Abstract
Hypoxia inducible factor 1α (HIF-1α) is a basic helix-loop-helix-PAS (bHLH-PAS) transcription factor that mediates certain cellular responses to low oxygen tension, iron chelators, Co2+, Ni2+, Mg2+, and low intracellular glucose concentration. Upon exposure to the above conditions, HIF-1α is upregulated and heterodimerizes with the Ah receptor nuclear translocator (ARNT, also known as HIF-1β), the heterodimeric complex binds TACGTG-containing genomic enhancer elements, and activates transcription of target genes. As a first step in developing genetic models to study the biology related to cellular hypoxia, we have cloned the murine HIF-1α cDNA, determined the tissue-specific expression of its mRNA, functionally analyzed its protein product, and characterized its promoter and its genomic structure. A comparison between the murine and human HIF-1α protein sequence reveals 95%, 99%, and 83% identity in the bHLH, PAS, and variable domains, respectively. RNAse protection assays demonstrate that in adult mice, the mHIF-1α mRNA is expressed at high levels in kidney, heart, brain, thymus, and placenta, with moderate expression in liver, spleen, testis, and lung and much lower expression in skeletal muscle testis. Northern blot analysis indicates that the mRNA of the murine HIF-1α is transcribed in two forms, a major 4-kb species and a minor 5-kb species; both are present in all tissues examined. The Hif-1α promoter is GC rich, does not have a TATA element near its transcriptional start site, and does not respond to hypoxia or Co2+. The mHIF-1α structural gene is composed of 15 exons. The splice junction sites within the bHLH and the PAS domains of HIF-1α gene are highly conserved with respect to a number of previously characterized members of the bHLH-PAS superfamily. However, unlike other bHLH-PAS genes, where the variable domain is encoded by 2 exons, the variable region of the mHIF-1α gene is encoded by 7 exons. Furthermore, most of these splice junction sites in the variable region are conserved with that of HIF-2α, a recently cloned hypoxia-responsive bHLH-PAS protein (also known as MOP2, EPAS1, and HLF). These data suggest that HIF-1α, along with HIF-2α, represents a new subclass of the bHLH-PAS superfamily.
Keywords: Murine Hif-1α, Hypoxia inducible factor 1α, bHLH-PAS superfamily, Molecular characterization
THE basic helix–loop–helix (bHLH) domain is common to a variety of transcription factors that regulate xenobiotic metabolism (8,27), neurogenesis (6), myogenesis (26,38), cellular proliferation (1), and hematopoiesis (37). The bHLH domain directs homotypic interactions between protein pairs, thus influencing the DNA binding specificity generated by the dimeric complex. An emerging superfamily of bHLH proteins also shares homology in a second region termed the PAS domain. The PAS domain is named for the first three proteins that were shown to contain it: PER, the Drosophila protein that controls circadian rhythm (5), ARNT, the dimeric partner of the AHR (15), and SIM, the Drosophila protein that controls specification of cell fate during midline cell differentiation (6,24). The PAS domain is a region of 200–300 amino acids containing two 60-amino acid degenerate repeats (24) that mediates protein dimerization (17). In the AHR, the PAS domain is required for dimerization with ARNT and is also necessary for interactions with the 90-kDa heat shock protein (Hsp90), as well as ligand binding (7,13).
HIF-1α is a newly identified member of the bHLH-PAS superfamily (37). This transcription factor responds to conditions of low oxygen tension and activates hypoxia-responsive genes such as erythropoietin (EPO), vascular endothelial growth factor (VEGF) (12), and certain glycolytic enzymes (22,34). Under normal physiological conditions, HIF-1α protein levels are very low or un-detectable in cells. Upon exposure to low oxygen tension, or treatment with Co2+ or desferroxamine, the HIF-1α protein is substantially increased in nuclear fractions of cells (37). The observed responsiveness of HIF-1α to low 02 tension, Co2+, Ni2+, Mg2+, and desferroxamine treatment suggests that a heme sensor regulates the intracellular level of HIF-1α (14). A model supported by evidence from a number of laboratories suggests that HIF-1α concentration drives the formation of HIF-1α/ARNT complexes in vivo, which in turn activates the expression of genes that increase erythrocyte production, and energy production via glycolysis. In an effort to develop genetic models to study the biology related to HIF-1α gene, we have characterized the murine HIF-1α locus and its encoded mRNA and protein.
MATERIALS AND METHODS
cDNA Cloning
Polymerase chain reaction (PCR) was employed to obtain the murine HIF-1α cDNA using an Uni-ZAP C57BL/6 mouse kidney phage cDNA library as a template (Stratagene, La Jolla, CA). Typical amplification reactions employed 109 plaque forming units of the phage library and 2.5 units of Taq polymerase in 50 μl reactions buffered with 50 mM KC1, 10 mM Tris-HCl, and 2.0 mM MgCl2, at pH 8.3. Initially, a fragment of the murine HIF-lα cDNA was obtained using the human HIF-lα-specific oligonucleotides OL358 and OL365. Once murine-specific sequences were obtained in this initial PCR, succeeding fragments of the murine cDNA were amplified using one murine-specific oligonucleotide and one human-specific oligonucleotide (bold) in the following combinations: OL404 vs. OL412, OL394 vs. OL363, OL413 vs. OL430, and OL433 vs. OL431. Sequences corresponding to the 5′ and 3′ UTRs were obtained by PCR amplification with primer pairs corresponding to the T3 promoter site of the Uni-ZAP vector plus OL395 and primer pairs corresponding to the T7 promoter site of the Uni-ZAP vector plus OL458, respectively. PCR products were purified by gel electrophoresis, sub-cloned into pGEM-T vector (Promega, Madison, WI), and sequenced using the dideoxy chain termination method (31).
PCR was employed to construct an HIF-1α cDNA that contained the entire ORF. Oligonucleotide primers OL404 and OL363 were used to amplify a 1475-bp fragment containing the initial methionine and the 5′ half of the ORF and primers OL411 and OL431 were used to amplify a 1464-bp fragment containing the 3′ half of the ORF up to the stop codon. These two cDNA fragments were subcloned into separate pGEM-T vectors in the T7 orientation and then digested with Csp45I and NotI. The resulting cDNA fragments were ligated to generate an ORF-containing plasmid, termed pGmHIF-1α. To obtain a vector with greater expression levels for in vitro experiments, the SacII and NotI of pGmHIF-1α was subcloned into the corresponding sites of pBSK. The NcoI/EcoRI fragment of this plasmid was subcloned into the corresponding sites of pSPUTK (Stratagene, La Jolla, CA) and designated pSmHIF-1α. In vitro transcription and translation of the murine HIF-1α protein were performed using the TNT coupled reticulocyte lysate as described previously (7).
Antibody Generation and Western Blot
The human HIF-1α antibody was generated as described previously (16). The ARNT-specific antisera was a generous gift of Dr. Alan Poland of the McArdle Laboratory for Cancer Research, University of Wisconsin, Madison (28). Western blots were performed as described previously (3).
Gel Shift Assays
Synthetic oligonucleotides containing core TACGTG (OL414) or CACGTG (OL445) sequences were end-labeled with [γ-32P]ATP by T4 nucleotide kinase. The radiolabeled oligonucleotide probes were annealed with a 10-fold molar excess of the complementary oligonucleotides OL415 and OL446, respectively. The clone pm-ARNT, containing the complete ORF of the C57BL/6J mouse ARNT cDNA, was a generous gift from Alan Poland (University of Wisconsin-Madison, WI). Approximately 0.2 fmol of in vitro expressed mHIF-1α and mARNT proteins were incubated for 10 min at 30°C, followed by the addition of the nonspecific competitor, poly(dI-dC) (100 ng, 10 min at RT) and 32P-labeled complementary oligonucleotides (1 ng, 70,000 cpm, 10 min at RT). For the antibody blocking experiments, antisera was added to this reaction and incubated for 10 min at room temperature. In oligonucleotide competition experiments, equal amounts of mHIF-1α and mARNT were incubated with an unlabeled complementary pair of synthetic oligonucleotides and poly(dI-dC) for 10 min at room temperature. Following this incubation, 32P-labeled oligonucleotides were added for 10 min at room temperature prior to analysis by PAGE (3).
RNA Analysis
Northern blots were performed using 2 μg of poly(A)+ RNA per lane from eight different mouse tissues (Clontech, Palo Alto, CA). A probe was constructed from the Apa1-Sac1 fragment of PL428 and random primed in the presence of [α-32P]CTP (10). Hybridization was performed overnight at 65 °C in a solution containing 5 × SSPE, 2 × Denhardt’s, 0.5% SDS, and 100 mg/ml heat-denatured salmon sperm DNA (Life Technologies, Gaithersburg, MD). Following overnight hybridization, the blot was washed four times with 2 × SSC (0.3 M NaCl, 0.03 M sodium citrate), 0.5% SDS at room temperature for 5 min, and then once with 0.1 × SSC, 0.1% SDS at 65°C for 15 min. The blot was exposed to film for 24 h at –80°C.
Ribonuclease protection assays were performed with 10 μg of total RNA, prepared from 10-month-old C57BL/6 mouse tissues using the TRIzol reagent (Life Technologies, Gaithersburg, MD). The riboprobe for ribonuclease protection was generated from the plasmid PL428. This plasmid was linearized with HindIII and used as template for synthesis of a 370-nucleotide antisense riboprobe with SP6 polymerase in the presence of [α-32P]UTP (10 mCi/ml, 3000 Ci/mmol). This riboprobe was designed to protect a 291-bp fragment of mHIF-1α mRNA. The riboprobe was added to total RNA in molar excess such that the signals of all protected fragments were within the linear range of the assay. Hybridization was performed using Hybspeed Buffer (Ambion Inc., Austin,TX) at 68°C for 1 h. After hybridization, the RNA was subjected to RNAse A1/T1 for 30 min at 37°C. The protected bands were quantified on BAS2000 phosphoimaging system. For presentation, the dried gel was exposed to film for 24 h at –70°C. A 109-nucleotide riboprobe from the conserved region of human 18S subunit of ribosomal RNA was used as an internal control (Ambion Inc., Austin, TX). The 18S riboprobe often results in multiple bands ranging from 66 to 80 nucleotides (data not shown) (2).
Identification of the Transcriptional Start Site
The RNAse protection assay was performed essentially as described above. PL529 containing 1.7 kb PstI fragment upstream of the translation starting codon ATG was cut with XbaI to make a riboprobe of approximately 1.4 kb using T7 polymerase in the presence of [α-32P]UTP (3000 Ci/ mmol). UTP (1 mM) was added to ensure generation of full-length riboprobe. Riboprobe (450 pg) (45,000 cpm; S.A. = 1 × 108 cpm/μl) and 15 μg of total RNA from E17.5d mouse placenta were used for the assay.
Cell Culture and Transient Transfection
Human Hep3B cells were maintained in MEM medium supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 2.5 μg/ml fungizone, 1 mM nonessential amino acids, 1 mM sodium pyruvate, and 2 mM l-gluta-mine at 37°C in 5% CO2. For transient transfection experiments, a 1.9-kb SacI fragment containing the 5′ UTR of the murine HIF-1α cDNA was subcloned into the SacI site of the pGL2-basic plasmid (Promega, Madison, WI) to produce a construct that contained the HIF-1α promoter fused to the luciferase gene. This fragment was also subcloned in the reverse orientation using the XbaI/BglII sites of pGL-2 basic plasmid. Transient transfections were performed by the lipofectamine method (Life Technologies, Gaithersburg, MD) in six-well polystyrene tissue culture. Cells were incubated with the DNA liposome complex for 5 h in OPTI-MEM I reduced serum medium. Following incubation, fresh media were added with or without 75 μM CoCl2 and incubated at 37°C overnight. To control for transfection efficiency, 0.1 μg of β-galactosidase control plasmid pCH110 was cotransfected in each plate. To quantify activity, cells were harvested and the luciferase and the β-galactosidase activities were determined using the luciferase and Galacto-Light™ protocols according to manufacturer’s instructions (Promega, TROPIX, Inc.). All luciferase values were normalized to β-galactosidase activity (32).
Determination of the HIF-1α Gene Structure
Two overlapping P1 phage clones (P1mHIF-1α 5635 and P1mHIF-1α 5636) containing the murine HIF-1α gene were identified from pools of phage by PCR using the primer pair OL406 and OL407 (Genome Systems, St. Louis, MO). Subclones were obtained by either restriction digest or PCR. Exon–intron splice junctions and their corresponding locations were identified by nucleotide sequencing. The PCR was typically performed using 103 colony forming units of P1 phage and various combinations of mHIF-1α-specific oligonucleotide primers. The PCR products were analyzed by Southern blot and subcloned into the pGEM-T vector as described above. PCR fragments typically contained at least one exon–intron boundary. Intron size was estimated from the size difference of the PCR products obtained using the genomic and cDNA as template. The plasmids were sequenced by the dideoxy chain termination method (31).
Oligonucleotide primers used (from 5′ to 3′) are given below:
OL145 AGCTCGAAATTAACCCTCACTAAAGG,
OL146 GAATTGTAATACGACTCACTATAGGG,
OL358 AACTCAGTTTGAACTAACTGG,
OL363 CCAGTGACTCTGGATTTGGTTC,
OL365 GACAGTTGCTTGAGTTTCAACC,
OL394 CAGCAACGTGGAAGGTGCTT,
OL404 GCGGTACCGGGACCGATTCACCATGGAG,
OL411 CGGCGACACCATCATCTCTCTG,
OL412 AATATGGCCCGTGCAGTGAAGC,
OL413 GCAATGTCTCCTTTACCTTCAT,
OL430 TAGGGCTTCTTGGATGAGATTT,
OL431 AAGTTTGTGCAGTATTGTAG,
OL433 CCAGTTACAGAAACCTACCATC,
OL406 CATGACGTGCTTGGTGCTGAT,
OL407 GAGGCTGTGTCGACTGAGAAATGT.
Complementary pairs of oligonucleotides used in gel shift assays are given below. Only the 5′ oligonucleotide is shown. Note constant flanking sequences, with variable core sequences underlined:
OL316/317 TCGAGCTGGGCAGGTCATGTGGCAAGGC,
OL323/324 TCGAGCTGGGCAGGTCAGCTGGCAAGGC,
OL396/397 TCGAGCTGGGCAGGTAACGTGGC
OL398/399 TCGAGCTGGGCAGGTGACGTGGCAAGGC
OL414/415 TCGAGCTGGGCAGGGTACGTGGCAAGGC,
OL445/446 TCGAGCTGGGCAGGTCACGTGGCAAGGC.
RESULTS AND DISCUSSION
Cloning of the Murine HIF-1α cDNA
To develop genetic models of HIF-1α function, we cloned the murine HIF-1α cDNA using sequence information from the human cDNA (16, 37). Figure 1 provides the murine HIF-1 cDNA sequence obtained by PCR. The ORF contains 2511 nucleotides that encode a protein of 836 amino acids with a predicated molecular weight of 95 kDa. Nucleotide number one is defined as the first A of the initiation codon (ATG). The initiation codon is defined by the presence of an upstream in-frame termination codon (found at –201). The cloned 5′ and 3′ UTRs of mHIF-1α contains 257 and 1205 nucleotides, respectively. A consensus polyadenylation signal is located at nucleotide 3674, 24 nucleotides upstream of a poly(A) sequence. A comparison of the murine and human HIF-1α amino acid sequences revealed that the murine HIF-1α protein shared with its human homologue 95% identity in the bHLH domain, 99% identity in the PAS domain, and 83% identity in the C-terminal variable region (data not shown). While this work was in progress, two independent groups also reported cDNA sequences for murine HIF-1α (22,39). The open reading frame for the sequence we report here is essentially identical to that reported by Li et al. (22). Although these two sequences are in agreement, their prediction of initiation codon is in contrast to that reported by Wenger et al. (39), where the initiation codon is placed at a position corresponding to our nucleotide at position 37. At the present time, we cannot rule out either a true polymorphism, the existence of an additional exon, or a sequencing error as the underlying cause of this difference.
FIG. 1.

cDNA sequence and the deduced protein sequence of the murine HIF-1α gene. Numbers on the right indicate the nucleotide numbering with the A in the initiation codon ATG defined as +1. Numbers on the left side indicate the amino acid position of the deduced mHIF-1α protein. The star at the end of the protein sequence indicates the stop codon. Positions of the in-frame termination codon (TAG) upstream of the initiation codon ATG and the polyadenylation signal are underlined in the 5′ and 3′ ends, respectively. To obtain the entire cDNA, fragments of the murine cDNA were amplified and subcloned into pGEM-T vectors: OL358 and OL365 generated plasmid PL426, OL404 vs. OL412 generated PL430, OL394 vs. OL363 generated PL428, OL413 vs. OL430 generated PL431, OL433 vs. OL431 generated PL432, a T3 promoter oligo vs. OL395 generated PL505, and a T7 promoter oligo vs. OL458 generated PL506.
Tissue-Specific Expression of Murine HIF-1α mRNA
Northern blot analysis was first performed to determine the size of HIF-1α mRNA transcript. RNAse protection assays were performed to determine the level of HIF-1α mRNA expression in different tissues (Fig. 2A, B). Northern blots revealed a major 4-kb transcript and a minor 5-kb transcript in all tissues examined (Fig. 2A). The size of the major transcript is consistent with the 3973-bp size of the full-length murine HIF-1α cDNA that we obtained by PCR. Given the rare nature of the 5-kb transcript, this species was not pursued further. RNAse protection assays were performed on 10 different tissues from the C57BL/6 mouse (Fig. 2B) and a 109-nucleotide riboprobe from the conserved region of human 18S subunit of ribosomal RNA was use as the internal control [(2), data not shown]. The results show that the mHIF-1α mRNA was abundantly expressed in placenta, kidney, heart, brain, and thymus. Moderate expression of mHIF-1α was found in liver, spleen, testis, and lung, and low expression was found in skeletal muscle (Fig. 2B). Given that HIF-1α protein expression has recently been reported to be regulated by a posttranscriptional mechanism [(18); our unpublished observations], these data identify tissues where HIF-1α has the potential to play an important physiological role.
FIG. 2.
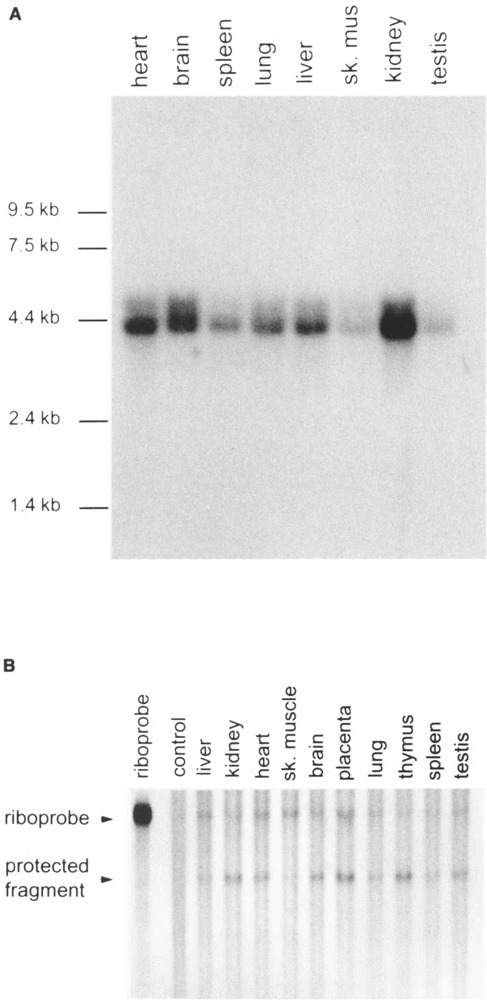
Analysis of the murine HIF-1α expression in mouse tissues. (A) Northern blot. Approximately 2 μg of poly(A) RNA per lane from eight different mouse tissues was hybridized with a random primed fragment of murine HIF-1α cDNA. (B) RNAse protection assay. Approximately 10 μg of total RNA per lane from 10 different C57BL/6 mouse tissues was hybridized at 68°C for 1 h with a 370-bp [α-32P]UTP-labeled riboprobe synthesized from a HindIII linearized murine HIF-1α cDNA (PL428). The protected mRNA fragment is 290 nucleotides. The amount of total RNA in each lane was normalized using a 109-nucleotide riboprobe from the conserved region of human 18S subunit of ribosomal RNA. The film was exposed for 24 h at –70°C.
Functional Expression of mHIF-1α
To investigate the biochemical properties of the mHIF-1α protein, mHIF-1α cDNA was expressed using a reticulocyte lysate system (Promega, Madison, WI). Quantitiation of protein expression was performed by monitoring incorporation of [35S]methionine by autoradiography and by Western blot with antisera raised against the recombinant human protein (Fig. 3). The results of these analyses revealed that the expressed mHIF-1α protein has an apparent molecular weight of ∼86 kDa, which is within 10% of the 95 kDa predicted from its open reading frame. The human HIF-1α antibody displays much greater sensitivity at detecting the human HIF-1α than the murine HIF-1α (Fig. 3). This is not surprising given that the antigen used is from the variable region of the hHIF-1α (residues 329 to 531), where the amino acid sequence identity between the HIF-1α orthologues is only 85%.
FIG. 3.
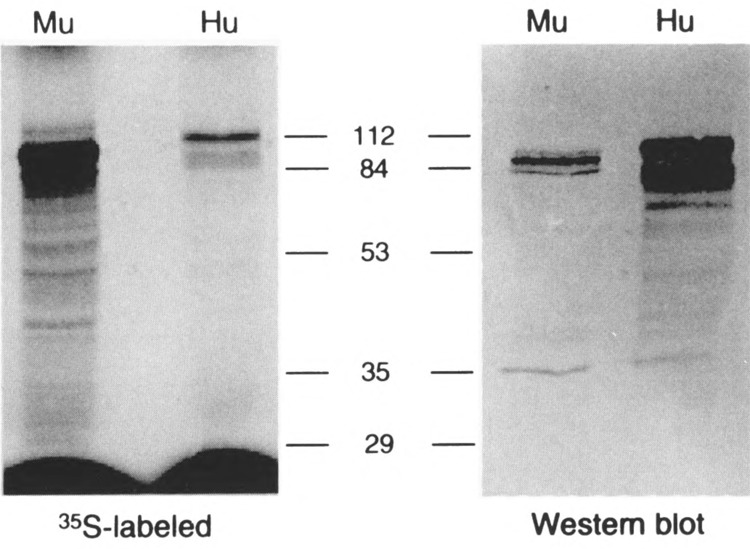
In vitro expression of the murine HIF-1α protein and the Western Blot analysis. Left: The murine HIF-1α was expressed in TNT reticulocyte lysate using 35S-labeled methionine and resolved in 7.5% SDS-PAGE. Right: The same gel (left) was analyzed by Western Blot using the human HIF-1α specific antibody. Hu: human; Mu: murine.
To determine the DNA binding specificity of the mHIF-1α protein, a gel shift assay was performed using double-stranded oligonucleotides containing the known hypoxia-responsive element found within the human EPO gene enhancer (TACGTG). In the presence of equimolar amounts of mHIF-1α and mARNT, specific interactions were obtained with the radiolabled TACGTG (OL414/415) sequence (Fig. 4). In the presence of mHIF-1α or mARNT alone, no reactivity was observed. The binding specificity to the TACGTG sequence was demonstrated by the observation that oligonucleotides containing AACGTG (OL396/397), GACGTG (OL398/399), or CAGCTG (OL323/324) did not compete for mHIF-1α/ARNT binding. Like its human orthologues, the mHIF-1α/ARNT complex also showed specific binding for the core sequences of CACGTG (OL445/446) and CATGTG (OL316/317) (16,20). The presence of both mouse HIF-1α and ARNT proteins in the complex was demonstrated by the observation that both the hHIF-1α antibody and the ARNT antibody completely abolished the DNA-protein interaction (Fig. 4).
FIG. 4.
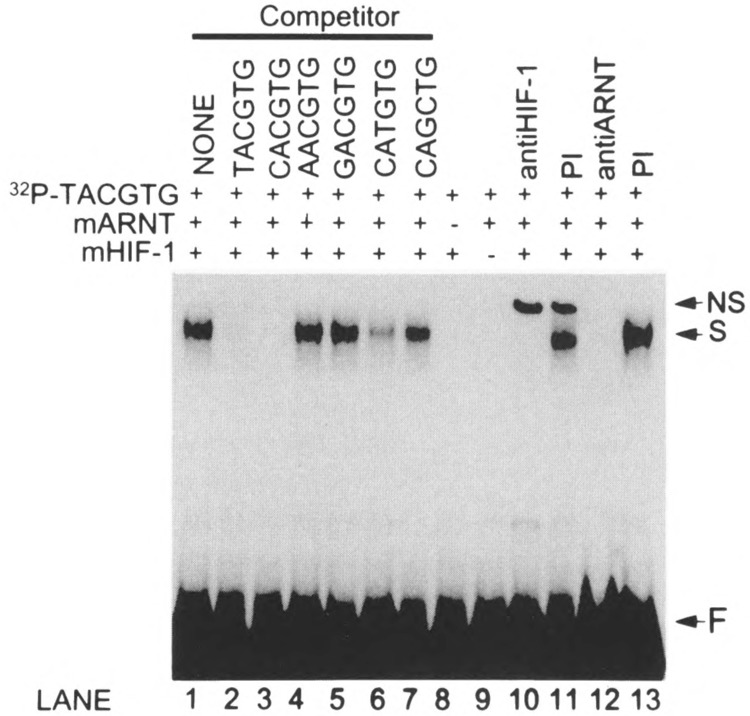
DNA binding specificity of the murine HIF-1α /ARNT complex. Approximately 0.2 fmol of reticulocyte lysate-expressed murine HIF-1α and ARNT were analyzed by gel shift assays using 32P-end labeled double-strand oligonucleotide corresponding to the consensus HRE (TACGTG, OL415/OL416). The murine HIF-1α and ARNT complex specifically binds to TACGTG (lane 1). Competition assays with divergent core sequences were performed as described in the text (lanes 2–7). No specific binding was seen with the murine HIF-1α or ARNT alone (lane 8 and 9). The anti-HIF-1α or anti-ARNT immuglobin abolished the specific binding, but the nonspecific IgG does not (lanes 10–13). NS: nonspecific binding; S: specific binding; F: free 32P-end labeled oligonucleotides.
Characterization of the mHIF-1α Gene
To determine the complete structure of the mHIF-1α gene, nine genomic DNA clones were obtained by either PCR or standard subcloning from P1 phage clones (Fig. 5). Sequence analysis of these clones revealed that the mHIF-1α gene contains 14 introns and 15 exons. Table 1 shows the intron-exon splice junction of the murine HIF-1α gene. All the splice junctions have the consensus GT . . . AG donor and acceptor dinucleotides. Figure 5 illustrates the structural organization of the murine HIF-1α genomic DNA and relates the 15 exons to the functional domains of the encoded protein. Exon 2 contains the bHLH domain, exons 3 through 8 comprise the PAS domain, and exons 9 through 15 encode the variable region. Recently, two hypoxia-responsive domains (HRD) have been characterized (22,30). HRD1 is within exons 11 and 12 whereas HRD2 is within exons 14 and 15.
FIG. 5.
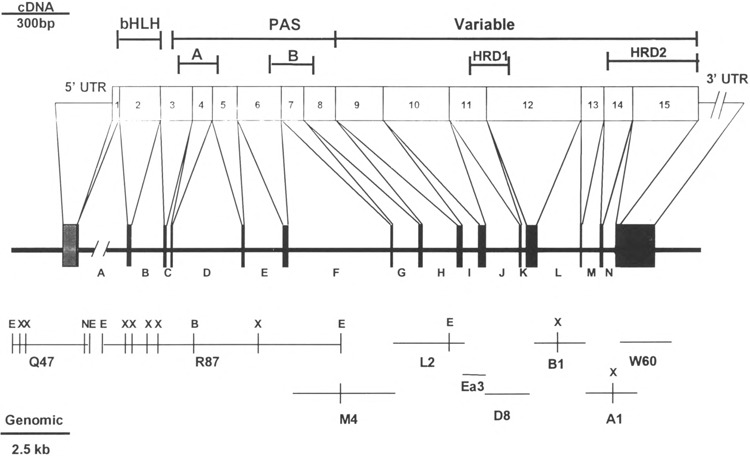
The murine HIF-1α gene structure. The exons of the murine HIF-1α cDNA are represented by numbers. Boxes represent the coding regions (exons) of the genomic DNA. Shaded boxes respresent the 5′ and 3′ untranslated regions (UTRs). Letters represent the introns. Scale bars are shown for the cDNA (top left) and for the genomic DNA (bottom left), respectively. Restriction sites of the murine HIF-1α gene are indicated as follows: B: BamHI; E: EcoRI; N: NotI; X: XbaI. This restriction map does not include a 9-kb fragment of the first intron. The clone comprising the region between exon 1 and intron A was obtained by digesting the P1 phage clone 5635 with EcoRI and identifying the fragment by Southern blot. The corresponding band was subcloned into the EcoRI site of pBSK and referred to as PLQ47. Similarly, an 8.5-kb fragment was obtained by Southern blot with a random primed exon 2 cDNA fragment (PLR87).
TABLE 1.
INTRON-EXON SPLICE JUNCTIONS OF THE MURINE HIF-lα GENE
| Position | Exon No. | Exon Seq. | Intron Size (bp) | Intron Seq. | Exon No. | Exon Seq. |
|---|---|---|---|---|---|---|
| 35/36 | 1 | G/AAA/AA | >10000 | GTAAG . . . TGCAG | 2 | G/ATG/AG |
| 226/227 | 2 | AT/GCC/G | 1100 | GTGAG . . . TGCAG | 3 | GT/GGT/C |
| 372/373 | 3 | ACT/CAG/ | 82 | GTAAA . . . TGCAG | 4 | TTT/GAA/ |
| 457/458 | 4 | GA/AAT/G | 2500 | GTGAG . . . TTCAG | 5 | GC/CCA/G |
| 570/571 | 5 | TGG/AAG/ | 1300 | GTAGC . . . TTCAG | 6 | GTG/CTT/ |
| 773/774 | 6 | T/GAA/AG | 3700 | GTAAA . . . AACAG | 7 | A/ATT/AC |
| 880/881 | 7 | AT/GAT/A | 900 | GTAAG . . . CCCAG | 8 | TG/TCT/A |
| 1028/1029 | 8 | TT/GTA/A | 1250 | GTAAT . . . gacag | 9 | GT/GGT/A |
| 1249/1250 | 9 | GC/GAT/G | 500 | GTCAG . . . TGCAG | 10 | AC/ACA/G |
| 1533/1534 | 10 | CCT/GAG/ | 1100 | GTTGG . . . AACAG | 11 | AGA/CTT/ |
| 1698/1699 | 11 | ACT/CAG | 108 | GTATG . . . GAAAG | 12 | GAC/ACT/ |
| 2123/2124 | 12 | T/CAA/AG | 1500 | GTATT . . . TCCAG | 13 | A/AAT/AC |
| 2232/2233 | 13 | CCA/ATT/ | 600 | GTAAG . . . TTTAG | 14 | GGA/ACA/ |
| 2359/2360 | 14 | CC/TCC/G | 400 | GTTAG . . . TTTAG | 15 | AT/TTA/G |
| 3740 | 15 | AATAAA |
The positions of the murine HIF-lα intron–exon splice junctions and their adjacent sequences are shown. The consensus splice donor and acceptor dinucleotides of each intron are underlined. The genetic codons are separated by slashes.
Using RNAse protection assay, we identified two transcriptional start sites that mapped to approximately 137 and 130 bp upstream of the PstI site (Fig. 6A, B). In support of the conclusion that this region comprises bona fide transcriptional start sites, we offer two supporting observations. First, the boundaries of this region are within 18 nucleotides of the 5′ end of the HIF-1α cDNA obtained by RACE (Fig. 6B) and within 4 nucleotides of the 5′ end of a murine HIF-1α cDNA sequence independently obtained by another group (22). Second, the boundaries of the predicted transcriptional start sites are proximal to two 8-nucleotide sequences that have high homology to the consensus for transcriptional initiation (21). Based upon the relative intensity of the two protected species in the RNAse protection assay, we conclude that the HIF-1α promoter uses two nearby independent transcriptional start sites with equal efficiency (Fig. 6B). Analysis of the regions upstream of these start sites indicated that the promoter is extremely GC rich (68% GC in the 500 bp upstream of the start site region), similar to promoters found in many “housekeeping” genes. Although consensus recognition sequences for a number of well-characterized transcription factors were identified, no TATA or CCAAT boxes were found in the sequences proximal to the transcriptional start sites. In keeping with many TATA-less promoters, two SP1 sites were found in a region 20–40 nucleotides upstream of the proposed transcriptional starts (21). Interestingly, these characteristics of the promoter region of the murine HIF-1α gene are similar to that of the murine AHR gene, another member of the bHLH-PAS superfamily (33).
FIG. 6.
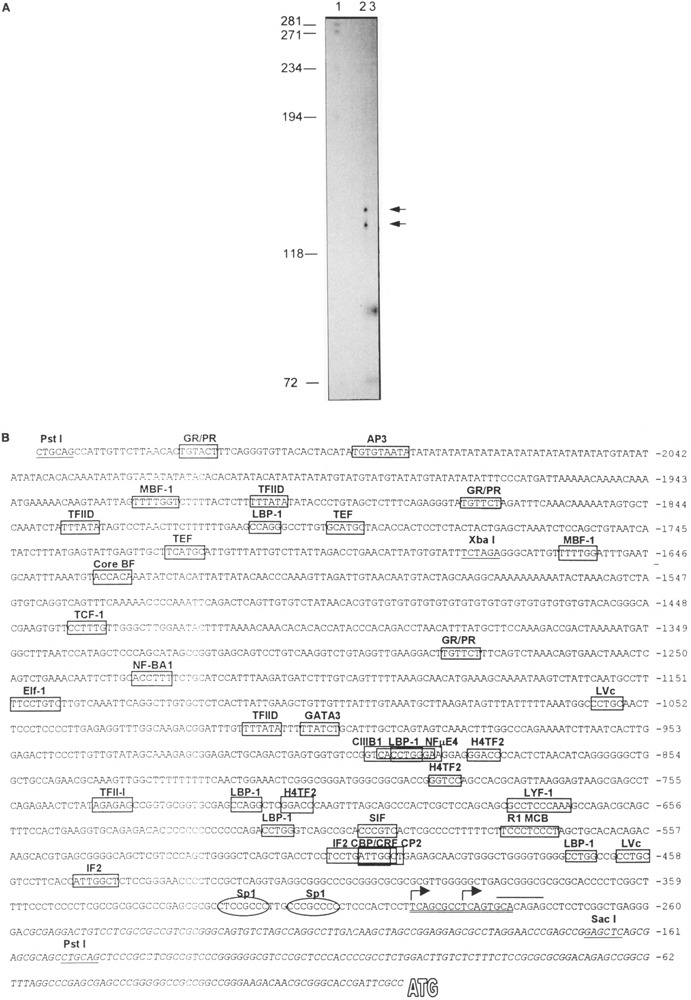
Identification and characterization of the murine HIF-1α promoter. (A) RNAse protection assay to identify transcriptional start sites (see text). Left: Size in bp of DNA molecular weight markers. Lane 1: radiolabeled, double-stranded DNA markers. Lane 2: 15 μg of total RNA from 17.5-day mouse placenta. Lane 3: 15 μg of yeast RNA as a control. The sizes of the two protected fragments were calculated from a linear regression analysis of the migration of the DNA standards. As a result, we estimate that the sizes of the protected fragments are approximately 130 and 137 nucleotides in length. (B) DNA sequence of the murine Hif-1α gene 5′ region. The PstI and XbaI sites used to generate the plasmid for the riboprobe for RNAse protection assays are underlined. Putative transcriptional start sites are depicted as follows: Italics: 5′ UTR of cDNA as described in Fig. 1. Overline: approximate boundaries of region predicted to contain two independent start sites by RNAse protection. Double underline: sequences closely conforming to a reported consensus initiation site, KCABHYBY, where K = G/T, B = C/G/T, and Y = C/T (21). Arrows: Predicted transcriptional start sites based on all of the above information. To identify potential binding sites for known transcription factors, the “Matrix Search” program (http://bimas.dcrt.nih.gov/molbio/signal) was used and sites with a “match ratio” score of greater than 1 are depicted by a box (4). Two consensus Sp1 sites were also identified with scores of 0.77 and 0.75. Given the importance of these sites in initiation from TATA-less promoters, these sites are also identified by circles.
Once the genomic region harboring transcriptional start sites was mapped, we subcloned the corresponding 1.9-kb fragment upstream of a luciferase reporter gene (pGL2-basic by Promega, WI) in both the forward and the reverse orientations. These constructs were used in transient transfection of human Hep3B hepatoma cell lines and the activity of each reporter construct was measured in the absence or presence of either 75 μM CoCl2 or hypoxia treatment (Fig. 7A, B). The results indicate that the mHIF-1α promoter greatly activates reporter gene activity in human hepatoma cells and the promotor is only active in the forward orientation. However, this promoter was not induced by either CoCl2 or hypoxia treatment (Fig. 7A, B). This observation is consistent with the notion that HIF-1α protein levels are regulated by a posttranscriptional mechanism [(18); unpublished observations].
FIG. 7.
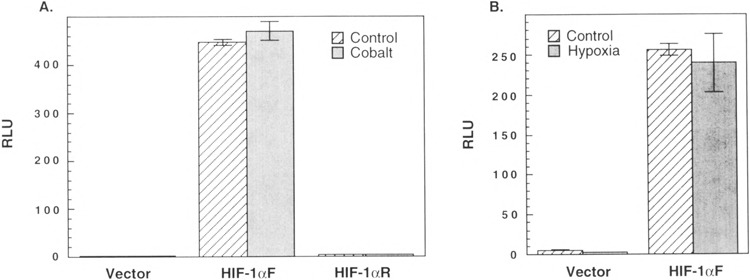
Functional analysis of the of the murine HIF-1α promoter. A 1.9-kb fragment upstream of the SacI site of the murine HIF-1α promoter (Fig. 6B) was subcloned in either forward (HIF-1αF) or reverse (HIF-1αR) orientation into the luciferase expression vector pGL2-basic (vector). (A) 2 μg or (B) 1 μg of each expression plasmid along with 0.1 μg of the β-galactosidase control plasmid was transiently transfected into Hep3B cells. The cells were incubated in the absence or presence of either 75 μM CoCl2 (A) or 1 % 02 (B) for 48 h before being harvested. The value of luciferase activity was normalized to that of β-galactosidase control plasmid and the data represent mean ± SE of triplicate samples. RLU: relative luciferase units. Parallel experiments to demonstrate induction of HIF-1 protein under these conditions were performed (data not shown).
PAS Gene Family Splice Junction Comparison
Recent evidence has demonstrated that a super-family of PAS-encoding genes exist within the mammalian genome (9,16,29,36,40). We are interested in a comparison of the structural organization of these genes as a first step in understanding evolutionary relationships and to help guide our efforts to generate informative chimeric molecules to explore related signaling pathways. We have previously provided a complete characterization of the murine AHR gene and partial characterization of the murine ARNT gene (23,32). Among other members of the bHLH-PAS proteins, the genomic DNA structures of Drosophila Per and Sim were also previously characterized (19,25). Recently, our laboratory and others have characterized a homologue of HIF-1α, that we now refer to as HIF-2α (9,16,36). [HIF-2α had also been referred to as MOP2, EPAS1, and HLF (9,16,36). The fact that this homologue has 50% sequence identity with HIF-1α, that it also dimerizes with ARNT, and that it is also hypoxia responsive leads us to suggest that this gene product is more appropriately referred to as HIF-2α.] In an effort to better understand the evolutionary relationship among bHLH-PAS proteins, we compared the structural organization of the mouse Hif-1α gene with the available information of other members of the bHLH-PAS superfamily, mouse Ahr, Drosophila sim, human HIF-2α, and Drosophila per (19,25,32,36) (Fig. 8). This comparison demonstrates that the bHLH domain is always encoded by a single exon and that the PAS domains are commonly encoded by 5–7 exons with highly conserved splice junction sites. However, the variable region is composed of 1 or 2 exons in Sim or Ahri and 7 and 8 exons in the Hif-1α and HIF-2α, respectively. The splice junctions in the variable region are highly conserved between Hif-1α and HIF-2α. These results suggest that Hif-1α and HIF-2α are derived from a common primordial gene and belong to a new subclass of the bHLH-PAS gene superfamily. Similarly, Ahr and Sim belong to a separate subclass in the superfamily (16,32). The poor splice site conservation in Drosophila per protein, coupled with the observation that this gene does not encode a bHLH motif, suggests that per represents another subclass of the bHLH-PAS superfamily.
FIG. 8.
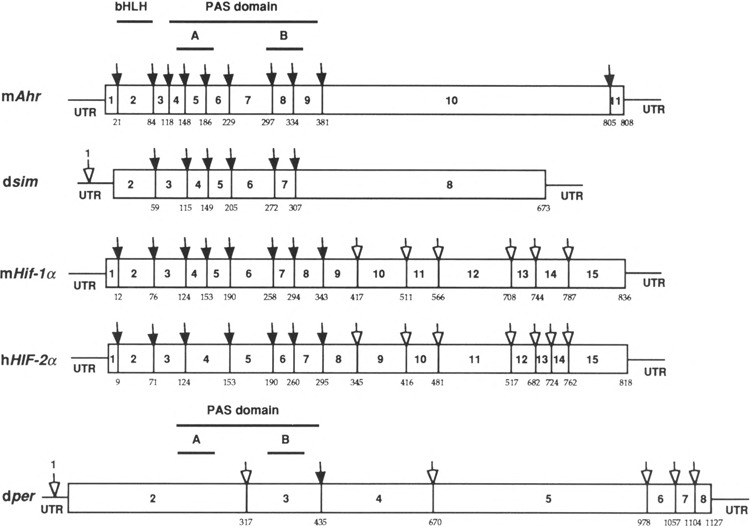
Schematic comparison of splice junction sites of mouse AHR, Drosophila Sim, mouse HIF-1α, human HIF-2α, and Drosophila Per. The protein sequences of mAHR (33), dSIM (25), mHIF-1α, hHIF-2α (36), and dPER (19) were aligned by the clustal method using the DNASTAR program (DNA Star, Madison, WI). The numbers of amino acids at which the splicing occurs were marked underneath each protein. The filled arrows represent splice junction sites that are conserved compared with that of mAHR. The hollow arrows represent those splice junction sites that are not conserved. The numbers of exons were labeled either between splice junctions or on top of the arrows above 5′ UTR.
Conclusion
The murine Hif-1α locus will undoubtedly be the focus of continued scientific investigation in coming years. This expanding interest arises from its role in the regulation of EPO, VEGF, and glycolysis under conditions of low oxygen and low glucose (12,23,34,35). The observation that an endothelial-specific homologue, Hif-2α, also exists suggests that the corresponding promoters are likely to utilize distinct tissue-specific regulatory elements to control promoter activity. Our characterization of the Hif-1α promoter and its structural gene is a first step in understanding how these HIFs are regulated under normal condition and physiological stress. Description of the structural gene is also a requisite step in attempts to generate informative murine strains harboring null or mutant alleles at this locus. Such mutant strains have already provided great insights into the biology of other bHLH-PAS proteins (11,23,33).
ACKNOWLEDGMENTS
We thank Drs. Peggy Farnham and Tom Gilmore for their help and Alan Poland for the gift of pmARNT.
REFERENCES
- 1. Amati B.; Land H. Myc-Max-Mad: A transcription factor network controlling cell cycle progression, differentiation and death. Curr. Opin. Gen. Dev. 4:102–108; 1994. [DOI] [PubMed] [Google Scholar]
- 2. Carver L. A.; Hogenesch J. B.; Bradfield C. A. Tissue specific expression of the rat Ah-receptor and ARNT mRNAs. Nucleic Acids Res. 22:3038–3044; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan W. K.; Chu R.; Jain S.; Reddy J. K.; Bradfield C. A. Baculovirus expression of the Ah receptor and Ah receptor nuclear translocator. Evidence for additional dioxin responsive element-binding species and factors required for signaling. J. Biol. Chem. 269:26464–26471; 1994. [PubMed] [Google Scholar]
- 4. Chen Q. K.; Hertz G. Z.; Stormo G. D. MATRIX SEARCH 1.0: A computer program that scans DNA sequences for transcriptional elements using a database of weight matrices. Comput. Appl. Biosci. 11:563–566; 1995. [DOI] [PubMed] [Google Scholar]
- 5. Citri Y.; Colot H. V.; Jacquier A. C.; Yu Q.; Hall J. C.; Baltimore D.; Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature 326:42–47; 1987. [DOI] [PubMed] [Google Scholar]
- 6. Crews S. T.; Thomas J. B.; Goodman C. S. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell 52:143–151; 1988. [DOI] [PubMed] [Google Scholar]
- 7. Dolwick K. M.; Swanson H. I.; Bradfield C. A. In vitro analysis of Ah receptor domains involved in ligand-activated DNA recognition. Proc. Natl. Acad. Sci. USA 90:8566–8570; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eisen H. J.; Hannah R. R.; Legraverend C.; Okey A. B.; Nebert D. W. The Ah-receptor: Controlling factor in the induction of drug-metabolizing enzymes by certain chemical carcinogens and other environmental pollutants. Biochem. Actions Horm. X:227–257; 1983. [Google Scholar]
- 9. Ema M.; Taya S.; Yokotanii N.; Sogawa K.; Matsuda Y.; Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1-alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA 94:4273–4278; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Feinberg A. P.; Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6–13; 1983. [DOI] [PubMed] [Google Scholar]
- 11. Fernandez-Salguero P.; Pineau T.; Hilbert D. M.; McPhail T.; Lee S. S. T.; Kimura S.; Nebert D.; Rudikoff S.; Ward J. M.; Gonzalez F. J. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science 268:722–726; 1995. [DOI] [PubMed] [Google Scholar]
- 12. Forsythe J. A.; Jiang B. H.; Iyer N. V.; Agani F.; Leung S. W.; Koos R. D.; Semenza GL Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16:4604–4613; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fukunaga B. N.; Probst M. R.; Reisz-Porszasz S.; Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor. J. Biol. Chem. 270:29270–29278; 1995. [DOI] [PubMed] [Google Scholar]
- 14. Goldberg M. A.; Dunning S. P.; Bunn H. F. Regulation of the erythropoietin gene: Evidence that the oxygen sensor is a heme protein. Science 242:1412–1415; 1988. [DOI] [PubMed] [Google Scholar]
- 15. Hoffman E. C.; Reyes H.; Chu F. F.; Sander F.; Conley H.; rooks A.; Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252:954–958; 1991. [DOI] [PubMed] [Google Scholar]
- 16. Hogenesch J. B.; Chan W. C.; Jackiw V. H.; Brown R. C.; Gu Y. Z.; Pray-Grant M.; Perdew G. H.; Bradfield C. A. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interact with components of the dioxin signaling pathway. J. Biol. Chem. 272:8581; 1997. [DOI] [PubMed] [Google Scholar]
- 17. Huang Z. J.; Edery I.; Rosbash M. PAS is a dimerLtion domain common to Drosophila period and several transcription factors. Nature 364:259–262; 1993. [DOI] [PubMed] [Google Scholar]
- 18. Huang L. E.; Arany Z.; Livingston D. M.; Bunn H. F. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 271:32253–32259; 1996. [DOI] [PubMed] [Google Scholar]
- 19. Jackson F. R.; Bargiello T. A.; Yun S. H.; Young M. W. Product of per locus of Drosophila shares homology with proteoglycans. Nature 320:185–188; 1986. [DOI] [PubMed] [Google Scholar]
- 20. Jiang B. H.; Rue E.; Wang G. L.; Roe R.; Semenza G. L. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 271:17771–17778; 1996. [DOI] [PubMed] [Google Scholar]
- 21. Kollmar R.; Farnham P. J. Site-specific initiation of transcription by RNA polymerase II. Proc. Soc. Exp. Biol. Med. 203:127–139; 1993. [DOI] [PubMed] [Google Scholar]
- 22. Li H.; Ko H. P.; Whitlock J. P. Induction of phosphoglycerate kinase 1 gene expression by hypoxia. Roles of Arnt and HIF1alpha. J. Biol. Chem. 271:21262–21267; 1996. [DOI] [PubMed] [Google Scholar]
- 23. Maltepe E.; Schmidt J. V.; Baunoch D.; Bradfield C. A.; Simon M. C. ARNT is essential for responses to oxygen and glucose deprivation and developmental angiogenesis in the mouse. Nature 386:403–407; 1997. [DOI] [PubMed] [Google Scholar]
- 24. Nambu J. R.; Lewis J. O.; Wharton K. A. Jr.; Crews S. T. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67:1157–1167; 1991. [DOI] [PubMed] [Google Scholar]
- 25. Nambu J. R.; Franks R. G.; Hu S.; Crews S. T. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell 63:63–75; 1990. [DOI] [PubMed] [Google Scholar]
- 26. Olson E. N. MyoD family: A paradigm for development? Genes Dev. 4:1454–1461; 1990. [DOI] [PubMed] [Google Scholar]
- 27. Poland A.; Knutson J. C. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: Examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 22:517–554; 1982. [DOI] [PubMed] [Google Scholar]
- 28. Pollenz R. S.; Sattler C. A.; Poland A. The aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator protein show distinct sub-cellular localizations in Hepa 1c1c7 cells by immunofluorescence microscopy. Mol. Pharmacol. 45:428–438; 1994. [PubMed] [Google Scholar]
- 29. Probst M. R.; Fan C. M.; Tessier-Lavigne M.; Hankinson O. Two murine homologs of the Drosophila single-minded protein that interact with the mouse aryl hydrocarbon receptor nuclear translocator protein. J. Biol. Chem. 272:4451–4457; 1997. [DOI] [PubMed] [Google Scholar]
- 30. Pugh C. W.; O’Rourke J. F.; Nagao M.; Gleadle J. M.; Ratcliffe P. J. Activation of hypoxi-inducible factor-1; definition of regulatory domains within the α subunit. J. Biol. Chem. 272:11205–11214; 1997. [DOI] [PubMed] [Google Scholar]
- 31. Sanger F.; Nicklen S.; Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463–5467; 1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt J. V.; Carver L. A.; Bradfield C. A. Molecular characterization of the murine Ahr gene. Organization, promoter analysis, and chromosomal assignment. J. Biol. Chem. 268:22203–22209; 1993. [PubMed] [Google Scholar]
- 33. Schmidt J. V.; Su G. H.-T.; Reddy J. K.; Simon M. C.; Bradfield C. A. Characterization of a murine Ahr null allele: Animal model for the toxicity of halogenated dioxins and biphenyls. Proc. Natl. Acad. Sci. USA 93:6731–6736; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Semenza G. L.; Roth P. H.; Fang H. M.; Wang G. L. Transcriptional legulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269:23757–23763; 1994. [PubMed] [Google Scholar]
- 35. Semenza G. L.; Nejfelt M. K.; Chi S. M.; Antonarakis S. E. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc. Natl. Acad. Sci. USA 88:5680–5684; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tian H.; McKnight S. L.; Russell D. W. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72–82; 1997. [DOI] [PubMed] [Google Scholar]
- 37. Wang G. L.; Jiang B. H.; Rue E. A.; Semenza G. L Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510–5514; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weintraub H.; Davis R.; Tapscott S.; Thayer M.; Krause M.; Benezra R.; Blackwell T. K.; Turner D.; Rupp R.; Hollenberg S.; Zhuang Y.; Lassar A. The myoD gene family: Nodal point during specification of the muscle cell lineage. Science 251:761–766; 1991. [DOI] [PubMed] [Google Scholar]
- 39. Wenger R. H.; Rolfs A.; Marti H. H.; Guenet J.-L.; Gassmann M. Nucleotide sequence, chromosomal assignment and mRNA expression of mouse hypoxia-inducible factor-1α. Biochem. Biophys. Res. Commun. 223:54–59; 1996. [DOI] [PubMed] [Google Scholar]
- 40. Zhou Y. D.; Barnard M.; Tian H.; Li X.; Ring H. Z.; Francke U.; Shelton J.; Richardson J.; Russell D. W.; McKnight S. L. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. USA 94:713–718; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


