Abstract
The initial report of the mcr-1 (mobile colistin resistance) gene has led to many reports of mcr-1 variants and other mcr genes from different bacterial species originating from human, animal and environmental samples in different geographical locations. Resistance gene nomenclature is complex and unfortunately problems such as different names being used for the same gene/protein or the same name being used for different genes/proteins are not uncommon. Registries exist for some families, such as bla (β-lactamase) genes, but there is as yet no agreed nomenclature scheme for mcr genes. The National Center for Biotechnology Information (NCBI) recently took over assigning bla allele numbers from the longstanding Lahey β-lactamase website and has agreed to do the same for mcr genes. Here, we propose a nomenclature scheme that we hope will be acceptable to researchers in this area and that will reduce future confusion.
A plasmid-borne gene encoding a phosphoethanolamine transferase conferring resistance to colistin was reported in 2015 in China and named mcr-1 (mobile colistin resistance).1 An identical gene has now been reported in several different bacterial species isolated from as far back as the 1980s, from different hosts and in at least 30 countries in six continents.2–5 This gene is associated with one copy, two copies or no copies of ISApl1 on different plasmids, including I2-, X4- and HI-type plasmids.6 Several publications have now reported minor variants of the original mcr-1, as well as more divergent genes also predicted to encode phosphoethanolamine transferases.7
Various registries have been responsible for keeping track of and assigning names to some families of resistance genes, e.g. http://www.lahey.org/Studies/ for β-lactamases, now managed by the National Center for Biotechnology Information (NCBI); https://www.ncbi.nlm.nih.gov/pathogens/submit_beta_lactamase/.8 Other resistance gene families have remained relatively neglected and it has not been uncommon for the same gene/protein to be assigned multiple names or for different genes/proteins to be given the same name. One contributing factor is that many researchers prefer that the sequences they deposit in International Nucleotide Sequence Database Collaboration (INSDC) databases are given ‘hold-until-publication’ (HUP) status until any associated manuscripts are accepted. This means that researchers who find a potential new allele may not be aware of all the other alleles that have been identified but which are not yet publicly available. When there is no clearing house to ‘reserve’ and keep track of allele numbers this leads to nomenclature collisions and a system similar to the one in place for β-lactamases is needed to avoid confusion. NCBI has agreed to assign allele numbers to and keep track of mcr genes, in consultation with the antibiotic resistance research community.
The first variant of mcr-1 with a single non-synonymous nucleotide change was reported in a Klebsiella pneumoniae ST512 clinical isolate from Italy, on an X4-type plasmid in a genetic context also seen for mcr-1, and was named mcr-1.2.9 Following on from this, we propose that mcr gene and allele numbers are assigned in the same manner as for β-lactamases, i.e. based on amino acid sequence identity rather than on nucleotide sequence identity, and that the original allele is effectively mcr-#.1, with subsequent alleles then assigned mcr-#.2, mcr-#.3, mcr-#.4 etc. (e.g. mcr-1.2, mcr-1.3, mcr-1.4 etc.). The terms ‘mcr-1-family/MCR-1-family’ etc. can then be used to refer more generally to genes/proteins within each group and for related genes/proteins where a specific allele number has not been requested. Each full-length MCR protein will also be assigned a WP_ number, a non-redundant protein accession number used to annotate the same protein sequence in different RefSeq entries from one or more species. In response to requests from researchers, allele numbers have already been assigned by NCBI to additional MCR-1 variants (Table 1), all of which have one or two amino acid changes from MCR-1 (Table 2).
Table 1.
mcr names assigned by NCBI and/or available in GenBank
This citation does not correspond to the first report in GenBank.
Originally called MCR-2.1, but renamed here with the agreement of the authors (AbuOun et al.13). An identical protein was also published as MCR-2.2.14 A protein originally called MCR-2.213 almost as closely related to MCR-1, has been renamed MCR-6.1.
Also reported by Roer et al.,38 but no GenBank entry.
The original mcr-4.2 sequence (MG581979.1/AUE2202934) is missing the last three nucleotides and stop codon, so details of the first complete version published are given.
Originally also named mcr-4.2.
Originally named mcr-4.3, but initially published without an INSDC entry.37
Table 2.
Nucleotide/amino acid changes in mcr-1/MCR-1 alleles from species other than Moraxellaa
| Alleleb | Identified inc | Nucleotide differences from mcr-1.1 | Amino acid differences from MCR-1.1 |
|---|---|---|---|
| 1.1 | E. coli, Escherichia fergusonii, S. enterica, Shigella, Klebsiella, Citrobacter, Enterobacter, Kluyvera ascorbata, Cronobacter sakazakii, Providencia alcalifaciens | — | — |
| 1.2 | E. coli, K. pneumoniae | A8T | Gln3Leu |
| 1.3 | E. coli | AA111-2GG | Ile38Val |
| 1.4 | E. coli | G1318A | Asp440Asn |
| 1.5 | E. coli | C1354T | His452Tyr |
| 1.6 | S. enterica | G1263A, G1607A | Arg536His |
| 1.7 | E. coli | G643A | Ala215Thr |
| 1.8 | E. coli | A8G | Gln3Arg |
| 1.9d | E. coli | T1238C | Val413Ala |
| 1.11e | E. coli | GTG19-21dup | Val7dup |
| 1.12 | E. coli | G9C | Gln3His |
| 1.13 | E. coli | G465A | Met155Ile |
mcr-1.10 is found in Moraxella spp. and has 36 nucleotide differences from mcr-1.1 and MCR-1.10 has 7 amino acid differences from MCR-1.1.
Allele numbers assigned by NCBI are in bold.
Species/genera in which each allele has been detected to date.
Using start codon that matches other mcr-1 genes rather than the one in the original INSDC entry.
dup, duplication of nucleotides/amino acids at positions indicated.
The mcr-2 gene, identified in Escherichia coli isolated from food animals in Belgium, is 77% identical to mcr-1. It encodes a phosphoethanolamine transferase significantly different from MCR-1 (81% amino acid identity, 89% similarity) that confers colistin resistance and is carried on an X4-type plasmid,4 but in a different genetic context from mcr-1.10 The initial reports of MCR-11 and MCR-2,1,4 and subsequent identification of genes encoding proteins with ∼60% identity to both MCR-1 and MCR-2 in various Moraxella species,11 suggested that this genus may be the original source of genes related to mcr-1 and mcr-2. These genes were apparently mobilized by different ISs, ISApl16,12 in the case of mcr-1 and ISEc69 in the case of mcr-2,10 and transferred to other bacterial species relevant to human and veterinary medicine.
A gene from Moraxella spp. that is 97.6% identical to mcr-1, encoding a protein with seven amino acid changes from the original MCR-1, was then identified as a closer ancestor by AbuOun et al.13 and named mcr-1.10. This group also reported a gene from Moraxella pluranimalium that is 96.1% identical to the original mcr-2, encoding a protein with only eight amino acid differences from MCR-2 that was named MCR-2.1.13 Our suggested scheme would assign ‘MCR-2.1’ to the original MCR-2 protein and as the name MCR-2.2 has already been used for an identical protein (MG545606),14 we propose referring to this protein as MCR-2.2 and the gene as mcr-2.2 (Table 1). The name MCR-2.2 has also been used for a third gene from Moraxella spp. reported by AbuOun et al.,13 which encodes a protein 87.9% identical to the original MCR-2. As this protein is also 82.8% identical to MCR-1 (Figure 1) we suggest that it becomes part of a new group and is assigned the name MCR-6.1. Both of these changes have been agreed by AbuOun et al.13 and updated in the respective GenBank entries. We have not proposed mcr-# numbers for the genes encoding proteins designated MCR-POR (from Moraxella porci; MF432696) MCR-LIN (Moraxella lincolnii; MF432697), mcr-OSL (Moraxella osloensis, MF432698) or MCR-CAT (Moraxella catarrhalis, e.g. CP000205) by Kieffer et al.11 at this time, as closely related, mobilized versions are yet to be identified outside Moraxella.
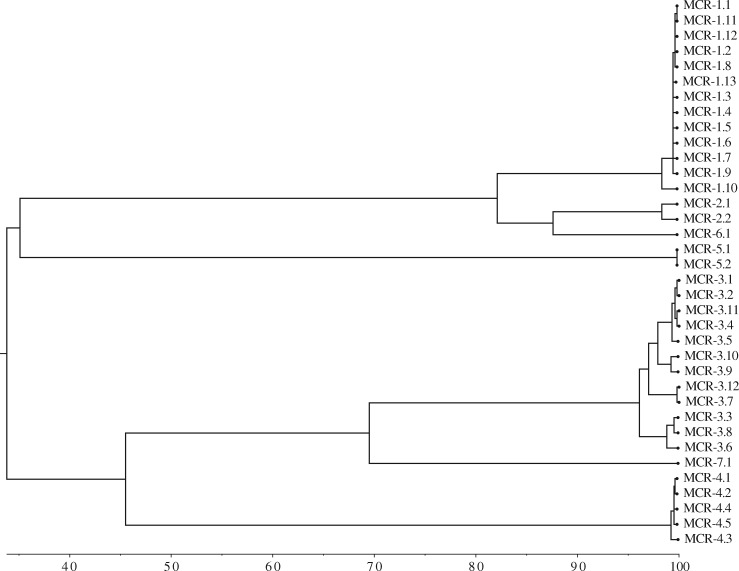
Figure 1.
Relationships between proposed MCR subgroups and alleles. UPGMA tree constructed from a MUSCLE alignment of protein sequences obtained from accession numbers listed in Table 1. Numbers on the bottom axis represent percentage identity among aligning amino acids, but do not reflect differences due to insertions and/or deletions.
The name mcr-3 was given to a gene reported as 45% identical to mcr-1 and 47% identical to mcr-2, encoding a protein that is ∼32% identical to MCR-1 and MCR-2, found on an HI2-type plasmid in E. coli isolated from a pig in China in 2015.15 Genes encoding proteins identical to or with one amino acid change from MCR-3 were also identified in contigs from E. coli, K. pneumoniae, Salmonella enterica subsp. enterica serovar Typhimurium available in the INSDC database.15 Genes encoding proteins up to 94% identical to MCR-3 were also identified in various Aeromonas spp., leading to the suggestion that members of this genus may be the source of mcr-3-family genes.15
Multiple examples of MCR-3 and a number of MCR-3 variants have now been identified from both Aeromonas and other species [Table 3 and Figure S1 (available as Supplementary data at JAC Online)] and have been assigned MCR-3.# numbers by NCBI (Tables 1 and 3). These mcr-3-family variant genes and the corresponding MCR-3-family proteins all show >93% nucleotide and >94% amino acid identity to one another, but the relationships between members of this group appear to be complex. The publication reporting mcr-3.3 in Aeromonas veronii also identified a downstream gene encoding another phosphoethanolamine transferase 84.8% identical to MCR-3.1.16 As it was found not to confer colistin resistance the encoded protein was designated ‘MCR-3.3-like’ by the authors.16 Examination of nucleotide alignments of mcr-3.1, mcr-3.3 and this mcr-3-like gene suggest that mcr-3.1 could be a hybrid made up of the start of mcr-3.3 and the end of the mcr-3-like gene, perhaps generated by homologous recombination (Figure S1). Proteins designated MCR-3.2, MCR-3.4, MCR-3.5 and MCR-3.11, all found in species other than Aeromonas, are minor variants of MCR-3.1 (Table 3).
Table 3.
Nucleotide/amino acid changes in selected mcr-3/MCR-3 allelesa
| Alleleb | Identified inc | Nucleotide differences from mcr-3.1 or mcr-3.7 | Amino acid differences from MCR-3.1 or MCR-3.7 |
|---|---|---|---|
| 3.1 | E. coli, K. pneumoniae, Salmonella Typhimurium, Shigella sonnei | — | — |
| 3.2 | E. coli, Shigella | C1463T | Thr488Ile |
| 3.4 | K. pneumoniae, S. sonnei | G1118T | Gly373Val |
| 3.5 | E. coli, Shigella | A67G, C1370A, C1463T | Met23Val, Ala457Glu, Thr488Ile |
| 3.11 | E. coli | G1118T, CA1402-3AC | Gly373Val, Gln468Thr |
| 3.7 | Aeromonas media | — | — |
| 3.12 | E. coli | T380C + 9 other differences | Val127Ala |
mcr-3.3 and mcr-3.6–mcr-3.9 have been found in various Aeromonas spp. (see Figure S1a) and have 34–100 nucleotide changes from mcr-3.1, while the MCR-3.3 and MCR-3.6–MCR-3.9 proteins have 11–27 amino acid differences from MCR-3.1. mcr-3.10 has been found in Aeromonas caviae, E. coli and Proteus mirabilis and has 19 nucleotide changes from mcr-3.1 and MCR-3.10 has 7 amino acid changes from MCR-3.1.
Allele numbers assigned by NCBI are in bold.
Species/genera in which each allele has been detected to date.
A gene encoding a protein 34%, 35% and 49% identical to MCR-1, MCR-2 and MCR-3, respectively, and 82%–99% identical to genes encoded by Shewanella spp. was named mcr-417 and several variants have now been identified (Table 4). A gene identified in a d-tartrate fermenting S. enterica subsp. enterica serovar Paratyphi B isolate from chicken meat in Germany was named mcr-5.18 The mcr-5 gene is part of the Tn21-family transposon Tn6452, located on a non-conjugative plasmid. The MCR-5 protein shows low identities (33%–36%) to MCR-1, MCR-2, MCR-3 and MCR-4.18 The minor variant MCR-5.2 differs by deletion of Glu234 (698–700AAGΔ).19 A gene designated mcr-7.1 encoding a protein ∼70% identical to MCR-3.1, also possibly originating from Aeromonas and ∼30%–45% identical to other MCR types, has now been reported.20
Table 4.
Nucleotide/amino acid changes in mcr-4/MCR-4 alleles from species other than Shewanella
| Allelea | Identified inb | Nucleotide differences from mcr-4.1 | Amino acid differences from MCR-4.1 |
|---|---|---|---|
| 4.1 | E. coli, Salmonella spp. | — | — |
| 4.2 | E. coli, Salmonella Typhimurium | A992G | Gln331Arg |
| 4.3c | Enterobacter cloacae | T536G, G706T | Val179Gly, Val236Phe |
| 4.4 | E. coli | C613A, A992G | His205Asn, Gln331Arg |
| 4.5 | E. coli | C329T, A992G | Pro110Leu, Gln331Arg |
| 4.6d | Salmonella Kedougou | not available | Val236Phe |
Other subgroups of MCR-type enzymes encoded by mobilized genes are likely to be identified in the future and deciding when an mcr gene/MCR protein should be assigned a new ‘family’ number is potentially more problematic than numbering alleles with minor variations. Existing family numbers were mostly assigned to newly identified MCR proteins by the authors that discovered them. We suggest generally retaining these numbers (apart from redesignating ‘MCR-2.2’ as MCR-6.1, above) as there are clear distinctions between current groups (Figure 1). While various percentage identities have been put forward to define resistance gene subgroups (summarized in Hall and Schwarz21), avoiding fixed numerical cut-offs has also been suggested.22 Current MCR groupings suggest a cut-off between ∼88% and ∼96%, but we think it is better not to define a precise cut-off, to enable re-evaluation of boundaries between families as new data become available. Such flexibility would allow discussions between authors, reviewers, editors, other representatives from the antibiotic resistance community and NCBI to arrive at the best solution if members of potential new subgroups are identified, while assignment of a number by NCBI, if required, avoids possible nomenclature clashes.
Information in Tables 1–4 and Figure 1 represents the current situation (May 2018) for unambiguous mcr-# family and allele numbers designated according to the principles described here. All definitive numbers will be available in the continually updated resistance gene database under BioProject PRJNA313047. MCR variants identified by NCBI staff for which allele numbers have not been requested have been assigned WP_ numbers and are identified as ‘MCR-# family phosphoethanolamine–lipid A transferase’ (and could be assigned allele numbers if requested by submitting authors).
Analysis of currently available sequences indicates that no synonymous nucleotide changes are present in genes encoding MCR-1.2–MCR-1.13 and all alleles of MCR-2–MCR-7 (May 2018). In the case of MCR-1.1 a few different synonymous nucleotide variants, some affecting the first of two ATG codons at the start of the gene and only two found in more than one to two sequences (Table S1), are found amongst the >600 examples in INSDC databases. If such nucleotide differences do become important for epidemiological tracking of mcr genes then the use of automated allele assignments based on nucleotide sequences can be explored.
We hope that the suggestions presented here are acceptable to researchers working in this area. To avoid confusion, we would encourage authors to submit all new allele sequences to INSDC and to approach NCBI (pd-help@ncbi.nlm.nih.gov) for assignment and registration of mcr allele numbers prior to submitting sequences and/or manuscripts, and journals to recommend this and initiate discussions if manuscripts describing related genes that may fall into new subgroups are received.
Supplementary Material
Acknowledgements
We thank Muna Anjum, Rene Hendriksen, Ana Rita Rebelo and Jeanette Teo for agreeing to change published numbers for mcr genes/proteins, authors of papers/GenBank entries listed in Table 1 for agreeing to make changes prior to publication and Laurent Poirel for providing information on sequences prior to release of GenBank entries.
Funding
This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Library of Medicine (M. F., D. H. H., W. K., A. P.) and Medical Research Council/National Natural Science Foundation of China grant DETER-XDRE-CHINA (MR/P007295/1 and 81661138002; J. S., T. W., Y. W.).
Transparency declarations
Y. D. has served on advisory boards for Shionogi, Meiji Seika Pharma, Tetraphase Pharmaceuticals and Merck, and has received research funding from Merck and The Medicines Company for studies unrelated to this work. S. K.-S. has received a research grant from Merck for unrelated studies. S. M.-K. has received funding from Pfizer, Merck, Opgen Inc. and Huvepharma for studies unrelated to the current work. G. M. R. has served on advisory boards or provided consultancies for Achaogen, Angelini, AstraZeneca, Elitech, Menarini, Merck, Nordic Pharma, Pfizer, Rempex/The Medicines Company, Roche and Thermofisher, has received congress lecture fees from AstraZeneca, Basilea, Biotest, Pfizer and Zambon, and has received research grants from Accelerate, Alifax, Angelini, AstraZeneca, Basilea, Becton-Dickinson, bioMérieux, Biotest, Cepheid, Checkpoints, Elitech, Liofilchem, Merck, Nordic Pharma, Novartis, Pfizer, Rempex/The Medicines Company, Seegene, Zambon, VenatorX and Symcel for studies unrelated to this work. All other authors: none to declare.
Supplementary data
Figure S1 and Table S1 are available as Supplementary data at JAC Online.
References
- 1. Liu YY, Wang Y, Walsh TR. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 2. Skov RL, Monnet DL.. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 2016; 21: pii=30155. [DOI] [PubMed] [Google Scholar]
- 3. Schwarz S, Johnson AP.. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 2016; 71: 2066–70. [DOI] [PubMed] [Google Scholar]
- 4. Xavier BB, Lammens C, Ruhal R. et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 2016; 21: pii=30280. [DOI] [PubMed] [Google Scholar]
- 5. Ellem JA, Ginn AN, Chen SC. et al. Locally acquired mcr-1 in Escherichia coli, Australia, 2011 and 2013. Emerg Infect Dis 2017; 23: 1160–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Snesrud E, He S, Chandler M. et al. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother 2016; 60: 6973–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kluytmans J. Plasmid-encoded colistin resistance: mcr-one, two, three and counting. Euro Surveill 2017; 22: pii=30588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jacoby GA, Bonomo RA, Bradford PA. et al. Comment on: Resistance gene naming and numbering: is it a new gene or not? J Antimicrob Chemother 2016; 71: 2677–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Di Pilato V, Arena F, Tascini C. et al. mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of sequence type 512. Antimicrob Agents Chemother 2016; 60: 5612–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Partridge SR. mcr-2 in the IncX4 plasmid pKP37-BE is flanked by directly-oriented copies of ISEc69. J Antimicrob Chemother 2017; 72: 1533–5. [DOI] [PubMed] [Google Scholar]
- 11. Kieffer N, Nordmann P, Poirel L.. Moraxella species as potential sources of MCR-like polymyxin resistance determinants. Antimicrob Agents Chemother 2017; 61: e00129-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poirel L, Kieffer N, Nordmann P.. In-vitro study of ISApl1-mediated mobilization of the colistin resistance gene mcr-1. Antimicrob Agents Chemother 2017; 61: e00127-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. AbuOun M, Stubberfield EJ, Duggett NA. et al. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother 2017; 72: 2745–9. Erratum in: J Antimicrob Chemother 2018; 73: 2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poirel L, Kieffer N, Fernandez-Garayzabal JF. et al. MCR-2-mediated plasmid-borne polymyxin resistance most likely originates from Moraxella pluranimalium. J Antimicrob Chemother 2017; 72: 2947–9. [DOI] [PubMed] [Google Scholar]
- 15. Yin W, Li H, Shen Y. et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio 2017; 8: e00543–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ling Z, Yin W, Li H. et al. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob Agents Chemother 2017; 61: e01272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carattoli A, Villa L, Feudi C. et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 2017; 22: pii=30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Borowiak M, Fischer J, Hammerl JA. et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 2017; 72: 3317–24. [DOI] [PubMed] [Google Scholar]
- 19. Hammerl JA, Borowiak M, Schmoger S. et al. mcr-5 and a novel mcr-5.2 variant in Escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J Antimicrob Chemother 2018; 73: 1433–5. [DOI] [PubMed] [Google Scholar]
- 20. Yang YQ, Li YX, Lei CW. et al. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother 2018; 73: 1791–5. [DOI] [PubMed] [Google Scholar]
- 21. Hall RM, Schwarz S.. Resistance gene naming and numbering: is it a new gene or not? J Antimicrob Chemother 2016; 71: 569–71. [DOI] [PubMed] [Google Scholar]
- 22. Evans BA. Comment on: Resistance gene naming and numbering: is it a new gene or not? J Antimicrob Chemother 2016; 71: 1742–3. [DOI] [PubMed] [Google Scholar]
- 23. Yang YQ, Li YX, Song T. et al. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother 2017; 61: e01204-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao F, Feng Y, Lü X. et al. Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front Microbiol 2017; 8: 2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tijet N, Faccone D, Rapoport M. et al. Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS One 2017; 12: e0180347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu X, Hu Y, Luo M. et al. MCR-1.6, a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica serovar Typhimurium isolate from a healthy individual. Antimicrob Agents Chemother 2017; 61: e02632-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishino Y, Shimojima Y, Suzuki Y. et al. Detection of the mcr-1 gene in colistin-resistant Escherichia coli from retail meat in Japan. Microbiol Immunol 2017; 61: 554–7. [DOI] [PubMed] [Google Scholar]
- 28. Hernández M, Iglesias MR, Rodríguez-Lázaro D. et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill 2017; 22: pii=30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu L, Feng Y, Zhang X. et al. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother 2017; 61: e01757-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eichhorn I, Feudi C, Wang Y. et al. Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GERM-Vet and from diagnostic submissions. J Antimicrob Chemother 2018; 73: 1217–21. [DOI] [PubMed] [Google Scholar]
- 31. Wang X, Zhai W, Li J. et al. Presence of mcr-3 variant in Aeromonas caviae, Proteus mirabilis, and Escherichia coli from one domestic duck. Antmicrob Agents Chemother 2018; 62: e02106-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiang R, Liu BH, Zhang AY. et al. Colocation of the polymyxin resistance gene mcr-1 and a variant of mcr-3 on a plasmid in an Escherichia coli isolate from a chicken farm. Antimicrob Agents Chemother 2018; 62: e00501-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kieffer N, Nordmann P, Micke Moreno A. et al. Genetic and functional characterization of an MCR-3-like enzyme-producing Escherichia coli isolate recovered from swine in Brazil. Antimicrob Agents Chemother 2018; 62: e00278-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carretto E, Brovarone F, Nardini P. et al. Detection of mcr-4 positive Salmonella enterica serovar Typhimurium in clinical isolates of human origin, Italy, October to November 2016. Euro Surveill 2018; 23: pii=17-00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garcia V, Garcia-Menino I, Mora A. et al. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006-2017). Int J Antimicrob Agents 2018; 52: 104–8. [DOI] [PubMed] [Google Scholar]
- 36. Teo JWP, Kalisvar M, Venkatachalam I. et al. mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J Clin Microbiol 2018; 56: e01562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rebelo AR, Bortolaia V, Kjeldgaard JS. et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 2018; 23: pii=17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roer L, Hansen F, Stegger M. et al. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Euro Surveill 2017; 22: pii=30584. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.