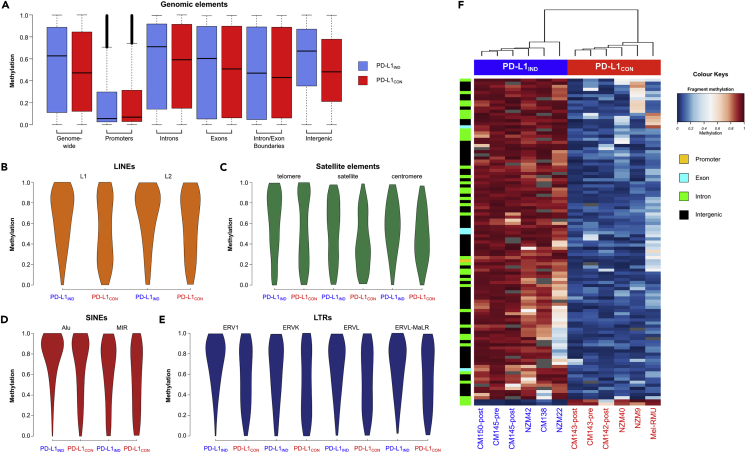
Figure 2.
Whole-Genome-Scale and Element-Wise Methylation Profiles in PD-L1IND and PD-L1CON Cell Lines
(A) Boxplots showing genome-wide and genomic element RRBS methylation profiles for PD-L1IND (blue) and PD-L1CON (red) cell lines; black bars indicate the median methylation.
(B–E) Equal-area violin plots of PD-L1CON and PD-L1IND DNA methylation levels for different classes of repeat elements. (B) LINE elements (L1 and L2), (C) Satellite elements (satellite, telomeric, and centromeric repeats), (D) SINE elements (Alu and MIR), and (E) LTRs (ERV1, ERVK, ERVL, and ERVL-MaLR). In all cases the y axis represents the methylation level on a 0–1 scale. Annotations for repeat elements were downloaded from the UCSC repeat masker database.
(F) Methylation levels for the 105 differentially methylated fragments (DMFs) showing >70% methylation difference between the PD-L1IND and PD-L1CON cell lines (blue = unmethylated, red = fully methylated).
See also Figures S4–S9 and Tables S2–S4. The methylation data are available at Database: NCBI GEO, accession number GSE107622.