Pathogens that infect the gastrointestinal and respiratory tracts are subjected to intense pressure due to the environmental conditions of the surroundings. This pressure has led to the development of mechanisms of bacterial tolerance or persistence which enable microorganisms to survive in these locations.
KEYWORDS: persistence, resistance, respiratory infection, tolerance, treatments, gastrointestinal infection
SUMMARY
Pathogens that infect the gastrointestinal and respiratory tracts are subjected to intense pressure due to the environmental conditions of the surroundings. This pressure has led to the development of mechanisms of bacterial tolerance or persistence which enable microorganisms to survive in these locations. In this review, we analyze the general stress response (RpoS mediated), reactive oxygen species (ROS) tolerance, energy metabolism, drug efflux pumps, SOS response, quorum sensing (QS) bacterial communication, (p)ppGpp signaling, and toxin-antitoxin (TA) systems of pathogens, such as Escherichia coli, Salmonella spp., Vibrio spp., Helicobacter spp., Campylobacter jejuni, Enterococcus spp., Shigella spp., Yersinia spp., and Clostridium difficile, all of which inhabit the gastrointestinal tract. The following respiratory tract pathogens are also considered: Staphylococcus aureus, Pseudomonas aeruginosa, Acinetobacter baumannii, Burkholderia cenocepacia, and Mycobacterium tuberculosis. Knowledge of the molecular mechanisms regulating the bacterial tolerance and persistence phenotypes is essential in the fight against multiresistant pathogens, as it will enable the identification of new targets for developing innovative anti-infective treatments.
INTRODUCTION
The survival of bacteria is at least partly associated with the capacity of these microorganisms to detect and react to changes in environmental conditions. The machinery required to respond to environmental features is universally present in prokaryotic and eukaryotic cells. Several response mechanisms in bacteria are activated under stress conditions and controlled by proteins whose expression is associated with regulator genes. Interestingly, interactions between these mechanisms enable an efficient, coordinated response to multiple stressors.
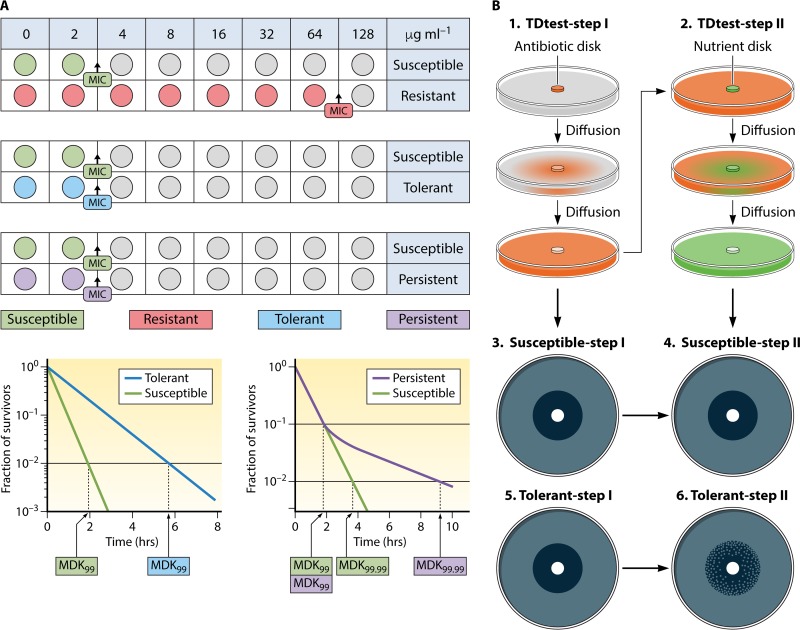
Antimicrobial resistance is one of the main problems of the 21st century. The rapid spread of multidrug-resistant (MDR) pathogens has been described as a global crisis that may lead to an era without effective antibiotics (1). Failure of antibiotic treatment is typically attributed to resistance. However, it has long been realized that other mechanisms, such as tolerance and persistence, can also help bacteria to survive antibiotic exposure (2). Resistant bacterial populations (resistance phenotype) have the following three main characteristics: (i) the use of active, mutation-associated defense mechanisms to withstand drug-induced stress; (ii) growth of the surviving cells under drug pressure; and (iii) an inherited phenotype. The cellular changes that result as effects of the mutations include inactivation of antibiotics by increasing efflux, modifying targets, and directly modifying the antibiotic (3–5). Tolerant bacterial populations (tolerance phenotype) are bacterial populations which can outlive exposure to raised concentrations of an antibiotic, without any modification of the MIC, by slowing down essential bacterial processes. Tolerance may be acquired through exposure to environmental stress conditions (6) and applies only to bactericidal compounds (2, 5). Persister bacterial subpopulations (persistence phenotype) are persister cells that exhibit an epigenetic trait whereby they are tolerant to antibiotics but remain dormant and are not metabolically active (3). The following are characteristic of persistent subpopulations: (i) cessation of cellular activity (dormancy), (ii) no growth or change in concentration in the presence of drug, (iii) no inherited persistence phenotype, and (iv) cells revert quickly to wild-type growth once the drug pressure is eliminated and nutrients are administered (3). The relationships between resistant and tolerant populations and persistent subpopulations are complex (2, 7).
In this review, we use the definitions of resistant and tolerant populations and persistent subpopulations described by several authors (3, 8) (Fig. 1). These researchers followed the experimental evidence showing that persister cells do not grow (3, 9–13).
FIG 1.
Illustration of resistant (green), tolerant (purple), and persistent (brown) subpopulations of bacteria interacting with the immune system (police) and antimicrobial treatment (superman) defense agents.
GASTROINTESTINAL AND RESPIRATORY ENVIRONMENTS
The environmental conditions in the gastrointestinal and respiratory environments differ in relation to their function in humans. Thus, the gastrointestinal environment is characterized by the presence of nutrients, gastric and pancreatic enzymes, bile salts, pH and temperature conditions, anaerobiosis, and bacterial competition. Moreover, the gut is the epicenter of antibiotic resistance (14).
On the other hand, the conditions in the respiratory environment are associated with its function, including variable levels of oxygen, nitrogen, carbon dioxide, and water vapor, pH and temperature conditions, external factors, and viral infections. The different characteristics largely determine which pathogens are capable of infecting these locations.
In this work, we analyzed the importance of the molecular machinery of tolerance and persistence in clinical pathogens that inhabit the gastrointestinal and respiratory environments. Gastrointestinal tract pathogens form part of the gastrointestinal microbiota (both commensal and opportunistic) (14) and include Escherichia coli, Salmonella spp., Vibrio spp., Klebsiella spp., Helicobacter spp., Enterococcus spp., Campylobacter jejuni, Shigella spp., Yersinia spp., and Clostridium difficile. Respiratory tract pathogens (both commensal and opportunistic) (15) include Staphylococcus aureus, Pseudomonas aeruginosa, Acinetobacter baumannii, Burkholderia cenocepacia, and Mycobacterium tuberculosis.
RELATIONSHIPS BETWEEN MOLECULAR MECHANISMS OF TOLERANCE AND PERSISTENCE
The molecular mechanisms involved in the formation of tolerant and/or persistent bacterial cells include the following: RpoS and the general stress response, oxidant tolerance (response to reactive oxygen species [ROS]), energy metabolism or drug efflux pumps, the SOS response, the quorum sensing (QS) system or bacterial communication, (p)ppGpp signaling, and toxin-antitoxin (TA) modules (7, 16, 17). In the following, we discuss these molecular mechanisms in gastrointestinal and respiratory bacteria.
General Stress Response (RpoS-Mediated Response)
RpoS and the other general stress responses are important molecular mechanisms whereby bacteria survive stress conditions (7). The rpoS gene encodes the sigma factor (S), which regulates the response (18) to conditions of stress, causing the accumulation of RpoS as cells enter the stationary phase and a rise in the number of related bacteria. RpoS-dependent gene expression leads to global bacterial stress resistance. Specific small RNAs (sRNAs) that encourage RpoS translation or the induction of antiadaptors that make this protein stable are induced in response to several stressors (19).
In isolates of the aforementioned gastrointestinal tract pathogens E. coli and Salmonella enterica serovar Typhimurium, RpoS and the general stress response have important roles in virulence, biofilm development, and bacterial survival. Under conditions of environmental stress or as cells enter the stationary phase, E. coli causes accumulation of RpoS (18, 19). RpoS mainly regulates genes and structural proteins associated with the formation and degradation of biofilms in response to stress (18). Several conditions can induce the general stress response in this pathogen, including nutrient deprivation, variations in temperature, biofilm production, high pH, oxidative stress, and other signals (20). In addition, the RpoS response and TA systems interact in E. coli (Fig. 2); for example, the antitoxins MqsA of the MqsR/MqsA TA module and DinJ of the YafQ/DinJ TA repress rpoS transcription and translation, respectively, until stress is encountered and the antitoxins are degraded (21, 22). In relation to S. Typhimurium, RpoS and the general stress response system also have a role in virulence or biofilm formation (23). RpoS levels are higher during the stationary phase or whenever bacteria are exposed to stress conditions (24).
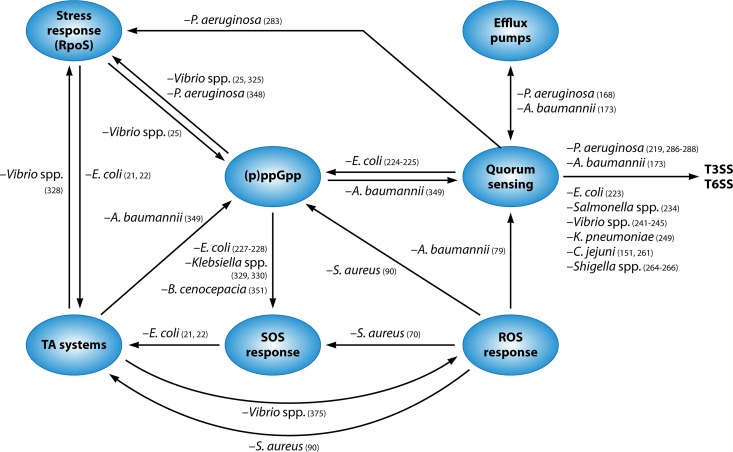
FIG 2.
Links between the different mechanisms of tolerance and persistence described for gastrointestinal and respiratory pathogens.
On the other hand, when Vibrio cholerae colonizes the human gastrointestinal tract, RpoS regulates a system known as the “mucosal escape response.” In this pathogen, RpoS expression is linked to an increase in the (p)ppGpp alarmone (Fig. 2), managing to improve motility and chemotaxis and presumably contributing to evasion of the mucosal response (25).
In clinical isolates of Klebsiella pneumoniae, a c-di-GMP phosphodiesterase protein controls the oxidative stress response and in vivo virulence, which is decreased by rpoS and/or by soxRS deletion, implying RpoS- or SoxRS-dependent control (26).
In Shigella flexneri and Shigella boydii, acid and base resistances are dependent on pH and are controlled by RpoS under stress conditions. For both types of resistance, the RpoS demand can be overcome by growth under anaerobic and moderately acidic conditions (27–30).
Interestingly, although RpoS is vital for the general stress response in numerous bacteria, it appears to be lacking in some pathogens, and it is not known whether other proteins carry out the same functions. Helicobacter pylori possesses alternative regulatory systems that were not observed until now because of the global response under stress conditions in isolates lacking the classic regulators. Proteins such as Fur and HspR compensate for the absence of RpoS (31). Three proteins associated with the stress response have been described for Enterococcus faecalis: (i) a general stress protein (gsp65), encoded by the hydroperoxide resistance ohr gene, which is induced in response to hydrogen peroxide, heat shock, acid pH, detergents, ethanol, sodium chloride, and tert-butylhydroperoxide (tBOOH) (32); (ii) the gsp62 protein, associated with the reaction to heat shock, acid pH, detergents (i.e., SDS or bile salts), ethanol, tBOOH, sodium chloride, and, to a lesser extent, hydrogen peroxide (33); and (iii) the gls24 protein, implicated in resistance to bile salts and virulence (34–36). Nevertheless, the RpoS protein did not seem to have a role in expression of the genes associated with the pathogenesis of Yersinia enterocolitica infection; however, the RpoS protein was required by Y. enterocolitica grown at 37°C to survive a variety of types of environmental stress (37) and hostile environments (38). Analysis of the C. jejuni NCTC 11168 genome sequence demonstrated that RpoS is absent in this organism. In addition, in a study searching for an RpoS homologue in the NCTC 11351 strain of C. jejuni, the authors concluded that no such homologue was found (39). The absence of an RpoS homologue was confirmed for C. jejuni NCTC 11351 by the lack of induction of stress resistance in the stationary phase (39). The gastrointestinal pathogen Clostridium also lacks RpoS. Proteins such as HSP and those comprising the GroESL and DnaKJ systems have been associated with the reaction to chemical stress, whether from autologous metabolites or allogeneic toxic chemicals (e.g., derived carboxylic acids), high H+ concentrations (low pH), antibiotics, or solvents (ethanol and butanol), all of which can have a major role in survival of the cells (40).
The important role of RpoS has also been described for respiratory pathogens, e.g., P. aeruginosa. In this bacterium, RpoS has been shown to positively affect the pls locus, which encodes enzymes that generate an extracellular polysaccharide involved in biofilm formation/expression (41).
However, other factors have been analyzed in pathogens such as S. aureus and B. cenocepacia. For S. aureus, the σB factor has been shown to be involved in the survival of cells under heat as well as electric and hydrostatic pressure (42). For B. cenocepacia, the importance of two sigma factors (other than RpoS), RpoN (AK34_313) and RpoE (AK34_2044), which are upregulated in the fixLJ deletion mutant relative to those in Burkholderia strain AU0158, has been studied. These sigma factors are critical for the survival of B. cenocepacia inside macrophages, and RpoN has been found to be essential for biofilm production (43–45).
Oxidant Tolerance (ROS Response)
Reactive oxygen species (ROS) are chemically reactive chemical species with oxygen (hydrogen peroxide [H2O2], superoxide [O2−], and hydroxyl radical [OH−]). In a biological context, ROS are produced as a natural response to the normal metabolism of oxygen and have important functions in cell signaling and homeostasis. Nevertheless, during times of environmental pressure (e.g., UV, heat, or drug exposure), ROS levels can rise. This can produce DNA damage, lipids, and proteins that cause cell death (46–48). Superoxide dismutase (SOD) and catalase enzymes or other antioxidant agents, such as glutathione and vitamin C, can eliminate ROS. When an imbalance between the mechanisms of production and elimination of ROS occurs, with an increase in the former, cells are subjected to oxidative stress (49).
In drug-tolerant E. coli cells, SOD and catalase have been shown to have a protective function (50). In other gastrointestinal pathogens, such as S. Typhimurium, many genes must be expressed in order to inactivate ROS, in a process controlled by regulons, such as SoxRS, OxyR, σS, σE, SlyA, and RecA, as well as the Dps protein, which halts bacterial growth under the control of the σ factor RpoS (51) (Fig. 2). The ROS response favors intestinal inflammation, thus allowing this microorganism to spread in the gut (52).
Interestingly, V. cholerae can generate two types of catalases, KatB and KatG, to promote ROS homeostasis (53). Moreover, in V. cholerae, the transcriptional regulator OxyR is critical for antioxidant defense and enables the microorganism to scavenge environmental ROS to facilitate population growth (54). For this pathogen, the importance of the role of the ROS response in mediating the cholix toxin, a virulence factor that displays the action of ADP-ribosyltransferase on eukaryotic elongation factor 2 of host cells and leads to cell death, was recently demonstrated (55).
On the other hand, K. pneumoniae pyogenic abscess isolates often contain heavy capsular polysaccharides (CPS) and escape phagocytosis or death due to the action of serum factors (56, 57), oxidative stress, and the ROS response (58, 59). The thick, viscous CPS also control bacterial colonization and biofilm production at the infection location (60).
H. pylori induces chronic inflammation of the stomach epithelium and evades clearance via numerous factors, such as adhesion, cell motility, and detoxification of ROS and toxins (61). All H. pylori strains encode catalase and SOD proteins to detoxify ROS, and H. pylori arginase limits NO production via macrophage-, neutrophil- and epithelial cell-derived nitric oxide synthase (62, 63).
Characterization of ROS detoxification enzymes, such as KatA, SodB, AhpC, Tpx, and Bcp, in C. jejuni has demonstrated the value of these cellular defense systems for the survival of this pathogen against ROS (64). During host colonization, C. jejuni is subjected to damage caused by ROS produced by the host immune system and the gut microbiota. However, C. jejuni possesses important ROS detoxification methods that allow it to outlive and colonize the host (64).
Some interesting features of the ROS response have also been described for other gastrointestinal bacteria, such as Shigella dysenteriae, Yersinia enterocolitica, and Clostridium difficile. The Shigella dysenteriae 1 toxin produces intestinal infection via a decrease in the endogenous intestinal protection against ROS (65). Two new SODs associated with survival in acidic environments, including that in intraphagocytic vesicles, have been described for Yersinia enterocolitica (66). The siderophore yersiniabactin increases the virulence of Y. enterocolitica and blocks the development of the ROS response through eukaryotic cells (leukocytes, monocytes, and macrophages) (67). Finally, in a study of the distribution of the main enzymes in Clostridium involved in antioxidative defense, i.e., SOD and catalase, the physiological responses (induction of SOD and catalase) to factors triggering oxidative stress in the cells of strict anaerobes were shown to be responsible for the ability of the bacteria to remain viable under aerobic conditions (68). Moreover, studies of the induction of ROS by TcdA and TcdB toxins have dissected pathways contributing to this event, and there is speculation about the role of ROS in mediating pathogenesis (69). Clostridium difficile manages oxidative stress efficiently, and survival of this anaerobic microorganism is therefore consistent with ROS being mediated by TcdA and TcdB and thus favoring inflammation (69). On the other hand, the enzyme glutamate dehydrogenase (essential for growth of C. difficile) participates in the production of alpha-ketoglutarate, which contributes to H2O2 tolerance associated with the ROS response (69).
It was recently demonstrated that heterogeneous respiratory bacteria, such as methicillin-resistant S. aureus (MRSA) strains, can exist as two populations, with a heteroresistant phenotype (HeR-MRSA) and a homoresistant phenotype (HoR-MRSA), which are induced by β-lactam antibiotics via oxidative stress and mediated by the ROS response and DNA damage (Fig. 2) (70). The production of ROS during β-lactam treatment seems to be regulated by catalases and dismutases (SODs), which protect organisms with the heteroresistant phenotype from cell death and encourage cell survival. Moreover, disabling the tricarboxylic acid (TCA) cycle activity has a negative effect on ROS production, showing the role of metabolic modifications in the homoresistant phenotype (70). Finally, mutagenesis stimulated by β-lactams in cells with the heteroresistant phenotype demonstrates the conjunction of ROS production and the SOS-induced response (70). On the other hand, conservation of the persistent state in biofilms produced by several pathogens has been associated with adaptation of metabolic processes, mainly the processes associated with ROS formation and the TCA cycle. In addition to the characteristics of cells associated with biofilm production, metabolic changes are also important, as confirmed by survival studies with planktonic S. aureus cells. The appearance of mutants without the TCA cycle enzymes succinate dehydrogenase and aconitase suggests an improved level of survival in the stationary phase (71, 72). The decrease in ROS formation was determined to be a fundamental characteristic at this point. The ROS response appears to be associated with programmed cell death in S. aureus strains (73).
SOD and catalase have been analyzed in P. aeruginosa, and the sodB gene has been connected with an important protective role against UV-C radiation (74). The same researchers also found that levels of the POX enzyme increased under stress tolerance (74). According to in vitro evidence, the environmental factors that trigger the selection of mucoid colonies are oxidative stress, low oxygen concentrations, and high osmolarity, i.e., conditions commonly found in the lungs of infected patients. Alginate overproduction confers resistance to phagocytosis but also appears to increase the sensitivity toward β-lactams and other types of antibiotics (75, 76) and contributes to the persistent inflammation of the airways (77). The parameters associated with mucoid transition in patients include persistence of bacteria in the sputum and the use of inhaled bronchodilators and inhaled antibiotics, such as colistin (78). A connection between the QS system and the expression of catalase and dismutase proteins in the ROS response of this pathogen has been described (Fig. 2) (79).
Four catalases (KatA, KatE, KatG, and KatX) have been described for Acinetobacter spp., among which KatE is the most effective in the resistance to H2O2 (80). In total, 107 differentially expressed proteins have been identified in Acinetobacter baumannii in relation to oxidative stress and are mainly involved in signaling, supposed virulence factors, and the stress response (including oxidative tolerance) (81). Interestingly, oxidant-tolerant cells of this pathogen showed a higher survival rate in response to several bactericidal antibiotics (41, 81).
Persistence is decreased in B. cenocepacia mutants lacking catalase or SOD proteins (82). The results obtained by Van Acker and colleagues have contributed greatly to our comprehension of the molecular machinery controlling antimicrobial tolerance in biofilms formed by the Burkholderia cepacia complex, revealing that these biofilms carry tolerant and persistent cells (83). The TCA cycle was downregulated in the remaining persistent cells, which thus prevented ROS production (detoxification). At the same time, the persistent cells switched on a different pathway, the glyoxylate shunt, which may become a new target for therapy. The aforementioned study also demonstrated that treatment of B. cenocepacia with tobramycin induces ROS production and that persister cells depend on ROS detoxification mechanisms, while processes responsible for ROS production (e.g., the TCA cycle, processes resulting in the production of NAD or flavin adenine dinucleotide, and the electron transport chain) are downregulated in persister cells (83).
Mycobacterial persister development under isoniazid pressure was related to the stochastic difference in the pulsatory expression of KatG, a catalase peroxidase needed for processing and activation of the drug. Slow-pulsing cells processed smaller amounts of the drug and therefore survived longer than fast-pulsing bacteria (84). The persister subpopulation of mycobacteria shows a different type of sensitivity to hydroxyl radicals due to their tolerance to antimicrobials, unlike the larger population, which is susceptible to them. There is increasing agreement on the importance of ROS and oxidative damage in antimicrobial tolerance in other populations resistant to antimicrobial elimination, and the crucial function of ROS in a stochastic persistent subpopulation pattern has been demonstrated for this pathogen (85). However, stress promotes other metabolic pathways in mycobacteria, leading to reduced levels of ROS and increasing the limit for antibiotic-mediated death (86).
Finally, several compounds induce the ROS response in gastrointestinal pathogens, leading to the development of tolerant populations and thus favoring colonization and bacterial pathogenesis, as follows. (i) Salicylate can induce tolerance in E. coli by generation of ROS. Salicylate-induced ROS cause a decrease in the membrane potential, reduce metabolism, and lead to increased tolerance of ROS (87). (ii) Iron is important for pathogenic bacteria, such as S. Typhimurium. Free cytoplasmic iron is used in the formation of radicals and can stimulate the antimicrobial actions of ROS. It has been shown that mice with deficient levels of this metal have a lower risk of developing S. Typhimurium infection (88). (iii) Although Salmonella does not produce indole, when it uses the indole produced by other bacteria, its tolerance of antibiotics increases (52). (iv) Antibiotics such as vancomycin and penicillin may have an impact. The dependence of Enterococcus spp. on SOD for tolerance of vancomycin and penicillin is usual for antimicrobial-susceptible enterococci (89). (v) A demonstration of the response to ethanol-induced stress (EIS) in S. aureus strains was provided by the expression of 1,091 genes, of which 291 were upregulated (90). The EIS caused upregulation of genes that promote stress response networks (90), including the ROS response, (p)ppGpp, and TA modules (Fig. 2). The transcriptional profile of MRSA pathogens indicated that they reacted to EIS by entering a state of dormancy and modifying the expression of elements from cross-protective stress response systems in an effort to preserve the preexisting proteins (90).
Energy Metabolism
Regarding energy metabolism, we highlight two mechanisms in relation to tolerant populations. First, cytochrome bd is a prokaryotic respiratory quinol:O2 oxidoreductase that enhances the tolerance of cells to oxidative stress (ROS response) and nitrosative stress conditions. When aerobic metabolism is restricted by oxygen limitation in the cells, the cytochrome d complex (encoded by the CydAB operon), one of the terminal oxidases in the respiratory network, predominates (91, 92). Overexpression of this operon has been implicated in chlorhexidine (biocide)-tolerant cells (93, 94) and in taurine metabolism (93).
Cytochrome bd complex.
The cytochrome bd complex (encoded by the CydAB operon) is the main element in the respiratory chain when aerobic metabolism is restricted by oxygen limitation (95). This mechanism has been analyzed in gastrointestinal bacteria, such as E. coli (95) and S. Typhimurium (96). When S. Typhimurium enters host tissue, it is subjected to an oxygen partial pressure (pO2) of 23 to 70 mm Hg (3% to 10% oxygen), which is significantly lower than the atmospheric pO2 of 160 mm Hg (21% oxygen) (97). The survival of S. Typhimurium in tissues during infection of mice is due to the presence of high-affinity cytochrome bd oxidase (98). Cytochrome bd-II produces an increase in epithelial oxygenation, which together with nitrate respiration drives expansion of this pathogen within the gut lumen (96, 99). This pathogenic strategy is acerbated by oral antibiotic therapy, since it enhances Clostridium depletion, which may clarify why treatment with oral antibiotics often fosters infection with antibiotic-sensitive S. enterica serovars producing human gastroenteritis (100). The cytochrome c peroxidases were also similar in E. coli strains described as H2O2 degraders and in anoxic Salmonella spp. (101).
The V. cholerae genome encodes four different respiratory oxygen reductases under limiting conditions, depending on the gastrointestinal localization (102). Three of these use ubiquinol as a natural electron donor (103), and the fourth (cbb3-type heme-copper oxygen reductase) (104) uses cytochrome c (105).
Little is known about the energy metabolism (i.e., cytochrome bd) in strains of Klebsiella pneumoniae. Nevertheless, the ability of K. pneumoniae to grow anaerobically with citrate as the unique carbon source is known (106). The presence in K. pneumoniae of the genes responsible for citrate fermentation has been described for a group of 13 kb (107), in which variation in several isolates helps with adaptation to nutritional characteristics (108).
A cytochrome bd-type oxidase has been described for the H. pylori H2-oxidizing membrane-associated respiratory chain (109), and differences relative to other pathogens have been reported (110). The possible relationship between the development of duodenal ulcers caused by H. pylori and its capacity to survive in the presence of bile acids conjugated with taurine has been contemplated (111).
Some authors have detected the presence in C. jejuni of a low-affinity oxidase resistant to cyanide, belonging to another type of cytochrome (112).
Functional studies of the cytochrome bd-type enzyme in E. faecalis V583 have been carried out (113). Interestingly, in relation to energy metabolism, the Clp ATP-dependent protease operon, implicated in stress survival, is highly conserved and enables growth at high temperatures in Gram-positive bacteria, including Enterococcus faecalis (114).
Cytochrome bd expression has been associated with normal intracellular survival and virulence in S. flexneri (115). Moreover, in a study involving an S. flexneri cydC strain lacking cytochrome bd, the bacteria were quickly cleared from the lungs of intranasally inoculated mice (116).
S. aureus and P. aeruginosa cause opportunistic infections and regularly infect the lungs of cystic fibrosis (CF) patients. S. aureus has the capacity to resist the action of pyocyanin and hydrogen cyanide, which are small respiratory inhibitors, because of the action of the cytochrome bd quinol oxidase (117). The cbb3-type cytochrome oxidase subunit (which catalyzes the final stage in respiration, displaying a strong affinity for oxygen) supports P. aeruginosa biofilm growth and bacterial pathogenesis (118). Hence, cbb3-type cytochrome oxidases may be important therapeutic targets (118). On the other hand, elevated quantities of the second messenger cyclic di-GMP (cdG), produced by several mutations, are linked to overproduction of Pel and Pls exopolysaccharides and fimbrial adhesins in P. aeruginosa and are consequently involved in small-colony variant (SCV) formation (119–122). In addition to the cdG signaling pathways, SCV generation is influenced by other regulatory systems, such as the components of the GacAS mechanism, in which GacA regulates the response and GacS is the phosphorylating transmembrane histidine protein kinase. This system controls the RsmAZY regulatory system, and the combined regulatory network influences the transition between acute and chronic infectious lifestyles of P. aeruginosa. Mechanistically, phosphorylated GacA fosters the transcription of RsmY, sRNAs, and RsmZ genes, thus blocking the activity of RsmA by participating in its binding and attenuating the inhibitory effects on the target mRNAs, including those related to the synthesis of the Psl exopolysaccharide (123, 124). The SCV phenotype is promoted by deletion of the rsmA gene in P. aeruginosa strain PAO1 (41). The deletion also increases infectious persistence in mouse lung infections (125). Moreover, mechanisms of specific tolerance, such as the production of periplasmic glucan molecules, which inactivate aminoglycosides, are preferentially expressed in P. aeruginosa biofilms rather than in planktonic cells (126, 127). Together these mechanisms promote the prevalence of P. aeruginosa in the airways of infected patients.
In Acinetobacter calcoaceticus, quinoprotein glucose dehydrogenase interacts with the b-type cytochrome(s) and with cytochrome o and cytochrome d (both cytochrome oxidases) under stress conditions (128).
Finally, in M. tuberculosis, the phoU gene controls phosphate uptake, thus regulating the pst operon. The phoU gene is well conserved among bacteria and has homologues in M. tuberculosis. Mutants of M. tuberculosis are influenced by the phoY2 gene, displaying low persistence both in vitro and in vivo (129–131).
Tau metabolism.
In relation to Tau metabolism, cysteine or sulfate deficiency leads to tauD (or an orthologue) being essential for growth of E. coli (132, 133). The presence of TauD leads to the production of sulfite for use as a source of sulfur by catalyzing the hydroxylation of taurine alpha-(2-aminoethanesulfonic acid) in the sulfonate. Several studies have focused on E. coli TauD, which is an important member of the huge, universal family of αKG-dependent nonheme iron oxygenases (134, 135).
Some studies have also been conducted with V. cholerae in relation to activation of the virulence factor associated with the metabolism of taurine and with bile salts, favoring colonization of the gastrointestinal tract (136–139). Studies of the role of taurine (enzymes and transporters) associated with sulfate metabolism in this pathogen are scarce.
However, when H. pylori colonizes the tissue, bile has been observed to influence cell gastric epithelial kinetics, promoting gastric cancer (140). Interestingly, bile chemotactic gradients (mainly taurocholic and taurodeoxycholic acids) across the gastric mucus layer may therefore contribute to directing H. pylori to the pyloric antrum and thus to enabling this important pathogen to attain high population densities on the mucous layer in the area of the gastric epithelium (141).
Taurine is another important source of energy in S. aureus, and CymR has been considered an important regulator of cysteine and taurine metabolism participating in biofilm formation (142).
Moreover, the important functional role of the tauRXYPI cluster, implicated in taurine metabolism in A. calcoaceticus, has been analyzed under nitrogen limitation conditions (143). The inhibitory effect of taurine on the formation of biofilm during alkane degradation was recently studied and shows that taurine probably affects the alkane-induced cell surface (144).
Finally, in B. cenocepacia, regulation of sulfur as an environmental source of energy has been associated with the tauABC operon (145).
Efflux Pumps
Efflux pumps are needed to eliminate toxic elements or to maintain the balance of compounds that are vital for bacterial survival (146). Various researchers recently demonstrated that increased expression of efflux systems is vital for active maintenance of a low intracellular antibiotic concentration, and thus specifically the persister state, in nongrowing, nondividing E. coli cells (147–149). Moreover, an active mechanism can also decrease the concentration of the toxic compound (drug), which raises the drug MIC and thus appears in populations with mixed resistance and tolerance phenotypes (5). The multidrug efflux pumps that participate in tolerance and resistance processes can be upregulated by several signals, such as oxidative stress (ROS response) and QS systems, including the type III secretion system (T3SS), T6SS, and other virulence factors (Fig. 2) (150, 151).
In vitro studies with E. coli have demonstrated that paraquat-induced tolerance is reliant on the AcrAB multidrug efflux machinery (152). Moreover, overexpression of this efflux pump was shown to be vital for the active maintenance of a low intracellular antibiotic concentration, and thus the tolerant persister state, specifically in nongrowing, nondividing E. coli cells, suggesting the coregulation of dormancy and active processes for the persistence phenotype (147, 153). In addition to the AcrAB system, Salmonella possesses at least 11 multidrug efflux pumps (11, 154). Among them, we highlight the following. (i) We first mention the AcrD efflux pump of S. Typhimurium (57). After inactivation of this pump, variations of expression of several genes involved in metabolism, stress responses, and virulence were detected. For example, a reduction in the expression of genes involved in pathogenesis or those that encode products of metabolism of tricarboxylic acid and purines was observed. The exact same effect was observed for the expression of virulence genes in Salmonella pathogenicity islands (SPI-1, SPI-2, SPI-3, SPI-10, and SPI-18). Levels of fumarate associated with swarming motility were also altered after inactivation of the AcrD efflux pump (11). (ii) The MdtD pump stimulates the efflux of citrate, an iron chelator generated during aerobic metabolism. When this efflux pump is induced by stress, iron is expelled from the cell, thus reducing bacterial growth. Expression of a citrate transporter (IceT) leads to a decrease in the vulnerability to ROS, nitrosative stress, and antimicrobial agents. Stress resistance and antibiotic tolerance are mediated by this protein (IceT) via regulation of metabolism, redox chemistry, and intracellular iron (88). (iii) Finally, in the presence of H2O2, the MacAB drug efflux pump protects S. Typhimurium against ROS by inducing the formation of a compound that confers resistance to extracellular H2O2. Another function of this protein is to facilitate the growth of S. Typhimurium within macrophages (154, 155). The functions of efflux pumps in multiresistance and in tolerance and/or persistence under stress conditions are not known for this pathogen.
A new multidrug efflux pump, EmrD-3, was recently discovered in V. cholerae O395 (156).
In K. pneumoniae, the AcrAB efflux pump promotes the development of pneumonia by acting as a virulence agent that fights the innate immune defense in the lung (157). Extreme virulence of K. pneumoniae isolates was recently found to be associated with increased expression of the AcrAB and OqxAB efflux pumps (158).
In H. pylori, an association between biofilm formation and loss of sensitivity to antibiotics was also detected through the action of two expulsion pumps in the process (159).
A contribution of the Cme ABC efflux pump to antibiotic resistance in isolates of Campylobacter jejuni has been reported (160). In addition, the cmeA gene, which encodes a multidrug efflux transporter, is upregulated in the presence of deoxycholic acid in C. jejuni wild-type strains relative to that in T6SS-deficient strains, suggesting a relationship between the suppression of proliferation and the role of this efflux pump, regulated by the T6SS (151). The relevance of the T6SS to the adhesion, invasion, and colonization of the host and also to the adaptation to deoxycholic acid was thus confirmed, demonstrating the key role of this system in the pathogenesis of C. jejuni (151).
Researchers in South Africa recently considered drug resistance, efflux pumps, and virulence characteristics in different species of Enterococcus that occur in surface waters (161). Most of the isolates had four efflux pump genes (mefA, tetK, tetL, and msrC) and virulence genes, such as the asa1, cylA, gel, and hyl genes (161).
Moreover, for Shigella, we highlight two efflux pumps, as follows. (i) The AcrAB efflux pump has been connected to resistance to bile salts as well as to survival and pathogenicity during host transit and later gastrointestinal infection (162). (ii) The MdtJI efflux pump, which is involved in the extrusion of toxic compounds and allows survival of bacteria within infected macrophages, has been described for Shigella (163).
Regarding the transporters, we can highlight the RosA/RosB efflux pump of Yersinia enterocolitica (164). For this pathogen, resistance to new cationic antimicrobial peptides has been described as being due to an ejection pump/potassium antiporter mechanism consisting of the RosA and RosB proteins (164).
Finally, CdeA, a multidrug efflux pump belonging to the MATE family and involved in Na+ transport, was identified in Clostridium difficile but has not been found to be associated with antibiotic resistance (165).
In respiratory bacteria, such as S. aureus, the expression of the Qac efflux pumps has been associated with increased tolerance to biocides (166). The predominant antibiotic resistance mechanism in P. aeruginosa CF isolates is the overexpression of efflux pumps, particularly those belonging to the RND superfamily. P. aeruginosa has genes for at least 12 of these systems, although we can highlight MexAB, which is involved in tolerance to colistin and biofilm formation (167). Moreover, this efflux pump is associated with the transport of 3-oxo-acyl-homoserine lactones, which are the signals used in cell-to-cell communication (QS) (168). The capacity of P. aeruginosa infections to persist in the lung regardless of antimicrobial treatment depends on the intrinsic tolerance of the bacterium to antibiotics and the readily acquired resistance to new drugs (169).
In Acinetobacter baumannii, increased biofilm production is associated with overexpression of two efflux pumps: AdeABC and AdeFGH (170–172). Under bile salt pressure, A. baumannii strain ATCC 17978 and A. baumannii clone ST79/PFGE-HUI-1 (a clinical strain lacking the AdeABC efflux pump) overexpress the glutamate/aspartate transporter as well as virulence components associated with activation of the QS system (biofilm, surface motility, and T6SS components) (173).
RND efflux pumps, such as BCAM1945-1947 (RND-9) and BCAM0925 (RND-8), protect the biofilm complex of B. cenocepacia from tobramycin, while the BCAL1672-1676 (RND-3) pump is important for resistance of biofilms to ciprofloxacin and tobramycin (174).
Finally, little information is available about efflux pumps in M. tuberculosis.
SOS Response
The SOS response is triggered by DNA damage, allowing repair of genetic material to enhance cell survival. The SOS system can be considered an important mechanism of bacterial survival under stress conditions, which are related to other stress responses (7, 50, 175). The SOS response involves genes that not only affect cellular processes, such as DNA recombination and repair, but also affect pathogenesis, antimicrobial resistance, and biofilm production (176). The proteins that make up the SOS system include a transcriptional repressor called LexA and a DNA-binding activating protein, RecA (177). However, other proteins may be involved.
As mentioned above, the RecA protein positively regulates the SOS system in E. coli (178, 179). The SOS system contributes to DNA repair but also induces the development of the type I TA module toxin TisB in a subpopulation of E. coli (180). Different studies have demonstrated that under these conditions (including treatment with fluoroquinolones), the SOS response encourages the production of persister cells through positive regulation of the expression of the TisB toxin in E. coli (152, 175, 181). In Salmonella spp., swarming motility, bacterial virulence, and antibiotic tolerance are related, and the bacteria are able to control their motility when DNA damage is present, by means of the SOS induction system (RecA) (176). The lack of the RecA protein in Salmonella enterica impairs the swarming ability and also reduces the capacity of the bacterium to cross the intestinal epithelium (182–184).
Stress response pathways that are crucial for the development and survival of V. cholerae persister cells, especially the SOS system (RecA), have been found to encourage horizontal gene transfer between microorganisms, thus enhancing resistance (185). Antibiotics such as quinolones activate the SOS response and thus increase the incidence of horizontal transfer in V. cholerae (177). Moreover, this pathogen responds to other antimicrobials, such as aminoglycosides, chloramphenicol, and tetracycline, by stimulating the SOS response (175).
Interestingly, several canonical heat and cold shock proteins in K. pneumoniae strains are upregulated under extreme temperatures (low [20°C] and high [50°C]). Among these proteins, we highlight RecA, although other proteins (GrapE, ClpX, and DeaD) may be involved in the response (186).
DNA damage repair in in vivo colonization by H. pylori requires enzymes, such as RecA and AddAB, which are involved in survival at low pH (187, 188).
The RecA protein has also been characterized in relation to DNA damage repair in Campylobacter jejuni (189). On the other hand, plasmids encoding UmuDC-like proteins with SOS function have been located in isolates of Streptococcus pneumoniae Rx1 and E. faecalis UV202 (190).
SOS proteins, such as topoisomerases, and histone-like DNA binding proteins, such as H-NS, HU, and IFH, which are essential for maintaining bacterial DNA organization, have been investigated in Shigella spp. (191). The involvement of H-NS in suppressing DNA repair in Shigella spp. after UV irradiation has been described (191). The role of H-NS in Yersinia enterocolitica has also been reported (192).
Finally, the features of the SOS response have been studied in C. difficile, and the authors reached the conclusion that the SOS system is related to C. difficile sporulation and that the induction of the SOS system can stimulate biofilm production in this pathogen (193). The same authors also showed that LexA controls the expression of toxin genes, metronidazole resistance, biofilm production, and sporulation (193). The relationship between sporulation and the SOS system was also found in Bacillus subtilis, through the sda gene (193). Although this gene is absent in C. difficile (194), Walter and colleagues studied how LexA interacted in vitro with the promoter region of another gene associated with sporulation, sspB (194).
The proportion of S. aureus mutants (i.e., SCVs that are very tolerant to antimicrobials and that can survive in host cells) is higher in cultures exposed to fluoroquinolones and mitomycin C than in nonexposed cultures, and this was found to be correlated with a larger proportion of mutations (indicated by resistance to rifampin) and followed by stimulation of the SOS DNA damage response (195). These findings indicate that environmental stimulation (e.g., with antimicrobial agents that lower replication fidelity) increases the formation of SCVs by activating the SOS response and thus boosts difficult-to-treat persistent infections (195). It has been found that Staphylococcus aureus strains can adapt to oxidative stress via a mechanism that produces subpopulations of H2O2-resistant SCVs with improved catalase production (196). This occurs through a mutagenic DNA repair pathway that includes RexAB, RecA, and polymerase V (Pol V) (196). The SOS response is more complex in P. aeruginosa than in E. coli due to the participation of two other chromosomal regulators (PrtR and PA0906), which are present in most P. aeruginosa isolates and other species of the genus, in addition to LexA (197–202). In P. aeruginosa, DNA damage causes autocleavage of LexA by RecA and the elimination of repression of the LexA regulon, as in E. coli (198, 201, 203). The induction of PrtR-managed genes adversely affects survival during genotoxic stress, decreases antimicrobial resistance, and reduces resistance to oxidant agents (204). In addition, the UmuDpR protein has been demonstrated to suppress expression of P. aeruginosa SOS genes regulated by LexA (205).
Regarding A. baumannii and Acinetobacter baylyi, several authors have examined the role of the RecA protein and UmuC proteins in repairing DNA damage, and therefore in the cellular defense against pressures produced by agents that damage DNA, antibiotics of various families (ciprofloxacin and tetracycline) (206), and oxidizing elements (207–209). Hemolytic Acb strains have been shown to be significantly more tolerant to UV treatment than nonhemolytic isolates and those belonging to other Acinetobacter species. This demonstrates the diversity of SOS responses in Acinetobacter strains and may partly explain the emergence of A. baumannii and Acinetobacter ursingii (210). Moreover, a DNA damage-inducible response has been described and found to promote drug resistance in A. baumannii, especially in stressful environments (211).
Little is known about the role of the SOS response in B. cenocepacia.
Finally, for M. tuberculosis, a regulon including Y-family DNA polymerases (ImuA′ and ImuB), which contributes greatly to damage tolerance (in conjunction with the C-family DNA polymerase DnaE2), has been described (212). Recently reported transcriptomic studies showed that various stress response regulons (e.g., the SOS response) and also different TA genes are positively regulated in M. tuberculosis persisters (213, 214). However, for this pathogen, an alternative RecA-independent DNA repair mechanism controlled by a ClpR-like factor has also been reported (215).
QS and Secretion Systems
QS is a bacterial communication network that allows cells to modify their collective behavior through signaling molecules, known as autoinducers, according to changes in the environment favoring the formation of persister cells (7, 216, 217). This process affects bacterial populations and determines the expression of genes regulating virulence, toxin production, motility, chemotaxis, biofilm production, and bacterial competition (secretion systems [T3SS and T6SS]) (Fig. 2), which may contribute to bacterial adaptation and colonization (218). In this review, we consider proteins from the QS system and secretion systems (T3SS and T6SS) due to their association with the development of persister cells, as described for pathogens such as P. aeruginosa and Mycobacterium spp. (219, 220). We hypothesize the importance in those pathogens of the relationship between the secretion systems (T3SS and T6SS) and the QS system regarding the development of persister cells. However, further studies of these secretion systems are required to clarify these relationships.
The QS systems described for E. coli include the LuxR homolog (SdiA receptor), LuxS (synthetase), autoinducer-2 (AI-2) and autoinducer-3 (AI-3) systems, and an indole-mediated signaling system (221). A QS system was related to the induction of E. coli persister cells through an indole molecule that overexpresses OxyR and phage shock regulons, preparing the subset of cells for future stress (222, 223). In contrast, other researchers have demonstrated that indole and indole analogs reduce persistence (224–226). Several researchers have investigated the connection between tolerance and persistence mechanisms and phenotypic factors controlled by the QS system. It has been shown that biofilms are often associated with intestinal infections caused by E. coli (18). As the main agent of urinary tract infections (UTIs), E. coli can form biofilms within the bladder epithelial cells and thus evade antibiotic activity (180). The bacterial growth rate determines the sensitivity to some antibiotics, as occurs with ciprofloxacin, and therefore the biofilm cells are protected from the action of these antibiotics when they grow at lower rates (20). Furthermore, biofilm formation in E. coli causes cells to have limited access to nutrients, thus increasing the levels of (p)ppGpp involved in tolerance to multiple drugs (Fig. 2) (227, 228). Finally, the QS system in enteropathogenic E. coli (EPEC) controls the activation of the type III secretion system, which participates in the modulation of virulence (229).
Three QS systems have been identified in Salmonella, all of which are formed by a synthase, a receptor, and a signal (221), as follows: (i) the unknown synthase, SdiA, and 3OC8HSL system, which has a motility function and promotes resistance to acids (230); (ii) the LuxS, LsrB, and AI-2 system, involved in expression of the Lsr gene cluster (uptake of AI-2) (231); and (iii) the QseB, QseC, and AI-3 system, which is involved in virulence features, motility ability, and biofilm production (232).
S. Typhimurium produces acyl-homoserine lactone (AHL) and the signaling molecule AI-2. Both molecules are produced and released mainly during exponential-phase growth and are implicated in biofilm formation (52). A recently published study showed that the presence of in vitro bile salts increases the killing of other bacteria by S. Typhimurium in the gut. This antibacterial activity is not efficient against all commensal bacteria. While S. Typhimurium outcompetes Klebsiella oxytoca or Klebsiella variicola, commensal species, such as Enterococcus cloacae, Bacteroides fragilis, Bifidobacterium longum, Parabacteroides distasonis, and Prevotella copri, are not overcome (233). Interestingly, Salmonella enterica and other pathogens present T6SS in association with T3SS, quorum sensing (QS), flagellum production, and QS regulators, which is essential for bacterial pathogenesis (234).
The different types of behavior controlled by QS in Vibrio spp., such as bioluminescence, T6SS and T3SS, biofilm production, and motility, make this bacterium an ideal model for studying quorum sensing. The following QS systems have been described for V. cholerae: (i) LuxS, LuxP, and AI-2; and (ii) CqsA, CqsS, and CAI-1. Both of these are related to biofilm production, extracellular polysaccharide formation, and other virulence agents (221, 235). QS systems are also involved in the V. cholerae persister phenotype (236). It is well known that V. cholerae can occur either as planktonic cells or adhered to a biofilm matrix that forms aggregates. As in other pathogens, biofilm production in V. cholerae is regulated by QS. Recent evidence suggests that biofilms are formed during the aquatic and intestinal phases of the V. cholerae life cycle and perform an essential function in environmental and intestinal survival and also in the transmission of infection (237). V. cholerae AlsR (quorum sensing-regulated activator) drives the expression of the acetoin gene cluster in response to glucose, acetate, or another activating signal (238). Furthermore, a model describing the regulation of the acetoin biosynthetic gene cluster by AlsR and AphA according to environmental conditions has been established for V. cholerae (238). In many species of Vibrio, the T3SS and T6SS are strongly associated with QS (239–242). V. cholerae uses the T6SS to compete with the different prokaryotic and eukaryotic cells that it finds in several environments and human hosts. Novel research on the expression of the T6SS indicates that this system may promote the persistence phenotype and the development of V. cholerae infection through direct opposition of bacterial competitors (243). In vitro studies have demonstrated that the V. cholerae T6SS is expressed by bacteriocins (mucins) and modulated by bile salts, which are modified by the microbiota (244). In relation to these findings, an intact T6SS is necessary for V. cholerae to become established in the guts of infant rabbits (245).
Interestingly, QS in K. pneumoniae is LuxS dependent, and AI-2 autoinducers (246, 247) participate in biofilm formation (248). In a study involving genomic extraction and data analysis for isolates of K. pneumoniae, three conserved regions were distinguished and found to contain T6SS genes, which are controlled by the QS system (249).
H. pylori generates extracellular signaling molecules associated with AI-2, which depends on LuxS function. In turn, this is determined by the growth phase, and production is higher in the mid-exponential phase (250–252). Its genetic variability and the capacity of H. pylori to develop biofilm, and thus to protect itself from environmental stressors, are responsible for the resistance of this pathogen to the usual treatments, and also for its persistence in human tissues (253). In vitro biofilm production by H. pylori is reflected in several studies (254–257). Moreover, this pathogen can even produce biofilms on the human gastric mucosa (257–260).
The LuxS-homologous protein which participates in the synthesis of AI-2 (a key participant in biofilm formation) has been located for C. jejuni. This bacterium can join to and produce biofilms on different surfaces (160). However, a luxS mutant did not show important changes in the virulence phenotype (CmeABC multidrug efflux pump, cell morphology, or mucin penetration). As mentioned above, C. jejuni possesses an operational T6SS encoded by a complete T6SS gene cluster that forms part of an integration factor located in the genomes of some C. jejuni isolates (261). Moreover, the T6SS participates in tolerance of bile salts and deoxycholic acid (DCA) and in the pathogenicity characteristics of bacteria, such as adhesion and invasion (151).
E. faecalis readily forms biofilms. It is capable of acquiring resistance determinants, such as elements of the QS system (fsrA, fsrC, and gelE) and two glycosyltransferase genes (GTF genes), via horizontal gene transfer encoded by epaI and epaOX, which promote biofilm formation (262). Moreover, the fsr QS component and the GelE protease regulated by QS are involved in gentamicin, daptomycin, and linezolid resistance in E. faecalis biofilms but not planktonic cells (262).
Regarding Shigella spp., many authors have searched for multiple factors related to the QS system, including the production of the AI-2 signal, in Shigella flexneri (263). Moreover, the connection between T3SS and the QS activation process was described in 2011 (264, 265). More recently, it was shown that Shigella sonnei, but not S. flexneri, encodes a T6SS, providing a higher capacity for survival in the intestine (266).
The QS of Yersinia enterocolitica has also been investigated (267). Homologues of the LuxI (AHL synthase) and LuxR (response regulator) protein families have been analyzed for several species of Yersinia. Although Y. enterocolitica has a LuxRI pair (YenRI), other species have two pairs (267). Moreover, the same researchers demonstrated the role of QS in swimming- and swarming-type motilities of Yersinia (268). Other investigators have shown that biofilm formation may be inherent in Y. enterocolitica. The presence of biofilms greatly increased the minimum inhibitory concentration for bacterial regrowth (MICBR) for all antimicrobials (269).
In Clostridium difficile, QS plays a role in toxin synthesis (270) as well as biofilm formation (271). Along with LuxS and SpoOA, flagella and the cysteine protease Cwp84 are important for biofilm formation in C. difficile (271, 272).
In vivo experiments have examined the function of quorum detection systems in Staphylococcus aureus (SarA and Agr) in the development of persister cells in various types of infections (221). Agr has been shown to be associated with the formation of the persistence phenotype in S. aureus (273). Mutations of either agrCA or agrD, but not RNAIII, showed a rise in persister cell formation in stationary-phase cultures (273) (Fig. 3). In S. aureus, a connection between the modulation of AI-2 and different phenotypes displaying capsule formation, biofilm production, antibiotic resistance, and virulence has been observed (274–276). These findings have been corroborated in both laboratory experiments and animal models infected with S. aureus, in which luxS controlled biofilm formation by regulating the icaR locus. This regulator is a repressor of the ica operon (responsible for producing a polysaccharide composed of β-1,6-linked N-acetylglucosamine), which is necessary for biofilm formation (275). However, the function of LuxS in regulating the QS in Staphylococcus spp. remains controversial.
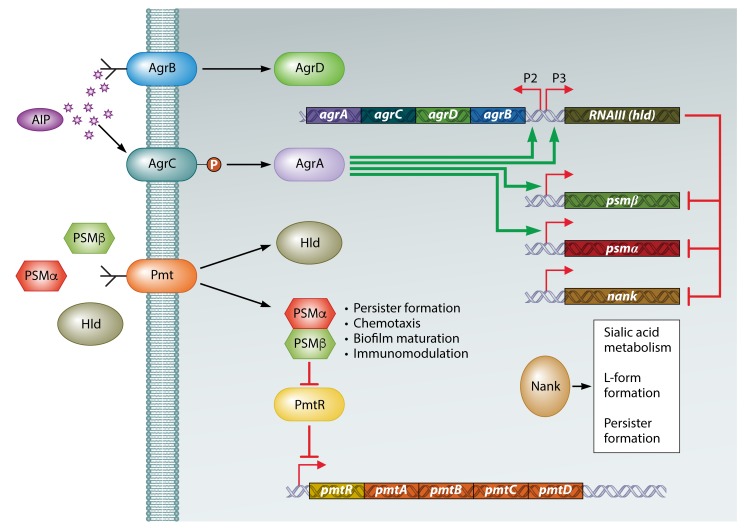
FIG 3.
Role of the Agr quorum sensing regulatory network in the formation of persisters. The agr operon is activated by an autoinducer peptide (AIP) encoded by agrD, modified and transported by AgrB, and processed by AgrC (histidine kinase) and AgrA (response regulator). AgrA positively regulates P2 and P3 of the agr operon, activating AIP production from P2 and RNAIII from P3. In addition, AgrA promotes the expression of the psm genes, which encode phenol-soluble modulins (PSMs). The PSMs are transported by the Pmt system, encoded by the pmt operon. PSMs join PmtR (repressor of the pmt operon) to activate the production of PSM transporters. RNAIII negatively regulates the psm and nank genes. PSMs inhibit the formation of persisters, while Nank promotes their formation. (Adapted from reference 273.)
Quorum sensing in P. aeruginosa involves at least three functional QS circuits, two of which are controlled by N-acyl-homoserine lactone (HSL) signals (LasI/LasR and RhlI/RhiR) and the other of which is controlled by quinolones (221). Five signals have been identified: AI-2, Pseudomonas quinolone signal (PQS), autoinducer peptides (AIPs), AHLs, and diffusible signal factors (DSFs) (277, 278). This system is vital for the colonization and later survival of bacteria during infection, as it coordinates phenotypic changes, especially at the beginning of the infection and during binding to the host cell (279). Expression of QS genes is important in determining the progress of infection (acute or chronic). More than 10% of the genes in P. aeruginosa are controlled by QS, and all of these genes are associated with the production of virulence factors, as well as with biofilm formation, antibiotic resistance, surface motility, and stress response-produced adjustments of the metabolic routes (280–282). Moreover, the rpoS gene is known to control the Las system (Fig. 2), which is probably involved in the generation of tolerance to ofloxacin in P. aeruginosa (283). Small molecules spread in the environment (QS signals) can trigger biofilm disintegration. One of the most important factors contributing to the survival of P. aeruginosa, by generation of resistance or tolerance, is the capacity to develop biofilms both in vivo and in vitro. Biofilms are more tolerant (up to 4 orders of magnitude) than planktonic cells to several antibiotics, as the antibiotics are less able to infiltrate the deepest parts of the structure and because the low oxygen and nutrient concentrations available to the bacteria at those locations render them metabolically inactive. Moreover, up to 1% of bacterial cells in biofilms are persister cells, which are dormant cells that are not affected by antibiotics (8). This type of cell is abundant in bacterial isolates from the lungs of chronic CF patients (284). In addition to mucoid colonies, SCVs, also known as dwarf colonies, are also commonly isolated from P. aeruginosa infections. SCVs are small (1 to 3 mm in diameter), form biofilms, attach strongly to surfaces, and display autoaggregative properties due to increased exopolysaccharide production (mainly Pel and Psl polysaccharides but sometimes also alginate) and high rates of production of pili (285). Moreover, these SCVs are usually nonmotile and resistant to several different classes of antibiotics (285). In vitro tests have shown that exposure to sublethal concentrations of antibiotics, such as aminoglycosides, selects for the formation of SCVs; hence, exposure to antibiotics may also trigger the selection of SCVs in vivo. In CF patients, prolonged persistent infections, deterioration of pulmonary function, and increased antibiotic resistance are all correlated with the presence of SCVs in sputum. For Pseudomona aeruginosa, the regulators and growth states involved in H2- or H3-T6SS expression, including quorum sensing and iron depletion, have been analyzed (286, 287). Analysis of RsmA-mRNA complexes of P. aeruginosa by gel shift assay as well as translational and transcriptional fusion led to the identification of 40 genes, distributed in six operons, which are translationally controlled by RsmA (288). RsmA is a negative regulator of these clusters as well as of the coding genes of T6SS-HIS-I related to chronic P. aeruginosa disease. In a process generally overlooked until now, RsmA has been demonstrated to act on most known T6SS genes, indicating that the type VI secretion system is also regulated by AmrZ (288).
The QS system in Acinetobacter spp. comprises the AbaR (receptor) and AbaI (synthase) proteins. These proteins are related to some virulence factors, such as motility, antibiotic resistance, survival properties, and biofilm establishment, in Acinetobacter baumannii and other Acinetobacter spp. (289, 290). In Acinetobacter strain M2 (originally named A. baumannii and later reclassified as Acinetobacter nosocomialis, due to genomic differences), synthesis of 3-hydroxy-C12-HSL requires AbaI (221). More than one AHL has been found in 63% of Acinetobacter isolates. Nonetheless, there is no correlation between different AHLs and particular species of Acinetobacter (291). Deletion of the abaI gene, which is associated with AHL formation, reduced biofilm formation by 30 to 40% relative to that in the wild-type strain (290). However, addition of an exogenous AHL obtained from Acinetobacter restored biofilm formation in the mutant (292). Moreover, the important role of a new QS enzyme, AidA, in bacterial defense against 3-oxo-C12-homoserine-lactone, an inhibitor of the quorum sensing system (293), was recently analyzed (294). Finally, in A. baumannii clinical strains from clone ST79/PFGE-HUI-1 (which lacks the AdeABC efflux pump) and a modified strain (named ATCC 17978 ΔadeB), the presence of bile salts (a stress condition) induced overexpression of genes involved in biofilm production, the T6SS, and surface motility, which are associated with QS (173).
One or more quorum sensing systems containing a synthase and an acyl-homoserine lactone receptor have been identified for all species of Burkholderia (295). Two complete quorum sensing systems, known as CciIR and CepIR, were discovered in B. cenocepacia J2315, in addition to a gene encoding a regulator known as CepR2 but lacking a synthase and, finally, a system based on Burkholderia diffusible signal factor (BDSF), known as RpfFBC (296–298). Biofilm formation in B. cenocepacia H111 is highly dependent on BapA, which is a surface protein, and BapR, a regulatory protein. Both the bapA and bapR genes need QS for high levels of expression (299). Moreover, in their study, Aguilar and collaborators reported that BapR is an important protein in relation to the development of persister cells, indicating that this regulator may be a useful target in the production of drugs to prevent the formation of biofilms and persister cells (299).
The complex M. tuberculosis biofilms can produce a subpopulation of drug-tolerant persister cells (300). In addition to persistence against antibiotics, biofilms can also be envisioned as being part of a key persistence strategy of M. tuberculosis against the host immune system in chronic infections, particularly those that do not display clinical symptoms (301). The QS system of M. tuberculosis is largely unknown, and we highlight expression of the WhiB3 protein in response to environmental signals present in vivo, which is consistent with a model of QS-mediated regulation (302).
(p)ppGpp Network
The (p)ppGpp response involves the enzymes guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp). Under starvation conditions (amino acid starvation) and other types of environmental pressure, the “alarmone” molecule is produced. The (p)ppGpp network includes Rel/SpoT homolog (RSH) proteins with a nucleotidyltransferase domain, some of which display only synthetic functions, only hydrolytic functions, or both (303, 304). Other proteins, such as the RelV (p)ppGpp synthase of Vibrio (305) and the RelQ (p)ppGpp synthase of Gram-positive bacteria, have an important role in the (p)ppGpp network (48). Cell processes such as replication, transcription, and translation are influenced by the (p)ppGpp network. Also, (p)ppGpp binds to RNA polymerase, which modifies the transcriptional profile and alters the translational machinery (such as rRNA and tRNA) until nutritional conditions improve (306–308). Interestingly, bacteria deficient in (p)ppGpp production usually display massive defects in persister cell formation and survival (7).
Two different (p)ppGpp-dependent (RSH proteins and amino acid starvation conditions) (309) and -independent pathways have been studied in relation to salt tolerance in E. coli (310). In E. coli, the generation of the abnormal amino acid isoaspartate, which can be repaired by isoaspartyl protein carboxyl methyltransferase (PCM), is a specific type of protein damage that is significant for bacterial survival. The relationship between unrepaired isoaspartyl protein damage and the development of E. coli persisters through activation of the (p)ppGpp network was recently investigated (310, 311). Moreover, Harms and collaborators confirmed the important role of (p)ppGpp and Lon in the development of persister cells in E. coli (312).
Several pathways are involved in the (p)ppGpp network (RSH proteins) in S. Typhimurium (313), including (i) osmoregulation of periplasmic glucan content by expression of the opgGH operon (314); (ii) enhancement of the expression of stress-dependent genes incorporating genetic material acquired by horizontal transfer (315); (iii) regulation of virulence proteins required for intracellular survival in macrophages (316); (iv) aminoglycoside resistance (317); (v) a defense mechanism against reactive nitrogen species (RNS) encountered in the host through the RNA polymerase regulatory protein DksA (318); (vi) regulated expression of the virulence elements (motility and biofilm production) from pathogenicity island 1, which is necessary to facilitate processes such as invasion and intracellular replication in host cell lines, such as human epithelial cells, as well as uptake by macrophages (319–321); and (vii) development of persister cells by inhibition of bacterial growth interfering with FtsZ assembly (322).
Several functions have been associated with the (p)ppGpp network (amino acid starvation) in V. cholerae. In 2012, a study of V. cholerae investigated how biofilm formation is controlled by a tangled regulatory mechanism, with QS acting as a negative regulator, the restrictive response mediated by (p)ppGpp synthases (RelA, SpoT, and RelV) acting as a positive agent, and both interacting to coordinate the production of biofilm with the environmental conditions (323). Moreover, several studies have associated this system with the development of virulence elements, such as cholera toxin (CT) and toxin-coregulated pilus (TCP) (323–325). However, it has also been found that production of hemagglutinin (HA)/protease, which influences the formation of biofilm and motility in V. cholerae, is independent of the (p)ppGpp network but requires HapR, RpoS, and cyclic AMP receptor protein (CRP) (326). It was recently shown that (p)ppGpp positively controls the production of acetoin and that this specific role of (p)ppGpp in V. cholerae enables the pathogen to survive in environments with considerable concentrations of glucose, as in the human intestine (327). Finally, the (p)ppGpp network in this microorganism has been linked to RpoS, which manages the “mucosal escape response” by establishing a specific response and executing chemotaxis and motility through intracellular proteolysis (25, 328).
However, the extensive literature shows that different intracellular stress responses, such as the SOS response, ROS, and (p)ppGpp (RSH proteins), can transform a subset of K. pneumoniae cells into persister cells with antibiotic tolerance (Fig. 2) (329). In addition, ROS generation is induced by multiple factors, such as suboptimal concentrations of aminoglycosides (which also activate the SOS response), paraquat, and H2O2 (control by RpoS, SoxRS, and YjcC). It can thus be concluded that antimicrobial treatment can promote the persistence phenotype in K. pneumoniae (330).
H. pylori must withstand the inhospitable conditions of the human stomach and does so via a minimum number of transcriptional regulators which control the stringent response, as analyzed in different studies (331, 332). A rel/spoT homolog (RSH) gene-deleted mutant was incapable of surviving under extreme conditions of acidity and oxygenation, including during infection and transmission (332). Moreover, the Rel protein has been demonstrated to be vital for the persistence phenotype in H. pylori inside macrophages during phagocytosis in the gastric environment (333). In addition, CO2 restriction considerably raises the level of (p)ppGpp and ATP inside this bacterium, although it does not reduce the mRNA level, indicating activation of a stringent response (334).
In C. jejuni strains, the (p)ppGpp network (RSH proteins), together with the phosphohydrolases (PPX/GPPA) and polyphosphates [poly(P)], has been related to motility, biofilm production, and the ability to survive under stress conditions, such as nutrient deficiency, as well as in the process of invasion and the persistence phenotype within the host cell (335, 336).
In E. faecalis, the lack of (p)ppGpp production results in a lower ability to maintain biofilm development (337). In this pathogen, (p)ppGpp production is regulated by the Rel/SpoT (RSH) proteins RelA and RelQ synthetase (338). RSH activates a strict response so that changes in (p)ppGpp levels affect survival under stress conditions as well as the virulence capacity (339). In a vancomycin-resistant Enterococcus faecium (VRE) subpopulation, a mutation in the stringent response (SR) pathway was recently reported that caused an increase in reference (p)ppGpp levels, which triggered antibiotic tolerance inside the biofilm (340). Finally, alarmone levels regulate the responses against environmental stress, tolerance to antibiotic treatment, and virulence elements in E. faecalis (341, 342).
In Shigella spp., with the stringent response, the RSH proteins together with the DksA protein showed activity (343).
Little is know about the role of the (p)ppGpp network in bacteria such as Yersinia enterocolitica and Clostridium difficile.
Persistence phenotype infections produced by S. aureus are nutrient restriction reactions that directly depend on the (p)ppGpp mechanism (344). The involvement of (p)ppGpp signaling in the development of persister cells as well as in the acquisition of antibiotic tolerance by S. aureus cells has been described (344). Nevertheless, more detailed analysis is required in relation to molecular principles, as recent studies did not find any connection between (p)ppGpp signaling and the development of persister cells in S. aureus (345, 346). CodY24, a repressor, has been shown to regulate expression of S. aureus gene homologues of Rel/SpoT proteins (RSH proteins), i.e., rsh genes, which constitute a distinct class of (p)ppGpp synthase genes (347). Mutations in codY or in rsh were not found to have any consequences on the number of persister cells during growth (346). It has also been noted that S. aureus has putative GTPases that are capable of recognizing guanosine tetraphosphate and guanosine pentaphosphate with a high affinity (345). Further analysis confirmed that these are active GTPases regulated by the presence of ribosomes (activation) and (p)ppGpp (inhibition). Once these molecules were characterized, it became clear that bacteria have a mechanism whereby cell growth can be halted under stress conditions by activation of (p)ppGpp, which blocks the correct formation of 70S ribosomes (345).
In P. aeruginosa strains, the RelA/SpoT protein RSH (stringent response) and the RpoS protein (stress conditions) promote the tolerance of P. aeruginosa biofilms to ciprofloxacin but not to tobramycin (Fig. 2) (348). RSH proteins have been described for this pathogen (349). Interestingly, in Acinetobacter oliveivorans DR1, QS controls (p)ppGpp synthase (RSH proteins), AHL, and histidine kinase proteins during participation in biofilm production and hexadecane metabolism (350).
As for cis-2-dodecenoic acid factor (discovered in B. cenocepacia), it belongs to the diffusible signal family and has been associated with increased levels of RecA (SOS response) and (p)ppGpp (RSH proteins) and a reduction in biofilm formation (Fig. 2) (351).
M. tuberculosis contains homologues of Rel proteins that react to low nutrient levels via activation of (p)ppGpp (352, 353). These proteins are necessary for growth in aerobic and anaerobic environments as well as for long-term survival in response to starvation (352), and they induce drug tolerance (354). In guinea pigs, cells which had lost Rel proteins were associated with a remarkable lack of tubercle lesions as well as an absence of caseous granulomas in histological sections (355). Interestingly, the success of the M. tuberculosis pathogen depends on cell growth in the host, regulated by (p)ppGpp (RSH proteins) and CarD (356). The lack of the CarD protein led to killing of M. tuberculosis due to DNA damage, starvation, and oxidative stress, all of which led to a reduction of rRNA transcription (Fig. 2) (356).
Toxin-Antitoxin Systems
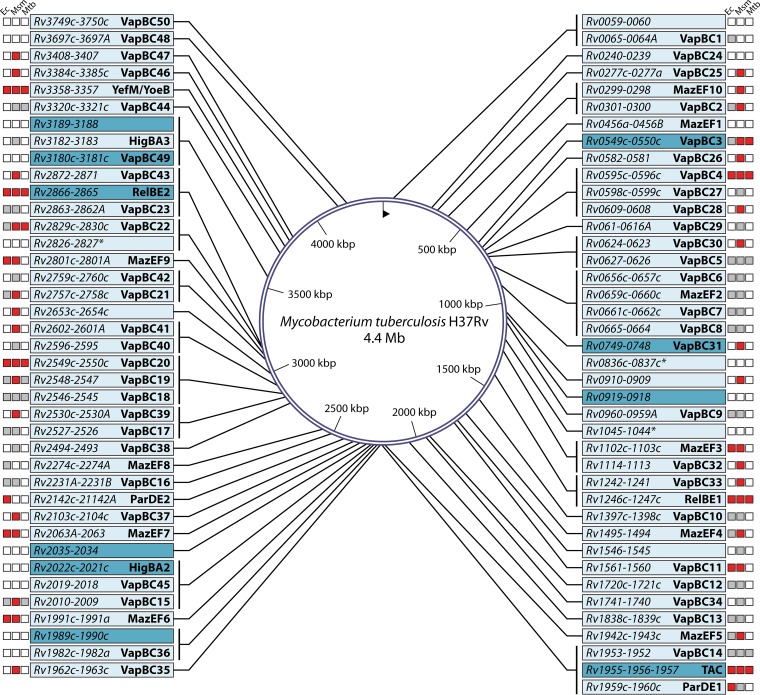
Finally, one of the best-studied mechanisms of formation of persister cells involves toxin-antitoxin (TA) systems, which trigger a state of bacterial dormancy to evade the effects of drugs or stress conditions (8). TA systems are small genetic systems located on bacterial plasmids as well as on chromosomes. TA loci usually are comprised of two genes, which encode a stable toxin and an unstable antitoxin that inhibits the toxin. TAs are currently divided into six distinct classes on the basis of the proteomic nature of the corresponding antitoxin (1, 16). See Fig. 4 for an explanation of the six best-studied types of TA modules. Critically, deletion of a single TA system that reduces persistence under certain conditions has been shown for the mqsR/mqsA locus (12, 357), the tisB/istR locus (181), and the dinJ/yafQ locus (358). Finally, several TA systems are triggered by the SOS system and (p)ppGpp to drive the development of persister cells (Fig. 2) (181, 359).
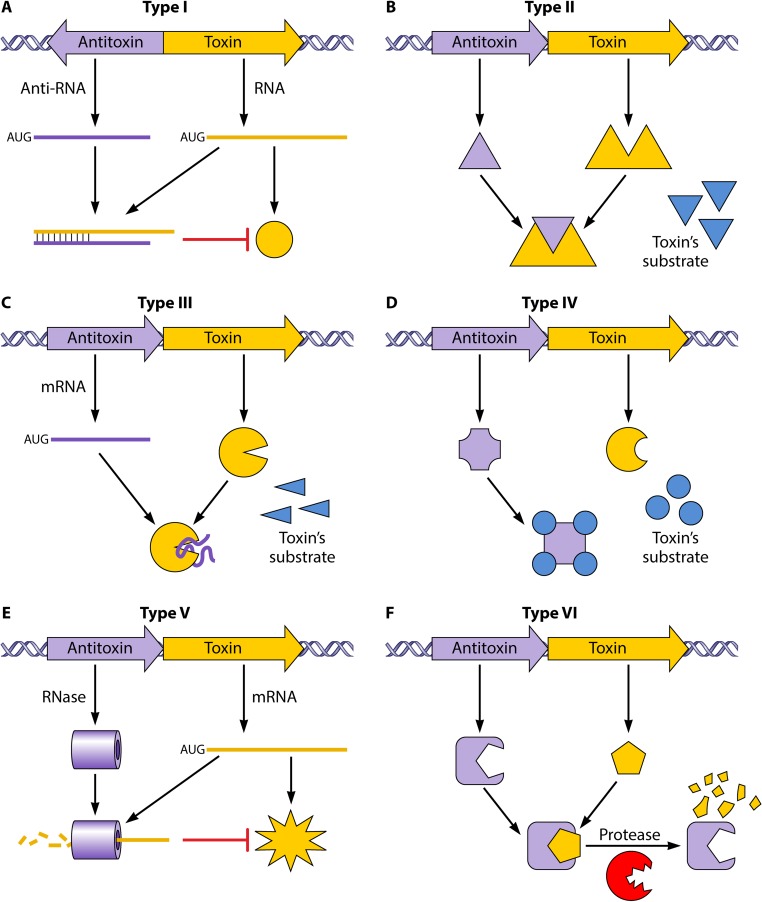
FIG 4.
Graphical representations of antitoxin-toxin interactions and their classification in the different types of TA systems in clinical pathogens. (A) Type I. There is an interaction between mRNAs that blocks toxin protein formation. (B) Type II. The antitoxin protein forms a complex with the toxin protein, inhibiting its function. (C) Type III. The antitoxin mRNA generates a complex with the toxin protein, inhibiting its function. (D) Type IV. The antitoxin protein competes with the toxin for its substrates. (E) Type V. The toxin mRNA is degraded by an RNase encoded by the antitoxin gene. (F) Type VI. A TA complex is formed by the union of toxin and antitoxin proteins, producing decomposition of the toxin by a cellular protease. (Adapted from reference 16.)
Research on the TA modules of Escherichia coli has provided most of the information that exists regarding persister formation (360). Also, type I, type II, and type IV TA loci are localized in the cryptic prophages of E. coli (361). In 2011, a model of E. coli persister formation based on the expression of 10 mRNAs for type II TA endonuclease modules was proposed (362). However, we must point out that that study was recently retracted due to the discovery of inadvertent artifacts (312). The following class II TA modules have been described: (i) the HipBA TA module, (ii) the TisB/IstR TA module, (iii) the HokB/SokB TA module, (iv) the YafQ/DinJ TA module, (v) the MazEF TA module, and (vi) the MqsRA TA module. The hipA toxin gene was the first gene to be associated with the production of persister cells (50, 363). TA loci encoding mRNases and persistence development are closely related (364, 365). In addition, an increment of the expression of HipA toxin in E. coli has been reported to halt growth by boosting (p)ppGpp production via RelA, a signal commonly related to amino acid requirements (365). By use of chloramphenicol to inhibit (p)ppGpp production in HipA-arrested cells, effects such as halting the initiation of replication and RNA synthesis were reduced, thus enabling recovery of susceptibility to β-lactam antibiotics (365). The TisB/IstR TA module is activated by the SOS system. TisB works as an ion channel which reduces proton motive force and the amount of ATP, enhancing the formation of persistent cells and causing antimicrobial tolerance (180). The HokB/SokB TA module is regulated by the (p)ppGpp system (181, 359). For the YafQ/DinJ TA module, the toxin shows an association with increasing persistence by reducing indole, thus establishing a relationship between the TA systems and the cell signaling QS system (224). The MazF toxin, which generally breaks down cellular RNAs, halts cell growth for some time and promotes formation of persister cells (366). The MqsR toxin influences persister cell formation (357). Moreover, the MqsRA TA in E. coli is physiologically vital for survival of bacterial cells in the gallbladder and upper intestinal tract, where concentrations of bile are generally high (367). As previously mentioned, TA systems promote persister states in bacteria by repressing bacterial metabolism or inhibiting cellular growth by the toxin. This state leads to bacterial tolerance of environmental stressors (368).
The presence of Salmonella inside macrophages creates stress conditions for the bacterium and induces the production of a subpopulation of persister bacteria by class II TA modules (369). Fourteen putative class II TA modules have been described for this pathogen, all of which are related to the development of persister cells inside macrophages. Among these, the toxin (TacT) contributes to the formation of these persisters in Salmonella populations by transiently blocking translation (370). The TacT toxin is an acetyltransferase that inhibits translation of tRNA molecules by disrupting the primary amine group of the amino acids, also favoring the formation of persister cells by the pathogen (370).
For V. cholerae, TA loci have been described as being present in a superintegron. V. cholerae TAs were assigned to the Phd/Doc, HigBA, RelBE, and HigBA families (371). However, only RelBE-family TA systems in V. cholerae are involved in biofilm and ROS generation (Fig. 2) (371).
Klebsiella pneumoniae isolates have a major role in antibiotic resistance, and the genes for several type I and II TA systems (Hok/Sok, PemK/PemI, and CcdA/CccB) (16) have previously been identified on resistance plasmids carried by this bacterium (372). A bioinformatics approach was used to analyze the distribution of the locus of the type II system and to determine the variability in the TA loci from 10 complete sequenced genomes of K. pneumoniae, revealing numerous putative type II TA loci (373). Some RelBE-like TA systems were also distributed in a manner different from that for the K. pneumoniae RelBE systems (59). Thus, the distributions of RelBE_1kp and RelBE_2kp loci differ in plasmids and chromosomes, although they were found in the same K. pneumoniae isolate (16, 373). However, a detailed distribution and the implications for the development of a persistence phenotype in K. pneumoniae are unknown (373).
A new type I TA system, the AapA1/IsoA1 locus, present on the chromosome of H. pylori, was recently characterized (374). Several type II TA systems associated with the bacterial persistence phenotype have also been localized in this pathogen, including (i) the HP0894-HP0895 proteins (375, 376), (ii) the HP0892-HP0893 proteins (RelE-family TA system), and (iii) the HP0967-HP0968 proteins (Vap family) (374).
CjrA/CjpT (type I) and VirA/VirT (type II) TA systems were identified in Campylobacter spp. (377). These systems were encoded by the pVir plasmid, which is involved in virulence and natural transformation. VirT belongs to the RelE family, one of the best-studied TA systems (374, 377).
Several plasmids encode toxin-antitoxin systems in strains of Enterococcus spp., as follows: (i) the plasmid pAD1-encoded Fst toxin (type I toxin-antitoxin) in E. faecalis affects the permeability of the membrane, thus altering the cellular responses to antibiotics (378–382); and (ii) plasmids encoding vancomycin resistance and harboring genes for the ω-ε-ζ Par toxin-antitoxin and Axe-Txe toxin-antitoxin have been located, but their involvement in the presence of persister cells has not been analyzed (383–386). Moreover, mazEF, mazEG, and higBA loci have often been found in E. faecalis and Enterococcus faecium clinical strains (387).
On the other hand, type II TA systems are related to bacterial persistence in Shigella sp. isolates and include YeeUT (388, 389), VapBC (390, 391), GmvAT, and CcdAB (392), while the only system described for Yersinia enterocolitica is the CcdA/B TA system, and its function has not been analyzed (393).
Only one type II TA system has been described for C. difficile, namely, MazEF (endoribonuclease), which is involved in C. difficile sporulation (68). However, by means of the RASTA-Bacteria TA database, additional putative TA systems, the COG2856-Xre and Fic systems, are assumed to exist in strain 630 of C. difficile (394).
S. aureus harbors some annotated chromosomal TA systems, and their function in persister formation is of interest (395). Several type II toxin-antitoxin systems have been identified in S. aureus, including MazEF and Axe1/Twe1 as well as Axe2/Twe2, both of which are RelBE homologues. A recent study showed that the two open reading frames directly over the sigB operon region in the S. aureus genome, herein designated masES and mazFS, represent a TA system (396). However, further studies of the involvement of these TA systems in the development of persister cells must be performed. Although the findings of bioinformatics studies of P. aeruginosa suggest that TA systems are abundant in the genome of this bacterium (397), the functions of these systems have not been established (16), except for the HigBA system, which downregulates virulence by the effect of the HigB toxin, which reduces the production of pyocyanin and pyochelin and surface motility (398).
The plasmid p3ABAYE is the most frequently found plasmid among the toxin-antitoxin plasmidic systems in A. baumannii. This plasmid (94 kb) probably encodes the following five TA systems: (i) RelBE; (ii) two HigBA systems, encoded in opposing directions; (iii) SplTA (DUF497/COG3514 domain proteins); and (iv) CheTA (HTH/GNAT domain proteins). These were found in a group of A. baumannii clinical strains from Lithuanian hospitals. The HigBA and SplTA TA systems were particularly common (88.6% predominance, taking into account the 476 clinical samples). Remarkably, in 46 of the A. baumannii isolates analyzed, expression of the HigBA toxin-antitoxin system was not observed (399, 400). All of these toxin-antitoxin systems were found in most clinical isolates of A. baumannii belonging to the ECI and ECII groups worldwide. The function of SplTA was found to be associated with the persistence phenotype (399). Moreover, the AbkB/AbkA toxin-antitoxin system, also known as SplTA, was found to be encoded by the most frequent resistance plasmid of A. baumannii, which carries an OXA24/40 β-lactamase (carbapenem resistance) gene (401). The toxin of this system has the ability to prevent translation when overexpressed in E. coli, through the scission of lpp mRNA plus the transfer of mRNA, all of which indicate that the AbkB toxin acts as an endoribonuclease. The stability of the plasmid in the absence of selection pressure can be explained by the presence of the AbkB/AbkA system, especially in the case of small plasmids pAC30a and pAC29a, which lack a blaOXA24/blaOXA40-like gene (401). Moreover, in a recent study of isolates of A. baumannii with resistance to carbapenem due to the presence of a plasmid encoding OXA24 β-lactamase and the AbkAB TA module, we analyzed the presence of a link between molecular mechanisms associated with the development of tolerance to a biocidal compound (chlorhexidine) and later formation of a subgroup of persister cells in the presence of an antibiotic (imipenem). These persister cells showed overexpression of the abkB toxin gene as well as downregulated expression of the abkA antitoxin gene (349).
Expression of most of the toxin genes (from TA systems) in biofilm cells of B. cenocepacia is upregulated relative to that in planktonic cells (82, 402). The high levels of expression of these bacterial toxins involve an increase in cellular survival after treatment with ciprofloxacin or tobramycin, confirming the importance of the toxins in the development of persister cells as well as the generation of antibiotic tolerance in biofilms (82, 402). The FixL toxin was recently identified in Burkholderia dolosa, which belongs to the B. cepacia complex (BCC). This toxin is homologous to FixL, which forms part of the two-component FixL/FixJ system of the Rhizobiales. In B. dolosa, the fixL gene acts as a general regulator of motility, biofilm formation, persistence, virulence, and intracellular invasion (45).
Finally, the genome of M. tuberculosis, a particular persistence phenotype pathogen, has an abundance of toxin-antitoxin systems. Most of these modules have been found to be functional in vivo in animal models, and they are involved in virulence and antibiotic tolerance (403–406). To date, numerous toxin-antitoxin systems (confirmed and putative) have been identified in the H37Rv isolate of M. tuberculosis; the most frequent are type II TAs (VapBC, MazEF, YefM/YoeB, RelBE, HigBA, and ParDE) (Fig. 5) (404). Interestingly, no other type of TA system has been detected in this bacterium (407, 408). Most M. tuberculosis systems (409) have been analyzed empirically, in Mycobacterium smegmatis and E. coli, with the aim of determining the toxin function in growth restriction as well as how to neutralize this function. Thus, researchers found that 37 of the TAs were functional under at least one condition (404, 405, 410–413). The articles cited summarize the current knowledge of the M. tuberculosis toxin-antitoxin modules. Moreover, analysis of the M. tuberculosis persister transcriptome shows positive regulation of the toxin-antitoxin systems (213, 414). Although the involvement of different TA modules in this repertoire is not fully understood, functional specialization can be considered due to the synergy observed under certain stress conditions (405, 406).
FIG 5.
Chromosomal map of M. tuberculosis H37Rv TA systems. TA systems are annotated according to the information in the Tuberculist database, except for VapBC45 (Rv2018-Rv2019), VapBC49 (Rv3181c-Rv3180c), VapBC50 (Rv3750c-Rv3749c), HigBA2 (Rv2022c-Rv2021c), HigBA3 (Rv3182-Rv3183), YefM/YoeB (Rv3357-Rv3358), and MazEF10 (Rv0298-Rv0299). Most of the TA systems depicted here probably belong to type II, except for those marked with an asterisk, which are putative type IV systems. For each system, the functionality in E. coli (Ec), M. smegmatis (Msm), and M. tuberculosis (Mtb) is depicted as follows: red indicates inhibition of growth, gray indicates no inhibition of growth, and white indicates that the system was not tested. The 10 most commonly induced TA systems in drug-tolerant persister cells are highlighted by a dark blue background. (Reproduced from reference 404 with permission.)
We summarize the molecular mechanisms of tolerance and persistence previously described for each pathogen in Table 1.
TABLE 1.
Targets of different mechanisms associated with bacterial persistencea
| Environment and pathogen | Target(s) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Stress response | ROS response | Energy metabolism | Efflux pump | SOS response | Quorum sensing (QS) | (p)ppGpp signaling | Toxin-antitoxin (TA) system | |
| Gastrointestinal tract | ||||||||
| E. coli | RpoS (21, 22) | Superoxide dismutase (SOD)/catalase (50) | Cytochrome bd oxidases (95, 98), cytochrome c peroxidases (101), TauD protein (132, 133) | AcrAB (147, 152) | RecA/RecBCD (178, 179), TisB toxin (180) | SdiA/LuxS (AI-2/AI-3 autoinducers/indole) (221), OxyR/phage shock (222, 223) | RSH proteins (309), PCMb (310, 311) | HipBA (363–365), TisB-IstR (179), HokB-SokB (181, 359), YafQ/DinJ (224), MazEF (366), MqsRA (357, 367) |
| Salmonella spp. | RpoS (23) | SoxRS, OxyR, σS and σE factors, SlyA protein, dps gene (51) | Cytochrome bd oxidases (98) | AcrAB (11, 154), AcrD (57), MdtD (88), MacAB (154) | RecA (176) | Unknown synthase/SdiA (3OC8HSL signal) (230); LuxS, LsrB (AI-2 signal) (231); QseB, QseC (AI-3 signal) (232); T3SS/T6SS (234) | RSH proteins (313), opgGH operon/stress-dependent genes/DksA/FtsZ interference (314–319, 322) | Putative class II TA systems/TacT (370) |
| Vibrio spp. | RpoS (25) | Catalases (KatB-KatG)/PhoB-PhoR system (53), OxyR (54), cholix (55) | Oxygen/reductases (102–105), taurine (136–139) | EmrD-3 (156) | RecA (185) | LuxS, LuxP (AI-2 signal)/CqsA, CqsS (CAI-1 signal)/AlsR/AphA (235–238); T3SS/T6SS (241–245) | RSH proteins (323), cholera toxin (CT)/toxin-coregulated pilus (TCP)/acetoin (324–327) | Phd-Doc/RelBE/HigBA/ParDE (371) |
| Klebsiella spp. | RpoS/SoxRS (26) | CPS (58) | AcrAB/OqxAB (158) | RecA/Viz/GrapE/ClpX/Dead proteins (186) | LuxS (AI-2 signal) (246, 247), T6SS (249) | RSH proteins (329), SoxRS/YjcC (330) | Hok-Sok/PemK-PemI/CcdA-CccB/RelBE (372, 373) | |
| Helicobacter spp. | Fur/HspR (31) | Catalase/SOD/arginase (62, 63) | Cytochrome bd oxidases (109–111), taurine (140) | Two efflux pumps (159) | RecA/AddAb (187, 188) | LuxS (AI-2 signal) (250–252) | RSH proteins (332, 333) | AapA1-IsoA1/RelE family (HP0894-HP0895)/Vap family (HP0967-HP0968) (374–376) |
| C. jejuni | KatA-SodB/AhpC-Tpx/Bcp (64) | Cytochrome bd oxidases (112) | CmeABC (151, 160) | RecA (189) | LuxS (AI-2 signal) (160), T6SS (151, 261) | RSH proteins/PPX/GPPA/PolP (335, 336) | CjrA-CjpT/VirA-VirT (374, 377) | |
| Enterococcus spp. | Gsp65 (ohr gene) (32), Gsp62 (33), Gls24 (34–36) | Cytochrome bd oxidases (113), Clp ATP protease (114) | MefA/TetK/TetL/MsrC (161) | RecA/UmuDC (190) | fsrA/fsrC/gelE/GTF genes (262) | RSH proteins (338) | Fst toxin (pAD1) (378–382), ω-ε-ζ (pw9-2)/Axe-Txe modules (pRUM) (383–386), MazEF/MazEG/HigBA (387) | |
| Shigella spp. | Toxin (65) | Cytochrome bd oxidases/CydC (115, 116) | AcrAB (162), MdtJI (163) | RecA/topoisomerases/histones (191) | AI-2 (signal) (263), T3SS/T6SS (264–266) | RSH proteins/DksA (343) | YeeUT/VapBC/GmvAT/CcdAB (388–392) | |
| Yersinia spp. | New SODs (66), yersiniabactin (67) | RosA/RosB (164) | RecA/histone-like proteins (192) | LuxI/LuxR-like proteins (267) | CcdAB (393) | |||
| C. difficile | HSP proteins (GroESL/DnaKJ) (40) | Catalase/SOD (68), TcdA-TcdB/glutamate dehydrogenase (GDH) (69) | CdeA (165) | RecA/LexA (193, 194) | LuxS/SpoOA (271, 272) | MazEF (68), COG2856-Xre/Fic (394) | ||
| Respiratory tract | ||||||||
| S. aureus | σB factor (42) | Catalase/SOD (70), aconitase/succinate dehydrogenase-TCA cycle enzymes (71) | Cytochrome bd oxidases (117), CymR (taurine) (118, 142) | Qac efflux (166) | RecA/LexA, RexAB/PolV (195, 196) | SarA/Agr (AI-2 signal) (273–276), LuxS/IcaR (275) | RSH proteins, rsh genes (344–346), CodY24 (347), GTPases (345) | MazEF/RelBE (395, 396), TisB-GhoT toxins (346) |
| P. aeruginosa | RpoS/pls locus (41) | Catalase/SOD/POX (74), mucoid phenotype (75, 76) | cbb3-type enzyme (118), GacAS system (123, 124) | MexAB (167, 168, 172) | RecA/LexA/PrtR/PA0906 (198, 201, 203), UmuDpR (205) | Las/Rhl QS systems (AI-2/PQS, AIP/AHL/DSF signals) (277, 278, 283), T6SS (RsmA/AmrZ) (286–288) | RSH proteins (348) | HigBA (398) |
| A. baumannii | SOD (79), catalases (KatA/KatE/KatG/KatX) (80) | Cytochrome bd oxidases (128), tauRXYPI operon (143) | AdeABC, AdeFGH (170–173) | RecA/LexA, UmuC (207–209) | AbaR/AbaI/AidA (N-[3-OH-C12] signal) (289–291, 294), T6SS system (173) | RSH proteins (349) | RelBE/HigBA/CheTA/AbkAB (SpltA) (349, 399–401) | |
| B. cepacia | RpoN/RpoE (fixLJ) (43–45) | SOD/catalase, glyoxylate (TCA cycle) (83) | tauABC operon (145) | BCAM0925 to -0927, BCAM1945 to -1947, BCAL1672 to -1676 (RND efflux) (174) | CepIR/CciIR/CepR2/RpfF (BapA and BapR) (296–298) | RSH proteins/DSF signals (351) | FixLJ (45) | |
| M. tuberculosis | Catalase (KatG) (84) | pst operon (129–131) | ImuA-ImuB/DnaE2 (212), ClpR-like protein (215) | WhiB3 regulator protein (302) | RSH proteins/CarD protein (352, 353) | VapBC/MazEF/YefM/YoeB/RelBE/HigBA/ParDE (193, 403–415, 480) | ||
Numbers in parentheses are reference numbers.
Isoaspartyl protein carboxyl methyltransferase.
MEASUREMENT OF LEVELS OF BACTERIAL TOLERANCE AND PERSISTENCE
Survival of bacteria in natural environments depends directly on the capacity of the microorganisms to sense and accommodate to the surroundings. Bacteria can recognize and modify themselves in different situations by monitoring the environment and also by creating signals and modifying gene expression accordingly.
In a recently published opinion article, Brauner and collaborators highlighted the importance of distinguishing between resistant, tolerant, and persistent bacterial cells before selecting the appropriate antibiotic treatment (5). With the aim of establishing differences in the diverse survival strategies under stress conditions, they propose measurement of the minimum duration of killing (MDK) in batch cultures of bacteria. The MDK, which is based on the concept of effective killing, is used to provide a quantitative measure of tolerance and indicates that tolerant strains must be exposed to the condition under consideration for a longer time than that for susceptible strains. The MDK is described as the usual time required by an antimicrobial to eliminate a certain proportion of the bacterial culture (5, 415). The MICs of antibiotics are similar for tolerant and susceptible isolates. However, the MDK99 (time to kill 99% of the cells in the culture) is generally higher for tolerant than for susceptible isolates. Furthermore, the MIC and MDK99 for persister cells in bacterial populations are also similar to those observed for susceptible bacteria; however, for cells in culture, the MDK99.99 is considerably higher for persister cells than for susceptible cells (Fig. 5) (5). A recently described novel method, the TDtest, would be capable of detecting distinct levels of antibiotic tolerance in clinical isolates. This technique was analyzed with E. coli isolates, which were also used to study the antimicrobials with the highest activity against tolerant populations as well as persister subpopulations (Fig. 6) (416, 417).
FIG 6.
(A) Characteristic drug responses for resistance, tolerance, and persistence phenotypes. The MIC value is useful for analyzing bacterial populations that are susceptible and resistant to antibiotics, while the MDK (minimal duration of bacterial killing) is a quantitative measure that is suitable for studying tolerant and persistent bacterial cells. Among susceptible bacterial cells, the MDK99 (time to kill 99% of culture cells) is higher for tolerant bacterial cells, while the MDK99.99 (time to kill 99.99% of culture cells) is higher for persister bacterial cells. (Adapted from reference 5 with permission from Springer Nature.) (B) Analysis of bacterial tolerance and persistence in clinical strains by use of a modified disk diffusion test (TDtest). (1) A disk with drug is added to the agar plate. (2) The drug-infused disk is replaced by a glucose-infused disk. The drug diffuses from the disk. (3 and 4) Susceptible bacteria show growth inhibition around the drug-infused disk. (5 and 6) Tolerant bacteria show growth inhibition around the drug-infused disk (5), and colonies inside the inhibition zone after addition of glucose show late-growing bacteria of this isolate (6). (Adapted from reference 416.)
NEW APPROACHES TO TREATMENT OF BACTERIAL PERSISTENCE
The development of new bacterial treatments must include tolerant and persister bacterial cells as targets. Hence, knowledge of the mechanisms of bacterial tolerance or persistence may provide targets for development of new anti-infective treatments for combating tolerant and persister cells (61), including antimicrobial peptides (synthetic and pyocins/bacteriocins), antivirulence compounds, phage therapy, anticancer drugs, and new molecules (Table 2).
TABLE 2.
Experimental treatments against persister cells
| Treatment type | Molecule(s) | Mechanism of action | Bacterial target(s) | Reference(s) |
|---|---|---|---|---|
| Antimicrobial peptides | Peptide 1018 | Blocking (p)ppGpp | Gram-negative pathogens: P. aeruginosa, E. coli, A. baumannii, K. pneumoniae, S. Typhimurium, B. cenocepacia | 418 |
| Gram-positive pathogens: methicillin-resistant S. aureus | ||||
| Peptide analogue | Blocking (p)ppGpp | M. smegmatis | 419 | |
| RelA proteins | Antibiofilm activity | |||
| Pyrazinoic acid (POA) | Blocking (p)ppGpp | M. tuberculosis | 420 | |
| Peptide SAAP-148 | Biofilm inhibition | S. aureus, A. baumannii | 421 | |
| R-type pyocins | Bacterial lysis | P. aeruginosa, Haemophilus spp., Neisseria spp., Campylobacter spp. | 423–434 | |
| Curvacin A (bacteriocin) | Bacterial lysis | L. monocytogenes | 439 | |
| Enterocidin B3A-B3B (bacteriocin) | Bacterial lysis | L. monocytogenes | 440 | |
| Enterocidin | Bacterial lysis | L. monocytogenes | 440 | |
| B3A-B3B/nisin (bacteriocin) | ||||
| Antivirulence compounds (QS inhibitor molecules) | QS inhibitory molecules | Inhibition of the MvfR virulence regulon | P. aeruginosa | 441 |
| Halogenated indoles | LuxR inhibition | Gram-negative pathogen: E. coli | 226 | |
| Gram-positive pathogen: S. aureus | ||||
| (Z)-4-Bromo-5-(bromomethylene)-3-methylfuran-2-(5H)-one | Quorum sensing inhibition | E. coli | 442 | |
| Benzimidazole derivative M64 | Blocking PqsR | P. aeruginosa | 441 | |
| RNAIII-inhibiting peptide (RIP) and its analogues | Inhibition of phosphorylation of target of RNAIII-activating protein (TRAP) | S. aureus | 447–449 | |
| Synthetic autoinducer of AIP and analogues (I to IV) | Inhibition of RNAIII | S. aureus | 447 | |
| Nonfunctional AIP analogues | Repression of many AgrC receptors (I to IV) | S. aureus | 450 | |
| Halogenated compounds | Quorum sensing inhibition | P. aeruginosa | 221, 451 | |
| AHL antagonists | P. aeruginosa | |||
| Cell extracts and secretion products | P. aeruginosa | |||
| QS inhibitors from food and plant sources | P. aeruginosa | |||
| Acylases and lactonases | P. aeruginosa | |||
| OmpA inhibitor | Inhibition of pathogenesis | A. baumannii, P. aeruginosa, E. coli | 453 | |
| Phage therapy | Phage cocktails | Antibiofilm activity | Coagulase-negative staphylococci, S. aureus, E. faecalis, E. faecium, E. coli, P. mirabilis, K. pneumoniae, P. aeruginosa, Acinetobacter spp. | 454–459 |
| Lytic phage phiIPLA-RODI | S. aureus | 460 | ||
| Endolysins | Antibacterial effect | S. aureus | 461 | |
| Gram-positive bacteria other than S. aureus | 462 | |||
| PlyE146 endolysin | Antibacterial effect | A. baumannii, P. aeruginosa, E. coli | 463 | |
| LysAB2 endolysin | Antibacterial effect | Methicillin-resistant S. aureus, A. baumannii, E. coli, and other bacteria (with modifications) | 464, 465 | |
| CF-301 lysin | Biofilm agent | Staphylococcus spp. | 481 | |
| Anticancer drugs | 5-Fluorouracil, gallium compounds, mitomycin C, cisplatin | Inhibition of persister cells | P. aeruginosa | 466–468 |
| New molecules | DG70 (biphenyl benzamide) | Inhibition of respiration | M. tuberculosis | 469 |
| Suramin | Inhibitor of RecA protein and SOS response | M. tuberculosis | 471 | |
| 3-(4-[4-Methoxyphenyl] piperazin-1-yl) piperidin-4-yl biphenyl-4-carboxylate | Waking of persister cells by unknown mechanism | E. coli | 472 | |
| P. aeruginosa | ||||
| Antibiotic acyldepsipeptide ADEP4 | ClpP protease activation | S. aureus | 474 | |
| Sytox Green NH125 (1-hexadecyl-2-methyl-3-[phenylmethyl]-1H-imidazolium iodide) | Permeabilization of the membrane | S. aureus | 475 | |
| Pyrazinamide (analogue of nicotinamide) | trans-Translation inhibition | M. tuberculosis, B. burgdorferi | 476 | |
| Daptomycin | Disruption of multiple aspects of bacterial cell membrane function | B. burgdorferi | 417 | |
| cis-2-Decenoic acid | Antibiofilm activity | P. aeruginosa | 478 | |
| Itaconate plus tobramycin | Inhibitor of ICL (isocitrate lyase) | B. cepacia | 83 | |
| Morin, pyrrolidine, quercetin, quinine, reserpine | Antibiofilm activity | S. aureus | 479 | |
| Small molecules | LexA autoproteolysis | 203 |
Use of a peptide prevented biofilm development and also yielded removal of mature biofilms of Gram-positive and Gram-negative pathogens, such as A. baumannii, P. aeruginosa, E. coli, methicillin-resistant S. aureus, K. pneumoniae, B. cenocepacia, and S. Typhimurium (418). Interestingly, the activity of inhibitory compounds analogous to Rel proteins [synthetic (p)ppGpp] has been described for Mycobacterium spp. (419). The impacts of these molecules on long-term persistence, biofilm disruption, and downregulation of (p)ppGpp have been analyzed in vivo in M. smegmatis (419). Another compound, pyrazinoic acid (POA), has been described for M. tuberculosis and showed significant inhibition of persister cells (420). Finally, we highlight the peptide SAAP-148, which has shown an important level of activity against persister cells of methicillin-resistant S. aureus and MDR A. baumannii (421). In contrast, P. aeruginosa protects itself from other strains of its species by use of the R-type pyocin (422), which is structurally similar to the contractile tail of the Myoviridae bacteriophage family (423) and is encoded by a unique cluster in the genome; although the pyocin mainly kills P. aeruginosa, it can also kill members of other genera, such as Neisseria, Campylobacter, and Haemophilus (424–429). Bacteriocins, such as pyocins, have been reported for other Gram-positive and Gram-negative bacteria (430–434). Bacteriocins are a diverse group of ribosomally produced antimicrobial peptides. Some bacteriocins undergo extensive posttranslational modifications, which, together with their mode of action, have been used for classification purposes (435). Moreover, bacteriocins are toxic bacterial peptides that are released in order to inhibit bacterial growth and biofilm production (18, 436, 437). The most recent classification scheme (438) suggests three classes based on the mechanisms of biosynthesis and biological activity of lactic acid bacterium (LAB) bacteriocins, although it may also be applied to bacteriocins of other microorganisms. The use of bacteriocins in antimicrobial packaging is particularly well suited for foods at risk of surface contamination (435). We highlight two studies of the use of bacteriocins as an antibiofilm strategy. In the first of these, researchers used curvacin A-producing Lactobacillus sakei CRL1862 to reduce the biofilm formation of Listeria monocytogenes (439). In the second study, Al-Seraih and collaborators analyzed the capacity of enterocidin B3A-B3B alone and in combination with nisin (another bacteriocin) to inhibit biofilm formation in Listeria monocytogenes (440).
Antivirulence treatments aim to inhibit bacterial virulence without affecting growth of the bacteria (150). In 2014, in a study analyzing persister cell strains of P. aeruginosa, QS molecules that inhibited the MvfR virulence regulon (LysR-type transcriptional regulator) limited lethal effects in mice (441). Moreover, halogenated indoles have been observed to remove bacterial biofilms and persistent cells formed by Gram-positive and Gram-negative microorganisms, such as S. aureus and E. coli (226). Pan et al. demonstrated that the QS inhibitor BF8 decreases the persistence of growing E. coli cultures and throwback antibiotic tolerance in the persisters (442). The benzimidazole derivative M64 is another QS inhibitor that has been described in relation to preventing the development of persister cells, by blocking the PqsR QS/virulence system of P. aeruginosa (441). However, resistance to these compounds is common (443–445) and increasing (446). Other inhibitory QS S. aureus peptides have also been evaluated and include the following. (i) The RNAIII-inhibiting peptide (RIP) and its analogues, which inhibit phosphorylation of a target protein (target of RNAIII-activating protein [TRAP]), leading to suppression of virulence characteristics in vitro (447, 448), are also effective in vivo. These compounds are more effective in combination with cefazolin, imipenem, or vancomycin (449). (ii) A synthetic autoinducer of AIP and its derivatives (I to IV) also inhibit RNAIII (447). (iii) Finally, nonfunctional AIP analogues can repress many AgrC receptors (I to IV). These compounds are the most potent described, to date (450). In addition, numerous inhibitors can be highlighted in relation to inhibitory QS in P. aeruginosa (221, 451), such as (i) halogenated compounds; (ii) AHL antagonists; (iii) cell extracts and secretion products; (iv) quorum quenchers obtained from plants and food; (v) acylases and lactonases (452); and (vi) an inhibitor of the OmpA protein, which is an important virulence factor (453).
Several authors have studied the use of lytic phage cocktails to prevent biofilm formation by bacteria, such as S. aureus and coagulase-negative staphylococci (CoNS), Enterococcus faecalis, E. faecium, E. coli, Proteus mirabilis, K. pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. (454–459). Fernández et al. demonstrated that S. aureus biofilms formed at nonlethal concentrations of phage phiIPLA-RODI present a unique physiological state that may benefit both the host and the predator (460). Thus, biofilms may be denser and contain more DNA, depending on phage pressure. Significantly, transcriptome sequencing (RNA-seq) data showed (p)ppGpp response expression, which may slow the movement of the bacteriophage within the biofilm. The result would be an equilibrium that would help bacterial cells to survive environmental pressures while maintaining a reservoir of sensitive bacterial cells available to the phage on reactivation of the latent carrier subpopulation. The study of lysins from phages and bacterial lytic proteins is also of great interest. Several endolysins have been found to exert a lytic function against Gram-positive bacteria, such as S. aureus (461) and other bacteria (462). In relation to Gram-negative bacteria, we highlight the endolysin PlyE146, which displays lytic activity against E. coli, P. aeruginosa, and A. baumannii (463). Finally, the LysAB2 endolysin, first described in 2011, shows activity against bacteria, such as methicillin-resistant S. aureus, A. baumannii, and E. coli (464). Interestingly, peptide-induced modification of this endolysin extended the range of lytic activity (465).
Recently, Wood et al. reported the effective use of anticancer drugs, including 5-fluorouracil (5-FU), gallium (Ga) compounds, mitomycin C, and cisplatin, to treat persistent bacterial infections (466–468).
New antibacterial agents against bacterial persisters have been described and include the following. (i) Specific inhibitors of respiration (MenG inhibitors, such as DG70 in Mycobacterium tuberculosis) are accepted antitubercular agents. Nevertheless, further studies will be necessary to optimize the route toward the proposal of candidates for validation of in vivo efficiency (469). In addition, analysis of physiological, biochemical, and pharmacological data shows that cytochrome bd plus the biosynthetic pathways of menaquinone, fumarate dehydrogenase, hydrogenase, and ubiquinone dehydrogenase are potential targets for the next generation of drugs (470). (ii) Suramin is an inhibitor of the RecA protein and the SOS response in M. tuberculosis (471). (iii) 3-(4-[4-Methoxyphenyl] piperazin-1-yl) piperidin-4-yl biphenyl-4-carboxylate wakes persisters, although the procedure has not been determined (472, 473). (iv) The antibiotic acyldepsipeptide ADEP4 eliminates ATP demands by ClpP protease activation, producing the death of persister cells (474). (v) Sytox Green NH125 acts against MRSA persisters by permeabilizing the bacterial membrane (475). (vi) Pyrazinamide (an analogue of nicotinamide) is used to treat M. tuberculosis and Borrelia burgdorferi infections. This compound acts by disrupting trans-translation during retrieval of the halted ribosome process (476). (vii) Daptomycin has also been associated with the killing of B. burgdorferi persisters (477). (viii) The compound cis-2-decenoic acid (CDA) reduces biofilm-derived P. aeruginosa persister cells (478). (ix) Pretreatment of Burkholderia cenocepacia biofilms with itaconic acid, an isocitrate lyase inhibitor, produced lower survival of persisters in a Burkholderia cenocepacia biofilm in response to a treatment including tobramycin (83). (x) The use of subinhibitory concentrations of 2′,3,4′,5,7-pentahydroxyflavone, tetrahydropyrrole, and quercetin, alone or in combination with antibiotics, can prevent or control biofilm formation. Synergetic interactions with antibiotics have been observed to affect biofilms of S. aureus for strain SA1199B overexpressing NorA. Culture of this strain with ciprofloxacin at subinhibitory concentrations led to acquisition of tolerance to the antibiotic. However, this was reversed when 2′,3,4′,5,7-pentahydroxyflavone and quinine were added, demonstrating that the inclusion of phytochemicals in combined therapies improves treatments and decreases antibiotic resistance, strongly affecting S. aureus in biofilm and planktonic states (479). (xi) Finally, small molecules which participate in the LexA autoproteolysis step in the SOS system may be used as SOS inhibitors and administered as adjuvants to current antibiotics (204).
CONCLUSIONS
Obtaining information about the metabolism of cells with tolerance and persistence phenotypes is challenging but also provides valuable tools for the development of antitolerance and/or antipersistence compounds. Interestingly, in-depth analysis of these mechanisms has revealed a larger number of data for gastrointestinal pathogens than for respiratory ones, possibly due to differences in environmental and antimicrobial pressures. The knowledge of the molecular mechanisms of tolerant populations and/or persistent subpopulations is key to the fight against multidrug-resistant (MDR) bacteria. The current lack of effective antibiotics against MDR pathogens drives the need to develop new bacterial treatments, which must include tolerant and persister bacterial cells as targets due to the relationships among these bacterial populations.
Moreover, phenotypic detection of tolerant populations and persistent subpopulations in microbiological clinical practice through MDK and TDtest measurements would help to (i) improve the guidelines for anti-infective treatments (selection as well as duration of treatment), (ii) avoid the evolution or maintenance of resistant bacterial populations, (iii) allow for study of the antitolerance and antipersistence ability of antimicrobials used in clinical practice, and (iv) allow for analysis of the efficiency of new anti-infective treatments (including antitolerance and/or antipersistence ability).
In conclusion, phenotypic, molecular, and clinical studies of these bacterial populations are important for the development of new anti-infective treatments efficient in the fight against MDR pathogens. Research on the diagnosis and treatment of infection according to bacterial populations, host environments, and patient features may become essential for the development of personalized medicine.
ACKNOWLEDGMENTS
This study was funded by grants PI13/02390 and PI16/01163, awarded to M. Tomás within the State Plan for R+D+I 2013–2016 (National Plan for Scientific Research, Technological Development and Innovation 2008–2011) and cofinanced by the ISCIII-Deputy General Directorate of Evaluation and Promotion of Research-European Regional Development Fund “A Way of Making Europe” and the Instituto de Salud Carlos III FEDER, Spanish Network for Research in Infectious Diseases (REIPI) (grants RD16/0016/0001 and RD16/0016/0006), as well as the Study Group on Mechanisms of Action and Resistance to Antimicrobials (GEMARA; SEIMC). M. Tomás was financially supported by the Miguel Servet Research Programme (SERGAS and ISCIII). R. Trastoy and L. Fernández-García were financially supported by a postspeciality from the Fundación Novo Santos (CHUAC-SERGAS, Galicia, Spain) and a predoctoral fellowship from the Xunta de Galicia (GAIN, Axencia de Innovación), respectively.
Biographies

R. Trastoy is a Clinical Microbiologist and obtained her degree in biology from the University of Santiago de Compostela (USC), Galicia, Spain (2007 to 2012), specializing in molecular biology. Since then, she has specialized in clinical microbiology and parasitology at the USC University Hospital Complex (CHUS), Galicia, Spain (2013 to 2017). During this period, she began conducting research in the field of microbiology. She is currently developing a line of research on hepatitis B virus, undertaking doctoral research within the framework of a national project. In addition, she is collaborating on different research projects, such as “Clinical Phage Therapy: New Challenges,” “Antimicrobial Persistence and/or Tolerance,” and “Molecular Diagnostic Tools: Development of Molecular Kits,” within the INIBIC-CHUAC microbiology research group, A Coruña, Spain, directed by M. Tomás. She also has some experience in clinical practice as a microbiologist.

T. Manso is currently a Ph.D. student in microbiology, in the odontologic sciences group, led by I. Tomás. She obtained her degree in chemistry from the University of Santiago de Compostela (USC), Spain, in 2013. Afterwards, she carried out training as a clinical microbiologist at the USC University Hospital Complex, completing her specialist training in May 2018. She has conducted research in microbiology from 2014 onwards. Since 2016, she has collaborated with M. Tomas on assignments related to resistance and virulence mechanisms of different pathogens. She has reviewed studies in journals, including Frontiers.

L. Fernández-García obtained her degree in biology from Oviedo University (2007 to 2012) and her master's degree in cellular, molecular, and genetic biology from A Coruña University (2014 to 2015). She is currently undertaking Ph.D. research at the Institute for Biomedical Investigation-A Coruña University Hospital Complex (INIBIC-CHUAC) as a member of the microbiology group, under the supervision of M. Tomás. She is currently the recipient of a Xunta de Galicia Ph.D. student contract. Her Ph.D. research focuses on mechanisms of persistence and tolerance in nosocomial pathogens. She has published nine articles in scientific journals and has also published one book chapter and has another in press.

L. Blasco obtained her Ph.D. in biotechnology from the University of Santiago de Compostela (USC) in 2011. She began her research career while undertaking a master's degree program in biotechnology at the USC. She has participated as a member of the biotechnology group of the Microbiology and Parasitology Department of the USC, working on several projects within the field of industrial microbiology. Since 2015, she has been carrying out postdoctoral research as part of the microbiology group led by M. Tomás at the Institute for Biomedical Research-A Coruña University Hospital Complex (INIBIC-CHUAC), Spain. Her main research interest is the search for new treatments for use in the fight against multiresistant bacteria (bacteriophages, endolysins, and quorum sensing inhibition).

A. Ambroa is currently pursuing a Ph.D. degree at the University of A Coruña (UDC) and has been undertaking research at the Institute for Biomedical Research, A Coruña (INIBIC), Spain, since September 2017. He obtained his degree in biotechnology from the Rovira i Virgili University (Tarragona, Spain) in 2016. He has completed internships at the Pontevedra Provincial Hospital (Pontevedra, Spain; June 2015), the Sant Joan de Reus University Hospital (Reus, Spain; January to March 2016), and the Faculty of Medicine and Health Science of the Rovira i Virgili University (Reus and Tarragona, Spain; March to June 2016). In 2017, he was awarded a master's degree in clinical investigation (with a specialty in clinical microbiology) from the University of Barcelona. During the master's degree course, he also completed another internship, at the Vall d'Hebron Hospital (Barcelona, Spain; January to June 2017). He is currently investigating the role of the type VI secretion system (T6SS) in multidrug-resistant bacteria in relation to virulence and resistance mechanisms, under the supervision of M. Tomás.

M. L. Pérez del Molino obtained a Ph.D. in the area of chemical physics from the University of Santiago de Compostela (USC), Galicia, Spain, with a specialty in clinical microbiology and parasitology at the University Clinical Hospital of Santiago de Compostela (1980 to 1983). Since 2016, she has been Head of the Microbiology and Parasitology Department of the University Clinical Hospital of Santiago de Compostela. She previously worked as an Assistant Professor in the Department of Microbiology and Parasitology of the USC (1984 to 1986). Subsequently, she worked as a Clinical Microbiologist in the area of diagnosis of respiratory pathology, focusing on tuberculosis disease (1988 to 2016), and as Head of the reference laboratory of mycobacteria in Galicia (1998 to 2018). She has collaborated with the WHO on studies of resistance to antituberculosis drugs; her work features over 60 publications about microbiological diagnosis, epidemiology, and antimicrobial resistance of mycobacteria and other respiratory pathogens.

G. Bou received his Ph.D. from the Molecular Biology Center (CSIC)-Autonoma University, Madrid, Spain. He also completed a residence in clinical microbiology at the Ramon y Cajal Hospital. Afterwards, he was granted a postdoctoral position with the Fulbright Scholarship program to work in the Laboratory of Medicine at Mayo Clinic, Rochester, MN. Dr. Bou then served as an Investigator at the National Health System in Spain (2001 to 2005) and as a consultant in clinical microbiology (2005 to 2010), and at present, he is the Head of the Microbiology Department of the University Hospital A Coruña (CHUAC). Since 2009, he has been an Associate Professor of medical microbiology at the University of Santiago de Compostela. Dr. Bou's research focuses on understanding the molecular basis for antimicrobial resistance in human pathogens, developing rapid tests for detecting resistant organisms, and designing and developing bacterial vaccines. So far, he has published more than 200 international peer-reviewed papers on these topics, as well as obtaining 7 related patents. Recently, he was conferred the honorary title of ESCMID Fellow for professional excellence and service to society.

R. García-Contreras has been an Associate Professor in the Microbiology and Parasitology Department of the Medicine Faculty of the National Autonomous University of Mexico (UNAM) since 2014. From 2010 to 2014, he was an Associate Professor at the National Institute of Cardiology. In 2005, he obtained his Ph.D. at UNAM. He completed two postdoctoral positions, the first in the Department of Chemical Engineering at Texas A&M University, in the group of Thomas K. Wood, working on the genetic basis of biofilm formation in Escherichia coli, and the second in the Molecular Cell Physiology Department at the VU University of Amsterdam, with Fred Boogerd, working on E. coli central metabolism. Currently, his research is centered on the study of the resistance mechanisms of Pseudomonas aeruginosa against antivirulence compounds and novel antimicrobials, the influence of quorum sensing in virulence and bacterial physiology, and the repurposing of drugs to treat multidrug-resistant bacteria.

T. K. Wood is the Endowed Biotechnology Chair and a Professor in the Department of Chemical Engineering at Pennsylvania State University. He was formerly the Northeast Utilities Endowed Chair in Environmental Engineering at the University of Connecticut (1998 to 2005) and the O'Connor Endowed Chair at Texas A&M University (2005 to 2012). He obtained his Ph.D. in chemical engineering from North Carolina State University in 1991 by studying heterologous protein production and obtained his B.S. from the University of Kentucky in 1985. His current research pursuits include understanding the genetic basis of biofilm formation in order to prevent disease and to utilize biofilms for beneficial biotransformations, including remediation, green chemistry, and energy production. He also uses systems biology approaches to understand cell resistance, specifically discerning the roles of toxin-antitoxin systems and cryptic prophages in antibiotic resistance and persistence. He also has utilized protein engineering to control biofilm formation as well as for bioremediation and green chemistry.

M. Tomás, M.D., Ph.D. (and Clinical Microbiologist), has investigated different mechanisms of resistance to antimicrobials in nosocomial MDR pathogens in various research centers, within the framework of the Rio Hortega and Miguel Servet program (ISCIII-SERGAS). She is currently working as the Molecular Microbiology Coordinator in the Microbiology Department of CHUAC and as a Principal Investigator (PI) in the Institute for Biomedical Research (INIBIC-CHUAC), initiating new research on the relationships between resistance, tolerance, and persistence mechanisms in microbial pathogens to improve new anti-infective treatments, such as phage and antivirulence therapies against persister cells. She has over 70 publications and 3 patents related to new anti-infective treatments and molecular techniques and has completed more than 10 projects as PI (5 research projects and 5 innovation projects) (http://www.mariatomas.me/). Finally, she is Guest Editor for several international journals (Frontiers in Cellular and Infection Microbiology and Marine Drugs) and is a member of ASM, ECCMID, SEIMC, and the REIPI network.
REFERENCES
- 1.Yang QE, Walsh TR. 2017. Toxin-antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol Rev 41:343–353. doi: 10.1093/femsre/fux006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 3.Kim JS, Wood TK. 2017. Tolerant, growing cells from nutrient shifts are not persister cells. mBio 8:e00354-17. doi: 10.1128/mBio.00354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKeegan KS, Borges-Walmsley MI, Walmsley AR. 2002. Microbial and viral drug resistance mechanisms. Trends Microbiol 10:S8–S14. doi: 10.1016/S0966-842X(02)02429-0. [DOI] [PubMed] [Google Scholar]
- 5.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 6.McDermott W. 1958. Microbial persistence. Yale J Biol Med 30:257–291. [PMC free article] [PubMed] [Google Scholar]
- 7.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 8.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilization. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 10.Hobby G. 1942. Observations on the mechanism of action of penicillin. Proc Soc Exp Biol Med 50:281–285. doi: 10.3181/00379727-50-13773. [DOI] [Google Scholar]
- 11.Kwan BW, Valenta JA, Benedik MJ, Wood TK. 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother 57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luidalepp H, Joers A, Kaldalu N, Tenson T. 2011. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J Bacteriol 193:3598–3605. doi: 10.1128/JB.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol 6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlet J. 2012. The gut is the epicentre of antibiotic resistance. Antimicrob Resist Infect Control 1:39. doi: 10.1186/2047-2994-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanshew AS, Jette ME, Rosen SP, Thibeault SL. 2017. Integrating the microbiota of the respiratory tract with the unified airway model. Respir Med 126:68–74. doi: 10.1016/j.rmed.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Garcia L, Blasco L, Lopez M, Bou G, Garcia-Contreras R, Wood T, Tomas M. 2016. Toxin-antitoxin systems in clinical pathogens. Toxins (Basel) 8:E227. doi: 10.3390/toxins8070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prax M, Bertram R. 2014. Metabolic aspects of bacterial persisters. Front Cell Infect Microbiol 4:148. doi: 10.3389/fcimb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma G, Sharma S, Sharma P, Chandola D, Dang S, Gupta S, Gabrani R. 2016. Escherichia coli biofilm: development and therapeutic strategies. J Appl Microbiol 121:309–319. doi: 10.1111/jam.13078. [DOI] [PubMed] [Google Scholar]
- 19.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, Tarone AM, Benedik MJ, Peti W, Page R, Wood TK. 2011. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol 7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Benedik MJ, Wood TK. 2012. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ Microbiol 14:669–679. doi: 10.1111/j.1462-2920.2011.02618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lago M, Monteil V, Douche T, Guglielmini J, Criscuolo A, Maufrais C, Matondo M, Norel F. 2017. Proteome remodelling by the stress sigma factor RpoS/sigmaS in Salmonella: identification of small proteins and evidence for post-transcriptional regulation. Sci Rep 7:2127. doi: 10.1038/s41598-017-02362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiroda M, Pratt ZL, Dopfer D, Wong AC, Kaspar CW. 2014. RpoS impacts the lag phase of Salmonella enterica during osmotic stress. FEMS Microbiol Lett 357:195–200. doi: 10.1111/1574-6968.12523. [DOI] [PubMed] [Google Scholar]
- 25.Wurm P, Tutz S, Mutsam B, Vorkapic D, Heyne B, Grabner C, Kleewein K, Halscheidt A, Schild S, Reidl J. 2017. Stringent factor and proteolysis control of sigma factor RpoS expression in Vibrio cholerae. Int J Med Microbiol 307:154–165. doi: 10.1016/j.ijmm.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Huang CJ, Wang ZC, Huang HY, Huang HD, Peng HL. 2013. YjcC, a c-di-GMP phosphodiesterase protein, regulates the oxidative stress response and virulence of Klebsiella pneumoniae CG43. PLoS One 8:e66740. doi: 10.1371/journal.pone.0066740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol 176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterman SR, Small PL. 1996. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol 21:925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 29.Chan YC, Blaschek HP. 2005. Comparative analysis of Shigella boydii 18 foodborne outbreak isolate and related enteric bacteria: role of rpoS and adiA in acid stress response. J Food Prot 68:521–527. doi: 10.4315/0362-028X-68.3.521. [DOI] [PubMed] [Google Scholar]
- 30.Jennison AV, Verma NK. 2007. The acid-resistance pathways of Shigella flexneri 2457T. Microbiology 153:2593–2602. doi: 10.1099/mic.0.2007/006718-0. [DOI] [PubMed] [Google Scholar]
- 31.de Vries N, van Vliet AHM, Kusters JG. 2001. Gene regulation, p 321–334. In Mobley HLT, Mendz GL, Hazell SL (ed), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [PubMed] [Google Scholar]
- 32.Rince A, Giard JC, Pichereau V, Flahaut S, Auffray Y. 2001. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J Bacteriol 183:1482–1488. doi: 10.1128/JB.183.4.1482-1488.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rince A, Uguen M, Le Breton Y, Giard JC, Flahaut S, Dufour A, Auffray Y. 2002. The Enterococcus faecalis gene encoding the novel general stress protein Gsp62. Microbiology 148:703–711. doi: 10.1099/00221287-148-3-703. [DOI] [PubMed] [Google Scholar]
- 34.Teng F, Nannini EC, Murray BE. 2005. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J Infect Dis 191:472–480. doi: 10.1086/427191. [DOI] [PubMed] [Google Scholar]
- 35.Choudhury T, Singh KV, Sillanpaa J, Nallapareddy SR, Murray BE. 2011. Importance of two Enterococcus faecium loci encoding Gls-like proteins for in vitro bile salts stress response and virulence. J Infect Dis 203:1147–1154. doi: 10.1093/infdis/jiq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rince A, Le Breton Y, Verneuil N, Giard JC, Hartke A, Auffray Y. 2003. Physiological and molecular aspects of bile salt response in Enterococcus faecalis. Int J Food Microbiol 88:207–213. doi: 10.1016/S0168-1605(03)00182-X. [DOI] [PubMed] [Google Scholar]
- 37.Badger JL, Miller VL. 1995. Role of RpoS in survival of Yersinia enterocolitica to a variety of environmental stresses. J Bacteriol 177:5370–5373. doi: 10.1128/jb.177.18.5370-5373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto T, Hanawa T, Ogata S. 1994. Induction of Yersinia enterocolitica stress proteins by phagocytosis with macrophage. Microbiol Immunol 38:295–300. doi: 10.1111/j.1348-0421.1994.tb01779.x. [DOI] [PubMed] [Google Scholar]
- 39.Kelly AF, Park SF, Bovill R, Mackey BM. 2001. Survival of Campylobacter jejuni during stationary phase: evidence for the absence of a phenotypic stationary-phase response. Appl Environ Microbiol 67:2248–2254. doi: 10.1128/AEM.67.5.2248-2254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venkataramanan KP, Jones SW, McCormick KP, Kunjeti SG, Ralston MT, Meyers BC, Papoutsakis ET. 2013. The Clostridium small RNome that responds to stress: the paradigm and importance of toxic metabolite stress in C. acetobutylicum. BMC Genomics 14:849. doi: 10.1186/1471-2164-14-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Schaik W, Abee T. 2005. The role of sigmaB in the stress response of Gram-positive bacteria—targets for food preservation and safety. Curr Opin Biotechnol 16:218–224. doi: 10.1016/j.copbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Saldias MS, Lamothe J, Wu R, Valvano MA. 2008. Burkholderia cenocepacia requires the RpoN sigma factor for biofilm formation and intracellular trafficking within macrophages. Infect Immun 76:1059–1067. doi: 10.1128/IAI.01167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flannagan RS, Valvano MA. 2008. Burkholderia cenocepacia requires RpoE for growth under stress conditions and delay of phagolysosomal fusion in macrophages. Microbiology 154:643–653. doi: 10.1099/mic.0.2007/013714-0. [DOI] [PubMed] [Google Scholar]
- 45.Schaefers MM, Liao TL, Boisvert NM, Roux D, Yoder-Himes D, Priebe GP. 2017. An oxygen-sensing two-component system in the Burkholderia cepacia complex regulates biofilm, intracellular invasion, and pathogenicity. PLoS Pathog 13:e1006116. doi: 10.1371/journal.ppat.1006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renggli S, Keck W, Jenal U, Ritz D. 2013. Role of autofluorescence in flow cytometric analysis of Escherichia coli treated with bactericidal antibiotics. J Bacteriol 195:4067–4073. doi: 10.1128/JB.00393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X, Drlica K. 2014. Reactive oxygen species and the bacterial response to lethal stress. Curr Opin Microbiol 21:1–6. doi: 10.1016/j.mib.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van den Bergh B, Fauvart M, Michiels J. 2017. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev 41:219–251. doi: 10.1093/femsre/fux001. [DOI] [PubMed] [Google Scholar]
- 51.Farizano JV, Torres MA, Pescaretti ML, Delgado MA. 2014. The RcsCDB regulatory system plays a crucial role in the protection of Salmonella enterica serovar Typhimurium against oxidative stress. Microbiology 160:2190–2199. doi: 10.1099/mic.0.081133-0. [DOI] [PubMed] [Google Scholar]
- 52.Gart EV, Suchodolski JS, Welsh TH Jr, Alaniz RC, Randel RD, Lawhon SD. 2016. Salmonella Typhimurium and multidirectional communication in the gut. Front Microbiol 7:1827. doi: 10.3389/fmicb.2016.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goulart CL, Barbosa LC, Bisch PM, von Kruger WM. 2016. Catalases and PhoB/PhoR system independently contribute to oxidative stress resistance in Vibrio cholerae O1. Microbiology 162:1955–1962. doi: 10.1099/mic.0.000364. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Chen S, Zhang J, Rothenbacher FP, Jiang T, Kan B, Zhong Z, Zhu J. 2012. Catalases promote resistance of oxidative stress in Vibrio cholerae. PLoS One 7:e53383. doi: 10.1371/journal.pone.0053383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogura K, Terasaki Y, Miyoshi-Akiyama T, Terasaki M, Moss J, Noda M, Yahiro K. 2017. Vibrio cholerae cholix toxin-induced HepG2 cell death is enhanced by tumor necrosis factor-alpha through ROS and intracellular signal-regulated kinases. Toxicol Sci 156:455–468. doi: 10.1093/toxsci/kfx009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campos MA, Vargas MA, Regueiro V, Llompart CM, Alberti S, Bengoechea JA. 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun 72:7107–7114. doi: 10.1128/IAI.72.12.7107-7114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo Y, Wang S, Zhan L, Jin Y, Duan J, Hao Z, Lv J, Qi X, Chen L, Kreiswirth BN, Wang L, Yu F. 2017. Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front Cell Infect Microbiol 7:24. doi: 10.3389/fcimb.2017.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CT, Wu CC, Chen YS, Lai YC, Chi C, Lin JC, Chen Y, Peng HL. 2011. Fur regulation of the capsular polysaccharide biosynthesis and iron-acquisition systems in Klebsiella pneumoniae CG43. Microbiology 157:419–429. doi: 10.1099/mic.0.044065-0. [DOI] [PubMed] [Google Scholar]
- 59.Sahly H, Podschun R, Oelschlaeger TA, Greiwe M, Parolis H, Hasty D, Kekow J, Ullmann U, Ofek I, Sela S. 2000. Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect Immun 68:6744–6749. doi: 10.1128/IAI.68.12.6744-6749.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu MC, Lin TL, Hsieh PF, Yang HC, Wang JT. 2011. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS One 6:e23500. doi: 10.1371/journal.pone.0023500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fisher RA, Gollan B, Helaine S. 2017. Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 62.Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, Wilson KT. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci U S A 98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang G, Alamuri P, Maier RJ. 2006. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol 61:847–860. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- 64.Flint A, Sun YQ, Butcher J, Stahl M, Huang H, Stintzi A. 2014. Phenotypic screening of a targeted mutant library reveals Campylobacter jejuni defenses against oxidative stress. Infect Immun 82:2266–2275. doi: 10.1128/IAI.01528-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaur T, Singh S, Dhawan V, Ganguly NK. 1998. Shigella dysenteriae type 1 toxin induced lipid peroxidation in enterocytes isolated from rabbit ileum. Mol Cell Biochem 178:169–179. doi: 10.1023/A:1006826829687. [DOI] [PubMed] [Google Scholar]
- 66.Dhar MS, Gupta V, Virdi JS. 2013. Detection, distribution and characterization of novel superoxide dismutases from Yersinia enterocolitica biovar 1A. PLoS One 8:e63919. doi: 10.1371/journal.pone.0063919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paauw A, Leverstein-van Hall MA, van Kessel KP, Verhoef J, Fluit AC. 2009. Yersiniabactin reduces the respiratory oxidative stress response of innate immune cells. PLoS One 4:e8240. doi: 10.1371/journal.pone.0008240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brioukhanov AL, Netrusov AI. 2004. Catalase and superoxide dismutase: distribution, properties, and physiological role in cells of strict anaerobes. Biochemistry (Mosc) 69:949–962. doi: 10.1023/B:BIRY.0000043537.04115.d9. [DOI] [PubMed] [Google Scholar]
- 69.Fradrich C, Beer LA, Gerhard R. 2016. Reactive oxygen species as additional determinants for cytotoxicity of Clostridium difficile toxins A and B. Toxins (Basel) 8:E25. doi: 10.3390/toxins8010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosato RR, Fernandez R, Paz LI, Singh CR, Rosato AE. 2014. TCA cycle-mediated generation of ROS is a key mediator for HeR-MRSA survival under beta-lactam antibiotic exposure. PLoS One 9:e99605. doi: 10.1371/journal.pone.0099605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun 70:6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaupp R, Schlag S, Liebeke M, Lalk M, Gotz F. 2010. Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms. J Bacteriol 192:2385–2394. doi: 10.1128/JB.01472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas VC, Sadykov MR, Chaudhari SS, Jones J, Endres JL, Widhelm TJ, Ahn JS, Jawa RS, Zimmerman MC, Bayles KW. 2014. A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog 10:e1004205. doi: 10.1371/journal.ppat.1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghorbal SK, Maalej L, Chourabi K, Khefacha S, Ouzari HI, Chatti A. 2016. Antioxidant defense mechanisms in Pseudomonas aeruginosa: role of iron-cofactored superoxide dismutase in response to UV-C radiations. Curr Microbiol 73:159–164. doi: 10.1007/s00284-016-1043-7. [DOI] [PubMed] [Google Scholar]
- 75.Ciofu O, Fussing V, Bagge N, Koch C, Hoiby N. 2001. Characterization of paired mucoid/non-mucoid Pseudomonas aeruginosa isolates from Danish cystic fibrosis patients: antibiotic resistance, beta-lactamase activity and RiboPrinting. J Antimicrob Chemother 48:391–396. doi: 10.1093/jac/48.3.391. [DOI] [PubMed] [Google Scholar]
- 76.Owlia P, Nosrati R, Alaghehbandan R, Lari AR. 2014. Antimicrobial susceptibility differences among mucoid and non-mucoid Pseudomonas aeruginosa isolates. GMS Hyg Infect Control 9:Doc13. doi: 10.3205/dgkh000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beaudoin T, Lafayette S, Nguyen D, Rousseau S. 2012. Mucoid Pseudomonas aeruginosa caused by mucA mutations result in activation of TLR2 in addition to TLR5 in airway epithelial cells. Biochem Biophys Res Commun 428:150–154. doi: 10.1016/j.bbrc.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 78.Martha B, Croisier D, Fanton A, Astruc K, Piroth L, Huet F, Chavanet P. 2010. Factors associated with mucoid transition of Pseudomonas aeruginosa in cystic fibrosis patients. Clin Microbiol Infect 16:617–623. doi: 10.1111/j.1469-0691.2009.02786.x. [DOI] [PubMed] [Google Scholar]
- 79.Bhargava N, Sharma P, Capalash N. 2014. Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect Immun 82:3417–3425. doi: 10.1128/IAI.01600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, Fernando DM, Mason KM, Santana E, Loewen PC, Kumar A, Liu Y. 2016. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci 148:31–40. doi: 10.1016/j.lfs.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soares NC, Cabral MP, Gayoso C, Mallo S, Rodriguez-Velo P, Fernandez-Moreira E, Bou G. 2010. Associating growth-phase-related changes in the proteome of Acinetobacter baumannii with increased resistance to oxidative stress. J Proteome Res 9:1951–1964. doi: 10.1021/pr901116r. [DOI] [PubMed] [Google Scholar]
- 82.Van Acker H, Sass A, Dhondt I, Nelis HJ, Coenye T. 2014. Involvement of toxin-antitoxin modules in Burkholderia cenocepacia biofilm persistence. Pathog Dis 71:326–335. doi: 10.1111/2049-632X.12177. [DOI] [PubMed] [Google Scholar]
- 83.Van Acker H, Sass A, Bazzini S, De Roy K, Udine C, Messiaen T, Riccardi G, Boon N, Nelis HJ, Mahenthiralingam E, Coenye T. 2013. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS One 8:e58943. doi: 10.1371/journal.pone.0058943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. 2013. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 85.Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. 2012. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci U S A 109:12147–12152. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baek SH, Li AH, Sassetti CM. 2011. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol 9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang T, El Meouche I, Dunlop MJ. 2017. Bacterial persistence induced by salicylate via reactive oxygen species. Sci Rep 7:43839. doi: 10.1038/srep43839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frawley ER, Crouch ML, Bingham-Ramos LK, Robbins HF, Wang W, Wright GD, Fang FC. 2013. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc Natl Acad Sci U S A 110:12054–12059. doi: 10.1073/pnas.1218274110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ladjouzi R, Bizzini A, Lebreton F, Sauvageot N, Rince A, Benachour A, Hartke A. 2013. Analysis of the tolerance of pathogenic enterococci and Staphylococcus aureus to cell wall active antibiotics. J Antimicrob Chemother 68:2083–2091. doi: 10.1093/jac/dkt157. [DOI] [PubMed] [Google Scholar]
- 90.Pando JM, Pfeltz RF, Cuaron JA, Nagarajan V, Mishra MN, Torres NJ, Elasri MO, Wilkinson BJ, Gustafson JE. 2017. Ethanol-induced stress response of Staphylococcus aureus. Can J Microbiol 63:745–757. doi: 10.1139/cjm-2017-0221. [DOI] [PubMed] [Google Scholar]
- 91.Pereira CS, Thompson JA, Xavier KB. 2013. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev 37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 92.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta 1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rema T, Medihala P, Lawrence JR, Vidovic S, Leppard GG, Reid M, Korber DR. 2016. Proteomic analyses of chlorhexidine tolerance mechanisms in Delftia acidovorans biofilms. mSphere 1:e00017-15. doi: 10.1128/mSphere.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Z, Yang M, Peterfreund GL, Tsou AM, Selamoglu N, Daldal F, Zhong Z, Kan B, Zhu J. 2011. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc Natl Acad Sci U S A 108:810–815. doi: 10.1073/pnas.1014640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Korshunov S, Imlay JA. 2010. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol Microbiol 75:1389–1401. doi: 10.1111/j.1365-2958.2010.07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Baumler AJ. 2016. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C, Kieda C. 2011. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med 15:1239–1253. doi: 10.1111/j.1582-4934.2011.01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Craig M, Sadik AY, Golubeva YA, Tidhar A, Slauch JM. 2013. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol Microbiol 89:887–902. doi: 10.1111/mmi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jones-Carson J, Husain M, Liu L, Orlicky DJ, Vazquez-Torres A. 2016. Cytochrome bd-dependent bioenergetics and antinitrosative defenses in Salmonella pathogenesis. mBio 7:e02052-16. doi: 10.1128/mBio.02052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pavia AT, Shipman LD, Wells JG, Puhr ND, Smith JD, McKinley TW, Tauxe RV. 1990. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J Infect Dis 161:255–260. doi: 10.1093/infdis/161.2.255. [DOI] [PubMed] [Google Scholar]
- 101.Khademian M, Imlay JA. 2017. Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc Natl Acad Sci U S A 114:E6922–E6931. doi: 10.1073/pnas.1701587114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McGregor A, Klartag M, David L, Adir N. 2008. Allophycocyanin trimer stability and functionality are primarily due to polar enhanced hydrophobicity of the phycocyanobilin binding pocket. J Mol Biol 384:406–421. doi: 10.1016/j.jmb.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 104.Hemp J, Christian C, Barquera B, Gennis RB, Martinez TJ. 2005. Helix switching of a key active-site residue in the cytochrome cbb3 oxidases. Biochemistry 44:10766–10775. doi: 10.1021/bi050464f. [DOI] [PubMed] [Google Scholar]
- 105.Chang HY, Ahn Y, Pace LA, Lin MT, Lin YH, Gennis RB. 2010. The diheme cytochrome c(4) from Vibrio cholerae is a natural electron donor to the respiratory cbb(3) oxygen reductase. Biochemistry 49:7494–7503. doi: 10.1021/bi1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meyer M, Dimroth P, Bott M. 2001. Catabolite repression of the citrate fermentation genes in Klebsiella pneumoniae: evidence for involvement of the cyclic AMP receptor protein. J Bacteriol 183:5248–5256. doi: 10.1128/JB.183.18.5248-5256.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bott M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch Microbiol 167:78–88. doi: 10.1007/s002030050419. [DOI] [PubMed] [Google Scholar]
- 108.Chen YT, Liao TL, Wu KM, Lauderdale TL, Yan JJ, Huang IW, Lu MC, Lai YC, Liu YM, Shu HY, Wang JT, Su IJ, Tsai SF. 2009. Genomic diversity of citrate fermentation in Klebsiella pneumoniae. BMC Microbiol 9:168. doi: 10.1186/1471-2180-9-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maier RJ, Fu C, Gilbert J, Moshiri F, Olson J, Plaut AG. 1996. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol Lett 141:71–76. doi: 10.1111/j.1574-6968.1996.tb08365.x. [DOI] [PubMed] [Google Scholar]
- 110.Smith MA, Finel M, Korolik V, Mendz GL. 2000. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch Microbiol 174:1–10. doi: 10.1007/s002030000174. [DOI] [PubMed] [Google Scholar]
- 111.Han SW, Evans DG, el-Zaatari FA, Go MF, Graham DY. 1996. The interaction of pH, bile, and Helicobacter pylori may explain duodenal ulcer. Am J Gastroenterol 91:1135–1137. [PubMed] [Google Scholar]
- 112.Jackson RJ, Elvers KT, Lee LJ, Gidley MD, Wainwright LM, Lightfoot J, Park SF, Poole RK. 2007. Oxygen reactivity of both respiratory oxidases in Campylobacter jejuni: the cydAB genes encode a cyanide-resistant, low-affinity oxidase that is not of the cytochrome bd type. J Bacteriol 189:1604–1615. doi: 10.1128/JB.00897-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Winstedt L, Frankenberg L, Hederstedt L, von Wachenfeldt C. 2000. Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J Bacteriol 182:3863–3866. doi: 10.1128/JB.182.13.3863-3866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Derre I, Rapoport G, Msadek T. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol 31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 115.Way SS, Sallustio S, Magliozzo RS, Goldberg MB. 1999. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J Bacteriol 181:1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Way SS, Borczuk AC, Goldberg MB. 1999. Adaptive immune response to Shigella flexneri 2a cydC in immunocompetent mice and mice lacking immunoglobulin A. Infect Immun 67:2001–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, Gotz F. 2006. Microevolution of cytochrome bd oxidase in staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol 188:8079–8086. doi: 10.1128/JB.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jo J, Cortez KL, Cornell WC, Price-Whelan A, Dietrich LE. 2017. An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. Elife 6:e30205. doi: 10.7554/eLife.30205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kirisits MJ, Prost L, Starkey M, Parsek MR. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meissner A, Wild V, Simm R, Rohde M, Erck C, Bredenbruch F, Morr M, Romling U, Haussler S. 2007. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ Microbiol 9:2475–2485. doi: 10.1111/j.1462-2920.2007.01366.x. [DOI] [PubMed] [Google Scholar]
- 122.Ueda A, Wood TK. 2009. Connecting quorum sensing, c-di-GMP, Pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chambers JR, Sauer K. 2013. Small RNAs and their role in biofilm formation. Trends Microbiol 21:39–49. doi: 10.1016/j.tim.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mulcahy H, O'Callaghan J, O'Grady EP, Macia MD, Borrell N, Gomez C, Casey PG, Hill C, Adams C, Gahan CG, Oliver A, O'Gara F. 2008. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect Immun 76:632–638. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 127.Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. 2010. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1→3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904. doi: 10.1093/glycob/cwq047. [DOI] [PubMed] [Google Scholar]
- 128.Beardmore-Gray M, Anthony C. 1986. The oxidation of glucose by Acinetobacter calcoaceticus: interaction of the quinoprotein glucose dehydrogenase with the electron transport chain. J Gen Microbiol 132:1257–1268. [DOI] [PubMed] [Google Scholar]
- 129.Shi W, Zhang Y. 2010. PhoY2 but not PhoY1 is the PhoU homologue involved in persisters in Mycobacterium tuberculosis. J Antimicrob Chemother 65:1237–1242. doi: 10.1093/jac/dkq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Namugenyi SB, Aagesen AM, Elliott SR, Tischler AD. 2017. Mycobacterium tuberculosis PhoY proteins promote persister formation by mediating Pst/SenX3-RegX3 phosphate sensing. mBio 8:e00494-17. doi: 10.1128/mBio.00494-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tischler AD, Leistikow RL, Kirksey MA, Voskuil MI, McKinney JD. 2013. Mycobacterium tuberculosis requires phosphate-responsive gene regulation to resist host immunity. Infect Immun 81:317–328. doi: 10.1128/IAI.01136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McCusker KP, Klinman JP. 2009. Modular behavior of tauD provides insight into the origin of specificity in alpha-ketoglutarate-dependent nonheme iron oxygenases. Proc Natl Acad Sci U S A 106:19791–19795. doi: 10.1073/pnas.0910660106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Javaux C, Joris B, De Witte P. 2007. Functional characteristics of TauA binding protein from TauABC Escherichia coli system. Protein J 26:231–238. doi: 10.1007/s10930-006-9064-x. [DOI] [PubMed] [Google Scholar]
- 134.Price JC, Barr EW, Hoffart LM, Krebs C, Bollinger JM Jr. 2005. Kinetic dissection of the catalytic mechanism of taurine:alpha-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry 44:8138–8147. doi: 10.1021/bi050227c. [DOI] [PubMed] [Google Scholar]
- 135.Bollinger JM Jr, Krebs C. 2006. Stalking intermediates in oxygen activation by iron enzymes: motivation and method. J Inorg Biochem 100:586–605. doi: 10.1016/j.jinorgbio.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 136.Hay AJ, Zhu J. 2015. Host intestinal signal-promoted biofilm dispersal induces Vibrio cholerae colonization. Infect Immun 83:317–323. doi: 10.1128/IAI.02617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sanchez LM, Cheng AT, Warner CJ, Townsley L, Peach KC, Navarro G, Shikuma NJ, Bray WM, Riener RM, Yildiz FH, Linington RG. 2016. Biofilm formation and detachment in Gram-negative pathogens is modulated by select bile acids. PLoS One 11:e0149603. doi: 10.1371/journal.pone.0149603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xue Y, Tu F, Shi M, Wu CQ, Ren G, Wang X, Fang W, Song H, Yang M. 2016. Redox pathway sensing bile salts activates virulence gene expression in Vibrio cholerae. Mol Microbiol 102:909–924. doi: 10.1111/mmi.13497. [DOI] [PubMed] [Google Scholar]
- 139.Hay AJ, Yang M, Xia X, Liu Z, Hammons J, Fenical W, Zhu J. 2017. Calcium enhances bile salt-dependent virulence activation in Vibrio cholerae. Infect Immun 85:e00707-16. doi: 10.1128/IAI.00707-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Loogna P, Franzen L, Sipponen P, Domellof L. 2001. Helicobacter pylori, N-methyl-N′-nitro-N′-nitrosoguanidine, and bile modulate gastric cell kinetics in experimental cancer. Virchows Arch 439:653–660. doi: 10.1007/s004280100411. [DOI] [PubMed] [Google Scholar]
- 141.Lapierre H, Petitclerc D, Dubreuil P, Pelletier G, Gaudreau P, Morisset J, Couture Y, Brazeau P. 1987. Synergism between a human growth hormone-releasing factor and thyrotropin-releasing factor on growth hormone release in relation with the stage of lactation of Holstein dairy cows. Reprod Nutr Dev 27:605–607. doi: 10.1051/rnd:19870421. [DOI] [PubMed] [Google Scholar]
- 142.Soutourina O, Poupel O, Coppee JY, Danchin A, Msadek T, Martin-Verstraete I. 2009. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol Microbiol 73:194–211. doi: 10.1111/j.1365-2958.2009.06760.x. [DOI] [PubMed] [Google Scholar]
- 143.Krejcik Z, Schleheck D, Hollemeyer K, Cook AM. 2012. A five-gene cluster involved in utilization of taurine-nitrogen and excretion of sulfoacetaldehyde by Acinetobacter radioresistens SH164. Arch Microbiol 194:857–863. doi: 10.1007/s00203-012-0806-1. [DOI] [PubMed] [Google Scholar]
- 144.Eom HJ, Park W. 2017. Inhibitory effect of taurine on biofilm formation during alkane degradation in Acinetobacter oleivorans DR1. Microb Ecol 74:821–831. doi: 10.1007/s00248-017-1010-2. [DOI] [PubMed] [Google Scholar]
- 145.Lochowska A, Iwanicka-Nowicka R, Zielak A, Modelewska A, Thomas MS, Hryniewicz MM. 2011. Regulation of sulfur assimilation pathways in Burkholderia cenocepacia through control of genes by the SsuR transcription factor. J Bacteriol 193:1843–1853. doi: 10.1128/JB.00483-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blanco P, Hernando-Amado S, Reales-Calderon JA, Corona F, Lira F, Alcalde-Rico M, Bernardini A, Sanchez MB, Martinez JL. 2016. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4:E14. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pu Y, Zhao Z, Li Y, Zou J, Ma Q, Zhao Y, Ke Y, Zhu Y, Chen H, Baker MA, Ge H, Sun Y, Xie XS, Bai F. 2016. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell 62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gerdes K, Semsey S. 2016. Microbiology: pumping persisters. Nature 534:41–42. doi: 10.1038/nature18442. [DOI] [PubMed] [Google Scholar]
- 149.Du Toit A. 2017. Bacterial physiology: efflux pumps, fitness and virulence. Nat Rev Microbiol 15:512–513. doi: 10.1038/nrmicro.2017.97. [DOI] [PubMed] [Google Scholar]
- 150.Beceiro A, Tomas M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev 26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lertpiriyapong K, Gamazon ER, Feng Y, Park DS, Pang J, Botka G, Graffam ME, Ge Z, Fox JG. 2012. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One 7:e42842. doi: 10.1371/journal.pone.0042842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu Y, Vulic M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kim J-S, Wood TK. 2016. Persistent persister misperceptions. Front Microbiol 7:2134. doi: 10.3389/fmicb.2016.02134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bogomolnaya LM, Andrews KD, Talamantes M, Maple A, Ragoza Y, Vazquez-Torres A, Andrews-Polymenis H. 2013. The ABC-type efflux pump MacAB protects Salmonella enterica serovar Typhimurium from oxidative stress. mBio 4:e00630-13. doi: 10.1128/mBio.00630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Andersen JL, He GX, Kakarla P, KC R, Kumar S, Lakra WS, Mukherjee MM, Ranaweera I, Shrestha U, Tran T, Varela MF. 2015. Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int J Environ Res Public Health 12:1487–1547. doi: 10.3390/ijerph120201487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Varela MF, Kumar S, He G. 2013. Potential for inhibition of bacterial efflux pumps in multidrug-resistant Vibrio cholerae. Indian J Med Res 138:285–287. [PMC free article] [PubMed] [Google Scholar]
- 157.Padilla E, Llobet E, Domenech-Sanchez A, Martinez-Martinez L, Bengoechea JA, Alberti S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bialek-Davenet S, Lavigne JP, Guyot K, Mayer N, Tournebize R, Brisse S, Leflon-Guibout V, Nicolas-Chanoine MH. 2015. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 70:81–88. doi: 10.1093/jac/dku340. [DOI] [PubMed] [Google Scholar]
- 159.Attaran B, Falsafi T, Ghorbanmehr N. 2017. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J Gastroenterol 23:1163–1170. doi: 10.3748/wjg.v23.i7.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bezek K, Kurincic M, Knauder E, Klancnik A, Raspor P, Bucar F, Smole Mozina S. 2016. Attenuation of adhesion, biofilm formation and quorum sensing of Campylobacter jejuni by Euodia ruticarpa. Phytother Res 30:1527–1532. doi: 10.1002/ptr.5658. [DOI] [PubMed] [Google Scholar]
- 161.Molale LG, Bezuidenhout CC. 2016. Antibiotic resistance, efflux pump genes and virulence determinants in Enterococcus spp. from surface water systems. Environ Sci Pollut Res Int 23:21501–21510. doi: 10.1007/s11356-016-7369-7. [DOI] [PubMed] [Google Scholar]
- 162.Nickerson KP, Chanin RB, Sistrunk JR, Rasko DA, Fink PJ, Barry EM, Nataro JP, Faherty CS. 2017. Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect Immun 85:e01067-16. doi: 10.1128/IAI.01067-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Leuzzi A, Di Martino ML, Campilongo R, Falconi M, Barbagallo M, Marcocci L, Pietrangeli P, Casalino M, Grossi M, Micheli G, Colonna B, Prosseda G. 2015. Multifactor regulation of the MdtJI polyamine transporter in Shigella. PLoS One 10:e0136744. doi: 10.1371/journal.pone.0136744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bengoechea JA, Skurnik M. 2000. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol Microbiol 37:67–80. doi: 10.1046/j.1365-2958.2000.01956.x. [DOI] [PubMed] [Google Scholar]
- 165.Dridi L, Tankovic J, Petit JC. 2004. CdeA of Clostridium difficile, a new multidrug efflux transporter of the MATE family. Microb Drug Resist 10:191–196. [DOI] [PubMed] [Google Scholar]
- 166.Smith K, Gemmell CG, Hunter IS. 2008. The association between biocide tolerance and the presence or absence of qac genes among hospital-acquired and community-acquired MRSA isolates. J Antimicrob Chemother 61:78–84. doi: 10.1093/jac/dkm395. [DOI] [PubMed] [Google Scholar]
- 167.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 168.Minagawa S, Inami H, Kato T, Sawada S, Yasuki T, Miyairi S, Horikawa M, Okuda J, Gotoh N. 2012. RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol 12:70. doi: 10.1186/1471-2180-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.He X, Lu F, Yuan F, Jiang D, Zhao P, Zhu J, Cheng H, Cao J, Lu G. 2015. Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob Agents Chemother 59:4817–4825. doi: 10.1128/AAC.00877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yoon EJ, Chabane YN, Goussard S, Snesrud E, Courvalin P, De E, Grillot-Courvalin C. 2015. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 6:e00309-15. doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Richmond GE, Evans LP, Anderson MJ, Wand ME, Bonney LC, Ivens A, Chua KL, Webber MA, Sutton JM, Peterson ML, Piddock LJ. 2016. The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio 7:e00430-16. doi: 10.1128/mBio.00430-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lopez M, Blasco L, Gato E, Perez A, Fernandez-Garcia L, Martinez-Martinez L, Fernandez-Cuenca F, Rodriguez-Bano J, Pascual A, Bou G, Tomas M. 2017. Response to bile salts in clinical strains of Acinetobacter baumannii lacking the AdeABC efflux pump: virulence associated with quorum sensing. Front Cell Infect Microbiol 7:143. doi: 10.3389/fcimb.2017.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Buroni S, Matthijs N, Spadaro F, Van Acker H, Scoffone VC, Pasca MR, Riccardi G, Coenye T. 2014. Differential roles of RND efflux pumps in antimicrobial drug resistance of sessile and planktonic Burkholderia cenocepacia cells. Antimicrob Agents Chemother 58:7424–7429. doi: 10.1128/AAC.03800-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 38:1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 176.Irazoki O, Mayola A, Campoy S, Barbe J. 2016. SOS system induction inhibits the assembly of chemoreceptor signaling clusters in Salmonella enterica. PLoS One 11:e0146685. doi: 10.1371/journal.pone.0146685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 178.Fuchs RP. 2016. Tolerance of lesions in E. coli: chronological competition between translesion synthesis and damage avoidance. DNA Repair (Amst) 44:51–58. doi: 10.1016/j.dnarep.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 179.Sutherland JH, Cheng B, Liu IF, Tse-Dinh YC. 2008. SOS induction by stabilized topoisomerase IA cleavage complex occurs via the RecBCD pathway. J Bacteriol 190:3399–3403. doi: 10.1128/JB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K. 2017. ATP-dependent persister formation in Escherichia coli. mBio 8:e02267-16. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dorr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gomez-Gomez JM, Manfredi C, Alonso JC, Blazquez J. 2007. A novel role for RecA under non-stress: promotion of swarming motility in Escherichia coli K-12. BMC Biol 5:14. doi: 10.1186/1741-7007-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Medina-Ruiz L, Campoy S, Latasa C, Cardenas P, Alonso JC, Barbe J. 2010. Overexpression of the recA gene decreases oral but not intraperitoneal fitness of Salmonella enterica. Infect Immun 78:3217–3225. doi: 10.1128/IAI.01321-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mayola A, Irazoki O, Martinez IA, Petrov D, Menolascina F, Stocker R, Reyes-Darias JA, Krell T, Barbe J, Campoy S. 2014. RecA protein plays a role in the chemotactic response and chemoreceptor clustering of Salmonella enterica. PLoS One 9:e105578. doi: 10.1371/journal.pone.0105578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tripathy S, Sen R, Padhi SK, Mohanty S, Maiti NK. 2014. Upregulation of transcripts for metabolism in diverse environments is a shared response associated with survival and adaptation of Klebsiella pneumoniae in response to temperature extremes. Funct Integr Genomics 14:591–601. doi: 10.1007/s10142-014-0382-3. [DOI] [PubMed] [Google Scholar]
- 187.Amundsen SK, Fero J, Salama NR, Smith GR. 2009. Dual nuclease and helicase activities of Helicobacter pylori AddAB are required for DNA repair, recombination, and mouse infectivity. J Biol Chem 284:16759–16766. doi: 10.1074/jbc.M109.005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Thompson SA, Blaser MJ. 1995. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect Immun 63:2185–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gaasbeek EJ, van der Wal FJ, van Putten JP, de Boer P, van der Graaf-van Bloois L, de Boer AG, Vermaning BJ, Wagenaar JA. 2009. Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J Bacteriol 191:3785–3793. doi: 10.1128/JB.01817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Munoz-Najar U, Vijayakumar MN. 1999. An operon that confers UV resistance by evoking the SOS mutagenic response in streptococcal conjugative transposon Tn5252. J Bacteriol 181:2782–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Palchaudhuri S, Tominna B, Leon MA. 1998. H-NS regulates DNA repair in Shigella. J Bacteriol 180:5260–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bladel I, Wagner K, Beck A, Schilling J, Alexander Schmidt M, Heusipp G. 2013. The H-NS protein silences the pyp regulatory network of Yersinia enterocolitica and is involved in controlling biofilm formation. FEMS Microbiol Lett 340:41–48. doi: 10.1111/1574-6968.12073. [DOI] [PubMed] [Google Scholar]
- 193.Walter BM, Cartman ST, Minton NP, Butala M, Rupnik M. 2015. The SOS response master regulator LexA is associated with sporulation, motility and biofilm formation in Clostridium difficile. PLoS One 10:e0144763. doi: 10.1371/journal.pone.0144763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Walter BM, Rupnik M, Hodnik V, Anderluh G, Dupuy B, Paulič N, Žgur-Bertok D, Butala M. 2014. The LexA regulated genes of the Clostridium difficile. BMC Microbiol 14:88. doi: 10.1186/1471-2180-14-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vestergaard M, Paulander W, Ingmer H. 2015. Activation of the SOS response increases the frequency of small colony variants. BMC Res Notes 8:749. doi: 10.1186/s13104-015-1735-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Painter KL, Strange E, Parkhill J, Bamford KB, Armstrong-James D, Edwards AM. 2015. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect Immun 83:1830–1844. doi: 10.1128/IAI.03016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 198.Cirz RT, O'Neill BM, Hammond JA, Head SR, Romesberg FE. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol 188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Brazas MD, Hancock RE. 2005. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3222–3227. doi: 10.1128/AAC.49.8.3222-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chang W, Small DA, Toghrol F, Bentley WE. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. doi: 10.1186/1471-2164-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sanders LH, Rockel A, Lu H, Wozniak DJ, Sutton MD. 2006. Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. J Bacteriol 188:8573–8585. doi: 10.1128/JB.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mo CY, Birdwell LD, Kohli RM. 2014. Specificity determinants for autoproteolysis of LexA, a key regulator of bacterial SOS mutagenesis. Biochemistry 53:3158–3168. doi: 10.1021/bi500026e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Penterman J, Singh PK, Walker GC. 2014. Biological cost of pyocin production during the SOS response in Pseudomonas aeruginosa. J Bacteriol 196:3351–3359. doi: 10.1128/JB.01889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Diaz-Magana A, Alva-Murillo N, Chavez-Moctezuma MP, Lopez-Meza JE, Ramirez-Diaz MI, Cervantes C. 2015. A plasmid-encoded UmuD homologue regulates expression of Pseudomonas aeruginosa SOS genes. Microbiology 161:1516–1523. doi: 10.1099/mic.0.000103. [DOI] [PubMed] [Google Scholar]
- 206.Jara LM, Cortes P, Bou G, Barbe J, Aranda J. 2015. Differential roles of antimicrobials in the acquisition of drug resistance through activation of the SOS response in Acinetobacter baumannii. Antimicrob Agents Chemother 59:4318–4320. doi: 10.1128/AAC.04918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Aranda J, Bardina C, Beceiro A, Rumbo S, Cabral MP, Barbe J, Bou G. 2011. Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J Bacteriol 193:3740–3747. doi: 10.1128/JB.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hare JM, Ferrell JC, Witkowski TA, Grice AN. 2014. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi. PLoS One 9:e93861. doi: 10.1371/journal.pone.0093861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Witkowski TA, Grice AN, Stinnett DB, Wells WK, Peterson MA, Hare JM. 2016. UmuDAb: an error-prone polymerase accessory homolog whose N-terminal domain is required for repression of DNA damage inducible gene expression in Acinetobacter baylyi. PLoS One 11:e0152013. doi: 10.1371/journal.pone.0152013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hare JM, Bradley JA, Lin CL, Elam TJ. 2012. Diverse responses to UV light exposure in Acinetobacter include the capacity for DNA damage-induced mutagenesis in the opportunistic pathogens Acinetobacter baumannii and Acinetobacter ursingii. Microbiology 158:601–611. doi: 10.1099/mic.0.054668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Norton MD, Spilkia AJ, Godoy VG. 2013. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J Bacteriol 195:1335–1345. doi: 10.1128/JB.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Warner DF, Ndwandwe DE, Abrahams GL, Kana BD, Machowski EE, Venclovas C, Mizrahi V. 2010. Essential roles for imuA′- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 107:13093–13098. doi: 10.1073/pnas.1002614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Keren I, Minami S, Rubin E, Lewis K. 2011. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2:e00100-11. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Smollett KL, Smith KM, Kahramanoglou C, Arnvig KB, Buxton RS, Davis EO. 2012. Global analysis of the regulon of the transcriptional repressor LexA, a key component of SOS response in Mycobacterium tuberculosis. J Biol Chem 287:22004–22014. doi: 10.1074/jbc.M112.357715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang Y, Huang Y, Xue C, He Y, He ZG. 2011. ClpR protein-like regulator specifically recognizes RecA protein-independent promoter motif and broadly regulates expression of DNA damage-inducible genes in mycobacteria. J Biol Chem 286:31159–31167. doi: 10.1074/jbc.M111.241802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ali L, Goraya MU, Arafat Y, Ajmal M, Chen JL, Yu D. 2017. Molecular mechanism of quorum-sensing in Enterococcus faecalis: its role in virulence and therapeutic approaches. Int J Mol Sci 18:E960. doi: 10.3390/ijms18050960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Golz G, Sharbati S, Backert S, Alter T. 2012. Quorum sensing dependent phenotypes and their molecular mechanisms in Campylobacterales. Eur J Microbiol Immunol 2:50–60. doi: 10.1556/EuJMI.2.2012.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li M, Long Y, Liu Y, Chen R, Shi J, Zhang L, Jin Y, Yang L, Bai F, Jin S, Cheng Z, Wu W. 2016. HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the type III secretion system in ciprofloxacin induced persister cells. Front Cell Infect Microbiol 6:125. doi: 10.3389/fcimb.2016.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Troselj V, Treuner-Lange A, Søgaard-Andersen L, Wall D. 2018. Physiological heterogeneity triggers sibling conflict mediated by the type VI secretion system in an aggregative multicellular bacterium. mBio 9:e01645-17. doi: 10.1128/mBio.01645-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Castillo-Juarez I, Maeda T, Mandujano-Tinoco EA, Tomas M, Perez-Eretza B, Garcia-Contreras SJ, Wood TK, Garcia-Contreras R. 2015. Role of quorum sensing in bacterial infections. World J Clin Cases 3:575–598. doi: 10.12998/wjcc.v3.i7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lee JH, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 223.Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat Chem Biol 8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hu Y, Kwan BW, Osbourne DO, Benedik MJ, Wood TK. 2015. Toxin YafQ increases persister cell formation by reducing indole signalling. Environ Microbiol 17:1275–1285. doi: 10.1111/1462-2920.12567. [DOI] [PubMed] [Google Scholar]
- 225.Kwan BW, Osbourne DO, Hu Y, Benedik MJ, Wood TK. 2015. Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol Bioeng 112:588–600. doi: 10.1002/bit.25456. [DOI] [PubMed] [Google Scholar]
- 226.Lee J-H, Kim Y-G, Gwon G, Wood TK, Lee J. 2016. Halogenated indoles eradicate bacterial persister cells and biofilms. AMB Express 6:123. doi: 10.1186/s13568-016-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppee JY, Ghigo JM, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sircili MP, Walters M, Trabulsi LR, Sperandio V. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect Immun 72:2329–2337. doi: 10.1128/IAI.72.4.2329-2337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Soares JA, Ahmer BM. 2011. Detection of acyl-homoserine lactones by Escherichia and Salmonella. Curr Opin Microbiol 14:188–193. doi: 10.1016/j.mib.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kendall MM, Sperandio V. 2007. Quorum sensing by enteric pathogens. Curr Opin Gastroenterol 23:10–15. doi: 10.1097/MOG.0b013e3280118289. [DOI] [PubMed] [Google Scholar]
- 232.Moreira CG, Weinshenker D, Sperandio V. 2010. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect Immun 78:914–926. doi: 10.1128/IAI.01038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 113:E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Leung KY, Siame BA, Snowball H, Mok YK. 2011. Type VI secretion regulation: crosstalk and intracellular communication. Curr Opin Microbiol 14:9–15. doi: 10.1016/j.mib.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 235.Silva AJ, Parker WB, Allan PW, Ayala JC, Benitez JA. 2015. Role of methylthioadenosine/S-adenosylhomocysteine nucleosidase in Vibrio cholerae cellular communication and biofilm development. Biochem Biophys Res Commun 461:65–69. doi: 10.1016/j.bbrc.2015.03.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Jubair M, Morris JG Jr, Ali A. 2012. Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS One 7:e45187. doi: 10.1371/journal.pone.0045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Teschler JK, Cheng AT, Yildiz FH. 2017. The two-component signal transduction system VxrAB positively regulates Vibrio cholerae biofilm formation. J Bacteriol 199:e00139-17. doi: 10.1128/JB.00139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kovacikova G, Lin W, Skorupski K. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol Microbiol 57:420–433. doi: 10.1111/j.1365-2958.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- 239.Swartzman E, Silverman M, Meighen EA. 1992. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J Bacteriol 174:7490–7493. doi: 10.1128/jb.174.22.7490-7493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Waters CM, Bassler BL. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ruwandeepika HA, Karunasagar I, Bossier P, Defoirdt T. 2015. Expression and quorum sensing regulation of type III secretion system genes of Vibrio harveyi during infection of gnotobiotic brine shrimp. PLoS One 10:e0143935. doi: 10.1371/journal.pone.0143935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. 2010. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A 107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Joshi A, Kostiuk B, Rogers A, Teschler J, Pukatzki S, Yildiz FH. 2017. Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol 25:267–279. doi: 10.1016/j.tim.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bachmann V, Kostiuk B, Unterweger D, Diaz-Satizabal L, Ogg S, Pukatzki S. 2015. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl Trop Dis 9:e0004031. doi: 10.1371/journal.pntd.0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Fu Y, Waldor MK, Mekalanos JJ. 2013. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Zhu H, Liu HJ, Ning SJ, Gao YL. 2011. A luxS-dependent transcript profile of cell-to-cell communication in Klebsiella pneumoniae. Mol Biosyst 7:3164–3168. doi: 10.1039/c1mb05314k. [DOI] [PubMed] [Google Scholar]
- 247.De Araujo C, Balestrino D, Roth L, Charbonnel N, Forestier C. 2010. Quorum sensing affects biofilm formation through lipopolysaccharide synthesis in Klebsiella pneumoniae. Res Microbiol 161:595–603. doi: 10.1016/j.resmic.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 248.Anderl JN, Zahller J, Roe F, Stewart PS. 2003. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother 47:1251–1256. doi: 10.1128/AAC.47.4.1251-1256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Sarris PF, Zoumadakis C, Panopoulos NJ, Scoulica EV. 2011. Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect Genet Evol 11:157–166. doi: 10.1016/j.meegid.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 250.Doherty NC, Shen F, Halliday NM, Barrett DA, Hardie KR, Winzer K, Atherton JC. 2010. In Helicobacter pylori, LuxS is a key enzyme in cysteine provision through a reverse transsulfuration pathway. J Bacteriol 192:1184–1192. doi: 10.1128/JB.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Shen F, Hobley L, Doherty N, Loh JT, Cover TL, Sockett RE, Hardie KR, Atherton JC. 2010. In Helicobacter pylori auto-inducer-2, but not LuxS/MccAB catalysed reverse transsulphuration, regulates motility through modulation of flagellar gene transcription. BMC Microbiol 10:210. doi: 10.1186/1471-2180-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. 2015. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio 6:e00379. doi: 10.1128/mBio.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Cammarota G, Sanguinetti M, Gallo A, Posteraro B. 2012. Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection. Aliment Pharmacol Ther 36:222–230. doi: 10.1111/j.1365-2036.2012.05165.x. [DOI] [PubMed] [Google Scholar]
- 254.Stark RM, Gerwig GJ, Pitman RS, Potts LF, Williams NA, Greenman J, Weinzweig IP, Hirst TR, Millar MR. 1999. Biofilm formation by Helicobacter pylori. Lett Appl Microbiol 28:121–126. doi: 10.1046/j.1365-2672.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 255.Cole SP, Harwood J, Lee R, She R, Guiney DG. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol 186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cellini L, Grande R, Di Campli E, Di Bartolomeo S, Di Giulio M, Traini T, Trubiani O. 2008. Characterization of an Helicobacter pylori environmental strain. J Appl Microbiol 105:761–769. doi: 10.1111/j.1365-2672.2008.03808.x. [DOI] [PubMed] [Google Scholar]
- 257.Yonezawa H, Osaki T, Kamiya S. 2015. Biofilm formation by Helicobacter pylori and its involvement for antibiotic resistance. Biomed Res Int 2015:914791. doi: 10.1155/2015/914791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Carron MA, Tran VR, Sugawa C, Coticchia JM. 2006. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg 10:712–717. doi: 10.1016/j.gassur.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 259.Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. 2006. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg 10:883–889. doi: 10.1016/j.gassur.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 260.Cellini L, Grande R, Di Campli E, Traini T, Di Giulio M, Lannutti SN, Lattanzio R. 2008. Dynamic colonization of Helicobacter pylori in human gastric mucosa. Scand J Gastroenterol 43:178–185. doi: 10.1080/00365520701675965. [DOI] [PubMed] [Google Scholar]
- 261.Bleumink-Pluym NM, van Alphen LB, Bouwman LI, Wosten MM, van Putten JP. 2013. Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog 9:e1003393. doi: 10.1371/journal.ppat.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Dale JL, Cagnazzo J, Phan CQ, Barnes AM, Dunny GM. 2015. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob Agents Chemother 59:4094–4105. doi: 10.1128/AAC.00344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Xu P, Yang J, Lu LL, Feng EL, Wang HL, Lu Y, Zhu L. 2015. The effect of quorum sensing system for growth competitiveness on Shigella flexneri. Yi Chuan 37:487–493. doi: 10.16288/j.yczz.15-002. [DOI] [PubMed] [Google Scholar]
- 264.Day WA Jr, Maurelli AT. 2001. Shigella flexneri LuxS quorum-sensing system modulates virB expression but is not essential for virulence. Infect Immun 69:15–23. doi: 10.1128/IAI.69.1.15-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Dorman CJ. 29 December 2004, posting date Virulence gene regulation in Shigella. EcoSal Plus 2004. doi: 10.1128/ecosalplus.8.9.3. [DOI] [PubMed] [Google Scholar]
- 266.Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ. 2017. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21:769.e3–776.e3. doi: 10.1016/j.chom.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 267.Atkinson S, Sockett RE, Camara M, Williams P. 2006. Quorum sensing and the lifestyle of Yersinia. Curr Issues Mol Biol 8:1–10. [PubMed] [Google Scholar]
- 268.Atkinson S, Chang CY, Sockett RE, Camara M, Williams P. 2006. Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility. J Bacteriol 188:1451–1461. doi: 10.1128/JB.188.4.1451-1461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ioannidis A, Kyratsa A, Ioannidou V, Bersimis S, Chatzipanagiotou S. 2014. Detection of biofilm production of Yersinia enterocolitica strains isolated from infected children and comparative antimicrobial susceptibility of biofilm versus planktonic forms. Mol Diagn Ther 18:309–314. doi: 10.1007/s40291-013-0080-1. [DOI] [PubMed] [Google Scholar]
- 270.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. 2015. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 6:e02569. doi: 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Janoir C. 2016. Virulence factors of Clostridium difficile and their role during infection. Anaerobe 37:13–24. doi: 10.1016/j.anaerobe.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 272.Maldarelli GA, Piepenbrink KH, Scott AJ, Freiberg JA, Song Y, Achermann Y, Ernst RK, Shirtliff ME, Sundberg EJ, Donnenberg MS, von Rosenvinge EC. 2016. Type IV pili promote early biofilm formation by Clostridium difficile. Pathog Dis 74:ftw061. doi: 10.1093/femspd/ftw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Xu T, Wang XY, Cui P, Zhang YM, Zhang WH, Zhang Y. 2017. The Agr quorum sensing system represses persister formation through regulation of phenol soluble modulins in Staphylococcus aureus. Front Microbiol 8:2189. doi: 10.3389/fmicb.2017.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhao L, Xue T, Shang F, Sun H, Sun B. 2010. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect Immun 78:3506–3515. doi: 10.1128/IAI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Yu D, Zhao L, Xue T, Sun B. 2012. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol 12:288. doi: 10.1186/1471-2180-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Xue T, Zhao L, Sun B. 2013. LuxS/AI-2 system is involved in antibiotic susceptibility and autolysis in Staphylococcus aureus NCTC 8325. Int J Antimicrob Agents 41:85–89. doi: 10.1016/j.ijantimicag.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 277.Kaplan JB. 2010. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res 89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2011. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 279.Gonzalez JE, Keshavan ND. 2006. Messing with bacterial quorum sensing. Microbiol Mol Biol Rev 70:859–875. doi: 10.1128/MMBR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Venturi V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev 30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 281.Williams P, Camara M. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol 12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 282.Barr HL, Halliday N, Camara M, Barrett DA, Williams P, Forrester DL, Simms R, Smyth AR, Honeybourne D, Whitehouse JL, Nash EF, Dewar J, Clayton A, Knox AJ, Fogarty AW. 2015. Pseudomonas aeruginosa quorum sensing molecules correlate with clinical status in cystic fibrosis. Eur Respir J 46:1046–1054. doi: 10.1183/09031936.00225214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kayama S, Murakami K, Ono T, Ushimaru M, Yamamoto A, Hirota K, Miyake Y. 2009. The role of rpoS gene and quorum-sensing system in ofloxacin tolerance in Pseudomonas aeruginosa. FEMS Microbiol Lett 298:184–192. doi: 10.1111/j.1574-6968.2009.01717.x. [DOI] [PubMed] [Google Scholar]
- 284.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Malone JG. 2015. Role of small colony variants in persistence of Pseudomonas aeruginosa infections in cystic fibrosis lungs. Infect Drug Resist 8:237–247. doi: 10.2147/IDR.S68214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Lesic B, Starkey M, He J, Hazan R, Rahme LG. 2009. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155:2845–2855. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S. 2012. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem 287:27095–27105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Allsopp LP, Wood TE, Howard SA, Maggiorelli F, Nolan LM, Wettstadt S, Filloux A. 2017. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114:7707–7712. doi: 10.1073/pnas.1700286114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Kang YS, Jung J, Jeon CO, Park W. 2011. Acinetobacter oleivorans sp. nov. is capable of adhering to and growing on diesel-oil. J Microbiol 49:29–34. doi: 10.1007/s12275-011-0315-y. [DOI] [PubMed] [Google Scholar]
- 290.Gaddy JA, Actis LA. 2009. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol 4:273–278. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Gonzalez RH, Dijkshoorn L, Van den Barselaar M, Nudel C. 2009. Quorum sensing signal profile of Acinetobacter strains from nosocomial and environmental sources. Rev Argent Microbiol 41:73–78. [PubMed] [Google Scholar]
- 292.Niu C, Clemmer KM, Bonomo RA, Rather PN. 2008. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol 190:3386–3392. doi: 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Stacy DM, Welsh MA, Rather PN, Blackwell HE. 2012. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-acyl homoserine lactones. ACS Chem Biol 7:1719–1728. doi: 10.1021/cb300351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lopez M, Mayer C, Fernandez-Garcia L, Blasco L, Muras A, Ruiz FM, Bou G, Otero A, Tomas M. 2017. Quorum sensing network in clinical strains of A. baumannii: AidA is a new quorum quenching enzyme. PLoS One 12:e0174454. doi: 10.1371/journal.pone.0174454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Suppiger A, Schmid N, Aguilar C, Pessi G, Eberl L. 2013. Two quorum sensing systems control biofilm formation and virulence in members of the Burkholderia cepacia complex. Virulence 4:400–409. doi: 10.4161/viru.25338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. 2008. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- 297.Coenye T. 2010. Social interactions in the Burkholderia cepacia complex: biofilms and quorum sensing. Future Microbiol 5:1087–1099. doi: 10.2217/fmb.10.68. [DOI] [PubMed] [Google Scholar]
- 298.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 299.Aguilar C, Schmid N, Lardi M, Pessi G, Eberl L. 2014. The IclR-family regulator BapR controls biofilm formation in B. cenocepacia H111. PLoS One 9:e92920. doi: 10.1371/journal.pone.0092920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Fauvart M, De Groote VN, Michiels J. 2011. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol 60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 301.Richards JP, Ojha AK. 2014. Mycobacterial biofilms. Microbiol Spectr 2:MGM2-0004-2013. doi: 10.1128/microbiolspec.MGM2-0004-2013. [DOI] [PubMed] [Google Scholar]
- 302.Banaiee N, Jacobs WR Jr, Ernst JD. 2006. Regulation of Mycobacterium tuberculosis whiB3 in the mouse lung and macrophages. Infect Immun 74:6449–6457. doi: 10.1128/IAI.00190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, Rubin H. 2005. Functional regulation of the opposing(p) ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44:9913–9923. doi: 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- 304.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.He H, Cooper JN, Mishra A, Raskin DM. 2012. Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol 194:2962–2972. doi: 10.1128/JB.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Cashel M. 1969. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem 244:3133–3141. [PubMed] [Google Scholar]
- 307.Chatterji D, Fujita N, Ishihama A. 1998. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 308.Chatterji D, Ojha AK. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol 4:160–165. doi: 10.1016/S1369-5274(00)00182-X. [DOI] [PubMed] [Google Scholar]
- 309.Carneiro S, Lourenço A, Ferreira EC, Rocha I. 2011. Stringent response of Escherichia coli: revisiting the bibliome using literature mining. Microb Inform Exp 1:14. doi: 10.1186/2042-5783-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Tarusawa T, Ito S, Goto S, Ushida C, Muto A, Himeno H. 2016. (p)ppGpp-dependent and -independent pathways for salt tolerance in Escherichia coli. J Biochem 160:19–26. doi: 10.1093/jb/mvw008. [DOI] [PubMed] [Google Scholar]
- 311.Kudrin P, Varik V, Oliveira SR, Beljantseva J, Del Peso Santos T, Dzhygyr I, Rejman D, Cava F, Tenson T, Hauryliuk V. 2017. Subinhibitory concentrations of bacteriostatic antibiotics induce relA-dependent and relA-independent tolerance to β-lactams. Antimicrob Agents Chemother 61:e02173-16. doi: 10.1128/AAC.02173-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Harms A, Fino C, Sørensen MA, Semsey S, Gerdes K. 2017. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8:e01964-17. doi: 10.1128/mBio.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Spector MP. 1998. The starvation-stress response (SSR) of Salmonella. Adv Microb Physiol 40:233–279. doi: 10.1016/S0065-2911(08)60133-2. [DOI] [PubMed] [Google Scholar]
- 314.Costa CS, Pizarro RA, Anton DN. 2009. Influence of RpoS, cAMP-receptor protein, and ppGpp on expression of the opgGH operon and osmoregulated periplasmic glucan content of Salmonella enterica serovar Typhimurium. Can J Microbiol 55:1284–1293. doi: 10.1139/W09-086. [DOI] [PubMed] [Google Scholar]
- 315.Song M, Kim HJ, Ryu S, Yoon H, Yun J, Choy HE. 2010. ppGpp-mediated stationary phase induction of the genes encoded by horizontally acquired pathogenicity islands and cob/pdu locus in Salmonella enterica serovar Typhimurium. J Microbiol 48:89–95. doi: 10.1007/s12275-009-0179-6. [DOI] [PubMed] [Google Scholar]
- 316.Haneda T, Sugimoto M, Yoshida-Ohta Y, Kodera Y, Oh-Ishi M, Maeda T, Shimizu-Izumi S, Miki T, Kumagai Y, Danbara H, Okada N. 2010. Comparative proteomic analysis of Salmonella enterica serovar Typhimurium ppGpp-deficient mutant to identify a novel virulence protein required for intracellular survival in macrophages. BMC Microbiol 10:324. doi: 10.1186/1471-2180-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Koskiniemi S, Pranting M, Gullberg E, Nasvall J, Andersson DI. 2011. Activation of cryptic aminoglycoside resistance in Salmonella enterica. Mol Microbiol 80:1464–1478. doi: 10.1111/j.1365-2958.2011.07657.x. [DOI] [PubMed] [Google Scholar]
- 318.Henard CA, Vazquez-Torres A. 2012. DksA-dependent resistance of Salmonella enterica serovar Typhimurium against the antimicrobial activity of inducible nitric oxide synthase. Infect Immun 80:1373–1380. doi: 10.1128/IAI.06316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Lim S, Yoon H, Kim M, Han A, Choi J, Ryu S. 2013. Hfq and ArcA are involved in the stationary phase-dependent activation of Salmonella pathogenicity island 1 (SPI1) under shaking culture conditions. J Microbiol Biotechnol 23:1664–1672. doi: 10.4014/jmb.1305.05022. [DOI] [PubMed] [Google Scholar]
- 320.Rice CJ, Ramachandran VK, Shearer N, Thompson A. 2015. Transcriptional and post-transcriptional modulation of SPI1 and SPI2 expression by ppGpp, RpoS and DksA in Salmonella enterica sv Typhimurium. PLoS One 10:e0127523. doi: 10.1371/journal.pone.0127523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Azriel S, Goren A, Rahav G, Gal-Mor O. 2016. The stringent response regulator DksA is required for Salmonella enterica serovar Typhimurium growth in minimal medium, motility, biofilm formation, and intestinal colonization. Infect Immun 84:375–384. doi: 10.1128/IAI.01135-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Yamaguchi T, Iida K, Shiota S, Nakayama H, Yoshida S. 2015. Elevated guanosine 5′-diphosphate 3′-diphosphate level inhibits bacterial growth and interferes with FtsZ assembly. FEMS Microbiol Lett 362:fnv187. doi: 10.1093/femsle/fnv187. [DOI] [PubMed] [Google Scholar]
- 323.Haralalka S, Nandi S, Bhadra RK. 2003. Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J Bacteriol 185:4672–4682. doi: 10.1128/JB.185.16.4672-4682.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Pal RR, Bag S, Dasgupta S, Das B, Bhadra RK. 2012. Functional characterization of the stringent response regulatory gene dksA of Vibrio cholerae and its role in modulation of virulence phenotypes. J Bacteriol 194:5638–5648. doi: 10.1128/JB.00518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Oh YT, Park Y, Yoon MY, Bari W, Go J, Min KB, Raskin DM, Lee KM, Yoon SS. 2014. Cholera toxin production during anaerobic trimethylamine N-oxide respiration is mediated by stringent response in Vibrio cholerae. J Biol Chem 289:13232–13242. doi: 10.1074/jbc.M113.540088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Silva AJ, Benitez JA. 2006. A Vibrio cholerae relaxed (relA) mutant expresses major virulence factors, exhibits biofilm formation and motility, and colonizes the suckling mouse intestine. J Bacteriol 188:794–800. doi: 10.1128/JB.188.2.794-800.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Oh YT, Lee KM, Bari W, Raskin DM, Yoon SS. 2015. (p)ppGpp, a small nucleotide regulator, directs the metabolic fate of glucose in Vibrio cholerae. J Biol Chem 290:13178–13190. doi: 10.1074/jbc.M115.640466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Lery LM, Goulart CL, Figueiredo FR, Verdoorn KS, Einicker-Lamas M, Gomes FM, Machado EA, Bisch PM, von Kruger WM. 2013. A comparative proteomic analysis of Vibrio cholerae O1 wild-type cells versus a phoB mutant showed that the PhoB/PhoR system is required for full growth and rpoS expression under inorganic phosphate abundance. J Proteomics 86:1–15. doi: 10.1016/j.jprot.2013.04.038. [DOI] [PubMed] [Google Scholar]
- 329.Riesenberg D, Erdei S, Kondorosi E, Kari C. 1982. Positive involvement of ppGpp in derepression of the nif operon in Klebsiella pneumoniae. Mol Gen Genet 185:198–204. doi: 10.1007/BF00330786. [DOI] [PubMed] [Google Scholar]
- 330.Ren H, He X, Zou X, Wang G, Li S, Wu Y. 2015. Gradual increase in antibiotic concentration affects persistence of Klebsiella pneumoniae. J Antimicrob Chemother 70:3267–3272. doi: 10.1093/jac/dkv251. [DOI] [PubMed] [Google Scholar]
- 331.Wells DH, Gaynor EC. 2006. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J Bacteriol 188:3726–3729. doi: 10.1128/JB.188.10.3726-3729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Mouery K, Rader BA, Gaynor EC, Guillemin K. 2006. The stringent response is required for Helicobacter pylori survival of stationary phase, exposure to acid, and aerobic shock. J Bacteriol 188:5494–5500. doi: 10.1128/JB.00366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zhou YN, Coleman WG Jr, Yang Z, Yang Y, Hodgson N, Chen F, Jin DJ. 2008. Regulation of cell growth during serum starvation and bacterial survival in macrophages by the bifunctional enzyme SpoT in Helicobacter pylori. J Bacteriol 190:8025–8032. doi: 10.1128/JB.01134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Park SA, Ko A, Lee NG. 2011. Stimulation of growth of the human gastric pathogen Helicobacter pylori by atmospheric level of oxygen under high carbon dioxide tension. BMC Microbiol 11:96. doi: 10.1186/1471-2180-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Malde A, Gangaiah D, Chandrashekhar K, Pina-Mimbela R, Torrelles JB, Rajashekara G. 2014. Functional characterization of exopolyphosphatase/guanosine pentaphosphate phosphohydrolase (PPX/GPPA) of Campylobacter jejuni. Virulence 5:521–533. doi: 10.4161/viru.28311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Jeon B. 2014. A tangle of poly-phosphate in Campylobacter. Virulence 5:449–450. doi: 10.4161/viru.28690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Chavez de Paz LE, Lemos JA, Wickstrom C, Sedgley CM. 2012. Role of (p)ppGpp in biofilm formation by Enterococcus faecalis. Appl Environ Microbiol 78:1627–1630. doi: 10.1128/AEM.07036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Hauryliuk V, Atkinson GC. 2017. Small alarmone synthetases as novel bacterial RNA-binding proteins. RNA Biol 14:1695–1699. doi: 10.1080/15476286.2017.1367889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Gaca AO, Kajfasz JK, Miller JH, Liu K, Wang JD, Abranches J, Lemos JA. 2013. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. mBio 4:e00646-13. doi: 10.1128/mBio.00646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Honsa ES, Cooper VS, Mhaissen MN, Frank M, Shaker J, Iverson A, Rubnitz J, Hayden RT, Lee RE, Rock CO, Tuomanen EI, Wolf J, Rosch JW. 2017. RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. mBio 8:e02124-16. doi: 10.1128/mBio.02124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Raj NB, Engelhardt J, Au WC, Levy DE, Pitha PM. 1989. Virus infection and interferon can activate gene expression through a single synthetic element, but endogenous genes show distinct regulation. J Biol Chem 264:16658–16666. [PubMed] [Google Scholar]
- 342.Abranches J, Martinez AR, Kajfasz JK, Chavez V, Garsin DA, Lemos JA. 2009. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol 191:2248–2256. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Sharma AK, Payne SM. 2006. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol Microbiol 62:469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- 344.Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee HW, Hong JI, Hartland EL, Stinear TP, Howden BP. 2010. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Corrigan RM, Bellows LE, Wood A, Grundling A. 2016. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc Natl Acad Sci U S A 113:E1710–E1719. doi: 10.1073/pnas.1522179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 347.Geiger T, Goerke C, Fritz M, Schafer T, Ohlsen K, Liebeke M, Lalk M, Wolz C. 2010. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun 78:1873–1883. doi: 10.1128/IAI.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Stewart PS, Franklin MJ, Williamson KS, Folsom JP, Boegli L, James GA. 2015. Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 59:3838–3847. doi: 10.1128/AAC.00433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Fernandez-Garcia L, Fernandez-Cuenca F, Blasco L, Lopez-Rojas R, Ambroa A, Lopez M, Pascual A, Bou G, Tomas M. 2018. Relationship between tolerance and persistence mechanisms in Acinetobacter baumannii strains with AbkAB toxin-antitoxin system. Antimicrob Agents Chemother 62:e00250-18. doi: 10.1128/AAC.00250-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Kang YS, Park W. 2010. Contribution of quorum-sensing system to hexadecane degradation and biofilm formation in Acinetobacter sp. strain DR1. J Appl Microbiol 109:1650–1659. doi: 10.1111/j.1365-2672.2010.04793.x. [DOI] [PubMed] [Google Scholar]
- 351.Dean SN, Chung MC, van Hoek ML. 2015. Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Appl Environ Microbiol 81:7057–7066. doi: 10.1128/AEM.02165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol 182:4889–4898. doi: 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Sajish M, Tiwari D, Rananaware D, Nandicoori VK, Prakash B. 2007. A charge reversal differentiates (p)ppGpp synthesis by monofunctional and bifunctional Rel proteins. J Biol Chem 282:34977–34983. doi: 10.1074/jbc.M704828200. [DOI] [PubMed] [Google Scholar]
- 354.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 355.Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC. 2010. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202:1397–1404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. 2009. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kim Y, Wood TK. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun 391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Harrison JJ, Wade WD, Akierman S, Vacchi-Suzzi C, Stremick CA, Turner RJ, Ceri H. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob Agents Chemother 53:2253–2258. doi: 10.1128/AAC.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versees W, Hofkens J, Jansen M, Fauvart M, Michiels J. 2015. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 360.Norton JP, Mulvey MA. 2012. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog 8:e1002954. doi: 10.1371/journal.ppat.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Wang X, Wood TK. 2016. Cryptic prophages as targets for drug development. Drug Resist Updat 27:30–38. doi: 10.1016/j.drup.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 362.Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 363.Schumacher MA, Balani P, Min J, Chinnam NB, Hansen S, Vulić M, Lewis K, Brennan RG. 2015. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524:59–64. doi: 10.1038/nature14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Germain E, Roghanian M, Gerdes K, Maisonneuve E. 2015. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci U S A 112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 365.Bokinsky G, Baidoo EE, Akella S, Burd H, Weaver D, Alonso-Gutierrez J, Garcia-Martin H, Lee TS, Keasling JD. 2013. HipA-triggered growth arrest and beta-lactam tolerance in Escherichia coli are mediated by RelA-dependent ppGpp synthesis. J Bacteriol 195:3173–3182. doi: 10.1128/JB.02210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Cho J, Carr AN, Whitworth L, Johnson B, Wilson KS. 2017. MazEF toxin-antitoxin proteins alter Escherichia coli cell morphology and infrastructure during persister formation and regrowth. Microbiology 163:308–321. doi: 10.1099/mic.0.000436. [DOI] [PubMed] [Google Scholar]
- 367.Kwan BW, Lord DM, Peti W, Page R, Benedik MJ, Wood TK. 2015. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ Microbiol 17:3168–3181. doi: 10.1111/1462-2920.12749. [DOI] [PubMed] [Google Scholar]
- 368.Silva-Herzog E, McDonald EM, Crooks AL, Detweiler CS. 2015. Physiologic stresses reveal a Salmonella persister state and TA family toxins modulate tolerance to these stresses. PLoS One 10:e0141343. doi: 10.1371/journal.pone.0141343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Ban GH, Kang DH, Yoon H. 2015. Transcriptional response of selected genes of Salmonella enterica serovar Typhimurium biofilm cells during inactivation by superheated steam. Int J Food Microbiol 192:117–123. doi: 10.1016/j.ijfoodmicro.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 370.Cheverton AM, Gollan B, Przydacz M, Wong CT, Mylona A, Hare SA, Helaine S. 2016. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol Cell 63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wang Y, Wang H, Hay AJ, Zhong Z, Zhu J, Kan B. 2015. Functional RelBE-family toxin-antitoxin pairs affect biofilm maturation and intestine colonization in Vibrio cholerae. PLoS One 10:e0135696. doi: 10.1371/journal.pone.0135696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Duprilot M, Decre D, Genel N, Drieux L, Sougakoff W, Arlet G. 2017. Diversity and functionality of plasmid-borne VagCD toxin-antitoxin systems of Klebsiella pneumoniae. J Antimicrob Chemother 72:1320–1326. doi: 10.1093/jac/dkw569. [DOI] [PubMed] [Google Scholar]
- 373.Wei YQ, Bi DX, Wei DQ, Ou HY. 2016. Prediction of type II toxin-antitoxin loci in Klebsiella pneumoniae genome sequences. Interdiscip Sci 8:143–149. doi: 10.1007/s12539-015-0135-6. [DOI] [PubMed] [Google Scholar]
- 374.Arnion H, Korkut DN, Masachis Gelo S, Chabas S, Reignier J, Iost I, Darfeuille F. 2017. Mechanistic insights into type I toxin antitoxin systems in Helicobacter pylori: the importance of mRNA folding in controlling toxin expression. Nucleic Acids Res 45:4782–4795. doi: 10.1093/nar/gkw1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Han KD, Matsuura A, Ahn HC, Kwon AR, Min YH, Park HJ, Won HS, Park SJ, Kim DY, Lee BJ. 2011. Functional identification of toxin-antitoxin molecules from Helicobacter pylori 26695 and structural elucidation of the molecular interactions. J Biol Chem 286:4842–4853. doi: 10.1074/jbc.M109.097840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Pathak C, Im H, Yang YJ, Yoon HJ, Kim HM, Kwon AR, Lee BJ. 2013. Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity. Biochim Biophys Acta 1834:2579–2590. doi: 10.1016/j.bbapap.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 377.Shen Z, Patil RD, Sahin O, Wu Z, Pu XY, Dai L, Plummer PJ, Yaeger MJ, Zhang Q. 2016. Identification and functional analysis of two toxin-antitoxin systems in Campylobacter jejuni. Mol Microbiol 101:909–923. doi: 10.1111/mmi.13431. [DOI] [PubMed] [Google Scholar]
- 378.Weaver KE, Weaver DM, Wells CL, Waters CM, Gardner ME, Ehli EA. 2003. Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responses to lantibiotics. J Bacteriol 185:2169–2177. doi: 10.1128/JB.185.7.2169-2177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. 2009. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 155:2930–2940. doi: 10.1099/mic.0.030932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Weaver KE. 2012. The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs. RNA Biol 9:1498–1503. doi: 10.4161/rna.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Weaver KE. 2015. The type I toxin-antitoxin par locus from Enterococcus faecalis plasmid pAD1: RNA regulation by both cis- and trans-acting elements. Plasmid 78:65–70. doi: 10.1016/j.plasmid.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Weaver KE, Chen Y, Miller EM, Johnson JN, Dangler AA, Manias DA, Clem AM, Schjodt DJ, Dunny GM. 2017. Examination of Enterococcus faecalis toxin-antitoxin system toxin Fst function utilizing a pheromone-inducible expression vector with tight repression and broad dynamic range. J Bacteriol 199:e00065-17. doi: 10.1128/JB.00065-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Sletvold H, Johnsen PJ, Hamre I, Simonsen GS, Sundsfjord A, Nielsen KM. 2008. Complete sequence of Enterococcus faecium pVEF3 and the detection of an omega-epsilon-zeta toxin-antitoxin module and an ABC transporter. Plasmid 60:75–85. doi: 10.1016/j.plasmid.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 384.Liu Y, Wang Y, Schwarz S, Li Y, Shen Z, Zhang Q, Wu C, Shen J. 2013. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob Agents Chemother 57:42–48. doi: 10.1128/AAC.01605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Grady R, Hayes F. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol Microbiol 47:1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- 386.Bjorkeng EK, Hjerde E, Pedersen T, Sundsfjord A, Hegstad K. 2013. ICESluvan, a 94-kilobase mosaic integrative conjugative element conferring interspecies transfer of VanB-type glycopeptide resistance, a novel bacitracin resistance locus, and a toxin-antitoxin stabilization system. J Bacteriol 195:5381–5390. doi: 10.1128/JB.02165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Soheili S, Ghafourian S, Sekawi Z, Neela VK, Sadeghifard N, Taherikalani M, Khosravi A, Ramli R, Hamat RA. 2015. The mazEF toxin-antitoxin system as an attractive target in clinical isolates of Enterococcus faecium and Enterococcus faecalis. Drug Des Devel Ther 9:2553–2561. doi: 10.2147/DDDT.S77263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Arbing MA, Handelman SK, Kuzin AP, Verdon G, Wang C, Su M, Rothenbacher FP, Abashidze M, Liu M, Hurley JM, Xiao R, Acton T, Inouye M, Montelione GT, Woychik NA, Hunt JF. 2010. Crystal structures of Phd-Doc, HigA, and YeeU establish multiple evolutionary links between microbial growth-regulating toxin-antitoxin systems. Structure 18:996–1010. doi: 10.1016/j.str.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Al-Hasani K, Rajakumar K, Bulach D, Robins-Browne R, Adler B, Sakellaris H. 2001. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb Pathog 30:1–8. doi: 10.1006/mpat.2000.0404. [DOI] [PubMed] [Google Scholar]
- 390.Dienemann C, Boggild A, Winther KS, Gerdes K, Brodersen DE. 2011. Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J Mol Biol 414:713–722. doi: 10.1016/j.jmb.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Xu K, Dedic E, Cob-Cantal P, Dienemann C, Boggild A, Winther KS, Gerdes K, Brodersen DE. 2013. Protein expression, crystallization and preliminary X-ray crystallographic analysis of the isolated Shigella flexneri VapC toxin. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:762–765. doi: 10.1107/S1744309113014012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.McVicker G, Tang CM. 2016. Deletion of toxin-antitoxin systems in the evolution of Shigella sonnei as a host-adapted pathogen. Nat Microbiol 2:16204. doi: 10.1038/nmicrobiol.2016.204. [DOI] [PubMed] [Google Scholar]
- 393.Lepka D, Kerrinnes T, Skiebe E, Hahn B, Fruth A, Wilharm G. 2009. Adding to Yersinia enterocolitica gene pool diversity: two cryptic plasmids from a biotype 1A isolate. J Biomed Biotechnol 2009:398434. doi: 10.1155/2009/398434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Gil F, Pizarro-Guajardo M, Alvarez R, Garavaglia M, Paredes-Sabja D. 2015. Clostridium difficile recurrent infection: possible implication of TA systems. Future Microbiol 10:1649–1657. doi: 10.2217/fmb.15.94. [DOI] [PubMed] [Google Scholar]
- 395.Donegan NP, Thompson ET, Fu Z, Cheung AL. 2010. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J Bacteriol 192:1416–1422. doi: 10.1128/JB.00233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Fu Z, Donegan NP, Memmi G, Cheung AL. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J Bacteriol 189:8871–8879. doi: 10.1128/JB.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Andersen SB, Ghoul M, Griffin AS, Petersen B, Johansen HK, Molin S. 2017. Diversity, prevalence, and longitudinal occurrence of type II toxin-antitoxin systems of Pseudomonas aeruginosa infecting cystic fibrosis lungs. Front Microbiol 8:1180. doi: 10.3389/fmicb.2017.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Wood TL, Wood TK. 2016. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen 5:499–511. doi: 10.1002/mbo3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Jurenaite M, Markuckas A, Suziedeliene E. 2013. Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J Bacteriol 195:3165–3172. doi: 10.1128/JB.00237-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Ghafourian S, Good L, Sekawi Z, Hamat RA, Soheili S, Sadeghifard N, Neela V. 2014. The mazEF toxin-antitoxin system as a novel antibacterial target in Acinetobacter baumannii. Mem Inst Oswaldo Cruz 109:502–505. doi: 10.1590/0074-0276130601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Mosqueda N, Gato E, Roca I, Lopez M, de Alegria CR, Fernandez Cuenca F, Martinez-Martinez L, Pachon J, Cisneros JM, Rodriguez-Bano J, Pascual A, Vila J, Bou G, Tomas M. 2014. Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J Antimicrob Chemother 69:2629–2633. doi: 10.1093/jac/dku179. [DOI] [PubMed] [Google Scholar]
- 402.Hall CW, Mah TF. 2017. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 403.Winther K, Tree JJ, Tollervey D, Gerdes K. 2016. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res 44:9860–9871. doi: 10.1093/nar/gkw781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Sala A, Bordes P, Genevaux P. 2014. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 6:1002–1020. doi: 10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Tiwari P, Arora G, Singh M, Kidwai S, Narayan OP, Singh R. 2015. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat Commun 6:6059. doi: 10.1038/ncomms7059. [DOI] [PubMed] [Google Scholar]
- 407.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. 2010. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res 38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Blower TR, Short FL, Rao F, Mizuguchi K, Pei XY, Fineran PC, Luisi BF, Salmond GP. 2012. Identification and classification of bacterial type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res 40:6158–6173. doi: 10.1093/nar/gks231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, Clarke K, Cronin A, Davis P, Goodhead I, Holroyd N, Jagels K, Lord A, Moule S, Mungall K, Norbertczak H, Quail MA, Rabbinowitsch E, Walker D, White B, Whitehead S, Small PL, Brosch R, Ramakrishnan L, Fischbach MA, Parkhill J, Cole ST. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Gupta A. 2009. Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol Lett 290:45–53. doi: 10.1111/j.1574-6968.2008.01400.x. [DOI] [PubMed] [Google Scholar]
- 411.Huang F, He ZG. 2010. Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic Acids Res 38:8219–8230. doi: 10.1093/nar/gkq737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Singh R, Barry CE III, Boshoff HI. 2010. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J Bacteriol 192:1279–1291. doi: 10.1128/JB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Zhu L, Sharp JD, Kobayashi H, Woychik NA, Inouye M. 2010. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J Biol Chem 285:39732–39738. doi: 10.1074/jbc.M110.163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Walter ND, Dolganov GM, Garcia BJ, Worodria W, Andama A, Musisi E, Ayakaka I, Van TT, Voskuil MI, de Jong BC, Davidson RM, Fingerlin TE, Kechris K, Palmer C, Nahid P, Daley CL, Geraci M, Huang L, Cattamanchi A, Strong M, Schoolnik GK, Davis JL. 2015. Transcriptional adaptation of drug-tolerant Mycobacterium tuberculosis during treatment of human tuberculosis. J Infect Dis 212:990–998. doi: 10.1093/infdis/jiv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Regoes RR, Wiuff C, Zappala RM, Garner KN, Baquero F, Levin BR. 2004. Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob Agents Chemother 48:3670–3676. doi: 10.1128/AAC.48.10.3670-3676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Gefen O, Chekol B, Strahilevitz J, Balaban NQ. 2017. TDtest: easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci Rep 7:41284. doi: 10.1038/srep41284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Mascio CT, Alder JD, Silverman JA. 2007. Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob Agents Chemother 51:4255–4260. doi: 10.1128/AAC.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.de la Fuente-Nunez C, Reffuveille F, Haney EF, Straus SK, Hancock RE. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog 10:e1004152. doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Syal K, Flentie K, Bhardwaj N, Maiti K, Jayaraman N, Stallings CL, Chatterji D. 2017. Synthetic (p)ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob Agents Chemother 61:e00443-17. doi: 10.1128/AAC.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Njire M, Wang N, Wang B, Tan Y, Cai X, Liu Y, Mugweru J, Guo J, Hameed HMA, Tan S, Liu J, Yew WW, Nuermberger E, Lamichhane G, Zhang T. 2017. Pyrazinoic acid inhibits a bifunctional enzyme in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e00070-17. doi: 10.1128/AAC.00070-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.de Breij A, Riool M, Cordfunke RA, Malanovic N, de Boer L, Koning RI, Ravensbergen E, Franken M, van der Heijde T, Boekema BK, Kwakman PHS, Kamp N, El Ghalbzouri A, Lohner K, Zaat SAJ, Drijfhout JW, Nibbering PH. 2018. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci Transl Med 10:eaan4044. doi: 10.1126/scitranslmed.aan4044. [DOI] [PubMed] [Google Scholar]
- 422.Williams SR, Gebhart D, Martin DW, Scholl D. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Kageyama M. 1964. Studies of a pyocin. I. Physical and chemical properties. J Biochem 55:49–53. [DOI] [PubMed] [Google Scholar]
- 424.Blackwell CC, Law JA. 1981. Typing of non-serogroupable Neisseria meningitidis by means of sensitivity to R-type pyocines of Pseudomonas aeruginosa. J Infect 3:370–378. doi: 10.1016/S0163-4453(81)91996-4. [DOI] [PubMed] [Google Scholar]
- 425.Blackwell CC, Winstanley FP, Telfer Brunton WA. 1982. Sensitivity of thermophilic campylobacters to R-type pyocines of Pseudomonas aeruginosa. J Med Microbiol 15:247–251. doi: 10.1099/00222615-15-2-247. [DOI] [PubMed] [Google Scholar]
- 426.Campagnari AA, Karalus R, Apicella M, Melaugh W, Lesse AJ, Gibson BW. 1994. Use of pyocin to select a Haemophilus ducreyi variant defective in lipooligosaccharide biosynthesis. Infect Immun 62:2379–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Filiatrault MJ, Munson RS Jr, Campagnari AA. 2001. Genetic analysis of a pyocin-resistant lipooligosaccharide (LOS) mutant of Haemophilus ducreyi: restoration of full-length LOS restores pyocin sensitivity. J Bacteriol 183:5756–5761. doi: 10.1128/JB.183.19.5756-5761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Morse SA, Jones BV, Lysko PG. 1980. Pyocin inhibition of Neisseria gonorrhoeae: mechanism of action. Antimicrob Agents Chemother 18:416–423. doi: 10.1128/AAC.18.3.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Morse SA, Vaughan P, Johnson D, Iglewski BH. 1976. Inhibition of Neisseria gonorrhoeae by a bacteriocin from Pseudomonas aeruginosa. Antimicrob Agents Chemother 10:354–362. doi: 10.1128/AAC.10.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Birmingham VA, Pattee PA. 1981. Genetic transformation in Staphylococcus aureus: isolation and characterization of a competence-conferring factor from bacteriophage 80 alpha lysates. J Bacteriol 148:301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Coetzee HL, De Klerk HC, Coetzee JN, Smit JA. 1968. Bacteriophage-tail-like particles associated with intra-species killing of Proteus vulgaris. J Gen Virol 2:29–36. doi: 10.1099/0022-1317-2-1-29. [DOI] [PubMed] [Google Scholar]
- 432.Jabrane A, Sabri A, Compere P, Jacques P, Vandenberghe I, Van Beeumen J, Thonart P. 2002. Characterization of serracin P, a phage-tail-like bacteriocin, and its activity against Erwinia amylovora, the fire blight pathogen. Appl Environ Microbiol 68:5704–5710. doi: 10.1128/AEM.68.11.5704-5710.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Strauch E, Kaspar H, Schaudinn C, Dersch P, Madela K, Gewinner C, Hertwig S, Wecke J, Appel B. 2001. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl Environ Microbiol 67:5634–5642. doi: 10.1128/AEM.67.12.5634-5642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Zink R, Loessner MJ, Scherer S. 1995. Characterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/endolysin gene. Microbiology 141:2577–2584. doi: 10.1099/13500872-141-10-2577. [DOI] [PubMed] [Google Scholar]
- 435.Naghmouchi K, Baah J, Hober D, Jouy E, Rubrecht C, Sane F, Drider D. 2013. Synergistic effect between colistin and bacteriocins in controlling Gram-negative pathogens and their potential to reduce antibiotic toxicity in mammalian epithelial cells. Antimicrob Agents Chemother 57:2719–2725. doi: 10.1128/AAC.02328-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Benmechernene Z, Fernandez-No I, Kihal M, Bohme K, Calo-Mata P, Barros-Velazquez J. 2013. Recent patents on bacteriocins: food and biomedical applications. Recent Pat DNA Gene Seq 7:66–73. doi: 10.2174/1872215611307010010. [DOI] [PubMed] [Google Scholar]
- 437.Shin JM, Gwak JW, Kamarajan P, Fenno JC, Rickard AH, Kapila YL. 2016. Biomedical applications of nisin. J Appl Microbiol 120:1449–1465. doi: 10.1111/jam.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Alvarez-Sieiro P, Montalban-Lopez M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Perez-Ibarreche M, Castellano P, Leclercq A, Vignolo G. 2016. Control of Listeria monocytogenes biofilms on industrial surfaces by the bacteriocin-producing Lactobacillus sakei CRL1862. FEMS Microbiol Lett 363:fnw118. doi: 10.1093/femsle/fnw118. [DOI] [PubMed] [Google Scholar]
- 440.Al-Seraih A, Belguesmia Y, Baah J, Szunerits S, Boukherroub R, Drider D. 2017. Enterocin B3A-B3B produced by LAB collected from infant faeces: potential utilization in the food industry for Listeria monocytogenes biofilm management. Antonie Van Leeuwenhoek 110:205–219. doi: 10.1007/s10482-016-0791-5. [DOI] [PubMed] [Google Scholar]
- 441.Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He J, Kitao T, Righi V, Milot S, Tzika A, Rahme L. 2014. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog 10:e1004321. doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Pan J, Xie X, Tian W, Bahar AA, Lin N, Song F, An J, Ren D. 2013. (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5H)-one sensitizes Escherichia coli persister cells to antibiotics. Appl Microbiol Biotechnol 97:9145–9154. doi: 10.1007/s00253-013-5185-2. [DOI] [PubMed] [Google Scholar]
- 443.Garcia-Contreras R, Martinez-Vazquez M, Velazquez Guadarrama N, Villegas Paneda AG, Hashimoto T, Maeda T, Quezada H, Wood TK. 2013. Resistance to the quorum-quenching compounds brominated furanone C-30 and 5-fluorouracil in Pseudomonas aeruginosa clinical isolates. Pathog Dis 68:8–11. doi: 10.1111/2049-632X.12039. [DOI] [PubMed] [Google Scholar]
- 444.Garcia-Contreras R, Maeda T, Wood TK. 2013. Resistance to quorum-quenching compounds. Appl Environ Microbiol 79:6840–6846. doi: 10.1128/AEM.02378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Garcia-Contreras R, Nunez-Lopez L, Jasso-Chavez R, Kwan BW, Belmont JA, Rangel-Vega A, Maeda T, Wood TK. 2015. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J 9:115–125. doi: 10.1038/ismej.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Garcia-Contreras R, Maeda T, Wood TK. 2016. Can resistance against quorum-sensing interference be selected? ISME J 10:4–10. doi: 10.1038/ismej.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Balaban N, Goldkorn T, Gov Y, Hirshberg M, Koyfman N, Matthews HR, Nhan RT, Singh B, Uziel O. 2001. Regulation of Staphylococcus aureus pathogenesis via target of RNAIII-activating protein (TRAP). J Biol Chem 276:2658–2667. doi: 10.1074/jbc.M005446200. [DOI] [PubMed] [Google Scholar]
- 448.Yang G, Cheng H, Liu C, Xue Y, Gao Y, Liu N, Gao B, Wang D, Li S, Shen B, Shao N. 2003. Inhibition of Staphylococcus aureus pathogenesis in vitro and in vivo by RAP-binding peptides. Peptides 24:1823–1828. doi: 10.1016/j.peptides.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 449.Giacometti A, Cirioni O, Ghiselli R, Dell'Acqua G, Orlando F, D'Amato G, Mocchegiani F, Silvestri C, Del Prete MS, Rocchi M, Balaban N, Saba V, Scalise G. 2005. RNAIII-inhibiting peptide improves efficacy of clinically used antibiotics in a murine model of staphylococcal sepsis. Peptides 26:169–175. doi: 10.1016/j.peptides.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 450.Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. 2013. Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J Am Chem Soc 135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- 451.López M, Barbosa B, Gato E, Bou G, Tomás M. 2014. Patents on antivirulence therapies. World J Pharmacol 3:97–109. doi: 10.5497/wjp.v3.i4.97. [DOI] [Google Scholar]
- 452.Zhang ZH, Wu CC, Chen XW, Li X, Li J, Lu MJ. 2016. Genetic variation of hepatitis B virus and its significance for pathogenesis. World J Gastroenterol 22:126–144. doi: 10.3748/wjg.v22.i1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Vila-Farres X, Parra-Millan R, Sanchez-Encinales V, Varese M, Ayerbe-Algaba R, Bayo N, Guardiola S, Pachon-Ibanez ME, Kotev M, Garcia J, Teixido M, Vila J, Pachon J, Giralt E, Smani Y. 2017. Combating virulence of Gram-negative bacilli by OmpA inhibition. Sci Rep 7:14683. doi: 10.1038/s41598-017-14972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Abedon ST. 2015. Ecology of anti-biofilm agents. II. Bacteriophage exploitation and biocontrol of biofilm bacteria. Pharmaceuticals (Basel) 8:559–589. doi: 10.3390/ph8030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Pires DP, Oliveira H, Melo LD, Sillankorva S, Azeredo J. 2016. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol 100:2141–2151. doi: 10.1007/s00253-015-7247-0. [DOI] [PubMed] [Google Scholar]
- 456.Motlagh AM, Bhattacharjee AS, Goel R. 2016. Biofilm control with natural and genetically-modified phages. World J Microbiol Biotechnol 32:67. doi: 10.1007/s11274-016-2009-4. [DOI] [PubMed] [Google Scholar]
- 457.Bardy P, Pantucek R, Benesik M, Doskar J. 2016. Genetically modified bacteriophages in applied microbiology. J Appl Microbiol 121:618–633. doi: 10.1111/jam.13207. [DOI] [PubMed] [Google Scholar]
- 458.Gutierrez D, Rodriguez-Rubio L, Martinez B, Rodriguez A, Garcia P. 2016. Bacteriophages as weapons against bacterial biofilms in the food industry. Front Microbiol 7:825. doi: 10.3389/fmicb.2016.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Kaistha SD, Umrao PD. 2016. Bacteriophage for mitigation of multiple drug resistant biofilm forming pathogens. Recent Pat Biotechnol 10:184–194. doi: 10.2174/1872208310666160919122155. [DOI] [PubMed] [Google Scholar]
- 460.Fernández L, Gonzalez S, Campelo AB, Martinez B, Rodriguez A, Garcia P. 2017. Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus. Sci Rep 7:40965. doi: 10.1038/srep40965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Gutierrez D, Fernandez L, Rodriguez A, Garcia P. 2018. Are phage lytic proteins the secret weapon to kill Staphylococcus aureus? mBio 9:e01923-17. doi: 10.1128/mBio.01923-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Gilmer DB, Schmitz JE, Euler CW, Fischetti VA. 2013. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57:2743–2750. doi: 10.1128/AAC.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Larpin Y, Oechslin F, Moreillon P, Resch G, Entenza JM, Mancini S. 2018. In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS One 13:e0192507. doi: 10.1371/journal.pone.0192507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Lai MJ, Lin NT, Hu A, Soo PC, Chen LK, Chen LH, Chang KC. 2011. Antibacterial activity of Acinetobacter baumannii phage varphiAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Appl Microbiol Biotechnol 90:529–539. doi: 10.1007/s00253-011-3104-y. [DOI] [PubMed] [Google Scholar]
- 465.Peng SY, You RI, Lai MJ, Lin NT, Chen LK, Chang KC. 2017. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci Rep 7:11477. doi: 10.1038/s41598-017-11832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Soo VW, Kwan BW, Quezada H, Castillo-Juarez I, Perez-Eretza B, Garcia-Contreras SJ, Martinez-Vazquez M, Wood TK, Garcia-Contreras R. 2017. Repurposing of anticancer drugs for the treatment of bacterial infections. Curr Top Med Chem 17:1157–1176. doi: 10.2174/1568026616666160930131737. [DOI] [PubMed] [Google Scholar]
- 467.Kwan BW, Chowdhury N, Wood TK. 2015. Combatting bacterial infections by killing persister cells with mitomycin C. Environ Microbiol 17:4406–4414. doi: 10.1111/1462-2920.12873. [DOI] [PubMed] [Google Scholar]
- 468.Chowdhury N, Wood TL, Martínez-Vázquez M, García-Contreras R, Wood TK. 2016. DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol Bioeng 113:1984–1992. doi: 10.1002/bit.25963. [DOI] [PubMed] [Google Scholar]
- 469.Sukheja P, Kumar P, Mittal N, Li SG, Singleton E, Russo R, Perryman AL, Shrestha R, Awasthi D, Husain S, Soteropoulos P, Brukh R, Connell N, Freundlich JS, Alland D. 2017. A novel small-molecule inhibitor of the Mycobacterium tuberculosis demethylmenaquinone methyltransferase MenG is bactericidal to both growing and nutritionally deprived persister cells. mBio 8:e02022-16. doi: 10.1128/mBio.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Cook GM, Greening C, Hards K, Berney M. 2014. Energetics of pathogenic bacteria and opportunities for drug development. Adv Microb Physiol 65:1–62. doi: 10.1016/bs.ampbs.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 471.Nautiyal A, Patil KN, Muniyappa K. 2014. Suramin is a potent and selective inhibitor of Mycobacterium tuberculosis RecA protein and the SOS response: RecA as a potential target for antibacterial drug discovery. J Antimicrob Chemother 69:1834–1843. doi: 10.1093/jac/dku080. [DOI] [PubMed] [Google Scholar]
- 472.Wood TK. 2016. Combatting bacterial persister cells. Biotechnol Bioeng 113:476–483. doi: 10.1002/bit.25721. [DOI] [PubMed] [Google Scholar]
- 473.Kim JS, Heo P, Yang TJ, Lee KS, Cho DH, Kim BT, Suh JH, Lim HJ, Shin D, Kim SK, Kweon DH. 2011. Selective killing of bacterial persisters by a single chemical compound without affecting normal antibiotic-sensitive cells. Antimicrob Agents Chemother 55:5380–5383. doi: 10.1128/AAC.00708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Kim W, Conery AL, Rajamuthiah R, Fuchs BB, Ausubel FM, Mylonakis E. 2015. Identification of an antimicrobial agent effective against methicillin-resistant Staphylococcus aureus persisters using a fluorescence-based screening strategy. PLoS One 10:e0127640. doi: 10.1371/journal.pone.0127640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE III, Wang H, Zhang W, Zhang Y. 2011. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333:1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Feng J, Auwaerter PG, Zhang Y. 2015. Drug combinations against Borrelia burgdorferi persisters in vitro: eradication achieved by using daptomycin, cefoperazone and doxycycline. PLoS One 10:e0117207. doi: 10.1371/journal.pone.0117207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Marques CN, Morozov A, Planzos P, Zelaya HM. 2014. The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl Environ Microbiol 80:6976–6991. doi: 10.1128/AEM.01576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Abreu AC, Saavedra MJ, Simoes LC, Simoes M. 2016. Combinatorial approaches with selected phytochemicals to increase antibiotic efficacy against Staphylococcus aureus biofilms. Biofouling 32:1103–1114. doi: 10.1080/08927014.2016.1232402. [DOI] [PubMed] [Google Scholar]
- 480.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 481.Schuch R, Khan BK, Raz A, Rotolo JA, Wittekind M. 2017. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent. Antimicrob Agents Chemother 61:e02666-16. doi: 10.1128/AAC.02666-16. [DOI] [PMC free article] [PubMed] [Google Scholar]