Abstract
Bcl-2 family proteins play a crucial role in regulating apoptosis, a process critical for development, eliminating damaged or infected cells, host-pathogen interactions and in disease. Dysregulation of Bcl-2 proteins elicits an expansive cell survival mechanism promoting cell migration, invasion and metastasis. Through a network of intra-family protein–protein interactions Bcl-2 family members regulate the release of cell death factors from mitochondria. NRZ is a novel zebrafish pro-survival Bcl-2 orthologue resident on mitochondria and the endoplasmic reticulum (ER). However, the mechanism of NRZ apoptosis inhibition has not yet been clarified. Here we examined the interactions of NRZ with pro-apoptotic members of the Bcl-2 family using a combination of isothermal calorimetry and mutational analysis of NRZ. We show that NRZ binds almost all zebrafish pro-apoptotic proteins and displays a broad range of affinities. Furthermore, we define the structural basis for apoptosis inhibition of NRZ by solving the crystal structure of both apo-NRZ and a holo form bound to a peptide spanning the binding motif of the pro-apoptotic zBad, a BH3-only protein orthologous to mammalian Bad. The crystal structure of NRZ revealed that it adopts the conserved Bcl-2 like fold observed for other cellular pro-survival Bcl-2 proteins and employs the canonical ligand binding groove to bind Bad BH3 peptide. NRZ engagement of Bad BH3 involves the canonical ionic interaction between NRZ R86 and Bad D104 and an additional ionic interaction between NRZ D79 and Bad R100, and substitution of either NRZ R86 or D79 to Ala reduces the binding to Bad BH3 tenfold or more. Our findings provide a detailed mechanistic understanding for NRZ mediated anti-apoptotic activity in zebrafish by revealing binding to both Bad and Noxa, suggesting that NRZ is likely to occupy a unique mechanistic role in zebrafish apoptosis regulation by acting as a highly promiscuous pro-apoptotic Bcl-2 binder.
Introduction
Multicellular organisms have evolved a multitude of mechanisms to remove superfluous cells1. Pivotal among the mechanisms for cell removal is programmed cell death or apoptosis, a process that maintains tissue homeostasis, removes damaged, infected in response to pathogen invasion2, or otherwise unwanted cells, such as during embryonic development, where it plays a critical role in shaping body and tissue structures3,4. Members of the B-cell lymphoma-2 (Bcl-2) family of proteins are key players of cellular life and death decisions and regulate the intrinsic or mitochondrial associated cell death3,4. Consisting of ~20 proteins the Bcl-2 family is characterized by the presence of conserved sequence motifs referred to as Bcl-2 homology or BH motifs. Structurally, the Bcl-2 proteins are organized into two major sub-families, those that share the Bcl-2 fold (Bcl-2, Bcl-xL, Bcl-w, Mcl-1, A1, Bcl-B, Bax, Bak Bok) and a distantly related group, the BH3-only proteins that bear only a BH3-motif (Bim, Bad, Bmf, Bid, Bik, Hrk, Puma and Noxa) that with the exception of Bid are disordered5. Those with pro-survival activity in mammals comprise Bcl-2, Bcl-xL, Bcl-w, Mcl-1, A1 and Bcl-B. Their primary function is to antagonize the activation of caspases by directly interacting and inhibiting pro-apoptotic Bcl-2 proteins4,6 of the BH3-only group or Bax, Bak and Bok. It is ultimately the balance and specificity7 of interactions between pro-apoptotic and pro-survival Bcl-2 proteins that regulates apoptosis and determines cellular fate8.
The pro-apoptotic Bcl-2 proteins promote apoptosis via mitochondrial outer membrane permeabilization (MOMP)4,5. Critical to the execution of MOMP are Bax and Bak9,10 as they trigger formation of oligomeric pores that breach the mitochondrial outer membrane and release pro-apoptogenic molecules such as SMAC/DIABLO and cytochrome-c11 into the cytoplasm to activate the caspase cascade that proteolyses key cellular components and ultimately demolishes the cell4. In contrast to Bax and Bak, the BH3-only proteins induce apoptosis by utilizing their BH3 motif via two mechanisms: either indirectly by neutralizing pro-survival Bcl-2 by binding to a conserved ligand binding groove or directly by interacting with Bax and Bak via an alternative interaction site5,12. In healthy cells, BH3-only proteins act as sentinels of cellular well-being and are up-regulated in response to cellular insults including growth factor deprivation, exposure to cytotoxic drugs or viral infections, leading to the activation of cell death mechanisms6. Other less well described functions have also been attributed to the Bcl-2 family. For example, Bcl-2 family members may regulate or monitor intracellular calcium13,14 in the unfolded protein response (UPR) and trigger apoptosis through activation of the BH3-only proteins15 when the unfolded protein levels in the endoplasmic reticulum (ER) become excessive16. While elements of the intrinsic apoptotic pathway are highly conserved from sponges17 to mammals6 there are also significant differences between organisms18.
Many Bcl-2 family members are conserved between fish and mammals19–21, but some notable differences exist between the apoptotic machineries in mammals and fish. For instance, overexpression of the zebrafish (Danio rerio) pro-apoptotic Bcl-2 proteins zBmf1, zBik, zPuma and zNoxa triggered dose-dependent caspase activation and subsequent cell death, whereas overexpression of zBad and zBok did not lead to cell death compared to overexpression of human Bad22. Also, expression of zebrafish bad gene did not result in embryonic lethality23. Furthermore, zBad pro-apoptotic activity is regulated through phosphorylation of conserved serine residues23,24, in a manner similar to mouse Bad25. One significant difference from mammals is that zebrafish lack orthologues of the pro-survival Bcl-2 proteins Bcl-w and A1, as well as pro-apoptotic Bak and Hrk20, but contain a novel pro-survival protein called NRZ, that was initially identified as an orthologue of the avian pro-survival Bcl-2 protein NR-1326–28. Also present is a recently described pro-apoptotic Bcl-2 protein Bcl-wav29 that is without a mammalian orthologue. Thus, there are a number of significant differences between Bcl-2-regulated apoptosis in zebrafish and mammals.
NRZ is a 19 kDa protein expressed in the epiboly of the extra-maternal yolk syncytial layer (YSL) of zebrafish eggs28. In vivo, NRZ has been shown to be a potent inhibitor of apoptosis after serum withdrawal, and controls development during somatogenesis and gastrulation28. Mechanistically, NRZ was shown to inhibit apoptosis by antagonizing zBax-BH328. Nrz knockout triggers intracellular Ca2+ increase in YSL, which results in the blockade of development prior to gastrulation13. Additionally, NRZ appears to arrest Ca2+ release from endoplasmic reticulum (ER) by direct interaction with the inositol triphosphate type 1 receptor, IP3R1, calcium channel13. Knockdown of NRZ is lethal in zebrafish embryos, and intriguingly, some of this activity appears to be independent of its role in apoptosis28. Loss of NRZ prevents embryonic development via upregulation of the transcription factor Snail-1, a cell adhesion regulator to arrest gastrulation at the shield stage28,30. Other activities ascribed to NRZ include inhibition of the apoptosis accelerating function of Bcl-wav, a pro-apoptotic Bcl-2 member found in teleost fish29.
Although there are significant differences between the apoptosis machinery of zebrafish and mammals there remains a high degree of conservation with many direct orthologues of mammalian apoptotic genes present in the genome of the zebrafish19,20. Coupled with the presence of many orthologous mammalian genes are the advantages of zebrafish as a model organism, such as their rapid development, embryo transparency and genetic accessibility20. Analysis of zebrafish genetics is providing a better understanding of the fundamental interactions governing apoptosis and is of significant interest in deciphering human disease, including cancer20 and host-pathogen interactions31. Here we report the first systematic biochemical analysis and high-resolution structure determination of a zebrafish pro-survival Bcl-2 protein, NRZ. Our findings suggest that NRZ is a unique pro-survival Bcl-2 protein with an unusual pro-apoptotic Bcl-2 binding profile unlike its counterparts in mammalian systems.
Materials and methods
Protein expression and purification
Synthetic cDNA encoding for codon-optimized NRZ (Uniprot Accession number Q8UWD5) lacking the 28 C-terminal residues (Bioneer, Melbourne, Australia) was cloned into the bacterial expression vector pCoofy432 Recombinant NRZ was expressed in BL21-CodonPlus cells using the auto-induction method33 for 24 h at 25 °C with shaking. Bacterial cells were collected by centrifugation at 4000 rpm (JLA 9.1000 rotor, Avanti J-E Beckman Coulter, Mount Waverly, Australia) for 20 min and re-suspended in 50 ml lysis buffer A (50 mM Tris, pH 8.5, 300 mM NaCl and 2 mM BME (β-Mercaptoethanol) supplemented with lysozyme and DNaseI. The cells were lysed using sonication (programme 7, Model 705 Sonic Dismembrator, Fisher Scientific, Hampton, New Hampshire, US) and the resultant lysate was transferred into SS34 tubes for further centrifugation at 16,000 rpm (JA-25.50 rotor, Beckman Coulter Avanti J-E) for 30 min. The supernatant was loaded onto a HisTrap HP, 5 ml (GE Healthcare, Little Chalford, UK) equilibrated with buffer A. After sample application, the column was washed with 100 ml of buffer B (50 mM Tris, pH 8.5, 300 mM NaCl, 25 mM imidazole and 2 mM BME (β-Mercaptoethanol) and the target protein was eluted with buffer C (50 mM Tris, pH 8.5, 300 mM NaCl, 300 mM imidazole and 2 mM BME (β-Mercaptoethanol)) followed by HRV 3C protease cleavage while dialyzed overnight into buffer A at 4 °C. The cleaved protein was passed again through the column to remove the cleaved His-MBP tag, with the remaining protein being concentrated using a centrifugal concentrator with 10 kDa molecular weight cutoff (Amicon® Ultra 15) to a final volume of 4 ml. Concentrated NRZ was subjected to size-exclusion chromatography using a Superdex S75 16/600 column mounted on an ÄKTAXpress system (GE Healthcare) equilibrated in 25 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM BME where it eluted as a single peak. The final sample purity was estimated to be higher than 95% based on SDS–PAGE analysis. Appropriate fractions were pooled and concentrated using a centrifugal concentrator with 10 kDa molecular weight cutoff (Amicon ® Ultra 15) to final concentration of 28 mg ml−1.
Expression and purification of NRZ mutants R86A and D79A
NRZ mutants D79A and R86A were codon-optimized and synthesized (Genscript) and subsequently cloned into the pGEX-6P-3 vector (Invitrogen). Expression and purification were performed using the same protocol as for wild-type NRZ.
Measurement of dissociation constants
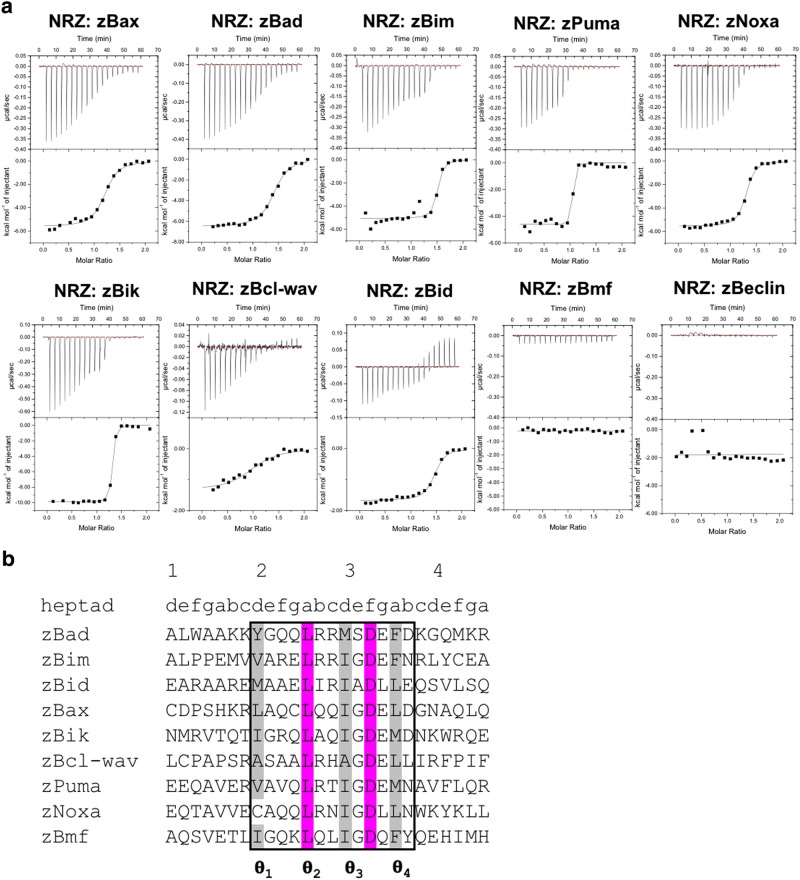
Binding affinities were measured by isothermal titration calorimetry (ITC) employing a MicroCal iTC200 system (GE Healthcare) at 25 °C using wt-NRZ, and NRZ mutants D79A and R86A in 25 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM BME at a final concentration of 30 μM as previously described34. BH3 motif peptide ligands were used at a concentration of 300 μM and titrated using 19 injections of 2.0 μl of ligand. All affinity measurements were performed in triplicate. Protein concentrations were measured using a Nanodrop UV spectrophotometer (Thermo Scientific, Scoresbury, Australia) at a wavelength of 280 nm. Peptide concentrations were calculated based on the dry peptide weight after synthesis. The zebrafish BH3-motif peptides used were commercially synthesized and were purified to a final purity of 95% (GenScript, Piscataway, New Jersey, US). zBim: ALPPEMVVARELRRIGDEFNRLYCEA (UniProt accession code B2KKY9, residues 117–142), zPuma: EEQAVERVAVQLRTIGDEMNAVFLQR (accession code Q0GKC9, residues 123–148), zBik: NMRVTQTIGRQLAQIGDEMDNKWRQE (accession code Q5RGV6, residues 30–55), zBax: ELCDPSHKRLAQCLQQIGDELDGNAQLQ (accession code Q9I9N4, residues 52–79), zBid: EARAAREMAAELIRIADLLEQSVLSQAA (accession code Q0GKC5, residues 85–112), zBad: ALWAAKKYGQQLRRMSDEFDKGQMKR (accession code A7MCM4, 88–113 residues), zBmf: AQSVETLIGQKLQLIGDQFYQEHIMH (accession code Q0GKC7, residues 89–114), zNoxa: EQTAVVECAQQLRNIGDLLNWKYKLL (accession code Q0GKC8, residues 5–30), zBok: PRGVLVDVSVVLLKLGDELECMRPYV (accession code Q7T381, residues 60–85), zBeclin: DGGTMENLSRRLKVTSNLFDIMSGQT (accession code F1RCP1, residues 102–127), zBcl-wav: LCPAPSRASAALRHAGDELLIRFPIF (accession code D2Y5Q2, residues 42–67).
Crystallization and data collection
Crystals of apo NRZ were obtained at a protein concentration of 28 mg/ml in 1.0 M magnesium sulphate hydrate, 0.1 M sodium acetate trihydrate pH 4.6. The crystals were cryo-protected in mother liquor supplemented with 30% glucose and flash cooled in liquid nitrogen. The apo NRZ crystals in this condition appeared as thick needles belonging to the P43 space group of the tetragonal crystal system.
All diffraction data were collected on the MX2 beamline at the Australian Synchrotron using an Eiger detector (Dectris, Baden-Dättwil, Switzerland) with an oscillation range of 0.1° per frame using wavelength 0.9537 Å. The diffraction data were integrated using XDS35 and scaled using AIMLESS36. The crystals of apo-NRZ contained one molecule of NRZ in the asymmetric unit with a calculated solvent content of 47.0%. The structure of apo-NRZ was solved by molecular replacement using PHASER37 with previously solved structure of NRZ: Bad BH3 (PDB ID: 6FBX) as a search model. The final TFZ and LLG values were 15.4 and 541.6, respectively. The final apo-NRZ structure was built manually over multiple cycles using Coot38 and refined using PHENIX39 to a final Rwork/Rfree of 0.194/0.222 with 96.3% of residues in Ramachandran favoured region of the plot and no outliers. All data collection and refinement statistics are summarized in Table 2.
Table 2.
X-ray crystallographic data collection and refinement statistics
| Native NRZ Apo | Native NRZ: Bad BH3 | |
|---|---|---|
| Data collection | ||
| Space group | P43 | P63 |
| Cell dimensions | ||
| a, b, c (Å) | 48.18, 48.18, 75.33 | 87.62, 87.62, 36.77 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 |
| Wavelength (Å) | 0.9537 | 0.9537 |
| Resolution (Å) | 48.18–2.0 (2.07–2.0) * | 43.81–1.639 (1.68–1.64) * |
| Rsym or Rmerge | 0.051 (1.16) | 0.092 (1.92) |
| I/σI | 18.3 (1.7) | 15.4 (1.3) |
| Completeness (%) | 99.93 (99.57) | 100 (100) |
| Redundancy | 6.9 (6.9) | 18.6 (9.9) |
| Refinement | ||
|---|---|---|
| Resolution (Å) | 48.18–2.0 (2.07–2.0) | 43.81–1.639 (1.68–1.64) |
| No. of reflections | 11,653 | 19,885 |
| Rwork/Rfree | 0.194/0.222 | 0.187/0.206 |
| No. of atoms | ||
| Protein | 1194 | 1361 |
| Ligand/ion | 0 | 0 |
| Water | 39 | 105 |
| B-factors | ||
| Protein | 54.98 | 39.6 |
| Ligand/ion | 0 | 0 |
| Water | 52.42 | 44.2 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.003 |
| Bond angles (°) | 0.84 | 0.51 |
* Values in parentheses are for the highest resolution shell.
Complexes of NRZ with zBad BH3 were prepared as previously described40. Briefly, NRZ: zBad BH3 complexes were reconstituted by adding zBad BH3 peptides at a 1:1.25 molar ratio to NRZ. The reconstituted complex was concentrated to 28 mg ml−1 using a 3 kDa molecular weight cutoff centrifugal concentrator (Millipore), flash-cooled and stored under liquid nitrogen. High-throughput sparse matrix screening was carried out using 96-well sitting-drop trays (Swissci, Neuheim, Switzerland) and the vapour-diffusion method at 20 °C. Crystals of NRZ: zBad BH3 were obtained at 28 mg ml−1 using the sitting-drop method at 20 °C in 0.2 M sodium fluoride, 0.1 M Bis-Tris propane, pH 6.5, 20% (W/V) PEG 3350. The crystals were flash-cooled at −173 °C in mother liquor supplemented with 30% (w/v) glucose. The NRZ: zBad BH3 complex formed single rod-shaped crystals belonging to space group P63 in the hexagonal crystal system.
Diffraction data for NRZ: zBad BH3 complex were collected on the MX2 beamline at the Australian Synchrotron using a with an Eiger detector with an oscillation range of 0.1° per frame using wavelength 0.9537. Collected diffraction data were integrated using XDS35 and scaled using AIMLESS36. Molecular replacement was performed using PHASER37 with the structure of Mcl-1 (PDB ID: 5KU9)41 as a search model. NRZ: zBad BH3 crystals contain one molecule of NRZ and 1 molecule of zBad BH3 in the asymmetric unit, with a 43.7% solvent content and final TFZ and LLG values of 9.2 and 63.76, respectively. The final model of NRZ: zBad BH3 was built manually over several cycles using Coot38 and refined using PHENIX39 with a final Rwork/Rfree of 0.187/0.206, with 98.7% of residues in Ramachandran favoured region of the plot and no outliers.
All images for NRZ apo and NRZ: zBad BH3 complex were generated using PyMOL molecular graphic system, version 1.8.6 (Schrödinger, LLC, New York, USA). All software was accessed through the SBGrid suite42.
Sequence alignments
Sequence alignments were performed using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/) with the default settings.
Results
In order to determine the molecular basis for apoptosis control mediated by NRZ in zebrafish we systematically examined the ability of NRZ to bind to peptides spanning the BH3 motif of zebrafish encoded pro-apoptotic Bcl-2 proteins. Analysis of the D. rerio genome indicated that genes are present for orthologues of the mammalian pro-apoptotic Bcl-2 family members Bid, Bim, Bax, Bad, Bik, Bmf, Puma and Noxa4,23,24,44, as well as Bcl-wav, a pro-apoptotic paralogue unique to fish29. In addition a Beclin 1 orthologue, a protein that harbours a BH3-like motif that is involved in autophagy45 and was previously shown to interact with pro-survival Bcl-2 proteins46, is also present. Isothermal titration calorimetry (ITC) was used to determine the affinity of NRZ for peptides that span the BH3 motifs of zBid, zBim, zBax, zBad, zBik, zBmf, zPuma, zNoxa, zBcl-wav and zBeclin (Fig 1, Table 1). The BH3 motifs were chosen through sequence alignment with the known mammalian pro-apoptotic Bcl-2 proteins by identifying the signature sequence LXXXGDE of the BH3 motif5, where X is any amino acid. Interestingly, our ITC data showed that NRZ binds most BH3 motifs with the exception of those from Bmf and Beclin, which displayed no detectable affinity. Several BH3-only proteins interacted with NRZ with high affinities, including zBik (KD 12 nM), zPuma (KD 36 nM) and zBim (KD 41 nM), while zNoxa (KD 142 nM), zBad (KD 343 nM), zBid (KD 409 nM) and zBax (KD 688 nM) were bound with more modest affinities. In contrast, Bcl-wav, a recently discovered novel pro-apoptotic Bcl-2 family member of zebrafish29 engaged NRZ with only micromolar affinity (KD 3570 nM).
Fig. 1. Titration curves showing the raw heats of titration for ITC measurements of NRZ: BH3 motif interactions.
NRZ interacts with Bax as well as all other BH3-only proteins but not Bmf or Beclin-1. Affinities are summarized in Table 1
Table 1.
ITC affinity measurements of NRZ: BH3 motif interactions
| Peptide | WT NRZ KD (nM) |
NRZ R86A KD (nM) |
NRZ D79A KD (nM) |
|---|---|---|---|
| Bax | 688 ± 111 | 4600 ± 440 | 264 ± 42 |
| Bim | 41 ± 5 | 168 ± 8 | 13 ± 4 |
| Bad | 343 ± 48 | 4800 ± 88 | 3330 ± 41 |
| Puma | 36 ± 4 | 2770 ± 162 | 117 ± 5 |
| Bik | 12 ± 2 | 17 ± 3 | 14 ± 3 |
| Noxa | 142 ± 16 | 510 ± 41 | 335 ± 31 |
| Bcl-wav | 3570 ± 162 | NB | NB |
| Bid | 409 ± 55 | NB | 220 ± 16 |
| Bmf | NB | NB | NB |
| Beclin | NB | NB | NB |
26-mer peptides spanning the BH3-motyif of D. rerio pro-apoptotic Bcl-2 family members or Beclin-1 from zebrafish were employed. All KD values (in nM) are the means of three replicates with standard error
NB no binding
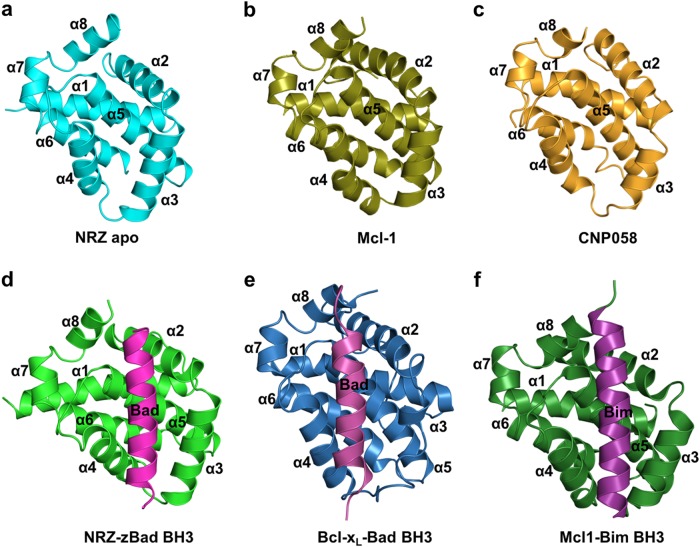
To determine the structural basis of NRZ interaction with BH3 motifs of pro-apoptotic Bcl-2 proteins, we determined the crystal structures of apo-NRZ and its complexed holo form bound to the BH3-motif of zBad (Fig. 2a, d, Table 2). Similar to other pro-survival Bcl-2 proteins, NRZ adopts a conserved Bcl-2-like fold consisting of eight α-helices that form a globular helical bundle. Helices α2-5 form the canonical hydrophobic ligand binding groove observed in other Bcl-2 family proteins47 that is utilized to accommodate the zBad BH3 peptide (Fig. 2d). An analysis using DALI48 showed that complexes of Mcl-1 (PDB ID 2NL9)49 (Fig. 2b, f) and Bcl-xL (PDB ID 4QNQ)50 bound to Bim and Bad BH3 peptides are the closest structural Bcl-2 homologs with R.M.S.D. value 1.7 Å and 1.9 Å over 136 and 132 Cα atoms, respectively, with sequence identities of 15% and 18%, respectively. The closest viral Bcl-2 homolog to NRZ is CNP05851 (R.M.S.D. of 2.0 Å over 133 Cα atoms, Fig. 2c) with a sequence identity of 16%.
Fig. 2. Crystal structures of NRZ, NRZ: Bad BH3 complex and comparison with closely related complexes.
Cartoon representation of (a) apo NRZ (light blue). Helices are labelled as α1–8, with the view being into the hydrophobic groove formed by α2–5. b Cartoon representation of Mcl-1 (olive)49. c CNP058 (orange)51, the closest structural homolog from viral pro-survival Bcl-2 proteins for NRZ. d NRZ (green) in complex with zBad BH3 motif (hot pink). e Bcl-xL (blue) in complex with Bad BH3 motif (yellow)50, f Mcl-1 (brown) in complex with Bim (magenta) BH3 motif49. All views in b–f are as in a
NRZ: BH3 motif interactions
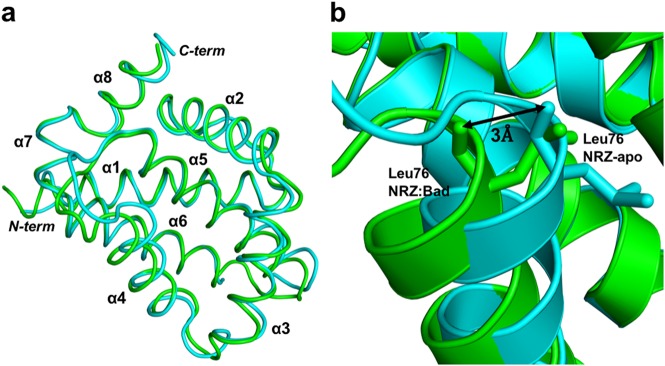
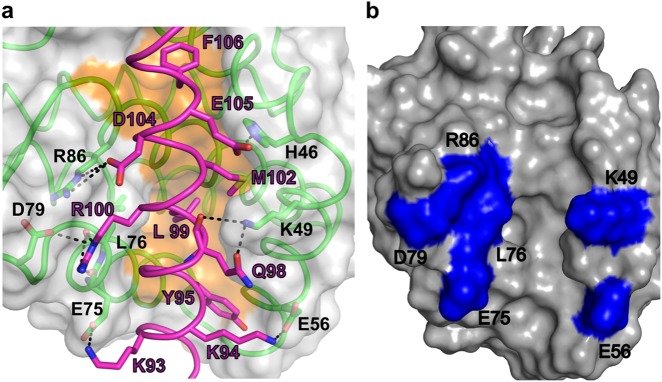
NRZ utilizes the canonical hydrophobic binding groove that is also found in other pro-survival Bcl-2 proteins4 to accommodate the zBad BH3-motif (Fig. 3a) using a combination of hydrophobic and ionic interactions as well as hydrogen bonds. To accommodate the zBad BH3-peptide NRZ undergoes localized conformational changes (Fig. 3a, d). Upon binding of the BH3-motif, the C-terminal end of α4-helix moves by 3.0 Å (Fig. 4a, b) relative to apo-NRZ, thus enlarging the binding groove to accommodate the Bad BH3 motif. Detailed inspection of the NRZ: zBad BH3 complex interface reveals five salt bridges between Lys93Bad and Glu75NRZ, Lys94Bad and Glu56NRZ, Glu105Bad and His46NRZ, Arg100Bad with Asp79NRZ and Asp104Bad with Arg86NRZ. In addition to ionic interactions, the NRZ: zBad interface also features three hydrogen bonds between Arg100Bad–Leu76NRZ, Arg100Bad–Glu75NRZ, Gln98Bad–Lys49NRZ. Finally, the four highly conserved hydrophobic residues Y95, 99L, M102 and F106 from the zBad BH3 motif protrude into the ligand binding groove of NRZ and are accommodated in four hydrophobic pockets at the floor of the binding groove.
Fig. 3. Superimposition of apo NRZ with NRZ: zBad BH3 complex.
Comparison of the backbone structures of NRZ and NRZ: zBad BH3 complex. a Cartoon representation of apo NRZ (light blue) superimposed onto NRZ: zBad BH3 complex (green). The view is into the canonical hydrophobic binding groove formed by α2–5. b Close up view of NRZ helix α4, which is shifted by 3 Å from its original position in the apo NRZ structure upon zBad BH3 motif binding, thus enlarging the binding groove
Fig. 4. Binding of BH3 motif peptides to NRZ result in local reorganization of the canonical NRZ binding groove.
a Detailed view of the NRZ: zBad BH3 motif interface. The NRZ surface, backbone and floor of the binding groove are shown in grey, green and orange, respectively, while zBad BH3 is shown in hot pink. The four key hydrophobic residues of zBad (Y95, L99, M102, F106) protruding into the binding groove, and the conserved salt-bridge formed by NRZ D79 and zBad BH3 R100 are labelled, as well as all other residues involved in additional ionic interactions and hydrogen bonds. Interactions are denoted as dashed black lines. b Surface view of the conserved residues that are involved in NRZ-zBad BH3 interactions. Residues shown in blue are residues within canonical binding groove of NRZ (K49, E56, E75, L76, D79 and R86) that are conserved in NRZ-like proteins amongst different fish species
To validate the structure of NRZ: Bad BH3 we mutated two key NRZ residues involved in ionic interactions, Asp79 and Arg86 to Ala, and examined the ability of these mutants to bind BH3-motif peptides (Table 1). Both mutants showed substantially reduced binding to Bad, with D79A displaying a tenfold reduction in Bad binding, whereas NRZ R86A displayed a 14-fold reduction in affinity. However, the contributions to binding BH3-motif peptides from these residues are not uniform across all BH3-motifs, indicating differences in the specific importance of these contacts. For example, Bik is not affected strongly by these two mutations and Bid binding is only impacted by R86A, whereas Bcl-wav binding is ablated for both R86A and D79A.
Discussion
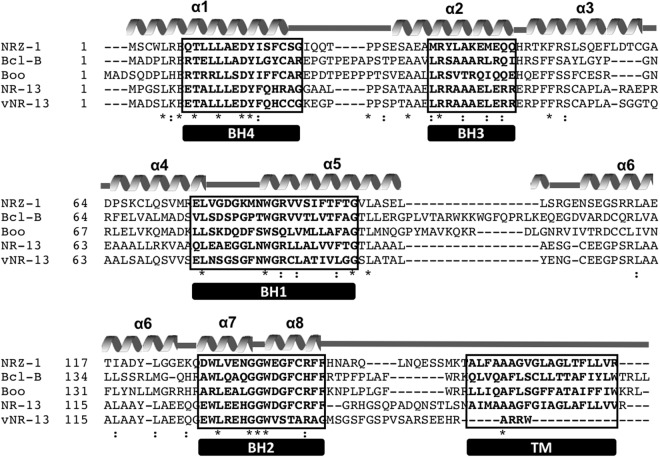
Developing a detailed understanding of Bcl-2 family function in apoptosis regulation is not only important for identifying their biological roles but, is crucial in the design of new therapeutic strategies directed against this family4,52. Indeed, as a major arbiter of programmed cell death, there is a significant interest in resolving the function of Bcl-2 family proteins at a molecular level with the aim of targeting them for their role in cancer6,53. Zebrafish are proving a valuable model system in this context as the mechanisms of apoptosis activation and function appear to be similar to those found in mammals28. Although there are many similarities between the Bcl-2 families in mammals and fish there are also significant differences that require clarification20. NRZ was initially identified as a D. rerio Bcl-2 orthologue of avian NR-1326 via database searches26,28. Sequence alignment revealed that NRZ shares 40 and 39% identity with chicken NR-13 and turkey herpes virus NR-13 with respectively. Significantly lower sequence identity is shared with the mammalian orthologues of NRZ where only 25 and 23% sequence identity are observed for the human pro-survival Bcl-2 protein Bcl-B (also known as Bcl-2 like protein 10 or NrH) and the mouse orthologue Boo (or Diva), respectively54. Zebrafish NRZ features significant sequence differences from other Bcl-2 proteins with only three residues identical between NRZ, Bcl-B and Boo in the region spanning helices α3–5 that constitute the canonical ligand binding groove (Fig. 5), thus potentially providing a basis for a unique ligand binding profile for this pro-survival Bcl-2 family member4,28. Here we examined the structure and interactions of NRZ by determining the structures of apo-NRZ and its complex with zBad BH3, the orthologue of mammalian Bad and measuring the binding affinities for BH3-motifs. The structures revealed the conformational changes in NRZ after binding of BH3 motif ligand and provide a structural basis for NRZ mediated apoptosis inhibition.
Fig. 5. Sequence alignment of NRZ with Bcl-2 homologs from other organisms.
The sequence alignment of Bcl-2 family proteins was generated with MUSCLE66 using sequences from zebrafish NRZ (Uniprot Accession number Q8UWD5), human Bcl-B (Q9HD36), mouse Boo (Q9Z0F3), chicken NR-13 (Q90ZN1) and herpes virus vNR-13 (Q9DH00). The α-helical secondary structure elements (α1–8) are marked as grey helices and loop regions are indicated as grey lines based on the crystal structure of NRZ. The boxed regions of the sequences are denoting the Bcl-2 homology motifs (BH motifs 1–4) and trans-membrane domains (TM) at the end of the sequences. Conserved identical residues between sequences are denoted as “*”, similar residues are denoted as “:” and semi conserved residues denoted as “.”
Surprisingly, our structural search and comparison using DALI48 revealed that the closest structural homolog of NRZ is in fact Mcl-155 with an R.M.S.D value of 1.7 Å over 136 Cα atoms, and a sequence identity of 15%. Sequence alignment of NRZ (Fig. 5) showed that NRZ shares sequence features of other multi-domain members of Bcl-2 family. However, the BH regions show considerable sequence variation and these sequence variations that are located in the binding groove account in part for the selectivity differences observed for BH3-ligands47 compared to NRZ’s mammalian counterparts. Interestingly, NRZ shares only 18% sequence identity with the two zebrafish encoded Mcl-1 homologs zMcl-1a and zMcl-1b, suggesting that they may not be functionally redundant and differentially interact with pro-apoptotic Bcl-2 proteins in zebrafish.
The overall fold of the NRZ: zBad BH3 complex is very similar to that observed in other Bcl-2 complexes. Despite the overall similarity in fold, several interesting differences are observed in the crystal structures and protein:peptide interfaces of the NRZ: zBad and Mcl-1:Bim complexes. The difference in peptide binding mode of these complexes were calculated56 as solvent accessible surface and associated thermodynamic properties of Gibb’s free energy change (ΔG) of interface formation and dissociation. The binding of zBad to NRZ buries a total of 2366 Å2 solvent accessible surface and solvation energy of isolated structure −10.1 kcal/mol and ΔG of interface formation and dissociation of −3.9 kcal/mol. In contrast, binding of human Bim to human Mcl-1 buries a total of 2665 Å2 and solvent accessible surface and solvation energy of isolated structure −10.4 kcal/mol and ΔG of interface formation and dissociation of −4.8 kcal/mol. However, the human Bcl-xL:Bad complex forms a larger (total of 3268 Å2 and solvent accessible surface and solvation energy of isolated structure −14.4 kcal/mol and ΔG of interface formation and dissociation of −2.2 kcal/mol) ligand binding interface than that of NRZ: zBad and human Mcl-1:Bim. Structurally, NRZ features a more ordered α3 helix compared to human Bcl-xL, which upon Bim binding unravels the α3 helix57, and leads to an opening of the canonical ligand binding groove of ~9 Å due to an outward movement of α3 and a pivoting of α4. In contrast, NRZ maintains the ordered α3 helix on binding Bad, which leads to the C-terminal end of α4 helix moving by 3 Å relative to that in apo-NRZ (Fig. 4b).
Similar to other multi-domain Bcl-2 family proteins, including pro-apoptotic proteins Bax, Bak and Bok, NRZ also contains the highly conserved sequence motif “NWGR” as part of the BH1 motif at the N-terminal end of α5 (Fig. 2). In addition to forming a helix cap4 this region plays a vital role for recognition of the BH3-only proteins58. A hallmark of BH3 motif interactions with mammalian pro-survival Bcl-2 proteins is the formation of an ionic bond between the conserved Arginine of the “NWGR” sequence motif of pro-survival Bcl-2 proteins with the absolutely conserved Aspartate residue of the BH3 motif58. In the human Bcl-xL:Bad complex (1G5J50)(Fig. 3e), the corresponding R139 residue in the NWGR motif interacts with D119 of the Bad BH3 peptide50. Previous studies revealed that a R139Q mutation in Bcl-xL results in loss of pro-survival function and ability to interact with Bax59. Surprisingly, this highly conserved interaction between the Arginine in the NWGR motif that is present in all other mammalian Bcl-2:BH3 complexes solved to date4,60 is very weak in the NRZ: zBad BH3 complex, instead an additional ionic interaction between Asp79NRZ-Arg100Bad is observed that may compensate for the weaker and longer range canonical ionic interaction between Arg86NRZ-Asp104Bad. Mutagenesis of both Asp79 and Arg86 in NRZ indicated that indeed both contribute to the binding of Bad, with Arg86 still playing an important role in binding BH3-motif peptides despite being more distant from the partner Asp104 in Bad when compared to other pro-survival Bcl-2: BH3 peptide complexes (Table 1). Notably, the individual mutations affected binding to several BH3 motif peptides differentially, suggesting that careful mutagenesis may be utilized to probe the role of individual NRZ interactions with pro-apoptotic Bcl-2 proteins61.
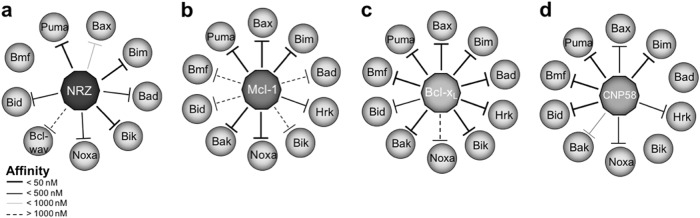
NRZ displays a very distinct ligand interaction profile when compared to its most structurally related proteins, Mcl-1, Bcl-xL and CNP058. Intriguingly the sole viral Bcl-2 member encoded by a fish virus, grouper iridovirus GIV66, only binds Bim, thus displaying a radically different ligand binding profile compared to NRZ62. Among mammalian pro-survival Bcl-2 proteins a distinct Bad/Noxa dyad is observed, with Bcl-2, Bcl-xL and Bcl-w binding Bad, but not Noxa, whereas Mcl-1 and A1 bind Noxa but not Bad7. In contrast, NRZ binds both Bad and Noxa with 340 nM and 140 nM affinity, respectively, a feature not previously seen outside of virus encoded pro-survival Bcl-2, with African swine fever virus encoded A179L and fowlpox virus encoded FPV039 the only known pro-survival Bcl-2 proteins that are Bad and Noxa binders63,64. NRZ shows no affinity for Bmf, which is bound by both human Mcl-1 and Bcl-xL (Fig. 6)7, and overall displays a ligand binding profile that most closely resembles A1, albeit with different affinities for individual ligands65. Overall, the affinity measurements suggest that NRZ is unlikely to be a functional Mcl-1 homolog, as might be expected as there are two Mcl-1 orthologues in D. rerio, Mcl-1a and Mcl-1b26, and it is also unlikely to be a functional Bcl-B homolog, considering that human Bcl-B is only able to engage Bax and Bim54. NRZ also does not bind the BH3 motif of the autophagy regulator Beclin-1, a feature previously observed for both mammalian Bcl-2 and Bcl-xL50, suggesting that NRZ does not harbour a dual role in regulating apoptosis and autophagy.
Fig. 6. Comparison of the BH3 motif binding profile of NRZ with Mcl-1, Bcl-xL and CNP058.
a Binding profile of zebrafish NRZ with BH3 motif peptides (b) binding profile of human Mcl-1 (c) binding profile of human Bcl-xL and (d) binding profile of canarypox virus CNP058. BH3 motif peptides used in (b) and (c) are of human origin, whereas peptides in (d) are from chicken. Bars indicate binding affinity ranges from <50 nM, <500 nM, <1000 nM, and >1000 nM as shown in inset
In summary, like other pro-survival Bcl-2 protein structures solved to-date, NRZ adopts a Bcl-2 like fold and its most closely related structural homologs are the cellular apoptosis inhibitor Mcl-1 and the canarypox viral Bcl-2 protein CNP058. Furthermore, we demonstrated that NRZ harbours a unique BH3 motif binding profile. However, while NRZ is a close structural homolog of Mcl-1 it seems unlikely to be a functional orthologue based on its different binding profile, in particular the ability to engage both Bad and Noxa, a feature that has not been previously observed in mammalian pro-survival Bcl-2 proteins. This study suggests that NRZ likely occupies a unique mechanistic role in zebrafish apoptosis regulation. Thus, further functional studies are required in vivo to delineate the role of NRZ in apoptosis signalling. Our findings demonstrate the complexities of delineating Bcl-2 family function and the pitfalls of assumed functional and evolutionary similarity based on sequence and structure alone.
Data availability
The raw X-ray diffraction data were deposited at the SBGrid Data Bank (http://data.sbgrid.org)43 as dataset entries doi: 10.15785/SBGRID/6H1N and 10.15785/SBGRID/6FBX. The coordinates have been deposited in the Protein Data Bank (accession code 6H1N and 6FBX).
Acknowledgements
We would like to thank staff at the MX beamlines at the Australian Synchrotron for help with X-ray data collection, and the Comprehensive Proteomics Platform at La Trobe University for core instrument support. This work was supported by the Australian Research Council (Fellowship FT130101349 to M.K.) and La Trobe University (Scholarship to C.D.S.).
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by: P. Bouillet
Contributor Information
Mark G. Hinds, Email: mhinds@unimelb.edu.au
Marc Kvansakul, Email: m.kvansakul@latrobe.edu.au.
References
- 1.Green Douglas R., Llambi Fabien. Cell Death Signaling. Cold Spring Harbor Perspectives in Biology. 2015;7(12):a006080. doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kvansakul Marc, Caria Sofia, Hinds Mark. The Bcl-2 Family in Host-Virus Interactions. Viruses. 2017;9(10):290. doi: 10.3390/v9100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–3683. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kvansakul M, Hinds MG. The Bcl-2 family: structures, interactions and targets for drug discovery. Apoptosis. 2015;20:136–150. doi: 10.1007/s10495-014-1051-7. [DOI] [PubMed] [Google Scholar]
- 5.Kvansakul M, Hinds MG. The structural biology of BH3-only proteins. Methods Enzymol. 2014;544:49–74. doi: 10.1016/B978-0-12-417158-9.00003-0. [DOI] [PubMed] [Google Scholar]
- 6.Luna-Vargas MP, Chipuk JE. The deadly landscape of pro-apoptotic BCL-2 proteins in the outer mitochondrial membrane. FEBS J. 2016;283:2676–2689. doi: 10.1111/febs.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Kalkavan H, Green DR. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018;25:46–55. doi: 10.1038/cdd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czabotar PE, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Lindsten T, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol. Cell. 2000;6:1389–1399. doi: 10.1016/S1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaux DL. Apoptogenic factors released from mitochondria. Biochim. Biophys. Acta. 2011;1813:546–550. doi: 10.1016/j.bbamcr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Shamas-Din A, Kale J, Leber B, Andrews DW. Mechanisms of action of Bcl-2 family proteins. Cold Spring Harb. Perspect. Biol. 2013;5:a008714. doi: 10.1101/cshperspect.a008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popgeorgiev N, et al. The apoptotic regulator Nrz controls cytoskeletal dynamics via the regulation of Ca2+ trafficking in the zebrafish blastula. Dev. Cell. 2011;20:663–676. doi: 10.1016/j.devcel.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Prudent J, Popgeorgiev N, Bonneau B, Gillet G. Bcl-2 proteins, cell migration and embryonic development: lessons from zebrafish. Cell Death Dis. 2015;6:e1910. doi: 10.1038/cddis.2015.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonneau B, Prudent J, Popgeorgiev N, Gillet G. Non-apoptotic roles of Bcl-2 family: The calcium connection. Biochim. Et. Biophys. Acta. 2013;1833:1755–1765. doi: 10.1016/j.bbamcr.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 16.Szegezdi E, MacDonald DC, Ní Chonghaile T, Gupta S, Samali A. Bcl-2 family on guard at the ER. Am. J. Physiol. 2009;296:C941–C953. doi: 10.1152/ajpcell.00612.2008. [DOI] [PubMed] [Google Scholar]
- 17.Caria S, Hinds MG, Kvansakul M. Structural insight into an evolutionarily ancient programmed cell death regulator – the crystal structure of marine sponge BHP2 bound to LB-Bak-2. Cell Death Dis. 2017;8:e2543. doi: 10.1038/cddis.2016.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaux DL, Haecker G, Strasser A. An evolutionary perspective on apoptosis. Cell. 1994;76:777–779. doi: 10.1016/0092-8674(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 19.Santos NMSd, Vale Ad, Reis MIR, Silva MT. Fish and apoptosis: molecules and pathways. Curr. Pharm. Des. 2008;14:148–169. doi: 10.2174/138161208783378743. [DOI] [PubMed] [Google Scholar]
- 20.Eimon PM, Ashkenazi A. The zebrafish as a model organism for the study of apoptosis. Apoptosis. 2010;15:331–349. doi: 10.1007/s10495-009-0432-9. [DOI] [PubMed] [Google Scholar]
- 21.Yuan Z, et al. Expression of Bcl-2 genes in channel catfish after bacterial infection and hypoxia stress. Dev. Comp. Immunol. 2016;65:79–90. doi: 10.1016/j.dci.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Krumschnabel G, Podrabsky JE. Fish as model systems for the study of vertebrate apoptosis. Apoptosis. 2009;14:1–21. doi: 10.1007/s10495-008-0281-y. [DOI] [PubMed] [Google Scholar]
- 23.Jette CA, et al. BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ. 2008;15:1063. doi: 10.1038/cdd.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW. BH3-only proteins: orchestrators of apoptosis. Biochim. Et. Biophys. Acta. 2011;1813:508–520. doi: 10.1016/j.bbamcr.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Akhtar RS, Ness JM, Roth KA. Bcl-2 family regulation of neuronal development and neurodegeneration. Biochim. Et. Biophys. Acta. 2004;1644:189–203. doi: 10.1016/j.bbamcr.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Inohara N, Nunez G. Genes with homology to mammalian apoptosis regulators identified in zebrafish. Cell Death Differ. 2000;7:509–510. doi: 10.1038/sj.cdd.4400679. [DOI] [PubMed] [Google Scholar]
- 27.Lee Rm, Gillet G, Burnside J, Thomas SJ, Neiman P. Role of Nr13 in regulation of programmed cell death in the bursa of Fabricius. Genes Dev. 1999;13:718–728. doi: 10.1101/gad.13.6.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnaud E, et al. The zebrafish bcl-2 homologue Nrz controls development during somitogenesis and gastrulation via apoptosis-dependent and -independent mechanisms. Cell Death Differ. 2005;13:1128. doi: 10.1038/sj.cdd.4401797. [DOI] [PubMed] [Google Scholar]
- 29.Prudent J, et al. Bcl-wav and the mitochondrial calcium uniporter drive gastrula morphogenesis in zebrafish. Nat. Commun. 2013;4:2330. doi: 10.1038/ncomms3330. [DOI] [PubMed] [Google Scholar]
- 30.Vega S, et al. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torraca V, Mostowy S. Zebrafish infection: from pathogenesis to cell biology. Trends Cell Biol. 2018;28:143–156. doi: 10.1016/j.tcb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scholz J, Besir H, Strasser C, Suppmann S. A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC Biotechnol. 2013;13:12. doi: 10.1186/1472-6750-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Burton DR, Caria S, Marshall B, Barry M, Kvansakul M. Structural basis of Deerpox virus-mediated inhibition of apoptosis. Acta Crystallogr. D Biol. Crystallogr. 2015;71:1593–1603. doi: 10.1107/S1399004715009402. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 37.McCoy A. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kvansakul M, Czabotar PE. Preparing Samples for Crystallization of Bcl-2 Family Complexes. In: Puthalakath H, Hawkins CJ, editors. Programmed Cell Death: Methods and Protocols. New York, NY: Springer New York; 2016. pp. 213–229. [DOI] [PubMed] [Google Scholar]
- 41.Johannes JW, et al. Structure based design of non-natural peptidic macrocyclic Mcl-1 inhibitors. ACS Med. Chem. Lett. 2017;8:239–244. doi: 10.1021/acsmedchemlett.6b00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morin A, et al. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer PA, et al. Data publication with the structural biology data grid supports live analysis. Nat. Commun. 2016;7:10882. doi: 10.1038/ncomms10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Priault M, et al. Differential dependence on Beclin 1 for the regulation of pro-survival autophagy by Bcl-2 and Bcl-xL in HCT116 colorectal cancer cells. PLoS ONE. 2010;5:e8755. doi: 10.1371/journal.pone.0008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kvansakul M, Hinds MG. Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis. 2013;4:e909. doi: 10.1038/cddis.2013.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holm L, Rosenström Pi. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czabotar PE, et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl Acad. Sci. USA. 2007;104:6217–6222. doi: 10.1073/pnas.0701297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petros AM, et al. Rationale for Bcl-XL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anasir M, Baxter A, Poon I, Hulett M, Kvansakul M. Structural and functional insight into canarypox virus CNP058 mediated regulation of apoptosis. Viruses. 2017;9:305. doi: 10.3390/v9100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cory S, Roberts AW, Colman PM, Adams JM. Targeting BCL-2-like proteins to kill cancer cells. Trends Cancer. 2016;2:443–460. doi: 10.1016/j.trecan.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Fuchs Y, Steller H. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat. Rev. Mol. Cell Biol. 2015;16:329–344. doi: 10.1038/nrm3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rautureau GJP, et al. The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis. 2012;3:e443. doi: 10.1038/cddis.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fire E, Gullá SV, Grant RA, Keating AE. Mcl-1–Bim complexes accommodate surprising point mutations via minor structural changes. Protein Sci. 2010;19:507–519. doi: 10.1002/pro.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 57.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–352. doi: 10.1016/S1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 58.Day CL, et al. Structure of the BH3 domains from the p53-inducible bh3-only proteins noxa and puma in complex with Mcl-1. J. Mol. Biol. 2008;380:958–971. doi: 10.1016/j.jmb.2008.05.071. [DOI] [PubMed] [Google Scholar]
- 59.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, et al. Structure of BcI-X$_L$-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 60.Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim. Et. Biophys. Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 61.Campbell S, et al. Structural insight into BH3 domain binding of vaccinia virus antiapoptotic F1L. J. Virol. 2014;88:8667–8677. doi: 10.1128/JVI.01092-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banjara S, Mao J, Ryan TM, Caria S, Kvansakul M. Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim. J. Biol. Chem. 2018;293:5464–5477. doi: 10.1074/jbc.RA117.000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banjara S, Caria S, Dixon LK, Hinds MG, Kvansakul M. Structural Insight into African Swine Fever Virus A179L-mediated inhibition of apoptosis. J. Virol. 2017;91:e02228–16. doi: 10.1128/JVI.02228-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anasir MI, Caria S, Skinner MA, Kvansakul M. Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039. J. Biol. Chem. 2017;292:9010–9021. doi: 10.1074/jbc.M116.768879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smits C, Czabotar PE, Hinds MG, Day CL. Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure. 2008;16:818–829. doi: 10.1016/j.str.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 66.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw X-ray diffraction data were deposited at the SBGrid Data Bank (http://data.sbgrid.org)43 as dataset entries doi: 10.15785/SBGRID/6H1N and 10.15785/SBGRID/6FBX. The coordinates have been deposited in the Protein Data Bank (accession code 6H1N and 6FBX).