Summary
Modern research technologies, including DNA, protein, and antibody microarrays identify a steadily growing number of clues that are useful in molecular disease classification, drug development, and the prediction of response to treatment. Subsequent validation of the clinical importance of such candidate genes or proteins requires large-scale analysis of human tissues. To date, this analysis constitutes an important bottleneck in the process of discovery because tissue analysis by the conventional slide-by-slide strategy is slow and expensive. To overcome these limitations, tissue microarray (TMA) technology has been developed. TMA allows for the simultaneous analysis of up to 1,000 tissue samples in a single experiment, using all types of in-situ analyses including immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), and RNA in situ hybridization (RNA-ISH). TMA technology has the potential to greatly facilitate the translation of basic research into clinical practice. Potential applications include the establishment of associations between molecular changes and clinical endpoints, testing of potential therapeutic targets using tissue samples from specific cancer patients, standardization of molecular detection of targets, and rapid translation of results from cell lines and animal models to human cancer. Because of its beneficial economic aspects and ability to differentiate ethnic differences in tumor biology, TMA applications may become particularly important in developing countries.
The sequence of the human genome has recently been unraveled.1,2 At the same time, new methods have been introduced that allow for a comprehensive expression analysis of the thousands of genes on the RNA or protein level in a given tissue. As a result, a rapidly increasing number of genes have been identified that may play a role in cancer or other diseases. It is hoped that these findings will eventually lead to clinically useful applications. However, the number of potentially useful candidate genes poses significant problems in the evaluation of their potential utility. Large-scale functional gene analyses and molecular epidemiologic studies are needed for this purpose. Collecting data on molecular epidemiology is often a significant challenge for basic researchers due to limited availability of human tissues with clinical information. Tissue microarray (TMA) technology greatly facilitates large-scale tissue analyses. In this method, up to 1,000 different minute tissue samples can be brought onto one microscope glass slide, and be simultaneously analyzed by all types of in-situ methods.
Tissue microarray methodology
In tissue microarray (TMA) methodology, minute tissue cylinders (typical diameter: 0.6 mm) are taken from various primary tumor blocks (the “donor” blocks) and subsequently assembled in an array-like format into one empty “recipient” block (Figure 1). Simple and inexpensive arraying instruments can be used for this purpose. Regular microtomes can be used to cut sections from these TMA blocks. Virtually all types of in situ tissue analyses can be done on TMA sections without significant modifications of existing protocols. These include immunohistochemistry (IHC) for protein detection, fluorescence in situ hybridization (FISH) for detection of copy number changes of genes or chromosomes as well as RNA in situ hybridization (RNA-ISH) for quantification of RNA expression levels (Figure 2). Importantly, the small diameter of the specimen taken from each donor block minimizes the damage to patient tissue that might be needed for additional diagnostic procedures. This allows for extensive use of diagnostic tissue samples for research purposes, and at the same time retains sufficient tissue material for additional diagnostic procedures if needed.
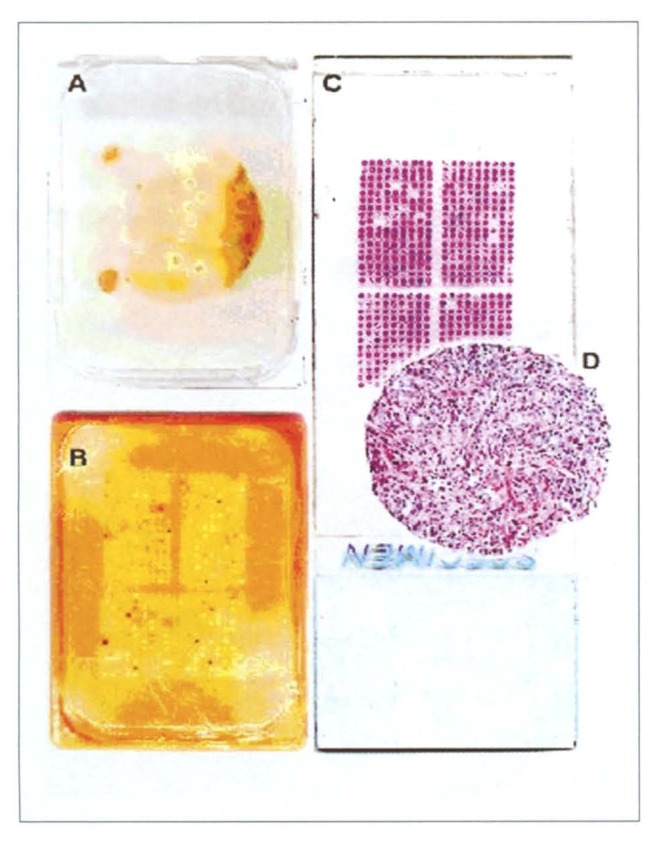
Figure 1.
Tissue microarray (TMA) manufacturing. A) Donor block from which several 0.6 mm tissue cores have been removed. Note that the original tissue block remains fully interpretable. B) Recipient block with the completed TMA. C) Hematoxylin & eosin stained tissue section of the TMA. D) Magnification of a tissue spot.
Figure 2.
Tissue microarray applications. Overview of TMA sections investigated by A) immunohistochemistry and B) RNA-ISH. C) Tissue spot showing focal expression (black granular staining) of BTG2 mRNA as investigated by RNA-ISH. The TMA section was coated with a photographic emulsion for detection of a radioactively labeled antisense-RNA probe. D) Tissue spot from a breast cancer array showing strong (3+) membranous staining of the HER2 protein by immunohistochemistry. E) Sector from the same tissue spot as in D) analyzed by FISH, using probes detecting HER2 gene amplification (clusters of red signals) and centromere 17 copy numbers (green signals).
TMAs are optimally suited for large-scale expression profiling projects. More than 400 punches can theoretically be obtained from a tissue with a diameter of 25 mm if the entire block can be used for research. All these cores can be placed on 400 different copies of a particular TMA block. As 200 to 300 sections can be taken per TMA block, between 80 000 and 120 000 analyses could be done from a set of TMAs. This means that TMAs would allow the expression analysis of all human genes in one set of tissues. TMAs can be made not only from paraffin embedded tissues, but also from frozen tissue samples (Figure 3).3,4 Frozen TMAs are well suited for RNA-ISH detection of low-level transcripts, or for IHC if antibodies that work on formalin fixed tissues are unavailable.
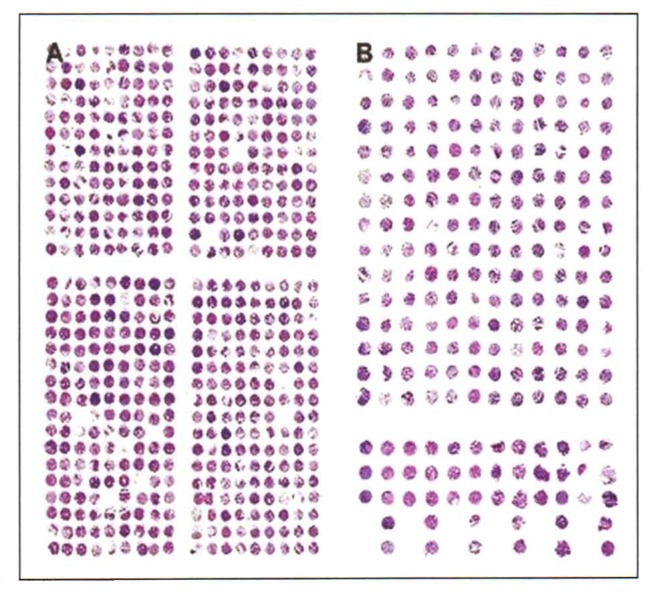
Figure 3.
Tissue microarrays from paraffin (A) and frozen tissues (B). TMAs from frozen tissues become more irregular and distorted than TMAs from formalin fixed material because the commercially available arrayers have not been designed for frozen array making. Therefore a larger space between samples is recommended (e.g., 1 mm).
General applications
TMAs can be used for many different applications. Most research requiring in-situ tissue analysis can be done in a TMA format. More than 150 publications using the TMA approach have now been published. Most have used TMAs in cancer research to investigate the prevalence of molecular changes and their associations with tumor progression and/or prognosis. However, even simple TMAs containing tissues without any clinical background information can be used for collecting significant new information on the epidemiology of molecular changes in different tumor types.
In one study a “multitumor” TMA was used to analyze 4788 different samples from more than 130 different tumor categories for cyclin E amplification and overexpression.5 More information on the importance of a specific change in one particular tumor type can be collected by using better-characterized tissues with information on the pathology and/or the clinical outcome. A multitude of studies used TMAs to find associations between molecular alterations and tumor phenotype. For example, associations between cyclin E,6 FGFR1, RAF1,7 MDM2 or CDK48 amplification or MAGE-A4 expression9 and stage and grade were found in urinary bladder cancer. CK7 and CK20 expression were associated with histologic grade in colorectal carcinoma.10 Several clinico-pathological associations were observed in prostate cancer studies. For example, IGFBP2 expression was related to a hormone-refractory state,11 and EIF3S3 amplification was linked to advanced tumor stage.12 TMAs were also successfully used to investigate potential heterogeneity between primary tumors and their metastases. Simon et al. previously used a TMA composed of paired primary tumors and metastases from 196 breast carcinomas to demonstrate a high concordance in the HER2 amplification/overexpression between primary tumors and their nodal metastases.13 Various molecular features were analyzed for their prognostic significance on TMAs with samples from patients with clinical follow up information, for example in bladder,6,8,14 breast,15–20 prostate,21–23 brain,24–26 liver,27 kidney,28 and colorectal tumors,29–31 Hodgkin’s lymphoma,32 and malignant melanoma.33 Studies using prognosis TMAs have yielded a considerable amount of novel information. For example, significant associations were found between 17q23 amplifications18 or COX2 expression15 and breast cancer prognosis, between TOP2A expression and prognosis in glioblastoma,24 between MYC and AIB1 expression and prognosis in hepatocellular carcinoma,27 and between IGFBP2 and prostate cancer prognosis.11
In all these studies, maximal standardization of the procedure is a major strongpoint of the TMA method. Very often all tissues analyzed are located on one glass section and treated under absolutely identical conditions, including reagent concentrations, temperature, pretreatment, antigen retrieval, slide age, and section thickness. This eliminates the many possible pitfalls of molecular diagnostics and assures maximal comparability of results within one study. We see great applicability of TMA technology not only to basic cancer research but also to anatomical pathology (Table 1).
Table 1.
Potential applications of tissue microarray technology.
|
Tissue microarray applications in developing countries
So far, TMA technology has predominantly been used in European and U.S. laboratories. However, the TMA method may have even greater importance in developing countries. Recently, studies have suggested significant genetic differences between cancers occurring in different ethnic groups. For example, significant differences were found in the pattern of gene amplification between European and Arabian breast cancers (Al Kuraya, unpublished), and between the frequency of HER2 and nm23 expression between British and Japanese gastric cancers.34 As the management and treatment of cancer patients is increasingly based on molecular tumor characteristics, ethnic variations may be of practical significance.
Currently, our knowledge of the prevalence and clinical significance of molecular tumor features is based on data collected on Western tumors. It now appears possible that not all conclusions drawn from these studies are applicable to patients from other ethnic backgrounds. Large-scale surveys on the molecular epidemiology of clinically important molecular changes are necessary to better define the dimension of ethnic differences in tumor biology. TMAs represent an ideal tool to rapidly determine the epidemiology of important molecular features in different ethnic groups. Once the necessary tissues have been collected and the TMAs manufactured, all these studies could easily be done in a short period of time.
Economical factors are another strong reason for using TMAs. Studies have confirmed that TMAs help to save reagents, manpower and money because one can investigate up to 1000 tumors in one glass slide section with the same amount of reagent that was previously necessary to analyze one tumor (in the large section format). So, in laboratories with limited financial resources one of the major advantages of TMA is the small amount of reagents needed for scientific studies. For example, more than 10 000 tumors have been analyzed in the University of Basel for amplification. If regular sections were used for this purpose, the reagent costs would have exceeded US$200,000 per 10,000 tumors. In the TMA setting, we stained 30 slides, containing 10 000 tumor sections for about $600 (Table 2).
Table 2.
Cost analysis of 10,000 tumors.
| Large section method | Tissue microarray method |
|---|---|
| 1 tumor per slide @ $20 per slide | 334 tumors per slide @ $20 per slide |
| 10 000 tumors = $200 000 | 10 000 tumors = $600 |
We hope that strong collaborative projects can be initiated in the future that involve research laboratories in different developing countries. Such collaborations are highly important to clarify whether patients from different ethnic backgrounds are equally prone to benefit from modern targeted cancer therapies. TMAs will be optimally suited to highlight such potentially important ethnic differences in a cost effective manner.
Is representativity a limitation of tissue microarray technology?
The most significant and obvious limitation to the TMA technique is the size of the arrayed tissues, which measure only 0.6 mm in diameter. Initially, the method was criticized because it was questioned whether such small tumor samples could be representative and give meaningful information for an entire tumor. To address this question, at least 20 studies have compared IHC findings on TMAs with their corresponding traditional “large” sections—the current gold standard for molecular tumor tissue analysis.16,20,26,28,32,35–49 A high level of concordance has been found in most of these studies.16,20,26,28,35–42,44,45,47,48 Some results suggest that two or three samples may provide more representative information than a single sample,38,41,44,45,48 and that adding more than four or five samples does not improve the concordance level to any important extent.38,44 However, it is necessary to understand that the general assumption of classical large sections, that they are representative of an entire tumor, may be wrong. In the optimal case, a “large” section will contain about 0.0018 cm3 of tumor tissue (3 cm × 2 cm × 3 μm). This is only about 1/7500 of 14 cm3, which represents the approximate volume of a tumor measuring 3 centimeter in diameter. Considering these numbers, the representativity problem is obviously much greater between the entire tumor and a traditional “large” section than between a TMA sample and a “large” section.
Rather than comparing results obtained on large sections with TMA sections, studies validating the TMA technique should determine whether associations between molecular features and tumor phenotype or clinical outcome can be found in a TMA setting. In fact, numerous studies have confirmed previously well-established associations between molecular findings and clinical outcome.18,28,39,50–53 For example, significant associations were found between estrogen or progesteron expression52 and HER-2 alterations18 and survival in breast cancer patients, between vimentin expression and prognosis in kidney cancer28 as well as between Ki67 labeling index and prognosis in urinary bladder cancer,39 soft tissue sarcoma,50 and Hurthle cell carcinoma.51 Studies that successfully used tissues from Hodgkin’s lymphoma for analyses on TMAs provide further evidence that highly heterogeneous tumors can be analyzed in a TMA format.32,37,47
Future developments
Currently, the availability of TMA sections is limited. TMAs that are available from commercial sources or from non-profit organizations are often small and have little clinical data attached. Academic sources that have large TMA repositories are overloaded with requests for collaboration and cannot meet all demands. However, the number of institutions with TMA facilities is increasing. Virtually all institutions dealing with tissue analyses will be using TMAs in the future. Also the quality of TMAs will improve as more are made from particularly precious tissue resources like rare tumor types at referral centers or tissues from patients included in clinical trials. In fact, multiple clinical trial groups are now implementing TMAs as a part of their trial protocols.
Automation of TMA analysis is another major need since manual TMA reading is cumbersome, slow and subjective. TMAs are optimally suited for automated IHC analysis because the most critical step for automation—the selection of the area to be analyzed—has already been accomplished. In a “low tech” approach, the automated analysis would more or less be limited to measurement of the total signal intensity per tissue spot. Although this approach cannot distinguish neoplastic from non-neoplastic epithelial cells or from stroma cells, our home-made system revealed the expected associations with outcome information in all cases. More sophisticated analyses could include multicolor fluorescent detection systems for IHC or intelligent pattern recognition software solutions. In contrast to TMA analysis, automation of tissue arraying devices has little importance because the availability of tissues and their pathological characterization, and not the arraying process itself, is the rate-limiting step in TMA manufacturing.
Large databases that include the results of in situ analyses obtained on large and well-documented TMA resources constitute another expected development. Such databases may not only contain the staining results of all tissues and related clinico-pathological information, but potentially images of each individually stained spot. The total size of such databases will be enormous, as they will include millions of images. Implementation is largely dependent on the development of suitable data storage solutions.
TMAs will significantly contribute to an accelerated transition of laboratory observations to clinical applications. Once a gene product is measurable by an in situ test, it is possible, in one examination, to analyze all normal tissues, and all different tumor types, as well as a variety of tissues from non-neoplastic diseases to immediately determine where the gene of interest might have clinical utility.
References
- 1.Venter JC, Adams MD, Myers EW, Li P, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Fejzo MS, Slamon DJ. Frozen tumor tissue microarray technology for analysis of tumor RNA, DNA, and proteins. Am J Pathol. 2001;159:1645–1650. doi: 10.1016/S0002-9440(10)63011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon R, Sauter G. Tissue microarrays for miniaturized high-throughput molecular profiling of tumors. Exp Hematol. 2002;30:1365–1372. doi: 10.1016/s0301-472x(02)00965-7. [DOI] [PubMed] [Google Scholar]
- 5.Andersen CL, Monni O, Wagner U, et al. A. High-throughput copy number analysis of 17q23 in 3520 tissue specimens by fluorescence in situ hybridization to tissue microarrays. Am J Pathol. 2002;161:73–79. doi: 10.1016/S0002-9440(10)64158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter J, Wagner U, Kononen J, Fijan A, et al. High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am J Pathol. 2000;157:787–794. doi: 10.1016/s0002-9440(10)64592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon R, Richter J, Wagner U, Fijan A, et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res. 2001;61:4514–4519. [PubMed] [Google Scholar]
- 8.Simon R, Struckmann K, Schraml P, et al. Amplification pattern of 12q13-q15 genes (MDM2, CDK4, GLI) in urinary bladder cancer. Oncogene. 2002;21:2476–2483. doi: 10.1038/sj.onc.1205304. [DOI] [PubMed] [Google Scholar]
- 9.Kocher T, Zheng M, Bolli M, Simon R, et al. Prognostic relevance of MAGE-A4 tumor antigen expression in transitional cell carcinoma of the urinary bladder: a tissue microarray study. Int J Cancer. 2002;100:702–705. doi: 10.1002/ijc.10540. [DOI] [PubMed] [Google Scholar]
- 10.Park SY, Kim HS, Hong EK, Kim WH. Expression of cytokeratins 7 and 20 in primary carcinomas of the stomach and colorectum and their value in the differential diagnosis of metastatic carcinomas to the ovary. Hum Pathol. 2002;33:1078–1085. doi: 10.1053/hupa.2002.129422. [DOI] [PubMed] [Google Scholar]
- 11.Bubendorf L, Kolmer M, Kononen J, Koivisto P, Mousses S, Chen Y, Mahlamaki E, Schraml P, Moch H, Willi N, Elkahloun AG, Pretlow TG, Gasser TC, Mihatsch MJ, Sauter G, Kallioniemi OP. Hormone therapy failure in human prostate cancer: analysis by complementary DNA and tissue microarrays. J Natl Cancer Inst. 1999;91:1758–1764. doi: 10.1093/jnci/91.20.1758. [DOI] [PubMed] [Google Scholar]
- 12.Saramaki O, Willi N, Bratt O, Gasser TC, Koivisto P, Nupponen NN, Bubendorf L, Visakorpi T. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am J Pathol. 2001;159:2089–2094. doi: 10.1016/S0002-9440(10)63060-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon R, Nocito A, Hubscher T, Bucher C, Torhorst J, Schraml P, Bubendorf L, Mihatsch MM, Moch H, Wilber K, Schotzau A, Kononen J, Sauter G. Patterns of her-2/neu amplification and overexpression in primary and metastatic breast cancer. J Natl Cancer Inst. 2001;93:1141–1146. doi: 10.1093/jnci/93.15.1141. [DOI] [PubMed] [Google Scholar]
- 14.Rao J, Seligson D, Visapaa H, Horvath S, Eeva M, Michel K, Pantuck A, Belldegrun A, Palotie A. Tissue microarray analysis of cytoskeletal actin-associated biomarkers gelsolin and E-cadherin in urothelial carcinoma. Cancer. 2002;95:1247–1257. doi: 10.1002/cncr.10823. [DOI] [PubMed] [Google Scholar]
- 15.Ristimaki A, Sivula A, Lundin J, Lundin M, Salminen T, Haglund C, Joensuu H, Isola J. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res. 2002;62:632–635. [PubMed] [Google Scholar]
- 16.Ginestier C, Charafe-Jauffret E, Bertucci F, Eisinger F, Geneix J, Bechlian D, Conte N, Adelaide J, Toiron Y, Nguyen C, Viens P, Mozziconacci MJ, Houlgatte R, Birnbaum D, Jacquemier J. Distinct and complementary information provided by use of tissue and DNA microarrays in the study of breast tumor markers. Am J Pathol. 2002;161:1223–1233. doi: 10.1016/S0002-9440(10)64399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poremba C, Heine B, Diallo R, Heinecke A, Wai D, et al. Telomerase as a prognostic marker in breast cancer: high-throughput tissue microarray analysis of hTERT and hTR. J Pathol. 2002;198:181–189. doi: 10.1002/path.1191. [DOI] [PubMed] [Google Scholar]
- 18.Barlund M, Forozan F, Kononen J, Bubendorf L, Chen Y, Bittner ML, Torhorst J, Haas P, Bucher C, Sauter G, Kallioniemi OP, Kallioniemi A. Detecting activation of ribosomal protein S6 kinase by complementary DNA and tissue microarray analysis. J Natl Cancer Inst. 2000;92:1252–1259. doi: 10.1093/jnci/92.15.1252. [DOI] [PubMed] [Google Scholar]
- 19.Bucher C, Torhorst J, Kononen J, Haas P, Askaa J, et al. Automated, High-Throughput Tissue Microarray Analysis for Assessing the Significance of HER-2 Involvement in Breast Cancer. Proceedings of the ASCO annual meeting, 2000; Abstr. #2388; New Orleans, LA. [Google Scholar]
- 20.Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol. 2001;159:2249–2256. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bubendorf L, Kononen J, Koivisto P, Schraml P, et al. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res. 1999;59:803–806. [PubMed] [Google Scholar]
- 22.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 23.Mousses S, Bubendorf L, Wagner U, Hostetter G, Kononen J, et al. Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res. 2002;62:1256–1260. [PubMed] [Google Scholar]
- 24.Miettinen HE, Jarvinen TA, Kellner U, et al. High topoisomerase llalpha expression associates with high proliferation rate and and poor prognosis in oligodendrogliomas. Neuropathol Appl Neurobiol. 2000;26:504–512. doi: 10.1046/j.1365-2990.2000.00282.x. [DOI] [PubMed] [Google Scholar]
- 25.Miettinen HE, Paunu N, Rantala I, Kalimo H, et al. Cell cycle regulators (p21, p53 pRb) in oligodendrocytic tumors: a study by novel tumor microarray technique. J Neurooncol. 2001;55:29–37. doi: 10.1023/a:1012961918848. [DOI] [PubMed] [Google Scholar]
- 26.Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, Kallioniemi OP, Kononen J. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000;60:6617–6622. [PubMed] [Google Scholar]
- 27.Wang Y, Wu MC, Sham JS, Zhang, et al. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer. 2002;95:2346–2352. doi: 10.1002/cncr.10963. [DOI] [PubMed] [Google Scholar]
- 28.Moch H, Schraml P, Bubendorf L, Mirlacher M, Kononen J, et al. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Pathol. 1999;154:981–986. doi: 10.1016/S0002-9440(10)65349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung GG, Provost E, Kielhorn EP, Charette LA, Smith BL, Rimm DL. Tissue microarray analysis of beta-catenin in colorectal cancer shows nuclear phospho-beta-catenin is associated with a better prognosis. Clin Cancer Res. 2001;7:4013–4020. [PubMed] [Google Scholar]
- 30.Hoos A, Nissan A, Stojadinovic A, Shia J, Hedvat CV, et al. Tissue Microarray Molecular Profiling of Early, Node-negative Adenocarcinoma of the Rectum: A Comprehensive Analysis. Clin Cancer Res. 2002;8:3841–3849. [PubMed] [Google Scholar]
- 31.Otsuka M, Kato M, Yoshikawa T, Chen H, et al. Differential expression of the L-plastin gene in human colorectal cancer progression and metastasis. Biochem Biophys Res Commun. 2001;289:876–881. doi: 10.1006/bbrc.2001.6047. [DOI] [PubMed] [Google Scholar]
- 32.Garcia JF, Camacho FI, Morente M, et al. Hodgkin’s and Reed-Stemberg cells harbor alterations in the major tumor suppressor pathways and cell-cycle checkpoints: analyses using tissue-microarrays. Blood. 2002;12:12. doi: 10.1182/blood-2002-04-1128. [DOI] [PubMed] [Google Scholar]
- 33.Kielhorn E, Provost E, Olsen D, D’Aquila TG, Smith BL, Camp RL, Rimm DL. Tissue microarray-based analysis shows phospho-beta-catenin expression in malignant melanoma is associated with poor outcome. Int J Cancer. 2003;103:652–656. doi: 10.1002/ijc.10893. [DOI] [PubMed] [Google Scholar]
- 34.Livingstone JI, Yasui W, Tahara E, Wastell C. Are Japanese and European gastric cancer the same biological entity? An immunohistochemical study. Br J Cancer. 1995;72:976–980. doi: 10.1038/bjc.1995.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yosepovich A, Kopolovic J. Tissue microarray technology--a new and powerful tool for the molecular profiling of tumors. Harefuah. 2002;141:1039–1041. 1090. [PubMed] [Google Scholar]
- 36.Hendriks Y, Franken P, Dierssen JW, et al. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol. 2003;162:469–477. doi: 10.1016/S0002-9440(10)63841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzankov A, Zimpfer A, Lugli A, Krugmann J, Went P, Schraml P, Maurer R, Ascani S, Pileri S, Geley S, Dirnhofer S. High-throughput tissue microarray analysis of G1-cyclin alterations in classical Hodgkin’s lymphoma indicates overexpression of cyclin E1. J Pathol. 2003;199:201–207. doi: 10.1002/path.1279. [DOI] [PubMed] [Google Scholar]
- 38.Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab Invest. 2000;80:1943–1949. doi: 10.1038/labinvest.3780204. [DOI] [PubMed] [Google Scholar]
- 39.Nocito A, Bubendorf L, Maria Tinner E, Suess K, et al. Microarrays of bladder cancer tissue are highly representative of proliferation index and histological grade. J Pathol. 2001;194:349–357. doi: 10.1002/1096-9896(200107)194:3<349::AID-PATH887>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 40.Rassidakis GZ, Jones D, Thomaides A, Sen F, Lai R, Cabanillas F, McDonnell TJ, Medeiros LJ. Apoptotic rate in peripheral T-cell lymphomas. A study using a tissue microarray with validation on full tissue sections. Am J Clin Pathol. 2002;118:328–334. doi: 10.1309/HKMV-VMPP-0CH8-3DPQ. [DOI] [PubMed] [Google Scholar]
- 41.Hoos A, Urist MJ, Stojadinovic A, Mastorides S, Dudas ME, Leung DH, Kuo D, Brennan MF, Lewis JJ, Cordon-Cardo C. Validation of tissue microarrays for immunohistochemical profiling of cancer specimens using the example of human fibroblastic tumors. Am J Pathol. 2001;158:1245–1251. doi: 10.1016/S0002-9440(10)64075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mucci NR, Akdas G, Manely S, Rubin MA. Neuroendocrine expression in metastatic prostate cancer: evaluation of high throughput tissue microarrays to detect heterogeneous protein expression. Hum Pathol. 2000;31:406–414. doi: 10.1053/hp.2000.7295. [DOI] [PubMed] [Google Scholar]
- 43.Natkunam Y, Warnke RA, Montgomery K, Falini B, van De Rijn M. Analysis of mum1/irf4 protein expression using tissue microarrays and immunohistochemistry. Mod Pathol. 2001;14:686–694. doi: 10.1038/modpathol.3880373. [DOI] [PubMed] [Google Scholar]
- 44.Rubin MA, Dunn R, Strawderman M, Pienta KJ. Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am J Surg Pathol. 2002;26:312–319. doi: 10.1097/00000478-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 45.Engellau J, Akerman M, Anderson H, et al. Tissue microarray technique in soft tissue sarcoma: immunohistochemical Ki-67 expression in malignant fibrous histiocytoma. Appl Immunohistochem Mol Morphol. 2001;9:358–363. doi: 10.1097/00129039-200112000-00012. [DOI] [PubMed] [Google Scholar]
- 46.Gulmann C, Butler D, Kay E, Grace A, Leader M. Biopsy of a biopsy: validation of immunoprofiling in gastric cancer biopsy tissue microarrays. Histopathology. 2003;42:70–76. doi: 10.1046/j.1365-2559.2003.01556.x. [DOI] [PubMed] [Google Scholar]
- 47.Hedvat CV, Hegde A, Chaganti, et al. Application of tissue microarray technology to the study of non-Hodgkin’s and Hodgkin’s lymphoma. Hum Pathol. 2002;33:968–974. doi: 10.1053/hupa.2002.127438. [DOI] [PubMed] [Google Scholar]
- 48.Fernebro E, Dictor M, Bendahl PO, Ferno M, Nilbert M. Evaluation of the tissue microarray technique for immunohistochemical analysis in rectal cancer. Arch Pathol Lab Med. 2002;126:702–705. doi: 10.5858/2002-126-0702-EOTTMT. [DOI] [PubMed] [Google Scholar]
- 49.Merseburger AS, Kuczyk MA, et al. Limitations of tissue microarrays in the evaluation of focal alterations of bcl-2 and p53 in whole mount derived prostate tissues. Oncol Rep. 2003;10:223–228. [PubMed] [Google Scholar]
- 50.Hoos A, Stojadinovic A, Mastorides S, Urist MJ, Polsky D, Di Como CJ, Brennan MF, Cordon-Cardo C. High Ki-67 proliferative index predicts disease specific survival in patients with high-risk soft tissue sarcomas. Cancer. 2001;92:869–874. doi: 10.1002/1097-0142(20010815)92:4<869::aid-cncr1395>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 51.Hoos A, Stojadinovic A, Singh B, Dudas ME, et al. Clinical significance of molecular expression profiles of Hurthle cell tumors of the thyroid gland analyzed via tissue microarrays. Am J Pathol. 2002;160:175–183. doi: 10.1016/S0002-9440(10)64361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torhorst J, Bucher C, Kononen J, Haas P, Schraml P, Zuber M, Köchli O, Mross F, Dieterich H, Moch H, Mihatsch M, Kallioniemi O, Sauter G. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol. 2001;159:2249–2256. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schraml P, Kononen J, Bubendorf L, Moch H, Bissig H, Nocito A, Mihatsch M, Kallioniemi O, Sauter G. Tissue microarrays for gene amplification surveys in many different tumor types. Clin Cancer Res. 1999;5:1966–1975. [PubMed] [Google Scholar]