Coinfections involving viruses are being recognized to influence the disease pattern that occurs relative to that with single infection. Classically, we usually think of a clinical syndrome as the consequence of infection by a single virus that is isolated from clinical specimens.
KEYWORDS: bystander protection, diverse TCR repertoire, attrition, coinfection, cross reactivity, exclusion, persistence, virus
SUMMARY
Coinfections involving viruses are being recognized to influence the disease pattern that occurs relative to that with single infection. Classically, we usually think of a clinical syndrome as the consequence of infection by a single virus that is isolated from clinical specimens. However, this biased laboratory approach omits detection of additional agents that could be contributing to the clinical outcome, including novel agents not usually considered pathogens. The presence of an additional agent may also interfere with the targeted isolation of a known virus. Viral interference, a phenomenon where one virus competitively suppresses replication of other coinfecting viruses, is the most common outcome of viral coinfections. In addition, coinfections can modulate virus virulence and cell death, thereby altering disease severity and epidemiology. Immunity to primary virus infection can also modulate immune responses to subsequent secondary infections. In this review, various virological mechanisms that determine viral persistence/exclusion during coinfections are discussed, and insights into the isolation/detection of multiple viruses are provided. We also discuss features of heterologous infections that impact the pattern of immune responsiveness that develops.
INTRODUCTION
It is common to attribute a viral disease to infection by a single agent. However, under natural circumstances hosts may be infected by multiple agents with the outcome influenced by contributions from more than the incriminated virus, but rarely in diagnostic laboratories do we consider the input of multiple agents. Regarding terminology, infection by more than one variety of microorganism (viruses, bacteria, protozoa, etc.) is termed mixed infection. In virology, coinfection is used to describe simultaneous infection of a cell or organism by separate viruses (1). The term superinfection is used if one virus infects the host some time before infection by the second virus. However, in the literature, the definitions of coinfection and mixed infection have been used interchangeably (2–5). The meaning of these terms depends on the context, whether applied to a single cell, a cell line, part of a host, or a whole host (1, 4). In an infected cell, viruses can interact with a large number of cellular proteins (virus-host interactome) that may either support or inhibit virus replication. As with virus-host protein interactions, protein-protein interactions between unrelated viruses are also possible (6, 7). Coinfections may result in genetic exchange between agents to generate recombinant viruses. Chimeric viruses (mixed nucleic acid) observed in metagenomic studies have suggested the possibility of genetic exchange even among heterologous viruses, but this issue needs further evaluation (8). Recombination effects can influence viral evolution, disease dynamics, sensitivity to antiviral therapy, and eventually the fate of the host (9).
Coinfections may play a pivotal role in reducing or augmenting disease severity (10–13). However, because of the high specificity of diagnostic assays, they usually miss detection of additional relevant agents. When individual cells are coinfected, one virus usually influences replication of the other, a phenomenon termed viral interference. The result can be clearance (exclusion) of one virus but persistence of the other (14). Viral interference may be mediated by factors such as interferons (IFNs), defective interfering (DI) particles, production of trans-acting proteases, cellular factors, and nonspecific double-stranded RNA (dsRNA) (1). Besides virus-virus interactions, the nature of the host also plays an important role in shaping coinfection patterns. For example, bacterial isolates from a particular geographical region are usually infected more efficiently by bacteriophages isolated from the same niche (15).
The response of the host immune system also influences the outcome of viral coinfections. Upon antigen exposure, naive T cells convert into activated effector T cells and eventually long-term memory T cells. Memory responses generated against one infection may influence the quantity and quality of the immune response to subsequent secondary infection. This influence of immunity to primary infection on a subsequent unrelated infection is known as heterologous immunity. Heterologous immunity can occur between very closely related infectious agents such as multiple variants of a particular virus type, among different viruses, or between viruses, bacteria, protozoa, or different parasites (2). A variety of immune cells participate in heterologous immunity, and these may induce either a protective or immunopathological response (2). Finally, studying coinfections in short-lived laboratory animal systems can be misleading since the outcome of coinfections in clean containment facilities does not replicate what occurs in natural environments in hosts exposed often for decades to multiple pathogens.
DETECTION OF COINFECTIONS
Multiple viruses are capable of causing disease syndromes, though we usually consider the outcome of infection by a single virus. However, almost invariably under natural circumstances, hosts may be infected by multiple agents, with the outcome influenced by contributions from more than a single agent. In diagnostic laboratories, we rarely consider the input of multiple-agent infections. Current understanding of mixed infections is biased and is targeted on the culturable or presumed disease-causing agents. The laboratory investigation of disease is usually directed to correlate the clinical symptoms with a particular pathogen, with the aim of establishing that agent as the etiology. In reality, the disease could be associated with multiple agents. Therefore, the clinical implication, diagnosis, and therapeutic management of such viral infections are of considerable importance. Unlike bacteria, where individual organisms can be rapidly purified from a mixed culture by colony purification, multiple viruses cannot be easily purified directly from clinical specimens. For virus isolation, the clinical specimens need to be detected in an appropriate host; this approach permits amplification of the divergent viruses present in the clinical specimens. Unfortunately, divergent viruses in a specimen may block replication of the target virus (viral interference) and hence result in a misdiagnosis. Classically, the detection of the coinfection has been based on serology and virus isolation, both of which may be compromised by inadequate sensitivity and specificity. The advent of PCR in the 1990s enhanced the specificity and sensitivity of coinfection detection, but because PCR amplification needs prior sequence information on the target genome, PCR encounters problems when amplifying for divergent viruses from clinical specimens. Next-generation sequencing (NGS) platforms have completely revolutionized virus diagnostics and novel virus discovery. NGS does not need prior sequence information about the target genome and allows detection of most potential genomes present in the clinical specimens, and therefore it is considered highly effective for the detection of multiple agents (16–19). However, isolating multiple viruses in a purified form is cumbersome and is rarely achieved. Viruses have variable host range/tropism. Consequently, in a particular cell type, one virus usually replicates faster, eventually resulting in the elimination of other coinfecting viruses upon long-term culture.
Virus Isolation
Compared to the use of embryonated eggs and laboratory animals, employment of cell culture in laboratories in the 1960s provided a less expensive and more convenient tool for virus isolation. Besides the diagnostic utility, virus isolation is essential for product development (vaccines and diagnostic agents) and is also crucial for clinical decisions such as discriminating disease from subclinical infections (20) and deciding when to implement, change, continue, or discontinue drug therapy (21). Isolating multiple viruses in a purified form represents a major bottleneck in cases of coinfections. The presence of a viral genome or antigen in a clinical specimen does not always warrant virus isolation (22, 23). During cell culture adaptation of a virus (virus isolation), several blind passages are usually required before appearance of cytopathic effects (CPE) (24). It is likely that due to a difference in the rates of replication or due to viral interference, one of the viruses will be eliminated before appearance of CPE. If the culture conditions are more permissible for the adventitious virus, it is likely that it will exclude the targeted agent on high passage, thereby resulting in failure of the targeted isolation of a known virus. Even under conditions where both the coinfecting viruses are able to persist until the appearance of CPE, it is not mandatory that both of them will participate in the formation of CPE (14). However, in such instances, at least one of the viruses can be purified by plaque assay (14). Moreover, we have witnessed conditions where despite formation of CPE (in mixed culture), none of the coinfecting viruses formed plaques (14), though subsequent higher passage of the mixed culture allowed plaque formation by one of the viruses (14).
Depending on the nature of coinfecting viruses, strategies for virus purification from mixed culture vary (Table 1) and may include (i) elimination of the enveloped viruses by treatment with the organic solvents (25), (ii) hemagglutination to separate a hemagglutinating virus, (iii) endpoint dilution assay to purify multiple agents, (iv) antibody (Ab) neutralization to eliminate other confecting viruses (26), (v) acid/alkali treatment if one of the viruses is more susceptible to extreme pH, (vi) plaque assay to purify single or multiple viruses, and (vii) transfection of the viral RNA mixture into target cells, which allows amplification (production) of only positive-sense RNA viruses, thereby eliminating negative-sense RNA viruses from the mixed culture (14).
TABLE 1.
Strategies for purification of multiple viruses from mixed culture
| Strategy | Remark |
|---|---|
| Treatment with organic solvents to eliminate enveloped viruses | Unsuccessful if the concn of the organic solvents required for complete inactivation of the virus particles is toxic to the target cells |
| Removal of hemagglutinating viruses | Complete adsorption of hemagglutinating virus is difficult to achieve |
| Plaque assay | Not all viruses form plaques |
| Limiting-dilution assay | Quite cumbersome, as testing so many replicates by PCR is labor-intensive |
| Neutralization with antiserum | Considered an ideal strategy for targeted elimination of a known virus; however, at lower passage levels, virus may not form CPE, and at higher passage, when it starts forming CPE, defective interfering particles may appear that interfere with plaque formation as well as facilitate extinction of standard viral genome |
| Passage in cell types that do not support growth of divergent viruses | Depends on virus(es) and cell types used for coinfection |
| Treatment with acid/alkali | One of the coinfecting viruses may be sensitive toward extreme pH; therefore, it can be eliminated by exposure to extreme pH |
| Viral RNA transfection | Most efficient method for the elimination of RNA viruses with negative-sense genomes |
Complications in Isolation of Multiple Viral Agents
Inability to produce CPE.
Isolation/purification of multiple viral agents from natural infection is quite cumbersome (Table 1). In the beginning of cell culture adaptation, viruses usually do not show cytopathic effects (CPE) (are noncytolytic), and so plaque purification is not feasible. Later, when CPE is evident, all coinfecting viruses may not contribute to CPE formation, thereby allowing purification of only CPE-forming virus. Under such circumstances, antibody neutralization of the cytolytic virus may allow purification of the noncytolytic viruses. However, further blind passages may be required until noncytolytic virus does become cytolytic (14).
DI particles.
Defective interfering (DI) particles are produced following high-multiplicity-of-infection (high-MOI) passage of a virus in cell culture (27, 28). DI particles have a defective or deleted genome, replicate quite rapidly compared to the wild-type (WT) virus, and generally require another helper virus (wild type) for effective replication (29–31). Two defective RNA genomes may also act synergistically to produce cytopathology (32). DI particles may hamper the plaque-forming ability of WT virus (14, 33). The presence of DI particles progressively reduces levels of standard viral genome such that at higher passage levels, the wild-type viral genome may not be detectable by PCR (14, 32, 34, 35). DI particles also produce rapid CPE, and this may prematurely terminate the life cycles of other coinfecting (homo- or heterologous) viruses, eventually resulting in their extinction. However, little is known about direct interaction of DI particle with a heterologous virus.
Rescue of positive-sense RNA virus directly from clinical specimens.
Viruses with positive-sense RNA genomes can generate infectious virus upon delivery of their viral RNA into host cells. This property may be exploited to eliminate negative-sense RNA viruses from mixed cultures. However, in most instances, viral RNA derived only from the cell culture-adapted viruses, but not that from clinical specimens, produces CPE in the established cell lines (14). Transfecting viral RNA (derived from clinical specimens) into primary cells may sometimes show rapid CPE (36, 37), although the reduced amount of viral RNA may require additional passages until CPE becomes observable in primary cells (36). The RNA delivery method, which allows elimination of the DI genome (14), is considered more suitable than antiserum treatment for purification of positive-sense RNA viruses from mixed culture (Table 1).
Improved Virus Isolation
The selection of appropriate body sites and the proper collection, transport, processing, and preservation (freezing conditions) of specimens all contribute to enhance the success of virus isolation. Specimens with large amounts of virus (24, 38) and centrifugation-enhanced inoculation also increase the chances of isolating viruses from clinical specimens (39). A single cell line is not always suitable for isolating multiple viruses, but cocultured and genetically modified cell lines have made it possible to simultaneously isolate multiple viruses.
Cocultured cells.
As a consequence of isolation in cell culture, viruses may undergo genetic changes (40). The success of virus isolation may also depend on the nature of cells used for infection, and a single cell type may not always be appropriate for isolation of multiple viral agents (5, 14, 41). Cocultured cells, where multiple cell types are cultured together in a single monolayer, may solve the problem of isolating multiple viruses (42, 43), and to this end, a variety of mixed cell cultures have been recommended for detection/isolation of multiple viruses. A mixture of MRC-5 and A549 cells is useful to detect cytomegalovirus (CMV), herpes simplex virus (HSV), and adenovirus in the same specimen and can be as sensitive as immunofluorescence or isolation in a single cell type (43). Similarly, a coculture of mink lung and human adenocarcinoma cells (R-Mix cells) is useful for the rapid isolation of respiratory viruses (parainfluenza virus types 1, 2, and 3, influenza A and B viruses, rouse sarcoma virus [RSV], adenovirus, HSV, CMV and enteroviruses) (44–48). R-Mix cells also facilitate the isolation of highly pathogenic respiratory viruses such as severe acute respiratory syndrome coronavirus (SARS-CoV), which cannot be grown without a containment laboratory. Therefore, there might be a risk associated with use of R-Mix cells for virus isolation. An alternative approach being used is the R-MixToo cell line (consisting of MDCK and A549 cells), which does not support SARS-CoV infection (49) and is more sensitive than R-Mix cells for detection of influenza B viruses and adenovirus (50). Both R-Mix and R-MixToo cells facilitate growth of diverse strains of influenza viruses (51, 52) and provide a faster and sensitive cell culture system for isolation of respiratory viral agents. The times needed for positive cultures are 1.4 and 5.2 days, respectively, for R-Mix and single culture (46, 52). Additionally, a mixture of MRC-5 and CV1 cells facilitates multiplex detection of HSV-1, HSV-2, and varicella-zoster virus (VZV) (53, 54). The CPE formed in these cocultured cell lines is as sensitive as fluorescence-based assays (54). Finally, Vero/BHK-21 cocultured cells are adequate for concurrent isolation of peste des petits ruminants virus (PPRV) and foot-and-mouth disease virus (FMDV) (14). These cocultured cell lines are also quite sensitive for the detection/isolation of viral agents with a very low virus titer and those which grow slowly (42). However, their cost is usually higher than that of a single-cell culture (42).
Transgenic cell lines.
Some genetically engineered cell lines (transgenic cell lines) have been developed to enhance the efficiency of virus detection (41, 55–58). A genetically modified cell line named BHKICP6lacZ-5 (enzyme-linked virus-inducible system [trade name ELVIS]; Diagnostics Hybrids, Inc.) which uses an HSV promoter sequence (UL39 gene) in association with Escherichia coli lacZ was developed. Within a few hours of HSV-1/HSV-2 infection, virus-associated transactivators strongly activate the promoter (55) to induce β-galactosidase that can be detected with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (a chromogenic substrate) (55). Whereas single-cell systems detect virus (CPE) in 48 h, BHKICP6 transgenic cell lines can detect virus within 16 to 24 h (59). The original ELVIS approach detected only HSV, but it has now been modified to distinguish HSV-1 and HSV-2 (42, 60, 61) and is less expensive but less specific than PCR (42).
The field needs a cell line system to detect multiple enterovirus strains. Human embryonic lung fibroblasts and primary monkey kidney, A549, and BGMK cells are generally used for enterovirus isolation, and these produce CPE within 5 days (62). Compared to use of a single cell type, coculturing these cells has enhanced the possibility of virus isolation (63, 64). Compared with wild-type BGMK cells, BGMK-hDAF, a genetically engineered cell line expressing human decay-accelerating factor (hDAF) and with an expanded host range, can enhance enterovirus detection (64, 65). The sensitivity of these cell lines was further increased by coculturing BGMK-hDAF with CaCo-2 (BGMK-hDAF/CaCo-2 [marketed as Super E-Mix cell; Diagnostic Hybrid Inc.]) (65).
Hemadsorption.
Hemadsorption is useful approach to detect viruses which produce slow or no CPE in cultured cells (5, 42). Hemadsorption is applicable to those viruses that express hemagglutinin proteins on the plasma membrane of infected cells. Examples include members of the family Orthomyxoviridae and Paramyxoviridae. Hemagglutination testing is usually performed in virus-infected cells by replacing cell culture medium with a suspension of red blood cells. Hemadsorbing foci can be seen as early as 12 h following infection with influenza A and B viruses (39).
Nucleic Acid-Based Tests
Multiplex PCR.
PCR directly targets viral genomes and is more specific than enzyme immunoassays. However, PCR is labor-intensive and expensive, particularly for the detection of multiple viral agents. Quantitative real-time PCR (qRT-PCR) for concurrent detection of heterogeneous viruses in a single reaction has reduced the overall cost (66–81). Although the qRT-PCR system is quite sensitive, adsorption and fluorescence spectra for different fluorophores used in the fluorescent-labeled probe systems tend to interfere with each other, which limits reliable detection to a maximum 4 or 5 different viruses (82).
Chemiluminescence and magnetic separation.
Detection of a limited number of fluorophores, which is a drawback of qRT-PCR, may be overcome by employing a chemiluminescent label-based assay (83, 84). Optical labels such as colorimetric nanoparticles (85–87), fluorescent tags (88), and chemiluminescent labels (89) are increasingly being used for DNA hybridization assays. However, due to simple instrumentation, increased sensitivity, and low background, chemiluminescent label-based techniques are preferred over fluorescence-based detection (90, 91). Because of their easy manipulation under an external magnetic field, surface-modified magnetic particles can be used for enrichment of the target molecules, and this permits high-throughput and automated detection platforms (92–95). Based on these advancements, Ali et al. combined magnetic separation technology (for nucleic acid purification) with a chemiluminescence technique for more sensitive (as low as 10 viral RNA copies) detection of multiple viral agents (82). The technique involved simultaneous extraction of the viral nucleic acid and amplification of the viral genomes in a single tube by qRT-PCR (with biotin-11-dUTP being incorporated into the amplified products during amplification). This was followed by capture of the virus-specific gene segments by different amino-modified probes attached with carboxyl-coated magnetic nanoparticles (82).
NGS.
Despite the availability of a wide range of sensitive and specific diagnostic assays, profiling of microbial species has not been possible. Microarray-based methods, such as ViroChip (96–100) and PathChip (101, 102), that allowed detection of multiple agents but did not support detection of a divergent virus were developed. Sequence-independent amplification techniques (next-generation sequencing [NGS] platforms) have been successfully employed for rapid detection of novel (16–19) and multiple (103) viruses in clinical settings (104–106) and have allowed whole-virus genome organization (107) and analyses of minority variants (108, 109). NGS detects sequences from almost all potential organisms in an unbiased manner. It also allows concurrent genetic characterization of diverse groups of known viruses as well as divergent viruses that evade conventional testing (110, 111). For example, transcriptome analyses of >220 invertebrates identified 1,445 RNA viruses, including those that represented new virus families (112).
Conventional NGS systems omit detection of single-stranded DNA (ssDNA) viruses, although modified library preparation has now made it possible to amplify/detect ssDNA viruses (113–117). The main disadvantage of NGS is the high cost and unsuitability for high-throughput application to detect viruses in multiple clinical samples. Moreover, guidelines that allow interpretation of viral sequences with clinical relevance are lacking (103). In a given clinical specimen, NGS reveals both viral and cellular sequences, but patient privacy must be maintained before transmitting the data from research into clinic settings (103). Nevertheless, the cost of NGS is sharply declining, and in the future it may be competitive with current diagnostic assays (107).
ViroCap (probe enrichment).
NGS often fails to detect viruses detectable by PCR (118) and may not produce sufficient data for comprehensive analysis of the viral genomes, particularly in specimens that contain minimal virus. Several strategies can be used to increase the virus-specific sequence reads. For example, low-speed centrifugation and filtration to remove host/bacterial cells, treatment with nucleases to remove free nucleic acids (not encapsidated by virus), and ultracentrifugation to increase the concentration of virus particles improve the approach. However, employment of these enrichment strategies is not sufficient to capture all viral sequences present in clinical specimens.
ViroCap is a test system developed recently (119) that is based on a targeted sequence panel to enrich viral genomes and includes 190 viral genera and 337 species. To define a unique set of reference sequences, ∼1 billion bp of annotated viral genome sequences was reduced to <200 million bp of targets. This probe enrichment process involves hybridization of DNA/RNA probes to the cDNA fragments in a shotgun library. This is followed by 10 to 15 cycles of PCR prior to sequencing. Besides comprehensively detecting most vertebrate viruses, this system can detect divergent viruses having low sequence similarity (∼50%) to the known vertebrate viruses (119). Compared to NGS, ViroCap increases virus detection by >50%. Because the targeted sequence enrichment increases the percentage of virus-associated sequence reads, it yields better viral coverage and needs fewer of total sequence reads. ViroCap has the potential to reduce sequencing cost and is flexible, since new viral sequences may be periodically added to increase representation of viruses in the shotgun library. However, ViroCap is incapable of detecting novel viruses (that do not share any nucleotide sequence similarity with known viruses), and the technology is still in the validation phase. It may take a few years until it is available for clinical use.
Heteroduplex mobility analysis (HMA).
If multiple strains/subtypes of a virus are present in a clinical specimen, PCR amplification results in two heterologous double-stranded DNA products of similar size. When these heterologous DNA fragments are denatured and allowed to anneal, they form homo- and heteroduplexes, which are derived from identical and nonidentical strains, respectively. The formation of these homo- and heteroduplexes (nucleotide mismatches) results in altered migration in agarose gel electrophoresis. This method has been utilized to illustrate divergent sequences present in torque teno virus (TTV) and hepatitis C virus (HCV) (120). Likewise, amino acid alterations in cytopathic and noncytopathic form of bovine viral diarrhea virus (BVDV) could be analyzed by distinct polypeptide profiles in virus-infected cells (121).
Multicolor Imaging with Self-Assembled Quantum Dot Probes
Multicolor quantum dot (QD) probes allow simultaneous detection and evaluation of coinfection of a cell by multiple viral agents. The process involves conjugation of quantum dot probes with Staphylococcus aureus protein A (SpA) and virus-specific antibodies (Abs). The application of a cocktail of multicolored QD-SpA-Ab probes to coinfected cells generates multiple fluorescence. This method has allowed simultaneous detection of influenza A virus (IAV) subtypes H1N1, H3N2, and H9N2 and human adenovirus in coinfected cells (122).
Laboratory Viral Stocks Contaminated with Unknown Viruses
Unlike for bacteria, where mixed cultures can be rapidly purified by plating on agar, virus purification from mixed culture remains a challenge. Whereas some of the viruses may be plaque purified, those which do not form CPE are cumbersome to purify. The clinical specimens may also contain cryptic viral agents. If the cell line is equally susceptible and the life cycle of the cryptic agent is shorter, the target virus is likely to be eliminated (viral interference) after few passages, even before its adaptation (CPE formation) in the cell culture system. Such divergent viruses may also be acquired accidently during in vitro propagation of the clinical specimens, although their presence is difficult to realize unless examined. Our laboratory is part of a culture collection center (repository). We faced such a problem when a parvovirus isolate came to our repository for deposition. We authenticated the virus deposit by observing CPE in MDCK cells and amplification of parvovirus-specific genome by PCR, and thereafter an accession number was assigned. Four years later, the virus isolate was distributed to another laboratory, where it was grown in A72 cells. After a few passages, the culture was found to be negative for the parvovirus genome. Upon further investigation, it was found to be positive for canine adenovirus. When the original virus stock which came to us for deposition was examined, it was found to be positive for both parvovirus and adenovirus, suggesting coinfection of these viruses in the original culture. The A72 cells favored the growth of adenovirus over parvovirus, and the latter was eventually eliminated. It is not possible to detect such divergent (unknown) viruses by virus species-specific assays, although NGS has made it possible to detect most potential genomes (pathogen/host) in clinical specimens (16, 123).
VIROLOGICAL OUTCOMES OF COINFECTIONS
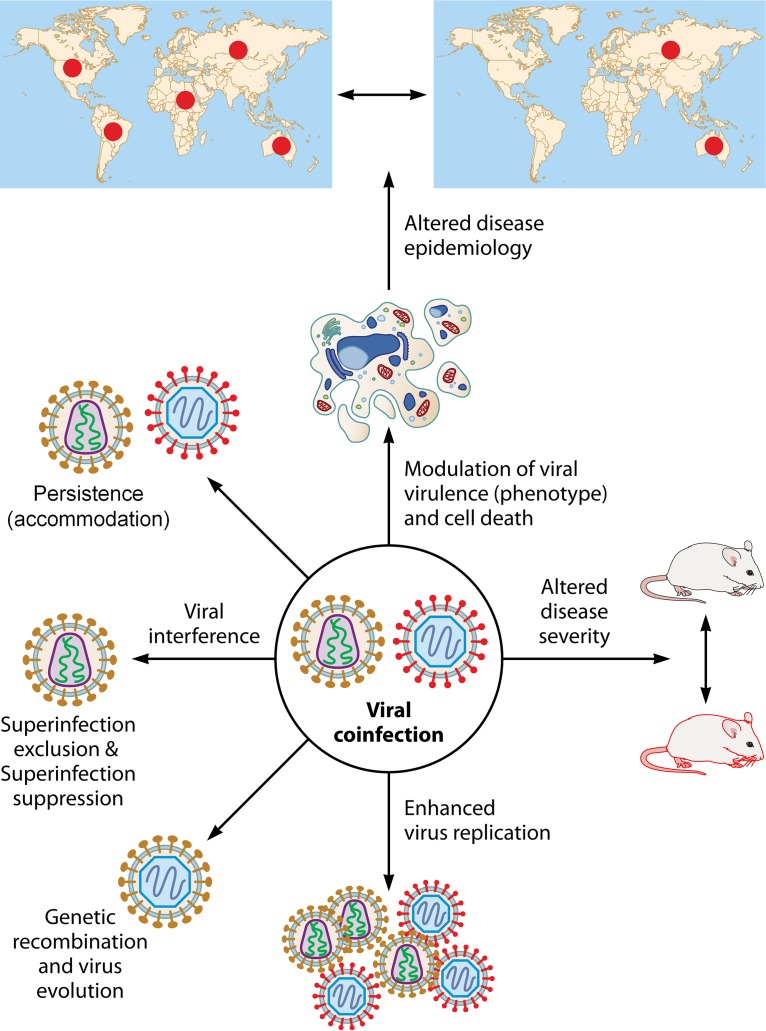
Coinfections are increasingly being reported (Table 2). However, little is known about their effect on other coinfecting agents and the host. The most common outcome of coinfection is viral interference, where one virus competitively suppresses replication of the other confecting viruses. Besides interference, coinfections of certain viruses may also promote an increase in viral replication. In several other cases, coinfections have no effect on virus replication, and thus all the coinfecting viruses can coexist (accommodation). Coinfections are generally believed to exert a negative effect on health (124). They may modulate viral virulence and cell death, thereby altering disease severity and epidemiology. Establishing the outcome of coinfections requires integrated monitoring and research on multiple pathogens. However, there is a dearth of such data.
TABLE 2.
Viral coinfections, detection, and outcomesa
| Confecting viruses | Outcome | Method(s) of detection | Remark(s) | Reference(s) |
|---|---|---|---|---|
| DNV and CHIKV | Accommodation | Nucleic acid | CHIKV neither triggered nor suppressed DNV replication; mosquitoes with DNV infection were equally susceptible to infection by CHIKV | 145 |
| DNV and DENV | Interference/enhancement | Nucleic acid | Reduced DENV replication concomitant with increased DNV replication | 146, 147 |
| DNV and DENV | Accommodation | Nucleic acid | Persistent DENV and DNV coinfection | 148 |
| DNV, DENV, and JEV | Accommodation | Immunofluorescence | Stable infection of all three viruses without any CPE | 149 |
| CHIKV and JEV | NA | Antibody | Prevalence of antibodies against dual infection | 150 |
| IBV and avian pneumovirus | Interference | Viral titers, nucleic acid, antibody | IBV interfered with replication of pneumovirus vaccine strain in fowl | 151 |
| IBV and NDV | Interference | Viral titers, nucleic acid | IBV interfered with NDV replication | 152–162 |
| Sylvatic and endemic DENV strains | Interference | Viral titers | Primary virus suppressed secondary virus infection | 141 |
| NDV and HPAIV | Interference | Viral titers | NDV blocked HPAIV replication | 163 |
| WMV and ZYMV | Interference | Nucleic acid | ZYMV inhibited WMV replication | 164 |
| Henipavirus and rubulavirus | NA | Nucleic acid | Evidence of dual virus infection in bats | 165 |
| SINV and LACV | Interference | Viral titers | Replication of both viruses suppressed | 166 |
| SINV and LACV | Interference | Viral titers | LACV titers suppressed but no effect on SINV titers if SINV infection was 2 h before LACV infection | 166 |
| DENV2 and DENV4 | Interference | Viral titers | Suppression of both viruses but greater suppression of DENV2 | 142 |
| SINV, SFV and SINV, RRV | Exclusion | Viral titers | Persistently SINV-infected cells excluded superinfecting heterologous alphaviruses | 167 |
| SINV and YFV | Accommodation | Viral titers | Persistently SINV-infected cells did not impair YFV replication | 167 |
| CxFV and WNV | Enhancement | Viral titers/nucleic acid | Enhanced WNV transmission in mosquitoes | 168 |
| CxFV and WNV | Accommodation | Viral titers/nucleic acid | CxFV had no impact on WNV replication | 168 |
| IPNV and VHSV | Interference/accommodation | mRNA | Accommodation on coinfection and primary VHSV and secondary IPNV infection but interference on primary IPNV and secondary VHSV infection | 169 |
| NDV and LPAIV | Interference | Viral titers | Coinfection decreased LPAIV shedding and transmission but had no impact on clinical signs. | 170 |
| NDV and HPAIV | Interference | Viral titers | Coinfected ducks survived for shorter duration | 170 |
| HSV and VZV | Exclusion | Immunofluorescence | Exclusion of each other | 171 |
| BHV-1 gD and HSV-1/BHV-1/PRV | Interference | Viral titers | Expression of BHV-1 gD inhibited HSV-1, PRV, and BHV-1 replication | 172 |
| TTSuV1a and ASFV | NA | Nucleic acid | NA | 173 |
| hMPV and hRSV | NA | NA | Increased hospitalization rates in humans | 174 |
| HCV and TTV | NA | Nucleic acid (HMA) | NA | 120 |
| Fowlpox virus and ILTV | NA | Viral titers/nucleic acid | NA | 175 |
| WSSV and IIHNV | NA | Nucleic acid/histopathology | Increased mortality in Pacific white shrimp | 176 |
| Influenza A/H1N1 and A/H3N2 viruses | NA | Nucleotide sequencing | Demonstrated ability of these two influenza virus subtypes to coinfect humans and a potential risk of influenza virus reassortment | 177 |
| CIAV and IBDV | Interference/enhancement | Nucleic acid/cytofluorometric analysis | Enhanced CIAV titers in bursa and thymus but diminished IBDV-induced lymphocyte disorder | 178 |
| Cytopathic and noncytopathic BVDV | NA | Radioimmunoprecipitation/polypeptide profile | Induced different polypeptide profiles | 121 |
| Multiple coronaviruses (BtCoV HKU2, BtCoV HKU8, BtCoV HKU1, BtCoV HKU7, BtCoV HKU10) | NA | Nucleotide sequencing | Evidence of multiple coronavirus infections in bats (zoonoses) | 179 |
| SINV and DENV4 | Interference | Viral titers/nucleotide sequencing | SINV infection resisted DENV infection | 180 |
| ZIKV, CHIKV, and DENV | NA | Nucleic acid/antibody | Cross-reactivated antibodies against ZIKV/DENV may lead to misleading serological conclusions | 181 |
| CHIKV and DENV | NA | Nucleic acid/antibody | CHIKV and DNV were successfully purified by plaque purification and antibody neutralization, respectively | 26, 182–187 |
| DENV3 and CHIKV | Interference | Cell culture/nucleic acid | Interference depended on virus dose | 143 |
| DENV and CHIKV | NA | Antibody | NA | 188 |
| DENV1 and DENV3 | Interference | Antibody | Higher DENV3 prevalence | 189 |
| GPV and ORFV | NA | Nucleic acid | NA | 190 |
| Human adenovirus, human enterovirus, RSV, and human rhinovirus | NA | Nucleic acid | NA | 191 |
| HIV-1 and influenza virus | Enhancement | NA | High risk of influenza infection in HIV-1-infected individuals | 192 |
| PCV2 and CSFV | NA | NA | Induction of stress response and apoptotic signaling pathways | 193 |
| DENV and HIV-1 | NA | Nucleic acid | NA | 194 |
| DENV, CHIKV, and ZIKV | NA | Nucleic acid | Concurrent circulation of DENV, CHIKV, and ZIKV and their coinfection | 195 |
| PPRV and FMDV | Interference | Viral titers/nucleic acid | Reciprocal replicative suppression in BHK-21 and Vero cells, respectively | 14 |
| Aura virus, SFV, RRV, and flaviviruses | Interference | Viral titers/nucleic acid | Insect cells persistently infected with SINV resisted infection of both homologous (SINV) and heterologous (Aura virus, SFV, and RRV) alphaviruses but had no effect on flaviviruses | 167 |
| IAV and RSV | Interference | Viral titers/immunofluorescence | IAV competitively suppressed RSV at the level of viral protein synthesis and budding | 196 |
| IAV and hPIV2 | Enhancement | Viral titers/immunofluorescence | hPIV2 facilitated IAV replication | 197 |
| Wild-type and oseltamivir-resistant IAV strains | Interference | Viral titers/nucleotide sequencing | H3N2 and H1N1 differed in their ability to suppress replication and in transmissibility of the respective drug-resistant viral mutants | 198 |
| Swine influenza virus and PRRV | Interference | Viral titers/nucleic acid | Primary virus infection interfered with replication of the secondary virus | 199 |
| Group 1 and group 2 Brazilian vaccinia viruses | Interference/enhancement | Viral titers/nucleic acid | Higher titers in lungs, lower titers in spleen; greater disease severity in mice | 200 |
| HBV and HCV | Coexistence | Viral titers/nucleic acid | Coexistence in Huh-7 cells without any interference | 201 |
| WNV (different strains) | Interference | Viral titers | Block in transmission of superinfecting virus | 202 |
| WNV. SLEV | Interference | Viral titers/nucleic acid | Block in transmission of superinfecting virus | 203 |
| HBV and HCV | Enhanced disease severity | Antibody | HBV-exposed individuals experienced enhanced HCV-associated disease severity | 204 |
Abbreviations: ASFV, African swine fever virus; BHV, bovine herpesvirus; BVDV, bovine viral diarrhea virus; CHIKV, chikungunya virus; CIAV, chicken infectious anemia virus; CSFV, classical swine fever virus; CxFV, culex flavivirus; DENV, dengue virus; DNV, densovirus; FMDV, foot-and-mouth disease virus; GPV, goatpox virus; HCV, hepatitis virus; HDA, heteroduplex mobility analysis; HIV-1, human immunodeficiency virus type 1; HMA, heteroduplex mobility analysis; hMPV, human metapneumovirus; HPAIV, highly pathogenic avian influenza virus; hRSV, human respiratory syncytial virus; HPIV, human parainfluenza virus; HSV, herpes simplex virus; IBDV, infectious bursal disease virus; IBV, infectious bronchitis virus; IIHNV, infectious hypodermal and hematopoietic necrosis virus; ILTV, infectious laryngotracheitis virus; IAV, influenza A virus; IPNV, infectious pancreatic necrosis virus; JEV, Japanese encephalitis virus; LACV, La Crosse virus; LPAIV, low-pathogenic avian influenza virus; NDV, Newcastle disease virus; ORFV, Orf virus; PCV, porcine circo virus; PIV, parainfluenza virus; PPRV, peste des petits ruminants virus; PRV, pseudorabies virus; RRV, Ross River virus; RSV, respiratory syncytial virus; SFV, Semliki Forest virus; SINV, Sindbis virus; SLEV, St. Louis encephalitis virus; TTSuV1a, torque teno sus virus strain 1; TTV, TT virus; VHSV, viral hemorrhagic septicemia virus; VZV, varicella-zoster virus; WMV, watermelon mosaic virus; WNV, West Nile virus; WSSV, white spot syndrome virus; YFV, yellow fever virus; ZIKV, Zika virus; ZYMV, zucchini yellow mosaic virus; NA, not applicable.
Viral Interference (Competitive Suppression)
A phenomenon whereby one virus interferes with the replication of other viruses so as to become resistant towards a second superinfecting virus is termed viral interference (1). Innate viral interference mediated via interferons (IFNs) is the most common form of viral interference (125, 126). Upon binding with their cognate receptors, IFNs induce multiple so-called interferon-stimulated genes (ISGs), many of which activate numerous cell signaling pathways (127–137). These ISGs regulate the activity of numerous innate immune mediators that nonspecifically block virus replication.
Non-interferon-mediated viral interference, also called intrinsic interference, is a virus-induced cellular state of resistance to subsequent viral infection. Initially it was observed in Newcastle disease virus (NDV) superinfection where the refractory state against NDV emerged exclusively in cells that experienced prior viral infection. The effect was due to molecules encoded by the virus (viral genome/proteins) and not to the intrinsic capacity of cells (138). Later this was also observed in FMDV, where the attenuated A24 Cruzeiro strain interfered with the multiplication of a homologous wild-type strain as well as that of heterologous wild-type strains. The interference occurred intracellularly without any role of DI particles or interferons and was directed exclusively against FMDV (139). Intrinsic interference may occur between similar, closely related, or unrelated viruses (140–144) (Table 2).
In non-interferon-mediated viral interference, competition between two viruses exists for the metabolites, replication sites (205), or those host factors that support virus replication (148, 167, 206–217). One virus modulates the host machinery in its favor, thereby interfering with the replication of other coinfecting viruses. A requirement for common cellular factors for unrelated viruses indicates that heterologous viral interference can also occur (1). Besides competition for cellular factors, several other mediators of viral interference are also known. These include DI particles (218), RNA interference (RNAi) (219–223), trans-acting viral proteins (224–226), and nonspecific dsRNA (227, 228), and these are listed in Table 3.
TABLE 3.
Mediators of viral interference
| Mediator(s) | Remark(s) or virus(es) involved | Reference(s) |
|---|---|---|
| Defective interfering particles | FMDV | 14 |
| trans-Acting proteases | Primary virus (SINV) nonstructural protein (NSP2) rapidly degraded uncleaved P123 protein of superinfecting virus | 224–226 |
| Interference due to individual viral proteins | ||
| BHV-1 | Expression of BHV-1 glycoprotein D in MDCK cells interfered with replication of BHV-1, pseudorabies virus, and HSV-1 | 172 |
| Poxviruses | Heterodimers formed by viral A56 and K2 proteins at the cell surface resisted superinfection | 229, 230 |
| WNV | Long-term incubation of superinfecting virus with primary virus-containing cells generated variant viruses that could overcome superinfection exclusion | 231 |
| Competition for cellular factors | DENV2 and DENV4 coinfection of mosquito cells resulted in reduced replication of both viral strains | 142 |
| Nonspecific dsRNA | Administration of both sequence-specific and non-sequence-specific dsRNA in bees resulted in lower viral titers; treatment with nonspecific dsRNA in adult bees resulted in enhanced survival following deformed wing virus infection | 227, 228 |
| RNAi | ||
| DENV | DENV NS4B protein exerted a suppressive effect on RNAi response | 232 |
| FHV | FHV B2 protein prevented Dicer-2-mediated cleavage of long dsRNA as well as loading of siRNA into RISC | 233, 234 |
| Dicistro viruses | Encode protein 1A, which interacts with Dicer-2 or AGO2 | 235 |
| Multiple flavivirus infection in insects | Generate cDNAs from the defective genome that are eventually transcribed by host transcription machinery to produce small dsRNAs, the source that induces the Dicer-2/RISC apparatus (RNAi pathway) that eventually regulates virus replication | 236, 237 |
| Interference by temp-sensitive mutants | Viral mutants that acquire dominance over wild-type virus | 238–245 |
Viral interference usually occurs at specific steps of the virus replication cycle. These include attachment (144, 246–259), entry (217, 260–263), genome replication (167, 217, 231, 264–268), and budding (269). However, the infection may also invoke inhibition of multiple steps. For example, infection with recombinant Semliki Forest virus (SFV) inhibited attachment, entry, and uncoating in the subsequent secondary infection (217).
Superinfection exclusion.
Superinfection exclusion is a phenomenon by which an established viral infection interferes with a second, closely related virus infection (159). This phenomenon occurs in both plant and animal viruses (270–272) and has important consequences for virus replication, pathogenesis, and evolution. It affects genome diversification, antiviral drug resistance, and evasion of vaccine-mediated immune responses. The members of a particular virus family may differ in their ability to exclude a superinfecting virus (217, 231, 267, 273–276) (Table 4). For example, infection with the Old World arenavirus results in downregulation of its receptors (α-dystroglycan) and thus induces resistance to superinfection. To distinguish coinfecting viruses at the level of transcription and translation, Gaudin and Kirchausen (286) developed a dual-reporter assay. They observed that, in contrast to infection with the Old World arenaviruses, infection with New World arenavirus (Junin virus [JUNV]) in A549 and Vero cells did not downregulate transferring receptors, and thus the cells were unable to resist superinfection. In contrast, persistently infected (with JUNV) cells did exclude superinfecting homologous or antigenically related arenavirus (277). Likewise, Env-, Vpu-, and Nef-mediated downregulation of the CD4 receptor resulted in HIV-1 superinfection exclusion (278). In vivo evidence of superinfection exclusion is rare (279–281). Examples include pigs persistently infected with classical swine fever virus (CSFV), which exclude vaccine strains upon immunization (282). Moreover, persistently infected pigs may also exclude highly virulent CSFV upon challenge infection (279). Viral and cellular factors that mediate superinfection exclusion in diverse groups of viruses are summarized in Table 4.
TABLE 4.
Superinfection exclusion
| Virus | Mechanism of exclusion | Reference(s) |
|---|---|---|
| Bovine viral diarrhea virus | Primary virus blocked entry and RNA synthesis of the superinfecting virus | 283 |
| Hepatitis C virus | Presence of primary virus RNA/proteins resisted superinfection | 266, 275 |
| Rubella virus | Exclusion occurred after entry but before accumulation of detectable amt of viral RNA | 267 |
| Semliki Forest virus | Superinfection exclusion occurs at the level of binding and endosomal fusion | 217 |
| Sindbis virus | Superinfection exclusion is mediated via viral nonstructural proteins (proteases) | 167, 284 |
| Measles virus | Superinfection exclusion is mediated via downregulation of CD46 (cellular receptor) | 255, 285 |
| Borna disease virus | Selective inhibition of polymerase activity of incoming viruses | 264 |
| HIV-1 | HIV-1 Nef interferes with the superinfecting virus at the level of viral entry by downregulating CCR5, the major HIV-1 coreceptors | 248, 254 |
| Vaccinia virus | Superinfecting virus is unable to carry out its DNA synthesis and early gene transcription | 271 |
| West Nile virus | Competition for the cellular factors that are required for synthesis of the viral genome | 231 |
| Junin virus | No superinfection exclusion observed (virus failed to downregulate transferring receptor and thus was unable to resist superinfection) | 286 |
| Persistently infected K3 cells excluded homologous arenavirus and antigenically related Tacaribe virus with an unknown mechanism | 287 | |
| Primary virus blocked protein synthesis by the superinfecting virus | 277 | |
| Classical swine fever virus | Dysregulation of innate immune response with an unknown mechanism | 279 |
| Influenza A virus | Expression of neuraminidase by primary virus blocked secondary virus attachment to the host cells | 288 |
| Vaccinia virus | Primary virus infection blocked fusion of the viral (secondary virus) and cellular membranes | 229 |
| Expression of A33 and A36 proteins in infected cells pushes the superinfecting virus particles on actin tails toward neighboring cells. | 289 | |
| Primary virus gene products inhibit early gene expression by the superinfecting virus | 271, 290–292 | |
| Heterodimer formed by the primary viral proteins (A56 and K2) at the cell surface inhibited secondary virus entry | 293–296 | |
| Alphaherpesviruses (HSV, PRV, and EHV) | Glycoprotein D-mediated receptor interference | 172, 297–299 |
| Alphaherpesviruses (HSV and PRV) | Expression of immediate early viral genes (ICP0, ICP4, ICP22, and ICP27) resists superinfection with unknown mechanisms | 300 |
| PIV3 and NDV | Primary virus hemagglutinin-neuraminidase protein inhibited attachment of the superinfecting virus | 301, 302 |
| Alphabaculoviruses | Primary virus infection inhibited budding of the superinfecting virus | 269 |
| Citrus tristeza virus | Viral protein p33 mediates superinfection exclusion with an unknown mechanism | 272, 273 |
| Deformed wing virus (lethal and nonlethal types) | Lethal infection results in death of honey bees; prior infection with nonlethal DNV resists superinfection by a lethal deformed wing virus | 303 |
Superinfection suppression.
The instance where persistently infected cells withstand challenge of a heterologous virus is termed superinfection suppression. Superinfection suppression has been observed between densovirus (DNV) and dengue virus (DENV). Persistently DNV-infected cells resist DENV challenge (with a reduced rate of DENV-2 infection, decreased DENV-2 production, and reduced mortality) in insect cells (147, 304). However, the superinfection suppression between DNV and DENV, as well as that between infectious hypodermal and hematopoietic necrosis virus or (IHHNV) and white spot syndrome virus (WSSV) (176), should be referred to as superinfection disease suppression because the most prominent outcome is decreased disease severity rather than decreased viral infection (147).
Interference due to vaccination (live-attenuated viruses).
The poliovirus vaccine strain is known to restrict the growth of standard (WT) virus (305). Later, following vaccination, this interference is achieved by stimulating antibody production that restricts the growth of the secondary virus. This evidence was derived primarily from field trials in which large-scale immunization campaigns against polio were found to displace antigenically unrelated enteroviruses (306). In addition, enteroviruses also interfered with poliovirus vaccines and led to vaccine failure (306). A similar phenomenon has been experienced with diverse groups of viruses, such as NDV (307), IAV (308), and DENV (268). However, the interference varies with the cell types and virus prototypes involved. Consequently, understanding viral interference is of utmost importance for the formulation and recommendation of any combination of vaccines (159).
Enhanced Virus Replication
Competitive inhibition is not the only outcome of coinfection (Fig. 1). Compared to single infection, CMV/HSV coinfection results in enhanced virus replication and virulence (309). Likewise, La Crosse virus (LACV) and Sindbis virus (SINV) coinfection in C6/36 cells resulted in enhanced SINV replication (166). In a study by Goto et al., human parainfluenza virus 2 (hPIV2) infection-associated cell fusion facilitated IAV replication and modulated pathological consequences (197). In another study, the simultaneous inoculation of culex flavivirus (CxFV) and West Nile virus (WNV) facilitated WNV transmission (168), although prior infection with CxFV had no effect on WNV replication.
FIG 1.
Virological outcomes of coinfections. Coinfections involving viruses may have several virological consequences. The most common outcome of coinfection is viral interference, where one virus competitively suppresses replication of the other confecting viruses. Interference between closely related viruses eventually results in elimination of the secondary coinfecting virus and is referred to as superinfection exclusion. The instances where persistently infected cells withstand the challenge of a heterologous virus are termed superinfection suppression. Besides diminished viral replication (interference), coinfections with certain viruses may also trigger enhancement of the replication of one or both of the confecting viruses. In several other cases, coinfection has no effect on the virus replication, and thus all the coinfecting viruses can coexist (accommodation). Coinfection may modulate viral virulence and cell death, thereby altering disease severity and epidemiology. However, genetic recombination between coinfecting viruses depends on the similarity between the coinfecting viruses.
Persistence
Contrary to the case in acute lethal infections, where virus particles are eventually cleared either by the immune system or by elimination of the host, persistently infected cells harbor virus for long times without clearance (1), thereby facilitating viral transmission to new hosts (236). Viruses isolated from persistent infections usually impede growth of the standard virus (242, 310–317). Since these viruses have managed to outgrow wild-type virus, they dominate over the parental virus in acute infections (245). Due to the inability to shut off the host cell machinery, persistent viruses have a reduced ability to kill infected cells.
DNV persistently infects mosquito populations, and this serves as a good model to study susceptibility to other viral coinfections in persistently DNV-infected cells (145). Compared to naive cells, persistently DNV-infected cells resist CPE formation upon DENV challenge (146, 147). The molecular mechanism underlying viral persistence is not completely understood. One potential mechanism is the activity of DI particles (318). Studies on flock house virus (FHV) suggest involvement of both host and viral factors in the maintenance of viral persistence (319–321). During establishment of in vitro persistent infection, the FHV genome remains unaltered; the mutations in the viral genome start accumulating after several successive passages (319), suggesting that a modified cellular environment, rather than virus itself, is crucial in establishing persistent infection. Following infection, ongoing virus replication is blocked either by the elimination of infected cell or by a host-directed RNAi response. Studies by Goic et al. suggest that FHV persistence in Drosophila melanogaster cells is accomplished through combined use of the RNAi and reverse transcriptase activity (237). Diverse RNA segments of FHV genome are reverse transcribed by long terminal repeat (LTR) retrotransposons. The resulting DNA molecules integrate with the host genome (322). Alternatively, the viral genome may be maintained as extrachromosomal circular DNA molecules (323). In both the cases, the viral DNA is steadily transcribed and produces dsRNA. These dsRNA structures are eventually sensed by the RNAi machinery to block viral replication. Blocking of reverse transcription prevents the emergence of chimeric DNA, hence interrupting viral persistence (237).
Active, persistent infection by multiple viruses without any apparent signs of illness is referred to as viral accommodation and is commonly observed in arthropods (148) and shrimp (147, 304, 324, 325). There is little evidence that shrimp or other arthropods possess an immune system (326), but exposure of the shrimp to inactivated virions or envelope proteins can result in short-lived resistance to viral challenge (327). However, persistently infected shrimp only resist viral challenge until they remain infected (328), and there is no system equivalent to immune memory. In shrimp, mortality from viral diseases is an outcome of virus-triggered apoptosis (147, 329–331), and the viral accommodation that prevents triggering of apoptosis is not understood. The phenomenon of viral accommodation suggests that multiple viruses can stably coexist in the same cell and that the possibility of genetic exchange between them depends on the degree of similarity between coinfecting viruses.
Modulation of Cell Death
In retroviruses, viral DNA integration in the host genome is catalyzed by both viral (integrase) and cellular (DNA-dependent protein kinase [DNA-PK]) factors. The initial events during viral DNA integration are sensed as a DNA damage response by the host, and this results in cell death (apoptosis). By promoting aggregation of the unintegrated viral DNA, superinfection exclusion in retroviruses may be employed to prevent cell death (332). In HIV-1, superinfection of primary T cells results in an increased level of apoptosis (274). One potential reason for HIV-1 inhibition of superinfection is to block premature cell death so that the virus may get sufficient time for replication.
Change in Virus Phenotype
Plaque assay is one of the most common methods to quantify virus particles. It is generally believed that a plaque represents a single infectious unit. As such, the number of plaques is believed to have a linear correlation with virus dilutions. However, a recent study has demonstrated that a plaque may contain multiple parental viruses. This possibly occurs due to the formation of virus aggregates, because even at an extremely low MOI, 5 to 7% of the poliovirus plaque population was found to be associated with multiple parental viruses (333). Coinfection with heterologous viruses (separate virus stock) may also result in altered plaque morphology, as seen with IAV and cowpox virus coinfection in BHK-21 cells (334). Likewise, plaques were small and opaque when persistently rubella virus-infected Vero cells were superinfected with another homologous virus (267).
Altered Disease Severity
In most instances, the contribution of coinfection at increasing or decreasing disease severity is difficult to determine. For example, in a PPRV/FMDV dual infection in goats, we noticed ∼50% fatality (14). The fatality rate in PPR-affected sheep or goat flocks varies between 10 and 90% (335). Except in some young animals, FMDV usually does not cause any fatality in sheep and goats (336), so any role of FMDV/PPRV coinfection in fatality in goats could not be determined (14). Several other reports also suggest unaltered disease severity in mixed infections (337–342). Conversely, compared to the case for monoinfection, a higher rate of hospitalization/admission to the intensive care unit has been reported following multiple infections in humans, for example, coinfections with TTV, norovirus, and adenovirus (343), RSV and human metapneumovirus (hMPV) (174), IAV (344), and multiple respiratory viral agents (345). HIV-1-infected individuals, especially those with diminished CD4+ counts, also have a high risk of influenza virus (192) and HCV (346) infection. In contrast, a less severe clinical impact of viral coinfections has also been reported (341, 347).
Hepatitis B virus (HBV) and HCV coinfections are quite common due to their shared mode of transmission. Compared to monoinfection, HBV/HCV coinfection results in more severe fibrosis and cirrhosis as well excess liver-related mortality (348, 349). Moreover, previous HBV infection (based on antibody detection) has also been also shown to significantly enhance the risk of decompensated cirrhosis (204). Clinical examination of HBV/HCV-coinfected patients suggests reciprocal replicative suppression (interference) (350). However, in an in vitro model (Huh-7 cells) of coinfection, both HBV and HCV could propagate in the same cell without any interference (201). Therefore, it was concluded that viral interference observed clinically in HBV/HCV-coinfected patients is mediated via host immune responses.
Experimental studies on viral coinfections are rare. In one study, reovirus and SARS-CoV infection in guinea pigs resulted in rapid death of the animals (351). Another experimental viral coinfection was described for vaccinia viruses (VVs). Based on plaque size and virulence in mice, two distinct groups of vaccinia viruses (group I and group II) that are associated with the exanthematous outbreaks in cattle are known (352, 353). Coinfection of these two vaccinia viruses was reported in a natural outbreak in horses (353, 354). A mouse model of infection demonstrated more severe disease in coinfected (with vaccinia virus subtypes) than in monoinfected mice (200).
Altered Disease Epidemiology
By influencing disease severity and transmissibility and vaccine effectiveness, mixed infections may impact disease epidemiology. For a competition to succeed among multiple viral strains, they must be prevalent in the same geographical region, infect corresponding hosts, and target the same cells within that host. Viruses such as the DENV, with multiple variants and circulation across wide geographic regions, meet all these criteria (355–358). In nature, both humans and vectors (insects) are infected by multiple DENV subtypes (359–362). One major discrepancy between humans and vectors is that in the former, virus is cleared by the immune response, whereas in insects, it may persist for a long time. Therefore, vectors serve as a mixing vessel for any competition to take place between diverse viral strains. Since DENV2 and DENV4 coinfection results in competitive suppression, colonization of new viral strains may be blocked in those areas where mosquitoes are infected with multiple endemic DENV strains (142, 363–365).
Natural coinfection of rhinovirus and influenza virus does occur frequently in humans, but the situation is transitory (366), because rhinovirus negatively affects influenza virus replication (366). However, depending on the nature of the virus prototypes involved, coinfected hosts may shed more transmissible molecules than the singly infected host, and this can result in a higher disease prevalence (367). To comprehensively understand the significance of viral coinfections in epidemiology, further studies in natural populations are needed.
Genetic Recombination and Virus Evolution
Coinfection of a single cell with multiple viral strains allows genetic recombination, a major event driving viral diversity and escape from available antiviral drugs and vaccine-induced immunity (368–370). With segmented viruses, reassortment of the viral segments is a major source for the generation of novel viruses (371–373). The major influenza A pandemics in 1957, 1968, and 2009 all emerged from reassortment of viral segments (374). In influenza virus-infected cells, the efficiency with which a given neuraminidase (NA) removes sialic acid receptors determines reassortment between two or more viruses. However, the addition of an NA inhibitor in virus-infected cells can reduce viral titers and enhance superinfection and hence reassortment events (288).
Novel strains of poliovirus have been identified during early periods of excretion, and these appear to be generated due to recombination between poliovirus types 2 and 3 (375). Similarly, recombination between a persistently infected bovine viral diarrhea virus (BVDV) (noncytopathic form) and a vaccine strain resulted in the formation of a cytopathic form of BVDV. This led to lethal mucosal disease (376). Superinfection exclusion thus prevents the generation of viral diversity and detrimental recombination events, as well as maintaining cellular resources for primary virus infection. However, viruses such as HIV-1 generally replicate in short-lived T cells, and resistance to superinfection, which occurs primarily due to downregulation of CD4 receptors, barely reduces the recombination frequency (274).
Importin-α7, a cellular factor, is critically required for efficient IAV replication. Upon IAV challenge, importin-α7-knockout mice developed more severe disease than wild-type mice. In addition, virus recovered from the challenged mice was more virulent. This might have occurred due to more frequent recombination events and increased probability of superinfection in knockout mice (377). This evidence suggests that host-directed antiviral therapy may also result in the generation of more virulent viral phenotypes and hence should be considered carefully.
The impact of interstrain competition must be quantified in the epidemiological settings, as these may eventually influence long-term virus persistence and emergence of virus variants. It is worth studying these effects in vivo in connection with the host immune system.
Factors Influencing Outcome of Coinfections
Virus dose and the time lag between coinfecting viruses.
The time gap between coinfecting viruses is a major factor which influences viral interference. When wild-type and mutant SFV (SFV-tr) strains were added together at an MOI of 5, all the cells became infected, but if wild-type SFV was added 15 min after SFV-tr, fewer than 30% of cells were infected. Consequently, 15 min was enough to establish interference in most cell types (217). Similarly, instead of coinfection, infection with FMDV at 12 h after PPRV infection induced viral interference (14). In vaccinia virus superinfection, the secondary virus could not replicate at all if it was applied 4 h later (271).
The efficiency of viral interference also depends on the virus dose. Thus, when secondary virus was infected at a 10-fold larger amount (MOI = 50), the resistance to superinfection was overcome in a majority of the cells even when the secondary virus was applied 30 min later (217). At equal multiplicities of initial infection, DENV-3 and chikungunya virus (CHIKV) coinfection resulted in copersistence, with a similar result at higher CHIKV and lower DENV-3 infection levels. However, at lower CHIKV and higher DENV-3 infection levels, DENV suppressed CHIKV replication (143).
Cell types.
A major factor which influences viral interference is the cell type used for coinfection. For example, Vero and BHK 21 are permissive cell lines for PPRV and FMDV, respectively. During PPRV and FMDV coinfection, a reciprocal competitive suppression (interference) occurs in BHK21 and Vero cells, respectively (14). In HIV-1 superinfection, Vpu and Env were found to more significantly affect downmodulation of the CD4+ in peripheral blood mononuclear cells (PBMCs) than in the Jurkat T cells (274). Likewise, CD4+ downmodulation by HIV-1 is more profound in Jurkat T cells than in PBMCs (378), suggesting a role of cell type in viral interference (379). Sperm proteins, human T-lymphotrophic virus (HTLV), Epstein-Barr virus (EBV), and CMV share similarity with the HIV-1 cellular receptor CD4+ present on T helper lymphocytes (380). The binding of HIV-1 to these additional CD4+ homologues on sperm or other coinfecting viruses allows it to infect additional cell types which are not infected normally (380).
Route of infection.
The route of infection also impacts the consequences of viral coinfections. For example, LACV-infected mosquitoes (Aedestriseriatus) remained sensitive to secondary heterologous infection with bunyaviruses if the primary virus was administered transovarially (381). However, when inoculation of the primary virus was by the intrathoracic route, the mosquitoes resisted superinfection (382). In CxFV/WNV coinfection, when the mosquitoes were inoculated by the intrathoracic route, both CxFV and WNV were present in the saliva (168). However, CxFV was not detectable in the saliva of singly infected mosquitoes, suggesting that CxFV infects the salivary glands by “piggybacking” on WNV (168).
Age.
In humans, coinfections are more commonly observed in children than in adults (11). A study carried out in a population ranging from 0 to 105 years suggested that children <5 years showed an increased rate of viral coinfection than older persons (345). In another study, it was observed that the propensity for viral coinfection was greater in children age 6 to 24 months than in infants (0 to 6 months) (347). A study by Zhang et al. (383) demonstrated that among children <3 years of age, the 13- to 24-month age group had relatively higher rates of viral coinfections than the 8-to 12-month or 25- to 36-month age group.
Rate of virus replication and CPE formation.
Cytolytic viruses rapidly deplete cellular resources and induce cell death. If coinfecting viruses significantly vary in their replication cycle length, the one with the shorter life cycle will persist because other coinfecting viruses with longer cycles will be prematurely terminated. For example, in PPRV/NDV coinfection, the relatively long replication cycle of PPRV (∼24 h, compared to 8 h for NDV) led to its removal upon long-term cell culture passage in Vero cells (14).
In buffalopox virus (BPXV) infection, evidence of both peak virus titers and CPE formation is observed at ∼48 h postinfection (hpi) (14). In PPRV infection, although evidence of the new progeny virus particle is observed quite early, at ∼24 hpi (but with very low titers), virus titers progressively increase until 7 days postinfection (dpi), and the CPE cannot be observed until 4 to 5 dpi. Therefore, in BPXV/PPRV coinfection, faster CPE formation by BPXV eliminated PPRV on long-term passage (14) in Vero cells.
IMMUNOLOGY OF VIRAL COINFECTIONS
Animals and humans are exposed to a variety of environmental pathogens and thus have a different infection history than mice grown in a containment (specific-pathogen-free) facility. Upon encountering an antigen following viral infection, naive T cells become activated effector cells, ultimately leading to memory T-cell formation. Memory responses elicited as a result of previously encountered pathogens play a significant role in deciding the type and magnitude of immune responses mounted against the subsequent infections (384). Immune responses to previously experienced pathogens can modify responses made against unrelated pathogens. This is referred to as heterologous immunity, and it can occur between related or unrelated viruses or between viruses or other types of pathogens (2).
The heterologous immune response may provide protective immunity or may lead to immunopathology, depending upon multiple factors. These include the type of viral infection (385), dose of virus (386), stage of infection (387), and, in some circumstances, age of the host (388). Diverse arrays of mechanisms are involved, and almost entire immune cell types are known to participate and modify the outcome of infections. Altered immunodominance hierarchies, a remodeled T-cell receptor (TCR) repertoire, and cross-reactivity are the major changes recorded during heterologous infections. In this section, we discuss current knowledge and recent developments in heterologous immunity related to concurrent as well as sequential infections. Several mechanisms and immune outcomes with specific examples are mentioned, and these are related to virus control measures, prevention of inflammatory consequences, and implications for diagnostic and vaccination strategies.
Immunity or Immunopathology
Certain heterologous viral infections result in protective immunity by employing mechanisms that include innate immune activation (389), bystander protection by activated CD4+ or CD8+ T cells (388), and cross-reactive CD8+ T cells (390). However, in some instances, heterologous infections result in an unaltered immune response, suggesting that the viral coinfection had no significant consequences. Indeed, work with heterologous viral infections such as IAV, murine cytomegalovirus (MCMV), lymphocytic choriomeningitis virus (LCMV), and vaccinia virus (VV) has convincingly demonstrated that IAV-immune mice were protected against VV but not MCMV. In fact, IAV suppressed the clearance of MCMV and LCMV. In addition, the cross talk was not reciprocal, with virus A affecting responses to virus B but not the reverse effect. For example, LCMV-immune mice may resist VV challenge, but VV-immune mice remained fully susceptible to LCMV challenge (391).
Immunopathology is yet another facet of heterologous immunity where viral coinfections culminate in severe and prolonged lesions. Following viral infection, an effector CD8+ T-cell response is generated, which helps in antigen clearance. Upon clearance of the antigen, regulatory responses are induced, which suppress effector responses, thereby preventing collateral damage due to the excessive cytokine production by the activated effector T cells. Thus, the infection is resolved with minimum tissue damage. However, heterologous infections under some circumstances could bring about an uncontrolled immune response and consequent development of immunopathology. This happens with DENV infection and in flu infections both of which involve development of a cytokine storm (392). When infected with MCMV, LCMV-immune mice may exhibit enhanced immunopathology and augmented viral loads, and MCMV-specific memory T-cell inflation was suppressed (390). Similarly, LCMV-immune VV infected mice experienced a more severe outcome (393). The memory T-cell response in IAV-immune LCMV-infected mice enhanced lung immunopathology.
In humans, cross-reactivity in EBV and IAV epitopes (394) resulted in the amplification of the cross-reactive T cells that had subsided affinity for the cells expressing viral antigen and thus were inefficient in clearing infection. Primary DENV infection generates a high-avidity CD8+ T-cell response. Upon a secondary DENV infection, the augmentation of the primary virus-specific CD8+ T cells occurs due to cross-reactivity, rather than the cells specific for secondary infection.
Whatever the mechanism, the particular sequence of infections, time interval between infecting viruses, and route of infection are the decisive factors that determine the outcome (pathological/protective) of heterologous viral infections. The dose of virus may also affect the extent of immunopathology. In a study with LCMV clone 13 infections in a mouse model, a low dose of the virus generated strong effector T-cell responses that efficiently cleared the virus. High viral doses, on the other hand, resulted in T-cell clonal exhaustion, viral persistence, and limited immunopathology. Interestingly, intermediate viral doses could elicit an immune response with a lower rate of exhaustion and provided sufficient time for profound collateral damage to occur in the lung and liver, often resulting in death (386).
Net Outcome of Viral Coinfections
The net outcome depends upon the stage at which the subsequent pathogen is encountered (Fig. 2). For instance, the second incoming pathogen may enter at a stage when prior infection has primed the innate immune responses or at a stage when it encounters a polarized helper T-cell subset. Preexisting primary virus-specific CD4+ and CD8+ T cells are also known in some cases to provide bystander protection against the subsequent pathogen, although the mechanisms involved are not understood (395).
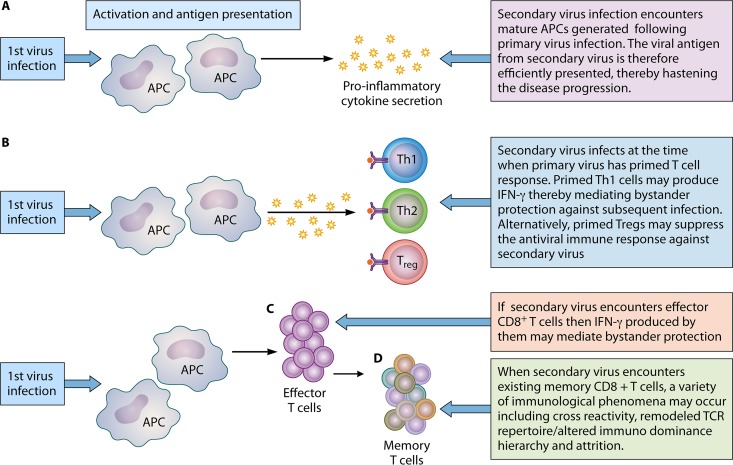
FIG 2.
Immunological outcomes of heterologous viral infections. The immunological outcome is dependent upon the stage at which a subsequent viral pathogen is encountered. (A) Primary virus infection activates APCs. Subsequent infection entering following maturation of APCs culminates in efficient antigen presentation that may hasten the disease progression (immunopathology or protective immune response, depending on the nature of the immune response mounted). (B) Upon encountering antigen, activated APCs secrete cytokines that ultimately influence the type of T-cell differentiation. When the new incoming pathogen encounters already polarized T helper cells, bystander protection is mediated by these polarized Th1 cells. However, encounter with a polarized regulatory T cell can suppress immune responses against new pathogen. (C) Bystander protection from IFN-γ production may also be mediated under conditions where subsequent heterologous infection occurs during an ongoing effector CD8+ T-cell response. (D) When a new virus infects the host with an established memory CD8+ T-cell pool as a result of prior viral infection, the outcome may be cross-reactivity (can be protective or pathological), a remodeled TCR repertoire, or an altered immunodominance hierarchy. Inversely, the incoming pathogen can result in attrition (type 1 IFN dependent) of preexisting memory CD8+ T cells.
Primed innate immune responses.
Innate immunity plays a crucial role in safeguarding against viral infections. Following pathogen recognition by specific receptors, various inflammatory cascades are triggered that eventually results in secretion of cytokines and chemokines, activation of antigen-presenting cells (APCs), and recruitment of innate immune cells. These can, in turn, modulate responses to subsequent viral infection. The extent of this modulation depends largely upon the time gap between the two infecting viruses. Simultaneous coinfection, for instance, may lead to higher viral loads and increased immunopathology. In an alternate situation, primary infection may lead to maturation of the APCs that eventually augment antigen presentation upon subsequent viral infection. For example, in one study, LCMV infection caused stimulation of Kupffer cells, recruitment of T cells and NK cells, and enhanced production of tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), and IFN-α/β. This scenario favored faster elimination of HBV in the coinfected animals (396). Similarly, herpesviruses are known to establish persistent infections and latency. Subsequent secondary infection of the persistently infected host may culminate in an altered immune response. For example, herpesvirus latency is known to enhance the basal activation status of innate immune compartments and thus protective immunity to subsequent secondary infections (389). However, this concept is not generally accepted.
Polarized T helper subset.
CD4+ T cells exert innumerable activities in antiviral immunity (397). These include mediating either a protective or immunopathological role in a setting of heterologous infection. Prior infection of the host may lead to APC activation. These activated APCs undergo cytokine secretion that eventually drives TH subset differentiation, i.e., TH1, TH2, TH17, or regulatory T cells (Treg), depending upon the cytokine milieu contributed by the innate immune cells that respond to the primary viral infection. A preexisting polarized T helper immune response generated against primary virus infection may induce bystander protective immunity (387) or an immunopathological response upon exposure to a subsequent secondary viral infection. Adoptive transfer of CD8+ and CD4+ T-cell subsets from LCMV-immune to naive mice revealed heterologous immunity to Pichinde virus (PICV) and VV in the recipients (398).
An exacerbated immune response to viral infections is controlled by several regulatory mechanisms, which otherwise might result in immunopathology and autoimmune disease. These regulatory mechanisms include the expansion of regulatory T cells (399, 400) and various inhibitory protein-mediated interactions. For example, the Tim3/galectin 9 (401–405), PD1-PDL1 axis (406), and CTLA-4 and CD80/86 (406) interactions strongly influence the outcome.
Regulatory T cells.
FoxP3+ CD4+ T cells (407) play a pivotal role in influencing the amplitude of T-cell responses to viral infections (2). Treg expand during viral infection, with an enhanced suppressive function (400). Such Treg can alter the magnitude and other features of effector T-cell responses and limit immunopathology upon exposure to subsequent heterologous infection. This Treg-mediated impact either may positively influence the outcome or can be detrimental to the host (387). However, the sequel of coinfection may be influenced by numerous other components such as the infection status, dosage of the pathogen, genetic makeup, and immunological condition of the host in addition to the presence of other concurrent infections (408).
Treg that have expanded during primary infection can suppress bystander responses (408) induced by exposure to a subsequent heterologous pathogen. Treg depletion at both the acute and memory stages of an antiviral immune response may lead to enhanced CD8+ T-cell responses (400). Viral infection can thus temporarily dampen immunity to subsequent viral infections. Upon IAV infection, the expanded Treg affect the nature and magnitude of the effector responses, as well as their contribution to lung immunopathology following heterologous infection. In line with this, Treg induced following IAV infection mitigate the T-cell responses following heterosubtypic IAV challenge and thus diminish pathology following heterologous IAV challenge (409).
Treg were also shown to regulate virus clearance and immunopathology in persistent viral infections (410, 411). Accordingly, with a heterologous infection model of persistent and nonpersistent viral infections, Treg generated as a result of past infections diminished subsequent immune responses and lung immunopathology upon exposure to heterologous virus infection (412). With an IAV and LCMV heterologous infection model, when Treg were depleted in IAV-immune mice and subsequently infected with LCMV, unexpectedly, lung pathology was reduced. The LCMV-specific CD8+ T-cell responses in the spleen were significantly reduced but not those in the mesenteric lymph nodes (mLNs). The explanation advocated was inefficient effector T-cell trafficking to lymph nodes due to the absence of Treg in both the naive mice and LCMV-infected IAV-immune mice. The study thus confirmed the established role of Treg in regulating effector T-cell exit from lymph nodes (413). Moreover, there was no enhancement of virus-specific effector responses when IAV-expanded Treg were depleted during LCMV infection (414, 415).
The observations above contrast with the report where PC61 (anti-CD25) treatment inhibited regulatory T cells expanded by IAV infection, which resulted in extensive lung pathology upon subsequent LCMV challenge (412). The drastic decrease in the degree of lung pathology upon Treg depletion was attributed to the fact that the LCMV-specific CD8+ T cells were overactivated and subsequently partially exhausted in mice immune to IAV and infected with LCMV.
In influenza virus-immune mice, infection with LCMV resulted in increased viral titers and lung pathology along with a modified cytokine profile in comparison to those in naive mice infected with LCMV. This was explained by the increased numbers of CD4+ Foxp3+ regulatory T cells in the lungs of influenza virus-immune mice compared to those in naive or LCMV-immune mice. Therefore, it is plausible that modulating the normal proportions of Treg and effector T-cell responses might have played a role in altering the responses in influenza virus-immune mice infected with LCMV. In this heterologous IAV/LCMV infection model, acute LCMV infection provided peak CD4+ cells, CD8+ cells, and Treg at day 3, which started decreasing at day 7, in the mLNs. However, in influenza virus-immune mice, the Treg persisted at elevated levels until day 9 following LCMV infection. Thus, in influenza virus-immune mice, heterologous LCMV infection resulted in altered Treg kinetics. This led to higher viral loads, increased proinflammatory cytokine and chemokine levels in the lungs, and ultimately immunopathology (409).
Preexisting CD8+ T cells that mediate bystander protection.
CD8+ T cells are known to confer protective heterologous immunity. In an adoptive transfer model, transfer of CD8+ and CD4+ T cells from LCMV-immune mice to naive mice provided protective heterologous immunity to PICV in the recipients (398). Furthermore, in a mouse model of coinfections, CMV infection conferred protection against IAV infection. In the same study, CMV-seropositive human adults displayed increased antibody responses to influenza vaccination compared to those in seronegative individuals. The enhanced responses included CD8+ T-cell activity and higher levels of IFN-γ. Thus, prior CMV infection has a beneficial effect on the immune system of young healthy humans. The mechanism of this CMV-mediated beneficial effect was attributed to the bystander protection offered by the IFN-γ (produced from CMV-specific CD8+ T cells) (388), and in a parallel mice study from the same group of researchers, it was demonstrated that this CD8+ T-cell-mediated protective immunity was completely abrogated in IFN-γ knockout mice. Alternatively, persistent infections may result in a modified inflammatory environment that leads to a change in dominance patterns. In a murine model of latent CMV infection, diminished CD8+ T-cell responses were observed upon secondary heterologous infection by WNV, IAV, or human herpesvirus 1 (416).
Superinfection of persistently infected host with acute heterologous infection such as with IAV results in persistent activation and proliferation of virus-specific cells (388). Similarly, simultaneous infection of gammaherpesvirus and influenza viruses resulted in enhanced numbers of lymphocytes in peripheral blood, as well as CD8+ CD4− T cells and CD19+ CD45+ B cells, in lungs of coinfected animals compared to singly infected mice. This in turn generated higher levels of IFN-γ and antibodies and ultimately a stimulated immune system (417).
Attrition of preexisting CD8+ T cells upon subsequent heterologous infection.
It has been confirmed in several animal model systems that an incoming heterologous infection may deplete the preexisting CD8+ T cells. This is referred to as attrition (418, 419). The finite space in the immune compartment supports the theory of attrition, where the subsequent viral infection induces interferon which in turn mediates apoptosis of memory T cells generated upon encountering primary virus infection. More than one class of interferons can also induce erosion of preexisting memory (420).
A cell surface receptor known as programmed cell death protein 1 (PD1) promotes apoptosis and serves as an immune checkpoint. It induces self-tolerance by inhibiting T-cell inflammatory activity. PD1 suppresses autoimmunity by depletion of autoreactive CD8+ T cells in mice (421). In a mouse model of heterologous CMV/HBV infection, PD1 suppression was found to be upregulated by CMV-specific CD8+ T cells, which was associated with enhanced apoptotic activity of these cells. Blockade of the PD1 pathway by anti-PDL1 antibody restored the proliferation and cytokine secretion by these CMV-specific CD8+ T cells (422). Thus, besides type 1 IFN, PD1-mediated apoptosis is yet another mechanism proposed for attrition.
Attrition could have a catastrophic consequence for vaccine-induced memory. However, contrasting data with a prime-boost vaccination strategy with VV suggested that immunological memory can grow in size while still preserving memory for previously encountered pathogens (423). VV induces poor interferon responses, and moreover, it generates effector memory cells that are present outside the lymph nodes, in contrast to central memory located within the lymphoid compartment. The possibility of central memory being more tightly regulated and the rapid erosion of immunological memory require further investigation. Measles virus provides long-lasting protective immunity in humans (65 years). Likewise, memory B cells persist for an indefinite period after smallpox immunization, suggesting that memory responses do not need to erode rapidly. In conclusion, some researchers support attrition because the space in the lymphoid compartment is finite; however, others do not support attrition, and various theories are proposed to explain both facets, thus presenting the idea of controversy.
It was argued whether virus-specific CD4+ memory T cells undergo attrition upon heterologous virus infection. However, data from mouse models indicate that LCMV-specific CD4+ T cells are relatively stable following various heterologous virus infections and protein immunization. However, in contrast, under the same circumstances, LCMV-specific CD8+ T cells were significantly reduced, suggesting that the T helper and cytotoxic memory cell pools are independently regulated (424).
Mathematical modeling studies indicate that upon each subsequent infection, viral clearance is challenged, and when the number of infections crosses a threshold, then viral control is completely abraded, thus supporting the loss of memory CD8+ T cells upon each new incoming infection (2).
Common Phenomena Observed in the Setting of Heterologous Infection
Altered immunodominance hierarchies.
Adaptive immunity is defined by the fact that a unique T-cell repertoire is established to a wide range of immunodominant epitopes. Immunodominance hierarchy depends upon the dose of antigen and numbers of T and B cells (425). Following a viral infection, multiple major histocompatibility complex class I (MHC-I) molecules are coexpressed along with the generation of numerous immunogenic peptides. However, the majority of the antiviral cytotoxic T lymphocyte subsets target only a few peptide/MHC class I complexes. This phenomenon where only limited peptide/MHC class I molecules are targeted by antiviral cytotoxic T lymphocytes is known as immunodominance. Among these peptide/MHC complexes, some epitopes are dominant and others are subdominant. For example, following IAV infection, although the primary CD8+ T-cell responses to DbNP366 and DbPA224 epitopes are of equivalent size, after secondary challenge, the DbNP366-specific T cells become the predominant responders. Naive hosts respond quite differently to antigen exposure than an antigen (heterologous)-experienced host. The presence of antigenically experienced cross-reactive CD8+ T cells which compete with the proliferation of naive CD8+ T cells partly contributes to altered epitope-specific hierarchies.
Remodeled TCR repertoire.
The response to an antigen can be represented as the number of T cells that are recruited and the structure of their antigen receptors. Quantifying an immune response at the repertoire level is now becoming very common. The array of individual clonotypes with TCRs specific for a distinct peptide MHC epitope is known as the TCR repertoire (426). The repertoire varies considerably in constituent TCR frequency and diversity. A diverse TCR repertoire benefits the host in combating a large number of pathogens. Tools such as TCRdist have recently been developed, which could be used to calculate the similarity and differences of key features of T-cell receptors and to identify those T-cell receptors that could recognize similar epitopes (427).
Heterologous infection can lead to the broadening of an otherwise narrow repertoire by recruiting the nondominant clones but at the same time could narrow the repertoire due to cross-reactivity. Primary infection-associated repertoires can be remarkably altered by a new, heterologous infection. For example, the NP205–212 epitope is encoded by both LCMV and PICV. This NP epitope elicits a TCR repertoire that is different in both infection types (LCMV and PICV) but is highly cross-reactive. Heterologous infection, i.e., infection of PICV-immune and LCMV-immune mice with LCMV and PICV, respectively, resulted in a narrow oligoclonal repertoire with clones having unpredictable TCR sequences. In this heterologous infection study, non-cross-reactive epitope-containing TCR repertoires were unaffected. However, cross-reactive CD8+ T cells proliferated after heterologous challenge. On the other hand, minimal alteration in the repertoire was observed in mice following homologous viral challenge, and the expected TCR motifs were observed (428). In another study, PICV infection followed by heterologous LCMV infection resulted in dominance of a subdominant NP epitope (429). Thus, discrepancies may result from challenging or vaccinating hosts with distinct immunological histories.
Alteration in repertoire diversity can also follow homologous challenge. In a study with bluetongue virus (BTV), virus-specific CD8+, but not CD4+, T cells expanded during the recall responses to BTV challenge. In addition, primary responses elicited a wider range of repertoire for MHC-I and MHC-II epitopes than the memory response, where a narrowed repertoire was induced in a dominant motif in VP7 (amino acid position 139 to 291) (430).
Cross-reactivity.
One of the several mechanisms proposed for the altered immunodominance hierarchies in heterologous infections is cross-reactivity. Cross-reactivity is the capacity of the TCR to recognize multiple peptide/MHC complexes, and this can occur in several different modes. These include the same TCR recognizing multiple peptide/MHC complexes or by molecular mimicry in which the TCR can bind to unrelated peptide/MHC complexes in a variable manner or may itself change the conformation within the flexible CDR3 loops.
Cross-reactivity may be advantageous as well as disadvantageous for the host. Cross-reactive immune responses in viral coinfections can either inhibit or augment the growth of new incoming pathogen (431). On the beneficial side, cross-reactivity could protect under conditions where a large number of pathogenic antigens and a limited TCR repertoire mounted by the host occur. On the harmful side, cross-reactivity may involve narrowing the TCR repertoire and consequently viral escape (428).
Cross-reactive CD8+ T-cell response were shown to occur during coinfection with multiple homologous strains, such as in IAV and DENV strains. It also occurs between completely unrelated viruses. In humans, the BMLF1280 antigen of EBV cross-reacts with IAV epitope M158.13, which is HLA-A*0201 restricted. This results in the induction of cross-reactive T cells with a reduced affinity for virus antigen-expressing cells and inefficient viral clearance (394). Upon secondary DENV infection in humans, the CD8+ T cells generated have a higher avidity to previously encountered DENV epitopes, and thus these cells expand compared to those T cells for the newer serotype expressed by the current infection (406). Another example includes HCV and IAV (394). HCV infection demonstrates a variety of symptoms varying from subclinical to clinical. The infection either is cleared from the host or may become persistent. One study reported that patients with acute HCV mount a T-cell response recognizing a diverse group of peptides; however, patients with chronic HCV mount a narrow T-cell response directed against cross-reactive influenza virus and HCV epitopes (432). Thus, cross-reactivity regulates disease severity in acute and chronically infected human patients. Cross-reactive memory cells elicited by past exposure to infection could influence immune response to other infectious agents, and this could impact the efficacy of vaccines (433).
ADE.
Virus-specific antibodies are well known to clear virus infection; however, a subneutralizing amount of antibodies can also augment virus infection and therefore disease severity. In antibody-dependent enhancement of infection (ADE), subneutralizing, cross-reactive antiviral antibodies bind to virion particles and facilitate infection of cells expressing Fc-γ receptors (Fc-γRs), including macrophages, monocytes, and some dendritic cell subsets. ADE usually occurs in patients who have preexisting antiviral immunity and are subsequently exposed to a heterologous virus. Alternatively, ADE can occur due to the presence of maternal antibodies in infants (434).
In DENV, the cross-reactive antibodies are hypothesized to promote DENV infection and antigenemia, which eventually result in severe DENV syndrome characterized by fever, hypotension, vascular leakage, thrombocytopenia, hemoconcentration, and end-organ damage (435). Besides ADE, cross-reactive, dysfunctional T-cell responses may also contribute to enhancing disease severity (435, 436). In addition, modifications on subtypes of cross-reactive IgG can also regulate interactions with specific Fc-γRs to influence disease severity (437).
Due to amino acid relatedness (nearly 43%) and cross-reactive antibodies between DENV and Zika virus (ZIKV), there is speculation that preexisting cross-reactive T cells and DENV antibodies can facilitate ZIKV infection. Bardina et al. (438) immunized STAT2−/− immunodeficient mice by injecting DENV- or WNV-immune plasma and subsequently inoculated them with ZIKV. The cross-reactive antibodies against WNV and DENV facilitated ZIKV replication and lethality. A higher incidence of ZIKV infection with more severe clinical manifestations (congenital malformations) was noticed in areas with a prior flavivirus infection, which could be explained by ADE. This, however, has not been substantiated.
Implications of Heterologous Immunity
Diagnostics and therapeutics.
Physicians and clinicians usually do not consider more than one viral etiological agent for diagnosis and therapy. Nevertheless, the association between the presence of coinfection and increased/decreased disease severity is still unclear. Indeed, an adequate diagnostic procedure investigating diverse groups of viral pathogens is important for appropriate therapy (439).
During an ongoing HCV infection, HIV coinfection hastens the development of hepatic fibrosis. Thus, therapy in coinfected individuals demands judicious implementation of the therapeutic regime (440). In another study, it was demonstrated that TNF is vital for VV control in naive mice, yet in LCMV-immune mice TNF is not essential for VV clearance (441). Thus, anti-TNF therapy could be safe in such cases. If an immunopathological outcome is expected in diagnosed coinfections, appropriate interventions (anticytokine therapy) can be employed to prevent severe immunopathology (398, 442, 443).
Transplantation.
Information regarding the previous infection history is also of vital importance in transplant recipients because successive heterologous viral infections result in increased numbers of alloantigen-specific T cells. These cells require tolerization before the graft transplant. Indeed, studies have shown that alloreactive immune responses elicited following viral infection could hamper tolerance induction (444). Several inhibitory mechanisms and costimulatory blockades (445–447) have also been performed, either singly or in combinations to enhance the rate of allograft survival. These attempts to disrupt T-cell activation could compromise T-cell-mediated antiviral immunity in a host with already ongoing persistent viral infection, such as infection with Epstein-Barr virus (EBV), HCV, and CMV, but also for a new incoming heterologous infection. Thus, the choice of costimulatory blockades could have critical consequences for transplant recipients. Therefore, a thorough knowledge of heterologous immunity helps guide successful engraftment and management of transplant recipients.
Vaccination.
Under the majority of circumstances, the infection history in humans and particularly animal species remains largely unknown. Thus, predicting the outcome of vaccination in heterologous infections in experienced hosts is challenging.
The efficacy of vaccines might be reduced due to immune-dominant alterations of undesired T-cell responses (443, 448) as a consequence of cross-reactivity. Cross-reactive T cells play a crucial role in pathogenic and protective immunity to heterologous infection. Hence, careful identification of such cross-reactive memory T cells could aid in vaccine designs. Thus, vaccines lacking cross-reactive epitopes (391) could be supplemented to formulate effective vaccination strategies (449). As discussed above, heterologous challenge can lead to erosion of the memory CD8+ T cells generated against the previously experienced antigen. This erosion could be explained by space restrictions within the immune compartment, which, if true, would have disastrous consequences for memory CD8+ T cells elicited in response to vaccination. This could mean that new incoming heterologous infection would displace the memory CD8+ T cells generated against the target pathogen by vaccination and vice versa, where vaccination could displace the preexisting memory CD8+ T cells elicited upon exposure to a previous heterologous infection.
With LCMV and vesicular stomatitis virus (VSV) prime-boost vaccination strategies, the enhancement of the memory CD8+ T-cell compartment was demonstrated to harbor newly developed clones of effector memory CD8+ T cells (423). In addition, attrition focused on secondary lymphoid tissue and the central memory population, whereas the prime-boost vaccination strategy mainly induced effector memory CD8+ T cells that resided within the nonlymphoid compartment (423). Moreover, attrition is induced by viruses that are strong interferon inducers. Thus, it is noteworthy that vaccines which generate a strong interferon response could end up in causing attrition of preexisting memory CD8+ T cells. However, this notion requires more study.
The induction of cross-reactivity and attrition are major concerns in vaccination. For instance, the VV vaccine is known to induce potent immune responses. Thus, such vaccines reduce the risk of infections and are likely to have heterologous impacts on the immune system. Smallpox immunization is advocated to lower the risk of asthma and malignant melanoma (450), due to heterologous effects of the vaccine on the immune system. However, we no longer vaccinate against smallpox, since the disease has been eradicated (451).
Similarly, the measles vaccine has an additional advantage; besides providing protection against measles, it could provide protective immunity against other, unrelated infections. Nevertheless, this wide-ranging beneficial effect could be abolished if the measles vaccine was followed by an inactivated diphtheria-tetanus-pertussis DTP vaccine (452).
MATHEMATICAL MODELS OF VIRAL COINFECTIONS
In order to better understand disease dynamics as well as to devise better therapeutic regimens, mathematical models of viral coinfections have also been developed (453). Most of the mathematical models involve HIV/HCV coinfection. Vickerman et al. first proposed a mathematical model for HCV/HIV transmission and concluded that sharing of needles/syringes is likely to increase HIV/HCV incidence in injecting drug users and that HCV infection indicates the risk of HIV infection (454). In another model, it was suggested that health care workers must be given sterile equipment (water filters and cookers) to prevent HCV infection (455). HIV loads impact the severity of HCV infection (456); therefore, treatment with highly active antiretroviral therapy (HAART) is specifically recommended to reduce the number of carriers (457). Mathematical models also suggest that the treatment efficacy influences the natural progression of HCV in HCV/HIV coinfection (456), and HAART is associated with a reduction in the transmission of HIV (458, 459). Moreover, HCV progressively induces a negative effect on human health, irrespective of the HIV status (460). In 2015, Birger et al. (461) refined a preexisting model of HCV infection by integrating dynamics of HIV and HCV coinfection as well as components of the immune system that clear infection. It was concluded that the propensity for HCV infection is greater in immunocompromised HIV-1 patients.
Rong et al. (462) presented a mathematical model for drug-sensitive and drug-resistant HCV. The model concluded that viral mutations acquired during the course of drug therapy have no major impact on the dynamics of different viral strains. Although low levels of HCV variants may be generated, they are liable to be completely suppressed due to fitness disadvantages (462, 463).
Pinky and Dobrovolny (464) developed a mathematical model to study the dynamics of IAV, RSV, rhinovirus, hPIV, and human metapneumovirus (hMPV) coinfections. The model suggested that one virus dominates over the other simply by being the first to infect, without involvement of viral interference or immune response. Rhinovirus, the most rapidly replicating virus, interferes with replication of other coinfecting viruses, while PIV, the most slowly replicating virus, is interrupted in the presence of other viral agents (464).
By considering that infection is cleared before initiation of the cellular regeneration, most of the prevailing mathematical models (for respiratory viruses) do not consider regeneration of the cells within the respiratory tract. In order to determine the effect of cellular regeneration on coinfection dynamics, Pinky and Debrovolny (465) investigated four mathematical models that incorporate distinct mechanisms of cellular regeneration. The models suggested that chronic illness is possible only with one viral species. Coexistence of multiple viruses in chronic conditions is unlikely to occur if the regeneration model is considered (465).
CONCLUDING REMARKS
Understanding the drivers of multiple infections and virus-virus interactions is an emerging field in virology. Classically, the laboratory examination of clinical specimens is biased to associate it with a single identified pathogen. Frequently, additional agents that contribute to the disease outcome are undetected. Due to viral interference, one virus infection may alter consequences of the other coinfecting viruses. Coinfection by related viruses may lead to genetic recombination/reassortment. This produces antigenic variants that can escape vaccine-induced immunity as well as the efficacy of antiviral drug therapy. Nonpathogenic divergent viruses present in clinical specimens may influence detection of pathogenic viruses, particularly when cell cultures are the diagnostic approach used. The advent of sequence-independent (nucleic acid-based) high-throughput technologies of microbial identification that enable microbial profiling (detection of both pathogenic and nonpathogenic microbes) in clinical specimens has permitted more precise diagnosis. In vitro and in vivo propagation of viruses also produces subgenomic viral particles (DI particles) that in some affect the phenotypic and virulence properties of heterologous viruses during coinfection, a topic still needing further investigation. Genome-wide transcriptomics and proteomics, coupled with small interfering RNA (siRNA) screens to analyze cellular factors required for virus replication, are likely to identify key molecules crucial for innate and adaptive viral interference. The approaches could also elucidate mechanisms of viral persistence, accommodation, enhancement, and superinfection exclusion/suppression during viral coinfections.
A well-adapted immune response is also critical for efficient control of pathogens involved in heterologous infections. Thus, memory responses to one infecting virus can markedly influence the type and magnitude of the immune response mounted against subsequent infections. In addition, the secondary infection may either deplete aspects of existing immune memory or generate additional effects which impact immune defense. The net outcome of heterologous immune responses could be either protection or immunopathology mediated by cross-reactivity, altered immunodominance hierarchies, or a remodeled TCR repertoire. The consequences of coinfections need to be better understood and the knowledge applied to improve diagnostics, preventative vaccines, and antiviral therapies.
ACKNOWLEDGMENTS
The Science and Engineering Research Board (India) supported this work (grant number SB/SO/AS-20/2014).
The funding agency had no role in design, data collection and interpretation, or the decision to submit this work for publication.
Biographies

Naveen Kumar, Ph.D., received his B.V.Sc. (2000) and M.V.Sc. (2002) degrees from the College of Veterinary Sciences, Bikaner, India, and his Ph.D. degree (2006) from the Friedrich Loeffler Institute, Insel Riems, Germany, and CCS Haryana Agricultural University, Hisar, India, under a Sandwich Model Scholarship program. He joined Emory University, Atlanta, GA, USA, as a as a postdoctoral fellow and worked (2006 to 2011) on understanding the interactions of influenza virus with host cell signaling pathways. He joined the Indian Council of Agricultural Research in 2011 as a Senior Scientist and is currently serving at the ICAR—National Centre for Veterinary Type Cultures, Hisar, India. He is a World Organisation for Animal Health (OIE)-designated member of the research group on PPR. Dr. Kumar's research interest is in the area of infectious diseases of animals and has focused on understanding virus-host interactions and cell biology of mixed infections.

Shalini Sharma, Ph.D., received her B.V.Sc. (2002) and M.V.Sc. (2004) degrees from the College of Veterinary Sciences, Bikaner, India, and her Ph.D. degree (2011) from the University of Tennessee, Knoxville, TN, USA. She worked on understanding immunity to and immunopathology of acute viral infections during her doctoral research work. Thereafter (2011 to 2014), she was a postdoctoral fellow at St. Jude Children's Research Hospital, Memphis, TN, USA, where she worked on immunity to and immunopathology of heterologous viral infections. Since 2014, she has been an Assistant Professor at Lala Lajpat Rai University of Veterinary and Animal Sciences, Hisar, Haryana, India. Her area of interest is infectious diseases with focus on understanding cellular immune responses against selected pathogens of domestic livestock.

Sanjay Barua, Ph.D., received his B.V.Sc. degree from the College of Veterinary Science, Guwahati, India, in 1991 and his M.V.Sc. (Veterinary Virology) from the Indian Veterinary Research Institute (IVRI), Izatnagar, India, in 1994. He joined the Central Institute for Research on Goats, Mathura, as a Scientist (Veterinary Microbiology) in 1997. He was awarded a Ph.D. degree in 2002 by IVRI, Izatnagar. He joined the National Centre for Veterinary Type Cultures (NCVTC), Hisar, in 2006 and is presently in charge of the national repository of animal microbes. He has more than 20 years of research experience in the field of virology. He is particularly interested in exploring viral diversity across various domestic animals in India.

Bhupendra N. Tripathi, Ph.D., obtained his B.V.Sc. and A.H. degrees from CSA University of Agriculture & Technology, Kanpur, India, and his M.V.Sc. (1987) and Ph.D. (1990) degrees from the Indian Veterinary Research Institute (IVRI), Izatnagar, India. He joined IVRI in 1993 and worked there in the capacities of Scientist, Senior Scientist, and Principal Scientist before moving in 2009 to CSWRI, Avikanagar, India, as Head, Animal Health Division. He also served as a postdoctoral fellow at the Institute of Animal Health, Compton, United Kingdom, and the Moredun Research Institute, Edinburgh, United Kingdom. Currently he is Director at the ICAR—National Research Centre on Equines and Project Coordinator of the National Centre for Veterinary Type Cultures. Dr. Tripathi is a veterinary pathologist. His area of interest is infectious diseases with focus on understanding mechanisms by which microbial pathogens cause disease, including molecular virology to in vivo pathogenesis.

Barry T. Rouse, Ph.D., obtained his B.V.Sc degree (1965) from the University of Bristol, England, and his M.Sc. (1967) and Ph.D. (1970) degrees from the University of Guelph (Canada). He worked as a postdoctoral fellow (1970 to 1972) at the Walter and Eliza Hall Institute of Medical Research, Australia, and also received a D.Sc. degree (1997) from the University of Bristol, England. Currently he is a Distinguished Professor at the Department of Biomedical and Diagnostic Sciences, University of Tennessee, Knoxville, TN, USA. He has been extensively involved in reviewing NIH grants since 1978 and has been a member of the Faculty of 1000 since its inception. Dr. Rouse's research interest is in the field of infectious disease and has focused on viral immunology and immunopathology. Dr. Rouse's group was the first to show the role of regulatory T cells (Treg) in the host response to a virus infection.
REFERENCES
- 1.Salas-Benito JS, De Nova-Ocampo M. 2015. Viral interference and persistence in mosquito-borne flaviviruses. J Immunol Res 2015:873404. doi: 10.1155/2015/873404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma S, Thomas PG. 2014. The two faces of heterologous immunity: protection or immunopathology. J Leukoc Biol 95:405–416. doi: 10.1189/jlb.0713386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghedin E, Fitch A, Boyne A, Griesemer S, DePasse J, Bera J, Zhang X, Halpin RA, Smit M, Jennings L, St George K, Holmes EC, Spiro DJ. 2009. Mixed infection and the genesis of influenza virus diversity. J Virol 83:8832–8841. doi: 10.1128/JVI.00773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moschidou P, Martella V, Lorusso E, Desario C, Pinto P, Losurdo M, Catella C, Parisi A, Banyai K, Buonavoglia C. 2011. Mixed infection by Feline astrovirus and Feline panleukopenia virus in a domestic cat with gastroenteritis and panleukopenia. J Vet Diagn Invest 23:581–584. doi: 10.1177/1040638711404149. [DOI] [PubMed] [Google Scholar]
- 5.Waner JL. 1994. Mixed viral infections: detection and management. Clin Microbiol Rev 7:143–151. doi: 10.1128/CMR.7.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendez-Rios J, Uetz P. 2010. Global approaches to study protein-protein interactions among viruses and hosts. Future Microbiol 5:289–301. doi: 10.2217/fmb.10.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durmus S, Ulgen KO. 2017. Comparative interactomics for virus-human protein-protein interactions: DNA viruses versus RNA viruses. FEBS Open Biol 7:96–107. doi: 10.1002/2211-5463.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roux S, Enault F, Bronner G, Vaulot D, Forterre P, Krupovic M. 2013. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat Commun 4:2700. doi: 10.1038/ncomms3700. [DOI] [PubMed] [Google Scholar]
- 9.Ojosnegros S, Beerenwinkel N, Domingo E. 2010. Competition-colonization dynamics: an ecology approach to quasispecies dynamics and virulence evolution in RNA viruses. Commun Integr Biol 3:333–336. doi: 10.4161/cib.3.4.11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Munoz SL. 2017. Viral coinfection is shaped by host ecology and virus-virus interactions across diverse microbial taxa and environments. Virus Evol 3:vex011. doi: 10.1093/ve/vex011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scotta MC, Chakr VC, de Moura A, Becker RG, de Souza AP, Jones MH, Pinto LA, Sarria EE, Pitrez PM, Stein RT, Mattiello R. 2016. Respiratory viral coinfection and disease severity in children: a systematic review and meta-analysis. J Clin Virol 80:45–56. doi: 10.1016/j.jcv.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cebey-Lopez M, Herberg J, Pardo-Seco J, Gomez-Carballa A, Martinon-Torres N, Salas A, Martinon-Sanchez JM, Justicia A, Rivero-Calle I, Sumner E, Fink C, Martinon-Torres F, GENDRES Network . 2016. Does viral co-infection influence the severity of acute respiratory infection in children? PLoS One 11:e0152481. doi: 10.1371/journal.pone.0152481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asner SA, Science ME, Tran D, Smieja M, Merglen A, Mertz D. 2014. Clinical disease severity of respiratory viral co-infection versus single viral infection: a systematic review and meta-analysis. PLoS One 9:e99392. doi: 10.1371/journal.pone.0099392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar N, Barua S, Riyesh T, Chaubey KK, Rawat KD, Khandelwal N, Mishra AK, Sharma N, Chandel SS, Sharma S, Singh MK, Sharma DK, Singh SV, Tripathi BN. 2016. Complexities in isolation and purification of multiple viruses from mixed viral infections: viral interference, persistence and exclusion. PLoS One 11:e0156110. doi: 10.1371/journal.pone.0156110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores CO, Valverde S, Weitz JS. 2013. Multi-scale structure and geographic drivers of cross-infection within marine bacteria and phages. ISME J 7:520–532. doi: 10.1038/ismej.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visser M, Bester R, Burger JT, Maree HJ. 2016. Next-generation sequencing for virus detection: covering all the bases. Virol J 13:85. doi: 10.1186/s12985-016-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bialasiewicz S, McVernon J, Nolan T, Lambert SB, Zhao G, Wang D, Nissen MD, Sloots TP. 2014. Detection of a divergent Parainfluenza 4 virus in an adult patient with influenza like illness using next-generation sequencing. BMC Infect Dis 14:275. doi: 10.1186/1471-2334-14-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho T, Tzanetakis IE. 2014. Development of a virus detection and discovery pipeline using next generation sequencing. Virology 471-473:54–60. doi: 10.1016/j.virol.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 19.Kumar N, Barua S, Thachamvally R, Tripathi BN. 2016. Systems perspective of morbillivirus replication. J Mol Microbiol Biotechnol 26:389–400. doi: 10.1159/000448842. [DOI] [PubMed] [Google Scholar]
- 20.Limaye AP, Bakthavatsalam R, Kim HW, Kuhr CS, Halldorson JB, Healey PJ, Boeckh M. 2004. Late-onset cytomegalovirus disease in liver transplant recipients despite antiviral prophylaxis. Transplantation 78:1390–1396. doi: 10.1097/01.TP.0000145989.22373.03. [DOI] [PubMed] [Google Scholar]
- 21.Lyall EG, Ogilvie MM, Smith NM, Burns S. 1994. Acyclovir resistant varicella zoster and HIV infection. Arch Dis Child 70:133–135. doi: 10.1136/adc.70.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooney MK. 1976. Selection of cell culture systems for virus isolation. Am J Med Technol 42:158–165. [PubMed] [Google Scholar]
- 23.Chua KB, Chua KH, Chua IL, Chen KF. 2004. A new cell culture tube in diagnostic virology for virus isolation. Malays J Pathol 26:69–71. [PubMed] [Google Scholar]
- 24.Kumar N, Barua S, Riyesh T, Tripathi BN. 2017. Advances in peste des petits ruminants vaccines. Vet Microbiol 206:91–101. doi: 10.1016/j.vetmic.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Engelenburg FA, Terpstra FG, Schuitemaker H, Moorer WR. 2002. The virucidal spectrum of a high concentration alcohol mixture. J Hosp Infect 51:121–125. doi: 10.1053/jhin.2002.1211. [DOI] [PubMed] [Google Scholar]
- 26.Chang SF, Su CL, Shu PY, Yang CF, Liao TL, Cheng CH, Hu HC, Huang JH. 2010. Concurrent isolation of chikungunya virus and dengue virus from a patient with coinfection resulting from a trip to Singapore. J Clin Microbiol 48:4586–4589. doi: 10.1128/JCM.01228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson KA, Yin J. 2010. Population dynamics of an RNA virus and its defective interfering particles in passage cultures. Virol J 7:257. doi: 10.1186/1743-422X-7-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frensing T, Heldt FS, Pflugmacher A, Behrendt I, Jordan I, Flockerzi D, Genzel Y, Reichl U. 2013. Continuous influenza virus production in cell culture shows a periodic accumulation of defective interfering particles. PLoS One 8:e72288. doi: 10.1371/journal.pone.0072288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radloff RJ, Young SA. 1983. Defective interfering particles of encephalomyocarditis virus. J Gen Virol 64:1637–1641. doi: 10.1099/0022-1317-64-7-1637. [DOI] [PubMed] [Google Scholar]
- 30.Roux L, Holland JJ. 1979. Role of defective interfering particles of Sendai virus in persistent infections. Virology 93:91–103. doi: 10.1016/0042-6822(79)90278-2. [DOI] [PubMed] [Google Scholar]
- 31.Treuhaft MW, Beem MO. 1982. Defective interfering particles of respiratory syncytial virus. Infect Immun 37:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Arriaza J, Domingo E, Escarmis C. 2005. A segmented form of foot-and-mouth disease virus interferes with standard virus: a link between interference and competitive fitness. Virology 335:155–164. doi: 10.1016/j.virol.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Janda JM, Davis AR, Nayak DP, De BK. 1979. Diversity and generation of defective interfering influenza virus particles. Virology 95:48–58. doi: 10.1016/0042-6822(79)90400-8. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Arriaza J, Manrubia SC, Toja M, Domingo E, Escarmis C. 2004. Evolutionary transition toward defective RNAs that are infectious by complementation. J Virol 78:11678–11685. doi: 10.1128/JVI.78.21.11678-11685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belsham GJ, Jamal SM, Tjornehoj K, Botner A. 2011. Rescue of foot-and-mouth disease viruses that are pathogenic for cattle from preserved viral RNA samples. PLoS One 6:e14621. doi: 10.1371/journal.pone.0014621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snowdon WA. 1966. Growth of foot-and mouth disease virus in monolayer cultures of calf thyroid cells. Nature 210:1079–1080. doi: 10.1038/2101079a0. [DOI] [PubMed] [Google Scholar]
- 38.Kumar N, Wadhwa A, Chaubey KK, Singh SV, Gupta S, Sharma S, Sharma DK, Singh MK, Mishra AK. 2014. Isolation and phylogenetic analysis of an orf virus from sheep in Makhdoom, India. Virus Genes 48:312–319. doi: 10.1007/s11262-013-1025-9. [DOI] [PubMed] [Google Scholar]
- 39.Phipps PH, McCulloch BG, Miller HR, Rossier E. 1989. Rapid detection of influenza virus infections in human fetal lung diploid cell cultures. J Infect 18:269–278. doi: 10.1016/S0163-4453(89)80063-5. [DOI] [PubMed] [Google Scholar]
- 40.Sanjuan R, Domingo-Calap P. 2016. Mechanisms of viral mutation. Cell Mol Life Sci 73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hematian A, Sadeghifard N, Mohebi R, Taherikalani M, Nasrolahi A, Amraei M, Ghafourian S. 2016. Traditional and modern cell culture in virus diagnosis. Osong Public Health Res Perspect 7:77–82. doi: 10.1016/j.phrp.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leland DS, Ginocchio CC. 2007. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev 20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brumback BG, Wade CD. 1994. Simultaneous culture for adenovirus, cytomegalovirus, and herpes simplex virus in same shell vial by using three-color fluorescence. J Clin Microbiol 32:2289–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinberg A, Brewster L, Clark J, Simoes E, consortium A. 2004. Evaluation of R-Mix shell vials for the diagnosis of viral respiratory tract infections. J Clin Virol 30:100–105. doi: 10.1016/j.jcv.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 45.Lim G, Park TS, Suh JT, Lee HJ. 2010. Comparison of R-mix virus culture and multiplex reverse transcriptase-PCR for the rapid detection of respiratory viruses. Korean J Lab Med 30:289–294. doi: 10.3343/kjlm.2010.30.3.289. [DOI] [PubMed] [Google Scholar]
- 46.Barenfanger J, Drake C, Mueller T, Troutt T, O'Brien J, Guttman K. 2001. R-Mix cells are faster, at least as sensitive and marginally more costly than conventional cell lines for the detection of respiratory viruses. J Clin Virol 22:101–110. doi: 10.1016/S1386-6532(01)00171-8. [DOI] [PubMed] [Google Scholar]
- 47.Dunn JJ, Woolstenhulme RD, Langer J, Carroll KC. 2004. Sensitivity of respiratory virus culture when screening with R-mix fresh cells. J Clin Microbiol 42:79–82. doi: 10.1128/JCM.42.1.79-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.St George K, Patel NM, Hartwig RA, Scholl DR, Jollick JA Jr, Kauffmann LM, Evans MR, Rinaldo CR Jr. 2002. Rapid and sensitive detection of respiratory virus infections for directed antiviral treatment using R-Mix cultures. J Clin Virol 24:107–115. doi: 10.1016/S1386-6532(01)00239-6. [DOI] [PubMed] [Google Scholar]
- 49.Gillim-Ross L, Taylor J, Scholl DR, Ridenour J, Masters PS, Wentworth DE. 2004. Discovery of novel human and animal cells infected by the severe acute respiratory syndrome coronavirus by replication-specific multiplex reverse transcription-PCR. J Clin Microbiol 42:3196–3206. doi: 10.1128/JCM.42.7.3196-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim DK, Poudel B. 2013. Tools to detect influenza virus. Yonsei Med J 54:560–566. doi: 10.3349/ymj.2013.54.3.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuen KY, Chan PK, Peiris M, Tsang DN, Que TL, Shortridge KF, Cheung PT, To WK, Ho ET, Sung R, Cheng AF. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471. doi: 10.1016/S0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 52.Choi WS, Noh JY, Baek JH, Seo YB, Lee J, Song JY, Park DW, Lee JS, Cheong HJ, Kim WJ. 2015. Suboptimal effectiveness of the 2011-2012 seasonal influenza vaccine in adult Korean populations. PLoS One 10:e0098716. doi: 10.1371/journal.pone.0098716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmutzhard J, Merete Riedel H, Zweygberg Wirgart B, Grillner L. 2004. Detection of herpes simplex virus type 1, herpes simplex virus type 2 and varicella-zoster virus in skin lesions. Comparison of real-time PCR, nested PCR and virus isolation. J Clin Virol 29:120–126. [DOI] [PubMed] [Google Scholar]
- 54.Huang YT, Hite S, Duane V, Yan H. 2002. CV-1 and MRC-5 mixed cells for simultaneous detection of herpes simplex viruses and varicella zoster virus in skin lesions. J Clin Virol 24:37–43. doi: 10.1016/S1386-6532(01)00230-X. [DOI] [PubMed] [Google Scholar]
- 55.Olivo PD. 1996. Transgenic cell lines for detection of animal viruses. Clin Microbiol Rev 9:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morabito KM, Erez N, Graham BS, Ruckwardt TJ. 2016. Phenotype and hierarchy of two transgenic T cell lines targeting the respiratory syncytial virus KdM282-90 epitope is transfer dose-dependent. PLoS One 11:e0146781. doi: 10.1371/journal.pone.0146781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabuchi Y, Arai Y, Ohta S, Shioya H, Takahashi R, Ueda M, Takeguchi N, Asano S, Obinata M. 2002. Development and characterization of conditionally immortalized gastric epithelial cell lines from transgenic rats harboring temperature-sensitive simian virus 40 large T-antigen gene. Cell Struct Funct 27:71–79. doi: 10.1247/csf.27.71. [DOI] [PubMed] [Google Scholar]
- 58.Beggs AH, Miner JH, Scangos GA. 1990. Cell type-specific expression of JC virus T antigen in primary and established cell lines from transgenic mice. J Gen Virol 71:151–164. doi: 10.1099/0022-1317-71-1-151. [DOI] [PubMed] [Google Scholar]
- 59.Stabell EC, O'Rourke SR, Storch GA, Olivo PD. 1993. Evaluation of a genetically engineered cell line and a histochemical beta-galactosidase assay to detect herpes simplex virus in clinical specimens. J Clin Microbiol 31:2796–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turchek BM, Huang YT. 1999. Evaluation of ELVIS HSV ID/Typing System for the detection and typing of herpes simplex virus from clinical specimens. J Clin Virol 12:65–69. doi: 10.1016/S0928-0197(98)00066-X. [DOI] [PubMed] [Google Scholar]
- 61.Patel N, Kauffmann L, Baniewicz G, Forman M, Evans M, Scholl D. 1999. Confirmation of low-titer, herpes simplex virus-positive specimen results by the enzyme-linked virus-inducible system (ELVIS) using PCR and repeat testing. J Clin Microbiol 37:3986–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kok TW, Pryor T, Payne L. 1998. Comparison of rhabdomyosarcoma, buffalo green monkey kidney epithelial, A549 (human lung epithelial) cells and human embryonic lung fibroblasts for isolation of enteroviruses from clinical samples. J Clin Virol 11:61–65. doi: 10.1016/S0928-0197(98)00026-9. [DOI] [PubMed] [Google Scholar]
- 63.She RC, Crist G, Billetdeaux E, Langer J, Petti CA. 2006. Comparison of multiple shell vial cell lines for isolation of enteroviruses: a national perspective. J Clin Virol 37:151–155. doi: 10.1016/j.jcv.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 64.Peaper DR, Landry ML. 2014. Laboratory diagnosis of viral infection. Handb Clin Neurol 123:123–147. doi: 10.1016/B978-0-444-53488-0.00005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang YT, Yam P, Yan H, Sun Y. 2002. Engineered BGMK cells for sensitive and rapid detection of enteroviruses. J Clin Microbiol 40:366–371. doi: 10.1128/JCM.40.2.366-371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shchelkunov SN, Shcherbakov DN, Maksyutov RA, Gavrilova EV. 2011. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. J Virol Methods 175:163–169. doi: 10.1016/j.jviromet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garg G, Kumar D, Asim M, Husain SA, Das BC, Kar P. 2016. Multiplex reverse transcriptase-PCR for simultaneous detection of hepatitis B, C, and E virus. J Clin Exp Hepatol 6:33–39. doi: 10.1016/j.jceh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leveque N, Van Haecke A, Renois F, Boutolleau D, Talmud D, Andreoletti L. 2011. Rapid virological diagnosis of central nervous system infections by use of a multiplex reverse transcription-PCR DNA microarray. J Clin Microbiol 49:3874–3879. doi: 10.1128/JCM.01214-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hole K, Clavijo A, Pineda LA. 2006. Detection and serotype-specific differentiation of vesicular stomatitis virus using a multiplex, real-time, reverse transcription-polymerase chain reaction assay. J Vet Diagn Invest 18:139–146. doi: 10.1177/104063870601800201. [DOI] [PubMed] [Google Scholar]
- 70.Hindson BJ, Reid SM, Baker BR, Ebert K, Ferris NP, Tammero LF, Lenhoff RJ, Naraghi-Arani P, Vitalis EA, Slezak TR, Hullinger PJ, King DP. 2008. Diagnostic evaluation of multiplexed reverse transcription-PCR microsphere array assay for detection of foot-and-mouth and look-alike disease viruses. J Clin Microbiol 46:1081–1089. doi: 10.1128/JCM.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pang Z, Li A, Li J, Qu J, He C, Zhang S, Li C, Zhang Q, Liang M, Li D. 2014. Comprehensive multiplex one-step real-time TaqMan qRT-PCR assays for detection and quantification of hemorrhagic fever viruses. PLoS One 9:e95635. doi: 10.1371/journal.pone.0095635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z, Qi X, Zhou M, Bao C, Hu J, Wu B, Wang S, Tan Z, Fu J, Shan J, Zhu Y, Tang F. 2013. A two-tube multiplex real-time RT-PCR assay for the detection of four hemorrhagic fever viruses: severe fever with thrombocytopenia syndrome virus, Hantaan virus, Seoul virus, and dengue virus. Arch Virol 158:1857–1863. doi: 10.1007/s00705-013-1677-8. [DOI] [PubMed] [Google Scholar]
- 73.Jiang Y, Fang L, Shi X, Zhang H, Li Y, Lin Y, Qiu Y, Chen Q, Li H, Zhou L, Hu Q. 2014. Simultaneous detection of five enteric viruses associated with gastroenteritis by use of a PCR assay: a single real-time multiplex reaction and its clinical application. J Clin Microbiol 52:1266–1268. doi: 10.1128/JCM.00245-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beuret C. 2004. Simultaneous detection of enteric viruses by multiplex real-time RT-PCR. J Virol Methods 115:1–8. doi: 10.1016/j.jviromet.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 75.You HL, Chang SJ, Yu HR, Li CC, Chen CH, Liao WT. 2017. Simultaneous detection of respiratory syncytial virus and human metapneumovirus by one-step multiplex real-time RT-PCR in patients with respiratory symptoms. BMC Pediatr 17:89. doi: 10.1186/s12887-017-0843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thonur L, Maley M, Gilray J, Crook T, Laming E, Turnbull D, Nath M, Willoughby K. 2012. One-step multiplex real time RT-PCR for the detection of bovine respiratory syncytial virus, bovine herpesvirus 1 and bovine parainfluenza virus 3. BMC Vet Res 8:37. doi: 10.1186/1746-6148-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liao RS, Tomalty LL, Majury A, Zoutman DE. 2009. Comparison of viral isolation and multiplex real-time reverse transcription-PCR for confirmation of respiratory syncytial virus and influenza virus detection by antigen immunoassays. J Clin Microbiol 47:527–532. doi: 10.1128/JCM.01213-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y, Cui D, Zheng S, Yang S, Tong J, Yang D, Fan J, Zhang J, Lou B, Li X, Zhuge X, Ye B, Chen B, Mao W, Tan Y, Xu G, Chen Z, Chen N, Li L. 2011. Simultaneous detection of influenza A, influenza B, and respiratory syncytial viruses and subtyping of influenza A H3N2 virus and H1N1 (2009) virus by multiplex real-time PCR. J Clin Microbiol 49:1653–1656. doi: 10.1128/JCM.02184-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bonroy C, Vankeerberghen A, Boel A, De Beenhouwer H. 2007. Use of a multiplex real-time PCR to study the incidence of human metapneumovirus and human respiratory syncytial virus infections during two winter seasons in a Belgian paediatric hospital. Clin Microbiol Infect 13:504–509. doi: 10.1111/j.1469-0691.2007.01682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beck ET, Jurgens LA, Kehl SC, Bose ME, Patitucci T, LaGue E, Darga P, Wilkinson K, Witt LM, Fan J, He J, Kumar S, Henrickson KJ. 2010. Development of a rapid automated influenza A, influenza B, and respiratory syncytial virus A/B multiplex real-time RT-PCR assay and its use during the 2009 H1N1 swine-origin influenza virus epidemic in Milwaukee, Wisconsin. J Mol Diagn 12:74–81. doi: 10.2353/jmoldx.2010.090095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arens MQ, Buller RS, Rankin A, Mason S, Whetsell A, Agapov E, Lee WM, Storch GA. 2010. Comparison of the Eragen Multi-Code Respiratory Virus Panel with conventional viral testing and real-time multiplex PCR assays for detection of respiratory viruses. J Clin Microbiol 48:2387–2395. doi: 10.1128/JCM.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ali Z, Wang J, Tang Y, Liu B, He N, Li Z. 2016. Simultaneous detection of multiple viruses based on chemiluminescence and magnetic separation. Biomater Sci 5:57–66. doi: 10.1039/C6BM00527F. [DOI] [PubMed] [Google Scholar]
- 83.Defoort JP, Martin M, Casano B, Prato S, Camilla C, Fert V. 2000. Simultaneous detection of multiplex-amplified human immunodeficiency virus type 1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA using a flow cytometer microsphere-based hybridization assay. J Clin Microbiol 38:1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mitterer G, Schmidt WM. 2006. Microarray-based detection of bacteria by on-chip PCR. Methods Mol Biol 345:37–51. [DOI] [PubMed] [Google Scholar]
- 85.Wang Q, Li RD, Yin BC, Ye BC. 2015. Colorimetric detection of sequence-specific microRNA based on duplex-specific nuclease-assisted nanoparticle amplification. Analyst 140:6306–6312. doi: 10.1039/C5AN01350J. [DOI] [PubMed] [Google Scholar]
- 86.Varshney M, Li Y. 2007. Interdigitated array microelectrode based impedance biosensor coupled with magnetic nanoparticle-antibody conjugates for detection of Escherichia coli O157:H7 in food samples. Biosens Bioelectron 22:2408–2414. doi: 10.1016/j.bios.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 87.Muangchuen A, Chaumpluk P, Suriyasomboon A, Ekgasit S. 2014. Colorimetric detection of Ehrlichia canis via nucleic acid hybridization in gold nano-colloids. Sensors (Basel) 14:14472–14487. doi: 10.3390/s140814472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thapa RK, Choi JY, Gupta B, Ramasamy T, Poudel BK, Ku SK, Youn YS, Choi HG, Yong CS, Kim JO. 2016. Liquid crystalline nanoparticles encapsulating cisplatin and docetaxel combination for targeted therapy of breast cancer. Biomater Sci 4:1340–1350. doi: 10.1039/C6BM00376A. [DOI] [PubMed] [Google Scholar]
- 89.Bronstein I, McGrath P. 1989. Chemiluminescence lights up. Nature 338:599–600. doi: 10.1038/338599a0. [DOI] [PubMed] [Google Scholar]
- 90.Lara FJ, Airado-Rodriguez D, Moreno-Gonzalez D, Huertas-Perez JF, Garcia-Campana AM. 2016. Applications of capillary electrophoresis with chemiluminescence detection in clinical, environmental and food analysis. A review. Anal Chim Acta 913:22–40. doi: 10.1016/j.aca.2016.01.046. [DOI] [PubMed] [Google Scholar]
- 91.Nakashima K, Ikeda R, Wada M. 2009. Analytical studies on the development of high-performance liquid chromatographic methods with fluorescence or chemiluminescence detections and their practical applications. Anal Sci 25:21–31. doi: 10.2116/analsci.25.21. [DOI] [PubMed] [Google Scholar]
- 92.Fuentes M, Mateo C, Rodriguez A, Casqueiro M, Tercero JC, Riese HH, Fernandez-Lafuente R, Guisan JM. 2006. Detecting minimal traces of DNA using DNA covalently attached to superparamagnetic nanoparticles and direct PCR-ELISA. Biosens Bioelectron 21:1574–1580. doi: 10.1016/j.bios.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 93.Min JH, Woo MK, Yoon HY, Jang JW, Wu JH, Lim CS, Kim YK. 2014. Isolation of DNA using magnetic nanoparticles coated with dimercaptosuccinic acid. Anal Biochem 447:114–118. doi: 10.1016/j.ab.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 94.Berensmeier S. 2006. Magnetic particles for the separation and purification of nucleic acids. Appl Microbiol Biotechnol 73:495–504. doi: 10.1007/s00253-006-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peterson RD, Chen W, Cunningham BT, Andrade JE. 2015. Enhanced sandwich immunoassay using antibody-functionalized magnetic iron-oxide nanoparticles for extraction and detection of soluble transferrin receptor on a photonic crystal biosensor. Biosens Bioelectron 74:815–822. doi: 10.1016/j.bios.2015.07.050. [DOI] [PubMed] [Google Scholar]
- 96.Liu Q, Bai Y, Ge Q, Zhou S, Wen T, Lu Z. 2007. Microarray-in-a-tube for detection of multiple viruses. Clin Chem 53:188–194. doi: 10.1373/clinchem.2006.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kostina EV, Riabinin VA, Agafonov AP, Ternovoi VA, Siniakov AN. 2011. Microarray for diagnostics of human pathogenic influenza A virus subtypes. Bioorg Khim 37:715–717. [DOI] [PubMed] [Google Scholar]
- 98.Martinez MA, Soto-Del Rio Mde L, Gutierrez RM, Chiu CY, Greninger AL, Contreras JF, Lopez S, Arias CF, Isa P. 2015. DNA microarray for detection of gastrointestinal viruses. J Clin Microbiol 53:136–145. doi: 10.1128/JCM.01317-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yadav BS, Pokhriyal M, Vasishtha DP, Sharma B. 2014. Animal Viruses Probe dataset (AVPDS) for microarray-based diagnosis and identification of viruses. Curr Microbiol 68:301–304. doi: 10.1007/s00284-013-0477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shieh B, Li C. 2004. Multi-faceted, multi-versatile microarray: simultaneous detection of many viruses and their expression profiles. Retrovirology 1:11. doi: 10.1186/1742-4690-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jaing CJ, Thissen JB, Gardner SN, McLoughlin KS, Hullinger PJ, Monday NA, Niederwerder MC, Rowland RR. 2015. Application of a pathogen microarray for the analysis of viruses and bacteria in clinical diagnostic samples from pigs. J Vet Diagn Invest 27:313–325. doi: 10.1177/1040638715578484. [DOI] [PubMed] [Google Scholar]
- 102.Kunze A, Dilcher M, Abd El Wahed A, Hufert F, Niessner R, Seidel M. 2016. On-chip isothermal nucleic acid amplification on flow-based chemiluminescence microarray analysis platform for the detection of viruses and bacteria. Anal Chem 88:898–905. doi: 10.1021/acs.analchem.5b03540. [DOI] [PubMed] [Google Scholar]
- 103.Prachayangprecha S, Schapendonk CM, Koopmans MP, Osterhaus AD, Schurch AC, Pas SD, van der Eijk AA, Poovorawan Y, Haagmans BL, Smits SL. 2014. Exploring the potential of next-generation sequencing in detection of respiratory viruses. J Clin Microbiol 52:3722–3730. doi: 10.1128/JCM.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Capobianchi MR, Giombini E, Rozera G. 2013. Next-generation sequencing technology in clinical virology. Clin Microbiol Infect 19:15–22. doi: 10.1111/1469-0691.12056. [DOI] [PubMed] [Google Scholar]
- 105.Lipkin WI. 2010. Microbe hunting. Microbiol Mol Biol Rev 74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mokili JL, Rohwer F, Dutilh BE. 2012. Metagenomics and future perspectives in virus discovery. Curr Opin Virol 2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yanagisawa H, Tomita R, Katsu K, Uehara T, Atsumi G, Tateda C, Kobayashi K, Sekine KT. 2016. Combined DECS analysis and next-generation sequencing enable efficient detection of novel plant RNA viruses. Viruses 8:70. doi: 10.3390/v8030070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Caraballo Cortes K, Bukowska-Osko I, Pawelczyk A, Perlejewski K, Ploski R, Lechowicz U, Stawinski P, Demkow U, Laskus T, Radkowski M. 2016. Next-generation sequencing of 5′ untranslated region of hepatitis C virus in search of minor viral variant in a patient who revealed new genotype while on antiviral treatment. Adv Exp Med Biol 885:11–23. doi: 10.1007/5584_2015_186. [DOI] [PubMed] [Google Scholar]
- 109.Molenaar N, Burger JT, Maree HJ. 2015. Detection of a divergent variant of grapevine virus F by next-generation sequencing. Arch Virol 160:2125–2127. doi: 10.1007/s00705-015-2466-3. [DOI] [PubMed] [Google Scholar]
- 110.Smits SL, Osterhaus AD. 2013. Virus discovery: one step beyond. Curr Opin Virol doi: 10.1016/j.coviro.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3:e00473-12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, Buchmann J, Wang W, Xu J, Holmes EC, Zhang YZ. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 113.Lecomte E, Tournaire B, Cogne B, Dupont JB, Lindenbaum P, Martin-Fontaine M, Broucque F, Robin C, Hebben M, Merten OW, Blouin V, Francois A, Redon R, Moullier P, Leger A. 2015. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol Ther Nucleic Acids 4:e260. doi: 10.1038/mtna.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gansauge MT, Gerber T, Glocke I, Korlevic P, Lippik L, Nagel S, Riehl LM, Schmidt A, Meyer M. 2017. Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res 45:e79. doi: 10.1093/nar/gkx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burnham P, Kim MS, Agbor-Enoh S, Luikart H, Valantine HA, Khush KK, De Vlaminck I. 2016. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep 6:27859. doi: 10.1038/srep27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wales N, Caroe C, Sandoval-Velasco M, Gamba C, Barnett R, Samaniego JA, Madrigal JR, Orlando L, Gilbert MT. 2015. New insights on single-stranded versus double-stranded DNA library preparation for ancient DNA. Biotechniques 59:368–371. doi: 10.2144/000114364. [DOI] [PubMed] [Google Scholar]
- 117.Gansauge MT, Meyer M. 2013. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc 8:737–748. doi: 10.1038/nprot.2013.038. [DOI] [PubMed] [Google Scholar]
- 118.Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. 2012. Sequence analysis of the human virome in febrile and afebrile children. PLoS One 7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wylie TN, Wylie KM, Herter BN, Storch GA. 2015. Enhanced virome sequencing using targeted sequence capture. Genome Res 25:1910–1920. doi: 10.1101/gr.191049.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.White PA, Li Z, Zhai X, Marinos G, Rawlinson WD. 2000. Mixed viral infection identified using heteroduplex mobility analysis (HMA). Virology 271:382–389. doi: 10.1006/viro.2000.0323. [DOI] [PubMed] [Google Scholar]
- 121.Donis RO, Dubovi EJ. 1987. Differences in virus-induced polypeptides in cells infected by cytopathic and noncytopathic biotypes of bovine virus diarrhea-mucosal disease virus. Virology 158:168–173. doi: 10.1016/0042-6822(87)90250-9. [DOI] [PubMed] [Google Scholar]
- 122.Fayyadh TK, Ma F, Qin C, Zhang X, Zhang Z, Cui Z. 2017. Simultaneous detection of multiple viruses in their co-infected cells using multicolour imaging with self-assembled quantum dot probes. Microchim Acta 184:2815–2824. doi: 10.1007/s00604-017-2300-6. [DOI] [Google Scholar]
- 123.Boonham N, Kreuze J, Winter S, van der Vlugt R, Bergervoet J, Tomlinson J, Mumford R. 2014. Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res 186:20–31. doi: 10.1016/j.virusres.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 124.Griffiths EC, Pedersen AB, Fenton A, Petchey OL. 2011. The nature and consequences of coinfection in humans. J Infect 63:200–206. doi: 10.1016/j.jinf.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haller O. 2015. Jean Lindenmann: from viral interference to interferon and beyond (1924-2015). J Interferon Cytokine Res 35:239–241. doi: 10.1089/jir.2015.1500. [DOI] [PubMed] [Google Scholar]
- 126.Dianzani F. 1975. Viral interference and interferon. Ric Clin Lab 5:196–213. [DOI] [PubMed] [Google Scholar]
- 127.Wang X, Hinson ER, Cresswell P. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 128.Tan KS, Olfat F, Phoon MC, Hsu JP, Howe JL, Seet JE, Chin KC, Chow VT. 2012. In vivo and in vitro studies on the antiviral activities of viperin against influenza H1N1 virus infection. J Gen Virol 93:1269–1277. doi: 10.1099/vir.0.040824-0. [DOI] [PubMed] [Google Scholar]
- 129.Seo JY, Yaneva R, Cresswell P. 2011. Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10:534–539. doi: 10.1016/j.chom.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Neil SJ. 2013. The antiviral activities of tetherin. Curr Top Microbiol Immunol 371:67–104. [DOI] [PubMed] [Google Scholar]
- 131.Lenschow DJ. 2010. Antiviral properties of ISG15. Viruses 2:2154–2168. doi: 10.3390/v2102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moldovan JB, Moran JV. 2015. The zinc-finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet 11:e1005121. doi: 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Carthagena L, Parise MC, Ringeard M, Chelbi-Alix MK, Hazan U, Nisole S. 2008. Implication of TRIM alpha and TRIMCyp in interferon-induced anti-retroviral restriction activities. Retrovirology 5:59. doi: 10.1186/1742-4690-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen K, Huang J, Zhang C, Huang S, Nunnari G, Wang FX, Tong X, Gao L, Nikisher K, Zhang H. 2006. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol 80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Birdwell LD, Zalinger ZB, Li Y, Wright PW, Elliott R, Rose KM, Silverman RH, Weiss SR. 2016. Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal Oas gene expression and independent of virus-induced interferon. J Virol 90:3160–3172. doi: 10.1128/JVI.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakayama M, Nagata K, Kato A, Ishihama A. 1991. Interferon-inducible mouse Mx1 protein that confers resistance to influenza virus is GTPase. J Biol Chem 266:21404–21408. [PubMed] [Google Scholar]
- 137.Streitenfeld H, Boyd A, Fazakerley JK, Bridgen A, Elliott RM, Weber F. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J Virol 77:5507–5511. doi: 10.1128/JVI.77.9.5507-5511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Marcus PI, Carver DH. 1967. Intrinsic interference: a new type of viral interference. J Virol 1:334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Polacino P, Kaplan G, Palma EL. 1985. Homologous interference by a foot-and-mouth disease virus strain attenuated for cattle. Arch Virol 86:291–301. doi: 10.1007/BF01309833. [DOI] [PubMed] [Google Scholar]
- 140.Hanley KA, Nelson JT, Schirtzinger EE, Whitehead SS, Hanson CT. 2008. Superior infectivity for mosquito vectors contributes to competitive displacement among strains of dengue virus. BMC Ecol 8:1. doi: 10.1186/1472-6785-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pepin KM, Hanley KA. 2008. Density-dependent competitive suppression of sylvatic dengue virus by endemic dengue virus in cultured mosquito cells. Vector Borne Zoonotic Dis 8:821–828. doi: 10.1089/vbz.2008.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pepin KM, Lambeth K, Hanley KA. 2008. Asymmetric competitive suppression between strains of dengue virus. BMC Microbiol 8:28. doi: 10.1186/1471-2180-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Potiwat R, Komalamisra N, Thavara U, Tawatsin A, Siriyasatien P. 2011. Competitive suppression between chikungunya and dengue virus in Aedes albopictus c6/36 cell line. Southeast Asian J Trop Med Public Health 42:1388–1394. [PubMed] [Google Scholar]
- 144.Steck FT, Rubin H. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology 29:642–653. [DOI] [PubMed] [Google Scholar]
- 145.Sivaram A, Barde PV, Gokhale MD, Singh DK, Mourya DT. 2010. Evidence of co-infection of chikungunya and densonucleosis viruses in C6/36 cell lines and laboratory infected Aedes aegypti (L.) mosquitoes. Parasit Vectors 3:95. doi: 10.1186/1756-3305-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wei W, Shao D, Huang X, Li J, Chen H, Zhang Q, Zhang J. 2006. The pathogenicity of mosquito densovirus (C6/36DNV) and its interaction with dengue virus type II in Aedes albopictus. Am J Trop Med Hyg 75:1118–1126. [PubMed] [Google Scholar]
- 147.Burivong P, Pattanakitsakul SN, Thongrungkiat S, Malasit P, Flegel TW. 2004. Markedly reduced severity of dengue virus infection in mosquito cell cultures persistently infected with Aedes albopictus densovirus (AalDNV). Virology 329:261–269. doi: 10.1016/j.virol.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 148.Kanthong N, Khemnu N, Sriurairatana S, Pattanakitsakul SN, Malasit P, Flegel TW. 2008. Mosquito cells accommodate balanced, persistent co-infections with a densovirus and dengue virus. Dev Comp Immunol 32:1063–1075. doi: 10.1016/j.dci.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 149.Kanthong N, Khemnu N, Pattanakitsakul SN, Malasit P, Flegel TW. 2010. Persistent, triple-virus co-infections in mosquito cells. BMC Microbiol 10:14. doi: 10.1186/1471-2180-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thenmozhi V, Paramasivan R, Samuel PP, Kamaraj T, Balaji T, Dhananjeyan KJ, Venkatasubramani K, Tyagi BK. 2013. Dual infection in human by Japanese encephalitis virus & chikungunya virus in Alappuzha district, Kerala, India. Indian J Med Res 138:362–363. [PMC free article] [PubMed] [Google Scholar]
- 151.Cook JK, Huggins MB, Orbell SJ, Mawditt K, Cavanagh D. 2001. Infectious bronchitis virus vaccine interferes with the replication of avian pneumovirus vaccine in domestic fowl. Avian Pathol 30:233–242. doi: 10.1080/03079450120054640. [DOI] [PubMed] [Google Scholar]
- 152.Beard CW. 1967. Infectious bronchitis virus interference with Newcastle disease virus in monolayers of chicken kidney cells. Avian Dis 11:399–406. doi: 10.2307/1588186. [DOI] [PubMed] [Google Scholar]
- 153.Bracewell CD, Dawson PS, Allan WH. 1972. Antibody responses to a live Newcastle disease vaccine when combined with a live infectious bronchitis vaccine. Vet Rec 90:248–249. doi: 10.1136/vr.90.9.248. [DOI] [PubMed] [Google Scholar]
- 154.Brown JL, Cunningham CH. 1971. Immunofluorescence of avian infectious bronchitis virus and Newcastle disease virus in singly and dually infected cell cultures. Avian Dis 15:923–934. doi: 10.2307/1588883. [DOI] [PubMed] [Google Scholar]
- 155.Hanson LE, White FH, Alberts JO. 1956. Interference between Newcastle disease and infectious bronchitis viruses. Am J Vet Res 17:294–298. [PubMed] [Google Scholar]
- 156.Raggi LG, Lee GG. 1963. Infectious bronchitis virus interference with growth of Newcastle disease virus. I. Study of interference in chicken embryos. Avian Dis 7:106–122. [PubMed] [Google Scholar]
- 157.Hidalgo H, Raggi LG. 1976. Identification of seven isolants of infectious bronchitis virus by interference with the B-1 isolant of Newcastle disease virus. Avian Dis 20:167–172. doi: 10.2307/1589486. [DOI] [PubMed] [Google Scholar]
- 158.Raggi LG, Pignattelli P. 1975. Identification of infectious bronchitis virus by interference with the B-1 isolant of Newcastle disease virus. Waxing and waning of interference. Avian Dis 19:334–342. doi: 10.2307/1588988. [DOI] [PubMed] [Google Scholar]
- 159.Gelb J Jr, Ladman BS, Licata MJ, Shapiro MH, Campion LR. 2007. Evaluating viral interference between infectious bronchitis virus and Newcastle disease virus vaccine strains using quantitative reverse transcription-polymerase chain reaction. Avian Dis 51:924–934. doi: 10.1637/7930-020807-REGR.1. [DOI] [PubMed] [Google Scholar]
- 160.Yachida S, Kuwahara E, Iritani Y, Hayashi Y. 1986. In ovo interference of embryo non-lethal avian infectious bronchitis viruses (IBV) with velogenic Newcastle disease virus and embryo adapted IBV. Res Vet Sci 40:1–3. [PubMed] [Google Scholar]
- 161.Hidalgo H, Gallardo R, Vivar J, Toro H. 1985. Identification of field isolates of infectious bronchitis virus by interference with the La Sota of strain of Newcastle disease virus. Avian Dis 29:335–340. doi: 10.2307/1590493. [DOI] [PubMed] [Google Scholar]
- 162.Beard CW. 1968. An interference type of serological test for infectious bronchitis virus using Newcastle disease virus. Avian Dis 12:658–665. doi: 10.2307/1588450. [DOI] [PubMed] [Google Scholar]
- 163.Costa-Hurtado M, Afonso CL, Miller PJ, Shepherd E, DeJesus E, Smith D, Pantin-Jackwood MJ. 2016. Effect of infection with a mesogenic strain of Newcastle disease virus on infection with highly pathogenic avian influenza virus in chickens. Avian Dis 60:269–278. doi: 10.1637/11171-051915-Reg. [DOI] [PubMed] [Google Scholar]
- 164.Salvaudon L, De Moraes CM, Mescher MC. 2013. Outcomes of co-infection by two potyviruses: implications for the evolution of manipulative strategies. Proc Biol Sci 280:20122959. doi: 10.1098/rspb.2012.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Barr J, Smith C, Smith I, de Jong C, Todd S, Melville D, Broos A, Crameri S, Haining J, Marsh G, Crameri G, Field H, Wang LF. 2015. Isolation of multiple novel paramyxoviruses from pteropid bat urine. J Gen Virol 96:24–29. doi: 10.1099/vir.0.068106-0. [DOI] [PubMed] [Google Scholar]
- 166.Bara JJ, Muturi EJ. 2014. Effect of mixed infections of Sindbis and La Crosse viruses on replication of each virus in vitro. Acta Trop 130:71–75. doi: 10.1016/j.actatropica.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 167.Karpf AR, Lenches E, Strauss EG, Strauss JH, Brown DT. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J Virol 71:7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kent RJ, Crabtree MB, Miller BR. 2010. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex Flavivirus Izabal. PLoS Negl Trop Dis 4:e671. doi: 10.1371/journal.pntd.0000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lopez-Vazquez C, Alonso MC, Dopazo CP, Bandin I. 2016. In vivo study of viral haemorrhagic septicaemia virus and infectious pancreatic necrosis virus coexistence in Senegalese sole (Solea senegalensis). J Fish Dis doi: 10.1111/jfd.12585. [DOI] [PubMed] [Google Scholar]
- 170.Pantin-Jackwood MJ, Costa-Hurtado M, Miller PJ, Afonso CL, Spackman E, Kapczynski DR, Shepherd E, Smith D, Swayne DE. 2015. Experimental co-infections of domestic ducks with a virulent Newcastle disease virus and low or highly pathogenic avian influenza viruses. Vet Microbiol 177:7–17. doi: 10.1016/j.vetmic.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sloutskin A, Yee MB, Kinchington PR, Goldstein RS. 2014. Varicella-zoster virus and herpes simplex virus 1 can infect and replicate in the same neurons whether co- or superinfected. J Virol 88:5079–5086. doi: 10.1128/JVI.00252-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dasika GK, Letchworth GJ. 2000. Homologous and heterologous interference requires bovine herpesvirus-1 glycoprotein D at the cell surface during virus entry. J Gen Virol 81:1041–1049. doi: 10.1099/0022-1317-81-4-1041. [DOI] [PubMed] [Google Scholar]
- 173.Luka PD, Erume J, Yakubu B, Owolodun OA, Shamaki D, Mwiine FN. 2016. Molecular detection of torque teno sus virus and coinfection with African swine fever virus in blood samples of pigs from some slaughterhouses in Nigeria. Adv Virol 2016:6341015. doi: 10.1155/2016/6341015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Semple MG, Cowell A, Dove W, Greensill J, McNamara PS, Halfhide C, Shears P, Smyth RL, Hart CA. 2005. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis 191:382–386. doi: 10.1086/426457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Diallo IS, Taylor J, Gibson J, Hoad J, De Jong A, Hewitson G, Corney BG, Rodwell BJ. 2010. Diagnosis of a naturally occurring dual infection of layer chickens with fowlpox virus and gallid herpesvirus 1 (infectious laryngotracheitis virus). Avian Pathol 39:25–30. doi: 10.1080/03079450903447412. [DOI] [PubMed] [Google Scholar]
- 176.Otta SK, Arulraj R, Ezhil Praveena P, Manivel R, Panigrahi A, Bhuvaneswari T, Ravichandran P, Jithendran KP, Ponniah AG. 2014. Association of dual viral infection with mortality of Pacific white shrimp (Litopenaeus vannamei) in culture ponds in India. VirusDisease 25:63–68. doi: 10.1007/s13337-013-0180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Myers CA, Kasper MR, Yasuda CY, Savuth C, Spiro DJ, Halpin R, Faix DJ, Coon R, Putnam SD, Wierzba TF, Blair PJ. 2011. Dual infection of novel influenza viruses A/H1N1 and A/H3N2 in a cluster of Cambodian patients. Am J Trop Med Hyg 85:961–963. doi: 10.4269/ajtmh.2011.11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vaziry A, Silim A, Bleau C, Frenette D, Lamontagne L. 2013. Dual infections with low virulent chicken infectious anaemia virus (lvCIAV) and intermediate infectious bursal disease virus (iIBDV) in young chicks increase lvCIAV in thymus and bursa while decreasing lymphocyte disorders induced by iIBDV. Avian Pathol 42:88–99. doi: 10.1080/03079457.2013.766306. [DOI] [PubMed] [Google Scholar]
- 179.Ge XY, Wang N, Zhang W, Hu B, Li B, Zhang YZ, Zhou JH, Luo CM, Yang XL, Wu LJ, Wang B, Zhang Y, Li ZX, Shi ZL. 2016. Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virol Sin 31:31–40. doi: 10.1007/s12250-016-3713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Muturi EJ, Bara J. 2015. Sindbis virus interferes with dengue 4 virus replication and its potential transmission by Aedes albopictus. Parasit Vectors 8:65. doi: 10.1186/s13071-015-0667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cabral-Castro MJ, Cavalcanti MG, Peralta RH, Peralta JM. 2016. Molecular and serological techniques to detect co-circulation of DENV, ZIKV and CHIKV in suspected dengue-like syndrome patients. J Clin Virol 82:108–111. doi: 10.1016/j.jcv.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 182.Rosso F, Pacheco R, Rodriguez S, Bautista D. 2016. Co-infection by Chikungunya virus (CHIK-V) and dengue virus (DEN-V) during a recent outbreak in Cali, Colombia: Report of a fatal case. Rev Chilena Infectol 33:464–467. doi: 10.4067/S0716-10182016000400013. [DOI] [PubMed] [Google Scholar]
- 183.Chahar HS, Bharaj P, Dar L, Guleria R, Kabra SK, Broor S. 2009. Co-infections with chikungunya virus and dengue virus in Delhi, India. Emerg Infect Dis 15:1077–1080. doi: 10.3201/eid1507.080638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hapuarachchi HA, Bandara KB, Hapugoda MD, Williams S, Abeyewickreme W. 2008. Laboratory confirmation of dengue and chikungunya co-infection. Ceylon Med J 53:104–105. doi: 10.4038/cmj.v53i3.252. [DOI] [PubMed] [Google Scholar]
- 185.Leroy EM, Nkoghe D, Ollomo B, Nze-Nkogue C, Becquart P, Grard G, Pourrut X, Charrel R, Moureau G, Ndjoyi-Mbiguino A, De-Lamballerie X. 2009. Concurrent chikungunya and dengue virus infections during simultaneous outbreaks, Gabon, 2007. Emerg Infect Dis 15:591–593. doi: 10.3201/eid1504.080664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nayar SK, Noridah O, Paranthaman V, Ranjit K, Norizah I, Chem YK, Mustafa B, Chua KB. 2007. Co-infection of dengue virus and chikungunya virus in two patients with acute febrile illness. Med J Malaysia 62:335–336. [PubMed] [Google Scholar]
- 187.Schilling S, Emmerich P, Gunther S, Schmidt-Chanasit J. 2009. Dengue and Chikungunya virus co-infection in a German traveller. J Clin Virol 45:163–164. doi: 10.1016/j.jcv.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 188.Brooks JB, Ruiz CA, Fragoso YD. 2016. Acute illness with neurological findings caused by coinfection of dengue and chikungunya viruses in a Brazilian patient. J Infect Public Health doi: 10.1016/j.jiph.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 189.Dittmar D, Castro A, Haines H. 1982. Demonstration of interference between dengue virus types in cultured mosquito cells using monoclonal antibody probes. J Gen Virol 59:273–282. doi: 10.1099/0022-1317-59-2-273. [DOI] [PubMed] [Google Scholar]
- 190.Chu Y, Yan X, Gao P, Zhao P, He Y, Liu J, Lu Z. 2011. Molecular detection of a mixed infection of Goatpox virus, Orf virus, and Mycoplasma capricolum subsp. capripneumoniae in goats. J Vet Diagn Invest 23:786–789. doi: 10.1177/1040638711407883. [DOI] [PubMed] [Google Scholar]
- 191.Esposito S, Zampiero A, Bianchini S, Mori A, Scala A, Tagliabue C, Sciarrabba CS, Fossali E, Piralla A, Principi N. 2016. Epidemiology and clinical characteristics of respiratory infections due to adenovirus in children living in Milan, Italy, during 2013 and 2014. PLoS One 11:e0152375. doi: 10.1371/journal.pone.0152375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gonzalez Alvarez DA, Lopez Cortes LF, Cordero E. 2016. Impact of HIV on the severity of influenza. Expert Rev Respir Med 10:463–472. doi: 10.1586/17476348.2016.1157474. [DOI] [PubMed] [Google Scholar]
- 193.Zhou N, Fan C, Liu S, Zhou J, Jin Y, Zheng X, Wang Q, Liu J, Yang H, Gu J, Zhou J. 2017. Cellular proteomic analysis of porcine circovirus type 2 and classical swine fever virus coinfection in porcine kidney-15 cells using isobaric tags for relative and absolute quantitation-coupled LC-MS/MS. Electrophoresis 38:1276–1291. doi: 10.1002/elps.201600541. [DOI] [PubMed] [Google Scholar]
- 194.Espinoza-Gomez F, Delgado-Enciso I, Valle-Reyes S, Ochoa-Jimenez R, Arechiga-Ramirez C, Gamez-Arroyo JL, Vazquez-Campuzano R, Guzman-Bracho C, Vasquez C, Lopez-Lemus UA. 2017. Dengue virus coinfection in human immunodeficiency virus-1-infected patients on the west coast of Mexico. Am J Trop Med Hyg doi: 10.4269/ajtmh.17-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Carrillo-Hernandez MY, Ruiz-Saenz J, Villamizar LJ, Gomez-Rangel SY, Martinez-Gutierrez M. 2018. Co-circulation and simultaneous co-infection of dengue, chikungunya, and zika viruses in patients with febrile syndrome at the Colombian-Venezuelan border. BMC Infect Dis 18:61. doi: 10.1186/s12879-018-2976-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shinjoh M, Omoe K, Saito N, Matsuo N, Nerome K. 2000. In vitro growth profiles of respiratory syncytial virus in the presence of influenza virus. Acta Virol 44:91–97. [PubMed] [Google Scholar]
- 197.Goto H, Ihira H, Morishita K, Tsuchiya M, Ohta K, Yumine N, Tsurudome M, Nishio M. 2016. Enhanced growth of influenza A virus by coinfection with human parainfluenza virus type 2. Med Microbiol Immunol 205:209–218. doi: 10.1007/s00430-015-0441-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hurt AC, Nor'e SS, McCaw JM, Fryer HR, Mosse J, McLean AR, Barr IG. 2010. Assessing the viral fitness of oseltamivir-resistant influenza viruses in ferrets, using a competitive-mixtures model. J Virol 84:9427–9438. doi: 10.1128/JVI.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dobrescu I, Levast B, Lai K, Delgado-Ortega M, Walker S, Banman S, Townsend H, Simon G, Zhou Y, Gerdts V, Meurens F. 2014. In vitro and ex vivo analyses of co-infections with swine influenza and porcine reproductive and respiratory syndrome viruses. Vet Microbiol 169:18–32. doi: 10.1016/j.vetmic.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Calixto R, Oliveira G, Lima M, Andrade AC, Trindade GS, de Oliveira DB, Kroon EG. 2017. A model to detect autochthonous group 1 and 2 Brazilian vaccinia virus coinfections: development of a qPCR tool for diagnosis and pathogenesis studies. Viruses 10:15. doi: 10.3390/v10010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bellecave P, Gouttenoire J, Gajer M, Brass V, Koutsoudakis G, Blum HE, Bartenschlager R, Nassal M, Moradpour D. 2009. Hepatitis B and C virus coinfection: a novel model system reveals the absence of direct viral interference. Hepatology 50:46–55. doi: 10.1002/hep.22951. [DOI] [PubMed] [Google Scholar]
- 202.Rozeboom LE, Kassira EN. 1969. Dual infections of mosquitoes with strains of West Nile virus. J Med Entomol 6:407–411. doi: 10.1093/jmedent/6.4.407. [DOI] [PubMed] [Google Scholar]
- 203.Pesko K, Mores CN. 2009. Effect of sequential exposure on infection and dissemination rates for West Nile and St. Louis encephalitis viruses in Culex quinquefasciatus. Vector Borne Zoonotic Dis 9:281–286. doi: 10.1089/vbz.2007.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wang H, Swann R, Thomas E, Innes HA, Valerio H, Hayes PC, Allen S, Barclay ST, Wilks D, Fox R, Bhattacharyya D, Kennedy N, Morris J, Fraser A, Stanley AJ, Gunson R, McLntyre PG, Hunt A, Hutchinson SJ, Mills PR, Dillon JF. 2018. Impact of previous hepatitis B infection on the clinical outcomes from chronic hepatitis C? A population-level analysis. J Viral Hepat doi: 10.1111/jvh.12897. [DOI] [PubMed] [Google Scholar]
- 205.Zebovitz E, Brown A. 1968. Interference among group A arboviruses. J Virol 2:1283–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Brinton MA. 2001. Host factors involved in West Nile virus replication. Ann N Y Acad Sci 951:207–219. doi: 10.1111/j.1749-6632.2001.tb02698.x. [DOI] [PubMed] [Google Scholar]
- 207.Riis B, Rattan SI, Clark BF, Merrick WC. 1990. Eukaryotic protein elongation factors. Trends Biochem Sci 15:420–424. doi: 10.1016/0968-0004(90)90279-K. [DOI] [PubMed] [Google Scholar]
- 208.Li D, Wei T, Abbott CM, Harrich D. 2013. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol Mol Biol Rev 77:253–266. doi: 10.1128/MMBR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Van Der Kelen K, Beyaert R, Inze D, De Veylder L. 2009. Translational control of eukaryotic gene expression. Crit Rev Biochem Mol Biol 44:143–168. doi: 10.1080/10409230902882090. [DOI] [PubMed] [Google Scholar]
- 210.De Nova-Ocampo M, Villegas-Sepulveda N, del Angel RM. 2002. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- 211.Davis WG, Blackwell JL, Shi PY, Brinton MA. 2007. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J Virol 81:10172–10187. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Blackwell JL, Brinton MA. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol 71:6433–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yocupicio-Monroy M, Padmanabhan R, Medina F, del Angel RM. 2007. Mosquito La protein binds to the 3′ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology 357:29–40. doi: 10.1016/j.virol.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 214.Garcia-Montalvo BM, Medina F, del Angel RM. 2004. La protein binds to NS5 and NS3 and to the 5′ and 3′ ends of dengue 4 virus RNA. Virus Res 102:141–150. doi: 10.1016/j.virusres.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 215.Gomila RC, Martin GW, Gehrke L. 2011. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS One 6:e16687. doi: 10.1371/journal.pone.0016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kenney JL, Solberg OD, Langevin SA, Brault AC. 2014. Characterization of a novel insect-specific flavivirus from Brazil: potential for inhibition of infection of arthropod cells with medically important flaviviruses. J Gen Virol 95:2796–2808. doi: 10.1099/vir.0.068031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Singh IR, Suomalainen M, Varadarajan S, Garoff H, Helenius A. 1997. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virology 231:59–71. doi: 10.1006/viro.1997.8492. [DOI] [PubMed] [Google Scholar]
- 218.Kim GN, Kang CY. 2005. Utilization of homotypic and heterotypic proteins of vesicular stomatitis virus by defective interfering particle genomes for RNA replication and virion assembly: implications for the mechanism of homologous viral interference. J Virol 79:9588–9596. doi: 10.1128/JVI.79.15.9588-9596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chotkowski HL, Ciota AT, Jia Y, Puig-Basagoiti F, Kramer LD, Shi PY, Glaser RL. 2008. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology 377:197–206. doi: 10.1016/j.virol.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. 2009. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog 5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sim S, Jupatanakul N, Ramirez JL, Kang S, Romero-Vivas CM, Mohammed H, Dimopoulos G. 2013. Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS Negl Trop Dis 7:e2295. doi: 10.1371/journal.pntd.0002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wu X, Hong H, Yue J, Wu Y, Li X, Jiang L, Li L, Li Q, Gao G, Yang X. 2010. Inhibitory effect of small interfering RNA on dengue virus replication in mosquito cells. Virol J 7:270. doi: 10.1186/1743-422X-7-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pijlman GP. 2014. Flavivirus RNAi suppression: decoding non-coding RNA. Curr Opin Virol 7:55–60. doi: 10.1016/j.coviro.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 224.Lemm JA, Rice CM. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol 67:1916–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lemm JA, Rumenapf T, Strauss EG, Strauss JH, Rice CM. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J 13:2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Shirako Y, Strauss JH. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol 68:1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Flenniken ML, Andino R. 2013. Non-specific dsRNA-mediated antiviral response in the honey bee. PLoS One 8:e77263. doi: 10.1371/journal.pone.0077263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nunes FM, Aleixo AC, Barchuk AR, Bomtorin AD, Grozinger CM, Simoes ZL. 2013. Non-target effects of green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) used in honey bee RNA interference (RNAi) assays. Insects 4:90–103. doi: 10.3390/insects4010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Laliberte JP, Moss B. 2014. A novel mode of poxvirus superinfection exclusion that prevents fusion of the lipid bilayers of viral and cellular membranes. J Virol 88:9751–9768. doi: 10.1128/JVI.00816-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Moss B. 2012. Poxvirus cell entry: how many proteins does it take? Viruses 4:688–707. doi: 10.3390/v4050688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zou G, Zhang B, Lim PY, Yuan Z, Bernard KA, Shi PY. 2009. Exclusion of West Nile virus superinfection through RNA replication. J Virol 83:11765–11776. doi: 10.1128/JVI.01205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kakumani PK, Ponia SS, S RK, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, Bhatnagar RK. 2013. Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J Virol 87:8870–8883. doi: 10.1128/JVI.02774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Lingel A, Simon B, Izaurralde E, Sattler M. 2005. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep 6:1149–1155. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. 2005. Dual modes of RNA-silencing suppression by flock house virus protein B2. Nat Struct Mol Biol 12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 235.Kingsolver MB, Huang Z, Hardy RW. 2013. Insect antiviral innate immunity: pathways, effectors, and connections. J Mol Biol 425:4921–4936. doi: 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Goic B, Saleh MC. 2012. Living with the enemy: viral persistent infections from a friendly viewpoint. Curr Opin Microbiol 15:531–537. doi: 10.1016/j.mib.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 237.Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh MC. 2013. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol 14:396–403. doi: 10.1038/ni.2542. [DOI] [PubMed] [Google Scholar]
- 238.Chakraborty PR, Ahmed R, Fields BN. 1979. Genetics of reovirus: the relationship of interference to complementation and reassortment of temperature-sensitive mutants at nonpermissive temperature. Virology 94:119–127. doi: 10.1016/0042-6822(79)90442-2. [DOI] [PubMed] [Google Scholar]
- 239.Pohjanpelto P, Cooper PD. 1965. Interference between polioviruses induced by strains that cannot multiply. Virology 25:350–357. doi: 10.1016/0042-6822(65)90054-1. [DOI] [PubMed] [Google Scholar]
- 240.Cooper PD. 1965. Rescue of one phenotype in mixed infections with heat-defective mutants of type 1 poliovirus. Virology 25:431–438. doi: 10.1016/0042-6822(65)90064-4. [DOI] [PubMed] [Google Scholar]
- 241.Jofre JT, Courtney RJ, Schaffer PA. 1981. A dominant lethal temperature-sensitive mutant of herpes simplex virus type 1. Virology 111:173–190. doi: 10.1016/0042-6822(81)90663-2. [DOI] [PubMed] [Google Scholar]
- 242.Norkin LC. 1980. Persistent infections of green monkey kidney cells initiated with temperature-sensitive mutants of simian virus 40. Virology 107:375–388. doi: 10.1016/0042-6822(80)90305-0. [DOI] [PubMed] [Google Scholar]
- 243.Keranen S. 1977. Interference of wild type virus replication by an RNA negative temperature-sensitive mutant of Semliki Forest virus. Virology 80:1–11. doi: 10.1016/0042-6822(77)90376-2. [DOI] [PubMed] [Google Scholar]
- 244.Youngner JS, Frielle DW, Whitaker-Dowling P. 1986. Dominance of temperature-sensitive phenotypes. I. Studies of the mechanism of inhibition of the growth of wild-type vesicular stomatitis virus. Virology 155:225–235. [DOI] [PubMed] [Google Scholar]
- 245.Youngner JS, Quagliana DO. 1976. Temperature-sensitive mutants of vesicular stomatitis virus are conditionally defective particles that interfere with and are rescued by wild-type virus. J Virol 19:102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Benson RE, Sanfridson A, Ottinger JS, Doyle C, Cullen BR. 1993. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J Exp Med 177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Le Guern M, Levy JA. 1992. Human immunodeficiency virus (HIV) type 1 can superinfect HIV-2-infected cells: pseudotype virions produced with expanded cellular host range. Proc Natl Acad Sci U S A 89:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Michel N, Allespach I, Venzke S, Fackler OT, Keppler OT. 2005. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr Biol 15:714–723. doi: 10.1016/j.cub.2005.02.058. [DOI] [PubMed] [Google Scholar]
- 249.Hrecka K, Swigut T, Schindler M, Kirchhoff F, Skowronski J. 2005. Nef proteins from diverse groups of primate lentiviruses downmodulate CXCR4 to inhibit migration to the chemokine stromal derived factor 1. J Virol 79:10650–10659. doi: 10.1128/JVI.79.16.10650-10659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Geleziunas R, Bour S, Wainberg MA. 1994. Cell surface down-modulation of CD4 after infection by HIV-1. FASEB J 8:593–600. doi: 10.1096/fasebj.8.9.8005387. [DOI] [PubMed] [Google Scholar]
- 251.Willey RL, Maldarelli F, Martin MA, Strebel K. 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol 66:7193–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Breiner KM, Schaller H, Knolle PA. 2001. Endothelial cell-mediated uptake of a hepatitis B virus: a new concept of liver targeting of hepatotropic microorganisms. Hepatology 34:803–808. doi: 10.1053/jhep.2001.27810. [DOI] [PubMed] [Google Scholar]
- 253.Breiner KM, Urban S, Glass B, Schaller H. 2001. Envelope protein-mediated down-regulation of hepatitis B virus receptor in infected hepatocytes. J Virol 75:143–150. doi: 10.1128/JVI.75.1.143-150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Nethe M, Berkhout B, van der Kuyl AC. 2005. Retroviral superinfection resistance. Retrovirology 2:52. doi: 10.1186/1742-4690-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Schneider-Schaulies J, Schnorr JJ, Brinckmann U, Dunster LM, Baczko K, Liebert UG, Schneider-Schaulies S, ter Meulen V. 1995. Receptor usage and differential downregulation of CD46 by measles virus wild-type and vaccine strains. Proc Natl Acad Sci U S A 92:3943–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Walters KA, Joyce MA, Addison WR, Fischer KP, Tyrrell DL. 2004. Superinfection exclusion in duck hepatitis B virus infection is mediated by the large surface antigen. J Virol 78:7925–7937. doi: 10.1128/JVI.78.15.7925-7937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Bratt MA, Rubin H. 1968. Specific interference among strains of Newcastle disease virus. II. Comparison of interference by active and inactive virus. Virology 35:381–394. [DOI] [PubMed] [Google Scholar]
- 258.Bratt MA, Rubin H. 1968. Specific interference among strains of Newcastle disease virus. 3. Mechanisms of interference. Virology 35:395–407. [DOI] [PubMed] [Google Scholar]
- 259.Steck FT, Rubin H. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology 29:628–641. [DOI] [PubMed] [Google Scholar]
- 260.Rikkonen M, Peranen J, Kaariainen L. 1992. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology 189:462–473. doi: 10.1016/0042-6822(92)90570-F. [DOI] [PubMed] [Google Scholar]
- 261.Ranki M, Ulmanen I, Kaariainen L. 1979. Semliki Forest virus-specific nonstructural protein is associated with ribosomes. FEBS Lett 108:299–302. doi: 10.1016/0014-5793(79)81232-6. [DOI] [PubMed] [Google Scholar]
- 262.Simon KO, Cardamone JJ Jr, Whitaker-Dowling PA, Youngner JS, Widnell CC. 1990. Cellular mechanisms in the superinfection exclusion of vesicular stomatitis virus. Virology 177:375–379. doi: 10.1016/0042-6822(90)90494-C. [DOI] [PubMed] [Google Scholar]
- 263.Whitaker-Dowling P, Youngner JS, Widnell CC, Wilcox DK. 1983. Superinfection exclusion by vesicular stomatitis virus. Virology 131:137–143. doi: 10.1016/0042-6822(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 264.Geib T, Sauder C, Venturelli S, Hassler C, Staeheli P, Schwemmle M. 2003. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J Virol 77:4283–4290. doi: 10.1128/JVI.77.7.4283-4290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol 77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Schaller T, Appel N, Koutsoudakis G, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J Virol 81:4591–4603. doi: 10.1128/JVI.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Claus C, Tzeng WP, Liebert UG, Frey TK. 2007. Rubella virus-induced superinfection exclusion studied in cells with persisting replicons. J Gen Virol 88:2769–2773. doi: 10.1099/vir.0.83092-0. [DOI] [PubMed] [Google Scholar]
- 268.Whitaker-Dowling P, Youngner JS. 1987. Viral interference-dominance of mutant viruses over wild-type virus in mixed infections. Microbiol Rev 51:179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Beperet I, Irons SL, Simon O, King LA, Williams T, Possee RD, Lopez-Ferber M, Caballero P. 2014. Superinfection exclusion in alphabaculovirus infections is concomitant with actin reorganization. J Virol 88:3548–3556. doi: 10.1128/JVI.02974-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.McAllister WT, Barrett CL. 1977. Superinfection exclusion by bacteriophage T7. J Virol 24:709–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Christen L, Seto J, Niles EG. 1990. Superinfection exclusion of vaccinia virus in virus-infected cell cultures. Virology 174:35–42. doi: 10.1016/0042-6822(90)90051-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bergua M, Zwart MP, El-Mohtar C, Shilts T, Elena SF, Folimonova SY. 2014. A viral protein mediates superinfection exclusion at the whole-organism level but is not required for exclusion at the cellular level. J Virol 88:11327–11338. doi: 10.1128/JVI.01612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Folimonova SY. 2012. Superinfection exclusion is an active virus-controlled function that requires a specific viral protein. J Virol 86:5554–5561. doi: 10.1128/JVI.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wildum S, Schindler M, Munch J, Kirchhoff F. 2006. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J Virol 80:8047–8059. doi: 10.1128/JVI.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Tscherne DM, Evans MJ, von Hahn T, Jones CT, Stamataki Z, McKeating JA, Lindenbach BD, Rice CM. 2007. Superinfection exclusion in cells infected with hepatitis C virus. J Virol 81:3693–3703. doi: 10.1128/JVI.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Huang C, Kolokoltsova OA, Yun NE, Seregin AV, Ronca S, Koma T, Paessler S. 2015. Highly pathogenic New World and Old World human arenaviruses induce distinct interferon responses in human cells. J Virol 89:7079–7088. doi: 10.1128/JVI.00526-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ellenberg P, Edreira M, Scolaro L. 2004. Resistance to superinfection of Vero cells persistently infected with Junin virus. Arch Virol 149:507–522. doi: 10.1007/s00705-003-0227-1. [DOI] [PubMed] [Google Scholar]
- 278.Thiel N, Zischke J, Elbasani E, Kay-Fedorov P, Messerle M. 2015. Viral interference with functions of the cellular receptor tyrosine phosphatase CD45. Viruses 7:1540–1557. doi: 10.3390/v7031540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Munoz-Gonzalez S, Perez-Simo M, Colom-Cadena A, Cabezon O, Bohorquez JA, Rosell R, Perez LJ, Marco I, Lavin S, Domingo M, Ganges L. 2016. Classical swine fever virus vs. classical swine fever virus: the superinfection exclusion phenomenon in experimentally infected wild boar. PLoS One 11:e0149469. doi: 10.1371/journal.pone.0149469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Meignier B, Norrild B, Roizman B. 1983. Colonization of murine ganglia by a superinfecting strain of herpes simplex virus. Infect Immun 41:702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Jarosinski KW. 2012. Dual infection and superinfection inhibition of epithelial skin cells by two alphaherpesviruses co-occur in the natural host. PLoS One 7:e37428. doi: 10.1371/journal.pone.0037428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Munoz-Gonzalez S, Perez-Simo M, Munoz M, Bohorquez JA, Rosell R, Summerfield A, Domingo M, Ruggli N, Ganges L. 2015. Efficacy of a live attenuated vaccine in classical swine fever virus postnatally persistently infected pigs. Vet Res 46:78. doi: 10.1186/s13567-015-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Lee YM, Tscherne DM, Yun SI, Frolov I, Rice CM. 2005. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J Virol 79:3231–3242. doi: 10.1128/JVI.79.6.3231-3242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Adams RH, Brown DT. 1985. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J Virol 54:351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ludlow M, McQuaid S, Cosby SL, Cattaneo R, Rima BK, Duprex WP. 2005. Measles virus superinfection immunity and receptor redistribution in persistently infected NT2 cells. J Gen Virol 86:2291–2303. doi: 10.1099/vir.0.81052-0. [DOI] [PubMed] [Google Scholar]
- 286.Gaudin R, Kirchhausen T. 2015. Superinfection exclusion is absent during acute Junin virus infection of Vero and A549 cells. Sci Rep 5:15990. doi: 10.1038/srep15990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Ellenberg P, Linero FN, Scolaro LA. 2007. Superinfection exclusion in BHK-21 cells persistently infected with Junin virus. J Gen Virol 88:2730–2739. doi: 10.1099/vir.0.83041-0. [DOI] [PubMed] [Google Scholar]
- 288.Huang IC, Li W, Sui J, Marasco W, Choe H, Farzan M. 2008. Influenza A virus neuraminidase limits viral superinfection. J Virol 82:4834–4843. doi: 10.1128/JVI.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Doceul V, Hollinshead M, van der Linden L, Smith GL. 2010. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science 327:873–876. doi: 10.1126/science.1183173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Joklik WK. 1964. The intracellular uncoating of poxvirus DNA. II. The molecular basis of the uncoating process J Mol Biol 8:277–288. [DOI] [PubMed] [Google Scholar]
- 291.Joklik WK. 1964. The intracellular uncoating of poxvirus DNA. I. The fate of radioactively-labeled rabbitpox virus. J Mol Biol 8:263–276. [DOI] [PubMed] [Google Scholar]
- 292.Moss B, Rosenblum EN, Grimley PM. 1971. Assembly of virus particles during mixed infection with wild-type vaccinia and a rifampicin-resistant mutant. Virology 45:135–148. doi: 10.1016/0042-6822(71)90120-6. [DOI] [PubMed] [Google Scholar]
- 293.Turner PC, Moyer RW. 2008. The vaccinia virus fusion inhibitor proteins SPI-3 (K2) and HA (A56) expressed by infected cells reduce the entry of superinfecting virus. Virology 380:226–233. doi: 10.1016/j.virol.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Wagenaar TR, Moss B. 2009. Expression of the A56 and K2 proteins is sufficient to inhibit vaccinia virus entry and cell fusion. J Virol 83:1546–1554. doi: 10.1128/JVI.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wagenaar TR, Moss B. 2007. Association of vaccinia virus fusion regulatory proteins with the multicomponent entry/fusion complex. J Virol 81:6286–6293. doi: 10.1128/JVI.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Wagenaar TR, Ojeda S, Moss B. 2008. Vaccinia virus A56/K2 fusion regulatory protein interacts with the A16 and G9 subunits of the entry fusion complex. J Virol 82:5153–5160. doi: 10.1128/JVI.00162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Campadelli-Fiume G, Qi S, Avitabile E, Foa-Tomasi L, Brandimarti R, Roizman B. 1990. Glycoprotein D of herpes simplex virus encodes a domain which precludes penetration of cells expressing the glycoprotein by superinfecting herpes simplex virus. J Virol 64:6070–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Meurens F, Schynts F, Keil GM, Muylkens B, Vanderplasschen A, Gallego P, Thiry E. 2004. Superinfection prevents recombination of the alphaherpesvirus bovine herpesvirus 1. J Virol 78:3872–3879. doi: 10.1128/JVI.78.8.3872-3879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Dutta SK, Myrup AC, Thaker SR. 1986. In vitro interference between equine herpesvirus types 1 and 2. Am J Vet Res 47:747–750. [PubMed] [Google Scholar]
- 300.Criddle A, Thornburg T, Kochetkova I, DePartee M, Taylor MP. 2016. gD-independent superinfection exclusion of alphaherpesviruses. J Virol 90:4049–4058. doi: 10.1128/JVI.00089-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Horga MA, Gusella GL, Greengard O, Poltoratskaia N, Porotto M, Moscona A. 2000. Mechanism of interference mediated by human parainfluenza virus type 3 infection. J Virol 74:11792–11799. doi: 10.1128/JVI.74.24.11792-11799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Morrison TG, McGinnes LW. 1989. Avian cells expressing the Newcastle disease virus hemagglutinin-neuraminidase protein are resistant to Newcastle disease virus infection. Virology 171:10–17. doi: 10.1016/0042-6822(89)90505-9. [DOI] [PubMed] [Google Scholar]
- 303.Mordecai GJ, Brettell LE, Martin SJ, Dixon D, Jones IM, Schroeder DC. 2016. Superinfection exclusion and the long-term survival of honey bees in Varroa-infested colonies. ISME J 10:1182–1191. doi: 10.1038/ismej.2015.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Flegel TW. 2001. The shrimp response to viral pathogens, 254–278. In Browdy CL, Jory DE (ed), The new wave: proceedings of a special session on sustainable shrimp farming. World Aquaculture Society, Sorrento, LA. [Google Scholar]
- 305.Sabin AB. 1959. Present position of immunization against poliomyelitis with live virus vaccines. Br Med J 1:663–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Wheelock EF, Larke RP, Caroline NL. 1968. Interference in human viral infections: present status and prospects for the future. Prog Med Virol 10:286–347. [PubMed] [Google Scholar]
- 307.Li X, Hanson RP. 1989. In vivo interference by Newcastle disease virus in chickens, the natural host of the virus. Arch Virol 108:229–245. doi: 10.1007/BF01310936. [DOI] [PubMed] [Google Scholar]
- 308.Whitaker-Dowling P, Lucas W, Youngner JS. 1990. Cold-adapted vaccine strains of influenza A virus act as dominant negative mutants in mixed infections with wild-type influenza A virus. Virology 175:358–364. doi: 10.1016/0042-6822(90)90420-V. [DOI] [PubMed] [Google Scholar]
- 309.Rinaldo CR Jr, Richter BS, Black PH, Callery R, Chess L, Hirsch MS. 1978. Replication of herpes simplex virus and cytomegalovirus in human leukocytes. J Immunol 120:130–136. [PubMed] [Google Scholar]
- 310.Norkin LC. 1976. Rhesus monkeys kidney cells persistently infected with simian virus 40: production of defective interfering virus and acquisition of the transformed phenotype. Infect Immun 14:783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Ahmed R, Chakraborty PR, Fields BN. 1980. Genetic variation during lytic reovirus infection: high-passage stocks of wild-type reovirus contain temperature-sensitive mutants. J Virol 34:285–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Elliott RM, Wilkie ML. 1986. Persistent infection of Aedes albopictus C6/36 cells by Bunyamwera virus. Virology 150:21–32. doi: 10.1016/0042-6822(86)90262-X. [DOI] [PubMed] [Google Scholar]
- 313.Kowal KJ, Stollar V. 1980. Differential sensitivity of infectious and defective-interfering particles of Sindbis virus to ultraviolet irradiation. Virology 103:149–157. doi: 10.1016/0042-6822(80)90133-6. [DOI] [PubMed] [Google Scholar]
- 314.Peleg J, Stollar V. 1974. Homologous interference in Aedes aegypti cell cultures infected with Sindbis virus. Arch Gesamte Virusforsch 45:309–318. doi: 10.1007/BF01242874. [DOI] [PubMed] [Google Scholar]
- 315.Shenk TE, Koshelnyk KA, Stollar V. 1974. Temperature-sensitive virus from Aedes albopictus cells chronically infected with Sindbis virus. J Virol 13:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ju G, Birrer M, Udem S, Bloom BR. 1980. Complementation analysis of measles virus mutants isolated from persistently infected lymphoblastoid cell lines. J Virol 33:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Frielle DW, Huang DD, Youngner JS. 1984. Persistent infection with influenza A virus: evolution of virus mutants. Virology 138:103–117. doi: 10.1016/0042-6822(84)90151-X. [DOI] [PubMed] [Google Scholar]
- 318.Perrault J. 1981. Origin and replication of defective interfering particles. Curr Top Microbiol Immunol 93:151–207. [DOI] [PubMed] [Google Scholar]
- 319.Dasgupta R, Selling B, Rueckert R. 1994. Flock house virus: a simple model for studying persistent infection in cultured Drosophila cells. Arch Virol Suppl 9:121–132. [DOI] [PubMed] [Google Scholar]
- 320.Rechavi O, Minevich G, Hobert O. 2011. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Dasgupta R, Free HM, Zietlow SL, Paskewitz SM, Aksoy S, Shi L, Fuchs J, Hu C, Christensen BM. 2007. Replication of flock house virus in three genera of medically important insects. J Med Entomol 44:102–110. doi: 10.1093/jmedent/41.5.102. [DOI] [PubMed] [Google Scholar]
- 322.Eickbush TH, Jamburuthugoda VK. 2008. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res 134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Wu Y, Marsh JW. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503–1506. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- 324.Flegel TW. 2007. Update on viral accommodation, a model for host-viral interaction in shrimp and other arthropods. Dev Comp Immunol 31:217–231. doi: 10.1016/j.dci.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 325.Flegel TW. 2012. Historic emergence, impact and current status of shrimp pathogens in Asia. J Invertebr Pathol 110:166–173. doi: 10.1016/j.jip.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 326.Johansson MW, Soderhall K. 1996. The prophenoloxidase activating system and associated proteins in invertebrates. Prog Mol Subcell Biol 15:46–66. doi: 10.1007/978-3-642-79735-4_3. [DOI] [PubMed] [Google Scholar]
- 327.Namikoshi A, Wu JL, Yamashita T, Nishizawa T, Nishioka T, Arimoto M, Muroga K. 2004. Vaccination trials with Penaeus japonicus to induce resistance to white spot syndrome virus. Aquaculture 229:25–35. doi: 10.1016/S0044-8486(03)00363-6. [DOI] [Google Scholar]
- 328.Venegas CA, Nonaka L, Mushiake K, Nishizawa T, Murog K. 2000. Quasi-immune response of Penaeus japonicus to penaeid rod-shaped DNA virus (PRDV). Dis Aquat Organ 42:83–89. doi: 10.3354/dao042083. [DOI] [PubMed] [Google Scholar]
- 329.Khanobdee K, Soowannayan C, Flegel TW, Ubol S, Withyachumnarnkul B. 2002. Evidence for apoptosis correlated with mortality in the giant black tiger shrimp Penaeus monodon infected with yellow head virus. Dis Aquat Organ 48:79–90. doi: 10.3354/dao048079. [DOI] [PubMed] [Google Scholar]
- 330.Sahtout AH, Hassan MD, Shariff M. 2001. DNA fragmentation, an indicator of apoptosis, in cultured black tiger shrimp Penaeus monodon infected with white spot syndrome virus (WSSV). Dis Aquat Organ 44:155–159. doi: 10.3354/dao044155. [DOI] [PubMed] [Google Scholar]
- 331.Wongprasert K, Khanobdee K, Glunukarn SS, Meeratana P, Withyachumnarnkul B. 2003. Time-course and levels of apoptosis in various tissues of black tiger shrimp Penaeus monodon infected with white-spot syndrome virus. Dis Aquat Organ 55:3–10. doi: 10.3354/dao055003. [DOI] [PubMed] [Google Scholar]
- 332.Daniel R, Katz RA, Skalka AM. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 333.Aguilera ER, Erickson AK, Jesudhasan PR, Robinson CM, Pfeiffer JK. 2017. Plaques formed by mutagenized viral populations have elevated coinfection frequencies. mBio 8:e02020-16. doi: 10.1128/mBio.02020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Hansen H, Okeke MI, Nilssen O, Traavik T. 2004. Recombinant viruses obtained from co-infection in vitro with a live vaccinia-vectored influenza vaccine and a naturally occurring cowpox virus display different plaque phenotypes and loss of the transgene. Vaccine 23:499–506. doi: 10.1016/j.vaccine.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 335.Baron MD, Diallo A, Lancelot R, Libeau G. 2016. Peste des petits ruminants virus. Adv Virus Res 95:1–42. doi: 10.1016/bs.aivir.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 336.Wernery U, Kaaden OR. 2004. Foot-and-mouth disease in camelids: a review. Vet J 168:134–142. doi: 10.1016/j.tvjl.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 337.Downham MA, Gardner PS, McQuillin J, Ferris JA. 1975. Role of respiratory viruses in childhood mortality. Br Med J 1:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Meissner HC, Murray SA, Kiernan MA, Snydman DR, McIntosh K. 1984. A simultaneous outbreak of respiratory syncytial virus and parainfluenza virus type 3 in a newborn nursery. J Pediatr 104:680–684. doi: 10.1016/S0022-3476(84)80943-9. [DOI] [PubMed] [Google Scholar]
- 339.Olson LC, Lexomboon U, Sithisarn P, Noyes HE. 1973. The etiology of respiratory tract infections in a tropical country. Am J Epidemiol 97:34–43. doi: 10.1093/oxfordjournals.aje.a121481. [DOI] [PubMed] [Google Scholar]
- 340.Rahman M, Huq F, Sack DA, Butler T, Azad AK, Alam A, Nahar N, Islam M. 1990. Acute lower respiratory tract infections in hospitalized patients with diarrhea in Dhaka, Bangladesh. Rev Infect Dis 12(Suppl 8):S899–S906. doi: 10.1093/clinids/12.Supplement_8.S899. [DOI] [PubMed] [Google Scholar]
- 341.Martin ET, Fairchok MP, Stednick ZJ, Kuypers J, Englund JA. 2013. Epidemiology of multiple respiratory viruses in childcare attendees. J Infect Dis 207:982–989. doi: 10.1093/infdis/jis934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Brand HK, de Groot R, Galama JM, Brouwer ML, Teuwen K, Hermans PW, Melchers WJ, Warris A. 2012. Infection with multiple viruses is not associated with increased disease severity in children with bronchiolitis. Pediatr Pulmonol 47:393–400. doi: 10.1002/ppul.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Tang MB, Yu CP, Chen SC, Chen CH. 2014. Co-Infection of adenovirus, norovirus and torque teno virus in stools of patients with acute gastroenteritis. Southeast Asian J Trop Med Public Health 45:1326–1336. [PubMed] [Google Scholar]
- 344.Goka E, Vallely P, Mutton K, Klapper P. 2013. Influenza A viruses dual and multiple infections with other respiratory viruses and risk of hospitalisation and mortality. Influenza Other Respir Viruses 7:1079–1087. doi: 10.1111/irv.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Goka EA, Vallely PJ, Mutton KJ, Klapper PE. 2015. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol Infect 143:37–47. doi: 10.1017/S0950268814000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Griesbeck M, Valantin MA, Lacombe K, Samri-Hassimi A, Bottero J, Blanc C, Sbihi Z, Zoorob R, Katlama C, Guiguet M, Altfeld M, Autran B, HepACT-VIH Study Group. 2017. Hepatitis C virus drives increased type I interferon-associated impairments associated with fibrosis severity in antiretroviral treatment-treated HIV-1-hepatitis C virus-coinfected individuals. AIDS 31:1223–1234. doi: 10.1097/QAD.0000000000001455. [DOI] [PubMed] [Google Scholar]
- 347.Martin ET, Kuypers J, Wald A, Englund JA. 2012. Multiple versus single virus respiratory infections: viral load and clinical disease severity in hospitalized children. Influenza Other Respir Viruses 6:71–77. doi: 10.1111/j.1750-2659.2011.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Amin J, Law MG, Bartlett M, Kaldor JM, Dore GJ. 2006. Causes of death after diagnosis of hepatitis B or hepatitis C infection: a large community-based linkage study. Lancet 368:938–945. doi: 10.1016/S0140-6736(06)69374-4. [DOI] [PubMed] [Google Scholar]
- 349.Alberti A, Pontisso P, Chemello L, Fattovich G, Benvegnu L, Belussi F, De Mitri MS. 1995. The interaction between hepatitis B virus and hepatitis C virus in acute and chronic liver disease. J Hepatol 22:38–41. [PubMed] [Google Scholar]
- 350.Sagnelli E, Coppola N, Messina V, Di Caprio D, Marrocco C, Marotta A, Onofrio M, Scolastico C, Filippini P. 2002. HBV superinfection in hepatitis C virus chronic carriers, viral interaction, and clinical course. Hepatology 36:1285–1291. doi: 10.1053/jhep.2002.36509. [DOI] [PubMed] [Google Scholar]
- 351.Liang L, He C, Lei M, Li S, Hao Y, Zhu H, Duan Q. 2005. Pathology of guinea pigs experimentally infected with a novel reovirus and coronavirus isolated from SARS patients. DNA Cell Biol 24:485–490. doi: 10.1089/dna.2005.24.485. [DOI] [PubMed] [Google Scholar]
- 352.Trindade GS, Lobato ZI, Drumond BP, Leite JA, Trigueiro RC, Guedes MI, da Fonseca FG, dos Santos JR, Bonjardim CA, Ferreira PC, Kroon EG. 2006. Isolation of two vaccinia virus strains from a single bovine vaccinia outbreak in rural area from Brazil: implications on the emergence of zoonotic orthopoxviruses. Am J Trop Med Hyg 75:486–490. [PubMed] [Google Scholar]
- 353.Oliveira G, Assis F, Almeida G, Albarnaz J, Lima M, Andrade AC, Calixto R, Oliveira C, Diomedes Neto J, Trindade G, Ferreira PC, Kroon EG, Abrahao J. 2015. From lesions to viral clones: biological and molecular diversity amongst autochthonous Brazilian vaccinia virus. Viruses 7:1218–1237. doi: 10.3390/v7031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Campos RK, Brum MC, Nogueira CE, Drumond BP, Alves PA, Siqueira-Lima L, Assis FL, Trindade GS, Bonjardim CA, Ferreira PC, Weiblen R, Flores EF, Kroon EG, Abrahao JS. 2011. Assessing the variability of Brazilian vaccinia virus isolates from a horse exanthematic lesion: coinfection with distinct viruses. Arch Virol 156:275–283. doi: 10.1007/s00705-010-0857-z. [DOI] [PubMed] [Google Scholar]
- 355.Chareonsook O, Foy HM, Teeraratkul A, Silarug N. 1999. Changing epidemiology of dengue hemorrhagic fever in Thailand. Epidemiol Infect 122:161–166. doi: 10.1017/S0950268898001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Gubler DJ. 1994. Perspectives on the prevention and control of dengue hemorrhagic fever. Gaoxiong Yi Xue Ke Xue Za Zhi 10(Suppl):S15–S18. [PubMed] [Google Scholar]
- 357.Gubler DJ. 1998. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 11:480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Nisalak A, Endy TP, Nimmannitya S, Kalayanarooj S, Thisayakorn U, Scott RM, Burke DS, Hoke CH, Innis BL, Vaughn DW. 2003. Serotype-specific dengue virus circulation and dengue disease in Bangkok, Thailand from 1973 to 1999. Am J Trop Med Hyg 68:191–202. [PubMed] [Google Scholar]
- 359.Aaskov J, Buzacott K, Thu HM, Lowry K, Holmes EC. 2006. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science 311:236–238. doi: 10.1126/science.1115030. [DOI] [PubMed] [Google Scholar]
- 360.Gubler DJ, Kuno G, Sather GE, Waterman SH. 1985. A case of natural concurrent human infection with two dengue viruses. Am J Trop Med Hyg 34:170–173. doi: 10.4269/ajtmh.1985.34.170. [DOI] [PubMed] [Google Scholar]
- 361.Lorono-Pino MA, Cropp CB, Farfan JA, Vorndam AV, Rodriguez-Angulo EM, Rosado-Paredes EP, Flores-Flores LF, Beaty BJ, Gubler DJ. 1999. Common occurrence of concurrent infections by multiple dengue virus serotypes. Am J Trop Med Hyg 61:725–730. doi: 10.4269/ajtmh.1999.61.725. [DOI] [PubMed] [Google Scholar]
- 362.Wang WK, Chao DY, Lin SR, King CC, Chang SC. 2003. Concurrent infections by two dengue virus serotypes among dengue patients in Taiwan. J Microbiol Immunol Infect 36:89–95. [PubMed] [Google Scholar]
- 363.Vasilakis N, Shell EJ, Fokam EB, Mason PW, Hanley KA, Estes DM, Weaver SC. 2007. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology 358:402–412. doi: 10.1016/j.virol.2006.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Rudnick A, Chan YC. 1965. Dengue type 2 virus in naturally infected Aedes albopictus mosquitoes in Singapore. Science 149:638–639. doi: 10.1126/science.149.3684.638. [DOI] [PubMed] [Google Scholar]
- 365.Vasilakis N, Holmes EC, Fokam EB, Faye O, Diallo M, Sall AA, Weaver SC. 2007. Evolutionary processes among sylvatic dengue type 2 viruses. J Virol 81:9591–9595. doi: 10.1128/JVI.02776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Linde A, Rotzen-Ostlund M, Zweygberg-Wirgart B, Rubinova S, Brytting M. 2009. Does viral interference affect spread of influenza? Euro Surveill 1414(40):pii=19354. [PubMed] [Google Scholar]
- 367.Susi H, Barres B, Vale PF, Laine AL. 2015. Co-infection alters population dynamics of infectious disease. Nat Commun 6:5975. doi: 10.1038/ncomms6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Blackard JT, Cohen DE, Mayer KH. 2002. Human immunodeficiency virus superinfection and recombination: current state of knowledge and potential clinical consequences. Clin Infect Dis 34:1108–1114. doi: 10.1086/339547. [DOI] [PubMed] [Google Scholar]
- 369.Malim MH, Emerman M. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469–472. doi: 10.1016/S0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 370.Lama J. 2003. The physiological relevance of CD4 receptor down-modulation during HIV infection. Curr HIV Res 1:167–184. doi: 10.2174/1570162033485276. [DOI] [PubMed] [Google Scholar]
- 371.Zhou J, Yang F, Yang J, Ma L, Cun Y, Song S, Liao G. 2017. Reassortment of high-yield influenza viruses in vero cells and safety assessment as candidate vaccine strains. Hum Vaccin Immunother 13:111–116. doi: 10.1080/21645515.2016.1231261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Urbaniak K, Markowska-Daniel I. 2014. In vivo reassortment of influenza viruses. Acta Biochim Pol 61:427–431. [PubMed] [Google Scholar]
- 373.Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73:8851–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Elderfield R, Barclay W. 2011. Influenza pandemics. Adv Exp Med Biol 719:81–103. doi: 10.1007/978-1-4614-0204-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Minor PD, John A, Ferguson M, Icenogle JP. 1986. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J Gen Virol 67:693–706. doi: 10.1099/0022-1317-67-4-693. [DOI] [PubMed] [Google Scholar]
- 376.Becher P, Orlich M, Thiel HJ. 2001. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J Virol 75:6256–6264. doi: 10.1128/JVI.75.14.6256-6264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Resa-Infante P, Paterson D, Bonet J, Otte A, Oliva B, Fodor E, Gabriel G. 2015. Targeting importin-alpha7 as a therapeutic approach against pandemic influenza viruses. J Virol 89:9010–9020. doi: 10.1128/JVI.00583-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Chen BK, Gandhi RT, Baltimore D. 1996. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J Virol 70:6044–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Cortez V, Odem-Davis K, McClelland RS, Jaoko W, Overbaugh J. 2012. HIV-1 superinfection in women broadens and strengthens the neutralizing antibody response. PLoS Pathog 8:e1002611. doi: 10.1371/journal.ppat.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Root-Bernstein RS, Hobbs SH. 1993. Does HIV “piggyback” on CD4-like surface proteins of sperm, viruses, and bacteria? Implications for co-transmission, cellular tropism and the induction of autoimmunity in AIDS. J Theor Biol 160:249–264. [DOI] [PubMed] [Google Scholar]
- 381.Borucki MK, Chandler LJ, Parker BM, Blair CD, Beaty BJ. 1999. Bunyavirus superinfection and segment reassortment in transovarially infected mosquitoes. J Gen Virol 80:3173–3179. doi: 10.1099/0022-1317-80-12-3173. [DOI] [PubMed] [Google Scholar]
- 382.Beaty BJ, Bishop DH, Gay M, Fuller F. 1983. Interference between bunyaviruses in Aedes triseriatus mosquitoes. Virology 127:83–90. doi: 10.1016/0042-6822(83)90373-2. [DOI] [PubMed] [Google Scholar]
- 383.Zhang G, Hu Y, Wang H, Zhang L, Bao Y, Zhou X. 2012. High incidence of multiple viral infections identified in upper respiratory tract infected children under three years of age in Shanghai, China. PLoS One 7:e44568. doi: 10.1371/journal.pone.0044568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Brehm KE, Kumar N, Thulke HH, Haas B. 2008. High potency vaccines induce protection against heterologous challenge with foot-and-mouth disease virus. Vaccine 26:1681–1687. doi: 10.1016/j.vaccine.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 385.Chen HD, Fraire AE, Joris I, Welsh RM, Selin LK. 2003. Specific history of heterologous virus infections determines anti-viral immunity and immunopathology in the lung. Am J Pathol 163:1341–1355. doi: 10.1016/S0002-9440(10)63493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Cornberg M, Kenney LL, Chen AT, Waggoner SN, Kim SK, Dienes HP, Welsh RM, Selin LK. 2013. Clonal exhaustion as a mechanism to protect against severe immunopathology and death from an overwhelming CD8 T cell response. Front Immunol 4:475. doi: 10.3389/fimmu.2013.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Page KR, Scott AL, Manabe YC. 2006. The expanding realm of heterologous immunity: friend or foe? Cell Microbiol 8:185–196. doi: 10.1111/j.1462-5822.2005.00653.x. [DOI] [PubMed] [Google Scholar]
- 388.Furman D, Jojic V, Sharma S, Shen-Orr SS, Angel CJ, Onengut-Gumuscu S, Kidd BA, Maecker HT, Concannon P, Dekker CL, Thomas PG, Davis MM. 2015. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med 7:281ra243. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW IV. 2007. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 390.Che JW, Daniels KA, Selin LK, Welsh RM. 2017. Heterologous immunity and persistent murine cytomegalovirus infection. J Virol 91:e01386-16. doi: 10.1128/JVI.01386-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Welsh RM, Che JW, Brehm MA, Selin LK. 2010. Heterologous immunity between viruses. Immunol Rev 235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.La Gruta NL, Kedzierska K, Stambas J, Doherty PC. 2007. A question of self-preservation: immunopathology in influenza virus infection. Immunol Cell Biol 85:85–92. doi: 10.1038/sj.icb.7100026. [DOI] [PubMed] [Google Scholar]
- 393.Yang HY, Joris I, Majno G, Welsh RM. 1985. Necrosis of adipose tissue induced by sequential infections with unrelated viruses. Am J Pathol 120:173–177. [PMC free article] [PubMed] [Google Scholar]
- 394.Welsh RM, Fujinami RS. 2007. Pathogenic epitopes, heterologous immunity and vaccine design. Nat Rev Microbiol 5:555–563. doi: 10.1038/nrmicro1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Stelekati E, Wherry EJ. 2012. Chronic bystander infections and immunity to unrelated antigens. Cell Host Microbe 12:458–469. doi: 10.1016/j.chom.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Guidotti LG, Borrow P, Hobbs MV, Matzke B, Gresser I, Oldstone MB, Chisari FV. 1996. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci U S A 93:4589–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Sant AJ, McMichael A. 2012. Revealing the role of CD4(+) T cells in viral immunity. J Exp Med 209:1391–1395. doi: 10.1084/jem.20121517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Selin LK, Varga SM, Wong IC, Welsh RM. 1998. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J Exp Med 188:1705–1715. doi: 10.1084/jem.188.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Belkaid Y, Piccirillo CA, Mendez S, Shevach EM, Sacks DL. 2002. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature 420:502–507. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- 400.Suvas S, Kumaraguru U, Pack CD, Lee S, Rouse BT. 2003. CD4+CD25+ T cells regulate virus-specific primary and memory CD8+ T cell responses. J Exp Med 198:889–901. doi: 10.1084/jem.20030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Sharma S, Sundararajan A, Suryawanshi A, Kumar N, Veiga-Parga T, Kuchroo VK, Thomas PG, Sangster MY, Rouse BT. 2011. T cell immunoglobulin and mucin protein-3 (Tim-3)/galectin-9 interaction regulates influenza A virus-specific humoral and CD8 T-cell responses. Proc Natl Acad Sci U S A 108:19001–19006. doi: 10.1073/pnas.1107087108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Kuchroo VK, Meyers JH, Umetsu DT, DeKruyff RH. 2006. TIM family of genes in immunity and tolerance. Adv Immunol 91:227–249. doi: 10.1016/S0065-2776(06)91006-2. [DOI] [PubMed] [Google Scholar]
- 403.Nagahara K, Arikawa T, Oomizu S, Kontani K, Nobumoto A, Tateno H, Watanabe K, Niki T, Katoh S, Miyake M, Nagahata S, Hirabayashi J, Kuchroo VK, Yamauchi A, Hirashima M. 2008. Galectin-9 increases Tim-3+ dendritic cells and CD8+ T cells and enhances antitumor immunity via galectin-9-Tim-3 interactions. J Immunol 181:7660–7669. doi: 10.4049/jimmunol.181.11.7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Kuchroo VK, Dardalhon V, Xiao S, Anderson AC. 2008. New roles for TIM family members in immune regulation. Nat Rev Immunol 8:577–580. doi: 10.1038/nri2366. [DOI] [PubMed] [Google Scholar]
- 405.Degauque N, Mariat C, Kenny J, Zhang D, Gao W, Vu MD, Alexopoulos S, Oukka M, Umetsu DT, DeKruyff RH, Kuchroo V, Zheng XX, Strom TB. 2008. Immunostimulatory Tim-1-specific antibody deprograms Tregs and prevents transplant tolerance in mice. J Clin Invest 118:735–741. doi: 10.1172/JCI32562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Rouse BT, Sehrawat S. 2010. Immunity and immunopathology to viruses: what decides the outcome? Nat Rev Immunol 10:514–526. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.La Cava A, Van Kaer L, Fu Dong S. 2006. CD4+CD25+ Tregs and NKT cells: regulators regulating regulators. Trends Immunol 27:322–327. doi: 10.1016/j.it.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 408.Belkaid Y, Rouse BT. 2005. Natural regulatory T cells in infectious disease. Nat Immunol 6:353–360. [DOI] [PubMed] [Google Scholar]
- 409.Brincks EL, Roberts AD, Cookenham T, Sell S, Kohlmeier JE, Blackman MA, Woodland DL. 2013. Antigen-specific memory regulatory CD4+Foxp3+ T cells control memory responses to influenza virus infection. J Immunol 190:3438–3446. doi: 10.4049/jimmunol.1203140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Reuter D, Sparwasser T, Hunig T, Schneider-Schaulies J. 2012. Foxp3+ regulatory T cells control persistence of viral CNS infection. PLoS One 7:e33989. doi: 10.1371/journal.pone.0033989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Tseng KC, Ho YC, Hsieh YH, Lai NS, Wen ZH, Li C, Wu SF. 2012. Elevated frequency and function of regulatory T cells in patients with active chronic hepatitis C. J Gastroenterol 47:823–833. doi: 10.1007/s00535-012-0544-9. [DOI] [PubMed] [Google Scholar]
- 412.Kraft AR, Wlodarczyk MF, Kenney LL, Selin LK. 2013. PC61 (anti-CD25) treatment inhibits influenza A virus-expanded regulatory T cells and severe lung pathology during a subsequent heterologous lymphocytic choriomeningitis virus infection. J Virol 87:12636–12647. doi: 10.1128/JVI.00936-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Lund JM, Hsing L, Pham TT, Rudensky AY. 2008. Coordination of early protective immunity to viral infection by regulatory T cells. Science 320:1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Haeryfar SM, DiPaolo RJ, Tscharke DC, Bennink JR, Yewdell JW. 2005. Regulatory T cells suppress CD8+ T cell responses induced by direct priming and cross-priming and moderate immunodominance disparities. J Immunol 174:3344–3351. doi: 10.4049/jimmunol.174.6.3344. [DOI] [PubMed] [Google Scholar]
- 415.Ruckwardt TJ, Bonaparte KL, Nason MC, Graham BS. 2009. Regulatory T cells promote early influx of CD8+ T cells in the lungs of respiratory syncytial virus-infected mice and diminish immunodominance disparities. J Virol 83:3019–3028. doi: 10.1128/JVI.00036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Cicin-Sain L, Brien JD, Uhrlaub JL, Drabig A, Marandu TF, Nikolich-Zugich J. 2012. Cytomegalovirus infection impairs immune responses and accentuates T-cell pool changes observed in mice with aging. PLoS Pathog 8:e1002849. doi: 10.1371/journal.ppat.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Ancicova L, Wagnerova M, Janulikova J, Chalupkova A, Hrabovska Z, Kostolansky F, Vareckova E, Mistrikova J. 2015. Simultaneous infection with gammaherpes and influenza viruses enhances the host immune defense. Acta Virol 59:369–379. doi: 10.4149/av_2015_04_369. [DOI] [PubMed] [Google Scholar]
- 418.Selin LK, Vergilis K, Welsh RM, Nahill SR. 1996. Reduction of otherwise remarkably stable virus-specific cytotoxic T lymphocyte memory by heterologous viral infections. J Exp Med 183:2489–2499. doi: 10.1084/jem.183.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Selin LK, Lin MY, Kraemer KA, Pardoll DM, Schneck JP, Varga SM, Santolucito PA, Pinto AK, Welsh RM. 1999. Attrition of T cell memory: selective loss of LCMV epitope-specific memory CD8 T cells following infections with heterologous viruses. Immunity 11:733–742. doi: 10.1016/S1074-7613(00)80147-8. [DOI] [PubMed] [Google Scholar]
- 420.Dudani R, Murali-Krishna K, Krishnan L, Sad S. 2008. IFN-gamma induces the erosion of preexisting CD8 T cell memory during infection with a heterologous intracellular bacterium. J Immunol 181:1700–1709. doi: 10.4049/jimmunol.181.3.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Lin SJ, Peacock CD, Bahl K, Welsh RM. 2007. Programmed death-1 (PD-1) defines a transient and dysfunctional oligoclonal T cell population in acute homeostatic proliferation. J Exp Med 204:2321–2333. doi: 10.1084/jem.20062150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Zhang JY, Zhang Z, Jin B, Zhang SY, Zhou CB, Fu JL, Wang FS. 2008. Programmed death-1 up-regulation is involved in the attrition of cytomegalovirus-specific CD8+ T cells in acute self-limited hepatitis B virus infection. J Immunol 181:3741–3744. doi: 10.4049/jimmunol.181.6.3741. [DOI] [PubMed] [Google Scholar]
- 423.Vezys V, Yates A, Casey KA, Lanier G, Ahmed R, Antia R, Masopust D. 2009. Memory CD8 T-cell compartment grows in size with immunological experience. Nature 457:196–199. doi: 10.1038/nature07486. [DOI] [PubMed] [Google Scholar]
- 424.Varga SM, Selin LK, Welsh RM. 2001. Independent regulation of lymphocytic choriomeningitis virus-specific T cell memory pools: relative stability of CD4 memory under conditions of CD8 memory T cell loss. J Immunol 166:1554–1561. doi: 10.4049/jimmunol.166.3.1554. [DOI] [PubMed] [Google Scholar]
- 425.La Gruta NL, Kedzierska K, Pang K, Webby R, Davenport M, Chen W, Turner SJ, Doherty PC. 2006. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc Natl Acad Sci U S A 103:994–999. doi: 10.1073/pnas.0510429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Kedzierska K, La Gruta NL, Stambas J, Turner SJ, Doherty PC. 2008. Tracking phenotypically and functionally distinct T cell subsets via T cell repertoire diversity. Mol Immunol 45:607–618. doi: 10.1016/j.molimm.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, Crawford JC, Clemens EB, Nguyen THO, Kedzierska K, La Gruta NL, Bradley P, Thomas PG. 2017. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547:89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Cornberg M, Chen AT, Wilkinson LA, Brehm MA, Kim SK, Calcagno C, Ghersi D, Puzone R, Celada F, Welsh RM, Selin LK. 2006. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest 116:1443–1456. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Brehm MA, Pinto AK, Daniels KA, Schneck JP, Welsh RM, Selin LK. 2002. T cell immunodominance and maintenance of memory regulated by unexpectedly cross-reactive pathogens. Nat Immunol 3:627–634. doi: 10.1038/ni806. [DOI] [PubMed] [Google Scholar]
- 430.Rojas JM, Rodriguez-Calvo T, Sevilla N. 2017. Recall T cell responses to bluetongue virus produce a narrowing of the T cell repertoire. Vet Res 48:38. doi: 10.1186/s13567-017-0444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Selin LK, Brehm MA, Naumov YN, Cornberg M, Kim SK, Clute SC, Welsh RM. 2006. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol Rev 211:164–181. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Urbani S, Amadei B, Fisicaro P, Pilli M, Missale G, Bertoletti A, Ferrari C. 2005. Heterologous T cell immunity in severe hepatitis C virus infection. J Exp Med 201:675–680. doi: 10.1084/jem.20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. 2001. Cross-reactivity between hepatitis C virus and influenza A virus determinant-specific cytotoxic T cells. J Virol 75:11392–11400. doi: 10.1128/JVI.75.23.11392-11400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Halstead SB. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476–481. [DOI] [PubMed] [Google Scholar]
- 435.Lum FM, Couderc T, Chia BS, Ong RY, Her Z, Chow A, Leo YS, Kam YW, Renia L, Lecuit M, Ng LFP. 2018. Antibody-mediated enhancement aggravates chikungunya virus infection and disease severity. Sci Rep 8:1860. doi: 10.1038/s41598-018-20305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, Yenchitsomanus PT, McMichael A, Malasit P, Screaton G. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med 9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 437.Tripathi S, Balasubramaniam VR, Brown JA, Mena I, Grant A, Bardina SV, Maringer K, Schwarz MC, Maestre AM, Sourisseau M, Albrecht RA, Krammer F, Evans MJ, Fernandez-Sesma A, Lim JK, Garcia-Sastre A. 2017. A novel Zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLoS Pathog 13:e1006258. doi: 10.1371/journal.ppat.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Bardina SV, Bunduc P, Tripathi S, Duehr J, Frere JJ, Brown JA, Nachbagauer R, Foster GA, Krysztof D, Tortorella D, Stramer SL, Garcia-Sastre A, Krammer F, Lim JK. 2017. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356:175–180. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Stefanska I, Romanowska M, Donevski S, Gawryluk D, Brydak LB. 2013. Co-infections with influenza and other respiratory viruses. Adv Exp Med Biol 756:291–301. doi: 10.1007/978-94-007-4549-0_36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Rockstroh JK, Spengler U. 2004. HIV and hepatitis C virus co-infection. Lancet Infect Dis 4:437–444. doi: 10.1016/S1473-3099(04)01059-X. [DOI] [PubMed] [Google Scholar]
- 441.Nie S, Cornberg M, Selin LK. 2009. Resistance to vaccinia virus is less dependent on TNF under conditions of heterologous immunity. J Immunol 183:6554–6560. doi: 10.4049/jimmunol.0902156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Chen HD, Fraire AE, Joris I, Brehm MA, Welsh RM, Selin LK. 2001. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat Immunol 2:1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- 443.Wlodarczyk MF, Kraft AR, Chen HD, Kenney LL, Selin LK. 2013. Anti-IFN-gamma and peptide-tolerization therapies inhibit acute lung injury induced by cross-reactive influenza A-specific memory T cells. J Immunol 190:2736–2746. doi: 10.4049/jimmunol.1201936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Adams AB, Pearson TC, Larsen CP. 2003. Heterologous immunity: an overlooked barrier to tolerance. Immunol Rev 196:147–160. doi: 10.1046/j.1600-065X.2003.00082.x. [DOI] [PubMed] [Google Scholar]
- 445.Li Y, Li XC, Zheng XX, Wells AD, Turka LA, Strom TB. 1999. Blocking both signal 1 and signal 2 of T-cell activation prevents apoptosis of alloreactive T cells and induction of peripheral allograft tolerance. Nat Med 5:1298–1302. doi: 10.1038/15256. [DOI] [PubMed] [Google Scholar]
- 446.Jones TR, Ha J, Williams MA, Adams AB, Durham MM, Rees PA, Cowan SR, Pearson TC, Larsen CP. 2002. The role of the IL-2 pathway in costimulation blockade-resistant rejection of allografts. J Immunol 168:1123–1130. doi: 10.4049/jimmunol.168.3.1123. [DOI] [PubMed] [Google Scholar]
- 447.Demirci G, Gao W, Zheng XX, Malek TR, Strom TB, Li XC. 2002. On CD28/CD40 ligand costimulation, common gamma-chain signals, and the alloimmune response. J Immunol 168:4382–4390. doi: 10.4049/jimmunol.168.9.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Chen AT, Cornberg M, Gras S, Guillonneau C, Rossjohn J, Trees A, Emonet S, de la Torre JC, Welsh RM, Selin LK. 2012. Loss of anti-viral immunity by infection with a virus encoding a cross-reactive pathogenic epitope. PLoS Pathog 8:e1002633. doi: 10.1371/journal.ppat.1002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Cornberg M, Clute SC, Watkin LB, Saccoccio FM, Kim SK, Naumov YN, Brehm MA, Aslan N, Welsh RM, Selin LK. 2010. CD8 T cell cross-reactivity networks mediate heterologous immunity in human EBV and murine vaccinia virus infections. J Immunol 184:2825–2838. doi: 10.4049/jimmunol.0902168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Sorup S, Benn CS, Poulsen A, Krause TG, Aaby P, Ravn H. 2014. Live vaccine against measles, mumps, and rubella and the risk of hospital admissions for nontargeted infections. JAMA 311:826–835. doi: 10.1001/jama.2014.470. [DOI] [PubMed] [Google Scholar]
- 451.Bhattacharya S. 2008. The World Health Organization and global smallpox eradication. J Epidemiol Community Health 62:909–912. doi: 10.1136/jech.2006.055590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Gil A, Kenney LL, Mishra R, Watkin LB, Aslan N, Selin LK. 2015. Vaccination and heterologous immunity: educating the immune system. Trans R Soc Trop Med Hyg 109:62–69. doi: 10.1093/trstmh/tru198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Esteva L, Vargas C. 2003. Coexistence of different serotypes of dengue virus. J Math Biol 46:31–47. doi: 10.1007/s00285-002-0168-4. [DOI] [PubMed] [Google Scholar]
- 454.Vickerman P, Martin NK, Hickman M. 2012. Understanding the trends in HIV and hepatitis C prevalence amongst injecting drug users in different settings—implications for intervention impact. Drug Alcohol Depend 123:122–131. doi: 10.1016/j.drugalcdep.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 455.Corson S, Greenhalgh D, Taylor A, Palmateer N, Goldberg D, Hutchinson S. 2013. Modelling the prevalence of HCV amongst people who inject drugs: an investigation into the risks associated with injecting paraphernalia sharing. Drug Alcohol Depend 133:172–179. doi: 10.1016/j.drugalcdep.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 456.Carvalho ARM, Pinto CMA, Baleanu D. 2018. HIV/HCV coinfection model: a fractional-order perspective for the effect of the HIV viral load. Adv Differ Equat 2:22. [Google Scholar]
- 457.Carvalho AR, Pinto CM. 2014. A coinfection model for HIV and HCV. Biosystems 124:46–60. doi: 10.1016/j.biosystems.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 458.Waziri AS, Massawe ES, Makinde OD. 2012. Mathematical modelling of HIV/AIDS dynamics with treatment of vertical transmission. Appl Math 2:77–79. doi: 10.5923/j.am.20120203.06. [DOI] [Google Scholar]
- 459.Nthiiri JK, Lavi GO, Mayonge A. 2015. Mathematical model of pneumonia and HIV/AIDS coinfection in the presence of protection. Int J Math Anal 9:2069–2085. doi: 10.12988/ijma.2015.55150. [DOI] [Google Scholar]
- 460.Bhunu CP, Mushayabasa S. 2013. Modelling the transmission dynamics of HIV/AIDS and hepatitis C virus coinfection. HIV AIDS Rev 12:37–42. doi: 10.1016/j.hivar.2013.03.001. [DOI] [Google Scholar]
- 461.Birger R, Kouyos R, Dushoff J, Grenfell B. 2015. Modeling the effect of HIV coinfection on clearance and sustained virologic response during treatment for hepatitis C virus. Epidemics 12:1–10. doi: 10.1016/j.epidem.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 462.Rong L, Riberio RM, Perelson AS. 2012. Modeling quasispecies and drug resistence in hepatitis C patients treated with a protease inhibitor. Bull Math Biol 4:1789–1817. doi: 10.1007/s11538-012-9736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Alexander HK, Bonhoeffer S. 2012. Pre-existence and emergence of drug resistance in a generalized model of intra-host viral dynamics. Epidemics 4:187–202. doi: 10.1016/j.epidem.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 464.Pinky L, Dobrovolny HM. 2016. Coinfections of the respiratory tract: viral competition for resources. PLoS One 11:e0155589. doi: 10.1371/journal.pone.0155589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Pinky L, Dobrovolny HM. 2017. The impact of cell regeneration on the dynamics of viral coinfection. Chaos 27:063109. doi: 10.1063/1.4985276. [DOI] [PubMed] [Google Scholar]