Summary
The transcriptional co-activator p300 is essential for p53 transactivation, although its precise role remains unclear. We report that p53 activates the acetyltransferase activity of p300 through the enhancement of p300 autoacetylation. Autoacetylated p300 accumulates near the transcription start sites accompanied by a similar enrichment of activating histone marks near those sites. Abrogation of p53-p300 interaction by a site-directed peptide inhibitor abolished p300-mediated histone acetylation, suggesting a crucial role played by the activation in p53-mediated gene regulation. Gain-of-function mutant p53, known to impart aggressive proliferative properties in tumor cells, also activates p300 autoacetylation. The same peptide abolished many of the gain-of-function properties of mutant p53 as well. Reversal of gain-of-function properties of mutant p53 suggests that molecules targeting the p53-p300 interface may be good candidates for anti-tumor drugs.
Subject Areas: Molecular Biology, Molecular Mechanism of Gene Regulation, Cancer
Graphical Abstract

Highlights
-
•
Wild-type and mutant p53 are potent inducers of p300 autoacetylation
-
•
p53 activates p300 catalytic activity by altering its structural conformation
-
•
Induction of p300 autoacetylation possibly enhances p53-targeted gene expression
-
•
Mutant-p53-induced p300 autoacetylation could be critical for tumorigenicity
Molecular Biology; Molecular Mechanism of Gene Regulation; Cancer
Introduction
Organismic complexity scales with the intricacies and sophistication of gene regulation. How this intricate regulation is achieved, however, is not fully understood. Specificity, both spatial and temporal, is at the heart of this extraordinarily elaborate and complex regulatory process (Levine and Tjian, 2003). Protein-protein interactions among multiple transcription factors bound to adjacent sequences, DNA-mediated cooperativity, and other factors have been found to enhance site selectivity and to initiate the assembly of multi-protein regulatory complexes. After assembly the regulatory complex initiates the recruitment of co-activators, chromatin modifiers, and other factors, thereby increasing chromatin accessibility and the initiation of RNA synthesis (Dulac, 2010, Schneider and Grosschedl, 2007). It is not known whether any steps beyond the assembly of regulatory complexes influence the spatial and temporal specificity of gene regulation.
p300 (KAT3B) is a transcriptional co-activator with intrinsic acetyltransferase activity. Being a large adaptor protein, p300 bridges the basal transcription machinery to DNA sequence-specific transcription factors (Chan and La Thangue, 2001, Ogryzko et al., 1996). By virtue of its structural plasticity, p300 can interact with the intrinsically disordered transactivation domains of a vast array of transcription factors, thereby functioning as a nodal integrator of signals terminating in the regulation of transcription (Bedford et al., 2010, Dyson and Wright, 2016, Lee et al., 1998, Perissi et al., 1999). Therefore, its recruitment via the interaction with sequence-specific transcription factors can regulate transcription from the chromatin template (Kundu et al., 2000). This protein is often recruited to assembled regulatory complexes, where it acetylates histone tails in the vicinity. In this way, p300 promotes localized chromatin accessibility and, subsequently, selective transcriptional initiation (Vo and Goodman, 2001).
The basal catalytic activity of p300 is enhanced by trans-autoacetylation of its lysine-rich regulatory loop (Thompson et al., 2004). It had been proposed that, in the absence of acetylation, the positively charged lysine-rich auto-inhibitory loop folds back onto the enzyme active site, thus hindering optimal substrate-enzyme interaction. Since p300 is involved in diverse physiological roles, the precise regulation of its function is essential. It has been demonstrated that some factors affect the levels of p300 autoacetylation in the cell. Intriguingly, these factors have distinct cellular functions and induce p300 autoacetylation under different cellular contexts. The Anaphase Promoting Complex/Cyclosome (APC/C) subunits APC5 and APC7 are capable of enhancing p300 autoacetylation, which is required for proper cell cycle progression (Turnell et al., 2005). Similarly, the transcription factor MAML1 has been shown to increase the levels of p300 autoacetylation, thereby inducing the transcription of Notch pathway genes (Hansson et al., 2009). During cellular stress, GAPDH associates with the E3 ligase Siah1 and shuttles to the nucleus, where it enhances p300 autoacetylation, which in turn activates the p53-mediated apoptotic pathway (Sen et al., 2008). p300 autoacetylation is negatively regulated by the class III lysine deacetylase SIRT2, and as expected, the suppression of p300 autoacetylation leads to the reduction of its enzymatic activity (Black et al., 2008). We have previously reported the presence of global histone hyperacetylation in oral cancer. Mechanistically, this abnormality is a consequence of enhanced p300 autoacetylation, which is induced by NPM1 and nuclear GAPDH in a nitric oxide (NO) signaling-dependent manner. Ac-p300 may be the driving force that causes alterations in the epigenetic landscape and, consequently, the global deregulation of transcription that is required for oral carcinogenesis (Arif et al., 2010). Thus, modulation of p300 autoacetylation may be a key factor in regulating cellular responses.
p300 is a co-activator of the tumor suppressor p53 and acetylates it on several lysine residues, mainly on its C-terminal domain (Gu and Roeder, 1997, Lill et al., 1997). Studies have attributed the enhancement of DNA sequence-specific binding ability of p53 to the p300-mediated acetylation, whereas contradictory findings have shown that p53 C-terminal acetylation may not have a profound effect on its promoter binding (Barlev et al., 2001, Espinosa and Emerson, 2001). Nevertheless, accumulation of acetylated p53 has been observed during cellular stresses (Reed and Quelle, 2014, Sakaguchi et al., 1998) and p53 acetylation has been studied extensively with respect to cell cycle arrest, apoptosis, senescence, and ferroptosis (Bieging et al., 2014, Jiang et al., 2015). Studies using the acetylation-defective mutants of p53 have shown that the acetylation of p53 is not required for its DNA binding, rather it is crucial for the recruitment of co-activators such as p300, which are essential for p53 transactivation function (Barlev et al., 2001), thereby reiterating the fact that the interaction between p300 and p53 is functionally important. p53 interacts with p300 in its tetrameric conformation, through its bipartite transactivation domain AD1 and AD2, whereas p300 interacts through several domains, namely, TAZ1, KIX, TAZ2, and IBiD (Krois et al., 2016, Teufel et al., 2007). Intriguingly, numerous factors such as E1A, DDX24, WTX, MYBB1A, and Skp2, can regulate p53 tumor suppressive functions in vivo by dictating its association with p300, thereby proving that the p300-p53 axis is an important cell fate determinant in stress responses (Kim et al., 2012, Kitagawa et al., 2008, Kumazawa et al., 2011, Lill et al., 1997, Shi et al., 2016, Vasudevarao et al., 2014). The p53 gene acquires several mutations that abolish its tumor suppressive functions and confer oncogenic gain-of-function (GOF) properties that contribute to tumorigenesis. Approximately 97% of the missense mutations map to the DNA-binding domain (DBD), of which a small number occur in high frequency in cancers (Freed-Pastor and Prives, 2012). GOF p53 mutants induce aberrant transactivation in cancers through the recruitment of p300, but the effect of mutant p53 on p300 catalytic activity is yet unknown. In the present study, we addressed the mechanism of factor-induced p300 autoacetylation and its importance in p53-dependent gene regulation.
We found that the master tumor suppressor protein p53 specifically induces p300 autoacetylation. Comparison of the genomic distribution of ac-p300 and p300 before and after stimulation by p53 indicates that ac-p300 is specifically and uniquely present around transcription start sites (TSS) of p53-regulated genes. This study establishes the mechanism of p53-mediated induction of p300 autoacetylation and its chromatin recruitment at transcriptional regulatory elements. Our data indicate that GOF mutant p53 may execute its downstream oncogenic functions through the modulation of p300/CBP autoacetylation and function. The implications of these observations are further explored.
Results
p53 Directly Modulates p300 Autoacetylation and Acetyltransferase Activity
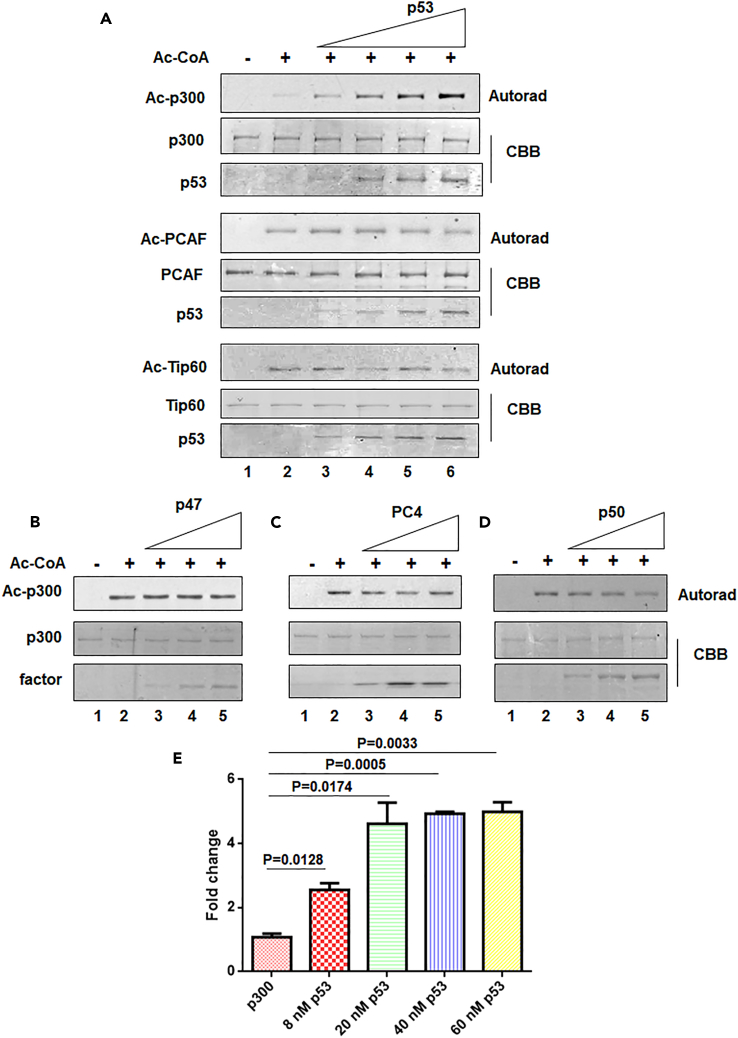
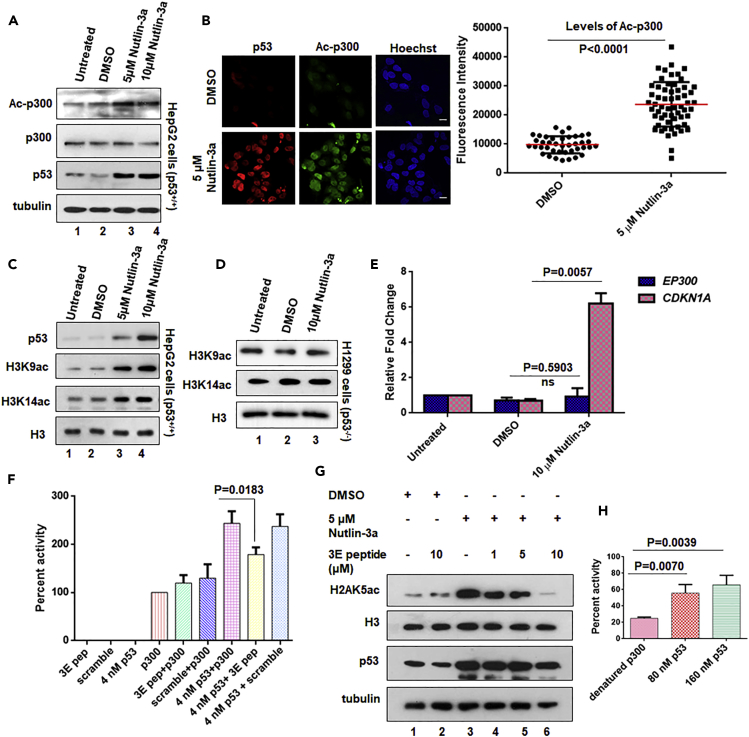
To explore whether p53 has an effect on the acetyltransferase activity of p300, an in vitro acetyltransferase assay was designed to detect the effect of p53 on the levels of p300 autoacetylation. p53 specifically induced the autoacetylation of 20 nM p300 in a concentration-dependent manner (ranging from 40 to 160 nM p53 monomers) but exhibited a negligible effect on the autoacetylation of the other two classes of lysine acetyltransferases (KATs), PCAF, and Tip60 (Figure 1A). An approximately 4.5-fold enhancement of p300 autoacetylation was observed in the presence of 160 nM p53 (2:1 ratio of p53 tetramers to p300). Because p53 forms a stable tetramer and may possibly interact with p300 as a tetramer (Teufel et al., 2007), a significant degree of p300 autoacetylation was reached only when p53 was present in a 1:1 ratio of tetrameric p53 to p300. At a suboptimal concentration (40 nM) of p53 (tetrameric p53 to p300 ratio of 0.5:1), a negligible increase in the autoacetylation was observed (Figure S1A). Moreover, p53 is a substrate of most major nuclear KATs, including p300/CBP (Gu and Roeder, 1997), PCAF (KAT2B) (Liu et al., 1999), TIP60 (KAT5) (Sykes et al., 2006, Tang et al., 2006), MOZ (KAT6A) (Rokudai et al., 2013), and MOF (KAT8) (Sykes et al., 2006). Therefore, the specificity exhibited by p53 toward the enhancement of p300 autoacetylation, with no effect seen for PCAF and Tip60, led us to conclude that p53 is not merely a substrate of p300 but a regulator of the intrinsic enzymatic activity of p300. This specific role of p53 was investigated further. Three different substrates of p300, p47 (N-terminal truncated [Δ40p53] isoform of p53, incapable of binding to p300), Positive Coactivator 4 [PC4], and p50 (a subunit of nuclear factor κB), were also tested and found not to effectively induce p300 autoacetylation (Figures 1B–1D). To investigate the effect of the p53-mediated induction of p300 autoacetylation, we performed an in vitro histone acetyltransferase assay using recombinant histones as the substrate. In agreement with the results of the autoacetylation assay, we observed an approximately 3-fold increase in p300 histone acetyltransferase activity at a p300:p53 molar ratio of 1:2. Notably, we observed an approximately 4.5-fold increase in p300 activity in the presence of p53, thus suggesting that p53 positively modulates p300 KAT activity through the induction of autoacetylation. Further increase in the p53 concentration did not affect the HAT activity beyond 4.5-fold (Figure 1E). The in vitro p53-mediated induction of p300 autoacetylation was further verified in the cellular context. Endogenous p53 levels were stabilized in HepG2 cells by using the small molecule inhibitor of the MDM2-p53 interaction, Nutlin-3a. The stabilization of p53 after Nutlin-3a treatment appeared to positively regulate the levels of p300 autoacetylation in the cells in comparison with cells treated or not treated with DMSO (Figure 2A). Furthermore, the fluorescence intensity due to p300 autoacetylation was significantly (p < 0.001) enhanced in HepG2 cells treated with Nutlin-3a expressing wild-type p53 (Figure 2B). We expected that hyper-autoacetylated p300 would induce p300-specific histone acetylation in cells. To address this possibility, histone acetylation at H3K9 and H3K14 were investigated in the presence and absence of Nutlin-3a (Figure 2C). It has been previously demonstrated that p53 acetylation and histone acetylation increase after Nutlin-3a treatment (Haaland et al., 2014). Therefore, to test for whether the enhancement in histone acetylation was due to the p53-mediated enhancement of p300 autoacetylation, we treated p53-null H1299 cells with Nutlin-3a and probed for the levels of different histone acetylation marks in treated versus control cells (Figure 2D). Interestingly, we found that the histone acetylation levels did not show any appreciable alteration in Nutlin-3a-treated H1299 cells, thereby signifying the importance of p53 in the activation of p300 KAT activity. To rule out the possibility that p300 might be transcriptionally regulated by p53 rather than post-translationally modulated, p300 transcript levels were compared between cells treated with Nutlin-3a and DMSO. No change in the levels of EP300 transcript was observed in the presence of p53 compared with CDKN1A (an early p53 target gene) transcripts, thus suggesting that p300 is not regulated by p53 at the transcriptional level (Figure 2E). Collectively, these results established p53 as a bona fide inducer of p300 autoacetylation and its acetyltransferase activity. To elucidate the mechanistic details of p53-mediated activation of p300, the importance of the direct interaction between p53 and p300 was investigated. p53 directly contacts p300 through four important residues (L22, W23, W53, F54) in its transactivation domain. Mutations in these residues severely compromise p53 transactivation (Teufel et al., 2007). After DNA damage, p53 is phosphorylated by several kinases, and phosphorylated p53 binds to its co-activator p300 with higher affinity (Lambert et al., 1998, Lee et al., 2010). Interestingly, the higher the number of phosphorylated sites on the p53 N-terminus, the higher its affinity for CBP/p300 (Lee et al., 2009, Lee et al., 2010). A triple-phospho-mimic peptide (S15E, T18E, S20E) comprising the 11th to 30th amino acid residue of the p53 (TAD1 domain) protein along with a cell penetrating sequence and nuclear localization sequence (NLS) was used to obstruct the interaction between p53 and p300 (Polley et al., 2008) (Figure S1B, Table S1). Using a filter-binding assay-based strategy, the importance of the p53-p300 interaction for the activation of p300 was assessed on the basis of the level of p300-mediated histone acetylation (Figure 2F). The peptides and p53 alone did not bind to the p81 phosphocellulose filters; therefore, the measured radioactivity was only from the tritium-labeled acetylated histones bound to the filters. The basal activity of p300 in the absence of any inducer was considered as 100% activity, and the peptides alone did not alter the activity of p300, thus signifying that any change in histone acetylation observed thereafter was a direct consequence of the p300-p53 interaction. In the presence of 4 nM p53, a 3-fold increase in the histone acetylation was observed. The presence of scrambled peptide did not alter the p53-mediated induction of p300 activity, but p53 phosphomimic peptides (3E pep) effectively decreased histone acetylation levels. These results indicated that the p53 phosphomimic peptide effectively interfered with the p53-p300 interaction, thus presumably leading to decreased p300 autoacetylation and activity, as indicated by a significant decrease in histone acetylation levels. The scrambled peptide exhibited a negligible effect on p53-mediated p300 activation, thus ensuring that the decrease observed in the case of the p53 phosphomimic peptide was indeed due to the disruption of the p53-p300 axis and was not a non-specific effect of the peptides (Figure 2F). Furthermore, to ascertain whether these peptides were effective in cells, HepG2 cells were treated with Nutlin-3a to stabilize the levels of p53. The cells were then treated with different concentrations of the p53 phosphomimic peptide, which has cell-penetrating sequences (Figure 2G). The levels of p53 increased after Nutlin-3a treatment, and the peptide treatment did not alter p53 protein levels. Therefore, the p53 peptide did not alter cellular p53 protein levels, and the observed alteration in histone acetylation is a direct consequence of the disruption of p53-p300 interaction. In agreement with the previous observation, H2AK5 acetylation increased on p53 stabilization with Nutlin-3a (Figures 2G and S1C). After treatment with the interfering peptide, the levels of H2AK5 acetylation gradually decreased in a concentration-dependent manner (Figure 2G), whereas H2AK5ac levels remained unchanged after scrambled peptide treatment (Figure S1C). The direct interaction between p300 and p53 appeared to be crucial for the induction of p300 catalytic activity; therefore, we wanted to investigate whether p53 could modulate the structure of p300. Since p300 is a large, intrinsically disordered protein of 300 kDa, it could be a possibility that p53 may act as a molecular chaperone for p300, thereby stabilizing its disordered structure. To address this, we designed an in vitro radioactivity-based histone acetyltransferase assay whereby we denatured p300 at 45°C and then incubated the denatured enzyme with p53 at 30°C. The refolding or recovery of p300 activity was scored by its ability to acetylate recombinant histones. Interestingly, we observed a p53 concentration-dependent rescue of p300 activity, suggesting that p53 could possibly act as a molecular chaperone for p300 (Figure 2H). Overall, we observed that p53 is a potent inducer of p300 autoacetylation and enzymatic activity, which may be through the modulation of p300 structure by p53.
Figure 1.
The Tumor Suppressor p53 Enhances p300 Autoacetylation
(A) An in vitro acetyltransferase assay was performed to determine the levels of autoacetylation of different lysine acetyltransferases (KATs) in the presence of 40, 80, 120, and 160 nM of recombinant wild-type p53 (Lanes 3-6), respectively. The autoradiograms indicate the levels of p300, PCAF, and Tip60 autoacetylation, respectively, whereas the Coomassie Brilliant Blue (CBB)-stained protein panels show the loading control. (See Figure S1A for densitometric quantification).
(B–D) An in vitro acetyltransferase assay was performed to determine the levels of p300 autoacetylation in the presence of different p300 substrates (B) p47, (C) PC4, and (D) p50. 20 nM p300 and 50, 100, and 150 nM of each protein factor were used.
(E) Enhancement of p300 catalytic activity in the presence of increasing concentrations of wild-type p53 as indicated was determined by filter-binding histone acetyltransferase assay, using recombinant histone H3 as substrate. The mean fold change was plotted ±SD; statistical analysis was performed using unpaired two-tailed Student's t test.
Figure 2.
p53 Modulates p300 Autoacetylation and Acetyltransferase Activity in Cells
(A) HepG2 cells were treated with DMSO (vehicle control) or Nutlin-3a as indicated (lane 2–4); levels of Ac-p300, p300, and p53 were analyzed by immunoblotting. Alpha-tubulin levels were considered as the loading control.
(B) HepG2 cells were treated with 5 μM Nutlin-3a for 24 hr, and the levels of Ac-p300 and p53 were assessed by co-immunofluorescence. Fluorescence intensity of Ac-p300 in control cells versus Nutlin-3a-treated cells have been quantified (represented as mean ± SD, unpaired two-tailed Student's t test, n = 50 of three independent experiments). Scale bar, 10 μm.
(C and D) HepG2 cells (C) and H1299 cells (D) were treated with DMSO (vehicle control) or Nutlin-3a as indicated. Histone acetylation levels were analyzed by immunoblotting using H3K9ac and H3K14ac antibodies. Total histone H3 levels were used as the loading control.
(E) The relative transcript levels of EP300 and CDKN1A (p53-responsive gene) were determined by qRT-PCR (mean ± SD). Unpaired two-tailed Student's t test statistical analysis was performed. Actin transcript level was used as the internal control. The relative p300 transcripts do not alter significantly on Nutlin-3a treatment.
(F) Results of filter-binding assay indicate that the p53 phosphomimic N-terminal peptide (3E pep) can effectively reduce the activity of p300 by interfering with its interaction to p53; 4 nM p53 phosphomimic and scrambled peptides were used. (Also see Figure S1B.)
(G) HepG2 cells treated with Nutlin-3a or DMSO, and p53 phosphomimic peptide (3E peptide) as indicated, and the levels of H2AK5ac, H3, p53, and tubulin were probed by immunoblotting. (Also see Figure S1C.)
(H) The rescue of heat-denatured 20 nM p300 activity (activity rescue assay), on addition of wild-type p53, at the indicated concentrations, was determined by the in vitro filter-binding assay, using recombinant histone H3 and [3H] acetyl-CoA.
Redistribution of Autoacetylated p300 upon p53-Mediated Activation
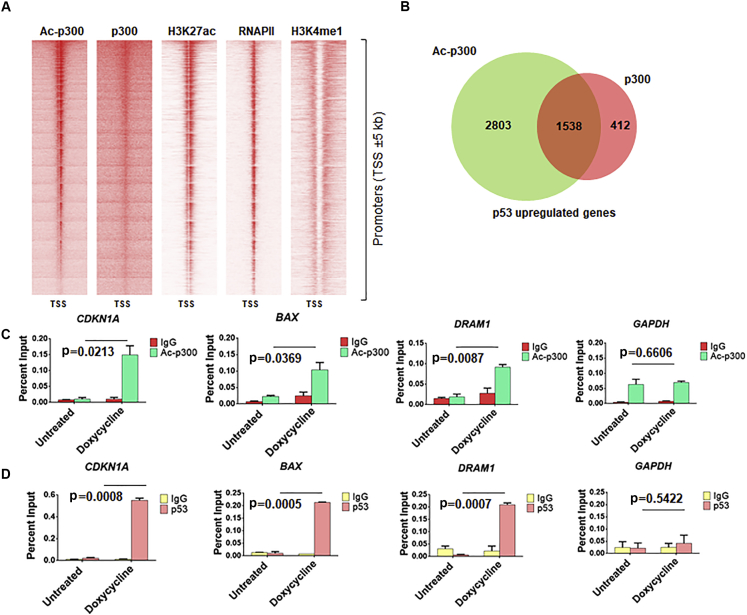
If the p53-mediated activation of p300 is important for p53-mediated regulation of targeted genes, then it is expected that activated forms should be present in the vicinity of the regulated genes. We have thus attempted to explore the effect of p53 expression on the presence of p300 in different genomic sites. The differential occupancy of ac-p300 and p300 on the promoters of genes that are expressed after p53 stimulation has been investigated. The enrichment patterns indicated a distinct preference of ac-p300 for the promoter proximal regions, after p53 expression, whereas only a weak enrichment pattern was observed for p300 (Figure S2A), thus reinforcing the possibility that the autoacetylated form of p300 is actively associated with p53-mediated transcriptional initiation (Figure 3A). We analyzed the differential occupancy of p300 and ac-p300 with publicly available datasets of RNAPII, H3K27ac (active promoter mark), and H3K4me1 (enhancer mark) distribution. The abundance of ac-p300 at the promoters correlated with the presence of RNA Pol II enrichment and active promoter marks such as H3K27ac (Figure 3A). These data indicated a specific role for ac-p300 within the entire pool cellular p300. The co-activator occupancy has been integrated with the microarray expression analysis of the p53-induced differentially expressed genes (Jiang et al., 2015), revealing a greater overlap between p53-driven transcription and ac-p300 (4,341 genes) recruitment (Figures 3C and 3D) in comparison with p300 (1950 genes) (Figure 3B).
Figure 3.
Redistribution of p300 after p53-Mediated Enhancement of p300 Autoacetylation
(A) Heatmaps depicting the enrichment of ac-p300, p300, H3K27ac, RNA Polymerase II (RNAPII), and H3K4me1 at promoters (TSS ±5 kb).
(B) Venn diagram showing the overlap between the up-regulated genes enriched at the promoters with both ac-p300 and p300 and genes unique to each set.
(C and D) ChIP-qPCR to determine the occupancy of (C) p53 and (D) autoacetylated p300 (Ac-p300) at p53-responsive gene promoters. (See Figure S2.)
The Gain-of-Function Mutant p53 Induces p300 Autoacetylation to Enhance Tumorigenesis
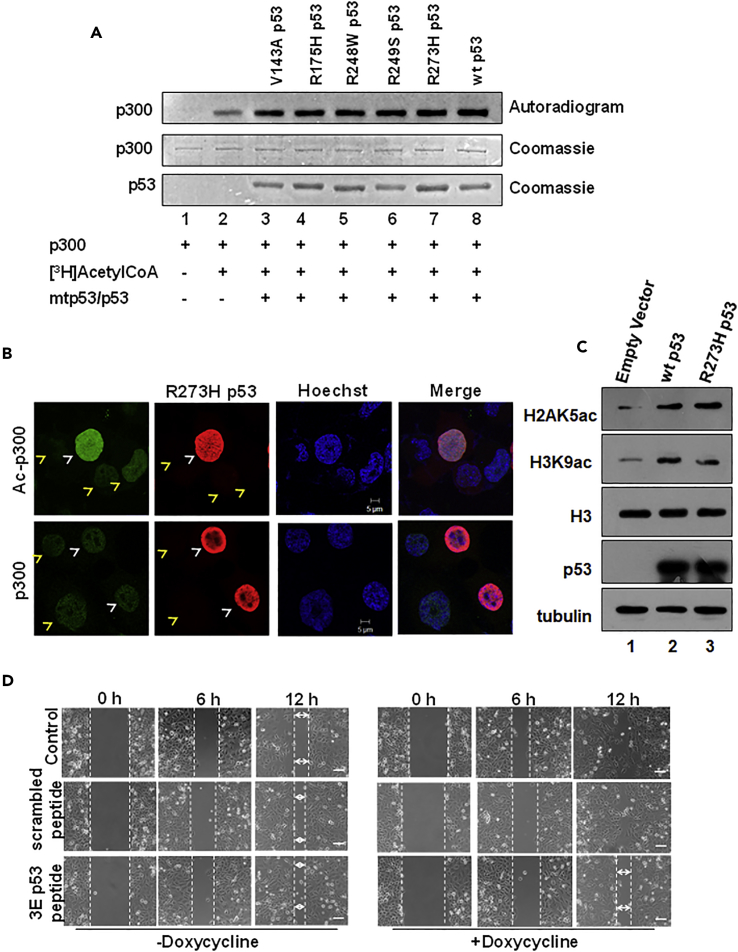
The TP53 gene is mutated in approximately 50% of malignancies, of which 75% are missense mutations (Freed-Pastor and Prives, 2012, Olivier et al., 2010). The missense mutations mapping to the DNA binding domain not only abrogate the tumor suppressive functions of p53 but also impart unique oncogenic functions to the mutant protein (Freed-Pastor and Prives, 2012). Since our previous results elucidated the mechanisms of p53-mediated induction of intermolecular p300 autoacetylation and the role of autoacetylated p300 in p53 downstream gene expression, we investigated whether these GOF mutants retained the ability to enhance p300 catalytic activity through the induction of autoacetylation. We tested different GOF hotspot mutants, three conformational mutants (R175H, V143A, and R249S), and two DNA contact mutants (R273H and R248W), in an in vitro autoacetylation assay. All the GOF mutants appeared to enhance p300 autoacetylation in vitro, which was comparable with the enhancement observed in the presence of wild-type p53 (Figure 4A). In H1299 cells ectopically expressing GOF p53 mutants, the levels of autoacetylated p300 were checked by immunofluorescence in the mutant p53 transfected cell versus the untransfected cells. The intensity of autoacetylated p300 immunofluorescence staining revealed that the cells expressing the GOF mutants, both DNA contact mutants (Figures S3A and S3B) and conformational mutants (Figures S3C and S3D), appeared to have significantly higher levels of autoacetylated p300 in comparison with the untransfected controls (Figure S3). To understand the pathophysiological relevance of GOF mutant p53-mediated enhancement of p300 autoacetylation, the R273H p53 mutant, which has one of the highest incidences in cancers (6.7%), was chosen for further investigation. To establish that R273H p53 mutant could alter the levels of p300 autoacetylation alone without affecting the overall p300 protein levels, immunofluorescence was performed in H1299 cells ectopically expressing R273H p53. As observed in the earlier immunofluorescence staining, the autoacetylation of p300 increased dramatically in the presence of R273H (Figure 4B, upper panel) while exhibiting a negligible change in the overall p300 protein levels (Figure 4B, lower panel). These data reinforce the fact that similar to wild-type p53, the GOF R273H mutant too, modulates p300 only at the autoacetylation level and does not appear to affect p300 protein stability or expression. As established earlier that increased KAT activity is a direct consequence of p300 autoacetylation, the levels of different p300-specific histone marks were checked, after ectopically expressing the mutant R273H and wild-type p53 as a control. It is evident from the western blotting analysis that histone acetylation on H2AK5 and H3K9 were up-regulated in cells transfected with the mutant and wild-type p53 in comparison with the vector-transfected cells (Figure 4C). To investigate the role of R273H mutant-mediated induction of p300 autoacetylation in regulating the tumorigenic potential of the mutant, an inducible Tet-ON H1299 cell line for R273H p53 expression was created. Earlier experiments have established that the direct interaction of p53 with p300 is essential for the induction of p300 autoacetylation. Therefore, the aim of the experiment was also to test whether the direct interaction of p300 and p53 mutant R273H played a role in the GOF of this mutant. In a wound healing assay, the cells were treated with doxycycline to induce the expression of R273H p53 and the untreated cells served as the experimental controls. Under both conditions, R273H p53 presence and absence, the cells were treated with either the p53-p300 interfering p53 phosphomimic peptide or the control scrambled peptide. On doxycycline treatment the wound created in the control cells healed faster, suggesting that the R273H p53 mutant indeed exhibited GOF properties, marked by higher proliferation and migration. The cells treated with the scrambled peptide showed similar closure time as the control cells, signifying that the scrambled peptide did not have any apparent pleiotropic effect. Interestingly, the cells treated with the phosphomimic p300-p53 interfering peptide migrated and proliferated slower than the control cells, implicating a possible role of p300 interaction in the tumorigenic potential of R273H p53 (Figure 4D). Moreover, the delay in wound closure observed on p53 phosphomimic peptide treatment was comparable with that of the untreated (doxycycline negative) cells, suggesting that the peptide treatment may have abrogated the effect of the p53 mutant expression. Furthermore, in the doxycycline-negative cells, the control, p53 phosphomimic peptide, and scrambled peptide-treated cells migrated and proliferated at similar rates, suggesting that the effect observed on p53 phosphomimic peptide treatment in the doxycycline-treated cells had a direct correlation with the presence of mutant p53 (Figure 4D). Collectively, these results implicate the direct role of p300 (and possible p300 autoacetylation) in the induction of R273H tumorigenic functions, creating a positive feedback loop between the two proteins. Western blotting analysis was done with the cells used in the assay to ascertain the expression status of R273H p53 in the presence and absence of doxycycline treatment (Figure 4A). Since in the above-mentioned experiment the mutant R273H was artificially expressed in H1299 cells, it was important to demonstrate a similar phenomenon in a cancerous cell line endogenously expressing the R273H p53 mutant. Therefore, a similar wound healing assay was performed in the AW13516 oral cancer cell line, which expresses high levels of the R273H p53 mutant. Similar to the previous experiment, the p53 phosphomimic peptide drastically impaired the rate of wound closure (Figure S4B, middle panel, 12 h) in comparison with the untreated and scrambled peptide-treated controls (Figure S4B, top and bottom panels, 12 h).
Figure 4.
p53 Gain-of-Function (GOF) Mutants Are Inducers of p300 Autoacetylation
(A) An in vitro HAT assay was performed to determine the levels of autoacetylation of p300 in the presence of recombinant GOF p53 mutants and wild-type p53.
(B) p53 null H1299 cells were transfected with R273H p53 mutant. The levels of Ac-p300 (autoacetylated p300) (top panel, green), p300 (bottom panel, green), and mutant p53 (red) were determined by immunofluorescence. Transfected cells and untransfected cells are indicated by white and yellow arrows, respectively. Scale bar, 5 μm. (See Figure S3.)
(C) Western blotting analysis was performed to determine the levels of p300-specific histone acetylation levels in H1299 cells transiently transfected with pCMV2 Empty Vector, wild-type p53, and the DNA contact mutant R273H p53. Histone H3 levels served as the loading control.
(D) Representative images (of two biological repeats) of a wound healing assay performed in doxycycline-inducible R273H p53 H1299 stable cell line. The cells were treated with either 10 μM phosphomimic p53 peptide or 10 μM scrambled peptide as indicated with or without doxycycline treatment. Scale bar, 100 μm. (See Figure S4.)
To establish further that the GOF mutant p53-mediated induction of p300 autoacetylation may be an oncogenic pathway through which mutant p53 can exert its tumorigenic potential, we tested whether loss-of-function (LOF) tetramerization-defective mutants could also induce p300 autoacetylation. Tetramerization-defective mutants were transfected into H1299 cells, and the levels of autoacetylated p300 was probed by immunofluorescence. It was found that the p53 tetramerization mutants failed to enhance p300 autoacetylation in the H1299 cells (Figures S5A and S5B). It has been shown earlier that p53 proteins that cannot form tetramers lose their ability to interact with p300 (Itahana et al., 2009). It is fair to assume that the physical interaction of p53 and p300 is a key requisite for the induction of p300 autoacetylation. The L344A p53 tetramerization-defective mutant was tested in an autoacetylation assay. The autoacetylation of p300 does not alter in the presence of the mutant (Figure S5C). When H1299 cells were transfected with the L344A p53 mutant, we observed no enhancement in the global levels of p300 autoacetylation or any alteration in p300 protein levels (Figures S5D and S5E). These experiments revealed the importance of p53 tetramerization and the requirement for the direct interaction between p53 and p300 for p300 autoacetylation and augmentation of its acetyltransferase activity (Figure 5).
Figure 5.
Model Depicting the Physiological Significance of p53-Mediated Enhancement of p300 Autoacetylation
The histone acetyltransferase is present in a “closed” inactive conformation when present in an unbound state, but in the presence of p53, a substantial rearrangement in the domain architecture of p300 is observed. In the p53-bound complex, p300 undergoes a conformational switch from an inactive to an altered conformation. This altered conformational state of p300 is more conducive to intermolecular autoacetylation, a phenomenon that enhances the catalytic activity of p300. When p300 is activated by wild-type p53, there is a re-distribution of autoacetylated p300 onto p53-responsive gene promoters, leading to the downstream tumor suppressive functions of p53, whereas when mutant p53 induces the autoacetylation of p300, we observe an increased tumorigenic potential. The gain-of-function effect of mutant p53 is abrogated when the interaction between mutant p53 and p300 is disrupted.
Discussion
The functional cross talk between p300 and p53 is well established; however, the role of p53 in the modulation of the enzymatic activity of p300 has not been addressed before. The chromatin immunoprecipitation sequencing (ChIP-seq) analysis of ac-p300 genome occupancy after p53 induction provided further insights into the transcriptional cascades triggered by effector (p53)-mediated induction of p300 autoacetylation. p300 autoacetylation can control its catalytic activity (Thompson et al., 2004), but the dynamics of p300 autoacetylation under physiological stimuli are not fully understood. The present findings provide experimental evidence that establishes p53 as a bona fide modulator of p300 autoacetylation. Remarkably, among all of the substrates of p300 tested, only p53 enhanced p300 autoacetylation, thus confirming the molecular specificity of this phenomenon. The preferential induction of p300 autoacetylation by p53 over that of other acetyltransferases further exhibited the specificity of p53-mediated induction of p300 autoacetylation. This result further suggested that the induction of autoacetylation is not merely a consequence of enzyme-substrate interaction. The structural plasticity of p300 not only increases its interactome but also provides an additional level of regulation for this essential epigenetic enzyme. Thompson et al have shown that the basal catalytic activity of p300 is stimulated through the autoacetylation of certain lysine residues present on the auto-inhibitory loop residing in the KAT domain. It was speculated that, on acetylation of these lysine residues, the loop would be dislodged thereby allowing enhanced catalytic activity (Thompson et al., 2004). Surface-enhanced Raman spectroscopy (SERS)-based structural studies on the minimal p300 HAT domain provided evidence to demonstrate that p300 when hyper-autoacetylated in the presence of acetyl-CoA exhibited a structural reorganization in the HAT domain (Arif et al., 2007). This finding was corroborated by another SERS-based structural study using small molecule activators of p300, CTB and CTPB, which showed a similar structural alteration in the HAT domain (Mantelingu et al., 2007). The 2.8 Å crystal structure of p300 catalytic core domain (bromodomain-RING-PHD-HAT (PDB: 4BHW)) revealed that the bromodomain-RING-PHD domains associate closely with the HAT domain, through several conserved contacts. In the crystal structure, the spatial arrangement of the domains suggests that p300 alone is present in a closed inactive conformation, which the authors speculated could be relieved by inter-molecular autoacetylation (Delvecchio et al., 2013). Studies have indicated that a conformational alteration may be a prerequisite for complete activation of p300 acetyltransferase activity (Delvecchio et al., 2013, Kaypee et al., 2018, Yi et al., 2017). In our recent study, we have shown that the decameric histone chaperone NPM1 could alter the structural conformation of p300, functioning as a molecular chaperone for the intrinsically disordered enzyme thereby inducing p300 autoacetylation and acetyltransferase activity, whereas the disruption of NPM1 oligomerization abrogates its ability to activate p300 (Kaypee et al., 2018). Drawing parallels to this study, we find that p53, in its tetrameric form, is a potent inducer of p300 autoacetylation. On disruption of its tetramers (in tetramerization-defective mutants), p53 appears to lose its ability to enhance p300 autoacetylation. Corroborating the model proposed by Teufel et al., we can conjecture that a single molecule of p300 can wrap around tetrameric p53 (Teufel et al., 2007), as a result of which p300 attains a conformation that may be conducive to intermolecular autoacetylation, as opposed to the closed inactive conformation observed in the crystal structure of the catalytic core of p300 (Delvecchio et al., 2013). However, the conformational alterations should be further validated through extensive structural studies on the full-length p300-p53 complex. This model may have further implications on the recruitment of p300/CBP to chromatin.
p300/CBP is recruited to gene promoters by several transcription factors (An et al., 2004, Kundu et al., 2000) where it can assemble transcription complexes that facilitate gene expression. The autoacetylation of p300 has been proposed to induce distinct structural changes that are critical for its activity. Black et al. have shown that the presence of p300 at the preinitiation complex (PIC) exerts an inhibitory effect of transcription in vitro. The release of p300 from the complex is attributed to the possible conformational switch in the p300 structure after autoacetylation (Black et al., 2006). It is clear from this study that p300 autoacetylation is essential for maximal transcription to proceed. The predominant occupancy of ac-p300 on the TSS of the promoters is a novel finding based on the obtained ChIP-seq analysis. These data are in agreement with the factor-dependent recruitment of the acetyltransferases p300/CBP and subsequent gene activation, as previously reported. Evident from our biochemical data, the direct interaction of p53 with its co-activator p300/CBP is functionally important in this process and serves as the mechanism of induction of autoacetylation. Ceschin et al. have shown that CBP is methylated at multiple sites by the arginine methyltransferase protein CARM1 and the arginine methylation of CBP stimulates CBP activity through the induction of CBP autoacetylation. Use of polyclonal antibodies recognizing specific CBP methylation species has indicated that each methylated class of CBP has differential HAT activities and distinct transcriptional effects, thereby diversifying the estrogen receptor response (Ceschin et al., 2011). The contextual recruitment of p300 has been noted in another study performed in the MCF7 cell line, in which it has been observed that, in resting cells, p300 appears to show a preference toward neural lineage gene promoters, whereas the enrichment of p300 shifts toward estrogen-regulated gene targets after estradiol treatment (Wang and Li, 2016). In a recent report, CBP has been shown to interact with RNAs. At active enhancers, CBP interacts with enhancer RNAs, which bind to the CBP catalytic domain, inducing CBP autoacetylation (Bose et al., 2017). The resultant enhancement in CBP activity leads to increased histone acetylation and expression of target genes. In the present work, the differential occupancy of p300 and ac-p300 was investigated after p53 activation. After autoacetylation, in the presence of p53, p300 appeared to show a strong preference for the TSS (Figure 3A and S2A), in agreement with the recruitment model discussed earlier. The binding of ac-p300 was strongly associated with RNA Pol II occupancy and the promoter acetylation mark H3K27ac. This finding suggested that ac-p300 is indeed involved in the assembly of the basal transcription machinery at promoter-proximal regions of genes. The overall pool of p300 did not show an appreciable alteration in chromatin occupancy in the presence of p53, whereas a distinct redistribution in ac-p300 enrichment was observed. Notably, the integration of the ChIP-seq data with the gene expression microarray analysis revealed that the enrichment of ac-p300 was a better determinant of p53-mediated gene expression than the presence of unmodified p300. This result was in agreement with previous studies in which p300 has been shown to occupy transcriptionally silent regions or the chromatin, whereas the co-occupancy of (presumably autoacetylated) p300 with its histone acetylation mark H3K27ac was observed at transcriptionally active genes (Holmqvist and Mannervik, 2013). With the current evidence, it can be speculated that the ability to induce structural alterations in p300, leading to enhancement of its acetyltransferase activity, may be a key determinant in p300 chromatin recruitment and downstream transcriptional programs. The ChIP-seq data suggest that the phenomenon of factor-induced p300 autoacetylation may play a pivotal role in the integration of stimulus-driven transcriptional pathways. Specifically, the recruitment of the catalytically active form of p300 may trigger a rapid transcriptional response to internal and external signaling cues. In this study, it is the p53-driven pathway that stimulates a burst in ac-p300 levels and an alteration of the epigenetic landscape, which may be essential for these p53 signaling pathways. We hypothesize that after p53 is stabilized in the cell or overexpressed, it associates with its ubiquitously expressed co-activator p300. This association triggers a conformational alteration in p300, thereby potentiating intermolecular autoacetylation. It is this catalytically active form of p300 that is subsequently enriched in p53-target gene regulatory chromatin regions via its interaction with p53. The active ac-p300 then acetylates histone tails at these loci, decompacting chromatin and leading to enhanced transcription p53-dependent and (possibly) independent gene networks (Figure 5, Table S2).
A large proportion of tumors harbor mutations in p53 that nullify specific DNA-binding properties. A subset of these mutants, the GOF mutants, also gain aggressive proliferation properties, and it is now known that this is achieved by relocation to regulatory regions of genes responsible for proliferation and drug resistance followed by activation of expression of those genes. The results presented here underline that, in spite of the loss of DNA-binding properties, these mutants retain their ability to activate the co-activator p300 through the induction of autoacetylation, thus promoting the activation of these growth-promoting genes in the new locale. Abolition of the GOF properties by a site-directed peptide inhibitor of p53-p300 interaction suggests that these classes of inhibitors may offer promise against tumors bearing aggressive GOF mutant p53 alleles (Figure 5).
The activation of p300 by p53 suggests that p300 may be preferentially activated when it is recruited to p53 at its target sites. This activation would suggest an enhanced activity of p300 when it is complexed with target-site-bound p53, thereby leading to localized transcriptional initiation. Thus, the augmented activity of p300 at genes to which it is recruited by p53 adds another element of specificity beyond that of the transcription factor binding. Moreover, p53 was able to modulate the levels of histone acetylation concomitant with the induction of p300 autoacetylation, thus signifying that the tumor suppressor p53 can alter the epigenetic landscape through the regulation of p300 acetyltransferase activity. We speculate that the activation of p300 by p53 demonstrated here may be a representative example of a general mechanism for attaining the high degree of spatiotemporal specificity required for attaining exquisite and intricate gene regulation.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by Jawaharlal Nehru Center for Advanced Scientific Research (JNCASR) (MBGU/TKK), Sir JC Bose Fellowship, Department of Science and Technology, Government of India (SR/S2/JCB-28/2010) and Virtual National Oral Cancer Institute, Department of Biotechnology, Government of India (BT/PR17576/MED/30/1690/2016). T.K.K. is a Sir JC Bose Fellow. We acknowledge Mr. Madavan Vasudevan and Ms. Madhura Tathode, Bionivid Technology Pvt. Ltd., for the ChIP-seq analysis. We acknowledge Dr. Dirk Gründemann (University of Cologne, Germany) for the pEBTetD SLC22A1 construct. We acknowledge Dr. Amit Dutt (ACTREC, Mumbai, India) for the AW13516 cell line. We acknowledge the Confocal Imaging Facility at JNCASR. We have used illustration templates from the website somersault1824 (http://www.somersault1824.com) available under a Creative Commons Attribution-Noncommercial-Share Alike license (CC BY-NC-SA 4.0) in the Model (Figure 5) and Graphical Abstract.
Author Contributions
T.K.K. and S.K. have conceived the project. T.K.K., S.K., and S.R. have written the manuscript. S.K., S.A.S., and S.P. have executed the molecular biology experiments. P.G. and N.S.R. have synthesized and characterized the peptides used in this study.
Declaration of Interests
The authors declare no competing interests.
Published: June 29, 2018
Footnotes
Supplemental Information includes Transparent Methods, five figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.isci.2018.06.002.
Supplemental Information
References
- An W., Kim J., Roeder R.G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Arif M., Kumar G.V.P., Narayana C., Kundu T.K. Autoacetylation induced specific structural changes in histone acetyltransferase domain of p300: probed by surface enhanced Raman spectroscopy. J. Phys. Chem. B. 2007;111:11877–11879. doi: 10.1021/jp0762931. [DOI] [PubMed] [Google Scholar]
- Arif M., Vedamurthy B.M., Choudhari R., Ostwal Y.B., Mantelingu K., Kodaganur G.S., Kundu T.K. Nitric oxide-mediated histone hyperacetylation in oral cancer: target for a water-soluble HAT inhibitor, CTK7A. Chem. Biol. 2010;17:903–913. doi: 10.1016/j.chembiol.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Barlev N.A., Liu L., Chehab N.H., Mansfield K., Harris K.G., Halazonetis T.D., Berger S.L. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Bedford D.C., Kasper L.H., Fukuyama T., Brindle P.K. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.C., Choi J.E., Lombardo S.R., Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Black J.C., Mosley A., Kitada T., Washburn M., Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol. Cell. 2008;32:449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose D.A., Donahue G., Reinberg D., Shiekhattar R., Bonasio R., Berger S.L. RNA binding to CBP stimulates histone acetylation and transcription. Cell. 2017;168:135–149.e22. doi: 10.1016/j.cell.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceschin D.G., Walia M., Wenk S.S., Duboé C., Gaudon C., Xiao Y., Fauquier L., Sankar M., Vandel L., Gronemeyer H. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 2011;25:1132–1146. doi: 10.1101/gad.619211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.M., La Thangue N.B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Delvecchio M., Gaucher J., Aguilar-Gurrieri C., Ortega E., Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat. Struct. Mol. Biol. 2013;20:1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. Role of intrinsic protein disorder in the function and interactions of the transcriptional coactivators CREB-binding protein (CBP) and p300. J. Biol. Chem. 2016;291:6714–6722. doi: 10.1074/jbc.R115.692020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa J.M., Emerson B.M. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- Freed-Pastor W.A., Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Roeder R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Haaland I., Opsahl J.A., Berven F.S., Reikvam H., Fredly H.K., Haugse R., Thiede B., McCormack E., Lain S., Bruserud Ø. Molecular mechanisms of nutlin-3 involve acetylation of p53, histones and heat shock proteins in acute myeloid leukemia. Mol. Cancer. 2014;13:116. doi: 10.1186/1476-4598-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson M.L., Popko-Ścibor A.E., Saint Just Ribeiro M., Dancy B.M., Lindberg M.J., Cole P.A., Wallberg A.E. The transcriptional coactivator MAML1 regulates p300 autoacetylation and HAT activity. Nucleic Acids Res. 2009;37:2996–3006. doi: 10.1093/nar/gkp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist P.-H., Mannervik M. Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription. 2013;4:18–23. doi: 10.4161/trns.22601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana Y., Ke H., Zhang Y. p53 Oligomerization is essential for its C-terminal lysine acetylation. J. Biol. Chem. 2009;284:5158–5164. doi: 10.1074/jbc.M805696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Kon N., Li T., Wang S.-J., Su T., Hibshoosh H., Baer R., Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaypee S., Sahadevan S.A., Sudarshan D., Halder Sinha S., Patil S., Senapati P., Kodaganur G.S., Mohiyuddin A., Dasgupta D., Kundu T.K. Oligomers of human histone chaperone NPM1 alter p300/KAT3B folding to induce autoacetylation. Biochim. Biophys. Acta. 2018;1862:1729–1741. doi: 10.1016/j.bbagen.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Kim W.J., Rivera M.N., Coffman E.J., Haber D.A. The WTX tumor suppressor enhances p53 acetylation by CBP/p300. Mol. Cell. 2012;45:587–597. doi: 10.1016/j.molcel.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Lee S.H., McCormick F. Skp2 Suppresses p53-dependent apoptosis by inhibiting p300. Mol. Cell. 2008;29:217–231. doi: 10.1016/j.molcel.2007.11.036. [DOI] [PubMed] [Google Scholar]
- Krois A.S., Ferreon J.C., Martinez-Yamout M.A., Dyson H.J., Wright P.E. Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein. Proc. Natl. Acad. Sci. USA. 2016;113:E1853–E1862. doi: 10.1073/pnas.1602487113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumazawa T., Nishimura K., Kuroda T., Ono W., Yamaguchi C., Katagiri N., Tsuchiya M., Masumoto H., Nakajima Y., Murayama A. Novel nucleolar pathway connecting intracellular energy status with p53 activation. J. Biol. Chem. 2011;286:20861–20869. doi: 10.1074/jbc.M110.209916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu T.K., Palhan V.B., Wang Z., An W., Cole P.A., Roeder R.G. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- Lambert P.F., Kashanchi F., Radonovich M.F., Shiekhattar R., Brady J.N. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- Lee C.W., Sørensen T.S., Shikama N., La Thangue N.B. Functional interplay between p53 and E2F through co-activator p300. Oncogene. 1998;16:2695–2710. doi: 10.1038/sj.onc.1201818. [DOI] [PubMed] [Google Scholar]
- Lee C.W., Arai M., Martinez-Yamout M.A., Dyson H.J., Wright P.E. Mapping the interactions of the p53 transactivation domain with the KIX domain of CBP. Biochemistry. 2009;48:2115–2124. doi: 10.1021/bi802055v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Ferreon J.C., Ferreon A.C.M., Arai M., Wright P.E. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc. Natl. Acad. Sci. USA. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Lill N.L., Grossman S.R., Ginsberg D., DeCaprio J., Livingston D.M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Liu L., Scolnick D.M., Trievel R.C., Zhang H.B., Marmorstein R., Halazonetis T.D., Berger S.L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantelingu K., Kishore A.H., Balasubramanyam K., Kumar G.V.P., Altaf M., Swamy S.N., Selvi R., Das C., Narayana C., Rangappa K.S. Activation of p300 histone acetyltransferase by small molecules altering enzyme structure: probed by surface-enhanced Raman spectroscopy. J. Phys. Chem. B. 2007;111:4527–4534. doi: 10.1021/jp067655s. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz R.L., Russanova V., Howard B.H., Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Olivier M., Hollstein M., Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Dasen J.S., Kurokawa R., Wang Z., Korzus E., Rose D.W., Glass C.K., Rosenfeld M.G. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc. Natl. Acad. Sci. USA. 1999;96:3652–3657. doi: 10.1073/pnas.96.7.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley S., Guha S., Roy N.S., Kar S., Sakaguchi K., Chuman Y., Swaminathan V., Kundu T., Roy S. Differential recognition of phosphorylated transactivation domains of p53 by different p300 domains. J. Mol. Biol. 2008;376:8–12. doi: 10.1016/j.jmb.2007.11.082. [DOI] [PubMed] [Google Scholar]
- Reed S.M., Quelle D.E. p53 acetylation: regulation and consequences. Cancers (Basel) 2014;7:30–69. doi: 10.3390/cancers7010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokudai S., Laptenko O., Arnal S.M., Taya Y., Kitabayashi I., Prives C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc. Natl. Acad. Sci. USA. 2013;110:3895–3900. doi: 10.1073/pnas.1300490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K., Herrera J.E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C.W., Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Sen N., Hara M.R., Kornberg M.D., Cascio M.B., Bae B.-I., Shahani N., Thomas B., Dawson T.M., Dawson V.L., Snyder S.H. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat. Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Dai C., Qin J., Gu W. Negative regulation of the p300-p53 interplay by DDX24. Oncogene. 2016;35:528–536. doi: 10.1038/onc.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes S.M., Mellert H.S., Holbert M.A., Li K., Marmorstein R., Lane W.S., McMahon S.B. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Luo J., Zhang W., Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Teufel D.P., Freund S.M., Bycroft M., Fersht A.R. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc. Natl. Acad. Sci. USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.R., Wang D., Wang L., Fulco M., Pediconi N., Zhang D., An W., Ge Q., Roeder R.G., Wong J. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- Turnell A.S., Stewart G.S., Grand R.J.A., Rookes S.M., Martin A., Yamano H., Elledge S.J., Gallimore P.H. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- Vasudevarao M.D., Mizar P., Kumari S., Mandal S., Siddhanta S., Swamy M.M.M., Kaypee S., Kodihalli R.C., Banerjee A., Naryana C. Naphthoquinone-mediated inhibition of lysine acetyltransferase KAT3B/p300, basis for non-toxic inhibitor synthesis. J. Biol. Chem. 2014;289:7702–7717. doi: 10.1074/jbc.M113.486522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N., Goodman R.H. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Wang X., Li S. [Corrigendum] Chromatin immunoprecipitation-sequencing predicts p300 binding sites in the MCF7 human breast cancer cell line. Int. J. Mol. Med. 2016;38:675. doi: 10.3892/ijmm.2016.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P., Wang Z., Feng Q., Chou C.-K., Pintilie G.D., Shen H., Foulds C.E., Fan G., Serysheva I., Ludtke S.J. Structural and functional impacts of ER coactivator sequential recruitment. Mol. Cell. 2017;67:733–743.e4. doi: 10.1016/j.molcel.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.