Abstract
A n in vitro differentiation system utilizing retinoic acid (RA) treatment of pluripotent murine P19 embryonal carcinoma (EC) cells, which can be induced to differentiate into various cell types, was optimized for maximal induction of cel type I collagen (Collal) gene expression. Differentiation was associated with apoptotic death of the majority of cells, indicating that this in vitro system faithfully mimics the in vivo differentiation process. Collal mRNA became detectable by RNase protection assay after 3 days of RA treatment and, after 6 days, reached a level comparable to that in NIH 3T3 fibroblasts. A fte r induction of differentiation the Collal gene remained transcriptionally active for extended periods of time even in the absence of RA. A minigene version of the murine Collal gene was constructed that contains all of the so far known Collal regulatory elements. This construct exhibited the correct expression pattern in stable transfection experiments: it was expressed in fibroblasts, but not in undifferentiated P19 EC cells, and it was transcriptionally activated after induction of differentiation. This experimental system should be a useful tool for dissecting the molecular mechanisms involved in the developmental activation and stageand tissue-specific expression of the murine Collal gene.
Keywords: Murine α1 type I collagen, Collal, In vitro differentiation, P19 EC cells, Retinoic acid
TYPE I collagen, the most abundant of the collagens, is expressed in a variety of cell types, at various stages of development, and under various physiological conditions, and its regulation is accordingly complex [for reviews, see (6,8,26,38,43)]. Numerous studies have addressed the molecular mechanisms involved in the regulation of the genes coding for the α1 and α2 chains of type I collagen in various species, including human, rat, mouse, and chicken. However, despite considerable efforts, the details of type I collagen gene regulation remain elusive. It has generally been reported that the α1 and α2 promoters confer basic tissue-specific expression to reporter gene or minigene constructs in transfection experiments and transgenic animals (31,34,37,40). In addition, cis-regulatory elements have been identified that mediate the effect of various modulators of type I collagen expression (15,32,39) or are required for high levels of expression in different collagen-producing cell types (3,4,33,34). However, the functional significance of additional regulatory elements, particularly in the first introns of the human and the murine al genes (5,17,20,24,31,39,41), but also other putative elements located within the coding or the 3′-flanking sequences (8), remains unclear and controversial and warrants further studies. Moreover, to our knowledge, none of the minigene or reporter gene constructs reported to date showed position-independent, copy number-dependent expression in transfection experiments or transgenic animals. It is conceivable that DNase-hypersensitive sites in the distal 5′-and 3′-flanking regions of the human (2) and murine (M. Breindl, unpublished observation) α1 genes contain elements with insulating or domain-opening functions, which are necessary for position independence and warrant further analysis. Furthermore, in addition to interactions between proximal and distal cis-regulatory elements and trans-acting factors, chromatin structure and DNA methylation constitute important mechanisms in the developmental activation and tissue-specific expression of mammalian genes. Whereas several previous studies have addressed the developmentally regulated changes in the chromatin structure and methylation pattern of the murine α1 type I collagen (Collal) promoter (7,9,29,30), the molecular mechanisms involved in mediating these changes are virtually unknown and require additional experimentation.
The most commonly used experimental approach to the identification and functional analysis of regulatory elements is to molecularly clone them into reporter or minigene constructs and introduce the constructs into tissue culture cells and, ultimately, into transgenic animals. However, transient or stable transfection assays frequently do not reveal the function of certain regulatory elements, specifically when they are not left in their normal context and spatial arrangement, and the production of transgenic animals is expensive and time consuming. Moreover, it is difficult to study the molecular mechanisms involved in developmental changes in chromatin structure and DNA methylation in vivo because of the limited availability of cellular material needed for biochemical analyses and, more importantly, because most tissues are mixtures of collagen-producing and nonproducing cells, which may exhibit heterogeneity in their chromatin structure and methylation pattern. We have therefore developed an alternative experimental approach to study the developmental activation and tissue-specific regulation of the Collal gene using in vitro differentiation of pluripotent P19 embryonal carcinoma (EC) cells and a murine Collal minigene construct that maintains all so far known Collal regulatory sequences in their normal spatial orientation. We show here that P19 EC cells can be induced by retinoic acid (RA) to differentiate into cells that transcriptionally activate the endogenous Collal gene and produce high levels of Collal mRNA for extended periods of time, and that the minigene shows the same differentiation-specific transcriptional activation as the endogenous gene.
MATERIALS AND METHODS
Cell Lines and In Vitro Differentiation
P19 EC cells (kindly provided by E. Adamson) and NIH 3T3 fibroblasts were grown as previously described (30). For in vitro differentiation, P19 EC cells were seeded into 100-mm plastic tissue culture plates at a density of 0.5 × 106 cells/plate and RA was immediately added at a concentration of 5 × 10–7 M. Plates were incubated for 48 h, at which time the medium was removed and fresh medium with RA was added. Medium was then changed every 24 h until all dead cells had been removed. After 4–5 days, the differentiated cells remaining on the plate were passed and grown at subconfluent density in the presence or absence of RA for further analysis.
Immunofluorescent Staining
NIH 3T3 fibroblasts and P19 cells that had been differentiated with RA for 6 days were fixed in 95% ethanol containing 1% acetic acid at –20°C for 20 min and then treated with 150 mM NH4C1 for 15 min. The cells were permeabilized with 0.2% Triton X-100 and blocked in a solution of 1% normal goat serum, 1% bovine serum albumin, 1% gelatin from cold water fish skin, and 0.05 M glycine in PBS. The primary antiserum was a rabbit anti-mouse type I collagen polyclonal antiserum (purchased from Chemicon International) and the secondary antibody was fluorescein-conjugated goat anti-rabbit IgG, F(ab′)2 fragment (Jackson ImmunoResearch Laboratories).
Stable Transfections
NIH 3T3 fibroblasts were transfected using standard calcium phosphate coprecipitation as described (30). Undifferentiated P19 EC cells were transfected using DOTAP (Boehringer). The Collal minigene plasmid was cotransfected in a 5:1 molar ratio with a plasmid carrying a neomycin resistance gene, and neomycin-resistant colonies were selected in the presence of 500 μg/ml geneticin (Gibco BRL) in the growth medium as previously described (9). To compensate for possible position effects, all experiments were performed with populations of stably transfected clones and only populations containing > 20 clones were included in the analyses. Pools of stably transfected clones were expanded and used for further analyses.
Preparation ofRNA and RNase Protection Assays
RNA was prepared by a modified acid phenol procedure (11), and the concentration and structural integrity of all samples were determined by agarose gel electrophoresis and ethidium bromide staining. Two different Collal-specific probes were used in RNase protection assays. In initial experiments (data shown in Fig. 3) a probe was used that protects 112 bp from the first exon of the Collal gene, as previously described (9). To analyze expression of the transfected Collal mini-gene we constructed a probe that can discriminate between transcripts from the endogenous Collal gene and the minigene as follows: RNA from NIH 3T3 fibroblasts was reverse transcribed with AMV reverse transcriptase (Promega) using a primer that spans the junction of exons 9 and 10 of the Collal gene (primer 1). The resulting cDNA was then amplified by PCR with Taq polymerase (Perkin Elmer) using a primer complementary to exon 5 (primer 2) and a second, nested primer also spanning the junction of exons 9 and 10 (primer 3) of the Collal gene. The latter two primers contained recognition sequences for restriction endonucleases that were subsequently used to molecularly clone the PCR product into the plasmid vector pGEM-4Z (Promega), which contains Sp6 and T7 bacteriophage promoters to produce the plasmid COLEX5-10. For probe preparation plasmid COLEX5-10 was linearized with Nhe I and transcribed with Sp6 polymerase. The resulting transcripts are 348 nucleotides long and protect 288 nucleotides of transcripts from the endogenous Collal gene and 124 nucleotides of transcripts from the minigene. In these latter experiments (data shown in Fig. 5) a GAPDH probe was included that was prepared using plasmid pTRI-GAPDH-Mouse (Ambion). This probe protects 316 nucleotides of GAPDH transcripts and was used as a loading control. All enzymatic reactions were performed following the recommendations of the suppliers and standard recombinant DNA procedures (36). The protected fragments were separated on a 6% acrylamide, 7 M urea denaturing gel and visualized and quantitated using a phosphoimager (Molecular Dynamics).
FIG. 5.
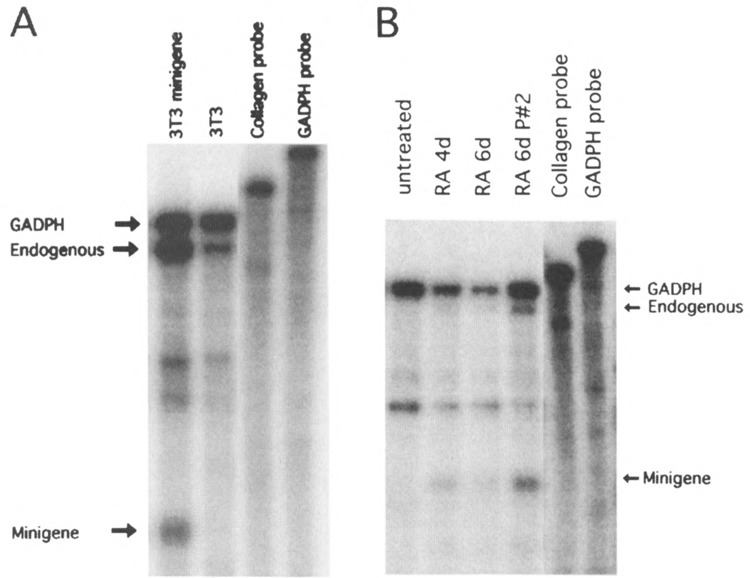
RA-induced differentiation of P19 EC cells results in transcriptional activation of the endogenous Collal gene and the minigene. Total RNA was isolated from P19 EC and NIH 3T3 fibroblasts and analyzed by RNase protection assay using a probe that differentiates between the endogenous and minigene transcripts and a GAPDH probe as loading control (see the Materials and Methods section). The position of the GAPDH signal and the Collal endogenous and minigene signals are indicated by arrows. The integrity of the probes used in these assays is shown in the two rightmost lanes. (A) Minigene-transfected and untransfected NIH 3T3 fibroblasts. (B) Minigene-transfected, undifferentiated, and RA differentiated P19 cells analyzed 4 and 6 days after induction of differentiation as indicated; the cells analyzed in the 6d P#2 lane had been passed twice after induction of differentiation and show a relative higher expression of both the endogenous Collal gene and the minigene.
FIG. 3.
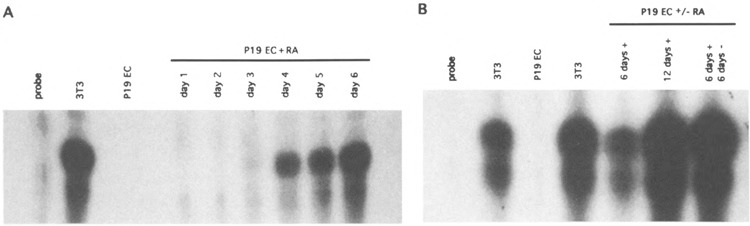
RA-induced differentiation results in transcriptional activation of the murine Collal gene. Total RNA was isolated from 3T3 cells, undifferentiated P19 EC cells, and P19 EC cells treated with RA for the indicated lengths of time in days and analyzed by RNase protection assay using a probe that protects 112 nucleotides of Collal mRNA as described (9). (A) P19 EC cells were differentiated with RA for 1–6 days as described in the Materials and Methods section. On the original autoradiogram Collal mRNA was first detectable after 3 days of RA treatment. (B) P19 EC cells were differentiated with RA for 6 days and grown for 6 more days in the absence or presence of RA as indicated and analyzed by RNase protection assay. The concentration and integrity of the RNA preparations were determined by agarose gel electrophoresis (not shown), and equal amounts of RNA were assayed in each lane. With this probe we usually see in RNase protection assays a main band at 112 nucleotides and a lower band or smear, the intensity of which varies in different experiments (9,29,30). The origin of this lower band has not been determined, but it is assumed to result from inaccurate initiation of transcription and/or technical artefacts such as breathing of the hybrid molecules or unspecific degradation during RNase digestion.
DNA Sequences
The DNA sequences referred to in this article have been published (19) or are accessible in the EMBL (accession number X54876) or Gene Bank data libraries (accession numbers U38307, U38544, and U50767). The nucleotide sequences of the primers were as follows:
primer 1: 5′-GCTTCCCCATCATCTCC-3′
primer 2: 5′-TCTAAGCTTACTTCCTGGTCCTCCTG-3′
primer 3: 5′-CTCCTGCAGCCCCATCATCTCCATTC-3′.
RESULTS
Optimization of an In Vitro Differentiation System for Maximal Collal Gene Transcription
Previous studies have shown that P19 EC cells can be induced to differentiate into a variety of different cell types (16,22,23,35). The differentiation protocols used in those studies involved the aggregation of the cells into embryoid bodies by culture in bacterial-grade plastic dishes to which the cells do not adhere prior to differentiation. In the presence of dimethyl sulfoxide (DMSO) aggregates of P19 cells formed cardiac and skeletal muscle, whereas in the presence of RA the cells differentiated into neuronal, glial, and fibroblast-like cells. However, it was not previously known whether the Collal gene is transcriptionally activated under these latter conditions. We therefore first optimized the differentiation process for our purpose (i.e., maximal induction of Collal gene transcription) by varying the concentrations of DMSO and RA and by differentiating the cells with and without prior aggregation. Expression of Collal mRNA was determined by RNase protection assay and immunostaining with a type I collagen-specific antiserum. The results of these initial experiments showed that maximal Collal expression was achieved when the cells were treated with 5 × 10–7 M RA without prior aggregation, and when cells were kept at subconfluent cell densities throughout the differentiation process (data not shown). In a typical experiment, P19 EC cells were seeded onto 100-mm tissue culture plates at a density of 0.5 × 106 cells per plate and RA was added. After 2–3 days the majority of the cells (between 40% and 70% in different experiments)
The Collal Gene Is Transcriptionally Activated and Stably Expressed at High Levels in the Differentiated Cells
We next monitored the transcriptional activation of the Collal gene throughout the differentiation. RNA was isolated from the living, attached portion of the RA-treated cells at different times after induction of differentiation, and Collal transcription was determined by RNase protection assay using a radiolabeled riboprobe that protects 112 nucleotides from the first exon of the Collal gene. Figure 3 shows that undifferentiated P19 EC cells did not express Collal mRNA, confirming previous results (9,13,29). However, after 3 days of RA treatment Collal transcripts became detectable. The amount of Collal mRNA increased with continued RA treatment and after 6 days reached a level comparable to that in NIH 3T3 fibroblasts (Fig. 3A). Similarly, the amount of type I collagen stainable with a type I-specific anti-serum was comparable to that in 3T3 fibroblasts (Fig. 1E, F). Additional experiments showed that the differentiated cells continued to express Collal mRNA at levels comparable to that in 3T3 cells for at least 2 weeks, even when RA treatment was discontinued after 6 days (Fig. 3B), although we did observe variation in the levels of Collal mRNA between experiments (see the Discussion section). These results show that the endogenous Collal gene is stably activated and expressed at high levels in the in vitro differentiated P19 EC cells.
A Collal Minigene Exhibits Correct Transcriptional Activation in Differentiating P19 Cells
The Collal minigene construct used in these studies is schematically shown in Fig. 4, and the design of this and several other Collal minigene versions containing different combinations of regulatory elements will be described in detail elsewhere (manuscript in preparation). Briefly, the minigene contains 4 kb of 5′-flanking sequence, exons 1–6 from the 5′ end, exons 49–52 from the 3′ end, and 3 kb of 3′-flanking sequence. The role of the different regulatory elements in the 5′-flanking region, first intron, and 3′-flanking region of the Collal gene has been discussed in several recent reviews [(6,8,26,38,43); manuscript submitted for publication]. The 5′ and 3′ portions of the minigene were joined at intron sequences (introns 6 and 48) to maintain correct splicing signals and translational reading frame. Unique restriction sites for Not 1 and Srf I were added at the 5′ and 3′ ends of the minigene, respectively, to allow cloning into the minigene additional regulatory elements (putative remote regulatory elements, nuclear matrix binding sites). A similar minigene construct of the human Collal gene has been described (24).
FIG. 4.
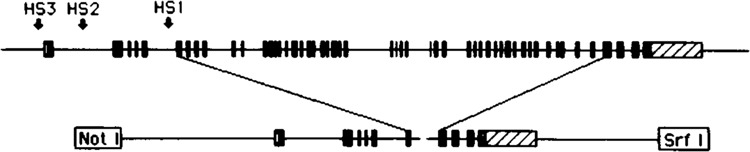
Schematic representation of the murine Collal minigene. The minigene used in these experiments contains 4 kb of 5′-flanking sequence, exons 1–6 from the 5′ end, exons 49–52 from the 3′ end, and 3 kb of 3′-flanking sequence. The 5′ and 3′ portions of the minigene were joined at intron sequences (a unique SnaB1 site in intron 6 and a blunted HindIII site in intron 48) to maintain correct splicing signals and translational reading frame. Unique restriction sites for Not 1 and Srf I were added at the 5′ and 3′ ends of the minigene, respectively, to allow subsequent addition of additional, putative remote regulatory elements as they are identified.
To determine whether the Collal minigene showed the same transcriptional repression in undifferentiated P19 cells and differentiation-dependent transcriptional activation as the endogenous Collal gene, we stably transfected the Collal minigene into undifferentiated P19 EC cells and analyzed its expression before and after in vitro differentiation. As a control, NIH 3T3 fibroblasts were transfected with the same minigene construct. Collal mRNA expression was analyzed by RNase protection assay using a probe that differentiated between transcripts from the endogenous and the minigene (see the Materials and Methods section). We also included a murine GAPDH probe in these assays as an RNA loading control. The results of a representative experiment are shown in Fig. 5. To compensate for possible position effects, all experiments were performed with populations of stably transfected clones and only populations containing > 20 clones were included in the analyses. Upon stable transfection, the Collal minigene exhibited the expected expression pattern: it was expressed at relatively high levels in fibroblasts (Fig. 5A), but at undetectable levels in undifferentiated P19 EC cells (Fig. 5B). Moreover, when the transfected P19 cells were induced to differentiate, the minigene showed the same time course of transcriptional activation as the endogenous gene: transcripts became detectable between 4 and 6 days after the induction of differentiation (Fig. 5B). Similar results were obtained with five independently transfected cell populations. In both 3T3 fibroblasts and differentiated P19 cells the level of minigene expression was considerably lower when normalized to copy numbers than that of the endogenous gene, presumably due to position effects (see the Discussion section). These results indicate that the minigene has the regulatory elements necessary for the developmental activation and cell-specific expression of the Collal promoter but that it lacks additional elements that are required for high level, position-independent expression of the Collal gene.
DISCUSSION
Embryonal carcinoma cells are an excellent model to study many biochemical and cell and molecular biological aspects of cell differentiation because they can be grown in large amounts in tissue culture in an undifferentiated state and can be induced to differentiate into various cell types (10,16,21–23,35,42,44). P19 EC cells have previously been shown to differentiate in the presence of RA into fibroblast-like cells (23,35), and we have used a modified differentiation protocol for these cells that allows us to analyze molecular mechanisms involved in the transcriptional derepression of the murine Collal gene. Our results show that after induction of differentiation a large fraction of the cells undergo apoptotic cell death, as characterized by cell shrinking and rounding (Fig. 1) and endogenous internucleosomal DNA degradation (Fig. 2). Because apoptotic cell death is a common event during normal embryonic development (25,27), this indicates that the in vitro differentiation faithfully mimics the in vivo differentiation process. This observation is in contrast to a previous report, which indicated that RA does not induce cell death in P19 EC cell cultures at the concentrations used (35). The reason for this discrepancy is not known but presumably reflects different properties of specific subpopulations of P19 EC cells or variations between the differentiation protocols used. More recent studies by others have confirmed our observation and have shown that induction of differentiation in embryonic cells (12), including the P19 EC cells used in this study (14), is frequently associated with apoptosis.
Under optimal conditions RA-induced differentiation of P19 EC cells without prior aggregation of the cells into embryoid bodies is sufficient to stably activate transcription of Collal mRNA, and the differentiated cells continue to transcribe the gene at levels comparable to, or even in excess of, that of murine 3T3 fibroblasts for extended periods of time (Fig. 3). However, we have observed some variation in the levels of Collal mRNA expressed in the differentiated P19 cells between experiments. Whereas early passage P19 cells (stored in liquid nitrogen) reproducibly activate Collal mRNA under the experimental conditions described here to levels comparable to those shown in Figs. 3 and 5, it seems that upon continuous and prolonged passage in culture P19 EC cells lose the ability to differentiate into type I collagen-producing cells but they can still be induced to differentiate into other cell types. The reason for this is not clear but may reflect changes in the DNA methylation pattern of the Collal gene or other genes that need to be expressed during the differentiation process. Such changes in the methylation pattern of nonessential genes have actually been observed to occur in many cell lines in culture (1). Alternatively, some so far unidentified serum factor may influence the differentiation process by promoting or inhibiting the growth of collagen-producing cells. The identity of such a putative regulatory factor is not known. Preliminary experiments have shown that addition of the fibrogenic cytokines TGF-β, bFGF, PDGF, or TNFα to the culture medium of late passage P19 cells during differentiation has no effect on the level of Collal mRNA in this system (K. Hall, unpublished observation).
The minigene construct used in these experiments maintains in a correct spatial orientation all of the Collal regulatory elements known so far (6,8,26,38,43). In addition to the well-studied elements in the promoter, 5′-flanking region, and first intron, it also contains intragenic sequences through exon 6, including the fifth intron, which harbors a constitutive DNase I-hypersensitive site of unknown function (7), as well as 3 kb of 3′-flanking sequence, which has recently been found to have a moderate stimulatory effect on the Collal promoter and to contain binding sites for several transcription factors, including USF and AP-1 (R. A. Rippe et al., manuscript submitted). Despite the presence of all of these regulatory elements the Collal minigene is expressed at levels lower than the endogenous Collal gene. Phosphoimager quantification of minigene expression in several independent transfections has shown that in 3T3 fibroblasts the level of minigene mRNA is between 10% and 40% of that of the endogenous gene when calculated per copy number (data not shown). This indicates that expression of the transfected minigene is subject to position effects and suggests that, although the construct contains cis-regulatory elements that confer basic cell and differentiation specificity to the Collal promoter, additional, so far unidentified, elements are necessary for a position independence and copy number dependence of the transgenes. Candidate sequences for such elements are DNase-hypersensitive sites in the distal 5′- and 3′-flanking regions, which have been identified in the human (2) and murine (M. Breindl, unpublished observation) Collal genes and may have insulating or domain-opening functions.
Previous studies have shown that the developmental activation of the murine Collal gene in vivo is accompanied by changes in its chromatin structure and DNA methylation pattern. Its promoter is transcriptionally repressed, in an inactive chromatin conformation, and methylated in early embryonic cells, whereas it is transcriptionally active in a hypersensitive chromatin conformation and unmethylated in collagen-producing cells (7,9,13,29,30). Although numerous studies in a wide variety of experimental systems underscore the important regulatory functions of chromatin structure and DNA methylation in mammalian gene expression (18,28), the molecular mechanisms involved in these changes are only poorly understood. In the case of stage- and tissue-specifically expressed genes such as the Collal gene it is difficult to study these events in vivo, mainly because tissues are usually mixtures of collagen-producing and nonproducing cells in which chromatin structure and methylation pattern of the Collal promoter differ and are therefore refractory to experimental analysis. These questions are nevertheless of considerable interest for understanding the complex mechanisms regulating Collal gene expression and are now amenable to experimental analysis using the in vitro differentiation system described here. Moreover, similar in vitro differentiation systems should prove useful for dissecting the molecular mechanisms involved in the developmental activation and stage- and tissue-specific expression of other mammalian genes.
ACKNOWLEDGEMENTS
We thank Dr. E. Adamson for her strain of P19 EC cells and helpful advice and A. Sreedhar for her help with the immunofluorescent staining. This work was supported by NIH research grant AR41909.
REFERENCES
- 1. Antequera F.; Boyes J.; Bird A. P. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 62:503–514; 1990. [DOI] [PubMed] [Google Scholar]
- 2. Barsh G. S.; Roush C. L.; Gelinas R. E. DNA and chromatin structure of the human alpha1(I) collagen Gene. J. Biol. Chem. 259:14906–14913; 1984. [PubMed] [Google Scholar]
- 3. Bedalov A.; Breault D. T.; Sokolov B. P.; Lichtler A. C.; Bedalov I.; Clark S. H.; Mack K.; Killan J. S.; Woody C. O.; Kream B. E.; Rowe D. W. Regulation of α1(I) collagen in vascular smooth muscle cells. J. Biol. Chem. 269:4903–4909; 1994. [PubMed] [Google Scholar]
- 4. Bedalov A.; Salvatori R.; Dodig M.; Kronenberg M. S.; Kapural B.; Bogdanovic Z.; Kream B. E.; Woody C. O.; Clark S. H.; Mack K.; Rowe D. W.; Lichtler A. L. Regulation of COL1A1 gene expression in type I collagen producing tissues: Identification of a 49 base pair region which is required for transgene expression in bone of transgenic mice. J. Bone Miner. Res. 10:1443–1452; 1995. [DOI] [PubMed] [Google Scholar]
- 5. Bornstein P. Regulation of expression of the α1(I) collagen gene: A critical appraisal of the role of the first intron. Matrix Biol. 15:3–10; 1996. [DOI] [PubMed] [Google Scholar]
- 6. Bornstein P.; Sage H. Regulation of collagen gene expression. Prog. Nucleic Acid Res. Mol. Biol. 37:67–106; 1989. [DOI] [PubMed] [Google Scholar]
- 7. Breindl M.; Harbers K.; Jaenisch R. Retrovirus-induced lethal mutation in collagen I gene of mice is associated with an altered chromatin structure. Cell 38:9–16; 1984. [DOI] [PubMed] [Google Scholar]
- 8. Brenner D. A.; Rippe R. A.; Rhodes K.; Trotter J. F.; Breindl M. Fibrogenesis and type I collagen gene regulation. J. Lab. Clin. Med. 124:755–760; 1994. [PubMed] [Google Scholar]
- 9. Chan H.; Hartung S.; Breindl M. Retrovirus-induced interference with collagen I gene expression in Movl3 fibroblasts is maintained in the absence of DNA methylation. Mol. Cell. Biol. 11:47–54; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiang P. K.; Burbelo P. D.; Brugh S. A.; Gordon R. K.; Fukuda K.; Yamada Y. Activation of collagen IV gene expression in F9 teratocarcinoma cells by 3-deazaadenosine analogs. J. Biol. Chem. 267:4988–4991; 1992. [PubMed] [Google Scholar]
- 11. Chomczynski P.; Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanatephenol-chloroform extraction. Anal. Biochem. 162:156–159; 1987. [DOI] [PubMed] [Google Scholar]
- 12. Coucouvanis E.; Martin G. R. Signals for death and survival: A two-step mechanism for cavitation in the vertebrate embryo. Cell 83:279–287; 1995. [DOI] [PubMed] [Google Scholar]
- 13. Hartung S.; Jaenisch R.; Breindl M. Retrovirus insertion inactivates mouse alphal(I) collagen gene by blocking initiation of transcription. Nature 320:365–367; 1986. [DOI] [PubMed] [Google Scholar]
- 14. Horn V.; Minucci S.; Ogrysko V.; Adamson E. D.; Howard B. H.; Levin A. A.; Ozato K. RAR and RXR selective ligands cooperatively induce apoptosis and neuronal differentiation in P19 embryonal carcinoma cells. FASEB J. 10:1071–1077; 1996. [DOI] [PubMed] [Google Scholar]
- 15. Inagaki Y.; Truter S.; Ramirez F. Transforming growth factor-beta stimulates alpha 2(I) collagen gene expression through a cis-acting element that contains an Sp1-binding site. J. Biol. Chem. 296:14828–14834; 1994. [PubMed] [Google Scholar]
- 16. Jones-Villeneuve E. M. V.; McBurney M. W.; Rogers K. A.; Kalnins V. I. Retinoic acid induces embryonal carcinoma cell to differentiate into neurons and glia. J. Cell Biol. 94:253–262; 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katai H.; Stephenson J. D.; Simkevich C. P.; Thompson J. P.; Raghow R. An AP-l-like motif in the first intron of human pro alpha 1(I) collagen gene is a critical determinant of its transcriptional activity. Mol. Cell. Biochem. 118:119–129; 1992. [DOI] [PubMed] [Google Scholar]
- 18. Lewin B. Chromatin and gene expression: Constant questions, but changing answers. Cell 79:397–406; 1994. [DOI] [PubMed] [Google Scholar]
- 19. Li S-W.; Khillan J.; Prockop D. J. The complete cDNA sequence for the mouse proαl(I) chain of type I procollagen. Matrix Biol. 14:593–595; 1994. [DOI] [PubMed] [Google Scholar]
- 20. Liska D.; Reed M. J.; Sage E. H.; Bornstein P. Cell-specific expression of αl(I) collagen-hGH minigenes in transgenic mice. J. Cell Biol. 125:695–704; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin G. R.; Evans M. J. Differentiation of clonal lines of teratocarcinoma cells: Formation of embryoid bodies in vitro. Proc. Natl. Acad. Sci. USA 72:1441–1445; 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McBurney M. W. Clonal lines of teratocarcinoma cells in vitro: Differentiation and cytogenetic characteristics. J. Cell. Physiol. 89:441–456; 1976. [DOI] [PubMed] [Google Scholar]
- 23. McBurney M. W.; Jones-Villeneuve E. M. V.; Edwards M. K. S.; Anderson P. J. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature 229:165–167; 1982. [DOI] [PubMed] [Google Scholar]
- 24. Olsen A. S.; Geddis A. E.; Prockop D. J. High levels of expression of a minigene version of the human proαl(I) collagen gene in stably transfected mouse fibroblasts. J. Biol. Chem. 266:1117–1121; 1991. [PubMed] [Google Scholar]
- 25. Oppenhein R. W. Cell death during development of the nervous system. Annu. Rev. Neurosci. 14:453–501; 1991. [DOI] [PubMed] [Google Scholar]
- 26. Prockop D. J.; Kivirikko K. I. Collagens: Molecular biology, diseases, and potentials for therapy. Annu. Rev. Biochem. 64:403–434; 1995. [DOI] [PubMed] [Google Scholar]
- 27. Raff M. Social controls on cell survival and cell death. Nature 356:397–400; 1992. [DOI] [PubMed] [Google Scholar]
- 28. Razin A.; Kafri T. DNA methylation from the embryo to the adult. Prog. Nucleic Acid Res. Mol. Biol. 48:53–81; 1994. [DOI] [PubMed] [Google Scholar]
- 29. Rhodes K.; Breindl M. Developmental changes in the methylation status of regulatory elements in the murine αl(I) collagen gene. Gene Expr. 2:59–69; 1992. [PMC free article] [PubMed] [Google Scholar]
- 30. Rhodes K.; Rippe R. A.; Umezawa A.; Nehls M.; Brenner D. A.; Breindl M. DNA methylation represses murine αl(I) collagen promoter by an indirect mechanism. Mol. Cell. Biol. 14:5950–5960; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rippe R. A.; Lorenzen S.-I.; Brenner D. A.; Breindl M. Regulatory elements in the 5'-flanking region and the first intron contribute to transcriptional control of the mouse alpha 1 type I collagen gene. Mol. Cell. Biol. 9:2224–2227; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ritzenthaler J. D.; Goldstein R. H.; Fine A.; Lichtler A.; Rowe D. W.; Smith B. D. Trans-forming-growth-factor-β activation elements in the distal promoter regions of the rat al type I collagen gene. Biochem. J. 280:157–162; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rossert J. A.; Chen S. S.; Eberspaecher H.; Smith C. D.; deCrombrugghe B. Identification of a minimal sequence of the mouse αl(I) collagen promoter that confers high-level osteoblast expression in transgenic mice and that binds a protein selectively present in osteoblasts. Proc. Natl. Acad. Sci. USA 93:1027–1031; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rossert J. A.; Eberspaecher H.; deCrombrugghe B. Separate cis-acting DNA elements of the mouse pro-αl(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell Biol. 129:1421–1432; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rudnicki M. A.; Sawtell N. M.; Reuhl K. R.; Berg R.; Craig J. C.; Jardine K.; Lessard J. L.; McBurney M. W. Smooth muscle actin expression during P19 embryonal carcinoma differentiation in cell culture. J. Cell. Physiol. 142:89–98; 1990. [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J.; Fritsch E. F.; Maniatis T. Molecular cloning. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37. Schmidt A.; Rossi P.; deCrombrugghe B. Transcriptional control of the mouse α2(I) collagen gene: Functional deletion analysis of the promoter and evidence for cell-specific expression. Mol. Cell. Biol. 6:347–354; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Slack J. D.; Liska D. A. J.; Bornstein P. Regulation of expression of the type I collagen genes. Am. J. Med. Genet. 45:140–151; 1993. [DOI] [PubMed] [Google Scholar]
- 39. Slack J. L.; Parker M. I.; Bornstein P. Transcriptional repression of the αl(I) collagen gene by ras is mediated in part by an intronic API site. J. Cell. Biochem. 58:380–392; 1995. [DOI] [PubMed] [Google Scholar]
- 40. Sokolov B. P.; Ala-Koko L.; Dhulipala R.; Arita M.; Khillan J. S.; Prockop D. J. Tissue-specific expression of the gene for type I procollagen (COL1A1) in transgenic mice. J. Biol. Chem. 270:9622–9629; 1995. [DOI] [PubMed] [Google Scholar]
- 41. Sokolov B. P.; Mays P. K.; Khillan J. S.; Prockop D. J. Tissue- and development-specific expression in transgenic mice of α type I procollagen (COL1A1) minigene construct with 2.3 kb of the promoter region and 2 kb of 3′-flanking region. Specificity is independent of the putative regulatory sequences in the first intron. Biochemistry 32:9242–9249; 1993. [DOI] [PubMed] [Google Scholar]
- 42. Strickland S.; Smith K. K.; Marotti K. R. Hormonal induction of differentiation in teratocarcinoma stem cells: Generation of parietal endoderm by retinoic acid and dibutyryl cAMP. Cell 21:347–355; 1980. [DOI] [PubMed] [Google Scholar]
- 43. Vuorio E.; deCrombrugghe B. The family of collagen genes. Annu. Rev. Biochem. 59:837–872; 1990. [DOI] [PubMed] [Google Scholar]
- 44. Watt F. M. Cell culture models of differentiation. FASEB J. 5:287–294; 1991. [DOI] [PubMed] [Google Scholar]
- 45. Wyllie A. H. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284:555–556; 1980. [DOI] [PubMed] [Google Scholar]


