Abstract
As the most commonly occurring cancer in women worldwide, breast cancer poses a formidable public health challenge on a global scale. Breast cancer consists of a group of biologically and molecularly heterogeneous diseases originated from the breast. While the risk factors associated with this cancer varies with respect to other cancers, genetic predisposition, most notably mutations in BRCA1 or BRCA2 gene, is an important causative factor for this malignancy. Breast cancers can begin in different areas of the breast, such as the ducts, the lobules, or the tissue in between. Within the large group of diverse breast carcinomas, there are various denoted types of breast cancer based on their invasiveness relative to the primary tumor sites. It is important to distinguish between the various subtypes because they have different prognoses and treatment implications. As there are remarkable parallels between normal development and breast cancer progression at the molecular level, it has been postulated that breast cancer may be derived from mammary cancer stem cells. Normal breast development and mammary stem cells are regulated by several signaling pathways, such as estrogen receptors (ERs), HER2, and Wnt/β-catenin signaling pathways, which control stem cell proliferation, cell death, cell differentiation, and cell motility. Furthermore, emerging evidence indicates that epigenetic regulations and noncoding RNAs may play important roles in breast cancer development and may contribute to the heterogeneity and metastatic aspects of breast cancer, especially for triple-negative breast cancer. This review provides a comprehensive survey of the molecular, cellular and genetic aspects of breast cancer.
Keywords: BRCA1/2, Breast cancer, Cancer stem cells, Estrogen receptors, HER2, Noncoding RNAs, Triple-negative breast cancer, Tumor heterogeneity
Introduction
For many years, breast cancer has had the highest incidence of all cancers in women worldwide.1, 2, 3, 4 Patients have better survival compared with more fatal cancers possibly because the breast tissue is physically not a necessary organ for human survival. Yet the mental and emotional disturbances from major surgeries as well as deaths by relapse or metastasis seriously endanger women's health. Since the earliest known descriptions of breast cancer originating in ancient Egypt, people have been dedicated to finding means of eradicating this disease. Leaps and bounds have been made in terms of this endeavor, especially in recent years. Mastectomy and chemotherapy have greatly improved the survival of breast cancer patients and more elegant forms of surgical procedures are now being applied to minimize the post-treatment psychological impact.5, 6, 7 However, without fully understanding the underlying mechanism and pathogenesis, the efficiency of prevention and treatment will always be limited.
Breast cancer is a compilation of distinct malignancies that manifests in the mammary glands. Carcinomas make up the majority of breast cancers while sarcomas such as phyllodes tumors and angiosarcomas are rarely seen. Thanks to the rapid progresses in molecular biology, systems biology and genome sciences in the past decades, our understanding about this disease has been dramatically expanded at cellular, molecular and genomic levels. Here, we intend to provide a comprehensive up-to-date overview of the basic biological aspects of breast cancer, including the risk factors, specific breast cancer classifications and subtypes, possible roles of mammary stem cells in breast cancer, major signaling pathways in breast cancer development, common gene mutations in breast cancer, the regulatory roles of epigenetics and noncoding RNAs in breast cancer, the molecular basis of triple-negative breast cancer, tumor heterogeneity of breast cancer, and the mechanism underlying breast cancer metastasis. It is our goal to present the aforementioned information in hopes of disseminating the present understanding of the molecular and genetic bases of breast cancer, which can be employed to assist in the development of novel and targeted therapies as a means of realizing the full potential of personalized medicine for breast cancer.
Disease landscape and risk factors of breast cancer
U.S. breast cancer statistics
It is estimated that about one in eight US women will develop invasive breast cancer over the course of her lifetime. In 2018, over 266,000 new cases of invasive breast cancer are expected to be diagnosed in women in the U.S., along with nearly 64,000 new cases of non-invasive breast cancer in US women.1, 8, 9 As of January 2018, there are already more than 3.1 million US women with a history of breast cancer, which includes the women currently being treated as well as women who have finished treatment. In addition to the statistics regarding women, approximately 2500 new cases of invasive breast cancer are expected to be diagnosed in US men in 2018. Even so, breast cancer incidence rates in the US have steadily declined since 2000.1 On account of treatment advances, increased awareness, and the possibility for earlier detection through screening, breast cancer death rates have been decreasing since 1989. Even with this marked progress, over 40,000 US women are expected to die in 2018 from breast cancer. In fact, for women in the US, breast cancer death rates are higher than those of any other cancer except lung cancer.1, 8, 9
In addition to having the second highest cancer-related death rate, breast cancer is among most commonly diagnosed cancer in US women. In total, about 30% of newly diagnosed cancers in women will be breast cancers. Among US women under the age of 45, breast cancer is more common in African-American women than white women. African-American women are more likely to die of breast cancer. On the other hand, the risk of developing and dying from breast cancer is lower for Asian, Hispanic, and Native-American women in the US1.
Genetic predispositions as important risk factors of breast cancer
At its most basic, a risk factor is defined as anything that affects individual's chance of getting a disease, in this case breast cancer. Certain major risk factors for breast cancer are beyond individual's control.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 For example, simply being a woman is the main risk factor for breast cancer as this disease is about 100 times more likely to occur in women than in men. Aging inevitably increases one's risk of breast cancer as evinced by the fact that most breast cancers are diagnosed in women age 55 and older. Beyond the inherent risks of gender and aging as they relate to breast cancer, it has been well documented that a woman's risk of developing breast cancer nearly doubles if she has a first-degree relative (mother, sister, or daughter) diagnosed with breast cancer. Close to 15% of US women who suffer from breast cancer also have a family member who has been diagnosed.8, 9, 22
Overall, about 5–10% of breast cancers are linked to gene mutations inherited from a parent. The most common cause of hereditary breast cancer is an inherited mutation in the BRCA1 or BRCA2 gene.4, 8, 9, 22, 23, 24 Statistically, women with a BRCA1 mutation have a 55–65% lifetime risk of developing breast cancer. For women with a BRCA2 mutation, the lifetime risk is 45%. On average, a woman with a BRCA1 or BRCA2 gene mutation has about 70% chance of getting breast cancer by age 80. The effect of the mutation is related to how many other family members have breast cancer, as breast cancer risk goes up if more family members are affected. In the US, BRCA mutations are more common in Jewish people of Ashkenazi (Eastern European) origin than in other racial and ethnic groups although anyone can have these mutations. Women with one of these two mutations are also more likely to be diagnosed with breast cancer at a younger age, as well as to have cancer in both breasts. The impact of the BRCA1 and BRCA 2 mutation expands beyond just breast cancer as having mutations in either of these genes is associated with an increased ovarian cancer risk as well. Conversely, BRCA1 mutations are found less frequently in breast cancers occurring in men while BRCA2 mutations are associated with a lifetime breast cancer risk of only about 6.8%.4, 8, 9, 22, 23, 24
Although less common and less drastic in their increase of breast cancer risk than the BRCA mutations, inherited mutations in many other genes can also lead to breast cancer development.4, 8, 9, 22, 23, 24 Some of the mutated genes include ATM (inheriting 2 abnormal copies of this gene causes the disease ataxia-telangiectasia), TP53 (inherited mutations of this gene cause Li-Fraumeni syndrome with an increased risk of breast cancer, as well as some other cancers such as leukemia, brain tumors, and sarcomas), CHEK2 (a CHEK2 mutation can increase breast cancer risk about 2-fold), PTEN (inherited mutations in this gene can cause Cowden syndrome which is accompanied by a higher risk for both non-cancerous and cancerous tumors in the breasts, as well as growths in the digestive tract, thyroid, uterus, and ovaries), CDH1 (inherited mutations cause hereditary diffuse gastric cancer with an increased risk of invasive lobular breast cancer), STK11 (mutations in this gene can lead to Peutz-Jeghers syndrome with a higher risk of many types of cancer, including breast cancer), and PALB2 (PALB2 gene makes a protein that interacts with the protein made by the BRCA2 gene, resulting in mutations in this gene causing a higher risk of breast cancer).4, 8, 9, 22, 23, 24
Properly and carefully consulted genetic testing of mutations in the BRCA1 and BRCA2 genes, as well as other less commonly mutated genes such as PTEN or TP53 in women in the high risk group can be beneficial for early detection and/or prevention of breast cancer development.4, 8, 9, 22, 23, 24 However, it is important to understand the limitations of genetic testing and what it can and can't tell an individual. In terms of practically making use of genetic testing for detection and prevention of breast cancer, it's also necessary to keep in mind that the testing is quite expensive and may not be covered by all health insurance plans. While genetic testing can be helpful in some cases, not every woman needs to be tested.
“Non-genetic” risk factors of breast cancer
Family history of breast cancer: While less than 15% of women with breast cancer have a family member with this disease, women who do have close blood relatives with breast cancer have a higher risk.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 25 For instance, having a first-degree relative (mother, sister, or daughter) with breast cancer almost doubles a woman's risk while having two first-degree relatives with the disease increases the woman's risk about 3-fold. Interestingly, women with a father or brother who have breast cancer also have a higher risk of breast cancer. Within the context on an individual, a woman with cancer in one breast has a higher risk of developing a new cancer in the other breast or in another part of the same breast.4, 8, 9, 22, 23, 24
Race and ethnicity: In general, Caucasian women are slightly more likely to develop breast cancer than African-American women although breast cancer is more common in African-American women under age 45. Furthermore, African-American women are more likely to die from breast cancer at any age. Other races such as Asian, Hispanic, and Native American women have a lower risk of developing and dying from breast cancer.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21
Certain benign breast conditions: Women with dense breasts on mammogram have a risk of breast cancer that is about 1.5–2 times that of women with average breast density even though multi factors play a role in determining breast density, such as age, menopausal status, the use of certain drugs (such as menopausal hormone therapy) and pregnancy. Certain non-proliferative lesions may marginally affect breast cancer risk. These non-proliferative lesions include fibrosis and/or simple cysts, mild hyperplasia, adenosis, phyllodes tumor, single papilloma, duct ectasia, periductal fibrosis, squamous and apocrine metaplasia, epithelial-related calcifications, other tumors (lipoma, hamartoma, hemangioma, neurofibroma, adenomyoepithelioma), or mastitis.26, 27
Certain proliferative breast lesions: Some proliferative lesions without atypia seem to raise a woman's risk of breast cancer slightly.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 Examples of such proliferative lesions are ductal hyperplasia, fibroadenoma, sclerosing adenosis, papillomatosis or radial scar. However, certain proliferative lesions with atypia in the ducts or lobules of the breast tissue will increase breast cancer risk 4–5-fold; and these include atypical ductal hyperplasia (ADH) and atypical lobular hyperplasia (ALH).28, 29
Lobular carcinoma in situ (LCIS) or lobular neoplasia: LCIS cells are cancer-like and grow in the lobules of the milk-producing glands of the breast, but are limited within the walls of the lobules.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 LCIS is traditionally grouped with ductal carcinoma in situ (DCIS) as a non-invasive breast cancer, while recent updates in the field consider LCIS to be benign. However, LCIS differs from DCIS in that it usually does progress to become invasive cancer if it is not treated. Women with LCIS also have a much higher risk of developing cancer in either breast.
Chest radiation therapy: Women, who were treated with radiation therapy to the chest for another cancer when they were younger, have higher risk for developing breast cancer.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 The impact of this factor on increasing risk is highest if the individual had radiation as a teen or young adult, when the breasts were still developing. Conversely, radiation treatment after age 40 does not seem to increase breast cancer risk.
Exposure to diethylstilbestrol (DES): From 1940s through early 1970s some pregnant women were given an estrogen-like drug DES because it was thought to lower the incidence of miscarriage.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 These women have a slightly increased risk of breast cancer, and women whose mothers took DES during pregnancy may also have a slightly higher risk of breast cancer.
Lifestyle and Personal Behavior-Related Risk Factors of Breast Cancer Vast majority (about 85%) of breast cancers occur in women without apparent family history of breast cancer. These cancers may be caused by genetic mutations that occur as a result of the aging process and lifestyle-related risk factors, rather than inherited mutations.
Birth control and contraceptives: Many birth control methods use hormones, which may increase breast cancer risk.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 Women using oral contraceptives have a slightly higher risk of breast cancer than women who have never used them, although the risk seems to go back to normal over time once the regimen is stopped. As an injectable form of progesterone, Depo-Provera has been shown to have an increase in breast cancer risk, but there is seemingly no increased risk in women five years after they have stopped receiving the shots. Birth control implants, intrauterine devices (IUDs), skin patches, and vaginal rings usually also use hormones and thus in theory may increase breast cancer risk. Consequently, whenever considering the use of hormonal birth control, women should discuss the coupling of this impact with any other risk factors for breast cancer with their health care providers.
Hormone replacement therapy (HRT) after menopause: The hormone estrogen (often combined with progesterone) has been used to relieve symptoms of menopause and to prevent osteoporosis.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 30 Combined hormone therapy is needed in most cases as use of estrogen alone can increase the risk of cancer of the uterus. However, for women who have had a hysterectomy, estrogen by itself can be used. Postmenopausal combined hormone therapy increases the risk of breast cancer, the chances of dying from breast cancer, and the likelihood that the cancer may be found only at a more advanced stage. This increase in risk is usually seen with as little as two years of use. However, the increased risk from combined HRT is reversible and its risk applies only to current and recent users, as a woman's breast cancer risk seemingly returns to that of the general population within five years of stopping HRT. The use of bioidentical or “natural” estrogen and/or progesterone is not necessarily safer or more effective, and thus should be considered to have the same health risks as any other type of HRT. Short term use of estrogen alone after menopause does not seem to increase the risk of breast cancer much. However, long-term use of estrogen therapy (e.g., >15 years) was reported to increase the risk of ovarian and breast cancer. Thus, the decision to use any forms of HRT should be made by a woman and her physician after weighing the possible risks and benefits, and considering her other risk factors for heart disease, breast cancer, and osteoporosis.
Excessive alcohol consumption: Drinking alcohol is clearly linked to an increased risk of breast cancer, and the increase in risk caused by this factor correlates with the amount of alcohol consumed.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 31 For example, women who have two to three drinks a day have approximately 20% higher risk of breast cancer compared to women who don't drink alcohol. Women who have only one alcoholic drink per day have a very small increase in risk.
Significant overweight or obese: Before menopause women's ovaries make most of the body's estrogen, while fat tissue makes only a small amount.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 However, when the ovaries stop making estrogen after menopause, most of a woman's estrogen comes from fat tissue. Thus, having more fat tissue after menopause will raise estrogen levels and increase breast cancer risk. Furthermore, being overweight tends to lead to higher blood insulin levels, and higher insulin levels are linked to certain cancers, including breast cancer. Nonetheless, the link between body weight and breast cancer risk is complex and remains to be fully understood.
Not having children or not breastfeeding: Women who have not had children or who have their first child after age 30 have a slightly higher overall risk for breast cancer. Conversely, having multiple pregnancies and/or becoming pregnant at an early age reduce breast cancer risk.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 Nonetheless, pregnancy seems to have different effects on different types of breast cancer, and pregnancy seems to increase risk for triple-negative breast cancer. It has been suggested that breastfeeding may slightly lower breast cancer risk, especially if it is continued for 1.5–2 years. A possible explanation for this effect is that breastfeeding reduces woman's total number of lifetime menstrual cycles.
Starting menstruation early or stopping menopause after age 55: Women will have more menstrual cycles if they start menstruating early, especially before age 12, and thus they will have a longer lifetime exposure to the hormones estrogen and progesterone, leading to a slightly higher risk of breast cancer.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 Similarly, women will have more menstrual cycles if they go through menopause later, especially after age 55, and also have a longer lifetime exposure to estrogen and progesterone with a higher risk of breast cancer.
Lack of physical activity: Growing evidence indicates that regular physical activity, especially in women past menopause, may reduce breast cancer risk.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 It is not completely clear how physical activity might reduce breast cancer risk, but it may be due to the fact that activity levels affect body weight, inflammation, hormones, and energy balance.
Classifications and stages of breast cancer
Types of breast cancer based on pathology, invasiveness and prevalence
There are many types of breast cancers as it can present in distinct areas of the breast, such as the ducts, the lobules, or the tissue in between. The type of breast cancer is determined by the specific cells that are affected. Based on which cell origin is involved, breast cancers can be divided into two broad classifications, carcinomas and sarcomas. Carcinomas are breast cancers arising from the epithelial component of the breast, which consists of the cells that line the lobules and terminal ducts responsible for making milk. Sarcomas are a much rarer form of breast cancer (<1% of primary breast cancer) arising from the stromal components of the breast, which include myofibroblasts and blood vessel cells. These groups are not always sufficient categories as, in some cases, a single breast tumor can be a combination of different cell types.4, 8, 9, 22, 23, 24
Most breast cancers are carcinomas. Within the large group of carcinomas, there are many different types of breast cancer identified based on their invasiveness relative to the primary tumor sites. Accurately being able to distinguishing between the various subtypes is vital as they each have different prognoses and treatment implications. Based on criteria of pathological features and invasiveness, common breast cancers can be divided into three major groups: non-invasive (or in situ), invasive, and metastatic breast cancers.4, 8, 9, 22, 23, 24
Non-invasive (or in situ) breast cancer
Ductal carcinoma in situ (DCIS; also called intraductal carcinoma): As one of the most common types of breast cancer, DCIS is a non-invasive or pre-invasive breast cancer, which develops inside of pre-existing normal ducts.4, 8, 9, 22, 23, 24 While DCIS is itself not invasive, in situ carcinomas have high potential to become invasive cancers, so early and adequate treatment is important in preventing the patient from developing an invasive cancer.
Invasive or infiltrating breast cancer Invasive breast cancers have cancer cells that invade and spread outside of the normal breast lobules and ducts, growing into the surrounding breast stromal tissue.4, 8, 9, 22, 23, 24 About two-thirds of women with an invasive form of breast cancer are 55 or older when they are diagnosed. Invasive carcinomas have the potential to spread to other sites of the body, such as the lymph nodes or other organs and to form metastases thus entering the classification of metastatic breast cancers.4, 8, 9, 22, 23, 24 Based on the tissue and cell types involved, invasive breast cancers are further divided into following two types:
Invasive Ductal Carcinoma (IDC): IDC is the most common type of breast cancer with about 80% of all breast cancers being constituted by invasive ductal carcinomas.4, 8, 9, 22, 23, 24 The IDC classification includes several subtypes: tubular carcinoma of the breast, medullary carcinoma of the breast, mucinous carcinoma of the breast, papillary carcinoma of the breast, and cribriform carcinoma of the breast.4, 8, 9, 22, 23, 24
Invasive Lobular Carcinoma (ILC): ILC is the second most common type of breast cancers and accounts for approximately 10–15% of all breast cancers.4, 8, 9, 22, 23, 24 Although ILC can affect women at any age, it is more common in older women. ILC tends to occur later in life than IDC, e.g. in the early 60s as opposed to the mid-to late 50s for IDC.4, 8, 9, 22, 23, 24
Together, 90–95% of all breast cancer cases fall into invasive subcategories. IDC and ILC cancers each exhibit distinct pathologic features. Lobular carcinomas grow as single cells arranged individually, in single file, or in sheets, and they have distinct molecular and genetic aberrations that distinguish them from ductal carcinomas. Ductal and lobular carcinomas may have different prognoses and treatment options and are thus important to clearly differentiate from one another.
Metastatic breast cancer
Metastatic breast cancers, also known as stage IV or advanced breast cancers, are late stage breast cancers, which have spread to other organs in the body.4, 8, 9, 22, 23, 24 Metastases from breast cancers can be found in lymph nodes in the armpit, and/or in distant sites such as the lung, liver, bone and brain. Even after the primary tumor is removed, microscopic tumor cells or micro-metastases may remain in the body, which allows the cancer to return and disseminate. Clinically, patients may initially be diagnosed with metastatic disease (or de novo metastatic breast cancers), or they may develop metastases months or years after receiving initial treatment. The risk of breast cancer returning and metastasizing is not clearly understood or predictable as it varies from person to person, largely depending on the unique molecular biology of the tumor and the stage at the time of the original diagnosis. Unfortunately, approximately 30% of the women diagnosed with early-stage breast cancer will develop a metastatic form of the disease.4, 8, 9, 22, 23, 24
Less common types of breast cancer
While IDC and ILC account for approximately 90–95% of all breast cancer cases, several rare types of breast cancers can observed in a clinical setting.4, 8, 9, 22, 23, 24
Inflammatory breast cancers (IBC): IBC is an uncommon type of invasive breast cancer that comprise 1%–5% of all breast cancers.4, 8, 9, 22, 23, 24 IBC differs from other types of breast cancers in its symptoms, prognosis and treatment. Typical IBC symptoms include inflammation-like breast swelling, purple or red color of the skin, and pitting or thickening of the skin of the breast, all of which are likely caused by cancer cells blocking lymph vessels in the skin. IBC often does not present with a breast lump and may not be identifiable on mammograms. IBC tends to occur in younger women, and is more common in African-American women as well as in women who are overweight or obese. Furthermore, IBC tends to be more aggressive, growing and spreading much more quickly than the common types of breast cancers. IBC is always first diagnosed at a locally advanced stage where the breast cancer cells have grown into the skin. In about one-third cases, IBCs are already metastasized to distant sites of the body at diagnosis, which makes it more difficulty to treat successfully.4, 8, 9, 22, 23, 24
Breast cancers in men and children & adolescents: Male breast carcinomas, which can be either in situ or invasive, account for <1% of all breast cancer cases.4, 8, 9, 22, 23, 24 Male breast cancers can look identical to those seen in women as most cases consist of invasive ductal carcinomas with estrogen receptor (ER) expression. The most common breast lesion in males is gynecomastia (or breast enlargement), which may involve either the unilateral breast or bilateral breasts. Breast lesions are also uncommon in children and adolescents, but do occur. These rare cases can include both benign lesions, such as juvenile fibroadenoma, and malignant lesions, such as secretory carcinoma. Pediatric patients are more likely to suffer from metastatic tumors to the breast from lymphoma or alveolar rhabdomyosarcoma.
Paget disease of the breast: This rare form of breast cancer starts in the breast ducts, spreads to the skin of the nipple and then expands to the areola (the dark circle around the nipple).4, 8, 9, 22, 23, 24 Paget disease makes up <3% of all breast cancer cases. Paget's cells look significantly different from normal cells and divide rapidly. About half of the cells are positive for estrogen and progesterone receptors, and most cells are positive for the HER2 protein. The cancer is typically diagnosed with a biopsy of the tissue, sometimes followed by a mammogram, sonogram or MRI to confirm the diagnosis. It should also be pointed out that Paget's disease of the breast is not related in any medical way to other conditions named after Sir James Paget, such as Paget's disease of the bone.
Papillary carcinoma: Papillary carcinoma is another rare form of breast cancer that accounts for <3% of all breast cancer cases.4, 8, 9, 22, 23, 24 Papillary carcinoma cancer cells are usually arranged in finger-like projections. In some cases, the cancer cells are quite small in size and form micropapillary. Most papillary carcinomas are invasive, and are treated in the same manner as IDCs. However, invasive papillary carcinoma usually has a better prognosis than other invasive breast cancer. It is noteworthy that papillary carcinomas may also be detected when they are still noninvasive. A noninvasive papillary carcinoma is usually considered a variety of DCIS.
Phyllodes tumor: Phyllodes tumors are rare breast tumors that develop in the stromal cells of the breast.4, 8, 9, 22, 23, 24 Most of these tumors are benign, but about one-quarter are malignant. Phyllodes tumors are most commonly found in women in their 40s, and in women with Li-Fraumeni syndrome who have an increased risk for this type of tumor.
Angiosarcoma of the breast: As a form of sarcoma, angiosarcoma is a rare cancer that originates the epithelial cells that line blood or lymph vessels.4, 8, 9, 22, 23, 24 Angiosarcomas make up <1% of all breast cancers and it can involve the breast tissue or the skin of the breast. Some arise from prior radiation therapy in that area. Although rare, angiosarcomas tend to grow and spread quickly and need to be treated accordingly.
Molecular or intrinsic subtypes of breast cancer
Breast cancer encompasses a heterogeneous and phenotypically diverse group of diseases. It is composed of several biological subtypes that have distinct behaviors and responses to therapy.4, 8, 9, 22, 23, 24 Gene expression studies have identified several distinct breast cancer subtypes that differ significantly in prognosis as well as in the therapeutic targets present in the cancer cells. With the advance of gene expression profiling techniques, the list of intrinsic genes that differentiate these subtypes is now made up of several clusters of genes relating to estrogen receptor (ER) expression (the luminal cluster), human epidermal growth factor 2 (HER2) expression, proliferation, and a unique cluster of genes called the basal cluster.4, 8, 9, 22, 23, 24 Through a utilization of these understandings, breast cancers are usually divided into five intrinsic or molecular subtypes that are based on the expression pattern of certain genes (Table 1).
Table 1.
Molecular/intrinsic subtypes of breast cancers.
| Subtypes | Molecular Signatures | Characteristics | Treatment optionsa |
|---|---|---|---|
| Luminal A | ER+, PR±, HER2-, Low Ki67 | ∼70%, Most common | Hormonal Therapy |
| Best prognosis | Targeted Therapy | ||
| Luminal B | ER+, PR±, HER2±, | 10%–20% | Hormonal Therapy |
| High Ki67 | Lower survival than Luminal A | Targeted Therapy | |
| HER2 | ER-, PR-, HER2+ | 5%–15% | Targeted Therapy |
| Triple Negative | ER-, PR-, HER2- | 15%–20% | Limited Targeted Therapy |
| More common in black women | |||
| Diagnosed at younger age | |||
| Worst prognosis | |||
| Normal-like | ER+, PR±, HER2-, Low Ki67 | Rare | Hormonal Therapy |
| Low proliferation gene cluster expression | Targeted Therapy |
Besides conventional surgical and non-surgical treatment.
Luminal A breast cancer: This subtype is estrogen-receptor (ER) and/or progesterone-receptor (PR) positive, HER2 negative, and has low levels of Ki-67. Luminal A cancers account for about 40% of all breast cancers. They are low-grade, slow growing, and tend to have the best prognosis. Treatment typically involves hormonal therapy.4, 8, 9, 22, 23, 24
Luminal B breast cancer: Accounting for <20% of all breast cancers, this subtype is ER and/or PR positive, either HER2 positive or HER2 negative, and presents high levels of Ki-67. Luminal B cancers grow slightly faster than luminal A cancers, and their prognosis is slightly worse.4, 8, 9, 22, 23, 24
HER2-enriched breast cancer: This subtype makes up 10%–15% of breast cancers and is characterized by the absence of ER and PR expression, the high expression of the HER2 and proliferation gene clusters, and the low expression of the luminal and basal clusters.4, 8, 9, 22, 23, 24 HER2-enriched cancers grow faster than luminal cancers and have a generally worse prognosis. However, they can be successfully treated with targeted therapies aimed at the HER2 protein, such as Herceptin (or trastuzumab), Perjeta (or pertuzumab), Tykerb (or lapatinib), and Kadcyla (or T-DM1 or ado-trastuzumab emtansine). It must be clarified that the HER2-enriched subtype is not synonymous with clinically HER2-positive breast cancer. While about 50% of clinical HER2-positive breast cancers are HER2-enriched, the remaining 50% can include any molecular subtype but are mostly HER2-positive luminal subtypes. However, about 30% of HER2-enriched tumors are clinically HER2-negative.4, 8, 9, 22, 23, 24
Triple-negative/basal-like breast cancer (TNBC): Accounting for approximately 20% of all breast cancers, the TNBC subtype is characterized as ER-negative, PR-negative and HER2-negative.4, 8, 9, 22, 23, 24 TNBC is more common in women with BRCA1 gene mutations as well as among women younger than 40 years of age and African-American women. TNBC usually behaves more aggressively than other types of breast cancer making it a high grade breast cancer. The most common histology seen in TNBC is infiltrating ductal carcinoma, although a rare histologic subtype, medullary carcinoma, is generally also triple negative. Unlike other breast cancer subtypes with an arsenal of targeted regimens such as ER antagonists and HER2 monoclonal antibodies, TNBC's non-surgical treatment has been limited to conventional chemotherapy, until the recent approval of the PARP inhibitor Olaparib for BRCA1 and BRCA2 mutation carriers, who are more likely to develop TNBC.4, 8, 9, 22, 23, 24
Normal-like breast cancer: This subtype is similar to luminal A disease. It is ER and/or PR-positive, HER2 negative, and has low levels of the protein Ki-67.4, 8, 9, 22, 23, 24 While normal-like breast cancer has a good prognosis, its prognosis is still slightly worse than luminal A cancer's prognosis.
The cellular and molecular heterogeneity of breast cancers mandates the analyses of multiple genetic alterations in concert, which has been made possible by the emergence of next-generation genomics and transcriptomics techniques.4, 8, 9, 22, 23, 24 Gene expression studies have thus identified several distinct breast cancer subtypes that differ markedly in prognosis and in their expression of therapeutic targets.32, 33, 34, 35, 36, 37, 38 In fact, the intrinsic subtypes bifurcate into two segregated groups that correspond to the expression of hormone receptor-related genes. This segregation is consistent with both literature and clinical experience which show ER-positive and ER-negative cancers to define biologically distinct phenotypes that may derive from different progenitor cells.35 Furthermore, the genomewide analyses and transcriptomic profiling have provided critical insights into the nuances of the molecular classification of breast cancers,34, 38 and have helped established several diagnostic and prognostic panels, such as the Oncotype Dx 21-gene Recurrence Score (RS),39 the Breast Cancer Index (BCI),40 the Predictor Analysis of Microarray 50 (PAM50) Risk of Recurrence (ROR) score,41, 42, 43, 44, 45 the Amsterdam 70-gene prognostic profile (Mammaprint),46, 47, 48, 49, 50 and the Genomic Grade Index (GGI).51
Clinical staging and survival rates of breast cancer
Once breast cancer is diagnosed, tests are performed to determine the stage of the disease, which will impact the treatment patients receive. The clinical staging of breast cancer is identical across breast cancer subtypes according to the American Joint Committee on Cancer (AJCC) and the International Union for Cancer Control (UICC) Tumor, Node, and Metastasis (TNM) breast cancer staging system: Stage 0, Stage I, Stage II, Stage III and Stage IV, as detailed in Table 2.
Table 2.
Anatomic stage groups of breast cancer.
| Stages | Definition | |
|---|---|---|
| Stage 0 | Ductal Carcinoma In Situ | |
| Stage I | IA | Primary invasive tumor with a size of ≤20 mm No nodal involvement |
| IB | Nodal micrometastases (>0.2 mm, <2.0 mm) with or without ≤ 20 mm primary tumor | |
| Stage II | IIA | Movable ipsilateral Level I, II lymph node metastases with ≤20 mm primary tumor; Or > 20 mm, ≤ 50 mm tumor with no nodal involvement |
| IIB | Movable ipsilateral Level I, II lymph node metastases with >20 mm, ≤ 50 mm tumor; Or > 50 mm tumor with no nodal involvement | |
| Stage III | IIIA | Movable ipsilateral Level I, II lymph node metastases with >50 mm tumor; Or any size primary tumor with fixed ipsilateral Level I, II or internal lymph node metastases |
| IIIB | Primary tumor with chest wall and/or skin invasion | |
| IIIC | Any size primary tumor with supraclavicular or ipsilateral Level III lymph node metastases; Or with ipsilateral Level I, II and internal lymph node metastases | |
| Stage IV | Any case with distant organ metastasis | |
Notes: 1). Lobular carcinoma in situ is now considered benign thus removed from the breast cancer staging system.
2). The Anatomic Stage Group is to be used when biomarker tests are not available.
Source: AJCC Cancer Staging Manual, Eighth Edition, The American College of Surgeons (ACS), Chicago, IL, USA. With reprint permission of ACS.
Cell origins of breast cancer
Mammary gland development and mammary stem cells
Human mammary gland development is a progressive process that is initiated during embryonic life.52, 53 The mammary gland consists of a highly branched network of epithelial tubes, embedded within a complex stroma (Fig. 1).53 The mammary epithelium originates during embryonic development from an epidermal placode.53 Regulated by epithelial–mesenchymal interactions, the placodes descend into the underlying mesenchyme and produce the rudimentary ductal structure of the gland present at birth.52, 53 At birth, the breast rudiment is formed by 10–12 primitive ductal elements located beneath the nipple–areola complex.52 The breast undergoes dramatic changes in size, shape, and function in association with puberty, pregnancy, and lactation, in response to steroid hormone and growth factor receptor signaling.53 One unique feature of the breast is that mammary epithelium is highly responsive to local and systemic signals and displays significant morphologic changes of the ductal tree during puberty and pregnancy.54
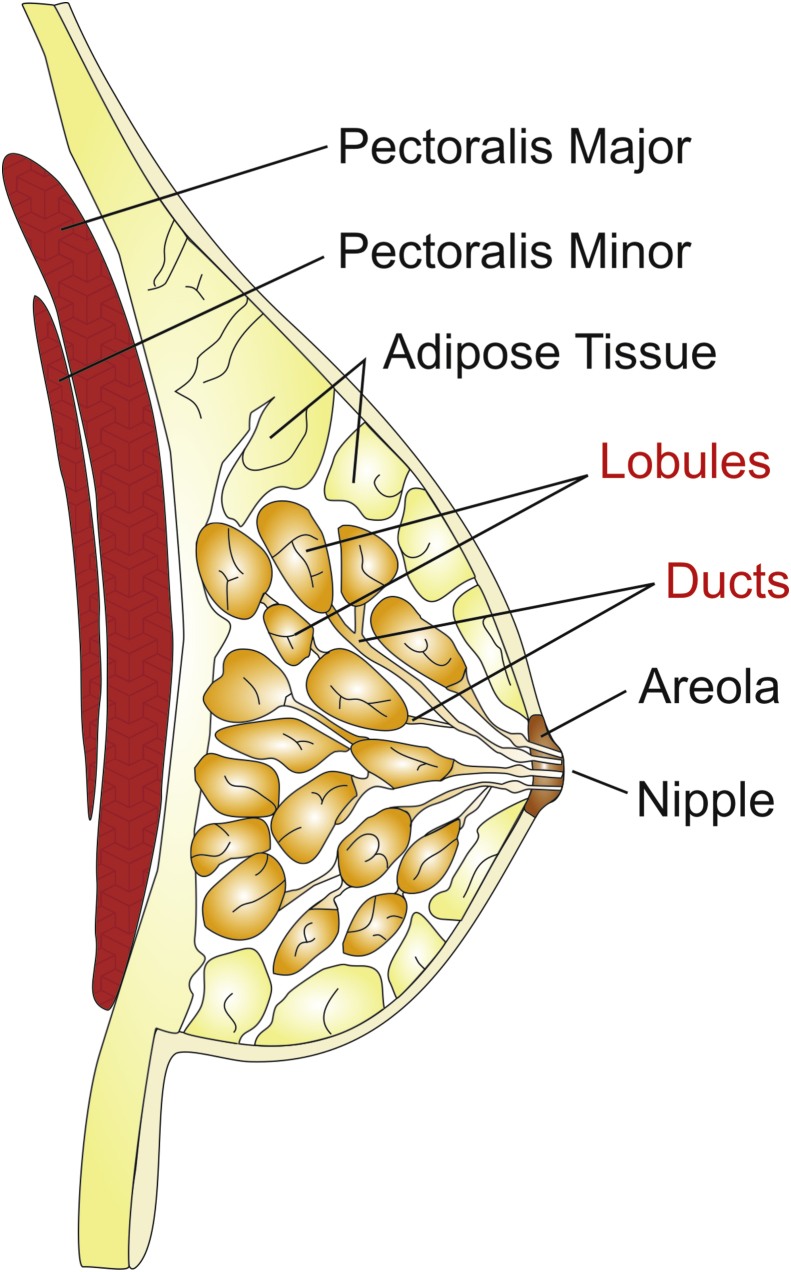
Figure 1.
Anatomical and histologic origins of breast cancer. Most breast cancers arise from the lobules or the ducts of the breast. In some cases, the tumor infiltrates the skin or components of the chest wall such as the pectoralis muscles. The tumor cells also are capable of converting the microenvironment into a tumor-friendly state to promote their growth and expansion.
Transplantation and lineage tracing studies indicated that a hierarchy of stem and progenitor cells exists among the mammary epithelium.54, 55 In fact, lineage tracing has highlighted the existence of bipotent mammary stem cells (MaSCs) in situ as well as long-lived unipotent cells that drive morphogenesis and homeostasis of the ductal tree.54, 56 Accumulating evidence suggests the existence of a heterogeneous MaSC compartment comprising fetal MaSCs, slow-cycling cells, and both long-term and short-term repopulating cells.54, 55, 56 Furthermore, diverse luminal progenitor subtypes have been identified in both normal mouse and normal human mammary tissue.56 This suggests that the elucidation of the normal cellular hierarchy is an important step toward understanding the “cells of origin” for breast cancer (Fig. 2).54, 55, 56, 57
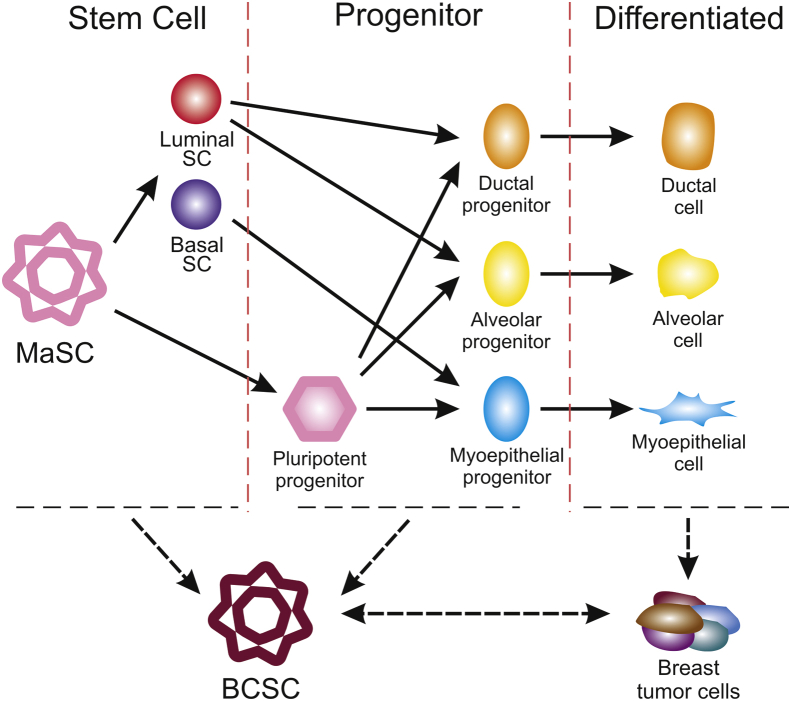
Figure 2.
Mammary cell hierarchy and breast cancer stem cells. Mammary stem cells (MaSCs) are multipotent self-renewing cells of great importance in the development and replenishment of mammary glands, as well as having implications in cellular origin of breast cancer stem cells (BCSCs), which has been extensively traced using various methods using human and mice samples. Two models have been established to define the relationship between MaSCs and breast cancer.
Mammary stem cells (MaSCs) exist as a very small proportion of cells in the mammary gland that are undifferentiated and can produce new MaSCs through self-renewal as well as to give rise to a variety of differentiated cells through symmetric and asymmetric divisions.57 Asymmetric divisions are hypothesized to give rise to progenitor cells.57, 58, 59, 60 In a healthy body, SCs are well understood in their longevity and self-renewal properties for their role in maintenance, repair, and tissue regeneration.60 It is accepted that MaSCs possess functions under normal circumstances involved in responding to cellular needs during the course of reproductive life. The function of MaSCs is thought to occur though close interaction with their specific cellular microenvironment which is also referred to as the mammary stem cell niche.57, 60, 61 The specific locations of these cells, with their unique self-renewal and differentiation characteristics, remain a topic for debate as does their role in breast cancer. MaSCs are broadly referred to as slow-cycling and long-lived stem cells, or as progenitor cells, which exist as a singular, role-committed population. There is still much debate over the cells' differentiation capabilities into one of both types of mammary epithelial cell lineages, as well as over the impact of cell plasticity.58
Roles of mammary stem cells in breast cancer development
One attempt to explain MaSCs in relationship to breast cancer, as these cells do have a normal role in the body, is the umbrella concept or cancer stem cell (CSC) theory.62, 63, 64, 65 While CSC theory has a fairly extensive history,65 the focus here will be on the most recent advancements and understandings of the theory as they relate to breast cancer (Fig. 2).54, 55, 56, 57 The primary bifurcation in this theory is whether CSCs are the cell of origin from which cancer cells develop, a hypothesis based on observations of the similarities between tissue replenishment and tumorigenesis, or if malignant cells attain stem cell potential.60, 66 Effectively, the debate centralizes around the importance of bipotent and unipotent luminal and myoepithelial stem cells during both normal cell development and carcinogenesis.54, 55, 56, 57
One challenge in accumulating data to either support or negate this theory is the inaccuracy and variability between human and mouse models of selected biomarkers some of which include CD49f, EpCAM, CD10 and ALDH1A1, SSEA4, CD44, CK12+/CK19+ 66. Gaining an understanding of cellular heterogeneity, which CSC theory attempts to explain, is imperative to the development of more specialized treatments for breast cancer, in which cell heterogeneity poses a major hurdle. CSC theory rests on a widely accepted structure of cellular hierarchy in both normal and carcinogenic cells with stem cells residing at the apex and cells differentiating from that point out.66 The consequence of this structure is that MaSCs, as distinct from mammary repopulating cells, are bi- or multi-potent, a conclusion that has been well supported. These mammary repopulating cells can be identified as adopting multipotent characteristics under regenerative conditions viewed through transplantation.67, 68
MaSCs are posited to exist in distinct microenvironment known as a stem cell niche, which places added importance on mutual feedback relationships between the epithelial cells that come from MaSCs, surrounding stromal cells, and the extracellular matrix.57 However, the signaling pathways and the ways in which they contribute to the normal and carcinogenic roles of MaSCs are not fully understood.54, 55, 56, 57 Two major cell lineages lie at the root of this argument in terms of breast CSC origin: basal cells and luminal cells.54, 55, 56, 57 Basal cells are one of the two main cell lineages in the in the mammary gland, and the other being luminal cells. Basal cells surround the luminal cell layer in the structure of the mammary gland. Luminal cells line the lumen of the mammary ducts and alveoli. Basal cells normally express cytokeratin-5, -14, and smooth muscle actin while luminal cells express cytokeratin-8 and can be either positive or negative for hormone receptors.54, 55, 56, 57 Recent studies have been able to exceed prior limitations to mouse models for cellular lineage-tracing to using patient-derived breast cancer organoids.69 The construction of a living breast cancer organoid biobank is one such project that may be employed for research on cancer cell heterogeneity and drug discovery as a more accurate model.69, 70, 71
Major signaling pathways in breast cancer development and progression
There are remarkable parallels between normal development and cancer progression at the molecular level.52, 53 Normal human development is tightly controlled by complex signaling pathways, which allow cells to communicate with each other and their surrounding environment.72, 73 Not surprisingly, many of these same signaling pathways are dysregulated or hijacked by cancer cells and CSCs.74 In essence, cancer is driven by genetic and epigenetic alterations that allow cells to escape the mechanisms that normally control their proliferation, survival and migration.74 Many of these alterations map to signaling pathways that govern cell proliferation and division, cell death, cell differentiation and fate, and cell motility.74 Thus, activating mutations of proto-oncogenes can cause hyperactivation of these signaling pathways, whereas inactivation of tumor suppressors eliminates critical negative regulators of signaling.74 Here, we will focus on the predominant signaling pathways that regulate normal mammary gland development and breast cancer stem cell functions, namely estrogen receptor (ER) signaling, HER2 signaling and canonical Wnt signaling.
ER signaling and ER-Positive breast cancer
Estrogen receptors (ERs) consist of membrane estrogen receptors (mostly G protein-coupled receptors) and nuclear estrogen receptors (ERα, ERβ).75, 76, 77, 78 Both ERα and ERβ are transcriptional factors that either activate or repress the expression of target genes upon ligand binding. ERα (coded by ESR1) and ERβ (coded by ESR2) share common structural features that serve their main functions while upholding receptor-specific signal transduction through exclusive elements.77, 78, 79 ERα and ERβ both contain six functional domains with various degrees of similarity and retain the ability of forming heterodimers.75, 77 The region with the highest identity of 96% is the DNA-binding domain (DBD), which mediates the interaction of ER dimers and estrogen response elements (EREs) of target genes (Fig. 3).77, 78, 79, 80
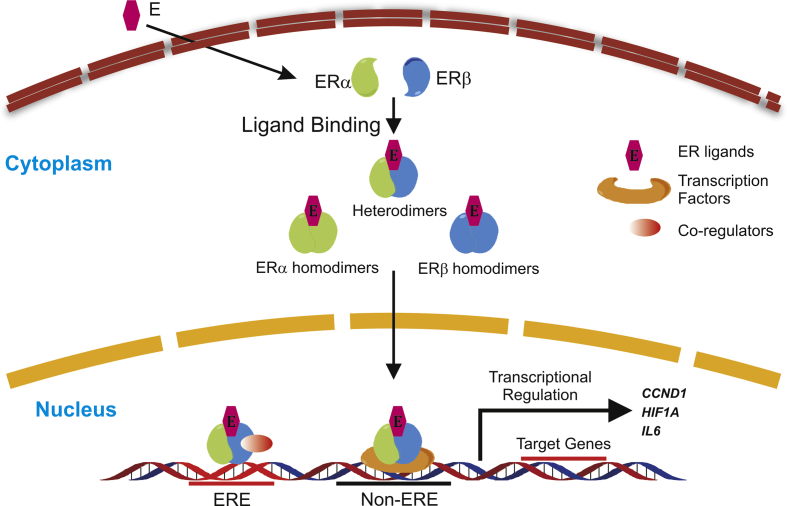
Figure 3.
ER signaling pathway. Breast cancer cells have relatively high ERα expression and low ERβ expression. These two types of nuclear hormone receptors form homo- or heterodimers upon ligand binding and translocate into the cell nucleus for transcriptional regulation, which is the main function of ERs. ER dimers bind to the ERE region of target genes and recruit co-regulators to achieve the regulation of transcriptional activity. Another mechanism by which ERs control the expression of target genes is acting as a co-regulator for other transcription factors.
Apart from the ERE-dependent pathway, ERs also have the capability of transcription regulation without the involvement of EREs.77, 78, 79 In ER-mediated transcription regulatory process, many co-activators and co-repressors,81, 82 including BRCA183 play important roles. BRCA1 acts as a tumor suppressor partially by inhibiting ERα signaling.84 The intricacy of ER-mediated signaling suggests its complex functions in many biological and pathological settings. ERα plays a major role in the pathogenesis of breast cancers as approximately 75% of breast cancers have positive expression of this specific type of hormonal receptor.85 The function of ERα in breast cancer biology is finely tuned by many factors and plays an active role in numerous occurrences of cross-talk.77, 80
One of the best characterized mechanisms by which ERα promotes the growth of breast tumor cells is through its interaction with cyclin D1.86, 87, 88 Cyclin D1 is an important activator of cyclin-dependent kinases (CDKs) 4 and 6, which coordinates the transition of cell cycle from G1 to S phase in many cancer cells.89 The synergism within the ERα and cyclin D1 feedback loop could moderately explain the mechanism of antiestrogen therapy resistance and provide rationale for the combined use of selective CDK4/6 inhibitors and hormonal therapy agents in ER + patients.90, 91, 92
Breast cancer biology and therapeutics does not only submit to the influence of the full-size ERα, but also many of its isoforms.93 Depending on the structural domains, these isoforms exhibit paradoxical effects in the regulation of ERα signaling.94, 95, 96 High expression of a truncated isoform of ERα, ERα36, was found to be correlated with metastatic phenotype and poor prognosis in breast cancer patients.97 The most commonly used ERα antagonist Tamoxifen had an adverse effect on ERα36, exacerbating stem cell qualities by upregulating the transcription of ALDH1A1, thus promoting the progression of the disease instead of suppressing it. This discovery could account for part of the reality that only about 70% of ER + patients respond to hormonal therapy and potentially increase the precision of personalized treatment.97
While ERα is the one of the mostly frequently examined parameters in breast cancer patients, ERβ has been gradually drawing attention. ERβ has partial overlap with ERα in the tissue expression profile. Normal breast tissue frequently expresses ERβ and the level decline along with the progression of breast tumors.98 Consistent with the pattern in expression, many studies provided both in vitro and in vivo evidence supporting its role as a breast cancer suppressor.99, 100 In a conditional p53 knockout mouse model, the formation of mammary tumors was sped up by the simultaneous knockout of Esr2 (mouse gene coding ERβ).101 Tumors in the double knockout mice also displayed more malignant phenotypes, suggesting ERβ execute its tumor-suppressing role at least partly by interacting with p53. ERβ signaling in breast cancer and other circumstances are not as well elucidated as ERα at the moment.101 However, since more studies are implying a significant position for ERβ in the mammary tumorigenesis, it will not be surprising if ERβ turn out to be just as crucial as ERα, if not more.80
HER2 signaling and HER2-Positive breast cancer
Human epidermal growth factor receptors (EGFRs, or HERs) 1 to 4 constitute a family of tyrosine kinase receptors expressed in normal tissues and in many types of cancer.102, 103, 104, 105, 106 Human epidermal growth factor receptor-2 (or HER2/NEU, c-ERBB2) is a member of the EGFRs.104, 106 Like the others, HER2 is a receptor tyrosine kinase that consists of an extracellular ligand-binding domain, a transmembrane domain, and an intracellular domain.105, 107 The constitutively active form makes HER2 the preferred component to form dimers with other molecules and grants HER2 the capability of affecting many cellular functions through various pathways.108, 109 Ligand binding and subsequent dimerization stimulate phosphorylation of tyrosine residues in the intracellular domain of HER2, leading to the activation of multiple downstream signaling pathways such as the mitogen-activated protein kinase (MAPK) and the phosphatidylinositol 4,5-bisphosphate 3-kinase (PI3K) pathways.110, 111, 112 These signaling pathways are heavily associated with breast tumorigenesis (Fig. 4).105, 106, 107, 113
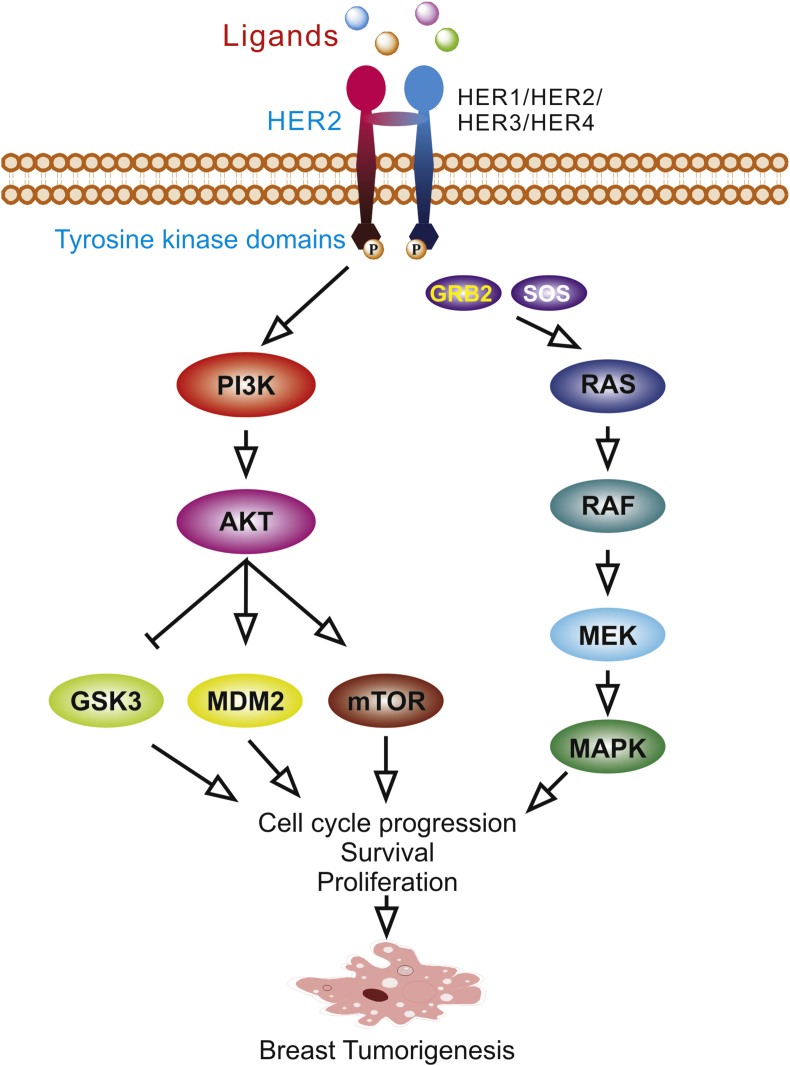
Figure 4.
HER2 signaling pathway. HER2 as well as the other members of the EGFR family are receptor tyrosine kinases which are located on the cell membrane and responds to a wide variety of ligands. Phosphorylation of the tyrosine kinase domain in the cytoplasm initiates downstream oncogenic signaling pathways such as PI3K/AKT pathway and Ras/MAPK pathway.
HER2 is amplified in various human breast cancer cell lines.114 HER2 signaling amplification results in HER2 protein overexpression which is linked to tumor cell proliferation and cancer progression.115 Targeted therapies are developed to bind specific molecules in signaling pathways important for cancer development and progression, providing most effective therapy in appropriately selected patients.105 Novel mechanisms underlying the relationship between HER2 and breast cancer have been uncovered recently.105, 116 While the interplay of HER2 and ERs has long been recognized,102 it was recently discovered that a new intermediary factor MED1 has significant impact on HER2-driven tumorigenesis.116 The precancerous effect of HER2 was also found to be linked to inflammation and the expansion of cancer stem-like cells (CSCs) in breast cancer.117 A newly identified enhancer located at the 3′ gene body of HER2 was reported to be the target locus of known HER2 regulator, TFAP2C.118 Other epigenetic mechanisms, such as DNA methylation and histone modifications, also affect this process.
HER2 has been widely used in generating animal models for investigating cancer biology and therapeutic efficiency, especially in breast tumor models because of its powerful transformation-inducing ability.119, 120, 121, 122, 123 A positive rate of 15%–20% in HER2 status (Overexpression/Amplification) (3 + by IHC or an amplified HER2 gene copy number by FISH)124 generally identifies those women who might benefit from HER2 targeted therapies, such as monoclonal antibodies or tyrosine kinase inhibitors (TKIs). Such novel targeted treatments have greatly improved the prognoses of patients with HER2+ neoplasms.125, 126
Breast cancer cells that express HER2 are more likely to progress to metastasis.127 Progesterone and progesterone-induced paracrine signals likely induce migration in early may induce migration in early primary tumor cells and, in this way, activate mammary stem cells. This proposed impact is consistent with HER2-stimulated stem-cell qualities that have been reported.123 Thus, HER2 testing is used to select patients appropriate for what is potentially resistant and expensive therapy.128 In pursuit of heightened specificity of treatment, HER2 molecular analysis has become an integral part of the diagnostic breast cancer patient work-up.128
Canonical Wnt/β-catenin signaling in breast cancer
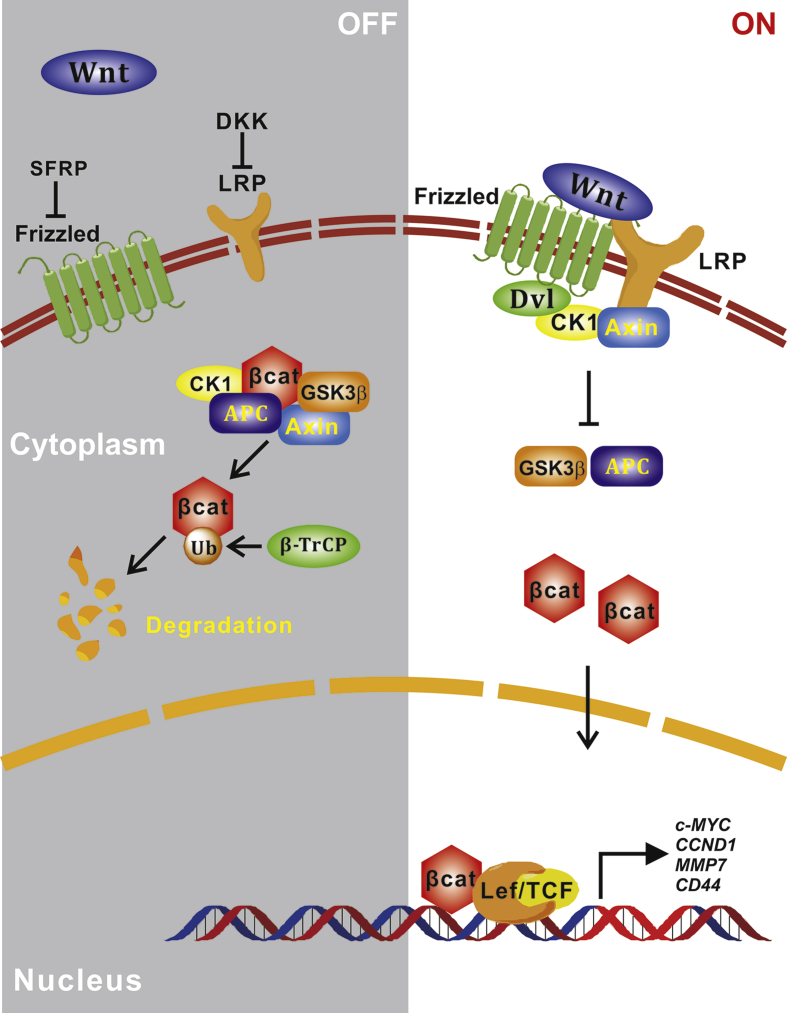
Wnt proteins are a family of highly glycosylated, secreted proteins with pivotal roles in various developmental processes including embryonic induction, generation of cell polarity, and cell fate specification, as well as in maintaining adult tissue homeostasis.129, 130, 131, 132, 133, 134, 135, 136 The canonical Wnt/β-catenin signaling is initiated by the binding of the secreted Wnt proteins, which is palmitoylated by Porcupin, to both co-receptors Frizzled and low-density lipoprotein receptor-related protein 5 and 6 (LRP5/6).131, 132, 134, 135 Wnt–receptor interaction leads to recruitment of Axin and Dishelved proteins to the cell membrane and induces inhibition of glycogen synthase kinase (GSK)-3β protein, which is a negative regulator of the Wnt pathway driven by β-catenin to proteasomal degradation that is induced by phosphorylation.131, 132, 134, 135 Inhibition of GSK-3β leads to β-catenin accumulation in the cytoplasm, and its subsequent translocation into the nucleus to act as co-transcriptional activator together with CREB binding protein (CBP) and T-cell factor/lymphoid enhancing factor (TCF/LEF) transcription factors and regulating oncogenes, such as MYC, CCND1 and other target genes (Fig. 5).131, 132, 133, 134, 135, 136, 137, 138
Figure 5.
Canonical Wnt/β-catenin signaling pathway. Canonical Wnt signaling plays significant roles in many biological and pathological processes such as mammary gland development and breast tumorigenesis. Wnt ligands bind with membrane receptors frizzled and LRPs, attenuating the ubiquitination of β-catenin by β-TrCP. Accumulation of β-catenin allows for nucleus translocation and downstream transcriptional activation. Inhibitors of Wnt signaling such as DKKs and SFRPs function as tumor suppressors by contributing to the degradation of β-catenin, thus impeding the transcription of β-catenin-targeting oncogenes.
The first clue about the role of Wnt signaling in mammary gland development came from the identification of the first mammalian Wnt molecule, Wnt1, as MMTV-induced mouse mammary oncogenesis involved an insertion at Wnt1 locus (or called MMTV int1).139 Soon after, Wnt3 was identified as another common insertion site for MMTV-induced mammary tumors in mouse.140, 141 Accordingly, mammary gland-specific expression of stabilized β-catenin has been shown to result in aggressive adenocarcinomas, consisting predominantly of glandular and undifferentiated cells.142 Interestingly, while neither Wnt1 nor Wnt3 is highly expressed in the adult mammary gland, other members of the Wnt family, such as Wnt-2, 4, 5a, 5b, 6, and 7, are expressed at various stages of mammary development.143, 144, 145 Wnt-2, 5a, and 7b are strongly expressed in the virgin mouse mammary gland, but are downregulated during pregnancy, while pregnancy strongly induces the expression of Wnt-4, 5b, and 6.144 Overexpression of Wnt-4 was shown to result in an increase in ductal branching,146 while the mammary tissue of Wnt-4−/− mice exhibited significantly reduced ductal branching compared to their wild-type counterparts.147 Mice lacking Lef-1 or Lrp6, and mice overexpressing the Wnt inhibitor Dickkopf all failed to develop an appropriate mammary bud.148, 149, 150 Studies using the Wnt reporter mice (Axin2-LacZ) showed positive staining in nearly all branches of the mammary ductal system, and Wnt activity was detected in cells located in the basal layer of the mammary ducts, which has been suggested to be the MaSC niche.151, 152
In breast cancer, Wnt signaling is likely constitutively activated through an autocrine mechanism. While mutations in Wnt signaling are not common in breast cancer, it has been shown that approximately 50% of clinical breast cancer cases exhibit high levels of stabilized β-catenin.153, 154 The positive regulator of Wnt signaling Dvl is amplified in 50% of breast cancers.155 Conversely, a secreted Wnt inhibitor, Frizzled-related protein 1 (FRP1) was reportedly lost in 78% of malignant breast cancers,156 and associated with poor prognosis.157 Down regulation of Wnt inhibitor Dickkopf 1 (DKK1) suggests the importance of Wnt regulation in the metastatic process in breast cancer.158 Furthermore, the expression of APC is lost in approximately 36–50% of breast cancers either by mutations, loss of heterozygosity, or hypermethylation.159, 160 The Wnt/β-catenin pathway was remarkably activated in basal-like breast tumors, and its nuclear localization was correlated with worse prognosis.161, 162 Not only does activated β-catenin promote triple-negative breast cancer, it also has a role in HER2-driven mammary tumors shown by in vivo data.163 With many of Wnt ligands being able to promote the progression of breast tumors, the restoration of many Wnt inhibitors that has been silenced by mechanisms such as DNA methylation and miRNAs in tumors has effectively attenuated tumor growth.164, 165, 166 Wnt pathway activation was shown to increase radiation resistance of progenitor cells in the mouse mammary gland and human breast cancer cell lines,167, 168 indicating that Wnt signaling is involved in resistance to current anticancer drugs potentially by regulating stem and progenitor cell populations.
Emerging evidence suggests that breast cancer initiation and maintenance may be regulated by a small population of cells within the tumor, either stem cells or cells that exhibit stem-like properties.169 The expression of Wnt1 in human mammary epithelial cells increased stem cell self-renewal, resistance to apoptosis and failure to senescence.170 Similarly, an expanded mammary stem cell pool was identified from a population of committed luminal progenitors in MMTV-Wnt1 mouse model,171 indicating that Wnt-1 activation may induce the appearance of aberrant progenitor cells. It was also found there was a 6.4-fold expansion of MaSCs in premalignant MMTV-Wnt1 transgenic mammary glands.152 The pre-neoplastic lesions and tumors of Wnt-1 mice were shown to have expanded stem/progenitor cell populations.172 Similar results were obtained from transgenic mice expressing β-catenin or c-myc, two downstream components of Wnt signaling, while mammary tumors in transgenic mice expressing Neu, H-Ras or plyoma middle T antigen did not exhibit a similar enrichment for MaSCs.172 Conditional deletion of a single APC allele in either the mammary stem/progenitor population or luminal cells of lactating mice revealed that mammary tumors only developed when APC was deleted in mammary progenitor cells, but not in luminal cells, suggesting Wnt-induced tumorigenesis targets the stem/progenitor population.173 Lastly, the inhibition of WNT1 was shown to alter the phenotype to CD44+CD24−ALDH1− and reduce tumor formation and cellular migration.174 Furthermore, suppression of GSK3/β-catenin signaling via the inhibitory activity of protein kinase D1 (PRKD1) was sufficient to reduce the stemness features of breast cancer cells.175 Thus, Wnt signaling may play an important role in maintaining mammary stem cell properties.
Other signaling pathways in breast cancer
Normal mammary stem cells and mammary development is controlled by a variety of hormones and signaling pathways.151 In addition to the three pathways discussed above, many other pathways and their crosstalk play important roles in regulating normal mammary development, as well as in breast cancer development if they are dysregulated. These include CDKs (Cyclin dependent kinase), Notch, SHH, PI3K/Akt/mTOR, and others.128
Cyclin dependent kinases (CDKs): Cell cycle progression is regulated by cyclins, CDKs, and CDK inhibitors (CDKIs).176 All cancers activate the cell cycle to sustain their survival. The overexpression of cyclin D1 and cyclin E as well as the decreased expression of CDKI p27Kip1 were found in human breast cancer.177 Cyclin D1 amplification is seen in nearly 60% of breast cancers. Furthermore, estrogen utilizes cyclin D1 to exert its mitogenic effects. It was reported that high tumor expression of cyclin D1 and overexpression of HER2 were associated with reduced recurrence-free survival and tamoxifen responsiveness.178 The oral CDK4/6 inhibitor Palbocyclib alone was shown to inhibit cell cycle progression ER-positive cell lines, including those with HER2 amplification, which were most sensitive to growth inhibition by palbocyclib while nonluminal/basal subtypes were most resistant.91 Thus, Palbocyclib is a promising therapeutic in breast cancer, and has been recently approved as a combination with fulvestrant for the treatment of hormone receptor-positive, HER2-negative metastatic breast cancer.128, 179
Notch signaling: Notch signaling is implicated in the pathogenesis of breast cancer and as such may represent a novel therapeutic target. Notch signaling consist of five Notch ligands, namely Delta-like (Dll) 1, 3, 4, and Jagged (JAG) 1, 2, which are single transmembrane proteins.180 There are four Notch receptors which act through the same basic signaling pathway and is activated by binding of Notch ligand on one cell to the extracellular domain of a Notch receptor on another neighboring cell. The Notch ligand–receptor complex then undergoes several key proteolytic cleavages, yielding the Notch extracellular domain and the Notch intracellular domain (NICD). NICD acts as a transcriptional factor and regulates downstream target genes.180 It was reported that high-level expression of Jag1 (Jag1High) and/or Notch1 (Notch1High) in tumors correlated with poor outcome and is an independent prognostic indicator in primary human breast cancers.181, 182, 183 Mammosphere self-renewal was inhibited by Notch 4 blocking antibody or an inhibitor of the γ-secretase enzyme,184 and the efficiency of DCIS-derived mammosphere production was significantly reduced when Notch signaling was inhibited.185 In primary breast cancer and breast cancer cell line-derived tumorspheres, Notch 3 and Jag1 have emerged as key regulators of cancer stem cell renewal and hypoxia survival.186 Furthermore, Notch signaling also interacts with HER2 signaling pathway, which is active in approximately 20% of breast cancers and associated with a more aggressive disease.
Sonic Hedgehog (SHH) signaling: SHH signaling plays a critical role in organizing cell growth and differentiation during embryonic tissue patterning,187, 188 and is important in mouse mammary gland development. Disruption of its downstream transcriptional targets, Patched homolog-1 (PTCH-1) or glioma-associated oncogene-2 (GLI-2) resulted in severe defects in ductal morphogenesis.189, 190 It was shown that disruptions of these genes also occurred in breast cancer,190, 191 suggesting a role for the SHH pathway in breast tumorigenesis. This data suggests that SHH activation may contribute to the relapse of breast cancer and may serve as a predictor of postoperative relapse since higher expression of SHH, PTCH-1, GLI-1, and SMOH in breast cancer was shown to correlate with breast cancer invasiveness.191
Breast tumor kinase (BRK): BRK is a non-receptor tyrosine kinase identified in breast tumors,192 and is overexpressed in more than 60% of breast cancer cases, but not in normal mammary glands or benign lesions.193, 194 Depletion of BRK in breast cancer cells was shown to impair EGFR-regulated signaling.195 Activation of BRK by stably expressing BRK-Y447F significantly increased MAPK activity, cell proliferation and migration in breast cancer cells, while a decreased migration was observed in breast cancer cells depleted of BRK.196 Thus, BRK may play an important role in promoting cell proliferation and migration in breast cancer cells.
PI3K/AKT/mTOR pathway: As discussed in the following section, oncogenic mutation of PI3K (i.e., PIK3CA) is a common one in human breast cancer that may lead to the dedifferentiation of luminal or basal mammary progenitor cells, allowing them to attain multi-lineage potential.197 Hyperactivation of AKT and the subsequent hyperactivation of downstream mTOR may underlie resistance to endocrine therapies.198, 199, 200 Clinically, activation of AKT is associated with a worse outcome among patients receiving endocrine therapy, with reduced clinical benefit in patients with positive expression of activated AKT.201 It was reported that there was an inverse correlation between AKT activation and partial response rates.202 Accordingly, the expression of phosphorylated S6 kinase (S6K), a downstream mediator of mTOR activation, can predict overall survival in patients with hormone receptor-positive breast cancer receiving adjuvant endocrine therapy.115 Thus, mTOR inhibition may be able to restore anti-estrogen sensitivity.
Cancer gene mutations in breast cancer
BRCA1/2 mutations in breast cancer
Approximately 10%–20% breast cancer patients have at least one first-degree relatives affected with breast cancer.8, 203 Among them, up to 20% of women with a family history of breast cancer have a mutation in the breast cancer susceptibility genes 1 or 2 (BRCA1 or BRCA2).204 Thus, the significance of BRCA1 and BRCA2 in breast cancer can be told from their names.205, 206 The prevalence of germline BRCA mutations is relatively high in women of Ashkenazi Jewish ethnicity, where the risk is estimated to be 30%–35%.207, 208, 209 In male breast cancer cases, up to 14% have a BRCA2 mutation although 4.5% of Ashkenazi Jewish men presenting with breast cancer have a BRCA1 mutation.210, 211 Moreover, among women with ovarian cancer, regardless of family history, about 15% are attributable to BRCA mutations,212 while the proportion with a germline BRCA mutation may be as high as approximately 40% in Ashkenazi Jewish women with epithelial ovarian cancer.213
The BRCA proteins share a similar, and cooperative, tumor suppressing mechanism by repairing DNA damage through homology-directed repair (HDR), which inhibits tumorigenesis.206, 214 Thus, deletion mutations and/or loss of function in the BRCA genes lead to decreased DNA repair efficiency and possibly give rise to the expansion of cancerous cells, elevating the risk of developing breast cancer by five to six fold.1, 215
Even though BRCA mutations can be inherited and responsible for a certain proportion of familial cases, investigators have found no difference in the incidence of breast cancer in BRCA mutation carriers with or without intimate family history.216 A recent study has shown that smoking also plays a minor role in increasing the likelihood of developing breast cancer, as well as other cancers for BRCA-mutation carriers.217 These results will increase the accuracy of identifying the high-risk population and the efficiency of preventative measures.
For more than two decades, the prevalence of genetic variations of the BRCA genes in breast cancer and other cancers has been well-investigated.218, 219, 220, 221, 222, 223 Interpretations that are more comprehensive were made available with the advancement of data collection and analysis. Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA; http://cimba.ccge.medschl.cam.ac.uk/) has made significant contributions to the characterization of the BRCA landscape. A latest update report of the CIMBA dataset summarized a total of 1650 and 1731 unique mutations in BRCA1 and BRCA2 respectively.224 Of all the types of BRCA1 and BRCA2 mutations, frameshift is the most common, mostly leading to the generation of premature stop codons and therefore decreasing the levels of mature RNAs and functional proteins. Enduring efforts made by researchers or organizations such as CIMBA demonstrate the ubiquitous perception of higher ratio of BRCA mutations in younger patients with more aggressive subgroups of breast cancer.223, 225, 226, 227 Nonetheless, the majority of these studies imply a comparable outcome among patients regardless of BRCA status, limiting the application of BRCA mutation status in prognosis prediction.
Clinical data suggest that BRCA1 mutation-related breast cancers that do not overexpress ER or ERBB2 are related to the expression of the basal epithelial markers, which are already associated with ER/ERBB2-negative tumors.228 While the cells are phenotypically basal-like, there is evidence that BRCA-1 basal-like breast cancer cells as well as sporadic basal-like breast tumors are not originated from basal stem cells, but rather from luminal epithelial progenitors.229 Cells with a potential for phenotypic plasticity do not necessarily reflect histology in tumor phenotypes.229
Oncogenic mutations of PIK3CA in breast cancer
Phosphatidylinositol 3-kinase (PI3K) is divided into three classes (I–III) based on their structure and substrate specificity. Class I PI3K is further categorized into class IA and IB, in which Class IA PI3K is the class most closely implicated in cancer.230 Structurally, PI3K is constituted of a p110 catalytic subunit and p85 regulatory subunit. There are three isoforms of p110, namely p110α (encoded by PIK3CA), p110β, and p110δ. While p110δ is expressed exclusively in leukocytes, p110α and p110β are ubiquitously expressed.230, 231 Conversely, human regulatory subunits p85α, p85β, and p55γ are encoded by PI3K regulatory subunit 1 (PIK3R1), PIK3R2, and PIK3R3, respectively.231 PI3K signaling is initiated by the growth factor activated receptor tyrosine kinase, or RAS protein, though direct interaction with p85 or via adaptor proteins, resulting PI3K being recruited to the membrane.230, 231 Activated PI3K subsequently activates critical downstream mediators AKT and mTOR, leading to enhanced growth, anti-apoptosis, cell-cycle progression, and translation.230, 231, 232
The PI3K/AKT/mTOR pathway is the most frequently enhanced oncogenic pathway in breast cancer.230 Among mechanisms of PI3K enhancement, PIK3CA mutations are most frequently (∼30%) observed, along with protein loss of PTEN,230, 233 although somatic mutations of PIK3CA coding p110α in various solid malignancies were first reported in 2004.234 The majority of PIK3CA somatic mutations are located in the two “hot spots”, E542K or E545K in exon 9, and H1047R or H1047L in exon 20, both of which are gain-of-function mutations and have transforming capacity.230, 234 Interestingly, PIK3CA gene amplification was reported even before PIK3CA mutations were identified, and was found in various malignancies, including approximately 10% of cases of breast cancer.235, 236
In addition to PIK3CA mutations, there are many other PI3K-enhancing mechanisms, such as HER2 amplification, dysfunction of PTEN, and AKT1 activating mutation. For example, PIK3R1 mutations were found in breast cancer with much lower occurrence (∼3%) as the PIK3R1 gene product p85α plays a tumor-suppressor role by stabilizing p110α.38, 230 AKT1 mutations (E17K) have been found in 1.4%–8% of breast cancers, especially in tumors expressing both ER and PR.237 PIK3CA mutations and gain of copy number of PIK3CA and PTEN loss and PTEN mutations have also been reported to coexist, although PIK3CA, PTEN, AKT1, and PIK3R1 mutations are reported to be mutually exclusive.230, 237, 238
Oncogenic PIK3CA mutations are thought to cause dedifferentiation of luminal or basal mammary progenitor cells, allowing them to attain multi-lineage potential.197 Mutations that upregulate the PI3K pathway in ER-positive breast cancers, such as downregulation of PTEN, overexpression of HER2 or IGF-1R, or the activation of mutant AKT1, can lead to the acquired resistance to hormone therapies in ER-positive tumors.233 Overall, PIK3CA mutations are most likely found in luminal-type (HR-positive/HER2-negative) tumors, in particular those with markers indicating less aggressive tumor characteristics.230 Furthermore, it was reported that treatments with PI3K inhibitors in ER-positive, PIK3CA-mutant breast cancers had to be continuous to achieve optimal therapeutic effects, and to overcome proliferative rebound and antiestrogen resistance; and yet the occurrence of such resistance observed in one third of patients in this subtype and resulted in an exceptionally poor prognosis.239 Nonetheless, it remains to be determined whether PIK3CA mutations are valid prognostic or predictive biomarkers for the clinical management of breast cancer patients.
Other gene mutations in breast cancer
Mutations of the BRCA1 and BRCA2 genes do not explain the occurrence of breast cancer in every breast cancer prone family.240, 241, 242 In fact, several breast cancer susceptibility genes, many of which are associated with rare genetic syndromes, have been identified and may account for less than 1% of all hereditary breast cancers.240, 241, 242, 243 These gene mutations are much less common and those representative of this group of genes include the following:
ATM and ataxia telangiectasia: The ATM gene product participates in damaged DNA repair. Most ataxia telangiectasia patients do not survive to an age at which breast cancer generally occurs. The risk of ataxia-telangiectasia carriers to develop breast cancer is estimated to be 11% by the age of 50 and 30% by the age of 70.244
TP53 and Li-Fraumeni syndrome: Inactivating mutations in the TP53 gene have been found in many cancer types including breast cancer.245 Li-Fraumeni syndrome is an autosomal dominant disorder, caused by germline mutations in the TP53 gene, leading to an increased risk of osteosarcomas, leukemia, brain tumors, adrenocortical carcinomas, and breast cancers.246 The risk of developing breast cancer before age 45 is18-fold higher for affected females, as compared to the general population.241 While germline mutations in the TP53 gene may account for <1% of breast cancer cases, somatic mutations in the TP53 gene are reported in 19–57% of human breast cancers, and LOH is found in 30–42%.241
PTEN and Cowden syndrome: Cowden syndrome is an autosomal dominant disorder, characterized by the development of hamartomas and benign tumors.241 Mutations in the PTEN gene are present in 80% of Cowden syndrome families.247 Truncating PTEN mutations in Cowden syndrome families cause a 25–50% lifetime breast cancer risk in women.241 In sporadic breast cancer patients, germline and somatic mutations in the PTEN gene are rare.
STK11 or LKB1 and Peutz-Jeghers syndrome: Peutz-Jeghers syndrome is caused by truncating germline mutations in the LKB1 gene and is an autosomal dominant disorder characterized by hamartomatous polyps in the small bowel and pigmented macules of the buccal mucosa, lips, fingers, and toes.241, 248, 249, 250
PALB2 (Partner and Localizer of BRCA2): PALB2 was originally identified as a BRCA2-interacting protein but subsequently also shown to interact with BRCA1.251, 252 Biallelic germline loss-of-function mutations in PALB2 (also known as FANCN) cause Fanconi's anemia, whereas monoallelic loss-of-function mutations are associated with an increased risk of breast cancer and pancreatic cancer.253 Loss-of-function mutations in PALB2 are an important cause of hereditary breast cancer, with respect both to the frequency of cancer-predisposing mutations and to the risk associated with them.254, 255, 256
CHEK2: Checkpoint kinase 2 (CHEK2) is a serine/threonine kinase, which is activated upon DNA damage and plays an important role in governing DNA repair, cell cycle arrest or apoptosis in response to the initial damage. Loss of kinase function due to mutation is correlated with several types of cancer, mainly breast cancer.257
CDH1: CDH1 encodes for the E-cadherin protein. Hereditary diffuse gastric cancer is an autosomal dominant inherited disease associated of CDH1 germline mutations. While the affected individuals develop a rare type of stomach cancer, invasive lobular breast cancer is the second most frequent type of neoplasia.258, 259
Epigenetics and non-coding RNAS in breast cancer
The important role of epigenetic changes such as aberrant DNA methylation and histone modification in cancer causation, progression and treatment has been increasingly recognized and exploited, as the enzymatic processes that control the epigenome present new opportunities for developing therapeutic strategies to target transcriptional abnormalities that are inherent to the cancer epigenome.260, 261 In addition, epigenetic factors may provide a mechanism for the development of cancer cell heterogeneity. It was reported that the analysis of DNA methylation and gene expression identifies conserved, reproducible patterns of association in three main breast cancer cohorts, which were related to two main branches of expressed qualities, tumor infiltrating immune cell signatures and ER signaling.262 The significant demethylation in the binding sites at the enhancers of three well-studied transcriptional factors, ERα, FOCA1, and GATA has been detected in ER + breast cancers in comparison to normal breast tissue.262, 263, 264 The elevated expression of these genes was also seen to be elevated in Luminal A and Luminal B types in contrast to Normal- and Basal-like breast cancers strongly linking this cluster of transcription factors to estrogen signaling.262 DNA methylation at the enhancers of ERα binding sites may also be a root cause of resistance to anti-estrogen treatments like tamoxifen for ER + breast cancers.97, 265, 266, 267
Recent rapid advances in large-scale whole genome sequencing and RNA sequencing techniques, in combinations with bioinformatics analyses have profoundly changed our understanding about human genome. Surprisingly, comprehensive global transcriptomic analyses indicated that up to 80% of the human genome while <2% of them encode proteins.268, 269, 270, 271 Thus, the vast majority of mammalian transcriptome encompass noncoding RNAs (ncRNAs) that play a variety of important regulatory roles in gene expression and other biological processes.271, 272 The ncRNAs are divided into two categories according to the size and structural or regulatory properties. The ncRNAs >200 nucleotides are called long noncoding RNAs or lncRNAs, whereas ncRNAs <200 nt include microRNAs (or miRNAs), small nucleolar RNAs (or snoRNAs), and piwi RNAs (or piRNAs).272, 273, 274 The miRNAs is the best studied short noncoding RNAs (sncRNAs), being extensively characterized for their biogenesis, function and importance in tumorigenesis.270, 271, 272 On the other hand, lncRNAs have emerged as key regulators of developmental processes, including mammary gland development.271, 272 In fact, lncRNA dysregulation is implicated in the development of various cancers, including breast cancer.272
A recent genome-wide transcriptional survey was carried out to explore the lncRNA landscape across 995 breast tissue samples was recently performed.275 A total of 215 lncRNAs were found aberrantly expressed in breast tumors, as compared to normal samples.275 On the basis of lncRNA expression, unsupervised hierarchical clustering of breast tumors identified four breast cancer subgroups that correlate tightly with PAM50-defined mRNA-based subtypes.275 Further analysis of the co-expression of lncRNA genes and protein-coding genes revealed a correlation, on one hand, between luminal A-specific lncRNAs and the activation of PI3K, FGF and TGF-β pathways, and on the other hand, between basal-like-specific lncRNAs and the activation of EGFR-dependent pathways and of the epithelial-to-mesenchymal transition, suggesting a wide range of biological functions associated with lncRNAs in breast cancer275
Recent studies have shown that ncRNAs are key players in the initiation and progression of cancer, and epigenetic mechanisms are deeply involved in their dysregulation. A growing list of miRNA genes aberrantly methylated in cancer, suggesting that many miRNAs may act as tumor suppressors or oncogenes.261 In addition, it has been shown that dysregulation of lncRNAs plays critical roles in tumorigenesis.261 Since ncRNAs are involved in regulating gene expression through interaction with epigenetic modifiers, their dysregulation may cause epigenetic alterations in cancer. Dissection of the interrelationships between ncRNAs and epigenetic alterations has the potential to reveal novel approaches to the diagnosis and treatment of cancer.261 It has been shown that lncRNAs can impact a variety of factors through mediated regulation of protein coding RNAs by acting cis and/or trans to guide epigenetic modifier complexes to targeted sites and protein stability, as well as by affecting miRNA binding capabilities.276, 277 LncRNAs, and other noncoding RNAs such as miRNAs, work in conjunction with RNA-binding proteins (RBPs), to alter both co- and post-transcriptional gene regulation.278 There are many cases in which, through epigenetic modifications, lncRNAs act as gene suppressors though lncRNAs may also act as gene activators.279, 280 MANCR (mitotically-associated noncoding RNA) is a lncRNA of particular interest, especially in its proposed association to TNBC, in the current search for therapeutic targets for its noted upregulation in breast cancer cells.281
Consistent with the genomic instability associated with TNBC, the mutation rate in both coding and noncoding regions were significantly higher in ER-negative/HER2-negative tumors.282 Substitution errors are the most frequent aberrations in noncoding regulatory regions, namely the promoters of cancer-associated genes including OBSCN and TP53. Additionally, identified genetic variations in promotors, introns, and other such regulatory noncoding regions may impact the phosphorylation, protein–protein interaction, and regulation of cancer-associated genes including ATM/ATR, FGFR1, FOXA1, IGF1R, NF1, NOTCH2, and TOP2A.282 It has been reported that the hypomethylation phenotype is observed at a higher rate in many lncRNAs in breast cancer and other cancers as opposed to the well-studied phenomenon of CpG island hypermethylation phenotype (CIMP) in tumors.262 The hypomethylation of lncRNA EPIC1 upregulates its expression, and EPIC1 overexpression is related to poor survival outcomes in independent patient cohorts, in particular, for luminal B breast cancer, and was shown to promote tumorigenesis through interacting directly with MYC to increase the occupation of MYC target genes and promote cell-cycle progression.265
The impacts of lncRNA epigenetic alterations can be studied using a repurposing of HM450 probes to analyze lncRNA promoters and comparing the lncRNA DNA methylation profile of lncRNA promotors to an existing database.265, 283 In fact, several studies identified lncRNA copy-number alterations and expression alterations, on account of which there is a point of reference for looking into methods for identifying and understanding potentially relevant lncRNAs.265, 277, 284 A recent pan-cancer analysis of S-phase enriched lncRNAs using nascent RNA capture assays has identified hundreds of potential biomarkers and oncogenic drivers, contributing to the process of illuminating the role of lncRNAs in S-phase regulation.277 Nonetheless, while herculean efforts have been in place to identify genetic driver mutations for breast cancer, the realm of noncoding RNA sequences and epigenetic factors remain largely unexplored by comparison.
Molecular basis of triple-negative breast cancer (TNBC)
Triple-negative breast cancer (TNBC) is broadly defined as tumors that lack expression of the estrogen receptor (ER), progesterone receptor (PR), and HER2.285, 286, 287 TNBC accounts for approximately 20% of breast cancers and is more commonly diagnosed in women younger than 40 years, as well as in African-American women.288 Genetically, <20% of patients with TNBC harbor a breast cancer gene (BRCA) mutation, particularly in BRCA1.289 Pathologically, TNBC is usually high grade and commonly infiltrating ductal carcinoma exhibiting geographic necrosis.290 TNBC Patients usually have a poorer outcome compared with those with other breast cancer subtypes owing to an inherently aggressive clinical behavior and a lack of effective targeted therapies.286, 291 The diagnosis of TNBC relies on the accurate determination of ER and PR protein levels by immunohistochemistry (IHC) and of HER2 by IHC and/or fluorescence in situ hybridization (FISH).286, 287 Such accurate assessment is crucial to avoid false diagnosis of ER-negative and/or HER2-negative disease in patients that would be benefited from endocrine therapy and/or HER2-targeted drugs.285, 286 TNBC clinical phenotype usually consists of the basal-like molecular subtype, although TNBC and basal-like breast cancers are not synonymous and yet there is substantial heterogeneity within TNBCs.
The molecular landscape of TNBC is complex and remains to be fully understood, largely due to the heterogeneous genetic and clinical features. An extensive IHC staining study of 172 TNBC samples revealed that only 71% of TNBCs were assigned the basal subtype.292 In a converse analysis of 160 tumors, in which subtype was identified and correlated with IHC staining, 77% of basal tumors were triple-negative by IHC.292 A single-cell sequencing study was attempted to dissect TNBC-specific tumor heterogeneity.293 On the basis of gene expression profiles, six TNBC subtypes were identified, including two basal-like-related subgroups (basal-like 1 and 2, or BL1, and BL2), two mesenchymal-related subgroups (mesenchymal or M, and mesenchymal stem-like, or MSL), one immunomodulatory subgroup (IM), and one luminal androgen receptor group (LAR).286, 293 Such TNBC subtypes were further shown to closely link to histological types, IM tumors overlapped with medullary breast cancer, M and MSL tumors with metaplastic breast cancer, and LAR tumors with apocrine tumors.286, 293 Interestingly, a separate study involving genomic and transcriptomic analyses of 2000 breast tumors identified 10 integrative clusters (IntClust) of breast cancers.294 It was shown that basal-like tumors were heterogeneously distributed among different groups, with IntClust 4 and 10 accounting for 80% of them.286, 294 Cancers classified as IntClust 4 had extensive lymphocytic infiltration with a strong immune and inflammatory signature and few copy-number aberrations (‘CNA-devoid’ subgroup) and patients within this subgroup had a favorable outcome,286, 294 further highlighting the clinical relevance of breast cancer heterogeneity. Conversely, the basal-like tumors within the IntClust 10 subtype had high genomic instability with major chromosomal aberrations, such as chromosome 5 loss, 8q gain, 10p gain or 12p gain.286, 294
A direct comparison between TNBC subtypes and intrinsic molecular (PAM50) subtypes also revealed that most TNBCs were classified as basal-like (80.6%), followed by HER2-enriched (10.2%), normal-like (4.7%), luminal B (3.5%) and luminal A (1.1%) using PAM50 subtyping.295 A direct overlap comparison between the PAM50 classification and the TNBC subtypes indicated that the majority of the TNBC subtypes were classified as basal-like tumors by PAM50 analysis (BL1 at 99%, BL2 at 95%, IM at 84% and M at 97%), with the exception of MSL and LAR.286, 295 Approximately 50% of MSL TNBCs were classified as basal-like, 28% as normal-like and 14% as luminal B tumors.295 Furthermore, tumors within the LAR subtype were mainly classified as HER2-enriched (74%) or luminal B (14%).286, 295 Nonetheless, a recent molecular profiling analysis of TNBCs also identified four stable subtypes, luminal androgen receptor, mesenchymal, basal-like immunosuppressed, and basal-like immune-activated.296
While it is critical to identify molecular and genetic landscapes of TNBC at genomewide level, it is also important to delineate the contributing roles of individual genes and/or pathways in TNBCs, as those genes and/or pathways may serve as potentially actionable targets for TNBC treatments.286, 297, 298 As forkhead box O (FOXO) transcription factors have a recognized role in tumor development and progression, it was shown that FOXO3a expression was highly expressed in TNBC tumors with negative clinical and pathological features, including lymph node metastasis and perineural invasion, and correlated with poor disease-free survival.299
Genome-wide mutational landscape analyses indicate that TNBC tumors on average carry 1.68 somatic mutations per Mb of coding regions and harbor ∼60 somatic mutations in per tumor.38, 300 However, the mutation burden is not uniform; and some TNBCs may have a high mutation burden of >4.68 somatic mutations per Mb.38, 286, 300 The overall consensus is that TNBCs have a frequent occurrence of multiple copy-number aberrations involving genes that lead to alterations in multiple signal pathways, which include the mutations/deletions of BRCA1/2 in the DNA repair pathway; the mutations and/or amplifications of PIK3CA and AKT3 and deletions/mutations of PTEN, TSC1 and INPP4B of the PI3K/mTOR pathway; the amplifications of FGFR1, EGFR and IGF1R, mutations of ERBB2, ERBB3 and ERBB4, and amplifications/mutations of BRAF, KRAS, and HRAS and the deletion of DUSP4 of the RAS/RAF/MEK pathway; the deletions of RB1 and amplifications of CDK6, CCND1and CCND2 of the cell-cycle checkpoints; the amplification of JAK2 in the JAK/STAT pathway; along with androgen receptor pathway, Notch pathway, JNK/AP-1 pathway, and HIF1-α/ARNT pathway.38, 286, 287, 294, 297, 298, 300 Many of these alterations may serve as novel targets for personalized targeted therapies in TNBCs. As nearly 90% of chemo-resistant TNBCs contain alterations in many of these pathways, some targeted agents such as PARP inhibitors, PI3K inhibitors, MEK inhibitors, heat shock protein 90 (HSP 90) inhibitors and histone deacetylase (HDAC) inhibitors, and BET bromodomain inhibitors, as well as immune checkpoint inhibitors, are currently under clinical investigation.286, 287, 297, 298, 301, 302, 303, 304
Tumor heterogeneity and evolution of breast cancer
Breast cancer tumor heterogeneity is one of the hallmarks of malignancy, which includes intertumor heterogeneity observed in breast cancers from different individuals and intratumor heterogeneity caused by the presence of heterogeneous cell populations within an individual tumor.23, 305, 306, 307, 308 Breast cancer intratumor heterogeneity is the main hurdle in the development of effective treatments and personalized medicine.306, 307, 308, 309, 310, 311, 312 The intratumor heterogeneity was first described by Rudolf Carl Virchow, one of the founders of modern pathology, in mid and late 19th century.313 For quite a while, breast cancer phenotypic heterogeneity was used to classify breast cancers based on histological types.314 The clinical implications of tumor heterogeneity were well-recognized early in the process, and breast cancer was one of the first solid tumor types, in which the clinical and treatment implications of heterogeneity for cellular phenotypes were established by analyzing the expression of the estrogen receptors.315 With the rapid advances in molecular biology and genomics techniques, intratumor heterogeneity at the functional, genetic, and cellular levels has begun to be appreciated, and the identification of intrinsic molecular subtypes based on global gene expression profiling studies in breast cancer was pioneered with a rather rapid translation of this knowledge into clinical management of breast cancer.34, 35, 311, 316
Breast cancer intertumor heterogeneity is best illustrated by clinical staging of the disease based on physical examination and imaging findings, while the morphologic heterogeneity of breast carcinoma constitutes the basis for the histopathologic classification of breast cancer.307 Clinical grade of breast cancer is also highly representative of tumor heterogeneity, which is a reliable prognostic factor. However, breast cancers of different grades exhibit distinct profiles by proteomic, genomic and transcriptomic analyses, which serve as the basis to classify breast cancer into the five major intrinsic molecular subtypes with prognostic and therapy implications.286, 297, 298, 301, 302, 303, 304 Furthermore, biomarker heterogeneity in breast cancers, especially the expression or lack of expression of ER, PR and HER2, is recognized as well-established prognostic and predictive factors as their expression in breast cancers is critical in guiding patient treatment. Nonetheless, numerous biomarkers have been investigated in breast cancer for potential diagnostic, prognostic, and therapeutic implications.307, 308, 310, 311
Breast cancer intratumor heterogeneity is likely caused by the phenotypic plasticity, clonal evolution of cancer stem cells and epithelial–mesenchymal transition (EMT) driven by epigenetic, genetic, and microenvironmental alterations in breast cancers.23, 305, 306, 307, 308, 309, 310, 311 Intratumor morphologic heterogeneity is usually appreciated as variability in different areas of tumor (or spatial heterogeneity) and as tumor progression over time (or temporal heterogeneity).307, 308 Morphologically distinct areas within individual tumors may indicate clonal evolution with specific genetic aberrations, while temporal heterogeneity may include evolution of an invasive tumor over time or in response to therapy, development of metastatic disease and progression from in situ to invasive carcinoma.301, 307, 317, 318, 319, 320
The clonal evolution of genetically heterogeneous tumor initiating cells or cancer stem cells may contribute significantly to intratumor heterogeneity of breast cancer, even though the establishment of tumor heterogeneity has been traditionally explained by the cancer stem cell hypothesis23, 305, 321 or the clonal evolution/selection model.322 Both hypotheses propose that tumors originate from single cells that have acquired multiple molecular alterations and developed indefinite proliferative potential under optimal microenvironment to form a cancer.323, 324 However, the cancer stem cell hypothesis believes heterogeneity is caused by aberrant differentiation programs at an assumption of the existence of a hierarchical organization of cancer cells.323, 324 On the other hand, the clonal evolution hypothesis attributes intra-tumor heterogeneity to speciation by natural selection that does not rely on a hierarchical model.323, 324 Furthermore, the cancer stem cell hypothesis speculates that only a small fraction of stem cells are responsible for tumor progression and inherently therapy-resistant.323, 324 In the clonal evolution model, tumor progression and resistance to therapy should follow Darwinian evolutionary rules, in which the emergence of clones able to progress or be resistant to a therapy should depend on the size of cell population, gene mutation rate, cell proliferation rate, and selective pressures from the microenvironment and/or external selective pressures.323, 324 While each hypothesis has its pros and cons, increasing evidence suggests that these two hypotheses may not be mutually exclusive but rather complementary.324, 325 Nonetheless, given that fact that a high degree of phenotypic and genetic intra-tumor heterogeneity exists in breast tumors, the molecular, genetic and epigenetic mechanisms underlying intratumor heterogeneity remain to be fully understood.23, 306, 309
The tumor microenvironment may also contribute significantly to tumor heterogeneity as the tumor microenvironment includes not only cancer cells but also, immune cells, inflammatory cells, vascular cells, lymphatic cells, fibroblasts, and fibrous tissue, all of which impact the cancer's response to therapy.304, 305, 326, 327 Thus, a comprehensive assessment of individual patient tumor microenvironment may be beneficial for personalized medicine. It was shown, by altering their own immunogenicity as well as by implementing immunosuppressive responses in the tumor microenvironment, cancer cells can evade destruction by host immune system and continue to proliferate.328 Thus, the recruitment of these normal cells into the tumor microenvironment coupled with the acquisition of the self-perpetuating qualities in cancer cells may play a major role in sustained tumor development.327, 329 As the PD-1/PD-L1 is the immune checkpoint pathway and plays an important role in immunosuppression in the tumor microenvironment, it has been suggested that antibody inhibition in this pathway may bolster the natural immune system and be able to combat cancer cells.329
Breast cancer metastasis
Cancer metastasis is a complex and yet inefficient process, which requires the regulation of a number of biological steps prior to the presentation of overt disease.330, 331, 332 While the detailed mechanisms underlying cancer metastasis are far from understood, the metastatic cascade likely involves sequential events of tumor cell dissociation, neoangiogenesis of the primary tumor, intravasation, survival and diffusion through the lymphatic and systemic circulation, adhesion to target tissues, extravasation, establishment of metastatic foci at the tissue parenchyma, and a final manifestation of clinically apparent metastasis at the secondary site.330, 331, 332
Like many other solid tumors, breast cancer starts as a local disease, but can spread to the lymph nodes and distant organs, namely metastatic breast cancer (MBC, also known as advanced or stage IV breast cancers).331 Breast cancer metastasis accounts for the majority of deaths from breast cancer. In fact, 10%–15% of breast cancer patients eventually develop distant metastases within three years after the initial detection of the primary tumor.331 However, it is not unusual to see the manifestation of micrometastases at distant sites >10 years after the initial diagnosis. Thus, patients with breast cancer are at risk of developing metastasis during their entire lifetime.331, 333 One salient feature of cancer metastasis is that many tumor types tend to preferentially colonize in certain tissues or organs. Breast cancers exhibit an organ-specific pattern of dissemination and preferentially metastasize to bone (occurrence rate at 47%–60%), the liver (occurrence rate at 19%–20%), lung (occurrence rate at 16%–34%) and brain (occurrence rate at 10%–16%).330, 331, 332, 334, 335, 336, 337, 338
As for its primary tumor, metastatic breast cancer is highly heterogeneous, rendering it difficult to assess risk factors for metastasis and to find effective treatments of this disease.331, 332, 333, 339 A recent study examined the genomic evolution of breast cancer metastasis and relapse of the 299 samples from 170 patients with locally relapsed or metastatic breast cancer, and found that metastasis or relapse disseminated late from primary tumors, and continued to acquire mutations.340 As a result, most distant metastases acquired driver mutations were not seen in the primary tumors, drawing from a wider repertoire of cancer genes than early drivers, many of which include a number of clinically actionable alterations and mutations inactivating SWI-SNF and JAK2-STAT3 pathways.340 Thus, detection of breast cancer metastasis at the earliest stage is important for the management and prediction of breast cancer progression. Emerging techniques using the analysis of circulating tumor cells show promising results in predicting and identifying the early stages of breast cancer metastasis in patients.333
Despite intense efforts, our understanding of the underlying mechanisms of metastatic cancer in general has progressed minimally.332, 341 Over a century ago, the English surgeon Stephen Paget developed the seminal “seed and soil” concept, largely based on the observations that breast cancer preferentially spread to the liver but not the spleen.342 The “seed and soil” hypothesis emphasizes that the selection of a site for secondary tumor development is not only made by the tumor cell (or the “seed”), but also influenced by the properties of the target organ (or “soil”).342 Emerging evidence suggests that the relationship between disseminating tumor cells (DTCs) and the microenvironment of the target tissues/organs they colonize is the cornerstone of metastasis and plays a pivotal role in determining whether DTCs can survive and grow to form metastatic tumors.331, 333, 339, 342
There are currently two prevailing and competitive models of metastasis, the early dissemination model, which hypothesizes that cancer cells with metastatic potential are seeded early in tumor formation and lay dormant for a period of time, and the late dissemination model, which suggests that metastasis may arise as a result of Darwinian selection during multistep tumor progression and the metastatic cells arise out of later stage tumor heterogeneity that produces more invasive, mesenchymal-like cancer cells.332, 341, 343, 344 The late dissemination model is seemingly supported by the fact that a successful metastatic cell must dissociate from the primary tumor, invade the surrounding tissues and penetrate the local vasculature, survive within the circulation, and be immobilized in the capillaries of target tissues/organs where it can extravasate to enter, survive and adapt to a novel and potentially inhospitable microenvironment.332, 342, 345
Extensive genomic studies have revealed that primary tumors and metastases of varied cancer types were closely related from a genetic standpoint, but failed to uncover genetic changes that are uniquely associated with progression to metastasis.346, 347, 348, 349, 350 These results suggest that the metastasis seeding cells may originate from a subpopulation within the primary tumor and possess the genetic alterations required for both primary and metastatic tumor growth.332 Such subpopulation must therefore rely on epigenetic and posttranscriptional programs superimposed on their genetic mutations to complete the metastatic process.332
Alternative to the clonal evolution model is that a small subpopulation of cells, or commonly referred to as cancer stem cells (CSCs), at the apex of the tumorigenic hierarchy may retain pluripotency and give rise to heterogeneous, differentiated progeny while maintaining its own numbers.332 Given the fact that CSCs display tumor-initiating properties, it is conceivable that CSCs may also have the ability to disseminate and seed metastases, serving as metastasis-initiating cells (MICs), although cells that initiate metastatic growth have been difficult to characterize, and no hard evidence exists to suggest that MICs are distinct from CSCs.332, 351, 352, 353, 354 Nonetheless, the CSC model should be compatible with the early dissemination hypothesis. It has been speculated that the latent phase of breast tumorigenesis may include the spread of breast cancer cells at an early, pre-symptomatic stage of primary tumor development when the disseminated cancer cells remain undetected in bone marrow for years before presenting clinically identifiable features.123, 355 Dormant cancer cells with stem-cell properties in bone marrow can be activated in response to injury induced by chemotherapy to promote new tumor formation.60 Furthermore, it is suggested that circulating tumor cells (CTCs) in the blood may play an active role of carcinoma metastasis, and the presence of CTCs correlates to poor patient prognosis and an increase in metastatic sites in patient data.356 Nonetheless, strong evidence to draw a mechanistic connection between CTCs and tumor metastasis is lacking clinically, although it was shown in mouse models that metastasis-initiating cells (MICs) may exist in breast cancer CTCs.356
It has long been recognized that epithelial-to-mesenchymal transition (EMT) is an important cellular feature of metastatic cancer cells.357, 358, 359 EMT is a critical pathway in the mesenchymal movement of single migratory cells, where the cancer cells can undergo changes from an epithelial phenotype to a mesenchymal-like phenotype.332 The process is reversible, allowing epithelial cells, such as metastatic cancer cells that have undergone EMT to regain their epithelial features through mesenchymal-to-epithelial transition (MET).357, 358, 359 It was shown that the EMT cells, while not found at secondary cancer sites in lineage-tracing studies, may be able to give neighboring non-EMT cells access to the secondary site thus influencing metastasis.360 Furthermore, EMT process involves a number of cell functions which contribute to the imposition of mesenchymal traits on epithelial cells and thus can play a role in metastasis.361, 362 As EMT is well associated with cancer metastasis, tumor heterogeneity, and consequent drug resistance, the intersection between EMT regulation and hypoxia in the tumor microenvironment have been recently investigated.360, 361, 363 MET may precede seeding at a secondary site in CTCs lodged in a capillary, which provides an advantage at this stage as they need no EMT to MET reprogramming to proliferate in capillaries or within secondary tissues.332 Nonetheless, recent studies on pancreatic carcinoma models refuted EMT as a prerequisite for metastasis, suggesting instead that EMT may be more relevant to resistance to therapy.364, 365 Thus, it remains to be determined whether EMT is required and necessary for breast cancer metastasis.
Given the deadly consequence of metastatic breast cancer, it is imperative to detect metastasis at its earliest stage.331, 366 Importantly, the detection of biomarkers is usually necessary steps to determine treatment options.366 The American Society of Clinical Oncology recommends 13 categories of breast tumor markers that showed evidence of clinical utility and were recommended for use in practice including ER, PR, HERs, carcinoembryonic antigen (CEA), CA 15-3, CA 27.29, urokinase plasminogen activator (UPA), plasminogen activator inhibitor 1 (PAI), and certain multi-parameter gene expression assays.366 Among these biomarkers HER2 has attracted much attention as a possible prognostic marker.331 With the rapid advances in genomic sequencing technologies, it is expected that new diagnostic and prognostic markers of breast cancer metastasis will be identified.367, 368 These markers will be likely in multi-gene panel formats. Furthermore, the drastic improvements over detection sensitivity and throughput of whole genome sequencing technologies will enable us to reliably detect genomic changes in circulating tumor cells.369, 370, 371, 372, 373 Till then the full potential of personalized precision medicine will be realized in the clinical management of breast cancers.
Concluding remarks
In this review, we provide a comprehensive survey on the basic biological aspects of breast cancer. As we have elucidated, breast cancer carries complex genetic, epigenetic, and environmental factors in how it manifests in the individual patient. While the most common inherited genetic factor, the BRCA1 and BRCA2 gene mutations, have rightfully been studied in depth, upwards of 85% of breast cancers occur in women without apparent family history of the disease which includes the inherited BRCA1/2 mutations. We hope to draw attention to the suggestion that naturally arises from this statistic that these cancers may be caused by genetic mutations that occur as a result of the aging process and lifestyle-related risk factors, rather than inherited mutations. We have reviewed how, while most breast cancers are carcinomas, common breast cancers can be divided into three major groups: non-invasive (or in situ), invasive, and metastatic breast cancers. Gene expression studies further identified several distinct molecular subtypes that differ significantly in prognosis as well as in the therapeutic targets present in the cancer cells. A continued specification of subtype identification is imperative in the development of individualized treatment. While it remains in its somewhat controversial nascence, the mammary stem cell theory may serve to shed light on the cellular origins of breast cancer. Normal breast development and mammary stem cells are regulated by some of the same signaling pathways including estrogen receptors (ERs), HER2, and Wnt/β-catenin signaling pathways, which control stem cell proliferation, cell death, cell differentiation, and cell motility. The heterogeneity and plasticity within breast cancers poses one of the most pressing issues for treatment. The recent evidence supports that epigenetic regulations and noncoding RNAs may play important roles in breast cancer development and may contribute to the heterogeneity and metastatic aspects of breast cancer, especially that of triple-negative breast cancer. Continuing progress in genomic and systems biology technologies will aid in understanding of the molecular and genetic bases of tumor heterogeneity and breast cancer metastasis, and will enable us to identify more reliable diagnostic and prognostic biomarkers for metastatic and recurrent breast cancers. We hope to encourage the further development of novel diagnostic and therapeutic measures to fully realize the best possible patient outcomes for those who suffer from breast cancer.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
The authors apologize to the investigators whose original work couldn't be cited due to space constraints. Research in the authors' laboratories was supported in part by research grants from the National Institutes of Health (CA226303 to TCH), the National Key Research and Development Program of China (2016YFC1000803 and 2011CB707906 to TCH) and the Natural Science Foundation of China (#30670811, #31171243, and #31420103915 to GR). YF was a recipient of pre-doctorate fellowship from the China Scholarship Council. Funding sources were not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Veronesi U., Boyle P., Goldhirsch A., Orecchia R., Viale G. Breast cancer. Lancet. 2005;365(9472):1727–1741. doi: 10.1016/S0140-6736(05)66546-4. [DOI] [PubMed] [Google Scholar]
- 5.Mathews F.S. The ten-year survivors of radical mastectomy. Ann Surg. 1933;98(4):635–643. doi: 10.1097/00000658-193310000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cliffton E.E., Young L.E. Carcinoma of the breast; five to twenty-year follow-up following radical mastectomy. Am J Surg. 1951;82(2):185–190. doi: 10.1016/0002-9610(51)90340-6. [DOI] [PubMed] [Google Scholar]
- 7.Figueiredo M.I., Cullen J., Hwang Y.T., Rowland J.H., Mandelblatt J.S. Breast cancer treatment in older women: does getting what you want improve your long-term body image and mental health? J Clin Oncol. 2004;22(19):4002–4009. doi: 10.1200/JCO.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 8.Collaborative Group on Hormonal Factors in Breast C Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001;358(9291):1389–1399. doi: 10.1016/S0140-6736(01)06524-2. [DOI] [PubMed] [Google Scholar]
- 9.Hulka B.S. Epidemiology of susceptibility to breast cancer. Prog Clin Biol Res. 1996;395:159–174. [PubMed] [Google Scholar]
- 10.Kaminska M., Ciszewski T., Lopacka-Szatan K., Miotla P., Staroslawska E. Breast cancer risk factors. Prz Menopauzalny. 2015;14(3):196–202. doi: 10.5114/pm.2015.54346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y.S., Zhao Z., Yang Z.N. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13(11):1387–1397. doi: 10.7150/ijbs.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singletary S.E. Rating the risk factors for breast cancer. Ann Surg. 2003;237(4):474–482. doi: 10.1097/01.SLA.0000059969.64262.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howell A., Anderson A.S., Clarke R.B. Risk determination and prevention of breast cancer. Breast Cancer Res. 2014;16(5):446. doi: 10.1186/s13058-014-0446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anothaisintawee T., Wiratkapun C., Lerdsitthichai P. Risk factors of breast cancer: a systematic review and meta-analysis. Asia Pac J Publ Health. 2013;25(5):368–387. doi: 10.1177/1010539513488795. [DOI] [PubMed] [Google Scholar]
- 15.Ozsoy A., Barca N., Dolek B.A. The relationship between breast cancer and risk factors: a single-center study. Eur J Breast Health. 2017;13(3):145–149. doi: 10.5152/tjbh.2017.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McTiernan A. Behavioral risk factors in breast cancer: can risk be modified? Oncologist. 2003;8(4):326–334. doi: 10.1634/theoncologist.8-4-326. [DOI] [PubMed] [Google Scholar]
- 17.Patterson R.E., Cadmus L.A., Emond J.A., Pierce J.P. Physical activity, diet, adiposity and female breast cancer prognosis: a review of the epidemiologic literature. Maturitas. 2010;66(1):5–15. doi: 10.1016/j.maturitas.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Rock C.L., Demark-Wahnefried W. Can lifestyle modification increase survival in women diagnosed with breast cancer? J Nutr. 2002;132(11 suppl l) doi: 10.1093/jn/132.11.3504S. 3504S-3507S. [DOI] [PubMed] [Google Scholar]
- 19.Yang X.R., Sherman M.E., Rimm D.L. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomark Prev. 2007;16(3):439–443. doi: 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 20.Barnard M.E., Boeke C.E., Tamimi R.M. Established breast cancer risk factors and risk of intrinsic tumor subtypes. Biochim Biophys Acta. 2015;1856(1):73–85. doi: 10.1016/j.bbcan.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Anderson W.F., Rosenberg P.S., Prat A., Perou C.M., Sherman M.E. How many etiological subtypes of breast cancer: two, three, four, or more? J Natl Cancer Inst. 2014;106(8) doi: 10.1093/jnci/dju165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colditz G.A., Kaphingst K.A., Hankinson S.E., Rosner B. Family history and risk of breast cancer: nurses' health study. Breast Cancer Res Treat. 2012;133(3):1097–1104. doi: 10.1007/s10549-012-1985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117(11):3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allison K.H. Molecular pathology of breast cancer: what a pathologist needs to know. Am J Clin Pathol. 2012;138(6):770–780. doi: 10.1309/AJCPIV9IQ1MRQMOO. [DOI] [PubMed] [Google Scholar]
- 25.Collins L.C., Baer H.J., Tamimi R.M., Connolly J.L., Colditz G.A., Schnitt S.J. The influence of family history on breast cancer risk in women with biopsy-confirmed benign breast disease: results from the Nurses' Health Study. Cancer. 2006;107(6):1240–1247. doi: 10.1002/cncr.22136. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Costantino J.P., Tan-Chiu E., Wickerham D.L., Paik S., Wolmark N. Lower-category benign breast disease and the risk of invasive breast cancer. J Natl Cancer Inst. 2004;96(8):616–620. doi: 10.1093/jnci/djhs105. [DOI] [PubMed] [Google Scholar]
- 27.Hartmann L.C., Sellers T.A., Frost M.H. Benign breast disease and the risk of breast cancer. N Engl J Med. 2005;353(3):229–237. doi: 10.1056/NEJMoa044383. [DOI] [PubMed] [Google Scholar]
- 28.Dupont W.D., Page D.L. Risk factors for breast cancer in women with proliferative breast disease. N Engl J Med. 1985;312(3):146–151. doi: 10.1056/NEJM198501173120303. [DOI] [PubMed] [Google Scholar]
- 29.Dupont W.D., Parl F.F., Hartmann W.H. Breast cancer risk associated with proliferative breast disease and atypical hyperplasia. Cancer. 1993;71(4):1258–1265. doi: 10.1002/1097-0142(19930215)71:4<1258::aid-cncr2820710415>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 30.Vogel V.G. Epidemiology, genetics, and risk evaluation of postmenopausal women at risk of breast cancer. Menopause. 2008;15(suppl 4):782–789. doi: 10.1097/gme.0b013e3181788d88. [DOI] [PubMed] [Google Scholar]
- 31.Byrne C., Webb P.M., Jacobs T.W. Alcohol consumption and incidence of benign breast disease. Cancer Epidemiol Biomark Prev. 2002;11(11):1369–1374. [PubMed] [Google Scholar]
- 32.Prat A., Parker J.S., Karginova O. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abd El-Rehim D.M., Pinder S.E., Paish C.E. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol. 2004;203(2):661–671. doi: 10.1002/path.1559. [DOI] [PubMed] [Google Scholar]
- 34.Perou C.M., Sorlie T., Eisen M.B. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 35.Sorlie T., Perou C.M., Tibshirani R. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sotiriou C., Neo S.Y., McShane L.M. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100(18):10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu K., Lee C.H., Tan P.H., Tan P. Conservation of breast cancer molecular subtypes and transcriptional patterns of tumor progression across distinct ethnic populations. Clin Cancer Res. 2004;10(16):5508–5517. doi: 10.1158/1078-0432.CCR-04-0085. [DOI] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paik S., Shak S., Tang G. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 40.Ma X.J., Wang Z., Ryan P.D. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5(6):607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Parker J.S., Mullins M., Cheang M.C. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen T.O., Parker J.S., Leung S. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16(21):5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chia S.K., Bramwell V.H., Tu D. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res. 2012;18(16):4465–4472. doi: 10.1158/1078-0432.CCR-12-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowsett M., Sestak I., Lopez-Knowles E. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31(22):2783–2790. doi: 10.1200/JCO.2012.46.1558. [DOI] [PubMed] [Google Scholar]
- 45.Laenkholm A.V., Jensen M.B., Eriksen J.O. PAM50 risk of recurrence score predicts 10-year distant recurrence in a comprehensive Danish cohort of postmenopausal women allocated to 5 Years of endocrine therapy for hormone receptor-positive early breast cancer. J Clin Oncol. 2018;36(8):735–740. doi: 10.1200/JCO.2017.74.6586. [DOI] [PubMed] [Google Scholar]
- 46.Krop I., Ismaila N., Andre F. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: american society of clinical Oncology clinical practice guideline focused update. J Clin Oncol. 2017;35(24):2838–2847. doi: 10.1200/JCO.2017.74.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Vijver M.J., He Y.D., van't Veer L.J. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 48.Buyse M., Loi S., van't Veer L. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 49.Mook S., Schmidt M.K., Weigelt B. The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann Oncol. 2010;21(4):717–722. doi: 10.1093/annonc/mdp388. [DOI] [PubMed] [Google Scholar]
- 50.Cardoso F., Van't Veer L., Rutgers E., Loi S., Mook S., Piccart-Gebhart M.J. Clinical application of the 70-gene profile: the MINDACT trial. J Clin Oncol. 2008;26(5):729–735. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- 51.Sotiriou C., Wirapati P., Loi S. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 52.Macias H., Hinck L. Mammary gland development. Wiley Interdiscip Rev Dev Biol. 2012;1(4):533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huebner R.J., Ewald A.J. Cellular foundations of mammary tubulogenesis. Semin Cell Dev Biol. 2014;31:124–131. doi: 10.1016/j.semcdb.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visvader J.E., Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28(11):1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inman J.L., Robertson C., Mott J.D., Bissell M.J. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142(6):1028–1042. doi: 10.1242/dev.087643. [DOI] [PubMed] [Google Scholar]
- 56.Fu N., Lindeman G.J., Visvader J.E. The mammary stem cell hierarchy. Curr Top Dev Biol. 2014;107:133–160. doi: 10.1016/B978-0-12-416022-4.00005-6. [DOI] [PubMed] [Google Scholar]
- 57.Lloyd-Lewis B., Harris O.B., Watson C.J., Davis F.M. Mammary stem cells: premise, properties, and perspectives. Trends Cell Biol. 2017;27(8):556–567. doi: 10.1016/j.tcb.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Mascre G., Dekoninck S., Drogat B. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489(7415):257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 59.Aragona M., Dekoninck S., Rulands S. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat Commun. 2017;8:14684. doi: 10.1038/ncomms14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soteriou D., Fuchs Y. A matter of life and death: stem cell survival in tissue regeneration and tumour formation. Nat Rev Cancer. 2018;18(3):187–201. doi: 10.1038/nrc.2017.122. [DOI] [PubMed] [Google Scholar]
- 61.Sreekumar A., Roarty K., Rosen J.M. The mammary stem cell hierarchy: a looking glass into heterogeneous breast cancer landscapes. Endocr Relat Cancer. 2015;22(6):T161–T176. doi: 10.1530/ERC-15-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 63.Kreso A., Dick J.E. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 64.Nassar D., Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- 65.Papaccio F., Paino F., Regad T., Papaccio G., Desiderio V., Tirino V. Concise review: cancer cells, cancer stem cells, and mesenchymal stem cells: influence in cancer development. Stem Cells Transl Med. 2017;6(12):2115–2125. doi: 10.1002/sctm.17-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brooks M.D., Burness M.L., Wicha M.S. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell. 2015;17(3):260–271. doi: 10.1016/j.stem.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Keymeulen A., Rocha A.S., Ousset M. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 68.van Amerongen R., Bowman A.N., Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11(3):387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 69.Sachs N., de Ligt J., Kopper O. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1-2) doi: 10.1016/j.cell.2017.11.010. 373–386.e310. [DOI] [PubMed] [Google Scholar]
- 70.Jabs J., Zickgraf F.M., Park J. Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol Syst Biol. 2017;13(11):955. doi: 10.15252/msb.20177697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pauli C., Hopkins B.D., Prandi D. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7(5):462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hunter T. Signaling–2000 and beyond. Cell. 2000;100(1):113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 73.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28(5):730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 74.Sever R., Brugge J.S. Signal transduction in cancer. Cold Spring Harb Perspect Med. 2015;5(4) doi: 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar V., Green S., Stack G., Berry M., Jin J.R., Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51(6):941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 76.Osborne C.K., Schiff R., Fuqua S.A., Shou J. Estrogen receptor: current understanding of its activation and modulation. Clin Cancer Res. 2001;7(suppl 12):4338s–4342s. discussion 4411s-4412s. [PubMed] [Google Scholar]
- 77.Renoir J.M., Marsaud V., Lazennec G. Estrogen receptor signaling as a target for novel breast cancer therapeutics. Biochem Pharmacol. 2013;85(4):449–465. doi: 10.1016/j.bcp.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 78.Cheskis B.J., Greger J.G., Nagpal S., Freedman L.P. Signaling by estrogens. J Cell Physiol. 2007;213(3):610–617. doi: 10.1002/jcp.21253. [DOI] [PubMed] [Google Scholar]
- 79.Bjornstrom L., Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 80.Saha Roy S., Vadlamudi R.K. Role of estrogen receptor signaling in breast cancer metastasis. Int J Breast Cancer. 2012;2012:654698. doi: 10.1155/2012/654698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klinge C.M. Estrogen receptor interaction with co-activators and co-repressors. Steroids. 2000;65(5):227–251. doi: 10.1016/s0039-128x(99)00107-5. [DOI] [PubMed] [Google Scholar]
- 82.Marino M., Galluzzo P., Ascenzi P. Estrogen signaling multiple pathways to impact gene transcription. Curr Genom. 2006;7(8):497–508. doi: 10.2174/138920206779315737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan S., Wang J., Yuan R. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284(5418):1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 84.Fan S., Ma Y.X., Wang C. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene. 2001;20(1):77–87. doi: 10.1038/sj.onc.1204073. [DOI] [PubMed] [Google Scholar]
- 85.Nadji M., Gomez-Fernandez C., Ganjei-Azar P., Morales A.R. Immunohistochemistry of estrogen and progesterone receptors reconsidered: experience with 5,993 breast cancers. Am J Clin Pathol. 2005;123(1):21–27. doi: 10.1309/4wv79n2ghj3x1841. [DOI] [PubMed] [Google Scholar]
- 86.Said T.K., Conneely O.M., Medina D., O'Malley B.W., Lydon J.P. Progesterone, in addition to estrogen, induces cyclin D1 expression in the murine mammary epithelial cell, in vivo. Endocrinology. 1997;138(9):3933–3939. doi: 10.1210/endo.138.9.5436. [DOI] [PubMed] [Google Scholar]
- 87.Cicatiello L., Addeo R., Sasso A. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol Cell Biol. 2004;24(16):7260–7274. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zwijsen R.M., Wientjens E., Klompmaker R., van der Sman J., Bernards R., Michalides R.J. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88(3):405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 89.Lundberg A.S., Weinberg R.A. Control of the cell cycle and apoptosis. Eur J Cancer. 1999;35(14):1886–1894. doi: 10.1016/s0959-8049(99)00292-0. [DOI] [PubMed] [Google Scholar]
- 90.Finn R.S., Aleshin A., Slamon D.J. Targeting the cyclin-dependent kinases (CDK) 4/6 in estrogen receptor-positive breast cancers. Breast Cancer Res. 2016;18(1):17. doi: 10.1186/s13058-015-0661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finn R.S., Dering J., Conklin D. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O'Leary B., Finn R.S., Turner N.C. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 93.Fuqua S.A., Cui Y. Estrogen and progesterone receptor isoforms: clinical significance in breast cancer. Breast Cancer Res Treat. 2004;87(suppl 1):S3–S10. doi: 10.1007/s10549-004-1577-4. [DOI] [PubMed] [Google Scholar]
- 94.Fuqua S.A., Fitzgerald S.D., Chamness G.C. Variant human breast tumor estrogen receptor with constitutive transcriptional activity. Cancer Res. 1991;51(1):105–109. [PubMed] [Google Scholar]
- 95.Flouriot G., Brand H., Denger S. Identification of a new isoform of the human estrogen receptor-alpha (hER-alpha) that is encoded by distinct transcripts and that is able to repress hER-alpha activation function 1. EMBO J. 2000;19(17):4688–4700. doi: 10.1093/emboj/19.17.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bollig A., Miksicek R.J. An estrogen receptor-alpha splicing variant mediates both positive and negative effects on gene transcription. Mol Endocrinol. 2000;14(5):634–649. doi: 10.1210/mend.14.5.0460. [DOI] [PubMed] [Google Scholar]
- 97.Wang Q., Jiang J., Ying G. Tamoxifen enhances stemness and promotes metastasis of ERalpha36(+) breast cancer by upregulating ALDH1A1 in cancer cells. Cell Res. 2018;28(3):336–358. doi: 10.1038/cr.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang B., Omoto Y., Iwase H. Differential expression of estrogen receptor alpha, beta1, and beta2 in lobular and ductal breast cancer. Proc Natl Acad Sci USA. 2014;111(5):1933–1938. doi: 10.1073/pnas.1323719111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paruthiyil S., Parmar H., Kerekatte V., Cunha G.R., Firestone G.L., Leitman D.C. Estrogen receptor beta inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res. 2004;64(1):423–428. doi: 10.1158/0008-5472.can-03-2446. [DOI] [PubMed] [Google Scholar]
- 100.Lin C.Y., Strom A., Li Kong S. Inhibitory effects of estrogen receptor beta on specific hormone-responsive gene expression and association with disease outcome in primary breast cancer. Breast Cancer Res. 2007;9(2):R25. doi: 10.1186/bcr1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bado I., Nikolos F., Rajapaksa G. Somatic loss of estrogen receptor beta and p53 synergize to induce breast tumorigenesis. Breast Cancer Res. 2017;19(1):79. doi: 10.1186/s13058-017-0872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oda K., Matsuoka Y., Funahashi A., Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100014. 2005 0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sergina N.V., Moasser M.M. The HER family and cancer: emerging molecular mechanisms and therapeutic targets. Trends Mol Med. 2007;13(12):527–534. doi: 10.1016/j.molmed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roskoski R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 105.Arteaga C.L., Engelman J.A. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wee P., Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 2017;9(5) doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wieduwilt M.J., Moasser M.M. The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol Life Sci. 2008;65(10):1566–1584. doi: 10.1007/s00018-008-7440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garrett T.P.J., McKern N.M., Lou M. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11(2):495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 109.Graus-Porta D., Beerli R.R., Daly J.M., Hynes N.E. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16(7):1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Burgess A.W. EGFR family: structure physiology signalling and therapeutic targets. Growth Factors. 2008;26(5):263–274. doi: 10.1080/08977190802312844. [DOI] [PubMed] [Google Scholar]
- 111.Mayer I.A., Arteaga C.L. The pi3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016;67:11–28. doi: 10.1146/annurev-med-062913-051343. [DOI] [PubMed] [Google Scholar]
- 112.Ono M., Kuwano M. Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to gefitinib and other EGFR-targeting drugs. Clin Cancer Res. 2006;12(24):7242–7251. doi: 10.1158/1078-0432.CCR-06-0646. [DOI] [PubMed] [Google Scholar]
- 113.Elizalde P.V., Cordo Russo R.I., Chervo M.F., Schillaci R. ErbB-2 nuclear function in breast cancer growth, metastasis and resistance to therapy. Endocr Relat Cancer. 2016;23(12):T243–T257. doi: 10.1530/ERC-16-0360. [DOI] [PubMed] [Google Scholar]
- 114.King C.R., Kraus M.H., Aaronson S.A. Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science. 1985;229(4717):974–976. doi: 10.1126/science.2992089. [DOI] [PubMed] [Google Scholar]
- 115.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 116.Yang Y., Leonard M., Zhang Y. HER2-Driven breast tumorigenesis relies upon interactions of the estrogen receptor with coactivator MED1. Cancer Res. 2018;78(2):422–435. doi: 10.1158/0008-5472.CAN-17-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu S., Lee J.S., Jie C. HER2 overexpression triggers an IL1alpha proinflammatory circuit to drive tumorigenesis and promote chemotherapy resistance. Cancer Res. 2018;78(8):2040–2051. doi: 10.1158/0008-5472.CAN-17-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu Q., Kulak M.V., Borcherding N. A novel HER2 gene body enhancer contributes to HER2 expression. Oncogene. 2018;37(5):687–694. doi: 10.1038/onc.2017.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goel S., Wang Q., Watt A.C. Overcoming therapeutic resistance in HER2-positive breast cancers with CDK4/6 inhibitors. Cancer Cell. 2016;29(3):255–269. doi: 10.1016/j.ccell.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li Z., Szabolcs M., Terwilliger J.D., Efstratiadis A. Prostatic intraepithelial neoplasia and adenocarcinoma in mice expressing a probasin-Neu oncogenic transgene. Carcinogenesis. 2006;27(5):1054–1067. doi: 10.1093/carcin/bgi324. [DOI] [PubMed] [Google Scholar]
- 121.Guy C.T., Webster M.A., Schaller M., Parsons T.J., Cardiff R.D., Muller W.J. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89(22):10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Muller W.J., Sinn E., Pattengale P.K., Wallace R., Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54(1):105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 123.Hosseini H., Obradovic M.M., Hoffmann M. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540(7634):552. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wolff A.C., Hammond M.E., Hicks D.G. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 125.Romond E.H., Perez E.A., Bryant J. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353(16):1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 126.Untch M., von Minckwitz G., Gerber B. Survival analysis after neoadjuvant chemotherapy with trastuzumab or lapatinib in patients with human epidermal growth factor receptor 2-positive breast cancer in the GeparQuinto (G5) study (GBG 44) J Clin Oncol. 2018 doi: 10.1200/JCO.2017.75.9175. JCO2017759175. [DOI] [PubMed] [Google Scholar]
- 127.Robert S., Schwartz M.D., John K., Erban M.D. Timing of metastasis in breast cancer. N Engl J Med. 2017:2486–2488. doi: 10.1056/NEJMcibr1701388. [DOI] [PubMed] [Google Scholar]
- 128.Nwabo Kamdje A.H., Seke Etet P.F., Vecchio L., Muller J.M., Krampera M., Lukong K.E. Signaling pathways in breast cancer: therapeutic targeting of the microenvironment. Cell Signal. 2014;26(12):2843–2856. doi: 10.1016/j.cellsig.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 129.Logan C.Y., Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 130.Luo J., Chen J., Deng Z.L. Wnt signaling and human diseases: what are the therapeutic implications? Lab Invest. 2007;87(2):97–103. doi: 10.1038/labinvest.3700509. [DOI] [PubMed] [Google Scholar]
- 131.Yang K., Wang X., Zhang H. The evolving roles of canonical WNT signaling in stem cells and tumorigenesis: implications in targeted cancer therapies. Lab Invest. 2016;96(2):116–136. doi: 10.1038/labinvest.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mohammed M.K., Shao C., Wang J. Wnt/beta-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumorigenesis and cancer chemoresistance. Genes Dis. 2016;3(1):11–40. doi: 10.1016/j.gendis.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Clevers H., Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 135.Nusse R., Clevers H. Wnt/beta-Catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 136.Krishnamurthy N., Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.He T.C., Chan T.A., Vogelstein B., Kinzler K.W. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99(3):335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.He T.C., Sparks A.B., Rago C. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 139.Nusse R., Varmus H.E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 140.Roelink H., Wagenaar E., Lopes da Silva S., Nusse R. Wnt-3, a gene activated by proviral insertion in mouse mammary tumors, is homologous to int-1/Wnt-1 and is normally expressed in mouse embryos and adult brain. Proc Natl Acad Sci USA. 1990;87(12):4519–4523. doi: 10.1073/pnas.87.12.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roelink H., Nusse R. Expression of two members of the Wnt family during mouse development–restricted temporal and spatial patterns in the developing neural tube. Genes Dev. 1991;5(3):381–388. doi: 10.1101/gad.5.3.381. [DOI] [PubMed] [Google Scholar]
- 142.Imbert A., Eelkema R., Jordan S., Feiner H., Cowin P. Delta N89 beta-catenin induces precocious development, differentiation, and neoplasia in mammary gland. J Cell Biol. 2001;153(3):555–568. doi: 10.1083/jcb.153.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gavin B.J., McMahon A.P. Differential regulation of the Wnt gene family during pregnancy and lactation suggests a role in postnatal development of the mammary gland. Mol Cell Biol. 1992;12(5):2418–2423. doi: 10.1128/mcb.12.5.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Weber-Hall S.J., Phippard D.J., Niemeyer C.C., Dale T.C. Developmental and hormonal regulation of Wnt gene expression in the mouse mammary gland. Differentiation. 1994;57(3):205–214. doi: 10.1046/j.1432-0436.1994.5730205.x. [DOI] [PubMed] [Google Scholar]
- 145.Buhler T.A., Dale T.C., Kieback C., Humphreys R.C., Rosen J.M. Localization and quantification of Wnt-2 gene expression in mouse mammary development. Dev Biol. 1993;155(1):87–96. doi: 10.1006/dbio.1993.1009. [DOI] [PubMed] [Google Scholar]
- 146.Bradbury J.M., Edwards P.A., Niemeyer C.C., Dale T.C. Wnt-4 expression induces a pregnancy-like growth pattern in reconstituted mammary glands in virgin mice. Dev Biol. 1995;170(2):553–563. doi: 10.1006/dbio.1995.1236. [DOI] [PubMed] [Google Scholar]
- 147.Brisken C., Heineman A., Chavarria T. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 2000;14(6):650–654. [PMC free article] [PubMed] [Google Scholar]
- 148.Chu E.Y., Hens J., Andl T. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131(19):4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- 149.Boras-Granic K., Chang H., Grosschedl R., Hamel P.A. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295(1):219–231. doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 150.Lindvall C., Zylstra C.R., Evans N. The Wnt co-receptor Lrp6 is required for normal mouse mammary gland development. PLoS One. 2009;4(6):e5813. doi: 10.1371/journal.pone.0005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stingl J., Eirew P., Ricketson I. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 152.Shackleton M., Vaillant F., Simpson K.J. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 153.Brennan K.R., Brown A.M. Wnt proteins in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2004;9(2):119–131. doi: 10.1023/B:JOMG.0000037157.94207.33. [DOI] [PubMed] [Google Scholar]
- 154.Bafico A., Liu G., Goldin L., Harris V., Aaronson S.A. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6(5):497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 155.Nagahata T., Shimada T., Harada A. Amplification, up-regulation and over-expression of DVL-1, the human counterpart of the Drosophila disheveled gene, in primary breast cancers. Cancer Sci. 2003;94(6):515–518. doi: 10.1111/j.1349-7006.2003.tb01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ugolini F., Adelaide J., Charafe-Jauffret E. Differential expression assay of chromosome arm 8p genes identifies Frizzled-related (FRP1/FRZB) and Fibroblast Growth Factor Receptor 1 (FGFR1) as candidate breast cancer genes. Oncogene. 1999;18(10):1903–1910. doi: 10.1038/sj.onc.1202739. [DOI] [PubMed] [Google Scholar]
- 157.Veeck J., Geisler C., Noetzel E. Epigenetic inactivation of the secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008;29(5):991–998. doi: 10.1093/carcin/bgn076. [DOI] [PubMed] [Google Scholar]
- 158.Zhou X.L., Qin X.R., Zhang X.D., Ye L.H. Downregulation of Dickkopf-1 is responsible for high proliferation of breast cancer cells via losing control of Wnt/beta-catenin signaling. Acta Pharmacol Sin. 2010;31(2):202–210. doi: 10.1038/aps.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Virmani A.K., Rathi A., Sathyanarayana U.G. Aberrant methylation of the adenomatous polyposis coli (APC) gene promoter 1A in breast and lung carcinomas. Clin Cancer Res. 2001;7(7):1998–2004. [PubMed] [Google Scholar]
- 160.Furuuchi K., Tada M., Yamada H. Somatic mutations of the APC gene in primary breast cancers. Am J Pathol. 2000;156(6):1997–2005. doi: 10.1016/s0002-9440(10)65072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Khramtsov A.I., Khramtsova G.F., Tretiakova M., Huo D., Olopade O.I., Goss K.H. Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176(6):2911–2920. doi: 10.2353/ajpath.2010.091125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin S.Y., Xia W., Wang J.C. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci USA. 2000;97(8):4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schade B., Lesurf R., Sanguin-Gendreau V. beta-Catenin signaling is a critical event in ErbB2-mediated mammary tumor progression. Cancer Res. 2013;73(14):4474–4487. doi: 10.1158/0008-5472.CAN-12-3925. [DOI] [PubMed] [Google Scholar]
- 164.Yin X., Xiang T., Li L. DACT1, an antagonist to Wnt/beta-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. 2013;15(2):R23. doi: 10.1186/bcr3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xiang T., Li L., Yin X. Epigenetic silencing of the WNT antagonist Dickkopf 3 disrupts normal Wnt/beta-catenin signalling and apoptosis regulation in breast cancer cells. J Cell Mol Med. 2013;17(10):1236–1246. doi: 10.1111/jcmm.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li M., Han Y., Zhou H. Transmembrane protein 170B is a novel breast tumorigenesis suppressor gene that inhibits the Wnt/beta-catenin pathway. Cell Death Dis. 2018;9(2):91. doi: 10.1038/s41419-017-0128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen M.S., Woodward W.A., Behbod F. Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J Cell Sci. 2007;120(Pt 3):468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- 168.Woodward W.A., Chen M.S., Behbod F., Alfaro M.P., Buchholz T.A., Rosen J.M. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci USA. 2007;104(2):618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kakarala M., Wicha M.S. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol. 2008;26(17):2813–2820. doi: 10.1200/JCO.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ayyanan A., Civenni G., Ciarloni L. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci USA. 2006;103(10):3799–3804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vaillant F., Asselin-Labat M.L., Shackleton M., Forrest N.C., Lindeman G.J., Visvader J.E. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68(19):7711–7717. doi: 10.1158/0008-5472.CAN-08-1949. [DOI] [PubMed] [Google Scholar]
- 172.Li Y., Welm B., Podsypanina K. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA. 2003;100(26):15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kuraguchi M., Ohene-Baah N.Y., Sonkin D., Bronson R.T., Kucherlapati R. Genetic mechanisms in Apc-mediated mammary tumorigenesis. PLoS Genet. 2009;5(2):e1000367. doi: 10.1371/journal.pgen.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jang G.B., Kim J.Y., Cho S.D. Blockade of Wnt/beta-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep. 2015;5:12465. doi: 10.1038/srep12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kim do Y., Park E.Y., Chang E. A novel miR-34a target, protein kinase D1, stimulates cancer stemness and drug resistance through GSK3/beta-catenin signaling in breast cancer. Oncotarget. 2016;7(12):14791–14802. doi: 10.18632/oncotarget.7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nurse P.M. Nobel Lecture. Cyclin dependent kinases and cell cycle control. Biosci Rep. 2002;22(5-6):487–499. doi: 10.1023/a:1022017701871. [DOI] [PubMed] [Google Scholar]
- 177.Sutherland R.L., Musgrove E.A. Cyclins and breast cancer. J Mammary Gland Biol Neoplasia. 2004;9(1):95–104. doi: 10.1023/B:JOMG.0000023591.45568.77. [DOI] [PubMed] [Google Scholar]
- 178.Gonzalez-Angulo A.M., Guarneri V., Gong Y. Downregulation of the cyclin-dependent kinase inhibitor p27kip1 might correlate with poor disease-free and overall survival in inflammatory breast cancer. Clin Breast Cancer. 2006;7(4):326–330. doi: 10.3816/CBC.2006.n.045. [DOI] [PubMed] [Google Scholar]
- 179.Walker A.J., Wedam S., Amiri-Kordestani L. FDA approval of palbociclib in combination with fulvestrant for the treatment of hormone receptor-positive, HER2-negative metastatic breast cancer. Clin Cancer Res. 2016;22(20):4968–4972. doi: 10.1158/1078-0432.CCR-16-0493. [DOI] [PubMed] [Google Scholar]
- 180.Radtke F., Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3(10):756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 181.Reedijk M., Odorcic S., Chang L. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65(18):8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 182.Dickson B.C., Mulligan A.M., Zhang H. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20(6):685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 183.Reedijk M., Pinnaduwage D., Dickson B.C. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008;111(3):439–448. doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- 184.Dontu G., Jackson K.W., McNicholas E., Kawamura M.J., Abdallah W.M., Wicha M.S. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6(6):R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Farnie G., Clarke R.B., Spence K. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99(8):616–627. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- 186.Sansone P., Storci G., Giovannini C. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cell. 2007;25(3):807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- 187.Ingham P.W., McMahon A.P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 188.Jiang J., Hui C.C. Hedgehog signaling in development and cancer. Dev Cell. 2008;15(6):801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Garcia-Zaragoza E., Perez-Tavarez R., Ballester A. Intraepithelial paracrine Hedgehog signaling induces the expansion of ciliated cells that express diverse progenitor cell markers in the basal epithelium of the mouse mammary gland. Dev Biol. 2012;372(1):28–44. doi: 10.1016/j.ydbio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 190.Fiaschi M., Rozell B., Bergstrom A., Toftgard R., Kleman M.I. Targeted expression of GLI1 in the mammary gland disrupts pregnancy-induced maturation and causes lactation failure. J Biol Chem. 2007;282(49):36090–36101. doi: 10.1074/jbc.M704280200. [DOI] [PubMed] [Google Scholar]
- 191.Jeng K.S., Sheen I.S., Jeng W.J., Yu M.C., Hsiau H.I., Chang F.Y. High expression of Sonic Hedgehog signaling pathway genes indicates a risk of recurrence of breast carcinoma. OncoTargets Ther. 2013;7:79–86. doi: 10.2147/OTT.S54702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mitchell P.J., Barker K.T., Martindale J.E. Cloning and characterisation of cDNAs encoding a novel non-receptor tyrosine kinase, brk, expressed in human breast tumours. Oncogene. 1994;9(8):2383–2390. [PubMed] [Google Scholar]
- 193.Barker K.T., Jackson L.E., Crompton M.R. BRK tyrosine kinase expression in a high proportion of human breast carcinomas. Oncogene. 1997;15(7):799–805. doi: 10.1038/sj.onc.1201241. [DOI] [PubMed] [Google Scholar]
- 194.Aubele M., Auer G., Walch A.K. PTK (protein tyrosine kinase)-6 and HER2 and 4, but not HER1 and 3 predict long-term survival in breast carcinomas. Br J Cancer. 2007;96(5):801–807. doi: 10.1038/sj.bjc.6603613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ludyga N., Anastasov N., Gonzalez-Vasconcellos I., Ram M., Hofler H., Aubele M. Impact of protein tyrosine kinase 6 (PTK6) on human epidermal growth factor receptor (HER) signalling in breast cancer. Mol Biosyst. 2011;7(5):1603–1612. doi: 10.1039/c0mb00286k. [DOI] [PubMed] [Google Scholar]
- 196.Miah S., Martin A., Lukong K.E. Constitutive activation of breast tumor kinase accelerates cell migration and tumor growth in vivo. Oncogenesis. 2012;1:e11. doi: 10.1038/oncsis.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Koren S., Reavie L., Couto J.P. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature. 2015;525(7567):114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- 198.Perez-Tenorio G., Stal O. Southeast Sweden Breast Cancer G. Activation of AKT/PKB in breast cancer predicts a worse outcome among endocrine treated patients. Br J Cancer. 2002;86(4):540–545. doi: 10.1038/sj.bjc.6600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kim E.K., Kim H.A., Koh J.S. Phosphorylated S6K1 is a possible marker for endocrine therapy resistance in hormone receptor-positive breast cancer. Breast Cancer Res Treat. 2011;126(1):93–99. doi: 10.1007/s10549-010-1315-z. [DOI] [PubMed] [Google Scholar]
- 200.deGraffenried L.A., Friedrichs W.E., Russell D.H. Inhibition of mTOR activity restores tamoxifen response in breast cancer cells with aberrant Akt Activity. Clin Cancer Res. 2004;10(23):8059–8067. doi: 10.1158/1078-0432.CCR-04-0035. [DOI] [PubMed] [Google Scholar]
- 201.Menard S., Fortis S., Castiglioni F., Agresti R., Balsari A. HER2 as a prognostic factor in breast cancer. Oncology. 2001;61(suppl 2):67–72. doi: 10.1159/000055404. [DOI] [PubMed] [Google Scholar]
- 202.Tokunaga E., Kimura Y., Mashino K. Activation of PI3K/Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006;13(2):137–144. doi: 10.2325/jbcs.13.137. [DOI] [PubMed] [Google Scholar]
- 203.Slattery M.L., Kerber R.A. A comprehensive evaluation of family history and breast cancer risk. The Utah Population Database. JAMA. 1993;270(13):1563–1568. [PubMed] [Google Scholar]
- 204.Couch F.J., Nathanson K.L., Offit K. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science. 2014;343(6178):1466–1470. doi: 10.1126/science.1251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Narod S.A. BRCA mutations in the management of breast cancer: the state of the art. Nat Rev Clin Oncol. 2010;7(12):702–707. doi: 10.1038/nrclinonc.2010.166. [DOI] [PubMed] [Google Scholar]
- 206.Narod S.A., Salmena L. BRCA1 and BRCA2 mutations and breast cancer. Discov Med. 2011;12(66):445–453. [PubMed] [Google Scholar]
- 207.King M.C., Marks J.H., Mandell J.B. New York Breast Cancer Study G. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 208.Gershoni-Baruch R., Dagan E., Fried G. Significantly lower rates of BRCA1/BRCA2 founder mutations in Ashkenazi women with sporadic compared with familial early onset breast cancer. Eur J Cancer. 2000;36(8):983–986. doi: 10.1016/s0959-8049(00)00045-9. [DOI] [PubMed] [Google Scholar]
- 209.Hodgson S.V., Heap E., Cameron J. Risk factors for detecting germline BRCA1 and BRCA2 founder mutations in Ashkenazi Jewish women with breast or ovarian cancer. J Med Genet. 1999;36(5):369–373. [PMC free article] [PubMed] [Google Scholar]
- 210.Couch F.J., Farid L.M., DeShano M.L. BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet. 1996;13(1):123–125. doi: 10.1038/ng0596-123. [DOI] [PubMed] [Google Scholar]
- 211.Struewing J.P., Coriaty Z.M., Ron E. Founder BRCA1/2 mutations among male patients with breast cancer in Israel. Am J Hum Genet. 1999;65(6):1800–1802. doi: 10.1086/302678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Alsop K., Fereday S., Meldrum C. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol. 2012;30(21):2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Moslehi R., Chu W., Karlan B. BRCA1 and BRCA2 mutation analysis of 208 Ashkenazi Jewish women with ovarian cancer. Am J Hum Genet. 2000;66(4):1259–1272. doi: 10.1086/302853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Trainer A.H., Lewis C.R., Tucker K., Meiser B., Friedlander M., Ward R.L. The role of BRCA mutation testing in determining breast cancer therapy. Nat Rev Clin Oncol. 2010;7(12):708–717. doi: 10.1038/nrclinonc.2010.175. [DOI] [PubMed] [Google Scholar]
- 215.Kuchenbaecker K.B., Hopper J.L., Barnes D.R. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 216.Metcalfe K.A., Lubinski J., Gronwald J. The risk of breast cancer in BRCA1 and BRCA2 mutation carriers without a first-degree relative with breast cancer. Clin Genet. 2018;93(5):1063–1068. doi: 10.1111/cge.13191. [DOI] [PubMed] [Google Scholar]
- 217.Ko K.P., Kim S.J., Huzarski T. The association between smoking and cancer incidence in BRCA1 and BRCA2 mutation carriers. Int J Cancer. 2018;142(11):2263–2272. doi: 10.1002/ijc.31257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ford D., Easton D.F., Bishop D.T., Narod S.A., Goldgar D.E. Risks of cancer in BRCA1-mutation carriers. Breast cancer linkage consortium. Lancet. 1994;343(8899):692–695. doi: 10.1016/s0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
- 219.Gayther S.A., Mangion J., Russell P. Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet. 1997;15(1):103–105. doi: 10.1038/ng0197-103. [DOI] [PubMed] [Google Scholar]
- 220.Chen S., Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25(11):1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nilsson M.P., Hartman L., Idvall I., Kristoffersson U., Johannsson O.T., Loman N. Long-term prognosis of early-onset breast cancer in a population-based cohort with a known BRCA1/2 mutation status. Breast Cancer Res Treat. 2014;144(1):133–142. doi: 10.1007/s10549-014-2842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li G., Guo X., Tang L. Analysis of BRCA1/2 mutation spectrum and prevalence in unselected Chinese breast cancer patients by next-generation sequencing. J Cancer Res Clin Oncol. 2017;143(10):2011–2024. doi: 10.1007/s00432-017-2465-8. [DOI] [PubMed] [Google Scholar]
- 223.Copson E.R., Maishman T.C., Tapper W.J. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): a prospective cohort study. Lancet Oncol. 2018;19(2):169–180. doi: 10.1016/S1470-2045(17)30891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rebbeck T.R., Friebel T.M., Friedman E. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat. 2018;39(5):593–620. doi: 10.1002/humu.23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Anders C.K., Hsu D.S., Broadwater G. Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. J Clin Oncol. 2008;26(20):3324–3330. doi: 10.1200/JCO.2007.14.2471. [DOI] [PubMed] [Google Scholar]
- 226.Brianese R.C., Nakamura K.D.M., Almeida F. BRCA1 deficiency is a recurrent event in early-onset triple-negative breast cancer: a comprehensive analysis of germline mutations and somatic promoter methylation. Breast Cancer Res Treat. 2018;167(3):803–814. doi: 10.1007/s10549-017-4552-6. [DOI] [PubMed] [Google Scholar]
- 227.Yadav S., Ladkany R., Yadav D. Impact of BRCA mutation status on survival of women with triple-negative breast cancer. Clin Breast Cancer. 2017 doi: 10.1016/j.clbc.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 228.Foulkes W.D. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. Cancer Spectr Knowl Environ. 2003;95(19):1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 229.Molyneux G., Geyer F.C., Magnay F.A. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7(3):403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 230.Mukohara T. PI3K mutations in breast cancer: prognostic and therapeutic implications. Breast Cancer (Dove Med Press) 2015;7:111–123. doi: 10.2147/BCTT.S60696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fruman D.A., Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13(2):140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Engelman J.A. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 233.Miller T.W., Rexer B.N., Garrett J.T., Arteaga C.L. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res. 2011;13(6):224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Samuels Y., Wang Z., Bardelli A. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 235.Liu P., Cheng H., Roberts T.M., Zhao J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gonzalez-Angulo A.M., Chen H., Karuturi M.S. Frequency of mesenchymal-epithelial transition factor gene (MET) and the catalytic subunit of phosphoinositide-3-kinase (PIK3CA) copy number elevation and correlation with outcome in patients with early stage breast cancer. Cancer. 2013;119(1):7–15. doi: 10.1002/cncr.27608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Stemke-Hale K., Gonzalez-Angulo A.M., Lluch A. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68(15):6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Millis S.Z., Ikeda S., Reddy S., Gatalica Z., Kurzrock R. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19784 diverse solid tumors. JAMA Oncol. 2016;2(12):1565–1573. doi: 10.1001/jamaoncol.2016.0891. [DOI] [PubMed] [Google Scholar]
- 239.Yang W., Hosford S.R., Dillon L.M. Strategically timing inhibition of phosphatidylinositol 3-kinase to maximize therapeutic Index in estrogen receptor alpha-positive, PIK3CA-mutant breast cancer. Clin Cancer Res. 2016;22(9):2250–2260. doi: 10.1158/1078-0432.CCR-15-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Easton D. Breast cancer genes–what are the real risks? Nat Genet. 1997;16(3):210–211. doi: 10.1038/ng0797-210. [DOI] [PubMed] [Google Scholar]
- 241.de Jong M.M., Nolte I.M., te Meerman G.J. Genes other than BRCA1 and BRCA2 involved in breast cancer susceptibility. J Med Genet. 2002;39(4):225–242. doi: 10.1136/jmg.39.4.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Nusbaum R., Vogel K.J., Ready K. Susceptibility to breast cancer: hereditary syndromes and low penetrance genes. Breast Dis. 2006;27:21–50. doi: 10.3233/bd-2007-27103. [DOI] [PubMed] [Google Scholar]
- 243.Hoskins K.F., Stopfer J.E., Calzone K.A. Assessment and counseling for women with a family history of breast cancer. A guide for clinicians. JAMA. 1995;273(7):577–585. [PubMed] [Google Scholar]
- 244.Easton D.F. The inherited component of cancer. Br Med Bull. 1994;50(3):527–535. doi: 10.1093/oxfordjournals.bmb.a072908. [DOI] [PubMed] [Google Scholar]
- 245.Harris C.C., Hollstein M. Clinical implications of the p53 tumor-suppressor gene. N Engl J Med. 1993;329(18):1318–1327. doi: 10.1056/NEJM199310283291807. [DOI] [PubMed] [Google Scholar]
- 246.Garber J.E., Goldstein A.M., Kantor A.F., Dreyfus M.G., Fraumeni J.F., Jr., Li F.P. Follow-up study of twenty-four families with Li-Fraumeni syndrome. Cancer Res. 1991;51(22):6094–6097. [PubMed] [Google Scholar]
- 247.Marsh D.J., Kum J.B., Lunetta K.L. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8(8):1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 248.Giardiello F.M., Welsh S.B., Hamilton S.R. Increased risk of cancer in the Peutz-Jeghers syndrome. N Engl J Med. 1987;316(24):1511–1514. doi: 10.1056/NEJM198706113162404. [DOI] [PubMed] [Google Scholar]
- 249.Hemminki A., Markie D., Tomlinson I. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391(6663):184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 250.Jenne D.E., Reimann H., Nezu J. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18(1):38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 251.Xia B., Sheng Q., Nakanishi K. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 252.Zhang F., Ma J., Wu J. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19(6):524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tischkowitz M., Xia B. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 2010;70(19):7353–7359. doi: 10.1158/0008-5472.CAN-10-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Antoniou A.C., Foulkes W.D., Tischkowitz M. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371(17):1651–1652. doi: 10.1056/NEJMc1410673. [DOI] [PubMed] [Google Scholar]
- 255.Lee A.S., Ang P. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371(17):1650–1651. doi: 10.1056/NEJMc1410673. [DOI] [PubMed] [Google Scholar]
- 256.Sopik V., Narod S.A. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371(17):1650. doi: 10.1056/NEJMc1410673#SA1. [DOI] [PubMed] [Google Scholar]
- 257.Apostolou P., Papasotiriou I. Current perspectives on CHEK2 mutations in breast cancer. Breast Cancer (Dove Med Press) 2017;9:331–335. doi: 10.2147/BCTT.S111394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Corso G., Intra M., Trentin C., Veronesi P., Galimberti V. CDH1 germline mutations and hereditary lobular breast cancer. Fam Cancer. 2016;15(2):215–219. doi: 10.1007/s10689-016-9869-5. [DOI] [PubMed] [Google Scholar]
- 259.Hansford S., Kaurah P., Li-Chang H. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1(1):23–32. doi: 10.1001/jamaoncol.2014.168. [DOI] [PubMed] [Google Scholar]
- 260.Baylin S.B., Jones P.A. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Suzuki H., Maruyama R., Yamamoto E., Niinuma T., Kai M. Relationship between noncoding RNA dysregulation and epigenetic mechanisms in cancer. Adv Exp Med Biol. 2016;927:109–135. doi: 10.1007/978-981-10-1498-7_4. [DOI] [PubMed] [Google Scholar]
- 262.Wahl G.M., Spike B.T. Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. NPJ Breast Cancer. 2017;3:14. doi: 10.1038/s41523-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Castoria G., Giovannelli P., Lombardi M. Tyrosine phosphorylation of estradiol receptor by Src regulates its hormone-dependent nuclear export and cell cycle progression in breast cancer cells. Oncogene. 2012;31(46):4868–4877. doi: 10.1038/onc.2011.642. [DOI] [PubMed] [Google Scholar]
- 264.Boidot R., Vegran F., Jacob D. The transcription factor GATA-1 is overexpressed in breast carcinomas and contributes to survivin upregulation via a promoter polymorphism. Oncogene. 2010;29(17):2577–2584. doi: 10.1038/onc.2009.525. [DOI] [PubMed] [Google Scholar]
- 265.Wang Z., Yang B., Zhang M. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018 doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Choi H.J., Joo H.S., Won H.Y. Role of RBP2-induced ER and IGF1R-ErbB signaling in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2018;110(4) doi: 10.1093/jnci/djx207. [DOI] [PubMed] [Google Scholar]
- 267.Stone A., Zotenko E., Locke W.J. DNA methylation of oestrogen-regulated enhancers defines endocrine sensitivity in breast cancer. Nat Commun. 2015;6:7758. doi: 10.1038/ncomms8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Djebali S., Davis C.A., Merkel A. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Meseure D., Drak Alsibai K., Nicolas A., Bieche I., Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. BioMed Res Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Elkon R., Agami R. Characterization of noncoding regulatory DNA in the human genome. Nat Biotechnol. 2017;35(8):732–746. doi: 10.1038/nbt.3863. [DOI] [PubMed] [Google Scholar]
- 271.Iyer M.K., Niknafs Y.S., Malik R. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lo P.K., Wolfson B., Zhou X., Duru N., Gernapudi R., Zhou Q. Noncoding RNAs in breast cancer. Brief Funct Genomics. 2016;15(3):200–221. doi: 10.1093/bfgp/elv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Taft R.J., Pang K.C., Mercer T.R., Dinger M., Mattick J.S. Non-coding RNAs: Regulators of disease. J Pathol. 2010;220(2):126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 274.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 275.Van Grembergen O., Bizet M., de Bony E.J. Portraying breast cancers with long noncoding RNAs. Sci Adv. 2016;2(9):e1600220. doi: 10.1126/sciadv.1600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Khorkova O., Hsiao J., Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24. doi: 10.1016/j.addr.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ali M.M., Akhade V.S., Kosalai S.T. PAN-cancer analysis of S-phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nat Commun. 2018;9(1):883. doi: 10.1038/s41467-018-03265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Janakiraman H., House R.P., Gangaraju V.K., Diehl J.A., Howe P.H., Palanisamy V. The long (lncRNA) and short (miRNA) of it: TGFbeta-mediated control of RNA-binding proteins and noncoding RNAs. Mol Cancer Res. 2018;16(4):567–579. doi: 10.1158/1541-7786.MCR-17-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Nakagawa S., Kageyama Y. Nuclear lncRNAs as epigenetic regulators-beyond skepticism. Biochim Biophys Acta. 2014;1839(3):215–222. doi: 10.1016/j.bbagrm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 280.Yoon J.H., Abdelmohsen K., Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425(19):3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Tracy K.M., Tye C.E., Ghule P.N. Mitotically-associated lncRNA (MANCR) affects genomic stability and cell division in aggressive breast cancer. Mol Cancer Res. 2018;16(4):587–598. doi: 10.1158/1541-7786.MCR-17-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Gyorffy B., Pongor L., Bottai G. An integrative bioinformatics approach reveals coding and non-coding gene variants associated with gene expression profiles and outcome in breast cancer molecular subtypes. Br J Cancer. 2018;118(8):1107–1114. doi: 10.1038/s41416-018-0030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15(4):272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 284.Athie A., Huarte M. FAL1ing inside an amplicon. Cancer Cell. 2014;26(3):303–304. doi: 10.1016/j.ccr.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 285.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 286.Bianchini G., Balko J.M., Mayer I.A., Sanders M.E., Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Denkert C., Liedtke C., Tutt A., von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389(10087):2430–2442. doi: 10.1016/S0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]
- 288.Trivers K.F., Lund M.J., Porter P.L. The epidemiology of triple-negative breast cancer, including race. Cancer Causes Control. 2009;20(7):1071–1082. doi: 10.1007/s10552-009-9331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Gonzalez-Angulo A.M., Timms K.M., Liu S. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17(5):1082–1089. doi: 10.1158/1078-0432.CCR-10-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Livasy C.A., Karaca G., Nanda R. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19(2):264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 291.Malorni L., Shetty P.B., De Angelis C. Clinical and biologic features of triple-negative breast cancers in a large cohort of patients with long-term follow-up. Breast Cancer Res Treat. 2012;136(3):795–804. doi: 10.1007/s10549-012-2315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bertucci F., Finetti P., Cervera N. How basal are triple-negative breast cancers? Int J Cancer. 2008;123(1):236–240. doi: 10.1002/ijc.23518. [DOI] [PubMed] [Google Scholar]
- 293.Navin N., Kendall J., Troge J. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Curtis C., Shah S.P., Chin S.F. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lehmann B.D., Pietenpol J.A. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232(2):142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Burstein M.D., Tsimelzon A., Poage G.M. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21(7):1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Costa R., Shah A.N., Santa-Maria C.A. Targeting Epidermal Growth Factor Receptor in triple negative breast cancer: new discoveries and practical insights for drug development. Cancer Treat Rev. 2017;53:111–119. doi: 10.1016/j.ctrv.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 298.Marotti J.D., de Abreu F.B., Wells W.A., Tsongalis G.J. Triple-negative breast cancer: next-generation sequencing for target identification. Am J Pathol. 2017;187(10):2133–2138. doi: 10.1016/j.ajpath.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 299.Rehman A., Kim Y., Kim H. FOXO3a expression is associated with lymph node metastasis and poor disease-free survival in triple-negative breast cancer. J Clin Pathol. 2018 doi: 10.1136/jclinpath-2018-205052. [Published online March 27, 2018] [DOI] [PubMed] [Google Scholar]
- 300.Kandoth C., McLellan M.D., Vandin F. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Balko J.M., Giltnane J.M., Wang K. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4(2):232–245. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Schadendorf D., Hodi F.S., Robert C. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol. 2015;33(17):1889–1894. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Shu S., Lin C.Y., He H.H. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529(7586):413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Asano Y., Kashiwagi S., Goto W. Prediction of treatment responses to neoadjuvant chemotherapy in triple-negative breast cancer by analysis of immune checkpoint protein expression. J Transl Med. 2018;16(1):87. doi: 10.1186/s12967-018-1458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121(10):3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Marusyk A., Almendro V., Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 307.Turashvili G., Brogi E. Tumor heterogeneity in breast cancer. Front Med (Lausanne) 2017;4:227. doi: 10.3389/fmed.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Ellsworth R.E., Blackburn H.L., Shriver C.D., Soon-Shiong P., Ellsworth D.L. Molecular heterogeneity in breast cancer: state of the science and implications for patient care. Semin Cell Dev Biol. 2017;64:65–72. doi: 10.1016/j.semcdb.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 309.Almendro V., Marusyk A., Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 310.McGranahan N., Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27(1):15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 311.Beca F., Polyak K. Intratumor heterogeneity in breast cancer. Adv Exp Med Biol. 2016;882:169–189. doi: 10.1007/978-3-319-22909-6_7. [DOI] [PubMed] [Google Scholar]
- 312.Remsik J., Fedr R., Navratil J. Plasticity and intratumoural heterogeneity of cell surface antigen expression in breast cancer. Br J Cancer. 2018;118(6):813–819. doi: 10.1038/bjc.2017.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Brown T.M., Fee E. Rudolf Carl Virchow: medical scientist, social reformer, role model. Am J Publ Health. 2006;96(12):2104–2105. doi: 10.2105/AJPH.2005.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Foote F.W., Jr., Stewart F.W. A histologic classification of carcinoma of the breast. Surgery. 1946;19:74–99. [PubMed] [Google Scholar]
- 315.Hawkins R.A., Roberts M.M., Forrest A.P. Oestrogen receptors and breast cancer: current status. Br J Surg. 1980;67(3):153–169. doi: 10.1002/bjs.1800670302. [DOI] [PubMed] [Google Scholar]
- 316.Perou C.M., Parker J.S., Prat A., Ellis M.J., Bernard P.S. Clinical implementation of the intrinsic subtypes of breast cancer. Lancet Oncol. 2010;11(8):718–719. doi: 10.1016/S1470-2045(10)70176-5. author reply 720–711. [DOI] [PubMed] [Google Scholar]
- 317.Nik-Zainal S., Van Loo P., Wedge D.C. The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Shah S.P., Morin R.D., Khattra J. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 319.Kuukasjarvi T., Karhu R., Tanner M. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57(8):1597–1604. [PubMed] [Google Scholar]
- 320.Simpson P.T., Reis-Filho J.S., Gale T., Lakhani S.R. Molecular evolution of breast cancer. J Pathol. 2005;205(2):248–254. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- 321.Meacham C.E., Morrison S.J. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Greaves M., Maley C.C. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Turner N.C., Reis-Filho J.S. Genetic heterogeneity and cancer drug resistance. Lancet Oncol. 2012;13(4):e178–e185. doi: 10.1016/S1470-2045(11)70335-7. [DOI] [PubMed] [Google Scholar]
- 324.Martelotto L.G., Ng C.K., Piscuoglio S., Weigelt B., Reis-Filho J.S. Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 2014;16(3):210. doi: 10.1186/bcr3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Campbell L.L., Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6(19):2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 326.Place A.E., Jin Huh S., Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13(6):227. doi: 10.1186/bcr2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Maley C.C., Aktipis A., Graham T.A. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer. 2017;17(10):605–619. doi: 10.1038/nrc.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 329.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 330.Chambers A.F., Groom A.C., MacDonald I.C. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 331.Weigelt B., Peterse J.L. Van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 332.Riggi N., Aguet M., Stamenkovic I. Cancer metastasis: a reappraisal of its underlying mechanisms and their relevance to treatment. Annu Rev Pathol. 2018;13:117–140. doi: 10.1146/annurev-pathol-020117-044127. [DOI] [PubMed] [Google Scholar]
- 333.Scully O.J., Bay B.H., Yip G., Yu Y. Breast cancer metastasis. Cancer Genomics Proteomics. 2012;9(5):311–320. [PubMed] [Google Scholar]
- 334.Coleman R.E., Rubens R.D. The clinical course of bone metastases from breast cancer. Br J Cancer. 1987;55(1):61–66. doi: 10.1038/bjc.1987.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Selzner M., Morse M.A., Vredenburgh J.J., Meyers W.C., Clavien P.A. Liver metastases from breast cancer: long-term survival after curative resection. Surgery. 2000;127(4):383–389. doi: 10.1067/msy.2000.103883. [DOI] [PubMed] [Google Scholar]
- 336.Minn A.J., Gupta G.P., Siegel P.M. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Lin N.U., Bellon J.R., Winer E.P. CNS metastases in breast cancer. J Clin Oncol. 2004;22(17):3608–3617. doi: 10.1200/JCO.2004.01.175. [DOI] [PubMed] [Google Scholar]
- 338.Chen W., Hoffmann A.D., Liu H., Liu X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precision Oncology. 2018;2(1) doi: 10.1038/s41698-018-0047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Santa-Maria C.A., Gradishar W.J. Changing treatment paradigms in metastatic breast cancer: lessons learned. JAMA Oncol. 2015;1(4):528–534. doi: 10.1001/jamaoncol.2015.1198. quiz 549. [DOI] [PubMed] [Google Scholar]
- 340.Yates L.R., Knappskog S., Wedge D. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32(2) doi: 10.1016/j.ccell.2017.07.005. 169–184 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Klein C.A. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9(4):302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 342.Fidler I.J. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 343.Fidler I.J. Selection of successive tumour lines for metastasis. Nat New Biol. 1973;242(118):148–149. doi: 10.1038/newbio242148a0. [DOI] [PubMed] [Google Scholar]
- 344.Klein C.A. Cancer. The metastasis cascade. Science. 2008;321(5897):1785–1787. doi: 10.1126/science.1164853. [DOI] [PubMed] [Google Scholar]
- 345.Talmadge J.E., Fidler I.J. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70(14):5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Naxerova K., Jain R.K. Using tumour phylogenetics to identify the roots of metastasis in humans. Nat Rev Clin Oncol. 2015;12(5):258–272. doi: 10.1038/nrclinonc.2014.238. [DOI] [PubMed] [Google Scholar]
- 347.Bozic I., Antal T., Ohtsuki H. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010;107(43):18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Garraway L.A., Lander E.S. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 349.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Jr., Kinzler K.W. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Yachida S., Jones S., Bozic I. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Oskarsson T., Batlle E., Massague J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14(3):306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Visvader J.E., Lindeman G.J. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10(6):717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 353.Celia-Terrassa T., Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev. 2016;30(8):892–908. doi: 10.1101/gad.277681.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Peitzsch C., Tyutyunnykova A., Pantel K., Dubrovska A. Cancer stem cells: the root of tumor recurrence and metastases. Semin Cancer Biol. 2017;44:10–24. doi: 10.1016/j.semcancer.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 355.Hartkopf A.D., Stefanescu D., Wallwiener M. Tumor cell dissemination to the bone marrow and blood is associated with poor outcome in patients with metastatic breast cancer. Breast Cancer Res Treat. 2014;147(2):345–351. doi: 10.1007/s10549-014-3113-5. [DOI] [PubMed] [Google Scholar]
- 356.Baccelli I., Schneeweiss A., Riethdorf S. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31(6):539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 357.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 358.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 359.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Neelakantan D., Zhou H., Oliphant M.U.J. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun. 2017;8:15773. doi: 10.1038/ncomms15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Tsai J.H., Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27(20):2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Sanchez-Tillo E., Liu Y., de Barrios O. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69(20):3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Ye X., Brabletz T., Kang Y. Upholding a role for EMT in breast cancer metastasis. Nature. 2017;547(7661):E1–E3. doi: 10.1038/nature22816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Fischer K.R., Durrans A., Lee S. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Zheng X., Carstens J.L., Kim J. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Harris L., Fritsche H., Mennel R. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 367.Desmedt C., Voet T., Sotiriou C., Campbell P.J. Next-generation sequencing in breast cancer: first take home messages. Curr Opin Oncol. 2012;24(6):597–604. doi: 10.1097/CCO.0b013e328359554e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Lips E.H., Michaut M., Hoogstraat M. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res. 2015;17(1):134. doi: 10.1186/s13058-015-0642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Beije N., Jager A., Sleijfer S. Circulating tumor cell enumeration by the CellSearch system: the clinician's guide to breast cancer treatment? Cancer Treat Rev. 2015;41(2):144–150. doi: 10.1016/j.ctrv.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 370.Hall C., Valad L., Lucci A. Circulating tumor cells in breast cancer patients. Crit Rev Oncog. 2016;21(1-2):125–139. doi: 10.1615/CritRevOncog.2016016120. [DOI] [PubMed] [Google Scholar]
- 371.Lee J.S., Magbanua M.J.M., Park J.W. Circulating tumor cells in breast cancer: applications in personalized medicine. Breast Cancer Res Treat. 2016;160(3):411–424. doi: 10.1007/s10549-016-4014-6. [DOI] [PubMed] [Google Scholar]
- 372.Boral D., Vishnoi M., Liu H.N. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat Commun. 2017;8(1):196. doi: 10.1038/s41467-017-00196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Chen Y.H., Hancock B.A., Solzak J.P. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer. 2017;3:24. doi: 10.1038/s41523-017-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]