Abstract
Objectives
4′-Ethnyl-2′-fluoro-2′-deoxyadenosine (EFdA) is a novel translocation-defective reverse transcriptase inhibitor. We investigated the virological and biochemical inhibitory potentials of EFdA against a broad spectrum of subtype-specific chimeric viruses and compared it with tenofovir alafenamide, nevirapine, efavirenz, rilpivirine and etravirine.
Methods
pNL4.3 chimeric viruses encoding gag-pol from treatment-naive patients (n = 24) and therapy-failure patients (n = 3) and a panel of reverse transcriptase inhibitor-resistant strains (n = 7) were used to compare the potency of reverse transcriptase inhibitor drugs. The phenotypic drug susceptibility assay was performed using TZM-bl cells. In vitro inhibition assays were done using patient-derived reverse transcriptase. IC50 values of NNRTIs were calculated using a PicoGreen-based spectrophotometric assay. Steady-state kinetics were used to determine the apparent binding affinity (Km.dNTP) of triphosphate form of EFdA (EFdA-TP) and dATP.
Results
Among the chimeric treatment-naive viruses, EFdA had an ex vivo antiretroviral activity [median (IQR) EC50 = 1.4 nM (0.6–2.1 nM)] comparable to that of tenofovir alafenamide [1.6 nM (0.5–3.6 nM)]. Subtype-specific differences were found for etravirine (P = 0.004) and rilpivirine (P = 0.017), where HIV-1C had the highest EC50 values. EFdA had a greater comparative efficiency [calculated by dividing the efficiency of monophosphate form of EFdA (EFdA-MP) incorporation (kcat.EFdA-TP/Km.EFdA-TP) over the efficiency of dATP incorporation (kcat.dATP/Km.dATP)] compared with the natural substrate dATP, with a fold change of between 1.6 and 3.2. Ex vivo analysis on reverse transcriptase inhibitor-resistant strains showed EFdA to have a higher potency. Despite the presence of rilpivirine DRMs, some non-B strains showed hypersusceptibility to rilpivirine.
Conclusions
Our combined virological and biochemical data suggest that EFdA inhibits both WT and reverse transcriptase inhibitor-resistant viruses efficiently in a subtype-independent manner. In contrast, HIV-1C is least susceptible to etravirine and rilpivirine.
Introduction
Combination ART (cART) has significantly reduced mortality and morbidity related to HIV-1 infection.1 Advancement of new drugs in clinical settings has been evidenced by their higher tolerability and less adverse effects. For example, the NRTI tenofovir alafenamide,2 which is a novel prodrug of tenofovir, and the NNRTIs etravirine3 and rilpivirine4 have lower toxicity than respective compounds of previous generations. In addition, several new highly efficacious and tolerable integrase strand transfer inhibitors (INSTIs), including dolutegravir, cabotegravir and bictegravir, have been developed.5–7 Importantly, NRTIs still serve as the backbone of cART. Given the increasing global trend of resistance associated with currently used NRTIs and NNRTIs,8 there is a continued need of new NRTIs that have efficacy against NRTI-resistant viruses, especially in low- and middle-income countries.
4′-Ethnyl-2′-fluoro-2′-deoxyadenosine (EFdA) is a new anti-HIV drug that inhibits HIV-1 reverse transcriptase (RT) via a novel mechanism, by acting as a chain-terminating agent that decreases translocation of RT. Thus, EFdA has been termed as translocation-defective reverse transcriptase inhibitor (TDRTI).9 In contrast to all approved NRTIs, EFdA retains a 3′OH group, which is expected to improve its phosphorylation efficiency in vivo by cellular kinases, making it a strong competitor with the natural (dATP) substrate during HIV-1 cDNA synthesis.10 It has been shown that EFdA has low toxicity and a high in vivo potency in Simian immunodeficiency virus (SIV)-infected rhesus macaques and humanized mice.11,12 The first Phase I study in humans reported that the drug suppressed HIV-1 replication for at least 7 days when administered as a single dose as low as 0.5 mg.13 Phase II studies in humans are currently underway. The extraordinary antiretroviral potency and pharmacokinetic properties of EFdA suggest that it is a strong candidate to serve as a long-acting drug and could serve in prophylaxis approaches. EFdA also has potential to be used against NRTI-resistant HIV strains.14
An additional potential problem associated with antiretroviral drugs is the subtype-specific differences in their susceptibility, which have not been evaluated systematically for most drugs. Furthermore, the effect of subtype-specific drug resistance mutations (DRMs) on second-generation reverse transcriptase inhibitors (RTIs) has not been well characterized. With the discovery of new and more potent RTIs that act through novel mechanisms, there is great need for the evaluation of phenotypic drug resistance and in vitro potency for diverse HIV-1 subtypes. This is especially relevant to low- and middle-income countries, where the subtype heterogeneity is very high, and both transmitted as well as acquired HIV drug resistances are increasing significantly.15
Earlier studies on HIV-1 subtype B (HIV-1B) have shown synergism between EFdA and rilpivirine, which may prove to be advantageous against specific RTI-resistant strains.16 In addition, strains containing the DRM K65R, which emerges among patients failing treatment with the first approved oral prodrug of tenofovir, tenofovir disoproxil fumarate, are hypersusceptible to EFdA.14 It should be noted that HIV-1C viruses develop the K65R mutation rapidly.17 Moreover, tenofovir alafenamide is associated with higher intracellular concentrations of tenofovir and may in vitro retain activity against tenofovir disoproxil fumarate-resistant viruses,18 although clinical relevance has not been shown. It is known that HIV-1 subtypes may affect the treatment response for PIs,19,20 but whether subtype-related differences exist for EFdA has not been studied, although it has potent efficacy against HIV-2. In view of the limited information available on the efficacies of different RTIs against different subtypes, the primary aim of this study was to identify the ex vivo (virological) and in vitro (biochemical) antiretroviral potential of all NNRTIs (nevirapine, efavirenz, etravirine and rilpivirine), the most commonly used NRTI (tenofovir alafenamide) and the novel TDRTI EFdA using diverse HIV-1 subtypes with or without resistance mutations.
Materials and methods
Cell lines, plasmids and antiretrovirals
TZM-bl21 cells were obtained from NIH AIDS Reagent Program, NIH, USA. 293 T cells were purchased from ATCC, USA. TZM-bl and 293 T cells were maintained in culture medium consisting of DMEM (Sigma, USA) supplemented with 10% FBS, penicillin/streptomycin antibiotics (100 IU/mL and 50 μg/mL, respectively) and 2 mM l-glutamine. MT-4 cells were maintained in RPMI medium containing 10% FBS, penicillin/streptomycin antibiotics (100 IU/mL and 50 μg/mL, respectively) and 2 mM l-glutamine. The plasmid pNL43,22 which is derived from a WT HIV-1B strain, and seven RT-resistant HIV-1B viruses containing major DRMs [plasmid clones 12227 (M41L, T215Y, K101P, K103N), 12229 (M41L, L74V, M184V, T215Y, L100I, K103N), 12233 (K70R, M184V, T215F, K101E, Y181V), 12235 (M41L, E44D, D67N, T69D, L74I, L210W, T215Y, K101E, Y181C, G190A), 12239 (M41L, T215D, K101E, E138K, Y181C), 12241 (K101E, G190S), 12243 (M41L, D67G, L74I, M184V, T215Y, L100I, M230L)] were obtained from the NIH AIDS Reagent Program, NIH, USA.23 The following drugs were purchased from Selleckchem, USA: nevirapine, efavirenz, etravirine, rilpivirine and tenofovir alafenamide.
Clinical specimens
Stored plasma samples from patients who were either therapy naive (n = 42) or had a virological treatment failure (n = 9) were randomly selected from the HIV cohort at Karolinska University Hospital, Stockholm, Sweden. Of these, 11 samples did not give any positive clones even after multiple efforts, and 13 were not replication competent. Finally we obtained 24 replication-competent viruses from treatment-naive individuals who were pure subtypes (HIV-1A1, n = 1; HIV-1B, n = 5; HIV-1C, n = 15) or circulating recombinant forms (CRFs) (01_AE, n = 1; 02_AG, n = 2), whilst the viruses extracted from patients with therapy failure (n = 3) were HIV-1B (n = 1), HIV-1D (n = 1) and HIV-1C (n = 1), as determined by our previously published near full-length sequencing protocol.24 The A1, 01_AE and 02_AG were grouped as A-like viruses as described earlier for statistical analysis.25
The three viruses extracted from therapy-failure patients had the following mutations: HIV-1D, K65R, T69del, L100I, Y181C, Y188L, H221Y; HIV-1C, K65R, M184V, V108I, Y181C, G190S; HIV-1B, M41L, D67N, K70R, M184V, L210W, T215Y, K219E, K103S and G190A. Ethics clearance for the study was obtained from the Regional Ethics Committee of Stockholm, Sweden (Dnr 2014/928-31/2 and 2013/1944-31/4). All participants gave informed consent.
Recombinant virus production
Briefly, viral RNA was extracted using the QIAmp viral RNA extraction kit (Qiagen, Hilden, Germany) from 140 μL of plasma. The gag-pol fragment (HXB2: 0702-5798) was cloned into pNL43 plasmid following digestion with BssHII and SalI (New England Biolabs, USA) and ligation using T4 DNA ligase (New England Biolabs), as described by us previously.26 The chimeric viruses were produced by transient transfection of the plasmids into the 293 T cell line using FuGENE® HD Transfection Reagent (Promega, USA) and harvested 72 h later by collection of the cell-free supernatant via centrifugation; aliquots were stored at −80°C.
Drug susceptibility assay (DSA)
DSA was performed by determining the extent to which EFdA, nevirapine, efavirenz, etravirine, rilpivirine and tenofovir alafenamide inhibited the replication of the reference virus (NL43) and the 24 treatment-naive patient-derived chimeric viruses (HIV-1B, 6; HIV-1C, 14; HIV-1A-like, 4). Drugs serially diluted in culture medium, spanning from 10 to 0.000001 μM, were added in triplicate in 96-well plates that had been seeded 24 h before the start with 10 000 TZM-bl cells/well. The viruses were added to each well at an moi of 0.05 IU/cell in the presence of 10 μg/mL DEAE. Virus replication was quantified by measuring luciferase activity (relative light units) using the Bright-Glo™ Luciferase Assay System (Promega, USA) 48 h post-infection. Drug concentrations required for inhibiting virus replication by 50% (EC50) were calculated from a dose–response curve using non-linear regression analysis (GraphPad Prism, version 6.07; GraphPad Software, La Jolla, CA, USA).
The DSA experiments were performed with three technical replicates for each virus with the specified dynamic concentration range of the drug, and at least two independent analyses (biological replicates) were performed. The reproducibility of the DSA was assessed based on the 95% CI obtained for the drug EC50 and the degree of correlation between technical replicates. The output for the drug EC50 results was used to compute the fold change value for each virus relative to NL4.3 before being exported to GraphPad Prism software for graphical representation and statistical analysis.
Cloning and purification of RT
Patient-derived HIV-1 RT enzymes were used in NNRTI susceptibility and kinetics assays. All enzymes were purified as described previously.27 Briefly, The RT genes of different subtypes were PCR amplified from DNA amplicons of viruses isolated from patients infected with diverse subtypes. For the cloning of the p66 subunit, NcoI and SalI restriction sites were added, except for 02_AG, for which primers were designed to add the SalI restriction site. For the cloning of the p51 subunit, BamHI and SalI restriction sites were added. The p66 subunit was cloned in the pCDFDuet-1 vector (EMD Millipore, Billerica, MA, USA), whereas for p51 the pRSFDuet-1 vector (EMD Millipore) was used. The p51 subunit contained a 6X histidine tag on its N terminus when expressed. The cloned genes on each plasmid are controlled by a T7 promoter and lac operator. The proteins were expressed and purified using an Ni-affinity column.
IC50 values of NNRTIs
The NNRTI susceptibility assays were conducted in 96-well plates by measuring the conversion of double-stranded DNA from Td100/Pd18 template-primer using a PicoGreen-based spectrophotometric assay in the presence of increasing concentrations of NNRTIs. The reactions, consisting of 20 nM RTs from different subtypes, 50 nm Td100/Pd18 and 10 μm dNTPs in a buffer containing 50 mM Tris, pH 7.8, and 60 mM KCl, were initiated by adding 5 mm MgCl2. The reactions were allowed to proceed for 30 min at 37°C followed by quenching with the addition of 100 mm EDTA. Quant-iTTM PicoGreen reagent (Invitrogen, USA) was added to quantify the amount of double-stranded DNA formed in TE buffer. The reaction mixtures were excited at 480 nm and fluorescence was monitored at 520 nm using an EnSpire Multilabel plate reader (PerkinElmer Life Sciences). Dose–response curves of triplicate samples were plotted using GraphPad Prism version 6.07 (GraphPad, Inc.) to determine the IC50 values of NNRTIs.
Steady-state kinetics of dATP and EFdA-TP incorporation
We used steady-state kinetics to determine the apparent binding affinity (Km.dNTP) of triphosphate form of EFdA (EFdA-TP) and dATP with enzyme/template-primer (31/18-mer) complex. In these assays, 10 nM RT and 100 nM T/P were incubated in a buffer containing 50 mM Tris-HCl, pH 7.8, and 60 mM KCl. The reactions were initiated by the addition of 5 mM MgCl2 and increasing concentrations of dATP (10 nM to 25 μM) or EFdA-TP (2.5 nM to 5 μM). The reactions were allowed to proceed for 2 min followed by quenching by the addition of a solution containing 95% formamide, 0.01% bromophonol blue and 25 mM EDTA. The reaction products were resolved on a 16% polyacrylamide/7 M urea gel. The bands corresponding to 19-mer product were visualized on Typhoon (Thermo Fisher, USA) and quantified by ImageQuant (GE Electronics, USA). The initial velocity of reaction at each dATP or EFdA-TP concentration was fitted to a hyperbolic function (Michaelis–Menten equation) to determine kcat and Km.substrate. The efficiency of nucleotide incorporation [monophosphate form of EFdA (EFdA-MP) or dAMP] was defined as the ratio of kcat to Km. The comparative efficiency of EFdA-TP over dATP was calculated by dividing the efficiency of EFdA-MP incorporation (kcat.EFdA-TP/Km.EFdA-TP) over the efficiency of dATP incorporation (kcat.dATP/Km.dATP).
Statistical analysis
Descriptive values for the EC50 in nM for each drug are represented as the median and IQR. The difference between two drugs was evaluated using the Wilcoxon matched-pairs signed-rank test. Subtype-specific differences were measured using the Kruskal–Wallis test. All statistical analysis were computed in GraphPad Prism version 6.07 (GraphPad, Inc.).
Results
Ex vivo antiretroviral potency on WT viruses
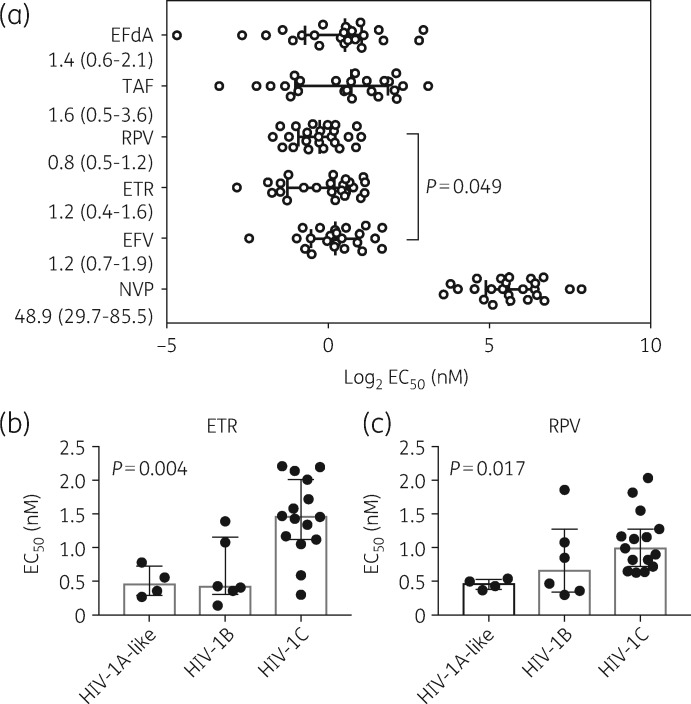
The chimeric viruses derived from therapy-naive patients (n = 24) were used to investigate the susceptibility to six RTI drugs: EFdA, nevirapine, efavirenz, etravirine, rilpivirine and tenofovir alafenamide (Figure 1a). EFdA had an ex vivo antiretroviral activity [median (IQR) EC50=1.4 nM (0.6–2.1 nM)] comparable to that of tenofovir alafenamide [1.6 nM (0.5–3.6 nM)] (Figure 1a). The NNRTIs rilpivirine [0.8 nM (0.5–1.2 nM)], etravirine [1.2 nM (0.4–1.6 nM)] and efavirenz [1.1 nM (0.7–1.9 nM)] all had higher potency than nevirapine [48.9 nM (29.7–85.5 nM)] (P < 0.001, for all comparisons). In addition, rilpivirine showed higher potency than efavirenz (P = 0.049).
Figure 1.
Effects of EFdA and other RTIs on 24 chimeric viruses representing HIV-1B (n = 5), HIV-1C (n = 15), HIV-1A1 (n = 1), HIV-1-01_AE (n = 1), HIV-1-02_AG (n = 2) and WT reference pNL4-3. (a) Intra-drug comparative analysis of the log2 EC50. Each virus is represented by a single circle on the graph. The log2 median EC50 value and IQR are indicated. Significant differences are shown as P values obtained from performing the Mann–Whitney test for each of two drugs. The actual EC50 values are presented as median (IQR) below each drug abbreviation. The G190A contacting viruses were excluded from the median EC50 analysis of nevirapine. Subtype-specific comparisons for (b) etravirine and (c) rilpivirine on HIV-1B, HIV-1C and HIV-1A-like viruses. The median EC50 value and IQR are indicated. Significant differences were found for etravirine (P = 0.004) and rilpivirine (P = 0.017). Statistical analyses were performed using the Kruskal–Wallis test. TAF, tenofovir alafenamide; RPV, rilpivirine; ETR, etravirine; EFV, efavirenz; NVP, nevirapine.
While comparing the specific potency for HIV-1B, HIV-1C and HIV-1A-like strains, no statistically significant difference was found for EFdA [median (IQR) EC50=1.69 nM (1.41–2.68 nM), 1.41 nM (0.38–1.67 nM) and 1.69 nM (1.41–2.68 nM); Kruskal–Wallis test P = 0.41], tenofovir alafenamide (Kruskal–Wallis test P = 0.23) and the first-generation NNRTI (nevirapine, P = 0.13; efavirenz, P = 0.64). However, subtype-specific differences were found for the second-generation NNRTIs etravirine (P = 0.004) (Figure 1b) and rilpivirine (P = 0.017) (Figure 1c), where HIV-1C had the highest EC50 values.
In vitro biochemical analysis of WT viruses
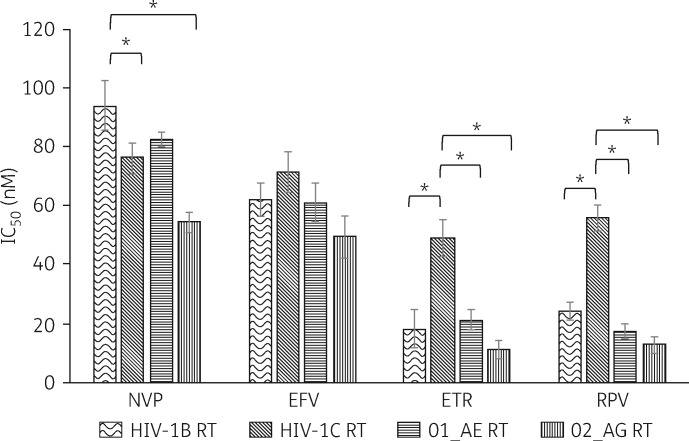
To confirm the relatively lower susceptibility to etravirine and rilpivirine for HIV-1C observed in cell-based assays, we determined IC50 values in cell-free biochemical assays. In vitro biochemical assays targeting nevirapine, etravirine and rilpivirine identified subtype-specific differences, but this was not observed for efavirenz (Figure 2). For nevirapine, non-B subtypes showed lower IC50 values compared with HIV-1B (HIV-1B versus HIV-1C, P = 0.02; HIV-1B versus HIV-102_AG, P = 0.005). In line with the cell-based assay, for both etravirine and rilpivirine, HIV-1C had higher IC50 values compared with HIV-1B (P = 0.03 and P = 0.02, respectively), 01_AE (P = 0.02 and P = 0.02, respectively) and 02_AG (P = 0.006 and P = 0.01, respectively). The steady-state kinetics of nucleotide incorporation showed that EFdA was preferred over natural substrate dATP, with a fold change of between 1.6 and 3.2 (Table 1). No statistical difference in comparative efficiency was observed between the subtypes. Intriguingly, the polymerase efficiency of the monophosphate form of EFdA (EFdA-MP) incorporation by HIV-1C RT was significantly less than those of the RT of the other subtypes.
Figure 2.
In vitro biochemical data on the IC50 values of different NNRTIs for different HIV-1 subtypes. Statistical analyses were performed using a paired t-test. P values <0.05 are indicated by asterisks. NVP, nevirapine; EFV, efavirenz; ETR, etravirine; RPV, rilpivirine.
Table 1.
Steady-state kinetic parameters (Km and kcat) for EFdA-MP and dAMP incorporation by HIV-1 RTs from different subtypes
| Subtype |
kcat (m−1) |
Km.dNTP (nM) |
kcat/Km.dNTP (nM−1 m−1) |
Comparative efficiencya | |||
|---|---|---|---|---|---|---|---|
| dATP | EFdA-TP | dATP | EFdA-TP | dATP | EFdA-TP | ||
| HIV-1B | 16 ± 2 | 19 ± 3 | 49 ± 4 | 21 ± 3 | 0.33 | 0.90 | 2.8 |
| HIV-1C | 8 ± 3 | 9 ± 2 | 39 ± 2 | 27 ± 3 | 0.21 | 0.33 | 1.6 |
| 01_AE | 19 ± 2 | 22 ± 3 | 44 ± 3 | 16 ± 4 | 0.43 | 1.38 | 3.2 |
| 02_AG | 22 ± 3 | 18 ± 4 | 55 ± 5 | 17 ± 3 | 0.40 | 1.01 | 2.5 |
Comparative efficiency is the ratio of the incorporation efficiency (kcat/Km) of EFdA-MP over that of dAMP [(kcat/Km)EFdA-MP/(kcat/Km)dAMP].
Ex vivo potency on drug-resistant viruses
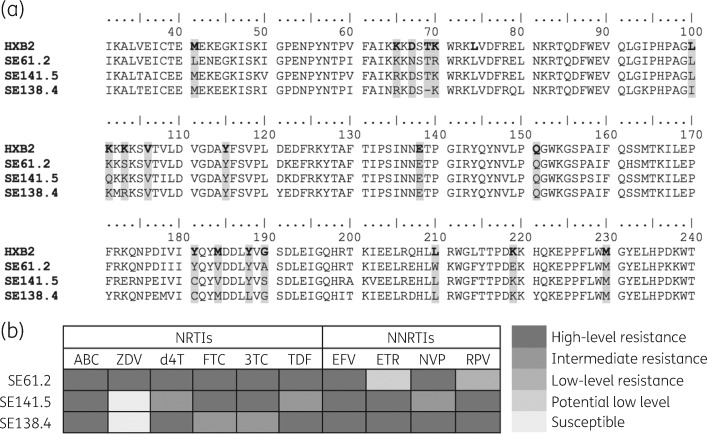
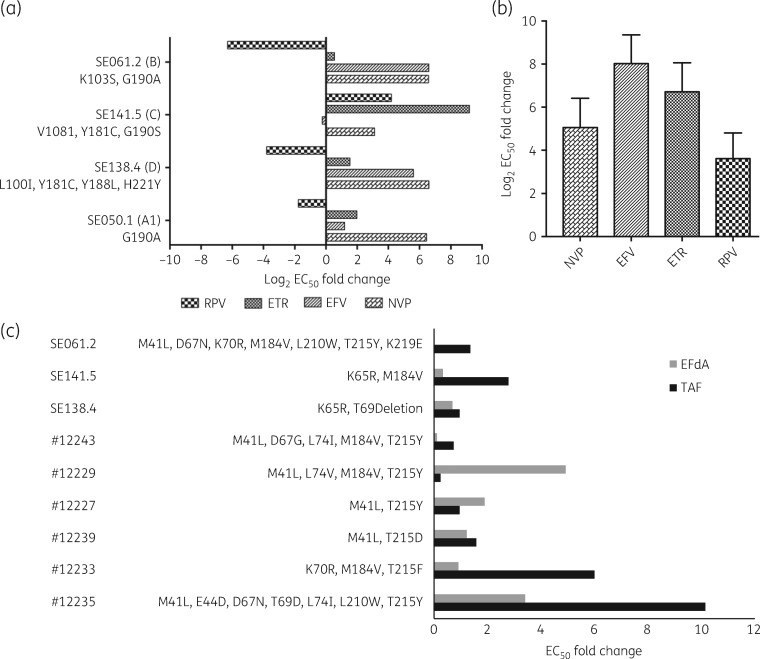
The multiple sequence alignment is presented in Figure 3(a). In SE138.4, a polymorphic mutation K103R was detected, whereas in SE141.5, K101Q, a relatively non-polymorphic mutation, was detected. No other accessory mutations were detected. The genotypic resistance prediction in the Stanford HIV drug resistance database identified all the strains as intermediate- to high-level resistant to all NRTIs except zidovudine and NNRTIs (Figure 3b). The DSA was performed on three resistant viruses from treatment-failure patients, one G190A-containing strain from a treatment-naive individual, and seven NNRTI-resistant viruses obtained from the NIH AIDS Reagent Program. The log2 fold changes in EC50 against WT pNL4-3 viruses in NNRTIs are shown in Figure 4(a). It is interesting to note that despite the presence of four rilpivirine mutations (L100I, Y181C, Y188L and H221Y), the HIV-1D strain (SE138.4) showed hypersusceptibility to rilpivirine, and another virus (SE141.5) with V108I, Y181C and G190S mutations showed 18 (4 log2) fold lower susceptibility to rilpivirine. The third HIV-1B virus (SE061.2) with the G190A mutation was hypersusceptible to rilpivirine. Similar data were obtained for the HIV-1A1 virus with the single G190A mutation obtained from a treatment-naive individual. As expected, the seven NNRTI-panel HIV-1B viruses showed reduced susceptibility to all the drugs (nevirapine, efavirenz, etravirine and rilpivirine) (Figure 4b).
Figure 3.
Multiple sequence alignment and genotypic resistance prediction. (a) Multiple sequence alignment of the clinical isolates from treatment-failure patients. HXB2 was used as reference. The DRM positions are shaded. (b) Genotypic resistance prediction of the clinical isolates in the Stanford HIV drug resistance database. ABC, abacavir; ZDV, zidovudine, d4T, stavudine; FTC, emtricitabine; 3TC, lamivudine; TDF, tenofovir disoproxil fumarate; EFV, efavirenz; ETR, etravirine; NVP, nevirapine; RPV, rilpivirine.
Figure 4.
Ex vivo DRM profile of the chimeric viruses with DRMs. (a) Fold resistance of the laboratory isolates with NNRTI mutations against the NNRTIs. (b) Log2 EC50 fold resistance on NNRTI-panel viruses from the NIH AIDS Reagent Program, NIH, USA in our assay against WT pNL4-3 indicated fold resistance. Data presented as median (IQR). (c) Ex vivo sensitivity analysis of fold resistance to EFdA and tenofovir alafenamide on viruses having NRTI mutations. EC50 fold change is shown. RPV, rilpivirine; ETR, etravirine; EFV, efavirenz; NVP, nevirapine; TAF, tenofovir alafenamide.
Among the seven NNRTI-resistant panel viruses, six also had NRTI mutations. EFdA suppressed seven out of nine viruses with NRTI mutations, of which hypersusceptibility was found in four viruses: 12243 (fold resistance = 0.091), SE138.4 (fold resistance = 0.69), SE141.5 (fold resistance = 0.33) and SE061.2 (fold resistance = 0.02). Interestingly two of the viruses, SE138.4 and SE141.5, had the K65R mutation. EFdA showed 3-fold reduced susceptibility in a virus with both thymidine analogue mutations pathway 1 (TAM-1) (M41L, T210W and T215Y) and pathway 2 (TAM-2) (D67N), along with other mutations (E44D, L74I and T69D), but had still better activity than tenofovir alafenamide (fold resistance = 10). Interestingly when both TAM-1 (M41L, T210W, T215Y) and TAM-2 (D67N, K70R, K219E) with the M184V mutation were present, EFdA showed hypersusceptibility. One virus with TAM-1 (M41L, T215Y) and L74V showed a high fold resistance to EFdA. Despite the presence of the K65R mutation, one virus (K65R and T69deletion) showed susceptibility to tenofovir alafenamide, and another (K65R and M184V) 3-fold resistance (Figure 4c).
Discussion
cART remains the most effective approach for managing the HIV pandemic. However, the introduction of novel antiretrovirals has created a need for phenotypic analysis to evaluate their potency and drug resistance profiles, not only for HIV-1B but also for the globally dominating non-B subtypes. In this study, we created a panel of non-B subtype chimeric viruses with or without DRMs and investigated the ex vivo and in vitro potency of first- and second-generation RTIs commonly used in low- and middle-income countries, and compared these with the novel drug EFdA. Our biochemical and virological study indicated that EFdA exhibited high efficiency against both WT viruses and DRM-containing viruses in a subtype-independent manner. In addition, EFdA showed equal efficacy or hypersusceptibility for most drug-resistant viruses when compared with WT viruses. In contrast, the second-generation NNRTIs etravirine and rilpivirine had a lower potency in HIV-1C strains.
In the DSA, EFdA displayed ex vivo potency for WT chimeric viruses comparable to that shown by other RTIs. The EC50 values for other RTIs used in our study were in the ranges reported previously. Thus, the reported median EC50 values are in the range of 5 nM28,29 to 10 nM18 for tenofovir alafenamide, between 1.5 nM18 and 2.0 nM30 for efavirenz, 0.1–2.0 nM for rilpivirine,31 and 1.4–4.0 nM32 for etravirine, and our value of 48.87 nM for nevirapine is close to the reported value of 84 nM found in enzymatic assays.33 The slight differences in published values compared with our values are most likely attributable to differences in the phenotypic assays used.
The results of our analysis of the EFdA susceptibility of the RTI-resistant strains confirm the strong potential EFdA has as an RTI, e.g. in a salvage regimen. Furthermore, no subtype-specific differences in EFdA susceptibility were found, supporting the use of EFdA in countries where non-HIV-1B viruses are dominating.
Moreover, tenofovir alafenamide, nevirapine and efavirenz had similar efficacy, independent of the subtype. In contrast, HIV-1C was less susceptible to both etravirine and rilpivirine, as compared with non-HIV-1C. This is a novel finding although an earlier study concluded that HIV-1C viruses containing DRMs against nevirapine and efavirenz will have decreased efficacy against both etravirine and rilpivirine in nearly half of the patients. However, no comparisons were done with non-HIV-1C in that study.34 Our finding adds further caution over the use of second-generation NNRTIs in HIV-1C-infected patients. Analysis of patients from the EARNEST trial35 failing first-line ART reported that non-B subtypes differ in DRMs, which impacts the residual NNRTI susceptibility. In particular, a higher rate of etravirine and rilpivirine resistance was predicted for HIV-1C, which limits their potential utility in further regimens.36,37 Another complementary possibility is that HIV-1C might harbour uncharacterized natural SNPs that possibly could reduce the fitness and delay the onset of viruses harbouring etravirine DRMs, resulting in a further accumulation of DRMs. Such an example is the emergence of the N348I mutation in the connection domain of HIV-1 RT, which has been shown to reduce the fitness of E138K-carrying HIV-1 strains known to be have a reduced etravirine susceptibility, delaying the detection of these resistant viruses.38 The same could be the case for rilpivirine.
The complex issue of subtype-specific differences in resistance is illustrated by our finding that rilpivirine DRMs, e.g. G190A, were present in viral strains that were hypersusceptible to rilpivirine. This could be due to the presence of other mutations as well. Thus, one of the HIV-1B-panel viruses (NIH1883), which had G190A along with Y181C and K101E, showed a 12-fold resistance, but, in contrast, an HIV-1D virus with four potential rilpivirine mutations was hypersusceptible to rilpivirine. This finding was further supported by biochemical data suggesting that HIV-1C had higher IC50 values of etravirine and rilpivirine, indicating a subtype-specific effect in the binding affinity of different HIV-1 RTs. This finding further supports our earlier report where we showed that HIV-1C RT had a 2-fold higher IC50 of rilpivirine.37
EFdA consistently suppressed both therapy-naive and resistant viruses. The K65R- and M184V-carrying strains were suppressed more effectively by EFdA than by tenofovir alafenamide. The superiority of EFdA is supported by mechanistic studies that have used single cycle cell-based assays showing that has tenofovir-resistant viruses with the K65R mutation have a 2.5-fold increased hypersusceptibility to EFdA-TP.14 The 2′fluoro group in EFdA is resistant to cellular degradation by deaminases and extends the half-life of EFdA more than 72 h in in vitro experiments,12 making EFdA a strong candidate for use as a long-acting drug. A recent study showed a significant increase in the proportion of patients with TAM at failure of tenofovir disoproxil fumarate-based first-line ART including the standard tenofovir disoproxil fumarate mutation K65R.8 Therefore, zidovudine may not be ideal for those patients as a second-line option. Therefore, EFdA can potentially fill the gap as viral strains with K65R are hypersusceptible to EFdA. These in vitro experiments give further support for the use of EFdA in all HIV-1 subtypes. Thus, the comparative efficiency determinations showed that EFdA-TP was selected better (by at least 2-fold) by RTs from all subtypes. The high binding efficacy of EFdA is mainly due to 4′ethynyl group, which binds to a hydrophobic pocket explicitly present in the structure of HIV-1 RT,9,10 and this binding site is conserved in HIV-1B, HIV-1C, 01_AE and 02_AG.
Our study has limitations that merit comment. First, the low numbers and unevenness of therapy-naive chimeric viruses could potentially result in a statistical bias. This is mainly due to the failure of cloning or non-functional viruses. Second, four HIV-1 RTs were used to study the efficacy of six drugs in our biochemical assays. To the best of our knowledge, this is the highest number of enzymes and drugs used in any study as most of the earlier studies were often restricted to one. It must also be emphasized that these experiments were conducted as a proof-of-concept.
In conclusion, our combined in vitro virological and biochemical analysis suggests that EFdA inhibits both WT and RTI-resistant viruses efficiently in a subtype-independent manner and is, therefore, a valid choice of drug for clinical trials involving both therapy-naive (as first-line drug) and therapy-failure (as second-line drug) individuals. In contrast, subtype-specific differences were observed for etravirine and rilpivirine. Rilpivirine DRMs in non-B subtypes need further investigation given that rilpivirine is part of a long-acting formulation under development. It can be claimed that these subtype-specific differences for etravirine and rilpivirine described by us in vitro and ex vivo have not been shown to be of relevance in vivo. However, when an antiretroviral is introduced on a large scale to patients outside well-controlled clinical trials, it is still possible that such differences may result in differences in clinical outcome, especially in patients with suboptimal adherence.
Acknowledgements
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells (cat. #8129) from Dr John C. Kappes and Dr Xiaoyun Wu, and IV-1 NL4-3 Infectious Molecular Clone (pNL4-3) (cat. #114) from Dr Malcolm Martin.
Funding
This work was supported by grants from the Swedish Research Council (grant numbers 2016-01675 to A. S. and 2017–01330 to U. N), a grant from Stockholm County Council (grant number ALF20160074 to A. S. and U. N.), a Bond Life Sciences Center grant (DU108) and a National Institutes of Health (NIH) CTSA grant (TR002345) (to K. S.) and NIH RO1 grants (grant numbers AI076119 to S. G. S. and GM118012 to K. S., U. N., S. G. S. and A. S.).
Transparency declarations
None to declare.
Author contributions
U. N. conceived and designed the studies and analysis plan. D. T. N. and S. G. A. performed the virological assays (supervised by U. N.). D. T. N., S. G. A. and U. N. are responsible for the virological data. D. T. N. performed all the statistical analysis for the virological assays and prepared the figures. K. S. and R. R. performed the biochemical assays. K. S. wrote the biochemical part of the study. K. S. and S. G. S. are responsible for the biochemical data. A. S. interpreted the clinical data and is responsible for the InfCare cohort. D. T. N. wrote the first draft of the manuscript, which was reviewed by U. N., K. S., A. S. and S. G. S. All the authors approved the final version of the manuscript.
References
- 1. Wang H. Estimates of global, regional, and national incidence, prevalence, and mortality of HIV, 1980–2015: the Global Burden of Disease Study 2015. Lancet HIV 2016; 3: e361–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. De Clercq E. Tenofovir alafenamide (TAF) as the successor of tenofovir disoproxil fumarate (TDF). Biochem Pharmacol 2016; 119: 1–7. [DOI] [PubMed] [Google Scholar]
- 3. Floris-Moore MA, Mollan K, Wilkin AM. et al. Antiretroviral activity and safety of once-daily etravirine in treatment-naive HIV-infected adults: 48-week results. Antivir Ther 2016; 21: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen C, Wohl D, Arribas JR. et al. Week 48 results from a randomized clinical trial of rilpivirine/emtricitabine/tenofovir disoproxil fumarate vs. efavirenz/emtricitabine/tenofovir disoproxil fumarate in treatment-naive HIV-1-infected adults. AIDS 2014; 28: 989–97. [DOI] [PubMed] [Google Scholar]
- 5. Clotet B, Feinberg J, van Lunzen J. et al. Once-daily dolutegravir versus darunavir plus ritonavir in antiretroviral-naive adults with HIV-1 infection (FLAMINGO): 48 week results from the randomised open-label phase 3b study. Lancet 2014; 383: 2222–31. [DOI] [PubMed] [Google Scholar]
- 6. Margolis DA, Gonzalez-Garcia J, Stellbrink H-J. et al. Long-acting intramuscular cabotegravir and rilpivirine in adults with HIV-1 infection (LATTE-2): 96-week results of a randomised, open-label, phase 2b, non-inferiority trial. Lancet 2017; 390: 1499–510. [DOI] [PubMed] [Google Scholar]
- 7. Sax PE, DeJesus E, Crofoot G. et al. Bictegravir versus dolutegravir, each with emtricitabine and tenofovir alafenamide, for initial treatment of HIV-1 infection: a randomised, double-blind, phase 2 trial. Lancet HIV 2017; 4: e154–60. [DOI] [PubMed] [Google Scholar]
- 8. Gregson J, Tang M, Ndembi N. et al. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis 2016; 16: 565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Michailidis E, Marchand B, Kodama EN. et al. Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-ethynyl-2-fluoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. J Biol Chem 2009; 284: 35681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salie ZL, Kirby KA, Michailidis E. et al. Structural basis of HIV inhibition by translocation-defective RT inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA). Proc Natl Acad Sci USA 2016; 113: 9274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murphey-Corb M, Rajakumar P, Michael H. et al. Response of simian immunodeficiency virus to the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine in vitro and in vivo. Antimicrob Agents Chemother 2012; 56: 4707–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stoddart CA, Galkina SA, Joshi P. et al. Oral administration of the nucleoside EFdA (4′-ethynyl-2-fluoro-2′-deoxyadenosine) provides rapid suppression of HIV viremia in humanized mice and favorable pharmacokinetic properties in mice and the rhesus macaque. Antimicrob Agents Chemother 2015; 59: 4190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matthews RP, Schürmann D, Rudd DJ. et al. Single doses as low as 0.5 mg of the novel NRTTI MK-8591 suppress HIV for at least seven days In: IAS 2017, Paris, France. Abstract number: TUPDB0202LB. [Google Scholar]
- 14. Michailidis E, Ryan EM, Hachiya A. et al. Hypersusceptibility mechanism of tenofovir-resistant HIV to EFdA. Retrovirology 2013; 10: 65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gupta RK, Gregson J, Parkin N. et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis 2018; 18: 346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hachiya A, Reeve AB, Marchand B. et al. Evaluation of combinations of 4’-ethynyl-2-fluoro-2’-deoxyadenosine with clinically used antiretroviral drugs. Antimicrob Agents Chemother 2013; 57: 4554–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coutsinos D, Invernizzi CF, Xu H. et al. Template usage is responsible for the preferential acquisition of the K65R reverse transcriptase mutation in subtype C variants of human immunodeficiency virus type 1. J Virol 2009; 83: 2029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Margot NA, Johnson A, Miller MD. et al. Characterization of HIV-1 resistance to tenofovir alafenamide in vitro. Antimicrob Agents Chemother 2015; 59: 5917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haggblom A, Svedhem V, Singh K. et al. Virological failure in patients with HIV-1 subtype C receiving antiretroviral therapy: an analysis of a prospective national cohort in Sweden. Lancet HIV 2016; 3: e166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sutherland KA, Ghosn J, Gregson J. et al. HIV-1 subtype influences susceptibility and response to monotherapy with the protease inhibitor lopinavir/ritonavir. J Antimicrob Chemother 2015; 70: 243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei X, Decker JM, Liu H. et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 2002; 46: 1896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adachi A, Gendelman HE, Koenig S. et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 1986; 59: 284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Balamane M, Varghese V, Melikian GL. et al. Panel of prototypical recombinant infectious molecular clones resistant to nevirapine, efavirenz, etravirine, and rilpivirine. Antimicrob Agents Chemother 2012; 56: 4522–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grossmann S, Nowak P, Neogi U.. Subtype-independent near full-length HIV-1 genome sequencing and assembly to be used in large molecular epidemiological studies and clinical management. J Int AIDS Soc 2015; 18: 20035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tebit DM, Arts EJ.. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 2011; 11: 45–56. [DOI] [PubMed] [Google Scholar]
- 26. Neogi U, Singh K, Aralaguppe SG. et al. Ex vivo antiretroviral potency of newer integrase strand transfer inhibitors cabotegravir and bictegravir in HIV-1 non-B subtypes. AIDS 2018; 32: 469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh K, Marchand B, Rai DK. et al. Biochemical mechanism of HIV-1 resistance to rilpivirine. J Biol Chem 2012; 287: 38110–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Callebaut C, Stepan G, Tian Y. et al. In vitro virology profile of tenofovir alafenamide, a novel oral prodrug of tenofovir with improved antiviral activity compared to that of tenofovir disoproxil fumarate. Antimicrob Agents Chemother 2015; 59: 5909–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chapman H, Kernan M, Rohloff J. et al. Purification of PMPA amidate prodrugs by SMB chromatography and x-ray crystallography of the diastereomerically pure GS-7340. Nucleosides Nucleotides Nucleic Acids 2001; 20: 1085–90. [DOI] [PubMed] [Google Scholar]
- 30. Petropoulos CJ, Parkin NT, Limoli KL. et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother 2000; 44: 920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janssen PA, Lewi PJ, Arnold E. et al. In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2, 6-dimethylphenyl]amino]-2- pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J Med Chem 2005; 48: 1901–9. [DOI] [PubMed] [Google Scholar]
- 32. Das K, Clark AD Jr, Lewi PJ. et al. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J Med Chem 2004; 47: 2550–60. [DOI] [PubMed] [Google Scholar]
- 33. Grob PM, Wu JC, Cohen KA. et al. Nonnucleoside inhibitors of HIV-1 reverse transcriptase: nevirapine as a prototype drug. AIDS Res Hum Retroviruses 1992; 8: 145–52. [DOI] [PubMed] [Google Scholar]
- 34. Basson AE, Rhee SY, Parry CM. et al. Impact of drug resistance-associated amino acid changes in HIV-1 subtype C on susceptibility to newer nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 2015; 59: 960–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kityo C, Thompson J, Nankya I. et al. HIV drug resistance mutations in non-B subtypes after prolonged virological failure on NNRTI-based first-line regimens in sub-Saharan Africa. J Acquir Immune Defic Syndr 2017; 75: e45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neogi U, Shet A, Shamsundar R. et al. Selection of nonnucleoside reverse transcriptase inhibitor-associated mutations in HIV-1 subtype C: evidence of etravirine cross-resistance. AIDS 2011; 25: 1123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neogi U, Haggblom A, Singh K. et al. Factors influencing the efficacy of rilpivirine in HIV-1 subtype C in low- and middle-income countries. J Antimicrob Chemother 2016; 71: 367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu HT, Colby-Germinario SP, Oliveira M. et al. The connection domain mutation N348I in HIV-1 reverse transcriptase enhances resistance to etravirine and rilpivirine but restricts the emergence of the E138K resistance mutation by diminishing viral replication capacity. J Virol 2014; 88: 1536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]