Abstract
The proposition of cancer cells in a tumor sample, named as tumor purity, is an intrinsic factor of tumor samples and has potentially great influence in variety of analyses including differential methylation, subclonal deconvolution and subtype clustering. InfiniumPurify is an integrated R package for estimating and accounting for tumor purity based on DNA methylation Infinium 450 k array data. InfiniumPurify has three main functions getPurity, InfiniumDMC and InfiniumClust, which could infer tumor purity, differential methylation analysis and tumor sample cluster accounting for estimated or user-provided tumor purities, respectively. The InfiniumPurify package provides a comprehensive analysis of tumor purity in cancer methylation research.
Keywords: Cancer subtype classification, Differential methylation analysis, DNA methylation, Tumor purity, InfiniumPurify
Availability
The R package InfiniumPurify is available from http://cran.r-project.org/web/packages.
Introduction
Tumor purity, defined as the percentage of cancer cells in a solid tumor sample, is an important characteristic that cannot be ignored in cancer genomics or epigenomics data analysis.1, 2, 3, 4 Due to the normal cell contamination in tumor tissue, high-throughput data obtained from tumor samples are mixed signals of cancer and normal cells. Thus the purity effect must be accounted for in various data analyses such as sample clustering/classification and differential expression/methylation.5, 6 Till now, a few methods and software tools are available for tumor purity estimation, mainly based on gene expression or copy number variation data. A comprehensive review is provided by.7
Here we present the InfiniumPurify, a comprehensive R package to evaluate and account for tumor purity in a series of cancer methylation researches based on Infinium 450 k array data. It includes the following functions: getPurity, which estimates tumor purities from beta value matrices of tumor and normal samples; InfiniumDMC, which performs differential methylation analysis accounting for tumor purities estimated from getPurify; InfiniumPurify, which infers purified tumor methylomes from tumor, normal samples and purities; InfiniumClust, which classified tumor samples into different methylation subtypes corrected by tumor purities.
Methods
InfiniumPurify takes beta value matrix of tumor and normal samples as input, which could be obtained from ChAMP,8 DMRcate,9 minfi10 or some related R packages. Note that if starting with raw CEL data of Infinium 450 k array, a normalization step is essential for data preparation. To be specific, two types of probes (type-I and type-II) are used in Infinium 450 k chip and they may have different beta distributions.11 Moreover, tumor samples exhibit a global different pattern with normal samples, i.e., hyper methylation in promoter regions and global hypo-methylation in the whole genome. So we prefer functional normalization12 in data preparation.
getPurity: estimate tumor purity from DNA methylation Infinium 450 k array data
The function getPurity is used to estimate tumor purities of tumor samples. It takes methylation beta value matrix of tumor (and optionally normal) samples and tumor type as inputs, and outputs a vector of tumor purities for all tumor samples. If normal data are available and numbers of tumor and normal samples are both sufficient large (≥20), the function first identifies a number of informative differentially methylated CpG sites (iDMCs) by comparing the methylation differences between tumor and normal samples and variation in tumor samples. Then methylation levels of the selected iDMCs are used to estimate tumor purity for each tumor sample by density evaluation of Gaussian kernel. When normal sample is unavailable or tumor/normal samples are too few to get reliable iDMCs, getPurity will load pre-selected iDMCs identified from public TCGA data to infer tumor purities. In such case, the tumor type needs to be specified by the user.
As an application, we calculated tumor purities for all tumor samples with methylation 450 k array data in TCGA, which are available from https://doi.org/10.5281/zenodo.253193. Comparison with purity estimated from other tools shows good correlation.13, 14
InfiniumDMC: differential methylation analysis accounting for tumor purity
Tumor purity could serious bias or weaken differential methylation analysis if not correctly accounting for. There are a few discussions on differential expression analysis with the consideration of tumor purity, and most of them simply add tumor purity as a covariate in regression model.6 However, as is showed in our work through rigorous data modeling, the tumor purity has multiplicative effect on differential methylation (as well as different expression), instead of additive.14
InfiniumDMC takes beta value matrix of tumor (and optionally normal)and purities for all tumor samples as inputs. Note that the purities can be the results from getPurity or other tools. The DM calling is performed under the following two scenarios. With normal sample size more than 20, InfiniumDMC tests the significance of differential methylation comparing tumor and normal data based on a generalized least square procedure.14 Otherwise when normal samples are too few or unavailable, InfiniumDMC will use data from tumor samples alone and test the association between tumor beta values and tumor purities.14 The latter control-free DM calling method provides an alternative way to DM analysis when normal controls are not available or of low quality.
InfiniumPurify: deconvolute pure tumor methylomes
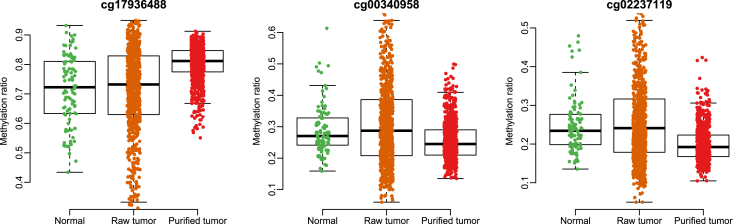
InfiniumPurify is to deconvolute pure tumor cellmethylomes from tumor samples, normal samples and tumor purity through a linear regression model. Intuitively, a CpG site is likely to be differentially methylated if it is highly correlated to tumor purities. In Figure 1, we show a CpG site with no significant methylation difference in tumor and normal samples by minfi. But its high correlation between tumor methylation and purity indicate that tumor methylations are seriously affected by tumor purity. After we corrected the purity effect by InfiniumPurify, its difference between purified tumor and normal methylomesis very significant.
Figure 1.
An example showing DMCs that are only detected by InfiniumPurify. Left panel shows their methylation level distributions in tumor and normal samples. Middle panel shows correlation between purities and methylation levels. Right panel shows methylation levels of normal and tumor samples after correcting for tumor purities.
InfiniumClust: cluster tumor sample accounting for tumor purity
DNA methylation plays an important role in tumorigenesis, thus clustering of tumor samples into different epigenetic subtypes is helpful in identifying diagnostic biomarker and therapeutic target in clinical practice. InfiniumClust is the first attempt to attribute tumor samples into subtypes after correcting tumor purity effect. It assumes pure normal methylome and tumor methylomes of different subtypes follow normal distribution after arcsine transformation. The clustering membership of a tumor sample is denoted as a latent variable that is optimized by Expectation-Maximization (EM) algorithm from the tumor-normal mixture model.15
InfiniumClust takes beta value matrix of and purities for a number of tumor samples and reports the probabilities of cluster membership. Given a user-specified number K of clusters, the function returns a list consisting of likelihood and membership matrix, where row corresponds to tumor samples and column corresponds to K clusters.
Conclusion
The R package InfiniumPurify contains a series of functions for DNA methylation analysis in cancer research accounting for tumor purity.
Conflict of interest
None declared.
Acknowledgements
The authors thank Weiwei Zhang and Yuzhen Sun for their help in R code and package debugging. This project was partially supported by the National Natural Science Foundation of China (61702325 and 61572327), Shanghai Science and Technology Innovation Action Plan (16391902900) and National Institute of Health (R01GM122083).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Carter S.L., Cibulskis K., Helman E. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. May 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshihara K., Shahmoradgoli M., Martinez E. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Loo P., Nordgard S.H., Lingjaerde O.C. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. Sep 28 2010;107(39):16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn J., Yuan Y., Parmigiani G. DeMix: deconvolution for mixed cancer transcriptomes using raw measured data. Bioinform. Aug 01 2013;29(15):1865–1871. doi: 10.1093/bioinformatics/btt301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaffe A.E., Irizarry R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. Feb 04 2014;15(2):R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aran D., Sirota M., Butte A.J. Systematic pan-cancer analysis of tumour purity. Nat Commun. Dec 04 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F., Zhang N., Wang J., Wu H., Zheng X. Tumor purity and differential methylation in cancer epigenomics. Brief Funct Genomics. Nov 2016;15(6):408–419. doi: 10.1093/bfgp/elw016. [DOI] [PubMed] [Google Scholar]
- 8.Morris T.J., Butcher L.M., Feber A. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. Feb 1 2014;30(3):428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters T.J., Buckley M.J., Statham A.L. De novo identification of differentially methylated regions in the human genome. Epigenet Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aryee M.J., Jaffe A.E., Corrada-Bravo H. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. May 15 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dedeurwaerder S., Defrance M., Bizet M., Calonne E., Bontempi G., Fuks F. A comprehensive overview of Infinium HumanMethylation450 data processing. Brief Bioinform. Nov 2014;15(6):929–941. doi: 10.1093/bib/bbt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortin J.P., Labbe A., Lemire M. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome biology. 2014;15(12):503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang N., Wu H.J., Zhang W., Wang J., Wu H., Zheng X. Predicting tumor purity from methylation microarray data. Bioinformatics. Nov 1 2015;31(21):3401–3405. doi: 10.1093/bioinformatics/btv370. [DOI] [PubMed] [Google Scholar]
- 14.Zheng X., Zhang N., Wu H.J., Wu H. Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies. Genome biology. 2017;18:183. doi: 10.1186/s13059-016-1143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W., Feng H., Wu H., Zheng X. Tumor purity improves cancer subtype classification from DNA methylation data. Bioinformatics. 2017;33(17):2651–2657. doi: 10.1093/bioinformatics/btx303. [DOI] [PMC free article] [PubMed] [Google Scholar]