Summary
Non-centrosomal Microtubule Organizing Centers (MTOCs) direct microtubule (MT) organization to exert diverse cell-type specific functions. In Drosophila spermatids, the giant mitochondria provide structural platforms for MT reorganization to support elongation of the extremely long sperm. However, the molecular basis for this mitochondrial MTOC and other non-centrosomal MTOCs have not been discerned. Here we report that Drosophila centrosomin (cnn) expresses two major protein variants: the centrosomal form (CnnC) and a non-centrosomal form in testes (CnnT). CnnC is established as essential for functional centrosomes, the major MTOCs in animal cells. We show that CnnT is expressed exclusively in testes by alternative splicing, and localizes to giant mitochondria in spermatids. In cell culture, CnnT targets to the mitochondrial surface, recruits the MT nucleator γ-TuRC, and is sufficient to convert mitochondria to MTOCs independent of core pericentriolar proteins that regulate MT assembly at centrosomes. We mapped two separate domains in CnnT. One that is necessary and sufficient to target it to mitochondria, and another that is necessary and sufficient to recruit γ-TuRCs and nucleate MTs. In elongating spermatids, CnnT forms speckles on the giant mitochondria that are required to recruit γ-TuRCs to organize MTs and support spermiogenesis. This molecular characterization of the mitochondrial MTOC defines a minimal molecular requirement for MTOC generation, and implicates the potent role of Cnn (or its related) proteins in the direct regulation of MT assembly and organization of non-centrosomal MTOCs.
Keywords: centrosome, non-centrosomal MTOC, microtubule, Centrosomin, mitochondria, spermatogenesis, spermatid, centriole, gamma tubulin, γ-TuRC
Graphical Abstract
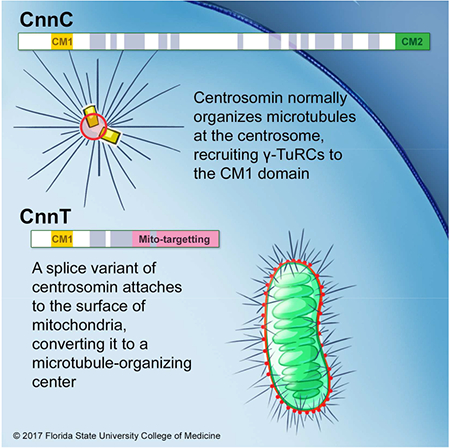
Introduction
Pericentriolar material (PCM) assembly at centrosomes regulates Microtubule-Organizing Center (MTOC) activity at centrosomes, the major MTOCs in animal cells. Non-centrosomal MTOCs also perform various essential functions in various cell types. In oocytes, they assemble in the cytoplasm and regulate bipolar spindle assembly [1, 2]. In syncytial myotubes, they organize at the nuclear periphery and Golgi apparatus to regulate nuclear positioning [3–5]. In developing Drosophila wing epithelia, they assemble at adherens junctions and regulate planar cell polarity [6]. In Drosophila testis, they form on the large fused mitochondria to promote sperm tail elongation [7]. Despite their importance in directing microtubule (MT) organization to exert cell-type-specific functions, the molecular composition and regulation of non-centrosomal MTOCs remain poorly understood.
Drosophila species have extremely long sperm; D. bifurca, in particular, produces the longest sperm known at nearly 6 cm [8]. During D. melanogaster spermiogenesis, the ~10 μm spherical spermatids elongate to ~1800 μm mature sperm, requiring an elaborate cytoskeleton network [7]. Four major structures run though the elongating spermatid during morphogenesis: the axoneme, cytoplasmic non-axonemal MTs, mitochondria, and F-actin cables [7]. The axoneme and actin are dispensable, but MTs and mitochondrial integrity are essential for spermatid elongation [7, 9, 10].
Mitochondria in Drosophila spermatids are specialized organelles with an important role in sperm morphogenesis. Following meiosis II, in the newly formed spermatid, mitochondria fuse into two giant mitochondrial derivatives that wrap around one another to produce the spherical “nebenkern”, a giant mitochondrial formation similar in size to the haploid nucleus [11]. During spermatid elongation, the two nebenkern derivatives unfurl and stretch with the elongating spermatid. The nebenkern is the main internal support that provides stiffness for elongating spermatids [7]; it also appears to provide a structural platform to organize MTs independently from centrosomes [7].
The molecular mechanisms by which the MTOCs assemble and function at the spermatid nebenkern are unknown. Here, we report that centrosomin (cnn) encodes two classes of proteins by alternative splicing. In addition to the well-characterized centrosomal form that is essential for functional centrosomes in various tissues [12–17], we show that the other form, CnnT, is expressed exclusively in testes and localizes to spermatid nebenkerns. CnnT recruits and activates the γ-Tubulin Ring Complex (γ-TuRC), the fundamental MT nucleator, generating unique MTOCs on the mitochondrial surface to support spermiogenesis. These data reveal how non-centrosomal MTOCs can be generated.
Results
CnnT is a Cnn variant expressed exclusively in testes
Drosophila melanogaster cnn expresses two classes of alternatively spliced protein variants: the long forms that include expression of the distal three 3’ coding exons, which encode the centrosomal form of Cnn (hereinafter CnnC, including variants PA, PB, PD, PE, PM and PN); and the short forms that don’t, but are instead expressed from a unique set of 3’ exons (all within a single intron that is spliced out when CnnC is produced), which we refer to as CnnT (Testis-specific Cnn, including PG, PH, PI and PK) (Figure 1A). CnnC is the well-characterized form that localizes to and is required for functional centrosomes [12–14, 16]. CnnC contains the conserved Centrosomin Motif 1 (CM1) near its N-terminus that is important for recruiting the MT nucleation factor γ-Tubulin (γ-Tub) [13]; and Centrosomin Motif 2 (CM2) at its C-terminus, required for centrosome localization and for furrow formation during embryonic cleavage cycles [18] (Figure 1A). CnnT shares the CM1 with CnnC, but has a unique C-terminus and lacks CM2 (Figure 1A). RT-PCR detects CnnT mRNA only in testes, and it was undetectable in ovaries, larval brains, or males with testes removed (Figure 1B). Additionally, all available cDNA clones and expressed sequence tags (ESTs) for CnnT came from testis libraries (flybase.org), suggesting that CnnT is expressed only in the male germline. In contrast, CnnC was expressed in testes and other tissues [12, 14, 19] (Figure 1B). A Cnn “conceptual form” that splices together the exons unique to both CnnC and CnnT was not detected by RT-PCR, consistent with the absence of such a cDNA clone in the EST library (flybase.org) (Figure 1B). The molecular weights of CnnC variants are ~150 kDa, and the four CnnT variants are predicted to be ~52-54 kDa. Western blotting of testis with an antibody that recognizes all Cnn variants (anti-Cnn(C+T); Figure 1A) showed that there was no detectable CnnC or CnnT in cnn null alleles cnnhk21 and cnn25cn1, while the cnnmfs3 and cnnmfs7 alleles, which truncate CnnC but leave CnnT intact, expressed CnnT but not CnnC (Figures 1A and C, S1A).
Figure 1. CnnT is expressed exclusively in testes.
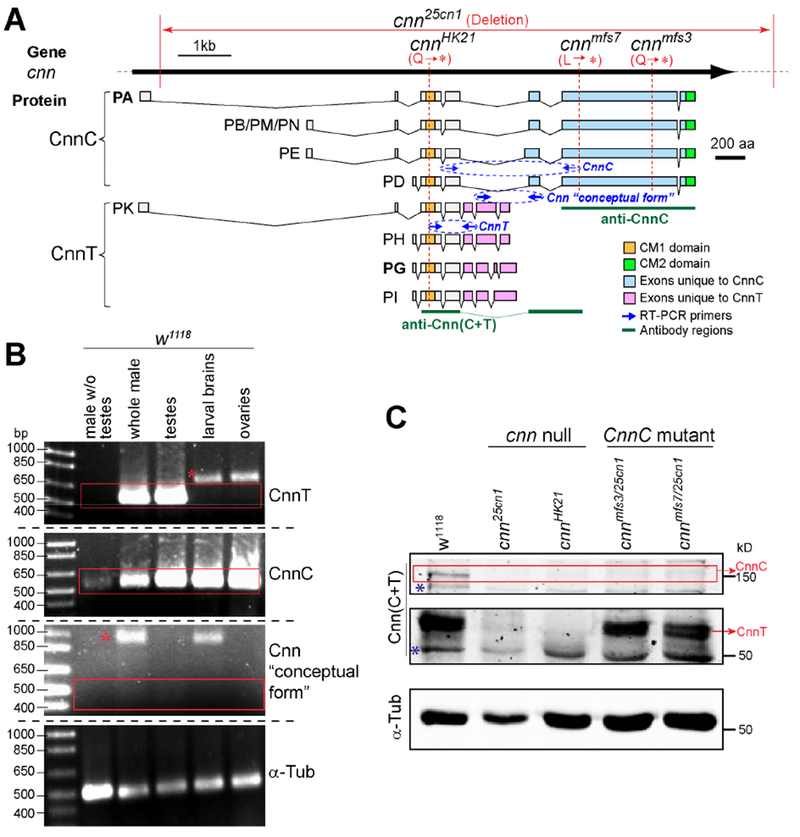
(A) Schematic showing the cnn gene, the CnnC and CnnT splice products, conserved domains CM1 and CM2, regions recognized by antibodies, primers used in RT-PCR, mutant alleles cnn25cn1, cnnHK21, cnnmfs3 and cnnmfs7.
(B) RT-PCR of cnn mRNAs. CnnT is predominantly expressed in testes. The “conceptual form “ with pink and blue exons (illustrated in A) spliced together is not expressed in the tissues examined. “*” indicates products from genomic DNA.
(C) Western blot of testes. WT (w1118) expresses CnnC and CnnT, cnn25cn1 and cnnHK21 express neither, cnnmfs3/25cnl and cnn mfs7/25cnl express CnnT but not CnnC. “*” indicates non-specific bands. See Figure S1A for whole blot.
CnnT localizes to mitochondria and recruits γ-TuRCs; its N-term and C-term are necessary and sufficient for γ-TuRC recruitment and mitochondrial targeting respectively
To study the function of CnnT, we expressed it in cultured cells (Cnn-PG; similar results were obtained with the other CnnT isoforms). Unlike its counterpart CnnC, CnnT did not localize to centrosomes, but instead localized at mitochondria in Drosophila Kc167 cells and various mammalian cell lines (Figures 2A, B). The non-centrosomal localization of CnnT is consistent with its lack of the CM2 domain (Figure 1A).
Figure 2. CnnT N-term recruits γ-TuRC and CnnT C-term targets to mitochondria; CnnT recruits γ-TuRCs to mitochondria independent of centrosome PCM proteins.
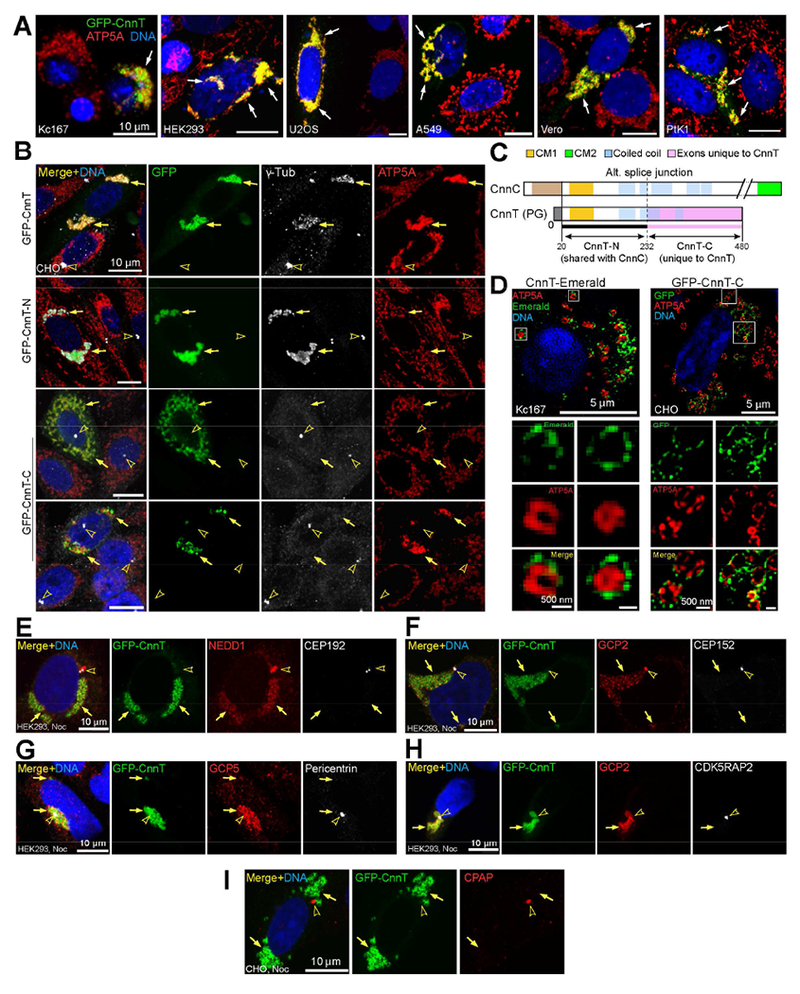
(A) Ectopic GFP-CnnT localizes to mitochondria marked by ATP5A in indicated cell lines.
(B) CnnT recruits γ-Tub to mitochondria; note clustering of mitochondria. CnnT-N forms aggregates that recruit γ-Tub but do not associate with mitochondria. CnnT-C localizes to mitochondria but does not recruit γ-Tub; mitochondria cluster upon high expression of CnnT-C (bottom panel).
(C) Schematic comparing CnnT to CnnC.
(D) 3D-SIM superresolution images showing CnnT and CnnT-C reside at the mitochondrial surface. ATP5A labels the inner mitochondrial membrane.
(E-I) Localization of indicated γ-TuRC-associated proteins and centrosomal PCM proteins. NEDD1 (E), GCP2 (F, H) and GCP5 (G) colocalize with CnnT at mitochondria, in addition to localizing to centrosomes. CEP192 (E), CEP152 (F), Pericentrin (G), CDK5RAP2 (H) and CPAP (I) localize to centrosomes, but do not colocalize with CnnT.
In each panel, arrows point to GFP-tagged proteins and arrowheads indicate centrosomes, “Noc” indicates nocodazole treatment before fixation. Scale bar as indicated. See also Figure S1.
Since CnnT contains the CM1 domain, which in CnnC recruits γ-TuRCs to centrosomes, we examined whether CnnT recruited γ-TuRC components to mitochondria. We found that CnnT recruited γ-Tub to mitochondria; in addition, Cnn-T induced clustering of mitochondria (Figure 2B). CnnT also recruited γ-TuRC-associated protein NEDD1/GCP-WD and γ-TuRC components GCP2 and GCP5 (Figure 2E-H).
To dissect the function of CnnT, we divided it into two parts: the N-term (CnnT-N), which contains CM1 but excludes ~20 N-terminal aa that are not essential for CnnT function (see below), and the C-term (CnnT-C) that is unique to CnnT (Figures 1A, 2C, S1B). CnnT-N formed aggregates in the cytoplasm that recruited γ-Tub but did not localize at mitochondria, whereas CnnT-C localized to mitochondria but did not recruit γ-Tub, and high expression of CnnT-C resulted in mitochondrial clustering (Figure 2B). Moreover, superresolution images from 3D-Structured Illumination Microscopy (3D-SIM) revealed that CnnT and CnnT-C reside on the surface of mitochondria (Figure 2D).
We conclude that the N-term of CnnT is necessary and sufficient to recruit γ-TuRCs, whereas the C-term is necessary and sufficient for mitochondrial targeting.
γ-TuRC recruitment by CnnT is independent of core centrosome components
We investigated whether γ-TuRC recruitment by CnnT involves other major centrosomal proteins, some of which, such as CDK5RAP2 (mammalian ortholog of Cnn), Pericentrin and CEP192/SPD-2, are key pericentriolar material (PCM) components and regulators of γ-TuRCs at mammalian centrosomes [20–24]. Aside from NEDD1, which associates with γ-TuRCs, the major PCM proteins CEP192, CEP152/Asl, Pericentrin, CDK5RAP2, and CPAP/Sas-4 localized to centrosomes but were not recruited by CnnT to mitochondria (Figure 2E-I). This indicates that CnnT may recruit γ-TuRCs directly, and independent of centrosome PCM proteins. We also showed that γ-TuRC proteins were recruited to mitochondria by CnnT following MT depolymerization by nocodazole treatment, indicating that γ-TuRC recruitment is not an indirect effect of microtubule association (Figure S1C, D).
CnnT converts mitochondria to MTOCs
Since CnnT recruits γ-TuRCs, which play a fundamental role in MT nucleation, we employed a MT regrowth assay to determine whether CnnT promotes MT assembly and can convert mitochondria to MTOCs. We treated cells with nocodazole to depolymerize MTs, and then recovered the cells in normal culture conditions following nocodazole removal to identify MTOCs as sites of immediate MT regrowth, which in most cells is the centrosome.
After 1 min of recovery from nocodazole treatment, prominent MTs projected from the mitochondria where CnnT localized (Figure 3). In cells expressing CnnT-N, MT nucleation sites formed in association with the CnnT-N aggregates throughout the cytoplasm (Figure 3). In contrast, CnnT-C, while retaining localization at mitochondria, did not nucleate MTs (Figure 3). Therefore, CnnT organizes functional MTOCs on mitochondria, and its N-term is necessary and sufficient for MT nucleation, consistent with its role in γ-TuRC recruitment.
Figure 3. CnnT converts mitochondria to microtubule-organizing centers.
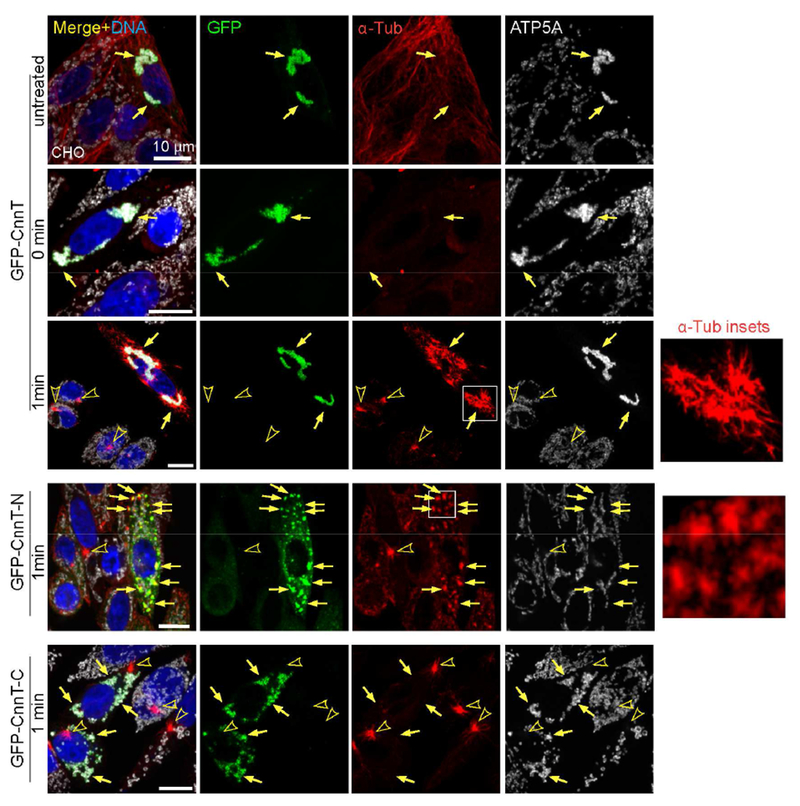
MT regrowth assay. At 1 min of regrowth, prominent MTs grow from mitochondria where CnnT localizes, in addition to growth from centrosomes. MT “asters” are organized from CnnT-N aggregates throughout the cytoplasm. CnnT-C on mitochondria does not nucleate MTs. Insets show zoom-in of MTs growing from nucleation sites. Arrows point to GFP-tagged proteins and arrowheads indicate centrosomes. ATP5A labels mitochondria, α-Tub marks MTs. Scale bars: 10 μm. See also Figures S1.
The CM1 domain is necessary and sufficient for MT nucleation from CnnT-organized mitochondrial MTOCs
The first 20 aa of CnnT, which are not included in CnnC, are dispensable for MT nucleation and mitochondrial localization of CnnT (Figures 4A, B). CnnC and CnnT both contain the CM1 domain that is important for MT nucleation [13, 20, 25–27]. To investigate CM1 function in CnnT, we generated CnnTΔ1, which harbors the Δ1 mutation that deletes 13 conserved amino acids in CM1 [13](Figures 4A, S2A). GFP-CnnTΔ1 localized to mitochondria but failed to recruit γ-Tub and to convert them to MTOCs (Figures 4B, S2E). GFP-CnnTΔ1 could not support MT regrowth even at longer time points (Figure S2B). We next examined the MTOC function of various deletions within CnnT, and with the homologous CM1 sequence from mouse CDK5RAP2 (MusCM1) (Figures 4A). These constructs contained or were fused with the CnnT mitochondrial targeting domain, which was sufficient to localize them to mitochondria (Figure S2C). We performed MT regrowth assays to determine whether these fusion proteins could convert mitochondria to MTOCs. When targeted to mitochondria, the 84 aa CM1 domain of CnnT, the MusCM1, and the CnnT N-term deletion fragments that include intact CM1 were sufficient to recruit γ-Tub and MT plus-end protein EB1 and convert mitochondria to MTOCs (Figures 4A, B, S2A, D, E). Fragments that disrupt CM1 were deficient in MTOC activity (Figures 4A, B, S2A, D, E). In Co-IP experiments, CnnT-N, CM1 and MusCM1 strongly associated with γ-Tub, whereas CnnT-N with the Δ1 mutation (CnnT-NΔ1) did not (Figure 4C). These results are consistent with previous findings that CM1 is essential for activating γ-TuRCs to nucleate MTs [20, 27, 28].
Figure 4. CM1 domain is necessary and sufficient to generate MTOCs on mitochondria.
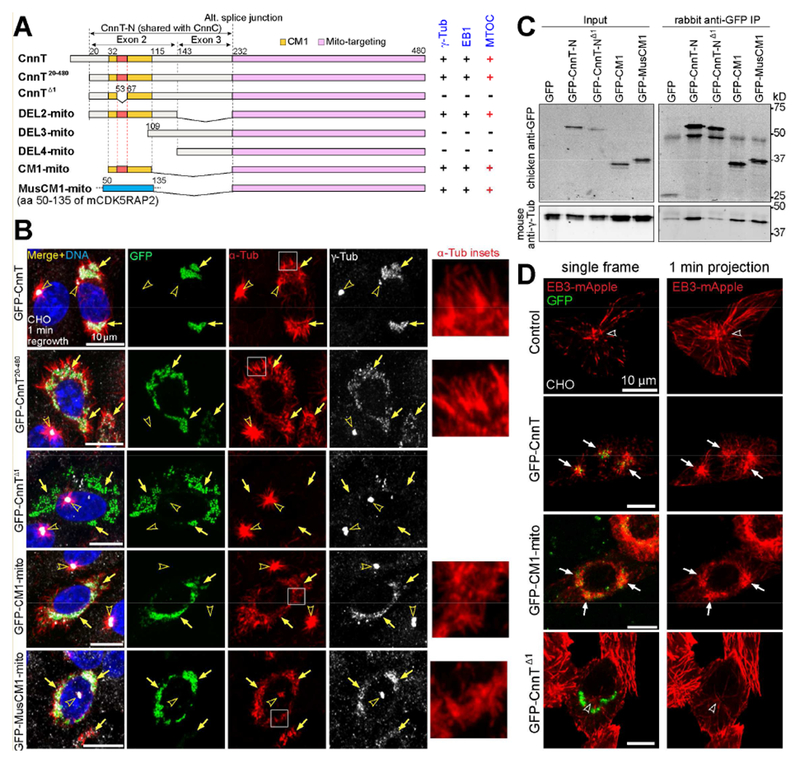
(A) Schematic of deletion constructs of CnnT and MusCM1 (mouse CDK5RAP2 CM1 domain). In some of the constructs, the mitochondrial-targeting domain is referred to as “mito”. A summary of γ-Tub and EB1 recruitment (see Figure S2E) and MTOC activity for each construct is included.
(B) γ-Tub localization and MT nucleation after 1 min of MT regrowth in cells expressing indicated constructs. All proteins are targeted to mitochondria through “mito” domain. Fusion proteins with intact CM1 from CnnT or MusCM1, but not a CM1 mutant (CnnTΔ1), recruit γ-Tub and convert mitochondria to MTOCs. Insets show zoom-in of MTs growing from nucleation sites at MTOCs on mitochondria.
(C) Western blot of Co-IP showing that CnnT-N, CM1, and MusCM1, but not CnnT-NΔ1, associate with γ-Tub.
(D) Plus-end MT assembly from MTOCs at CnnT or CM1-converted mitochondria. Control shows EB3-mApple comets emanating from the centrosome. GFP-CnnT and GFP-CM1-mito, but not GFP-CnnTΔ1, serve as sites for EB3 ejection from the mitochondria. Shown are single still frames and 1 min (15 frames) projections. See Movies S1–4.
Arrows point to GFP-tagged proteins and arrowheads indicate centrosomes. Scale bars: 10 μm. See also Figure S2.
MTs grow plus-end-outward from CnnT-organized mitochondrial MTOCs
We examined whether the MTs assembled at the mitochondrial MTOCs organized by CnnT grow with their plus ends outward, as centrosomes do, or with their minus ends pushed outward as kinetochores do. To detect MT plus-end growth, we co-expressed the MT plus-end tracking protein EB3 tagged with mApple fluorescent protein (EB3-mApple). In control cells, the centrosome was the major MTOC that ejected EB3 comets (Figure 4D; Movie S1). In CnnT-expressing cells, EB3 shot outwards from mitochondrial MTOCs (Figure 4D; Movie S2). In cells expressing GFP-CM1-mito, EB3 comets likewise emanated from mitochondria (Figure 4D; Movie S3). In contrast, cells expressing GFP-CnnTΔ1 did not produce EB3 comets from mitochondria (Figure 4D; Movie S4), further demonstrating the essential role of CM1 in MT assembly.
CnnT forms speckles that recruit γ-Tub to giant mitochondria in spermatids
To study the in vivo function of CnnT, we created transgenic flies carrying B2T-EGFP-FLAG-CnnT (hereafter B2T-GFP-CnnT), expressing GFP-CnnT under the control of the testis-specific β2 tubulin (B2T) promoter.
Immunostaining of B2T-GFP-CnnT adult testes showed that GFP-CnnT formed speckles on the surface of the spermatid nebenkern (Figures 5A, B). Nocodazole treatment before immunostaining revealed γ-Tub foci that colocalized with GFP-CnnT speckles on the nebenkern, in addition to residing at centrioles (Figure 5A). In wild-type (w1118) testes, endogenous CnnT localized at the nebenkern in early elongating spermatids, whereas endogenous CnnC localization at centrosomes was reduced after the spermatocyte stage and decreased to low levels in elongating spermatids (Figure S3A). Moreover, the anti-Cnn(C+T) antibody recognized both CnnC at the centrosome and CnnT on the nebenkern, but the anti-CnnC antibody only detected CnnC at centrosomes (Figures 1A, S3B).
Figure 5. CnnT forms speckles that recruit γ-Tub to spermatid nebenkerns.
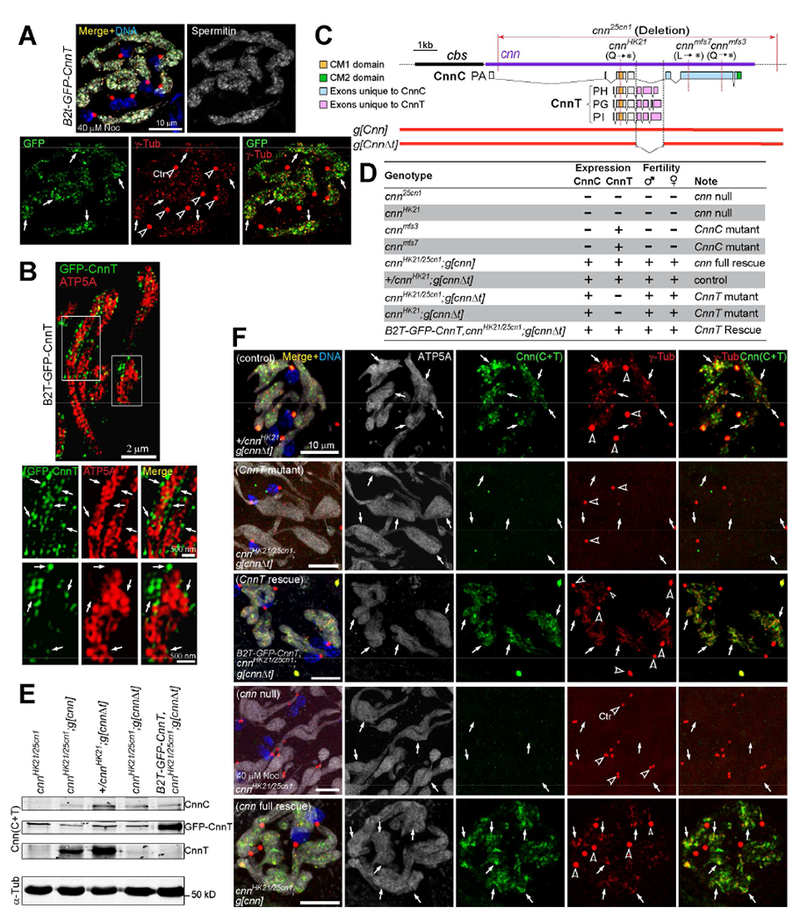
(A) GFP-CnnT forms speckles that colocalize with γ-Tub foci on giant mitochondria (nebenkern) in elongating spermatids. Spermitin labels nebenkerns.
(B) 3D-SIM superresolution images showing the majority of GFP-CnnT resides at the mitochondrial surface. ATP5A labels the inner mitochondrial membrane.
(C) Schematic of genomic constructs g[cnn] and g[cnnΔt]. g[cnnΔt] disrupts CnnT but leaves CnnC intact.
(D) Summary of CnnC and CnnT expression in testes and the adult fertility of indicated genotypes.
(E) Western blot of testes. See (D) for a summary of proteins expressed with each genotype. See Figure S3C for whole blot.
(F) CnnT is required for γ-Tub foci formation on the nebenkern in elongating spermatids. Endogenous CnnT is detected with the anti-Cnn(C+T) antibody. CnnT mutant abolishes the γ-Tub foci on nebenkerns, and the rescue construct restores it.
Panels A and F: arrows point to nebenkern locations and arrowheads indicate centrosomes (Ctr). Testes were treated with nocodazole before fixation. Scale bars as indicated.See also Figure S3.
CnnT mutant males show defects in MT organization and spermiogenesis
The transgenic construct g[cnn] carries the cnn locus on 15.3 kb of genomic DNA and rescues cnn mutants [14] (Figures 5C-E). We engineered g[cnn] to create g[cnnΔt] that disrupts CnnT but leaves CnnC intact (Figure 5C). We then introduced g[cnnΔt] into cnn null backgrounds to generate a mutant that specifically knocks out CnnT (Figures 5C-E). Western blot confirmed that the CnnT mutant expressed CnnC but not CnnT (Figures 5E, S3C). cnn nulls cnn25cn1 and cnnHK21, and CnnC mutants cnnmfs3 and cnn mfs7 are all male sterile and maternal effect lethal due to centrosome defects [12, 13, 17], which can be rescued with g[cnn] or CnnC transgenes [13, 14]. However, CnnT males and females were fertile (Figure 5D).
The CnnT mutant eliminated the γ-Tub foci on the nebenkern, while CnnC mutants retained them as expected (Figures 5F, S3D). Expression of GFP-CnnT restored γ-Tub localization to nebenkerns in CnnT (Figure 5F).
We investigated if CnnT foci on the nebenkern are required for MT organization in spermatids. In elongating spermatids, MT bundles were present in the cytoplasm and on nebenkerns, and CnnT foci were associated with the newly-formed MTs on nebenkerns following MT regrowth (Figure S4A). We next analyzed the MT plus-end binding protein EB1, which accumulates at functional MTOCs and at MT plus ends. CnnT and cnn null mutants showed dramatically decreased number of EB1-GFP comets/foci on nebenkerns after 1 min MT regrowth, additionally, they displayed significantly reduced MTs on nebenkerns; however, CnnC mutants did not show obvious difference from the control (Figures 6A, B). Therefore, CnnT but not CnnC is required for MT organization at nebenkerns in elongating spermatids, in accordance with organization of MTs on mitochondria by CnnT in cultured cells.
Figure 6. CnnT mutants show defects in MT organization and spermiogenesis.
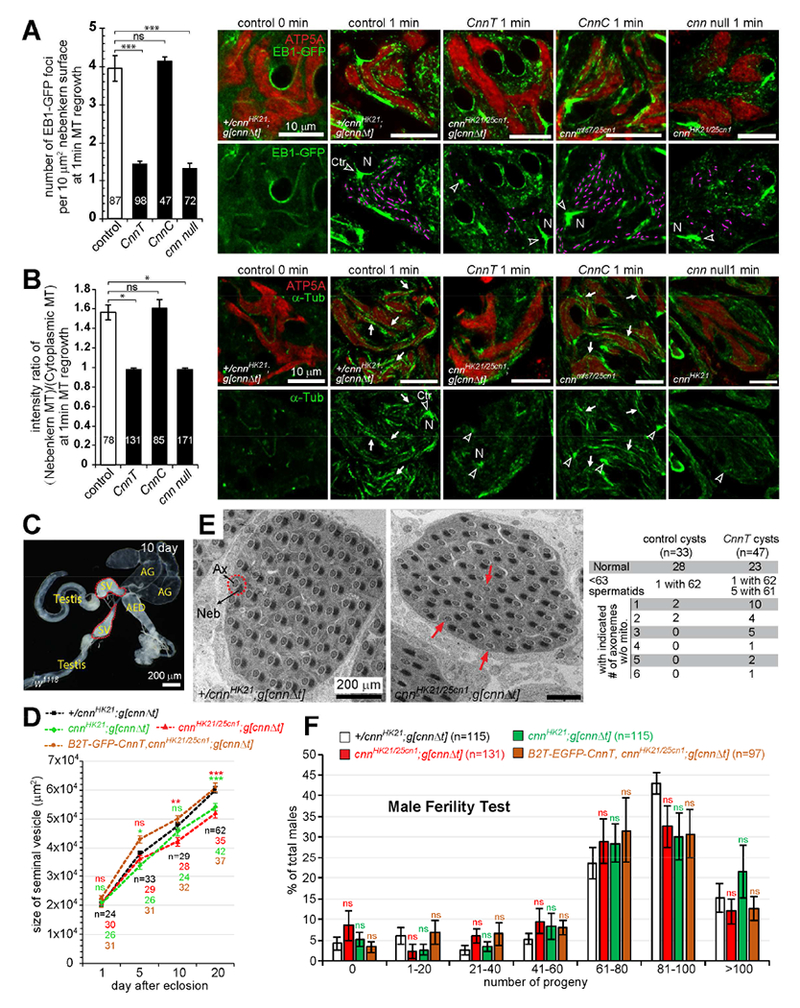
(A and B) Quantification of EB1-GFP foci (A) and MT intensity (B) on nebenkerns in elongating spermatids following 1 min MT regrowth. In control, EB1 accumulates at centrosomes (arrowheads), the nuclear (N) periphery, and as scattered foci on the nebenkern, and newly-formed MTs are associated with these locations. EB1 comets are highlighted with purple lines and arrows point to MTs associated with nebenkerns. CnnT and cnn null, but not CnnC mutants, showed significantly fewer EB1 foci and decreased MT signal on the nebenkern. Cytoplasmic MTs appear normal in all mutants. Numbers of nebenkerns assayed are indicated. At least 3 independent regrowth experiments were performed for each genotype. Error bars represent mean±SEM.
(C) The male reproductive system of a 10-day-old w1118 fly. AED, anterior ejaculatory duct; AG, accessory gland; SV, seminal vesicle.
(D) The size of the SV at indicated age. SV size was quantified as the area of the SV in the images (Figure S4C). Numbers of SVs measured are indicated, error bars represent mean±SE. Significance was measured between the control (+/cnnHK21;g[cnnΔt]) and CnnT mutants.
(E) TEM showing cross sections of late stage cysts. Control cysts normally have 64 spermatids, and thus 64 axoneme (Ax)-nebenkern (Neb) pairs (arrows). CnnT mutant cysts sometimes have fewer spermatids per cyst and spermatids often lack a nebenkern (red arrows). Cysts with 63-64 Ax-Neb pairs were classified as normal.
(F) CnnT males have overtly normal fertility. Shown is the percent of males that produced the indicated number of progeny. Numbers of flies tested are indicated, error bars represent mean±SEM.
Unpaired two-tailed Student’s test was used to determine the significance: ns, P > 0.05; *, P ≥ 0.05; **, P ≥ 0.01; ***, P ≥ 0.001. Scale bars as indicated. See also Figure S4.
In Drosophila, mature sperm are stored in the seminal vesicle (SV) (Figure 6C). After eclosion, the SV volume increases with age due to increased sperm production. cnn null and CnnC mutants had dramatically smaller SVs compared to the control (Figure S4B), because they have impaired meiosis and do not produce motile sperm [14]. CnnT mutants also had smaller SVs at 20 days post eclosion (Figures 6D, S4C), indicating that CnnT males may have reduced sperm production.
During spermatogenesis, cysts of 16 spermatocytes undergo two meiotic divisions to produce 64 haploid spermatids that remain joined in register as they elongate and individualize [11]. Transmission electron microscopy (TEM) showed that the CnnT mutant has higher incidence of containing fewer spermatids per cyst and, frequently, cysts contained one or more axonemes that were not accompanied with a mitochondrial derivative (Figure 6E). However, CnnT retained normal fertility (Figure 6F), and produced mature sperm with tail lengths comparable to the control (Figure S4D).
Ectopic expression of CnnT causes severe developmental defects
Since CnnT is expressed exclusively in the testes, we examined if ectopic CnnT expression impacts fly development. We generated transgenic flies carrying UAS-CnnT fusion proteins under the control of the UAS promoter and which can be manipulated by tissue-specific GAL4 “drivers” (UAS-GAL4 system, [29]). Ectopically expressed GFP-CnnT associated with mitochondria in brain and fat body cells in 3rd instar larvae (Figure S5A, B). When expressed ubiquitously (by tubP-GAL4), either CnnT or its N-term (CnnT-N), but not its mitochondrial targeting region CnnT-C, caused severe developmental impairments, including small and rough eyes, loss of bristles on the notum and abdomen, and defective wings (Figure 7A). Phenotypes from CnnT-N overexpression were more severe, and flies usually died soon after eclosion. However, when CnnT-N was targeted to the centriole with a PACT (pericentrin-AKAP-450 centrosomal targeting, [30]) domain, its overexpression caused no obvious defects (Figure 7A). Since both CnnT and CnnT-N have MTOC activity, it is likely that they compete with centrosomes for γ-TuRCs and redirect the MT organization in the cell, thus disrupting normal cell divisions and/or impairing cell physiology, leading to these defects.
Figure 7. Ectopic expression of CnnT causes severe developmental defects in flies; a model for divergent roles of Cnn variants.
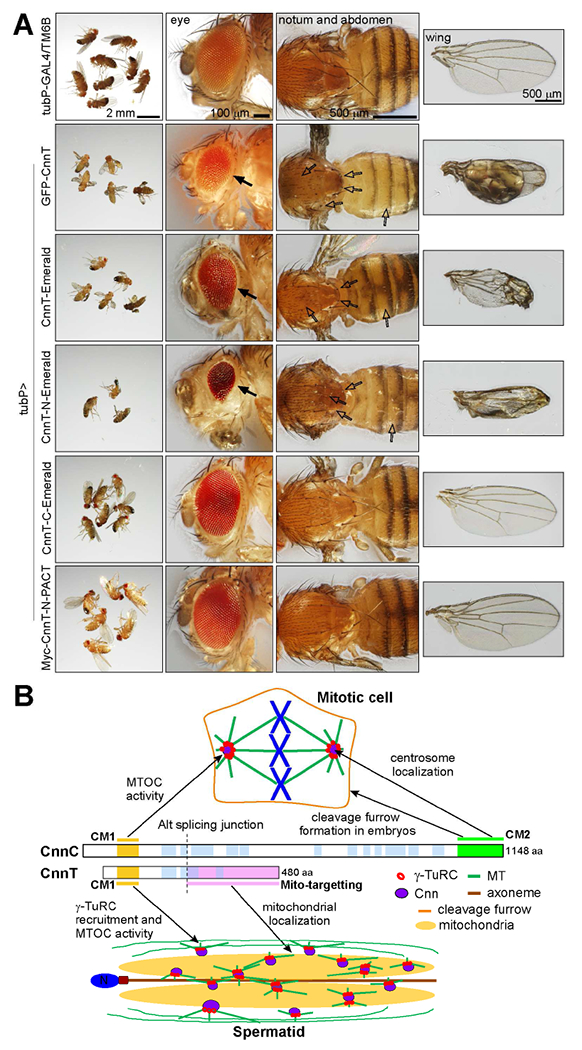
(A) Ectopic ubiquitous expression of indicated transgenes. Proteins were N-term tagged with GFP or 6xMyc or C-term tagged with Emerald. Overexpression of CnnT and CnnT-N, but not CnnT-C results in small and rough eyes (solid arrows), loss of bristles on the notum and abdomen (empty arrows), and defective wings. When CnnT-N is targeted to the centriole with a PACT domain (Myc-CnnT-N-PACT), its overexpression does not cause obvious defects. tubP-GAL4/TM6B is a control. Scale bars as indicated.
(B) A model for divergent roles of the two major Cnn variants. CnnC targets to centrosomes through the CM2 domain and is required for centrosome function. CnnT localizes to the giant mitochondria via its mito-targeting domain in elongating spermatids, where it recruits γ-TuRCs and nucleates MTs to promote spermiogenesis. See also Figure S5.
Discussion
In this study we show that CnnT is a testis-specific variant encoded by Drosophila cnn through alternative splicing. Unlike its centrosomal namesake, CnnT localizes to mitochondria, recruits γ-TuRCs, and converts mitochondria to MTOCs. The CM1 domain, which is shared with CnnC, is necessary and sufficient for MT nucleation; while the C-term of CnnT is necessary and sufficient for mitochondrial targeting. We discern divergent roles for Cnn variants in vivo: CnnC mainly localizes and promotes MTOC activity at centrosomes [12–14], and directs cleavage furrow organization during embryogenesis through its CM2 domain [18]; CnnT, on the other hand, localizes to the specialized mitochondria (nebenkern) in spermatids, where it recruits γ-TuRCs to organize MTs to promote spermiogenesis (Figure 7B). We show that CnnT is a bipartite organizer of unique MTOCs on mitochondria.
MT nucleation through γ-TuRCs
Because spontaneous nucleation of MTs is kinetically limited, γ-TuRCs or the minimal γ-Tub small complexes (γ-TuSCs) are essential to carry out robust MT nucleation in vivo, and they are involved at all MTOCs in eukaryotic cells. The majority (> 80%) of γ-Tub complexes are cytoplasmic, and not localized to centrosomes [31], yet centrosomes are the major sites of MT nucleation, indicating that MTOCs provide an activator for γ-TuRCs to polymerize MTs efficiently. Our study revealed that CM1 in Cnn is essential for activating γ-TuRC to initiate MT nucleation. Consistently, CM1 of other Cnn orthologs were shown to activate γ-TuRCs [20, 27, 32].
As the best-characterized MTOC, the centrosome is a supramolecular complex where the centrioles serve as a scaffold upon which sequential assembly of proteins such as CEP152/Asl, CPAP/Sas-4, Pericentrin/Plp, CEP192/Spd-2, and CDK5RAP2/Cnn are required for pericentriolar material (PCM) assembly and optimal MTOC activity [33, 34]. It is therefore remarkable that CnnT, with its simple composition of the CM1-containing γ-TuRC recruitment/binding domain fused to a mitochondrial targeting domain, is capable of organizing robust MTOCs at the mitochondrial surface without involvement of the key PCM proteins. The small protein MOZART (MZT) [35, 36] is a direct partner of CM1 [27] that also associates with GCP3 [37] and other GCP proteins [38], and cooperates with GCP6 [39]. GCP3, in particular, may directly control conformational changes of the γ-TuRC and thus regulate its MT nucleation activity [40, 41], and MZT1 appears to be a key link [27, 38]. The role of NEDD1, which was also recruited to CnnT MTOCs, is less clear. NEDD1 appears to be a γ-TuRC targeting factor [42, 43], associates with γ-TuRCs independently of CM1 in solution, and appears to anchor the γ-TuRC rather than activate it [28]. MZT1 is associated with both NEDD1-containing and CM1-containing γ-TuRCs [38].
Proteins with CM1 also have MTOC-targeting domains such that the γ-Tub complex (γ-TuC) is oligomerized at the site of action [44]. In S. pombe, a shortened “bonsai” version of the Cnn homolog Mto1p, that contains the CM1 domain and a binding site for Mto2p, allows for binding of γ-TuCs and oligomerization, enabling it to activate the γ-TuC without associating with an MTOC [45]. CM1 of human CDK5RAP2, however, is sufficient for γ-TuRC recruitment and MTOC activity [20]. Here we show that CM1 of Cnn is sufficient to recruit γ-TuRCs when tethered at mitochondria, and in solution associates with γ-Tub. Altogether, our findings indicate that, if appropriately targeted, CM1 is sufficient to generate an MTOC through the recruitment and activation of γ-TuRCs.
Cnn, Pericentrin, Spd-2, Ninein and their closely related proteins have been implicated to directly promote γ-TuRC-mediated MT nucleation. Among them, Cnn-related/CM1-containing proteins, including Drosophila Cnn, mammalian CDK5RAP2 and Myomegalin, fission yeast Mto1 and Pcp1, etc., appear to be the most potent stimulators for MT nucleation by γ-TuRCs, as shown by in vitro biochemical assays and/or by redirecting the protein (or protein fragments) to ectopic cellular locations to induce MT assembly at those sites (this study, and [20, 28, 32, 45]). Our data further support that Cnn (and orthologs) directly binds and stimulates γ-TuRCs to mediate MT assembly. With its simple bipartite molecular composition, CnnT targets to mitochondria, recruits and activates γ-TuRCs, and converts mitochondria to MTOCs.
Organizers and regulators of non-centrosomal MTOCs
Centrosomes and non-centrosomal MTOCs rely on γ-TuRCs for MT nucleation. Besides γ-TuRCs, the fundamental components in non-centrosomal MTOCs vary, but most of them are centrosome PCM proteins: for example, Pericentrin and CEP192 in mouse oocyte cytoplasmic MTOCs [1, 46]; CDK5RAP2/Cnn, Myomegalin, or Pericentrin/Plp at Golgi MTOCs; and Ninein, PCM-1, CDK5RAP2 and Pericentrin at the nuclear periphery MTOCs. How these proteins organize non-centrosomal MTOCs and regulate MT assembly at those sites remain largely unknown.
We show that Drosophila CnnT, a single small bipartite molecule consisting of a targeting domain and a microtubule nucleation-activating domain, can convert mitochondria to MTOCs. This is unique because CnnT does not regulate MT assembly at centrosomes, unlike the PCM proteins mentioned above that function both at centrosomes and non-centrosomal MTOCs. A previous report indicated that CnnT, described as the “short form” of Cnn, localized to centrosomes [47], however we were unable to detect CnnT at centrosomes. Furthermore, CnnT, or just the CM1 domain, is capable of interacting with γ-TuRC to drive MT nucleation in mammalian cells, and this function is independent of mammalian PCM proteins CDK5RAP2 and Pericentrin, indicating that CnnT (and CM1) has intrinsic and evolutionarily conserved capability of activating γ-TuRC-mediated MT assembly. These findings reveal significant insights in understanding how non-centrosomal MTOCs can be generated.
Mitochondrial MTOCs in Drosophila spermatids
Non-centrosomal MTOCs are assembled on the giant mitochondria in Drosophila spermatids [7], and here we show that CnnT is the major organizer of these mitochondrial MTOCs. Microtubules are essential for spermatid elongation [7]. Our work shows that loss of most of the MTOC activity on the nebenkern can be sufficiently compensated for by MT nucleation elsewhere in the cytoplasm. The mechanism for CnnT targeting to mitochondria is conserved, because it localizes to mitochondria across species. Since there are no obvious mitochondrial-targeting signals in CnnT, it is likely that CnnT is recruited to the mitochondrial surface by conserved mitochondrial protein(s). The mitochondrial-targeting domain of CnnT has no apparent homologs outside of Drosophilids. We also did not find a CDK5RAP2 or Myomegalin mRNA variant that contains CM1 but lacks the CM2 centrosome-localization domain among available genome data. Thus it is not clear if CnnT is unique to Drosophilids, or if mitochondrial MTOCs exist in other species.
Drosophila species have extremely long sperm with the current record being ~6 cm in D. bifurca [8], and they have evolved special cellular structures, including the nebenkern that stretch along the entire spermatid to support the dramatic morphological changes during sperm elongation. Non-centrosomal MTs were shown to be essential for spermatid morphogenesis [7]. However, our results indicate that diminished MT assembly on the nebenkern surface in the CnnT mutant does not greatly impede sperm morphogenesis; it is likely that other MT regulators may function in organizing cytoplasmic MTs, which may compensate for the loss of MTs on the nebenkern surface when CnnT is absent.
Why did Drosophila evolve such long sperm? It was suggested that postcopulatory sexual selection favors increased investment in sperm size rather than sperm number because fertilization is facilitated in the fly uterus where the egg is placed in juxtaposition to sperm housed in the seminal receptacle; and since female flies have a long seminal receptacle with length comparable to the sperm, it appears that longer sperm have advantages in passing through the reproductive tract to fertilize the egg [8, 48–50]. Thus, flies may have evolved CnnT to generate mitochondrial MTOCs to cope with the demands of morphogenesis of extremely long sperm.
Star Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Timothy Megraw (timothy.megraw@med.fsu.edu).
Experimental Model and Subject Details
Drosophila melanogaster strains
All fly strains were maintained at 25°C on standard fly food made from corn flour and yeast. B2t-GFP-CnnT was generated by cloning EGFP-FLAG-CnnT (isoform PH) into a male germ line expression vector (gift from Elizabeth Raff, Indiana University; [51]) under the control of B2t promoter [52]. UASt-GFP-CnnT was created by cloning EGFP-FLAG-CnnT into the insect expression vector pUAST [29] under the control of a UASt promoter. g[cnnΔt] has an 1187 bp deletion that covers all CnnT-specific exons, it was generated from g[cnn] [12, 14] by overlap extension PCR and “cut and paste” ligation processes utilizing the unique SrfI and SphI sites that flank the intron where all the alternatively spliced exons reside. B2t-GFP-CnnT, UASt-GFP-CnnT and g[cnnΔt] transgenic flies were created through standard germ line transformation methods and transgenes inserted onto the 3rd chromosome were recovered. UASp-CnnT-Emerald, UASp-CnnT-N-Emerald, UASp-CnnT-C-Emerald, UASp-Myc-CnnT-N-PACT transgenic flies were made by GenetiVision Inc. (Houston, TX) via PhiC31-mediated chromosome integration on the 3rd chromosome, with VK31:(3L)62E1 as the docking site for UASp-CnnT-Emerald, and VK20:(3R)99F8 for UASp-CnnT-N-Emerald, UASp-CnnT-C-Emerald, and UASp-Myc-CnnT-N-PACT. UAS transgene expression was undetectable in elongating spermatids when driven by various ubiquitous or testis-specific GAL4 drivers, though it could be detected in spermatogonia or other tissues. cnnhk21, cnn25cn1, cnnmfs3 and cnnmfs7 were described in [12–14]. Other strains include tubP-GAL4LL7 (chromosome 3; BDSC #5138) and hsp83-EB1-EGFP (chromosome 3; [7]).
Larval brains and fat bodies for RT-PCR and immunochemistry were dissected from mixed gender 3rd instar larvae. Testes for RT-PCR, western blotting and immunochemistry were dissected from newly eclosed male flies. For analysis of male fertility and the size of seminal vesicles, male flies aged 1-20 day post eclosion were used, with specific age reported in corresponding figures and figure legends. For electron microscopy, testes from male flies 3-5 days post eclosion were used.
Cell Culture
Drosophila melanogaster Kc167 cells (female, [53] FlyBase: FBtc0000001) were maintained in Hyclone CCM3 medium (Thermo Fisher Scientific) supplemented with 5% fetal bovine serum and Penicillin-Streptomycin (100 IU/mL Penicillin and 100 ug/mL Streptomycin) at room temperature. Cells were cotransfected with pUAS plasmid (pUASp-GFP-CnnT or pUASp-CnnT-Emerald) and pMT-GAL4 using lipofectamine 2000 and the protein expression was induced with 1 mM CuSO4 20–24 h later. Cells were prepared for immunostaining 20–24 h after induction.
Hamster CHO (female) cells, human HK293 (female), U2OS (female) and A549 (male) cells, Cercopithecus aethiops Vero (female) Cells, and Potorous tridactylus PtK1 (female) cells were maintained in DMEM (Dulbecco’s Modified Eagle Medium) (Cellgro) supplemented with 10% fetal bovine serum and Penicillin-Streptomycin (100 IU/mL Penicillin and 100 ug/mL Streptomycin). Cells were incubated at 37°C with 5% CO2. For transient protein expression, cells were transfected with plasmids using Lipofectamine 2000 and prepared for immunostaining 20–24 h after transfection. Cell lines were not authenticated; however, there were no overt differences among the cell lines regarding CnnT localization to mitochondria and their conversion to MTOCs, so authentication was not critical for this study.
Method Details
Accession numbers
Cnn-PG, ACZ94414.1; Cnn-PA, AAF58375.1; mouse CDK5RAP2, NP_666102.2
Molecular cloning
CnnT (full length PG, aa 1-480), CnnT20-480 (aa 20-480), CnnT-N (aa 20-231), CnnT-C (aa 232480), CM1 (aa 32-115), DEL2 (aa 20-143), DEL3-mito (aa 109-480), DEL4-mito (aa 143-480) were PCR-amplified from the Cnn-PG cDNA clone AT09084 (FBcl0015367). CnnT-NΔ1 (aa 20-480 with a deletion of aa 54-66) was amplified from the previously described construct EGFP-CnnΔ1 [13], MusCM1 was amplified from previously described construct pcDNA-GFP-mCDK5RAP2 [54]. CnnTΔ1, CM1-mito, DEL2-mito and MusCM1-mito were amplified by joining CnnT-NΔ1, CM1, DEL2 and MusCM1, respectively, to CnnT-C, using overlap extension PCR. EB3-mApple was human EB3 tagged with mApple at its C-terminus and it was amplified from plasmid mApple-EB3-7. DNA fragments were cloned into Gateway vector pENTR-D/TOPO (Invitrogen).
For expression in mammalian culture cells, DNA sequences were subcloned through Gateway cloning into pcDNA-DEST53 to generate the N-terminal GFP fusions. EB3-mApple was subcloned into pQCXP CMV/TO DEST to express in mammalian cell lines.
For expression in Drosophila, pUASp-GFP-CnnT, pUASp-CnnT-Emerald, pUASp-CnnT-N-Emerald, pUASp-CnnT-CM-Emerald and pUASp-Myc-CnnT-N-PACT were created through Gateway cloning into pPGW-attB, pPWEmerald-attB or pPMW-attB vectors. pPGW-attB and pPMW-attB generate an EGFP tag and a 6xMyc tag on the N terminus of the target protein respectively, and pPWEmerald-attB generates an Emerald tag on the C terminus of the target protein. pPGW-attB and pPMW-attB were engineered by cloning an attB sequence into pPGW and pPMW vectors respectively [55].
To construct pPWEmerald-attB, we first converted the unique SacII site of pPWH to a pair of SpeI and AscI sites by ligating in a double-stranded linker ( see Table S1). This generated a vector with this sequence and reading frame at the cloning site: ATG/GAT/CTC/CAC/CGC/ACT/AGT/ATA/TGG/CGC/GCC/AGC/GGT/GGA/GGC/CGC/ATC/TTT/TACCCATACGATGTTCCTGACTATGCGGGCTATCCCTATGACGTCCCGGACTATGCAGGATCCTATCCATATGACGTTCCAGATTACGCTGCTCATGGCGGA (The SpeI and AscI sites are underlined, the codon triplets are separated, and the three HA sequences are italicized). Next, we ligated a 368 bp fragment containing the attB sequence at the unique NsiI site in pPWH as described previously for other vectors [55]. Using this new vector, we ligated a PCR fragment that includes Emerald sequences flanked by an AscI site on the 5’ end, a SpeI site on the 3’ end, and a flexible protein linker at the 5’ end that encodes (Gly-Gly-Ser)×4 (PCR primers to amplify mEmerald listed in Table S1). The HA epitope originally found in pPWH is not expressed from pPWEmerald-attB.
pPGW, pPWH and pPMW were from Terence Murphy’s Drosophila Gateway Vector Collection at Carnegie Institution of Washington.
All primers for cloning are listed in Table S1.
Immunohistochemistry and Microscopy
For the staining of mammalian cells, cells were grown on 18 mm round coverslips, then fixed in −20°C methanol for 10min. After a few rinses in PBS, samples were incubated with antibodies. For nocodazole treatment, cells were incubated with 10 μM nocodazole in culture media for 2 h prior to fixation. For the staining of Drosophila Kc167 cells, cells were prepared according to the method described in [56]. Cells were incubated on poly-L-lysine-treated slides for 30 min, the slide was then rinsed briefly in PBS and then fixed in −20°C methanol for 10 min.
For the staining of larval brains, larval fat bodies and adult testes, tissues were dissected in Dulbecco’s PBS (DPBS), then transferred to a 4-μ1 drop of DPBS on a slide and then covered with a siliconized coverslip containing a 1 μL droplet of 18.5% formaldehyde in DPBS. After the tissue was allowed to flatten for 20–30 s under the weight of the coverslip, the slide was snap-frozen by plunging into liquid nitrogen. The slide was removed from liquid nitrogen and the coverslip was flipped off using a razor blade and then immersed into −20°C methanol and incubated for 10 min. The slides were then transferred to PBS. A Super PAP Pen (Immunotech) was used to draw a hydrophobic ring around the tissue to contain 50 uL antibody staining solutions. For nocodazole treatment of testes, dissected testes were broken apart and incubated with 40μM of Nocodazole in Shields and Sang M3 Insect Medium (Sigma) +10% FBS+ Penicillin-Streptomycin for 2hr. Afterwards, samples were prepared and fixed as stated above.
After fixation in methanol, cells or tissues were rinsed with PBS and then stained with antibodies in PBS solution containing 5 mg/mL bovine serum albumin and 0.1% saponin.
Guinea pig anti-Cnn(C+T) and rabbit anti-Cnn(C+T) were raised against aa 1-574 of Cnn PA [13], rabbit anti-CnnC was raised against last two 3’-most exons of CnnC [14]. Other antibodies used in this study are listed in the key resources table. Secondary goat antibodies conjugated to Alexa 488, 568, and 647 (1:1,000; Life Technologies) were used. DNA was stained with DAPI (1 μg/mL).
For confocal imaging, a Nikon A1 confocal microscope with a 60×/NA 1.49 oil immersion objective and NIS-Elements software was used. For superresolution imaging using 3D-structured illumination microscopy, DeltaVision OMX Blaze (GE Healthcare) with an Olympus 60×/NA1.42 oil immersion objective was used and images were processed with SoftWorx software.
Microtubule regrowth assay
For the regrowth in CHO cells, cells growing on coverslips were treated with 10 μM nocodazole in culture media for 2 h. After removal of nocodazole and then 5 quick rinses with ice-cold media, cells were fixed directly (as 0 min), or transferred immediately to 37°C media for the desired length of time and then fixed in −20°C methanol for 10 min and prepared for staining.
For MT regrowth in fly testes, dissected testes in M3 Insect Medium +10% FBS+ PenStrep and were placed in 0°C for 1 h. Samples were flattened and fixed directly (as 0 min) or transferred immediately to 23°C media for a desired length of time and then flattened and fixed in −20°C methanol for 10 min and prepared for staining.
Live cell imaging
For live imaging of EB3 comets in CHO cells. Cells growing in a 35 mm MatTek glass bottom petri dish were co-transfected with GFP fusions and EB3-mApple. 20-24 hrs after transfection, the time-lapse movies were captured by Nikon A1 confocal microscopy; frames were captured every 4 s for 4 min. More than five cells were imaged for each transfection and representative movies are shown in this study.
Co-immunoprecipitation and western blots
GFP-tagged proteins were expressed in CHO cells. Cells were treated with 10 μM nocodazole for 2 h before harvest. Cell extracts were incubated with ~1 μg of Rabbit anti-GFP antibodies for 3 h on ice, the mixtures were then incubated with Protein A Sepharose beads (GE Healthcare) for 1 h on ice. Beads were then washed, denatured and prepared for western blotting.
For western blotting, proteins were separated using an SDS-PAGE mini-gel electrophoresis system (Bio-Rad) and transferred to UltraCruz 0.45μm pore-size nitrocellulose membrane (Santa Cruz Biotechnology) using Trans-Blot SD Semi-Dry Transfer system (Bio-Rad). Membranes were probed with primary antibodies: rabbit anti-Cnn(C+T) serum (1:10,000), mouse anti-α-Tubulin (DM1A, 1:20,000), Chicken anti-GFP (1:10,000), and mouse anti-γ-Tubulin (GTU88, 1:10,000). For secondary antibodies, IRDye800CW Goat anti-mouse or antirabbit, IRDye680LT Goat anti-mouse or anti-rabbit, and IRDye800CW Goat anti-chicken antibodies (LI-COR Biosciences) were used (1:25,000). Membranes were scanned with an Odyssey Infrared Imaging System (LI-COR Biosciences). The Co-IP experiment was repeated three times.
RT-PCR
Total RNA from dissected tissues was isolated using TRIzol Reagent. cDNAs were then generated using SuperScript First-Strand Synthesis System (Invitrogen). 40 cycles were used in PCR amplification. For CnnT, CnnC, and Cnn “conceptual form“, primers used were illustrated in Figure 1A. For control α-tubulin, primers flanking intron 1 of α-tubulin-RA were used. Primers used are listed in the key resources table. This experiment was repeated at least four times.
Quantification of EB1-GFP comets on the nebenkern following microtubule regrowth
Dissected testes were subjected to 1 min MT regrowth, as described above for the MT regrowth assay, then fixed and stained. Images were taken with a Nikon A1 confocal microscope with a 60×/NA 1.49 oil immersion objective at a slice thickness of 0.45 μm. In a random region (> 20 μm2) of the nebenkern (labeled by ATP5A) that is away from the centrosome, the number of EB1-GFP comets with a length >0.3 μm in the region from each 0.45 μm image slice were counted and summed to reach a total number. The total surface area was approximated as twice the two dimensional area of the measured region. The number of EB1 comets per 10 μm2 of nebenkern surface was calculated as 10n/(2A), where n is the total number of EB1-GFP comets and A is the two dimensional area of the nebenkern. About 10-30 elongating nebenkerns were analyzed from each regrowth experiment, and at least 3 independent regrowth experiments were assayed for each genotype. No data were excluded from the measurements.
Quantification of MTs on the nebenkern following microtubule regrowth
Dissected testes were subjected to 1 min MT regrowth, as described above for the MT regrowth assay, then fixed and stained. Images were taken with a Nikon A1 confocal microscope with a 60×/NA 1.49 oil immersion objective at a slice thickness of 0.5 μm. Confocal stacks (1.0-2.0 μm total thickness) were processed into maximum intensity projection images. In a random region (> 20 μm2) of the nebenkern (labeled by ATP5A) not proximal to the centrosome, the mean MT intensity in the area was measured. Average mean cytoplasmic MT intensity was measured by averaging 5 random boxes (at least 5 μm2) in the spermatid cytoplasm and away from centrosomes and nebenkerns. The intensity ratio of (Nebenkern MT)/(Cytoplasmic MT) was calculated as (mean MT intensity on the nebenkern)/(average mean cytoplasmic MT intensity).
About 15-30 elongating nebenkerns were analyzed from each regrowth experiment, and at least 3 independent regrowth experiments were assayed for each genotype. No data were excluded from the measurements.
Measurements of adult male seminal vesicles
Newly eclosed male flies were collected and separated from females until the designated age. Seminal vesicles were dissected intact in PBS and placed in a drop of PBS on a glass slide. An Olympus MVX10 Macro Zoom Microscope equipped with MVPLAPO 1X/NA 0.25 and MVPLAPO 2XC/NA 0.5 objectives was used to capture images of seminal vesicles, and the size of seminal vesicles was measured as the area of the seminal vesicle in images, using the cellSens software. At least 25 seminal vesicles were measured for each genotype at designated age, standard error (SE) was calculated, and unpaired two-tailed Student’s test was used to determine the significance. No data were excluded from the measurements.
Tail length measurements of mature sperm
Seminal vesicles from males older than 10 days were dissected in media with 1 μg/ml Hoechst 33342 dye and were poked with fine forceps to release mature sperm. Individual sperm was imaged with an Eclipse TE2000-U inverted microscope equipped with a Plan Fluor 10× NA 0.30 phase contrast objective (Nikon), the NIS-Elements software (Nikon), and an ORCA-AG digital camera (Hamamatsu Photonics). The length of the tail was measured using the NIS-Elements software. Approximately 10 sperm were measured from each pair of testes, and at least 5 pairs of testes were assessed for each genotype. No data were excluded from the measurements.
Transmission electron microscopy
Dissected fly testes were fixed in Karnovsky’s fixative (Electron Microscopy Sciences) overnight at 4°C. Samples were postfixed in osmium tetroxide, dehydrated in a graded series of alcohol and propylene oxide, and then embedded in epoxy resin. Sections (~30 nm) of testis samples were cut in a Reichert-Jung Ultracut E Ultramicrotome, collected on copper grids, and stained with uranyl acetate and lead citrate. Samples were imaged with a Philips FEI BioTwin CM120 electron microscope.
Male fertility test
Virgin w1118 females and newly eclosed males were collected and held apart for 3–5 days before mating. In each test, a single male was mated with a single w1118 virgin female for 4 days. After 10 more days, the progeny from each pair mating were counted. For each genotype, at least 5 groups of 15-20 males each were assessed. The whole test was conducted at 25 °C. Neither the males nor the females were involved in previous procedures, and they were naive to the assay prior to its implementation. No data were excluded from the measurements.
Quantification and Statistical Analysis
Statistical analysis was performed using Microsoft Excel 2013. Two-tailed unpaired Student’s t tests were used to determine the significance. For quantification of EB1-GFP comets on the nebenkern following microtubule regrowth (Figure 6A), quantification of MTs on the nebenkern following microtubule regrowth (Figure 6B), measurements of adult male seminal vesicles (Figure 6D), measurements of sperm tail length (Figure S4), and male fertility test (Figure 6F), statistical details including “n”, data representation (SEM or SE) and significance measurements can be found in the figure and figure legends, as well as in their corresponding method details.
Supplementary Material
Movie S1 (related to Figure 4). Live imaging of EB3-mApple (red) in a control CHO cell
The centrosome is the major MTOC that ejects EB3-mApple comets. Movie was captured by time-lapse confocal microscopy (Nikon A1). Frames were captured every 4 s for 4 min. See also Figure 4D.
Movie S2 (related to Figure 4). Live imaging of EB3-mApple (red) in a CHO cell expressing GFP-CnnT (green)
EB3 comets eject outwards from mitochondrial MTOCs assembled by GFP-CnnT. Movie was captured by time-lapse confocal microscopy (Nikon A1). Frames were captured every 4 s for 4 min. See also Figure 4D.
Movie S3 (related to Figure 4). Live imaging of EB3-mApple (red) in a CHO cell expressing GFP-CM1-mito (green)
EB3 comets eject outwards from mitochondrial MTOCs assembled by GFP-CM1-mito. Movie was captured by time-lapse confocal microscopy (Nikon A1). Frames were captured every 4 s for 4 min. See also Figure 4D.
Movie S4 (related to Figure 4). Live imaging of EB3-mApple (red) in a CHO cell expressing GFP-CnnTM (green)
EB3 comets are only emitted from a major MTOC, which is presumably the centrosome. No EB3 comets eject from GFP-CnnTΔ1 foci. Movie was captured by time-lapse confocal microscopy (Nikon A1). Frames were captured every 4 s for 4 min. See also Figure 4D.
Acknowledgements
We thank Shigeo Hayashi for fly strains, Elizabeth Raff and Michael Davidson for plasmids, Andreas Merdes, Pavel Draber, Jens Luders, Brad Shuster and Jay Gopalakrishnan for antibodies, and Bloomington Drosophila Stock Center for fly stocks. We also thank Batory Foods (Atlanta, GA) for generously donating fly food. We thank Georgia Platt and the Biological Science Imaging Resource at FSU for help with electron microscopy. This work was supported by NIH grant GM068756.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schuh M, and Ellenberg J (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130, 484–498. [DOI] [PubMed] [Google Scholar]
- 2.Dumont J, and Desai A (2012). Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends in cell biology 22, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folker ES, and Baylies MK (2013). Nuclear positioning in muscle development and disease. Frontiers in physiology 4, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oddoux S, Zaal KJ, Tate V, Kenea A, Nandkeolyar SA, Reid E, Liu WH, and Ralston E (2013). Microtubules that form the stationary lattice of muscle fibers are dynamic and nucleated at Golgi elements. J Cell Biol 203, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tassin AM, Maro B, and Bornens M (1985). Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol 100, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matis M, Russler-Germain DA, Hu Q, Tomlin CJ, and Axelrod JD (2014). Microtubules provide directional information for core PCP function. eLife 3, e02893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noguchi T, Koizumi M, and Hayashi S (2011). Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr Biol 21, 805–814. [DOI] [PubMed] [Google Scholar]
- 8.Bjork A, and Pitnick S (2006). Intensity of sexual selection along the anisogamy-isogamy continuum. Nature 441, 742–745. [DOI] [PubMed] [Google Scholar]
- 9.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, and Raff JW (2006). Flies without centrioles. Cell 125, 1375–1386. [DOI] [PubMed] [Google Scholar]
- 10.Hoyle HD, and Raff EC (1990). Two Drosophila beta tubulin isoforms are not functionally equivalent. J Cell Biol 111, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabian L, and Brill JA (2012). Drosophila spermiogenesis: Big things come from little packages. Spermatogenesis 2, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Megraw TL, Li K, Kao LR, and Kaufman TC (1999). The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829–2839. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, and Megraw TL (2007). Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Molecular biology of the cell 18, 4037–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li K, Xu EY, Cecil JK, Turner FR, Megraw TL, and Kaufman TC (1998). Drosophila centrosomin protein is required for male meiosis and assembly of the flagellar axoneme. J Cell Biol 141, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerit DA, Jordan HA, Poulton JS, Fagerstrom CJ, Galletta BJ, Peifer M, and Rusan NM (2015). Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J Cell Biol 210, 79–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Megraw TL, Kao LR, and Kaufman TC (2001). Zygotic development without functional mitotic centrosomes. Curr Biol 11, 116–120. [DOI] [PubMed] [Google Scholar]
- 17.Vaizel-Ohayon D, and Schejter ED (1999). Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr Biol 9, 889–898. [DOI] [PubMed] [Google Scholar]
- 18.Kao LR, and Megraw TL (2009). Centrocortin cooperates with centrosomin to organize Drosophila embryonic cleavage furrows. Curr Biol 19, 937–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuer JG, Li K, and Kaufman TC (1995). The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development 121, 3861–3876. [DOI] [PubMed] [Google Scholar]
- 20.Choi YK, Liu P, Sze SK, Dai C, and Qi RZ (2010). CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol 191, 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmerman WC, Sillibourne J, Rosa J, and Doxsey SJ (2004). Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Molecular biology of the cell 15, 3642–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, and Doxsey SJ (1998). Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol 141, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fong KW, Choi YK, Rattner JB, and Qi RZ (2008). CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Molecular biology of the cell 19, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haren L, Stearns T, and Luders J (2009). Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One 4, e5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samejima I, Miller VJ, Groocock LM, and Sawin KE (2008). Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo. J Cell Sci 121, 3971–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin TC, Neuner A, Schlosser YT, Scharf AN, Weber L, and Schiebel E (2014). Cell-cycle dependent phosphorylation of yeast pericentrin regulates gamma-TuSC-mediated microtubule nucleation. eLife 3, e02208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin TC, Neuner A, Flemming D, Liu P, Chinen T, Jakle U, Arkowitz R, and Schiebel E (2016). MOZART1 and gamma-tubulin complex receptors are both required to turn gamma-TuSC into an active microtubule nucleation template. J Cell Biol 215, 823–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muroyama A, Seldin L, and Lechler T (2016). Divergent regulation of functionally distinct gamma-tubulin complexes during differentiation. J Cell Biol 213, 679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand AH, and Perrimon N (1993). Targeted Gene-Expression as a Means of Altering Cell Fates and Generating Dominant Phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Campos M, Basto R, Baker J, Kernan M, and Raff JW (2004). The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol 165, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moudjou M, Bordes N, Paintrand M, and Bornens M (1996). gamma-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci 109 (Pt 4), 875–887. [DOI] [PubMed] [Google Scholar]
- 32.Roubin R, Acquaviva C, Chevrier V, Sedjai F, Zyss D, Birnbaum D, and Rosnet O (2013). Myomegalin is necessary for the formation of centrosomal and Golgi-derived microtubules. Biology open 2, 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conduit PT, Wainman A, and Raff JW (2015). Centrosome function and assembly in animal cells. Nature reviews. Molecular cell biology 16, 611–624. [DOI] [PubMed] [Google Scholar]
- 34.Mennella V, Agard DA, Huang B, and Pelletier L (2014). Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends in cell biology 24, 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. (2010). Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teixido-Travesa N, Villen J, Lacasa C, Bertran MT, Archinti M, Gygi SP, Caelles C, Roig J, and Luders J (2010). The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Molecular biology of the cell 21, 3963–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhani DK, Goult BT, George GM, Rogerson DT, Bitton DA, Miller CJ, Schwabe JW, and Tanaka K (2013). Mzt1/Tam4, a fission yeast MOZART1 homologue, is an essential component of the gamma-tubulin complex and directly interacts with GCP3(Alp6). Molecular biology of the cell 24, 3337–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cota RR, Teixido-Travesa N, Ezquerra A, Eibes S, Lacasa C, Roig J, and Luders J (2017). MZT1 regulates microtubule nucleation by linking gammaTuRC assembly to adapter-mediated targeting and activation. J Cell Sci 130, 406–419. [DOI] [PubMed] [Google Scholar]
- 39.Masuda H, and Toda T (2016). Synergistic role of fission yeast Alp16GCP6 and Mzt1MOZART1 in gamma-tubulin complex recruitment to mitotic spindle pole bodies and spindle assembly. Molecular biology of the cell 27, 1753–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oakley BR, Paolillo V, and Zheng Y (2015). gamma-Tubulin complexes in microtubule nucleation and beyond. Molecular biology of the cell 26, 2957–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kollman JM, Merdes A, Mourey L, and Agard DA (2011). Microtubule nucleation by gamma-tubulin complexes. Nature reviews. Molecular cell biology 12, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luders J, Patel UK, and Stearns T (2006). GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol 8, 137–147. [DOI] [PubMed] [Google Scholar]
- 43.Haren L, Remy MH, Bazin I, Callebaut I, Wright M, and Merdes A (2006). NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol 172, 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin TC, Neuner A, and Schiebel E (2015). Targeting of gamma-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends in cell biology 25, 296–307. [DOI] [PubMed] [Google Scholar]
- 45.Lynch EM, Groocock LM, Borek WE, and Sawin KE (2014). Activation of the gamma-tubulin complex by the Mto1/2 complex. Curr Biol 24, 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clift D, and Schuh M (2015). A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat Commun 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eisman RC, Phelps MA, and Kaufman TC (2009). Centrosomin: a complex mix of long and short isoforms is required for centrosome function during early development in Drosophila melanogaster. Genetics 182, 979–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pitnick S, Markow T, and Spicer GS (1999). Evolution of multiple kinds of female sperm-storage organs in Drosophila. Evolution 53, 1804–1822. [DOI] [PubMed] [Google Scholar]
- 49.Lupold S, Manier MK, Puniamoorthy N, Schoff C, Starmer WT, Luepold SH, Belote JM, and Pitnick S (2016). How sexual selection can drive the evolution of costly sperm ornamentation. Nature 533, 535–538. [DOI] [PubMed] [Google Scholar]
- 50.Noguchi T, Koizumi M, and Hayashi S (2012). Mitochondria-driven cell elongation mechanism for competing sperms. Fly 6, 113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hutchens JA, Hoyle HD, Turner FR, and Raff EC (1997). Structurally similar Drosophila alpha-tubulins are functionally distinct in vivo. Molecular biology of the cell 8, 481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michiels F, Gasch A, Kaltschmidt B, and Renkawitzpohl R (1989). A 14-Bp Promoter Element Directs the Testis Specificity of the Drosophila-Beta-2 Tubulin Gene. Embo J 8, 1559–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cherbas P, Cherbas L, Lee SS, and Nakanishi K (1988). 26-[I-125]Iodoponasterone-a Is a Potent Ecdysone and a Sensitive Radioligand for Ecdysone Receptors. Proceedings of the National Academy of Sciences of the United States of America 85, 2096–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrera JA, Kao LR, Hammer RE, Seemann J, Fuchs JL, and Megraw TL (2010). CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell 18, 913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen JV, Kao LR, Jana SC, Sivan-Loukianova E, Mendonca S, Cabrera OA, Singh P, Cabernard C, Eberl DF, Bettencourt-Dias M, et al. (2015). Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila. J Cell Biol 211, 435–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kao LR, and Megraw TL (2004). RNAi in cultured Drosophila cells. Methods in molecular biology 247, 443–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen JV, and Megraw TL (2014). Spermitin: a novel mitochondrial protein in Drosophila spermatids. PLoS One 9, e108802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haren L, Remy MH, Bazin I, Callebaut I, Wright M, and Merdes A (2006). NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. Journal of Cell Biology 172, 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Draberova E, D’Agostino L, Caracciolo V, Sladkova V, Sulimenko T, Sulimenko V, Sobol M, Maounis NF, Tzelepis E, Mahera E, et al. (2015). Overexpression and Nucleolar Localization of gamma-Tubulin Small Complex Proteins GCP2 and GCP3 in Glioblastoma. Journal of neuropathology and experimental neurology 74, 723–742. [DOI] [PubMed] [Google Scholar]
- 60.Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, et al. (2017). Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell stem cell 20, 397–406 e395. [DOI] [PubMed] [Google Scholar]
- 61.Rodriguez D, Ramesh C, Henson LH, Wilmeth L, Bryant BK, Kadavakollu S, Hirsch R, Montoya J, Howell PR, George JM, et al. (2011). Synthesis and characterization of tritylthioethanamine derivatives with potent KSP inhibitory activity. Bioorganic & medicinal chemistry 19, 5446–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, and Kaufman PD (2009). A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klueg KM, Alvarado D, Muskavitch MAT, and Duffy JB (2002). Creation of a GAL4/UAS-coupled inducible gene expression system for use in Drosophila cultured cell lines. Genesis 34, 119–122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 (related to Figure 4). Live imaging of EB3-mApple (red) in a control CHO cell
The centrosome is the major MTOC that ejects EB3-mApple comets. Movie was captured by time-lapse confocal microscopy (Nikon A1). Frames were captured every 4 s for 4 min. See also Figure 4D.
Movie S2 (related to Figure 4). Live imaging of EB3-mApple (red) in a CHO cell expressing GFP-CnnT (green)
EB3 comets eject outwards from mitochondrial MTOCs assembled by GFP-CnnT. Movie was captured by time-lapse confocal microscopy (Nikon A1). Frames were captured every 4 s for 4 min. See also Figure 4D.
Movie S3 (related to Figure 4). Live imaging of EB3-mApple (red) in a CHO cell expressing GFP-CM1-mito (green)
EB3 comets eject outwards from mitochondrial MTOCs assembled by GFP-CM1-mito. Movie was captured by time-lapse confocal microscopy (Nikon A1). Frames were captured every 4 s for 4 min. See also Figure 4D.
Movie S4 (related to Figure 4). Live imaging of EB3-mApple (red) in a CHO cell expressing GFP-CnnTM (green)
EB3 comets are only emitted from a major MTOC, which is presumably the centrosome. No EB3 comets eject from GFP-CnnTΔ1 foci. Movie was captured by time-lapse confocal microscopy (Nikon A1). Frames were captured every 4 s for 4 min. See also Figure 4D.


