Abstract
Phosphorylation of the translation initiation factor eIF-2α downregulates protein synthesis by sequestering the guanylate exchange factor eIF-2B. The importance of this regulation has been demonstrated in the context of stress and virally induced repression of protein synthesis but has not been investigated relative to the control of protein synthesis during development. Transgenic Drosophila strains bearing aspartic acid or alanine substitutions at the presumed regulatory phosphorylation site (Ser50) of Drosophila eIF-2α were established. The expression of the eIF-2α mutant transgenes, under the transcriptional control of the hsp70 promoter, was induced at various times during development to assess the developmental and biochemical effects. Flies bearing the aspartic acid eIF-2α mutant (HD) transgene displayed a slow growth phenotype and small body size. Repeated induction of the HD transgene resulted in cessation of development. In contrast, flies bearing the alanine eIF-2α mutant (HA) displayed a fast growth phenotype and females were significantly larger than nontransgenic control sisters. The HD transgenic flies exhibit a relatively lower level of global protein synthesis than the HA transgenic flies, although the difference is statistically insignificant.
Keywords: Translation, eIF-2α, Development
EUKARYOTIC translation initiation factor eIF-2 plays a crucial role in the initiation process by tethering the initiator methionyl-tRNA to the 40S ribosomal subunit and is required for proper start site selection (9,14,15,17,24). eIF-2 is a heterotrimer composed of three polypeptides α, β, and γ. To begin the initiation cycle, eIF-2 forms a ternary complex with GTP and Met-tRNAi. This ternary complex then binds the 40S ribosomal sub-unit to form the 43S preinitiation complex, which scans along the mRNA in search of a suitable initiation codon. Concomitant with formation of the complete ribosome at the translation start site, eIF-2-GTP is hydrolyzed to eIF-2-GDP. Regulation of initiation is critically tied to the eIF-2-GDP to eIF-2-GTP exchange reaction, which is catalyzed by the multisubunit complex eIF-2B (also referred to as GEF). If the α-subunit of eIF-2 is phosphorylated, the GDP-GTP exchange reaction cannot occur (27,33). Instead, eIF-2B is sequestered in an inactive state with eIF-2-GDP. Because the amount of eIF-2B in the cell is relatively low compared to eIF-2, sequestration leads to insufficient eIF-2B to catalyze the exchange reaction of nonphosphorylated eIF-2. eIF-2B sequestration results in reduced levels of cellular eIF-2-GTP, thus diminishing the capacity for subsequent initiation events. The fate of this sequestered complex has not been studied in detail, but presumably type 1 phosphatases eventually dephosphorylate eIF-2α, thereby releasing GEF (34,37).
Biochemical analysis of eIF-2α (10) and site-directed mutagenesis of the mammalian and yeast (S. cerevisiae) eIF-2α genes have identified Ser51 as the regulatory phosphorylation site (12,19). Substitution of aspartic acid for Ser51, to mimic phosphoserine, resulted in a dominant repression of translation of mRNA encoded by genes cotransfected into mammalian cells. In contrast, substitution of alanine for Ser51, neutrally blocking phosphorylation, resulted in the stimulation of translation of transfected genes (19,28). Similar mutations of yeast eIF-2α Ser51 alter the translation of GCN4 (a transcriptional activator) in a manner consistent with the hypothesis that phosphorylation of Ser51 decreases the concentration of the ternary complex (12). Constitutive phosphorylation of eIF-2α in yeast results in more severe effects, including a dramatic reduction in growth rate (13) and in global protein synthesis (4).
Three protein kinases—yeast GCN2, mammalian double-stranded RNA-activated inhibitor (PKR), and mammalian hemin regulated inhibitor (HRI)—have been shown to phosphorylate specifically Ser51 of eIF-2α and thereby regulate global or mRNA-specific translation (5,12,17). A combination of genetic and biochemical experiments has demonstrated that the effects of these three eIF-2α kinases on translation are mediated through phosphorylation of Ser51 (10,12). Recent studies have implicated PKR in regulation of growth and differentiation (1,29).
Although eIF-2α phosphorylation has been shown to regulate translation at the cellular level, the importance of this control in regulating the development of a multicellular organism has not been addressed. As a prerequisite to investigating the developmental importance of eIF-2α, we have isolated the eIF-2α gene from Drosophila melanogaster (30). The Drosophila eIF-2α gene encodes a 341-amino acid protein with 57% and 44% identity to its human and yeast homologues, respectively. Human, yeast, and Drosophila eIF-2α share a block of 19 identical amino acids surrounding the critical phosphorylation site, Ser51 (Ser50 in Drosophila). Like yeast eIF-2α (13), Drosophila eIF-2α can be phosphorylated by mammalian HRI eIF-2α kinase (16,23) and its phosphorylation is elevated in heat-shocked cells (16). The homologous Ser residue in the Drosophila gene corresponds to Ser50 due to a deletion of one residue near the amino-terminus relative to yeast and human eIF-2α. Herein we report the developmental and biochemical effects of expressing alanine and aspartic acid substitutions of Ser50 in eIF-2α transgenes introduced into wild-type Drosophila.
MATERIALS AND METHODS
Drosophila Culture
Flies were reared on a standard cornmeal/molasses/agar media at 26°C. Larvae were heat shocked by immersing the bottom half of the culture vial in a 37°C water bath for 1 h. Adult flies were heat shocked by placing the flies in the culture vial in a 37°C dry incubator for 1 h.
Construction of the eIF-2α Mutant Plasmids
Site-directed mutagenesis after Kunkel and coworkers (21) was performed to create aspartic acid (GAC) or alanine (GCC) substitutions at Ser50 residue of Drosophila eIF-2α. In order to investigate the effects of eIF-2α mutants on Drosophila development, P-element-containing transformation vectors were used to reintroduce eIF-2α genes into flies. To achieve an appropriate temporal and spatial expression of transduced genes during Drosophila development, three different promoters were used: i) the Drosophila cytoplasmic actin 5C proximal promoter (7); ii) the Drosophila α-1 tubulin promoter (35); and iii) the Drosophila heat shock protein 70 promoter (22). The Drosophila actin 5C proximal promoter and a-l tubulin promoters are constitutively activated in most if not all cells whereas the hsp70 promoter is activated upon a brief heat shock (22). The pCaSpeR-act(Bam) vector, which contains the Drosophila actin 5C promoter and polyadenylation signal sequence, was provided by Carl Thummel (36). The transformation vector pCaSpeR is the parental vector in which a Drosophila white gene is flanked by P-element ends.
To construct the eIF-2α mutant genes under the control of the hsp70 promoter, the vector pWHSP216 was constructed using pCaSpeR-act(Bam), in which a 2.75-kb actin 5C promoter was replaced with the hsp70 promoter. Specifically, a 0.4-kb blunted NdeI-PstI fragment of pUChsneo-act (36), which contains hsp70 promoter sequences and a 86-bp untranslated leader sequence, was inserted into the SmaI site of the pBluescript II (KS+) vector. After identifying the orientation by XhoI digestion, a 0.4-kb EcoRI-BamHI fragment was used to replace the 2.75-kb EcoRI-BamHI fragment that contains the actin promoter in pCaSpeR-act. The resultant plasmid pWHSP216 contains a unique BamHI cloning site between the hsp70 promoter and actin polyadenylation signal sequence.
A 1.2-kb BamHI fragment, which contained either the wild-type eIF-2α gene (Ser50) or mutant type eIF-2α genes (Ala50 or Asp50), was inserted into the BamHI site of pCaSpeR-act (BamHI), pWTUB222, or pWHSP216 to create 18 constructs (3 promoters × 3 eIF-2α genes × 2 orientations). Nine constructs, which contained the eIF-2α gene in the same orientation as the promoter and polyadenylation signal sequence, were used to generate transgenic flies.
Isolation of eIF-2α Transgenic Strains
Transgenic flies were obtained by serially microinjecting the nine eIF-2α transformation constructs into 0–90 min w; Δ2-3 embryos. Trans-formants were identified in the second generation by screening for restoration of wild-type eye pigmentation. Insertion of P-elements into the Drosophila genome is facilitated by the Δ2-3 P-element transposase (32). Several crosses were performed to map the chromosomal location of the eIF-2α transgenes, make the transgenes homozygous, and remove the P-transposase so as to generate stable transgenic lines. Polymerase chain reaction (PCR) experiments using PCR primers that flanked an intron in eIF-2α were performed to confirm the presence of the eIF-2α transgene gene in the transgenic flies. As expected, two PCR products, a 640-bp product corresponding to the intronless eIF-2α transgene and a 900-bp product corresponding to the endogenous eIF-2α gene, were amplified from genomic DNA isolated from the transgenic flies. Only the 900-bp product was detected from nontransgenic flies.
Ribonuclease Protection Assays
Total RNA from Drosophila adult homogenates was isolated after procedures of Cavener and coworkers (3). An eIF-2α cRNA probe was derived from pCIAG200 and the rp49 cRNA probe was derived from pCR229. Total RNA (10 μg) was hybridized to 32P-labeled cRNA probes (about 150–600 pg for each probe) in hybridization buffer (80% deionized formamide, 100 mM sodium citrate, pH 6.4, 300 mM sodium acetate, pH 6.4, and 1 mM EDTA) at 95 °C for 3–4 min and 45 °C for overnight. The samples were digested with 200 μ1 of a diluted solution of RNase-A and RNase-Tl solution for 30 min at 37°C. A RNase inactivation/precipitation mixture (300 μ1) was added to the sample tubes and placed in a –20°C freezer for at least 15 min before pelleting the protected RNA fragments. The pellet was dissolved in 8 μl loading buffer and fractionated on a denaturing 8 M urea, 5% polyacrylamide gel.
Western Blot Analysis ofeIF-2α Protein
Female flies from hsp70-eIF-2α transgenic flies of approximately the same age were collected and placed in replicate vials with fresh yeast and allowed to feed overnight. Flies were subjected to a 1-h 37°C heat shock treatment and allowed to recover for 3 h in a 25°C water bath. Five flies were added to 35 μl of homogenization buffer (0.1 mM PMSF, 2 mg/aprotinin, 2 mg/leupeptin in phosphate-buffered saline) and were homogenized completely. Samples were microfuged at top speed at 4°C and the resulting supernatant was transferred to a fresh tube containing 30 μl of 2 × SDS sample buffer. The samples were boiled for 5 min and 1 fly equivalent per sample (11 μl) was run on a 9% SDS-PAGE gel with a 4.5% stacker. BI-ORAD Kaleidoscope prestained standard (30 μl) was used as a molecular weight marker, and in vitro transcribed/translated [35S]methionine-labeled eIF-2α (TNT Coupled Reticulocyte Lysate System, Promega) protein was used as a comigration control.
The gel and the nitrocellulose membrane were each incubated in transfer buffer (48 mM Tris, 39 mM glycine, 1.3 mM SDS, 20% methanol, pH 9.2) for 20 min at room temperature. Proteins were electrophoretically transferred to nitrocellulose at 15 V, 300 mA for 45 min in transfer buffer. Following transfer to nitrocellulose, the membrane was blocked in 10% milk in TBS-T (20 mM Tris base, 137 mM NaCl, 0.05% Tween-20, pH 7.6) for either 1 h at room temperature or at 4°C overnight. The membrane was then washed three times in TBS-T. The membrane was incubated for 1 h at room temperature with the primary antibody, α-Drosophila eIF-2α, in a 1:1000 dilution in TBS-T, followed by three washes in TBS-T. The secondary antibody, α-rabbit Ig-horseradish peroxidase linked (Amersham), was incubated with the membrane for 15 min at room temperature in a 1:5000 dilution in TBS-T. The membrane was washed three times in TBS-T (15, 10, and 5 min) followed by ECL detection (Amersham) according to the manufacturer’s protocol. At least two exposures were obtained for each nitrocellulose membrane to obtain the best film linearity. Autoradiograms were scanned and relative quantities of each sample were estimated.
Global Protein Synthesis
Five Drosophila third instar larvae per genotype were subjected to two cycles of a 1-h heat shock at 37°C and 1-h recovery at 25°C (heat shock larvae) whereas five other larvae were cultured continuously at 25 °C (non-heat shock larvae). Five larvae per samples were dissected along the anterior-posterior axis in 0.1 mM PMSF/PBS to expose the internal tissues. Dissected larvae were incubated for 3 h at 25°C in 10 μl of DMEM culture media lacking methionine and cysteine and 15 mCi of translation grade [35S]methionine. After the labeling period the larvae were washed three times in 100 μl of 0.1 mM PMSF/PBS. Larvae were homogenized in 50 μl 0.1 mM PMSF/PBS. The homogenates were centrifuged for 5 min at 25°C. Two replicate samples of 5 μl each of the larval supernatants were subjected to TCA precipitation followed by a determination of incorporated [35S]methionine into protein by scintillation counting. Total soluble protein of the larval supernatants was determined by the Bradford assay. The average value of the replicate TCA precipitation samples was divided by the average value of replicate samples from the Bradford assay (counts incorporated/total soluble protein). For each transgenic line tested, the value of the heat shock sample was divided by the non-heat shock value, producing a ratio indicative of protein synthesis rate following heat shock treatment. The average ratios from individual transgenic lines were then averaged together with ratios from other transgenic lines containing the same amino acid at the Ser50 position.
RESULTS
Isolation and Expression of Transgenic Strains of Drosophila Expressing Mutant and Wild-Type eIF-2α
Drosophila eIF-2α Ser50 was mutated to alanine (Ala50) or aspartic acid (Asp50). The alanine mutant is expected to block phosphorylation whereas the aspartic acid mutation is expected to partially mimic phosphorylation due to its negative charge. The Ala50 and Asp50 mutants and the wild-type eIF-2α genes were inserted into a P-element transformation vector under the transcriptional control of one of three different heterologous Drosophila promoters: actin 5C promoter, α-1 tubulin promoter, and the hsp70 heat shock promoter. The actin 5C and α-l tubulin promoters are expected to provide high constitutive expression, whereas the hsp70 provides low basal expression but can be induced by heat. Transgenic strains for all but two of the nine constructs were obtained. Several attempts to obtain transgenic strains of the Asp50 mutant genes under the control of either of the two constitutive promoters failed. We speculate that high constitutive expression of the Asp50 mutant gene results in lethality. All experiments described below were performed using the eIF-2α transgenes under hsp70 heat shock transcriptional control and are denoted HS (hsp70-Ser50), HA (hsp70-Ala50), and HD (hsp70-Asp50). Initially, seven HA lines, eight HD lines, and nine HS lines were isolated and mapped to one of the three major chromosomes.
To determine which of the strains were successfully expressing the transgene after a 1-h heat shock, RNase protection experiments were performed on RNA isolated from adult flies from all of the isolated HS, HA, and HD eIF-2α transgenic strains (Fig. 1). The RNase protection probe used distinguishes between the endogenous eIF-2α mRNA and the eIF-2α transgenic mRNA based upon differences in the 5′ UTR. Several independent transgenic strains for each of the three trans-genes exhibited a high level of transgenic eIF-2α mRNA upon heat shock induction. These strains were used in subsequent experiments.
FIG. 1.
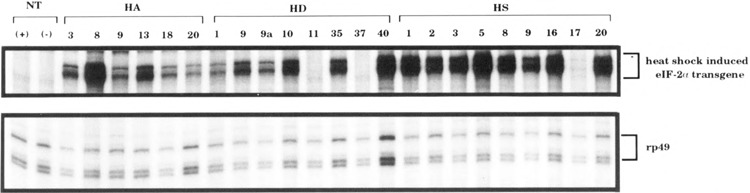
Expression of eIF-2α mRNA in eIF-2α transgenic adult flies assayed by ribonuclease protection assay (RPA). Total RNA was isolated from heat shock-treated animals; 10 mg total RNA was used for each RPA reaction. NT: nontransgenic control flies either heat shocked (+) or not heat shocked (–). HA: hsp70/Ser50Ala eIF-2α transgene. HD: hsp70/Ser50Asp eIF-2α transgene. HS: hsp70/Ser50 eIF-2α transgene. The number below each transgene represents the strain designation. The intense upper bands in the upper row corresponds to eIF-2α transgene mRNA. The reason for the presence of doublet bands is unknown but most likely reflects incomplete RNAse digestion very near the termini.
Polyclonal antisera was reared against a GST-eIF-2α fusion protein containing the carboxyl-terminal 95 amino acid residues of Drosophila eIF-2α. Western blot and immunoprecipitation experiments using the Drosophila eIF-2α antisera detects in vitro synthesized eIF-2α and eIF-2α from crude extracts of Drosophila tissues (Fig. 2A, B). In vitro and in vivo synthesized eIF-2α comigrate with an apparent molecular weight of approximately 40 kDa, consistent with the expected size based upon its inferred amino acid sequence (30).
FIG. 2.
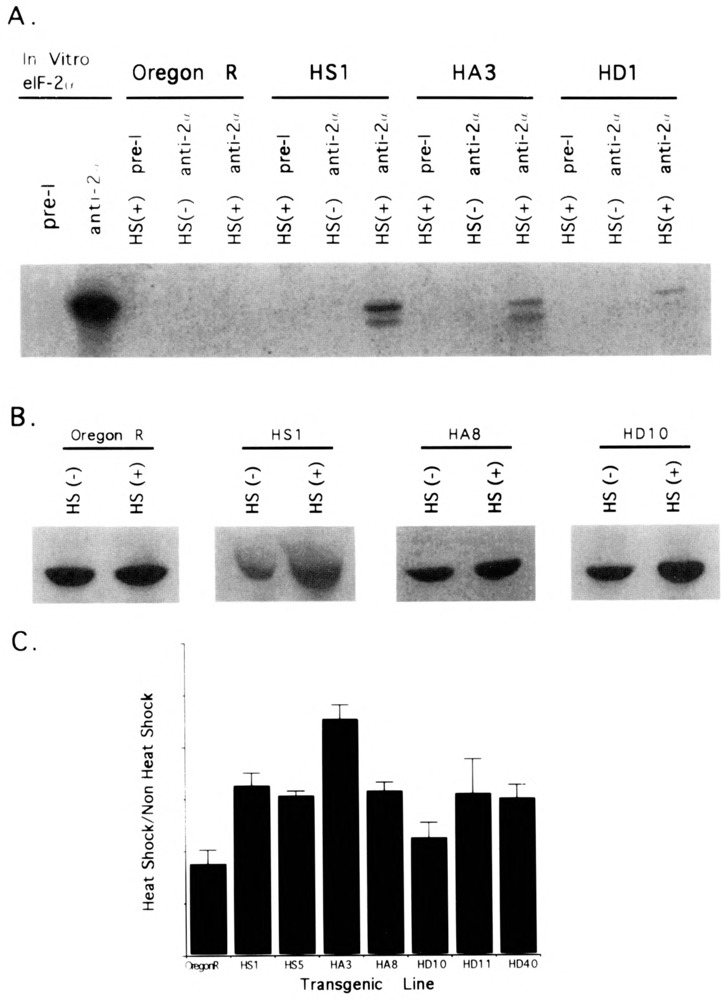
Expression of eIF-2α protein in heat-shocked and non-heat-shocked eIF-2α transgenic strains. HS (+): heat shocked for 1 h at 37°C; HS (–): non-heat shock control. (A) Immunoprecipitation of eIF-2α from in vitro synthesis of eIF-2α or from homogenates of wild-type (Oregon-R) larvae and eIF-2α transgenic larvae pulse labeled with 35S Met-Cys subjected to heat shock or non-heat shock treatment. Pre-I: preimmune serum. Anti-2α: eIF-2α polyclonal antisera. Two bands were consistently observed using anti-eIF-2α. The relationship of these two bands is currently unknown but may correspond to different phosphorylated forms of eIF-2α. Both bands are induced in the transgenic strains and both are observed after a longer exposure (not shown) in non-transgenic flies. Although this particular experiment seemed to indicate quantitative differences in eIF-2α protein among the three transgenic strains, subsequent experiments did not reveal consistent differences among them (not shown). (B) eIF-2α immuno-Western blots of total elF-2α present in homogenates of wild-type and eIF-2α transgenic adult flies subjected to a 1-h heat shock or not heat shocked. Although in this particular experiment only a single eIF-2α band is observed, adult tissues sometimes reveal doublet bands similar to larval tissues as seen in (A). (C) Quantitative estimates of the ratio of eIF-2α in homogenates of adult flies heat shocked vs. flies not heat shocked from densitometric scans of immuno-Western blots. Each bar represents the average ratio of two to four replicates.
Although the eIF-2α antisera cannot distinguish between endogenous and transgenic eIF-2α, we predicted that heat induction of the transgenic strains would elevate the level of eIF-2α in comparison with control, nontransgenic and non-heat-induced transgenic flies. Immunoprecipitation experiments of pulse-labeled third instar larvae (Fig. 2A) and adult flies (not shown) showed that the level of eIF-2α is elevated in the heat-shocked eIF-2α transgenic strains but not in the nontransgenic Oregon-R heat-shocked flies. We therefore posit that most of the newly synthesized eIF-2α in the heat-shocked transgenic strains is derived from the eIF-2α transgenes. Western blot analysis indicated that heat induction of the eIF-2α HS, HA, and HD transgenes increases the overall amount of eIF-2α by approximately 50% (Fig. 2B). Although significant differences were observed between individual lines, the average level of eIF-2α among the HS, HA, and HD strains was not significantly different (Fig. 2C). The nontransgenic control strain showed a slight decrease in eIF-2α protein consistent with the global repression of protein synthesis occurring as part of the normal heat shock response.
The eIF-2α HA and HD Transgenes Affect Developmental Rate and Body Weight
Developmental rate, body weight, and protein synthesis in Drosophila are highly interrelated (8). We speculated that expressing eIF-2α HD and HA transgenes may result in dominant effects on protein synthesis and ultimately lead to changes in developmental rate and body size. Nine HS, seven HA, and seven HD independent transgenic strains were crossed to a nontransgenic stock. The resultant transgenic/nontransgenic hemizygotes were then backcrossed singly to the nontransgenic stock. This genetic scheme should result in the generation of equal numbers of siblings bearing one copy of the transgene (as indicated by red-eyed flies) and no copies of the transgene (as indicated by white-eyed flies). These siblings, thus, would be reared in the same environment. Eggs were collected for a 24-h period for each cross. It is important to note that the total period of development will range between 10 and 20 days for eggs of nearly identical age. Developing animals were subjected to a single 1-h 37°C heat shock treatment at a specific time during development to express the eIF-2α transgene transiently. The heat shock was administered at a different time during development in separate experiments to determine the phenocritical period for the effects observed. Because the genetic scheme should result in a simple 1:1 ratio of transgenic to nontransgenic flies, the null hypothesis is that they will emerge from the puparium case in equal numbers each day over the 10-20-day period of development. Initially, eight developmental rate experiments were performed. Seven of the experiments entailed administering heat shock at different times during development and in one experiment no heat shock was done. A total of 4562 progeny were scored for the initial HD experiments, 4543 progeny scored for the HA experiments, and 5052 progeny scored for the HS experiments. The numbers of transgenic vs. nontransgenic progeny emerging each day (days 11–20) within each eIF-2α mutant were statistically homogeneous and were therefore pooled to simplify the presentation of the data. In addition, within each transgenic genotype, the data across all heat shock experiments were determined to be statistically homogeneous and therefore they were pooled for statistical analysis to test goodness of fit to the expected 1:1 genetic ratio using a chi-square test (Table 1). From the daily counts of transgenic and nontransgenic flies, the average developmental rates for each of the individual heat shock and non-heat shock experiments were determined and are presented in Table 2.
TABLE 1.
NUMBER OF eIF-2α TRANSGENIC AND NONTRANSGENIC SIBLINGS COMPLETING DEVELOPMENT EACH DAY
| Transgene | Sibling | T/S Ratio | Exp. | Chi-Square | ||
|---|---|---|---|---|---|---|
| HD | Day 11 | 273 | 539 | 0.5 | 406 | 87.1* |
| Day 12 | 614 | 752 | 0.8 | 683 | 13.9* | |
| Day 13 | 525 | 404 | 1.3 | 465 | 15.8* | |
| Day 14 | 384 | 216 | 1.8 | 300 | 47.0* | |
| Day 15 | 342 | 123 | 2.8 | 233 | 103.1* | |
| Day 16 | 178 | 56 | 3.2 | 117 | 63.6* | |
| Day 17 | 93 | 11 | 8.5 | 52 | 64.7* | |
| Day 18–20 | 43 | 3 | 14.3 | 23 | 34.8* | |
| HA | Day 11 | 418 | 347 | 1.2 | 383 | 6.6† |
| Day 12 | 783 | 749 | 1.0 | 766 | 0.8 | |
| Day 13 | 524 | 543 | 1.0 | 534 | 0.3 | |
| Day 14 | 283 | 321 | 0.9 | 302 | 2.4 | |
| Day 15 | 138 | 161 | 0.9 | 150 | 1.8 | |
| Day 16 | 62 | 108 | 0.6 | 85 | 12.4* | |
| Day 17–20 | 51 | 54 | 0.9 | 53 | 0.1 | |
| HS | Day 11 | 557 | 451 | 1.2 | 504 | 11.1* |
| Day 12 | 984 | 1008 | 1.0 | 996 | 0.3 | |
| Day 13 | 578 | 524 | 1.1 | 551 | 2.6 | |
| Day 14 | 277 | 246 | 1.1 | 262 | 1.8 | |
| Day 15 | 125 | 138 | 0.9 | 132 | 0.6 | |
| Day 16 | 48 | 55 | 0.9 | 52 | 0.5 | |
| Day 17–20 | 32 | 34 | 0.9 | 33 | 0.1 | |
| HD paternal | Day 11 | 87 | 213 | 0.4 | 150 | 52.9* |
| Day 12 | 170 | 297 | 0.6 | 234 | 34.5* | |
| Day 13 | 209 | 222 | 0.9 | 216 | 0.4 | |
| Day 14 | 131 | 51 | 2.6 | 91 | 35.2* | |
| Day 15 | 140 | 18 | 7.8 | 79 | 94.2* | |
| Day 16 | 39 | 0 | 20 | 39.0* | ||
| Day 17–20 | 6 | 0 | 3 | 6.0‡ | ||
| HD maternal | Day 11 | 44 | 255 | 0.2 | 150 | 148.9* |
| Day 12 | 160 | 405 | 0.4 | 283 | 106.2* | |
| Day 13 | 237 | 291 | 0.8 | 264 | 5.5‡ | |
| Day 14 | 202 | 112 | 1.8 | 157 | 25.8* | |
| Day 15 | 168 | 38 | 4.4 | 103 | 82.0* | |
| Day 16 | 109 | 4 | 27.3 | 57 | 97.6* | |
| Day 17–20 | 112 | 1 | 112.0 | 57 | 109.0* |
The expected ratio of transgenic to nontransgenic sibs is 1.0 for all days. Chi-square test for the goodness-of-fit of the observed numbers to the expected numbers was performed.
Significance levels: *0.001, †0.0l, ‡0.05.
TABLE 2.
TOTAL TIME OF DEVELOPMENT OF HD, HA, AND HS eIF-2α TRANSGENIC DROSOPHILA
| Stage of Heat Shock | HD | Sib | Δ | HA | Sib | Δ | HS | Sib | Δ |
|---|---|---|---|---|---|---|---|---|---|
| No heat shock | 13.09 | 12.46 | 0.63 | 12.97 | 13.12 | –0.15 | 12.86 | 12.86 | 0.00 |
| Embryo | 13.68 | 12.28 | 1.40 | 12.86 | 13.26 | –0.40 | 12.62 | 12.65 | –0.03 |
| First instar | 14.90 | 12.30 | 2.60 | 12.27 | 12.48 | –0.21 | 12.40 | 12.71 | –0.31 |
| Second instar | 14.62 | 12.40 | 2.22 | 12.97 | 13.47 | –0.50 | 12.70 | 12.65 | 0.05 |
| Third instar, early | 12.89 | 12.00 | 0.89 | 12.35 | 12.41 | –0.06 | 12.05 | 12.14 | –0.09 |
| Third instar, late | 12.54 | 12.07 | 0.47 | 12.34 | 12.25 | –0.09 | 11.99 | 11.90 | 0.09 |
| Pupa, early | 13.09 | 12.87 | 0.22 | 12.81 | 13.02 | –0.21 | 12.37 | 12.34 | 0.03 |
| Pupa, middle | 13.08 | 12.59 | 0.49 | 12.62 | 12.74 | –0.12 | 12.66 | 12.91 | –0.25 |
| Pupa, late | 13.52 | 12.99 | 0.53 | 13.17 | 13.33 | –0.16 | 12.78 | 12.86 | –0.08 |
| Heat shock ave. | 13.54 | 12.44 | 1.10 | 12.67 | 12.87 | –0.20 | 12.45 | 12.52 | –0.07 |
The HD transgenic flies took a significantly longer time to develop than their nontransgenic siblings in both the heat shock (Tables 1 and 2) and in the non-heat shock experiments (Table 2). During the first 2 days of the emergence period (days 11 and 12) a significantly smaller number of HD flies emerged compared to their nontransgenic sibs (Table 1). This trend is reversed in days 13–20 where a significantly larger number of HD flies emerged. Deviation from the expected 1:1 ratio are highly significant for all days of emergence. Although developmental rates differed between HD and their nontransgenic siblings, approximately equal numbers of each genotype completed development. The most pronounced developmental rate repression of the HD transgenic flies occurred when a heat shock treatment was administered early in larval development, prolonging development by an average of 2 days (Table 2).
In the absence of heat shock, the HD transgenic flies developed more slowly than their nontransgenic sibs, suggesting that the basal activity of the transgene’s hsp70 promoter was sufficient to retard developmental rate (Table 2). Thus, preloading eggs with a low level of the HD mutant protein, such as that provided by basal hsp70-driven expression, may magnify the developmental rate repression seen with subsequent heat shock induction of the transgene. In our original experiments we had established a nearly equal mixture of crosses using either male or female transgenic parents, and we pooled data across numerous single pair matings. That design would mask any maternal effect of preloading eggs with HD protein. We therefore decided to determine if the sex of the transgenic parent influences the degree of heat shock-induced HD-mediated repression. A second series of experiments was performed where a single heat shock was administered to embryos, first instar larvae, second instar larvae, or third instar larvae. We found a strong maternal effect of HD mothers in enhancing developmental rate repression compared with flies having HD fathers (Tables 1 and 3). A single heat shock administered at any of the four developmental stages prolonged the development of HD as much as an extra day when the HD transgene was maternally inherited compared to when it was paternally inherited.
TABLE 3.
MATERNAL EFFECT OF HD TRANSGENE ON TOTAL TIME OF DEVELOPMENT
| Stage of Heat Shock | HD Maternal | HD Paternal | Maternal vs. Paternal mHΔ-pHΔ | ||||
|---|---|---|---|---|---|---|---|
| HD | Sib | Δ | HD | Sib | Δ | ||
| Embryo | 14.05 | 12.51 | 1.54 | 13.28 | 12.27 | 1.01 | 0.77 |
| First instar | 15.32 | 12.30 | 3.02 | 14.28 | 12.11 | 2.18 | 1.04 |
| Second instar | 14.73 | 12.15 | 2.58 | 13.90 | 11.97 | 1.92 | 0.83 |
| Third instar, early | 14.15 | 12.27 | 1.87 | 13.31 | 12.09 | 1.22 | 0.84 |
| Average | 14.56 | 12.31 | 2.25 | 13.69 | 12.11 | 1.58 | 0.87 |
| SE | 0.29 | 0.08 | 0.24 | 0.06 | |||
| Paired t-test | 8.7 | 7.4 | 19.1 | ||||
| Significance level | 0.0005 < p < 0.005 | 0.0005 < p < 0.005 | p < 0.0005 | ||||
In contrast to the developmental rate repression of the HD transgenic strains, the development of the HA transgenic flies was more rapid than their wild-type siblings (Tables 1 and 2). For all 10 non-heat shock and heat shock experiments, the HA transgenic flies exhibited a more rapid developmental rate (Table 2). The difference in developmental rate is relatively small, but averaged over all experiments the difference (0.2 day) is significantly different (p < 0.025). Two other statistical tests confirm this interpretation: a) a nonparametric sign test indicates that the sign (+) of the difference in developmental rates (10 out of 10 experiments) deviates significantly from random expectation (5 out of 10 experiments), and b) chi-square analysis of data pooled for days 11 and 12 vs. days 13–20 indicates a significant difference (not shown). As was observed for the HD transgenic strains, induction of the HA transgene was most effective during early larval development. Induction of the HS wild-type control transgene did not affect developmental rate compared to nontransgenic siblings as there was no significant difference (p > 0.05) in developmental rate when averaged over all heat shock experiments. Also, a nonparametric sign test of developmental rate differences and chi-square test of pooled data (days 11–12 vs. days 13–20) did not reveal significant differences between HS transgenic and nontransgenic siblings.
Newly developed adult flies that had been previously heat shocked during development were sexed, counted, and weighed in mass (Table 4). Adult HD flies of both sexes weighed significantly less than their corresponding nontransgenic siblings. In contrast, HA females weighed significantly more than their nontransgenic sisters but the HA male body weight was approximately the same as their nontransgenic brothers. No significant differences in body weight were observed between the control HS transgenics and their nontransgenic siblings.
TABLE 4.
BODY WEIGHT OF HA, HD, AND HS eIF-2α TRANSGENIC ADULT DROSOPHILA
| Total Number of Flies | Total Weight (mg) | Average Weight of Single Fly (mg) | % Difference HX vs. Sib | Student’s t-Test | |
|---|---|---|---|---|---|
| HA female | 275 | 305.6 | 1.1112 | ||
| Sib female | 285 | 302.3 | 1.0607 | 4.54% | 7.5 |
| 0.0005 < p < 0.005 | |||||
| HA male | 256 | 205.8 | 0.8039 | ||
| Sib male | 245 | 195.7 | 0.7989 | 0.62% | 0.4 |
| 0.1 < p < 0.375 | |||||
| HD female | 152 | 158.8 | 1.0447 | ||
| Sib female | 192 | 212.2 | 1.1052 | –5.79% | –5.2 |
| 0.0005 < p < 0.005 | |||||
| HD male | 135 | 102.8 | 0.7614 | ||
| Sib male | 198 | 165.8 | 0.8373 | –9.97% | –5.8 |
| 0.0005 < p < 0.005 | |||||
| HS female | 259 | 280.3 | 1.0822 | ||
| Sib female | 244 | 261.8 | 1.0729 | 0.86% | 0.4 |
| 0.1 < p < 0.375 | |||||
| HS male | 252 | 199.8 | 0.7928 | ||
| Sib male | 219 | 176.9 | 0.8077 | –1.88% | –1.1 |
| 0.1 < p < 0.375 |
Repeated Heat Shock Treatments Completely Arrest Development of HD Transgenic Flies
Our inability to isolate transgenic strains bearing the Asp50-eIF-2α mutation controlled by the strong constitutive α-1 tubulin or actin 5C promoter suggested that high constitutive expression of the Asp50 mutant led to dominant lethality. To test this hypothesis, the HD transgenic strains were heat shocked for 2 h every day during development to elevate the expression of the HD mutant eIF-2α. The development and viability of the HA and HS transgenic strains was largely unaffected by the repeated heat shock inductions. In contrast, the development of the three independent HD transgenic lines tested were completely arrested. The development of almost all of the individuals was arrested at the second larval in-star. Some larvae progressed to the third larval instar stage where most died, but a few individuals continued to develop to the metamorphic stage. However, none of them developed to mature adults.
Global Protein Synthesis Rates in eIF-2α HA and HD Transgenic Strains Exhibit Small Differences
The pronounced developmental rate and viability differences observed between the HD and HA transgenic strains may have resulted from global changes in protein synthesis. To examine this possibility, transgenic flies were heat shocked, allowed to recover from the heat shock, dissected, and their proteins radiolabeled. TCA-precipitated radiolabeled proteins from whole fly homogenates from several replicate experiments were prepared and quantified. Pilot experiments were performed to maximize the induction of the transgenes while allowing for adequate recovery from heat shock. In general, global protein synthesis was not affected significantly by inducing the transgenes, nor were any significant differences observed among the HS, HD, and HA lines (Table 5). Although not statistically significant, the different transgenic strains exhibited the expected trend: the ratio of heat shock-induced to non-heat shock-induced level of global protein synthesis in the HA flies was higher than that observed for the HD flies. These small differences accumulated over several days of development may be sufficient to account for the developmental rate and body weight differences shown between the HD and HA transgenic strains.
TABLE 5.
RATIO OF TOTAL PROTEIN SYNTHESIS OF HEAT-SHOCKED VERSUS NON-HEAT-SHOCKED eIF-2α TRANSGENIC FLIES
| Transgenic Line | Number of Samples | Heat Shock/Non-Heat Shock | SE |
|---|---|---|---|
| HS | 4 | 1.14 | 0.290 |
| HA | 13 | 1.49 | 0.372 |
| HD | 10 | 0.96 | 0.081 |
We attempted to measure protein synthesis in transgenic larvae subjected to repeated heat shocks (not shown). However, the HD larvae stop feeding and become lethargic, thus invalidating any biochemical comparisons with the HA and HS larvae, which appear to be unaffected by the heat shock.
DISCUSSION
We propose that the eIF-2α encoded by the mutant transgenes replaces much of the wild-type eIF-2α in the holo-eIF-2 trimer, as was shown in similar experiments where eIF-2α mutant genes were transfected into mammalian cells (6), and that the observed developmental effects are due to alterations in the recycling eIF-2-GDP. In the case of the HD eIF-2α transgene, the aspartic acid substitution is predicted to mimic constitutive phosphorylation. In both yeast and mammalian cells, the homologous serine to aspartic acid substitution partially represses eIF-2-GDP/eIF-2-GTP recycling (12,31). Reducing the concentration of eIF-2-GTP in yeast results in the failure of ribosomes to initiate at specific ORFs present in the 5′ UTR of GCN4 mRNA, which would otherwise abrogate translation initiation at the GCN4 start codon. Although replacement of the endogenous eIF-2α gene in yeast with the Ser51 substituted with either aspartic acid or alanine affects the translation of GCN4, viability, growth rates, and global protein synthesis are not affected significantly (12). In contrast, transfection of mammalian cells with eIF-2α containing the homologous serine to aspartic acid substitution results in a dramatic reduction in translation of mRNAs encoded by transfected genes and ultimately to cell death. However, it is unclear as to whether global protein synthesis is affected (6,11,19,26,28,31). Thus, the dominant effects of expressing the Ser50Asp eIF-2α mutant transgene in Drosophila are similar in nature to that seen in mammalian cells. In addition to affecting viability, we observed that these mutations could alter developmental rate and body size of the whole organism. The expression of the Ser50Asp in mammalian cells and yeast cells has not been reported to alter developmental (cell cycle) rates; however, constitutive phosphorylation of eIF-2α in yeast retards growth rates (12).
We examined protein synthesis over a brief period of time (via a pulse labeling experiment) and found small but statistically insignificant differences between the HD and HA transgenic strains.
The significant body weight and developmental rate differences between these transgenic strains may be due to accumulative differences in protein synthesis over the entire developmental period. Developmental rate may be controlled translationally through regulation of global protein synthesis or through the regulation of specific mRNAs. The control of global protein synthesis could regulate developmental progression if the organism’s growth is dependent upon a threshold level of total protein. Alternatively, developmental progression could also be controlled by regulatory proteins that are under specific translational control. In a variety of studies, phosphorylation of eIF-2α has been shown to impact global protein synthesis and/or translation of specific mRNAs (18). The mechanism whereby phosphorylation of eIF-2α controls mRNA-specific translation is unknown except for GCN4 mRNA, where the concentration of the eIF-2-GTP determines the efficiency of termination-reinitiation.
The dominant negative repression of developmental rate exhibited by the HD transgenic strains does not necessarily mean that phosphorylation of Ser50 of eIF-2α plays a role in development. First, although we and others have shown that Drosophila eIF-2α is phosphorylated, the phosphorylated residue(s) has not been mapped. Secondly, the effects of dominant negative mutations do not reliably predict normal gene functions. The accelerated development of the HA transgenic flies is perhaps more suggestive that phosphorylation of eIF-2α has a developmental function, because the alanine substitution is presumably blocking phosphorylation, implying that the acceleration in development is the result of the inability to phosphorylate Ser51. Alternatively, the effects of the alanine and asparatic Ser50 eIF-2α mutations may due to other alterations in eIF-2α activity that are completely unrelated to phosphorylation at this site. We think that the former interpretation is much more likely because of the high degree of conservation of this region in eIF-2α and because the results obtained in this study are highly consistent with similar experiments in yeast and mammalian cells.
Controlling developmental rate could be a primary function of eIF-2α phosphorylation, or, more likely, the effect on developmental rate may be a secondary effect of some other developmental process that is regulated by eIF-2α phosphorylation. To address the role of eIF-2α phosphorylation during development directly, it will be necessary to examine the Ser50 mutations in Drosophila strains in which the endogenous wild-type eIF-2α is not present. Analogous experiments have been
performed in yeast (12), and it was shown that yeast development does not require the ability to phosphorylate eIF-2α at Ser51. However, the control of multicellular development of higher eukaryotes may require this form of translational regulation (2,20,25). Experiments are underway in our laboratory to isolate null eIF-2α mutants in Drosophila and identify eIF-2α kinases, which will allow us to examine critically the potential developmental role of eIF-2α phosphorylation. Recently, we have discovered a Drosophila homologue of yeast GCN2 (eIF-2α kinase) that appears to be expressed throughout development, suggesting that it may play a role in development beyond its role in the amino acid deprivation response seen in yeast.
ACKNOWLEDGEMENTS
We thank Xin Ye and DeAnne Olsen for valuable comments and criticism of this work. This work was supported by NSF grant MCB-9304983 to D.R.C.
REFERENCES
- 1. Barber G. N.; Jagus R.; Meurs E. F.; Hovanessian A. G.; Katze M. G. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J. Biol. Chem. 270:17423–17428; 1995. [DOI] [PubMed] [Google Scholar]
- 2. Cavener D. R.; Cavener B. A. Translation start sites and mRNA leaders. In: Maroni G., ed. An atlas of Drosophila genes. New York: Oxford University Press; 1993. [Google Scholar]
- 3. Cavener D.; Corbett G.; Cox D.; Whetten R. Isolation of the eclosion gene cluster and the developmental expression of the Gld gene in Drosophila melanogaster . EMBO J. 5:2939–2948; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chefalo P. J.; Yang J. M.; Ramaiah K. V.; Gehrke L.; Chen J. J. Inhibition of protein synthesis in insect cells by baculovirus-expressed heme-regulated eIF-2 alpha kinase. J. Biol. Chem. 269:25788–25794; 1994. [PubMed] [Google Scholar]
- 5. Chen J. J.; London I. M. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem. Sci. 20:105–108; 1995. [DOI] [PubMed] [Google Scholar]
- 6. Choi S. Y.; Scherer B. J.; Schnier J.; Davies M. V.; Kaufman R. J.; Hershey J. W. Stimulation of protein synthesis in COS cells transfected with variants of the alpha-subunit of initiation factor eIF-2. J. Biol. Chem. 267:286–293; 1992. [PubMed] [Google Scholar]
- 7. Chung Y.; Keller E. B. Positive and negative regulatory elements mediating transcription from the Drosophila melanogaster actin 5C distal promoter. Mol. Cell. Biol. 10:6172–6180; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Church R. B.; Robertson F. W. Biochemical analysis of genetic differences in the growth of Drosophila . Genet. Res. 7:383–407; 1966. [DOI] [PubMed] [Google Scholar]
- 9. Cigan A. M.; Pabich A. M.; Feng L.; Donahue T. F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc. Natl. Acad. Sci. USA 86:2784–2788; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colthurst D. R.; Campbell D. G.; Proud C. G. Structure and regulation of eukaryotic initiation factor eIF-2: Sequence of the site in the a subunit phosphorylated by the haem-controlled repressor and by the double-stranded RNA-activated inhibitor. Eur. J. Biochem. 166:357–363; 1987. [DOI] [PubMed] [Google Scholar]
- 11. Davies M. V.; Furtado M.; Hershey J. W.; Thimmappaya B.; Kaufman R. J. Complementation of adenovirus virus-associated RNA I gene deletion by expression of a mutant eukaryotic translation initiation factor. Proc. Natl. Acad. Sci. USA 86:9163–9167; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dever T. E.; Feng L.; Wek R. C.; Cigan A. M.; Donahue T. F.; Hinnebusch A. G. Phosphorylation of initiation factor 2a by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585–596; 1992. [DOI] [PubMed] [Google Scholar]
- 13. Dever T. E.; Chen J. J.; Barber G. N.; Cigan A. M.; Feng L.; Donahue T. F.; London I. M.; Katze M. G.; Hinnebusch A. G. Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc. Natl. Acad. Sci. USA 90:4616–4620; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donahue T. F.; Cigan A. M. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol. Cell. Biol. 8:2955–2963; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donahue T. F.; Cigan A. M.; Pabich E. K.; Valavicius B. C. Mutations at a Zn(II) finger motif in the yeast eIF-20 gene alter ribosomal start-site selection during the scanning process. Cell 54:621–632; 1988. [DOI] [PubMed] [Google Scholar]
- 16. Duncan R. F.; Cavener D. R.; Qu S. Heat shock effects on phosphorylation of protein synthesis initiation factor proteins eIF-4E and eIF-2 alpha in Drosophila . Biochemistry 34:2985–2997; 1995. [DOI] [PubMed] [Google Scholar]
- 17. Hershey J. W. Protein phosphorylation controls translation rates. J. Biol. Chem. 264:20823–20826; 1989. [PubMed] [Google Scholar]
- 18. Hershey J. W. B. Translational control in mammalian cells. Annu. Rev. Biochem. 60:717–756; 1991. [DOI] [PubMed] [Google Scholar]
- 19. Kaufman R. J.; Davies M. V.; Pathak V. K.; Hershey J. W. The phosphorylation state of eukaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol. Cell. Biol. 9:946–958; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kozak M. An analysis of vertebrate mRNA sequences: Intimations of translational control. J. Cell Biol. 115:887–903; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488–492; 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lis J. T.; Simon J. A.; Suttun C. A. New heat shock puffs and β-galactosidase activity resulting from transformation of Drosophila with an hsp70-lacZ hybrid gene. Cell 35:403–410; 1983. [DOI] [PubMed] [Google Scholar]
- 23. Mateu M. G.; Sierra J. M. Protein synthesis in Drosophila melanogaster embryos. Two mechanisms for guanine nucleotide exchange on eukaryotic initiation factor 2. Eur. J. Biochem. 165:507–513; 1987. [DOI] [PubMed] [Google Scholar]
- 24. Merrick W. C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 56:291–315; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mitchell H. K.; Petersen N. S. Rapid changes in gene expression in differentiating tissues of Drosophila . Dev. Biol. 85:233–242; 1981. [DOI] [PubMed] [Google Scholar]
- 26. Murtha-Riel P.; Davies M. V.; Scherer B. J.; Choi S. Y.; Hershey J. W.; Kaufman R. J. Expression of a phosphorylation-resistant eukaryotic initiation factor 2 alpha-subunit mitigates heat shock inhibition of protein synthesis. J. Biol. Chem. 268:12946–51; 1993. [PubMed] [Google Scholar]
- 27. Pain V. M. Initiation of protein synthesis in mammalian cells. Biochem. J. 235:625–637; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pathak V. K.; Schindler D.; Hershey J. W. Generation of a mutant form of protein synthesis initiation factor eIF-2 lacking the site of phosphorylation by eIF-2 kinases. Mol. Cell. Biol. 8:993–995; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petryshyn R.; Chen J.; London I. M. Growth-related expression of a double-stranded RNA-dependent protein kinase in 3T3 cells. J. Biol. Chem. 259:14736–14742; 1984. [PubMed] [Google Scholar]
- 30. Qu S.; Cavener D. R. Isolation and characterization of the Drosophila melanogaster eIF-2 alpha gene encoding the alpha subunit of translation initiation factor eIF-2. Gene 140:239–242; 1994. [DOI] [PubMed] [Google Scholar]
- 31. Ramaiah K. V.; Davies M. V.; Chen J. J.; Kaufman R. J. Expression of mutant eukaryotic initiation factor 2 alpha subunit (eIF-2 alpha) reduces inhibition of guanine nucleotide exchange activity of eIF-2B mediated by eIF-2 alpha phosphorylation. Mol. Cell. Biol. 14:4546–53; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robertson H. M.; Engels W. R. Modified P elements that mimic the P cytotype in Drosophila melanogaster . Genetics 123:815–824; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rowlands A. G.; Panniers R.; Henshaw E. C. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 263:5526–5533; 1988. [PubMed] [Google Scholar]
- 34. Szyszka R.; Kudlicki W.; Kramer G.; Hardesty B.; Galabru J.; Hovanessian A. A type 1 phosphoprotein phosphatase active with phosphorylated Mr = 68,000 initiation factor 2 kinase. J. Biol. Chem. 264:3827–3831; 1989. [PubMed] [Google Scholar]
- 35. Theurkauf W. E.; Baum H.; Bo J.; Wensink P. C. Tissue-specific and constitutive alpha-tubulin genes of Drosophila melanogaster code for structurally distinct proteins. Proc. Natl. Acad. Sci. USA 83:8417–8481; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thummel C. S.; Boulet A. M.; Lipshitz H. D. Vectors for Drosophila P element mediated transformation and tissue culture transfection. Gene 74:445–456; 1990. [DOI] [PubMed] [Google Scholar]
- 37. Wek R. C.; Cannon J. F.; Dever T. E.; Hinnebusch A. G. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2 alpha kinase GCN2. Mol. Cell. Biol. 12:5700–5710; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]


