Abstract
Growing evidence suggests that aberrant microRNA (miRNA) expression contributes to hepatocellular carcinoma (HCC) development and progression. However, the potential role and mechanism of miR-665 in the progression of liver cancer remains largely unknown. Our current study showed that miR-665 expression was upregulated in HCC cells and tissues. High expression of miR-665 exhibited more severe tumor size, vascular invasion and Edmondson grading in HCC patients. Gain- or loss-of-function assays demonstrated that miR-665 promoted cell proliferation, migration, invasion, and the epithelial–mesenchymal transition (EMT) of HCC cells in vitro and in vivo. Tyrosine phosphatase receptor type B (PTPRB) was downregulated in HCC tissues, and was negatively correlated with miR-665 expression. Through western blotting and luciferase reporter assay, PTPRB was identified as a direct downstream target of miR-665. Restoration of PTPRB reverses the effects of miR-665 on HCC migration, invasion, and cell proliferation. A mechanistic study showed that PTPTRB mediated the functional role of miR-665 through regulation of the Hippo signaling pathway. In conclusion, our results suggested that miR-665 was a negative regulator of the PTPRB and could promote tumor proliferation and metastasis in HCC through decreasing Hippo signaling pathway activity, which can be a potential target for HCC treatment.
Introduction
Hepatocellular carcinoma (HCC) is a common malignancy, particularly in China, due to the prevalence of hepatitis B virus1–3. In recent years, surgery and interventional therapy have made great progress, but the prognosis of patients with HCC remains poor4. It is well known that the main reasons for the poor prognosis of HCC patients are recurrence and metastasis5. Therefore, exploring the mechanisms of HCC progression is crucial to improve early diagnosis and treatment6,7.
MicroRNAs (miRNAs) are a class of small noncoding RNAs composed of ~22 nucleotides that can be combined with the 3′UTR of target mRNAs to provide post-transcriptional regulation8. Growing evidence confirms that dysregulated miRNAs are involved in various biological processes of HCC, including cell proliferation, cell cycle, apoptosis, invasion, and migration9–11. Recently, the role of miR-665 has been identified. Li et al.12 found that miR-665 aggravated inflammation and apoptosis in intestinal ischemia/reperfusion via regulating autophagy. Dong et al.13 confirmed the reduced miR-665 expression in patients with osteosarcoma, and miR-665 had an inhibitory effect on the proliferation and migration of osteosarcoma cells. However, the exact roles of miR-665 in HCC growth and metastasis as well as the molecular mechanisms involved remain unclear.
Genetic evidence has established inhibitory roles for Hippo signaling in the control of tumorigenesis in a variety of tissues, particularly the liver14. The Hippo signaling pathway activates LATS kinases, which phosphorylates YAP, leading to the cytoplasmic retention of YAP15. Tyrosine phosphatase receptor type B (PTPRB) is a potential target of miR-665(predicted by TargetScan and miRanda). Recent studies have also discovered that PTPRB may function as a tumor suppressor in carcinogenesis and cancer development16. However, a functional link between the miR-665/PTPRB axis and Hippo signaling pathway in association with HCC proliferation, migration, and invasion remains to be further studied.
In this study, we demonstrated a significant increase of miR-665 in HCC cells and tissues. We showed that miR-665 promoted tumor proliferation, migration, and invasion both in vitro and in vivo. We confirmed that miR-665 inhibited Hippo signaling activity through suppression of PTPRB. These findings indicated that miR-665 played a key role in the progression of liver cancer.
Methods and Materials
Tissue samples
Tissue samples were obtained from 50 patients who were undergoing liver resection in the Jiangsu Province Hospital. Approval was obtained from the ethics committee of the Jiangsu Province Hospital. All HCC and normal tissues were collected and restored in liquid nitrogen. The clinicopathological and demographic information of the patients is described in Table 1.
Table 1.
Association between miR-665 expression and clinicopathologic features of patients with hepatocellular carcinoma
| miR-665 levels | ||||
|---|---|---|---|---|
| Characteristics | Number(n = 50) | Low(n = 22) | High(n = 28) | P-value |
| Age (years) | 0.754 | |||
| ≥60 | 33 | 14 | 19 | |
| <60 | 17 | 8 | 9 | |
| Gender | 0.594 | |||
| Male | 36 | 15 | 21 | |
| Female | 14 | 7 | 7 | |
| HBV infection | 0.441 | |||
| Negative | 9 | 5 | 4 | |
| Positive | 41 | 17 | 24 | |
| Liver cirrhosis | 0.447 | |||
| Absent | 5 | 3 | 2 | |
| Present | 45 | 19 | 26 | |
| AFP (ng/ml) | 0.251 | |||
| ≤20 | 12 | 7 | 5 | |
| >20 | 38 | 15 | 23 | |
| Tumor size | 0.011a | |||
| ≤5 cm | 24 | 15 | 9 | |
| >5 cm | 26 | 7 | 19 | |
| Tumor multiplicity | 0.083 | |||
| Single | 32 | 17 | 15 | |
| Multiple | 18 | 5 | 13 | |
| Vascular invasion | 0.010a | |||
| No | 31 | 18 | 13 | |
| Yes | 19 | 4 | 15 | |
| Edmondson grade | 0.035a | |||
| I + II | 28 | 16 | 12 | |
| III + IV | 22 | 6 | 16 | |
a P < 0.05, statistically significant difference.
Cell culture
The human HCC cell lines and LO2 cells were obtained from the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) containing 10% fetal bovine serum within a humidified incubator containing 5% CO2 at 37 °C.
Fluorescence in situ hybridization (FISH)
The expression of miR-665 in HCC and adjacent non- HCC tissues was measured by FISH. The human miR-665 sequence is 3′-UCCCCGGAGUCGGAGGACCA-5′. LNA (locked-nucleic acid)-based probes against the mature miRNA sequence were used. The 5′-FAM-labeled miR-665 probe sequence was 5′-AGGGGCCTCAGCCTCCTGGT-3′. The probe was purchased from Servicebio (Wuhan, China).
Real-time quantitative polymerase chain reaction (PCR)
TRIzol (Invitrogen, USA) was used to extract RNA from tissues and cells. PrimeScript RT reagent Kit (Takara, China) was used to perform reverse transcription. Quantitative PCR were measured using SYBR Green Master(TaKaRa). The primers for PTPRB and β-actin were purchased from Realgene (Nanjing, China). The primers for miR-665 and U6 were obtained from RiboBio (Guangzhou, China).
The sequences of the primers are listed. PTPRB forward: 5′-ACAACACCACATACGGATGTAAC-3′, PTPRB reve-rse: 5′-CCTAGCAGGAGGTAAAGGATCT-3′; CTGF forward: 5′-CAGCATGGACGTTCGTCTG-3′, CTGF reverse: 5′-AACCACGGTTTGGTCCTTGG-3′; CYR61 forward: 5′-CAGCATGGACGTTCGTCTG-3′, CYR61 reverse: 5′-AA.CCACGGTTTGGTCCTTGG-3′; β-actin forward: 5′-TGA.CGTGGACATCCGCAAAG-3′, β-actin reverse 5′- CTG.GAAGGTGGACAGCGAGG-3′.
Establishment of stably transfected cells
We purchased commercially available LV3-has-miR-665-pre-microRNA vector (pre-miR-665), LV3-has-miR-665-sponge inhibitor vector (miR-665-inhibitor), vector containing the PTPRB DNA sequence (LV-PTPRB), and lentiviral vector containing PTPRB siRNA hairpin sequence (PTPRB-shRNA) constructs from GenePharma (Shanghai, China). After infecting the lentiviruses with HCC cells, 7 μg/mL puromycin (Sigma-Aldrich, USA) was used to select cells.
Cell migration and invasion assays
Transwell chambers (Millipore, USA) were used for cell migration and invasion assays. For migration assays, cells in the upper chamber were cultured with serum-free DMEM, and the lower chamber was filled with 10% serum-containing DMEM. For invasion assays, cells were seeded on Matrigel-coated membrane inserts. Twenty-four hours later, cells that migrated or invaded across the Transwell membrane were stained with 0.1% crystal violet for 30 min.
Wound-healing assay
Cells were seeded on six-well plates and were grown to confluence overnight. The cells were then scratched using a 200 μl tip and the wound recovery was observed after 0 and 48 h.
Cell counting Kit-8 assay
HCC cell growth was evaluated by a CCK-8 Kit (Dojindo, Japan). Two thousand cells per well were seeded in 96-well plates with 100 μL culture medium for 5 days.
5-Ethynyl-2′-deoxyuridine (EdU) incorporation assay
EdU proliferation assay was carried out according to the manufacturer’s instructions. Cells were incubated with 50 μM EdU (RiboBio) for 2 h. Apollo staining and DAPI staining were then used to observe the EdU-positive cells.
Soft agar growth assay
First, HCC cells were suspended in medium plus 0.7% agar, then HCC cells were plated on top of a bottom layer with 1.4% medium agar mixture. Viable colonies ( ≥ 0.1 mm) were counted and photographed after 10 days.
Flow cytometry analysis of the cell cycle
Cells were harvested and fixed in 75% cold ethanol. After one night, cells were resuspended in phosphate-buffered saline (PBS), and stained with propidium iodide for 30 min. Flow cytometry was used to analyze the cell cycle.
Immunofluorescence analysis
We used 4% paraformaldehyde to fix transfected cells and 0.3% Triton X-100 to permeabilize the cells. The cells were subjected to immunofluorescent staining with the primary antibody to E-cadherin, vimentin, and YAP (Cell Signaling Technology, USA) for 16 h. Then, cells were washed and incubated with fluorescence-labeled antibody for 30 min.
Luciferase reporter assay
The 3′-UTR and mutated sequence of PTPRB were synthesized and inserted into the pmiR-GLO dual-luciferase vector (GenePharma, Shanghai, China). Cells overexpressing miR-665 or its control were transfected with PTPRB–3′-UTR-Wt and PTPRB–3′-UTR-Mut. After 48 h transfection, cells were collected and lyzed by lysis buffer (Promega, Madison, WI, USA).
Western blotting
Proteins were electrophoresed by 10% SDS-PAGE and transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA). Next, the membranes were blocked in 5% skimmed milk and incubated with specific primary antibodies (1:1000) at 4 °C overnight. Rabbit anti-PTPRB (Biorbyt, Cambridge, UK), β-actin, E-cadherin, Vimentin, Slug, p-LATS1, p-YAP, LATS1, YAP, CTGF and CYR61 (Cell Signaling Technology) antibodies were used. After that, the membranes were incubated with the second antibody.
Immunohistochemistry
The paraffin was cut into 4-μm sections and were incubated with primary antibodies for PTPRB (Biorbyt), Ki67 (Cell Signaling Technology), and VEGF-A (Abcam, Cambridge, UK). After one night, the sections were then incubated with the second antibody and stained with 3,3-diaminobenzidine solution.
In vivo animal experiments
For tumor growth assay, nude mice were obtained from the Animal Model Institute of Nanjing University (Nanjing, China). Nude mice were randomly divided into six groups (n = 5 per group). HepG2-NC, HepG2-pre-miR-665, HepG2-miR-665, SMMC-7721-NC, SMMC-7721-pre-miR-665, and SMMC-7721-miR-665-inhibitor stable cells (2 × 106 cells) in 100 μL PBS were injected to nude mice by subcutaneous injection.
For tumor metastasis assay, nude mice and cell types were the same as above. PBS of 106 cells was injected into nude mice through the tail vein. The occurrence of metastases was observed using IVIS Imaging System (Caliper Life Sciences, Waltham, MA, USA).
Statistical analysis
Data are expressed as mean ± standard deviation from at least three separate experiments. The χ2 test was used to analyze the association of miR-665 expression with clinicopathological features. Independent t-tests were used to compare differences between two groups. A paired t-test was used to analyze miR-665 and PTPRB mRNA levels in tissue samples. Correlations were determined by Pearson’s correlation analysis. Statistical analyses were performed using SPSS, v. 22.0 (SPSS Inc., Chicago, IL, USA). * P < 0.05, ** P < 0.01, and *** P < 0.001 were defined as statistically significant.
Results
miR-665 is upregulated in HCC cell lines and tissues
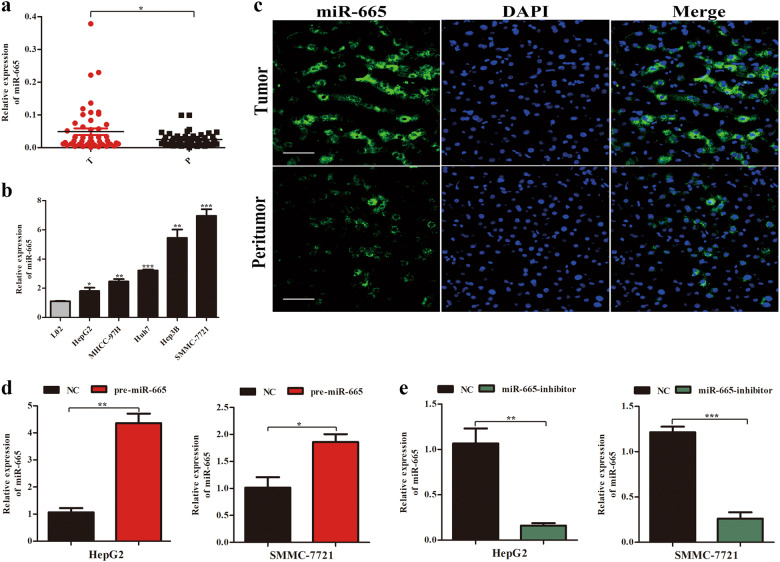
To explore whether miR-665 was abnormally regulated in HCC tissues, we performed real-time quantitative PCR (RT-qPCR) to detect miR-665 expression from 50 paired HCC tissues and adjacent peritumor tissues. Compared to peritumor samples, miR-665 expression was significantly increased in HCC tissues (Fig. 1a). To investigate the clinicopathological features of miR-665 in HCC, HCC patients were divided into miR-665 high- or low-group based on median value. Results showed that high levels of miR-665 were significantly associated with large tumor size (P = 0.011), vascular invasion (P = 0.010), and Edmondson grade (P = 0.035) (Table 1). Similarly, compared with the miR-665 expression in normal liver cell line LO2, expression of miR-665 was significantly increased in HCC cell lines, including HepG2, MHCC-97H, Huh7, Hep3B, and SMMC7721 (Fig. 1b). Next, we used three paired HCC and peritumoral tissues for FISH analysis of miR-665 and further confirmed the higher miR-665 expression in HCC tissues (Fig. 1c).
Fig. 1. miR-665 is upregulated in HCC tissues and cell lines.
a The mRNA levels of miR-665 in 50-paired human HCC and adjacent normal tissues. b The mRNA levels of miR-665 expression in HCC cell lines and normal L02 cells. c Expression of miR-665 in HCC and adjacent normal tissues was measured by FISH assay (scale bars, 50 μm). d, e HepG2 and SMMC-7721 cells were transfected with lentivirus overexpressing miR-665 (defined as pre-miR-665) or lentivirus with short hairpin RNA targeting miR-665 (defined as miR-665-inhibitor). Cells transfected with empty lentiviral vectors served as a negative control (NC). The miR-665 expression was analyzed by RT-qPCR after transfection; All experiments were performed three times and data were presented as mean ± SD. * P < 0.05, ** P < 0.01
To examine the biological function of miR-665 in HCC, we constructed both miR-665 overexpression and knockdown HCC cell lines using lentivirus-based approaches. As measured by RT-qPCR, we confirmed the transfection efficiency in HepG2 and SMMC-7721 cells (Fig. 1d, e).
miR-665 promotes HCC cell migration, invasion, and EMT in vitro
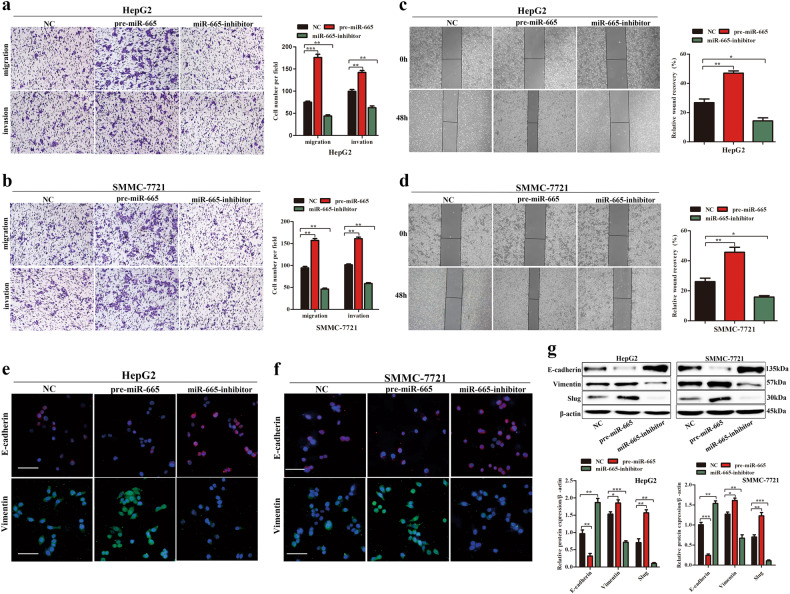
To investigate the influence of miR-665 on the migration and invasion of HCC cells in vitro, Transwell assay with or without matrigel and wound-healing assay were performed in HCC cells. The Transwell assays showed that miR-665 overexpression markedly enhanced cell migration and invasion in HepG2 and SMMC7721 cells, whereas knockdown of miR-665 significantly inhibited cell migration and invasion of HCC cells compared with controls (Fig. 2a, b). The wound-healing assay supported the conclusion that miR-665 promoted migration of HCC cells. The miR-665-overexpressing cells showed advanced wound healing while miR-665-silenced cells exhibited delayed wound healing (Fig. 2c, d). It is well known that the EMT process is related to the migration of cancer cells. Then we detected the EMT-related factors in HCC cells. Immunofluorescence results showed that overexpressing miR-665 resulted in reduced expression of E-cadherin but increased expression of vimentin in HepG2 and SMMC-7721 cells compared with controls and silencing miR-665 showed the opposite results (Fig. 2e, f). These results were also confirmed by western blotting. After miR-665 overexpression in HepG2 and SMMC-7721 cells, the protein level of E-cadherin was reduced and vimentin and slug levels were increased (Fig. 2g), while knockdown of miR-665 showed the opposite results (Fig. 2g). These data suggested that miR-665 promoted HCC cell migration and invasion.
Fig. 2. miR-665 promotes HCC cells migration, invasion, and EMT in vitro.
a, b Transwell assays of miR-665-overexpressing or -silencing HepG2 (a) and SMMC7721 (b) cells. c, d Wound-healing assays were performed in miR-665-overexpressing or -silencing HepG2 (c) and SMMC7721 (d) cells. e, f Expression of EMT markers in miR-665 -overexpressing or -silencing HepG2 (e) and SMMC7721 (f) cells was examined by immunofluorescence (red: E-cadherin, green: vimentin, blue: DAPI) (scale bar: 20 μm). g Western blotting was used to examine the levels of EMT-related protein. Three independent experiments were performed for each group. All data are represented as the means ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with NC
miR-665 promotes HCC cell proliferation and cell cycle progression in vitro
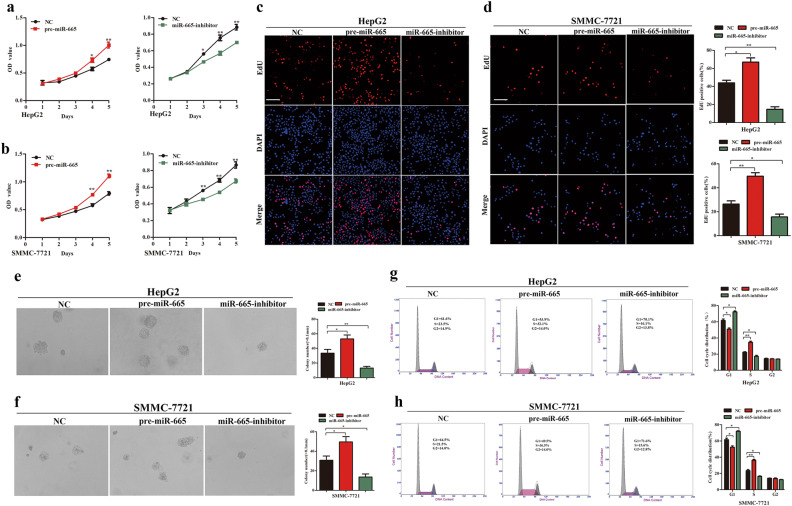
In the CCK8 assays, compared with the control groups, we found that upregulation of miR-665 significantly promoted proliferation of HepG2 and SMMC7721 cells, whereas downregulation of miR-665 significantly inhibited growth of HCC cells (Fig. 3a, b). Consistent with these results, EdU assays showed that stable ectopic overexpression of miR-665 increased the numbers of EdU-positive nuclei of HepG2 and SMMC-7721 cells compared with the controls, while stable knockdown of miR-665 reduced the numbers of EdU-positive nuclei (Fig. 3c, d). Soft agar growth assay also suggested that overexpression of miR-665 significantly increased anchorage-independent cell growth of HepG2 and SMMC-7721 cells, and downregulation of miR-665 attenuated anchorage-independent growth of HCC cells (Fig. 3e, f). The miR-665 promoted HCC cell proliferation; therefore we determined whether miR-665 affected cell cycle progression of HCC cells. As determined by flow cytometry, compared with the control group, overexpression of miR-665 led to a decrease in G1 phase and a significant increase in S phase in HepG2 and SMMC7721 cells (Fig. 3g, h). Silencing miR-665 expression showed the opposite results (Fig. 3g, h). These data demonstrated that miR-665 regulated HCC cell proliferation and cell cycle progression in vitro.
Fig. 3. miR-665 promotes HCC cell proliferation and cell cycle progression in vitro.
a, b CCK-8 assays in overexpressing or silencing miR-665 HepG2 (a) and SMMC7721 (b) cells. c, d EdU incorporation assays were performed to explore the cell proliferation in miR-665-overexpressing or -silencing HepG2 (c) and SMMC7721 (d) cells (red fluorescence represents Edu-positive cells; blue fluorescence from DAPI staining represents the total cells). e, f Soft agar growth assay in miR-665-overexpressing or -silencing HepG2 (e) and SMMC7721 (f) cells. g, h Cell cycle distribution of miR-665-overexpressing or -silencing HepG2 (g) and SMMC7721 (h) cells was measured by flow cytometry. Three independent experiments were performed for each group. All data are represented as the means ± SD. * P < 0.05, ** P < 0.01 compared with NC
PTPRB is a direct function target of miR-665 in HCC
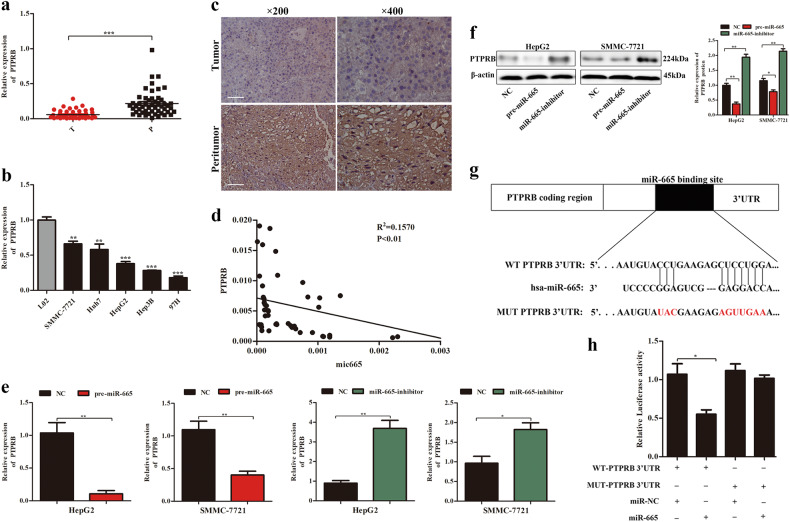
Targetscan and miRanda were used to analyze potential miR-665 targets. Among them, we were particularly interested in PTPRB, because the PTPR family is mostly regarded as tumor suppressors in carcinogenesis and cancer development16. RT-qPCR was used to analyze PTPRB expression in 50 paired HCC tissues and adjacent normal tissues. Compared to adjacent normal tissues, PTPRB expression was significantly lower in HCC tissues (Fig. 4a). Immunohistochemical analysis further confirmed the lower expression of PTPRB in HCC tissues (Fig. 4c). Consistent with the above results, mRNA level of PTPRB was also lower in HCC cells compared to LO2 cell (Fig. 4b). In addition, we found the PTPRB level was negatively correlated with miR-665 in HCC tissues (Fig. 4d).
Fig. 4. PTPRB is a direct target of miR-665 in HCC.
a The mRNA levels of PTPRB in 50 paired human HCC and adjacent normal tissues. b The mRNA levels of PTPRB expression in HCC cell lines and normal L02 cells. c Representative immunostaining images of PTPRB in HCC tissues and adjacent normal tissues. d A negative correlation between the levels of miR-665 and PTPRB expression in HCC specimens. e The mRNA level of PTPRB expression in overexpressing or silencing miR-665 HepG2 and SMMC7721 cells; cells transfected with empty lentiviral vectors were used as a negative control. f The protein levels of PTPRB in overexpressing or silencing miR-665 HepG2 and SMMC7721 cells; cells transfected with empty lentiviral vectors were used as a negative control. g The potential miR-665 binding site in the PTPRB 3′-UTR was predicted by TargetScan. h Dual-luciferase reporter assay was used to analyze the effects of miR-665 expression on the activities of WT and MUT PTPRB 3′-UTR. All data are represented as the means ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001
To verify that PTPRB was a bonafide downstream target of miR-665, how miR-665 expression changes affect the subsequent expression of PTPRB was determined by RT-qPCR and western blot. It was revealed that miR-665 overexpression was associated with decreased PTPRB mRNA and protein expression in HepG2 and SMMC-7721 cells, whereas silencing miR-665 led to the opposite results (Fig. 4e, f). Luciferase reporter assay was used to further confirm that PTPRB was a direct target of miR-665. The results showed that ectopic miR-665 expression markedly suppressed luciferase activity of WT 3′-UTR of PTPRB but not of the MUT 3′-UTR of PTPRB (Fig. 4g, h). These data suggested PTPRB was a direct target of miR-665.
Restoration of PTPRB reverses the effects of miR-665 on migration, invasion, and proliferation of HCC
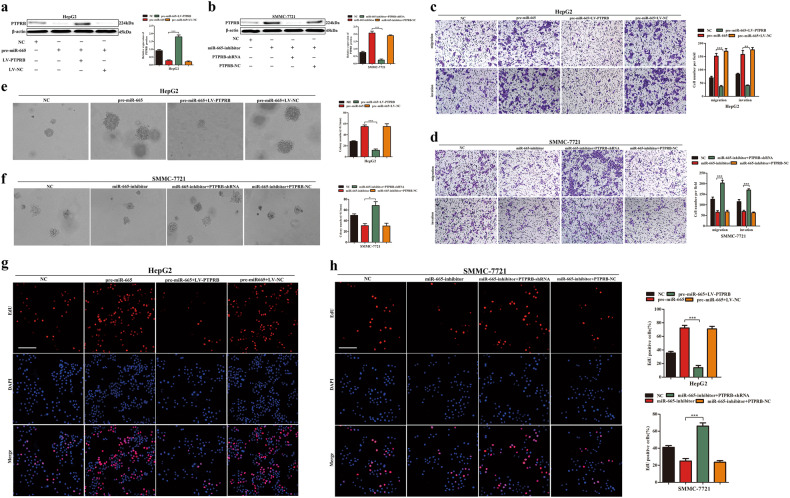
To confirm that miR-665 regulated migration, invasion, and proliferation of HCC cells through targeting PTPRB, HepG2 cells were transfected with pre-miR-665 and LV-PTPRB lentiviruses. Western blotting confirmed that the expression of PTPRB (Fig. 5a). SMMC-7721 cells were transfected with miR-665 inhibitor and PTPRB shRNA lentivirus. Western blotting also confirmed the expression of PTPRB (Fig. 5b). The results of Transwell assays showed that PTPRB overexpression reduced the promotive effects of migration and invasion in HepG2 cells, which was caused by miR-665 overexpression (Fig. 5c). In SMMC-7721 cells, similar rescue effects were observed; the inhibitory effects of migration and invasion caused by miR-665 silencing were reversed by PTPRB downregulation (Fig. 5d). Restoration of PTPRB also significantly reversed the effects of miR-665 on cell proliferation according to the soft agar growth assays and EdU assays in HepG2 and SMMC-7721 cells (Fig. 5e–h). Our results supported the findings that PTPRB was a functional mediator of miR-665 and played a key role in HCC proliferation, migration, and invasion.
Fig. 5. Rescue experiments performed to confirm that PTPRB is the functional target of miR-665 in HCC progression.
a Western blotting was conducted to examine PTPRB protein expression in the HepG2 cells transfected with miR-665 overexpression lentivirus (pre-miR-665) and PTPRB overexpression lentivirus (LV-PTPRB). b Western blotting was conducted to examine PTPRB protein expression in the SMMC-7721 cells transfected with lentivirus with short hairpin RNA targeting miR-665 (miR-665-inhibitor) and lentivirus with short hairpin RNA targeting PTPRB (PTPRB-shRNA). c, d Transwell assays of HepG2-pre-miR-665 cells transfected with LV-PTPRB or LV-NC (c), and SMMC-7721-miR-665-inhibitor cells transfected with PTPRB-shRNA or PTPRB-NC (d). e, f Soft agar growth assay of HepG2-pre-miR-665 cells transfected with LV-PTPRB or LV-NC (e), and SMMC-7721-miR-665-inhibitor cells transfected with PTPRB-shRNA or PTPRB-NC (f). g, h (EdU) incorporation analysis of HepG2-pre-miR-665 cells transfected with LV-PTPRB or LV-NC (g), and SMMC-7721-miR-665-inhibitor cells transfected with PTPRB-shRNA or PTPRB-NC (h). Three independent experiments were performed for each group. All data are represented as means ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001
miR-665 regulates the YAP-Hippo signaling pathway through PTPRB
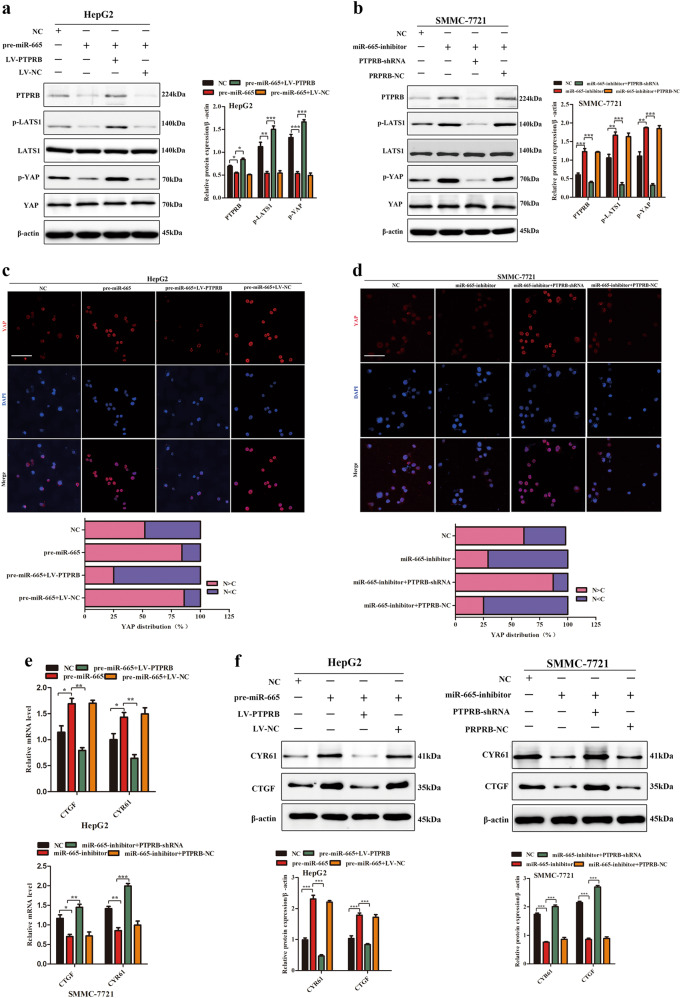
We further explored the mechanism by which the miR-665-PTPRB axis regulated the growth and metastasis of HCC. The recently discovered tumor suppressive Hippo signaling pathway had been involved in cancer progression17. It regulates diverse cellular functions in a broad range of tissues, including the hepatic systems18–20. Therefore, we speculated that the miR-665-PTPRB induced promotion of HCC proliferation and migration was associated with the Hippo pathway. Western blotting showed that overexpression of miR-665 in HepG2 cells decreased levels of phosphorylated-LATS1(p-LATS1) and its substrate, phosphorylated-YAP(p-YAP) (Fig. 6a). Conversely, downregulation of miR-665 in SMMC-7721 cells resulted in increased levels of p-LATS1 and p-YAP (Fig. 6b). More importantly, we also found that ectopic PTPRB expression reversed the inhibition of p-LATS1 and p-YAP caused by miR-665 overexpression in HepG2 cells (Fig. 6a). Similarly, in SMMC-7721 cells, the increased levels of p-LATS1 and p-YAP caused by miR-665 silencing were reversed by PTPRB downregulation (Fig. 6a).
Fig. 6. miR-665 inhibits the activity of Hippo signaling through PTPRB in HCC cells.
a Western blotting showed that overexpression of miR-665 in HepG2 cells decreased levels of p-LATS1 and its substrate, p-YAP, while ectopic PTPRB expression reversed the inhibition of p-LATS1 and p-YAP caused by miR-665 overexpression. b Western blotting showed that increased levels of p-LATS1 and p-YAP caused by miR-665 silencing were reversed by PTPRB downregulation. c Immunofluorescence analysis of nuclear retention and transcriptional activity of YAP in HepG2-pre-miR-665 cells transfected with LV-PTPRB or LV-NC. d Immunofluorescence analysis of nuclear retention and transcriptional activity of YAP in SMMC-7721-miR-665-inhibitor cells transfected with PTPRB-shRNA or PTPRB-NC (scale bar: 20 μm). e mRNA levels of CTGF and CYR61 in HepG2-pre-miR-665 cells and SMMC-7721-miR-665-inhibitor cells. f Protein levels of CTGF and CYR61 in HepG2-pre-miR-665 cells and SMMC-7721-miR-665-inhibitor cells. All data are represented as the means ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001
Hippo pathway controls of YAP transcription via the Hippo kinase cascade15. Phosphorylation of YAP promotes its cytoplasmic sequestration and degradation. In our study, immunofluorescence analysis revealed that miR-665 overexpression enhanced nuclear retention and transcriptional activity of YAP in HepG2 cells, while miR-665 silencing promoted YAP cytoplasmic sequestration and turned off its transcriptional activity in SMMC-7721cells (Fig. 6c, d). Furthermore, upregulation or downregulation of PTPRB reversed the above results in HepG2 and SMMC-7721cells (Fig. 6c, d).
Cysteine-rich angiogenic inducer 61 (CYR61) and connective tissue growth factor (CTGF) are the main downstream targets of YAP. We next investigated the levels of CTGF and CYR61. Results showed that overexpression of miR-665 in HepG2 cells increased expression of CTGF and Cyr61 at both the gene and protein levels. Conversely, downregulation of miR-665 in SMMC-7721 cells decreased levels of CTGF and Cyr61 (Fig. 6e, f). In addition, we also found that ectopic PTPRB expression reversed the promotion of CTGF and Cyr61 caused by miR-665 overexpression in HepG2 cells both at gene and protein levels. Similarly, in SMMC-7721 cells, the decreased levels of CTGF and Cyr61 caused by miR-665 silencing were reversed by PTPRB downregulation (Fig. 6e, f). These results suggested that miR-665 regulated the YAP–Hippo signaling pathway through PTPRB.
miR-665 promotes xenograft tumor growth in vivo
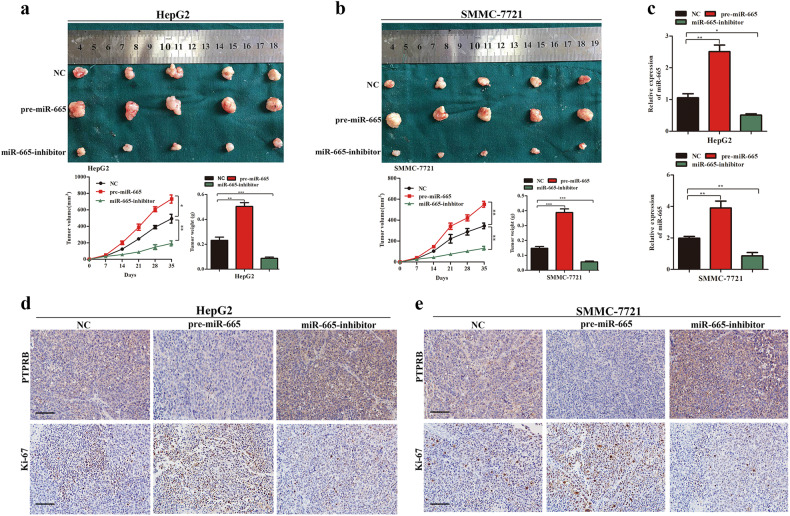
To examine the effect of miR-665 on cell tumorigenicity in vivo, HepG2 and SMMC-7721 cells with stable overexpression or knockdown of miR-665 were implanted into nude mice, and cells transfected with empty lentiviral vectors were used as a control. Each mouse was monitored once weekly, and the mice were euthanized after 5 weeks. Compared with controls, miR-665 overexpression promoted tumor growth of HepG2 and SMMC-7721 cells in mice, and downregulation of miR-665 inhibited the tumor growth of HCC cells (Fig. 7a, b). Next, we detected the expression of miR-665 in xenografts using RT-qPCR. As shown in the results, compared with the NC-treated group, higher miR-665 expression was observed in the pre-miR-665-treated mice group and lower miR-665 expression was found in the miR-665-inhibitor-treated mice group (Fig. 7c). Next, we detected PTPRB and Ki-67 expression of xenografts by immunohistochemistry. Results showed that PTPRB expression was decreased in the pre-miR-665-treated mice group and increased expression of PTPRB was found in the miR-665-inhibitor-treated mice group (Fig. 7d, e). In addition, miR-665 overexpression increased the number of Ki67-positive cells, while miR-665 knockdown decreased the number of Ki67-positive cells (Fig. 7d, e). These results demonstrated that miR-665 promoted tumor growth of HCC in vivo.
Fig. 7. miR-665 promotes tumor growth in vivo.
a Upper panel: representative pictures of HCC tumors from mice inoculated with HepG2-NC, HepG2-pre-miR-665, and HepG2-miR-665-inhibitor cells; Lower panel: growth curves for tumor volume and tumor weight in each group mice injected with HepG2 cells were calculated; n = 5 per group. b Representative pictures of HCC tumor from mice inoculated with SMMC-7721- NC, SMMC-7721-pre-miR-665, and SMMC-7721-miR-665-inhibitor cells; lower panel: growth curves for tumor volume and tumor weight in each group of mice injected with SMMC-7721 cells were calculated; n = 5 per group. c mRNA levels of miR-665 in the tumors derived from nude mice were measured by qPCR. d PTPRB and Ki67 staining of tumor tissues from each group of mice injected with HepG2 cells. e PTPRB and Ki67 staining of tumor tissues from each group of mice injected with SMMC7721 cells. All data are represented as the means ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001
miR-665 promotes HCC metastasis in in vivo
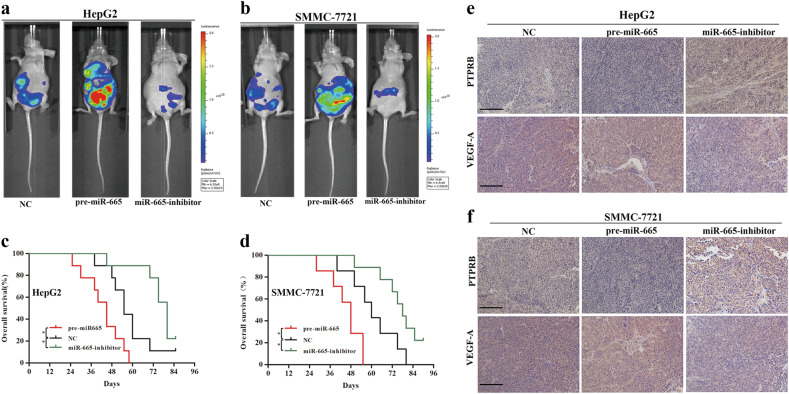
To investigate whether miR-665 promoted metastasis of HCC in vivo, SMMC-7721 and HepG2 cells with NC, miR-665 overexpression and miR-665 knockdown were injected into nude mice through the tail vein. Bioluminescence imaging analysis showed that nude mice injected with miR-665-overexpressing cells exhibited higher metastases compared to the controls (Fig. 8a, b). Conversely, mice injected with miR-665 knockdown cells showed lower metastases incidence compared to the controls (Fig. 8a, b). The HepG2- and SMMC7721-pre-miR-665 groups had a worse survival outcome compared to the control group, but the HepG2- and SMMC7721-miR-665-inhibitor groups had a longer overall survival time than the control group (Fig. 8c, d).
Fig. 8. miR-665 promotes HCC metastasis in vivo.
a, b Bioluminescent images of mice administered with HepG2 (a) and SMMC-7721 (b) cells via the tail veins. Images were taken after 6 weeks. n = 5 per group. c Overall survival of each group mice injected with HepG2 cells. d Overall survival of each group of mice injected with SMMC7721 cells. e PTPRB and VEGF-A staining was analyzed in tumor tissues from each group of mice injected with HepG2 cells. f PTPRB and VEGF-A staining in tumor tissues from each group of mice injected with SMMC-7721 cells. All data are represented as means ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001
We then isolated metastatic tumors from several organs and detected PTPRB and VEGF-A expression by immunohistochemistry. miR-665 overexpression decreased expression of PTPRB, while miR-665 knockdown increased expression of PTPRB (Fig. 8e, f). Expression of VEGF-A showed the opposite trends in tumors. The pre-miR-665 group increased expression of VEGF-A, but the miR-665-inhibitor group showed decreased expression of VEGF-A (Fig. 8e, f). Taken together, we demonstrated that miR-665 promoted HCC metastasis in vivo.
Discussion
miRNAs have been revealed as important regulators in progression of HCC21–23. Numerous studies have shown that dysregulation of miRNAs promotes the growth and metastasis of HCC cells24. Thus, illuminating the relevant mechanisms between miRNA and HCC is very important. In this study, we revealed that miR-665 was upregulated in HCC tissues and cell lines. Increased miR-665 level was correlated with malignant clinicopathological features, including large tumor size, vascular invasion, and Edmondson grade. These data suggested that miR-665 was involved in HCC progression and could be a potential novel target for HCC treatment.
Recently, the functional role of miR-665 in malignant tumors has been proposed. Dong et al.13 found that miR-665 expression was reduced in osteosarcoma tissue and miR-665 overexpression inhibited osteosarcoma cell proliferation and invasion through target Rab23. Chen et al.25 showed that miR-665 was upregulated in gastric adenocarcinoma but downregulated in signet ring cell carcinoma. Interestingly, levels of miR-665 were upregulated in some tumors but downregulated in others. This may suggest that miR-665 exerts different biological functions according to the different types of tumors. With regard to the progression of HCC, the exact biological functions of miR-665 in HCC are still poorly known. In this study, we demonstrated that miR-665 promoted cell proliferation, migration, and invasion by gain- and loss-of function experiment in vitro and in vivo. Epithelial-mesenchymal transition (EMT) plays a critical role in cancer cell progression and metastasis. The central feature of EMT is that epithelial cancer cells acquire molecular changes that promote the loss of epithelial characteristics and the acquisition of mesenchymal phenotypes, which promote the migration and invasion of cancer cells26. In addition, EMT is also characterized by decreased expression of E-cadherin and increased expression of vimentin27. In this study, we found that miR-665 overexpression decreased the level of E-cadherin but increased the levels of vimentin and slug, prompting that EMT was enhanced. The enhanced EMT progress was also consistent with our in vitro migration and invasion data. The above results confirmed the promoter role for miR-665 in HCC progression.
Furthermore, we provided sufficient evidence that the target gene of miR-665 was PTPRB. First, miR-665 negatively modulated PTPTB expression in HCC cells at the mRNA and protein levels. Second, miR-665 affected the luciferase activity of WT 3′-UTR but not of the MUT 3′-UTR of PTPRB. Third, miR-665 was inversely correlated with the expression of PTPRB in HCC tissues. PTPRB was reduced in HCC cells and regulating its expression could reverse the functions of miR-665 in HCC cells. PTPRB is a family member of protein tyrosine phosphatase receptor type (PTPRs). PTPRs are mostly regarded as tumor suppressors and are inactivated by genetic mutations in human cancer28,29. Recent studies have reported truncation mutations in metastatic melanoma and recurrent mutations in angiosarcoma of PTPRB30,31. We also confirmed that PTPRB was downregulated in HCC tissues compared to adjacent non-tumor tissues. These observations suggested that PTPRB may play a role as a tumor suppressor in tumorigenesis and progression.
The Hippo signaling is a central mechanism that inhibits tissue overgrowth and its activation pathways can suppress tumorigenesis32. Hippo pathway dysregulation has been involved in a wide range of human cancers including lung, colorectal, ovarian, liver, and prostate33–35. Central to the Hippo pathway is that Hippo signaling suppresses YAP transcription activities by activating LATS kinases, which directly phosphorylates YAP, causing YAP cytoplasmic retention and degradation15. Here, we further confirmed that miR-665 inhibited Hippo pathway activity through PTPRB. Western blotting showed that overexpression of miR-665 decreased levels of p-LATS1 and p-YAP, while downregulation of miR-665 resulted in increased levels of p-LATS1 and p-YAP in HCC cells. More importantly, we also found that modulating PTPRB expression reversed the effects on the Hippo signaling pathway of miR-665. Immunofluorescence analysis of YAP confirmed the above results. Although we confirmed that miR-665 affected the Hippo pathway through PTPRB, we were not able to confirm the specific mechanism of PTPRB on the Hippo pathway and this requires further investigation.
In this study, we reported a promoter role for miR-665 in HCC progression. We showed that miR-665 promoted tumor proliferation, migration, and invasion both in vivo and in vitro. We also confirmed that miR-665 inhibited Hippo signaling activity through direct suppression of PTPRB. In general, we have discovered the role and molecular mechanism of miR-665 in liver cancer, and miR-665 may be a potential target for HCC treatment in the future.
Acknowledgements
This work was partially supported by the National Natural Science Foundation of China (81273261 and 81270583), Priority Academic Program Development of Jiangsu Higher Education Institutions (814700901).
Ethics statement
All mice received humane care and experiments were performed according to the protocols approved by the Institutional Animal Care Committee of Nanjing Medical University.
Conflict of interest
The authors declare no competing interests.
Footnotes
Edited by G. Giannelli
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-023-05654-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuanchang Hu, Chao Yang.
Change history
2/9/2023
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41419-023-05654-x
Contributor Information
Jianhua Rao, Email: raojh@njmu.edu.cn.
Xuehao Wang, Email: wangxh@njmu.edu.cn.
References
- 1.Miller KD, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Wei W, Chua MS, Grepper S, So S. Small molecule antagonists of Tcf4/beta-catenin complex inhibit the growth of HCC cells in vitro and in vivo. Int. J. Cancer. 2010;126:2426–2436. doi: 10.1002/ijc.24810. [DOI] [PubMed] [Google Scholar]
- 4.Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut. 2015;64:842–848. doi: 10.1136/gutjnl-2014-307990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.He Y, et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Zou ZQ, Liu CX, Liu XZ. Immunotherapeutic interventions in chronic hepatitis B virus infection: a review. J. Immunol. Methods. 2014;407:1–8. doi: 10.1016/j.jim.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 9.Wei R, et al. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin. Cancer Res. 2013;19:4780–4791. doi: 10.1158/1078-0432.CCR-12-2728. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, et al. Methylation-mediated repression of microRNA-129-2 suppresses cell aggressiveness by inhibiting high mobility group box 1 in human hepatocellular carcinoma. Oncotarget. 2016;7:36909–36923. doi: 10.18632/oncotarget.9377. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Liu Z, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350–25365. doi: 10.18632/oncotarget.8129. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Li Z, et al. Targeting the miR-665-3p-ATG4B-autophagy axis relieves inflammation and apoptosis in intestinal ischemia/reperfusion. Cell Death Dis. 2018;9:483. doi: 10.1038/s41419-018-0518-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C, et al. MicroRNA-665 suppressed the invasion and metastasis of osteosarcoma by directly inhibiting RAB23. Am. J. Transl. Res. 2016;8:4975–4981. [PMC free article] [PubMed] [Google Scholar]
- 14.Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. J. Hepatol. 2015;63:1491–1501. doi: 10.1016/j.jhep.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim W, et al. Hepatic Hippo signaling inhibits protumoural microenvironment to suppress hepatocellular carcinoma. Gut. 2017;67:1692–1703. doi: 10.1136/gutjnl-2017-314061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi Y, Dai Y, Gui S. Protein tyrosine phosphatase PTPRB regulates Src phosphorylation and tumour progression in NSCLC. Clin. Exp. Pharmacol. Physiol. 2016;43:1004–1012. doi: 10.1111/1440-1681.12610. [DOI] [PubMed] [Google Scholar]
- 17.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 19.Jukam D, et al. The insulator protein BEAF-32 is required for Hippo pathway activity in the terminal differentiation of neuronal subtypes. Development. 2016;143:2389–2397. doi: 10.1242/dev.134700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poon CL, Mitchell KA, Kondo S, Cheng LY, Harvey KF. The Hippo pathway regulates neuroblasts and brain size in Drosophila melanogaster. Curr. Biol. 2016;26:1034–1042. doi: 10.1016/j.cub.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 21.He S, Zhang J, Lin J, Zhang C, Sun S. Expression and function of microRNA-27b in hepatocellular carcinoma. Mol. Med. Rep. 2016;13:2801–2808. doi: 10.3892/mmr.2016.4851. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, et al. A microRNA-based prediction model for lymph node metastasis in hepatocellular carcinoma. Oncotarget. 2016;7:3587–3598. doi: 10.18632/oncotarget.6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamori A, et al. MicroRNA expression in hepatocellular carcinoma after the eradication of chronic hepatitis virus C infection using interferon therapy. Hepatol. Res. 2016;46:E26–E35. doi: 10.1111/hepr.12518. [DOI] [PubMed] [Google Scholar]
- 24.Childs-Disney JL, Disney MD. Small molecule targeting of a microRNA associated with hepatocellular carcinoma. Acs. Chem. Biol. 2016;11:375–380. doi: 10.1021/acschembio.5b00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, et al. Screening of differential microRNA expression in gastric signet ring cell carcinoma and gastric adenocarcinoma and target gene prediction. Oncol. Rep. 2015;33:2963–2971. doi: 10.3892/or.2015.3935. [DOI] [PubMed] [Google Scholar]
- 26.Smith Bethany, Bhowmick Neil. Role of EMT in Metastasis and Therapy Resistance. Journal of Clinical Medicine. 2016;5(2):17. doi: 10.3390/jcm5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Yanyuan, Sarkissyan Marianna, Vadgama Jaydutt. Epithelial-Mesenchymal Transition and Breast Cancer. Journal of Clinical Medicine. 2016;5(2):13. doi: 10.3390/jcm5020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julien SG, Dube N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat. Rev. Cancer. 2011;11:35–49. doi: 10.1038/nrc2980. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 30.Behjati S, et al. Recurrent PTPRB and PLCG1 mutations in angiosarcoma. Nat. Genet. 2014;46:376–379. doi: 10.1038/ng.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding L, et al. Clonal architectures and driver mutations in metastatic melanomas. PLoS. One. 2014;9:e111153. doi: 10.1371/journal.pone.0111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong J, et al. Elucidation of a universal size-control mechanism in drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinhardt AA, et al. Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]