Strains of bacteria resistant to antibiotics, particularly those that are multiresistant, are an increasing major health care problem around the world. It is now abundantly clear that both Gram-negative and Gram-positive bacteria are able to meet the evolutionary challenge of combating antimicrobial chemotherapy, often by acquiring preexisting resistance determinants from the bacterial gene pool.
KEYWORDS: antibiotic resistance, insertion sequence, transposon, gene cassette, integron, plasmid, integrative conjugative element, resistance island
SUMMARY
Strains of bacteria resistant to antibiotics, particularly those that are multiresistant, are an increasing major health care problem around the world. It is now abundantly clear that both Gram-negative and Gram-positive bacteria are able to meet the evolutionary challenge of combating antimicrobial chemotherapy, often by acquiring preexisting resistance determinants from the bacterial gene pool. This is achieved through the concerted activities of mobile genetic elements able to move within or between DNA molecules, which include insertion sequences, transposons, and gene cassettes/integrons, and those that are able to transfer between bacterial cells, such as plasmids and integrative conjugative elements. Together these elements play a central role in facilitating horizontal genetic exchange and therefore promote the acquisition and spread of resistance genes. This review aims to outline the characteristics of the major types of mobile genetic elements involved in acquisition and spread of antibiotic resistance in both Gram-negative and Gram-positive bacteria, focusing on the so-called ESKAPEE group of organisms (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli), which have become the most problematic hospital pathogens.
INTRODUCTION
Antibiotic-resistant bacteria are a major cause of health care-associated infections around the world, and resistance has also emerged in infections in the wider community. Infections caused by multiresistant organisms significantly increase morbidity, mortality, and health care costs. Molecular analyses have revealed that widespread multiresistance has commonly been achieved by the acquisition of preexisting determinants followed by amplification in response to selection. The capture, accumulation, and dissemination of resistance genes are largely due to the actions of mobile genetic elements (MGE), a term used to refer to elements that promote intracellular DNA mobility (e.g., from the chromosome to a plasmid or between plasmids) as well as those that enable intercellular DNA mobility.
Insertion sequences (IS) and transposons (Tn) are discrete DNA segments that are able to move themselves (and associated resistance genes) almost randomly to new locations in the same or different DNA molecules within a single cell. Other elements, such as integrons (In), use site-specific recombination to move resistance genes between defined sites. As these types of MGE are often present in multiple copies in different locations in a genome, they can also facilitate homologous recombination (exchange of sequences between identical or related segments). Intercellular mechanisms of genetic exchange include conjugation/mobilization (mediated by plasmids and integrative conjugative elements [ICE]), transduction (mediated by bacteriophages), and transformation (uptake of extracellular DNA). Interactions between the various types of MGE underpin the rapid evolution of diverse multiresistant pathogens in the face of antimicrobial chemotherapy. Examples of these elements and processes are illustrated in Fig. 1.
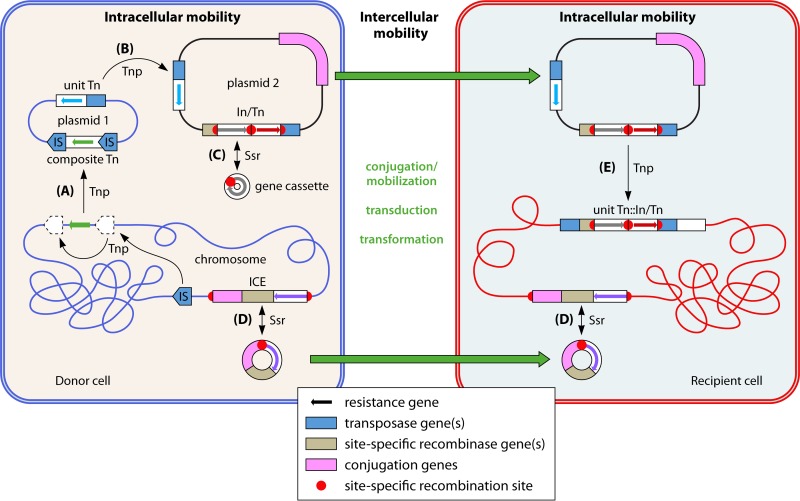
FIG 1.
Examples of mobile genetic elements (MGE) and processes involved in intracellular mobility or intercellular transfer of antibiotic resistance genes. Two cells of different strains or species are represented, with one acting as donor (envelope and chromosome shown in blue; contains two plasmids) and the other as recipient (shown in red). Various MGE are shown, with the functions of the genes they carry color coded as shown in the key. Different resistance genes associated with different MGE are represented by small arrows of various colors. Thin black arrows indicate intracellular processes, with those mediated by a transposase protein labeled Tnp and those mediated by a site-specific recombinase protein labeled Ssr. Thick green arrows represent intercellular (horizontal) transfer. Successive insertions of the same IS on both sides of a resistance gene may allow it to be captured and moved to another DNA molecule (e.g., from the chromosome to a plasmid) as part of a composite Tn (A). A unit Tn carrying a resistance gene may transpose between plasmids (B) or from a plasmid to the chromosome or vice versa. A gene cassette may move between In (a class 1 In/Tn structure is represented here) via a circular intermediate (C). An ICE can be integrated into the chromosome or excised as a circular element that can then conjugate into a recipient cell and integrate (reversibly) into the chromosome at a specific recombination site (D). A plasmid may be able to mediate its own intercellular transfer by conjugation or, if it lacks a conjugation region, be mobilized by another plasmid (or, alternatively, move horizontally by phage transduction or transformation). Tn and/or In and associated resistance genes on an incoming plasmid may move into the chromosome or other plasmid(s) in the recipient cell (E), as illustrated here for class 1 In/Tn, which are known to target unit Tn. See relevant sections of the text for further details.
It is not possible to cover all MGE involved in resistance in all bacterial species in a single review, so we have elected to focus primarily on the most important and/or topical elements in Gram-negative and Gram-positive bacterial species of particular concern clinically, namely, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. (the so-called ESKAPE group [1]) as well as Escherichia coli (giving the ESKAPEE group) (2); nonetheless, many of the MGE types described are of course broadly relevant to many other bacterial taxa. We aim to highlight the diversity and major characteristics of MGE associated with resistance in these organisms while citing other reviews that provide more detailed analyses of particular elements or biological processes.
IS AND COMPOSITE TRANSPOSONS
IS are generally small mobile elements that typically carry little more than one (sometimes two) transposase (tnp) gene, and their characteristics have been reviewed several times (see references 3 and 4 and references therein). They can be divided into groups based partly on active site motifs in Tnp, designated by key amino acids that come together in the active site, most commonly DDE (Asp, Asp, and Glu) but also DEDD and HUH (two His residues separated by a large hydrophobic amino acid) (5), and/or based on whether transposition is a conservative, cut-and-paste mechanism, where the IS is simply excised from the donor and inserted into the recipient, or replicative (6). Replicative transposition can occur by a copy-and-paste mechanism (the IS is replicated to join the donor and recipient in a cointegrate, which is then resolved to give the original donor plus the recipient with the IS [6]) or a copy-out-paste-in mechanism (the IS is replicated into a double-stranded circular intermediate that then integrates into the recipient [7]).
The ends of the most common (DDE) type of IS are generally defined by terminal inverted repeats (IR) that are designated left (IRL) and right (IRR) with respect to the direction of transcription of the tnp gene (Fig. 2A). Transposition involves binding of the IR by the Tnp protein, and as a result of repair of staggered cuts in different DNA strands during the transposition process, many IS create short flanking direct repeats (DR; typically ∼3 to 14 bp, but the length is characteristic for each IS) on insertion. These are also referred to as target site duplications (TSD), but many IS do not appear to target specific sequence motifs. Other types of IS may not have IR or create TSD. Because frequent transposition may be deleterious, expression of active transposase may be controlled by, for example, the need for a programmed frameshift to create a complete Tnp protein (8). The frameshift typically occurs within a “slippery codon” region, e.g., AAAAAAA in ISAba1 (9). While the mobility of some IS has been shown experimentally, such as by the detection of circular intermediates via inverse PCR, many have been defined only from the transposases that they encode, their IR, and/or their TSD. ISfinder (https://www-is.biotoul.fr/) provides a comprehensive database of IS and includes BLAST search tools (10). IS were originally assigned numbers, but ISfinder now assigns names that include a code for the species in which the IS was first identified (but this does not necessarily indicate that the IS originated in that species) and a number (e.g., ISAba1 for Acinetobacter baumannii).
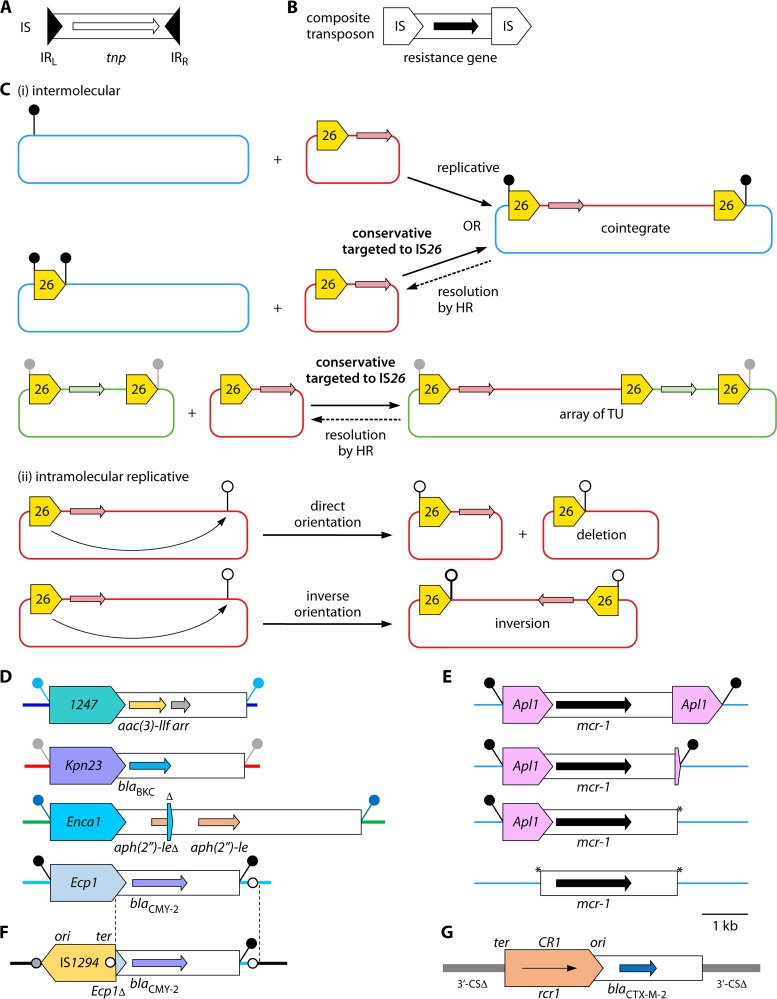
FIG 2.
Insertion sequences and composite transposons. (A) Components of a typical IS. (B) Composite transposon. IS are shown as block arrows, with the pointed end corresponding to IRR, and a captured resistance gene is shown as a black arrow. The two IS can also be oriented inversely. (C) Outcomes of transposition by IS26. (i) Intermolecular replicative transposition can insert a “translocatable unit” (TU; one copy of IS26 and an adjacent region) into a recipient that lacks IS26, while intermolecular conservative transposition targets an existing copy of IS26 (∼50× higher frequency). Both reactions create the same type of cointegrate, in which a “composite transposon”-like structure is flanked by 8-bp TSD (black lollipops) created during replicative transposition or preserved (if previously present) during conservative transposition. The cointegrate can be resolved by homologous recombination (HR), but not normally by the IS26 transposase. Intermolecular conservative transposition into a region that already contains two copies of IS26 flanking a resistance gene can give an array of TU. (ii) Intramolecular replicative transposition in direct orientation is another way of creating a TU, and in doing so deletes the region between the original IS26 element and the targeted position (white lollipop). Intramolecular replicative transposition in the inverse orientation inverts the region between the original IS26 element and the position targeted, so that TSD on the same strand are now reverse complements of one another (thick and thin lollipops). Diagrams are based on previously reported information (22, 25–28). (D) Transposition units (TPU) mediated by IS1380 family elements. △, 131 bp of IRR end of ISEnca1. Paired lollipops indicate different TSD sequences. Diagrams were drawn based on sequences from the following INSDC accession numbers: IS1247, AJ971344; ISKpn23, KP689347; ISEnca1, AY939911 (end of TPU found by alignment with Staphylococcus plasmids, e.g., pSTE1 [accession number HE662694]); and ISEcp1, FJ621588. (E) Different structures containing ISApl1 and mcr-1. Deletion of one or both copies of ISApl1 leaves “scars” (asterisks) (41, 45). Diagrams (from top to bottom) were drawn based on sequences from INSDC accession numbers CP016184, KY689635, KP347127, and KX084392. (F) IS1294 has captured part (delimited by dashed lines) of the ISEcp1-blaCMY-2 TPU plus 159 bp of the adjacent plasmid backbone (see the bottom diagram of panel D), ending with 4 bp matching its ter end (white circle), and targets a related 4-bp sequence (gray circle). The diagram was drawn based on sequences from INSDC accession number HG970648 (55). (G) By analogy with IS1294, ISCR1 captures regions adjacent to its ter end, but these are found adjacent to the ori end after insertion (by homologous recombination) (21, 59) between partial duplications of the 3′-CS of class 1 integrons (see Fig. 4B). The diagram was drawn based on the sequence from INSDC accession number AJ311891. The phenotypes conferred by resistance genes shown in the diagrams are given in Table 1 (panel E), Table 3 (panels D and F), and Table 4 (panel G).
Traditionally IS were not thought of as carrying “passenger” genes, but they can move resistance genes as part of a composite (also called compound) transposon, a region bounded by two copies of the same or related IS that can move as a single unit (Fig. 2B). Some of these have been given transposon numbers and are included in a transposon registry (11; http://transposon.lstmed.ac.uk/). More examples of a single IS mobilizing an adjacent region that includes one or more resistance genes are also being identified, particularly in Gram-negative bacteria. Many IS include a strong promoter that drives expression of the captured gene (12), and insertion upstream of an intrinsic chromosomal gene can also influence antibiotic resistance (e.g., ISAba1 with blaOXA-51-like genes in A. baumannii giving carbapenem resistance [13]). Alternatively, an IS may provide a −35 region only, which can combine with an adjacent −10-like sequence to create a hybrid promoter. These and other ways in which IS may influence antibiotic resistance phenotypes have been covered in a recent review (14). Here we concentrate on IS and composite transposons involved in movement of antibiotic resistance genes, listing examples from Gram-negative (Table 1) and Gram-positive (Table 2) bacteria and discussing some of the most important IS types in the following sections. For elements not discussed below, readers are referred to other reviews (4, 15–21).
TABLE 1.
Examples of IS and composite transposons associated with resistance genes in Gram-negative bacteria
| ISa | Tnb | Determinant | Resistance(s)c |
|---|---|---|---|
| IS1 | Tn9 | catA1 | Chloramphenicol |
| IS10 | Tn10 | tet(B) | Tetracycline |
| IS26d | Tn4352 | aphA1 | Kanamycin |
| Tn6020 | aphA1 | Kanamycin | |
| tet(C) | Tetracycline | ||
| tet(D) | Tetracycline | ||
| catA2 | Chloramphenicol | ||
| Tn2003 | blaSHV | β-Lactams | |
| cfr | Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | ||
| IS256e | cfr | Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | |
| IS50 | Tn5 | aph(3′)-IIa-ble-aph(6)-Ic | Kanamycin, bleomycin, streptomycin |
| IS903 | Tn903 | aphA1 | Kanamycin |
| IS1999 | Tn1999 | blaOXA-48-like | Carbapenems |
| ISApl1 | Tn6330 | mcr-1 | Colistin |
| ISEc69 | mcr-2 | Colistin | |
| ISAs2 | blaFOX-5 | BLBLI | |
| ISAba14 | TnaphA6 | aphA6 | Kanamycin |
| ISAba1 | Tn2006 | blaOXA-23 | Carbapenems |
| blaOXA-237 | Carbapenems | ||
| ISAba125 | Tn125 | blaNDM | Carbapenems |
See ISfinder (https://www-is.biotoul.fr/) for details of IS.
See the Tn registry (http://transposon.lstmed.ac.uk/) for further details, except for TnaphA6 (334).
SHV enzymes can be broad-spectrum or extended-spectrum β-lactamases; BLBLI, β-lactam–β-lactamase inhibitor combinations.
Regions flanked by two copies of IS26 and originally defined as composite transposons are listed, but recent findings suggest mobilization by a single copy of IS26 (see the text).
TABLE 2.
IS and composite transposons associated with resistance genes in staphylococci and enterococci
| ISa | Tn | Determinant | Associated resistance(s) | Hostb |
|---|---|---|---|---|
| IS16 | Tn1547 | vanB1 | Vancomycin | E |
| IS256 | cfr | Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | S | |
| Tn1547 | vanB1 | Vancomycin | E | |
| Tn4001 | aacA-aphD | Gentamicin/kanamycin/tobramycin | S | |
| Tn5281 | aacA-aphD | Gentamicin/kanamycin/tobramycin | E | |
| Tn5384 | aacA-aphD | Gentamicin/kanamycin/tobramycin | E | |
| Tn5384 | erm(B) | MLS antibiotics | E | |
| IS257c | aadD | Kanamycin/neomycin/paromomycin/tobramycin | S | |
| aphA-3 | Kanamycin/neomycin | S | ||
| bcrAB | Bacitracin | S | ||
| ble | Bleomycin | S | ||
| dfrK | Trimethoprim | S | ||
| erm(C) | MLS antibiotics | S | ||
| fosB5 | Fosfomycin | S | ||
| fusB | Fusidic acid | S | ||
| ileS2 (mupA) | Mupirocin | S | ||
| qacC | Antiseptics/disinfectants | S | ||
| sat4 | Streptothricin | S | ||
| tet(K) | Tetracycline | S | ||
| tet(L) | Tetracycline | S | ||
| vat(A) | Streptogramin A | S | ||
| vga(A) | Streptogramin A/pleuromutilins/lincosamides | S | ||
| vgb(A) | Streptogramin B | S | ||
| Tn924 | aacA-aphD | Gentamicin/kanamycin/tobramycin | E | |
| Tn4003 | dfrA | Trimethoprim | S | |
| Tn6072 | aacA-aphD | Gentamicin/kanamycin/tobramycin | S | |
| Tn6072 | spc | Spectinomycin | S | |
| IS1182 | Tn5405 | aadE | Streptomycin | S, E |
| Tn5405 | aphA-3 | Kanamycin/neomycin | S, E | |
| Tn5405 | sat4 | Streptothricin | S, E | |
| IS1216 | cfr | Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | E | |
| str | Streptomycin | E | ||
| Tn5385 | aacA-aphD | Gentamicin/kanamycin/tobramycin | E | |
| Tn5385 | aadE | Streptomycin | E | |
| Tn5385 | blaZ | Penicillins | E | |
| Tn5385 | erm(B) | MLS antibiotics | E | |
| Tn5385 | tet(M) | Tetracycline/minocycline | E | |
| Tn5482 | vanA | Vancomycin | E | |
| Tn5506 | vanA | Vancomycin | E | |
| IS1272 | TnSha1 | fabI | Triclosan | S |
| TnSha2 | fabI | Triclosan | S | |
| IS21-558 | cfr | Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | S | |
| lsa(B) | Lincosamides | S | ||
| ISEnfa4 | cfr | Phenicols/lincosamides/oxazolidinones/pleuromutilins/streptogramin A | S, E | |
| ISSau10 | aadD | Kanamycin/neomycin/paromomycin/tobramycin | S | |
| dfrK | Trimethoprim | S | ||
| erm(C) | MLS antibiotics | |||
| erm(T) | MLS antibiotics | S | ||
| tet(L) | Tetracycline | S |
IS26 and Related Elements
IS6 family elements IS26 (also known as IS6, IS15Δ, IS46, IS140, IS160, IS176, and IS1936) (22), IS257 (also known as IS431), and IS1216 have played a pivotal role in the dissemination of resistance determinants in Gram-negative (IS26) (Table 1) and Gram-positive (IS257 and IS1216) (Table 2) bacteria. These IS encode a single transposase, and the terminal IR of IS26 and IS257 both contain a −35 consensus (TTGCAA) that can create a hybrid promoter if fortuitously positioned (with an ∼17-bp spacer) near a −10 sequence upstream of the gene (14). Movement of these IS was originally demonstrated to occur by replicative transposition (15, 23, 24). This results in a cointegrate of the donor and recipient molecules with a directly repeated copy of the IS at each junction, creating a “composite transposon”-like structure flanked by characteristic 8-bp TSD (Fig. 2C, panel i). This may explain how small staphylococcal plasmids flanked by two directly oriented copies of IS257 have become incorporated into large plasmids or the chromosome (e.g., pUB110 within pSK41 or staphylococcal cassette chromosome mec [SCCmec] [see below]). RecA-dependent homologous recombination between the two IS copies can resolve such cointegrates, releasing the original donor and a modified recipient containing the IS flanked by TSD (25, 26).
Recently, a second mode of movement was described to explain the arrays of resistance genes separated by single copies of IS26 commonly seen in resistance plasmids and regions (22). The unit of mobility consists of one copy of IS26 and an adjacent region (which can be up to the next IS26 junction) and was termed a “translocatable unit” (TU) (22, 27). A TU preferentially inserts next to an existing copy of IS26 in a recipient molecule via a conservative process (no replication of IS26 and no creation of TSD, but any TSD already flanking the target IS26 are preserved), generating the same cointegrate structure as that created by replicative transposition (Fig. 2C, panel i). Importantly, this process, which is dependent on the IS26 transposase (Tnp26) and is recA independent, has been demonstrated to occur at a frequency ∼50 times higher than that of untargeted replicative transposition (22, 28). This means that once a chromosome or plasmid possesses a copy of IS26, it is predisposed to acquire further adjacent IS26 TU.
It appears that circular TU are not normally generated by a Tnp26-dependent mechanism but may occur following homologous recombination between IS26 copies (27) (Fig. 2C, panel i). Alternatively, intramolecular replicative transposition (Fig. 2C, panel ii) in direct orientation would release a TU-like structure (though the end would not be defined by a boundary with IS26 [Fig. 2C, panel ii] [25]). In doing so, the sequence between IS26 and the position targeted would be deleted; deletions flanking IS26-like elements have been described frequently. This is a way of streamlining resistance gene clusters by removing redundant or metabolically costly genes (25) and/or allowing expression of remaining genes to be modulated through creation of new hybrid promoters (29, 30). Intramolecular replicative transposition in inverse orientation reverses the segment between the original IS26 and the targeted site. Evidence for this comes from 8-bp sequences that are reverse complements of one another flanking the opposite ends of two IS26 copies (25) (Fig. 2C, panel ii). Further studies are beginning to unravel the details of the IS26 transposition process (31), which seem likely to also apply to other IS6 family members.
ISEcp1 and Related Elements
ISEcp1 (IS1380 family; encodes a DDE-type transposase), first identified in E. coli, has IR of about 14 bp and creates 5-bp TSD on transposition. ISEcp1 appears to be able to use IRL in combination with a sequence beyond its IRR end to move an adjacent region, creating 5-bp (or occasionally 6-bp) TSD flanking the whole “transposition unit” (32) (previously abbreviated TU [21], but TPU is used here to avoid confusion with the IS26 TU) (Fig. 2D). Insertion of ISEcp1 upstream of a chromosomal blaCTX-M-2 gene in Kluyvera and subsequent movement to a plasmid have been demonstrated (33), but the exact mechanism and any important characteristics of the sequences that can be used as alternatives to IRR have not been determined. ISEcp1 provides at least one promoter for captured genes (34) (and possibly a second [35]), and separation from this promoter results in reduced expression of blaCTX-M genes (36). ISEcp1 can also pick up regions of different lengths in different transposition events and thus can simultaneously move adjacent pieces of DNA with different origins (21). ISEcp1 appears to have been responsible for capturing many different resistance genes in this way (Table 3) in numerous cases from known source organisms (21).
TABLE 3.
Examples of resistance genes associated with ISEcp1 and related elements
| ISa | Determinant(s) | Resistance(s)b |
|---|---|---|
| ISEcp1c | blaCTX-M-1 group | 3GC |
| blaCTX-M-2 group | 3GC | |
| blaCTX-M-9 group | 3GC | |
| blaCTX-M-25 group | 3GC | |
| blaACC | 3GC, BLBLI | |
| blaCMY-2-like genes | 3GC, BLBLI | |
| blaOXA-181-like genes | Carbapenems | |
| blaOXA-204 | Carbapenems | |
| Some qnrB genes | Fluoroquinolones (low level) | |
| qnrE1 | Fluoroquinolones (low level) | |
| rmtC | Aminoglycosides (high level) | |
| IS1247 | aac(3)-IIf-arr | (Aminoglycosides, rifampind) |
| ISKpn23 | blaBKC | Carbapenems |
| aac(3)-IIb | GEN, TOB | |
| ISEnca1 | aph(2″)-Ie | GEN, TOB, KAN |
Information is available from references cited in the text as well as from other references (21, 253, 435, 508, 509) or references cited therein.
3GC, third-generation cephalosporins; BLBLI, β-lactam–β-lactamase inhibitor combinations; GEN, gentamicin; TOB, tobramycin; KAN, kanamycin.
Some ISEcp1 transposition units have been assigned Tn numbers. Search the Tn registry (http://transposon.lstmed.ac.uk/) with “ISEcp1” for details.
Predicted from homology to known genes.
Other IS1380 family elements, including ISKpn23 (37) and IS1247 (21), appear to have captured resistance genes (Table 3; Fig. 2D) in a fashion similar to that of ISEcp1. ISEnca1 (91% identical to ISEcp1) has been detected in the Gram-positive bacterium Enterococcus casseliflavus, associated with the aph(2″)-Ie gene (gentamicin resistance) (38).
ISApl1 and mcr-1
The IS30 family element ISApl1 (encodes a DDE-type transposase), first discovered in the pig pathogen Actinobacillus pleuropneumoniae (39), is involved in capture and mobilization of the recently identified mcr-1 (mobile colistin resistance) gene (40). ISApl1 is bounded by 27-bp IR and carries a single transposase gene. Like other IS30 family members, ISApl1 appears to use a “copy-out-paste-in” mechanism, via an intermediate containing 2 bp derived from the flanking sequence of the donor molecule between the abutted IRL and IRR ends, inserting in AT-rich sequences and generating 2-bp TSD (41). ISApl1 appears to be highly active (42).
mcr-1 is found as part of a segment apparently derived from Moraxella spp. (43) that also contains a gene usually annotated as a gene encoding PAP2 (a putative PAP family transmembrane protein). In the first plasmid characterized, a single copy of ISApl1 is present upstream of this segment (40). Other plasmids with either two complete ISApl1 elements or a complete ISApl1 element and a fragment of the IRR end flanking the mcr-1 segment, or completely lacking ISApl1, were then found (Fig. 2E). Examples of uninterrupted flanking sequences allowed confirmation that the ISApl1–mcr-1–pap2–ISApl1 arrangement is flanked by 2-bp TSD, suggesting movement of a composite transposon-type structure (41), and this was recently demonstrated (44). Identification of sequence changes and/or small deletions concentrated near the ends of the inserted mcr-1–pap2 segment led to the hypothesis that mcr-1 was first mobilized as part of an ISApl1-mediated composite transposon, with subsequent loss of one or both copies of ISApl1 by illegitimate recombination (41, 45). This may prevent subsequent movement and thus stabilize the mcr-1 gene in the plasmid, similar to loss of IS30 itself from a composite transposon (46). Inactivation of mcr-1 by insertion of IS10 (cut-and-paste mechanism) has been seen (47) and may be reversible, as precise excision of the IS10-flanked composite transposon Tn10 has been reported (48). Similarly, mcr-1 may be inactivated reversibly by insertion of IS1294b (49). This may be a way of dealing with fitness costs associated with modification of lipid A in the outer membrane by the MCR-1 phosphoethanolamine transferase in the absence of colistin (50, 51).
IS91-Like and ISCR Elements
Three related IS, IS91, IS801, and IS1294, lack conventional IR and move by rolling circle replication, which is catalyzed by the Y2 (two tyrosines in the active site) HUH-type enzyme that they encode (52). Replication proceeds from ori to ter (opposite to the direction of transcription of the internal gene), and these elements target a 4-bp sequence similar to the last 4 bp of the ter end (GAAC) and do not create TSD (53). In a proportion of transposition events (∼1 to 10% for IS1294 [54]), replication continues beyond ter into the adjacent sequence, which can then be transferred with the IS to new locations. IS91 and IS801 do not seem to have been involved in movement of known resistance genes, but IS1294 (Fig. 2F) and the variant IS1294b have transferred blaCMY-2-like genes originally associated with ISEcp1 between different plasmid types (55, 56).
An element first identified as a “common region” associated with different resistance genes in certain class 1 integrons (57) was renamed CR1 when related elements were identified (58), and the name ISCR has been used since recognition of their similarity to IS91-like IS (59). ISCR elements are assumed to move and pick up adjacent sequences by rolling circle replication (59), although this has not been demonstrated experimentally. The proteins that they encode (with the proposed name Rcr, for rolling circle replicase [60]) belong to the HUH Y1 family (single catalytic tyrosine). ISCR1 appears to have been responsible for capturing and moving a few different antibiotic resistance genes (Table 4) (21). These are found adjacent to the ori end of ISCR1 in “complex” class 1 integrons (Fig. 2G; see below), presumably as a result of incorporation of the circular molecule by recombination (58, 59). ISCR2 is associated with a few different resistance genes, particularly sul2 in the genomic island GIsul2 and its derivatives (61) (see below). Many other ISCR elements belong to the ISCR3 family (62), which includes hybrids presumably generated by recombination between related elements (21). One of these, ISCR27, may have been responsible for mobilization of a precursor of blaNDM from an unidentified source organism to A. baumannii (63), but ISCR1 may have contributed to subsequent movement (64).
TABLE 4.
Examples of resistance genes associated with ISCR elements
| ISa | Determinant(s) | Resistance(s)b |
|---|---|---|
| ISCR1 | dfrA10 | Trimethoprim |
| catA2 | Chloramphenicol | |
| armA | Aminoglycosides (high level) | |
| blaDHA | 3GC, BLBLI | |
| blaCMY/MOX-like genes | 3GC, BLBLI | |
| Some qnrB genes | Fluoroquinolones (low level) | |
| ISCR2 | sul2 | Sulfonamides |
| tet(31) | Tetracycline | |
| ISCR3 family elements | ||
| ISCR3 | floR | Florfenicol |
| ISCR4 | blaSPM-1 | Carbapenems |
| ISCR5 | blaOXA-45 | 3GC |
| ISCR6 | ant(4′)-IIb | TOB, AMK |
| ISCR14 | rmtB, rmtD | GEN, TOB, AMK |
| ISCR15 | blaAIM-1 | Carbapenems |
| ISCR27 | blaNDM | Carbapenems |
UNIT TRANSPOSONS
Unit transposons were traditionally thought of as elements larger than IS, bounded by IR rather than by a pair of IS, and including a transposase gene and an internal “passenger” gene(s), which may encode antibiotic resistance. This IS/Tn distinction is becoming more problematic, however, as there are now examples of relatives of well-known IS carrying passengers (transporter IS [tIS]) (65), which may have a Tn name/number, and cryptic relatives of transposons without any passengers, which may be given an IS name. The transposon registry (11) lists and provides numbers for unit transposons, and some Tn3 family transposons are included in ISfinder (10). Classes I, II, and III have been used as terms for different transposon types, but we have avoided these here, as definitions have changed over the years (66–68) and the same terms have different meanings for describing mobile elements in eukaryotes.
Antibiotic resistance genes are often associated with Tn3 family transposons, which were reviewed recently (68). Members of the broad Tn3 family are generally characterized by ∼38-bp terminal IR, with IRL and IRR named relative to the direction of transcription of the tnpA transposase gene, which is typically much larger than those of IS (∼3 kb). Tn3 family transposons also include a tnpR resolvase gene and a resolution (res) site, made up of two or three subsites, and may include passenger genes. Transposition occurs via a replicative mechanism in which TnpA catalyzes generation of a cointegrate structure, consisting of directly repeated copies of the transposon separating the original donor and recipient molecules (26). The cointegrate is then resolved into separate molecules, each containing a copy of the transposon, by site-specific recombination between the two directly oriented res sites, catalyzed by TnpR (68, 69). Transposition creates TSD of 5 bp (or occasionally 6 bp). Tn3 family members demonstrate transposition immunity, i.e., transposition of a second element into the same vicinity or the same DNA molecule is inhibited (68, 69), but homologous and/or res-mediated recombination between related elements can occur, creating hybrid elements.
Another transposon superfamily, referred to here as Tn7-like transposons, includes members associated with antibiotic resistance, such as Tn7 and Tn402-like elements in Gram-negative bacteria and Tn552 in Staphylococcus. Members of this group share some features, such as multiple genes encoding products (including a transposase regulator) involved in transposition rather than the single long tnpA gene found in the Tn3 family, but have different transposition mechanisms. Unlike Tn3 family transposons, members of this group may also target a particular site(s). The Tn3 family and Tn7-like transposons most relevant to antibiotic resistance in the species of interest are described below and/or listed in Table 5.
TABLE 5.
Unit transposons associated with resistance in staphylococci and enterococcia
| Transposon | Family | Determinant | Associated resistance(s) | Hostb |
|---|---|---|---|---|
| Tn551 | Tn3 | erm(B) | MLS antibiotics | S |
| Tn917 | erm(B) | MLS antibiotics | E | |
| Tn1546 | vanA | Vancomycin/teicoplanin | S, E | |
| Tn552 | Tn7 | blaZ | Penicillins | S, E |
| Tn5404 | aadE | Streptomycin | S | |
| aphA-3 | Kanamycin/neomycin | |||
| Tn554 | Other | erm(A) | MLS antibiotics | S, E |
| spc | Spectinomycin | |||
| Tn558 | fexA | Chloramphenicol/florfenicol | S | |
| Tn559 | dfrK | Trimethoprim | S | |
| Tn5406 | vga(A)v | Streptogramin A/pleuromutilins/lincosamides | S | |
| Tn6133 | erm(A) | MLS antibiotics | S | |
| spc | Spectinomycin | |||
| vga(E) | Streptogramin A/pleuromutilins/lincosamides |
Tn3 Family Transposons
Tn1, Tn2, and Tn3.
The archetype of the Tn3 family and the close relatives Tn1 and Tn2 were some of the earliest unit transposons to be identified in Gram-negative bacteria (70). In these elements, tnpA and tnpR are transcribed in opposite directions, and the res site lies between them (Fig. 3A). Tn1, Tn2, and Tn3 correspond to three named examples of a family of hybrid elements sharing ∼99% identity over most of their length but only ∼85% identity in short regions either side of res, which suggests homologous recombination followed by res-mediated recombination (70). Tn2 is the most common of the three in clinical isolates but is often annotated and referred to as Tn3 (71).
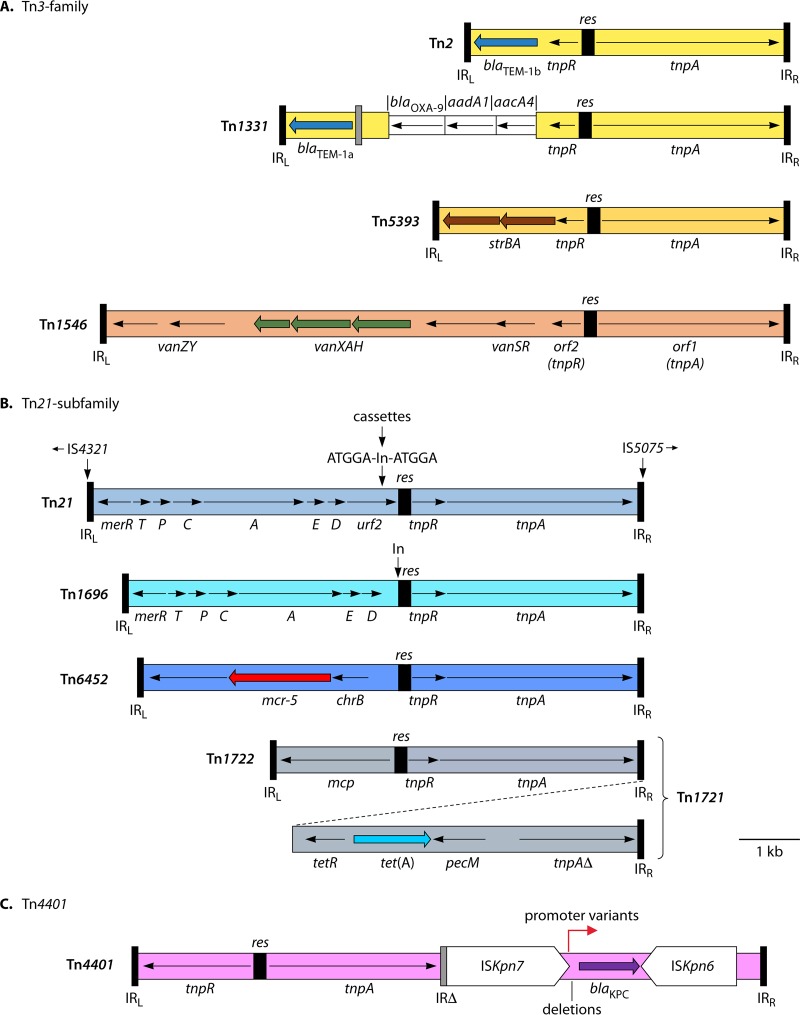
FIG 3.
Tn3 family transposons. The extents and orientations of various genes are shown by arrows, with thick arrows used to indicate antibiotic resistance genes (apart from those in gene cassettes). Terminal IR are indicated by black bars and putative ancestral IR relics by gray bars. res sites are shown as black boxes. (A) Tn3 family. Gene cassettes in Tn1331 are shown as narrower boxes. (B) Tn21 subfamily. For Tn21, the insertion site for class 1 In/Tn (see Fig. 4) and the 5-bp TSD are shown. Different integron structures and different cassettes may be present. IS4321 or IS5075 may be found inserted into IRL and/or IRR, in the indicated orientations. (C) Tn4401. The approximate position of deletions that lead to different promoter variants is indicated. Diagrams were drawn based on sequences from the following INSDC accession numbers: Tn2, AY123253; Tn1331, AF479774; Tn5393, AF262622; Tn1546, M97297; Tn21, AF071413; Tn1696, U12338; Tn6452; KY807920; Tn1721, X61367; and Tn4401, EU176011. The resistance genes shown confer resistance to the following antibiotics: blaTEM-1, broad-spectrum β-lactams; blaOXA-9, oxacillin; aadA1, streptomycin and spectinomycin; strAB, streptomycin; vanXAH, vancomycin/teicoplanin; mcr-5, colistin; tet(A), tetracycline; and blaKPC, carbapenems.
blaTEM genes, including those encoding extended-spectrum β-lactamases (ESBL) or inhibitor-resistant (IRT) variants, have always been found within Tn1, Tn2, Tn3, or variants, hybrids, or fragments of these transposons. A degenerate relic of an IR just upstream of blaTEM suggests capture following adjacent insertion of an ancestral cryptic transposon (68). A derivative of Tn3, named Tn1331, carries additional resistance genes in a region derived from a class 1 integron (Fig. 3A; see below) (72). Hybrid Tn1331-like elements with better matches to Tn1 or Tn2 in different segments are quite common, including in association with blaKPC genes (73). Recombination between different copies of Tn2 in different locations may also contribute to spread of resistance genes that have been inserted within this transposon by other mobile elements (74, 75).
Tn5393.
Tn5393 carries the strAB (streptomycin resistance) gene pair in the position equivalent to that of blaTEM in Tn3. Complete copies of Tn5393 with different insertions have been identified (76), but fragments of Tn5393 appear to be more common than the complete transposon in plasmids and genomic islands.
Tn1546.
The transposons discussed above are all associated with antibiotic resistance in Gram-negative bacteria, while the most notable member of the Tn3 family in Gram-positive bacterial species is Tn1546 (Fig. 3A). Similar to the tnpA and tnpR genes of Tn3, those of Tn1546 are transcribed in opposite directions and separated by the res site; it has 38-bp imperfect IR and creates 5-bp TSD on insertion (77). Tn1546 encodes resistance to vancomycin via the vanA gene cluster, whose expression is regulated by the vanRS gene products. Variants of Tn1546 display significant heterogeneity, including deletions and/or one or more IS inserted into the backbone structure (78). In some cases, these IS have also acquired additional resistance determinants, such as fosB3 (fosfomycin resistance) (79). Importantly, Tn1546 has been responsible for the spread of vancomycin resistance among enterococcal populations around the world, largely facilitated by its association with conjugative plasmids. Furthermore, Tn1546 has been delivered into methicillin-resistant S. aureus (MRSA) by plasmids on several occasions (see below).
Tn21 Subfamily Transposons
In members of the Tn21 subfamily of the Tn3 family, the tnpR and tnpA genes are in the same orientation and the res site is upstream of tnpR (80). This arrangement may give a more stable transposition module than the organization in the transposons described above, as the tnpA and tnpR genes are less likely to become separated by aberrant recombination in res (81). The 38-bp IR of Tn21 subfamily elements are the targets for the related IS4321 and IS5075 elements (IS110 family; encode a DDED transposase), which transpose via double-stranded circular intermediates and insert in one orientation at a specific position, presumably preventing further movement of the host transposon by transposition (82).
Tn21 and close relatives.
Tn21 (81) and related transposons (Fig. 3B) often carry a mercury resistance (mer) operon but are important in movement of antibiotic resistance genes, as they may also carry a class 1 integron (see below). Different members of this family have tnp regions that are ∼80% identical, and they carry different mer operons (e.g., Tn21 and Tn1696) (83) or other accessory genes (e.g., Tn1403) (84). Tn21 itself has an extra region between mer and the res site, and integrons with different structures and different cassette arrays are always found inserted at the same position in this extra sequence, flanked by the same TSD (81). In related transposons without this region, a class 1 integron may be inserted at different locations within the res site (see reference 21 for more details). Because such an interruption might affect resolution, this might help to explain why Tn21 is apparently more prevalent than other members of this subfamily (81).
A new mcr-type gene, mcr-5, was recently identified as part of a transposon designated Tn6452, identified in Salmonella, E. coli, and Cupriavidus gilardii (environmental Burkholderiaceae) (85, 86). The tnp region of Tn6452 is ∼80% identical to that of Tn21, and Tn6452 is bounded by identical 38-bp IR and creates the expected 5-bp TSD.
Tn1721.
Tn1721 consists of Tn1722 (tnpA, tnpR, and res) adjacent to a partial duplication of the IRR end of Tn1722 and the tet(A) tetracycline resistance determinant. The whole structure is flanked by 38-bp IR and has an extra internal copy of IRR (21). Tn1721 may have been created by internal deletion of an ancestral composite element flanked by two copies of Tn1722 (68). As in the case of Tn5393, fragments of Tn1721/Tn1722 are more common than the complete element in plasmids and resistance islands in Gram-negative bacteria.
Tn4401
Tn4401 (Fig. 3C), carrying blaKPC variants, also belongs to the broader Tn3 family, but the common description “Tn3 based” is not accurate, as the Tn3 and Tn4401 TnpA proteins are only about 39% similar/22% identical and the TnpR and nucleotide sequences are quite different. The organization is also different from that of Tn3, with blaKPC and the flanking ISKpn7 (upstream) and ISKpn6 (downstream) elements found between IRR of Tn4401 and the end of the tnpA gene. It appears that an ancestral transposon inserted upstream of blaKPC, with insertion of ISKpn6 disrupting the original IRR and forcing use of an alternative downstream sequence in subsequent transposition events (87).
Several variants of Tn4401 with different internal deletions have been distinguished by lowercase letters (88). The longest version, Tn4401b, has two experimentally confirmed promoters driving blaKPC expression: P2 (last 6 bp to 24 bp downstream of the ISKpn7 IRR) and P1 (74 to 46 bp upstream of the blaKPC start codon) (88). Deletions in Tn4401d (68 bp) and Tn4401a (99 bp; often incorrectly stated as 100 bp) remove regions between P1 and P2 that may form secondary structures but leave both promoters intact (88), as does the 188-bp deletion in Tn4401h (89). The most common form, Tn4401a, gives the highest levels of resistance (88), while Tn4401h gives higher levels than those with Tn4401b (89). Tn4401c and Tn4401e have deletions of 216 bp (incorrectly reported as 215 bp) and 255 bp, respectively, ending at the same place (27 bp upstream of the blaKPC start codon) and leaving P2 only, which was found to result in reduced blaKPC expression (88). Three other blaKPC contexts have either no deletion in this region (Tn4401f [90]) or the 216-bp (Tn4401g [91]) or 255-bp (another Tn4401h variant [92]) deletion, but regions upstream of blaKPC do not match the complete Tn4401 sequence, and these may better be considered “non-Tn4401 elements” (NTEKPC) that contain only part of Tn4401 (93).
Transposons Related to Tn7
Tn7.
The characteristics of Tn7 have been reviewed several times (see references 94 and 95 and references therein). Tn7 carries the tnsABCDE genes (Fig. 4A) and uses a “cut-and-paste” transposition mechanism. TnsB and TnsA together form a heteromeric transposase that excises Tn7 from its original site. IR of ∼28 bp are present at each extremity of Tn7, but there are also four 22-bp TnsB binding sites within 90 bp of the IRL end and three within 150 bp of the IRR end (95). TnsC is an adaptor for target capture that communicates between TnsA/B and either TnsD, directing insertion to a single chromosomal attTn7 site just downstream of the conserved glmS gene of Gram-negative bacteria, or TnsE, to target the lagging strands of replicating conjugative plasmids. This allows both vertical and horizontal transmission (95). Transposition generates 5-bp TSD, and Tn7 carries a class 2 integron (see below).
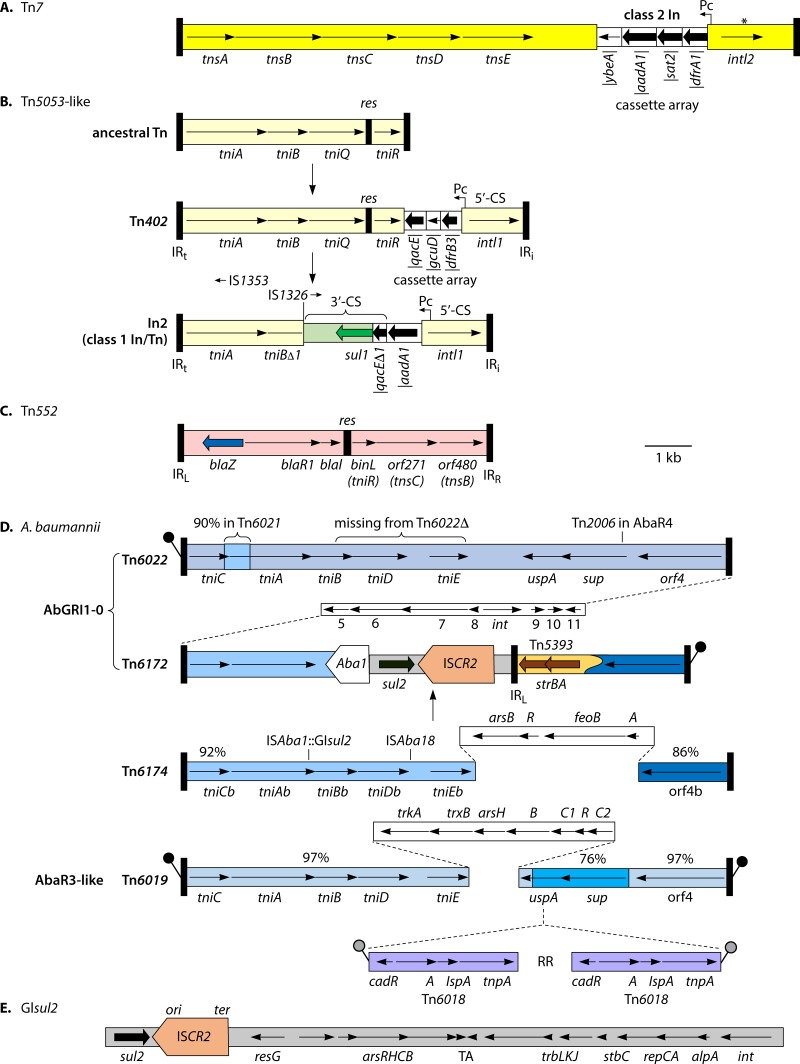
FIG 4.
Tn7-like transposons. Most features are shown as described in the legend to Fig. 3. (A) Tn7. The asterisk indicates the position of a common stop codon in intI2. The diagram was drawn based on the sequence from INSDC accession number AP002527. (B) Evolution of class 1 In/Tn. The diagrams show capture of intI1/attI1/Pc and gene cassettes, with qacE in the last position, by a Tn5053-like transposon. Subsequent deletion of parts of the final qacE cassette and tni region and insertion of sul1 create the 3′-CS, giving a typical “clinical” class 1 In/Tn which is not self-transposable, e.g., In2. Diagrams are based on information in reference 21 and sequences from INSDC accession numbers U67194 and AF071413. Different extents of tni and different IS may be present beyond the 3′-CS (see Fig. 5 in reference 21 for further details). (C) Tn552. Gene names shown in parentheses indicate relationships to those in Tn7/Tn5053 elements. The diagram was drawn based on the sequence from INSDC accession number X52734. (D) Transposons making up resistance islands in A. baumannii. The top diagram represents Tn6022; differences in minor variants Tn6021 (a short region with only 90% identity matches Tn6172) and Tn6022Δ are shown. The main part of the Tn6174 diagram corresponds to the hypothetical, ancestral Tn6173, which is also related to Tn6022 (percentage identities in different regions shown above) but has the ars/feo region replacing uspA and sup. Tn6174 itself has the two insertions shown above the diagram. Tn6172 was generated from Tn6174 by addition of Tn5393 (Fig. 3) and an internal deletion. In AbGRI1-0, a region flanked by Tn6022 and Tn6172 is inserted into the chromosomal comM gene. The backbone Tn6019 of AbaR3-like islands is related to Tn6022 (percent identities in different regions are shown) but contains an additional segment, shown above the relevant diagram. Various regions containing different antibiotic resistance genes are found between the two copies of Tn6018 (designated RR). Diagrams are based on previously published information (109, 110) and on sequences from the following INSDC accession numbers: Tn6022, CP012952; Tn6021 and Tn6164, CP012005; Tn6022Δ, JN247441; Tn6172, KU744946; and Tn6019, FJ172370. (E) GIsul2 (15.460 kb [188] rather than the initially reported 15.456 kb [61], apparently due to errors in the S. flexneri sequence). ars, arenite/arsenate resistance gene; TA, toxin-antitoxin system; alpA, regulation gene. The diagram is based on information in reference 188 and the sequence from INSDC accession number KX709966. The resistance genes shown confer resistance to the following antibiotics: aadA1, streptomycin and spectinomycin; sat2, streptothricin; dfrA1 and dfrB3, trimethoprim; qacE, quaternary ammonium compounds; sul1 and sul2, sulfonamides; blaZ, penicillins; and strAB, streptomycin.
Tn402-like transposons.
Tn402 (also called Tn5090) and other members of the Tn5053 family may carry a class 1 integron (see below) or a mer operon. They are bounded by 25-bp IR, create 5-bp TSD, and carry the tniABQR genes (Fig. 4B). TniA and TniB are related to TnsB and TnsC of Tn7, respectively (96), but transposition occurs via formation of a cointegrate, which also requires TniQ (also called TniD) and resolution by the TniR (TniC) resolvase acting at the adjacent res site (97). These transposons target the res site of Tn21 subfamily transposons but also resolution sites found on plasmids (98). Different Tn402-like tni regions, including hybrids, have been identified in association with class 1 integrons (99).
Tn552.
Strains of S. aureus resistant to penicillin emerged shortly after its therapeutic introduction, and Tn552-like elements are believed to be the origin of all β-lactamase genes in staphylococci (100). Tn552 itself carries genes encoding proteins related to TnsB (orf490) and TnsC (orf271) of Tn7 (94) as well as binL, encoding a serine recombinase, separated from the blaI, blaR1 (both encoding regulators [101]), and blaZ (β-lactamase) genes by a res site (Fig. 4C) (102). It is bounded by 116-bp IR and creates 6- or 7-bp TSD on transposition. Tn552-like transposons are sometimes found in the chromosome but are often carried by multiresistance plasmids and, like Tn5053-like elements, are usually inserted within the res site of the plasmid's resolution system (103–106). In many cases, genetic rearrangements are evident within or in the vicinity of these elements, presumably mediated by interactions between the transposon and plasmid resolution systems and repeated transposition events into them (15).
A. baumannii resistance islands.
Antibiotic resistance islands (AbaR and AbGRI1) found in global clones (GC) of A. baumannii are described in this section, as they are based on transposons related to Tn7 and Tn402 (95, 107, 108). Like Tn7 and Tn402, these transposons may target a specific site(s), as they are generally inserted into the chromosomal comM gene (encoding a protein of unknown function with an ATPase domain [109]), flanked by the same 5-bp TSD (ACCGC), but also on plasmids (110). These transposons are bounded by 25-bp IR and carry tniCAB, encoding Tn7 TnsA-, TnsB-, and TnsC-like proteins, as well as tniDE (orf2 and -3) and various downstream genes (Fig. 4D). Different resistance genes are inserted at different places in these transposon backbones.
Variants of the same basic transposon structure have been named Tn6022, Tn6022Δ (2.85-kb deletion), and Tn6021 (differences in part of tniCA) (Fig. 4D), while other variants are more complex. Tn6172 appears to have evolved from a hypothetical transposon, Tn6173, by addition of other elements to give Tn6174, followed by incorporation of Tn5393 and a subsequent large internal deletion (110). AbGRI1-0 consists of Tn6022 and Tn6172 flanking a region containing orf5 to orf11 and int (encoding a tyrosine recombinase) and may be derived from a plasmid-borne region. AbGRI1 variants may be derived from AbGRI1-0 by addition of resistance genes or deletions due to recombination between homologous transposon segments (110). In some A. baumannii isolates, a single Tn6022-like transposon is inserted into comM, e.g., AbaR4, which consists of Tn6022 with Tn2006 (Table 1) inserted.
The backbone of regions referred to as AbaR3-like is Tn6019, which is related to Tn6022 but has a different, longer region downstream of tniE (Fig. 4D) (109). A composite transposon-type structure consisting of two directly oriented copies of Tn6018 flanking different resistance regions is inserted in this backbone. Components of these resistance regions are apparently derived from a plasmid related to R1215 from Serratia marcescens, which is not stably maintained in A. baumannii (111).
GENE CASSETTES AND INTEGRONS
A gene cassette is a small mobile element (∼0.5 to 1 kb) consisting of a single gene (occasionally two), typically lacking a promoter, and an attC recombination site. Gene cassettes can exist in a free circular form but are nonreplicative and are usually found inserted into an integron (Fig. 1), characterized by an intI gene, an attI recombination site, and a promoter (Pc). intI encodes an atypical site-specific tyrosine recombinase, which has an extra domain compared to other members of this family (112), that catalyzes recombination between the attI site of the integron and the attC site of a cassette. This inserts the cassette into the integron in the orientation that allows expression of the cassette-borne gene from the Pc promoter. Multiple cassettes may be inserted into the same integron to create a cassette array (often incorrectly referred to as a “cassette”) that may confer multiresistance (Fig. 1). Different classes of integron have been defined based on the sequence of IntI (called IntI1, IntI2, IntI3, etc., with cognate attI1, attI2, and attI3 sites), with class 1 being the first reported and most common in antibiotic-resistant clinical isolates. Integrons and gene cassettes have been reviewed many times (e.g., see references 112–114).
Cassette Integration and Expression
attC sites associated with different cassettes differ in sequence, but all include two pairs of conserved 7- or 8-bp core sites at their outer ends (R″-L″ and R′-L′ [112] or 1L-2L and 2R-1R [114]). These are separated by a region of variable length that usually shows inverted repeatedness. Although the sequence similarity between different attC sites is low, single-stranded versions each form a conserved secondary structure, with two or three unpaired, protruding extrahelical bases. These are recognized by the IntI recombinase and are important in directing recombination to the bottom strand, ensuring that insertion occurs in one orientation only (112).
The most efficient IntI-mediated reaction is recombination between the double-stranded attI site and the single-stranded, folded attC site to insert a cassette into the first position in an array. IntI-mediated excision of cassettes typically occurs between two single-stranded, folded attC sites. IntI1 activity is regulated by LexA (SOS response master regulator) binding to a site overlapping the −10 box of the intI1 promoter, repressing expression to minimize unnecessary cassette shuffling. If the SOS response is triggered, repression is lifted, giving increased integrase activity when adaptation is required (112). Formation of single-stranded DNA during conjugation also favors both attC folding and recombination, as well as triggering of the SOS response, so that incoming cassettes are more likely to be integrated (112). intI2 expression is not regulated by the SOS response (115).
In class 1 integrons, the Pc promoter lies within the int1 gene, and minor sequence variations give an inverse relationship between Pc strength and IntI1 activity (116). In some class 1 integrons, insertion of three G's between potential −35 and −10 sites gives optimal 17-bp spacing, activating an additional promoter (P2) (116). attI2 of class 2 integrons contains two active Pc promoters, also with variants of different strengths (115). Expression of cassette genes is reduced with increasing distance from Pc and P2. Rather than being due to effects of attC secondary structure on transcription, as first proposed (117), this appears to be due to effects on translation (112). This means that cassettes can be carried at less cost at the “back” of an array but still have the potential to be shuffled to the “front” of the array. Some cassette genes lack a ribosome binding site (RBS), and ORF-11 (118) and the recently identified ORF-17 (119) in attI1 may contribute to expression if the cassette is the first in the array.
Class 1 Integrons
Tn402 (Fig. 4B) seems to have resulted from capture of the intI1/attI1/Pc combination, found on the chromosomes of betaproteobacteria in association with a qacE cassette (resistance to antiseptics), by a Tn5053 family transposon (120). In the more common “clinical” or “sul1-type” class 1 integrons, part of the tni region has been replaced by the 3′ conserved segment (3′-CS) (Fig. 4B). The longest versions of the 3′-CS include the qacEΔ1 gene, derived from the qacE cassette, and sul1 (encoding resistance to the early sulfonamide antibiotics), but only part of this region may be present. The term “class 1 In/Tn” has been suggested to encompass structures with intI1/attI1/Pc and either a full or truncated tni region (21). The 25-bp IR of class 1 In/Tn are known as IRi (at the integrase end) and IRt (at the tni end), and the region from IRi to the end of the attI1 site is called the 5′ conserved segment (5′-CS). While some class 1 In/Tn have lost tni transposition functions, there is evidence that they can be moved, presumably by compatible Tni proteins available in the same cell (121). Class 1 In/Tn may also move with an upstream ISPa17 element, which has IR related to IRi and IRt (122).
The first few class 1 In/Tn identified were given In numbers, In0 (no cassettes) to In6, intended to specify all components, including the cassettes, the length of the 3′-CS and tni region, and any additional elements, such as IS. “In2-like” (with IS1326 plus IS1353 inserted) and “In4-like” (with a shorter 3′-CS and inverted IRt ends of tni separated by IS6100) integrons seem to be the most common (114). INTEGRALL (123; http://integrall.bio.ua.pt/) now keeps a registry of In numbers, but these really correspond only to different cassette arrays. So-called “complex” class 1 integrons, usually with partial duplications of the 3′-CS, are created by insertion of circles containing ISCR1 and an associated resistance gene(s) by recombination into the 3′-CS or an existing ISCR1 element (Fig. 2G). The boundary with position 1,313 of the 3′-CS is used to define the ter end of ISCR1, although this may not be the original end (21).
Other Integron Classes
Class 2 integrons, associated with Tn7 (Fig. 4A) and variants, often have a nonfunctional IntI2 gene due to an internal stop codon and, probably as a consequence, house a limited variety of cassettes (124). Class 3 integrons are more similar to class 1 integrons and also appear to be associated with Tn402-like transposons (125). Only a few examples have been identified, mostly carrying cassettes that encode β-lactamases. Class 4 was previously used to refer to an integron found in the Vibrio cholerae chromosome. This and other “sedentary chromosomal integrons” (SCI; formerly called CI) may contain very large arrays of cassettes (>170 in V. cholerae), which all tend to have very similar attC sites. Although cassettes containing resistance genes make up a minority of those in SCI, they appear to be the source of cassettes found in “mobile” integrons (112). “Mobile” integron types, now designated class 4 and class 5 integrons (112), appear to be rare and have not been identified in the species of interest here.
Gene Cassettes and Antibiotic Resistance
A wide variety of gene cassettes containing resistance genes (named after the gene carried) have been identified (114; see http://app.spokade.com/rac/feature/list for updated lists). The most clinically relevant are those carrying genes encoding β-lactamases or aminoglycoside-modifying enzymes. The former include metallo-β-lactamases (MBL; class B), with the VIM and IMP types being the most common. Cassette-borne genes also encode class A GES enzymes, which are either ESBL or carbapenemases (with a mutation at amino acid 170), and class D OXA-10-like (which include ESBL variants) and OXA-1-like enzymes. Variants of the common aacA4/aac(6′)-Ib cassette may confer resistance to tobramycin plus gentamicin and/or amikacin or low-level resistance to fluoroquinolones due to different point mutations. Different fusions that compensate for the lack of an RBS in this cassette also create AacA4 proteins with different N-terminal ends (114). Certain cassette arrays (e.g., ∣dfrA17∣aadA5∣ and ∣dfrA12∣gcuF∣aadA2∣, giving resistance to trimethoprim [dfr] and to streptomycin and spectinomycin [aadA]; gcu indicates a gene cassette of unknown function) are very common in class 1 integrons (114).
Gene cassettes may be interrupted at a specific position in the attC site by an IS1111-attC element related to IS4321/IS5075 (see above) or by a group II intron (114). These small, mobile, site-specific elements encode a catalytic RNA (ribozyme) and a reverse transcriptase (126). A role for these introns in creation of gene cassettes has been suggested (127), but there are also arguments against this (112). Sometimes the partial attC site that follows an IS1111-attC element or an intron does not belong to the preceding cassette, suggesting IS- or intron-mediated deletion (114, 128), which may be a way of streamlining arrays. Group II introns, named using a combination of a species abbreviation and a number (129; http://webapps2.ucalgary.ca/~groupii), are also found inserted into conjugative plasmids (75), ICE, and pathogenicity islands (16).
Gene Cassettes and Integrons in Gram-Positive Bacteria
Gene cassettes and/or integrons have been reported for a few Gram-positive bacterial species, including Corynebacterium glutamicum (on a plasmid transferable to E. coli) (130), Staphylococcus (e.g., see reference 131), and Enterococcus (e.g., on a transferable plasmid [132]). However, many studies report only detection of intI1 by PCR, with sequencing of fragments in some cases. Searches with the class 1 integron 5′-CS or 3′-CS against sequences from Staphylococcus and Enterococcus species in GenBank (including the whole-genome shotgun contigs [WGS] database [accessed May 2018]) identified very few examples, most of which were fragments and none of which provided evidence of linkage to the chromosome or plasmids. Thus, there is presently no conclusive evidence demonstrating the existence of integrons in these genera.
MITEs AND TIMEs
MITEs are nonautonomous (i.e., incapable of self-transposition) derivatives of bacterial IS or transposons that retain the IR but which have lost central parts, including the transposase gene(s) (134). Pairs of MITEs, including Tn3-derived inverted-repeat miniature elements (TIMEs) (135), appear to have been involved in mobilization of resistance genes. For example, a composite transposon-like structure flanked by two copies of a 288-bp TIME (referred to as an integron mobilization unit [IMU]) was shown to transpose the intervening integron fragment when a Tn3 family transposase was provided (136). Two copies of the same 439-bp MITE were also identified flanking integron fragments carrying different cassette arrays in different Acinetobacter isolates (133, 137). MITEs and TIMEs may provide an explanation for movement of resistance genes if full-length IS or transposons cannot be found, but they can be difficult to identify.
RESISTANCE PLASMIDS
Plasmids are important vehicles for the carriage of other MGE and acquired antimicrobial resistance genes associated with these elements in both Gram-negative and Gram-positive genera, and they vary in size from less than a kilobase to several megabases (138). Their extrachromosomal existence stems from their ability to replicate and hence be inherited in a growing population of host cells, which often requires a cadre of gene systems dedicated to their efficient vertical inheritance. Conjugation or mobilization functions may also be present, allowing plasmids to spread horizontally. Together the genes encoding these functions form a “backbone” (139) that represents a core of plasmid housekeeping functions to which can be added “accessory” niche-adaptive activities that might benefit the host cell (and hence the plasmid itself) in a particular environment. In resistance plasmids, these accessory regions are typically made up of one or more resistance genes and associated mobile elements of the types described above (IS, Tn, and/or In). Closely related backbones may have different insertions and/or resistance regions, and conversely, different backbones may house the same resistance genes and associated mobile elements. In this section, we first provide a summary of the main functions encoded by plasmid backbones before going on to describe the basic characteristics of known plasmid groups that have played a major role in the spread of antibiotic resistance in the species that are the focus of this review.
Replication Initiation and Copy Number Control
Plasmid replication initiates at a defined region, the origin (ori), triggered either by an RNA transcript or, more commonly, by the binding of an initiation protein (Rep), encoded by a rep gene on the plasmid, to proximal iterated DNA repeat sequences termed iterons. The ori and the (typically colocated) initiator gene form the basic component of all plasmids, the minimal replicon. Thus, plasmids encode their own replication initiation but usually exploit the host's chromosomally encoded replication machinery (helicase, primase, polymerase, etc.) for DNA synthesis itself. Interactions with and dependence on host-encoded DNA replication proteins are among the factors that limit the host range of plasmids. Some plasmids are efficiently maintained only in closely related bacterial taxa and are hence termed narrow-host-range plasmids, whereas others are referred to as broad-host-range plasmids because they have been found or shown to replicate in quite diverse genera. Factors other than replication, particularly whether it is transmissible by conjugation or mobilization (see below), can also influence a plasmid's host range. Conjugation can be an extraordinarily promiscuous process, capable of even transkingdom genetic exchange (140). Transfer of resistance plasmids into hosts in which they cannot replicate is therefore likely to be commonplace, with other MGE (e.g., IS, Tn, and In) providing intracellular mobility mechanisms that give resistance genes an opportunity to “escape” to other functional replicons (the chromosome or other resident plasmids). Thus, even narrow-host-range plasmids can act as suicide vectors for the horizontal spread of resistance genes into divergent hosts.
Replication initiation proteins often possess one of several ancient conserved domains (141), which define the type of replication system. Three modes of plasmid replication have been described for circular plasmids (142). Rolling circle (RC) replication is commonly used by small plasmids in Gram-positive and, less commonly, Gram-negative bacteria (143). It relies on a Rep protein nicking one DNA strand at the double-stranded origin (dso), which provides a free 3′-OH to prime leading-strand DNA synthesis that displaces the remainder of the nicked strand. The displaced strand is then asymmetrically replicated from a second, distinct, single-stranded origin (sso). This mode of replication effectively limits plasmid size, so RC plasmids are usually cryptic or carry only a single resistance gene.
The other modes of plasmid replication rely on initiator-mediated localized melting of double-stranded DNA (dsDNA) at the origin to trigger replication based on RNA primers. Theta-mode replication resembles circular chromosome replication and is widely used by small to very large plasmids. DNA synthesis is continuous on the leading strand and discontinuous via Okazaki fragments on the lagging strand (144). IncQ plasmids utilize the third mode of replication, termed strand displacement, where both DNA strands are replicated continuously in opposite directions from the origin (144); these plasmids are also usually small. IncQ plasmids exhibit an extremely broad host range, as they encode their own helicase and primase proteins in addition to an initiator.
In order to balance the competing demands of effective plasmid inheritance and metabolic impost on the host, plasmids control their copy number. The details of plasmid copy number control systems vary greatly between plasmid types, but two basic strategies have been discerned. The first uses an antisense (countertranscript) RNA, constitutively expressed and hence proportional to plasmid copy number, which binds to the complementary rep mRNA to repress its transcription and/or translation; in plasmids that use an RNA initiator, such as ColE1, countertranscript binding inhibits maturation of the RNA primer (145, 146). In the second mechanism, the ori sites on two plasmid molecules are “handcuffed” together by interactions between Rep proteins bound to their iterons. This modulates Rep activity in response to the concentration of iterons within the cell, which is directly proportional to the plasmid copy number (147, 148).
Plasmids with multiple replication regions are quite common in both Gram-negative and Gram-positive bacteria, suggesting that fusions/cointegrations between plasmids occur frequently. It would be expected that the rep region with the highest intrinsic copy number would initiate replication of a multireplicon plasmid. Additional replicons may unduly increase the fitness cost of a cointegrate plasmid and can be eliminated by mutations or deletions, but they may also be advantageous, e.g., being able to use different replicons that can function in different host species may increase the plasmid host range. The presence of multiple replicons might also allow those that are not driving replication to diverge, potentially changing incompatibility (149) (see below).
Plasmid Maintenance
Once replicated, plasmids must be distributed between daughter cells when division takes place. For small plasmids maintained at a high copy number, efficient inheritance by both daughter cells can be achieved by random segregation. However, larger plasmids usually exist at a low copy number to minimize the burden on their hosts, which risk being outcompeted by plasmid-free counterparts in the environment. Large low-copy-number plasmids thus usually possess functional modules that contribute to plasmid maintenance (segregational stability) (150). These include multimer resolution (res), partitioning (par), and postsegregational killing systems.
Resolution systems convert plasmid multimers, which arise due to homologous recombination, into monomers that can be segregated independently into daughter cells. They usually comprise a gene encoding a site-specific recombinase and a cognate DNA site at which the recombinase acts, although some plasmids possess only a site that is recognized by a chromosomally encoded resolvase (151). Partitioning systems actively distribute plasmid copies to daughter cells and usually consist of two genes. The first encodes a DNA-binding “adaptor” protein that interacts with both a “centromere-like” DNA site and a “motor” protein encoded by the second gene; most par systems belong to one of three types, based on the class of motor protein which they encode (152, 153). Postsegregational killing systems, sometimes called plasmid addiction systems, kill progeny cells that fail to inherit a copy of the plasmid (i.e., if replication, resolution, and/or partitioning fails). They include toxin-antitoxin (TA) systems that encode a toxic polypeptide and an antitoxin component that inhibits the expression or activity of the toxin. A number of different TA system types have been described, distinguished primarily by the nature of the antitoxin (RNA or protein) and its mechanism of action (154), but plasmid TA systems all rely on an abundant antitoxin that is more labile than the longer-lived toxin component (either the toxic protein itself or the mRNA that encodes it) that it counteracts (155). Thus, in daughter cells that fail to inherit a copy of the plasmid, the antitoxin cannot be replenished and inhibition of toxin activity is eventually released, resulting in cell death. Restriction-modification systems, often found on plasmids and other mobile elements, can also act as postsegregational killing systems (156).
Conjugation and Mobilization
Plasmid propagation is facilitated not only through vertical transmission via cell division but also via horizontal transmission to other bacterial cells. Conjugative (self-transmissible) plasmids possess genetically complex systems for horizontal plasmid transfer, which significantly increase the size of their conserved backbone. The transfer (tra) regions of conjugative plasmids encode proteins for mating pair formation (MPF; classified into 8 types) (157) that function as a specialized type IV secretion system (T4SS) pore, as well as DNA transfer replication (DTR) proteins that process the plasmid DNA. The DTR proteins include a relaxase that specifically nicks the origin of transfer (oriT) of the DNA strand that is exported to the recipient cell (158). In Gram-negative bacteria, the T4SS assembles a conjugative pilus, a filamentous surface appendage that mediates interactions with recipient cells. Within the donor cell, the nucleoprotein complex, comprised of DTR proteins and nicked oriT (termed the relaxosome), is linked to the MPF pore (transferosome) by a coupling protein (T4CP), a multimeric ATPase belonging to the FtsK/SpoIIIE superfamily (159, 160). Conjugative plasmids also often carry genes encoding entry (surface) exclusion proteins that prevent the host from acting as a recipient cell for the same or related plasmids (161).
Some nonconjugative plasmids can be transferred horizontally by exploiting the MPF apparatus provided by a conjugative plasmid present in the same cell. Such mobilizable plasmids carry only a subset of the DTR functions (usually termed mob), including oriT and a gene for a corresponding relaxase. However, there is emerging evidence for both Gram-positive (162–164) and Gram-negative (165) organisms that plasmids that were assumed to be nontransmissible due to the lack of a relaxase gene may nonetheless actually be mobilizable (166) (see below).
Plasmid Classification
Originally, plasmid classification commonly relied on the phenomenon of incompatibility, based on the observation that closely related plasmids cannot coexist stably in the same cell. This is usually due to cross talk between the replication initiation systems of the two plasmids that “confuses” copy number control (the two different plasmids are perceived as the same), leading to a reduced copy number and hence to segregational instability in the absence of direct selection (17, 167). Thus, incompatible plasmids are likely related and are classified in the same Inc group. Extensive plasmid Inc typing schemes were established for both Gram-negative and Gram-positive bacteria (160), but the laborious nature of incompatibility testing resulted in it being superseded by hybridization (167), then PCR-based replicon typing (PBRT) (168, 169), and ultimately sequencing-based approaches (170). Nonetheless, the historical Inc groupings underpin the widely used PBRT and PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/) in silico replicon classification systems for plasmids from the Enterobacteriaceae (170). A contemporary replicon classification system for plasmids from Gram-positive genera was also devised, with groups rep1 to rep19 (171), but unfortunately the established Inc groupings were not incorporated. Additional rep families were subsequently added separately by the same author group (rep7b and rep20 to rep24, corresponding to the set used currently in the PlasmidFinder Enterococcus, Streptococcus, and Staphylococcus database) (172) and another group (173), resulting in discordant classification of some plasmids. Mobility typing (MOB typing), based on conjugative and mobilization relaxase genes, was also devised to extend plasmid identification/classification and to facilitate epidemiological tracking (174). These and other methods are summarized in a recent paper, which also provides discussion of the challenges of classifying plasmids from whole-genome sequence data (175).
Resistance Plasmids in the Enterobacteriaceae
Known resistance plasmids in the Enterobacteriaceae include large (up to at least 200 kb), usually conjugative, and small, often mobilizable, plasmids. PBRT (168) is commonly used to type plasmids in these organisms, and MOB typing (176, 177) to some extent, but the results of these methods are not always concordant (178). PlasmidFinder uses a database of amplicon sequences from PBRT and additional variants (170) and is a useful starting point for identifying plasmid types in whole-genome sequences. PCR methods for detecting different partitioning systems (179) or TA systems (180) are also available.
Plasmid multilocus sequence typing (pMLST) schemes (https://pubmlst.org/plasmid/) for some Inc groups assign allele numbers and sequence types (pST) (cf. MLST for strain typing). These schemes were often designed when few plasmid sequences were available and are based on the sequences of 2 to 5 gene fragments, so they can obviously reflect differences in just those few short regions (Fig. 5). While in some cases these schemes have been useful in identifying relationships between plasmids for epidemiological purposes, examples of plasmids of the same pST with differences outside the pMLST targets and different insertions of the same resistance gene have been identified (55). As it is now more economical and informative to sequence genomic DNA rather than to amplify and sequence individual pMLST targets, these schemes are being superseded. Identifying pMLST types from whole-genome sequences (e.g., by using the pMLST tool at https://cge.cbs.dtu.dk/services/pMLST/) may still be useful for comparison with historic data. For this reason, pMLST schemes are mentioned in the relevant sections below, but there is now a need to use WGS to understand the strengths and weaknesses of available pMLST schemes and to develop better means for comparison of entire plasmid backbones to identify relationships and evidence of recombination.
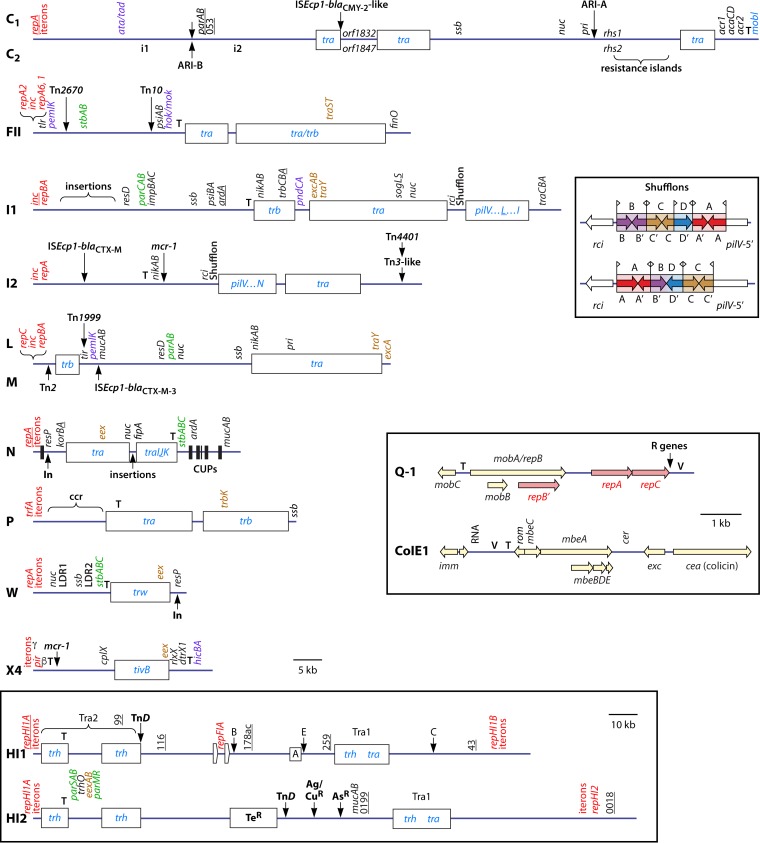
FIG 5.
Representative diagrams of the backbone organization of major plasmid types associated with antibiotic resistance in Enterobacteriaceae. Plasmid types are indicated on the left. Diagrams are approximately to scale, with those in boxes at a different scale (see scale bars). Selected genes/gene regions involved in various functions are shown by the following colors: red, replication/oriV; blue, conjugation; green, maintenance; brown, entry exclusion; and purple, TA. Additional features may be shown for different plasmid types, with most explained further in the text, except for the following: ssb, single-stranded DNA binding protein gene; pri/sog, primase gene; resD/resP, resolvase gene; stb, stability/partitioning gene; psiAB, plasmid SOS inhibition gene; impABC/mucAB, mutagenic DNA polymerase gene; ardA, antirestriction gene; korAB, kill override gene (involved in regulation of tra); ccr, central control region; LDR, long direct repeats. Origins of transfer (oriT) are indicated by “T,” if they have been defined. Insertion points for resistance regions common to plasmids of the same type are also indicated, in some cases, by labeled vertical arrows. C backbones are represented by a single line, with differences (presence/absence of ARI-A, orf1832 versus orf1847, rhs1 versus rhs2, and presence/absence of i1 and i2) shown above (C1) and below (C2). L and M backbones are also represented by a single line, with different insertions in common plasmids shown above (L) and below (M) (modified versions of Tn2 with additional resistance genes are also found at the site indicated for Tn2). These two plasmid types differ mainly in traY/excA (entry exclusion) and traX (relaxase), with differences in inc distinguishing the M1 and M2 types. For HI1 plasmids, the type 1 backbone is shown, with insertions found in type 2 plasmids indicated above (A to E; region D from reference 211 was recognized as a transposon, TnD, in reference 208). Insertions that give resistance to various heavy metals are indicated as follows; Te, tellurite; Ag, silver; Cu, copper; and As, arsenic. Targets for pMLST schemes are underlined (for C plasmids, repA, parA, parB, and 053; for I1 plasmids, repA, ardA, trbA, sogS, and pilL; for N plasmids, repA, korA, and traJ; for HI1 plasmids, repA [HCM1.64] as well as HCM1.99, HCM1.116, HCM1.178ac, HCM1.259, and HCM143 [abbreviated “99,” etc.]; and for HI2 plasmids, 0199 and 0018). Shufflons in I1 plasmid R64 (above) and I2 plasmid R721 (below) are shown in a separate box. Segment A contains partial open reading frames A and A′, etc. sfx repeats are represented by flags. Diagrams are based on information in previous publications and/or sequences from INSDC accession numbers for prototype plasmids, as follows: C1 and C2, references 181 and 193; FII, accession number AP000342; I1, references 55 and 514; I2, reference 236 and accession number KP347127; I1 and I2 shufflons, reference 493; L/M, references 239, 245, and 515; N, reference 250 and accession number AY046276; P, reference 260 and accession number U67194; W, reference 280 and accession number BR000038; X, reference 298; HI1, references 205, 208, and 211 and accession numbers AF250878 and AL513383; HI2, references 205 and 213 and accession number BX664015; Q-1, reference 307; and ColE1, reference 311 and accession number J01566.
Although plasmids are now often assigned to a group on the basis of sequence homology rather than information about true incompatibility, known resistance plasmids given the same Inc designation do mainly share backbones with similar organizations/functions. Given that similar backbone types may be associated with a number of different resistance genes and associated MGE, here we have focused mostly on the main characteristics of the plasmid backbones themselves (summarized in Table 6 and Fig. 5). The following sections group plasmids by the original Inc categories, although we removed “Inc” from the names to indicate that true incompatibility has not always been determined (as proposed for A/C plasmids [181, 182]). Plasmids of some of these groups are also found in P. aeruginosa, where they may have been given an alternative P-number designation, and those that carry resistance genes are mentioned in the relevant sections below. Another recent review of resistance plasmids in Enterobacteriaceae (183) includes information about the geographic distribution of plasmids from different sources (human, animal, and environmental) and associations with resistance genes.
TABLE 6.
Main characteristics of known resistance plasmids in Enterobacteriaceae
| Inca | Replicon(s) | Rep domainb | Copy no.c | MOBd | Host range | Conjugation or pilus descriptione |
|---|---|---|---|---|---|---|
| A/C (P-3) | A/C | — | L | MOBH12 | Broad | Thick and flexible |
| F | FII | FII | L | MOBF12 | Enterobacteriaceae | Thick and flexible |
| FIA | Rep_3 | |||||
| FIB | Rep_3 | |||||
| G (P-6) | G | — | L | MOBP14 | Broad, γ | Mobilizable |
| HI1 | HI1A | —e | L | MOBH11 | Enterobacteriaceae | Thick and flexibleg |
| HI1B | — | |||||
| FIA-like replicon | Rep_3 | |||||
| HI2 | HI1A | —f | L | MOBH11 | Enterobacteriaceae | Thick and flexibleg |
| HI2 | —f | |||||
| I complex | I1/Iγ/B/O/K/Z | FII | L | MOBP12 | Enterobacteriaceae | Rigid plus thin and flexibleh |
| I2 | I2 | FII | L | MOBP6 | Enterobacteriaceae | Rigid plus thin and flexibleh |
| J (ICE) | J | — | MOBH12 | Thick and flexible | ||
| L/M | L/M | FII | L | MOBP13 | Broad, α, β, γ | Rigid |
| N | N | Rep_3 | L | MOBF11 | Broad | Rigid |
| P (P-1) | P | Rep_3 | L | MOBP111 | Broad, α, β, γ | Rigid |
| Q-1 | Q-1 | RepC | H | MOBQ1 | Gram-negative and -positive bacteria | Mobilizable |
| Q-3 | Q-3 | RepC | H | ? | Broad | Mobilizable |
| R | R | Rep_3 | L | ? | ||
| T | T | —f | L | MOBH12 | ? | Thick and flexibleg |
| U | U | — | L | MOBP4 | Broad, α, β, γ | Rigid |
| W | W | Rep_3 | L | MOBF11 | Broad, α, β, γ | Rigid |
| X | X | Rep_3 | L | MOBP3 | Enterobacteriaceae | Thin and flexible |
| Y | Y | —f | L | Enterobacteriaceae | Plasmid-like prophage | |
| ColE1 | ColE1 | RNA II | H | MOBP5/HENi | Mobilizable |
P-numbers show designations used for Pseudomonas.
All plasmid types use a θ replication mechanism, except for Q plasmids, which use a strand displacement mechanism. Rep domains (see text on plasmids in Gram-positive bacteria for more details) from the conserved domains database (CDD) (141) were identified using BLASTp searches. —, no Rep domain identified in BLASTp searches. ColE1-like plasmids encode an RNA primer rather than a replication initiation protein.
H, high; L, low.
MOB type usually associated with the replicon(s) in known resistance plasmids. For details of MOB classification, see reference 174.
From reference 512.
RepHI1A, HI2, T, and Y replicons seem to belong to the same protein family.
Conjugation is temperature sensitive (209), and host range appears to be broader at lower temperatures (513).
The PilV tip adhesin of the thin pilus is varied by the shufflon recombination system.
HEN stands for amino acids H97, E104, and N106, whereas most relaxase active sites have three histidines (311).
A/C plasmids.
The main features of A/C plasmids were reviewed relatively recently (181) and are thus summarized and updated here. IncC plasmids were first reported in the 1960s, the compatible but related plasmid RA1 was then assigned to IncA, and these groups were subsequently combined (181, 182). More recently, it was suggested that A/C plasmids be divided into A/C1 and A/C2 groups due to differences in the repA initiator gene target used for PBRT (184). The A/C2 group, now equated with IncC (185), was further split into type 1 (called C1 here) and type 2 (C2) (186). Recently, RA1 (A) and a C1 plasmid were confirmed to be compatible but showed strong mutual entry exclusion (although the determinant has not yet been identified) (182). The same paper also recommends that the term IncA/C be avoided and suggests using “A/C,” “A-C complex,” or “RepA/C” when the two types have not been distinguished, e.g., identified by PBRT (182).
The backbones of RA1 (A), C1, and C2 plasmids have similar organizations. tra genes have been identified from homology to other systems and are not well studied (181). mobI, located upstream of repA, is also essential for conjugative transfer (187), and different conjugation frequencies have been reported for different A/C plasmids (181). The master activator complex AcaCD, essential for conjugative transfer, binds upstream of and positively regulates selected tra and other genes, and production of AcaCD itself is controlled by acr1- and acr2-encoded repressors (187). A/C plasmids can mobilize Salmonella genomic islands (SGI), which carry A/C tra gene homologs, but simultaneous transfer of both an A/C plasmid and an SGI appears to be rare (187).
The C1 and C2 types differ mainly in substitutions generating orf1832 versus orf1847 and rhs1 versus rhs2 (encoding Rhs proteins of unknown function and with different C termini), respectively, and in two insertions (i1 and i2) in C2 (Fig. 5) (181). Both C1 and C2 lineages may carry an antibiotic resistance island (ARI-B) (181) derived from GIsul2 (Fig. 4E and see below), which targets a specific site in these plasmids, following independent acquisition events (188). ARI-B regions mostly carry genes conferring resistance to older antibiotics (sul2, strAB, tet, and/or floR) between IS26 elements and partial duplications of ISCR2 (181). Most sequenced C1 plasmids also carry ARI-A, apparently derived from a complex multitransposon insertion (21, 189). ARI-A is always inserted in the same position upstream of rhs, flanked by the same TSD (unless part of rhs has been deleted), and may carry blaNDM and rmtC (190). C1 plasmids may also have an ISEcp1 TPU carrying blaCMY-2 (or a minor variant) inserted just upstream of traC, flanked by 5-bp TSD, or two copies of this TPU that may be rearranged (21). Resistance regions in C2 plasmids vary in organization and exact insertion points (although these are still in the rhs region), indicating multiple acquisition events, and they may carry different resistance genes, including blaKPC (181). Until recently, RA1 was the only plasmid of the A type to be reported, but a few additional sequences are now available (182).
PlasmidFinder (18-05-02 version) includes targets for A plasmids (reported as IncA/C) and C plasmids (reported as IncA/C2) (170). An early PCR scheme amplifying 12 A/C backbone regions was based on few plasmid sequences (191). Four genes (repA, parA, parB, and “orf053”) experimentally identified as being important for plasmid maintenance (and with similar expression patterns [192]) are used in a pMLST scheme that distinguishes C1 and C2 plasmids (193). A more recent PCR strategy discriminates C1 and C2 plasmids by using amplicon sizes (using primers targeting orf1832/orf1847, linking rhs1/rhs2 to the adjacent sequence, and flanking i1 and i2) and detects the presence/absence of ARI-B (194). Primers have also been designed to distinguish A and C plasmids (182). Phylogenetic analysis of 28 genes fully conserved in 82 C1 plasmids (including the four pMLST targets) identified five groups, and these genes are used in a more comprehensive cgpMLST (193) scheme available at https://pubmlst.org/plasmid/.
F plasmids.
The F (“fertility factor”) plasmid was the first example of a conjugative plasmid found in bacteria and is the basis of the designation IncF for plasmids with common sensitivity to specific phages and serological cross-reactivity, reflecting a common conjugation system (MPFF). This F-type mating apparatus may be associated with different replicons, with incompatibility testing originally defining subgroups FI to FVII (195). Combinations of three replicons (FIIA/FIC, FIA, and FIB) are commonly found together in multireplicon plasmids, including the F plasmid. Expression of the FII initiator RepA1 requires translation of the RepA6 (TAP) leader peptide and is regulated mainly by the countertranscript CopA (inc) RNA, but also by the transcriptional repressor protein CopB (repA2) (146). The FIA RepE initiator is regulated by handcuffing, with monomers bound to iterons on two different plasmids bridged by a dimer (148). In addition to multiple replicons, many F plasmids also carry different partitioning and TA systems and quite different sets of resistance and/or “virulence” genes, giving a diverse group of mosaic plasmids.
The original PBRT scheme included FIA, FIB, and FIC primers and a general IncF (FrepB) primer pair as well as primers for Salmonella enterica pSLT-type virulence plasmids (FIIS) (168). In addition to and probably partly as a consequence of having multiple replicons, F-type plasmids are highly mosaic, with few components in common, precluding development of a pMLST scheme (149). Additional/updated replicon primers were included in a replicon typing scheme (RST) based on diversity in the replicon regions (149). This scheme distinguishes FII replicons commonly found in E. coli (FII), Klebsiella (FIIK), Salmonella (FIIS), and Yersinia (FIIY) and uses a FAB formula (FII:FIA:FIB, e.g., F1:A2:B2). PlasmidFinder includes targets for FIA and FIC(FII) from plasmid F, plus different FIB and FII types, generally differentiated by plasmid names (rather than FAB numbers).
The functions of the proteins encoded by the ∼40-kb tra operon involved in formation of the F-type pilus have been well studied (196, 197). Analysis of available F conjugation regions identified five major groups, apparently all derived from a common ancestral system (MPFF) (157). Groups correlate strongly with bacterial host species, suggesting different adaptations, with four groups relevant to the Enterobacteriaceae (195). Group A includes most plasmids typeable by RST and currently has the most members (but this may be due at least partly to sequencing bias). This is the only group to have an easily identifiable finOP system (fertility inhibition FinO protein and finP antisense RNA) that regulates tra expression and conjugation (198). Group C plasmids appear to be rarer and are similar to plasmids originally defined as FV, which have a distinctive regulatory system. Group D plasmids, with differences from group A in their operon structures and regulatory genes, are mainly associated with Enterobacter. Group B includes plasmids from Yersinia and also a few from other species carrying blaNDM (pKOX_NDM1 and pRJF866 [195]), all defined as FIIY by RST.
F plasmids were among the earliest to be associated with antibiotic resistance and appear to be the most abundant plasmid type found in Enterobacteriaceae (199). The classical FII plasmid R100 (also called NR1; isolated from Shigella flexneri in Japan in the 1950s) carries a class 1 In/Tn (In2) inside Tn21 (Fig. 3), itself inside an IS1-mediated composite Tn carrying catA1 (chloramphenicol resistance gene), with the whole structure called Tn2670 (81). Some contemporary F plasmids appear to carry a resistance region derived from this structure (200). F plasmids often carry a blaCTX-M gene (201), especially blaCTX-M-15 (or increasingly blaCTX-M-27) in E. coli ST131, with these plasmids likely contributing to the success of this ST (202). FIIK plasmids are associated with blaKPC in ST258 (203) and other sequence types, and IncFIIY plasmids may carry blaNDM (64). F plasmids carrying mcr-1 have also been reported (204).
HI plasmids.
HI plasmids encode serologically related pili similar to the F pilus, are larger than most of the other conjugative plasmids discussed here, and may encode heavy metal, phage, and/or colicin resistance in addition to antibiotic resistance (205). DNA hybridization, restriction analysis, and incompatibility testing resulted in division into HI1, HI2, and HI3 groups, but only one HI3 (heavy metal resistance) plasmid (206) seems to have been found, and the sequence is not available. HI1 (archetype R27; isolated from S. enterica serovar Typhimurium in the United Kingdom in 1961) (207) and HI2 (archetype R478; isolated from Serratia marcescens in the United States in 1969) (205) both have multiple replicons, with a common RepHI1A replicon responsible for incompatibility. RepHI1B is unique to HI1 plasmids, which also have a RepFIA-like replicon (flanked by two copies of IS1 [208]) that confers one-way incompatibility with F plasmids. RepHI2 is unique to HI2 plasmids.
HI1 and HI2 plasmids have similar backbone organizations, with higher identities between equivalent proteins that are essential (205). Conjugation genes are in two separate regions: Tra1 (or Trh1; carries oriT and genes encoding the relaxosome and some MPF components) and Tra2 (Trh2; encodes most MPF proteins). The MPF system is related to that of F plasmids, while the relaxosome and pilin genes are more closely related to those of P plasmids. Optimal pilus synthesis occurs at 22 to 30°C, and although the pili remain stable at 37°C, formation of mating aggregates is inhibited (209). This thermosensitive conjugation may contribute to spread in the environment (209).
HI1 plasmids are mostly found in Salmonella but can also be found in E. coli (210). An analysis of available sequences led to a six-locus pMLST scheme and identified regions of difference (A to E) (Fig. 5) suggesting two different lineages (211), later called type 1 and type 2 (208). Another analysis suggested a slightly different classification (212). For HI2 plasmids, a two-locus typing scheme (open reading frames [ORFs] smr0018 and smr0199) was proposed, and primers to detect the presence/absence of three additional genes were also used (213). Fourteen and 12 ST have been assigned so far (as of May 2018) for HI1 and HI2 plasmids, respectively.
Additional groups of HI-like plasmids have now also been identified. pNDM-MAR encodes RepHI1B-like and RepFIB-like proteins (214), while pNDM-CIT encodes two different Rep proteins corresponding to RepHI1A and RepHI1B but only ~92% identical in each case (215). A phylogenetic tree derived from a concatenated core (mostly rep and trh regions) for available HI plasmids gave four groups (215). These correspond to groups HI1, HI2, HI3 (different from the original HI3 group; includes pNDM-MAR), and HI4 (includes pNDM-CIT) defined in a recent paper, apparently from analysis of only traI and trhC, which also identified an HI5 group (216). The original PBRT scheme included primers differentiating HI1 and HI2, and primers to detect pNDM-MAR-like (214) and pNDM-CIT-like (215) plasmids have been added. PlasmidFinder (18-05-02 version) includes three targets for RepHI1A, two for RepHI1B, and a single target for each of RepHI2 and the FIA-like rep found in HI1 plasmids, as detailed in the original paper (170), plus an additional [IncFIB(Mar)] target for pNDM-MAR.
Various resistance genes have been identified on HI plasmids, including blaIMP and blaCTX-M on HI2 (201). At least one plasmid in each of the HI3 to HI5 groups carries blaNDM-1. More recently, mcr-1 was identified on different HI1 plasmids (217, 218), and mcr-1 (219) and mcr-3 (220) on HI2 plasmids, all in E. coli.
I-complex plasmids.
Plasmids classified as Inc types I1 (Iα), Iγ, B/O, K, and Z were grouped into the I complex due to the similar serologies and morphologies of their pili. Replication (copy number) of these I-complex plasmids is regulated by an antisense inc RNA (also called rnaI) that inhibits translation of repA mRNA (also called repZ), encoding the RepA initiator protein. Translation of RepA requires translation of the upstream and overlapping repB gene (also called repY), encoding a short peptide, and formation of a pseudoknot secondary structure (146, 221). Incompatibility results from the interaction between inc RNA and a stem-loop (SL1) formed from the repAB mRNA. Plasmids classed as types B and Z are actually incompatible, suggesting that stable hybrid inhibitory complexes are formed (221).
The original PBRT scheme includes primers in and upstream of inc that detect both I1 and Iγ replicons (I1 FW/RV primers). K/B FW PBRT primers were stated to detect both K and B/O plasmids when paired with a K RV primer and B/O plasmids only when paired with a B/O RV primer (168). B/O primers were found to detect Z but to miss some I-complex plasmids, and different inc sequences were identified among plasmids classed in the Z group (222). Division of K plasmids into compatible K1 and K2 lineages was also proposed recently (223, 224), with new primers in inc and repB to detect and distinguish them (223). PlasmidFinder (18-05-02 version) includes one I1 target sequence and four sequences to cover B/O, K, and Z plasmids, all reported as ”B/O/K/Z.”
A pMLST scheme for I1 plasmids (225) uses five targets (Fig. 5) with 14 to 47 alleles, and nearly 300 pMLST profiles have been identified (as of May 2018; https://pubmlst.org/bigsdb?db=pubmlst_plasmid_seqdef), indicating extensive variation. Two main types of repA gene, namely, repABKI, found in plasmids classed as B/O, I1 (99% identical), and K (and Iγ; 92% identical), and repAZ (∼50% identical to repABKI), found in plasmids classed as Z, were also distinguished, and primers were designed for each type (222). Sequence comparisons indicate that repA of plasmids classed as K2 is more like repAZ (95% identical) and should be detected by the repAZ primers (there is one mismatch in the 3′ end of the reverse primer). Classification within the I complex may need to be revisited given that the effects of minor changes in inc and/or SL1 on incompatibility are not really known and that different inc types are apparently associated with the same repA type, and vice versa.
I-complex plasmids generate both a thick pilus (tra genes) for DNA transfer and a thin pilus (pil genes) that appears to stabilize the mating apparatus in liquid media but not on solid surfaces (226). A shufflon site-specific recombination system, consisting of rci (encoding a recombinase) and an adjacent region where 19-bp sfx repeat sequences separate segments containing partial reading frames, is present in I1, Iγ, and some other I-complex plasmids. This region overlaps pilV, encoding the tip adhesin of the thin pilus, and Rci-mediated recombination between sfx repeats causes rearrangements/deletions that create PilV variants with different C termini. This is reported to result in different conjugation efficiencies and biofilm and/or adherence properties (227).
The shufflons of archetypal plasmids R64 (I1) (227) and R621a (Iγ) (228) both have four segments separated by seven shf repeats. Segments denoted A, B, and C each contain two oppositely oriented partial reading frames that can form the 3′ end of pilV, while segment D has only one (Fig. 5). The K1 plasmid pCT has three segments flanked by six shf repeats, while up to eight shf repeats were reported for sequenced K2 plasmids (224), separating four segments. Comparison of the segments in K plasmids reveals one common to K1 and K2 plasmids, part of which is related to the I1 A segment, one related segment (∼85% identical) also related (80%/77% identical) to the I1 C segment, and a third common segment (∼74% identical) that matches parts of the I1 B and D segments. Available B-type plasmids appear to have a single shf repeat adjacent to part of the I1 C-like segment in K plasmids and no rci gene, giving only one PilV variant. Two related, sequenced plasmids typed as Z have one shufflon segment related to parts of the I1 B and C segments, flanked by three shf repeats (229).
Most examples of I1 plasmids are from E. coli or Salmonella, and many carry resistance genes, commonly blaCMY-2 and variants, blaCTX-M-15, or blaCTX-M-1 (mainly in animals) (230). Insertions tend to be in the same region of the plasmid, between a patch of genes of unknown function and stability genes (230) (Fig. 5). K1 plasmids from various locations carry blaCTX-M-14 (231), and available K2 plasmids carry blaCMY-2 or its variants (223, 224) or mcr-1 (232). B/O replicons have been detected by PCR in isolates carrying resistance genes, but at least some may correspond to Z-type plasmids (222), and few fully sequenced B plasmids seem to be available. Reported Z plasmids carry “older” resistance genes (222, 229).
I2 plasmids.
I2 (originally Iδ) plasmids have many features in common with the I-complex plasmids, including encoding thick and thin pili and possessing a shufflon, but the organization and sequences are different (Fig. 5). The archetypal I2 plasmid R721 (INSDC accession number AP002527) has three shufflon segments: A and C, equivalent to those of I1, and BD, the two ends of which are homologous to B and D of R64 (233) (Fig. 5). Some recently identified I2 plasmids have an additional segment (e.g., INSDC accession number KY795978, with the suggested designation “E”), and I2 shufflons seem to be actively rearranging (234).
I2 primers were not included in the original PBRT scheme, so this plasmid type was somewhat neglected until recent examples carrying blaCTX-M genes, including blaCTX-M-1/9/1 hybrids, were identified (235, 236). blaKPC in K. pneumoniae ST258 clade b (also called clade 2 or II) may be carried on an I2 plasmid (203). More recently, I2 plasmids have received attention as the vehicle of the first mcr-1 gene identified (40), with some carrying both a blaCTX-M gene and mcr-1. Different lineages of I2 plasmids were proposed based on a limited number of sequences (235), and mcr-1 genes have been found on at least two distinct I2 plasmid types (237). Preliminary analysis of over 100 I2 plasmid sequences now available suggests that these two types dominate among plasmids carrying an mcr-1 gene, that different resistance genes are carried by different I2 lineages, and that there may be extensive recombination in I2 backbones (S. R. Partridge, N. L. Ben Zakour, M. Kamruzzaman, and J. R. Iredell, unpublished data).
L/M plasmids.
Known L/M plasmids associated with resistance genes have a conserved backbone organization. The replication region consists of repA (initiation protein gene), repB, and repC genes regulated by inc antisense RNA, similar to that of I-complex plasmids (146, 238). The conjugation genes of L/M plasmids are split between a larger tra region and a smaller trb region and are also related to those of I plasmids (239).
After initially being defined as two separate groups (240), IncL and IncM were subsequently merged (241), but division into L, M1, and M2 groups was suggested recently, based on differences in inc, relaxase (traX), and entry exclusion genes (traY, excA) (242). Similar groups were also identified using a core genome of 20 genes in 20 available sequences (243). L and M plasmids can be distinguished using additional PBRT primers (242), while PlasmidFinder (18-05-02 version) has three targets for these plasmids (170), reported as IncL/M(pOXA-48), corresponding to L; IncL/M(pMU407), corresponding to M1; and IncL/M, corresponding to M2.
Although these plasmids are reported to have a broad host range, BLAST searches with L or M replicons reveal that almost all fully assembled plasmids are from Enterobacteriaceae. Most sequenced L plasmids are closely related and have variants of Tn1999, carrying a blaOXA-48-like gene, inserted into the tir (transfer inhibition) gene, resulting in a higher conjugation frequency (244). M2 plasmids often carry ISEcp1-blaCTX-M-3 (239) and/or clinically important genes, including armA, blaNDM, or blaIMP-4, within variants of the same Tn2-derived resistance region (245), while M1 plasmids carrying blaKPC, blaSHV (ESBL), or blaFOX (ampC) genes have been reported (242).
N plasmids.
N plasmids are relatively small conjugative plasmids. The N conjugation region is split into two parts, one encoding entry exclusion functions and pilus components and the other carrying oriT and some tra genes. These are separated by fipA, encoding a fertility inhibition protein that inhibits conjugation of coresident IncP1 plasmids by interacting with IncP1 TraG (246), and nuc, encoding a nuclease. Part of the N backbone is occupied by the conserved upstream repeat (CUP)-controlled regulon. The archetypal IncN plasmid R46 has six CUPs, which contain a strong promoter, separating several ccg (CUP-controlled genes), ard (antirestriction/regulatory genes), and other genes. Some N plasmids have fewer CUP repeats and only subsets of these genes, which may be explained by recombination between repeats (247).
Backbones of reported N plasmids appear to be well conserved. A pMLST scheme includes only three targets (248), and 20 ST are listed on the pMLST website (May 2018). Insertions are commonly a class 1 In/Tn (res site hunter) upstream of resP (resolvase) and/or other insertions in/close to fipA (such insertions may have a beneficial effect [249]). Genes including blaKPC, blaIMP, and blaCTX-M have been identified on N plasmids (201).
Plasmids related to the original N type (now called N1) have also been identified. Those designated N2 (249) have closely related backbones that share a similar organization, but limited nucleotide sequence identity, with N1 plasmids, with a different rep region (249, 250). Known examples carry blaNDM (249), blaCTX-M-62 (250), or various blaIMP genes (251) in insertions near fipA. The prototype N3 plasmid (note that a plasmid called N3 [252] is an N1 plasmid) has a backbone with an organization similar to that of the N2 backbone and encodes a RepA initiator ∼80% identical to N2 RepA (253). A recent paper identified two other N3 plasmids and placed a fourth in a separate group, called IncN3β, even though it encodes a distinct RepA initiator (254). PlasmidFinder (18-05-02 version) includes one target for N1 and one called N2 (170), but an additional target named N3 appears to detect some plasmids classed as N2 plasmids.
P/P-1 plasmids.
Plasmids called IncP in Enterobacteriaceae and IncP-1 in Pseudomonas were originally discovered in clinical isolates in the late 1960s (255). Representatives of the Pα (e.g., RP4/RK2 [256]) and Pβ (e.g., R751 [257]) subgroups have been well studied. The replication protein is encoded by trfA, and replication is controlled by a handcuffing mechanism similar to that for RepFIA (148). P plasmids have two conjugation regions: tra and trb. P plasmids are among the most stably maintained plasmids due to tight regulation of replication, conjugation, and maintenance by a central control region. IncC and KorB are partitioning proteins, but KorB also regulates gene expression, potentiated by IncC and in conjunction with KorA (139, 255). The broad host range of P plasmids may be due to a combination of accommodating differences in host factors needed for replication, the MPF apparatus being able to successfully interact with different cell types, and a lack of restriction sites (258). P plasmids can mobilize IncQ plasmids into Gram-positive bacteria.
The original PBRT primers detect Pα plasmids only, but P plasmids have now been divided into at least eight named clades, i.e., α, β1, β2, γ, δ, ε, ξ (259), and η (260), plus an unnamed clade from Neisseria (261) and a recently proposed new clade (262). About three quarters of backbone genes are shared by these clades, and hybrid plasmids have been found but generally have components from within the same clade (259, 260). Insertions tend to occur between ori and trfA and between the tra and trb operons, and many P plasmids carry antibiotic resistance genes (263). These do not generally confer resistance to the most clinically important antibiotics, although plasmids from the newest clade carry mcr-1 genes (262, 264). PlasmidFinder (18-05-02 version) includes targets for Pα, Pβ1, and the P plasmid carrying mcr-1, all reported as “IncP1.”
R plasmids.
The designation IncR was first given to pK245, carrying qnrS1, and primers to detect the repB gene of pK245 were published (265). PlasmidFinder (18-05-02 version) includes the PBRT amplicon from pK245 repB as the only IncR target (170). pK245 has two additional rep genes, named repE and repA (266). Plasmids with the R repB gene alone are apparently nonconjugative, lacking tra genes, and no relaxase gene has been identified (267). This may explain why complete plasmid sequences with R repB often have an additional replicon including FIIK, A/C, or untyped rep genes (267, 268). The original report of pK245 (266) also noted that repB is closely related to the β replicon of pGSH500 (isolated from K. pneumoniae in/prior to 1991), which also carries an FII-like (α) replicon (269). Given these considerations, it is not clear that plasmids that carry this type of replicon should really be considered a separate group. Plasmids with R repB have mostly been reported from K. pneumoniae, but also from Enterobacter cloacae and E. coli, carrying genes including blaNDM (sometimes with a 16S rRNA methylase gene [e.g., see reference 270]), blaKPC, blaVIM, and blaCTX-M-15 (201).
T plasmids.
The prototype IncT plasmid is Rts1, isolated from Proteus vulgaris. Rts1 replication (repA) and partitioning genes are most related to the Y plasmid P1 (see below), but replication is inhibited at 42°C. Conjugation genes are found in two clusters and encode proteins most related to those expressed from F and HI1 plasmids (271). Conjugation was reported to be efficient at 25°C but not at 37°C, but it was found later that this applies to liquid but not solid mating and may not be the same for all T plasmids (272). The higAB TA system causes temperature-sensitive postsegregational killing at 42°C (271). Rts1 may also be atypical, in that it contains two copies of an ∼50-kb region that share the same gene organization but limited identity (271).
Primers in repA of Rts1 were part of the original PBRT primer set, and PlasmidFinder (18-05-02 version) includes the amplicon sequence from Rts1 as the IncT target (170). Few T plasmids have been reported recently, but they were associated with blaCTX-M-2 in Proteus mirabilis strains from Japan by PBRT (273), and the complete sequence of a plasmid from Providencia rettgeri with a T repA gene and carrying blaNDM-1 is available (274). A T-type plasmid carrying blaOXA-181 from Citrobacter freundii (but with a partially truncated tra region) (253) has also been reported, but searches of GenBank, including the WGS database (May 2018), did not identify the T-like repA gene in any of the species of particular interest here.
U and G/P-6 plasmids.
The IncG (E. coli)/IncP-6 (P. aeruginosa) and IncU groups were assigned in the early 1980s. It was suggested that these groups could be merged, since IncP-6 iterons cloned in high copy number gave strong incompatibility with IncU and the replicons of the two groups are related (275), but MOB typing places them in different clades (Table 6) (174). Known U plasmids are mostly associated with environmental isolates and Aeromonas spp.
Rms149, the archetypal G plasmid, consists of a small backbone which appears to be made up of modules related to those found in different plasmid types, with multiple insertions that occupy about 80% of the sequence (276). It has characteristics of both smaller, mobilizable plasmids and larger, low-copy-number plasmids. IncU plasmids were not included in the original PBRT scheme, but primers for the repA gene were added later (265). The single PlasmidFinder (18-05-02 version) IncU target corresponds to the PBRT amplicon, and an IncP-6 target is also included (170). blaKPC has been reported to be present on P-6 plasmids smaller than Rms149 in P. aeruginosa and on a U plasmid that appears to lack mobilization genes (277, 278).
W plasmids.
The first IncW plasmid, pSa from Shigella, was described by T. Watanabe (hence IncW) in the late 1960s, and this group was reviewed relatively recently (279, 280). W plasmids are the smallest conjugative plasmids found in the Enterobacteriaceae, and most of the few available sequenced examples show a conserved backbone consisting of typical plasmid modules with different insertions (279, 280). W plasmids include a master regulation system similar to that of P plasmids (281). A new IncWβ group was recently suggested for a related plasmid, but it has a different replication module (254).
The original PBRT scheme includes primers to detect W plasmids, and the single IncW target in PlasmidFinder (18-05-02 version) corresponds to the amplicon produced (170). Three W plasmids identified some time ago (280), plus one carrying blaIMP-1 reported more recently (282), have a truncated class 1 In/Tn in the same position, suggesting a single insertion (280). The integron in pSa is one of the earliest examples to carry ISCR1 (57). There are reports of detection (by PCR) of W plasmids carrying related cassette arrays containing blaVIM-1/4 (283, 284) or blaKPC (285) in Tn4401, but sequences are not available.
X plasmids.
X plasmids were originally divided into X1 (e.g., R485) and X2 groups, typified by R6K, which has been well studied and whose sequence was reported recently (286). The π replication protein is encoded by pir (protein for initiation of replication), and X plasmids have three ori regions: γ (part of the minimal replicon), α, and β (287). Replication is regulated by handcuffing mediated by π dimers coupling two oriγ regions (148, 287). The conjugation region consists of genes for pilus synthesis and assembly (originally named pilX1 to -11 but renamed tivB1 to -11) plus taxC/rlxX (relaxase gene), taxB/cplX (coupling protein gene), and taxA/dtrX1 (auxiliary relaxosome protein gene) (286). Various other genes are conserved across the backbones of X plasmids, and the TA systems that they carry were surveyed recently (288).
X2 replicon primers were included in the original PBRT scheme (168), and then primers for taxC/rlxX1 were added for X1, X2, and the new X3 and X4 types (289). X5 (290) and X6 (291) types were proposed based on variation in taxC/rlxX1, but PlasmidFinder initially defined different X5 and X6 types based on pir (288). These have now been redesignated X7 and X8, respectively, and PlasmidFinder (18-05-02 version) includes multiple targets for some X plasmid types (four for X1 and two each for X3 to X5), but all are reported as IncX1, IncX4, etc. [except for IncX3(pEC14)] (170). An X3/X4 hybrid plasmid, apparently generated by formation and resolution of an X3-X4 cointegrate (292), and an X1/X2 hybrid (293) have also been identified.
X1 plasmids may carry oqxAB, encoding an efflux pump (289). X2 plasmids have been reported only rarely, but they may carry qnr genes (294). blaOXA-181 (295) and blaSHV-12, alone or with a blaNDM-4-like variant (296) or with blaKPC (297), have been seen on X3 plasmids. Different blaCTX-M genes are carried on related X4 plasmids (298, 299), almost identical X4 plasmids carry mcr-1 or variants (300), a different X4 plasmid carries mcr-2 (301), and the X3/X4 hybrid carries blaNDM-5 and mcr-1 (292). Known X5 and X6 plasmids carry a blaKPC gene (290, 291).
Y plasmids.
The Y group of plasmids corresponds to the prophage form of generalized transducing phages related to the P1 phage, which infects and lysogenizes E. coli and some other Enterobacteriaceae. P1 exists stably as a low-copy-number plasmid that replicates independently rather than integrating into the host chromosome and can transfer between bacterial cells as virus particles whose production it encodes. P1 replication is regulated by a handcuffing mechanism (148), and the partitioning, maintenance, and other systems have been well studied (302, 303).
The PBRT amplicon and the PlasmidFinder target (170) correspond to the same internal fragment of P1 repA, and detection of Y plasmids by PBRT has been reported for isolates carrying a few different resistance genes. A few plasmids with this replicon have been sequenced fully, including one that also includes F components and carries blaCTX-M-15 (304), one carrying mcr-1 (305), and a multireplicon plasmid carrying mcr-1 and other resistance genes (306).
Q plasmids.
IncQ plasmids are small and mobilizable, and their properties are covered in several reviews (307–309). In addition to repC (initiator protein gene), Q plasmids carry their own repA (helicase gene) and repB (primase gene; fused to the mobA relaxase gene) genes, giving them a broad host range, as they are not dependent on the host bacterium for these functions (308). Q plasmids have been split into groups Q1 to Q4, based on differences in Rep proteins and association with different lineages of mobilization proteins (307). Q1 plasmids carry mainly genes conferring resistance to “older” antibiotics, but one encodes GES-5 (a carbapenemase). Q3 plasmids carrying blaGES-1 (ESBL gene) and qnrS2 (low-level quinolone resistance gene) have been identified (307). Q plasmids often lack MGE normally associated with the resistance genes that they carry. These may have been lost after depositing their load to minimize plasmid size, which may be limited by the strand-displacement replication mechanism used (307).
Q plasmids were not included in the original PBRT scheme, but PlasmidFinder (18-05-02 version) includes IncQ1 and IncQ2 targets. A fragment of the archetypal Q1 plasmid, RSF1010, including repC and part of repA, is found in resistance regions on many large plasmids (21) as part of a rearranged, IS26-flanked region known as Tn6029 (310). The original PlasmidFinder IncQ1 target corresponded to only part of the Q replicon and lay wholly within Tn6029. It was recently replaced (in the PlasmidFinder update of 28 November 2017) by an expanded target that covers the start of repA and the adjacent region, with a match over the whole length (796 bp) suggesting a separate Q plasmid and shorter matches indicating truncated copies (e.g., 529 bp in Tn6029).
ColE1 and related plasmids.
ColE1 is a small plasmid that encodes colicin E1 (cea) and colicin immunity (imm). Replication requires a plasmid-encoded RNA primer, RNAII, rather than a replication protein. Replication and copy number are controlled by the rate of binding of antisense RNAI to RNAII, which prevents correct folding and thus primer formation. This interaction is also modulated by Rom (RNA one inhibition modulator; also called Rop, for repressor of primer), a small protein that stabilizes the RNAI-RNAII complex (146). ColE1 can be mobilized by different conjugative plasmids (including I1, F, P, and W plasmids) and requires oriT, mbeA (encoding the relaxase), and mbeBCD, while mbeE is not essential (311). ColE1 also carries a cer site for conversion of plasmid dimers to monomers by site-specific recombination catalyzed by host-encoded XerCD.
PlasmidFinder includes targets to detect ColE1-type plasmids (but the closest to ColE1 itself is <90% identical), and most of the targets with names starting with “Col” correspond to small plasmids that have a repA gene (170). qnrB19 has been detected in ColE1-like plasmids of two types, which appear to differ due to Xer-mediated events, and ISEcp1 may have been lost following insertion of qnrB19 (312). Tn1331 (Fig. 3) was originally identified in pJHCMW1 from K. pneumoniae, which replicates by a mechanism similar to that for ColE1 but lacks a rom equivalent and includes active (mrw) and defective (dxs) Xer-specific recombination sites (72). Plasmids related to ColE1 and carrying a colicin gene appear to be common in K. pneumoniae isolates (313), including ST258 isolates (203), and may carry a Tn1331 derivative and, in some cases, Tn4401 and blaKPC (314).
Other small plasmids with a ColE1-type replication system were recently recognized as having I1-like oriT and nikA (encoding relaxase accessory factor [RAF]) (166), but they lack an equivalent of the I1 nikB relaxase gene (165). One example (NTP16) was previously shown to be mobilizable by I1 plasmid R64 (315), similar to relaxase-in-trans mobilization demonstrated for plasmids with oriT only (165). NTP16-like plasmids have been identified in different locations and from a few different species (including E. coli ST131) from as early as the 1970s (165). XerCD-mediated recombination at the cer-like nmr recombination site may explain their different accessory regions (165). Several of these plasmids carry Tn2 or a modified version with a blaTEM gene, in one case an ESBL variant (165).
Resistance Plasmids in P. aeruginosa
Many resistance genes in P. aeruginosa are found on various resistance islands (see below) rather than on plasmids, and little information is available about the normal plasmid content of P. aeruginosa strains. Unlike the P/P-1 and G/P-6 types mentioned above, many plasmids from Pseudomonas spp. were not readily transferable to E. coli (narrow host range) and were classified using a separate incompatibility typing system (IncP-1 to IncP-13) (see the references in reference 316). These Inc types are found in various Pseudomonas species and are common in the environment, but many do not carry resistance genes. Relatively few resistance plasmids from P. aeruginosa have been sequenced, with a 2015 in silico study retrieving only 10 P. aeruginosa plasmids from GenBank, six of which were classified as IncP-1, IncP-2, or IncP-6 (138). Recently, more plasmids from P. aeruginosa, particularly from carbapenem-resistant isolates, have become available. These mostly carry cassette-borne carbapenemase genes found in class 1 In/Tn inserted into Tn21 subfamily transposons that may differ from those commonly found in plasmids in Enterobacteriaceae.
IncP-2.
Historically, IncP-2 plasmids were the most common transferable plasmids in P. aeruginosa (317). The examples identified were very large and typically encoded tellurite resistance in addition to antibiotic resistance (317). Several large (∼300 to 500 kb), related IncP-2 plasmids from P. aeruginosa, carrying cassette-borne carbapenemase genes (including blaIMP-9, its variant blaIMP-45, or blaVIM-2) in class 1 In/Tn with different structures and with identifiable replication, maintenance, and conjugation functions, have been sequenced (316, 318, 319).
P. aeruginosa plasmids carrying carbapenemase genes.
A large plasmid carrying the rare blaSIM-2 gene cassette in a class 1 In/Tn (320) and a smaller plasmid carrying blaKPC-2, different from the P-6/U plasmids with blaKPC mentioned above (321), were both sequenced recently. A group of related plasmids that carry a cassette-borne blaVIM-1 or blaVIM-2 carbapenemase gene in different class 1 In/Tn structures were sequenced (318, 322–324). They are also related to TNCP23, a region carrying a class 1 integron bounded by two copies of IS6100 (IS6 family) that seems to correspond to a plasmid inserted into the larger plasmid pKLC102, which is itself known to integrate as a genomic island (see below) (325). Other, unrelated plasmids carry blaVIM-1 (duplicate copies giving high-level carbapenem resistance) (323), blaVIM-2 (326), or blaVIM-7 (327).
Resistance Plasmids in A. baumannii
Plasmids found in A. baumannii are also less well studied than those in the Enterobacteriaceae, and as observed for P. aeruginosa, many resistance genes have been found on resistance islands (see above and Fig. 4). A PBRT scheme (AB-PBRT) with 19 types (GR1 to -19) has been proposed, based on 18 plasmid sequences available at the time (328), but there does not appear to be an associated Web resource, and these plasmids are not included in PlasmidFinder. An additional group, GR20, was recently proposed (329). Prior to the development of this PBRT scheme, no extensive surveys of the normal plasmid content of A. baumannii had been published (330), although plasmid typing had been used as part of epidemiological studies of resistant A. baumannii strains (331). A study of 96 isolates from various sources, using the AB-PBRT scheme, identified one to four plasmid types per isolate, mainly associated with blaOXA carbapenemase genes, most of which were nontransferable (330). An in silico study using the PBRT scheme identified at least one member of the GR1 to -19 group in 77% of the 70 A. baumannii plasmids obtained from GenBank, mostly GR10 plus GR2 and GR6 plasmids (138), but this is likely to be a biased set.
Most small plasmids in A. baumannii encode replicase proteins belonging to the Rep_3 superfamily (328, 329). Partitioning and TA systems have been identified, as well as a relaxase gene in many. pRAY-like plasmids (∼6 kb) have mobA (MOBHEN) and mobC genes, but a rep gene has not been identified. They carry the aadB gene cassette (encoding gentamicin and tobramycin resistance) outside an integron, in a secondary site, and are widely distributed (329, 332). A few larger, conjugative plasmids have been identified in A. baumannii. Closely related RepAci6 plasmids pAb-G7-2 (333) and pACICU2 (334) carry aphA6 (encoding kanamycin and amikacin resistance) in TnaphA6 (Table 1). Another RepAci6 plasmid carries blaOXA-23 in Tn2006, inserted into AbaR4 (Table 1 and Fig. 4) (335). Plasmids related to pNDM-BJ01, which are not classified by AB-PBRT (336), carry blaNDM-1 in Tn125 (Table 1), with aphA6 and ISAba4 regions upstream, and have been identified in A. baumannii and other Acinetobacter spp. but also in Enterobacter aerogenes (337).
Resistance Plasmids in Staphylococci
Clinical strains of staphylococci frequently harbor one or more plasmids that confer resistance to various classes of antibiotics, heavy metal ions, and/or antiseptics and disinfectants (15, 338, 339). Historically, the following three broad classes of staphylococcal resistance plasmids have been recognized: (i) small plasmids (<1 to 10 kb) that replicate by an asymmetric rolling-circle (RC) mechanism; (ii) multiresistance plasmids (>15 kb); and (iii) larger, conjugative multiresistance plasmids (15, 17, 18, 339).
RC-replicating plasmids.
Staphylococcal plasmids smaller than 10 kb usually employ an RC replication mechanism. They most often encode a single resistance determinant and are multicopy (10 to 60 copies per cell) (340). Four families of staphylococcal RC-replicating plasmids have been described and are exemplified by the tetracycline resistance plasmid pT181 [tet(K)] (341), the chloramphenicol resistance plasmid pC194 (cat) (342), the erythromycin resistance plasmid pE194 [erm(C)] (343), and the cryptic plasmid pSN2 (17, 344). Each of these plasmid families utilizes an evolutionarily distinct Rep protein, and these proteins are differentiated by the presence of conserved domains (Rep_trans, Rep_1, Rep_2, and RepL, respectively).
While RC plasmids are grouped according to the replication systems they carry, plasmids across the groups often share highly similar DNA segments that can carry resistance genes and/or mobilization functions. Thus, RC plasmids are considered mosaic structures composed of interchangeable functional modules (17, 345). In addition to the resistances described above, RC plasmids that confer resistance to streptomycin (str) (346), lincomycin [linA; now called lnu(A)] (347), fosfomycin (fosB) (348), quaternary ammonium compounds (qacC and smr) (349), aminoglycosides (aadD), or bleomycin (ble) (350) are known. Some RC plasmids, such as pC221 of the pT181 family, contain a mobCAB operon and oriT (nicked by the MobA relaxase) that can facilitate their mobilization by a coresident conjugative plasmid (351, 352). Likewise, other RC plasmids, including pT181, possess a pre gene and an RSA site, originally identified as a site-specific recombination function (353), which are now known to represent a distinct mobilization system homologous to the well-studied mobM relaxase/oriT system of the streptococcal plasmid pMV158 (354–356).
Multiresistance plasmids.
Staphylococcal multiresistance plasmids utilize theta replication and are maintained at approximately 5 copies per cell (357). Two groups of multiresistance plasmids have previously been described based on structural and function characteristics: the β-lactamase/heavy metal resistance plasmids (e.g., pI258) and a family of plasmids related to the prototype pSK1 (Fig. 6) (15, 358, 359). However, it is now clear that these groupings do not encompass the diversity/heterogeneity of multiresistance plasmids revealed through large-scale sequencing (338). In multiresistance plasmids, resistance genes are often interspersed with IS (most commonly IS257) and/or located within transposons or transposon-like elements, such as Tn551, Tn552 (Table 5), Tn4001, or Tn4003 (Table 2), conferring resistance to macrolide-lincosamide-streptogramin B (MLS) antibiotics (360), penicillin (102), aminoglycosides (361), or trimethoprim (362), respectively (15).
FIG 6.
S. aureus multiresistance plasmids. Representative multiresistance plasmids (pI258, pSK1, and pUSA300-HOU-MR) and pSK41-, pWBG749-, and pWBG4-family conjugative multiresistance plasmids (pLW1043, pBRZ01, and pWBG4, respectively) are shown (15, 104, 359, 384, 390, 398, 505, 516). IS, transposons, cointegrated plasmids, and resistance genes are shown, with resistances conferred by the latter listed in Table 2 or as follows: arsBC, arsenic resistance; bcrAB, bacitracin resistance; cadA and cadD, cadmium resistance; merAB, mercury resistance; msrA and mphC, macrolide resistance; and qacA, antiseptic/disinfectant resistance. The following plasmid maintenance genes/systems are also shown: par, novel partitioning system; parAB, type I partitioning system; parMR, type II partitioning system; rep, initiation of replication; res and sin, multimer resolution; TA, Fts-like toxin-antitoxin system. The conjugation-associated genes of pLW1043, pBRZ01, and pWBG4 are denoted tra, smp, and det, respectively.
A survey of 280 geographically and epidemiologically diverse staphylococci (n = 251 S. aureus strains) revealed that three plasmid lineages, represented by pIB485, pMW2, and pUSA300-HOU-MR (Fig. 6), encompassed more than half of all the multiresistance plasmids detected (338). pIB485- and pMW2-like plasmids were widely distributed geographically, whereas pUSA300-HOU-MR-like plasmids were found only in isolates from the United States. All three lineages usually carry Tn552-derived β-lactamase genes and genes for cadmium resistance; pUSA300-HOU-MR-like plasmids often also carry genes for resistance to macrolides, aminoglycosides, and bacitracin, while enterotoxin genes are a common feature of pIB485-like plasmids.
Most staphylococcal multiresistance plasmids utilize an evolutionarily common antisense RNA-controlled replication initiation system (363–365) encoding a replication initiator protein that contains a conserved RepA_N domain (338, 366); rep genes encoding this domain are prevalent on plasmids in many low-G+C Gram-positive genera (367). Other rep genes are sometimes evident in multiresistance plasmid sequences and appear to have been incorporated via cointegration of small RC plasmids, but these are usually inactivated by mutations/truncations of the coding sequence or corresponding ori. Exceptions to this are plasmids that possess both a repA_N-type gene and a distinct rep gene encoding an initiator with a Rep_3 domain. A rep_3-type gene in staphylococci was first detected in the small dfrA-containing Staphylococcus epidermidis trimethoprim resistance plasmid pSK639 (15, 368) but is increasingly being found on S. aureus multiresistance plasmids. In at least some cases, such as in pMW2, rep_3 appears to be responsible for replication, since only a remnant of a repA_N gene is evident (15, 338).
Multiresistance plasmids often carry a multimer resolution system incorporating a gene (usually annotated sin or bin3) that encodes a serine recombinase (103, 105, 338). The majority of plasmids bear a gene adjacent to and transcribed divergently from the repA_N gene, which is related to the pSK1 par locus (338). This is thought to represent a partitioning system, since it increases plasmid segregational stability (369), but it is unusual in that it encodes one rather than two Par proteins. However, more conventional two-gene type I and type II partitioning systems (153) are carried by a minority of multiresistance plasmids. Postsegregational killing systems, in the form of Fst-like type I toxin-antitoxin systems (370), have been found on some multiresistance plasmids but are often not detected/annotated due to their small size (359, 371).
Conjugative multiresistance plasmids.
Conjugative multiresistance plasmids are the largest plasmids found in staphylococci (>30 kb) and are defined by their ability to transfer from donor to recipient cells at low frequencies (372, 373). They can also promote the conjugative transmission of some smaller plasmids by mobilization or, if cointegration occurs, via conduction (351, 352, 372). Conjugative plasmids have also been found integrated into the chromosome (374).
Until recently, only one family of conjugative multiresistance plasmids had been characterized for staphylococci, as exemplified by plasmids such as pSK41 (375), pGO1, and pLW1043 (376, 377). Plasmids of this type were initially associated with the emergence of gentamicin resistance and were first isolated in North America in the mid-1970s (378–380), but they have also been identified in Europe and Japan (381–383) and, more recently, in community-acquired MRSA strains in the United States (384, 385).
Resistance to gentamicin and other aminoglycosides is mediated by derivatives of Tn4001 (Table 2) (aacA-aphD) that are truncated by copies of IS257 (386). This IS is usually present in multiple copies (up to nine) in pSK41-like plasmids, flanking diverse resistance genes in different members of this plasmid family (akin to IS26-associated gene arrays in Gram-negative species). These confer resistance to antiseptics and disinfectants (qacC) (349), mupirocin (mupA/ileS2) (29, 387), MLS antibiotics [erm(C)] (384), trimethoprim (dfrA) (381), tetracycline [tet(K)] (338), and linezolid (cfr) (388). In several cases, the resistance segments correspond to small plasmids, such as the RC plasmid pUB110 (encoding aminoglycoside [aadD] and bleomycin [ble] resistance) (389), that have been incorporated through IS257-mediated cointegrative capture (104). Some pSK41-like plasmids, such as pLW1043 (Fig. 6), also carry unit transposons, including Tn552-like β-lactamase transposons (Fig. 4) and, notably, the vanA glycopeptide resistance transposon Tn1546 (Table 5 and Fig. 3) (390), which is thought to have transposed from a transiently coresident enterococcal Inc18 plasmid (see below) (390, 391). Indeed, there is some evidence that intergeneric transfer of Inc18 vanA plasmids from enterococcal donor cells is enhanced by the presence of a pSK41-like plasmid in S. aureus recipient cells, but the mechanistic basis for this has not been elucidated (392). Although Inc18 vanA plasmids have occasionally been detected in S. aureus isolates, their rarity suggests a limitation to the establishment of these plasmids, which may be due to low replication efficiency, restriction-modification barriers, and/or high metabolic costs associated with vanA carriage in S. aureus (391).
Like most staphylococcal multiresistance plasmids (described above), pSK41-like plasmids utilize an antisense RNA-controlled repA_N replication initiation system (365, 393). They ubiquitously carry a multimer resolution (res) system (394) and a type II partitioning (par) locus (395), but an Fst-like toxin-antitoxin system is evident only on some members of the family (371). The transfer (tra) genes of these plasmids show similarity and synteny with those of the streptococcal/enterococcal Inc18 plasmid pIP501 and the lactococcal plasmid pMRC01, and several of their deduced products show distant homology to T4SS components encoded by conjugation systems of plasmids from Gram-negative bacteria (104, 396, 397).
An additional two types of staphylococcal conjugative plasmids, distinct from the pSK41 family, have been recognized only in the last few years and are represented by the prototype plasmids pWBG749 (163) and pWBG4 (398) (Fig. 6). pWBG749 was identified in an S. aureus isolate from Australia and harbors no antimicrobial resistance determinants, but related plasmids from around the world have been found to encode resistance to penicillin (blaZ), aminoglycosides (aacA-aphD on a Tn4001-like element), and vancomycin (vanA on a truncated Tn1546-like element) (163, 399). pWBG4 carries a Tn554-like element (Table 5) encoding resistance to MLS antibiotics [erm(A)] and spectinomycin (spc), and related plasmids have been found to confer resistance to aminoglycosides, trimethoprim (dfrD), or linezolid (cfr and fexA) (398, 400). pWBG749-like plasmids encode a RepA_N-type initiation protein, whereas pWBG4 encodes a protein with a PriCT_1 domain (like enterococcal Inc18 plasmids [see below]).
Bioinformatic analysis has indicated that only about 20% of S. aureus plasmids possess some form of mob relaxase gene to make them potentially mobilizable by a coresident conjugative plasmid (398). However, pWBG749 family plasmids (e.g., pBRZ01) (Fig. 6) were recently shown to mobilize both small RC and larger multiresistance plasmids that lack mob genes via a previously unidentified relaxase-in-trans mobilization mechanism (162, 163, 398). Short oriT “mimic” sequences on these plasmids, which closely resemble the oriT sequences of pWBG749 family plasmids, are sufficient to serve as substrates for the conjugative relaxase and conjugation machinery (162, 163). Equivalent mimics corresponding to the oriT sequence of pSK41-like conjugative plasmids have likewise been found on numerous plasmids that lack mob genes, although mobilization of such plasmids has not been demonstrated (164). About half (56%) of nonconjugative S. aureus plasmids were found to possess at least one pWBG749- or pSK41-like oriT mimic (many have both), including 89% of multiresistance plasmids, which rarely possess a mob gene (398). Moreover, conjugative mobilization of some RC plasmids, mediated by ICE, based on the relaxase activity of their Rep initiation protein nicking at their replicative ori, has also been described (401, 402). Together these observations have led to the suggestion that nearly all S. aureus plasmids might be mobilizable in the presence of a suitable coresident conjugation system (166).
Resistance Plasmids in Enterococci
Antibiotic resistance in enterococci is largely encoded (and transferred) by theta-replicating plasmids. Based on conserved domains in their replication initiators, these plasmids can be divided into the Rep_3, Inc18, and RepA_N families (171), but note that plasmids sometimes encode multiple replication initiators and that such mosaicism can confound classification. Characterized RC plasmids of enterococci encode Rep proteins containing the Rep_trans, Rep_1, or Rep_2 conserved domain and generally do not encode resistance, with an exception being the promiscuous plasmid pMV158, which confers tetracycline resistance via the tet(L) determinant (403). Similarly, the theta-replicating Rep_3 family plasmids (generally less than 10 kb) rarely encode resistance, with an exception being pAMα1, which also has tet(L) (404).
Inc18 plasmids.
Initially based on the incompatibility of plasmids such as pAMβ1 and pIP501, the so-called Inc18 family now includes plasmids with distantly related replication initiators that may in fact be compatible with original members of the family (16). These conjugative plasmids generally range in size from 25 to 50 kb and can be found in a wide variety of bacterial genera. Their replication initiation proteins contain a PriCT_1 domain and bind to an origin region located downstream of the rep gene, whose expression is tightly controlled by an antisense RNA and a transcriptional repressor, Cop (405). Furthermore, in addition to their T4SS-like conjugation machinery, encoded by the trs genes, they also encode a multimer resolution protein (Res) and a type I partitioning system (parAB) (406).
In enterococci, Inc18 plasmids typically confer resistance to MLS antibiotics [erm(B)] but can also encode resistance to multiple antibiotics, as in the case of pRE25 (406) (Fig. 7). They have also contributed to the spread of vancomycin resistance (vanA) and, in this regard, have been responsible (in most cases) for the delivery of Tn1546-like transposons to MRSA via conjugative transfer of pRE25-like plasmids, such as pWZ909 (407) (see above and Fig. 7). Furthermore, the Inc18-like mosaic plasmid pEF-01 (Fig. 7) was the first plasmid identified in Enterococcus faecalis to carry the cfr gene, which confers resistance to multiple antimicrobial classes, including phenicols, lincosamides, and oxazolidinones (408). Note that pMG1-like plasmids are related to those of the Inc18 family, as the replication initiation protein of pMG1 shares 32% amino acid sequence identity with the pRE25 initiator and contains a PriCT_1 domain (409). These conjugative plasmids can transfer into a variety of Gram-positive bacterial species and have contributed significantly to the spread of resistance to gentamicin (aacA-aphD) and vancomycin (vanA) among enterococci.
FIG 7.
Enterococcal multiresistance plasmids. Representatives of the Inc18 and RepA_N families are shown (15, 104, 359, 384, 390, 516; see the text for additional references). IS, transposons, and resistance genes are shown, with resistances conferred by the latter listed in Table 2 or as follows: cat (chloramphenicol resistance) and fexB (chloramphenicol/florfenicol resistance). The following plasmid maintenance genes/systems are also shown: cop, copy number control; parAB, type I partitioning system; rep, repA, and repB, initiation of replication; res, multimer resolution; txe-axe, toxin-antitoxin system. Note that pS177 is a pRUM-like plasmid.
RepA_N plasmids.
In general, RepA_N plasmids encode a replication initiation protein (RepA) that belongs to the RepA_N family, and they are broadly divided into the pheromone-responsive plasmids, the pRUM-like plasmids, and the so-called megaplasmids. However, unlike the case for their staphylococcal counterparts, little is known about how replication of these clinically important plasmids is regulated.
Pheromone-responsive conjugative plasmids, such as pAD1, are narrow-host-range enterococcal plasmids that have been studied extensively, particularly with respect to their pheromone-induced conjugation mechanism. In brief, potential recipient cells produce sex pheromones (encoded by the chromosome) that ultimately induce plasmid transfer via formation of a mating channel with donor cells. The conjugation machinery is encoded by plasmid tra genes, which display homology to those for T4SSs (397). In addition, these plasmids also encode a type I partitioning system (repBC) and the well-characterized RNA-regulated Fst toxin-antitoxin system (410). In the context of antimicrobial resistance, pheromone-responsive plasmids have largely been associated with dissemination of the vanA determinant (i.e., glycopeptide resistance) but can also variably confer resistance to multiple antibiotics, including streptomycin (aadE), kanamycin/neomycin (aphA-3), and MLS antibiotics [erm(B)], as in the case of pSL1 (411).
The pRUM-like plasmids are prevalent in E. faecium strains belonging to hospital-adapted clades (clonal complex 17 [CC17] related) and can confer resistance to chloramphenicol (cat), kanamycin/neomycin (aphA-3), MLS antibiotics [erm(B)], streptomycin (aadE), streptothricin (sat4), and/or vancomycin (vanA) (412). With respect to the latter, it is important that these plasmids have also been responsible for delivery of the Tn1546-like transposon to MRSA, as in the case of pS177 (413) (Fig. 7). In this regard, transfer was likely mediated via integration into a coresident conjugative plasmid (414), as pRUM-like plasmids generally do not carry conjugation or mobilization genes, although the prototype plasmid pRUM has a mob gene on a cointegrated plasmid (415). However, in addition to carrying repA, they do encode a type I partitioning system (parAB), a multimer resolution system (sin), and a proteic toxin-antitoxin system (axe-txe) (415). Interestingly, detailed sequence analysis of the pRUM-like plasmid pJEG40 (416) revealed that the repA gene may in fact be regulated via an antisense RNA-inhibited pseudoknot activation mechanism (S. M. Kwong, N. Firth, and S. O. Jensen, unpublished data).
The so-called megaplasmids range in size from 150 to 375 kb and are associated with the spread of both virulence and antibiotic resistance determinants among E. faecium clinical isolates (16). Sequencing of the prototypical plasmid pGL1 revealed that it encodes resistance to heavy metals, MLS antibiotics [erm(B)], and glycopeptides (vanA). These plasmids also encode a proteic toxin-antitoxin system, a type I partitioning system, and conjugation machinery related to T4SSs (417).
GENOMIC ISLANDS
A genomic island (GI) is a distinct region of a bacterial chromosome that has been acquired via horizontal transfer; in many cases, GIs are flanked by DR. GIs vary in size (they may be composed of several hundred genes) and can be classified based on the phenotype(s) that they encode. For example, GIs that contain multiple resistance determinants are referred to as resistance islands, whereas those that contain virulence factors are often called pathogenicity islands. A broad definition of GIs (418) encompasses elements with mobility functions, such as ICE, integrative mobilizable elements (IME), which require helper functions to conjugate, and elements that are excised from the chromosome and may be transferred horizontally via phage-mediated mechanisms (419), such as staphylococcal cassette chromosome elements (SCCmec) and S. aureus pathogenicity islands (SaPI).
Integrative Conjugative Elements
ICE (420) constitute a diverse group of mobile elements found in both Gram-negative and Gram-positive bacteria and have been reviewed recently (418). Like plasmids, ICE are self-transmissible by conjugation, but they integrate into the host chromosome and are replicated as part of it, although replication of excised ICE has now been demonstrated (421). ICE typically consist of a backbone (containing phage-like integration/excision functions, plasmid-like conjugation/maintenance components, and a regulation module) into which accessory genes are inserted. Excision of the ICE as a circular form and integration of this circle (at low frequency), usually into a unique attB site in the host chromosome, are catalyzed by the ICE-encoded site-specific integrase (Int). For some ICE, these processes, as well as in some cases conjugation, have been shown to be subject to complex regulation by ICE-encoded functions (418). Target sites are often at the 3′ ends of tRNA genes, and integration creates DR at the ends of the ICE, called attL and attR (418). The ICEberg website (http://db-mml.sjtu.edu.cn/ICEberg) collated information about ICE in an integrated database (422) but appears to be out of date (418).
ICE in Gram-negative bacteria.
Plasmid R391, originally called IncJ, is now classified as one of the archetypes of the SXT/R391 family of ICE. These elements integrate into an attB site in the 5′ end of the chromosomal prfC gene by site-specific recombination with their attP site, catalyzed by the IntSXT tyrosine recombinase. They have an IncA/C-related conjugation region, encoding a MOBHI family relaxase, and the regulation region includes a mobI gene and allows activation by the SOS response (418). The excised form of these elements is able to replicate, and they carry a partitioning system (421, 423). Different insertions are found at certain positions in a shared 47-kb backbone, and hybrid elements have also been found (424). SXT/R391 family elements are also able to mobilize adjacent sequences, including some genomic islands that have an oriT (418). SXT carries resistance genes and can be found in E. coli but is mainly associated with Vibrio spp.
GI/ICE appear to be particularly important in relation to resistance in P. aeruginosa, including high-risk clones (425), often carrying cassette-borne genes in class 1 In/Tn, sometimes inserted within Tn21 subfamily transposons. pKLC102/PAPI-1 and PAGI-2/PAGI-3 (P. aeruginosa pathogenicity/genomic island)-type ICE are integrated into tRNALys and tRNAGly genes, respectively (426, 427). pKLC102 was so named because it can exist as a free multicopy plasmid as well as a GI in some P. aeruginosa strains (325), and part of it seems to correspond to a smaller plasmid carrying a class 1 integron (see above). Three related PAGI-2-like islands carry a carbapenemase cassette in class 1 In/Tn in a transposon: GI2 (different from PAGI-2 itself; blaGES-5) (428), PAGI-15, and PAGI-16 (blaGES-24, blaIMP-6, or minor variant blaIMP-10) (429). The PAGI-2-like island PAGI-13 carries a class 1 integron and rmtD between ISCR3 family elements (430). ICEEc2, from this family but found in E. coli, carries a class 2 integron in Tn7 (431). It is not clear whether a GI designated GI1 (different from PAGI-1), inserted into the endA gene, would also be classified as an ICE. Variants with different class 1 In/Tn structures associated with Tn1403, or remnants of it, carry blaVIM-1 and other cassettes (425, 428, 432–434).
Tn4371 family ICE, which have IncP-like conjugation functions and target plasmids as well as the chromosome (435), have also been found in P. aeruginosa. Some carry resistance genes, including ICETn43716061, carrying the blaSPM-1 metallo-β-lactamase gene (436), which is found in some isolates that also carry PAGI-13 (430).
ICE in staphylococci and enterococci.
Tn916 (originally termed a conjugative transposon [CTn]) (11) exemplifies a group of related ICE that are found in a diverse range of bacteria. In most cases, the closely related Tn916-like elements encode resistance to tetracycline/minocycline [via tet(M)]; however, due to insertion of additional DNA elements into the basic backbone structure, they can also encode resistance to other antibiotics, such as MLS [erm(B)] and kanamycin/neomycin (aphA-3) in the case of Tn1545 (437).
Other Tn916-like elements include Tn6000, Tn5397, Tn5801, and Tn1549 (438–441). These elements have the same modular structure (same as that of Tn916) and confer resistance to tetracycline [via tet(M) or tet(S)] or, in the case of Tn1549, vancomycin (vanB2). Tn1549-like elements have made a significant contribution to the global spread of vancomycin resistance among enterococci (442). Interestingly, Tn6000, Tn5397, and Tn5801 have different integration/excision genes, likely due to recombination between different MGE, and additional regions of DNA. For example, Tn6000 also contains a restriction-modification system, a virulence locus, a group II intron, and a tyrosine integrase gene that is more closely related to those of staphylococcal pathogenicity islands (438).
ICE6013 represents a family of ICE in staphylococci that are not related to Tn916. ICE6013 was initially identified in human S. aureus ST239 isolates, in which it sometimes carries a Tn552 insertion (443); however, multiple subfamilies have now been identified in different staphylococcal species (444). While ICE6013 displays some sequence similarity to ICEBs1 of Bacillus subtilis (443), its encoded functions are largely uncharacterized. Interestingly, ORFs encoding an IS30-like DDE transposase were recently shown to be required for excision of ICE6013 prior to conjugative transfer (444). In this regard, the mechanism of recombination is clearly different from that mediated by the tyrosine integrases of Tn916-like elements, which target specific AT-rich sequences (445). It is therefore somewhat ironic/confusing that the term ICE has largely superseded the name conjugative transposon when elements such as Tn916 utilize an integrase and ICE6013 encodes a transposase-like enzyme.
Other Resistance Islands in Gram-Negative Bacteria
AbaR and AGR1 resistance islands were described in the Tn7-like transposon section above, and another, GIsul2, has also already been mentioned. GIsul2 (Fig. 4E) carries int (tyrosine site-specific recombinase) and resG (resolvase) genes and genes encoding potential replication, conjugation, and TA proteins, plus putative arsenate/arsenite resistance protein genes and ISCR2-sul2 (61, 188, 446). The conjugation genes are related to trb genes of IncPα plasmids, suggesting an interaction with these plasmids (188). GIsul2 was reported to be present in the chromosomes of S. flexneri and E. cloacae isolates (61) and as the progenitor of the ARI-B resistance region on C plasmids (188). Searches now identify a complete GIsul2 in E. coli and other Enterobacteriaceae. GIsul2 seems to be stably integrated, within the guaA (GMP synthetase) gene.
A. baumannii may also carry AGI1 (Acinetobacter genomic island 1), an IME with a backbone related to that of SGI, found in Salmonella, P. mirabilis, and recently Morganella morganii (447), and PGI-1 (P. mirabilis). Like these other IME, AGI1 is inserted at the 3′ end of the chromosomal trmE gene. It includes a resolvase (resG) gene and an adjacent large class 1 integron structure with three cassette arrays and two copies of the 3′-CS (448). Integrative mobile elements designated IMEX, which use chromosomally encoded XerC/D recombinases to integrate at chromosomal XerC/D binding sites (449), are beginning to be identified as vehicles for blaNMC-A/blaIMI carbapenemase genes in Enterobacter spp. (450–452).
Regions found in the chromosome of A. baumannii GC2 isolates, designated AbGRI2 (453) and AbGRI3 (454), are bounded by IS26 and can be thought of as equivalent to the resistance regions found in plasmids, presumably inserted and modified in the same way, or possibly transferred en bloc from a plasmid. Examples of similar chromosomal regions in other species include regions apparently derived from Tn2670 of R100 (21).
Staphylococcal Cassette Chromosome
SCCmec is a type of resistance island carried on the chromosome of MRSA isolates that confers resistance to methicillin, penicillin, and other β-lactam antibiotics. This element contains the mecA gene or the more recently identified mecC gene, both of which encode related low-affinity penicillin-binding proteins called PBP2a, and the divergently transcribed regulatory genes, mecR1 (for the signal transducer protein MecR1) and mecI (for the repressor protein MecI) (455–457). In the presence of a β-lactam antibiotic, MecR1 cleaves MecI bound to the operator region of the mecA/mecC promoter, resulting in derepression of transcription (458). This consequently enables cell wall synthesis to proceed via PBP2a production; most β-lactam antibiotics cannot bind PBP2a, with an exception being the fifth-generation cephalosporin ceftaroline fosamil (459). In some cases, both mecR1 and mecI are truncated by IS257 or IS1272, which results in constitutive expression of the mecA gene (460); however, the β-lactamase regulators BlaI and BlaR1 (if present) have also been shown to regulate mecA expression (101). Collectively, the mecA/C gene, the regulatory mec genes, and the associated IS are referred to as the mec gene complex. Several classes of the mec gene complex have been defined (A, B, C1, C2, D, and E) on the basis of the regulatory genes (i.e., truncated or not), the type and location/orientation of associated IS, and the hypervariable region located between the mecA gene and the downstream IS257 element (http://www.sccmec.org/).
In addition to mec genes, SCCmec elements also contain the ccr (cassette chromosome recombinase) gene complex, which is composed of the ccr gene(s) and surrounding open reading frames that, until recently, have not been assigned functions (discussed below). Three distinct ccr genes (ccrA, ccrB, and ccrC) share less than 50% nucleotide sequence identity with each other; different allotypes for ccrA, ccrB, and ccrC also exist. Each individual ccr gene encodes a recombinase that mediates the integration and excision of SCCmec elements at a specific site located at the 3′ end of the rlmH gene (previously referred to as orfX) (456, 461). Several types of ccr gene complex have also been defined, and these are comprised of either a combination of ccrA and ccrB allotypes (one of each; types 1 to 4 and 6 to 8) or single ccrC allotypes only (types 5 and 9) (http://www.sccmec.org) (462). Recently, a conserved gene (cch or cch2), located directly upstream of the ccr gene(s), was shown to encode an active DNA helicase (463); cch2 is also preceded by a putative primase gene (polA). The presence of such genes implies that SCC elements have the ability to replicate postexcision and that multiple circular copies would likely facilitate the horizontal transfer process. Furthermore, a gene downstream of the ccr gene(s) encodes a uracil-DNA glycosylase inhibitor (SAUGI), indicating that phages may facilitate SCCmec transfer, similar to the mechanism utilized by staphylococcal pathogenicity islands (398, 463) (see below).
At present, 12 SCCmec allotypes have been recognized based on structural diversity (Fig. 8) (464). In this regard, SCCmec types are defined by the mec and ccr gene complexes, while subtypes are based on the structure of the three joining (J) regions, located between ccr and the chromosomal region flanking SSCmec (J1), between mec and ccr (J2), and between rlmH and mec (J3). It is important to note that the J regions appear to act as a chromosomal reservoir for the accretion of antimicrobial resistance determinants, which are often associated with transposable elements (465). These include Tn554-like elements encoding resistance to cadmium (cad) or MLS antibiotics [erm(A)] and spectinomycin (spc), as well as segments flanked by IS257 that confer resistance to mercury (merAB), aminoglycosides (aadD) and bleomycin (ble), or tetracycline [tet(K)]; the last two segments represent integrated copies of the RC-replicating plasmids pUB110 and pT181, respectively (Fig. 8) (455). Recently, based on the structural diversity described above, a new online tool (SCCmecFinder; https://cge.cbs.dtu.dk/services/SCCmecFinder) was developed for rapid, sequence-based SCCmec typing of MRSA (466).
FIG 8.
Representative SCCmec elements (464, 478; see the text for additional references). IS, transposons, cointegrated plasmids, and resistance genes are shown, with resistances conferred by the latter listed in Table 2 or as follows: arsBC, arsenic resistance; cadA, cadmium resistance; fusC (previously known as far), fusidic acid resistance (517); and mecA/mecC, β-lactam resistance. Cassette recombinase genes (ccrA1 to -4, ccrB1 to -4, and ccrC1 and -2), mecA/C regulatory genes (mecI and mecR1), and an arginine catabolic mobile element (ACME) (441) are also shown; mec classes and ccr types are denoted by colored shading. Note that cch genes, polA, and SAUGI are not shown.
Prior to 1990, hospital-associated MRSA isolates predominantly contained SCCmec allotypes I to III; however, SCCmecIV is increasingly being identified in contemporary isolates (467). Interestingly, community-associated MRSA (CA-MRSA) isolates almost exclusively contain SCCmecIV (and SCCmecV, to a lesser extent), and this has now emerged as the most widely distributed SCCmec element (468, 469). Fitness experiments have shown that S. aureus strains containing SCCmecIV are indistinguishable from their isogenic methicillin-sensitive parents (470, 471). This may indicate that the prevalence of SCCmecIV is related (at least in part) to the apparently lesser burden it imposes on its hosts than those by other SCCmec elements, which is suggested to be due to differences in gene expression rather than DNA size (470). However, how efficiently it is transferred between strains may also be a key contributing factor.
It should be noted that SCCmec elements are not exclusive to S. aureus and, in fact, are more frequently carried by coagulase-negative staphylococci (CNS); the prevalence of methicillin resistance has been reported to be higher in CNS than in S. aureus, with global rates ranging from 75 to 90% for clinical isolates during the 1990s (472, 473). Interestingly, it has been suggested that the animal-related CNS Staphylococcus fleurettii and Staphylococcus vitulinus contributed to mec gene evolution (474, 475) and that assembly of the SCCmec element may have occurred in Staphylococcus sciuri (475). In any case, the CNS SCCmec elements display more diversity than those of S. aureus, and it is believed that CNS act as a reservoir from which methicillin-sensitive S. aureus can acquire SCCmec, contributing to the emergence of new MRSA clones, which appears to occur more frequently than originally thought, at least with respect to SCCmec allotypes IV and V (476, 477). However, not all SCC elements encode resistance to methicillin. Non-mec elements have been found to variably contain genes that contribute to the virulence or fitness/survival of the host strain. Two examples are SCCfus (also known as SCC476) (Fig. 8) and SCCHg (also known as SCCmercury), which encode resistance to fusidic acid and mercuric chloride, respectively (478, 479); these elements have been renamed in accordance with the nomenclature system proposed by the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (464). An additional example is the arginine catabolic mobile element (ACME), which has been identified in association with some SCCmec allotypes, mainly type IV (Fig. 8). This element utilizes SCCmec-encoded cassette recombinases (and thus integrates into the same site), carries the opp-3 and/or arc gene cluster, the latter of which encodes a complete arginine deiminase pathway, and has been shown to enhance fitness and skin colonization (471, 480, 481).
Staphylococcal Pathogenicity Islands
SaPIs are genomic islands that integrate at specific sites, are flanked by DR, and utilize the capsids of helper phages for their movement. They encode phage-like proteins that facilitate this process, including a master repressor (Stl) that controls SaPI excision via interaction with helper phage antirepressor proteins; these interactions are SaPI and phage specific (482). In most cases, SaPIs also encode one or more virulence factors, such as superantigens, but they rarely carry resistance determinants. However, some exceptions are SaPIj50, which encodes resistance to penicillin (bla) (483), and SaRIfusB, which encodes resistance to fusidic acid (fusB) (484). Note that while SaRIfusB is related to classical SaPIs, it does not encode any known virulence factors and is therefore referred to as a resistance island (484).
USING WGS TO BETTER UNDERSTAND MOBILE ANTIBIOTIC RESISTANCE
The increasing availability of next-generation DNA sequencing methods, including their use for tracking outbreaks of resistant organisms, has led to an explosion in the number of bacterial whole-genome sequences. Analysis of these data has underscored the significance of MGE, and the increasing use of this technology to characterize bacterial pathogens presents opportunities for understanding the evolution of resistance but also creates challenges. It is increasingly apparent that the boundaries between types of elements historically viewed as distinct are becoming blurred (e.g., IS with passenger genes resembling unit transposons, excised ICE replicating as plasmids do, etc.), and this needs to be borne in mind when naming new elements. Using existing resources to obtain names/numbers for novel IS (ISfinder [https://www-is.biotoul.fr]) (10), transposons (Transposon Registry [http://transposon.lstmed.ac.uk]) (11), gene cassettes (INTEGRALL [http://integrall.bio.ua.pt] [123] and RAC [http://app.spokade.com/rac/feature/list] [485]), and SCCmec elements (http://www.sccmec.org) (464) and submitting elements to these sites help to reduce confusion.
While use of long-read sequencing methods is becoming more common, many plasmid sequences in International Nucleotide Sequence Database Collaboration (INSDC) databases (GenBank, EMBL-EBI, and DDBJ) were derived from short-read methods, as these were more widely available and are more economical. However, complex resistance regions in plasmids or genomic islands present challenges for assembly from short-read data, as multiple copies of the same mobile element in different locations constitute repeats that are generally significantly longer than read lengths. Most assembly programs will collapse reads covering these repeats down to a single contig, while regions between repeats are usually found as separate contigs, potentially with fragments of repeats on each end. Methods such as plasmidSPAdes (486) may help to identify contigs derived from plasmids, and those such as PLACNET (487) may help to group contigs from a particular plasmid, but it can still be difficult to correctly assign resistance regions to particular plasmids (175, 488).
These difficulties mean that a significant number of plasmid sequences in INSDC databases may be misassembled, as suggested by, e.g., the presence of fragments of mobile elements that are not explained by truncation by another mobile element, which is quite unusual (489). Use of methods such as Bandage to visualize links between contigs (490) and correctly annotating the boundaries of MGE (using resources such as BLAST searches in ISfinder, ISMapper [491], information in the transposon registry, IntegronFinder [492], and MARA [http://mara.spokade.com] [489]) can help to identify assembly problems. These methods may also assist in designing strategies for PCR and Sanger sequencing across contig boundaries to ensure correct assembly.
Certain types of plasmids also pose particular assembly problems. For example, shufflons in I1 and I2 plasmids appear to be quite active, and different arrangements within the plasmid population used for sequencing may result in multiple contigs that cannot be assembled unambiguously (493) even using long-read data (234). One solution is to check reads for missing shufflon segments and then to order the shufflon segments as in a reference plasmid and note this in the INSDC entry (e.g., see the comment under accession number AP005147, for I1 plasmid R64). Plasmid backbones may also include multiple copies of other regions with sufficient identity to cause assembly issues, e.g., a repeated region (∼530 bp; 95% identity) in IncI2 plasmids carrying mcr-1 (237). Even with long-read methods, care must be taken to check for and remove one copy of any artifactual long repeats present at the end of plasmid contigs before circularizing, as minor differences between repeats due to errors may prevent this from happening automatically.
Many MGE sequences and plasmid backbone segments are also very highly conserved, so likely sequence errors, such as those in homopolymeric regions, may be quite obvious and can be checked and corrected as appropriate. Careful checking of plasmid sequences and assembly before submission to databases or publication, which is feasible due to their small size relative to that of whole chromosomes, will help to allow better identification of real differences between similar plasmids that are actually functionally or epidemiologically important. It is also helpful to standardize the start point and orientation of related plasmid sequences when submitting them to INSDC databases, to simplify comparative analysis. Such start points are already fairly well established for some plasmid types, e.g., in the replication region for A/C plasmids (499 bp upstream of the start codon of mobI) and at the start codon of tir in L/M plasmids.
Classification of plasmids also needs to move on from typing based on experimentally defined incompatibility, which may not be reflected adequately in sequence identity, and expanded to consider the whole backbone rather than just the replicon and/or relaxase region. At present, different types of schemes exist for different plasmid types, based on analyses by different groups of researchers, and some plasmid types have been neglected. Developing a more universal system will require cooperation between experts in pertinent fields, including plasmid biology and bioinformatics, as well as consultation and broad acceptance from the relevant research community. Recent suggestions to improve the annotation of plasmid backbone genes will hopefully stimulate discussion (286).
CONCLUSIONS
Horizontal gene transfer plays an important role in the acquisition of new properties, such as pathogenicity and antibiotic resistance, underpinning the formidable adaptive potential of bacteria. The previous sections of this review outline the diverse toolkit of MGE that the species of interest exploit to access an extended gene pool in order to overcome the evolutionary challenge that antimicrobial chemotherapy represents. We hope that this review also illustrates how the various mobile element types interact with each other, since it is largely the synergistic amalgamation of their differing properties that underpins the adaptive capacity of these bacteria. Although there are some notable differences between the elements important in Gram-negative and Gram-positive bacteria, such as the significant roles of integrons/gene cassettes in the former and of small RC plasmids in the latter, there are many more similarities. Transposable and integrative elements mediate the insertion of resistance genes into chromosomes and plasmids, with the latter serving as key vehicles of intercellular transmission. Nonetheless, recent findings remind us that there is still a lot to learn about MGE. For example, the realization that oriT-like sequences, which facilitate relaxase-in-trans mobilization, are commonplace in staphylococcal plasmids and the detection of analogous sequences in plasmids from other genera have implications for the possible importance of plasmids that have previously been considered nontransferable, and hence for the way we look at the spread of resistance genes.
Although the individual elements differ between organisms, evolutionary relationships reveal commonality in the element types found and the roles that they play in disparate bacterial hosts. The prevalence of Tn3- and Tn7-type transposons and the ubiquity of IS6 family IS are obvious examples of this, and likewise, there are equivalent collections of homologous functional modules for replication initiation, partitioning, transfer, etc., within the backbones of theta-replicating plasmids. The extensive sequence divergence evident between members of the same MGE family and between modules that share the same function reveals the evolutionarily ancient nature of these molecules and their extensive coevolution with their respective hosts.
Improved analysis methods for the annotation and classification of MGE will also be needed to obtain full value from the vast quantities of sequence data that are being generated, but the increasing diversity of sequenced elements means that there are rarely obvious approaches, and effective changes to nomenclature require broad consultation to ensure that these changes are taken up. Likewise, continued research into the basic biology of mobile elements will be needed for meaningful understanding of the properties of known and yet-to-be-discovered elements. Finally, it needs to be recognized that the current knowledge is based largely on analysis of clinical strains, representing a very limited snapshot of relevant bacterial ecology. It is becoming increasingly feasible to investigate intra- and interspecies linkages between organisms within health care settings, and to other niches in the broader environment, which should provide a more comprehensive understanding of the ecological pathways that ultimately lead to resistance in bacterial pathogens.
ACKNOWLEDGMENTS
Research on plasmids in Enterobacteriaceae was supported by National Health and Medical Research Council (Australia) project grant GNT1046886 to S.R.P. and Centre of Research Excellence grant GNT1001021. Research on staphylococcal plasmid biology was supported by National Health and Medical Research Council (Australia) project grant GNT1081412 to N.F., S.M.K., and S.O.J. and grant GNT1145697 to N.F.
We thank numerous colleagues, particularly Mick Chandler, Laura Frost, Ruth Hall, Jon Iredell, Josh Ramsay, Julian Rood, Ron Skurray, and Chris M. Thomas, for stimulating discussions over the years.
Biographies

Sally R. Partridge is a Principal Research Fellow at the Centre for Infectious Diseases and Microbiology at The Westmead Institute for Medical Research, Westmead Hospital, Sydney, Australia, and an Honorary Associate Professor, Sydney Medical School, University of Sydney. She received a B.A. in Biochemistry and a D.Phil. (focused on sporulation in Bacillus subtilis) from Oxford University. Since 1997, her main interest has been the genetics of mobile antibiotic resistance in Gram-negative bacteria, particularly the Enterobacteriaceae, including improving resistance gene nomenclature and sequence annotation, epidemiological aspects of resistance, and novel approaches to treatment of antibiotic-resistant infections.

Stephen M. Kwong obtained his Ph.D. from the School of Medicine at the National University of Singapore in 2001 before commencing postdoctoral studies at the University of Sydney. He is currently a Research Academic in the School of Life and Environmental Sciences at the University of Sydney. His current research is focused on the molecular biology of staphylococcal resistance plasmids, including their mechanisms of replication, maintenance, and transfer, and understanding the roles of plasmids and other mobile elements in facilitating the movement and spread of resistance genes.

Neville Firth is an Honorary Associate Professor in the School of Life and Environmental Sciences at the University of Sydney. He obtained a Ph.D. from Monash University, working on DNA transfer in Escherichia coli, and then undertook postdoctoral research at the University of Sydney, focusing on Staphylococcus aureus. He has studied the roles of mobile genetic elements in the evolution of antimicrobial resistance for over 20 years. His research seeks to understand the molecular biology of plasmids and transposable elements that facilitate the acquisition, maintenance, and dissemination of resistance genes in staphylococci.

Slade O. Jensen is an Associate Professor in the School of Medicine, Western Sydney University, and Head of the Antibiotic Resistance and Mobile Elements Group (ARMEG), based at the Ingham Institute for Applied Medical Research. He obtained a Ph.D. from the University of Sydney, focusing on the role of horizontal gene transfer in bacterial evolution. His current research interests include the development of novel antimicrobials and the evolution of antibiotic resistance in ESKAPE pathogens, particularly Staphylococcus aureus and Enterococcus faecium.
REFERENCES
- 1.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 2.Llaca-Díaz JM, Mendoza-Olazarán S, Camacho-Ortiz A, Flores S, Garza-González E. 2012. One-year surveillance of ESKAPE pathogens in an intensive care unit of Monterrey, Mexico. Chemotherapy 58:475–481. doi: 10.1159/000346352. [DOI] [PubMed] [Google Scholar]
- 3.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2015. Everyman's guide to bacterial insertion sequences. Microbiol Spectr 3:MDNA3-0030-2014. doi: 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 4.Chandler M, Mahillon J. 2002. Insertion sequences revisited, p 305–366. In Craig NL, Craigie R, Gellert M, Lambowitz AM (ed), Mobile DNA II. ASM Press, Washington, DC. [Google Scholar]
- 5.Hickman AB, Dyda F. 2015. Mechanisms of DNA transposition. Microbiol Spectr 3:MDNA3-0034-2014. doi: 10.1128/microbiolspec.MDNA3-0034-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallet B, Sherratt DJ. 1997. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol Rev 21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 7.Chandler M, Fayet O, Rousseau P, Ton Hoang B, Duval-Valentin G. 2015. Copy-out-paste-in transposition of IS911: a major transposition pathway. Microbiol Spectr 3:MDNA3-0031-2014. doi: 10.1128/microbiolspec.MDNA3-0031-2014. [DOI] [PubMed] [Google Scholar]
- 8.Chandler M, Fayet O. 1993. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol 7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 9.Mugnier PD, Poirel L, Nordmann P. 2009. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J Bacteriol 191:2414–2418. doi: 10.1128/JB.01258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts AP, Chandler M, Courvalin P, Guedon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, Berg DE. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167–173. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamruzzaman M, Patterson JD, Shoma S, Ginn AN, Partridge SR, Iredell JR. 2015. Relative strengths of promoters provided by common mobile genetic elements associated with resistance gene expression in Gram-negative bacteria. Antimicrob Agents Chemother 59:5088–5091. doi: 10.1128/AAC.00420-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 14.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. 2017. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol 43:709–730. doi: 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 15.Firth N, Skurray RA. 2006. The Staphylococcus—genetics: accessory elements and genetic exchange, p 413–426. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-positive pathogens, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 16.Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F. 2014. Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology, p 309–320. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston, MA. [PubMed] [Google Scholar]
- 17.Novick RP. 1989. Staphylococcal plasmids and their replication. Annu Rev Microbiol 43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- 18.Jensen SO, Lyon BR. 2009. Genetics of antimicrobial resistance in Staphylococcus aureus. Future Microbiol 4:565–582. doi: 10.2217/fmb.09.30. [DOI] [PubMed] [Google Scholar]
- 19.Haniford DB, Ellis MJ. 2015. Transposons Tn10 and Tn5. Microbiol Spectr 3:MDNA3-0002-2014. doi: 10.1128/microbiolspec.MDNA3-0002-2014. [DOI] [PubMed] [Google Scholar]
- 20.Pagano M, Martins AF, Barth AL. 2016. Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz J Microbiol 47:785–792. doi: 10.1016/j.bjm.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 22.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mollet B, Clerget M, Meyer J, Iida S. 1985. Organization of the Tn6-related kanamycin resistance transposon Tn2680 carrying two copies of IS26 and an IS903 variant, IS903B. J Bacteriol 163:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Needham C, Noble WC, Dyke KGH. 1995. The staphylococcal insertion sequence IS257 is active. Plasmid 34:198–205. doi: 10.1006/plas.1995.0005. [DOI] [PubMed] [Google Scholar]
- 25.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762-15. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siguier P, Gourbeyre E, Chandler M. 2017. Known knowns, known unknowns and unknown unknowns in prokaryotic transposition. Curr Opin Microbiol 38:171–180. doi: 10.1016/j.mib.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Harmer CJ, Hall RM. 2015. IS26-mediated precise excision of the IS26-aphA1a translocatable unit. mBio 6:e01866-15. doi: 10.1128/mBio.01866-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harmer CJ, Hall RM. 2016. IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1:e00038-16. doi: 10.1128/mSphere.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Roth E, Kwong SM, Alcoba-Florez J, Firth N, Mendez-Alvarez S. 2010. Complete nucleotide sequence and comparative analysis of pPR9, a 41.7-kilobase conjugative staphylococcal multiresistance plasmid conferring high-level mupirocin resistance. Antimicrob Agents Chemother 54:2252–2257. doi: 10.1128/AAC.01074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leelaporn A, Firth N, Byrne ME, Roper E, Skurray RA. 1994. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob Agents Chemother 38:2238–2244. doi: 10.1128/AAC.38.10.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harmer CJ, Hall RM. 2017. Targeted conservative formation of cointegrates between two DNA molecules containing IS26 occurs via strand exchange at either IS end. Mol Microbiol 106:409–418. doi: 10.1111/mmi.13774. [DOI] [PubMed] [Google Scholar]
- 32.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother 48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lartigue MF, Poirel L, Aubert D, Nordmann P. 2006. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring β-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob Agents Chemother 50:1282–1286. doi: 10.1128/AAC.50.4.1282-1286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother 47:2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurpiel PM, Hanson ND. 2012. Point mutations in the inc antisense RNA gene are associated with increased plasmid copy number, expression of blaCMY-2 and resistance to piperacillin/tazobactam in Escherichia coli. J Antimicrob Chemother 67:339–345. doi: 10.1093/jac/dkr479. [DOI] [PubMed] [Google Scholar]
- 36.Dhanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, Livermore DM, Woodford N. 2011. Variation in the genetic environments of blaCTX-M-15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 66:1005–1012. doi: 10.1093/jac/dkr041. [DOI] [PubMed] [Google Scholar]
- 37.Partridge SR. 2016. Mobilization of blaBKC-1 by ISKpn23? Antimicrob Agents Chemother 60:5102–5104. doi: 10.1128/AAC.00785-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YG, Qu TT, Yu YS, Zhou JY, Li LJ. 2006. Insertion sequence ISEcp1-like element connected with a novel aph(2″) allele [aph(2″)-Ie] conferring high-level gentamicin resistance and a novel streptomycin adenylyltransferase gene in Enterococcus. J Med Microbiol 55:1521–1525. doi: 10.1099/jmm.0.46702-0. [DOI] [PubMed] [Google Scholar]
- 39.Tegetmeyer HE, Jones SC, Langford PR, Baltes N. 2008. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet Microbiol 128:342–353. doi: 10.1016/j.vetmic.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 40.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 41.Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, Dyda F. 2016. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother 60:6973–6976. doi: 10.1128/AAC.01457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snesrud E, Ong AC, Corey B, Kwak YI, Clifford R, Gleeson T, Wood S, Whitman TJ, Lesho EP, Hinkle M, McGann P. 2017. Analysis of serial isolates of mcr-1-positive Escherichia coli reveals a highly active ISApl1 transposon. Antimicrob Agents Chemother 61:e00056-17. doi: 10.1128/AAC.00056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, Randall LP, Lemma F, Crook DW, Teale C, Smith RP, Anjum MF. 2017. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother 72:2745–2749. doi: 10.1093/jac/dkx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poirel L, Keiffer N, Nordmann P. 2017. In vitro study of ISApl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob Agents Chemother 61:e00127-17. doi: 10.1128/AAC.00127-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snesrud E, McGann P, Chandler M. 2018. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio 9:e02381-17. doi: 10.1128/mBio.02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabo M, Kiss J, Kotany G, Olasz F. 1999. Importance of illegitimate recombination and transposition in IS30-associated excision events. Plasmid 42:192–209. doi: 10.1006/plas.1999.1425. [DOI] [PubMed] [Google Scholar]
- 47.Terveer EM, Nijhuis RHT, Crobach MJT, Knetsch CW, Veldkamp KE, Gooskens J, Kuijper EJ, Claas ECJ. 2017. Prevalence of colistin resistance gene (mcr-1) containing Enterobacteriaceae in feces of patients attending a tertiary care hospital and detection of a mcr-1 containing, colistin susceptible E. coli. PLoS One 12:e0178598. doi: 10.1371/journal.pone.0178598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foster TJ, Lundblad V, Hanley-Way S, Halling SM, Kleckner N. 1981. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell 23:215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- 49.Zhou K, Luo Q, Wang Q, Huang C, Lu H, John RWA, Xiao Y, Li L. 2018. Silent transmission of an IS1294b-deactivated mcr-1 gene with inducible colistin resistance. Int J Antimicrob Agents 51:822–828. doi: 10.1016/j.ijantimicag.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Pham Thanh D, Thanh Tuyen H, Nguyen Thi Nguyen T, Chung The H, Wick RR, Thwaites GE, Baker S, Holt KE. 2016. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J Antimicrob Chemother 71:2314–2317. doi: 10.1093/jac/dkw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Q, Li M, Spiller OB, Andrey DO, Hinchliffe P, Li H, MacLean C, Niumsup P, Powell L, Pritchard M, Papkou A, Shen Y, Portal E, Sands K, Spencer J, Tansawai U, Thomas D, Wang S, Wang Y, Shen J, Walsh T. 2017. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat Commun 8:2054. doi: 10.1038/s41467-017-02149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandler M, de la Cruz F, Dyda F, Hickman AB, Moncalian G, Ton-Hoang B. 2013. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat Rev Microbiol 11:525–538. doi: 10.1038/nrmicro3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcillán-Barcia M, Bernales I, Mendiola M, de la Cruz F. 2002. IS91 rolling circle transposition, p 891–904. In Craig NL, Craigie R, Gellert M, Lambowitz AM (ed), Mobile DNA II. ASM Press, Washington, DC. [Google Scholar]
- 54.Tavakoli N, Comanducci A, Dodd HM, Lett MC, Albiger B, Bennett P. 2000. IS1294, a DNA element that transposes by RC transposition. Plasmid 44:66–84. doi: 10.1006/plas.1999.1460. [DOI] [PubMed] [Google Scholar]
- 55.Tagg KA, Iredell JR, Partridge SR. 2014. Complete sequencing of IncI1 sequence type 2 plasmid pJIE512b indicates mobilization of blaCMY-2 from an IncA/C plasmid. Antimicrob Agents Chemother 58:4949–4952. doi: 10.1128/AAC.02773-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yassine H, Bientz L, Cros J, Goret J, Bebear C, Quentin C, Arpin C. 2015. Experimental evidence for IS1294b-mediated transposition of the blaCMY-2 cephalosporinase gene in Enterobacteriaceae. J Antimicrob Chemother 70:697–700. doi: 10.1093/jac/dku472. [DOI] [PubMed] [Google Scholar]
- 57.Stokes HW, Tomaras C, Parsons Y, Hall RM. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39–50. doi: 10.1006/plas.1993.1032. [DOI] [PubMed] [Google Scholar]
- 58.Partridge SR, Hall RM. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob Agents Chemother 47:342–349. doi: 10.1128/AAC.47.1.342-349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levings RS, Djordjevic SP, Hall RM. 2008. SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob Agents Chemother 52:2529–2537. doi: 10.1128/AAC.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nigro SJ, Hall RM. 2011. GIsul2, a genomic island carrying the sul2 sulphonamide resistance gene and the small mobile element CR2 found in the Enterobacter cloacae subspecies cloacae type strain ATCC 13047 from 1890, Shigella flexneri ATCC 700930 from 1954 and Acinetobacter baumannii ATCC 17978 from 1951. J Antimicrob Chemother 66:2175–2176. doi: 10.1093/jac/dkr230. [DOI] [PubMed] [Google Scholar]
- 62.Toleman MA, Walsh TR. 2008. Evolution of the ISCR3 group of ISCR elements. Antimicrob Agents Chemother 52:3789–3791. doi: 10.1128/AAC.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toleman M, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera, likely constructed in Acinetobacter baumannii. Antimicrob Agents Chemother 56:2773–2776. doi: 10.1128/AAC.06297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wailan AM, Sidjabat HE, Yam WK, Alikhan NF, Petty NK, Sartor AL, Williamson DA, Forde BM, Schembri MA, Beatson SA, Paterson DL, Walsh TR, Partridge SR. 2016. Mechanisms involved in acquisition of blaNDM genes by IncA/C2 and IncFIIY plasmids. Antimicrob Agents Chemother 60:4082–4088. doi: 10.1128/AAC.00368-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siguier P, Gagnevin L, Chandler M. 2009. The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. Res Microbiol 160:232–241. doi: 10.1016/j.resmic.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 66.Brown NL, Evans LR. 1991. Transposition in prokaryotes: transposon Tn501. Res Microbiol 142:689–700. doi: 10.1016/0923-2508(91)90082-L. [DOI] [PubMed] [Google Scholar]
- 67.Nigro SJ, Hall RM. 2016. Loss and gain of aminoglycoside resistance in global clone 2 Acinetobacter baumannii in Australia via modification of genomic resistance islands and acquisition of plasmids. J Antimicrob Chemother 71:2432–2440. doi: 10.1093/jac/dkw176. [DOI] [PubMed] [Google Scholar]
- 68.Nicolas E, Lambin M, Dandoy D, Galloy C, Nguyen N, Oger CA, Hallet B. 2015. The Tn3-family of replicative transposons. Microbiol Spectr 3:MDNA3-0060-2014. doi: 10.1128/microbiolspec.MDNA3-0060-2014. [DOI] [PubMed] [Google Scholar]
- 69.Chandler M. 15 September 2016. Transposons: prokaryotic. eLS doi: 10.1002/9780470015902.a0000591.pub2. [DOI] [Google Scholar]
- 70.Partridge SR, Hall RM. 2005. Evolution of transposons containing blaTEM genes. Antimicrob Agents Chemother 49:1267–1268. doi: 10.1128/AAC.49.3.1267-1268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bailey J, Pinyon J, Abnantham S, Hall R. 2011. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother 66:745–751. doi: 10.1093/jac/dkq529. [DOI] [PubMed] [Google Scholar]
- 72.Sarno R, McGillivary G, Sherratt DJ, Actis LA, Tolmasky ME. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob Agents Chemother 46:3422–3427. doi: 10.1128/AAC.46.11.3422-3427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Partridge SR. 2015. What's in a name? ISSwi1 corresponds to transposons related to Tn2 and Tn3. mBio 6:e01344-15. doi: 10.1128/mBio.01344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ, Modernising Medical Microbiology Informatics Group, Sifri CD, Walker AS, Peto TE, Crook DW, Mathers AJ. 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zong Z, Ginn AN, Dobiasova H, Iredell JR, Partridge SR. 2015. Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements. Plasmid 80:118–126. doi: 10.1016/j.plasmid.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 76.Cain AK, Hall RM. 2011. Transposon Tn5393e carrying the aphA1-containing transposon Tn6023 upstream of strAB does not confer resistance to streptomycin. Microb Drug Resist 17:389–394. doi: 10.1089/mdr.2011.0037. [DOI] [PubMed] [Google Scholar]
- 77.Arthur M, Molinas C, Depardieu F, Courvalin P. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol 175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lopez M, Saenz Y, Alvarez-Martinez MJ, Marco F, Robredo B, Rojo-Bezares B, Ruiz-Larrea F, Zarazaga M, Torres C. 2010. Tn1546 structures and multilocus sequence typing of vanA-containing enterococci of animal, human and food origin. J Antimicrob Chemother 65:1570–1575. doi: 10.1093/jac/dkq192. [DOI] [PubMed] [Google Scholar]
- 79.Chen C, Xu X, Qu T, Yu Y, Ying C, Liu Q, Guo Q, Hu F, Zhu D, Li G, Wang M. 2014. Prevalence of the fosfomycin-resistance determinant, fosB3, in Enterococcus faecium clinical isolates from China. J Med Microbiol 63:1484–1489. doi: 10.1099/jmm.0.077701-0. [DOI] [PubMed] [Google Scholar]
- 80.Grinsted J, de la Cruz F, Schmitt R. 1990. The Tn21 subgroup of bacterial transposable elements. Plasmid 24:163–189. doi: 10.1016/0147-619X(90)90001-S. [DOI] [PubMed] [Google Scholar]
- 81.Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Partridge SR, Hall RM. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J Bacteriol 185:6371–6384. doi: 10.1128/JB.185.21.6371-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Partridge SR, Brown HJ, Stokes HW, Hall RM. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob Agents Chemother 45:1263–1270. doi: 10.1128/AAC.45.4.1263-1270.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stokes HW, Elbourne LD, Hall RM. 2007. Tn1403, a multiple-antibiotic resistance transposon made up of three distinct transposons. Antimicrob Agents Chemother 51:1827–1829. doi: 10.1128/AAC.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 86.Hammerl JA, Borowiak M, Schmoger S, Shamoun D, Grobbel M, Malorny B, Tenhagen BA, Kasbohrer A. 12 February 2018. mcr-5 and a novel mcr-5.2 variant in Escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J Antimicrob Chemother doi: 10.1093/jac/dky1020. [DOI] [PubMed] [Google Scholar]
- 87.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Naas T, Cuzon G, Truong HV, Nordmann P. 2012. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 56:4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheruvanky A, Stoesser N, Sheppard AE, Crook DW, Hoffman PS, Weddle E, Carroll J, Sifri CD, Chai W, Barry K, Ramakrishnan G, Mathers AJ. 2017. Enhanced Klebsiella pneumoniae carbapenemase expression from a novel Tn4401 deletion. Antimicrob Agents Chemother 61:e00025-17. doi: 10.1128/AAC.00025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bryant KA, Van Schooneveld TC, Thapa I, Bastola D, Williams LO, Safranek TJ, Hinrichs SH, Rupp ME, Fey PD. 2013. blaKPC-4 is encoded within a truncated Tn4401 in an IncL/M plasmid pNE1280 isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob Agents Chemother 57:37–41. doi: 10.1128/AAC.01062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chmelnitsky I, Shklyar M, Leavitt A, Sadovsky E, Navon-Venezia S, Ben Dalak M, Edgar R, Carmeli Y. 2014. Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn Microbiol Infect Dis 79:255–260. doi: 10.1016/j.diagmicrobio.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 92.Martinez T, Martinez I, Vazquez GJ, Aquino EE, Robledo IE. 2016. Genetic environment of the KPC gene in Acinetobacter baumannii ST2 clone from Puerto Rico and genomic insights into its drug resistance. J Med Microbiol 65:784–792. doi: 10.1099/jmm.0.000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Craig NL. 2002. Tn7, p 424–456. In Craig NL, Craigie R, Gellert M, Lambowitz AM (ed), Mobile DNA II. ASM Press, Washington, DC. [Google Scholar]
- 95.Peters JE. 2014. Tn7. Microbiol Spectr 2:MDNA3-0010-2014. doi: 10.1128/microbiolspec.MDNA3-0010-2014. [DOI] [PubMed] [Google Scholar]
- 96.Rådström P, Sköld O, Swedberg G, Flensburg J, Roy PH, Sundström L. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J Bacteriol 176:3257–3268. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kholodii GY, Mindlin SZ, Bass IA, Yurieva OV, Minakhina SV, Nikiforov VG. 1995. Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol Microbiol 17:1189–1200. doi: 10.1111/j.1365-2958.1995.mmi_17061189.x. [DOI] [PubMed] [Google Scholar]
- 98.Minakhina S, Kholodii G, Mindlin S, Yurieva O, Nikiforov V. 1999. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol Microbiol 33:1059–1068. doi: 10.1046/j.1365-2958.1999.01548.x. [DOI] [PubMed] [Google Scholar]
- 99.Labbate M, Chowdhury PR, Stokes HW. 2008. A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments. J Bacteriol 190:5318–5327. doi: 10.1128/JB.00199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gregory PD, Lewis RA, Curnock SP, Dyke KG. 1997. Studies of the repressor (BlaI) of β-lactamase synthesis in Staphylococcus aureus. Mol Microbiol 24:1025–1037. doi: 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 101.Hackbarth CJ, Chambers HF. 1993. blaI and blaR1 regulate β-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 37:1144–1149. doi: 10.1128/AAC.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rowland SJ, Dyke KG. 1990. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol 4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 103.Paulsen IT, Gillespie MT, Littlejohn TG, Hanvivatvong O, Rowland SJ, Dyke KG, Skurray RA. 1994. Characterisation of sin, a potential recombinase-encoding gene from Staphylococcus aureus. Gene 141:109–114. doi: 10.1016/0378-1119(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 104.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol 180:4350–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rowland SJ, Stark WM, Boocock MR. 2002. Sin recombinase from Staphylococcus aureus: synaptic complex architecture and transposon targeting. Mol Microbiol 44:607–619. doi: 10.1046/j.1365-2958.2002.02897.x. [DOI] [PubMed] [Google Scholar]
- 106.Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist Updat 6:41–52. doi: 10.1016/S1368-7646(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 107.Hamidian M, Hall RM. 2011. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother 66:2484–2491. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 108.Rose A. 2010. TnAbaR1: a novel Tn7-related transposon in Acinetobacter baumannii that contributes to the accumulation and dissemination of large repertoires of resistance genes. Biosci Horiz 3:40–48. doi: 10.1093/biohorizons/hzq006. [DOI] [Google Scholar]
- 109.Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:1162–1170. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 110.Hamidian M, Hall RM. 2017. Origin of the AbGRI1 antibiotic resistance island found in the comM gene of Acinetobacter baumannii GC2 isolates. J Antimicrob Chemother 72:2944–2947. doi: 10.1093/jac/dkx206. [DOI] [PubMed] [Google Scholar]
- 111.Blackwell GA, Hamidian M, Hall RM. 2016. IncM plasmid R1215 is the source of chromosomally located regions containing multiple antibiotic resistance genes in the globally disseminated Acinetobacter baumannii GC1 and GC2 clones. mSphere 1:e00117-16. doi: 10.1128/mSphere.00117-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Escudero JA, Loot C, Nivina A, Mazel D. 2015. The integron: adaptation on demand. Microbiol Spectr 3:MDNA3-0019-2014. doi: 10.1128/microbiolspec.MDNA3-0019-2014. [DOI] [PubMed] [Google Scholar]
- 113.Hall RM. 2012. Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann N Y Acad Sci 1267:71–78. doi: 10.1111/j.1749-6632.2012.06588.x. [DOI] [PubMed] [Google Scholar]
- 114.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev 33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 115.Jove T, Da Re S, Tabesse A, Gassama-Sow A, Ploy MC. 2017. Gene expression in class 2 integrons is SOS-independent and involves two Pc promoters. Front Microbiol 8:1499. doi: 10.3389/fmicb.2017.01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jové T, Da Re S, Denis F, Mazel D, Ploy MC. 2010. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet 6:e1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Collis C, Hall R. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother 39:155–162. doi: 10.1128/AAC.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hanau-Berçot B, Podglajen I, Casin I, Collatz E. 2002. An intrinsic control element for translational initiation in class 1 integrons. Mol Microbiol 44:119–130. doi: 10.1046/j.1365-2958.2002.02843.x. [DOI] [PubMed] [Google Scholar]
- 119.Papagiannitsis CC, Tzouvelekis LS, Tzelepi E, Miriagou V. 2017. attI1-located small open reading frames ORF-17 and ORF-11 in a class 1 integron affect expression of a gene cassette possessing a canonical Shine-Dalgarno sequence. Antimicrob Agents Chemother 61:e02070-16. doi: 10.1128/AAC.02070-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, Stokes HW. 2008. The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol 190:5095–5100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Petrovski S, Stanisich VA. 2010. Tn502 and Tn512 are res site hunters that provide evidence of resolvase-independent transposition to random sites. J Bacteriol 192:1865–1874. doi: 10.1128/JB.01322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Di Pilato V, Pollini S, Rossolini GM. 2014. Characterization of plasmid pAX22, encoding VIM-1 metallo-β-lactamase, reveals a new putative mechanism of In70 integron mobilization. J Antimicrob Chemother 69:67–71. doi: 10.1093/jac/dkt311. [DOI] [PubMed] [Google Scholar]
- 123.Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 124.Ramirez MS, Pineiro S, Argentinian Integron Study Group, Centron D. 2010. Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob Agents Chemother 54:699–706. doi: 10.1128/AAC.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Collis CM, Kim MJ, Partridge SR, Stokes HW, Hall RM. 2002. Characterization of the class 3 integron and the site-specific recombination system it determines. J Bacteriol 184:3017–3026. doi: 10.1128/JB.184.11.3017-3026.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Toro N, Jimenez-Zurdo JI, Garcia-Rodriguez FM. 2007. Bacterial group II introns: not just splicing. FEMS Microbiol Rev 31:342–358. doi: 10.1111/j.1574-6976.2007.00068.x. [DOI] [PubMed] [Google Scholar]
- 127.Léon G, Roy PH. 2009. Potential role of group IIC-attC introns in integron cassette formation. J Bacteriol 191:6040–6051. doi: 10.1128/JB.00674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Quiroga C, Roy PH, Centron D. 2008. The S.ma.I2 class C group II intron inserts at integron attC sites. Microbiology 154:1341–1353. doi: 10.1099/mic.0.2007/016360-0. [DOI] [PubMed] [Google Scholar]
- 129.Candales MA, Duong A, Hood KS, Li T, Neufeld RA, Sun R, McNeil BA, Wu L, Jarding AM, Zimmerly S. 2012. Database for bacterial group II introns. Nucleic Acids Res 40:D187–D190. doi: 10.1093/nar/gkr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nesvera J, Hochmannova J, Patek M. 1998. An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol Lett 169:391–395. doi: 10.1111/j.1574-6968.1998.tb13345.x. [DOI] [PubMed] [Google Scholar]
- 131.Nandi S, Maurer JJ, Hofacre C, Summers AO. 2004. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc Natl Acad Sci U S A 101:7118–7122. doi: 10.1073/pnas.0306466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Clark NC, Olsvik O, Swenson JM, Spiegel CA, Tenover FC. 1999. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob Agents Chemother 43:157–160. doi: 10.1093/jac/43.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gillings MR, Labbate M, Sajjad A, Giguere NJ, Holley MP, Stokes HW. 2009. Mobilization of a Tn402-like class 1 integron with a novel cassette array via flanking miniature inverted-repeat transposable element-like structures. Appl Environ Microbiol 75:6002–6004. doi: 10.1128/AEM.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Delihas N. 2011. Impact of small repeat sequences on bacterial genome evolution. Genome Biol Evol 3:959–973. doi: 10.1093/gbe/evr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Szuplewska M, Ludwiczak M, Lyzwa K, Czarnecki J, Bartosik D. 2014. Mobility and generation of mosaic non-autonomous transposons by Tn3-derived inverted-repeat miniature elements (TIMEs). PLoS One 9:e105010. doi: 10.1371/journal.pone.0105010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Poirel L, Carrer A, Pitout JD, Nordmann P. 2009. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob Agents Chemother 53:2492–2498. doi: 10.1128/AAC.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zong Z. 2014. The complex genetic context of blaPER-1 flanked by miniature inverted-repeat transposable elements in Acinetobacter johnsonii. PLoS One 9:e90046. doi: 10.1371/journal.pone.0090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shintani M, Sanchez ZK, Kimbara K. 2015. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol 6:242. doi: 10.3389/fmicb.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Thomas CM. 2000. Paradigms of plasmid organization. Mol Microbiol 37:485–491. [DOI] [PubMed] [Google Scholar]
- 140.Lacroix B, Citovsky V. 2016. Transfer of DNA from bacteria to eukaryotes. mBio 7:e00863-16. doi: 10.1128/mBio.00863-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. 1998. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev 62:434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khan SA. 2005. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 144.Lilly J, Camps M. 2015. Mechanisms of theta plasmid replication. Microbiol Spectr 3: PLAS-0029-2014. doi: 10.1128/microbiolspec.PLAS-0029-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.del Solar G, Espinosa M. 2000. Plasmid copy number control: an ever-growing story. Mol Microbiol 37:492–500. doi: 10.1046/j.1365-2958.2000.02005.x. [DOI] [PubMed] [Google Scholar]
- 146.Brantl S. 2014. Plasmid replication control by antisense RNAs. Microbiol Spectr 2:PLAS-0001-2013. doi: 10.1128/microbiolspec.PLAS-0001-2013. [DOI] [PubMed] [Google Scholar]
- 147.Chattoraj DK. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol 37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 148.Konieczny I, Bury K, Wawrzycka A, Wegrzyn K. 2014. Iteron plasmids. Microbiol Spectr 2:PLAS-0026-2014. doi: 10.1128/microbiolspec.PLAS-0026-2014. [DOI] [PubMed] [Google Scholar]
- 149.Villa L, García-Fernández A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 150.Sengupta M, Austin S. 2011. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect Immun 79:2502–2509. doi: 10.1128/IAI.00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Crozat E, Fournes F, Cornet F, Hallet B, Rousseau P. 2014. Resolution of multimeric forms of circular plasmids and chromosomes. Microbiol Spectr 2:PLAS-0025-2014. doi: 10.1128/microbiolspec.PLAS-0025-2014. [DOI] [PubMed] [Google Scholar]
- 152.Baxter JC, Funnell BE. 2014. Plasmid partition mechanisms. Microbiol Spectr 2:PLAS-0023-2014. doi: 10.1128/microbiolspec.PLAS-0023-2014. [DOI] [PubMed] [Google Scholar]
- 153.Salje J. 2010. Plasmid segregation: how to survive as an extra piece of DNA. Crit Rev Biochem Mol Biol 45:296–317. doi: 10.3109/10409238.2010.494657. [DOI] [PubMed] [Google Scholar]
- 154.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 156.Mruk I, Kobayashi I. 2014. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res 42:70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guglielmini J, Neron B, Abby SS, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2014. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res 42:5715–5727. doi: 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gomis-Rüth FX, Solà M, de la Cruz F, Coll M. 2004. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des 10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- 160.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 161.Garcillán-Barcia MP, de la Cruz F. 2008. Why is entry exclusion an essential feature of conjugative plasmids? Plasmid 60:1–18. doi: 10.1016/j.plasmid.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 162.O'Brien FG, Ramsay JP, Monecke S, Coombs GW, Robinson OJ, Htet Z, Alshaikh FA, Grubb WB. 2015. Staphylococcus aureus plasmids without mobilization genes are mobilized by a novel conjugative plasmid from community isolates. J Antimicrob Chemother 70:649–652. doi: 10.1093/jac/dku454. [DOI] [PubMed] [Google Scholar]
- 163.O'Brien FG, Yui Eto K, Murphy RJ, Fairhurst HM, Coombs GW, Grubb WB, Ramsay JP. 2015. Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res 43:7971–7983. doi: 10.1093/nar/gkv755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pollet RM, Ingle JD, Hymes JP, Eakes TC, Eto KY, Kwong SM, Ramsay JP, Firth N, Redinbo MR. 2016. Processing of nonconjugative resistance plasmids by conjugation nicking enzyme of staphylococci. J Bacteriol 198:888–897. doi: 10.1128/JB.00832-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moran RA, Hall RM. 2017. Analysis of pCERC7, a small antibiotic resistance plasmid from a commensal ST131 Escherichia coli, defines a diverse group of plasmids that include various segments adjacent to a multimer resolution site and encode the same NikA relaxase accessory protein enabling mobilisation. Plasmid 89:42–48. doi: 10.1016/j.plasmid.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 166.Ramsay JP, Firth N. 2017. Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr Opin Microbiol 38:1–9. doi: 10.1016/j.mib.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 167.Couturier M, Bex F, Bergquist PL, Maas WK. 1988. Identification and classification of bacterial plasmids. Microbiol Rev 52:375–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 169.Carloni E, Andreoni F, Omiccioli E, Villa L, Magnani M, Carattoli A. 2017. Comparative analysis of the standard PCR-based replicon typing (PBRT) with the commercial PBRT-kit. Plasmid 90:10–14. doi: 10.1016/j.plasmid.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 170.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jensen LB, Garcia-Migura L, Valenzuela AJ, Lohr M, Hasman H, Aarestrup FM. 2010. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods 80:25–43. doi: 10.1016/j.mimet.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 172.Lozano C, Garcia-Migura L, Aspiroz C, Zarazaga M, Torres C, Aarestrup FM. 2012. Expansion of a plasmid classification system for Gram-positive bacteria and determination of the diversity of plasmids in Staphylococcus aureus strains of human, animal, and food origins. Appl Environ Microbiol 78:5948–5955. doi: 10.1128/AEM.00870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.McCarthy AJ, Lindsay JA. 2012. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol 12:104. doi: 10.1186/1471-2180-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Garcillán-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 175.Orlek A, Stoesser N, Anjum MF, Doumith M, Ellington MJ, Peto T, Crook D, Woodford N, Walker AS, Phan H, Sheppard AE. 2017. Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front Microbiol 8:182. doi: 10.3389/fmicb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alvarado A, Garcillan-Barcia MP, de la Cruz F. 2012. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One 7:e40438. doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Compain F, Poisson A, Le Hello S, Branger C, Weill FX, Arlet G, Decre D. 2014. Targeting relaxase genes for classification of the predominant plasmids in Enterobacteriaceae. Int J Med Microbiol 304:236–242. doi: 10.1016/j.ijmm.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 178.Orlek A, Phan H, Sheppard AE, Doumith M, Ellington M, Peto T, Crook D, Walker AS, Woodford N, Anjum MF, Stoesser N. 2017. Ordering the mob: insights into replicon and MOB typing schemes from analysis of a curated dataset of publicly available plasmids. Plasmid 91:42–52. doi: 10.1016/j.plasmid.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bousquet A, Henquet S, Compain F, Genel N, Arlet G, Decre D. 2015. Partition locus-based classification of selected plasmids in Klebsiella pneumoniae, Escherichia coli and Salmonella enterica spp: an additional tool. J Microbiol Methods 110:85–91. doi: 10.1016/j.mimet.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 180.Mnif B, Vimont S, Boyd A, Bourit E, Picard B, Branger C, Denamur E, Arlet G. 2010. Molecular characterization of addiction systems of plasmids encoding extended-spectrum β-lactamases in Escherichia coli. J Antimicrob Chemother 65:1599–1603. doi: 10.1093/jac/dkq181. [DOI] [PubMed] [Google Scholar]
- 181.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 182.Ambrose SJ, Harmer CJ, Hall RM. 2018. Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid 96–97:7–12. doi: 10.1016/j.plasmid.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 183.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 23 January 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 184.Carattoli A, Miriagou V, Bertini A, Loli A, Colinon C, Villa L, Whichard JM, Rossolini GM. 2006. Replicon typing of plasmids encoding resistance to newer β-lactams. Emerg Infect Dis 12:1145–1148. doi: 10.3201/eid1207.051555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Harmer CJ, Partridge SR, Hall RM. 2016. pDGO100, a type 1 IncC plasmid from 1981 carrying ARI-A and a Tn1696-like transposon in a novel integrating element. Plasmid 86:38–45. doi: 10.1016/j.plasmid.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 186.Harmer CJ, Hall RM. 2014. pRMH760, a precursor of A/C2 plasmids carrying blaCMY and blaNDM genes. Microb Drug Resist 20:416–423. doi: 10.1089/mdr.2014.0012. [DOI] [PubMed] [Google Scholar]
- 187.Carraro N, Matteau D, Burrus V, Rodrigue S. 2015. Unraveling the regulatory network of IncA/C plasmid mobilization: when genomic islands hijack conjugative elements. Mob Genet Elements 5:1–5. doi: 10.1080/2159256X.2015.1006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Harmer CJ, Hamidian M, Hall RM. 2017. pIP40a, a type 1 IncC plasmid from 1969 carries the integrative element GIsul2 and a novel class II mercury resistance transposon. Plasmid 92:17–25. doi: 10.1016/j.plasmid.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 189.Partridge SR, Hall RM. 2004. Complex multiple antibiotic and mercury resistance region derived from the r-det of NR1 (R100). Antimicrob Agents Chemother 48:4250–4255. doi: 10.1128/AAC.48.11.4250-4255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Partridge SR, Iredell JR. 2012. Genetic contexts of blaNDM-1. Antimicrob Agents Chemother 56:6065–6067. doi: 10.1128/AAC.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, Leclerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lang KS, Danzeisen JL, Xu W, Johnson TJ. 2012. Transcriptome mapping of pAR060302, a blaCMY-2-positive broad-host-range IncA/C plasmid. Appl Environ Microbiol 78:3379–3386. doi: 10.1128/AEM.07199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hancock SJ, Phan MD, Peters KM, Forde BM, Chong TM, Yin WF, Chan KG, Paterson DL, Walsh TR, Beatson SA, Schembri MA. 2017. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother 61:e01740-16. doi: 10.1128/AAC.01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Harmer CJ, Hall RM. 2016. PCR-based typing of IncC plasmids. Plasmid 87–88:37–42. doi: 10.1016/j.plasmid.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 195.Fernandez-Lopez R, de Toro M, Moncalian G, Garcillan-Barcia MP, de la Cruz F. 2016. Comparative genomics of the conjugation region of F-like plasmids: five shades of F. Front Mol Biosci 3:71. doi: 10.3389/fmolb.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Frost LS, Koraimann G. 2010. Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol 5:1057–1071. doi: 10.2217/fmb.10.70. [DOI] [PubMed] [Google Scholar]
- 197.Arutyunov D, Frost LS. 2013. F conjugation: back to the beginning. Plasmid 70:18–32. doi: 10.1016/j.plasmid.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 198.Glover JNM, Chaulk SG, Edwards RA, Arthur D, Lu J, Frost LS. 2015. The FinO family of bacterial RNA chaperones. Plasmid 78:79–87. doi: 10.1016/j.plasmid.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 199.de Toro M, Garcillaon-Barcia MP, de la Cruz F. 2014. Plasmid diversity and adaptation analyzed by massive sequencing of Escherichia coli plasmids. Microbiol Spectr 2:PLAS-0031-2014. doi: 10.1128/microbiolspec.PLAS-0031-2014. [DOI] [PubMed] [Google Scholar]
- 200.He L, Partridge SR, Yang X, Hou J, Deng Y, Yao Q, Zeng Z, Chen Z, Liu JH. 2013. Complete nucleotide sequence of pHN7A8, an F33:A−:B− type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J Antimicrob Chemother 68:46–50. doi: 10.1093/jac/dks369. [DOI] [PubMed] [Google Scholar]
- 201.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 202.Johnson TJ, Danzeisen JL, Youmans B, Case K, Llop K, Munoz-Aguayo J, Flores-Figueroa C, Aziz M, Stoesser N, Sokurenko E, Price LB, Johnson JR. 2016. Separate F-type plasmids have shaped the evolution of the H30 subclone of Escherichia coli sequence type 131. mSphere 1:e00121-16. doi: 10.1128/mSphere.00121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wright MS, Perez F, Brinkac L, Jacobs MR, Kaye K, Cober E, van Duin D, Marshall SH, Hujer AM, Rudin SD, Hujer KM, Bonomo RA, Adams MD. 2014. Population structure of KPC-producing Klebsiella pneumoniae isolates from midwestern U.S. hospitals. Antimicrob Agents Chemother 58:4961–4965. doi: 10.1128/AAC.00125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gilmour MW, Thomson NR, Sanders M, Parkhill J, Taylor DE. 2004. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52:182–202. doi: 10.1016/j.plasmid.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 206.Alonso G, Vilchez G, Bruzual I, Rodriguez-Lemoine V. 2002. Characterization of plasmid MIP233 (IncHI3) of the H complex. Res Microbiol 153:149–153. doi: 10.1016/S0923-2508(02)01300-1. [DOI] [PubMed] [Google Scholar]
- 207.Sherburne CK, Lawley TD, Gilmour MW, Blattner FR, Burland V, Grotbeck E, Rose DJ, Taylor DE. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res 28:2177–2186. doi: 10.1093/nar/28.10.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cain AK, Hall RM. 2013. Evolution of IncHI1 plasmids: two distinct lineages. Plasmid 70:201–208. doi: 10.1016/j.plasmid.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 209.Taylor DE. 2009. Thermosensitive nature of IncHI1 plasmid transfer. Antimicrob Agents Chemother 53:2703. doi: 10.1128/AAC.00230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Phan MD, Wain J. 2008. IncHI plasmids, a dynamic link between resistance and pathogenicity. J Infect Dev Ctries 2:272–278. [DOI] [PubMed] [Google Scholar]
- 211.Phan MD, Kidgell C, Nair S, Holt KE, Turner AK, Hinds J, Butcher P, Cooke FJ, Thomson NR, Titball R, Bhutta ZA, Hasan R, Dougan G, Wain J. 2009. Variation in Salmonella enterica serovar Typhi IncHI1 plasmids during the global spread of resistant typhoid fever. Antimicrob Agents Chemother 53:716–727. doi: 10.1128/AAC.00645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kubasova T, Cejkova D, Matiasovicova J, Sekelova Z, Polansky O, Medvecky M, Rychlik I, Juricova H. 2016. Antibiotic resistance, core-genome and protein expression in IncHI1 plasmids in Salmonella Typhimurium. Genome Biol Evol 8:1661–1671. doi: 10.1093/gbe/evw105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Garcia-Fernandez A, Carattoli A. 2010. Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum β-lactamase and quinolone resistance genes. J Antimicrob Chemother 65:1155–1161. doi: 10.1093/jac/dkq101. [DOI] [PubMed] [Google Scholar]
- 214.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. 2012. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother 67:1645–1650. doi: 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 215.Dolejska M, Villa L, Poirel L, Nordmann P, Carattoli A. 2013. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J Antimicrob Chemother 68:34–39. doi: 10.1093/jac/dks357. [DOI] [PubMed] [Google Scholar]
- 216.Liang Q, Yin Z, Zhao Y, Liang L, Feng J, Zhan Z, Wang H, Song Y, Tong Y, Wu W, Chen W, Wang J, Jiang L, Zhou D. 2017. Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int J Antimicrob Agents 49:709–718. doi: 10.1016/j.ijantimicag.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 217.Zurfluh K, Klumpp J, Nuesch-Inderbinen M, Stephan R. 2016. Full-length nucleotide sequences of mcr-1-harboring plasmids isolated from extended-spectrum-β-lactamase-producing Escherichia coli isolates of different origins. Antimicrob Agents Chemother 60:5589–5591. doi: 10.1128/AAC.00935-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yu CY, Ang GY, Chong TM, Chin PS, Ngeow YF, Yin WF, Chan KG. 2017. Complete genome sequencing revealed novel genetic contexts of the mcr-1 gene in Escherichia coli strains. J Antimicrob Chemother 72:1253–1255. doi: 10.1093/jac/dkw541. [DOI] [PubMed] [Google Scholar]
- 219.Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, Zheng Z, Chan EW, Chen S. 2017. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 220.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Praszkier J, Pittard AJ. 2005. Control of replication in I-complex plasmids. Plasmid 53:97–112. doi: 10.1016/j.plasmid.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 222.Moran RA, Anantham S, Pinyon JL, Hall RM. 2015. Plasmids in antibiotic susceptible and antibiotic resistant commensal Escherichia coli from healthy Australian adults. Plasmid 80:24–31. doi: 10.1016/j.plasmid.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 223.Rozwandowicz M, Brouwer MS, Zomer AL, Bossers A, Harders F, Mevius DJ, Wagenaar JA, Hordijk J. 2017. Plasmids of distinct IncK lineages show compatible phenotypes. Antimicrob Agents Chemother 61:e01954-16. doi: 10.1128/AAC.01954-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Seiffert SN, Carattoli A, Schwendener S, Collaud A, Endimiani A, Perreten V. 2017. Plasmids carrying blaCMY-2/4 in Escherichia coli from poultry, poultry meat, and humans belong to a novel IncK subgroup designated IncK2. Front Microbiol 8:407. doi: 10.3389/fmicb.2017.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Garcia-Fernandez A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, Carattoli A. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother 61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 226.Bradley DE. 1984. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K and Z. J Gen Microbiol 130:1489–1502. [DOI] [PubMed] [Google Scholar]
- 227.Komano T. 1999. Shufflons: multiple inversion systems and integrons. Annu Rev Genet 33:171–191. doi: 10.1146/annurev.genet.33.1.171. [DOI] [PubMed] [Google Scholar]
- 228.Takahashi H, Shao M, Furuya N, Komano T. 2011. The genome sequence of the incompatibility group Iγ plasmid R621a: evolution of IncI plasmids. Plasmid 66:112–121. doi: 10.1016/j.plasmid.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 229.Venturini C, Hassan KA, Roy Chowdhury P, Paulsen IT, Walker MJ, Djordjevic SP.. 2013. Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS One 8:e78862. doi: 10.1371/journal.pone.0078862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Johnson TJ, Shepard SM, Rivet B, Danzeisen JL, Carattoli A. 2011. Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid 66:144–151. doi: 10.1016/j.plasmid.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 231.Cottell JL, Webber MA, Coldham NG, Taylor DL, Cerdeno-Tarraga AM, Hauser H, Thomson NR, Woodward MJ, Piddock LJ. 2011. Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerg Infect Dis 17:645–652. doi: 10.3201/eid1704.101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Dona V, Bernasconi OJ, Pires J, Collaud A, Overesch G, Ramette A, Perreten V, Endimiani A. 2017. Heterogeneous genetic location of mcr-1 in colistin-resistant Escherichia coli isolated from humans and retail chicken meat in Switzerland: emergence of mcr-1-carrying IncK2 plasmids. Antimicrob Agents Chemother 61:e01245-17. doi: 10.1128/AAC.01245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kim SR, Komano T. 1992. Nucleotide sequence of the R721 shufflon. J Bacteriol 174:7053–7058. doi: 10.1128/jb.174.21.7053-7058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sekizuka T, Kawanishi M, Ohnishi M, Shima A, Kato K, Yamashita A, Matsui M, Suzuki S, Kuroda M. 2017. Elucidation of quantitative structural diversity of remarkable rearrangement regions, shufflons, in IncI2 plasmids. Sci Rep 7:928. doi: 10.1038/s41598-017-01082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lv L, Partridge SR, He L, Zeng Z, He D, Ye J, Liu JH. 2013. Genetic characterization of IncI2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China. Antimicrob Agents Chemother 57:2824–2827. doi: 10.1128/AAC.02155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Liu L, He D, Lv L, Liu W, Chen X, Zeng Z, Partridge SR, Liu JH. 2015. blaCTX-M-1/9/1 hybrid genes may have been generated from blaCTX-M-15 on an IncI2 plasmid. Antimicrob Agents Chemother 59:4464–4470. doi: 10.1128/AAC.00501-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ellem JA, Ginn AN, Chen SC, Ferguson J, Partridge SR, Iredell JR. 2017. Locally acquired mcr-1 in Escherichia coli, Australia, 2011 and 2013. Emerg Infect Dis 23:1160–1163. doi: 10.3201/eid2307.161638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mierzejewska J, Kulinska A, Jagura-Burdzy G. 2007. Functional analysis of replication and stability regions of broad-host-range conjugative plasmid CTX-M3 from the IncL/M incompatibility group. Plasmid 57:95–107. doi: 10.1016/j.plasmid.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 239.Gołêbiewski M, Kern-Zdanowicz I, Zienkiewicz M, Adamczyk M, Żyliǹska J, Baraniak A, Gniadkowski M, Bardowski J, Cegłowski P. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob Agents Chemother 51:3789–3795. doi: 10.1128/AAC.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hedges RW, Datta N, Kontomichalou P, Smith JT. 1974. Molecular specificities of R factor-determined β-lactamases: correlation with plasmid compatibility. J Bacteriol 117:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Richards H, Datta N. 1979. Reclassification of incompatibility group L (IncL) plasmids. Plasmid 2:293–295. doi: 10.1016/0147-619X(79)90048-9. [DOI] [PubMed] [Google Scholar]
- 242.Carattoli A, Seiffert SN, Schwendener S, Perreten V, Endimiani A. 2015. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. doi: 10.1371/journal.pone.0123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Adamczuk M, Zaleski P, Dziewit L, Wolinowska R, Nieckarz M, Wawrzyniak P, Kieryl P, Plucienniczak A, Bartosik D. 2015. Diversity and global distribution of IncL/M plasmids enabling horizontal dissemination of β-lactam resistance genes among the Enterobacteriaceae. Biomed Res Int 2015:414681. doi: 10.1155/2015/414681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Potron A, Poirel L, Nordmann P. 2014. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob Agents Chemother 58:467–471. doi: 10.1128/AAC.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Partridge SR, Ginn AN, Paulsen IT, Iredell JR. 2012. pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother 56:6029–6032. doi: 10.1128/AAC.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Santini JM, Stanisich VA. 1998. Both the fipA gene of pKM101 and the pifC gene of F inhibit conjugal transfer of RP1 by an effect on traG. J Bacteriol 180:4093–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Delver EP, Belogurov AA. 1997. Organization of the leading region of IncN plasmid pKM101 (R46): a regulon controlled by CUP sequence elements. J Mol Biol 271:13–30. doi: 10.1006/jmbi.1997.1124. [DOI] [PubMed] [Google Scholar]
- 248.Garcia-Fernandez A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, Carattoli A. 2011. Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother 66:1987–1991. doi: 10.1093/jac/dkr225. [DOI] [PubMed] [Google Scholar]
- 249.Poirel L, Bonnin RA, Nordmann P. 2011. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli by high-throughput genome sequencing. Antimicrob Agents Chemother 55:4224–4229. doi: 10.1128/AAC.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Partridge SR, Paulsen IT, Iredell JR. 2012. pJIE137 carrying blaCTX-M-62 is closely related to p271A carrying blaNDM-1. Antimicrob Agents Chemother 56:2166–2168. doi: 10.1128/AAC.05796-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Jiang X, Yin Z, Yin X, Fang H, Sun Q, Tong Y, Xu Y, Zhang D, Feng J, Chen W, Song Y, Wang J, Chen S, Zhou D. 2017. Sequencing of blaIMP-carrying IncN2 plasmids, and comparative genomics of IncN2 plasmids harboring class 1 integrons. Front Cell Infect Microbiol 7:102. doi: 10.3389/fcimb.2017.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Humphrey B, Thomson NR, Thomas CM, Brooks K, Sanders M, Delsol AA, Roe JM, Bennett PM, Enne VI. 2012. Fitness of Escherichia coli strains carrying expressed and partially silent IncN and IncP1 plasmids. BMC Microbiol 12:53. doi: 10.1186/1471-2180-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Villa L, Carattoli A, Nordmann P, Carta C, Poirel L. 2013. Complete sequence of the IncT-type plasmid pT-OXA-181 carrying the blaOXA-181 carbapenemase gene from Citrobacter freundii. Antimicrob Agents Chemother 57:1965–1967. doi: 10.1128/AAC.01297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fernandez-Lopez R, Redondo S, Garcillan-Barcia MP, de la Cruz F. 2017. Towards a taxonomy of conjugative plasmids. Curr Opin Microbiol 38:106–113. doi: 10.1016/j.mib.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 255.Adamczyk M, Jagura-Burdzy G. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim Pol 50:425–453. [PubMed] [Google Scholar]
- 256.Pansegrau W, Lanka E, Barth PT, Figurski DH, Guiney DG, Haas D, Helinski DR, Schwab H, Stanisich VA, Thomas CM. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids. Compilation and comparative analysis. J Mol Biol 239:623–663. [DOI] [PubMed] [Google Scholar]
- 257.Thorsted PB, Macartney DP, Akhtar P, Haines AS, Ali N, Davidson P, Stafford T, Pocklington MJ, Pansegrau W, Wilkins BM, Lanka E, Thomas CM. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J Mol Biol 282:969–990. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- 258.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 259.Norberg P, Bergstrom M, Jethava V, Dubhashi D, Hermansson M. 2011. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat Commun 2:268. doi: 10.1038/ncomms1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sen D, Brown CJ, Top EM, Sullivan J. 2013. Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees. Mol Biol Evol 30:154–166. doi: 10.1093/molbev/mss210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pachulec E, van der Does C. 2010. Conjugative plasmids of Neisseria gonorrhoeae. PLoS One 5:e9962. doi: 10.1371/journal.pone.0009962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zhao F, Feng Y, Lu X, McNally A, Zong Z. 2017. IncP plasmid carrying colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob Agents Chemother 61:e02229-16. doi: 10.1128/AAC.02229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Popowska M, Krawczyk-Balska A. 2013. Broad-host-range IncP-1 plasmids and their resistance potential. Front Microbiol 4:44. doi: 10.3389/fmicb.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lu X, Hu Y, Luo M, Zhou H, Wang X, Du Y, Li Z, Xu J, Zhu B, Xu X, Kan B. 2017. MCR-1.6, a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica serovar Typhimurium isolate from a healthy individual. Antimicrob Agents Chemother 61:e02632-16. doi: 10.1128/AAC.02632-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Garcia-Fernandez A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 266.Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, Yan JJ, Su IJ, Tsai SF, Lauderdale TL. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 50:3861–3866. doi: 10.1128/AAC.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Compain F, Frangeul L, Drieux L, Verdet C, Brisse S, Arlet G, Decre D. 2014. Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob Agents Chemother 58:4207–4210. doi: 10.1128/AAC.02773-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Guo Q, Spychala CN, McElheny CL, Doi Y. 2016. Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J Antimicrob Chemother 71:882–886. doi: 10.1093/jac/dkv444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.da Silva-Tatley FM, Steyn LM. 1993. Characterization of a replicon of the moderately promiscuous plasmid, pGSH5000, with features of both the mini-replicon of pCU1 and the ori-2 of F. Mol Microbiol 7:805–823. doi: 10.1111/j.1365-2958.1993.tb01171.x. [DOI] [PubMed] [Google Scholar]
- 270.Khong WX, Marimuthu K, Teo J, Ding Y, Xia E, Lee JJ, Ong RT, Venkatachalam I, Cherng B, Pada SK, Choong WL, Smitasin N, Ooi ST, Deepak RN, Kurup A, Fong R, Van La M, Tan TY, Koh TH, Lin RT, Tan EL, Krishnan PU, Singh S, Pitout JD, Teo YY, Yang L, Ng OT, Carbapenemase-Producing Enterobacteriaceae in Singapore Study Group. 2016. Tracking inter-institutional spread of NDM and identification of a novel NDM-positive plasmid, pSg1-NDM, using next-generation sequencing approaches. J Antimicrob Chemother 71:3081–3089. doi: 10.1093/jac/dkw277. [DOI] [PubMed] [Google Scholar]
- 271.Murata T, Ohnishi M, Ara T, Kaneko J, Han CG, Li YF, Takashima K, Nojima H, Nakayama K, Kaji A, Kamio Y, Miki T, Mori H, Ohtsubo E, Terawaki Y, Hayashi T. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J Bacteriol 184:3194–3202. doi: 10.1128/JB.184.12.3194-3202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bradley DE, Whelan J. 1985. Conjugation systems of IncT plasmids. J Gen Microbiol 131:2665–2671. [DOI] [PubMed] [Google Scholar]
- 273.Kato K, Matsumura Y, Yamamoto M, Nagao M, Takakura S, Ichiyama S. 2017. Regional spread of CTX-M-2-producing Proteus mirabilis with the identical genetic structure in Japan. Microb Drug Resist 23:590–595. doi: 10.1089/mdr.2016.0148. [DOI] [PubMed] [Google Scholar]
- 274.Mataseje LF, Peirano G, Church DL, Conly J, Mulvey M, Pitout JD. 2016. Colistin-nonsusceptible Pseudomonas aeruginosa sequence type 654 with blaNDM-1 arrives in North America. Antimicrob Agents Chemother 60:1794–1800. doi: 10.1128/AAC.02591-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Haines AS, Cheung M, Thomas CM. 2006. Evidence that IncG (IncP-6) and IncU plasmids form a single incompatibility group. Plasmid 55:210–215. doi: 10.1016/j.plasmid.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 276.Haines AS, Jones K, Cheung M, Thomas CM. 2005. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J Bacteriol 187:4728–4738. doi: 10.1128/JB.187.14.4728-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Naas T, Bonnin RA, Cuzon G, Villegas MV, Nordmann P. 2013. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J Antimicrob Chemother 68:1757–1762. doi: 10.1093/jac/dkt094. [DOI] [PubMed] [Google Scholar]
- 278.Dai X, Zhou D, Xiong W, Feng J, Luo W, Luo G, Wang H, Sun F, Zhou X. 2016. The IncP-6 plasmid p10265-KPC from Pseudomonas aeruginosa carries a novel ΔISEc33-associated blaKPC-2 gene cluster. Front Microbiol 7:310. doi: 10.3389/fmicb.2016.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Fernández-López R, Garcillán-Barcia MP, Revilla C, Lázaro M, Vielva L, de la Cruz F. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol Rev 30:942–966. doi: 10.1111/j.1574-6976.2006.00042.x. [DOI] [PubMed] [Google Scholar]
- 280.Revilla C, Garcillan-Barcia MP, Fernandez-Lopez R, Thomson NR, Sanders M, Cheung M, Thomas CM, de la Cruz F. 2008. Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob Agents Chemother 52:1472–1480. doi: 10.1128/AAC.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Fernandez-Lopez R, de la Cruz F. 2014. Rebooting the genome: the role of negative feedback in horizontal gene transfer. Mob Genet Elements 4:1–6. doi: 10.4161/2159256X.2014.988069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Aoki K, Harada S, Yahara K, Ishii Y, Motooka D, Nakamura S, Akeda Y, Iida T, Tomono K, Iwata S, Moriya K, Tateda K. 2018. Molecular characterization of IMP-1-producing Enterobacter cloacae complex isolates in Tokyo. Antimicrob Agents Chemother 62:e02091-17. doi: 10.1128/AAC.02091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Miriagou V, Douzinas EE, Papagiannitsis CC, Piperaki E, Legakis NJ, Tzouvelekis LS. 2008. Emergence of Serratia liquefaciens and Klebsiella oxytoca with metallo-β-lactamase-encoding IncW plasmids: further spread of the blaVIM-1-carrying integron In-e541. Int J Antimicrob Agents 32:540–541. doi: 10.1016/j.ijantimicag.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 284.Shevchenko OV, Mudrak DY, Skleenova EY, Kozyreva VK, Ilina EN, Ikryannikova LN, Alexandrova IA, Sidorenko SV, Edelstein MV. 2012. First detection of VIM-4 metallo-β-lactamase-producing Escherichia coli in Russia. Clin Microbiol Infect 18:E214–E217. doi: 10.1111/j.1469-0691.2012.03827.x. [DOI] [PubMed] [Google Scholar]
- 285.Almeida AC, de Sa Cavalcanti FL, Vilela MA, Gales AC, de Morais MA Jr, Camargo de Morais MM. 2012. Escherichia coli ST502 and Klebsiella pneumoniae ST11 sharing an IncW plasmid harbouring the blaKPC-2 gene in an intensive care unit patient. Int J Antimicrob Agents 40:374–376. doi: 10.1016/j.ijantimicag.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 286.Thomas CM, Thomson NR, Cerdeno-Tarraga AM, Brown CJ, Top EM, Frost LS. 2017. Annotation of plasmid genes. Plasmid 91:61–67. doi: 10.1016/j.plasmid.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 287.Rakowski SA, Filutowicz M. 2013. Plasmid R6K replication control. Plasmid 69:231–242. doi: 10.1016/j.plasmid.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bustamante P, Iredell JR. 2017. Carriage of type II toxin-antitoxin systems by the growing group of IncX plasmids. Plasmid 91:19–27. doi: 10.1016/j.plasmid.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 289.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 290.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Du H, Chen L, Chavda KD, Pandey R, Zhang H, Xie X, Tang YW, Kreiswirth BN. 2016. Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 carbapenemases. Antimicrob Agents Chemother 60:2519–2523. doi: 10.1128/AAC.03053-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Sun J, Yang RS, Zhang Q, Feng Y, Fang LX, Xia J, Li L, Lv XY, Duan JH, Liao XP, Liu YH. 2016. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol 1:16176. doi: 10.1038/nmicrobiol.2016.176. [DOI] [PubMed] [Google Scholar]
- 293.Guo Q, Su J, McElheny CL, Stoesser N, Doi Y, Wang M. 2017. IncX2 and IncX1-X2 hybrid plasmids coexisting in FosA6-producing Escherichia coli. Antimicrob Agents Chemother 61:e00536-17. doi: 10.1128/AAC.00536-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Dobiasova H, Dolejska M. 2016. Prevalence and diversity of IncX plasmids carrying fluoroquinolone and β-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. J Antimicrob Chemother 71:2118–2124. doi: 10.1093/jac/dkw144. [DOI] [PubMed] [Google Scholar]
- 295.Liu Y, Feng Y, Wu W, Xie Y, Wang X, Zhang X, Chen X, Zong Z. 2015. First report of OXA-181-producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrob Agents Chemother 59:5022–5025. doi: 10.1128/AAC.00442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Espedido BA, Dimitrijovski B, van Hal SJ, Jensen SO. 2015. The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: identifying the role of IncX3 plasmids and the spread of blaNDM-4-like genes in the Enterobacteriaceae. J Clin Pathol 68:835–838. doi: 10.1136/jclinpath-2015-203044. [DOI] [PubMed] [Google Scholar]
- 297.Kassis-Chikhani N, Frangeul L, Drieux L, Sengelin C, Jarlier V, Brisse S, Arlet G, Decre D. 2013. Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid pKpS90 from Klebsiella pneumoniae. Antimicrob Agents Chemother 57:618–620. doi: 10.1128/AAC.01712-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Partridge SR, Ellem JA, Tetu SG, Zong Z, Paulsen IT, Iredell JR. 2011. Complete sequence of pJIE143, a pir-type plasmid carrying ISEcp1-blaCTX-M-15 from an Escherichia coli ST131 isolate. Antimicrob Agents Chemother 55:5933–5935. doi: 10.1128/AAC.00639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Stokes MO, Abuoun M, Umur S, Wu G, Partridge SR, Mevius DJ, Coldham NG, Fielder MD. 2013. Complete sequence of pSAM7, an IncX4 plasmid carrying a novel blaCTX-M-14b transposition unit isolated from Escherichia coli and Enterobacter cloacae from cattle. Antimicrob Agents Chemother 57:4590–4594. doi: 10.1128/AAC.01157-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Matamoros S, van Hattem JM, Arcilla MS, Willemse N, Melles DC, Penders J, Vinh TN, Thi Hoa N, COMBAT Consortium, de Jong MD, Schultsz C. 2017. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep 7:15364. doi: 10.1038/s41598-017-15539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 302.Lobocka MB, Rose DJ, Plunkett G III, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. 2004. Genome of bacteriophage P1. J Bacteriol 186:7032–7068. doi: 10.1128/JB.186.21.7032-7068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Yarmolinsky MB. 2004. Bacteriophage P1 in retrospect and in prospect. J Bacteriol 186:7025–7028. doi: 10.1128/JB.186.21.7025-7028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Shin J, Ko KS. 2015. A plasmid bearing the blaCTX-M-15 gene and phage P1-like sequences from a sequence type 11 Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6608–6610. doi: 10.1128/AAC.00265-15. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 305.Zhang C, Feng Y, Liu F, Jiang H, Qu Z, Lei M, Wang J, Zhang B, Hu Y, Ding J, Zhu B. 2017. A phage-like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob Agents Chemother 61:e02035-16. doi: 10.1128/AAC.02035-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Bai L, Wang J, Hurley D, Yu Z, Wang L, Chen Q, Li J, Li F, Fanning S. 2017. A novel disrupted mcr-1 gene and a lysogenized phage P1-like sequence detected from a large conjugative plasmid, cultured from a human atypical enteropathogenic Escherichia coli (aEPEC) recovered in China. J Antimicrob Chemother 72:1531–1533. doi: 10.1093/jac/dkw564. [DOI] [PubMed] [Google Scholar]
- 307.Loftie-Eaton W, Rawlings DE. 2012. Diversity, biology and evolution of IncQ-family plasmids. Plasmid 67:15–34. doi: 10.1016/j.plasmid.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 308.Meyer R. 2009. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rawlings DE, Tietze E. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev 65:481–496. doi: 10.1128/MMBR.65.4.481-496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Cain AK, Liu X, Djordjevic SP, Hall RM. 2010. Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb Drug Resist 16:197–202. doi: 10.1089/mdr.2010.0042. [DOI] [PubMed] [Google Scholar]
- 311.Francia MV, Varsaki A, Garcillán-Barcia MP, Latorre A, Drainas C, de la Cruz F. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev 28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 312.Pallecchi L, Riccobono E, Sennati S, Mantella A, Bartalesi F, Trigoso C, Gotuzzo E, Bartoloni A, Rossolini GM. 2010. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob Agents Chemother 54:678–682. doi: 10.1128/AAC.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Ramirez MS, Traglia GM, Lin DL, Tran T, Tolmasky ME. 2014. Plasmid-mediated antibiotic resistance and virulence in Gram-negatives: the Klebsiella pneumoniae paradigm. Microbiol Spectr 2:PLAS-0016-2013. doi: 10.1128/microbiolspec.PLAS-0016-2013. [DOI] [PubMed] [Google Scholar]
- 314.Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob Agents Chemother 53:1998–2004. doi: 10.1128/AAC.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Cannon PM, Strike P. 1992. Complete nucleotide sequence and gene organization of plasmid NTP16. Plasmid 27:220–230. doi: 10.1016/0147-619X(92)90024-5. [DOI] [PubMed] [Google Scholar]
- 316.Xiong J, Alexander DC, Ma JH, Deraspe M, Low DE, Jamieson FB, Roy PH. 2013. Complete sequence of pOZ176, a 500-kilobase IncP-2 plasmid encoding IMP-9-mediated carbapenem resistance, from outbreak isolate Pseudomonas aeruginosa 96. Antimicrob Agents Chemother 57:3775–3782. doi: 10.1128/AAC.00423-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Jacoby GA, Sutton L, Knobel L, Mammen P. 1983. Properties of IncP-2 plasmids of Pseudomonas spp. Antimicrob Agents Chemother 24:168–175. doi: 10.1128/AAC.24.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Botelho J, Grosso F, Quinteira S, Mabrouk A, Peixe L. 2017. The complete nucleotide sequence of an IncP-2 megaplasmid unveils a mosaic architecture comprising a putative novel blaVIM-2-harbouring transposon in Pseudomonas aeruginosa. J Antimicrob Chemother 72:2225–2229. doi: 10.1093/jac/dkx143. [DOI] [PubMed] [Google Scholar]
- 319.Liu J, Yang L, Chen D, Peters BM, Li L, Li B, Xu Z, Shirtliff ME. 2018. Complete sequence of pBM413, a novel multidrug resistance megaplasmid carrying qnrVC6 and blaIMP-45 from Pseudomonas aeruginosa. Int J Antimicrob Agents 51:145–150. doi: 10.1016/j.ijantimicag.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 320.Sun F, Zhou D, Wang Q, Feng J, Feng W, Luo W, Zhang D, Liu Y, Qiu X, Yin Z, Chen W, Xia P. 2016. The first report of detecting the blaSIM-2 gene and determining the complete sequence of the SIM-encoding plasmid. Clin Microbiol Infect 22:347–351. doi: 10.1016/j.cmi.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 321.Shi L, Liang Q, Feng J, Zhan Z, Zhao Y, Yang W, Yang H, Chen Y, Huang M, Tong Y, Li X, Yin Z, Wang J, Zhou D. 4 October 2017. Coexistence of two novel resistance plasmids, blaKPC-2-carrying p14057A and tetA(A)-carrying p14057B, in Pseudomonas aeruginosa. Virulence doi: 10.1080/21505594.2017.1372082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bonnin RA, Poirel L, Nordmann P, Eikmeyer FG, Wibberg D, Puhler A, Schluter A. 2013. Complete sequence of broad-host-range plasmid pNOR-2000 harbouring the metallo-β-lactamase gene blaVIM-2 from Pseudomonas aeruginosa. J Antimicrob Chemother 68:1060–1065. doi: 10.1093/jac/dks526. [DOI] [PubMed] [Google Scholar]
- 323.San Millan A, Toll-Riera M, Escudero JA, Canton R, Coque TM, MacLean RC. 2015. Sequencing of plasmids pAMBL1 and pAMBL2 from Pseudomonas aeruginosa reveals a blaVIM-1 amplification causing high-level carbapenem resistance. J Antimicrob Chemother 70:3000–3003. doi: 10.1093/jac/dkv222. [DOI] [PubMed] [Google Scholar]
- 324.Botelho J, Grosso F, Peixe L. 2017. Characterization of the pJB12 plasmid from Pseudomonas aeruginosa reveals Tn6352, a novel putative transposon associated with mobilization of the blaVIM-2-harboring In58 integron. Antimicrob Agents Chemother 61:e02532-16. doi: 10.1128/AAC.02532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Klockgether J, Reva O, Larbig K, Tummler B. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol 186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Vilacoba E, Quiroga C, Pistorio M, Famiglietti A, Rodriguez H, Kovensky J, Deraspe M, Raymond F, Roy PH, Centron D. 2014. A blaVIM-2 plasmid disseminating in extensively drug-resistant clinical Pseudomonas aeruginosa and Serratia marcescens isolates. Antimicrob Agents Chemother 58:7017–7018. doi: 10.1128/AAC.02934-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Li H, Toleman MA, Bennett PM, Jones RN, Walsh TR. 2008. Complete sequence of p07-406, a 24,179-base-pair plasmid harboring the blaVIM-7 metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob Agents Chemother 52:3099–3105. doi: 10.1128/AAC.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. 2010. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Lean SS, Yeo CC. 2017. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: good, bad, who knows? Front Microbiol 8:1547. doi: 10.3389/fmicb.2017.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Towner KJ, Evans B, Villa L, Levi K, Hamouda A, Amyes SG, Carattoli A. 2011. Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D β-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 55:2154–2159. doi: 10.1128/AAC.01661-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Seifert H, Boullion B, Schulze A, Pulverer G. 1994. Plasmid DNA profiles of Acinetobacter baumannii: clinical application in a complex endemic setting. Infect Control Hosp Epidemiol 15:520–528. [DOI] [PubMed] [Google Scholar]
- 332.Hamidian M, Nigro SJ, Hall RM. 2012. Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J Antimicrob Chemother 67:2833–2836. doi: 10.1093/jac/dks318. [DOI] [PubMed] [Google Scholar]
- 333.Hamidian M, Holt KE, Pickard D, Dougan G, Hall RM. 2014. A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J Antimicrob Chemother 69:955–958. doi: 10.1093/jac/dkt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Hamidian M, Hall RM. 2014. pACICU2 is a conjugative plasmid of Acinetobacter carrying the aminoglycoside resistance transposon TnaphA6. J Antimicrob Chemother 69:1146–1148. doi: 10.1093/jac/dkt488. [DOI] [PubMed] [Google Scholar]
- 335.Hamidian M, Kenyon JJ, Holt KE, Pickard D, Hall RM. 2014. A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J Antimicrob Chemother 69:2625–2628. doi: 10.1093/jac/dku188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. A novel plasmid and its variant harboring both blaNDM-1 gene and T4SS in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Chen Z, Li H, Feng J, Li Y, Chen X, Guo X, Chen W, Wang L, Lin L, Yang H, Yang W, Wang J, Zhou D, Liu C, Yin Z. 2015. NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front Microbiol 6:294. doi: 10.3389/fmicb.2015.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O'Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 1:581–591. doi: 10.1534/g3.111.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Malachowa N, DeLeo FR. 2010. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci 67:3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Khan SA. 1997. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev 61:442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Mojumdar M, Khan SA. 1988. Characterization of the tetracycline resistance gene of plasmid pT181 of Staphylococcus aureus. J Bacteriol 170:5522–5528. doi: 10.1128/jb.170.12.5522-5528.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Horinouchi S, Weisblum B. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol 150:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Horinouchi S, Weisblum B. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol 150:804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Walters JA, Dyke KG. 1990. Characterization of a small cryptic plasmid isolated from a methicillin-resistant strain of Staphylococcus aureus. FEMS Microbiol Lett 59:55–63. doi: 10.1111/j.1574-6968.1990.tb03798.x. [DOI] [PubMed] [Google Scholar]
- 345.Projan SJ, Novick R. 1988. Comparative analysis of five related staphylococcal plasmids. Plasmid 19:203–221. doi: 10.1016/0147-619X(88)90039-X. [DOI] [PubMed] [Google Scholar]
- 346.Projan SJ, Moghazeh S, Novick RP. 1988. Nucleotide sequence of pS194, a streptomycin-resistance plasmid from Staphylococcus aureus. Nucleic Acids Res 16:2179–2187. doi: 10.1093/nar/16.5.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Brisson-Noel A, Courvalin P. 1986. Nucleotide sequence of gene linA encoding resistance to lincosamides in Staphylococcus haemolyticus. Gene 43:247–253. doi: 10.1016/0378-1119(86)90213-1. [DOI] [PubMed] [Google Scholar]
- 348.Zilhao R, Courvalin P. 1990. Nucleotide sequence of the fosB gene conferring fosfomycin resistance in Staphylococcus epidermidis. FEMS Microbiol Lett 56:267–272. [DOI] [PubMed] [Google Scholar]
- 349.Littlejohn TG, DiBerardino D, Messerotti LJ, Spiers SJ, Skurray RA. 1991. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene 101:59–66. doi: 10.1016/0378-1119(91)90224-Y. [DOI] [PubMed] [Google Scholar]
- 350.McKenzie T, Hoshino T, Tanaka T, Sueoka N. 1986. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid 15:93–103. doi: 10.1016/0147-619X(86)90046-6. [DOI] [PubMed] [Google Scholar]
- 351.Projan SJ, Archer GL. 1989. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol 171:1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Caryl JA, Smith MCA, Thomas CD. 2004. Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro. J Bacteriol 186:3374–3383. doi: 10.1128/JB.186.11.3374-3383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Gennaro ML, Kornblum J, Novick RP. 1987. A site-specific recombination function in Staphylococcus aureus plasmids. J Bacteriol 169:2601–2610. doi: 10.1128/jb.169.6.2601-2610.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Priebe SD, Lacks SA. 1989. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J Bacteriol 171:4778–4784. doi: 10.1128/jb.171.9.4778-4784.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Grohmann E, Guzman LM, Espinosa M. 1999. Mobilisation of the streptococcal plasmid pMV158: interactions of MobM protein with its cognate oriT DNA region. Mol Gen Genet 261:707–715. doi: 10.1007/s004380050014. [DOI] [PubMed] [Google Scholar]
- 356.Lorenzo-Diaz F, Fernandez-Lopez C, Garcillan-Barcia MP, Espinosa M. 2014. Bringing them together: plasmid pMV158 rolling circle replication and conjugation under an evolutionary perspective. Plasmid 74:15–31. doi: 10.1016/j.plasmid.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Novick RP. 1990. The Staphylococcus as a molecular genetic system, p 1–37. In Novick RP. (ed), Molecular biology of the staphylococci. VCH Publishers, New York, NY. [Google Scholar]
- 358.Shalita Z, Murphy E, Novick RP. 1980. Penicillinase plasmids of Staphylococcus aureus: structural and evolutionary relationships. Plasmid 3:291–311. doi: 10.1016/0147-619X(80)90042-6. [DOI] [PubMed] [Google Scholar]
- 359.Jensen SO, Apisiridej S, Kwong SM, Yang YH, Skurray RA, Firth N. 2010. Analysis of the prototypical Staphylococcus aureus multiresistance plasmid pSK1. Plasmid 64:135–142. doi: 10.1016/j.plasmid.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 360.Khan SA, Novick RP. 1980. Terminal nucleotide sequences of Tn551, a transposon specifying erythromycin resistance in Staphylococcus aureus: homology with Tn3. Plasmid 4:148–154. doi: 10.1016/0147-619X(80)90004-9. [DOI] [PubMed] [Google Scholar]
- 361.Rouch DA, Byrne ME, Kong YC, Skurray RA. 1987. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J Gen Microbiol 133:3039–3052. [DOI] [PubMed] [Google Scholar]
- 362.Rouch DA, Messerotti LJ, Loo LS, Jackson CA, Skurray RA. 1989. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol 3:161–175. doi: 10.1111/j.1365-2958.1989.tb01805.x. [DOI] [PubMed] [Google Scholar]
- 363.Firth N, Apisiridej S, Berg T, O'Rourke BA, Curnock S, Dyke KG, Skurray RA. 2000. Replication of staphylococcal multiresistance plasmids. J Bacteriol 182:2170–2178. doi: 10.1128/JB.182.8.2170-2178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Kwong SM, Lim R, Lebard RJ, Skurray RA, Firth N. 2008. Analysis of the pSK1 replicon, a prototype from the staphylococcal multiresistance plasmid family. Microbiology 154:3084–3094. doi: 10.1099/mic.0.2008/017418-0. [DOI] [PubMed] [Google Scholar]
- 365.Kwong SM, Skurray RA, Firth N. 2004. Staphylococcus aureus multiresistance plasmid pSK41: analysis of the replication region, initiator protein binding and antisense RNA regulation. Mol Microbiol 51:497–509. doi: 10.1046/j.1365-2958.2003.03843.x. [DOI] [PubMed] [Google Scholar]
- 366.Kwong SM, Ramsay JP, Jensen SO, Firth N. 2017. Replication of staphylococcal resistance plasmids. Front Microbiol 8:2279. doi: 10.3389/fmicb.2017.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Weaver KE, Kwong SM, Firth N, Francia MV. 2009. The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61:94–109. doi: 10.1016/j.plasmid.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Apisiridej S, Leelaporn A, Scaramuzzi CD, Skurray RA, Firth N. 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 38:13–24. doi: 10.1006/plas.1997.1292. [DOI] [PubMed] [Google Scholar]
- 369.Simpson AE, Skurray RA, Firth N. 2003. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J Bacteriol 185:2143–2152. doi: 10.1128/JB.185.7.2143-2152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Weaver KE, Jensen KD, Colwell A, Sriram S. 1996. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol Microbiol 20:53–63. doi: 10.1111/j.1365-2958.1996.tb02488.x. [DOI] [PubMed] [Google Scholar]
- 371.Kwong SM, Jensen SO, Firth N. 2010. Prevalence of Fst-like toxin-antitoxin systems. Microbiology 156:975–977. doi: 10.1099/mic.0.038323-0. [DOI] [PubMed] [Google Scholar]
- 372.Macrina FL, Archer GL. 1993. Conjugation and broad host range plasmids in streptococci and staphylococci, p 313–329. In Clewell DB. (ed), Bacterial conjugation. Plenum Press, New York, NY. [Google Scholar]
- 373.Climo M, Sharma V, Archer G. 1996. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol 178:4975–4983. doi: 10.1128/jb.178.16.4975-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.McElgunn CJ, Zahurul M, Bhuyian A, Sugiyama M. 2002. Integration analysis of pSK41 in the chromosome of a methicillin-resistant Staphylococcus aureus K-1. J Basic Microbiol 42:190–200. doi:. [DOI] [PubMed] [Google Scholar]
- 375.Liu MA, Kwong SM, Jensen SO, Brzoska AJ, Firth N. 2013. Biology of the staphylococcal conjugative multiresistance plasmid pSK41. Plasmid 70:42–51. doi: 10.1016/j.plasmid.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 376.Morton TM, Eaton DM, Johnston JL, Archer GL. 1993. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J Bacteriol 175:4436–4447. doi: 10.1128/jb.175.14.4436-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Caryl JA, O'Neill AJ. 2009. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the staphylococci. Plasmid 62:35–38. doi: 10.1016/j.plasmid.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 378.Jaffe HW, Sweeney HM, Weinstein RA, Kabins SA, Nathan C, Cohen S. 1982. Structural and phenotypic varieties of gentamicin resistance plasmids in hospital strains of Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob Agents Chemother 21:773–779. doi: 10.1128/AAC.21.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Archer GL, Johnston JL. 1983. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob Agents Chemother 24:70–77. doi: 10.1128/AAC.24.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Goering RV, Ruff EA. 1983. Comparative analysis of conjugative plasmids mediating gentamicin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 24:450–452. doi: 10.1128/AAC.24.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Evans J, Dyke KG. 1988. Characterization of the conjugation system associated with the Staphylococcus aureus plasmid pJE1. J Gen Microbiol 134:1–8. [DOI] [PubMed] [Google Scholar]
- 382.Perez-Roth E, Lopez-Aguilar C, Alcoba-Florez J, Mendez-Alvarez S. 2006. High-level mupirocin resistance within methicillin-resistant Staphylococcus aureus pandemic lineages. Antimicrob Agents Chemother 50:3207–3211. doi: 10.1128/AAC.00059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Sasatsu M, Shima K, Shibata Y, Kono M. 1989. Nucleotide sequence of a gene that encodes resistance to ethidium bromide from a transferable plasmid in Staphylococcus aureus. Nucleic Acids Res 17:10103. doi: 10.1093/nar/17.23.10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 385.McDougal LK, Fosheim GE, Nicholson A, Bulens SN, Limbago BM, Shearer JES, Summers AO, Patel JB. 2010. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob Agents Chemother 54:3804–3811. doi: 10.1128/AAC.00351-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Byrne ME, Gillespie MT, Skurray RA. 1990. Molecular analysis of a gentamicin resistance transposonlike element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob Agents Chemother 34:2106–2113. doi: 10.1128/AAC.34.11.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Morton TM, Johnston JL, Patterson J, Archer GL. 1995. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother 39:1272–1280. doi: 10.1128/AAC.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Bender J, Strommenger B, Steglich M, Zimmermann O, Fenner I, Lensing C, Dagwadordsch U, Kekule AS, Werner G, Layer F. 2015. Linezolid resistance in clinical isolates of Staphylococcus epidermidis from German hospitals and characterization of two cfr-carrying plasmids. J Antimicrob Chemother 70:1630–1638. doi: 10.1093/jac/dkv025. [DOI] [PubMed] [Google Scholar]
- 389.Byrne ME, Gillespie MT, Skurray RA. 1991. 4′,4″ adenyltransferase activity on conjugative plasmids isolated from Staphylococcus aureus is encoded on an integrated copy of pUB110. Plasmid 25:70–75. doi: 10.1016/0147-619X(91)90008-K. [DOI] [PubMed] [Google Scholar]
- 390.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 391.Perichon B, Courvalin P. 2009. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:4580–4587. doi: 10.1128/AAC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Zhu W, Clark N, Patel JB. 2013. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob Agents Chemother 57:212–219. doi: 10.1128/AAC.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Kwong SM, Skurray RA, Firth N. 2006. Replication control of staphylococcal multiresistance plasmid pSK41: an antisense RNA mediates dual-level regulation of Rep expression. J Bacteriol 188:4404–4412. doi: 10.1128/JB.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.LeBard RJ, Jensen SO, Arnaiz IA, Skurray RA, Firth N. 2008. A multimer resolution system contributes to segregational stability of the prototypical staphylococcal conjugative multiresistance plasmid pSK41. FEMS Microbiol Lett 284:58–67. doi: 10.1111/j.1574-6968.2008.01190.x. [DOI] [PubMed] [Google Scholar]
- 395.Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, Skurray RA, Firth N. 2007. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature 450:1268–1271. doi: 10.1038/nature06392. [DOI] [PubMed] [Google Scholar]
- 396.Firth N, Berg T, Skurray RA. 1999. Evolution of conjugative plasmids from gram-positive bacteria. Mol Microbiol 31:1598–1600. [PubMed] [Google Scholar]
- 397.Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev 67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Ramsay JP, Kwong SM, Murphy RJ, Yui Eto K, Price KJ, Nguyen QT, O'Brien FG, Grubb WB, Coombs GW, Firth N. 2016. An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob Genet Elements 6:e1208317. doi: 10.1080/2159256X.2016.1208317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Rossi F, Diaz L, Wollam A, Panesso D, Zhou Y, Rincon S, Narechania A, Xing G, Di Gioia TS, Doi A, Tran TT, Reyes J, Munita JM, Carvajal LP, Hernandez-Roldan A, Brandao D, van der Heijden IM, Murray BE, Planet PJ, Weinstock GM, Arias CA. 2014. Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med 370:1524–1531. doi: 10.1056/NEJMoa1303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Shore AC, Lazaris A, Kinnevey PM, Brennan OM, Brennan GI, O'Connell B, Fessler AT, Schwarz S, Coleman DC. 2016. First report of cfr-carrying plasmids in the pandemic sequence type 22 methicillin-resistant Staphylococcus aureus staphylococcal cassette chromosome mec type IV clone. Antimicrob Agents Chemother 60:3007–3015. doi: 10.1128/AAC.02949-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Showsh SA, Andrews RE Jr. 1999. Analysis of the requirement for a pUB110 mob region during Tn916-dependent mobilization. Plasmid 41:179–186. doi: 10.1006/plas.1999.1398. [DOI] [PubMed] [Google Scholar]
- 402.Lee CA, Thomas J, Grossman AD. 2012. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol 194:3165–3172. doi: 10.1128/JB.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Burdett V. 1980. Identification of tetracycline-resistant R-plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother 18:753–760. doi: 10.1128/AAC.18.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Francia MV, Clewell DB. 2002. Amplification of the tetracycline resistance determinant of pAMα1 in Enterococcus faecalis requires a site-specific recombination event involving relaxase. J Bacteriol 184:5187–5193. doi: 10.1128/JB.184.18.5187-5193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Brantl S, Wagner EG. 1997. Dual function of the copR gene product of plasmid pIP501. J Bacteriol 179:7016–7024. doi: 10.1128/jb.179.22.7016-7024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Schwarz FV, Perreten V, Teuber M. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170–187. doi: 10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
- 407.Zhu W, Murray PR, Huskins WC, Jernigan JA, McDonald LC, Clark NC, Anderson KF, McDougal LK, Hageman JC, Olsen-Rasmussen M, Frace M, Alangaden GJ, Chenoweth C, Zervos MJ, Robinson-Dunn B, Schreckenberger PC, Reller LB, Rudrik JT, Patel JB. 2010. Dissemination of an Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:4314–4320. doi: 10.1128/AAC.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Liu Y, Wang Y, Wu C, Shen Z, Schwarz S, Du XD, Dai L, Zhang W, Zhang Q, Shen J. 2012. First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob Agents Chemother 56:1650–1654. doi: 10.1128/AAC.06091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Tanimoto K, Ike Y. 2008. Complete nucleotide sequencing and analysis of the 65-kb highly conjugative Enterococcus faecium plasmid pMG1: identification of the transfer-related region and the minimum region required for replication. FEMS Microbiol Lett 288:186–195. doi: 10.1111/j.1574-6968.2008.01342.x. [DOI] [PubMed] [Google Scholar]
- 410.Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL. 2009. Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 155:2930–2940. doi: 10.1099/mic.0.030932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Lim SK, Tanimoto K, Tomita H, Ike Y. 2006. Pheromone-responsive conjugative vancomycin resistance plasmids in Enterococcus faecalis isolates from humans and chicken feces. Appl Environ Microbiol 72:6544–6553. doi: 10.1128/AEM.00749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect 16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 413.Halvorsen EM, Williams JJ, Bhimani AJ, Billings EA, Hergenrother PJ. 2011. Txe, an endoribonuclease of the enterococcal Axe-Txe toxin-antitoxin system, cleaves mRNA and inhibits protein synthesis. Microbiology 157:387–397. doi: 10.1099/mic.0.045492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, Godfrey P, Palmer KL, Bodi K, Mongodin EF, Wortman J, Feldgarden M, Lawley T, Gill SR, Haas BJ, Birren B, Gilmore MS. 2012. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. mBio 3:e00112-12. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Grady R, Hayes F. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol Microbiol 47:1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- 416.van Hal SJ, Espedido BA, Coombs GW, Howden BP, Korman TM, Nimmo GR, Gosbell IB, Jensen SO. 2017. Polyclonal emergence of vanA vancomycin-resistant Enterococcus faecium in Australia. J Antimicrob Chemother 72:998–1001. doi: 10.1093/jac/dkw539. [DOI] [PubMed] [Google Scholar]
- 417.Laverde Gomez JA, van Schaik W, Freitas AR, Coque TM, Weaver KE, Francia MV, Witte W, Werner G. 2011. A multiresistance megaplasmid pLG1 bearing a hylEfm genomic island in hospital Enterococcus faecium isolates. Int J Med Microbiol 301:165–175. doi: 10.1016/j.ijmm.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 418.Delavat F, Miyazaki R, Carraro N, Pradervand N, van der Meer JR. 2017. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev 41:512–537. doi: 10.1093/femsre/fux008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Novick RP, Christie GE, Penades JR. 2010. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol 8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Burrus V, Pavlovic G, Decaris B, Guedon G. 2002. Conjugative transposons: the tip of the iceberg. Mol Microbiol 46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 421.Carraro N, Burrus V. 2015. The dualistic nature of integrative and conjugative elements. Mob Genet Elements 5:98–102. doi: 10.1080/2159256X.2015.1102796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou HY. 2012. ICEberg: a web-based resource for integrative and conjugative elements found in bacteria. Nucleic Acids Res 40:D621–D626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Carraro N, Poulin D, Burrus V. 2015. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet 11:e1005298. doi: 10.1371/journal.pgen.1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Burrus V, Marrero J, Waldor MK. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173–183. doi: 10.1016/j.plasmid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 425.Roy Chowdhury P, Scott MJ, Djordjevic SP.. 2017. Genomic islands 1 and 2 carry multiple antibiotic resistance genes in Pseudomonas aeruginosa ST235, ST253, ST111 and ST175 and are globally dispersed. J Antimicrob Chemother 72:620–622. doi: 10.1093/jac/dkw471. [DOI] [PubMed] [Google Scholar]
- 426.Kung VL, Ozer EA, Hauser AR. 2010. The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev 74:621–641. doi: 10.1128/MMBR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Klockgether J, Wurdemann D, Reva O, Wiehlmann L, Tummler B. 2007. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J Bacteriol 189:2443–2459. doi: 10.1128/JB.01688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Martinez E, Marquez C, Ingold A, Merlino J, Djordjevic SP, Stokes HW, Chowdhury PR. 2012. Diverse mobilized class 1 integrons are common in the chromosomes of pathogenic Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 56:2169–2172. doi: 10.1128/AAC.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Hong JS, Yoon EJ, Lee H, Jeong SH, Lee K. 2016. Clonal dissemination of Pseudomonas aeruginosa sequence type 235 isolates carrying blaIMP-6 and emergence of blaGES-24 and blaIMP-10 on novel genomic islands PAGI-15 and -16 in South Korea. Antimicrob Agents Chemother 60:7216–7223. doi: 10.1128/AAC.00640-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Silveira MC, Albano RM, Asensi MD, Carvalho-Assef AP. 2016. Description of genomic islands associated to the multidrug-resistant Pseudomonas aeruginosa clone ST277. Infect Genet Evol 42:60–65. doi: 10.1016/j.meegid.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 431.Roche D, Flechard M, Lallier N, Reperant M, Bree A, Pascal G, Schouler C, Germon P. 2010. ICEEc2, a new integrative and conjugative element belonging to the pKLC102/PAGI-2 family, identified in Escherichia coli strain BEN374. J Bacteriol 192:5026–5036. doi: 10.1128/JB.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Di Pilato V, Pollini S, Rossolini GM. 2015. Tn6249, a new Tn6162 transposon derivative carrying a double-integron platform and involved with acquisition of the blaVIM-1 metallo-β-lactamase gene in Pseudomonas aeruginosa. Antimicrob Agents Chemother 59:1583–1587. doi: 10.1128/AAC.04047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Roy Chowdhury P, Merlino J, Labbate M, Cheong EY, Gottlieb T, Stokes HW. 2009. Tn6060, a transposon from a genomic island in a Pseudomonas aeruginosa clinical isolate that includes two class 1 integrons. Antimicrob Agents Chemother 53:5294–5296. doi: 10.1128/AAC.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Roy Chowdhury P, Scott M, Worden P, Huntington P, Hudson B, Karagiannis T, Charles IG, Djordjevic SP. 2016. Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol 6:150175. doi: 10.1098/rsob.150175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Toleman MA, Walsh TR. 2011. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev 35:912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 436.Fonseca EL, Marin MA, Encinas F, Vicente AC. 2015. Full characterization of the integrative and conjugative element carrying the metallo-β-lactamase blaSPM-1 and bicyclomycin bcr1 resistance genes found in the pandemic Pseudomonas aeruginosa clone SP/ST277. J Antimicrob Chemother 70:2547–2550. doi: 10.1093/jac/dkv152. [DOI] [PubMed] [Google Scholar]
- 437.Cochetti I, Tili E, Mingoia M, Varaldo PE, Montanari MP. 2008. erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob Agents Chemother 52:1285–1290. doi: 10.1128/AAC.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Brouwer MS, Mullany P, Roberts AP. 2010. Characterization of the conjugative transposon Tn6000 from Enterococcus casseliflavus 664.1H1 (formerly Enterococcus faecium 664.1H1). FEMS Microbiol Lett 309:71–76. doi: 10.1111/j.1574-6968.2010.02018.x. [DOI] [PubMed] [Google Scholar]
- 439.Roberts AP, Johanesen PA, Lyras D, Mullany P, Rood JI. 2001. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology 147:1243–1251. doi: 10.1099/00221287-147-5-1243. [DOI] [PubMed] [Google Scholar]
- 440.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 441.Tsvetkova K, Marvaud JC, Lambert T. 2010. Analysis of the mobilization functions of the vancomycin resistance transposon Tn1549, a member of a new family of conjugative elements. J Bacteriol 192:702–713. doi: 10.1128/JB.00680-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Launay A, Ballard SA, Johnson PD, Grayson ML, Lambert T. 2006. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob Agents Chemother 50:1054–1062. doi: 10.1128/AAC.50.3.1054-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Smyth DS, Robinson DA. 2009. Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J Bacteriol 191:5964–5975. doi: 10.1128/JB.00352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Sansevere EA, Luo X, Park JY, Yoon S, Seo KS, Robinson DA. 2017. Transposase-mediated excision, conjugative transfer, and diversity of ICE6013 elements in Staphylococcus aureus. J Bacteriol 199:e00629-16. doi: 10.1128/JB.00629-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Mullany P, Williams R, Langridge GC, Turner DJ, Whalan R, Clayton C, Lawley T, Hussain H, McCurrie K, Morden N, Allan E, Roberts AP. 2012. Behavior and target site selection of conjugative transposon Tn916 in two different strains of toxigenic Clostridium difficile. Appl Environ Microbiol 78:2147–2153. doi: 10.1128/AEM.06193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Hamidian M, Hall RM. 2017. Acinetobacter baumannii ATCC 19606 carries GIsul2 in a genomic island located in the chromosome. Antimicrob Agents Chemother 61:e01991-16. doi: 10.1128/AAC.01991-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Schultz E, Barraud O, Madec JY, Haenni M, Cloeckaert A, Ploy MC, Doublet B. 2017. Multidrug resistance Salmonella genomic island 1 in a Morganella morganii subsp. morganii human clinical isolate from France. mSphere 2:e00118-17. doi: 10.1128/mSphere.00118-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Hamidian M, Holt KE, Hall RM. 2015. Genomic resistance island AGI1 carrying a complex class 1 integron in a multiply antibiotic-resistant ST25 Acinetobacter baumannii isolate. J Antimicrob Chemother 70:2519–2523. doi: 10.1093/jac/dkv137. [DOI] [PubMed] [Google Scholar]
- 449.Das B, Martinez E, Midonet C, Barre FX. 2013. Integrative mobile elements exploiting Xer recombination. Trends Microbiol 21:23–30. doi: 10.1016/j.tim.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 450.Antonelli A, D'Andrea MM, Di Pilato V, Viaggi B, Torricelli F, Rossolini GM. 2015. Characterization of a novel putative Xer-dependent integrative mobile element carrying the blaNMC-A carbapenemase gene, inserted into the chromosome of members of the Enterobacter cloacae complex. Antimicrob Agents Chemother 59:6620–6624. doi: 10.1128/AAC.01452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Boyd DA, Mataseje LF, Davidson R, Delport JA, Fuller J, Hoang L, Lefebvre B, Levett PN, Roscoe DL, Willey BM, Mulvey MR. 2017. Enterobacter cloacae complex isolates harboring blaNMC-A or blaIMI-type class A carbapenemase genes on novel chromosomal integrative elements and plasmids. Antimicrob Agents Chemother 61:e02578-16. doi: 10.1128/AAC.02578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Koh TH, Rahman NBA, Teo JWP, La MV, Periaswamy B, Chen SL. 2018. Putative integrative mobile elements that exploit the Xer recombination machinery carrying blaIMI-type carbapenemase genes in Enterobacter cloacae complex isolates in Singapore. Antimicrob Agents Chemother 62:e01542-17. doi: 10.1128/AAC.01542-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Blackwell GA, Nigro SJ, Hall RM. 2015. Evolution of AbGRI2-0, the progenitor of the AbGRI2 resistance island in global clone 2 of Acinetobacter baumannii. Antimicrob Agents Chemother 60:1421–1429. doi: 10.1128/AAC.02662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Blackwell GA, Holt KE, Bentley SD, Hsu LY, Hall RM. 2017. Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. J Antimicrob Chemother 72:1031–1039. doi: 10.1093/jac/dkw542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother 48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, Daum RS, Hiramatsu K. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother 46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF. 2001. A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science 291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 459.Ishikawa T, Matsunaga N, Tawada H, Kuroda N, Nakayama Y, Ishibashi Y, Tomimoto M, Ikeda Y, Tagawa Y, Iizawa Y, Okonogi K, Hashiguchi S, Miyake A. 2003. TAK-599, a novel N-phosphono type prodrug of anti-MRSA cephalosporin T-91825: synthesis, physicochemical and pharmacological properties. Bioorg Med Chem 11:2427–2437. doi: 10.1016/S0968-0896(03)00126-3. [DOI] [PubMed] [Google Scholar]
- 460.Katayama Y, Ito T, Hiramatsu K. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob Agents Chemother 45:1955–1963. doi: 10.1128/AAC.45.7.1955-1963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Katayama Y, Ito T, Hiramatsu K. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Wu Z, Li F, Liu D, Xue H, Zhao X. 2015. Novel type XII staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob Agents Chemother 59:7597–7601. doi: 10.1128/AAC.01692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Mir-Sanchis I, Roman CA, Misiura A, Pigli YZ, Boyle-Vavra S, Rice PA. 2016. Staphylococcal SCCmec elements encode an active MCM-like helicase and thus may be replicative. Nat Struct Mol Biol 23:891–898. doi: 10.1038/nsmb.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements. 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Hiramatsu K, Cui L, Kuroda M, Ito T. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 9:486–493. doi: 10.1016/S0966-842X(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 466.Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesoe RL, Lemvigh CK, Aarestrup FM, Lund O, Larsen AR. 2018. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3:e00612-17. doi: 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Chambers HF, DeLeo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Fang H, Hedin G, Li G, Nord CE. 2008. Genetic diversity of community-associated methicillin-resistant Staphylococcus aureus in southern Stockholm, 2000–2005. Clin Microbiol Infect 14:370–376. doi: 10.1111/j.1469-0691.2007.01941.x. [DOI] [PubMed] [Google Scholar]
- 469.Lina G, Durand G, Berchich C, Short B, Meugnier H, Vandenesch F, Etienne J, Enright MC. 2006. Staphylococcal chromosome cassette evolution in Staphylococcus aureus inferred from ccr gene complex sequence typing analysis. Clin Microbiol Infect 12:1175–1184. doi: 10.1111/j.1469-0691.2006.01548.x. [DOI] [PubMed] [Google Scholar]
- 470.Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bachi B, Cook GM. 2007. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother 51:1497–1499. doi: 10.1128/AAC.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, Etages SA, Jones A, Palazzolo-Ballance AM, Perdreau-Remington F, Sensabaugh GF, Deleo FR, Chambers HF. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis 197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 472.Garza-Gonzalez E, Morfõn-Otero R, Llaca-Dõaz JM, Rodriguez-Noriega E. 2010. Staphylococcal cassette chromosome mec (SCCmec) in methicillin-resistant coagulase-negative staphylococci. A review and the experience in a tertiary-care setting. Epidemiol Infect 138:645–654. doi: 10.1017/S0950268809991361. [DOI] [PubMed] [Google Scholar]
- 473.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M, SENTRY Participants Group. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY antimicrobial surveillance program, 1997–1999. Clin Infect Dis 32(Suppl 2):S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 474.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. 2010. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother 54:4352–4359. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Rolo J, Worning P, Nielsen JB, Bowden R, Bouchami O, Damborg P, Guardabassi L, Perreten V, Tomasz A, Westh H, de Lencastre H, Miragaia M. 2017. Evolutionary origin of the staphylococcal cassette chromosome mec (SCCmec). Antimicrob Agents Chemother 61:e02302-16. doi: 10.1128/AAC.02302-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Nubel U, Roumagnac P, Feldkamp M, Song J-H, Ko KS, Huang Y-C, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 105:14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A 101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother 50:1001–1012. doi: 10.1128/AAC.50.3.1001-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Joshi GS, Spontak JS, Klapper DG, Richardson AR. 2011. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol 82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Novick RP, Ram G. 2016. The floating (pathogenicity) island: a genomic dessert. Trends Genet 32:114–126. doi: 10.1016/j.tig.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Iwao Y, Ishii R, Tomita Y, Shibuya Y, Takano T, Hung WC, Higuchi W, Isobe H, Nishiyama A, Yano M, Matsumoto T, Ogata K, Okubo T, Khokhlova O, Ho PL, Yamamoto T. 2012. The emerging ST8 methicillin-resistant Staphylococcus aureus clone in the community in Japan: associated infections, genetic diversity, and comparative genomics. J Infect Chemother 18:228–240. doi: 10.1007/s10156-012-0379-6. [DOI] [PubMed] [Google Scholar]
- 484.O'Neill AJ, Larsen AR, Skov R, Henriksen AS, Chopra I. 2007. Characterization of the epidemic European fusidic acid-resistant impetigo clone of Staphylococcus aureus. J Clin Microbiol 45:1505–1510. doi: 10.1128/JCM.01984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Tsafnat G, Copty J, Partridge SR. 2011. RAC: repository of antibiotic resistance cassettes. Database (Oxford) 2011:bar054. doi: 10.1093/database/bar054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. doi: 10.1093/bioinformatics/btv688. [DOI] [PubMed] [Google Scholar]
- 487.Lanza VF, de Toro M, Garcillan-Barcia MP, Mora A, Blanco J, Coque TM, de la Cruz F. 2014. Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet 10:e1004766. doi: 10.1371/journal.pgen.1004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Arredondo-Alonso S, van Schaik W, Willems RJ, Schürch AC. 2017. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom 3:e000128. doi: 10.1099/mgen.0.000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Partridge SR, Tsafnat G. 2018. Automated annotation of mobile antibiotic resistance in Gram-negative bacteria: the Multiple Antibiotic Resistance Annotator (MARA) and database. J Antimicrob Chemother 73:883–890. doi: 10.1093/jac/dkx513. [DOI] [PubMed] [Google Scholar]
- 490.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, Hall RM, Holt KE. 2015. ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics 16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Cury J, Jove T, Touchon M, Neron B, Rocha EP. 2016. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res 44:4539–4550. doi: 10.1093/nar/gkw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Brouwer MS, Tagg KA, Mevius DJ, Iredell JR, Bossers A, Smith HE, Partridge SR. 2015. IncI shufflons: assembly issues in the next-generation sequencing era. Plasmid 80:111–117. doi: 10.1016/j.plasmid.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 494.Firth N, Skurray RA. 1998. Mobile elements in the evolution and spread of multiple-drug resistance in staphylococci. Drug Resist Updat 1:49–58. doi: 10.1016/S1368-7646(98)80214-8. [DOI] [PubMed] [Google Scholar]
- 495.Furi L, Haigh R, Al Jabri ZJ, Morrissey I, Ou HY, Leon-Sampedro R, Martinez JL, Coque TM, Oggioni MR. 2016. Dissemination of novel antimicrobial resistance mechanisms through the insertion sequence mediated spread of metabolic genes. Front Microbiol 7:1008. doi: 10.3389/fmicb.2016.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Kehrenberg C, Schwarz S. 2005. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob Agents Chemother 49:813–815. doi: 10.1128/AAC.49.2.813-815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Kadlec K, Schwarz S. 2010. Identification of a plasmid-borne resistance gene cluster comprising the resistance genes erm(T), dfrK, and tet(L) in a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 54:915–918. doi: 10.1128/AAC.01091-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Li D, Wu C, Wang Y, Fan R, Schwarz S, Zhang S. 2015. Identification of multiresistance gene cfr in methicillin-resistant Staphylococcus aureus from pigs: plasmid location and integration into a staphylococcal cassette chromosome mec complex. Antimicrob Agents Chemother 59:3641–3644. doi: 10.1128/AAC.00500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Liu Y, Wang Y, Schwarz S, Li Y, Shen Z, Zhang Q, Wu C, Shen J. 2013. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob Agents Chemother 57:42–48. doi: 10.1128/AAC.01605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Gómez-Sanz E, Kadlec K, Fessler AT, Zarazaga M, Torres C, Schwarz S. 2013. Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob Agents Chemother 57:3275–3282. doi: 10.1128/AAC.00171-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob Agents Chemother 51:483–487. doi: 10.1128/AAC.01340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Kadlec K, Schwarz S. 2009. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob Agents Chemother 53:776–778. doi: 10.1128/AAC.01128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Chen L, Mediavilla JR, Smyth DS, Chavda KD, Ionescu R, Roberts RB, Robinson DA, Kreiswirth BN. 2010. Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob Agents Chemother 54:3347–3354. doi: 10.1128/AAC.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Allignet J, El Solh N. 1999. Comparative analysis of staphylococcal plasmids carrying three streptogramin-resistance genes: vat-vgb-vga. Plasmid 42:134–138. doi: 10.1006/plas.1999.1412. [DOI] [PubMed] [Google Scholar]
- 505.Highlander SK, Hulten KG, Qin X, Jiang H, Yerrapragada S, Mason EO Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.O'Neill AJ, Chopra I. 2006. Molecular basis of fusB-mediated resistance to fusidic acid in Staphylococcus aureus. Mol Microbiol 59:664–676. doi: 10.1111/j.1365-2958.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 507.de Vries LE, Christensen H, Agersø Y. 2012. The diversity of inducible and constitutively expressed erm(C) genes and association to different replicon types in staphylococci plasmids. Mob Genet Elements 2:72–80. doi: 10.4161/mge.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Naseer U, Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist 17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- 509.Albornoz E, Tijet N, De Belder D, Gomez S, Martino F, Corso A, Melano RG, Petroni A. 2017. qnrE1, a member of a new family of plasmid-located quinolone resistance genes, originated from the chromosome of Enterobacter species. Antimicrob Agents Chemother 61:e02555-16. doi: 10.1128/AAC.02555-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Kadlec K, Schwarz S. 2010. Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 54:3475–3477. doi: 10.1128/AAC.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob Agents Chemother 55:4900–4904. doi: 10.1128/AAC.00528-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Frost LS. 1993. Conjugative pili and pilus-specific phages, p 189–221. In Clewell DB. (ed), Bacterial conjugation. Springer, Boston, MA. [Google Scholar]
- 513.Suzuki H, Yano H, Brown CJ, Top EM. 2010. Predicting plasmid promiscuity based on genomic signature. J Bacteriol 192:6045–6055. doi: 10.1128/JB.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Sampei G, Furuya N, Tachibana K, Saitou Y, Suzuki T, Mizobuchi K, Komano T. 2010. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 64:92–103. doi: 10.1016/j.plasmid.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 515.Borrell L, Yang J, Pittard AJ, Praszkier J. 2006. Interaction of initiator proteins with the origin of replication of an IncL/M plasmid. Plasmid 56:88–101. doi: 10.1016/j.plasmid.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 516.Holden MTG, Lindsay JA, Corton C, Quail MA, Cockfield JD, Pathak S, Batra R, Parkhill J, Bentley SD, Edgeworth JD. 2010. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J Bacteriol 192:888–892. doi: 10.1128/JB.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Lannergard J, Norstrom T, Hughes D. 2009. Genetic determinants of resistance to fusidic acid among clinical bacteremia isolates of Staphylococcus aureus. Antimicrob Agents Chemother 53:2059–2065. doi: 10.1128/AAC.00871-08. [DOI] [PMC free article] [PubMed] [Google Scholar]