Abstract
Few studies have compared the performances of those reported miRNAs as biomarkers for hypertension in a same cohort, we aimed to comprehensively examine the performances of those reported miRNAs as biomarkers for hypertension and identify the genes and pathways targetted by these miRNAs. Serum samples were collected from patients hospitalized for hypertension in Zhongshan Hospital. Gene expressions of 25 miRNAs were compared between hypertension and normal groups. Receiver operating characteristic (ROC) curves were used to evaluate the accuracy of those miRNAs as biomarkers for hypertension. miRWALK2.0 and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed to predict the target genes and pathways of selected miRNAs. A total of 164 participants were enrolled, amongst which 53 were patients with hypertension, 111 were normal population. MiR-122-5p (area under curve (AUC): 0.750), miR-199a-3p (AUC: 0.744), miR-208a-3p (AUC: 0.743), miR-423-5p (AUC: 0.740), and miR-223-5p (AUC: 0.718) showed better performance than others, and the best performance was the combination of miR-199a-3p, miR-208a-3p, miR-122-5p, and miR-223-3p (AUC: 0.80). Pathway analysis revealed that 94 pathways enriched with genes targetted by miR-199a-3p, miR-208a-3p, miR-122-5p, miR-223-5p. FoxO signaling was enriched with genes targetted by all the three miRNAs (miR-199a-3p, miR-208a-3p, miR-122-5p). The combination of miR-199a-3p, miR-208a-3p, miR-122-5p, and miR-223-3p has a good diagnostic performance for hypertension, and multitudes of possible mechanisms/pathways through which dysregulation of these miRNAs may impact risk of hypertension.
Keywords: biomarkers, hypertension, KEGG analysis
Introduction
Hypertension is the leading global risk factor for cardiovascular diseases [1], and has been long recognized as the most common risk factor for cardiovascular disease [1,2], chronic renal failure [3], stroke [1], and so on [4]. The adverse effects and unsatisfactory control rate of hypertension calls for earlier detection and more accurate therapy [5,6].
The understandings of miRNA [7–9] have advanced the diagnosis and treatment of hypertension from proteomics [10,11] and genomics [12] to epigenetics [13]. MiRNAs not only play a role in the pathogenesis of hypertension [14], but also can be used as biomarkers for hypertension [15]. Amount of circulating miRNAs have been identified as biomarkers for hypertension in different populations [15], however, few studies have compared those miRNAs in a same cohort of hypertension patients. These studies were limited in the used variable normalization methods and their results remain controversial. Hence, we cannot get a picture of the strength of those different miRNAs as biomarkers for hypertension.
In addition, miRNA profiles associated with hypertension in humans have not yet been studied in a comprehensive way. Many of the previous publications aimed at studying single miRNAs and focussed on limited downstream effects [16,17], and very few studies looked into the systematic interactions between miRNA and gene signaling pathways.
Thus, we aimed to comprehensively examine the performance of those reported miRNAs as biomarkers for hypertension in a same cohort and identify the genes and pathways targetted by these miRNAs.
Materials and methods
Patients and control subjects
Serum samples were collected from patients enrolled in the China National Heart Failure Registry (CN-HF). The CN-HF is a national, multicentered, prospective, and observational registry study, led by Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital (head unit) with 50–100 secondary and tertiary hospitals involved. All subjects signed an informed written consent to participate in the study that was approved by Ethical Committee of Zhongshan Hospital, Fudan University, China, which is according to the principles stated in the Declaration of Helsinki (approval number: B2012-140(2)).
Hypertension is defined as systolic blood pressure ≥ 140 mmHg, or diastolic blood pressure ≥ 90 mmHg, or on antihypertensive medications. Normal population enrolled the subjects who have no cardiovascular disease or any other disease. The levels of brain natriuretic peptide (BNP) and cTNT were obtained from the results of biochemical test, left ventricular ejection fraction (LVEF) was obtained from echocardiography in our hospital.
Serum preparation and RNA isolation
Blood samples (approximately 5 ml) were collected from each donor and placed in a serum separator tube. Samples were processed within 1 h. Separation of the serum was accomplished by centrifugation at 800×g for 10 min at room temperature, followed by a 15-min high-speed centrifugation at 10000×g at room temperature to completely remove the cell debris. The supernatant serum was recovered and stored at −80°C until analysis.
For RNA isolation, miRNA was extracted from 200 μl of each serum sample and eluted in 30 μl of RNase-free water using miRcutes serum/plasma miRNA isolation kit (Tiangen, Beijing, China) according to the manufacturer’s instructions.
cDNA synthesis and RT-qPCR
The miRNA isolated from blood sample was polyadenylated and reverse transcribed to cDNA in a final volume of 20 μl using miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China). Real-time PCR was performed in duplicate measurements using miRcute Plus miRNA qPCR Detection Kit (SYBR Green) (Tiangen, Beijing, China). The miRNA-specific primer sequences were designed by a biologics company (Tiangen, Beijing, China). Each amplification reaction was performed in a final volume of 20 μl containing 1 μl of the cDNA, 0.2 mM of each primer and 1× miRcute Plus miRNA Premix (with SYBR ROX). At the end of the PCR cycles, melting curve analyses as well as electrophoresis of the products on 3.0% agarose gels were performed in order to validate the specific generation of the expected PCR product. Each sample was run in duplicate for analysis.
Pathway analysis
To predict the target genes for the candidate miRNAs, we used the miRWALK2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2) [18,19], an open-access webserver that integrates 12 target prediction algorithms. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed for overlapped genes using DAVID (https://david.ncifcrf.gov/) [20,21]. A Fisher’s exact test P-value <0.05 was used to identify significantly targetted pathways in KEGG and enriched gene target pathways obtained from these databases.
Data analysis
Data were expressed in terms of mean ± S.D. or median (interquartile range) for numeric variables and as number (percent) for categorical variables. Comparisons of continuous variables amongst groups were performed by the Student’s t test or Mann–Whitney test, if appropriate. For comparison of categorical variable, chi-square test was used. The correlationship between miRNAs and biochemical indicators in hypertension patients was evaluated by Pearson correlation coefficient. Statistic analysis was performed with SPSS 22.0 (SPSS Inc., Chicago, Illinois, U.S.A.).
Results
Characteristics of the enrolled individuals
A total of 164 participants were enrolled, amongst which 53 were patients with hypertension, 111 were normal population. As was shown in Table 1, the age of hypertension patients was significantly higher than normal population (64.43 ± 9.60 compared with 46.29 ± 9.09, P<0.001), while the percentage of male gender has no difference (69.8 compared with 73.9%, P=0.581). In addition, the levels of α2-macroglobulin, β2-microglobulin, triglyceride in hypertension patients were significantly higher than normal population, while the levels of hemoglobin, glomerular filtration rate, high-density lipoprotein were significantly lower than normal population (Table 1).
Table 1. Characteristics of the enrolled individuals.
| Index | Hypertension n=53 | Control n=111 | P-value |
|---|---|---|---|
| Male gender, n (%) | 37 (69.8%) | 82 (73.9%) | 0.581 |
| SBP | 132.17 ± 13.29 | 115.70 ± 15.50 | <0.001 |
| Age | 64.43 ± 9.60 | 46.29 ± 9.09 | <0.001 |
| α2-macroglobulin | 1.62 (1.40–2.22) | 1.55 (1.23–1.76) | 0.006 |
| β2-microglobulin | 1.99 (1.70–2.51) | 1.62 (1.41–1.86) | <0.001 |
| HB | 133.17 ± 13.40 | 146.14 ± 18.38 | <0.001 |
| WBC | 6.12 ± 1.41 | 6.44 ± 1.66 | 0.228 |
| GFR | 79.66 ± 20.70 | 91.82 ± 15.99 | <0.001 |
| TC | 5.09 ± 0.83 | 4.41 ± 1.05 | <0.001 |
| HDL | 1.22 ± 0.28 | 1.44 ± 0.37 | 0.001 |
Abbreviations: GFR, glomerular filtration rate; HB, hemoglobin; HDL, high-density lipoprotein; SBP, systolic blood pressure; TC, triglyceride; WBC, white blood cell.
Selection of the reference genes for circulating miRNA
Considering that few studies evaluated the stability and superiority of serum miR-16, U6 snRNA (U6), 5S ribosomal RNA (5S), miR-19b, miR-15b, miR-24, let-7i as reference genes in cardiovascular disease, we first assess the suitability of those seven miRNAs as normalizers in cardiovascular disease using BestKeeper, NormFinder, and comparative ΔCq analysis. Our results showed that miR-16 and let-7i have the best performance, and 5S is not suitable as reference gene. Thus, in the present study miR-16 was used as the reference gene [22].
The predictive value of the selected miRNAs for hypertension
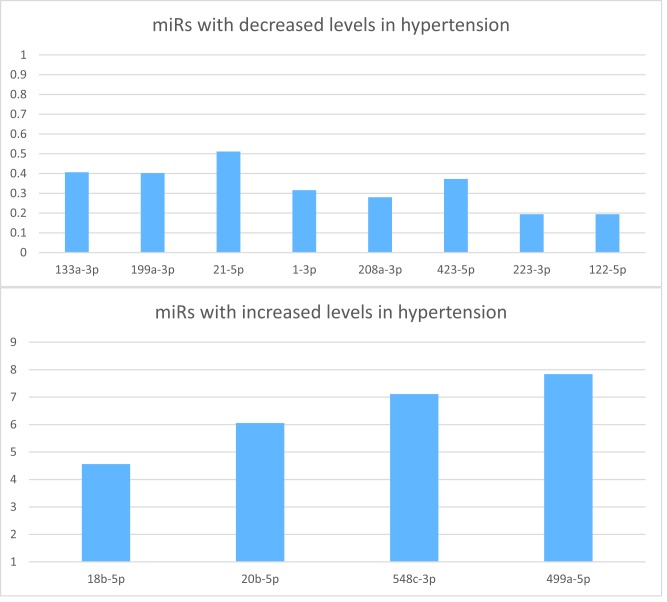
Significant negative correlations of miR-133a-3p (r: 0.175, P=0.031), miR-199a-3p (r: 0.293, P<0.001), miR-1-3p (r: 0.276, P=0.001), miR-208a-3p (r: 0.299, P<0.001), miR-423-5p (r: 0.299, P<0.001), miR-223-3p (r: 0.337, P<0.001), miR-122-5p (r: 0.368, P<0.001) were observed with hypertension. (Table 2 and Figure 1A). In contrast, statistically increased levels of miR-18b-5p (r: −0.259, P=0.001), miR-20b-5p (r: −0.306, P=0.001), miR-548c-3p (r: −0.231, P=0.007), miR-499a-5p (r: −0.244, P=0.005) were observed in hypertension patients (Table 2 and Figure 1B). The comparative ΔCt levels of miRNAs between normal group and hypertension group were shown in Supplementary Table S1.
Table 2. The correlation of levels of miRNAs with hypertension.
| MiRNA | RQ | Pearson correlation | P-value |
|---|---|---|---|
| 1-5p | 0.582 | 0.132 | 0.094 |
| 133a-3p | 0.406 | 0.175 | 0.031 |
| 23a-5p | 0.544 | 0.140 | 0.124 |
| 199a-3p | 0.403 | 0.293 | <0.001 |
| 20b-3p | 1.964 | −0.136 | 0.111 |
| 155-5p | 0.559 | 0.141 | 0.084 |
| 195-5p | 0.716 | 0.094 | 0.231 |
| 21-5p | 0.511 | 0.193 | 0.013 |
| 320a | 0.782 | 0.073 | 0.376 |
| 1-3p | 0.316 | 0.276 | 0.001 |
| 208a-3p | 0.280 | 0.299 | <0.001 |
| 423-5p | 0.372 | 0.299 | <0.001 |
| 106b-5p | 0.940 | 0.015 | 0.852 |
| 21-3p | 0.706 | 0.090 | 0.291 |
| 23a-3p | 0.542 | 0.153 | 0.053 |
| 126-5p | 0.578 | 0.138 | 0.107 |
| 133b | 0.595 | 0.096 | 0.235 |
| 675-3p | 2.035 | −0.193 | 0.020 |
| 223-3p | 0.194 | 0.337 | <0.001 |
| Let-7i-5p | 0.768 | 0.066 | 0.479 |
| 208b | 1.024 | −0.005 | 0.952 |
| 19b-3p | 1.253 | −0.064 | 0.952 |
| 122-5p | 0.194 | 0.368 | <0.001 |
| 18b-5p | 4.562 | −0.259 | 0.001 |
| 20b-5p | 6.057 | −0.306 | 0.001 |
| 548c-3p | 7.109 | −0.231 | 0.007 |
| 499a-5p | 7.838 | −0.244 | 0.005 |
Figue 1. 2 ΔΔCt of the selected miRNAs.
ΔΔCt of the selected miRNAs.
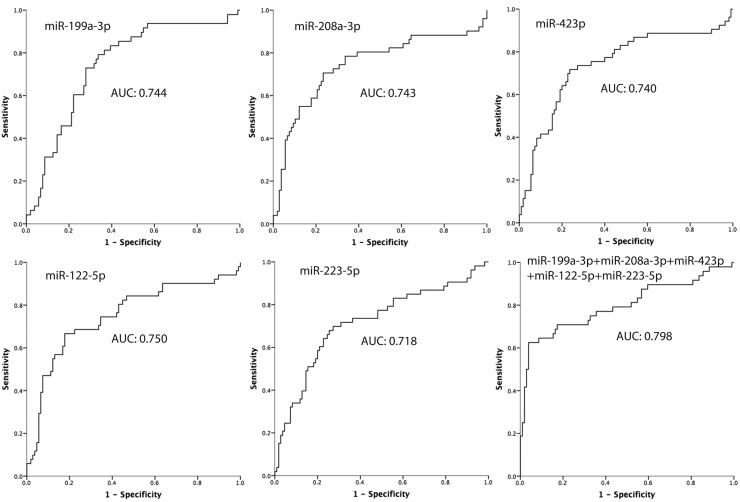
After identification of those miRNAs that were differentially expressed in a discovery phase, we further evaluated the diagnostic performances of those selected miRNAs by receiver operating characteristic (ROC) analysis. MiR-122-5p (area under curve (AUC): 0.750), miR-199a-3p (AUC: 0.744), miR-208a-3p (AUC: 0.743), miR-423-5p (AUC: 0.740), miR-223-5p (AUC: 0.718) showed better performance than others (Figure 2).
Figure 2. ROC curve for the first four miRNAs which have a good prediction ability for hypertension.
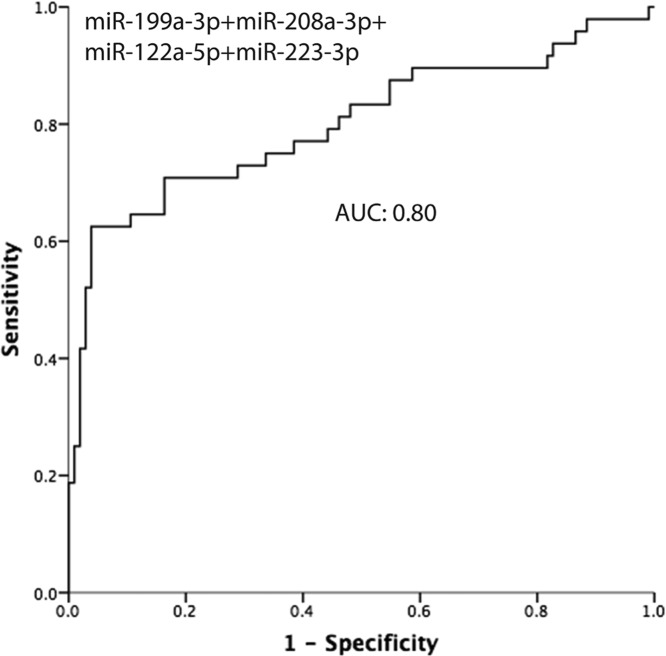
A combined analysis using those five miRNAs were used to explore the value of the combination, and the AUC was 0.798 (Figure 2). We also calculated the AUC of the combined analysis using the four from the five miRNAs, and the best performance was the combination of miR-199a-3p, miR-208a-3p, miR-122-5p, and miR-223-3p (AUC: 0.80) (Figure 3).
Figure 3. The predictive value of the combination of miR-199a-3p, miR-208a-3p, miR-122-5p, and miR-223-3p.
Correlation of the selected miRNAs with the biochemical indexes
Several miRNAs not only showed significant correlation with hypertension, but also had high association with indicators reflecting myocardial injury and cardiac function. For myocardial injury, ten of the selected miRNAs were highly related with cTNT, five of which were significantly associated with both hypertension and myocardial injury (miR-1-3p (r: 0.238, P:0.003), miR-208a-3p (r: −0.169, P:0.033), miR-20b-5p (r: −0.328, P<0.001), miR-548c-3p (r: −0.175, P=0.042), miR-499a-5p (r: −0.208, P=0.016). For cardiac function, miR-20b-5p was also related with BNP (r: −0.266, P=0.003) and LVEF (r: 0.219, P=0.015) (Table 3).
Table 3. Correlation of the selected miRNAs with the biochemical indexes.
| Indicator | miRNA | Pearson correlation | P-value |
|---|---|---|---|
| BNP | miR-1-3p | 0.231 | 0.004 |
| miR-20b-5p | −0.266 | 0.003 | |
| miR-548c-3p | −0.207 | 0.016 | |
| miR-499a-5p | −0.261 | 0.002 | |
| cTNT | miR-1-3p | 0.238 | 0.003 |
| miR-208a-3p | −0.169 | 0.033 | |
| miR-20b-5p | −0.328 | <0.001 | |
| miR-548c-3p | −0.175 | 0.042 | |
| miR-499a-5p | −0.208 | 0.016 | |
| LVEF | miR-208a-3p | −0.212 | 0.007 |
| miR-20b-5p | 0.219 | 0.015 | |
| miR-548c-3p | 0.189 | 0.028 |
Targetted genes and pathways for the four candidate miRNAs
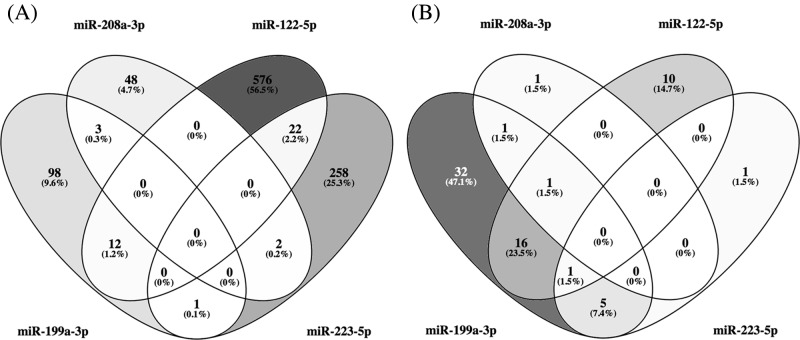
Using miRWALK2.0 software, 1060 predicted genes were identified to be targetted by the four candidate miRNAs. The number of genes targetted by each of the miRNAs are shown in boxes in Figure 4A. Since many of the genes were targetted by multiple miRNAs, the total of unique genes was only 1020. The multiple color Venn diagram presented in Figure 4A showed the distribution of these genes according to the candidate miRNAs that targetted them.
Figure 4. GO and KEGG analysis for the four candidate miRNAs.
(A) Venn’s diagram of genes targetted by each of the four candidate miRNAs. (B) Venn’s diagram of pathways enriched with genes targetted by the four candidate miRNAs with P<0.05. Pathways related to cancer were eliminated.
To identify pathways enriched with these genes, a KEGG pathway analysis was performed. This analysis identified 94 pathways enriched with genes targetted by miR-199a-3p, miR-208a-3p, miR-122-5p, and miR-223-5p. Many of the pathways, however, overlapped, so in total there were 68 unique pathways enriched at statistical significance of P<0.05.
Figure 4B shows distribution of the enriched pathways as a Venn diagram according to the candidate miRNAs, and Table 3 provides the name of the pathways with information about degree of enrichment with targetted genes. There was no pathway enriched with genes targetted by all four miRNAs with statistical significance. Pathways of FoxO signaling were enriched with genes targetted by three miRNAs (miR-199a-3p, miR-208a-3p, miR-122-5p). Twenty five pathways were enriched with genes that were targetted by two miRNAs (15 pathways when excluded the cancer pathways). The remaining 45 pathways were enriched with genes targetted only by one miRNA. We further analyzed the pathways enriched with genes targetted by the combination of four miRNAs, the most statistically significant of which were focal adhesion (P<3.0 × 10−6), glioma (P<1.0 × 10−5), endocytosis (P<2.0 × 10−4), MAPK signaling pathway (P<4 × 10−4) (Table 4).
Table 4. KEGG pathways enriched by genes targetted by the four candidate miRNAs (P<0.05).
| miR-199a-3p | miR-208a-3p | miR-122-5p | miR-223-5p | ||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA(s) | KEGG pathways | Target genes | P-value | Target genes | P-value | Target genes | P-value | Target genes | P-value |
| All four miRNAs | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| miR-199a-3p, miR-122-5p, miR-223-5p | Glioma | 5 | 0.0014 | — | — | 7 | 0.017 | 5 | 0.014 |
| miR-199a-3p, miR-208a-3p, miR-122-5p | FoxO signaling pathway | 8 | <6.0 × 10−5 | 4 | 0.016 | 10 | 0.027 | — | — |
| miR-199a-3p, miR-208a-3p | Epstein–Barr virus infection | 7 | 0.003 | 4 | 0.04 | — | — | — | — |
| miR-199a-3p, miR-223-5p | Focal adhesion | 14 | <3.0 × 10−6 | — | — | — | — | 8 | 0.03 |
| Pathways in cancer | 15 | <8.0 × 10−7 | 12 | 0.009 | |||||
| Melanoma | 7 | <2.0 × 10−5 | 6 | 0.004 | |||||
| Prostate cancer | 5 | 0.004 | 6 | 0.010 | |||||
| Endocytosis | 6 | 0.04 | 8 | 0.020 | |||||
| miR-199a-3p, miR-122-5p | Proteoglycans in cancer | 12 | 2.0 × 10−9 | — | — | 14 | 0.01 | — | — |
| Renal cell carcinoma | 6 | 1.2 × 10−4 | 8 | 0.004 | |||||
| Pancreatic cancer | 6 | 1.2 × 10−4 | 9 | 0.001 | |||||
| Sphingolipid signaling pathway | 7 | 2.6 × 10−4 | 10 | 0.01 | |||||
| Osteoclast differentiation | 7 | 4.2 × 10−4 | 12 | 0.003 | |||||
| Central carbon metabolism in cancer | 5 | <0.002 | 7 | 0.02 | |||||
| Toxoplasmosis | 6 | <0.002 | 9 | 0.03 | |||||
| Choline metabolism in cancer | 5 | 0.07 | 8 | 0.04 | |||||
| Insulin resistance | 5 | 0.009 | 10 | 0.007 | |||||
| Colorectal cancer | 4 | 0.01 | 8 | 0.004 | |||||
| Neurotrophin signaling pathway | 5 | 0.01 | 9 | 0.04 | |||||
| Epithelial cell signaling in Helicobacter pylori infection | 4 | 0.01 | 7 | 0.02 | |||||
| Pertussis | 4 | 0.01 | 7 | 0.03 | |||||
| Insulin signaling pathway | 5 | 0.02 | 11 | 0.01 | |||||
| Hepatitis B | 5 | 0.02 | 10 | 0.04 | |||||
| Tuberculosis | 5 | 0.04 | 15 | 0.002 | |||||
Discussion
Our study comprehensively examined the performance of those previously reported miRNAs in a same population. Our study showed that the combination of 199a-3p, 208a-3p, 122-5p, and 223-3p (AUC: 0.80) has a satisfactory diagnostic performance. Besides, some of those miRNAs also have correlation with myocardial injury (miR-1-3p, miR-208a-3p, miR-20b-5p, miR-548c-3p, miR-499a-5p), cardiac function (miR-20b-5p). Bioinformatics analysis showed that these four candidate miRNAs target the expression of more than 1020 genes that may impact 68 KEGG pathways. Although no pathway enriched with genes targetted by all four miRNAs with a statistical significance, pathways of FoxO signaling was enriched with genes targetted by three miRNAs (miR-199a-3p, miR-208a-3p, and miR-122-5p, respectively). Overall, our study identified four circulating miRNAs which target thousands of genes and many dozens of pathways. The interpretation and experimental validation of our findings creates a formidable challenge considering that our study measured circulating miRNAs and not intracellular in humans. Furthermore, not all human miRNAs are present in animals. Following this, we discuss our findings in the context of the limited literature regarding the pathways and miRNAs involved.
FoxO signaling pathway contains 132 genes/proteins, and in our study, 10 of these genes were predicted to be targetted by miR-122-5p, eight and four of these genes targetted by miR-199a-3p and miR-208a-3p, respectively. This pathway regulates the expression of genes in cellular physiological events including apoptosis, cell-cycle control, glucose metabolism, oxidative stress resistance, and longevity. Especially, FoxO signaling pathway was reported to play an important role in both vascular smooth muscle cells and endothelial cells, mediating the process of cell proliferation [23], vascular homeostasis [24], and age-related vascular changes [25]. Savai et al. [26] reported that FoxO1 in pulmonary artery smooth muscle cells (PASMCs) are a critical integrator of multiple signaling pathways driving pulmonary hypertension (PH), and reconstitution of FoxO1 activity offers a potential therapeutic option for PH. In addition, expression of p-FoxO/FoxO was elevated in both in spontaneously hypertensive rat (SHR), indicating the important role of FoxO signaling pathway in hypertension.
Except for FoxO signaling pathway, there were 15 pathways enriched with genes that were targetted by two miRNAs (excluding the cancer pathway). Epstein–Barr virus infection pathways targetted by both miR-199a-3p and miR-208a-3p, have been reported to be associated with secondary pulmonary arterial hypertension [27], while the mechanism remains to be discovered. Focal adhesion pathway plays an essential role in important biological processes including cell motility, cell proliferation, cell differentiation, regulation of gene expression, and cell survival. And total focal adhesion kinase was proved to be increased in renovascular hypertension, indicating the role of focal adhesion pathway in hypertension [28]. Endocytosis is a mechanism for cells to remove ligands, nutrients, and plasma membrane (PM) proteins, and lipids from the cell surface, bringing them into the cell interior and was associated with portal hypertension [29].
For pathways targetted by both miR-199a-3p, miR-122-5p, sphingolipid signaling pathway (targetted by) is known to have second messenger functions in a variety of cellular signaling pathways, and recently sphingosine-1-phosphate receptor 1 signaling was proved to regulate blood flow and pressure [30]. Insulin resistance and insulin signaling pathway have been suggested to be associated with hypertension in both clinical (metabolic syndrome) and basic trials. However, little has been done to identify the association of osteoclast differentiation and neurotrophin signaling pathway with hypertension. There were also three ways participating in infection, such as toxoplasmosis [31], pertussis [32], epithelial cell signaling in Helicobacter pylori infection [33], which have been proved to be related with hypertension, while pathways involving hepatitis B and tuberculosis have not been proved to be associated with hypertension. However, mechanisms of those pathways interacted with hypertension remains to be investigated.
In contrast with the results of our pathway analysis, which showed that those circulating miRNAs might target a large number of genes in multiple pathways, many of the previous publications aimed on studying single miRNAs and focussed on limited downstream effects. For example, studies on miR-223 have focussed its impact on the regulation of specific downstream gene target. miR-223 was down-regulated in human PAH lungs, in both the right heart and lungs from rodent models of PH. Down-regulation of miR-223 triggers PARP-1 and insulin-like growth factor 1 receptor (IGF-1R) overexpression, subsequent pathologic DNA damage repair, and increased proliferation [16,17]. The relationship between miRNAs and target genes in vivo is a complex network. Thus far, very few studies looked into the systematic interactions between miRNA and gene signaling pathways. In the present study, we performed pathway enrichment analysis and have identified top canonical pathways highly relevant to the pathogenesis of hypertension, including the FoxO signaling pathway.
Our study showed that in hypertension patients, the levels of miR-320a [34], miR-199a-3p [35,36] were lower than control group, while the levels of miR-20b-3p/5p [37], miR-208b [38], miR-499a-5p [38] increased, which kept the same with previous studies, indicating that the expression levels of these miRNAs were stable. However, the trend of many selected miRNAs in hypertension remains controversial in different studies, such as miR-1-3p/5p [39,40], miR-23a-3p/5p [34,35], miR-155-5p [41–43], miR-195-5p [34,43,44], miR-133a-3p [36,39], miR-223-3p [16,45], miR-122-5p [43,46]. In addition, the expression level of miR-423-5p and miR-106b-5p in our study was contrary to the previous reports [37]. There are several reasons that can explain these inconsistencies. First, the population in the previous studies include hypertension of different types (such as gestational hypertension, PH, renal vascular hypertension) or hypertension combined with different complication, which can effect the levels of hypertension. Second, miRNAs have many pathways to participate in the pathophysiology of hypertension, and other factors may affect the levels of hypertension (such as dietary structure, coffee consumption, exercise, pathogenesis, and so on). Those results suggested that the pathogenesis and complication of hypertension and the pathways of miRNAs should be taken into consideration when we identify miRNA as biomarkers for hypertension.
Our study found the best four biomarkers for hypertension (miR-122-5p, miR-199a-3p, 208a-3p, miR-423-5p, and miR-223-5p), and identified the best combination of those miRNAs (199a-3p, 208a-3p, 122-5p, and 223-3p), giving more information on miRNAs as biomarkers for hypertension. Furthermore, four of those miRNAs (miR-1-3p, miR-20b-5p, miR-548c-3p, miR-499a-5p) were significantly correlated with cTNT, indicating that those miRNAs maybe used as indicators for myocardial infarction. Those five miRNAs have also been shown to be correlated with myocardial infarction in previous studies [47]. In addition, miR-20b-5p was also related with BNP (relative risk (RR): −0.266, P=0.003) and LVEF (RR: 0.219, P=0.015), indicating the association between miR-20b-5p and cardiac function. In consistence with our results, miR-20b-5p had been proved to be significantly increased in response to hypertension-induced heart failure, and correlated with the levels of BNP.
The limitations of our study are: (i) because of amounts of miRNAs have been reported to be related with hypertension, we can only select those important, representative miRNAs instead of detecting them one by one. (ii) Considering that the the expressing levels of miRNAs may be different in different types of hypertension, we did not carry out a subgroup analysis according to the types of hypertension.
Conclusion
The combination of 199a-3p, 208a-3p, 122-5p, and 223-3p has a good diagnostic performance for hypertension, and multitudes of possible mechanisms/pathways through which dysregulation of these miRNAs may impact risk of hypertension.
Clinical perspectives
Few studies have comprehensively examined the performances of those reported miRNAs as biomarkers for hypertension and identify the genes and pathways targetted by these miRNAs.
The combination of 199a-3p, 208a-3p, 122-5p, and 223-3p has a good diagnostic performance for hypertension, and multitudes of possible mechanisms/pathways through which dysregulation of these miRNAs may impact risk of hypertension.
Our results give some information on the systematic interactions between miRNA and gene signaling pathways and provide some clues for mechanism research on miRNA and hypertension.
Supporting information
Supplemental Table 1. Levels of delta ct of microRNAs between normal group and hypertension group.
Abbreviations
- AUC
area under curve
- BNP
brain natriuretic peptide
- CN-HF
China National Heart Failure Registry
- Cq
quantification cycle
- Ct
threshold cycle
- cTNT
cardiac Troponin T
- FoxO
The forkhead box O
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LVEF
left ventricular ejection fraction
- MAPK
mitogen activated protein kinase
- PAH
pulmonary arterial hypertension
- PH
pulmonary hypertension
- PARP-1
poly(ADP-ribose) polymerase 1
- ROC
receiver operating characteristic
- RR
relative risk
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- 5S
5S ribosomal RNA
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 81730009, 81521001, 31430039]; (to Y.Z.) and the Shanghai Natural Science Foundation [grant number 18ZR1406900].
Author contribution
X.W., X.Z., J.Y., and Y.Z. were resonsible for the conception and design. Y.Z. and J.Y. provided administrative support. X.Z., J.P., X.D., Y.S., and C.Y. were responsible for provision of study materials or patients. X.Z., X.W., J.P., X.D., Y.S., C.Y., and J.Y. were responsible for collection and assembly of data. X.W., J.Y., Y.Z. were responsible for data analysis and interpretation. All the authors participated in manuscript writing and in final approval of manuscript.
References
- 1.Lawes C.M.M., Hoorn S.V. and Rodgers A. (2008) Global burden of blood-pressure-related disease, 2001. Lancet 371, 1513–1518 10.1016/S0140-6736(08)60655-8 [DOI] [PubMed] [Google Scholar]
- 2.Conen D.B.F. (2008) Noninvasive 24-h ambulatory blood pressure and cardiovascular disease: a systematic review and meta-analysis. J. Hypertens. 26, 1290–1299 10.1097/HJH.0b013e3282f97854 [DOI] [PubMed] [Google Scholar]
- 3.Harjutsalo V. and Groop P.H. (2014) Epidemiology and risk factors for diabetic kidney disease. Adv. Chronic Kidney Dis. 21, 260–266 10.1053/j.ackd.2014.03.009 [DOI] [PubMed] [Google Scholar]
- 4.Vidal-Petiot E., Ford I., Greenlaw N., Ferrari R., Fox K.M., Tardif J.-C. et al. (2016) Cardiovascular event rates and mortality according to achieved systolic and diastolic blood pressure in patients with stable coronary artery disease: an international cohort study. Lancet 388, 2142–2152 10.1016/S0140-6736(16)31326-5 [DOI] [PubMed] [Google Scholar]
- 5.Mills K.T., Bundy J.D., Kelly T.N., Reed J.E., Kearney P.M., Reynolds K. et al. (2016) Global disparities of hypertension prevalence and control:a systematic analysis of population-based studies from 90 countries. Circulation 134, 441–450 10.1161/CIRCULATIONAHA.115.018912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whelton P.K., Carey R.M., Aronow W.S., Casey D.E. Jr, Collins K.J., Dennison Himmelfarb C. et al. (2018) 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension, 71, 1269–1324, 10.1161/HYP.0000000000000066 [DOI] [PubMed] [Google Scholar]
- 7.Bartel D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauersachs J. and Thum T. (2011) Biogenesis and regulation of cardiovascular microRNAs. Circ. Res. 109, 334–347 10.1161/CIRCRESAHA.110.228676 [DOI] [PubMed] [Google Scholar]
- 9.Small E.M. and Olson E. (2011) Pervasive roles of microRNAs in cardiovascular biology. Nature 469, 336–342 10.1038/nature09783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrian A.D., Schriver S.D. and Prewitt R.L. (2001) Role of angiotensin II and free radicals in blood pressure regulation in a rat model of renal hypertension. Hypertension 38, 361–366 10.1161/01.HYP.38.3.361 [DOI] [PubMed] [Google Scholar]
- 11.Wang B., Li C., Huai R. and Qu Z. (2015) Overexpression of ANO1/TMEM16A, an arterial Ca2+-activated Cl- channel, contributes to spontaneous hypertension. J. Mol. Cell Cardiol. 82, 22–32 10.1016/j.yjmcc.2015.02.020 [DOI] [PubMed] [Google Scholar]
- 12.Trudu M., Janas S., Lanzani C., Debaix H., Schaeffer C., Ikehata M. et al. (2013) Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat. Med. 19, 1655–1660 10.1038/nm.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heggermont W.A. and Heymans S. (2012) MicroRNAs are involved in end-organ damage during hypertension. Hypertension 60, 1088–1093 10.1161/HYPERTENSIONAHA.111.187104 [DOI] [PubMed] [Google Scholar]
- 14.Caruso P., Dunmore B.J., Schlosser K., Schoors S., Dos Santos C., Perez-Iratxeta C. et al. (2017) Identification of microrna-124 as a major regulator of enhanced endothelial cell glycolysis in pulmonary arterial hypertension via ptbp1 (polypyrimidine tract binding protein) and pyruvate kinase m2. Circulation 136, 2451–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romaine S.P., Charchar F.J., Samani N.J. and Tomaszewski M. (2016) Circulating microRNAs and hypertension–from new insights into blood pressure regulation to biomarkers of cardiovascular risk. Curr. Opin. Pharmacol. 27, 1–7 10.1016/j.coph.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 16.Meloche J., Le Guen M., Potus F., Vinck J., Ranchoux B., Johnson I. et al. (2015) miR-223 reverses experimental pulmonary arterial hypertension. Am. J. Physiol. Cell Physiol. 309, C363–C372 10.1152/ajpcell.00149.2015 [DOI] [PubMed] [Google Scholar]
- 17.Shi L., Kojonazarov B., Elgheznawy A., Popp R., Dahal B.K., Böhm M. et al. (2016) miR-223-IGF-IR signalling in hypoxia- and load-induced right-ventricular failure: a novel therapeutic approach. Cardiovasc. Res. 111, 184–193 10.1093/cvr/cvw065 [DOI] [PubMed] [Google Scholar]
- 18.Dweep H. and Gretz N. (2015) miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods 12, 697 10.1038/nmeth.3485 [DOI] [PubMed] [Google Scholar]
- 19.Dweep H., Sticht C., Pandey P. and Gretz N. (2011) miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847 10.1016/j.jbi.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 20.Huang da W., Sherman B.T. and Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 21.Huang da W., Sherman B.T. and Lempicki R.A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Zhan X., Yuan J., Wu J., Deng X., Peng J. et al. (2018) Evaluation of the performance of serum miRNAs as normalizers in microRNA studies focused on cardiovascular disease. J. Thorac. Dis., 10, 2599–2607, 10.21037/jtd.2018.04.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahajan S.G., Fender A.C., Meyer-Kirchrath J., Kurt M., Barth M., Sagban T.A. et al. (2012) A novel function of FoxO transcription factors in thrombin-stimulated vascular smooth muscle cell proliferation. Thromb. Haemost. 108, 148–158 10.1160/TH11-11-0756 [DOI] [PubMed] [Google Scholar]
- 24.Paik J.H. (2006) FOXOs in the maintenance of vascular homoeostasis. Biochem. Soc. Trans. 34, 731–734 10.1042/BST0340731 [DOI] [PubMed] [Google Scholar]
- 25.Oellerich M.F. and Potente M. (2012) FOXOs and sirtuins in vascular growth, maintenance, and aging. Circ. Res. 110, 1238–1251 10.1161/CIRCRESAHA.111.246488 [DOI] [PubMed] [Google Scholar]
- 26.Savai R., Al-Tamari H.M., Sedding D., Kojonazarov B., Muecke C., Teske R. et al. (2014) Pro-proliferative and inflammatory signaling converge on FoxO1 transcription factor in pulmonary hypertension. Nat. Med. 20, 1289–1300 10.1038/nm.3695 [DOI] [PubMed] [Google Scholar]
- 27.Fukuda Y., Momoi N., Akaihata M., Nagasawa K., Mitomo M., Aoyagi Y. et al. (2015) Pulmonary arterial hypertension associated with chronic active Epstein-Barr virus infection. Pediatr. Int. 57, 731–734 10.1111/ped.12578 [DOI] [PubMed] [Google Scholar]
- 28.Pereira S.C., Parente J.M., Belo V.A., Mendes A.S., Gonzaga N.A., do Vale G.T. et al. (2018) Quercetin decreases the activity of matrix metalloproteinase-2 and ameliorates vascular remodeling in renovascular hypertension. Atherosclerosis 270, 146–153 10.1016/j.atherosclerosis.2018.01.031 [DOI] [PubMed] [Google Scholar]
- 29.Sandahl T.D., Støy S.H., Laursen T.L., Rødgaard-Hansen S., Møller H.J., Møller S. et al. (2017) The soluble mannose receptor (sMR) is elevated in alcoholic liver disease and associated with disease severity, portal hypertension, and mortality in cirrhosis patients. PLoS ONE 12, e0189345 10.1371/journal.pone.0189345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantalupo A., Gargiulo A., Dautaj E., Liu C., Zhang Y., Hla T. et al. (2017) S1PR1 (sphingosine-1-phosphate receptor 1) signaling regulates blood flow and pressure. Hypertension 70, 426–434 10.1161/HYPERTENSIONAHA.117.09088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westfall A.C., Lauer A.K., Suhler E.B. and Rosenbaum J.T. (2005) Toxoplasmosis retinochoroiditis and elevated intraocular pressure: a retrospective study. J. Glaucoma 14, 3–10 10.1097/01.ijg.0000146373.51495.c1 [DOI] [PubMed] [Google Scholar]
- 32.Domico M., Ridout D., MacLaren G., Barbaro R., Annich G., Schlapbach L.J. et al. (2018) Extracorporeal membrane oxygenation for pertussis: predictors of outcome including pulmonary hypertension and leukodepletion. Pediatr. Crit. Care Med. 19, 254–261 10.1097/PCC.0000000000001454 [DOI] [PubMed] [Google Scholar]
- 33.Wan Z., Hu L., Hu M., Lei X., Huang Y. and Lv Y. (2018) Helicobacter pylori infection and prevalence of high blood pressure among Chinese adults. J. Hum. Hypertens. 32, 158–164 10.1038/s41371-017-0028-8 [DOI] [PubMed] [Google Scholar]
- 34.Karolina D.S., Tavintharan S., Armugam A., Sepramaniam S., Pek S.L., Wong M.T. et al. (2012) Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 97, E2271–E2276 10.1210/jc.2012-1996 [DOI] [PubMed] [Google Scholar]
- 35.Sarrion I., Milian L., Juan G., Ramon M., Furest I., Carda C. et al. (2015) Role of circulating miRNAs as biomarkers in idiopathic pulmonary arterial hypertension: possible relevance of miR-23a. Oxid. Med. Cell Longev. 2015, 792846 10.1155/2015/792846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hromadnikova I., Kotlabova K., Hympanova L. and Krofta L. (2016) Gestational hypertension, preeclampsia and intrauterine growth restriction induce dysregulation of cardiovascular and cerebrovascular disease associated microRNAs in maternal whole peripheral blood. Thromb. Res. 137, 126–140 10.1016/j.thromres.2015.11.032 [DOI] [PubMed] [Google Scholar]
- 37.Dickinson B.A., Semus H.M., Montgomery R.L., Stack C., Latimer P.A., Lewton S.M. et al. (2013) Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur. J. Heart Fail. 15, 650–659 10.1093/eurjhf/hft018 [DOI] [PubMed] [Google Scholar]
- 38.Kontaraki J.E., Marketou M.E., Parthenakis F.I., Maragkoudakis S., Zacharis E.A., Petousis S. et al. (2015) Hypertrophic and antihypertrophic microRNA levels in peripheral blood mononuclear cells and their relationship to left ventricular hypertrophy in patients with essential hypertension. J. Am. Soc. Hypertens 9, 802–810 10.1016/j.jash.2015.07.013 [DOI] [PubMed] [Google Scholar]
- 39.Kontaraki J.E., Marketou M.E., Zacharis E.A., Parthenakis F.I. and Vardas P.E. (2014) Differential expression of vascular smooth muscle-modulating microRNAs in human peripheral blood mononuclear cells: novel targets in essential hypertension. J. Hum. Hypertens. 28, 510–516 10.1038/jhh.2013.117 [DOI] [PubMed] [Google Scholar]
- 40.Markus B., Grote K., Worsch M., Parviz B., Boening A., Schieffer B. et al. (2016) Differential expression of micrornas in endarterectomy specimens taken from patients with asymptomatic and symptomatic carotid plaques. PLoS ONE 11, e0161632 10.1371/journal.pone.0161632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DuPont J.J., McCurley A., Davel A.P., McCarthy J., Bender S.B., Hong K. et al. (2016) Vascular mineralocorticoid receptor regulates microRNA-155 to promote vasoconstriction and rising blood pressure with aging. JCI Insight 1, e88942 10.1172/jci.insight.88942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park M.Y., Herrmann S.M., Saad A., Widmer R.J., Tang H., Zhu X.Y. et al. (2015) Circulating and renal vein levels of microRNAs in patients with renal artery stenosis. Nephrol. Dial. Transplant. 30, 480–490 10.1093/ndt/gfu341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cengiz M., Karatas O.F., Koparir E., Yavuzer S., Ali C., Yavuzer H. et al. (2015) Differential expression of hypertension-associated microRNAs in the plasma of patients with white coat hypertension. Medicine 94, e693 10.1097/MD.0000000000000693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hromadnikova I., Kotlabova K., Ivankova K., Vedmetskaya Y. and Krofta L. (2017) Profiling of cardiovascular and cerebrovascular disease associated microRNA expression in umbilical cord blood in gestational hypertension, preeclampsia and fetal growth restriction. Int. J. Cardiol. 249, 402–409 10.1016/j.ijcard.2017.07.045 [DOI] [PubMed] [Google Scholar]
- 45.Gubrij I.B., Pangle A.K., Pang L. and Johnson L.G. (2016) Reversal of microRNA dysregulation in an animal model of pulmonary hypertension. PLoS ONE 11, e0147827 10.1371/journal.pone.0147827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hromadnikova I., Kotlabova K., Hympanova L. and Krofta L. (2015) Cardiovascular and cerebrovascular disease associated microRNAs are dysregulated in placental tissues affected with gestational hypertension, preeclampsia and intrauterine growth restriction. PLoS ONE 10, e0138383 10.1371/journal.pone.0138383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cortez-Dias N., Costa M.C., Carrilho-Ferreira P., Silva D., Jorge C., Calisto C. et al. (2016) Circulating miR-122-5p/miR-133b ratio is a specific early prognostic biomarker in acute myocardial infarction. Circ. J. 80, 2183–2191 10.1253/circj.CJ-16-0568 [DOI] [PubMed] [Google Scholar]