, Schwan RF. 2014. Impact of different cocoa hybrids (Theobroma cacao L.) and S. cerevisiae UFLA CA11 inoculation on microbial communities and volatile compounds of cocoa fermentation. Food Res Int 64:908–918. doi: 10.1016/j.foodres.2014.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Ferrocino I, Cocolin L. 2017. Current perspectives in food-based studies exploiting multi-omics approaches the effects of time pressure. Curr Opin Food Sci 13:10–15. doi: 10.1016/j.cofs.2017.01.002. [DOI] [Google Scholar]
- 8.Visintin S, Alessandria V, Valente A, Dolci P, Cocolin L. 2016. Molecular identification and physiological characterization of yeasts, lactic acid bacteria and acetic acid bacteria isolated from heap and box cocoa bean fermentations in West Africa. Int J Food Microbiol 216:69–78. doi: 10.1016/j.ijfoodmicro.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Visintin S, Ramos L, Batista N, Dolci P, Schwan F, Cocolin L. 2017. Impact of Saccharomyces cerevisiae and Torulaspora delbrueckii starter cultures on cocoa beans fermentation. Int J Food Microbiol 257:31–40. doi: 10.1016/j.ijfoodmicro.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Crafack M, Mikkelsen MB, Saerens S, Knudsen M, Blennow A, Lowor S, Takrama J, Swiegers JH, Petersen GB, Heimdal H, Nielsen DS. 2013. Influencing cocoa flavour using Pichia kluyveri and Kluyveromyces marxianus in a defined mixed starter culture for cocoa fermentation. Int J Food Microbiol 167:103–116. doi: 10.1016/j.ijfoodmicro.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 11.Pereira GVM, Alvarez JP, Neto DPDC, Soccol VT, Tanobe VOA, Rogez H, Góes-Neto A, Soccol CR. 2017. Great intraspecies diversity of Pichia kudriavzevii in cocoa fermentation highlights the importance of yeast strain selection for flavor modulation of cocoa beans. LWT-Food Sci Technol 84:290–297. doi: 10.1016/j.lwt.2017.05.073. [DOI] [Google Scholar]
- 12.Meersman E, Steensels J, Struyf N, Paulus T, Saels V, Mathawan M, Allegaert L, Vrancken G, Verstrepen KJ. 2016. Tuning chocolate flavor through development of thermotolerant Saccharomyces cerevisiae starter cultures with increased acetate ester. Appl Environ Microbiol 82:732–746. doi: 10.1128/AEM.02556-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arana-Sánchez A, Segura-García LE, Kirchmayr M, Orozco-Ávila I, Lugo-Cervantes E, Gschaedler-Mathis A. 2015. Identification of predominant yeasts associated with artisan Mexican cocoa fermentations using culture-dependent and culture-independent approaches. World J Microbiol Biotechnol 31:359–369. doi: 10.1007/s11274-014-1788-8. [DOI] [PubMed] [Google Scholar]
- 14.Fernández Maura Y, Balzarini T, Clapé Borges P, Evrard P, De Vuyst L, Daniel HM. 2016. The environmental and intrinsic yeast diversity of Cuban cocoa bean heap fermentations. Int J Food Microbiol 233:34–43. doi: 10.1016/j.ijfoodmicro.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 15.de Melo Pereira GV, Magalhaes KT, de Almeida EG, da Silva Coelho I, Schwan RF. 2013. Spontaneous cocoa bean fermentation carried out in a novel-design stainless steel tank: influence on the dynamics of microbial populations and physical-chemical properties. Int J Food Microbiol 161:121–133. doi: 10.1016/j.ijfoodmicro.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 16.da Veiga Moreira IM, Miguel MGDCP, Duarte WF, Dias DR, Schwan RF. 2013. Microbial succession and the dynamics of metabolites and sugars during the fermentation of three different cocoa (Theobroma cacao L.) hybrids. Food Res Int 54:9–17. doi: 10.1016/j.foodres.2013.06.001. [DOI] [Google Scholar]
- 17.Camu N, De Winter T, Addo SK, Takrama JS, Bernaert H, De Vuyst L. 2008. Fermentation of cocoa beans: influence of microbial activities and polyphenol concentrations on the flavour of chocolate. J Sci Food Agric 88:2288–2297. doi: 10.1002/jsfa.3349. [DOI] [Google Scholar]
- 18.Papalexandratou Z, Lefeber T, Bahrim B, Lee OS, Daniel HM, De Vuyst L. 2013. Hanseniaspora opuntiae, Saccharomyces cerevisiae, Lactobacillus fermentum, and Acetobacter pasteurianus predominate during well-performed Malaysian cocoa bean box fermentations, underlining the importance of these microbial species for a successful cocoa bean fermentation process. Food Microbiol 35:73–85. doi: 10.1016/j.fm.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH. 2007. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol 114:168–186. doi: 10.1016/j.ijfoodmicro.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Lefeber T, Papalexandratou Z, Gobert W, Camu N, De Vuyst L. 2012. On-farm implementation of a starter culture for improved cocoa bean fermentation and its influence on the flavour of chocolates produced thereof. Food Microbiol 30:379–392. doi: 10.1016/j.fm.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 21.Papalexandratou Z, De Vuyst L. 2011. Assessment of the yeast species composition of cocoa bean fermentations in different cocoa-producing regions using denaturing gradient gel electrophoresis. FEMS Yeast Res 11:564–574. doi: 10.1111/j.1567-1364.2011.00747.x. [DOI] [PubMed] [Google Scholar]
- 22.Lima LJ, Almeida MH, Nout MJ, Zwietering MH. 2011. Theobroma cacao L., “the food of the gods”: quality determinants of commercial cocoa beans, with particular reference to the impact of fermentation. Crit Rev Food Sci Nutr 51:731–761. doi: 10.1080/10408391003799913. [DOI] [PubMed] [Google Scholar]
- 23.Axelsson L. 2004. Lactic acid bacteria: classification and physiology. Marcel Dekker, New York, NY. [Google Scholar]
- 24.Deák T. 2007. Handbook of food spoilage yeasts. CRC Press, Boca Raton, FL. [Google Scholar]
- 25.De Vuyst L, Weckx S. 2016. The cocoa bean fermentation process: from ecosystem analysis to starter culture development. J Appl Microbiol 121:5–17. doi: 10.1111/jam.13045. [DOI] [PubMed] [Google Scholar]
- 26.Jespersen L, Nielsen DS, Hønholt S, Jakobsen M. 2005. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Res 5:441–453. doi: 10.1016/j.femsyr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Daniel HM, Vrancken G, Takrama JF, Camu N, De Vos P, De Vuyst L. 2009. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res 9:774–783. doi: 10.1111/j.1567-1364.2009.00520.x. [DOI] [PubMed] [Google Scholar]
- 28.Lefeber T, Janssens M, Camu N, De Vuyst L. 2010. Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media toward development of a starter culture for cocoa bean fermentation. Appl Environ Microbiol 76:7708–7716. doi: 10.1128/AEM.01206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camu N, De Winter T, Verbrugghe K, Cleenwerck I, Vandamme P, Takrama JS, Vancanneyt M, De Vuyst L. 2007. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl Environ Microbiol 73:1809–1824. doi: 10.1128/AEM.02189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouattara HD, Ouattara HG, Droux M, Reverchon S, Nasser W, Niamke SL. 2017. Lactic acid bacteria involved in cocoa beans fermentation from Ivory Coast: species diversity and citrate lyase production. Int J Food Microbiol 256:11–19. doi: 10.1016/j.ijfoodmicro.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Bartowsky EJ, Henschke PA. 2008. Acetic acid bacteria spoilage of bottled red wine—a review. Int J Food Microbiol 125:60–70. doi: 10.1016/j.ijfoodmicro.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Afoakwa EO, Paterson A, Fowler M, Ryan A. 2017. Flavor formation and character in cocoa and chocolate: a critical review. Crit Rev Food Sci Nutr 48:840–857. doi: 10.1080/10408390701719272. [DOI] [PubMed] [Google Scholar]
- 33.Koné MK, Guéhi ST, Durand N, Ban-koffi L, Berthiot L, Tachon AF, Brou K, Boulanger R, Montet D. 2016. Contribution of predominant yeasts to the occurrence of aroma compounds during cocoa bean fermentation. Food Res Int 89:910–917. doi: 10.1016/j.foodres.2016.04.010. [DOI] [Google Scholar]
- 34.Schwan RF, Wheals AE. 2004. The microbiology of cocoa fermentation and its role in chocolate quality. Crit Rev Food Sci Nutr 44:205–221. doi: 10.1080/10408690490464104. [DOI] [PubMed] [Google Scholar]
- 35.Kim B, Cho BR, Hahn JS. 2014. Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol Bioeng 111:115–124. doi: 10.1002/bit.24993. [DOI] [PubMed] [Google Scholar]
- 36.Molinari F, Gandolfi R, Aragozzini F, Leon R, Prazeres DMF. 1999. Biotransformations in two-liquid-phase systems: production of phenylacetaldehyde by oxidation of 2-phenylethanol with acetic acid bacteria. Enzyme Microb Technol 25:729–735. doi: 10.1016/S0141-0229(99)00107-6. [DOI] [Google Scholar]
- 37.Yakushi T, Matsushita K. 2010. Alcohol dehydrogenase of acetic acid bacteria: structure, mode of action, and applications in biotechnology. Appl Microbiol Biotechnol 86:1257–1265. doi: 10.1007/s00253-010-2529-z. [DOI] [PubMed] [Google Scholar]
- 38.Fleet GH. 1999. Microorganisms in food ecosystems. Int J Food Microbiol 50:101–117. doi: 10.1016/S0168-1605(99)00080-X. [DOI] [PubMed] [Google Scholar]
- 39.Cocolin L, Rantsiou K, Iacumin L, Urso R, Cantoni C, Comi G. 2004. Study of the ecology of fresh sausages and characterization of populations of lactic acid bacteria by molecular methods. Appl Environ Microbiol 70:1883–1894. doi: 10.1128/AEM.70.4.1883-1894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korabecná M, Liška V, Fajfrlík K. 2003. Primers ITS1, ITS2 and ITS4 detect the intraspecies variability in the internal transcribed spacers and 58S rRNA gene region in clinical isolates of fungi. Folia Microbiol (Praha) 48:233–238. doi: 10.1007/BF02930961. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Campos J, Escalona-Buendía HB, Orozco-Avila I, Lugo-Cervantes E, Jaramillo-Flores ME. 2011. Dynamics of volatile and non-volatile compounds in cocoa (Theobroma cacao L.) during fermentation and drying processes using principal components analysis. Food Res Int 44:250–258. doi: 10.1016/j.foodres.2010.10.028. [DOI] [Google Scholar]
- 42.Baayen RH, Davidson DJ, Bates DM. 2008. Mixed-effects modeling with crossed random effects for subjects and items. J Mem Lang 59:390–412. doi: 10.1016/j.jml.2007.12.005. [DOI] [Google Scholar]
- 43.Crawley MJ. 2007. The R book. John Wiley and Sons, Ltd., West Sussex, England. [Google Scholar]
- 44.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:1–11. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tedersoo L, Anslan S, Bahram M, Põlme S, Riit T, Liiv I, Kõljalg U, Kisand V, Nilsson H, Hildebrand F, Bork P, Abarenkov K. 2015. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10:1–43. doi: 10.3897/mycokeys.10.4852. [DOI] [Google Scholar]
- 46.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrocino I, Bellio A, Romano A, Macori G, Rantsiou K, Decastelli L, Cocolin L. 2017. RNA-based amplicon sequencing reveals microbiota development during ripening of artisanal versus industrial Lard d'Arnad. Appl Environ Microbiol 83:e00983-17. doi: 10.1128/AEM.00983-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.