Abstract
The upstream proximal region of the transthyretin (TTR) promoter and a distal enhancer are sufficient to drive liver-specific expression of the TTR gene, as demonstrated by experiments in transgenic mice. Previous analyses have characterized the binding of a number of liver-enriched transcription factors to the TTR promoter including hepatocyte nuclear factors one (HNF-1), HNF-4, and three distinct HNF-3 proteins (α, β, and γ), which are members of the winged helix (fork head) family. The TTR enhancer was shown to bind members of the CCAAT/enhancer binding protein (C/EBP) family at two distinct sites (TTR-2 and TTR-3), and an oligonucleotide containing the activation protein one (AP-1) binding sequence competed for recognition to a third enhancer site (TTR-1). In this study, we have carried out a detailed analysis of the transcription factors that recognize the TTR enhancer elements (TTR-1, TTR-2, and TTR-3 oligonucleotide sequences). Analysis of the TTR-1 site demonstrates that the putative AP-1 site in the TTR enhancer binds a ubiquitously expressed factor that is distinct from the AP-1 family of proteins. Next we demonstrate, via gel shift analysis, that the TTR-3 site is recognized by the C/EBP family in liver nuclear extracts. We also show that whereas the TTR-2 enhancer site is capable of binding recombinant C/EBP proteins, it does not bind C/EBP proteins from liver nuclear extracts. The TTR-2 site does, however, contain a variant HNF-3 recognition sequence that exclusively binds the HNF-3β isoform. Mutation of this HNF-3β-specific recognition sequence caused reductions in TTR enhancer activity. We had previously observed a 95% decrease in HNF-3β expression and a 20% reduction in HNF-3β expression in acute phase livers, which correlated with a 60% decrease in TTR gene transcription. We propose that the HNF-3β-specific binding site in the TTR enhancer may play a role in maintaining TTR gene expression during the acute phase response in spite of the dramatic reduction in HNF-3α protein levels.
Keywords: Hepatocyte nuclear factor 3, Fork head, Winged helix, Liver-enriched transcription factor, Hepatocyte-specific gene transcription, Isoform-specific DNA binding site
CELLULAR differentiation results in transcriptional induction of distinct sets of tissue-specific genes whose expression is vital for organ function (19). Deciphering transcriptional control mechanisms is thus critical to understanding the events involved in cellular differentiation. Toward this goal, functional dissection of numerous hepatocyte-specific promoter and enhancer regions has determined that such regulatory regions are structurally complex, consisting of multiple DNA binding sites recognized by distinct families of liver-enriched transcription factors (66). The combinatorial action of these factors on multiple DNA sites is required for the activation of transcription and plays a role in maintaining hepatocyte-specific gene expression (13). Structural similarities in the DNA binding and/or dimerization domains define five distinct families of liver-enriched factors: the winged helix/fork head family members hepatocyte nuclear factor (HNF)-3α, -3β and -3γ proteins (37,38); the steroid hormone receptor family members HNF-4 (59) and apolipoprotein AI regulatory protein-1 (ARP-1) (36); the POU-homeodomain family members HNF-1α and -1β proteins (8,23); the basic region leucine zipper (bZIP) family members CCAAT/enhancer binding protein α (C/EBPα) (39,40), C/EBPβ, and C/EBPδ (1,10,20,33,50,64); and the proline and acidic amino acid-rich (PAR) bZIP subfamily members albumin D-box binding protein (DBP) (45), chicken vitellogenin gene-binding protein (VBP)/mouse thyrotroph embryonic factor (TEF) (22,31), and hepatic leukemia factor (HLF) (28,29).
The HNF-3 proteins were originally identified as factors mediating hepatocyte-specific transcription of the transthyretin (TTR; encodes a serum carrier protein of thyroxine and vitamin A) and al-antitrypsin genes (15). The HNF-3 proteins share homology within a 100-amino acid winged helix DNA binding domain (12,38), which directs monomeric recognition of the DNA consensus sequence 5′-A(A/T)TRTT(G/T)RYTY-3′ (48). Subsequent studies showed that HNF-3 activates transcription of numerous liver genes encoding proteins important for liver function, including: the serum carrier proteins transferrin, albumin, a-fetoprotein, apolipoprotein B (ApoB), ApoAl, and insulin-like growth factor binding protein-1 (IGFBP-1) (6,9,15,21,26,41,60), and the enzymes tyrosine aminotransferase (TAT), cholesterol 7α -hydroxylase, phosphoenolpyruvate carboxykinase, phosphofructokinase, and aldolase B (30,43,46,54), and the human hepatitis B virus (11,47,53). In support of HNF-3’s role in regulating the expression of these hepatocyte-specific genes, a hepatoma cell line that expresses a dominant negative HNF-3 mutant extinguished transcription of several HNF-3 target genes (61). Furthermore, cytokines reduce hepatic HNF-3α and C/EBPα expression levels during the acute phase response, which correlates with a decrease in the expression of several of its target genes (52). The HNF-3 proteins also serve as accessory factors to the glucocorticoid receptor protein and provide hepatocyte-specific hormone induction of the TAT and IGFBP-1 genes (46,60). Furthermore, in vivo footprinting studies of the –10 kb albumin enhancer region have demonstrated that the HNF-3 proteins are involved in organizing the nucleosome pattern of the albumin enhancer exclusively in hepatocytes (42). The HNF-3 proteins thus not only play a role in transcriptional activation, but may also be involved in establishing hepatocyte-specific accessibility within these genes’ regulatory regions.
Mammalian HNF-3 (37,38) and the Drosophila homeotic gene fork head (fkh) (62) are prototypes of a family of transcription factors that share homology in their winged helix motif. To date, the mammalian winged helix protein family consists of over 30 distinct members, which are involved in the differentiation of diverse cellular lineages [for review see (13,27)]. In situ hybridization studies of stage-specific embryos demonstrate that the HNF-3α and HNF-3β genes also participate in pattern formation of the embryo. The HNF-3β genes begin expression during the primitive streak stage of the mouse embryo in the node that gives rise to endoderm, notochordal mesoderm, and neuroepithelium (4,44,55,57). HNF-3 expression is restricted to neuroepithelium in the floorplate of the neurotube, in the notochord, and throughout the gut endoderm. In support of the importance of HNF-3 expression in these developing structures, HNF-3β mutant embryos lack node and notochord, leading to defects in neurotube formation, somite organization, and gut endoderm invagination (3,63). Furthermore, ectopic hindbrain expression of the HNF-3β gene in trans-genic mice mediates the conversion of hindbrain to floorplate as evidenced by activation of the endogenous HNF-3α and HNF-3β genes as well as several additional floorplate marker genes (58). The HNF-3a and -3/3 genes are also induced during retinoic acid (RA)-mediated differentiation of embryonic carcinoma F9 cells to visceral endoderm (18,32). HNF-3 expression in differentiated F9 cells precedes activation of its target genes and results in decreased cellular proliferation. These embryonic studies suggest that the HNF-3 proteins play an important regulatory role in cellular commitment events.
We have used the TTR proximal promoter and distal enhancer regions (Fig. 1) as a model to understand hepatocyte-specific gene expression. These regulatory sequences are necessary and sufficient to elicit a normal hepatic expression pattern in transgenic mice (14,15,17,18,52,65). In TTR enhancer sequences required for transcriptional activation we previously identified two DNase I protected regions with recombinant C/EBPα protein (Fig. 1), designated TTR-2 and TTR-3 (16,17). We now demonstrate that liver-derived C/EBP proteins form specific complexes with the TTR-3 sequence only and not with TTR-2. We also identify a variant HNF-3 site in the TTR-2 sequence, which selectively binds the HNF-30 isoform, and show that this sequence is important for transcriptional activation. We suggest that the HNF-3β -selective site may be important for maintenance of TTR gene expression during the acute phase response when expression of HNF-3α and C/EBPα transcription factors drop precipitously (2,52). A third functional TTR enhancer site, TTR-1, was proposed to be a binding site for the AP-1 proteins on the basis of competition by an AP-1 binding site from the cd-antitrypsin (l-AT-B site) enhancer (17,25). In this study we demonstrate that the ubiquitously expressed factor binding to the TTR-1 sequence is not induced by TPA and is distinct from the AP-1 proteins, whereas the αl-AT-B sequence binds both the TTR ubiquitous factor and the AP-1 protein.
FIG. 1.
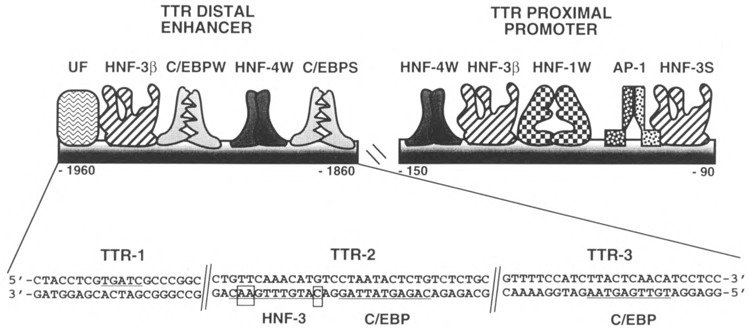
Transcription factors binding to the transthyretin (TTR) regulatory region. Schematically shown on the TTR promoter and enhancer region are the weak affinity protein binding sites for the hepatocyte nuclear factor-1 (HNF-1W), HNF-4W, activating protein-1 (AP-1) (15,52,59), and ubiquitous factor (UF; this study). There are weak and strong affinity sites for the CCAAT/enhancer binding protein (C/EBPW and C/EBPS) [(16,17) and this study] and two HNF-3β binding sites that are recognized exclusively by the HNF-30 isoform and a strong affinity HNF-3S site that binds the HNF-3α, HNF-3β, and HNF-γ proteins [(38) and this study]. The sequences of three oligonucleotides in the TTR enhancer are shown (TTR-1, TTR-2, and TTR-3) and indicated on the sequences are the positions of the C/EBP (16,17,25) and HNF-3 binding sites (boxed residues indicate mismatches with the HNF-3 consensus) (48). The TTR-1 and TTR-2 oligonucleotide sequences are separated by two nucleotides whereas the TTR-2 and TTR-3 oligonucleotide sequences are separated by 14 nucleotides that contain the HNF-4 binding site.
MATERIALS AND METHODS
Transfections to Generate Protein for Gel Shift Analysis
The cytomegalovirus (CMV) promoter-driven expression vectors containing the HNF-3α, HNF-3β, HNF-3γ, C/EBPα, or C/EBPβ cDNAs were transfected into human epithelial HeLa cells via calcium phosphate precipitation and nuclear extracts prepared 36 h later. Gel mobility shift assay was performed as described previously (52,56). A 100-fold molar excess of unlabeled oligonucleotide was included in the reaction when competition experiments were performed, unless otherwise indicated. Oligonucleotides used in gel mobility shift studies include the following: sites in the TTR enhancer comprising TTR-1, TTR-2, and TTR-3 [(17), Fig. 1], and the strong affinity HNF-3 (HNF-3S) site from the TTR promoter [–111/–88 base pair (bp)] (15), the HNF-3β promoter (–97/–67 bp) sequence containing both a weak affinity HNF-3 (HNF-3W) site and a strong affinity C/EBP site (49,56), a strong affinity C/EBP site derived from the hemopexin promoter (Hpx) (50), the αl-AT-B sequence containing an AP-1 consensus (5′-TKASTCA-3r where K = G or T and S = C or G) from the a1-antitrypsin enhancer (25), and the AP-1 site from the SV40 virus enhancer (5). Where indicated, HepG2 cells were treated with 160 nM of 12-O-tetradecanoyl-phorbol-13-acetate (TPA; dissolved in DMSO) or mock treated with DMSO in DMEM containing 0.5% fetal calf serum and nuclear extracts were prepared 4 h thereafter as previously described (52).
Generation and Analysis of Site-Directed Mutants via CAT Assay
Site-directed mutagenesis was performed via a PCR protocol previously described (52), with the following oligonucleotides (DNA International) and their opposite strand correlates: 5′ -TCG CCC GGC CCC TGG AAT TCT AGA TCC TAA TAC TCT-3′ for conversion of the HNF-3/3-specific site to a nonbinding site, and 5′-TCG CCC GGC CCC TAA ACA AAC ATT GCC TAA TAC TCT-3′ for conversion of the HNF-3/3-specific site to an HNF-β strong site. Human hepatoma HepG2 cells (34) were transfected with plasmid DNA using 10 μl of lipofectin as per manufacturer protocol (Bethesda Research lab) and then harvested 3 days later for preparation of cytoplasmic extracts. Quantitation of CAT enzyme levels was performed via butyrylation (Pharmacia) of [l4C]chloramphenicol (ICN) followed by xylene extraction of the product and liquid scintillation counting (Promega Technical Bulletin, 1994). The CMV-driven β-galactosidase plasmid was included in each transfection to normalize extracts for differences in transfection efficiency, as described previously (51,52,56).
RESULTS
The TTR-3 Site Possesses a Greater Affinity for Recombinant C/EBP Protein Than the TTR-2 Site
The TTR enhancer is critical for hepatic expression of the TTR minigene in transgenic mice (18,65). This enhancer stimulates activity of the proximal TTR promoter region by 5-to 10-fold in HepG2 cell transfections (14,15,17,52). Furthermore, transcriptional activation by the TTR enhancer requires two C/EBP binding sites designated TTR-2 and TTR-3 [(16,17), Fig. 1]. To determine the relative C/EBP binding affinity of these sites, we carried out gel mobility shift assays with recombinant C/EBPα and C/EBPβ proteins and the TTR enhancer sequences (Fig. 2A). Competition studies showed that binding of recombinant C/EBP protein to the TTR-3 sequence was competed by unlabeled TTR-3 oligonucleotide (100-fold molar excess) as well as by a HNF-3β promoter oligonucleotide that contains binding sites for C/EBP and HNF-3 proteins (49,56). The C/EBP binding site from the hemopexin (Hpx) promoter (50) was a more effective competitor for C/EBP binding to the TTR-3 oligonucleotide. By contrast, the TTR-2 sequence possessed low binding affinity for the C/EBP protein because it was unable to compete efficiently for C/EBP complex formation with itself or the TTR-3 sequence (Fig. 2A). These results suggest that the TTR-3 sequence possesses greater affinity for the C/EBP family of proteins than the TTR-2 sequence.
FIG. 2.
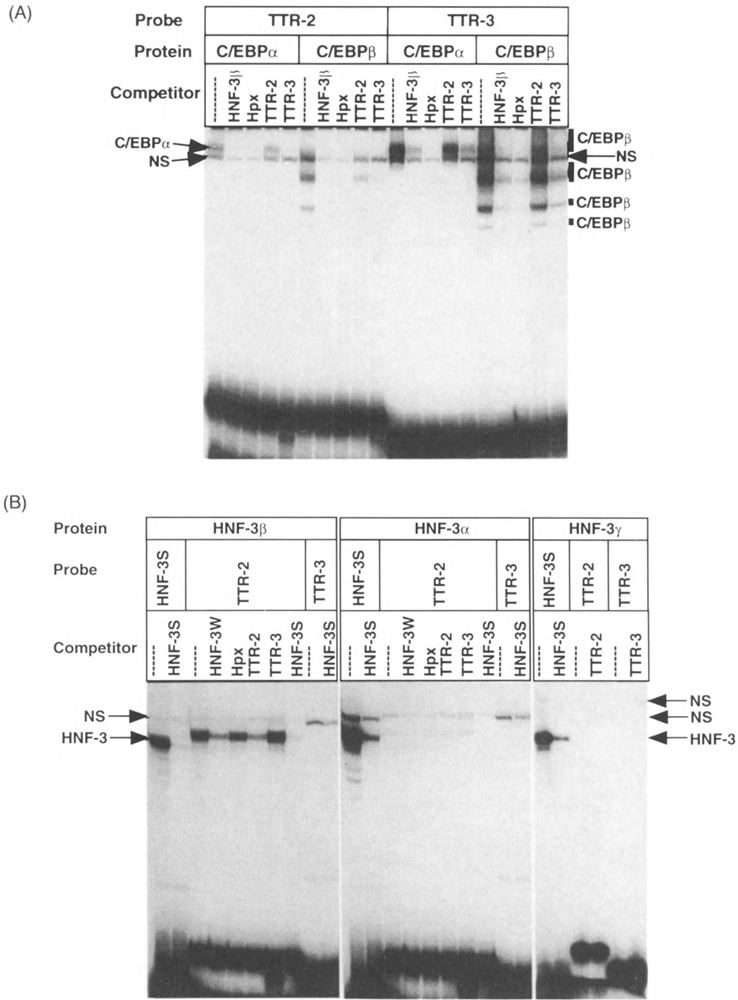
The TTR-2 sequence contains a divergent HNF-3 site that exclusively binds the HNF-3β isoform. (A) Recombinant C/EBPα and C/EBPβ proteins synthesized in HeLa cells were used for complex formation with the TTR-2 and TTR-3 oligonucleotides, and a 100-fold molar excess of the indicated competitors was included to examine binding affinities (see Fig. 1). This includes the HNF-3β promoter oligonucleotide (–97/–67), which binds both HNF-3 and C/EBP proteins (49,56), and the strong affinity C/EBP binding site from the hemopexin (Hpx) promoter (50). The position of the C/EBPα and C/EBPβ protein/DNA complexes and a nonspecific (NS) band that did not compete with C/EBP binding sites are also indicated. Control HeLa cell nuclear extracts did not contain any binding activity to the TTR-2 or TTR-3 oligonucleotides. (B) The TTR-2 sequence selectively binds the HNF-3/3 isoform. Recombinant HNF-3α, HNF-3β, and HNF-3γ proteins were used for complex formation with the TTR-2 and TTR-3 oligonucleotides and a 100-fold molar excess of the indicated competitors was included [HNF-3W is the same as HNF-3β in (A)]. The strong affinity HNF-3 site (HNF-3S) from the TTR promoter (15) was included to demonstrate that all of the recombinant HNF-3 isoforms were active for DNA binding activity and TTR-3 was used as a negative control. The position of the HNF-3 protein/DNA complexes (HNF-3) and several nonspecific complexes (NS) that were not inhibited by competition with HNF-3 binding sites are also indicated.
The TTR-2 Enhancer Sequence Selectively Binds the HNF-3β Isoform
Because of the weak C/EBP binding affinity exhibited by the functionally important TTR-2 sequence, we performed computer searches for other transcription factor binding sites and found homology with the HNF-3 binding site consensus (5′-AWTRTTKRYTY-3′, where R = A or G; W = A or T; K = G or T; Y = T or C) (48). We carried out gel mobility shift assays with the TTR-2 sequence and recombinant HNF-3α, HNF-3β, and HNF-3γ proteins (Fig. 2B). Each of the recombinant HNF-3 protein preparations was active as evidenced by complex formation with the strong affinity HNF-3S binding site from the TTR promoter (15). To our surprise, the TTR-2 sequence was exclusively bound by recombinant HNF-3β protein and not recognized by either HNF-3α or HNF-3γ proteins (Fig. 2B). We attribute the selective recognition of the TTR-2 sequence to several nucleotide differences at both ends of the HNF-3 consensus sequence (Fig. 1, boxed residues). Such nucleotide differences have been shown to influence recognition by winged helix family members (48). Furthermore, similar inhibition of HNF-3β complex formation was observed with either self-competition or cross-competition with a weak affinity HNF-3 site (HNF-3W) from the HNF-3/3 promoter region (49,56), whereas an oligonucleotide corresponding to the HNF-3S site was a more effective competitor (Fig. 1 and 2B). Moreover, the HNF-3 proteins did not bind to the TTR-3 sequence nor did C/EBP binding sites compete for HNF-3β complex formation with the TTR-2 oligonucleotide (Fig. 2B, TTR-3, and Hpx). These gel shift studies identify an HNF-3 site in the TTR-2 enhancer sequence that exclusively binds the HNF-3β isoform. It is interesting to note that a second HNF-3 binding site in the TTR promoter (–140/–130, see Fig. 1) has also been shown to bind the HNF-3β protein preferentially (38).
HNF-3β Protein Is the Major TTR-2 Binding Protein in Mouse Liver Nuclear Extracts
To examine whether liver-derived C/EBP and HNF-3β proteins could form complexes with the TTR-2 oligonucleotide, protein extracts were prepared from mouse liver nuclei. Analysis of this liver nuclear extract in DNA binding assays demonstrated recognition of the TTR-2 sequence by the HNF-3β isoform, because the TTR-2 protein/ DNA complex was disrupted with HNF-3β antibody but not with HNF-3α or HNF-3γ antisera (Fig. 3A). The HNF-3β protein complex with the TTR-2 sequences was not disrupted by the C/EBP binding sites but was effectively competed by itself and HNF-3 binding sites from the HNF-3/3 and TTR promoters (Fig. 3B). These gel shift assays also demonstrate that the TTR-2 oligonucleotide did not form detectable C/EBP complexes with liver nuclear extracts as evidenced by lack of competition with the Hpx or TTR-3 oligonucleotides (Fig. 3B).
FIG. 3.
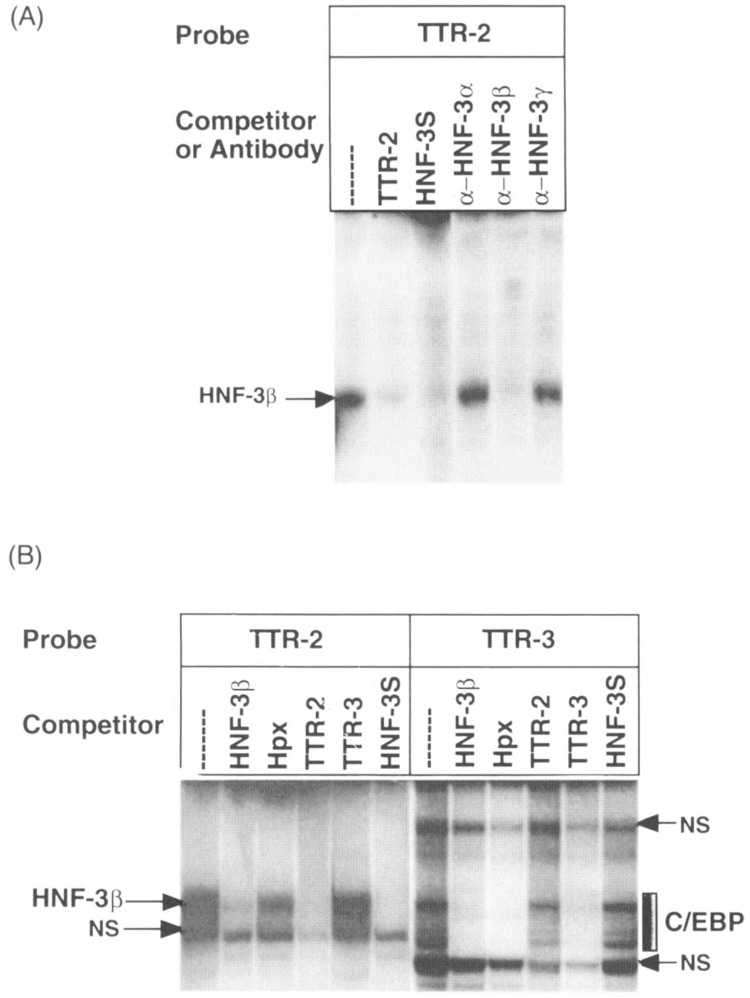
The HNF-3β protein is the major TTR-2 binding protein in mouse liver nuclear extracts. (A) Only the HNF-3β complex is disrupted when the TTR-2 oligonucleotide is used as the gel shift probe with HNF-3 isoform-specific antibodies. Shown is the top portion of the gel depicting the protein/DNA complexes with liver nuclear extracts. Included in the binding reactions are either competitions with the binding site oligonucleotides described in Fig. 2 legend or HNF-3 isoform-specific antisera. (B) Liver-derived C/EBP forms complexes with the TTR-3 site but not TTR-2. Gel shift competition assays using the TTR-2 and TTR-3 oligonucleotides and mouse liver nuclear extracts. Note that the HNF-3S site competes all of the TTR-2 protein complexes and it is not inhibited by the C/EBP site from the Hpx promoter. In contrast, TTR-3 is inhibited by C/EBP sites but not by HNF-3 sites.
Consistent with the recombinant C/EBP binding studies, the TTR-3 oligonucleotide formed abundant protein complexes that were effectively competed by unlabeled TTR-3 and with C/EBP binding sites derived from the Hpx and HNF-3β promoters. The multiple complexes are consistent with the recognition by homo- and heterodimers of the C/EBP protein family, which differ in their gel mobility (10,64). Furthermore, the TTR-2 and HNF-3S oligonucleotides were not effective competitors of C/EBP complex formation with the TTR-3 sequence. These studies suggest that TTR-3 is an authentic C/EBP binding site whereas the TTR-2 site is a selective binding site for HNF-3β protein. These results are consistent with the fact that the TTR-3 sequence contains the C/EBP consensus binding sequence (1) 5′-TKNNGNAAK-3′ (5′-TTGAGTAAG-3′ Fig. 1, bottom strand) whereas the TTR-2 sequence does not match the C/EBP consensus.
Disruption of the HNF-3β-Specific Site Results in Reduced TTR Enhancer Activity
To assess the contribution of the HNF-3β site in the TTR enhancer, we made site-directed mutations in the TTR enhancer, which either disrupted HNF-3/3 recognition or converted it so that it binds all of the HNF-3 isoforms (48). These TTR enhancer mutations were fused to the TTR promoter region driving the expression of the CAT reporter gene and used for transfections into HepG2 cells to determine the importance of the HNF-3β-specific site in TTR gene transcription. Disruption of the HNF-3β-specific site caused a 40% decrease in reporter gene activity compared to the wild-type TTR promoter and enhancer construct (Fig. 4). The TTR-2 enhancer mutant still demonstrates greater transcriptional levels than that of the TTR proximal promoter construct lacking the TTR enhancer, which exhibits a 70% reduction in transcriptional activity (Fig. 4), suggesting that disruption of the HNF-3β-specific site does not completely eliminate TTR enhancer activity. We then altered the HNF-3β-specific site in the TTR enhancer to a sequence that has previously been shown to bind all three HNF-3 isoforms (HNF-3 #4 site) (48). Conversion of the TTR-2 sequence to a site that also binds HNF-3a and HNF-37 causes a slight increase in transcriptional activity, suggesting that recognition by the other HNF-3 proteins potentiates transactivation function (Fig. 4).
FIG. 4.
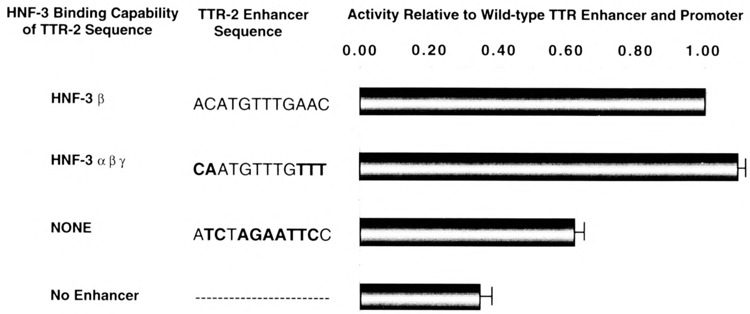
The HNF-3β-specific site is required for transcriptional activation by the TTR enhancer. Constructs consisting of the TTR enhancer and –202 promoter driving the expression of the CAT gene were transfected into HepG2 cells to determine the importance of the HNF-3β-specific site in the TTR-2 region of the TTR enhancer. Transient expression levels (average from three separate transfection experiments) of the wild-type TTR enhancer containing an HNF-3β-specific site were compared with mutations that allowed recognition of all of the HNF-3 isoforms or that eliminated HNF-3 binding (None). Also included for comparison is the activity of the TTR promoter constructs that lacked the TTR enhancer region. The CMV-driven β-galactosidase plasmid was included in each transfection to normalize cellular protein extracts for differences in transfection efficiency, as described previously (51,52,56).
A Ubiquitous Factor Binds Upstream of the HNF-3(3-Specific Binding Site
Analysis of the 5′ end of the TTR enhancer identified a functional sequence, designated TTR-1, that bound a ubiquitous factor (UF, Fig. 1), which was widely expressed in a number of tissue nuclear extracts (16,25). We proposed that TTR-1 was recognized by the AP-1 protein because it was competed by an α1-antitrypsin enhancer sequence (α1-AT-B) that contained a perfect AP-1 binding site [(54); see Fig. 1]. To explore this hypothesis, we carried out gel shift assays with these oligonucleotides and nuclear extracts prepared from TPA-treated HepG2 cells (Fig. 5). The al-AT-B site bound TPA-inducible AP-1 protein, as evidenced from cross-competition with the SV40 AP-1 site; the α1-AT-B site also inhibited AP-1 complex formation (Fig. 5, white dot). Nuclear extracts prepared from untreated HepG2 cells allowed us to identify the position of the UF/αl-AT-B complexes, which were not diminished by SV40 AP-1 competition (Fig. 5). In contrast to the αl-AT-B sequence, the TTR-1 probe did not form any TPA-inducible complexes and the UF complexes were cross-competed by the al-AT-B site but not by the AP-1, SP-1, and UF1-H3β sequences [(55,56); Fig. 3 and data not shown]. These results demonstrate that TTR-1 binds a ubiquitous factor (UF), which is distinct from the AP-1 protein/DNA complex. They also show that the al-AT-B sequence binds both the TTR UF and the TPA-inducible AP-1 proteins.
FIG. 5.
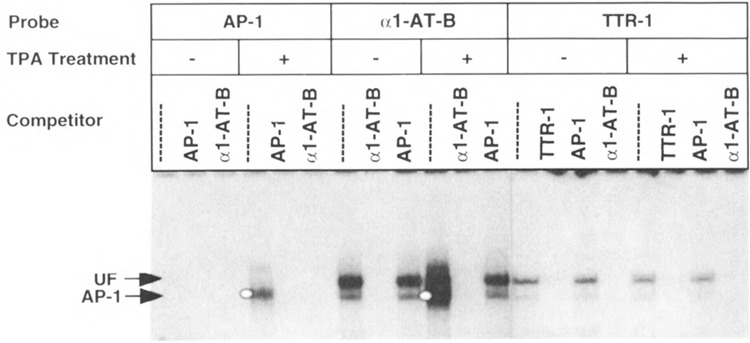
The TTR-1 site binds a ubiquitous factor that is distinct from AP-1. We carried out gel mobility shift assays with the indicated oligonucleotides (see Fig. 1) and nuclear extracts prepared from the human hepatoblastoma (HepG2) cell line that were either treated with TPA for 4 h or not treated (see the Materials and Methods section). The AP-1 oligonucleotide is the AP-1 sequence from the SV40 enhancer region (5) and the αl-AT-B oligonucleotide contains an AP-1 consensus site derived from the α1-antitrypsin (αl-AT) enhancer (25). Shown is the position of the TPA-induced AP-1 protein/DNA complex (white dot) and the ubiquitous factor (UF) complex on the gel. Note that the al-AT-B oligonucleotide binds both AP-1 and UF proteins. Competition lanes include a 100-fold molar excess of indicated unlabeled oligonucleotide.
DISCUSSION
Characterization of a Ubiquitous Factor Recognizing the TTR-1 Enhancer Site
In a previous report, three functional sequences were identified in the TTR enhancer region (TTR-1, TTR-2, and TTR-3), one of which (TTR-1) bound a nuclear factor found in a variety of tissue nuclear extracts [(16,25), see Fig. 1]. We had previously suggested that AP-1 protein recognizes TTR-1 because the al-AT-B enhancer sequence, which contains a perfect AP-1 consensus site, competed for binding (25). In this study, we demonstrate that TTR-1 sequence is not recognized by AP-1 but binds a distinct ubiquitously expressed factor (UF) (Figs. 1 and 5). The al-AT-B sequence, however, is recognized by both the UF proteins and the TPA-inducible AP-1 proteins (Fig. 5). Recent studies have identified an AP-1 site that partially overlaps the HNF-3S site in the TTR proximal promoter and confers TPA stimulation of the TTR promoter region (52). Therefore, as in the case of the hepatocyte-specific regulatory sequences of the αl-AT, metallothionein, and HNF-1 genes (5,25,35), the TTR promoter has evolved with an AP-1 sequence conferring transcrip-tional induction via the protein kinase C pathway.
The TTR-2 Site Exclusively Binds the HNF-3α Isoform and the TTR-3 Site Binds C/EBP Proteins in Mouse Nuclear Extracts
Two adjacent C/EBP binding sites (TTR-2 and TTR-3) were identified in the TTR enhancer that were required for transcriptional activation (16,17). In this study, we demonstrate that the TTR-3 sequence is recognized by both recombinant C/EBP proteins and authentic C/EBP protein found in liver nuclear extracts. In contrast, the TTR-2 sequence does not form detectable C/EBP protein complexes in liver nuclear extracts and does not specifically compete for recombinant C/EBP protein binding. These results are consistent with the fact that the TTR-3 sequence matches the C/EBP DNA binding consensus sequence (1) and that the TTR-2 sequence lacks significant homology to this consensus site. These data suggest that the TTR enhancer contains only one high-affinity binding sequence for the C/EBP protein, which is present within the TTR-3 enhancer site. In this study, we also show that the TTR-2 enhancer site contains a variant HNF-3 consensus sequence that is selectively binding the HNF-3β isoform. Disruption of this HNF-3β-specific enhancer site demonstrates that this sequence contributes significantly to TTR enhancer function (Figs. 2–4). These studies demonstrate the existence of variant HNF-3 sites that distinguish among the three HNF-3 isoforms. Analysis of promoter regions for this HNF-3β-specific site will allow identification of liver target genes that are primarily regulated by the HNF-3β isoform.
The TTR gene is a negatively responsive acute phase gene (24) whose decreased expression in the acute phase response contributes to the maintenance of a constant serum protein concentration by allowing for an increase in newly synthesized acute phase plasma proteins (7). Previous studies of the TTR promoter region identified a weak affinity HNF-3 site that exhibited a higher binding affinity for the HNF-3β isoform (38). The discovery of an additional HNF-3β-selective site in the TTR enhancer region suggests that these sites may play an important role under particular physiological conditions where the levels of the other HNF-3 proteins are reduced. We have previously shown that the induction of the acute phase response in the liver causes a transient 95% reduction in HNF-3a expression levels which coincides with a 60% decrease in expression levels of its target gene TTR (52). In contrast to HNF-3α expression, the HNF-3β protein is only slightly diminished during the acute phase response. The decrease in TTR gene expression during the acute phase response also coincides with a dramatic reduction of C/EBPα expression and increases in C/EBPβ and C/EBPδ isoforms (2). We have previously shown that the TTR-3 sequence is not an efficient competitor for C/EBPδ protein recognition (56). The decrease in both C/EBPα and HNF-3α expression during the acute phase response may therefore mediate the decrease in TTR gene transcription. The existence of two discriminating binding sites in the TTR DNA regulatory region suggests that HNF-3β recognition may play a role in maintaining TTR gene transcription during the acute phase response when transient reduction of other transcription factors occurs.
ACKNOWLEDGEMENTS
We thank Pradip Raychaudhuri for critically reading the manuscript and E. Lai for his generous gift of the HNF-3γ antisera. This work was supported by Public Health Service grant R01 GM43241-06 from the National Institute of General Medical Sciences. R.H.C. is an Established Investigator of the American Heart Association/Bristol-Myers Squibb. U.S. is a Charlotte Webster, Helen T. Barnes, and Broda O. Barnes Scholar in Molecular Medicine.
REFERENCES
- 1. Akira S.; Isshiki H.; Sugita T.; Tanabe O.; Kinoshita S.; Nishio Y.; Nakajima T.; Hirano T.; Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9:1897–1906; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alam T.; An M. R.; Papoconstantinou J. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 267:5021–5024; 1992. [PubMed] [Google Scholar]
- 3. Ang S. L.; Rossant J. HNF-3/3 is essential for node and notochord formation in mouse development. Cell 78:561–574; 1994. [DOI] [PubMed] [Google Scholar]
- 4. Ang S. L.; Wierda A.; Wong D.; Stevens K. A.; Cascio S.; Rossant J.; Zaret K. S. The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF3/forkhead proteins. Development 119:1301–1315; 1993. [DOI] [PubMed] [Google Scholar]
- 5. Angel P.; Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129–157; 1991. [DOI] [PubMed] [Google Scholar]
- 6. Auge-Gouillou C; Petropoulos I.; Zakin M. M. Liver-enriched HNF-3α and ubiquitous factors interact with the human transferrin gene enhancer. FEBS Lett. 323:4–10; 1993. [DOI] [PubMed] [Google Scholar]
- 7. Baumann H.; Gauldie J. The acute phase response. Immunol. Today 15:74–80; 1994. [DOI] [PubMed] [Google Scholar]
- 8. Baumhueter S.; Mendel D. B.; Conley P. B.; Kuo C. J.; Turk C.; Graves M. K.; Edwards C. A.; Courtois G.; Crabtree G. R. HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev. 4:372–379; 1990. [DOI] [PubMed] [Google Scholar]
- 9. Brooks A. R.; Blackhart B. D.; Haubold K.; Levy-Wilson B. Characterization of tissue-specific enhancer elements in the second intron of the human apolipoprotein B gene. J. Biol. Chem. 266:7848–7859; 1991. [PubMed] [Google Scholar]
- 10. Cao Z.; Umek R. M.; McKnight S. L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538–1552; 1991. [DOI] [PubMed] [Google Scholar]
- 11. Chen M.; Hieng S.; Qian X.; Costa R.; Ou J. Regulation of hepatitis B virus ENI enhancer activity by hepatocyte-enriched transcription factor HNF3. Virology 205:127–132; 1994. [DOI] [PubMed] [Google Scholar]
- 12. Clark K. L.; Halay E. D.; Lai E.; Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412–420; 1993. [DOI] [PubMed] [Google Scholar]
- 13. Costa R. H. Hepatocyte nuclear factor 3/fork head protein family: Mammalian transcription factors that possess divergent cellular expression patterns and binding specificities. In: Tronche F.; Yaniv M., eds. Liver gene transcription. Austin, TX: R. G. LandesCo.; 1994:183–206. [Google Scholar]
- 14. Costa R. H.; Grayson D. R. Site-directed muta-genesis of hepatocyte nuclear factor (HNF) binding sites in the mouse transthyretin (TTR) promoter reveal synergistic interactions with its enhancer region. Nucleic Acids Res. 19:4139–4145; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Costa R. H.; Grayson D. R.; Darnell J. Jr. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and α 1-antitrypsin genes. Mol. Cell. Biol. 9:1415–1425; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Costa R. H.; Grayson D. R.; Xanthopoulos K. G.; Darnell J. Jr. A liver-specific DNA-binding protein recognizes multiple nucleotide sites in regulatory regions of transthyretin, α 1-antitrypsin, albumin, and simian virus 40 genes. Proc. Natl. Acad. Sci. USA 85:3840–3844; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Costa R. H.; Lai E.; Grayson D. R.; Darnell J. Jr. The cell-specific enhancer of the mouse transthyretin (prealbumin) gene binds a common factor at one site and a liver-specific factor(s) at two other sites. Mol. Cell. Biol. 8:81–90; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Costa R. H.; Van Dyke T. A.; Yan C. ; Kuo F.; Darnell J. Jr. Similarities in transthyretin gene expression and differences in transcription factors: Liver and yolk sac compared to choroid plexus. Proc. Natl. Acad. Sci. USA 87:6589–6593; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Derman E.; Krauter K.; Walling L.; Weinberger C.; Ray M.; Darnell J. Jr. Transcriptional control in the production of liver-specific mRNAs. Cell 23:731–739; 1981. [DOI] [PubMed] [Google Scholar]
- 20. Descombes P.; Chojkier M.; Lichtsteiner S.; Fal-vey E.; Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 4:1541–1551; 1990. [DOI] [PubMed] [Google Scholar]
- 21. DiPersio C. M.; Jackson D. A.; Zaret K. S. The extracellular matrix coordinately modulates liver transcription factors and hepatocyte morphology. Mol. Cell. Biol. 11:4405–4414; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drolet D. W.; Scully K. M.; Simmons D. M.; Wegner M.; Chu K. T.; Swanson L. W.; Rosenfeld M. G. TEF, a transcription factor expressed specifically in the anterior pituitary during embryo-genesis, defines a new class of leucine zipper proteins. Genes Dev. 5:1739–1753; 1991. [DOI] [PubMed] [Google Scholar]
- 23. Frain M.; Swart G.; Monaci P.; Nicosia A.; Stampfli S.; Frank R.; Cortese R. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell 59:145–157; 1989. [DOI] [PubMed] [Google Scholar]
- 24. Fung W.; Thomas T.; Dickerson P.; Aldred A. R.; Milland J.; Dziadek M.; Powers B.; Hudson P.; Schreiber G. Structure and expression of rat transthyretin (prealbumin) gene. J. Biol. Chem. 263:480–488; 1988. [PubMed] [Google Scholar]
- 25. Grayson D. R.; Costa R. H.; Xanthopoulos K. G.; Darnell J. E. One factor recognizes the liver-specific enhancers in α 1-antitrypsin and transthyretin genes. Science 239:786–788; 1988. [DOI] [PubMed] [Google Scholar]
- 26. Harnish D. C.; Malik S.; Karathanasis S. K. Activation of apolipoprotein AI gene transcription by the liver-enriched factor HNF-3. J. Biol. Chem. 269:28220–28226; 1994. [PubMed] [Google Scholar]
- 27. Hromas R.; Costa R. The hepatocyte nuclear factor-3/forkhead transcription regulatory family in development, inflammation, and neoplasia. Crit. Rev. Oncol. Hematol. 20:129–140; 1995. [DOI] [PubMed] [Google Scholar]
- 28. Hunger S. P.; Ohyashiki K.; Toyama K.; Cleary M. L. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 6:1608–1620; 1992. [DOI] [PubMed] [Google Scholar]
- 29. Inaba T.; Roberts W. M.; Shapiro L. H.; Jolly K. W. Raimondii S. C. Smith S. D.; Look A. T. Fusion of the leucine zipper gene HLF to the H2A gene in human acute B-lineage leukemia. Science 257:531–534; 1992. [DOI] [PubMed] [Google Scholar]
- 30. Ip Y. T.; Poon D.; Stone D.; Granner D. K.; Chalkley R. Interaction of a liver-specific factor with an enhancer 4.8 kilobases upstream of the phosphoenolpyruvate carboxykinase gene. Mol. Cell. Biol. 10:3770–3781; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iyer S. V.; Davis D. L.; Seal S. N.; Burch J. B. Chicken vitellogenin gene-binding protein, a leucine zipper transcription factor that binds to an important control element in the chicken vitellogenin II promoter, is related to rat DBP. Mol. Cell. Biol. 11:4863–4875; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jacob A.; Budhiraja S.; Qian X.; Clevidence D.; Costa R. H.; Reichel R. R. Retinoic acid-mediated activation of HNF-3a during EC stem cell differentiation. Nucleic Acids Res. 22:2126–2133; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kinoshita S.; Akira S.; Kishimoto T. A member of the C/EBP family, NF-IL6/3, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc. Natl. Acad. Sci. USA 89:1473–1476; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Knowles B.; Howe C. C .; Arden D. P. Human hepatocellular carcinoma cell lines secrete the major plasma protein and hepatitis B surface antigen. Science 209:497–499; 1980. [DOI] [PubMed] [Google Scholar]
- 35. Kuo C. J.; Conley P. B.; Chen L.; Sladek F. M.; Darnell J. Jr.; Crabtree G. R. A transcriptional hierarchy involved in mammalian cell-type specification. Nature 355:457–461; 1992. [DOI] [PubMed] [Google Scholar]
- 36. Ladias J. A.; Karathanasis S. K. Regulation of the apolipoprotein AI gene by ARP-1, a novel member of the steroid receptor superfamily. Science 251:561–565; 1991. [DOI] [PubMed] [Google Scholar]
- 37. Lai E.; Prezioso V. R.; Smith E.; Litvin O.; Costa R. H.; Darnell J. E. Jr. HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 4:1427–1436; 1990. [DOI] [PubMed] [Google Scholar]
- 38. Lai E.; Prezioso V. R.; Tao W. F.; Chen W. S.; Darnell J. E. Jr. Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 5:416–427; 1991. [DOI] [PubMed] [Google Scholar]
- 39. Landschulz W. H.; Johnson P. F.; Adashi E. Y.; Graves B. J.; McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 2:786–800; 1988. [DOI] [PubMed] [Google Scholar]
- 40. Landschulz W. H.; Johnson P. F.; McKnight S. L. The DNA binding domain of the rat liver nucllar protein C/EBP is bipartite. Science 243:1681–1688; 1989. [DOI] [PubMed] [Google Scholar]
- 41. Liu J. K.; DiPersio C. M.; Zaret K. S. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol. Cell. Biol. 11:773–784; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McPherson C. E.; Shim E. Y.; Friedman D. S.; Zaret K. S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75:387–398; 1993. [DOI] [PubMed] [Google Scholar]
- 43. Molowa D. T.; Chen W. S.; Cimis G. M.; Tan C. P. Transcriptional regulation of the human cholesterol 7α-hydroxylase gene. Biochemistry 31:2539–2544; 1992. [DOI] [PubMed] [Google Scholar]
- 44. Monaghan A. P.; Kaestner K. H.; Grau E.; Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development 119:567–578; 1993. [DOI] [PubMed] [Google Scholar]
- 45. Mueller C. R.; Maire P.; Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell 61:279–291; 1990. [DOI] [PubMed] [Google Scholar]
- 46. Nitsch D.; Boshart M.; Schutz G. Activation of the tyrosine aminotransferase gene is dependent on synergy between liver-specific and hormone-responsive elements. Proc. Natl. Acad. Sci. USA 90:5479–5483; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ori A.; Shaul Y. Hepatitis B virus enhancer binds and is activated by the hepatocyte nuclear factor 3. Virology 207:98–106; 1995. [DOI] [PubMed] [Google Scholar]
- 48. Overdier D. G.; Porcella A.; Costa R. H. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell. Biol. 14:2755–2766; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pani L.; Qian X. B.; Clevidence D.; Costa R. H. The restricted promoter activity of the liver transcription factor hepatocyte nuclear factor 3/3 involves a cell-specific factor and positive autoactivation. Mol. Cell. Biol. 12:552–562; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Poli V.; Mancini F. P.; Cortese R. IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/EBP. Cell 63:643-653; 1990. [DOI] [PubMed] [Google Scholar]
- 51. Qian X.; Costa R. H. Analysis of HNF-3/3 protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res. 23:1184–1191; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Qian X.; Samadani U.; Porcella A.; Costa R. H. Decreased expression of Hepatocyte Nuclear Fac-tor-3a during the acute phase response influences transthyretin gene transcription. Mol. Cell. Biol. 15:1364–1376; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Raney A. K.; Zhang P.; McLachlan A. Regulation of transcription from the hepatitis B virus large surface antigen promoter by hepatocyte nuclear factor 3. J. Virol. 69:3265–3272; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raymondjean M.; Pichard A. L.; Gregori C. ; Ginot F.; Kahn A. Interplay of an original combination of factors: C/EBP, NFY, HNF-3 and HNF-1 in the rat adolase B gene promoter. Nucleic Acids Res. 19:6145–6153; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ruiz i Altaba A.; Prezioso V. R.; Darnell J. E.; Jessell T. M. Sequential expression of HNF-3β and HNF-3α by embryonic organizing centers: The dorsal lip/node, notochord and floor plate. Mech. Dev. 44:91–108; 1993. [DOI] [PubMed] [Google Scholar]
- 56. Samadani U.; Porcella A.; Pani L.; Johnson P. F.; Burch J.; Pine R.; Costa R. H. Cytokine regulation of the liver transcription factor HNF-3β is mediated by the C/EBP family and interferon regulatory factor 1. Cell Growth Differen. 6:879–890; 1995. [PubMed] [Google Scholar]
- 57. Sasaki H.; Hogan B. L. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development 118:47–59; 1993. [DOI] [PubMed] [Google Scholar]
- 58. Sasaki H.; Hogan B. L. HNF-3/3 as a regulator of floor plate development. Cell 76:103–115; 1994. [DOI] [PubMed] [Google Scholar]
- 59. Sladek F. M.; Zhong W. M.; Lai E.; Darnell J. E. Jr. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 4:2353–2365; 1990. [DOI] [PubMed] [Google Scholar]
- 60. Unterman T. G.; Goswami R. G.; Fareeduddin A.; Harris M. A.; Porcella A.; Costa R. H.; Lacson P. G. Hepatocyte nuclear factor-3 binds to the insulin response sequence in IGF binding protein-1 (IGFBP-1) promoter and enhances promoter function. Biochem. Biophys. Res. Commun. 203:1835–1841; 1994. [DOI] [PubMed] [Google Scholar]
- 61. Vallet V.; Antoine B.; Chafey P.; Vandewalle A.; Kahn A. Overproduction of a truncated hepatocyte nuclear factor 3 protein inhibits expression of liver-specific genes in hepatoma cells. Mol. Cell. Biol. 15:5453–5460; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Weigel D.; Jackie H. The fork head domain: A novel DNA binding motif of eukaryotic transcription factors? Cell 63:455–456; 1990. [DOI] [PubMed] [Google Scholar]
- 63. Weinstein D. C.; Ruiz i Altaba A.; Chen W. S.; Hoodless P.; Prezioso V. R.; Jessell T. M.; Darnell J. Jr. The winged-helix transcription factor HNF-3β is required for notochord development in the mouse embryo. Cell 78:575–588; 1994. [DOI] [PubMed] [Google Scholar]
- 64. Williams S. C.; Cantwell C. A.; Johnson P. F. A family of C/EBP-related proteins capable of forming covalent linked leucine zipper dimers in vitro. Genes Dev. 5:1553–1567; 1991. [DOI] [PubMed] [Google Scholar]
- 65. Yan C.; Costa R. H.; Darnell J. Jr.; Chen J. D.; Van Dyke T. A. Distinct positive and negative elements control the limited hepatocyte and choroid plexus expression of transthyretin in transgenic mice. EMBO J. 9:869–878; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zaret K. S. Control of hepatocyte differentiation by liver-enriched transcription factors. In: Tavoloni N.; Berk P. D., eds. Hepatic transport and bile secretion: Physiology and pathophysiology. New York: Raven Press, Ltd.; 1993:135–143. [Google Scholar]


