Abstract
Background
The molecular pathways involved in the transition from uterine quiescence to overt labour are mapped and form the currently established pharmacological targets for both the induction and inhibition of human labour. However, both spontaneous premature labour and functional dystocia occur and are difficult to treat adequately. The identification of upstream regulators involved in the onset and orchestration of labour pathways is essential to develop additional therapies that will contribute to the regulation of the timing of birth.
Objectives
To define uterine biological processes and their upstream activators involved in the transition from uterine quiescence to overt labour.
Study design
The uterus of non-pregnant and pregnant FVB M. musculus is collected at embryonic days (E) 6.5, 8.5, 10.5, 12.5, 15.5 and 17.5 and the uterine transcriptome is determined using the Illumina mouse Ref8v2 micro-array platform. K-means clustering and Ingenuity Pathway Analysis are applied to further dissect the transcriptome data.
Results
From E6.5 to E17.5, 5405 genes are significantly differentially expressed and they segregate into 7 unique clusters. Five of the 7 clusters are enriched for genes involved in specific biological processes that include regulation of gene-expression, T-cell receptor activation, Toll-like receptor signalling and steroid metabolism. The identification of upstream activators for differentially expressed genes between consecutive time points highlights the E10.5 to E12.5 window during which the role from progesterone switches from an activated state to the inhibited state reflecting the process of functional progesterone withdrawal essential for the transgression from myometrial quiescence to synchronized contractions. For this time window in which 189 genes are differentially expressed we define 22 putative upstream activators of which NUPR1 and TBX2 are the most significant with respectively an activated and an inhibited status.
Conclusions
Gene expression profiling of mice uterus from E6.5 to E17.5 results in 7 unique gene expression clusters from early to late pregnancy that define the landscape of molecular events in ongoing pregnancy. In the current dataset progesterone is predicted as an activated upstream regulator and maintainer of myometrial quiescence and is active till E10.5. Progesterone is predicted as an inhibited upstream regulator at E12.5. We identify 22 upstream regulators in the E10.5 to E12.5 time window where the switch to progesterone withdrawal occurs. They are putative relevant upstream activators of labour.
Introduction
The transition from uterine quiescence to overt labour in placental mammals is marked by functional progesterone withdrawal, induction of prostaglandins, increase of pro-inflammatory cytokines and interleukins, activation of the oxytocin receptor by oxytocin and increase of voltage gated Ca2+ channels facilitating contractions [1]. These processes have been investigated in detail and are rational and established therapeutical targets to intervene in the process of human labour when it either occurs preterm and/or does not advance properly as in functional labour dystocia.
Functional dystocia is defined as inadequate uterine activity where delivery fails to progress during the active phase of labour [2, 3]. Functional dystocia highly contributes to caesarean rates in nulliparous women [4, 5]. early interventions such as the administration of oxytocin only result in a clinically modest reduction in the caesarean section rate [6]. This implies that further activation of the oxytocin receptor only is not sufficient to generate strong synchronized contractions during the active phase of labour.
Preterm birth, defined as delivery before the 37th week of gestation, is the most common cause of neonatal mortality and the second leading cause of death in children under five years of age [7]. Preterm birth is associated with immediate and long term morbidity as well as growth and developmental delay [8, 9]. Progesterone treatment, counteracting the functional progesterone withdrawal, or administration of cyclooxygenase inhibitors to inhibit prostaglandin synthesis, do not substantially reduce the risk of preterm birth [10–13]. Although oxytocin receptor antagonists and calcium channel blockers are effective in delaying preterm delivery (~7 days) [14–16] the majority of the women (~65%) receiving this treatment still deliver preterm [17]. Research on understanding mechanisms governing parturition initiation has recently escalated owing to the increasing rates of preterm birth around the globe over the last 2 decades [18].
In the current study we aim to identify upstream regulators with a putative role in triggering molecular pathways relevant for the synchronized contractions necessary to attain a timely vaginal delivery.
In humans, it is unethical and thus impossible to track the uterine gene expression changes throughout gestation in relation to the initiation of parturition, making it necessary to use animal models. All animal models used to study human parturition have their limitations [19]. Because of their relative short gestation varying from 18.5 to 20.5 days, their fully available genome sequence and the opportunity to harvest uterine tissue during gestation, mice are an ideal species to perform gestational gene expression studies. It has been demonstrated that mice can mimic human pathologies with respect to the infection induced preterm birth [20–23].
Functional withdrawal of progesterone is essential for the progression from quiescence to overt labour. While maternal progesterone levels in rodents decline near term, in humans they remain high till birth and is it the switch of the highly bioactive B isoform of the progesterone receptor to a less bioactive A isoform that contributes to functional progesterone withdrawal [24]. This classic distinction between man and mice is less absolute then sometimes reported. The declined maternal progesterone levels in mice towards term remain well above the dissociation constant for binding to the progesterone receptor and the switch in progesterone receptor isoform expression also occurs in mice [25, 26]. In addition, functional progesterone withdrawal is in part achieved by increased local progesterone metabolism, a process similar in mice and human [26].
The downstream effects of functional progesterone withdrawal are induction of prostaglandins, pro-inflammatory mediators, expression of gap junction protein alpha 1 (connexin-43) and the oxytocin receptor. Although these processes in mice parturition are also not fully identical to human parturition there are demonstrable similarities [1, 19, 25–29]. Uterine specific knockout of Trp53 in mice results in both preterm birth and dystocia [30]. Previous RNA microarray studies on mouse uterus have mostly focused on selective time points during late gestation establishing pathways relevant to labour that are similar to those also relevant for human parturition [23, 26, 31–35].
Gene expression related time course studies on a prolonged earlier time periods of mouse gestation have not yet been reported and they are essential to define novel upstream triggers for labour associated pathways. The utilization of varying gestational time points, mice strains, experimental setups, interventions and array platforms used makes the merging of existing gene expression data extremely difficult and hampers the identification of potential upstream regulators.
In the current study, we define the gene expression patterns in FVB mouse uterus across gestation from E6.5 to E17.5. We identify gene expression clusters and define novel putative upstream regulators for labour.
Materials & methods
Tissue collection and sample preparation
All experimental procedures in this study were approved by the Animal Ethics Committee of the Academic Medical Center, Amsterdam, The Netherlands (Permit Number: DVF102563). All methods were performed in accordance with relevant guidelines and regulations. FVB/N mice were purchased from Harlan Laboratories. Staged embryos were obtained by crossing FVB/N mice and checking the next morning for a vaginal plug. The morning of discovery of the vaginal plug was designated as embryonic day (E) 0.5.
Mice samples were collected after termination by carbon dioxide inhalation from non-pregnant (NP)and pregnant FVB mice at E6.5, E8.5, E10.5, E12.5, E15.5 and E17.5 respectively. NP mice were taken at random. Median ages were 81 days (range 56–146) for non-pregnant and 79 days (range 61–99) for pregnant mice. Uterine tissue from pregnant mice was collected at implantation sites. Embryonic/placental tissue was removed and decidua was scraped of using a sterile blade. Uterine tissue was immediately placed into RNAlater and processed according to the instruction of the manufacturer. Samples were stored at -80°C until further use. Total RNA from uterine tissue samples was isolated using Trizol reagent (Thermo Fischer Scientific; 15596026) after lysing the tissues using the MagNA lyser (Roche). Isolated total RNA was cleaned up using RNA easy mini kit (Qiagen; 74104). The integrity of the RNA was checked using the Bioanalyser (Agilent). Because of a poor RIN score (RIN <7), one tissue sample from E6.5 and one tissue sample from E8.5 were excluded for further downstream use. For each individual mouse, total RNA samples with a RIN score greater than 7 were pooled. In total 32 total RNA samples, NP (n = 4), E6.5 (n = 4), E8.5 (n = 4), E10.5 (n = 5), E12.5 (n = 5) E15.5 (n = 5) and E17.5 (n = 5) were labelled using the Illumina total prep RNA amplification kit (AMIL1791) with an input of 200ng/μl.
Microarray hybridization
Illumina Mouse Ref8 platform was used to establish gene expression patterns of the current dataset. The hybridization of the microarray was performed by Service XS (Leiden, The Netherlands). For array hybridization 1 ug of total RNA of the uterus samples was used and samples were randomized over arrays.
Microarray data preprocessing and analysis
The data analysis was performed using the statistical software package R (version 3.3.2). The quality of the dataset was checked using arrayQualityMetrics (version 3.30.0) for 3 main parameters: comparison between arrays, array intensity distributions, and individual array quality. No outlying samples were identified. Probes (5528) with a non-significant detection P value of >0.05 on all 32 arrays were filtered out and not used for subsequent analysis. Normalization was performed on 32 samples starting from the Illumina sample and control probe profiles using the normexp-by-control background correction, quantile normalization, and log2 transformation from the limma package (v3.32.2). If multiple probes were mapped to the same Entrez Gene identifier according to the illuminaMousev2.db package (v1.26), the probe with the highest standard deviation was chosen. Separate contrasts were constructed for calculating differential expression between consecutive time points (NP vs E6.5, E6.5 vs E8.5, E8.5 vs E10.5, E10.5 vs E12.5, E12.5 vs E15.5 and E15.5 vs E17.5). Differential expression between the experimental conditions (time points) was assessed with a moderated t test using the linear model framework from the limma package. Resulting P values were corrected for multiple testing using the Benjamini-Hochberg false discovery rate. Corrected P values ≤0.05 were considered statistically significant. The resulting significant genes were used for prediction of upstream regulators. The gap statistic method (clusGap R function-cluster package v.2.0.5) was used to determine the optimal n number of clusters based on the firstSEmax value. Subsequently, K-means clustering (default algorithm) was used to obtain gene clusters using the scaled normalized relative gene expression data on a log2 scale for each expressed gene using packages clValid (v0.6–6) and cluster (v2.0.2).
Gene ontology and pathway enrichment analysis
The gene ontology (GO) Biological Process (2017) enrichment function provided by the Enrichr program [36, 37] was used for GO enrichment of all genes in each cluster. The GO enrichment was considered significant if it had a Benjamini-Hochberg adjusted P value less than 0.05. Pathway enrichment analysis for consecutive time point comparisons was done using the Enrichr program [36, 37] web based tool and the WikiPathways 2016 module.
Prediction of upstream regulators
Ingenuity pathway analysis (IPA) [38], an approach considering experimental observations in any species, tissue or cell type, was utilized to predict upstream regulators of the DEGs between consecutive embryonic time points. The significant (differential genes across consecutive time points (DEGct) from every comparison (FDR<0.05) was imported into IPA as separate text files with gene symbols and log2 fold change values. Upstream regulators prediction was based on the direction of fold change of the genes using MouseRef8 as gene background. The upstream regulators were considered significant if they had a significant prediction Z score greater than 2. The program gives a P value of overlap with the downstream targets. The Z scores can independently predict the activation or inhibition of an upstream regulator independent of the p value of overlap.
Data repository
The microarray data have been deposited in the NCBI Gene Expression. Accession number GSE85934.
Results
Quality control and normalization
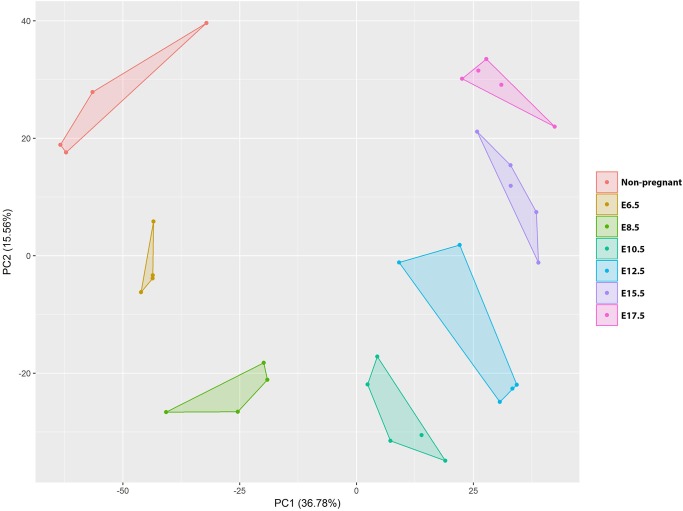
The expression profiles of in total 32 samples (with a minimum of 4 different mice per time point) were analysed on Illumina mouse Ref8v2 micro-array platform. Principal component analysis (PCA) shows that uterine tissue samples harvested during same embryonic time points cluster together, indicating significant gestational age based gene expression differences between time points (Fig 1).
Fig 1. Principal component analysis (PCA).
PCA of the quantile normalized dataset consisting of 32 mouse uterine RNA samples. The colours represent different embryonal days at which the tissue was harvested. % of total variance for PC1 and PC2 is indicated between brackets.
Gene expression clusters during gestation from E6.5 to E17.5
To define patterns of gene expression during gestation, differential gene expression was calculated between a minimum of two consecutive or non-consecutive time points across gestation (DEGots). Applying an adjusted p value filtering <0.05, 5405 DEGots are identified. With a 1.5 log fold change (lfc) cut-off this reduces to 971 DEGots and with a 2 lfc cut off that number further decreases to 520. To comprehend the entirety of gene expression networks and associated molecular pathways, the 5405 significant DEGots with no additional lfc were used for clustering and subsequent gene ontology enrichment analysis.
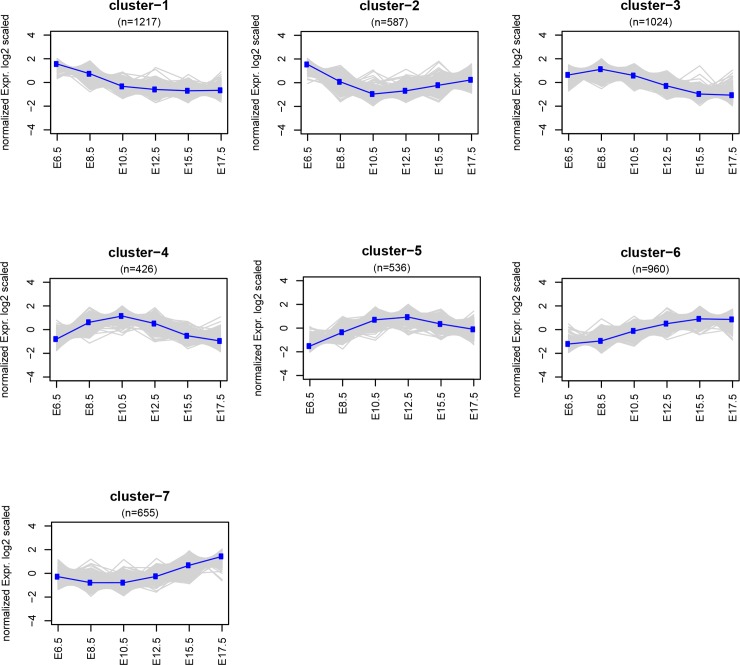
The gap statistic method [39] shows that the highest fraction of variance between differential gene expression patterns can be explained by 7 clusters (S1 Fig and Fig 2, S1 Table). Cluster 1 and cluster 2 show relatively high expression at E6.5. Cluster 2 shows a slight increase of expression towards later pregnancy, which differentiates it from cluster 1. Both clusters 1 and 2 do not display any significant (adjusted P-value < 0.05) enrichment of gene ontology (GO) terms. Cluster 3 gradually decreases from E8.5 to late pregnancy and is significantly enriched in processes involved in cytoplasmic translation and ribosome biogenesis. Cluster 4 and 5 have an arc shaped pattern with an increase from early-to mid-pregnancy and decrease from mid-to-late pregnancy. Cluster 4 is enriched in T cell receptor signalling and regulation of steroid biosynthesis with decreasing expression after E10.5. In cluster 5 expression levels drops after E12.5 and it is enriched in neutrophil activation. Cluster 6 increases from early to late pregnancy and is enriched in neutrophil activation, protein metabolism, Toll-like receptor signalling pathway/activation of immune response and apoptotic cell clearance. Cluster 7 shows an increase from mid to late pregnancy and is enriched in regulation of protein phosphorylation, protein-lipid complex remodelling and sterol transport (S2 Table). Clusters were evaluated with respect to the presence of genes well established as having a role in the transition from uterine quiescence to synchronized labour relevant for cortisol bioactivity (Hsd11b1 and Hsd11b2) [40], functional progesterone withdrawal (Stat5b, Zeb1, Zeb2,) [35, 41], the oxytocin receptor (Oxtr) and the Gja1 gene encoding the gap junction protein connexin 43 [42]. Cluster 3 with decreasing expression from early to late pregnancy contains Gja1. Cluster 1, with relatively high expression during early pregnancy that decreases as pregnancy advances, contains the Zeb1 and Hsd17b1 genes. Cluster 6, with overall increasing expression levels from E6.5 to E17.5, contains Hsd11b1 and the transcription factor Elf3 that has previously been shown to increase transcription from the Ptgs1 promoter [33].
Fig 2. K-means cluster analysis.
K-means clustering of differentially expressed genes changing over time (DEGots) from E6.5 to E17.5. The 5405 DEGots are clustered by K-means algorithm. n: number of genes in each cluster.
Identification of upstream regulators using DEGcts as downstream gene targets
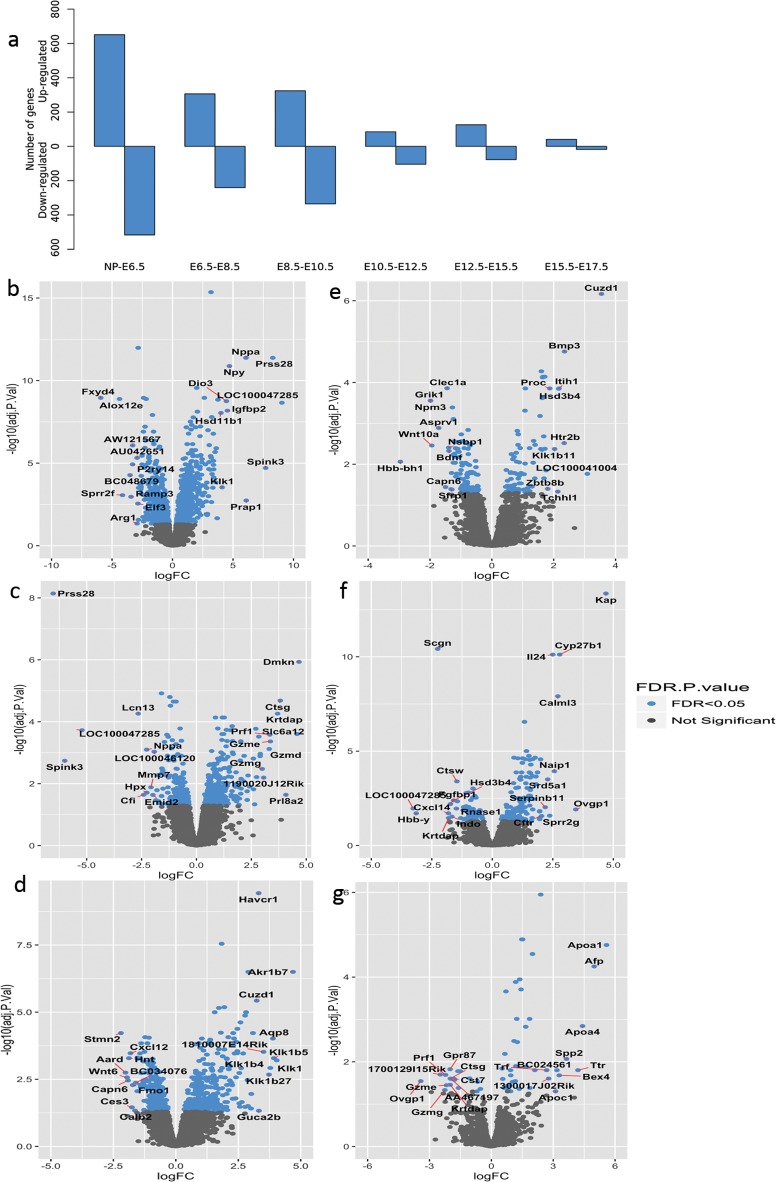
Identification of upstream regulators is based on the differential expression between consecutive time points (DEGcts) of progressing gestation. The number of significant DEGcts are illustrated in Fig 3A (S3 Table, pathway enrichment analysis for consecutive time point comparisons are present in S4 Table). The NP to E6.5 comparison yields the highest number of DEGcts. The volcano plots (Fig 3B–3G) visualize the spread of DEGcts, with higher lfc observed in earlier stages compared to the later time points. When comparing gene expression of the non-pregnant uterus to that at E6.5 LOC100047285, Prss28, Spink3 and Nppa with a lfc change >5 are highly upregulated at E6.5 compared to NP, and are subsequently down regulated at E8.5 compared to E6.5 (Fig 3C).
Fig 3. Differentially expressed genes between consecutive time points (DEGcts).
a: Number of significant DEGcts. b-g: Volcano plots of DEGcts with the log2 fold change (logFC) on the x-axis and adjusted P value on the y-axis. Please note that axis scales are not uniform. (b) Non-pregnant vs E6.5 (n = 1167), c) E6.5 vs E8.5 (n = 546), d) E8.5 vs E10.5 (n = 659), e) E10.5 vs E12.5 (n = 189), f) E12.5 vs E15.5 (n = 203) and g) E15.5 vs E17.5 (n = 59). The significantly differentially expressed genes are depicted by blue dots. (Benjamini- Hochberg adjusted pvalue <0.05). The top ten most up-regulated and down-regulated genes (by log2 fold change) in each comparison are labelled by gene name. Volcano plots of DEGcts in high resolution are present in supplementary figures (S2–S7 Figs).
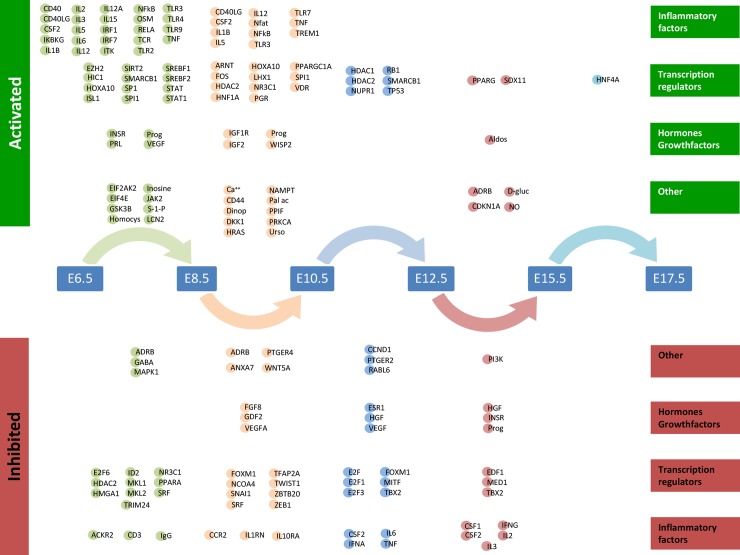
Ingenuity Pathway Analysis (IPA) [38] was applied to define upstream regulators relevant for each set of DEGcts (S5 Table). The upstream regulators identified for each DEGct comparison represent 3 main categories of functionality as illustrated in Fig 4. The top 5 upstream regulators for each comparison based on their Z-scores is illustrated in Table 1. From E15.5 to E17.7 the number of differentially expressed genes is at its nadir with only HNF4A predicted as an active upstream regulator.
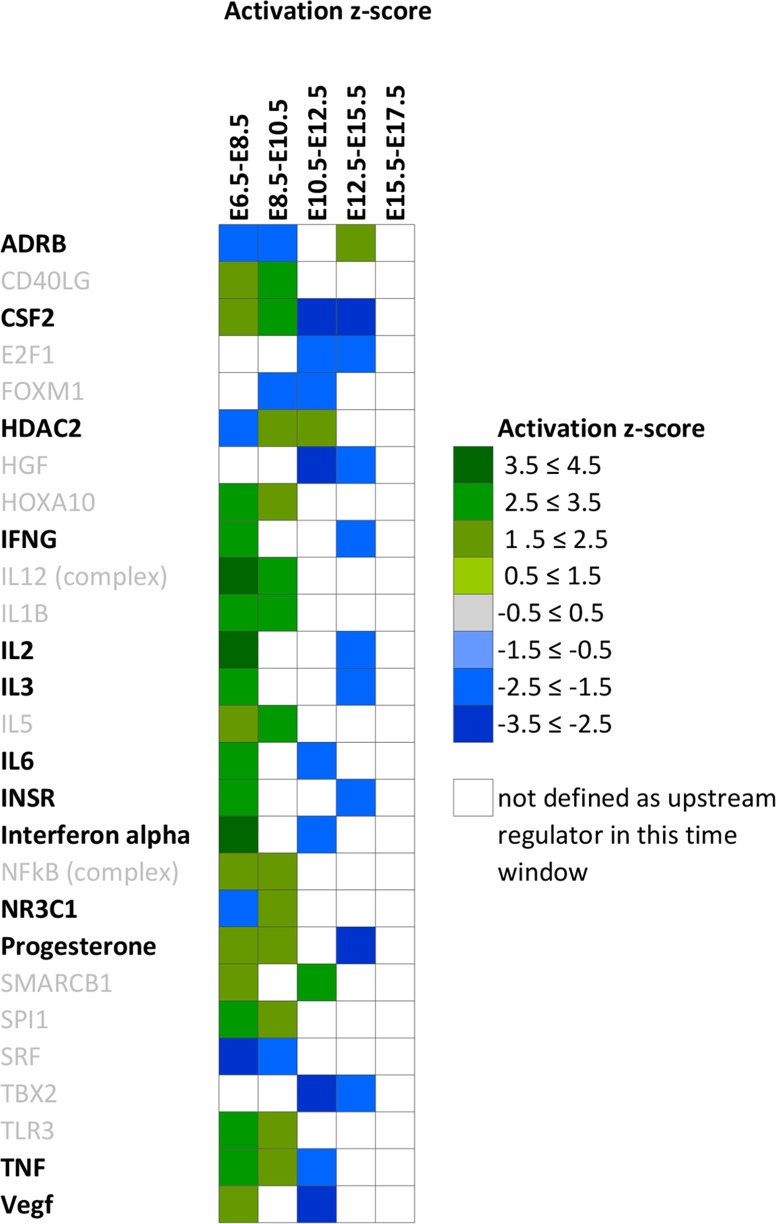
Fig 4. Predicted upstream regulators of differentially expressed genes between consecutive time points (DEGcts).
Time window specific identification of upstream analysis of the DEGcts using Ingenuity Pathway Analysis (IPA). The coloured circles are upstream regulators identified for each comparison of consecutive time points with an activation or inhibition Z score of respectively > 2.0 or < (-2.0) (S5 Table). Circle colours correspond to colours of the arrows linking the consecutive time points. Active upstream regulators are displayed at the top of the Figure and the inhibited upstream regulators are at the bottom of the Figure. The upstream regulators are grouped into four different categories: ‘Inflammatory factors’, ‘Transcription regulators’, ‘Hormones and growth factors’ and ‘Other’. Aldos = Aldosterone, D-gluc = D-glucose, Dinop = Dinoprost, Homocys = Homocysteine, NO = Nitric oxide, Pal ac = Palmitic acid, Prog = Progesterone, S-1-P = Shingosine-1-phosphate, Urso = Ursodeoxycholic acid.
Table 1. Activated and inhibited upstream regulators for each consecutive time point comparison.
| Compared embryonal time points | Activated upstream regulators |
Z-score | Inhibited upstream regulators |
Z-score |
|---|---|---|---|---|
| E6.5-E8.5 | IL2 | 4.156 | TRIM24 | -3.359 |
| IL12(complex) | 3.663 | PPARA | -2.618 | |
| Interferon alpha | 3.538 | MKL1 | -2.571 | |
| TNF | 3.430 | SRF | -2.554 | |
| SREBF1 | 3.347 | ADRB | -2.490 | |
| E8.5-E10.5 | IL5 | 3.347 | PTGER4 | -3.000 |
| CD40LG | 2.757 | IL10RA | -2.846 | |
| IL1B | 2.715 | SNAI1 | -2.577 | |
| CSF2 | 2.709 | TWIST1 | -2.514 | |
| PGR | 2.688 | GDF2 | -2.456 | |
| E10.5-E12.5 | NUPR1 | 3.771 | TBX2 | -3.450 |
| TP53 | 3.301 | HGF | -3.267 | |
| SMARCB1 | 2.917 | PTGER2 | -3.000 | |
| RB1 | 2.613 | CSF2 | -2.819 | |
| HDAC1 | 2.236 | Vegf | -2.704 | |
| E12.5-E15.5 | PPARG | 2.506 | CSF2 | -2.926 |
| D-glucose | 2.412 | progesterone | -2.547 | |
| ADRB | 2.236 | HGF | -2.402 | |
| aldosterone | 2.207 | IFNG | -2.317 | |
| CDKN1A | 2.168 | TBX2 | -2.236 | |
| E15.5-E17.5 | HNF4A | 2.208 | ||
Twenty-seven putative upstream regulators were predicted for more than one time window and 13 of them switch their direction of regulation during the gestational period investigated (Fig 5).
Fig 5. Predicted repetitive upstream activators.
Depicted are upstream activators with an activation or inhibition Z score of respectively > 2.0 or < (-2.0) present in at least 2 time window comparisons. Positive Z-scores are depicted in green, negative Z-scores are depicted in blue.
The important role of progesterone in the establishment of pregnancy and its functional withdrawal essential to the transition from uterine quiescence to overt synchronized contractions is well known. In the current dataset, we identify progesterone as one of the active hormonal upstream regulators both at E6.5 to E8.5 and E8.5-E10.5. Progesterone is not present as predicted upstream regulator from E10.5 to E12.5 and comes up again at E12.5-E15.5 with a negative Z score predictive value, showing the switch of this hormonal system to an inhibited state.
This finding draws attention to the E10.5-E12.5 window for which 22 upstream regulators are predicted, 6 of which the function is predicted as activated and 16 are predicted as inhibited. NUPR1, has the highest positive predictive Z score at E10.5 to E12.5. TBX2, with the highest negative predictive Z score at E10.5 to E12.5 is also listed as an inhibited upstream regulator for the E12.5 to 15.5 window of differential gene expression.
Discussion
Parturition is a natural phenomenon at the end of mammalian pregnancy when fetal maturation is compatible with extra-uterine life. Currently, both preterm labour and functional labour dystocia are clinical challenging obstetrical problems for which only limited therapeutics are available. Functional progesterone withdrawal, prostaglandin synthesis, oxytocin receptor activity and the expression of connexin-43 are all essential to activate voltage gated calcium channels and induce the high connectivity between uterine myocytes required for functional labour. The main goal of the current study is to identify upstream regulators that play a role in the initiation of parturition and the timely regulation of all pathways essential to attain synchronized contractions.
In the current study we collected uterine tissues from E6.5 to E17.5 in pregnant FVB mouse as well as from control non-pregnant mice and determined gestational time-point specific gene expression profiles using micro-array. Time points after E17.5 were not investigated as they have been previously well studied [32, 33, 35] and do not contribute to the aim of the current paper.
PCA shows proper clustering of gene expression profiles for the different sample time points.
Although the data from the non-pregnant mice uterus are not directly relevant for the identification of putative upstream regulators of labour they do demonstrate the validity of our approach. LOC100047285, Prss28, Spink3 and Nppa that are upregulated (lfc >5) at E6.5 compared to non-pregnant, are down regulated at E8.5 compared to E6.5 (Fig 3C) indicating a rather stage specific flip of gene expression in early pregnancy.
Prss28 (also known as Isp1 implantation serine protease1) and Spink3 (Secretory peptidase inhibitor) have both been previously implicated in implantation related processes[43–45]. Also Proline rich acidic protein 1 (Prap1) that is upregulated at E6.5 compared to non-pregnant has well-established functions in implantation [46].
Implantation requires an adaptive immune response where the maternal immune system should accept the fetus as a partial allograft. As elucidated in the results section, progesterone and inflammatory mediators (NFKB, STAT1, TLR2, TLR4, TNF, IL2, IL3, and IL6) are active upstream regulators at E6.5-E8.5–10.5 (Fig 4 and Table 1). Coinciding with progesterone withdrawal, activation of the inflammatory response is a well-known mechanism in the activation of labour. In the current dataset the activating inflammatory mediators between E6.5 and E10.5 disappear at E10.5 after which a subset reappears as inhibited upstream activators.
The immune related role of CSF2 (Predictive Z-score -2.82) in early pregnancy has been well described [47]. In the current study, CSF2 is predicted as an active upstream regulator from E6.5 to E10.5 after which it comes up as inhibited, suggesting a possible role in prevention of inflammatory response in mid-pregnancy. At the same time, active upstream regulators at E12.5-E15.5 such as Nitric oxide (Z-score 2.00) [48, 49], beta-adrenergic receptor complex ADRB (Z-score 2.24) [50, 51], PPARG (Z-score 2.51) [52–54] are established mediators of uterine quiescence.
The gene expression clusters during gestation shed light on the complexity of gene networks and the molecular cascades required for the on-going pregnancy and in preparation for parturition. Particularly, clusters 3, 4, 5, 6 & 7 that show significant enrichment in specific biological processes. Cluster 3 enriched in processes involving cellular mechanisms of regulation of gene expression and protein synthesis decreases from early to late pregnancy.
Cluster 4 & Cluster 5 indicate a suppression of T-cell receptor signaling/T cell activation during mid-pregnancy. On the other hand cluster 6 which increases from early to late pregnancy shows an enrichment in Toll-like receptor signaling some of which have been known to play an active part in labour [55, 56].
Functional progesterone withdrawal and induction of prostaglandins is one of the known key regulators to the transition from quiescence to overt labour in both mice and humans [57–59]. Previous studies in mice have shown that the increased expression of the oxytocin receptor and connexin-43 after E15.5 are preceded by the downregulation of the zinc finger transcription factors Zeb1 and Zeb2 evoked by the upregulation of the miR-200 family that perform a central role in the process of progesterone mediated uterine quiescence [26, 35]. We observe Zeb1 in cluster 1 which decreases from early to mid-pregnancy.
IPA defines progesterone as active upstream regulator in both the E6.5 to E8.5 and the E8.5 to E10.5 time windows fitting its well established role in the maintenance of pregnancy. Progesterone switches to an inhibited upstream regulator in the E12.5 to E15.5 window indicating functional progesterone withdrawal and highlighting the interest for the E10.5 to E12.5 time window.
From studies in C57Bl/6 mice it is evident that the transcription factor E47-like factor 3(Elf3) increases from E13.5 to E16.5 [33]. Elf3 is able to regulate the promoter activity of Ptgs1 (also known as Cox-1) that compared to E13.5 is upregulated at E15.5 and is responsible for the production of prostaglandins. Similarly, we observe Elf3 in Cluster 6 and Ptgs1 in Cluster 7 with a 1.7 lfc in expression during the E12.5 to E15.5 window in FVB mouse uterus. This again highlights the importance of the E10.5 to E12.5 time window for the identification of novel upstream regulators.
Previous studies on tumor protein p53 (Trp53), one of the active upstream regulators at E10.5 to E12.5 identified in the current study, have shown that mice with a conditional deletion of uterine Trp53 have an increased incidence of preterm birth. In addition, 75% of dams with preterm birth also show signs of dystocia with prolonged labour over a period of 12 hours [30].
NUPR1 predicted as an active upstream regulator, (Nupr1) present in cluster 6 gradually increases from early to late pregnancy and TBX2 predicted as an inhibited upstream regulator (Tbx2) is present in cluster 1 decreases from early to late pregnancy. TP53 (Trp53) present in cluster 6 decreases towards late pregnancy. In addition to TP53, we also observe NUPR1, TBX2, HGF and PTGER2 as upstream regulators with an absolute predictive Z-score above 3 identified at E10.5 to E12.5. They are strong candidates to play an important role in regulating the molecular events that are necessary for the transition from uterine quiescence to initiation of parturition.
Limitations of the study
The limitations of the study are the probe coverage and the fact that gene expression is unable to quantify regulation at the translational or posttranslational level. Given the homogeneity of gene expression within samples from a particular gestational time point and the profound positive correlations with previous experimental observations this study is a valuable asset of genes playing a role in gestation and parturition.
Supporting information
Each worksheet contains the mouse genes expressed in that specific cluster with scaled normalized relative gene expression data.
(XLS)
Each worksheet displays GO terms (Column A) of all biological processes per gene expression cluster. Adjusted P values <0,05 are considered statistically significant and are highlighted.
(XLS)
Each worksheet contains the DEGcts for the indicated specific time point comparison. Lfc expression level and adjusted P value are indicated. Lists are ranked by adjusted P-value. Data are used to construct Fig 3.
(XLS)
Each worksheet contains enriched pathways specific the indicated time point comparison.
(XLSX)
Upstream regulators corresponding to the differentially expressed genes defined for each consecutive time point comparison are listed. For each upstream regulator, the functional role and functional direction (either activating or inhibiting their downstream targets) as well as their downstream reported (human) genes are indicated.
(XLSX)
Weighted gap statistic measures within-clusters homogeneity and determining the minimum of clusters (x-axis) that explains the highest fraction of variance between differential gene expression patterns (y-axis). The optimal number of clusters was defined based on the saturation point of the curve.
(PDF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Acknowledgments
We thank P.D. Moerland and R. Keijser for helpful discussions regarding bio-informatics analysis. This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative.
Data Availability
The microarray data have been deposited in the NCBI Gene Expression Database (Accession number GSE85934).
Funding Statement
This work was supported by a grant from ZonMw the Netherlands grant (number 91210050)(CRS) and by the Netherlands Heart Institute (CRS, FF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vannuccini S, Bocchi C, Severi FM, Challis JR, Petraglia F. Endocrinology of human parturition. Ann Endocrinol (Paris). 2016;77(2):105–13. [DOI] [PubMed] [Google Scholar]
- 2.Baxi LV, Petrie RH. Pharmacologic effects on labor: effects of drugs on dystocia, labor, and uterine activity. Clin Obstet Gynecol. 1987;30(1):19–32. [DOI] [PubMed] [Google Scholar]
- 3.Garfield RE. Cellular and molecular bases for dystocia. Clin Obstet Gynecol. 1987;30(1):3–18. [DOI] [PubMed] [Google Scholar]
- 4.Karacam Z, Walsh D, Bugg GJ. Evolving understanding and treatment of labour dystocia. Eur J Obstet Gynecol Reprod Biol. 2014;182:123–7. 10.1016/j.ejogrb.2014.09.011 [DOI] [PubMed] [Google Scholar]
- 5.Neal JL, Lowe NK, Schorn MN, Holley SL, Ryan SL, Buxton M, et al. Labor Dystocia: A Common Approach to Diagnosis. J Midwifery Womens Health. 2015;60(5):499–509. 10.1111/jmwh.12360 [DOI] [PubMed] [Google Scholar]
- 6.Wei S, Wo BL, Qi HP, Xu H, Luo ZC, Roy C, et al. Early amniotomy and early oxytocin for prevention of, or therapy for, delay in first stage spontaneous labour compared with routine care. Cochrane Database Syst Rev. 2012;(9):CD006794 10.1002/14651858.CD006794.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. 10.1016/S0140-6736(08)60074-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saigal S, Doyle LW. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet. 2008;371(9608):261–9. 10.1016/S0140-6736(08)60136-1 [DOI] [PubMed] [Google Scholar]
- 9.Linsell L, Malouf R, Morris J, Kurinczuk JJ, Marlow N. Prognostic Factors for Poor Cognitive Development in Children Born Very Preterm or With Very Low Birth Weight: A Systematic Review. JAMA Pediatr. 2015;169(12):1162–72. 10.1001/jamapediatrics.2015.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norman JE, Marlow N, Messow CM, Shennan A, Bennett PR, Thornton S, et al. Vaginal progesterone prophylaxis for preterm birth (the OPPTIMUM study): a multicentre, randomised, double-blind trial. Lancet. 2016;387(10033):2106–16. 10.1016/S0140-6736(16)00350-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su LL, Samuel M, Chong YS. Progestational agents for treating threatened or established preterm labour. Cochrane Database Syst Rev. 2014;(1):CD006770 10.1002/14651858.CD006770.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanprakob T, Laopaiboon M, Lumbiganon P, Sangkomkamhang US. Cyclo-oxygenase (COX) inhibitors for preventing preterm labour. Cochrane Database Syst Rev. 2012;10:CD007748 10.1002/14651858.CD007748.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinebrant HE, Pileggi-Castro C, Romero CL, Dos Santos RA, Kumar S, Souza JP, et al. Cyclo-oxygenase (COX) inhibitors for treating preterm labour. Cochrane Database Syst Rev. 2015;(6):CD001992 10.1002/14651858.CD001992.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saleh SS, Al-Ramahi MQ, Al Kazaleh FA. Atosiban and nifedipine in the suppression of pre-term labour: a comparative study. J Obstet Gynaecol. 2013;33(1):43–5. 10.3109/01443615.2012.721822 [DOI] [PubMed] [Google Scholar]
- 15.Flenady V, Wojcieszek AM, Papatsonis DN, Stock OM, Murray L, Jardine LA, et al. Calcium channel blockers for inhibiting preterm labour and birth. Cochrane Database Syst Rev. 2014;(6):CD002255 10.1002/14651858.CD002255.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Vliet EOG, Nijman TAJ, Schuit E, Heida KY, Opmeer BC, Kok M, et al. Nifedipine versus atosiban for threatened preterm birth (APOSTEL III): a multicentre, randomised controlled trial. Lancet. 2016;387(10033):2117–24. 10.1016/S0140-6736(16)00548-1 [DOI] [PubMed] [Google Scholar]
- 17.Papatsonis DN, Van Geijn HP, Ader HJ, Lange FM, Bleker OP, Dekker GA. Nifedipine and ritodrine in the management of preterm labor: a randomized multicenter trial. Obstet Gynecol. 1997;90(2):230–4. 10.1016/S0029-7844(97)00182-8 [DOI] [PubMed] [Google Scholar]
- 18.Chang HH, Larson J, Blencowe H, Spong CY, Howson CP, Cairns-Smith S, et al. Preventing preterm births: analysis of trends and potential reductions with interventions in 39 countries with very high human development index. Lancet. 2013;381(9862):223–34. 10.1016/S0140-6736(12)61856-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell BF, Taggart MJ. Are animal models relevant to key aspects of human parturition? Am J Physiol Regul Integr Comp Physiol. 2009;297(3):R525–45. 10.1152/ajpregu.00153.2009 [DOI] [PubMed] [Google Scholar]
- 20.Ratajczak CK, Fay JC, Muglia LJ. Preventing preterm birth: the past limitations and new potential of animal models. Dis Model Mech. 2010;3(7–8):407–14. 10.1242/dmm.001701 [DOI] [PubMed] [Google Scholar]
- 21.Cappelletti M, Della Bella S, Ferrazzi E, Mavilio D, Divanovic S. Inflammation and preterm birth. J Leukoc Biol. 2016;99(1):67–78. 10.1189/jlb.3MR0615-272RR [DOI] [PubMed] [Google Scholar]
- 22.Migale R, Herbert BR, Lee YS, Sykes L, Waddington SN, Peebles D, et al. Specific Lipopolysaccharide Serotypes Induce Differential Maternal and Neonatal Inflammatory Responses in a Murine Model of Preterm Labor. Am J Pathol. 2015;185(9):2390–401. 10.1016/j.ajpath.2015.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Migale R, MacIntyre DA, Cacciatore S, Lee YS, Hagberg H, Herbert BR, et al. Modeling hormonal and inflammatory contributions to preterm and term labor using uterine temporal transcriptomics. BMC Med. 2016;14(1):86 10.1186/s12916-016-0632-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merlino AA, Welsh TN, Tan H, Yi LJ, Cannon V, Mercer BM, et al. Nuclear progesterone receptors in the human pregnancy myometrium: evidence that parturition involves functional progesterone withdrawal mediated by increased expression of progesterone receptor-A. J Clin Endocrinol Metab. 2007;92(5):1927–33. 10.1210/jc.2007-0077 [DOI] [PubMed] [Google Scholar]
- 25.Wagner GP, Tong Y, Emera D, Romero R. An evolutionary test of the isoform switching hypothesis of functional progesterone withdrawal for parturition: humans have a weaker repressive effect of PR-A than mice. Journal of perinatal medicine. 2012;40(4):345–51. 10.1515/jpm-2011-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams KC, Renthal NE, Condon JC, Gerard RD, Mendelson CR. MicroRNA-200a serves a key role in the decline of progesterone receptor function leading to term and preterm labor. Proc Natl Acad Sci U S A. 2012;109(19):7529–34. 10.1073/pnas.1200650109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keirse MJNC. Endogenous prostaglandins in human parturition In: Anderson ABM, Bennebroek Gravenhorst J, editors. Human Parturition. The Hague: Leiden University Press; 1979. p. 101–42. [Google Scholar]
- 28.Winchester SK, Imamura T, Gross GA, Muglia LM, Vogt SK, Wright J, et al. Coordinate regulation of prostaglandin metabolism for induction of parturition in mice. Endocrinology. 2002;143(7):2593–8. 10.1210/endo.143.7.8926 [DOI] [PubMed] [Google Scholar]
- 29.Doring B, Shynlova O, Tsui P, Eckardt D, Janssen-Bienhold U, Hofmann F, et al. Ablation of connexin43 in uterine smooth muscle cells of the mouse causes delayed parturition. J Cell Sci. 2006;119(Pt 9):1715–22. 10.1242/jcs.02892 [DOI] [PubMed] [Google Scholar]
- 30.Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest. 2010;120(3):803–15. 10.1172/JCI40051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhle RA, Pavlidis P, Grundy WN, Hirsch E. A high-throughput study of gene expression in preterm labor with a subtractive microarray approach. American journal of obstetrics and gynecology. 2001;185(3):716–24. 10.1067/mob.2001.117183 [DOI] [PubMed] [Google Scholar]
- 32.Salomonis N, Cotte N, Zambon AC, Pollard KS, Vranizan K, Doniger SW, et al. Identifying genetic networks underlying myometrial transition to labor. Genome Biol. 2005;6(2):R12 10.1186/gb-2005-6-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bethin KE, Nagai Y, Sladek R, Asada M, Sadovsky Y, Hudson TJ, et al. Microarray analysis of uterine gene expression in mouse and human pregnancy. Mol Endocrinol. 2003;17(8):1454–69. 10.1210/me.2003-0007 [DOI] [PubMed] [Google Scholar]
- 34.McConaha ME, Eckstrum K, An J, Steinle JJ, Bany BM. Microarray assessment of the influence of the conceptus on gene expression in the mouse uterus during decidualization. Reproduction. 2011;141(4):511–27. 10.1530/REP-10-0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci U S A. 2010;107(48):20828–33. 10.1073/pnas.1008301107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016;44(W1):W90–7. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics (Oxford, England). 2014;30(4):523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan M, Ye K. Determining the number of clusters using the weighted gap statistic. Biometrics. 2007;63(4):1031–7. 10.1111/j.1541-0420.2007.00784.x [DOI] [PubMed] [Google Scholar]
- 40.Damiani F, Makieva S, Rinaldi SF, Hua L, Marcolongo P, Petraglia F, et al. 11beta-hydroxysteroid dehydrogenase type 1 and pregnancy: Role in the timing of labour onset and in myometrial contraction. Mol Cell Endocrinol. 2017;447:79–86. 10.1016/j.mce.2017.02.034 [DOI] [PubMed] [Google Scholar]
- 41.Piekorz RP, Gingras S, Hoffmeyer A, Ihle JN, Weinstein Y. Regulation of progesterone levels during pregnancy and parturition by signal transducer and activator of transcription 5 and 20alpha-hydroxysteroid dehydrogenase. Mol Endocrinol. 2005;19(2):431–40. 10.1210/me.2004-0302 [DOI] [PubMed] [Google Scholar]
- 42.Mendelson CR. Minireview: fetal-maternal hormonal signaling in pregnancy and labor. Mol Endocrinol. 2009;23(7):947–54. 10.1210/me.2009-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma N, Liu S, Tang L, Irwin J, Meng G, Rancourt DE. Implantation Serine Proteinases heterodimerize and are critical in hatching and implantation. BMC Dev Biol. 2006;6:61 10.1186/1471-213X-6-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W, Han BC, Wang RC, Xiong GF, Peng JP. Role of secretory protease inhibitor SPINK3 in mouse uterus during early pregnancy. Cell and tissue research. 2010;341(3):441–51. 10.1007/s00441-010-1013-5 [DOI] [PubMed] [Google Scholar]
- 45.Tang L, Rancourt DE. Murine implantation serine proteinases 1 and 2: structure, function and evolution. Gene. 2005;364:30–6. 10.1016/j.gene.2005.07.041 [DOI] [PubMed] [Google Scholar]
- 46.Xiong GF, Zhang YS, Han BC, Chen W, Yang Y, Peng JP. Estradiol-regulated proline-rich acid protein 1 is repressed by class I histone deacetylase and functions in peri-implantation mouse uterus. Mol Cell Endocrinol. 2011;331(1):23–33. 10.1016/j.mce.2010.06.003 [DOI] [PubMed] [Google Scholar]
- 47.Hansen PJ, Dobbs KB, Denicol AC. Programming of the preimplantation embryo by the embryokine colony stimulating factor 2. Anim Reprod Sci. 2014;149(1–2):59–66. 10.1016/j.anireprosci.2014.05.017 [DOI] [PubMed] [Google Scholar]
- 48.Natuzzi ES, Ursell PC, Harrison M, Buscher C, Riemer RK. Nitric oxide synthase activity in the pregnant uterus decreases at parturition. Biochem Biophys Res Commun. 1993;194(1):1–8. 10.1006/bbrc.1993.1776 [DOI] [PubMed] [Google Scholar]
- 49.Sladek SM, Roberts JM. Nitric oxide synthase activity in the gravid rat uterus decreases a day before the onset of parturition. American journal of obstetrics and gynecology. 1996;175(6):1661–7. [DOI] [PubMed] [Google Scholar]
- 50.Breuiller M, Doualla-Bell F, Litime MH, Leroy MJ, Ferre F. Disappearance of human myometrial adenylate cyclase activation by prostaglandins at the end of pregnancy. Comparison with beta-adrenergic response. Adv Prostaglandin Thromboxane Leukot Res. 1991;21B:811–4. [PubMed] [Google Scholar]
- 51.Grammatopoulos D, Stirrat GM, Williams SA, Hillhouse EW. The biological activity of the corticotropin-releasing hormone receptor-adenylate cyclase complex in human myometrium is reduced at the end of pregnancy. J Clin Endocrinol Metab. 1996;81(2):745–51. 10.1210/jcem.81.2.8636298 [DOI] [PubMed] [Google Scholar]
- 52.Froment P, Gizard F, Defever D, Staels B, Dupont J, Monget P. Peroxisome proliferator-activated receptors in reproductive tissues: from gametogenesis to parturition. J Endocrinol. 2006;189(2):199–209. 10.1677/joe.1.06667 [DOI] [PubMed] [Google Scholar]
- 53.Borel V, Gallot D, Marceau G, Sapin V, Blanchon L. Placental implications of peroxisome proliferator-activated receptors in gestation and parturition. PPAR Res. 2008;2008:758562 10.1155/2008/758562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kowalewski MP, Meyer A, Hoffmann B, Aslan S, Boos A. Expression and functional implications of peroxisome proliferator-activated receptor gamma (PPARgamma) in canine reproductive tissues during normal pregnancy and parturition and at antiprogestin induced abortion. Theriogenology. 2011;75(5):877–86. 10.1016/j.theriogenology.2010.10.030 [DOI] [PubMed] [Google Scholar]
- 55.Patni S, Flynn P, Wynen LP, Seager AL, Morgan G, White JO, et al. An introduction to Toll-like receptors and their possible role in the initiation of labour. BJOG. 2007;114(11):1326–34. 10.1111/j.1471-0528.2007.01488.x [DOI] [PubMed] [Google Scholar]
- 56.Wahid HH, Dorian CL, Chin PY, Hutchinson MR, Rice KC, Olson DM, et al. Toll-Like Receptor 4 Is an Essential Upstream Regulator of On-Time Parturition and Perinatal Viability in Mice. Endocrinology. 2015;156(10):3828–41. 10.1210/EN.2015-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell MD, Flint AP. Progesterone withdrawal: effects on prostaglandins and parturition. Prostaglandins. 1977;14(3):611–4. [DOI] [PubMed] [Google Scholar]
- 58.Zakar T, Hertelendy F. Progesterone withdrawal: key to parturition. American journal of obstetrics and gynecology. 2007;196(4):289–96. 10.1016/j.ajog.2006.09.005 [DOI] [PubMed] [Google Scholar]
- 59.Mesiano S, Chan EC, Fitter JT, Kwek K, Yeo G, Smith R. Progesterone withdrawal and estrogen activation in human parturition are coordinated by progesterone receptor A expression in the myometrium. J Clin Endocrinol Metab. 2002;87(6):2924–30. 10.1210/jcem.87.6.8609 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each worksheet contains the mouse genes expressed in that specific cluster with scaled normalized relative gene expression data.
(XLS)
Each worksheet displays GO terms (Column A) of all biological processes per gene expression cluster. Adjusted P values <0,05 are considered statistically significant and are highlighted.
(XLS)
Each worksheet contains the DEGcts for the indicated specific time point comparison. Lfc expression level and adjusted P value are indicated. Lists are ranked by adjusted P-value. Data are used to construct Fig 3.
(XLS)
Each worksheet contains enriched pathways specific the indicated time point comparison.
(XLSX)
Upstream regulators corresponding to the differentially expressed genes defined for each consecutive time point comparison are listed. For each upstream regulator, the functional role and functional direction (either activating or inhibiting their downstream targets) as well as their downstream reported (human) genes are indicated.
(XLSX)
Weighted gap statistic measures within-clusters homogeneity and determining the minimum of clusters (x-axis) that explains the highest fraction of variance between differential gene expression patterns (y-axis). The optimal number of clusters was defined based on the saturation point of the curve.
(PDF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Please note that axis scales are not uniform.
(TIF)
Data Availability Statement
The microarray data have been deposited in the NCBI Gene Expression Database (Accession number GSE85934).