Abstract
Hypoxia is beneficial for the differentiation of stem cells transplanted for myocardial injury, but mechanisms underlying this benefit remain unsolved. Here, we report the impact of hypoxia-induced Jagged1 expression in cardiomyocytes (CMs) for driving the differentiation of cardiac stem cells (CSCs). Forced hypoxia-inducible factor 1α (HIF-1α) expression and physical hypoxia (5% O2) treatment could induce Jagged1 expression in neonatal rat CMs. Pharmacological inhibition of HIF-1α by YC-1 attenuated hypoxia-promoted Jagged1 expression in CMs. An ERK inhibitor (PD98059), but not inhibitors of JNK (SP600125), Notch (DAPT), NF-κB (PTDC), JAK (AG490), or STAT3 (Stattic) suppressed hypoxia-induced Jagged1 protein expression in CMs. c-Kit+ CSCs isolated from neonatal rat hearts using a magnetic-activated cell sorting method expressed GATA4, SM22α or vWF, but not Nkx2.5 and cTnI. Moreover, 87.3% of freshly isolated CSCs displayed Notch1 receptor expression. Direct co-culture of CMs with BrdU-labeled CSCs enhanced CSCs differentiation, as evidenced by an increased number of BrdU+/Nkx2.5+ cells, while intermittent hypoxia for 21 days promoted co-culture-triggered differentiation of CSCs into CM-like cells. Notably, YC-1 and DAPT attenuated hypoxia-induced differentiation. Our results suggest that hypoxia induces Jagged1 expression in CMs primarily through ERK signaling, and facilitates early cardiac lineage differentiation of CSCs in CM/CSC co-cultures via HIF-1α/Jagged1/Notch signaling.
Abbreviations: BMSCs, bone marrow stem cells; BrdU, 5-bromo-2′-deoxyuridine; CMs, cardiomyocytes; CSCs, cardiac stem cells; DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; ERK, extracellular signal-regulated kinase; FBS, fetal bovine serum; FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; HIF-1α, hypoxia-inducible factor 1α; HRE, hypoxia responsive element; JAK, Janus kinase; JNK, c-Jun N-terminal kinase; MACS, magnetic-activated cell sorting; MI, myocardial infarction; MOI, multiplicity of infection; NF-κB, nuclear factor κB; N-ICD, notch intracellular domain; PBS, phosphate buffer saline; PE, phycoerythrin; qPCR, quantitative PCR; RT-PCR, reverse transcription PCR; STAT3, signal transducer and activator of transcription 3; vWF, von Willebrand factor; YC-1, 3-(5′-hydroxymethyl-2′-furyl)-1-benzyl-indazole
KEY WORDS: Cardiac stem cell; Cardiomyocyte, Co-culture; Hypoxia; Notch1 signaling; Cell differentiation
Graphical abstract
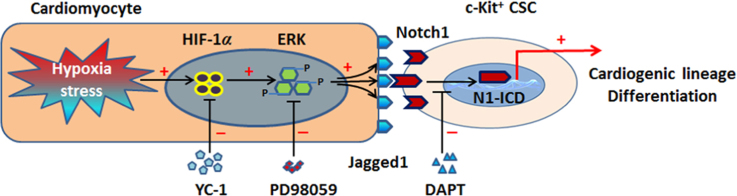
Hypoxia stress provokes Jagged1 expression in cardiomyocytes mainly through ERK signaling, subsequently leads to activation of Notch1 signaling in c-Kit+ cardiac stem cells (CSCs) via direct cell contact, which in turn favors cardiogenic lineage differentiation in CSCs.
1. Introduction
Stem cell transplantation has gradually become a promising therapeutic strategy for ischemic heart diseases such as myocardial infarction (MI). Despite some negative reports, accumulating data from preclinical and phases I and II clinical trials has provided evidence that stem cell transplantation is beneficial for the recovery of cardiac function following MI1, 2, 3, 4. However, the molecular mechanisms underlying cardiac functional recovery by stem cells suggest mostly paracrine and angiogenic effects, and only a small ratio of direct cell differentiation5, 6, 7. Considering the complexity of the regional myocardial tissue microenvironment after MI, mechanisms underpinning the contributions of stem cells to heart repair still remain unresolved, especially considering the low rate of cell differentiation.
Lineage negative (Lin−)/c-Kit positive (c-Kit+) cardiac stem cells (CSCs) are a resident adult stem cell population present in heart tissue8. Depletion of Lin+ cells is not necessary for enrichment of Lin−/c-Kit+ CSCs9. c-Kit+ CSCs, which are mainly localized in the atria, apex, and atrioventricular junction area8, display potential for self-renewal, clonogenicity, and differentiation10. c-Kit+ CSCs exhibited chemotaxis towards stem cell factor, and thereafter participated in repair of the injured heart11, 12, 13. Results of Stem Cell Infusion in Patients with Ischemic cardiOmyopathy (SCIPIO) clinical trials investigating c-Kit+ CSCs showed potential for treatment of MI3. Under a physiological state, CSCs localize to niches within heart tissues14, which are comprised by lineage-committed cardiomyocytes (CMs) and fibrocytes that act as the main supporting cells15, 16. By neighboring non-CMs, CSCs maintain their stemness in response to the balance of complex signals in their microenvironment. After stress such as MI, the hemostasis of niches is destroyed and certain signaling is activated to determine the fate of activated stem cells.
Notch signaling is a conserved pathway that mediates cellular processes such as proliferation, development, and differentiation17; the Notch pathway is retained in the CSC niche. Four receptors (Notch1–4) and five ligands (Jagged1–2; Delta-like 1, 3, and 4) of Notch signaling have been discovered in mammalian cells. After binding of the Notch receptor to its ligand, Notch signaling is activated, the Notch receptor is catalyzed, and the notch intracellular domain (N-ICD) is released. The N-ICD is thereafter translocated into the nucleus, where it binds with a co-activator to mediate downstream gene expression18. As a dogma, the canonical activation of Notch signaling reportedly limits neighboring cells. Abnormalities of Notch1 signaling lead to physiological defects in heart development19, as well as pathological deterioration of cardiac function20, 21, supporting a cardioprotective role for Notch signaling in the cardiovascular system. Our previous report found that activation of Notch1 signaling is beneficial for the differentiation of c-Kit+/Nkx2.5+ bone marrow stem cells (BMSCs) into CM-like cells in vitro22. The Sussman laboratory also reported Notch signaling to be pivotal for the fate of CSCs in vitro and in vivo23. However, it is still unclear how Notch signaling in CSC niches is activated under pathological conditions such as MI.
Hypoxia-inducible factor 1α (HIF-1α), a nuclear translational factor, is reportedly crucial for cardioprotection post-MI24. By mediating target genes harboring a hypoxia responsive element (HRE), HIF-1α plays an important biological role in hypoxic-ischemic diseases25. One mechanism of HIF-1α-mediated cardioprotection following MI is the activation of endogenous CSCs26; however, how HIF-1α activates endogenous CSCs remains largely unknown.
In this study, we found that hypoxia-impacted CMs could promote c-Kit+ CSCs differentiation via cell contact-triggered HIF-1α/Jagged1/Notch1 signaling. Our results provide new insight into the role of HIF-1α in CSCs differentiation after heart injury.
2. Methods and methods
2.1. Animals and ethics statement
Neonatal SD rat pups (within 72 h after birth) were purchased from Guangdong Medical Laboratory Animal Center (Guangzhou, China). All animal procedures were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Guangdong Medical University.
2.2. Isolation of CMs and c-Kit+ CSCs from neonatal rats
Neonatal CMs were isolated according to previously described standard protocols27, 28. CMs were maintained in DMEM/F-12 (Hyclone, Beijing, China) supplemented with 0.1 mmol/L 5-bromo-2′-deoxyuridine (BrdU) (B5002, Sigma, Shanghai, China), 10% fetal bovine serum (FBS, Gibco, Invitrogen, Carlsbad, CA), 100 U/mL penicillin, and 0.1 μg/mL streptomycin for 3 days, after which medium was changed to complete DMEM/F-12 without BrdU. c-Kit+ CSCs were isolated using a previously described magnetic-activated cell sorting (MACS) method protocol12, 22. CSCs were suspended in stem cell media (RASMX-90011, Cyagen, Guangzhou, China) supplemented with 100 U/mL leukemia inhibitory factor (LIF; LIF3010, Millipore, Burlington, MA, USA), 10% FBS, 100 U/mL penicillin, and 0.1 μg/mL streptomycin at 37 °C with saturated humidity. CSCs at passage 1 (P1) were subjected to identification of c-Kit and lineage markers by flow cytometry, reverse transcription-quantitative PCR (RT-PCR), and immunofluorescence. P3–P5 CSCs were used for associated experiments.
2.3. Forced expression of HIF-1α in CMs
Green fluorescent protein (GFP)-tagged HIF-1α-overexpressing lentivirus (HIF-1α-lent) and negative control lentivirus (NC-lent) were obtained from GeneCopoeia (FulenGen, Guangzhou, China). CMs were infected with virus at multiplicity of infection (MOI) of 100 and 30. Three days post-infection, GFP signals were observed under a microscope to judge the efficiency of viral infection. Cells post-viral infection for 1 day and 5 days were harvested and subjected to analysis.
2.4. Hypoxia and inhibitor treatment of CMs
Neonatal rat CMs were plated in 6-well plates. To mimic the hypoxic injury of myocardium, cells were maintained in a 5% O2 incubator (Galaxy 48R, Eppendorf, Germany) for various periods of time, and then harvested for associated experiments. Before hypoxic treatment, hypoxia (5% O2)-balanced DMEM/F-12 medium was changed. To further understand the mechanisms underlying hypoxia-induced Jagged1 expression in CMs, the following inhibitors were used: YC-1 (HIF-1α inhibitor, 10 μmol/L; Y102, Sigma), PD98059 [extracellular-regulated signal kinase (ERK) inhibitor, 50 nmol/L; sc-3532, Santa Cruz Biotechnology, Dallas, TX, USA], PTDC [nuclear factor κB (NF-κB) inhibitor, 10 μmol/L; P8765, Sigma), DAPT (Notch inhibitor, 10 μmol/L; D5942, Sigma), SP600125 [c-Jun N-terminal kinase (JNK) inhibitor, 25 μmol/L; S1460, Selleck Chemicals, Houston, TX, USA], AG490 [Janus kinase (JAK) inhibitor, 25 μmol/L; T3434, Sigma), and Stattic [signal transducer and activator of transcription 3 (STAT3) inhibitor,10 μmol/L; S7949, Sigma]. Dimethyl sulfoxide (DMSO; 0.1%, v/v) was used as a solvent control.
2.5. Co-culture of CSCs and CMs
BrdU (20 μmol/L) was added to medium for 48 h to label c-Kit+ CSCs, then 1×106 labeled CSCs were plated in 6-well plates on coverslips. After one night of recovery, CMs (ratio to CSCs, 10:1) were added to wells to allow direct interaction of CSCs and CMs. Cells were divided into four groups: (1) co-culture of CSCs and CMs (CSC+CM) under normoxia; (2) CSC+CM under hypoxia; (3) co-culture of 10 μmol/L YC-1-treated CSCs and CMs (CSC+CM+YC-1) under hypoxia; (4) Co-culture of 10 μmol/L DAPT-treated CSCs and CMs (CSC+CM+DAPT) under hypoxia. According to the pathophysiology of heart injury such as MI, both the transplanted exogenous CSCs and CMs in marginal zone of injury are under hypoxic conditions, we thus cultured the cell mixture in an atmosphere with 5% O2 as the hypoxic model. For hypoxic treatment, mixed cells were maintained with intermittent hypoxic cycles of 5% O2 for 12 h and 21% O2 for 24 h, for a total 21 days. Medium was changed every 48 h. After 21 days, coverslips were washed and fixed in cold paraformaldehyde for 15 min to prepare for immunofluorescence staining. For flow cytometrical analysis of cardiogenic lineage differentiation of CSCs, grouped cells were seeded on 6-cm dishes and subjected to hypoxic treatment mentioned above.
2.6. Flow cytometry
Flow cytometry was used to detect the expression of c-Kit, BrdU, Notch1, and Nkx2.5. CSCs were collected, labeled with antibodies for 30 min at room temperature, and washed twice with cold phosphate buffer saline (PBS). For c-Kit and Notch1 detection, phycoerythrin (PE)-coupled IgGs were added to allow further binding. For BrdU and Nkx2.5 detection, fluorescein (FITC)-BrdU and PE-Nkx2.5 antibodies were used. Non-immune IgG was used as a control. Cold PBS-washed cells were then subjected to flow cytometrical analysis. Data were collected and analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Three repeated wells were for each target genes.
2.7. Immunofluorescence staining
Cells grown on coverslips were subjected to indirect immunofluorescence to identify the expression and location of c-Kit, BrdU, and differentiation markers [Nkx2.5, cardiac troponin I (cTnI)]. Coverslips were washed in PBS three times and then treated with 0.1% Triton-PBS for 10 min. For BrdU staining, cells were incubated in 1 mol/L HCl for an additional 15 min. After washing with PBS, 0.3% bovine serum albumin was added to block non-immunological antigens. Diluted primary antibodies were then added to coverslips, which were allowed to incubate overnight at 4 °C. Antigen sites were localized with PE- and FITC-coupled IgGs (ProteinTech, Wuhan, China). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, 5 μg/mL) at room temperature for 15 min. Images were captured under a laser-scanning confocal microscope (TCS SP5 II, Leica, Germany). Details of primary antibodies are listed in Supplementary information Table S1.
2.8. RNA isolation and RT-PCR
Total RNA from cells and heart tissues were extracted with Trizol (15596-026, Invitrogen). To generate cDNA, 500 ng of total RNA was reverse transcribed using an RT kit (DRR047A, Takara, Kusatsu, Japan) with oligo(dT18) primers. Quantitative PCR (qPCR) was conducted using a LightCycler480 II instrument (Roche China, Guangzhou, China), and semi-quantitative PCR was performed using a Mastercycler® Gradient Thermal Cycler (Eppendorf, Germany). Primers were synthesized by Sangon Biotech (Shanghai, China) and are listed in Supplementary information Table S2. The values of melting temperatures were set as 60 °C. For all PCR analyses, the expression level of β-actin served as a loading control. The 2−∆∆Ct method was used to determine levels of target gene expression in qPCR.
2.9. Western blotting
CMs were homogenized with RIPA buffer (P0013B, Beyotime, Jiangsu, China) containing protease inhibitors. Protein homogenates were separated by SDS-PAGE and then transferred to nitrocellulose membranes, which were incubated with primary antibodies against Jagged1, HIF-1α, HIF-2α, ERK1/2, p-ERK1/2 and GAPDH at 4 °C overnight. Membranes were washed twice and incubated with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature. Protein bands were visualized using enhanced chemiluminescent reagents (Pierce, Rockford, IL) and analyzed using an InGenius LHR Gel Documentation System (Syngene, Frederick, MD).
2.10. Statistical analysis
Statistical analyses were conducted using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA). Data are expressed as means±SEM. Differences between two groups were analyzed using Student׳s t-test. For comparison of multiple groups, analysis of variance (ANOVA) tests were used followed by the Student-Newman-Keuls test. P<0.05 was considered to be statistically significant.
3. Results
3.1. Forced HIF-1α expression upregulated Jagged1 expression in CMs
To assess the functional role of HIF-1α on Jagged1 expression, we infected CMs with HIF-1α-overexpressing lentivirus (HIF-1α-lent). Under the microscope, GFP signal was localized in the nuclei of CMs, indicating correct localization of HIF-1α (Fig. 1A). As CMs exhibited low efficiency for gene transfection, we infected CMs with lentivirus at high MOI. At a MOI of 100, we found that forced expression of HIF-1α effectively upregulated Jagged1 expression in CMs (Fig. 1B), and this HIF-1α-triggered Jagged1 expression was positive related to lentiviral titers (Fig. 1C and D). Thus, Jagged1 expression could be mediated by HIF-1α.
Figure 1.
Forced HIF-1α expression in cardiomyocytes led to Jagged1 upregulation. (A) Primary neonatal rat cardiomyocytes infected with GFP-tagged HIF-1α-overexpressing lentivirus exhibited nuclear GFP signals. (B) Quantitative RT-PCR was used to detect Jagged1 mRNA levels in HIF-1α-overexpressing lentivirus-infected cardiomyocytes at a MOI of 100 on days 1 and 5. (C) Quantitative RT-PCR and (D) western blot were used to examine changes in mRNA and protein of Jagged1, respectively, in cardiomyocytes infected with HIF-1α-overexpressing lentivirus at MOIs of 30 and 100 on day 5. For (B), (C) and (D), data represent mean±SEM of three independent experiments (n=3). *P<0.05, **P<0.01, ***P<0.001.
3.2. Hypoxic stimuli sufficiently enhanced Jagged1 expression in CMs
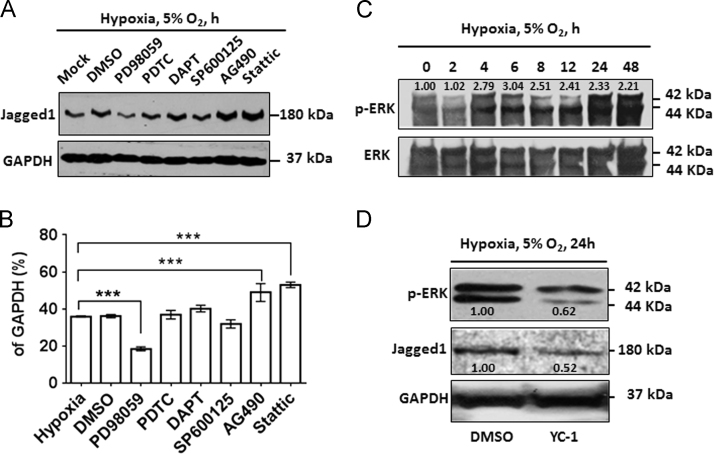
To further mimic the condition of MI, we next cultured CMs in a hypoxic incubator. According to Regula׳s report29, we used 5% O2 as our optimal hypoxic condition. When compared with normoxic (21% O2) controls, hypoxic stimuli significantly upregulated HIF-1α, but not HIF-2α, expression in CMs (Fig. 2A). Under the 5% O2 condition, hypoxia-mediated Jagged1 expression displayed a tendency to sustained increase after an initial transiently decline. At 8 h post-hypoxic treatment, the Jagged1 mRNA level reached a peak value (Fig. 2B), while the protein peak level lagged behind mRNA (Fig. 2C). To confirm that increased Jagged1 was HIF-1α-dependent, the HIF-1α inhibitor YC-1 [3-(5′-hydroxymethyl-2′-furyl)-1-benzyl-indazole] was used to block HIF-1α activity30, 31, 32. As expected, YC-1 significantly attenuated hypoxia-induced changes in Jagged1 mRNA levels (Fig. 2D).
Figure 2.
Physical hypoxia (5% O2) treatment increased Jagged1 expression in cardiomyocytes. (A) Physical hypoxia treatment of cardiomyocytes for 12 h led to significant upregulation of HIF-1α, but not HIF-2α. (B) Quantitative RT-PCR and (C) western blot were used to examine changes in Jagged1 mRNA and protein, respectively, in hypoxia-treated primary neonatal rat cardiomyocytes. (D) The HIF-1α inhibitor YC-1 (10 μmol/L) attenuated hypoxia-stimulated Jagged1 mRNA changes in cardiomyocytes. For all the panels, data represent mean±SEM of three independent experiments (n=3). *P<0.05, ***P<0.001.
3.3. Hypoxia enhanced Jagged1 expression in CMs mainly through ERK signaling
To understand the underlying signaling mediating hypoxia-induced Jagged1 expression in CMs, inhibitors of ERK (PD98059, 50 nmol/L), NF-κB (PTDC, 10 μmol/L), JNK (SP600125, 25 μmol/L), Notch (DAPT, 10 μmol/L), JAK (AG490, 25 μmol/L) and STAT3 (Stattic, 10 μmol/L) were applied to hypoxia-treated CMs. Results of western blotting indicated obvious attenuation of hypoxia-regulated Jagged1 protein expression by PD98059, while SP600125 had a slight effect, indicating ERK signaling was primarily involved in hypoxia-mediated Jagged1 expression. In contrast, NF-κB and Notch signaling showed no impact on Jagged1 expression under the hypoxic condition. Interestingly, although both AG490 and Stattic increased Jagged1 protein expression in hypoxia-treated CMs, JAK/STAT3 signaling appeared to downregulate Jagged1 protein expression in the current system (Fig. 3A and B). Further experimental results confirmed that hypoxia treatment activated ERK signaling, as evidenced by increased levels of 44 kDa phosphorylated ERK (p-ERK) (Fig. 3C). Notably, inhibition of HIF-1α by YC-1 led to a blockage of ERK signaling activation that was accompanied by downregulation of Jagged1 protein (Fig. 3D).
Figure 3.
Physical hypoxia (5% O2) induced Jagged1 expression in cardiomyocytes primarily through ERK signaling. (A) Jagged1 protein expression in hypoxia condition-maintained cardiomyocytes treated with DMSO (0.1%, v/v), PD98059 (50 nmol/L), PDTC (10 μmol/L), DAPT (10 μmol/L), SP600125 (25 μmol/L), AG490 (25 μmol/L), or Static (10 μmol/L) for 8 h detected by Western blot (B) was quantified relative to GAPDH levels. (C) Protein levels of total ERK and p-ERK were assessed by western blot in hypoxia-treated cardiomyocytes. (D) YC-1 inhibited hypoxia-activated ERK signaling and Jagged1 protein expression in cardiomyocytes. For (B), (C) and (D), data represent mean±SEM of three independent experiments (n=3). ***P<0.001.
3.4. Identification of rat CSCs
As young CSCs possess more biological advantages than old cells33, 34, and c-Kit+ CSCs are functionally competent for myocyte regeneration35, we isolated c-Kit+ CSCs from neonatal rats. Newly isolated neonatal rat CSCs were obviously positive for c-Kit (c-Kit+) and GATA4 (GATA4+), but negative for Nkx2.5 (Nkx2.5−) and cTnI(cTnI–). In addition, these cells were positive for makers of mature smooth muscle cells (SM22α+), epithelial cells (vWF+) (Fig. 4A, B and C). By flow cytometry analysis, 81.3% of isolated c-Kit+ CSCs expressed Notch1 receptor, while the purity of c-Kit+ CSCs following MACS reached 87.3% (Fig. 4D). The population-doubling time of c-Kit+ CSCs was 3.95±0.2 days (data not shown). Thus, freshly isolated CSCs were c-Kit+/Gata4+/Nkx2.5−/cTnI–/SM22α+/vWF+.
Figure 4.
Identification of c-Kit+ cardiac stem cells (CSCs). Neonatal rat c-Kit+ CSCs were isolated via a MACS method, and cells at Passage 1 were subjected to identification. (A) Cell morphology. (B) Detection of c-Kit, Nkx2.5 and cTnI expression by immunofluorescence. (C) Sub-quantitative RT-PCR was used to examine c-Kit and markers for cardiomyocyte (Nkx2.5, Gata4, cTnI), smooth muscle cell (SM22α), and epithelial cell (vWF) lineage differentiation. (D) Flow cytometry was used to analyze levels of c-Kit and Notch1 receptor in CSCs. For (D), data represent mean±SEM of three independent experiments (n=3).
3.5. Blocking HIF-1α/Notch signaling attenuates CSC-CM co-culture–increased CSC differentiation
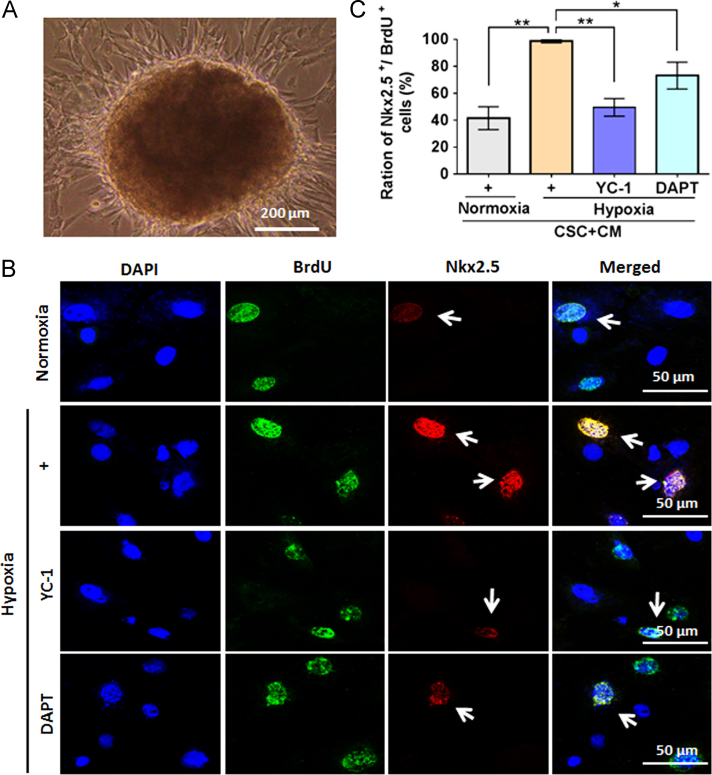
One report indicated that c-Kit was stably expressed in CSCs, even after 10 passages or a total of 40–50 days of culture9. To maintain the stability of CSCs in the present study, c-Kit+ CSCs at early passages (P3–P5) were used for associated experiments. When co-culturing c-Kit+ CSCs with CMs, these mixed cells could form cardiosphere-like structures (Fig. 5A). A small proportion of BrdU-tagged CSCs visualized by immunofluorescent staining were weakly positive for Nkx2.5, indicating co-culture of CMs and CSCs promoted CSCs differentiation (Fig. 5B, upper panels). Interestingly, this co-culture-induced CM-like differentiation was amplified under hypoxic conditions, while blocking HIF-1α (with YC-1) and Notch1 signaling (with DAPT) could suppress this differentiation tendency (Fig. 5B and C).
Figure 5.
Early cardiac differentiation of c-Kit+ cardiac stem cells (CSCs) after co-culture with cardiomyocytes. (A) Cardiosphere structures formed when c-Kit+ CSCs were co-cultured with cardiomyocytes. (B) Immunofluorescence staining was used to detect expression of Nkx2.5, an early marker for cardiac lineage in BrdU-labeled c-Kit+ CSCs. (C) Relative quantitative analysis of ratios of cells double-positive for Nkx2.5 and BrdU after co-culture of c-Kit+ CSCs with cardiomyocytes. For (C), data represent mean±SEM of 6 representative fluorescent fields of cells growth on the coverslips (n=6). *P<0.05, **P<0.01.
We further applied flow cytometry to quantify the efficiency of differentiation provoked by co-couture under hypoxia. As shown in Fig. 6, cells in the Q2 gate were positive for BrdU and Nkx2.5, indicating labeled CSCs in the early process of CM-like differentiation. The ratio of cells double-positive for BrdU and Nkx2.5 in the hypoxic condition was 7.14 times greater than in the normoxic condition. Again, YC-1 and DAPT decreased the ratio of BrdU and Nkx2.5 double-positive cells under hypoxia treatment. Cells in the Q4 gate were positive for Nkx2.5, representing a mixture of developing neonatal rat CMs and differentiating unlabeled CSCs. We observed that changes in the Q4 gate were similar to those of the Q2 gate for each groups.
Figure 6.
Quantitative analyses of Nkx2.5+ cells after co-culture BrdU-labeled c-Kit+ cardiac stem cells (CSCs) with cardiomyocytes. BrdU-labeled c-Kit+ CSCs were co-cultured with cardiomyocytes at a ratio of 1:6 under normoxic or hypoxic conditions for 21 days, and then (A) flow cytometry was used to analyze cells positive for both Nkx2.5 and BrdU (gate 2), and only positive for Nkx2.5 (gate 4). Cells in the normoxic condition were set as threshold and a total 2×104 cells were analyzed in each group. Fold-changes in cells (B) double-positive for Nkx2.5 and BrdU, and (C) only positive for Nkx2.5 were calculated. Data represent mean±SEM of three independent experiments (n=3). **P<0.01 vs. other groups.
4. Discussion
In the present investigation, we found that hypoxia (5% O2) treatment upregulated Jagged1 expression in CMs. Moreover, this hypoxia-dependent Jagged1 increase was mainly mediated through ERK signaling. When CSCs were co-cultured with CMs, their differentiation into a CM lineage under hypoxia was amplified compared with normoxia, indicating the effectiveness of hypoxic stimuli for CSCs differentiation. Interestingly, both YC-1 and DAPT could block early differentiation of hypoxia-amplified CSCs, suggesting a crucial role of HIF-1α/Notch1 signaling in hypoxia-induced CSCs differentiation. Our results reveal activation of Notch signaling through cell-to-cell contact as a new mechanism for determining the lineage commitment of CSCs.
Upregulation of HIF-1α is pivotal in cardioprotection following ischemic injury24. Although the underlying mechanisms are complex and as of yet unresolved, the molecule HIF-1α has attracted much attention for cardioprotection. Several reports have indicated that forced expression of HIF-1α in transplanted stem cells significantly favors their biological functions in vivo7, 36, 37, highlighting an important role for HIF-1α in stem cell transplantation after myocardial injury. We found that after hypoxia treatment, HIF-1α, but not HIF-2α, was most highly upregulated in CMs. Our current investigation focused on the role of HIF-1α/Notch1 signaling in the differentiation in cardiac stem cells.
The Notch pathway is an evolutionary conserved intercellular and intracellular mechanism that controls stem cell fate. In CSCs niches, the Notch1 receptor is presented on CSCs, while its ligand Jagged1 is presented on supporting cells16, 38. Activation of Notch1 signaling through its canonical mechanism favors the cardiogenic lineage commitment of stem cells22, 23, 39. In the current study, we successfully isolated c-Kit+ CSCs from neonatal rats that displayed near 81.3% purity for Notch1 receptor expression, positivity for GATA4, SM22α and vWF, and were negative for Nkx2.5 and cTnI. These results suggest c-Kit+ CSCs in the current study contained a low ratio of early cardiogenic, smooth muscle cell and epithelial cell lineage-committed cells.
As the primary supporting cells of the CSCs niche, CMs express the Notch ligand Jagged1, which exhibited enhanced expression in CMs after stress40. To further understand the mechanism underlying stress-induced Jagged1 expression, we first infected CMs with HIF-1α-overexpressing lentivirus. The results functionally demonstrated that Jagged1 expression was induced in CMs in a HIF-1α-dependent manner. Subsequently, physical hypoxia stimuli (5% O2) also effectively provoked Jagged1 expression in CMs; notably, the HIF-1α inhibitor YC-1 blocked this effect. We also found that physical hypoxia treatment of CMs led to a transient decline and then increase of Jagged1 expression. Due to the large demand of oxygen, CMs are very sensitive to the damage caused by low oxygen concentration. Thus, the trend of changes in Jagged1 expression in CMs may reflect the adaptive response of CMs to hypoxia treatment. As various signaling is activated post-MI in local injured myocardium, we sought to uncover the exact mechanism underpinning hypoxia-induced Jaggged1 expression. We focused on stress and inflammatory pathways, namely ERK, NF-κB, JAK, and JNK/Stat3 signaling. As a result of its function in heart development, Notch1 signaling was also included in this study. Our results showed that PD98059 displayed the most obvious Jagged1 decrease in hypoxia-treated CMs, while SP600125 produced only slight Jagged1 downregulation. However, PTDC and DAPT showed no effects on Jagged1 expression in hypoxia-treated CMs, while AG490 and Stattic increased Jagged1 expression. Thus, we concluded that hypoxia mediated Jagged1 expression mainly through ERK signaling. These results are consistent with previous reports indicating an effect of hypoxia on Notch signaling activation41, 42, 43. For example, forced HIF-1α expression or hypoxia could activate Notch signaling by upregulating Notch ligand Dll4 or Notch1 receptor in arterial differentiation of mouse embryonic stem cells41, 43. A very recent report also showed the functional association of HIF-1α with Jagged1 expression in BMSCs42. However, our current results provide further molecular evidence about the effect of hypoxia on Jagged1 expression, which shed more light on hypoxia-induced Notch signaling activation.
To mimic the contact between CMs and transplanted CSCs in vivo, we directly co-cultured CMs and CSCs in vitro. Although CSCs alone did not show Nkx2.5 expression, upon co-culture with CMs they formed 3-D cardiosphere structures with increased potential for cell contact, thus reforming the stem cell niche microenvironment in vitro. The CSCs we isolated were negative for Nkx2.5 and cTnI before co-culture. Co-culture of CMs and CSCs under normoxia increased the ratio of Nkx2.5+ CSCs, indicating cell contact could enhance early CSCs differentiation. This was consistent with previous reports suggesting CMs could support cardiac lineage commitment of stem cells44, 45. Under the hypoxic condition, this enhancement of cardiogenic lineage differentiation by co-culture was further amplified, as indicated by an increased ratio of Nkx2.5+ cells. An early report29 indicated that CMs maintained in hypoxic condition with oxygen concentrations of <2% for more than 12 h would undergo significant apoptosis. Thus, we cultured CMs in 5% O2 to maintain their viability. For the current co-culture system, we adopted a model of intermittent hypoxia, which involved the repetitive treatment of cells with cycles of 5% O2 for 12 hours and 21% O2 for 24 h over the course of 21 days. By not applying continuous hypoxia, the CMs maintained an uninjured status that can provide more efficient contacts with CSCs. Ultimately, our results indicated the functional success of our intermittent hypoxic model, as it substantially increased the early cardiac lineage differentiation of CSCs co-cultured with CMs compared with normoxic controls. Previous report had proposed the preliminary functional role of co-culture of CMs with CSCs in the cardiomyogenesis46, Kubo et al.47 also found that co-culture of CMs with c-Kit+ BMSCs favored BMSCs differentiation into functional CMs. This co-cultured CMs-facilitated CSCs differentiation ability was further observed by Kaushal laboratory33. Our current investigation provides novel evidences about detailed molecular mechanisms on co-cultured CMs-triggered CSCs differentiation post stresses.
The current investigation has some limitations. First, while the co-culture system definitely provided potential cell-to-cell contacts, paracrine effects should also be considered. Second, some reports have suggested that soluble Jagged1 is released from certain cells after stress; this should be examined in follow-up studies. Third, signaling pathways involved in hypoxia-treated CMs beyond those mentioned in this study should also be tested. Fourth, a more comprehensive panel of cardiogenic lineage differentiation markers is needed in the future. Finally, well-designed animal study would strengthen the current results.
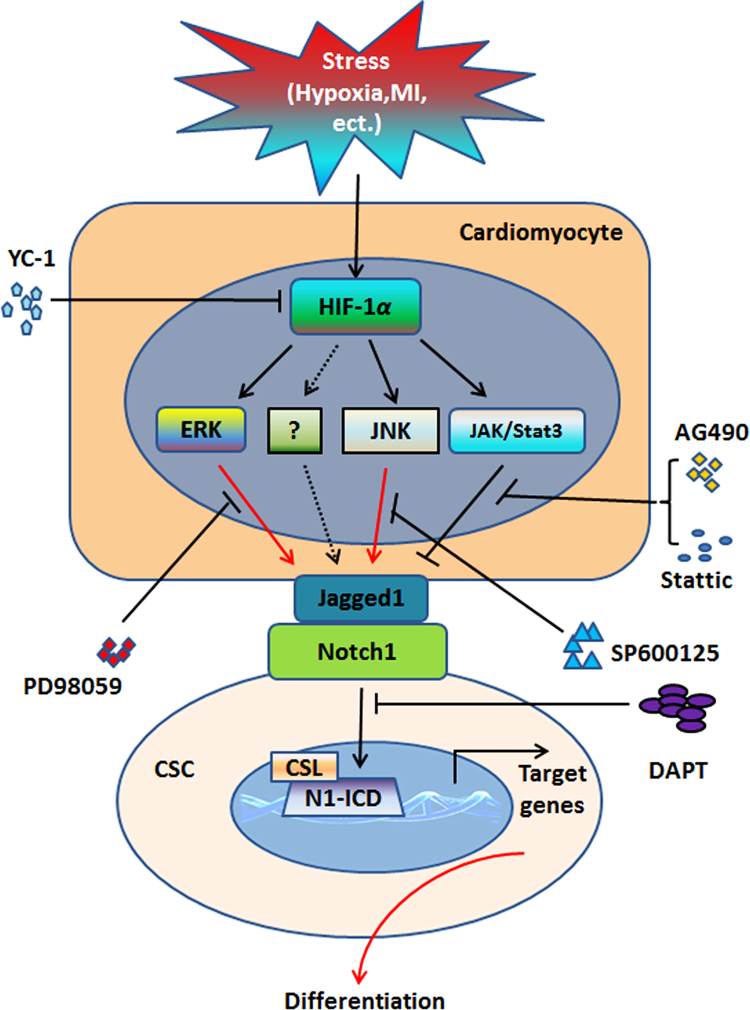
Taken together, our results indicate that hypoxia-stressed CMs promote early cardiac differentiation of CSCs through cell contacts and HIF-1α/Jagged1/Notch1 signaling, as illustrated in Fig. 7. The current investigation provides a new mechanism underlying hypoxia-facilitated stem cells differentiation, and sheds new light on the utility of hypoxia in stem cells-based translational medicine.
Figure 7.
Schematic illustration of mechanism by which cardiomyocytes promoted cardiac differentiation of c-Kit+ cardiac stem cells (CSCs) under hypoxic conditions. Under stress of hypoxia or other unfavorable conditions, enhanced HIF-1α may mediate Jagged1 expression in cardiomyocytes through ERK, JNK, or other unknown signaling pathways. Upregulated Jagged1 in cardiomyocytes binds with Notch1 receptors present on c-Kit+ CSCs, which directly contact cardiomyocytes, to activate Nocth1 signaling, thus mediating the cardiac differentiation of c-Kit+ CSCs. YC-1, HIF-1α inhibitor; PD98059, ERK inhibitor; SP600125, JNK inhibitor; AG490, JAK inhibitor; Stattic, Stat3 inhibitor; DAPT, Notch signaling inhibitor.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81170121, 81460042, 81541004 and 81670254), Science and Technology Project of Guangdong Province (2016A020214016), YangFan Plan of Guangdong Province (4YF16007G), and Excellent Graduate Student Training Program of Guangdong Medical University (YS2014013). We thank Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript. The authors declare no conflicts of interest in this work.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at 10.1016/j.apsb.2018.06.003.
Contributor Information
Junli Guo, Email: guojl79@163.com.
Wei Jie, Email: wei.jie@gdmu.edu.cn.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.Zwetsloot P.P., Vegh A.M., Jansen of Lorkeers S.J., van Hout G.P., Currie G.L., Sena E.S. Cardiac stem cell treatment in myocardial infarction: a systematic review and meta-analysis of preclinical studies. Circ Res. 2016;118:1223–1232. doi: 10.1161/CIRCRESAHA.115.307676. [DOI] [PubMed] [Google Scholar]
- 2.Makkar R.R., Smith R.R., Cheng K., Malliaras K., Thomson L.E., Berman D. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolli R., Chugh A.R., D׳Amario D., Loughran J.H., Stoddard M.F., Ikram S. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Guo J., Jie W., Kuang D., Ni J., Chen D., Ao Q. Ischaemia/reperfusion induced cardiac stem cell homing to the injured myocardium by stimulating stem cell factor expression via NF-kappaB pathway. Int J Exp Pathol. 2009;90:355–364. doi: 10.1111/j.1365-2613.2009.00659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang X.L., Li Q., Rokosh G., Sanganalmath S.K., Chen N., Ou Q. Long-term outcome of administration of C-Kit (Pos) cardiac progenitor cells after acute myocardial infarction: transplanted cells do not become cardiomyocytes, but structural and functional improvement and proliferation of endogenous cells persist for at least one year. Circ Res. 2016;118:1091–1105. doi: 10.1161/CIRCRESAHA.115.307647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao X., Liu Y., Gao J., Yang L., Mao D., Stefanitsch C. Nitric oxide releasing hydrogel enhances the therapeutic efficacy of mesenchymal stem cells for myocardial infarction. Biomaterials. 2015;60:130–140. doi: 10.1016/j.biomaterials.2015.04.046. [DOI] [PubMed] [Google Scholar]
- 7.Huang B., Qian J., Ma J., Huang Z., Shen Y., Chen X. Myocardial transfection of hypoxia-inducible factor-1alpha and co-transplantation of mesenchymal stem cells enhance cardiac repair in rats with experimental myocardial infarction. Stem Cell Res Ther. 2014;5:22. doi: 10.1186/scrt410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beltrami A.P., Barlucchi L., Torella D., Baker M., Limana F., Chimenti S. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 9.He J.Q., Vu D.M., Hunt G., Chugh A., Bhatnagar A., Bolli R. Human cardiac stem cells isolated from atrial appendages stably express c-kit. PLoS One. 2011;6:e27719. doi: 10.1371/journal.pone.0027719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bearzi C., Rota M., Hosoda T., Tillmanns J., Nascimbene A., De Angelis A. Human cardiac stem cells. Proc Natl Acad Sci U S A. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuang D., Zhao X., Xiao G., Ni J., Feng Y., Wu R. Stem cell factor/c-kit signaling mediated cardiac stem cell migration via activation of p38 MAPK. Basic Res Cardiol. 2008;103:265–273. doi: 10.1007/s00395-007-0690-z. [DOI] [PubMed] [Google Scholar]
- 12.Guo J., Jie W., Shen Z., Li M., Lan Y., Kong Y. SCF increases cardiac stem cell migration through PI3K/AKT and MMP2/9 signaling. Int J Mol Med. 2014;34:112–118. doi: 10.3892/ijmm.2014.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo K., Kuang D., Wang Y., Xia Y., Tong W., Wang X. SCF/c-kit transactivates CXCR4-serine 339 phosphorylation through G protein-coupled receptor kinase 6 and regulates cardiac stem cell migration. Sci Rep. 2016;6:26812. doi: 10.1038/srep26812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y., Yu X.Y., Wang Y. Recent concepts for the roles of progenitor/stem cell niche in heart repair. Am J Cardiovasc Dis. 2012;2:75–83. [PMC free article] [PubMed] [Google Scholar]
- 15.Urbanek K., Cesselli D., Rota M., Nascimbene A., De Angelis A., Hosoda T. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci U S A. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leri A., Rota M., Hosoda T., Goichberg P., Anversa P. Cardiac stem cell niches. Stem Cell Res. 2014;13:631–646. doi: 10.1016/j.scr.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borggrefe T., Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroeter E.H., Kisslinger J.A., Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 19.Luxan G., D׳Amato G., MacGrogan D., de la Pompa J.L. Endocardial Notch signaling in cardiac development and disease. Circ Res. 2016;118:e1–e18. doi: 10.1161/CIRCRESAHA.115.305350. [DOI] [PubMed] [Google Scholar]
- 20.Li Y., Hiroi Y., Liao J.K. Notch signaling as an important mediator of cardiac repair and regeneration after myocardial infarction. Trends Cardiovasc Med. 2010;20:228–231. doi: 10.1016/j.tcm.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin J., Hu H., Li X., Xue M., Cheng W., Wang Y. Inhibition of Notch signaling pathway attenuates sympathetic hyperinnervation together with the augmentation of M2 macrophages in rats post-myocardial infarction. Am J Physiol Cell Physiol. 2016;310:C41–C53. doi: 10.1152/ajpcell.00163.2015. [DOI] [PubMed] [Google Scholar]
- 22.Ding R., Jiang X., Ha Y., Wang Z., Guo J., Jiang H. Activation of Notch1 signalling promotes multi-lineage differentiation of c-Kit (POS)/NKX2.5 (POS) bone marrow stem cells: implication in stem cell translational medicine. Stem Cell Res Ther. 2015;6:91. doi: 10.1186/s13287-015-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gude N., Joyo E., Toko H., Quijada P., Villanueva M., Hariharan N. Notch activation enhances lineage commitment and protective signaling in cardiac progenitor cells. Basic Res Cardiol. 2015;110:29. doi: 10.1007/s00395-015-0488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tekin D., Dursun A.D., Xi L. Hypoxia inducible factor 1 (HIF-1) and cardioprotection. Acta Pharmacol Sin. 2010;31:1085–1094. doi: 10.1038/aps.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su H., Arakawa-Hoyt J., Kan Y.W. Adeno-associated viral vector-mediated hypoxia response element-regulated gene expression in mouse ischemic heart model. Proc Natl Acad Sci U S A. 2002;99:9480–9485. doi: 10.1073/pnas.132275299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang M., Nguyen P., Jia F., Hu S., Gong Y., de Almeida P.E. Double knockdown of prolyl hydroxylase and factor-inhibiting hypoxia-inducible factor with nonviral minicircle gene therapy enhances stem cell mobilization and angiogenesis after myocardial infarction. Circulation. 2011;124:S46–S54. doi: 10.1161/CIRCULATIONAHA.110.014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golden H.B., Gollapudi D., Gerilechaogetu F., Li J., Cristales R.J., Peng X. Isolation of cardiac myocytes and fibroblasts from neonatal rat pups. Methods Mol Biol. 2012;843:205–214. doi: 10.1007/978-1-61779-523-7_20. [DOI] [PubMed] [Google Scholar]
- 28.Louch W.E., Sheehan K.A., Wolska B.M. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol. 2011;51:288–298. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regula K.M., Baetz D., Kirshenbaum L.A. Nuclear factor-kappaB represses hypoxia-induced mitochondrial defects and cell death of ventricular myocytes. Circulation. 2004;110:3795–3802. doi: 10.1161/01.CIR.0000150537.59754.55. [DOI] [PubMed] [Google Scholar]
- 30.Segawa K., Fukuhara A., Hosogai N., Morita K., Okuno Y., Tanaka M. Visfatin in adipocytes is upregulated by hypoxia through HIF1alpha-dependent mechanism. Biochem Biophys Res Commun. 2006;349:875–882. doi: 10.1016/j.bbrc.2006.07.083. [DOI] [PubMed] [Google Scholar]
- 31.Zera K., Zastre J. Thiamine deficiency activates hypoxia inducible factor-1alpha to facilitate pro-apoptotic responses in mouse primary astrocytes. PloS One. 2017;12:e0186707. doi: 10.1371/journal.pone.0186707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masoud G.N., Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson D.L., Mishra R., Sharma S., Goh S.K., Deshmukh S., Kaushal S. A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation. 2012;126:S46–S53. doi: 10.1161/CIRCULATIONAHA.111.084699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castaldi A., Dodia R.M., Orogo A.M., Zambrano C.M., Najor R.H., Gustafsson A.B. Decline in cellular function of aged mouse c-kit+ cardiac progenitor cells. J Physiol. 2017;595:6249–6262. doi: 10.1113/JP274775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanada F., Kim J., Czarna A., Chan N.Y., Signore S., Ogorek B. c-Kit-positive cardiac stem cells nested in hypoxic niches are activated by stem cell factor reversing the aging myopathy. Circ Res. 2014;114:41–55. doi: 10.1161/CIRCRESAHA.114.302500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hnatiuk A.P., Ong S.G., Olea F.D., Locatelli P., Riegler J., Lee W.H. Allogeneic mesenchymal stromal cells overexpressing mutant human hypoxia-inducible factor 1-alpha (HIF1-alpha) in an ovine model of acute myocardial infarction. J Am Heart Assoc. 2016;5:e003714. doi: 10.1161/JAHA.116.003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerrada I., Ruiz-Sauri A., Carrero R., Trigueros C., Dorronsoro A., Sanchez-Puelles J.M. Hypoxia-inducible factor 1 alpha contributes to cardiac healing in mesenchymal stem cells-mediated cardiac repair. Stem Cells Dev. 2013;22:501–511. doi: 10.1089/scd.2012.0340. [DOI] [PubMed] [Google Scholar]
- 38.Boni A., Urbanek K., Nascimbene A., Hosoda T., Zheng H., Delucchi F. Notch1 regulates the fate of cardiac progenitor cells. Proc Natl Acad Sci U S A. 2008;105:15529–15534. doi: 10.1073/pnas.0808357105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Li H., Yu B., Zhang Y., Pan Z., Xu W., Li H. Jagged1 protein enhances the differentiation of mesenchymal stem cells into cardiomyocytes. Biochem Biophys Res Commun. 2006;341:320–325. doi: 10.1016/j.bbrc.2005.12.182. [DOI] [PubMed] [Google Scholar]
- 40.Croquelois A., Domenighetti A.A., Nemir M., Lepore M., Rosenblatt-Velin N., Radtke F. Control of the adaptive response of the heart to stress via the Notch1 receptor pathway. J Exp Med. 2008;205:3173–3185. doi: 10.1084/jem.20081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanner F., Lee K.L., Ortega G.C., Sohl M., Li X., Jin S. Hypoxia-induced arterial differentiation requires adrenomedullin and notch signaling. Stem Cells Dev. 2013;22:1360–1369. doi: 10.1089/scd.2012.0259. [DOI] [PubMed] [Google Scholar]
- 42.Ciria M., Garcia N.A., Ontoria-Oviedo I., Gonzalez-King H., Carrero R., De La Pompa J.L. Mesenchymal stem cell migration and proliferation are mediated by hypoxia-inducible factor-1alpha upstream of Notch and SUMO pathways. Stem Cells Dev. 2017;26:973–985. doi: 10.1089/scd.2016.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsang K.M., Hyun J.S., Cheng K.T., Vargas M., Mehta D., Ushio-Fukai M. Embryonic stem cell differentiation to functional arterial endothelial cells through sequential activation of ETV2 and NOTCH1 signaling by HIF1alpha. Stem Cell Rep. 2017;9:796–806. doi: 10.1016/j.stemcr.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubo H., Berretta R.M., Jaleel N., Angert D., Houser S.R. c-Kit+ bone marrow stem cells differentiate into functional cardiac myocytes. Clin Transl Sci. 2009;2:26–32. doi: 10.1111/j.1752-8062.2008.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeh Y.C., Wei H.J., Lee W.Y., Yu C.L., Chang Y., Hsu L.W. Cellular cardiomyoplasty with human amniotic fluid stem cells: in vitro and in vivo studies. Tissue Eng Part A. 2010;16:1925–1936. doi: 10.1089/ten.TEA.2009.0728. [DOI] [PubMed] [Google Scholar]
- 46.Belostotskaya G.B., Golovanova T.A. Characterization of contracting cardiomyocyte colonies in the primary culture of neonatal rat myocardial cells: a model of in vitro cardiomyogenesis. Cell Cycle. 2014;13:910–918. doi: 10.4161/cc.27768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kubo H., Berretta R.M., Jaleel N., Angert D., Houser S.R. c-Kit+ bone marrow stem cells differentiate into functional cardiac myocytes. Clin Transl Sci. 2009;2:26–32. doi: 10.1111/j.1752-8062.2008.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.