Summary
The endothelium first forms in the blood islands in the extra-embryonic yolk sac and then throughout the embryo to establish circulatory networks that further acquire organ-specific properties during development to support diverse organ functions. Here, we investigated the properties of endothelial cells (ECs), isolated from four human major organs—the heart, lung, liver, and kidneys—in individual fetal tissues at three months' gestation, at gene expression, and at cellular function levels. We showed that organ-specific ECs have distinct expression patterns of gene clusters, which support their specific organ development and functions. These ECs displayed distinct barrier properties, angiogenic potential, and metabolic rate and support specific organ functions. Our findings showed the link between human EC heterogeneity and organ development and can be exploited therapeutically to contribute in organ regeneration, disease modeling, as well as guiding differentiation of tissue-specific ECs from human pluripotent stem cells.
Subject Areas: Stem Cells Research, Developmental Biology, Biology of Human Development
Graphical Abstract
Highlights
-
•
Isolate, culture, and characterize four human organ-specific endothelial cells
-
•
Identify and validate heritable gene expression clusters in human organ-specific ECs
-
•
Uncover the contribution of organ-specific ECs on vascular and organ function
Stem Cells Research; Developmental Biology; Biology of Human Development
Introduction
Vascular development begins with the differentiation of endothelial cells (ECs) and the de novo formation of a primitive vascular network derived from angioblasts that first appear in the blood islands of the yolk sac and then migrate to the fetus where vascular networks are formed (Coffin et al., 1991, Hatzopoulos et al., 1998, Risau, 1997). As development proceeds, this primitive network remodels, presumably triggered by changes in hemodynamics, surrounding cell types and environment, to establish a hierarchical vessel tree with tissue-specific functionality important for the function of each organ (Adams and Alitalo, 2007, Carmeliet, 2003, Eichmann et al., 2005, le Noble et al., 2004). This development requires that ECs therefore deviate from an initial largely homogeneous embryonic population to acquire specific identities necessary to support the diverse needs of flow, transport, hormonal interactions, and cell trafficking across the endothelium of each organ (Atkins et al., 2011, Red-Horse et al., 2007). Recent studies also showed that ECs may be derived from local progenitors in different organs or tissues, which further enhances the complexity and diversity in the response of ECs to injury and regenerative capacity (Goldman et al., 2014, Mugford et al., 2008, Peng et al., 2013, Tian et al., 2015, Wang et al., 2010).
Although little is known about how each organ determines the functional properties of its endothelium, EC differences have been shown between arteries and veins, large and small vessels, and different microvascular beds in various organs (Aird, 2007a, Aird, 2007b, Chi et al., 2003, Nolan et al., 2013). Some of these properties depend on the tissue environment. Site-specific microenvironmental cues (i.e., cytokines, metabolites, biophysical signals, and direct cell-cell contact from parenchyma cells) communicate with ECs and induce posttranscriptional modification. In transplantation studies, ECs can be induced to gain other tissue-specific structural and morphologic phenotypes and gene expression patterns (Aird et al., 1997).
EC properties are also under epigenetic control. Epigenetic footprints that control basal expression of endothelial-specific genes in different organs are specified early during embryonic development and preserved during sequential mitotic cycles (Minami and Aird, 2005). When cells are removed from their in vivo microenvironment and grown in culture, most, but not all, gene expression patterns are lost upon passaging (Burridge and Friedman, 2010, Lacorre et al., 2004). Nevertheless, a previous analysis of messenger RNA (mRNA) from several human EC lines revealed heterogeneous signatures even in passaged cells, providing evidence that epigenetic modification mediates differential gene expression profiles of ECs (Chi et al., 2003).
A possible problem in the existing studies is that the ECs were isolated from different donors with various isolation methods for different tissues. Although this concern has been addressed in mouse species (Lim et al., 2003, Nolan et al., 2013), mouse ECs have different properties compared with human ECs and there are differences between human and mouse development (Xue et al., 2013). These studies also failed to examine the cells as they formed vascular networks, a critical issue for organ-specific EC heterogeneity, or to show that gene expression data correlated with differences in cell functions. The in vitro stability is particularly important, in that the preservation of expression with passaging is needed for the study of mechanisms in human ECs and vascular development in the future.
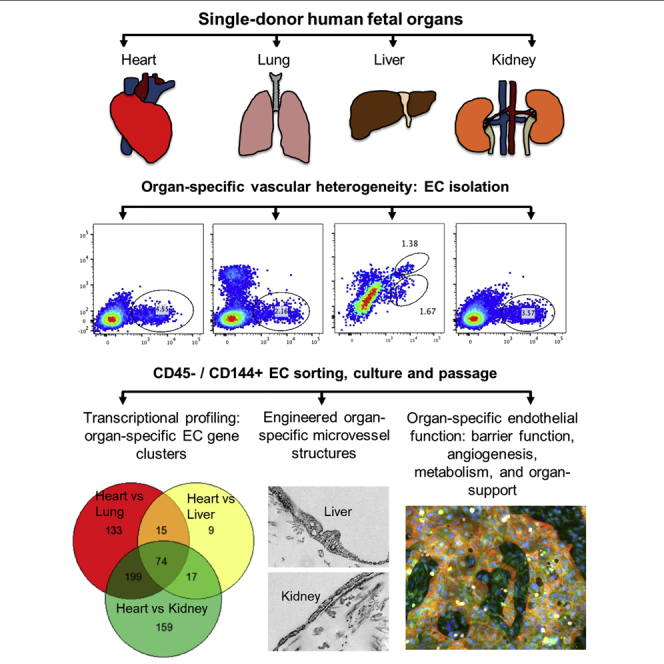
In the present study, we address these challenges and investigate human EC heterogeneity via the following different categories: (1) basal protein expression in ex vivo microvascular tissue beds; (2) organ-specific EC population in tissue; (3) morphology, structure, protein expression, transcriptional profiling, and vascular function of ECs after isolation, culture, and passaging; and (4) transcriptional signature validation in freshly isolated ECs, for four major developing organs—the heart, kidney, liver, and lung—obtained from individual human fetal donors. Together, our findings provide a comprehensive heterogeneity reference library after multiple passages in stabilized culture for human organ-specific ECs at the cellular, molecular, and transcriptional levels. This study will also contribute to understanding organ-specific vascular development, injuries, and potential development of targeted therapeutic interventions.
Results
Human Fetal ECs Show Organ-Specific Heterogeneity Ex Vivo
We examined the human fetal organ sets from three donors, constituting three biological replicates at 3 months' gestation (100–125 days). At this stage, all four major organs of interest—the heart, kidney, lung, and liver—have an established microvascular supply and exhibit organ-specific function. The heart beats at 120–160 bpm and is approximately 2 cm, the lungs have developed the entire air-conducting bronchial tree up to 20 generations with respiratory ducts and start to form barriers between alveoli and blood vessels, the liver is the major site of blood cell production and has also started to produce bile, and the kidneys have established nephrons and start to produce urine.
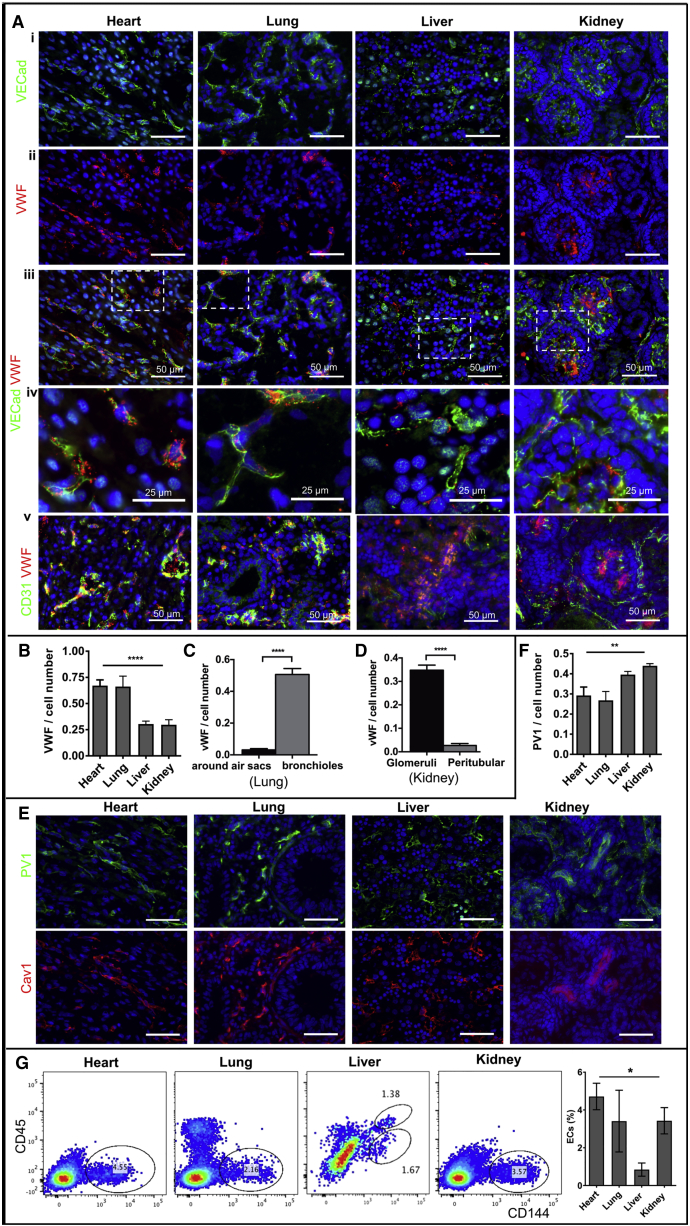
The vasculature of each organ displayed a heterogeneous organization, identified by the expression of EC markers, such as CD31, a transmembrane glycoprotein that constitutes endothelial intercellular junctions; CD144 (vascular endothelial [VE] cadherin), a major endothelial adhesion molecule; and von Willebrand factor (vWF), a glycoprotein that mediates platelet adhesion in the endothelium (Figure 1A). In the heart, ECs formed an organized network and contained a large amount of Weibel-Palade bodies with high expression of vWF (Figure 1A, ii–v). In the lungs, although the expression of vWF was overall high and comparable to that of the heart ECs (Figures 1A and 1B), it varied in different regions. Lower vWF expression was associated with pulmonary capillary ECs located near small air-sac-like structures, whereas a vWF mosaic pattern appeared in ECs neighboring the bronchioles and larger airways (Figures 1A and 1C). In the liver, the ECs displayed low vWF expression in VECad+ cells (Figures 1A and 1B), whereas there are clustered non-ECs containing high granular VWF in the interstitium (Figure 1A v), indicating the presence of hematopoietic progenitors. In the kidneys, two distinct microvascular structures were identified, with higher vWF expression in glomerular ECs and lower expression in peritubular microvascular ECs (Figures 1A, 1B, and 1D).
Figure 1.
Human Fetal Endothelial Cells Show Organ-Specific Heterogeneity Ex Vivo
(A) (A) Representative immunofluorescence images of frozen tissue sections from fetal human kidney, lung, liver, and heart stained with antibodies against CD144 (i) and vWF (ii) with merged (iii) and zoomed (iv) views, and (v) CD31 and vWF. Scale bar: 50 μm (i–iii, v) and 25 μm (iv).
(B–D) Quantification of vWF fluorescence area normalized to the number of CD31-positive cells for all four organs (B), within lungs (C), and kidneys (D). **** p ≤ 0.0001.
(E) Representative immunofluorescence images of frozen tissue sections stained with antibodies against PV1 and Cav1. Scale bar: 50 μm.
(F) Quantification of PV1 fluorescence area normalized to the number of CD31-positive cells for all four organs. ** p ≤ 0.01.
(G) Representative flow cytometry profiles of fresh total cell tissue suspension from fetal human heart, lung, liver, and kidney stained with antibodies against CD45 and CD144. Endothelial population is gated as CD144-positive and CD45-negative cells (left panels). The percentage of endothelial cells is compared among tissues against the total cell number (right panel). Bar plots were made from n = 3 images per donor for two donors (B–D, F) or three donor sets (G). Data are presented as mean ± SEM *p ≤ 0.05. SEM, standard error of the mean.
We then evaluated the structural differences of ECs in different fetal organs using the presence of caveolae, identified by the expression of caveolin-1 (Cav1), as well as stomatal and fenestral diaphragms, identified by the expression of plasmalemma vesicle-associated protein PLVAP (also called PV1)(Stan, 2007, Stan et al., 2012, Tkachenko et al., 2012). Our study showed that the heart and lung microvasculature has nearly complete Cav1 and PV1 colocalization, suggesting that the presence of PV1 was confined to stomatal diaphragms—a signature of continuous endothelium (Figures 1E and 1F). On the other hand, the kidney microvasculature, in particular, presented higher levels of and more discrete PV1 outside of Cav1 colocalization, indicating the presence of fenestral diaphragms—a signature of fenestrated endothelium (Figures 1E and 1F).
Human Fetal ECs Present as Diverse Organ-Specific Populations
To characterize the EC population within different fetal organ microvascular beds and distinguish them from hematopoietic cells, we used the pan-hematopoietic marker CD45 in combination with CD144. ECs were identified as CD144 positive and CD45 negative (CD144+/CD45-) by flow cytometry. Single cell suspensions, obtained by enzymatic digestion of mechanically minced fresh tissue followed by the filtered removal of large vessels and tissue chunks, showed well-demarcated CD144+/CD45- populations in each of the four fetal organs (Figures 1G and S1A). The highest percentage of ECs was from the heart (mean ± standard error [SE]: 4.7% ± 0.7 %), whereas the lowest percentage of ECs was found in the liver (0.8% ± 0.3%); the kidney and lung ECs represented 3.4% ± 1.6% and 3.4% ± 0.7%, respectively, of the entire cell suspension. The liver was the only tissue that showed a significant CD144+/CD45+ double-positive population, supporting the hematopoietic role of this organ during early stages of development. Kidney and lung ECs show distinct CD144+ population from the epithelial cell population (Epcam+) with no overlap (Figure S1B). Among the CD144+/CD45− cell population, the percentage for EphB4+ is 70%–90% for lung, liver, and kidney with no statistical significance among the three, whereas the heart cell suspension has significantly lower percentage for EphB4+ (∼25%). (Figure S1C). The majority of these ECs are CD34+ (85%–90% for heart and kidney ECs) (Figure S1C). Less than 2% ECs are GP38+ (podoplanin) for all four vascular beds, suggesting little lymphatic vascular development at this stage (Figure S1C). There is no detection of Robo1+ cells in this population (data not shown), which also suggests sufficient removal of large vessel tissue and minimal EC contaminations from large vessels (Figure S1D).
Human Fetal ECs Retain Ex Vivo Heterogeneity upon In Vitro Expansion
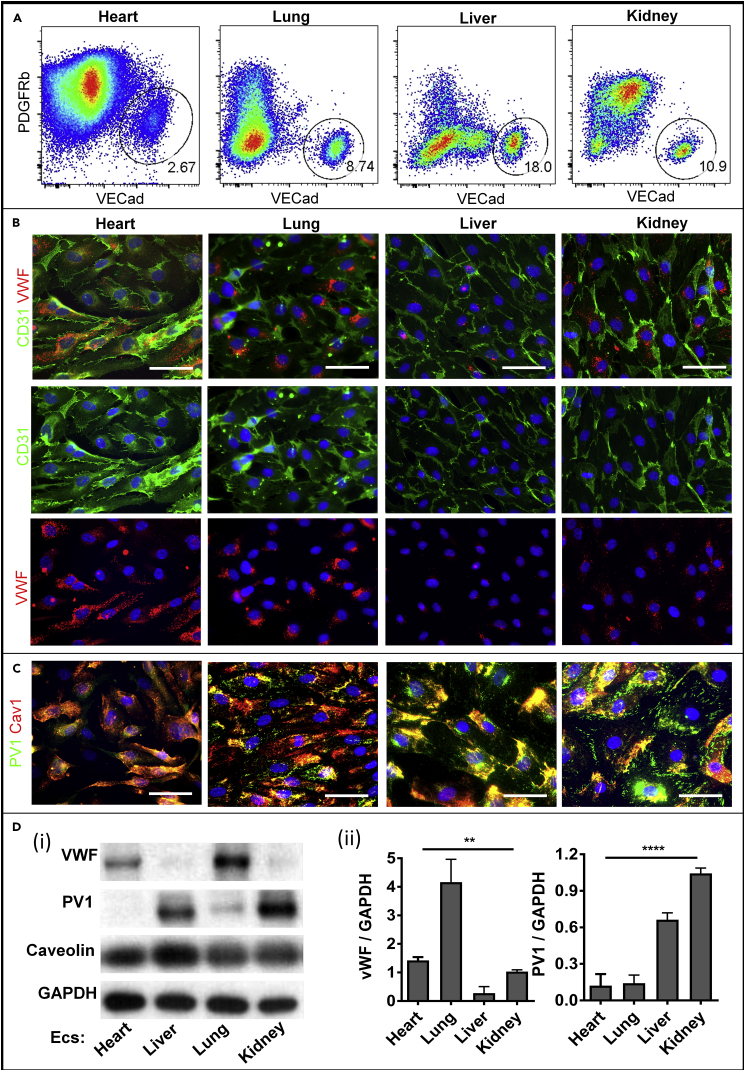
To study the persistence of EC heterogeneity in vitro and the epigenetic contribution to EC heterogeneity, we isolated and cultured the four human fetal organ-derived ECs through five passages (in each passage the cell number doubles). Important steps of the isolation procedure included the filtering and removal of large vessels and tissue chunks after enzymatic digestion, the removal of Epcam+ epithelial cell fraction from the whole tissue single cell suspension (particularly important for kidney and lung), the enrichment of the endothelial fraction through culture in low oxygen atmosphere with vascular endothelial growth factor (VEGF), the purification of the CD144+ endothelial fraction by flow cytometry, and the culture in VEGF-containing EC growth media for up to five passages (detailed in Methods). This isolation procedure also allows for the purification of other cell populations, important for further studies of regional organ heterogeneity and perivascular and parenchyma interactions (Figures 2A and S2A). The majority of cultured ECs are EphB4+ (>90%), with little contamination of lymphatic (GP38+ % < 4%) or large vascular (Robo1+ % < 1%) ECs; the CD34+ population in these cultured ECs ranges between 25% (liver) and 85% (kidney) (Figure S2B).
Figure 2.
Human Fetal ECs Retain the Ex Vivo Heterogeneity upon In Vitro Expansion
(A) Representative flow cytometry and sorting profiles of enriched cell suspension from fetal human heart, lung, liver, and kidney, stained with antibodies against VECad, PDGFRb.
(B and C) Representative immunofluorescence images of CD31 and vWF (B) and PV1 and Cav1 (C) fluorescence in cultured heart, lung, liver, and kidney ECs (scale bars: 50 μm).
(D) Representative western blot analysis (i) and densitometric quantification (ii) of vWF, PV1, and caveolin expression on cell lysates obtained from four ECs, using GAPDH as the loading control (n = 3 donors). Data are presented as mean ± SEM ** p ≤ 0.01, **** p ≤ 0.0001. SEM, standard error of the mean.
Isolated ECs displayed typical endothelial cobblestone appearance and maintained their morphologies through five passages in culture (Figure 2B). ECs also expressed CD31 with a high purity, greater than 99%, for all four fetal organs (Figure 2B). Similar to the ex vivo expression patterns, the isolated heart and lung ECs contain higher levels of vWF proteins, whereas the kidney and liver ECs have more PV1 protein expression (Figures 2B–2D). PV1 distributed largely at the peripheral cell surface membrane of the kidney and to a lesser extent in the liver ECs, whereas only a small fraction colocalized with Cav1, indicating the presence and persistence of fenestral diaphragms in these two EC types after isolation and expansion. In contrast, the heart and lung ECs expressed much more Cav1 but less PV1 than the kidney and liver ECs, with PV1 located mostly in the perinuclear region in the heart and lung ECs.
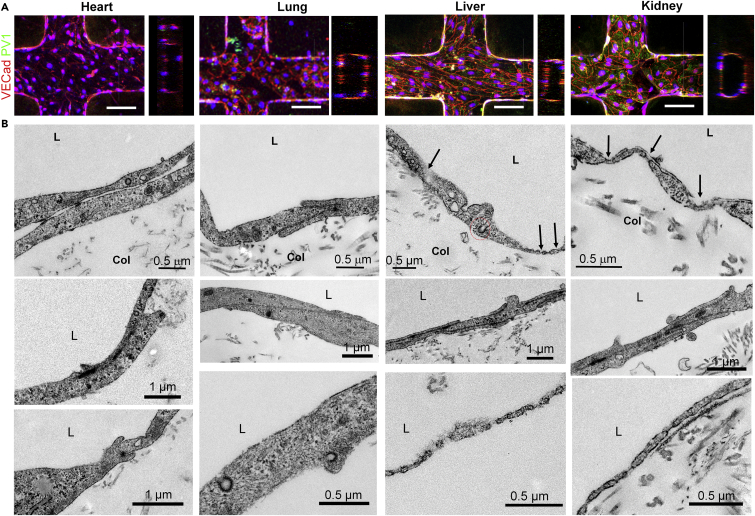
We used these ECs at passage 2 to engineer three-dimensional endothelialized microvascular networks. These were embedded in collagen microfluidic channels and cultured under gravity-driven flow, as previously described (Zheng et al., 2012). Cells from all four sources created similar networks of tubular endothelium (Figure 3A). Ultrastructural analysis via transmission electron microscopy revealed the formation of junctional complexes at cell-cell contacts and distinctive structures along the cell periphery in four types of microvessels (Figures 3B and S3). Engineered heart and lung microvessels showed focal contacts between adjacent cells and contained caveolae with stomatal diaphragms. Engineered kidney and liver microvessels showed complex and intercalated junctions between adjacent cells and presented numerous fenestrae (black arrows) along the peripheral regions of the cell membrane. The fenestrae in kidney endothelium contained diaphragms and had uniform morphology and size, whereas liver endothelium had discontinuous openings and contained fenestrae in varying sizes and structure along peripheral membranes. Not all fenestrae in the liver endothelium had diaphragms. In addition, the liver endothelium showed multivesicular bodies, lysosomes, and other distinct structures inside the cytoplasm (Figures 3B and S3). Although not directly compared with the microvascular ultrastructure in the four fetal tissues, these results establish the structural heterogeneity of organ-specific structural endothelium in vitro, which is consistent with known organotypic vasculature in vivo (Aird, 2007a, Augustin and Koh, 2017).
Figure 3.
Ultrastructural Heterogeneity of Human Fetal ECs upon In Vitro Expansion
(A) Projected immunofluorescence images and cross-sectional views of engineered perfusable, collagen-embedded, three-dimensional (3D) microvessel networks generated using the four types of organ-specific ECs (scale bar: 100 μm).
(B) Transmission electron microscopy of 3D microvessel networks showing distinctive structural signatures. Black arrows indicate the presence of fenestrae, and red circles denote clathrin-coated alveolae.
Global RNA Sequencing Reveals Heterogeneous Gene Expression Profiles in Cultured Organ-Specific ECs
We next sought to determine whether these cells have distinct molecular signatures after removal from the original microenvironment. We collected RNA for transcript profiling using RNAseq from single-donor derived heart, lung, liver, and kidney ECs obtained from three different donors after two passages in vitro. A total of 102 transcription factors and 93 co-factors were identified as highly expressed (top 20% transcript intensities) in all four types of cultured ECs (Figure S4A). These factors include known key regulators of vascular development such as ETS factor family (i.e., ETS1, FLI1, ELK3, ELF1 etc), ID proteins, KLF gene family (i.e., KLF3, KLF6, KLF10 etc), and genes important for vascular remodeling and VEGF signaling (i.e., SP1, HES1, SOX18, HIF1A, NFKB1, etc.) (De Val and Black, 2009, Lee et al., 2004, Park et al., 2013).
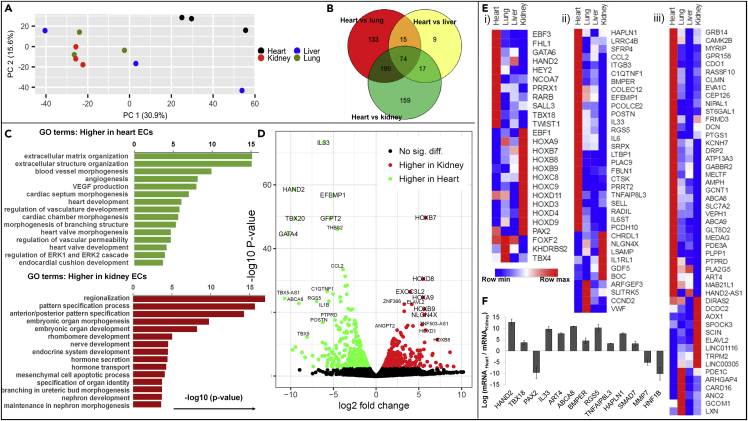
Principle component analysis showed distinct clustering of heart-derived ECs compared with the other three EC types (Figure 4A), and Venn diagrams defined the characteristic gene clusters for heart-specific ECs (Figure 4B). Differential expression analysis and Gene Ontology terminology analysis (Figures 4C, S4B, and S4C) showed that cultured heart ECs have higher gene expression levels, when compared with the other three types of ECs, with respect to extracellular matrix and structure organization, blood vessel morphogenesis, angiogenesis, VEGF production, heart chamber and valve morphogenesis, coronary vascular development and morphogenesis, branching morphogenesis, as well as regulation of vascular permeability. Compared with the heart ECs, lung ECs have higher expression for artery morphogenesis and development, developmental growth, and regionalization; liver ECs have higher expression for pattern specification, embryonic skeletal system development, regulation of leukocyte and homotypic cell-cell adhesion, regulation and activation of immune response, and bone marrow development; and kidney ECs in culture have higher expressions for regionalization, pattern specification process, embryonic organ morphogenesis/development/identity, hormone secretion and transport, and nephron development. These analyses are consistent with the corresponding stage of distinct organ development and function and indicate the important roles of different ECs in their corresponding organ development.
Figure 4.
Global RNA Sequencing Reveals Heterogeneous Gene Expression Profiles among Four EC Types (n = 3 Donors)
(A) 2D principle component analysis of RNA sequencing data for cultured organ-specific ECs showing clustered heart ECs compared with the kidney, lung, and liver ECs.
(B) Venn diagram of differentially expressed heart-specific ECs genes with respect to kidney, lung, and liver ECs.
(C and D) Gene Ontology terminology analysis (C) and volcano plot (D) showing different gene clusters for heart versus kidney ECs.
(E) Heatmaps of bona fide markers for the heart-, kidney-, and lung-specific EC transcription factors and co-factors (i), angiocrine and signaling factors (ii), and metabolism-related and other genes (iii).
(F) PCR validation of selected heart- and kidney-specific genes for additional three donor sets.
Differential expression analysis further revealed differentially expressed genes in paired comparison (Figures 4D and S4D). For example, genes such as GATA4, TBX5, TBX20, and HAND2 are expressed more than 200-fold higher in heart ECs, with high counts in RNAseq, compared with kidney ECs. These genes, although not studied in ECs, are known to be critical in heart development, heart valve formation, etc. (Rivera-Feliciano et al., 2006, Roche et al., 2013). Kidney ECs, on the other hand, highly and uniquely express many HOX genes that are known to be critical for kidney development. We further identified clustered genes as bona fide markers for organ-specific ECs by comparing one with the other three (fold change > 1.5 and false discovery rate [FDR] < 0.05). There are 74 heart-specific, 29 kidney-specific, 13 lung-specific highly expressed EC genes, whereas the numbers of liver-specific endothelial genes were not significant (Figures 4B and S4E). These organ-specific genes include transcription factors and regulation co-factors, angiocrine factors, extracellular matrix proteins, cell adhesion molecules, membrane transporters, signaling molecules, and metabolism-related genes (Figure 4E).
Heart-derived ECs showed upregulated expression of zinc finger, T-box, and basic-helix-loop-helix transcription factors involved in heart and embryonic vascular development (Brand, 2003, Fischer et al., 2004, Roche et al., 2013, Rutenberg et al., 2006, Xin et al., 2006) (GATA6, HAND2, HEY2, TBX18); transcription factors and co-factors involved in the heart valve and chamber development (Chakraborty et al., 2010, Chu et al., 2000, Lee and Yutzey, 2011) (FHL1); as well as factors involved in signaling regulation (Bergwerff et al., 2000, Chamberlain et al., 2014, Green and Vetter, 2011, Rhinn and Dolle, 2012) (EBF3, NCOA7, PRRX1, RARB, etc.). Kidney-derived ECs displayed upregulated expression of the helix-loop-helix transcription factor EBF1, which is critical in glomeruli development and maturation (Fretz et al., 2014, Schell et al., 2014), and the transcription factor PAX2, important for the regulation of branching morphogenesis and kidney development (Narlis et al., 2007). In addition, kidney ECs also expressed a large number of homeobox transcription factors, known as master developmental regulators in the kidney (Lechner and Dressler, 1997, Patterson and Potter, 2004) (HOXA9, HOXB7, HOXB8, HOXB9, HOXC8, HOXC9, HOXD3, HOXD4, HOXD9, HOXD11, etc.). Lung-derived ECs showed higher expression of the forkhead transcription factor FOXF2, known for the activation and regulation of pulmonary genes and embryonic development (Aitola et al., 2000); the T-box transcription factor TBX4, critical for lung branching (Arora et al., 2012); and the co-factor KHDRBS2, which regulates signal transduction during lung development (Lin et al., 2008). These organ-specific genes have not been found and studied in ECs previously, which may be important for the study of specific EC contribution during organ development.
Beside transcriptional regulators, heart-derived ECs highly expressed clusters of genes encoding for angiocrine factors, including annotated cytokines and growth factors (CCL2, IL6, IL33), signaling molecules, and extracellular matrix-remodeling proteins (BMPER, C1QTNF1, CTSK, HAPLN1, IL6ST, LTBP1, PCOLCE2, PLAC9, POSTN, RGS5, SFRP4, TNFAIP8L3). Other identified heart-derived EC-specific gene clusters included glycoproteins and surface receptors that mediate host defense, cell-cell adhesion and cell-matrix interaction (COLEC12, EFEMP1, ITGB3, LRRC4B, FBLN1, RADIL, PCDH10, PRRT2, SELL, SRPX), cytoplasm- and cytoskeleton-associated proteins (AMPH, CLMN, DRP2, FRMD3, MYRIP, GRB14), transporters (KCNH7, GRB14, ABCA8, ABCA9, EVA1C, GPR158, NIPAL1, RASSF10, SLC7A2), and metabolic regulators (ART4, ATP13A3, CAMK2B, GCNT1, GLT8D2, PDE3A, PLA2G5, PLPP1, PTGS1, ST6GAL1, etc.) (Figure 4F).
We validated the expression of selected genes from the kidney-specific and heart-specific EC gene lists by real-time polymerase chain reaction (PCR) analysis on both the RNA subjected to RNA sequencing and the three new RNA sets collected from single-donor organ-specific ECs (Figure 4F). The expression of EC genes identified as heart specific (i.e., HAND2, TBX18, IL33, ART4, ABCA8, BMPER, RGS5, TNFAIP8L3, HAPLN1, and SMAD7) were significantly upregulated in the heart-derived ECs compared with the kidney-derived ECs, and conversely kidney-specific genes (i.e., PAX2, HOXC10, MMP7, and HNF1B) were significantly upregulated in kidney-derived ECs. These organ-specific genes have minimal changes in the expression level through passage 5 (Figure S4F). We further used quantitative PCR (qPCR) analysis to validate the low expression of perivascular cell genes (i.e., NG2 and PDGFRb), confirming the absence of contaminating perivascular cells (Figure S4G).
Altogether, gene expression data show that ECs from fetal organs maintain organ-specific transcription profiles; in particular, heart-derived ECs display distinct clusters of transcription factors, angiocrine factors, surface glycoproteins, and metabolism that significantly distinguish these ECs from the other three EC types.
In Vitro-Ex Vivo Correlation of EC Heterogeneity
We then examined the EC heterogeneity ex vivo via RNAseq analysis and in situ immunofluorescence microscopy, to validate organ-specific gene clusters identified from cultured cells and further differentiate between the environmental and epigenetic contributions to EC heterogeneity. Freshly isolated ECs were collected via column purification with Epcam−CD144+/CD45−, and high-quality RNA were extracted for sequencing to assess gene expression in three single-donor organ sets. Column purification significantly enhanced cell survival during fresh isolation process. Fluorescence-activated cell sorting analysis for the column-purified ECs showed 70%–98% CD144+ cells and with no detectable perivascular cell contaminations for all four organs (Figure S5A). Principal component analysis revealed that ECs isolated from the four organs were distinct from each other, with organ replicates clustering tightly together (Figure S5B), suggesting higher degree of organ specificity when compared with the in vitro cultured ECs. Comparing heart and kidney ECs alone, there were more than 5,000 genes differentially expressed with significance in freshly isolated ECs, whereas only 867 genes were identified in culture (fold change >1.5 and FDR <0.1). This suggests that the majority of differentially expressed genes were likely dependent on the native microenvironment. However, a small portion of the expressed RNAs remained differentially expressed even in culture, suggesting that the differentiation has occurred at an epigenetic level. A potential epigenetic assay would help to confirm the epigenetic stability of ECs between in vivo and in vitro cultures.
Most top transcription factors and co-factors commonly expressed in culture were also highly expressed in freshly isolated ECs, whereas genes such as KLF2, KLF4, SOX4, SOX7, ID2, SNAI1, NR4A1, and EGR3, were significantly downregulated after in vitro expansion (Figures S5C and S5D). Among these highly expressed genes in freshly isolated sets, 32 transcription factors and 17 co-factors were differentially expressed in at least one type of EC and five transcription factors (ETS1, ELK3, SOX18, HMGA1, SOX7) were highly expressed with significant variation among all the ECs (Figure S5C), a difference not detected in cultured EC sets. Other top transcription factors, such as COUP-TFII (NR2F2) and ETS2, were differentially expressed in kidney versus heart, whereas ERF, ID1, E2F4, and SOX4 in the liver are different from those in the kidney, heart, and lung, which appear similar. These transcription factors have been recognized as important regulators of EC development and specification, and their differential expression suggests that vascular development occurs at a different pace in some of the four organs at the gestation time evaluated.
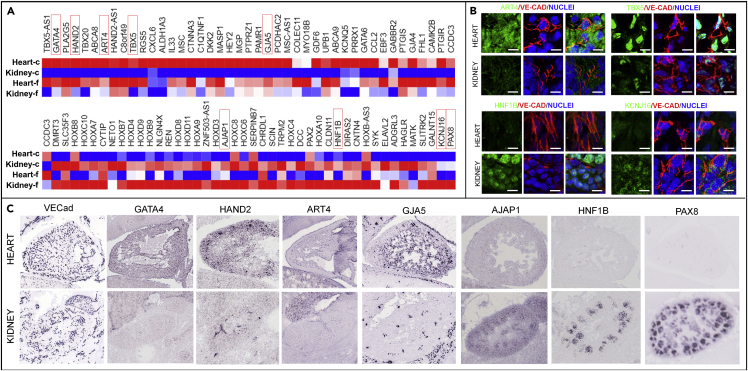
Next we validated the differentially expressed genes from cultured cells in freshly isolated cell sets, using heart and kidney paired comparison. Although the majority of differentially expressed genes in EC sets in vivo were lost after culture, over 100 highly upregulated genes (fold change >10) in either the heart or kidney ECs identified in culture remained elevated in freshly isolated ECs (Figure 5A). These differentially expressed gene clusters represent the epigenetic contribution to the organ-specific EC signature, which has the capacity to maintain in culture. Some of these identified genes were also verified in immunofluorescence staining in ex vivo tissues (Figure 5B: human and Figure 5C: mouse). The loss of differential expression of the highly expressed genes, from freshly isolated ECs to the cultured ECs, suggested the significant microenvironmental specifications for the developing ECs.
Figure 5.
Validation of Heterogeneous Gene Expression Profiles in Freshly Isolated ECs and Ex Vivo Tissue Sections (n = 3 Donors)
(A) Organ-specific genes identified from cultured sets (heart versus kidney) were validated in freshly isolated EC sets.
(B) Immunostaining of selected genes for heart and kidney in human fetal tissues at 120 days. Scale bar: 100 μm.
(C) Selected organ-specific gene expression in mouse embryo for heart and kidneys at E14.5 from Genepaint (genepaint.org).
Heart-Derived ECs Have the Highest Transendothelial Electrical Resistance, Angiogenic Potential, and Metabolic Rates among All Four ECs
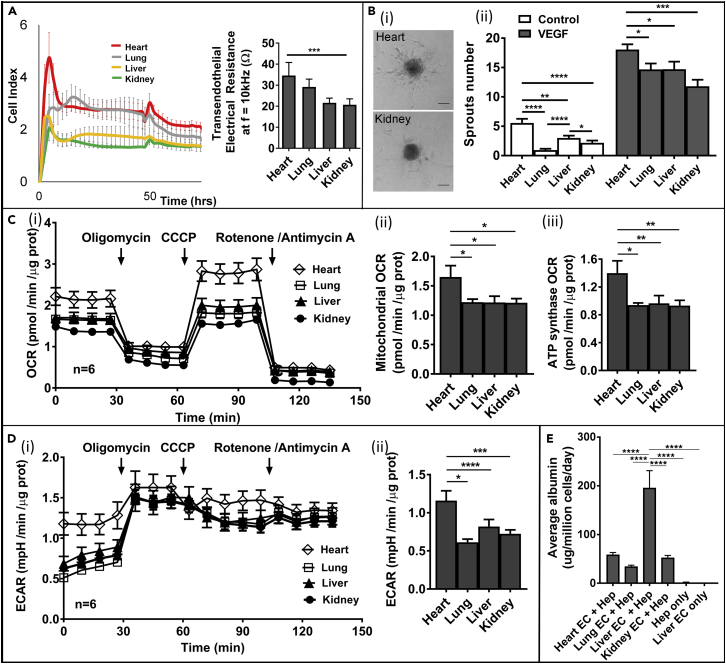
We next evaluated whether the variations in the transcriptional profiles of the four different EC types in culture led to distinct functions with respect to their barrier properties, angiogenic potential, and metabolic rates in vitro. To match the cell condition (passage 2) in RNAseq analysis, we used four ECs from single donor sets at the same passage number and culture media conditions for all the functional studies. Using xCELLigence RTCA SP instrumentation (ACEA Bioscience), we measured the real-time electrical impedance of EC monolayers at a particular frequency (f = 10 kHz). When EC monolayer reached confluency, the electrical impedance, recorded as the cell index, reached a plateau (Figure 6A), from which the transendothelial electrical resistance was measured. The monolayer of heart-derived ECs showed the highest electrical resistance (∼35 ± 1 Ω at f = 10 kHz) with statistical significance, whereas the electrical resistance of the kidney-derived ECs was the lowest among the four ECs (∼ 21 ± 2 Ω) (Figure 6A), supporting a better barrier function for the heart ECs when compared with cultured ECs from the fetal kidney.
Figure 6.
Cultured Organ-Specific Human Fetal ECs Display Distinct Vascular Functions and Bioenergetics and Support Specific Parenchyma Function
(A) Left panel: Raw data of transendothelial electrical resistance measurement versus time for four types of ECs in monolayer culture. Cell index is calculated as , where fn is the frequency at which the impedance measurement is carried out (fn = 10 kHz), R(fn,t) is the measured impedance at frequency fn at time point t, t0 is the time when the background is measured, and Zn is the corresponding frequency factor of fn (Zn = 15 Ω). Right panel: Electrical impedance measurements (10 kHz frequency) for confluent monolayers of organ-specific ECs (n = 5 cultures).
(B) Spheroid sprouting assay measurements of organ-specific EC angiogenic potential. (i) Representative bright-field images of heart and kidney EC sprouting spheroids, collagen-embedded, upon 24 hr of VEGF stimulation (40 ng/mL) (scale bar: 75 μm) and (ii) quantification of sprouts number at baseline and after VEGF stimulation (40 ng/mL) for organ-specific EC spheroids embedded in collagen matrix (24 hr) (n = 3 donors).
(C) Seahorse measurements of total cellular oxygen consumption rate (OCR) in organ-specific ECs: (i) time course of cellular oxygen consumption upon sequential addition of oligomycin (1 μM), CCCP (1 μM), and rotenone/antimycin A (1 μM each); (ii) quantification of mitochondrial OCR obtained upon electron transport chain inhibition with rotenone and antimycin A; and (iii) quantification of mitochondrial OCR due to ATP synthase activation obtained upon ATP synthase inhibition with oligomycin (n = 3 donors).
(D) Seahorse measurements of total extracellular acidification rate (ECAR) in organ-specific ECs (n = 3 donors): (i) time course of extracellular acidification upon sequential addition of oligomycin (1 μM), CCCP (1 μM), and rotenone/antimycin A (1 μM each) and (ii) ECAR quantification.
(E) ELISA measurement of albumin production from rat hepatocytes when cultured alone and when co-cultured with heart, lung, liver, and kidney ECs after 7 days (n = 4 replicates).
Data information: data are presented as mean ± SEM *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. SEM, standard error of the mean.
We then evaluated the ability of the ECs derived from four fetal organs to form new vessels in vitro using the spheroid sprouting assay (Heiss et al., 2015). Endospheroids were embedded in collagen and methylcellulose matrix and overlaid with endothelial growth medium, with or without 40 ng/mL VEGF, and grown for 24–48 hr to allow sprout formation (Figure 6B, i). Without VEGF exposure, most endospheroids were quiescent and devoid of any sprouting, except for the heart endospheroids. In the presence of 40 ng/mL VEGF, all endospheroids showed angiogenic activity, which was evident from the extensive capillary-like sprout formations. However, under VEGF exposure, heart endospheroids displayed a significantly higher number of sprouts (Figure 6B, ii).
Finally, we examined EC metabolism through the measurements of cellular oxygen consumption (respiration) and proton excretion (glycolysis) with a Seahorse Analyzer (Figures 6C and 6D). The oxygen consumption rate and extracellular acidification rate were measured at baseline, and in the presence of oligomycin, CCCP (carbonyl cyanide m-chlorophenylhydrazone), and rotenone/antimycin A, which were added sequentially (Figure 6C), and allowed for the calculation of respiration rates due to mitochondrial activity, ATP synthase, and spare capacities, as well as the glycolysis rate. Heart ECs showed the highest oxygen consumption rate due to an increase in both non-mitochondrial and mitochondrial respiration, which relied on enhanced ATP synthase activity, and also a significant spare respiratory capacity. A significantly higher glycolysis rate in addition to the mitochondrial respiration (Figure 6D) demonstrated that heart ECs possess the highest metabolic rate among the four types of ECs investigated, consistent with enhanced energetic support that can fuel a higher angiogenic potential.
Human Fetal Liver ECs Support Hepatocyte Function
We further examined whether organ-specific ECs can support specific parenchyma function. The four types of human fetal ECs at passage 2 were co-cultured with primary hepatocytes freshly isolated from rat liver. Each type of ECs was added on top of primary rat hepatocyte monolayers after their attachment for 24 hr following fresh isolation, at a ratio of 1 endo: 3 hepatocytes. Rat albumin was measured via enzyme-linked immunosorbent assay (ELISA) in media collected every 24 hr and compared after 7 days of culture. Co-culturing with ECs showed significant improvement in general compared with hepatocytes culture alone, in both albumin production and hepatocyte survival (Figures 6E and S6). Hepatocytes co-cultured with liver ECs showed significantly higher albumin production (∼200 μg/million cells per day) than those co-cultured with heart, lung, and kidney ECs (Figure 6E). Liver ECs appeared to provide additional signaling that promotes hepatocyte function (Figure S6C).
Discussion
Twenty years ago, studies in chick quail hybrids showed that the endothelium all over the body arises from blood islands that first appear in the embryonic yolk sac (DieterlenLievre et al., 1996, LeDouarin et al., 1996). Cells that arise from these early structures migrate throughout the body forming branches, and with time, they organize themselves into the arborized network of the adult arteries, veins, and capillaries. The initial formation of blood islands and tubes is called “vasculogenesis.” The lymphatic system develops in a similar fashion, originating from the venous branches. Blood vessels themselves can develop branches, primarily from the postcapillary venules. These branches form the vasculature of organs. More recently, it appears that endothelia can also arise locally within the mesenchyme of organs, including the heart and lungs. Thus diversity might arise both by interaction with parenchyma and by changes within the branched structure itself. Obviously, determining mechanisms that control local differentiation of ECs is difficult in whole tissue. We felt, therefore, that it would be desirable to develop cell lines. Because cells from fetuses usually have long replicative life spans, we chose to derive EC lines from four fetal tissues. We show that these human fetal ECs maintain stable organ-specific morphology and gene expression through five passages in culture. These fetal ECs express genes that are supportive to specific organ development and are different in metabolism, gene expression, and angiogenic responses to VEGF that may be relevant in vivo.
Among many possible organ-specific endothelial properties, we focused on three major categories: (1) vessel barrier, (2) angiogenic potential, and (3) metabolic functions. From a structural point of view, we showed that heart and lung ECs are continuous endothelium with focal close-contact junctions and no fenestrae formation, whereas kidney and liver ECs contain abundant fenestrae along the peripheral cell membrane but have complex intercalated junctions. These transcellular fenestrae would allow for large molecules to transport directly across the kidney and liver endothelium with low resistance, coinciding therefore with lower barrier strength. In addition, heart ECs have higher expression levels for adherent junctions, which not only contribute to higher barrier but also play important roles in signaling (Bazzoni and Dejana, 2004). Heart ECs also showed significantly higher transendothelial electrical resistance than kidney and liver ECs. Although not achieving physiological level of barrier in a monolayer culture (Kluger et al., 2013), these data suggest that at least one route of transport (transcellular or paracellular) (Komarova and Malik, 2010, Mehta and Malik, 2006) is higher in kidney and liver ECs compared with the heart ECs.
Our study specifically addressed ECs from the heart, liver, kidney, and lung, mainly owing to the tissue availability, and did not investigate ECs from other organs, such as the highly permeable high endothelial venules or the impermeable brain vasculature with unique distribution of tight junctions (Hirase et al., 1997, Schnitzer et al., 1994). Future studies of these ECs could provide more insights into the heterogeneous structure and molecular regulation of the endothelial barrier.
The second organ-specific property we demonstrated was the different angiogenic potential of ECs. Interestingly, heart ECs proved much more angiogenic than the other ECs derived from the other three organs under the same culture conditions and in the presence of identical stimuli. The higher angiogenic function in heart ECs is supported by a distinct cluster of genes categorized as angiocrine factors and signaling factors. These genes not only regulate angiogenesis but also provide important paracrine signaling for the maturation and function of developing cardiomyocytes. The different angiogenic potential may also be due to the specific origin of each EC type. Heart ECs are derived from at least three different sources, including the endocardium, sinus venous, and proepicardium (Zhang and Pu, 2013). Recent studies showed that ECs may even originate from different germ layers (del Monte and Harvey, 2012, Goldman et al., 2014, Mugford et al., 2008, Sims-Lucas et al., 2013, Wang et al., 2010). These varied origins could then lead to different capacities for angiogenesis via differential epigenetic control.
The endothelium has been shown critical to organ development and maturation, for example, in the pancreas, liver, and kidney (Abrahamson et al., 2009, Chen et al., 2016, Cleaver and Melton, 2003, Lammert et al., 2003, Schell et al., 2014, Tirziu and Simons, 2009, Villasenor and Cleaver, 2012), and can serve as progenitors for perivascular cells (Chen et al., 2016). It is possible that the endothelium is required to facilitate the differentiation of endoderm or mesoderm in the heart and to achieve mature function. Here, organ-specific EC identities, defined from the developing organs, could provide instructional signals toward the construction of organ-specific vascular microenvironments, and further the maturation of the developing parenchyma. This regulatory feedback system involving the endothelium is likely critical for successful organ regeneration, and for engineering functional parenchyma in vitro using stem cells.
We also observed heterogeneity in organ-specific EC metabolism. Remarkably, heart ECs showed the highest rates of oxygen consumption and glycolysis, supported by a distinct cluster of upregulated genes involved in metabolic regulation. ECs in mature vessel beds, despite their exposure to high oxygen levels, are known to be quiescent and to rely mostly on glycolysis (Eelen et al., 2015). During activation, ECs increase mitochondrial activity (Marcu et al., 2015), as well as glycolysis for energy production (De Bock et al., 2013a, De Bock et al., 2013b). However, little is known as to whether endothelial mitochondrial ATP generation is static or varies between vascular beds. Little is also known about the role of endothelial metabolism during development. Metabolism has been recently noted to be critical in regulating stem cell fate during early differentiation (Shyh-Chang and Daley, 2015, Shyh-Chang et al., 2013, Teslaa and Teitell, 2015). Our findings suggest that metabolism may also play an important role during the differentiation of endothelium toward organ-specific vasculature.
We further demonstrated the supportive role of these human fetal ECs in organ development and function by co-culturing with hepatocytes. Interestingly, these fetal ECs support hepatocyte survival and function, and liver-specific ECs also showed significantly superior supportive function in terms of albumin production from the hepatocytes. Although the mechanisms remain elusive and require investigation in depth, this functional study showed proof of principle that organ-specific ECs would be beneficial to support organ development, and engineer organ-on-a-chip to better recapitulate structure and function in vivo.
In summary, we have examined transcription profiles from fetal ECs cultured and passaged from four major human organs— the heart, lung, liver, and kidneys. We showed the distinct structural, and functional properties maintained over at least five passages in these four types of ECs. The results are still preliminary in several ways. First, we do not know how many more passages these cell lines can go through while preserving these properties. Longer replicative life spans could provide us with cells that would be reproducible reagents for mechanistic studies. Second, we do not know how closely the cultured cells themselves have diverse expression patterns that could represent the diversity of different EC phenotypes in vivo. For example, the high incidence in EphB4-positive cells in cultured sets suggest that endothelium form veins may be over-represented. Significant loss of differentially expressed genes from in vivo after culture suggest the importance of identifying environmental cues in vivo to understand and recapitulate the vascular and organ function. Third, although we did see some diversity in the structures of the EC-cell junctions, our measurement of electrical resistance suggests that none of these cell lines reproduce the functional impermeability of ECs with tight junctions. This is a general issue with cultured ECs with, as far as we know, only two reported exceptions. Aside from the cell junction, this may be important for other EC functions including control of endothelial differentiation by VE cadherin and shear-dependent transcription dependent on several cell junction proteins.
Nevertheless, our findings allowed us to identify for the first time the linkage between human EC heterogeneity and organ development, and differentiation between the intrinsic and microenvironment contribution to human EC heterogeneity. The availability of methods to culture organ-specific ECs makes it feasible to understand several organ-specific processes, including heart valve formation through the studies of endothelial-mesenchymal transition in vitro, formation and functional maturation of the kidney filtration barrier, and gas exchange interface in the lungs. Our work also provides important insights toward identifying precise organ-specific vascular targets and developing appropriate therapeutic interventions toward the treatment of organ-related vascular injuries and diseases.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We acknowledge the Lynn and Mike Garvey Imaging Laboratory in the Institute of Stem Cell and Regenerative Medicine, the Nanotech User Facility, and Flow Cytometry Facility, all at the University of Washington. We acknowledge the Electron Microscope Facility and Genomics Department in Fred Hutchinson Cancer Research Institute. We thank Drs. Marion Avril and Joseph Smith for their assistance in xCelligence analysis. We acknowledge the financial support of National Institute of Health (DP2DK102258, UH2/UH3 TR000504, UG3TR002158, R24HD000836, DP2HL137188, and UH2DK107343).
Author Contributions
Y.Z. conceived and designed the project. R.M., Y.J.C., J.X., C.L.F., R.N., and Y.Z. performed the experiments; Y.W., J.W.M., T.K.B., R.M., and Y.Z. performed RNAseq analysis. R.M., Y.J.C., K.M., C.E.M., K.R.S., J.H., S.M.S., and Y.Z. interpreted the data. R.M., Y.J.C., and Y.Z. wrote the manuscript. All authors edited and approved the manuscript.
Declaration of Interests
The authors declare that they have no conflict of interest.
Published: June 29, 2018
Footnotes
Supplemental Information includes Transparent Methods and six figures and can be found with this article online at https://doi.org/10.1016/j.isci.2018.05.003.
Supplemental Information
References
- Abrahamson D.R., Chen F., Chen M., Mecham R., Nguyen N., Jarad G. Development of kidney glomerular endothelial cells and their role in basement membrane assembly. Organogenesis. 2009;5:275–287. doi: 10.4161/org.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R.H., Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- Aird W.C. Phenotypic heterogeneity of the endothelium I. Structure, function, and mechanisms. Circ. Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- Aird W.C. Phenotypic heterogeneity of the endothelium II. Representative vascular beds. Circ. Res. 2007;100:174–190. doi: 10.1161/01.RES.0000255690.03436.ae. [DOI] [PubMed] [Google Scholar]
- Aird W.C., Edelberg J.M., WeilerGuettler H., Simmons W.W., Smith T.W., Rosenberg R.D. Vascular bed-specific expression of an endothelial cell gene is programmed by the tissue microenvironment. J. Cell Biol. 1997;138:1117–1124. doi: 10.1083/jcb.138.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitola M., Carlsson P., Mahlapuu M., Enerback S., Pelto-Huikko M. Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev. Dyn. 2000;218:136–149. doi: 10.1002/(SICI)1097-0177(200005)218:1<136::AID-DVDY12>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Arora R., Metzger R.J., Papaioannou V.E. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8:e1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins G.B., Jain M.K., Hamik A. Endothelial differentiation molecular mechanisms of specification and heterogeneity. Arterioscler. Thromb. Vasc. Biol. 2011;31:1476–1484. doi: 10.1161/ATVBAHA.111.228999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin H.G., Koh G.Y. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. 2017;357 doi: 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- Bazzoni G., Dejana E. Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiol. Rev. 2004;84:869–901. doi: 10.1152/physrev.00035.2003. [DOI] [PubMed] [Google Scholar]
- Bergwerff M., Gittenberger-de Groot A.C., Wisse L.J., DeRuiter M.C., Wessels A., Martin J.F., Olson E.N., Kern M.J. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000;436:12–19. doi: 10.1007/pl00008193. [DOI] [PubMed] [Google Scholar]
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- Burridge K.A., Friedman M.H. Environment and vascular bed origin influence differences in endothelial transcriptional profiles of coronary and iliac arteries. Am. J. Physiol. Heart Circ. Physiol. 2010;299:H837–H846. doi: 10.1152/ajpheart.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in health and disease. Nat. Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Wirrig E.E., Hinton R.B., Merrill W.H., Spicer D.B., Yutzey K.E. Twist1 promotes heart valve cell proliferation and extracellular matrix gene expression during development in vivo and is expressed in human diseased aortic valves. Dev. Biol. 2010;347:167–179. doi: 10.1016/j.ydbio.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain A.A., Lin M.Y., Lister R.L., Maslov A.A., Wang Y.D., Suzuki M., Wu B.R., Greally J.M., Zheng D.Y., Zhou B. DNA methylation is developmentally regulated for genes essential for cardiogenesis. J. Am. Heart Assoc. 2014;3:e000976. doi: 10.1161/JAHA.114.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhang H., Liu Y., Adams S., Eilken H., Stehling M., Corada M., Dejana E., Zhou B., Adams R.H. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat. Commun. 2016;7:12422. doi: 10.1038/ncomms12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J.T., Chang H.Y., Haraldsen G., Jahnsen F.L., Troyanskaya O.G., Chang D.S., Wang Z., Rockson S.G., Van de Rijn M., Botstein D. Endothelial cell diversity revealed by global expression profiling. Proc. Natl. Acad. Sci. USA. 2003;100:10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu P.H., Ruiz-Lozano P., Zhou Q., Cai C., Chen J. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech. Dev. 2000;95:259–265. doi: 10.1016/s0925-4773(00)00341-5. [DOI] [PubMed] [Google Scholar]
- Cleaver O., Melton D.A. Endothelial signaling during development. Nat. Med. 2003;9:661–668. doi: 10.1038/nm0603-661. [DOI] [PubMed] [Google Scholar]
- Coffin J.D., Harrison J., Schwartz S., Heimark R. Angioblast differentiation and morphogenesis of the vascular endothelium in the mouse embryo. Dev. Biol. 1991;148:51–62. doi: 10.1016/0012-1606(91)90316-u. [DOI] [PubMed] [Google Scholar]
- De Bock K., Georgiadou M., Carmeliet P. Role of endothelial cell metabolism in vessel sprouting. Cell Metab. 2013;18:634–647. doi: 10.1016/j.cmet.2013.08.001. [DOI] [PubMed] [Google Scholar]
- De Bock K., Georgiadou M., Schoors S., Kuchnio A., Wong B.W., Cantelmo A.R., Quaegebeur A., Ghesquiere B., Cauwenberghs S., Eelen G. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154:651–663. doi: 10.1016/j.cell.2013.06.037. [DOI] [PubMed] [Google Scholar]
- De Val S., Black B.L. Transcriptional control of endothelial cell development. Dev. Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Monte G., Harvey R.P. An endothelial contribution to coronary vessels. Cell. 2012;151:932–934. doi: 10.1016/j.cell.2012.11.004. [DOI] [PubMed] [Google Scholar]
- DieterlenLievre F., Duprat A.M., LeDouarin N. Developmental biology: origins and prospects. Med. Sci. 1996;12:67–75. [Google Scholar]
- Eelen G., de Zeeuw P., Simons M., Carmeliet P. Endothelial cell metabolism in normal and diseased vasculature. Circ. Res. 2015;116:1231–1244. doi: 10.1161/CIRCRESAHA.116.302855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann A., Yuan L., Moyon D., Lenoble F., Pardanaud L., Breant C. Vascular development: from precursor cells to branched arterial and venous networks. Int. J. Dev. Biol. 2005;49:259–267. doi: 10.1387/ijdb.041941ae. [DOI] [PubMed] [Google Scholar]
- Fischer A., Schumacher N., Maier M., Sendtner M., Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretz J.A., Nelson T., Velazquez H., Xi Y.G., Moeckel G.W., Horowitz M.C. Early B-cell factor 1 is an essential transcription factor for postnatal glomerular maturation. Kidney Int. 2014;85:1091–1102. doi: 10.1038/ki.2013.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman O., Han S.Y., Hamou W., de Villeroche V.J., Uzan G., Lickert H., Gouon-Evans V. Endoderm generates endothelial cells during liver development. Stem Cell Rep. 2014;3:556–565. doi: 10.1016/j.stemcr.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green Y.S., Vetter M.L. EBF proteins participate in transcriptional regulation of Xenopus muscle development. Dev. Biol. 2011;358:240–250. doi: 10.1016/j.ydbio.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzopoulos A.K., Folkman J., Vasile E., Eiselen G.K., Rosenberg R.D. Isolation and characterization of endothelial progenitor cells from mouse embryos. Development. 1998;125:1457–1468. doi: 10.1242/dev.125.8.1457. [DOI] [PubMed] [Google Scholar]
- Heiss M., Hellstrom M., Kalen M., May T., Weber H., Hecker M., Augustin H.G., Korff T. Endothelial cell spheroids as a versatile tool to study angiogenesis in vitro. FASEB J. 2015;29:3076–3084. doi: 10.1096/fj.14-267633. [DOI] [PubMed] [Google Scholar]
- Hirase T., Staddon J.M., Saitou M., AndoAkatsuka Y., Itoh M., Furuse M., Fujimoto K., Tsukita S., Rubin L.L. Occludin as a possible determinant of tight junction permeability in endothelial cells. J. Cell Sci. 1997;110:1603–1613. doi: 10.1242/jcs.110.14.1603. [DOI] [PubMed] [Google Scholar]
- Kluger M.S., Clark P.R., Tellides G., Gerke V., Pober J.S. Claudin-5 controls intercellular barriers of human dermal microvascular but not human umbilical vein endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2013;33:489–500. doi: 10.1161/ATVBAHA.112.300893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y., Malik A.B. Regulation of endothelial permeability via paracellular and transcellular transport pathways. Annu. Rev. Physiol. 2010;72:463–493. doi: 10.1146/annurev-physiol-021909-135833. [DOI] [PubMed] [Google Scholar]
- Lacorre D.A., Baekkevold E.S., Garrido I., Brandtzaeg P., Haraldsen G., Amalric F., Girard J.P. Plasticity of endothelial cells: rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood. 2004;103:4164–4172. doi: 10.1182/blood-2003-10-3537. [DOI] [PubMed] [Google Scholar]
- Lammert E., Cleaver O., Melton D. Role of endothelial cells in early pancreas and liver development. Mech. Dev. 2003;120:59–64. doi: 10.1016/s0925-4773(02)00332-5. [DOI] [PubMed] [Google Scholar]
- le Noble F., Moyon D., Pardanaud L., Yuan L., Djonov V., Matthijsen R., Breant C., Fleury V., Eichmann A. Flow regulates arterial-venous differentiation in the chick embryo yolk sac. Development. 2004;131:361–375. doi: 10.1242/dev.00929. [DOI] [PubMed] [Google Scholar]
- Lechner M.S., Dressler G.R. The molecular basis of embryonic kidney development. Mech. Dev. 1997;62:105–120. doi: 10.1016/s0925-4773(97)00667-9. [DOI] [PubMed] [Google Scholar]
- LeDouarin N., DieterlenLievre F., Teillet M.A. Quail-chick transplantations. Methods Cell Biol. 1996;51:23–59. [PubMed] [Google Scholar]
- Lee J.W., Bae S.H., Jeong J.W., Kim S.H., Kim K.W. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp. Mol. Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- Lee M.P., Yutzey K.E. Twist1 directly regulates genes that promote cell proliferation and migration in developing heart valves. PLoS One. 2011;6:e29758. doi: 10.1371/journal.pone.0029758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.C., Garcia-Cardena G., Allport J.R., Zervoglos M., Connolly A.J., Gimbrone M.A., Luscinskas F.W. Heterogeneity of endothelial cells from different organ sites in T-cell subset recruitment. Am. J. Pathol. 2003;162:1591–1601. doi: 10.1016/S0002-9440(10)64293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Ikegami M., Xu Y., Bosserhoff A.K., Malkinson A.M., Shannon J.M. Misexpression of MIA disrupts lung morphogenesis and causes neonatal death. Dev. Biol. 2008;316:441–455. doi: 10.1016/j.ydbio.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu R., Kotha S., Zhi Z., Qin W., Neeley C.K., Wang R.K., Zheng Y., Hawkins B.J. The mitochondrial permeability transition pore regulates endothelial bioenergetics and angiogenesis. Circ. Res. 2015;116:1336–1345. doi: 10.1161/CIRCRESAHA.116.304881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D., Malik A.B. Signaling mechanisms regulating endothelial permeability. Physiol. Rev. 2006;86:279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- Minami T., Aird W.C. Endothelial cell gene regulation. Trends Cardiovasc. Med. 2005;15:174–184. doi: 10.1016/j.tcm.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Mugford J.W., Sipila P., McMahon J.A., McMahon A.P. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev. Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlis M., Grote D., Gaitan Y., Boualia S.K., Bouchard M. Pax2 and Pax8 regulate branching morphogenesis and nephron differentiation in the developing kidney. J. Am. Soc. Nephrol. 2007;18:1121–1129. doi: 10.1681/ASN.2006070739. [DOI] [PubMed] [Google Scholar]
- Nolan D.J., Ginsberg M., Israely E., Palikuqi B., Poulos M.G., James D., Ding B.S., Schachterle W., Liu Y., Rosenwaks Z. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Kim T.M., Malik A.B. Transcriptional regulation of endothelial cell and vascular development. Circ. Res. 2013;112:1380–1400. doi: 10.1161/CIRCRESAHA.113.301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson L.T., Potter S.S. Atlas of Hox gene expression in the developing kidney. Dev. Dyn. 2004;229:771–779. doi: 10.1002/dvdy.10474. [DOI] [PubMed] [Google Scholar]
- Peng T., Tian Y., Boogerd C.J., Lu M.M., Kadzik R.S., Stewart K.M., Evans S.M., Morrisey E.E. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013;500:589–592. doi: 10.1038/nature12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K., Crawford Y., Shojaei F., Ferrara N. Endothelium-microenvironment interactions in the developing embryo and in the adult. Dev. Cell. 2007;12:181–194. doi: 10.1016/j.devcel.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Rhinn M., Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- Rivera-Feliciano J., Lee K.H., Kong S.W., Rajagopal S., Ma Q., Springer Z., Izumo S., Tabin C.J., Pu W.T. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–3618. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche P., Czubryt M.P., Wigle J.T. Molecular mechanisms of cardiac development. In: Ostadal B., Dhalla N.S., editors. Cardiac Adaptations: Molecular Mechanisms. Springer New York; 2013. pp. 19–39. [Google Scholar]
- Rutenberg J.B., Fischer A., Jia H.B., Gessler M., Zhong T.P., Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell C., Wanner N., Huber T.B. Glomerular development - shaping the multi-cellular filtration unit. Semin. Cell Dev. Biol. 2014;36:39–49. doi: 10.1016/j.semcdb.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Schnitzer J.E., Siflingerbirnboim A., Delvecchio P.J., Malik A.B. Segmental differentiation of permeability, protein glycosylation, and morphology of cultured bovine lung vascular endothelium. Biochem. Biophys. Res. Commun. 1994;199:11–19. doi: 10.1006/bbrc.1994.1185. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N., Daley G.Q. Metabolic switches linked to pluripotency and embryonic stem cell differentiation. Cell Metab. 2015;21:349–350. doi: 10.1016/j.cmet.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N., Daley G.Q., Cantley L.C. Stem cell metabolism in tissue development and aging. Development. 2013;140:2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims-Lucas S., Schaefer C., Bushnell D., Ho J., Logar A., Prochownik E., Gittes G., Bates C.M. Endothelial progenitors exist within the kidney and lung Mesenchyme. PLoS One. 2013;8:e65993. doi: 10.1371/journal.pone.0065993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan R.V. Endothelial stomatal and fenestral diaphragms in normal vessels and angiogenesis. J. Cell Mol. Med. 2007;11:621–643. doi: 10.1111/j.1582-4934.2007.00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan R.V., Tse D., Deharvengt S.J., Smits N.C., Xu Y., Luciano M.R., McGarry C.L., Buitendijk M., Nemani K.V., Elgueta R. The diaphragms of fenestrated endothelia: gatekeepers of vascular permeability and blood composition. Dev. Cell. 2012;23:1203–1218. doi: 10.1016/j.devcel.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslaa T., Teitell M.A. Pluripotent stem cell energy metabolism: an update. EMBO J. 2015;34:138–153. doi: 10.15252/embj.201490446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X.Y., Pu W.T., Zhou B. Cellular origin and developmental program of coronary angiogenesis. Circ. Res. 2015;116:515–530. doi: 10.1161/CIRCRESAHA.116.305097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirziu D., Simons M. Endothelium as master regulator of organ development and growth. Vasc. Pharmacol. 2009;50:1–7. doi: 10.1016/j.vph.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko E., Tse D., Sideleva O., Deharvengt S.J., Luciano M.R., Xu Y., McGarry C.L., Chidlow J., Pilch P.F., Sessa W.C. Caveolae, fenestrae and transendothelial channels retain PV1 on the surface of endothelial cells. PLoS One. 2012;7:e32655. doi: 10.1371/journal.pone.0032655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasenor A., Cleaver O. Crosstalk between the developing pancreas and its blood vessels: an evolving dialog. Semin. Cell Dev. Biol. 2012;23:685–692. doi: 10.1016/j.semcdb.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chadalavada K., Wilshire J., Kowalik U., Hovinga K.E., Geber A., Fligelman B., Leversha M., Brennan C., Tabar V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- Xin M., Davis C.A., Molkentin J.D., Lien C.L., Duncan S.A., Richardson J.A., Olson E.N. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc. Natl. Acad. Sci. USA. 2006;103:11189–11194. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Cai J.Y., Ma J., Huang Z., Guo M.X., Fu L.Z., Shi Y.B., Li W.X. Global expression profiling reveals genetic programs underlying the developmental divergence between mouse and human embryogenesis. BMC Genomics. 2013;14:568. doi: 10.1186/1471-2164-14-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pu W.T. The mysterious origins of coronary vessels. Cell Res. 2013;23:1063–1064. doi: 10.1038/cr.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Chen J., Craven M., Choi N., Totorica S., Diaz-Santana A., Kermani P., Hempstead B., Fischbach-Teschl C., Lopez J.A. In vitro microvessels for the study of angiogenesis and thrombosis. Proc. Natl. Acad. Sci. USA. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.