Abstract
Onion (Allium cepa L.) is an important bulbous vegetable crop that possesses important properties related to health as well as extraordinary colors. Naturally white onion bulbs were used in this study to reveal the complex metabolic mechanisms that underlie phenotypic traits, especially bulb pigmentation. Six libraries (three dark-red and three white) were constructed and analyzed to elucidate differences in cyanidin (Cy) metabolism between dark-red and white onion bulbs. Libraries were screened using RNA-sequencing (RNA-seq) to reveal the differentially expressed genes (DEGs) involved in anthocyanin biosynthesis at the transcriptional level. Comparison with the Kyoto Encyclopedia of Genes and Genomes (KEGG) database shows that a total of 27 unigenes participate in onion anthocyanin biosynthesis and 16 DEGs perform critical roles in flavonoid biosynthesis. Expression patterns of color-related flavonoid compounds associated with the onion anthocyanin biosynthesis pathway (ABP) show that flavonoid 3′,5′-hydroxylase (F3′5′H) and dihydroflavonol 4-reductase (DFR) genes play crucial roles in the biosynthesis of dark-red bulbs, the expression levels of flavonol synthase (FLS) and DFR genes may act to block blue pigmentation, and the loss of Cy from white onion bulbs might explain multibranching in the synthesis of this compound. Positive variation in the F3′5′H/F3′H ratio also affects onion bulb color diversity. The transcriptome presented here provides a basis for future onion molecular breeding based on variations in the diversity of ornamental plant pigmentation.
Introduction
Onion (Allium cepa L.) is classified within the family Alliaceae and is an important and well-known vegetable used for cooking. These plants are an important source of nutrients and antioxidants in human diets, are known to have numerous health-related benefits, and are popular because they can mitigate the effects of diabetes, bronchial asthma, and cardiovascular disease1–3. Onion bulb color varieties range from white, to pink, reddish-purple, and dark-red; the appeal of these different varieties depends on their colors and the fact that pigmentation imparts resistance to onion smudge, Colletotrichum circinans (Berk.). Thus, as purely white bulbs are prone to infection4, onion pigments are important not just for their benefits to human health, but because of resultant disease-resistance during breeding selection for novel varieties. Therefore, understanding the molecular mechanism (s) that underlie color inheritance in onion bulbs is critically important.
Flavonoids are common plant secondary metabolites belonging to the class of phenylpropanoids class and occur in various modified forms. One of the best known flavonoid functions is in pigmentation; the colors of various vegetables, flowers, and fruits are the result of specific flavonoids known as anthocyanin compounds. Specific flavonoids, the anthocyanins, essentially contribute to the coloration of fruits and vegetables in many species. The dominant pigments in onion bulbs are cyanidin (Cy) and delphinidin glycosides5–8. Anthocyanins, one class of flavonoids that have a basic C6-C3-C6 structure, are responsible for the orange-to-blue colors seen in many color plants, are water-soluble, and are stored in plant vacuoles. The three main branches (Cy, Pg and Del) comprise the anthocyanin biosynthesis pathway (ABP) and determine the orange-red, deep red, and blue hues of onions; These differences in the number of hydroxyl groups present in B-rings such that a higher quantity results in a blueish color. Similarly, a concomitant increase in the number of methyl groups results in a redder pigmentation9. Although a number of published reports have addressed anthocyanin metabolism in ornamental flowers and plants with colored bulbs10,11, the genetic mechanisms involved in the loss of this dark-red pigment in white onion bulbs have not so far been addressed.
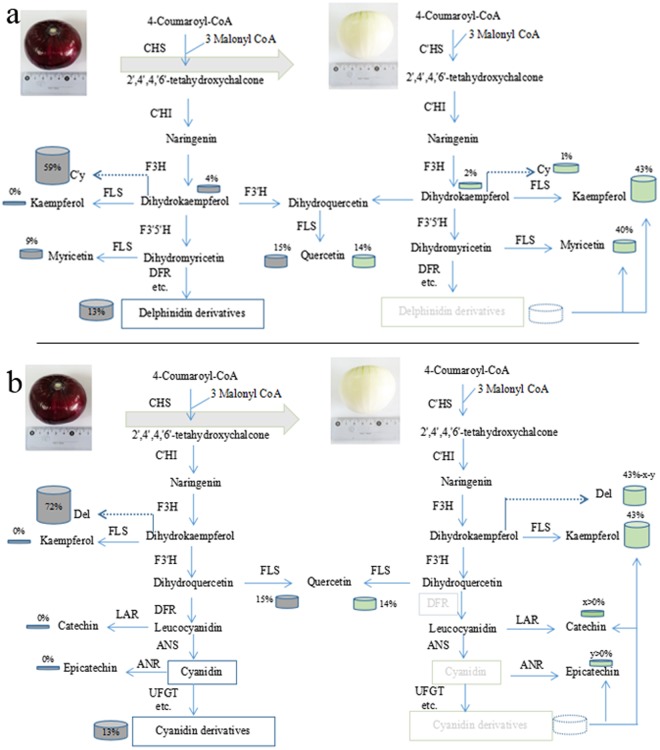
The level of onion bulb pigmentation is closely-related to anthocyanin metabolism2,12, a process that takes place in the cytosol and involves various enzymes. For example, chalcone synthase (CHS), the first committed enzyme, catalyzes the chalcone 2′,4′,4,6′-tetahydroxychalcone (THC) that is produced by one molecule of 4-coumaroyl CoA and three molecules of malonyl CoA. THC is then isomerized to colorless naringenin by chalcone isomerase (CHI), naringenin is hydroxylated by flavanone 3-hydroxylase (F3H) to produce dihydrokaempferol, while flavonoid 3′,5′-hydroxylase (F3′5′H) and flavonoid 3′-hydroxylase (F3′H) catalyze the hydroxylation of dihydrokaempferol (DHM) to produce dihydromyricetin and dihydroquercetin, respectively. These flavonoids are essential for the production of Del and Cy, respectively; thus, dihydroflavonols are reduced to lecoanthocyanidins via the effects of dihydroflavonol 4-reductase (DFR) and are converted to quercetin, kaempferol, and myricetin by flavonol synthase (FLS). At the same time, the leucoanthocyanidin dioxygenase anthocyanidin synthase (ANS) catalyzes correspondingly-colored anthocyanidins. These compounds are subsequently converted to anthocyanins by the UDP-flavonoid glucosyltransferase (UFGT) as well as some glucosyl/acyl/methyl-transferases, and are then transported into the vacuole via three different pathways (Fig. 1)9,13.
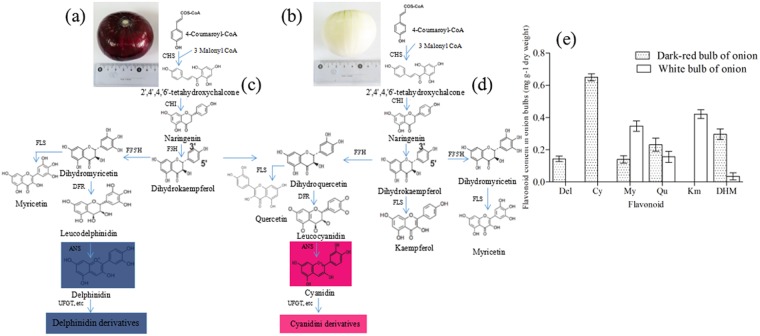
Figure 1.
Diagram to show the putative anthocyanin metabolic process in dark-red and white onion bulbs. (a,b) Mature dark-red and white onion bulbs; (c) The likely anthocyanin metabolic process in dark-red onion bulbs; (d) The likely anthocyanin metabolic process in white onion bulbs; (e) Flavonoid composition obtained by HPLC from dark-red and white onion bulbs. CHS: chalcone synthase; CHI: chalcone isomerase; F3H: flavanone 3-hydroxylase; F3′5′H: Flavonoid 3′,5′-hydroxylase; F3′H: flavonoid 3′-hydroxylase; DFR: dihydroflavonol 4-reductase; DHM: dihydrokaempferol; FLS, flavonol synthase; ANS: Anthocyanidin synthase; UFGT: UDP-flavonoid glucosyltransferase.
The inheritance of onion bulb colors is a complex pattern. Previous studies have revealed the presence of five major genes that are responsible for bulb colors14,15. The first gene, I, is a color-inhibiting factor which is not completely dominant over i; thus, when this gene occurs in a homozygous dominant condition, it inhibits the expression of onion bublb color, and all onion bulbs will be white regardless of the genotypes of the other genes. The second gene, C, is a basic color factor that is completely dominant over c and is required for the production of all colors. This means that homozygous recessive (cc) plants will have white bulbs which are referred to as recessive white to distinguish them from their dominant white counterparts. The third factor, G, causes a golden yellow color when present in either homozygous or heterozygous condition. Nevertheless, recessive homozygous (gg) plants appear chartreuse-colored bulbs. The last two genes, L and R, play complementary roles and produce red pigments when present in either homozygous or heterozygous conditions. Previous research has shown that the DFR gene transcript only accumulate in red onions while inactivation of this enzyme precludes the anthocyanin produced in the yellow onions15. It is nevertheless worth discussing which colors will be produced in onion bulbs by low level DFR expression.
Previous reports on colored bulbous plants, including potato (Solanum tuberosum L.)16, carrot (Daucus carota L.)17, and radish (Raphanus sativus L.)18, have emphasized anthocyanin metabolism. These vacuolar pigments are known to have important human health benefits and functions in plant stress responses. As in other bulb plants, onion pigments are known to be important in anthocyanin metabolism. Thus, RNA sequencing (RNA-seq) was performed in this study to further elucidate the metabolic pathways involving these compounds in dark-red onion bulbs. The RNA-seq method is an easy-to-use and efficient tool for detecting novel differentially expressed genes (DEGs)19,20. The aims of this study were therefore to: (1) Identify DEGs involved in the anthocyanin pathway of onion bulb pigmentation, and; (2) Determine candidate genes targeting the loss of pigmentation in onion bulbs.
Results and Discussion
Major color compounds in onion bulbs
We compared the metabolic profiles of onion bulb skin samples to evaluate phenotypes that lack color and to quantify the compounds involved in pigmentation. The major anthocyanin and flavonols composition of the purified in dark-red and white onions were identified by UPLC-PDA-Triple-TOF-MS. The results of this study predictably show that dark-red onion bulbs contain the anthocyanin delphindin and cyanidin, the flavonoids myricetin, quercetin, kaempferol and dihydrokaempferol (Fig. 1e and Supplementary Table S1). The antheocyanin and flavonoids compounds cyanidin 3-glucoside and cyanidin 3-malonoylglucoside as well as delphinidin 3-diglglucoside, delphinidin 3-glucoside and delphinidin aglycon and that this pigment is absent from white bulbs (Table 1 and Supplementary Fig. S1). MS data were collected in TOF-MS scan-Information dependent acquisition (IDA)-Product ion scan mode. Thus, to further understand the absence of color, we compared the intermediate products of the ABP metabolic process to determine the key enzymes present in dark-red and white onion bulbs (Fig. 1c,d). In this context, the presence of myricetin, quercetin, and kaempferol in white bulbs is indicative of an interrupted downstream ABP gene such as DFR, ANS, or UFGT.
Table 1.
Anthocyanins and flavonols identified from dark-red and white onion in this study.
| Variety | No. | Retention Time (min) | Extraction Mass (Da) | Found At Mass (Da) | M+ (m/z) | Error (ppm) | Peak area | Peak assignment |
|---|---|---|---|---|---|---|---|---|
| Dark-red onion | Ant1 | 9.51 | 535.1088 | 535.1083 | 535 | −0.73 | 23976666 | Cyanidin 3-malonoylglucoside |
| Ant2 | 20.52 | 449.1084 | 449.1081 | 449 | −0.75 | 2290833 | Cyanidin 3-glucoside | |
| Ant3 | 5.15 | 627.1561 | 627.1551 | 627 | −0.79 | 49188 | Delphinidin 3-diglucoside | |
| Ant4 | 27.63 | 465.1032 | 464.8732 | 465 | −5.53 | 54303 | Delphinidin 3-glucoside | |
| Ant5 | 34.54 | 356.2053 | 356.2033 | 303 | −2.13 | 43804 | Delphinidin aglycon | |
| Fla 1 | 11.02 | 310.3436 | 310.3406 | 289 | −2.01 | 37803 | Dihydrokaempferol | |
| Fla 2 | 26.31 | 348.9461 | 348.9441 | 319 | −1.73 | 3065458 | Myricetin | |
| Fla 3 | 36.82 | 364.5642 | 364.5636 | 303 | −1.65 | 3136864 | Quercetin | |
| White onion | Fla 1 | 11.03 | 36.5111 | 36.4353 | 289 | −2.78 | 3694 | Dihydrokaempferol |
| Fla 2 | 26.31 | 141.2461 | 141.673 | 319 | 2.13 | 11082 | Myricetin | |
| Fla 3 | 36.82 | 33.2043 | 33.0111 | 303 | −2.15 | 3132 | Quercetin | |
| Fla 4 | 45.23 | 230.3412 | 230.3398 | 287 | −1.93 | 23202 | Kaempferol |
Total anthocyanin and flavonols content
The total anthocyanin contents determined by the pH differential method, were (35.87 ± 0.54) mg 100 g−1 in fresh dark-red onion and (1.42 ± 0.87) mg 100 g−1 in fresh white onion, respectively. In this study, the anthocyanin content in dark-red was much higher than in white onion. The total flavonoid contents were (142.21 ± 2.46) mg 100 g−1 and (0.03 ± 0.32) mg 100 g−1, in fresh dark-red and white onion, respectively. What’s more, the flavonoid content in white onion was far lower than that in dark-red onion (Table 2).
Table 2.
Dark-red and white onion anthocyanin and flavonoids content (mean ± SE, n = 3).
| Variety | Anthocyanin content (mg 100g-1 FW) | Flavonoids content (mg 100g-1 FW) |
|---|---|---|
| Dark-red onion | 35.87 ± 0.54 | 142.21 ± 2.46 |
| White onion | 1.42 ± 0.87 | 0.03 ± 0.32 |
RNA-seq and de novo assembly
Six libraries were constructed in this study (three dark-red and three white) in order to gain further insights into the molecular mechanisms of dark-red onion bulb pigmentation. These comprised a total of 48.02 Gb clean reads, no less than 6.86 Gb for each sample, corresponding to an average percentage of bases with sequencing error rates lower than 1% (Q30) out of more than 92.49% (Supplementary Table S2). These values indicate that RNA-seq data quality was sufficient for further analysis.
Subsequent assembly generated 31,859,070 contigs with a mean length of 41.16 basis pairs (bp), including a length encompassing 50% of all nucleotide sequences of the largest 43 bp unigene length (N50). The overall assembly integrity of sequences is high; detailed results are presented in Supplementary Table S3.
Gene annotation and functional classification
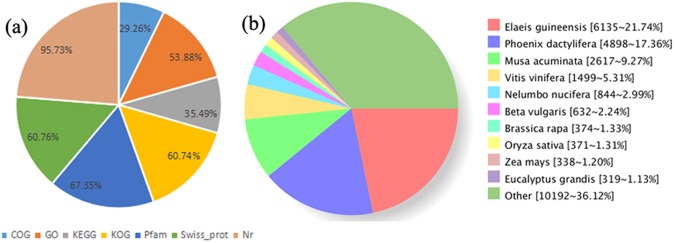
The BLASTX parameters E-value < 1e−5 and HMMER < 1e−10 were applied in this study, leading to the generation of 29,491 (31.29%) unigenes which were then annotated via a series of versus databases (i.e., non-redundant protein (Nr), Swiss-Rort, clusters of orthologous groups (COG), eukaryotic orthologous groups (KOG), Pfam, gene ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEEG)). Results show that 28,233 (95.73%) of annotated unigenes could be assigned to the Nr database, while the smallest proportion were assigned to the COG database (Fig. 2a). The distribution of major species is illustrated in Fig. 2b based on National Center for Biotechnology Information (NCBI) homology; data show that Elaeis guineensis (21.74%), Phoenix dactylifera (17.36%), and Musa acuminata (9.27%) all have high percentage homologies while the lowest value (1.13%) is seen in Brassica rapa and Eucalyptus grandis.
Figure 2.
Unigene characteristics. (a) Different databases percentage; (b) Numbers and percentages of annotated unigenes matching major species found in the Nr database using BLAST.
DEGs related to the development of dark-red color
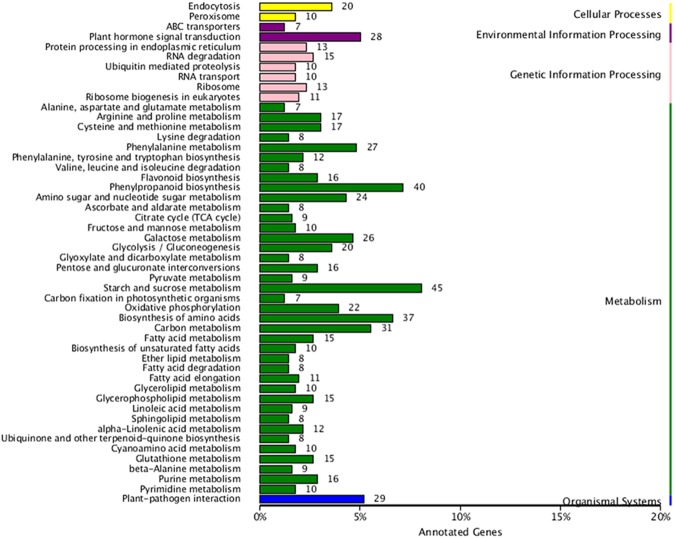
The unigenes obtained in this study were mapped against authoritative reference KEGG pathways to determine which are related to onion bulb colors. The results of this comparison revealed one secondary metabolic biosynthetic pathway (flavonoid biosynthesis) involved in color pigmentation; a total of 27 unigenes in total were assigned to this pathways (Table 3), while a total of 16 DEGs were shown to play key roles in flavonoid biosynthesis (Fig. 3 and Supplementary Table S4).
Table 3.
Bulb pigmentation candidate genes in onion (A. cepa L.).
| Function | Gene | Enzyme | KO id (EC no.) | No.All |
|---|---|---|---|---|
| Flavonoid biosynthesis | CHS | Chalcone synthase | K00660 (2.3.1.74) | 2 |
| CHI | Chalcone isomerase | K01859 (5.5.1.6) | 1 | |
| F3H | Flavanone 3-hydroxylase | K00475 (1.14.11.9) | 1 | |
| F3′H | Flavanone 3′-hydroxylase | K05280 (1.14.13.21) | 2 | |
| F3′5′H | Flavanone 3′5′-hydroxylase | K13083 (1.14.13.88) | 4 | |
| DFR | Dihydroflavonol 4-reductase | K13082 (1.1.1.219) | 1 | |
| ANS | Anthocyanidin synthase | K05277 (1.14.11.19) | 1 | |
| UFGT | Anthocyanidin 3-O-glucosyltransferase | K12930 (2.4.1.115) | 1 | |
| C3M | Coumaroylquinate(coumaroylshikimate) 3′-monooxygenase | K09754 (1.14.13.36) | 4 | |
| CoA | Caffeoyl-CoA O-methyltransferase | K00588 (2.1.1.104) | 3 | |
| C4H | Cinnamate-4-hydroxylase | K00487 (1.14.13.11) | 2 | |
| SOHT | Shikimate O-hydroxycinnamoyltransferase | K13065 (2.3.1.133) | 2 | |
| FLS | Flavonol synthase | K05278 (1.14.11.23) | 2 | |
| FOMT | Flavonol 3-O-methyltransferase | K05279 (2.1.1.76) | 1 |
Figure 3.
Comparison of KEGG enrichment DEG pathways between dark-red and white onion bulbs.
Comparisons of the transcriptional profiles of genes involved in anthocyanin metabolism in white and dark-red onion bulbs
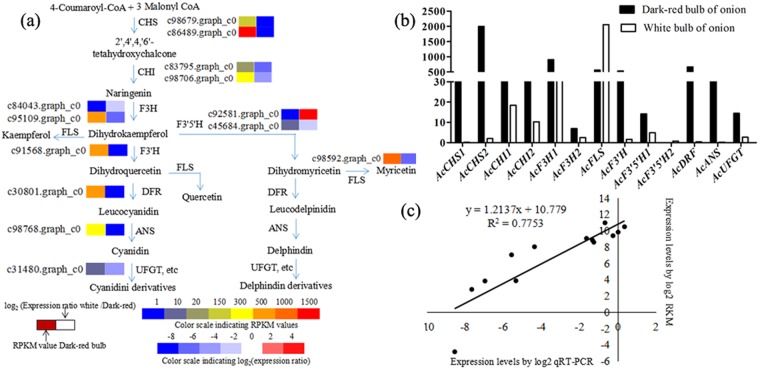
Previous studies on plant bulbs and flowers have shown that loss of color from dark-red to white is the result of major deficiencies in the anthocyanins (Cy and Del)8,11,17. This is also the case in onion bulbs; a color transition from dark-red to white is due to an ABP blockage preventing reactions that would normally lead to the anthocyanins Cy and Del. We therefore compared the transcriptomes of dark-red and white onion bulbs and their associated RNA-seq annotations to determine the transcripts involved in dark-red color metabolism. The 13 critical DEGs implicated in anthocyanin metabolism were then subjected to qRT-PCR analysis using the primers designed in this study in order to verify our RNA-seq results,. These qRT-PCR results reveal that the expression levels of selected DEGs expression levels were generally consistent with transcriptome sequencing data. The transcriptome sequencing data reported in this paper are therefore credible and are further corroborated by a high correlation coefficient (R2 = 0.775) (Fig. 4c). These comparisons reveal significant changes in the expression levels of 13 key uni-transcripts (Fig. 4a,b); while most key genes were expressed in anthocyanin biosynthesis, it is noteworthy that the level of the F3′5′H1 in dark-red bulbs was more than three times that seen in white one (14.16 versus 4.89), while the expression of the F3′5′H2 was far below in the dark-red onion (0.03 versus 0.75) (Fig. 4b). Data also show that the expression levels of DFR, ANS, and UFGT in the dark-red bulbs were up to 30 times higher than in white bulbs counterparts; this result demonstrate that F3′5′H and DFR genes play an important role in anthocyanin biosynthesis in dark-red onion bulbs and also provide an explanation for the absence of kaempferol in white bulbs (Fig. 1e).
Figure 4.
Physiological and metabolic data related to onion bulb pigmentation. (a) Detailed Cy and Del metabolic network showing subsets of nodes or metabolites that constitute this process. Enzyme and unigene names as well as expression patterns are listed next to each step, while expression patterns are summarized in two boxes; the PRKM values for dark-red onions are on the right, while the log2 expression ratio of white/dark-red are on the left; (b) Transcript accumulation of color-related genes involved in the anthocyanin metabolic process; (c) Gene expression correlation analysis for color-related genes in dark-red onion based on qRT-PCR and RNA-seq. Abbreviations as in Fig. 1.
Candidates which are responsible for the loss of Del and Cy in white onion bulbs
Although anthocyanidin metabolism in both white and dark-red onion bulbs involves the same enzymes prior to Del- and Cy-related reactions (Fig. 1c,d), the catalysis of subsequent specific reactions remains studied. It is generally acknowledged that CHS is the first catalyzing enzyme to produce the intermediate chalcones that are then processed in later metabolism21; thus, if this CHS enzyme is restricted, then both the production of anthocyanins and, subsequently, flavonoids will be limited and there will be no synergetic effect on the dark-red and white onion bulbs22,23. Previous studies have shown that F3′5′H plays an important role in the ABP pathway and is a prerequisite for the formation of Del (violet-to-blue) anthocyanins24; this implies that if the metabolism pathways for the minimal Del path of F3′5′H is cut, myricetin-related flavonols may not be determined in this study. However, as myricetin was detected in white onion bulbs anthocyanin at levels more than twice those in the dark-red onion (Fig. 1e), this explanation for the absence of Del is unsatisfactory. We therefore also addressed the question of whether, or not, the DFR gene which is crucial for anthocyanin formation can explain the absence of Del in the white onions; as this gene leads to colorless leucoanthocyanidins and no products of Del biosynthesis were found subsequent to dihydromyricetin in the white bulbs (Fig. 5a); DFR may be a candidate gene for this blocking process. At the same time, dihydroflavonols substrates represent branch points for the production of colourless flavonols (Km, My and Qu) through FLS; indeed, the expression levels of this gene are gradually up-regulated due to competing substrate (Fig. 4b). These results of this study therefore also reveal that both FLS and DFR genes catalyze the formation of dihydromyricetin and block Del synthesis, which is consistent with the result of earlier work25.
Figure 5.
Ideal model for Del and Cy elimination from white onion bulbs. (a) Loss of Del from white onion bulbs. In cases when DFR is suppressed, substrates used for Del synthesis become available for synthesis of Kaempferol and myricetin. (b) Ideal model for the loss of Cy from white onion bulbs. The fluxe due to Cy metabolism is limited in this case, and downstream reactions promote the turnover and degradation of this pigment in the white onion bulbs. The global output from the minimal anthocyanin subnetwork in onion was considered to be 100% and defined the relative content on each product. The dark grey and light green boxes on this figure denote the relative content of genes or compounds in the dark-red and white onion bulbs, respectively. Abbreviations: LAR: leucoanthocyanidin reductase; ANR: anthocyandin reductase. All other abbreviations as in Fig. 1.
Metabolism and expression patterns of dark-red and white onion bulbs were compared to determine candidates for the loss of Cy accumulation. In this context, both the enzymes FLS (c98592.graph_c0) and DFR (c30801.graph_c0) are likely to limit Cy accumulation, in agreement with the earlier work11; up-regulation of FLS occurs when substrates are available to catelyze with Km in white onion bulbs (Fig. 1e), while the down-regulation of DFR could be unable to completely block the Cy process. A number of recent studies have discussed the use of new approaches for detecting the loss of pigmentation phenotypes via blocking points in anthocyanin biosynthesis26,27; one report noted that when the anthocyandin reductase (ANR) gene in apple was introduced to tobacco, this limited expression of the DFR gene in apple blossoms. As a similar process might also explain the absence of anthocyanin28, we hypothesize that this might provide an ideal model for Cy elimination from white onion bulbs (Fig. 5b); even though, red Cy (which occurs in either very low concentrations or is present for only a very short time) is also determined via this mechanism. This result indicates that a complex metabolic mechanism underlies accumulation of this compound in the ABP that might be explained by a variety of factors (e.g., light, UV-A)29. It is clear that leucyanidin is formed before Cy and that this catalyzes two products (colourless catechin and red Cy) via leucocyanidin reductase (LAR) and ANS, respectively; this also means that Cy might be reduced to colorless epicatechin by ANR and so a stable form of this pigment might not exist. Ideally, catechin and epicatechin products should be detectable in white onion bulbs and absent from dark-red form, but further trials will be required to explore this transition in tuber plants. The loss of Cy from white onion bulbs might also provide an explanation multibranching of the Cy synthesis pathway.
The enzymes F3′5′H and F3′H play important roles in the accumulation of Del and Cy
Results show that the enzymes F3′5′H and F3′H both control crucial steps in the accumulation of Del and Cy anthocyanins and lead to significant changes in expression levels (Figs 1 and 4b). Previous work on a range of flower colors and plant bulbs has shown that the ratio between the expression of these genes controls the proportion and composition of flavonoids. In blue grape berries (Vitis vinifera), for example, a higher ratio of F3′5′H/F3′H leads to the production of more Del derivatives than in red or white cultivars25,30, while different ratios of these enzymes in Senecio cruentus determines Del and Cy accumulation and is the reason for their range of red and blue color varieties31. Similarly, roses (Roses hybrida) and carnations lack violet and blue color varieties because they do not produce Del, perhaps due to the loss of the F3′5′H gene during evolution. The high-performance liquid chromatography (HPLC) analysis presented here reveals a higher Cy content compared to Del (Fig. 1e) in dark-red onion bulbs and indicates that the F3′5′H/F3′H ratio is lower than that of F3′H/F3′5′H. This might indicated that both the genes F3′5′H and F3′H from onion (A. cepa L.) do not have a synergetic effect in regulating pigmentation.
Material and Methods
Plant material
The dark-red onion (A. cepa L.) cultivar ‘Xiu Qiu’ (fourth generation inbred lines from the dark-red ‘Shanxi’ variety) and the white onion cultivar ‘Ring Master’(fourth generation inbred lines from the white ‘Xinjiang’ variety) were used in this study. We performed a series of genetic tests to confirm whether white onions were dominant or recessive; as the F1 color of (‘Xiu Qiu’ × ‘Ring master’) was pink, the genotype of the white parent was i/i, and the phenotypes of the F2 segregating population were white, yellow, pink, and dark-red (Supplementary Fig. S2).
Fresh, undamaged 7 cm diameter samples weighing between 280 g and 320 g were collected on the 40th day after swelling from a test field at the Beijing Academy of Agriculture and Forestry Vegetable Research Center (Fig. 1a,b). Samples were washed, excess water was absorbed with blotting paper, and lengths between 1 cm and 2 cm from the top and the bottom of each were excised with a scalpel. Outer onion bulb layers were then divided into three parts for experiments; all materials were immediately frozen in liquid nitrogen (N) and stored at −80 °C prior to RNA extraction and flavonoid analyses.
Extraction of athocyanins and flavonoids
The extraction method used in this study follows previously reported protocols25,32 with slight modifications. Onion bulb peels frozen with liquid N were ground into fine powder and 50 mg subsamples were placed into 1.5 ml EP tubes. Extraction was then performed using 1 ml methanol containing 1% formic acid (v/v) before samples were sonicated in an ultrasonic bath at 4 °C for 24 hours. Samples were then centrifuged at 13,000 rpm for 10 minutes, supernatants were extracted into fresh tubes, and the whole protocol was repeated. Before the analysis, the extract was fltered by using 0.22 µm reinforced nylon membrane flters. Deionized water containing 0.01% hydrochloric acid were added up to 25 mL after the remain supernatants were combined and evaporated under vacuum to dryness at 35 °C to analyze the detailed anthocyanin and flavonoids composition and their antioxidant activities. A 3 µl supernatant volume from each sample was then injected for UPLC-MS analysis.
UPLC-PDA-Triple-TOF-MS analysis
Bulbs of differently-colored onions were subject to anthocyanin analysis using Shimadzu UHPLC LC-30A system (Shimadzu, Japan) coupled to an AB SCIEX 6600 Triple-TOF-MS (AB SCIEX, USA). Chromatographic separations were performed on an Acquity UPLC™ BEH C18 column (2.1 mm × 100 mm, i.d., 1.7 µm) (Waters Corp., MA, USA) at 40 °C Chromatographic separations were performed on an Acquity UPLC™ BEH C18 column (2.1 mm × 100 mm, i.d., 1.7 µm) (Waters Corp., MA, USA) at 40 °C. The solvent system consisted of water with 0.1% formic acid (mobile phase A) and acetonitrile with 0.1% formic acid (mobile phase B). The column was eluted with a linear gradient of 0–28% B over 0–22 min, 28–40% B over 22–22.5 min, 40–100% B over 22.5–23 min, the composition was held at 100% B for 2 min then returned to 0% B and re-equilibrated for an additional 3 min before injection of the next sample. The flow rate of mobile phase was 0.3 mL/min. PDA detector was set at 200~800 nm.
Mass spectrometry analysis was performed using an electrospray ionization source (ESI) in positive ion mode. The source parameters were set as follow: source temperature of 550 °C, ion spray voltage of 5.5 kV, declustering potential of 80 V, atomization gas pressure (GS1) of 0.34 MPa, air curtain gas (CUR) of 0.24 MPa and auxiliary air pressure (GS2) of 0.34 MPa. Nitrogen was used in all cases.
Anthocyanins (cyanidin 3-glucoside, cyanidin 3-malonylglucoside, delphinidin 3-diglycoside delphinidin 3-glucoside and delphinidin aglycon) and flavonols (quercetin, kaempferol, myricetin, and dihydromyricetin) standards were purchased from Sigma-Aldrich China (Shanghai); mean values and standard deviations from three biological replicates are reported in this study, expressed in milligrams per gram dry weight. Quantification of single compounds was achieved by peak area using corresponding standard samples33,34.
Total anthocyanins and flavonols content measurement
Total anthocyanin content in dark-red and white onion was measured by using the PH differential spectrophotometric method with some modifications35. Taking dark-red and white onion bulb 1.0 g, respectively, after grinding fully and adding precooling 10 ml 0.005% hydrochloric acid-methanol solution, storing at 4 °C for 12 h avoiding light and then collecting supernatant. The remaining residue was treated in the same way, combining with these supernatants, respectitvely. Taking the solution (1 ml) was dissolved with 0.025 mol L−1 potassium chloride buffer (PH = 1.0) and 0.4 mol L−1 sodium acetate buffer, respectivelly, and metered volume (25 ml). The absorbance was measured at 525 nm and 700 nm. Absorbance (A) of the diluted samples were then calculated as follows: anthocyanins content (mg L−1) = [(OD525 − OD700) PH1.0 − (OD525 − OD700) PH4.5] × 449.2 × 1000/26900 × DF, among of them, 449.2 is the relative molecular mass of cyanidin-3-glucoside; 26,900 is the molar absorptivity; DF is the dilution factor; 1000 is the factor to convert g to mg.
The total flavonols content in dark-red and white onion was measured by spectrophotometric method36. Rutin was used as a standard compound. Sample spectral absorbance measurements were read at 510 nm. The flavonols were calculated from the calibration curve: R (mg mL−1) = 0.1565OD750 – 0.0011 (R2 = 0.9925) and flavonols content (mg L−1) = R × DF/m. Among of them, DF is 2,500, m is the quality of the onion samples.
RNA extraction and RNA-seq library construction
Total RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions but incorporating a few modifications including precipitation with isovolumetric isopropanol and a high-salt solution (1.2 mol/L sodium chloride and 0.8 mol/L sodium citrate) rather than isopropanol precipitation when the supernatant fluid was removed. RNA concentration was initially characterized on a 1% agarose gel, and then examined using a NanoDrop 2100 spectrophotometer (Thermo Fisher Scientific, USA) to ensure RNA quality. Integrity RNA numbers were assessed using an Agilent 2100 system (Agilent Technologies, CA, USA) and were between 8.9 and 10.0 in all cases. Library construction and RNA-seq analyses were completed by the Biomarker Biotechnology Corporation (Beijing, China) utilizing RNA from each sample.
Two complementary DNA (cDNA) libraries were generated and sequenced in this study using an Illumina HiSeq. 4000 system. Messenger RNA was enriched using oligo(dT)-attached magnetic beads and sequences were randomly broken into short fragments via the addition of a fragmentation buffer. These short fragments were then used as templates for the synthesis of first-and second-strand cDNA using random hexamers and the addition of buffer, dNTPs, RHase H, and DNA polymerase I, respectively. Fragments of cDNA were then purified, subjected to end-repair, the addition of poly (A) tailing and ligation sequencing adapters, and size-selected using AMPure XP beads. Suitable fragments were then placed on 2% agarose gel for use as Polymerase Chain Reaction (PCR) templates for the amplification of cDNA libraries.
De novo transcriptome assembly and functional annotation
Raw pair-end (PE) 150 bp reads were initially filtered prior to assembly via the removal of adaptor and low quality reads as well as unknown nucleotides. The remaining high quality clean reads were then utilized for de novo transcriptome assembly using a Trinity platform (http://trinityrnaseq.sourceforge.net/) with the parameters ‘K-mer = 25’ and ‘group pairs distance = 300’ as well as other defaults and in the absence of a reference genome37. Thus, based on overlap regions, short reads were initially assembled into longer contigs which were then clustered to form components; the different constituent contigs of each were then used to build a De Bruijn diagram which was untangled using real reads to obtain transcriptional sequences that were clustered to obtain uni-transcripts using the TGI software clustering tool38.
The software BLAST (E-value ≤ le−5)39 was then used to compare resultant uni-transcript sequences against the non-redundant protein (Nr) (ftp://ftp.ncbi.nih.gov/blast/db/), Swiss-Prot (http://www.uniprot.org/), GO (http://www.geneontology.org/), COG (http://www.ncbi.nlm.nih.gov/COG/), KOG40, and KEGG (http://www.genome.jp/kegg/) databases. The software KOBAS2.041 was then used to determine KEGG unigene orthology results; after predicting the amino acid sequence of each unigene, we then used the software HMMER42 to make comparisons with the Pfam database43 to obtain unigene annotation information.
Differential expression analysis
The software Bowtie44 was used in this analysis to compare alignment results for the reads of each sample (i.e., three biological replicates in each case) versus the unigene library associated with RNA-seq utilizing expectation maximization (RSEM)45 to estimate expression level in each case. Thus, the expression abundance unigene differences among samples were represented by values of fragments per kilobase of transcript per million mapped reads (FPKM), and the software DESeq.46 was used to screen DEGs via pairwise comparisons. The universally recognized and effective Benjamini-Hochberg method was utilized to determine a significant P-value for the original hypothesis being tested in each case; corrected P-values incorporate false discovery rate (FDR) when screening DEG levels such that when the FDR was less than 0.001 and the log2 fold change (FC) was either less than −2 or greater than 2 in terms of FPKM between two libraries. FC values therefore denote the expression ratio between two samples.
Gene validation and expression analysis
Real-time-quantitative PCR (qRT-PCT) was performed using a Roche Light Cycler 480 machine (Bio-Rad, USA) incorporating a 96 real-time system in order to validate RNA-seq results and the roles of key enzymes related to in the anthocyanin biosynthetic pathway. SYBR® Premix Ex TaqTM (Tli RNaseH Plus) (TaKaRa, Dalian, China) was used for all PCR reactions, and primers were designed using the software Primer Premier 5.0 (Supplementary Table S5). Synthesis of cDNA and qRT-PCR were performed using previously described methods47; three technical qRT-PCT replicates of each sample as well as two biological replicates were performed to ensure reliability, and the inference gene β-actin48 was used to normalize gene expression levels.
Electronic supplementary material
Acknowledgements
This research was supported by the National Key Technology Research and Development Program of China (Grant No. 2014BAD01B08) and Technological Innovation Capacity Program of the Beijing Academy of Agricultural and Forestry Sciences (Grant No. KJCX201511, KJCX20170102, and KJCX20180401).
Author Contributions
Y.L. conceived the study and performed the research. while C.Z. and Z.Z. analyzed data, performed the experiments, created graphs, and wrote the manuscript. X.L. participated in data analysis and helped to draft the manuscript, while A.Z. and G.C. contributed analytical tools. L.C. helped to revise the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32472-5.
References
- 1.Havey MJ, Galmarini CR, Gökçe AF, Henson C. QTL affecting soluble carbohydrate concentrations in stored onion bulbs and their association with flavor and health-enhancing attributes. Genome. 2004;47:463–468. doi: 10.1139/g04-005. [DOI] [PubMed] [Google Scholar]
- 2.Kim S, Jones R, Yoo KS, Pike LM. The Llocus, one of complementary genes required for anthocyanin production in onions (Allium cepa), encodes anthocyanidin synthase. Theoretical & Applied Genetics. 2005;111(1):120–127. doi: 10.1007/s00122-005-2000-1. [DOI] [PubMed] [Google Scholar]
- 3.Nile SH, Park SW. Total phenolics, antioxidant and xanthine oxidase inhibitory activity of three colored onions (Allium cepa L.) Frontiers in Life Science. 2013;7:224–228. doi: 10.1080/21553769.2014.901926. [DOI] [Google Scholar]
- 4.Clarke AE, Jones HA, Little TM. Inheritance of bulb color in the onion. Genetics. 1944;29(6):569–575. doi: 10.1093/genetics/29.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhodes MJC, Price KR. Analytical problems in the study of flavonoid compounds in onions. Food Chemistry. 1996;57(1):113–117. doi: 10.1016/0308-8146(96)00147-1. [DOI] [Google Scholar]
- 6.Fossen T, et al. Characteristic Anthocyanin Pattern from Onions and other Allium spp. Journal of Food Science. 1996;61(4):703–706. doi: 10.1111/j.1365-2621.1996.tb12185.x. [DOI] [Google Scholar]
- 7.Donner H, Gao L, Mazza G. Separation and characterization of simple and malonylated anthocyanins in red onions. Allium cepa L. Food Research International. 1997;30(8):637–643. doi: 10.1016/S0963-9969(98)00011-8. [DOI] [Google Scholar]
- 8.Gennaro L, et al. Flavonoid and carbohydrate contents in Tropea red onions: effects of homelike peeling and storage. Journal of Agricultural & Food Chemistry. 2002;50(7):1904–10. doi: 10.1021/jf011102r. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant Journal. 2008;54(4):733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 10.Yuan YW, Sagawa JM, Frost L, Vela JP, Jr HDB. Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus) New Phytologist. 2014;204(4):1013–27. doi: 10.1111/nph.12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mano H, Ogasawara F, Sato K, Higo H, Minobe Y. Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiology. 2007;143(3):1252–68. doi: 10.1104/pp.106.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandwein BJ. The pigments in three cultivars of the common onion (Allium cepa) Journal of Food Science. 2010;30(4):680–685. doi: 10.1111/j.1365-2621.1965.tb01824.x. [DOI] [Google Scholar]
- 13.Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Science: An International Journal of Experimental Plant Biology. 2011;181(3):219–29. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, et al. Pink (P), a new locus responsible for a pink trait in onions (Allium cepa) resulting from natural mutations of anthocyanidin synthase. Molecular Genetics & Genomics. 2004;272(1):18–27. doi: 10.1007/s00438-004-1041-5. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, et al. Inactivation of DFR, (Dihydroflavonol 4-reductase) gene transcription results in blockage of anthocyanin production in yellow onions (Allium cepa) Molecular Breeding. 2004;14(3):253–263. doi: 10.1023/B:MOLB.0000047770.92977.04. [DOI] [Google Scholar]
- 16.Liu Y, et al. Comparative transcriptome analysis of white and purple potato to identify genes involved in anthocyanin biosynthesis. Plos One. 2015;10(6):e0129148. doi: 10.1371/journal.pone.0129148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu, Z. S. et al. Transcript profiling of structural genes involved in cyanidin-based anthocyanin biosynthesis between purple and non-purple carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biology, 14,1(2014-10-01), 14(1), 262 (2014). [DOI] [PMC free article] [PubMed]
- 18.Park NI, et al. Anthocyanin accumulation and expression of anthocyanin biosynthetic genes in Radish (Raphanus sativus) J Agric Food Chem. 2011;59(11):6034–9. doi: 10.1021/jf200824c. [DOI] [PubMed] [Google Scholar]
- 19.Blencowe BJ, Ahmad S, Lee LJ. Current-generation high throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. doi: 10.1101/gad.178800. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koes RE, Spelt CE, Elzen PJMV, Mol JNM. Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene. 1989;81:245–257. doi: 10.1016/0378-1119(89)90185-6. [DOI] [PubMed] [Google Scholar]
- 22.Clark ST, Verwoerd WS. A systems approach to identifying correlated gene targets for the loss of colour pigmentation in plants. BMC Bioinformatics. 2011;12:343. doi: 10.1186/1471-2105-12-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim W, Li J. Synergetic effect of the OnionCHIgene on the PAP1regulatory gene for enhancing the flavonoid profile of tomato skin. Scientific Reports. 2017;7:1. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka Y, Brugliera F. Flower colour and cytochromes p450. Philosophical Transactions of the Royal Society of London. 2013;368(1612):20120432. doi: 10.1098/rstb.2012.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lou Q, et al. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. Journal of Experimental Botany. 2014;65:12–3157. doi: 10.1093/jxb/eru168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellarin SD, Di GG. Transcriptional control of anthocyanin biosynthetic genes in extreme phenotypes for berry pigmentation of naturally occurring grapevines. BMC Plant Biology. 2007;7(1):46. doi: 10.1186/1471-2229-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SD, Rausher MD. Gene loss and parallel evolution contribute to species difference in flower color. Molecular Biology and Evolution. 2011;28:2799–2810. doi: 10.1093/molbev/msr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han YP, Vimolmangkang S, Soria-Guerra RE, Korban SS. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. Journal of Experimental Botany. 2012;63:2437–2447. doi: 10.1093/jxb/err415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YY, et al. Mdcop1 ubiquitin e3 ligases interact with mdmyb1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiology. 2012;160(2):1011–22. doi: 10.1104/pp.112.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castellarin SD. Colour variation in red grapevines (Vitis vinifera L.): Genomic organisation, expression of flavonoid 30-hydroxylase, flavonoid 3′, 5′-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genomics. 2006;7:12. doi: 10.1186/1471-2164-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin X, Huang H, Wang L, Sun Y, Dai S. Transcriptomics and metabolite analysis reveals the molecular mechanism of anthocyanin biosynthesis branch pathway in different Senecio cruentus cultivars. Frontiers in Plant Science. 2016;7(107):1307. doi: 10.3389/fpls.2016.01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang SL, et al. Quantification and analysis of anthocyanin and flavonoids compositions,and antioxidant activities in onions with three different colors. Journal of Integrative Agriculture. 2016;15(9):2175–2181. doi: 10.1016/S2095-3119(16)61385-0. [DOI] [Google Scholar]
- 33.Tomás-Barberán FA, et al. HPLC-DAD-ESIMS analysis of phenolic compounds in nectarines, peaches, and plums. Journal of Agricultural & Food Chemistry. 2001;49(10):4748. doi: 10.1021/jf0104681. [DOI] [PubMed] [Google Scholar]
- 34.Jaakola L, et al. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiology. 2002;130(2):729–39. doi: 10.1104/pp.006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Z, Wang B, Williams P, Pace RD. Identification of anthocyanins in muscadine grapes with hplc-esi-ms. LWT - Food Science and Technology. 2009;42(4):819–824. doi: 10.1016/j.lwt.2008.11.005. [DOI] [Google Scholar]
- 36.Wang LJ, et al. Variation of anthocyanins and flavonols in vaccinium uliginosum berry in lesser khingan mountains and its antioxidant activity. Food Chemistry. 2014;160(10):357–364. doi: 10.1016/j.foodchem.2014.03.081. [DOI] [PubMed] [Google Scholar]
- 37.Grabherr MG, Haas BJ, Yassour M. Full length transcriptome assembly from RNA Seq data without a reference genome. Nature Biotechnology Italic. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pertea G, Huang XQ, Liang F. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 39.Koonin EV, Fedorova ND, Jackson JD. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biology Italic. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schäffer AA. Gapped BLAST and PSI BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Research Italic. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie C, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddy SR. Profile hidden Markov models. Bioinformatics Italic. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 43.Finn, R. D., Bateman, A. & Clements, J. Pfam: the protein families database. Nucleic Acids Research Italic: gkt1223 (2013). [DOI] [PMC free article] [PubMed]
- 44.Langmead B, Trapnell C, Pop M. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology Italic. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Colin ND. RSEM: accurate transcript quantification from RNA Seq data with or without a reference genome. BMC Bioinformatics Italic. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leng N, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, C. et al. Transcriptome Analysis of Sucrose Metabolism during Bulb Swelling and Development in Onion (Allium cepa L.). Frontiers in Plant Science7 (2016). [DOI] [PMC free article] [PubMed]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.