Abstract
Multiple foci of morphologically and functionally differentiated hepatocytes are induced in the pancreas of adult rats subjected to a copper depletion-repletion regimen. Differentiation of hepatocytes in pancreas is preceded by irreversible depletion of over 90% of pancreatic acinar cells. Progressive acinar cell loss during 4–6 weeks of copper deficiency results in the proliferation of oval cells, some of which may serve as the hepatocyte precursor or stem cells. Albumin mRNA is detected in oval cells at 5 and 6 weeks by in situ hybridization at which time no morphologically identifiable hepatocytes are evident in the pancreas. Immunocytochemical analysis demonstrated the presence of stem cell factor (SCF) in proliferating oval cells during 6 weeks of copper depletion, and Northern blot analysis revealed the expression of liverenriched transcription factors in the rat pancreas during this 4–6-week period of copper deficiency. CCAAT/enhancer binding protein α (C/EBPα) mRNA was detected first at 4 weeks of copper deficiency. By 5 and 6 weeks of copper deficiency, the expression of mRNAs of C/EBPα, β, and δ, and hepatocyte nuclear factor-3/β (HNF-3β) was markedly enhanced. This enhanced expression of liver-enriched transcription factors and the SCF during oval cell proliferation in the pancreas preceding the expression of albumin mRNA and subsequent differentiation of hepatocyte phenotype further supports the identity of these oval cells as hepatocyte precursors or stem cells.
Keywords: Oval cells, Pancreatic hepatocytes, Stem cell factor, C/EBP, HNF
HEPATOCYTES develop in the pancreas, with constancy, in adult rats subjected to a copper depletion-repletion regimen (29,33). Rats maintained on a diet deficient in copper and supplemented with a copper-chelator reveal relentless pancreatic acinar cell apoptosis, beginning at 4 weeks, which culminates in global acinar cell loss by 8–9 weeks of deficiency (17,28,31,38). This acinar cell loss appears to serve as a trigger for the proliferation of ductular/periductular cells that resemble oval cells, similar to those proliferating in the liver under some experimental conditions (32). Sequential morphological observations suggested that some of these proliferating oval cells in the copper-depleted rat pancreas differentiate into hepatocytes (29,30). Furthermore, Northern analysis revealed the expression of albumin mRNA in the pancreas of rats fed a copper-deficient diet for 5 weeks, at which time period oval cells begin to proliferate, but morphologically discernible hepatocytes are not identifiable, implying that oval cells may be expressing albumin transcripts (29). Visualization of albumin mRNA trascripts in these oval cells by in situ hybridization further suggests that these cells could be the precursor (stem) cells of pancreatic hepatocytes (29,30).
Cell differentiation and expression of cell-specific genes are regulated by transcriptional factors (2). In mammals, the determination of fore-gut endodermal cells toward hepatic lineage is dependent, to a certain extent, on cues from mesenchymal cells and expression of liver-enriched transcription factors (4). In the adult liver, liver-specific gene expression is controlled by several transcriptional factors (9,16). Presently, four liver-enriched transcription factor families, namely CCAAT/enhancer binding protein (C/EBP), and hepatocyte nuclear factor (HNF)-l, -3, and -4 have been identified (18). Expression of HNF-1 family of transcription factors appears early during embryogenesis and these transcription factors appear to participate in hepatocyte differentiation and maintenance of the expression of tissue-specific genes (5,9). HNF-3 consists of three major DNA binding proteins, α, β and δ, that are expressed in different organs and play a role in the differentiation and development of structures derived from the primitive gut endo-derm and formation of mesoderm and neural axis (23). HNF-4, a member of the steroid hormone receptor superfamily, also expressed in several tissues, appears to be important in early murine development and organogenesis (10). C/EBP family of transcription factors consists of three proteins, α, β and δ, that play a role in maintaining the differentiated state of hepatocytes and inhibition of cell proliferation (21,40). We postulated the existence of stem cells in the adult pancreas and suggested that they are induced to proliferate only when there is a global loss of acinar cells (32). Evidence indicates that stem cell factor (SCF) and its receptor c-kit constitute an important signal transduction system implicated in cell proliferation and differentiation (26). Recent studies have shown that SCF may be important in liver stem cell maintenance and proliferation (14). To understand the early events that occur prior to the differentiation of morphologically identifiable hepatocytes in the pancreas, it is necessary to delineate the expression of liver-specific transcription factors in the pancreas of rats maintained on a copper-deficient diet. In the present study, we report that the expression of liver-enriched transcription factors and SCF occurs very early during the proliferation of oval cells in the pancreas of rats maintained on a copper-deficient diet, and the expression of these factors precedes the expression of albumin mRNA and hepatocyte differentiation. Once formed, the pancreatic hepatocytes proliferate and persist throughout the life span of these rats.
MATERIALS AND METHODS
Copper Depletion Procedure
Male Fischer 344 rats, weighing 80–90 g, were purchased from Charles River Breeding Laboratories (Wilmington, MA). After a week of acclimatization, rats were housed in individual wire mesh cages, allowed access to deionized water, and fed a copper-deficient diet (U.S. Biochemical Corp., Cleveland, OH) that was supplemented with 0.6% triethylene tetramine tetrahydrochloride (Aldrich Chemical Company, Milwaukee, WI) for a period of 4–6 weeks. Groups of four to six rats maintained on the copper-deficient diet were sacrificed at 4, 5, and 6 weeks. Four rats maintained on Purina rat chow served as controls and were sacrificed along with the 6-week copper-deficient batch. Portions of each pancreas were processed for light microscopy and the remainder used for RNA extraction.
In Situ Hybridization for Albumin mRNA
Portions of pancreas from rats maintained on a copper-deficient diet for 5 weeks were snap-frozen in liquid nitrogen and 6-μM-thick sections were cut using a cryostat and mounted on acid-cleaned slides. These sections were fixed in freshly prepared 4% paraformaldehyde in 0.1 M phosphate-buffered saline, pH 7.5, for 20 min, and dehydrated through graded series of ethanol. Rat albumin cDNA was labeled with [α-35S]dCTP (Amersham, Arlington, IL) to a specific activity of 3–5 × 108 cpm/μg DNA using the random priming method. The prehybridization, hybridization, and autoradiography were performed as described previously (37).
Immunohistochemistry
Sections of pancreas from 6-week copper-deficient rats were used for SCF immunostaining as described elsewhere (13). Briefly, cold acetone-fixed frozen sections were incubated with rabbit polyclonal antibody against mouse SCF (at 1:100 dilution), and the avidin-biotin complex method was followed as described by the manufacturer’s instructions (Vector, Burlingame, CA). Following immunohistochemical staining, the sections were counterstained with hematoxylin or methyl green.
RNA Extraction and Northern Hybridization
Total RNA was extracted from the pancreas of each experimental rat sacrificed at 4, 5, and 6 weeks and from the control rats according to the method described by Chomczynski and Sacchi (6). Because of marked atrophy, pancreases from two rats were pooled from the group fed copper-deficient diet for 6 weeks. In addition, RNA from livers of control rats was also extracted. Glyoxal denatured RNA (15 μg) was electrophoresed in 1% agarose gels and transferred to nylon membranes. Blots were prehybridized, and then hybridized with the following nick-translated 32P-labeled cDNAs: albumin, C/EBPα, β, and δ, and HNF-3β. For assessing equal loading of RNA in each lane, blots were hybridized for 18S ribosomal RNA or were stained with methylene blue for 18S and 28S RNA (15).
RESULTS
Morphological Changes
Sequential morphological alterations in the pancreas of rats maintained on a copper-deficient diet are similar to those previously described (31). At 4 weeks of copper deficiency, the lobular architecture appeared well preserved and changes in ac-inar cells were minimal. Apoptosis of an occasional acinar cell was encountered at 4 weeks; by 5 weeks the number of apoptotic cells increased significantly. Apoptotic changes in one or more cells in a given acinar lobule and atrophy of remaining acinar cells mainfested in a disarray of lobular architecure. Beginning at 5 weeks, the in-terlobular spaces became prominent with focal proliferation and streaming into the interstitial spaces of nondescript oval cells from the small pancreatic ductules. By 6 weeks of copper deficiency, a majority of acinar cells were lost and the pancreas at this period revealed marked proliferation of oval cells (Fig. 1).
FIG. 1.
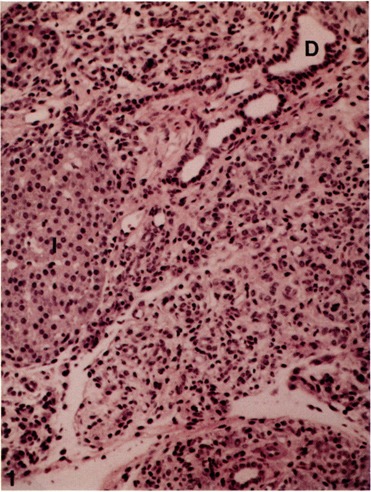
Pancreas from a rat maintained on a copper-deficient diet for 6 weeks showing marked acinar cell loss associated with oval cell proliferation. Islet of Langerhans (I) and ducts (D) are unaffected. Hematoxylin and eosin stain, × 100.
Expression of Albumin mRNA
In situ hybridization with 35S-labeled albumin cDNA revealed the expression in pancreas of albumin mRNA in some ductular and many interstitial oval cells at 5 weeks of copper deficiency (Fig. 2). No labeling was noted in the islets of Langerhans, in remaining acinar cells, and in the epithelium lining the larger ducts. No labeling was observed in normal pancreas and, as expected, hybridization with sense probe failed to reveal any signal. As previously reported, the expression of albumin mRNA during 5–8 weeks of copper depletion was most abundant in the oval cells (29,30). Although a majority of oval cells that proliferate during the copper depletion phase express albumin transcripts, it appears that only a small proportion of these cells make a commitment to become hepatocytes and differentiate accordingly. These differentiated hepatocytes then begin to proliferate to form large discrete islands of hepatocytes in the pancreas.
FIG. 2.
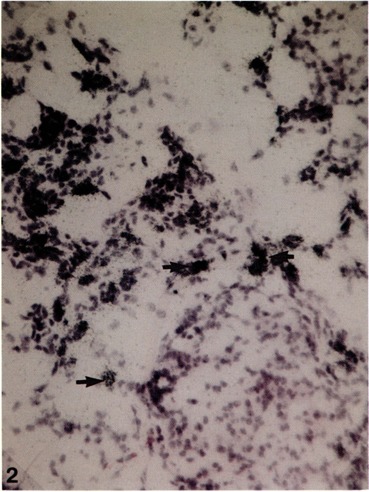
In situ hybridization of frozen section of pancreas from a rat maintained on a copper-deficient diet for 6 weeks for localization of albumin mRNA using 35S-labeled antisense RNA probe. Albumin transcripts are localized in periductular and oval cells. Hematoxylin and eosin stain, × 120.
Expression of Liver-Enriched Transcription Factors
In agreement with the in situ hybridization data, Northern blot analysis (Fig. 3) indicated that 5–6 weeks of feeding a copper-deficient diet caused a dramatic increase in the levels of the mRNA encoding albumin, a liver-specific gene product. Albumin mRNA was not detected in the pancreas of control animals and in those killed after 4 weeks of copper depletion. The size of albumin transcript was similar to that of the albumin mRNA found in the rat liver (Fig. 3A). To ascertain if the expression of albumin gene in the pancreas of copper-depleted rats is dependent upon on the transcriptional activation of liver-enriched transcription factors, we analyzed the expression of mRNAs encoding C/EBPα, β, and δ, and HNF-3/3 by Northern blotting. Pancreas from the 4-week experimental group showed faint signal for C/EBPα mRNA and at this time period there was no albumin expression. Pancreas from rats on the copper-deficient diet for 5 and 6 weeks showed increased levels of C/EBPα and δ mRNAs (Figs. 3 and 4). C/EBP/3 mRNA level was also increased at 5 weeks, but unlike the C/EBPα and δ, the C/EBPβ mRNA level decreased at 6 weeks. In the control pancreas, no C/EBP mRNAs were detected. HNF-3/3 expression in the pancreas of control rats, and from 5- and 6-week copper-deficient animals, was comparable (Fig. 3B). High levels of expression of C/EBPα and β, and HNF-3β mRNAs were seen in the livers, whereas the C/EBPδ expression was very low.
FIG. 3.
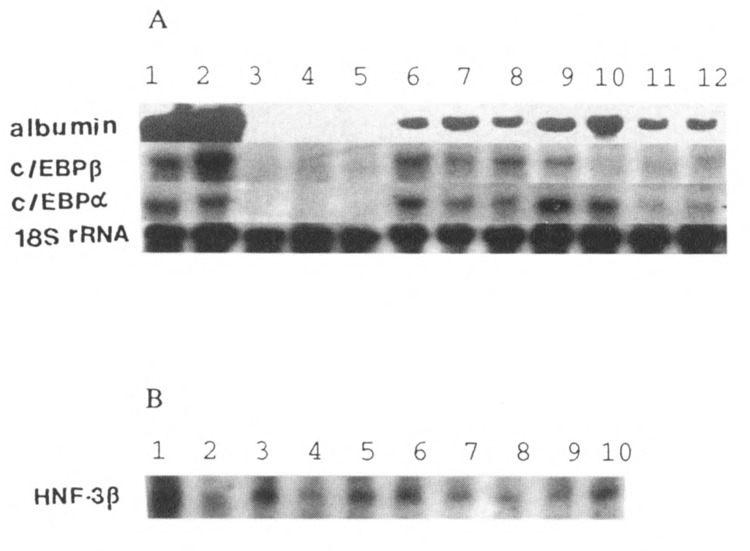
Northern analysis of total RNA isolated from pancreas of copper-deficient rats for mRNAs of albumin, and liver transcription factors C/EBPα and β, and HNF-3β. Fifteen micrograms of total RNA was hybridized with 32P-labeled cDNAs. (A) Lanes 1 and 2: liver; lanes 3–5: normal pancreas; lanes 6–9 and 10–12: pancreas from 5- and 6-week copper-deficient rats, respectively. (The sizes of cDNAs are: albumin, 2.3 kb; C/EBPβ, 1.5 kb; C/EBPα, 2.7 kb.) (B) HNF-3β mRNA. Lanes 1 and 2: liver; lanes 3–5: normal pancreas; lanes 6–8 and 9 and 10: pancreas from 5- and 6-week copper-deficient rats, respectively. (Size of HNF-3β is 2.1 kb.) The blot from albumin was stripped and rehybridized with 18S rRNA probe to show that similar quantities of RNA is loaded in each lane.
FIG. 4.
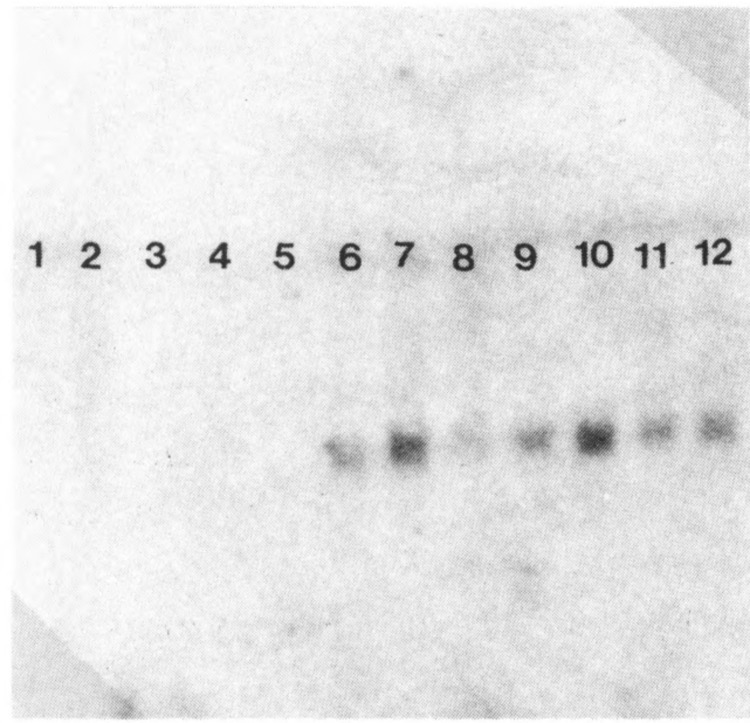
Expression of C/EBPa mRNA in the pancreas of control and copper-deficient rats. Lanes 1 and 2: liver; lanes 3–5: control pancreas; lanes 6–9 and 10–12: pancreas from 5- and 6-week copper-deficient diet, respectively. C/EBPα cDNA size is 1 kb.
SCE in Pancreatic Ovals Cells
Immunohistochemical analysis revealed the presence of SCF in clusters of oval cells scattered throughout the atrophic (acinar cell-depleted) pancreatic parenchyma of rats maintained on copper-deficient diet for 6 weeks (Fig. 5). Of interest is the remarkable intensity of SCF expression in oval cells surrounding the islets of Langerhans (i.e., in the peri-insular region) (Fig. 5). It is worth recaling that these peri-insular regions of islets of Langerhans exhibit a greater propensity for the differentiation of hepatocytes (34). The proliferation of oval cells in this region that were intensively positive for SCF is consistent with the later development of hepatocytes in the copper-deficient pancreas. Additional studies are essential to obtain data on the coexpression of SCF and its receptor c-kit at various intervals during copper depletion and repletion model of hepatocyte differentiation.
FIG. 5.
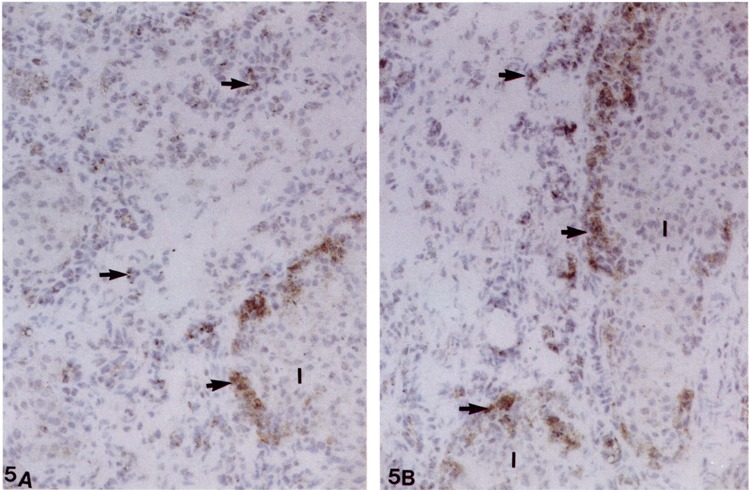
(A, B) Frozen section of pancreas from a rat maintained on a copper-deficient diet stained for stem cell factor by immunoperoxidase method. Oval cells (arrows) in the interstitium and around the islets of Langerhans (I) are strongly stained. Counterstained with hematoxylin.
DISCUSSION
Liver-enriched transcriptional factors, C/EBPα, β, δ, and HNF-1, -3, and -4, although detected in many tissues and organs, are abundantly expressed in the liver and play a crucial role in the transcription of many liver-specific genes and possibly in the liver organogenesis (4,7,18,22). In mammals, the liver develops from endoderm of the embryonic foregut. The foregut epithelial cells that are committed toward the liver cell lineage proliferate and migrate into the adjacent septum transversum and form liver. The intrahepatic bile ducts develop from hepatocytes (12). During development, the determination of stem cell fate, migration, and tissue morphogenesis are dependent on the interaction between extracellular matrix and cells (20). Differentiation of primitive gut endodermal cells into hepatocyte lineage has been shown to be coincident with interaction with the mesenchyme (4). Even after embryonic stage, the normal milieu appears necessary to maintain the differentiated state and has been well demonstrated under in vitro conditions. Hepatocytes lose phenotypic properties when grown on nonphysiological substratum (3), but when grown on basement membrane, they retain normal morphology and functions, possibly due to cell-matrix interactions resulting in the activation of specific transcription factors (24).
There is overwhelming evidence to support the existence of a stem cell compartment in the adult liver (39), but to date, various attempts to isolate, propagate, and induce differentiation of such stem cells into functionally viable hepatocytes have met with little success. Interestingly, in the adult rat liver, hepatocytes appear to develop from oval cells (39). Evidence for the participation of oval cells in hepatocyte differentiation is derived from the observation that these cells proliferate markedly under different experimental conditions, and they contain different transcription factors and hepatocyte-specific gene products (11,19,36). The observations of Nagy et al. (25) suggested that HNF-4 may be responsible for the final commitment of oval cells to differentiate into hepatocytes. Although the transition of oval cells in liver to hepatocytes has been suggested, the presence of preexisting mature hepatocytes in liver makes it difficult to unequivocally establish these lineage predictions. The rat pancreatic hepatocyte model we developed and optimized offers several advantages for the search for liver stem cell over the hepatocyte oval cell system (35). First, there are no preexisting hepatocytes within the pancreas to confound the observation of lineage transition and differentiation. Second, the copper deficiency model offers a window of opportunity to identify progenitor (stem) cells prior to the emergence of differentiated mature hepatocytes. The liver-enriched transcription factors and the earliest markers of liver specific transcription may be localized in these progenitor cells by in situ hybridization and by other methods. Third, the use of retrovirally driven β-galactosidase, as a marker gene for lineage analysis, should allow the fractionation of progenitor cells, and also permit analysis of their fate under in vivo and in vitro conditions. Finally, the ductal system of the pancreas is an effective conduit to transfect various genes and introduce growth factors at various intervals during copper depletion, and study their influence on the lineage and differentiation of the progenitor cells.
The results of the present study show that several liver-enriched transcriptional factors are expressed in the pancreas much earlier than the emergence of morphologically identifiable hepatocytes. The increased levels of mRNAs of these factors coincide with the proliferation of oval and ductular cells. The expression of C/EBPA mRNA, which was detectable in the pancreas after 4 weeks of copper deficiency, appears to precede the expression of albumin mRNA, indicating that this transcription factor may be necessary for the expression of albumin gene in oval cells. C/EBPβ and δ mRNAs appeared at the same time as albumin mRNA. The levels of C/EBPβ mRNA were high at 5 weeks, followed by a decrease at 6 weeks. C/EBPδ expression was very high in pancreas at 5 and 6 weeks and was not detectable in the normal liver. This is not surprising because it is known that C/EBPδ expression in the adult liver has been reported to be very low (1). The level of HNF-3β mRNA expression was comparable in the control pancreas and pancreas from 5- and 6-week copper-deficient animals. HNF-3/3 has been shown to be expressed in islets of Langerhans and proposed to participate in the expression of insulin gene (27). Nevertheless, in situ hybridization studies are necessary to discern the nature of cells that express these various transcrption factors during copper deficiency induced oval cell proliferation in the rat pancreas.
Although preliminary in nature, our observations on the expression of SCF in oval cells distributed randomly in the pancreatic parenchyma and more so in the proliferating oval cells in the peri-insular regions are of considerable interest. Pancreatic oval cell proliferation and their differentiation into hepatocytes may be dependent on the expression of SCF. In the rat liver, it has been shown that expansion of oval cell compartment in partial hepatectomy and 2-acetylaminoflourene model is associated with the expression of SCF and its receptor, c-kit (14).
In summary, these and other studies suggest that SCF and liver-enriched transcription factors play a potentially critical role in oval cell proliferation and in influencing these cells to make a commitment toward hepatocyte differentiation in the adult pancreas. During the past several years, we have presented ample evidence to show that these committed cells complete the hepatocyte differentiation program within the pancreatic melieu and these pancreatic hepatocytes respond to xenobiotics and to partial hepatectomy. A recent claim that this hepatocyte differentiation process is not completed in the pancreas is totally misleading and ignores a well-illustrated and well-documented body of evidence (8).
ACKNOWLEDGEMENTS
This research was supported by NIH grant DK37958, and the Adrian Mayer, M.D. Cancer Research Fund. We thank Dr. Robert Costa, University of Illinois School of Medicine, Chicago, for the cDNAs of liver-enriched transcrption factors used in this study. We thank Sujatha Pulikuri and V. Subbarao for excellent technical assistance.
REFERENCES
- 1. Alam T.; Ra An M.; Papaconstantinou J. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 267:5021–5024; 1992. [PubMed] [Google Scholar]
- 2. Alberts B.; Bray D.; Lewis J.; Raff M.; Roberts K.; Watson J. D. Molecular biology of the cell. New York: Garland Publishing Co.; 1989. [Google Scholar]
- 3. Bissell D. M.; Arenson D. M.; Maher J. J.; Roll F. J. Support of cultured hepatocytes by a lamininrich gel. Evidence for a functionally significant subendothelial matrix in normal rat liver. J. Clin. Invest. 79:801–812; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cascio S. C.; Zaret K. S. Hepatocyte differentiation initiates during endodermal-mesenchymal interactions prior to liver formation. Development 113:217–225; 1991. [DOI] [PubMed] [Google Scholar]
- 5. Cereghini S.; Otto M.; Power S.; Maury M. Expression patterns of vHNFl and HNF1 homeoproteins in early postimplantation embryos suggest distinct and sequential developmental roles. Development 116:783–797; 1992. [DOI] [PubMed] [Google Scholar]
- 6. Chomczynski P.; Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanatephenol-chloroform extraction. Ann. Biochem. 162:156–159; 1987. [DOI] [PubMed] [Google Scholar]
- 7. Clevidence D. E.; Overdier D. G.; Tao W.; Qian X.; Pani L.; Lai E.; Costa R. H. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc. Natl. Acad. Sci. USA 90:3948–3952; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dabeva M. D.; Hurston E.; Sharitz D. A. Transcription factor and liver-specific mRNA expression in facultative epithelial progenitor cells of liver and pancreas. Am. J. Pathol. 147:1633–1648; 1995. [PMC free article] [PubMed] [Google Scholar]
- 9. De Simone V.; Cortese R. Transcription factors and liver-specific genes. Biochim. Biophys. Acta 1132:119–126; 1992. [DOI] [PubMed] [Google Scholar]
- 10. Duncan S. A.; Manova K.; Chen W. S.; Hoodless P.; Weinstein D. C.; Bachvarova R. F.; Darnell J. E. Jr. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. USA 91:7598–7602; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evarts R. P.; Nagy P.; Marsden E.; Thorgeirsson S. S. A precursor-product relationship exists between oval cells and hepatocytes in rat liver. Carcinogenesis 8:1737–1740; 1987. [DOI] [PubMed] [Google Scholar]
- 12. Eyken P. V.; Sciot R.; Desmet V. Intrahepatic bile duct development in the rat: A cytokeratin-immunohistochemical study. Lab. Invest. 59:52–59; 1988. [PubMed] [Google Scholar]
- 13. Fujio K.; Hu Z.; Evarts R. P.; Marsden E. R.; Niu C.-H.; Thorgeirsson S. S. Coexpression of stem cell factor and c-kit in embryonic and adult liver. Exp. Cell Res. 224:243–250; 1996. [DOI] [PubMed] [Google Scholar]
- 14. Fujio K.; Evarts R. P.; Hu Z.; Marsden E. R.; Thorgeirsson S. S. Expression of stem cell factor and its receptor, c-kit, during liver regeneration from putative stem cells in adult rat. Lab. Invest. 70:511–516; 1994. [PubMed] [Google Scholar]
- 15. Hillis D.; Dixon T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 66:411–453; 1991. [DOI] [PubMed] [Google Scholar]
- 16. Johnson P. F. Transcriptional activators in hepatocytes. Cell Growth Differ. 1:47–51; 1990. [PubMed] [Google Scholar]
- 17. Kishimoto S.; Iwamoto S.; Masutani S.; Yamamoto R.; Jo T.; Saji F.; Terada N.; Sasaki Y.; Imaoka S.; Sugiyama T. Apoptosis of acinar cells in the pancreas of rats fed on a copper-depleted diet. Exp. Toxicol. Pathol. 45:489–495; 1994. [DOI] [PubMed] [Google Scholar]
- 18. Lai E.; Darnell J. E. Jr. Transcriptional control in hepatocytes: A window on development. Trends Biochem. Sci. 16:427–430; 1991. [DOI] [PubMed] [Google Scholar]
- 19. Lemire J. M.; Shiojiri N.; Fausto N. Oval cell proliferation and the origin of small hepatocytes in liver injury induced by D-galactosamine. Am. J. Pathol. 139:535–552; 1991. [PMC free article] [PubMed] [Google Scholar]
- 20. McDonald J. A. Matrix regulation of cell shape and gene expression. Curr. Opin. Cell Biol. 1:995–999; 1989. [DOI] [PubMed] [Google Scholar]
- 21. Mischoulon D.; Rana B.; Bucher N. L. R.; Farmer S. R. Growth-dependent inhibition of CCAAT enhancer-binding protein (C/EBPa) gene expression during hepatocyte proliferation in the renerating liver and in culture. Mol. Cell. Biol. 12:2553–2560; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miura N.; Iwai K.; Miyamoto I. Immunological characterization of hepatocyte nuclear factor 1 protein: Appearance of hepatocyte nuclear factor 1 protein in developing mouse embryos. Eur. J. Cell Biol. 60:376–382; 1993. [PubMed] [Google Scholar]
- 23. Monaghan A. P.; Kaestner K. H.; Grau E.; Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3α, β, and γ genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development 119:567–578; 1993. [DOI] [PubMed] [Google Scholar]
- 24. Nagaki M.; Shidoji Y.; Yamada Y.; Sugiyama A.; Tanaka M.; Akaike T.; Ohnishi H.; Moriwaki H.; Muto Y. Regulation of hepatic genes and liver transcription factors in rat hepatocytes by extracellular matrix. Biochem. Biophys. Res. Commun. 210:38–43; 1995. [DOI] [PubMed] [Google Scholar]
- 25. Nagy P.; Bisgaard H. C.; Thorgeirsson S. S. Expression of hepatic transcription factors during liver development and oval cell differentiation. J. Cell Biol. 126:223–233; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pawson T.; Bernstein A. Receptor tyrosine kinases: Genetic evidence for their role in Drosophila and mouse development. Trends Genet. 6:350–356; 1990. [DOI] [PubMed] [Google Scholar]
- 27. Philippe J.; Morel C; Prezioso V. R. Glucagon gene expression is negatively regulated by hepatocyte nuclear factor 3. Mol. Cell. Biol. 14:3514–3523; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rao M. S.; Subbarao V.; Yeldandi A. V.; Reddy J. K. Pancreatic acinar cell regeneration following copper deficiency-induced pancreatic necrosis. Int. J. Pancreatol. 2:71–85; 1987. [DOI] [PubMed] [Google Scholar]
- 29. Rao M. S.; Dwivedi R. S.; Yeldandi A. V.; Subbarao V.; Tan X.; Usman M. I.; Thangada S.; Nemali M. R.; Kumar S.; Scarpelli D. G.; Reddy J. K. Role of periductal and ductular epithelial cells of the adult rat pancreas in pancreatic hepatocyte lineage. Am. J. Pathol. 134:1069–1086; 1989. [PMC free article] [PubMed] [Google Scholar]
- 30. Rao M. S.; Yeldandi A. V.; Reddy J. K. Stem cell potential of ductular and periductular cells in the adult rat pancreas. Cell Differ. Dev. 29:155–163; 1990. [DOI] [PubMed] [Google Scholar]
- 31. Rao M. S.; Yeldandi A. V.; Subbarao V.; Reddy J. K. Role of apoptosis in copper deficiency-induced pancreatic involution in the rat. Am. J. Pathol. 142:1952–1957; 1993. [PMC free article] [PubMed] [Google Scholar]
- 32. Rao M. S.; Reddy J. K. Bipotential stem cells in the rat pancreas. Int. J. Pancreatol. 16:221–223; 1994. [Google Scholar]
- 33. Rao M. S.; Reddy J. K. Hepatic transdifferentiation in the pancreas. Semin. Cell Biol. 6:151–156; 1995. [DOI] [PubMed] [Google Scholar]
- 34. Reddy J. K.; Rao M. S.; Qureshi S. A.; Reddy M. K.; Scarpelli D. G.; Lalwani N. D. Induction and origin of hepatocytes in rat pancreas. J. Cell Biol. 98:2082–2090; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reddy J. K.; Rao M. S.; Yeldandi A. V.; Tan X.; Dwivedi R. S. Pancreatic hepatocytes: An in vivo model for cell lineage in pancreas of adult rat. Dig. Dis. Sci. 36:502–509; 1991. [DOI] [PubMed] [Google Scholar]
- 36. Sell S. Liver stem cells. Modern Pathol. 7:105–112; 1994. [PubMed] [Google Scholar]
- 37. Simmons D. L.; Lalley P. A.; Kasper C. B. Chromosomal assignments of genes coding for components of the mixed-function oxidase system in mice. J. Biol. Chem. 260:515–521; 1985. [PubMed] [Google Scholar]
- 38. Smith P. A.; Sunter J. P.; Case R. M. Progressive atrophy of pancreatic acinar tissue in rats fed a copper-deficient diet supplemented with D-penicil-lamine or triethylene tetramine: Morphological and physiological studies. Digestion 23:16–30; 1982. [DOI] [PubMed] [Google Scholar]
- 39. Thorgeirsson S. S. Hepatic stem cells. Am. J. Pathol. 142:1331–1333; 1993. [PMC free article] [PubMed] [Google Scholar]
- 40. Umek R. M.; Friedman A. D.; McKnight S. L. CCAAT-enhancer binding protein: A component of a differentiation switch. Science 251:288–292; 1991. [DOI] [PubMed] [Google Scholar]


