Abstract
Vertebrate small nuclear RNA (snRNA) gene promoters contain a distal, enhancer-like region that is composed of an octamer motif adjacent to at least one other element. Here we show that a human U6 snRNA distal region contains a SPH motif previously found in several chicken snRNA gene enhancers and the 5′-flanking region of vertebrate selenocysteine tRNA genes. SPH binding factor (SBF) was detected in either chicken or HeLa cell extracts that could bind SPH elements in a species-independent manner. Both human and chicken SBF required divalent cation to bind effectively to DNA. DNase I footprinting experiments indicated that human SBF specifically protected the human U6 SPH element. Furthermore, a SBF polypeptide of approximately 85 kDa was detected in both HeLa and chicken extracts following ultraviolet light-mediated cross-linking to human U6 or chicken U4 SPH elements. A part of the human U6 SPH element was quite sensitive to mutation, as demonstrated by both specific protein binding and transcription assays. From these data it is apparent that the distal regions of some RNA polymerase III-and RNA polymerase II-transcribed small RNA promoters are virtually identical in composition, and their mechanisms of transcriptional activation are possibly quite similar.
Keywords: Eukaryotic transcription, snRNA genes, U6 snRNA, SPH binding factor, Enhancer motifs
PROMOTERS for vertebrate small nuclear RNA (snRNA) genes consist of similar transcriptional control elements even though they are recognized by either RNA polymerase II (pol II) or pol III [reviewed in (5,12,25)]. Most genes for the abundant spliceosomal snRNAs are transcribed by pol II (Ul, U2, U4, and U5 snRNAs), whereas the U6 snRNA promoter is recognized by pol III. Both classes of promoters contain a snRNA-specific proximal sequence element (PSE) within 70 bp of the transcription start site. Vertebrate U6 promoters are distinguished by the presence of a TATA box in this region that is absent in the pol II snRNA gene promoters. Paradoxically, it is the presence of the TATA box that determines pol III specificity (14,15).
A distal region upstream of position −200 is present in both classes of vertebrate snRNA gene promoters and has some properties of a transcriptional enhancer. The distal region contains one copy of an octamer motif and an adjacent sequence element that varies among different snRNA promoters. The octamer motif is recognized by the well-studied ubiquitous transcription factor, Oct-1 [reviewed in (8)]. Some of the adjacent sequence elements that have been identified in pol II-transcribed snRNA gene distal regions include SPH motifs (3,19,20,27), a Spl binding site (2,10,22), an AP-2 binding site (23), a CCAAT motif (1), and a cyclic AMP response element (23). One of our laboratories has identified an octamer-adjacent element in a human U6 gene that we denoted as NONOCT, and we detected a factor from cultured human cell extracts that binds NONOCT (6). The relative structural simplicity and conservation of snRNA gene promoters makes them attractive for investigations into mechanisms of transcriptional activation.
In this report we demonstrate that the U6 NONOCT element is an example of the previously characterized snRNA SPH motif, well-studied in chicken genes by one of our laboratories (3,19,20,27). Thus, the organization of the human U6 (pol III) distal control region is remarkably similar to that of chicken U1 and U4B genes (pol II). Moreover, we detected a Mg2+-dependent SPH binding activity in HeLa cell extracts that is similar to chicken SPH binding factor (SBF). Furthermore, we identified a region of the U6 SPH element that is quite sensitive to point mutation as determined by effects on transcriptional activity and on efficiency of human SBF binding to several mutant templates.
MATERIALS AND METHODS
Plasmid Constructions
A plasmid containing the human U6 NONOCT-(SPH) and octamer motifs (pGEM/NPLUSO) was constructed by annealing the oligonucleotides 5′ -GATCCTATTTCCCATGATTCCTTCATAT TTGCATAT-3′ and 5′-GATCATATGCAAATA TGAAGGAATCATGGGAAATAG-3′, followed by ligation into the BamHI site of pGEM3Zf(-) (Promega). This plasmid was used for PCR to prepare probes for gel mobility shift and DNase I footprinting reactions as described below.
Plasmids containing random mutations within the human U6 NONOCT(SPH) motif were constructed using oligonucleotides containing randomized segments in a PCR-based protocol. The two oligonucleotides that were used separately for these constructions were:
RM1: 5′ -TCTAGAGGATCC(C/N)(C/N)(T/N)
(A/N)(T/N)(T/N)(T/N)(C/N)CCATGAT-3′
RM2: 5′-TCTAGAGGATCCCCTATTTC(C/N)
(C/N)(A/N)(T/N)(G/N)(A/N)(T/N)(T/N)-(C/N)CTT CATA
At each randomized position the DNA synthesizer inserted a mixture containing 62.5% of the wild-type base plus 12.5%, each, of the other three bases. Each randomized oligonucleotide was used for PCR with a bottom-strand oligonucleotide whose 5′ end corresponded to position −1 of the human U6 gene and the OCTCONMUT/maxiU6 plasmid (6). The PCR product was restricted with BamUl and Ndel (cuts at position −69 of human U6 flanking region) and gel purified. RM plasmids were constructed by ligation of this PCR fragment with an ∼410 bp Ndel-EcoRI fragment containing a human U6 maxigene (13) and the pGEM3Zf(−) vector (Promega) restricted with BamHI and EcoRI. Thus, all RM plasmids contained 244 bp of 5′-flanking sequence with a disrupted consensus octamer motif and random mutations within the NONOCT(SPH) region, and were in the context of a human U6 maxigene.
Sequences of all mutant U6 promoter templates were determined by the dideoxy method. Purified plasmids were prepared by alkaline lysis of bacterial cells, CsC1 gradient centrifugation, and chro-matography on Bio-Gel A5m resin (BioRad). DNA concentrations were determined spectropho-tometrically using absorbance at 260 nm and verified by visual examination of ethidium bromide-stained agarose gels.
Extracts and Protein Fractions
HeLa nuclear extract was purchased from Promega (cat. #E3110, lot #169301), divided into aliquots, and stored in liquid nitrogen until use. HeLa SI00 extract was prepared and fractionated on phosphocellulose and DEAE-cellulose as described previously (6).
Chicken nuclear extract was prepared from 8–10-day embryos, and SBF was partially purified for the experiments described in Figs. 4 and 7B by heparin agarose chromatography as previously described (19). For the experiment described in Fig. 1C, SBF was further purified by sequence-specific DNA affinity chromatography using an affinity resin prepared by the method of Wu et al. (26) with the following annealed oligonucleotides: 5 -GATCAAACCGCGCGCTGCATGCCGGGA GCACCAC-3′ and 5′-AGCATCGATAGCTGT GGTGCTCCCGGCATGCAGCGCGCGGTTTG ATC-3′. Compared to previous results, we observed that the SBF purified on this affinity resin contained an elevated amount of the species of SBF that forms the faster migrating cl complex on mobility shift gels [e.g., compare Fig. 1C, lanes 3, 5, and 6 of this report with Fig. 4 in (27)]. Because the SBF cl and c2 complexes have identical DNase I footprints (Miyake and Stumph, unpublished data), we believe that the cl complex may be due to a specific proteolysis of SBF that occurs during purification.
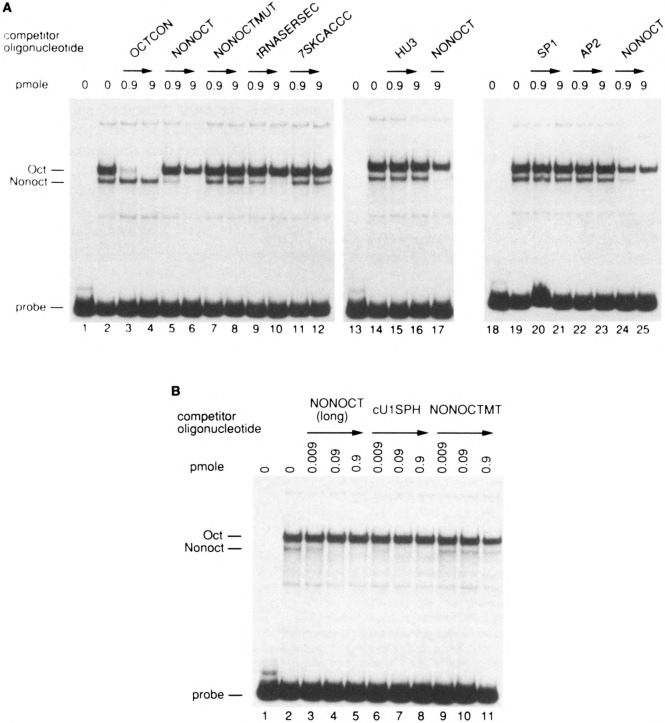
FIG. 4.
Competition of the chicken SBF/DNA complex by the human U6 NONOCT (SPH) element. Electrophoretic mobility shift reactions in lanes 2–21 contained chicken SBF [approximately 5 μg of the heparin agarose fraction (19)] and 5 mM MgCl2. Double-stranded oligonucleotides containing the chicken U1 SPH motif (lanes 1-11) or human NONOCT(SPH) element (lanes 12–22) were used as radiolabeled probes. Approximately 300 pg of radiolabeled probe was used in each binding reaction. Unlabeled competitor oligonucleotides corresponding to the chicken U1 or human U6 SPH motif (or a nonspecific oligonucleotide, NS) were added as indicated above the lanes in the following amounts: 5 ng in lanes 2, 5, 8, 13, 16, 19; 25 ng in lanes 3, 6, 9, 14, 17, 20; and 100 ng in lanes 4, 7, 10, 15, 18, 21. No competitor was added in lanes 11 and 12. The sequences of the cUlSPH and NONOCT(long) oligonucleotides used as probes and competitors are shown in Table 1. The nonspecific oligonucleotide competitor (NS) consisted of the following annealed sequences: 5′-GATCGGTTCAGGGAGCGCGCCGGCGCGCTGTGACGTAG-3′ and 5′-GA TCCTACGTCACAGCGCGCCGGCGCGCTCCCTGAACC-3′.
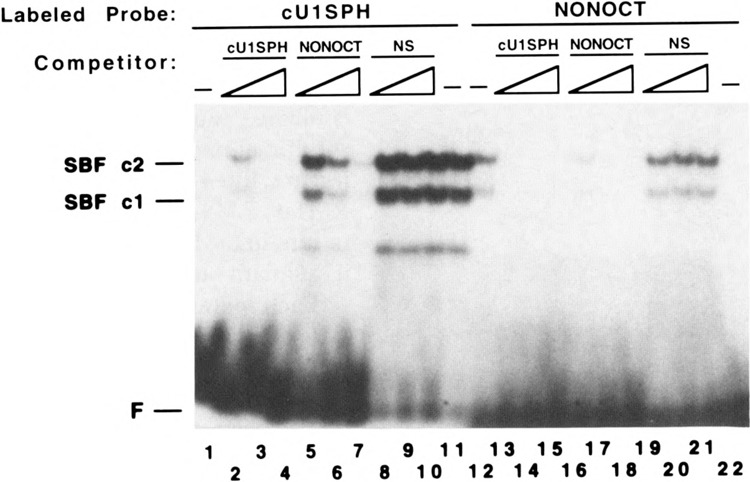
FIG. 7.
Ultraviolet light-mediated cross-linking to identify a DNA binding polypeptide of human and chicken SBF. An internally radiolabeled double-stranded oligonucleotide probe containing the human U6 NONOCT(SPH) and octamer motifs (A) or chicken U4B SPH motif (B) was prepared, incubated with protein, irradiated with ultraviolet light, treated with nucleases, and electrophoresed as described in the Materials and Methods section. (A) The human U6 SPH motif probe was incubated with HeLa extract protein fractionated over phosphocellulose as described (6). During the binding reactions, samples contained unlabeled double-stranded oligonucleotide competitors: lane 1: 3 pmol nonspecific DNA (N.S.); lane 2: 3 pmol OCTCON; lane 3: 3 pmol NONOCT(long). The sequences of OCTCON and NONOCT(long) are shown in Table 1. The N.S. oligonucleotide contained the following sequences that had been previously annealed: 5′-GATCCAGTCTGATCAGACTG-3′ and 5′-GATCCAGTCTGATCAGACTG-3′. Samples were electrophoresed on a 10% polyacrylamide gel. The arrow marked “hSBF” delineates the polypeptide that is specifically cross-linked to the NONOCT(SPH) element. Although this particular experiment did not include a lane of coelectrophoresed protein markers, the average apparent molecular weight of the hSBF polypeptide from four other experiments was approximately 85,000. (B) The chicken U4B SPH probe was incubated with the chicken SBF heparin agarose fraction (lanes 1-5) or with the HeLa P.35 fraction (lanes 6-10). Binding reactions for lanes 1, 5, 6, and 10 contained no added competitor oligonucleotides. Samples run in the other lanes contained competitor oligonucleotides as follows: lanes 2, 7: 3 pmol NONOCT(long); lanes 3, 8: 3 pmol cUlSPH; lanes 4, 9: 3 pmol of a nonspecific oligonucleotide with the sequence described in the legend to Fig. 4. Reactions loaded in lanes 5 and 10 were not irradiated with UV light, nor treated with nucleases. Samples were electrophoresed in an 8% polyacrylamide gel alongside protein markers whose mobilities are delineated to the right.
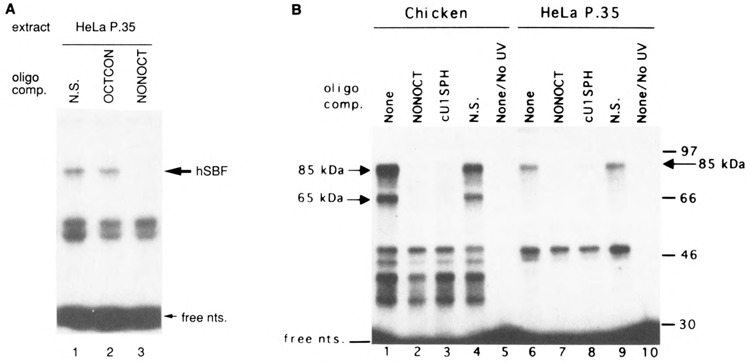
FIG. 1.
Identification of a DNA binding activity in HeLa cell extracts similar to chicken SBF. (A) Sequence of the enhancer region in the chicken U1ψ(O,S) construction. The octamer and SPH motifs are indicated by underlining. Lower-case letters indicate changes from the wild-type sequence, but these do not significantly affect enhancer activity (19). (B) Identification of a Mg2+-dependent DNA binding activity in HeLa cell extracts that interacts with the chicken U1 gene enhancer. A 118-bp 32P-labeled DNA fragment containing sequences of the U1 enhancer was incubated with increasing amounts of HeLa cell nuclear extract, and protein/DNA complexes were resolved by electrophoretic mobility shift assays. Complexes observed in the absence of MgCl2 (lanes 1-5) are indicated to the left of the autoradiograph, whereas complexes formed in the presence of 5 mM MgCl2 (lanes 6-10) are indicated to the right. (C) Similarity of electrophoretic mobilities and Mg2+-dependent DNA binding properties of chicken and human SBF/DNA complexes. Assays were performed as in (B), except that in lanes 2, 3, 5, and 6 the protein fraction used contained SBF purified from chicken embryo nuclear extracts by heparin agarose and DNA affinity chromatography. The SBF used in this experiment had been dialyzed against buffer containing 12.5 mM MgCl2. Thus, there is a minor contribution of MgCl2 from the added SBF (e.g., ∼0.4 mM MgCl2 final concentration in lane 6).
Electrophoretic Mobility Shift Assays
Experiments shown in Figs. 1, 2, and 4
FIG. 2.
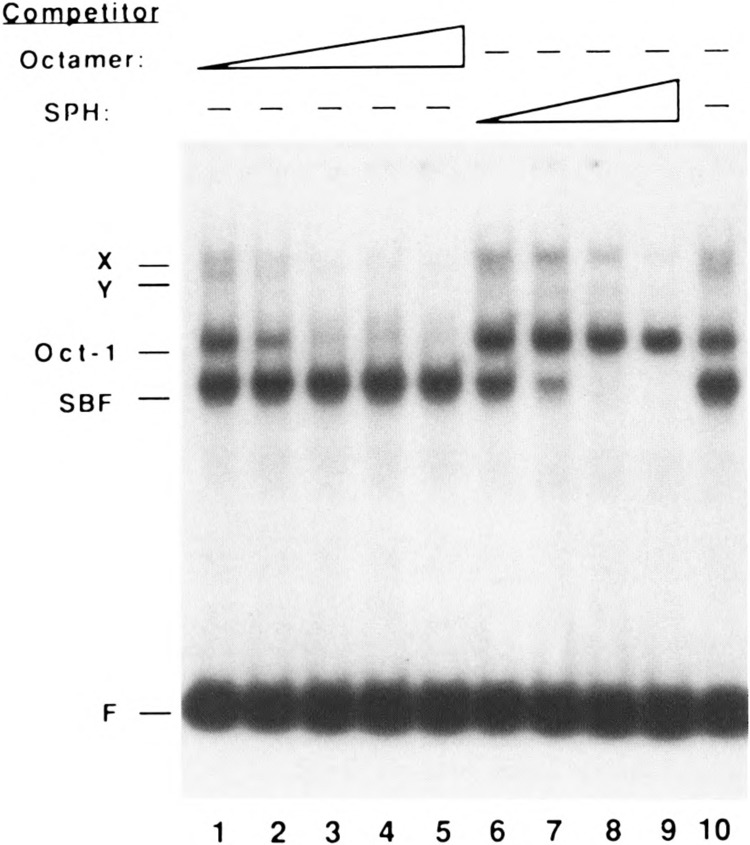
Competition of specific complexes by synthetic oligo-nucleotides containing octamer and SPH motifs. Electropho-retic mobility shift assays were performed as in Fig. 1 using a constant amount of HeLa nuclear extract (0.5 μl) in the presence of 5 mM MgCl2 and increasing amounts of octamer competitor (cUlOCT; 0.2, 1, 5, 25, and 125 ng, lanes 1-5), increasing amounts of SPH motif competitor (cU l SPH; 0.2, 1, 5, and 25 ng, lanes 6-9), or no competitor (lane 10). Cold competitor oligonucleotides were mixed with the labeled DNA fragment prior to adding the extract. Note that the band labeled Oct-1 was specifically competed by the octamer oligonucleotide, and the band labeled SBF was specifically competed by the SPH motif oligonucleotide. Although the molecular compositions of the X and Y complexes were not directly investigated in this study, the data are consistent with the possibility that the X complex contains two molecules of Oct-1, and that the Y complex contains both Oct-1 and SBF.
For the experiments shown in Figs. 1 and 2, a DNA fragment was generated by PCR using the plasmid U1ψ(O,S) (3) as a template and the oligonucleo-tides 5′-AAGTTCTAGATCGCCAGGGTTTTC CCAGTC-3′ and 5′-CTCCACGCCGAAGGTA CGCT-3′ as primers. The first oligonucleotide is identical to pUC19 vector DNA sequence located 250 to 279 bp upstream of the U1 transcription start site in U1Ψ (0,S), except for the underlined nucleotides that were altered to introduce XbaI and DpnII restriction sites. The second oligonucleotide is complementary to positions −118 to −137 in the U1 gene 5′-flanking DNA. PCR with these primers and U1Ψ(O,S) as a template generated a fragment 162 bp in length that contained the U1 enhancer and could be cut near each end with DpnII. Following DpnII digestion, the 118 bp central fragment was isolated by polyacryl-amide gel electrophoresis and labeled with 32P by filling in the 3′ recessed ends. For the experiment shown in Fig. 4 the cUlSPH and NONOCT(long) double-stranded oligonucleotides (Table 1) were labeled by a similar fill-in reaction.
TABLE 1.
OLIGONUCLEOTIDES USED FOR ELECTROPHORETIC MOBILITY SHIFT EXPERIMENTS
| Name | Length | Sequence |
|---|---|---|
| cUlSPH | 34 | GATCAAACCGCGCGCTGCATGCCGGGAGCACCACTTTGGCGCGCGACGTACGGCCCTCGTGGTGCTAG |
| cU1OCT | 22 | GATCGGAGCATGCAAATTAACTCCTCGTACGTTTAATTGACTAG |
| OCTCON | 17 | GATCCATATTTGCATATGTATAAACGTATACTAG |
| NONOCT (short) | 20 | GATCCTATTTCCCATGATTCGATAAAGGGTACTAAGCTAG |
| NONOCT (long) | 30 | GATCAGGGCCTATTTCCCATGATTCCTTCATCCCGGATAAAGGGTACTAAGGAAGTCTAG |
| NONOCTMUT* | 20 | GATCCTtcTagagATGATTCGAagAtctcTACTAAGCTAG |
| tRNASERSEC | 20 | CCGTTTCCCAGAATGCGCGG GGCAAAGGGTCTTACGCGCC |
| 7SKCACCC | 20 | CATGCCCCACCCATCTGCAA GTACGGGGTGGGTAGACGTT |
| HU3 | 20 | GTTTGTGATTGGCTGTCATT CAAACACTAACCGACAGTAA |
| SP1 | 22 | ATTCGATCGGGGCGGGGCGAGC TAAGCTAGCCCCGCCCCGCTCG |
| AP-2 | 26 | GATCGAACTGACCGCCCGCGGCCCGT CTAGCTTGACTGGCGGGCGCCGGGCA |
Lower-case letters indicate nucleotide changes between NONOCTMUT and NONOCT.
Various amounts of unfractionated HeLa nuclear extract or affinity-purified chicken SBF (Figs. 1 and 2) or heparin-agarose fractionated chicken SBF (Fig. 4) were incubated in a 20 μ1 reaction volume containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 11 mM dithiothreitol, 5% glycerol, 1 mM EDTA, 6 μg of poly(dA-dT) poly(dA-dT), and 20,000–30,000 cpm (∼0.5 ng) of 32P-labeled DNA. For the binding assays, MgCl2 was either omitted from the reactions or was included at various concentrations as indicated in the text and figures. For certain experiments, unlabeled competitor oligonucleotides, corresponding to the chicken U1 octamer motif or SPH motif or the human U6 NONOCT(long) sequence (Table 1), were added as described in the text and figure legends. The DNA/protein complexes were resolved from unbound DNA in native 4% polyacrylamide gels. The gels were electrophoresed in a cold room at 120 volts (22 mA) in 25 mM Tris-HCl (pH 8.3), 190 mM glycine, 1 mM EDTA as running buffer without circulation.
Experiments shown in Figs. 3 and 8D.
FIG. 3.
Competition of the human U6 NONOCT(SPH) complex by oligonucleotides containing vertebrate snRNA gene distal region elements. (A) Radiolabeled DNA probe containing wild-type human U6 NONOCT(SPH) and OCTCON sequences was used for electrophoretic mobility shift assays as described in the Materials and Methods section. Each sample contained approximately 3 fmol of probe DNA and 2 μg of protein from a HeLa cell SI00 extract. Competitor double-stranded oligonucleotides (sequences given in Table 1) were mixed into binding reactions prior to addition of the HeLa extract in amounts denoted above each lane. In (B), a longer version of the NONOCT-(SPH) region sequence was used [NONOCT(long) in Table 1]. Comparison of the efficiency of competition by the NONOCT(long) and NONOCT [“short”; (A) and (6)] oligonucleotides indicated a greater than 10-fold higher affinity for the longer oligonucleotide (results not shown).
FIG. 8.
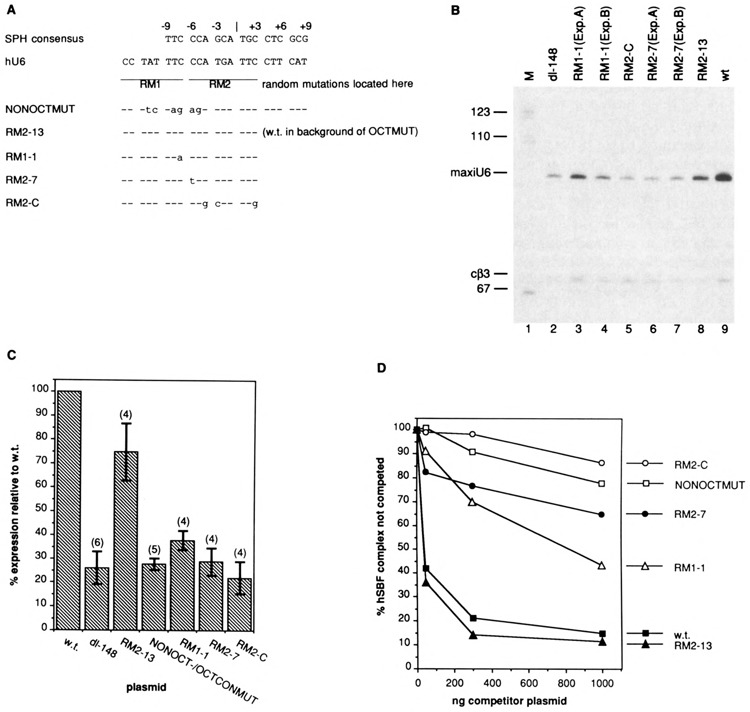
Mutations located in the human U6 NONOCT(SPH) region decrease U6 maxigene expression in transfected cells and are inefficient competitors in electrophoretic mobility shift assays. (A) Locations of mutations in templates used in these experiments. Sequences are aligned according to the SPH consensus from Fig. 5. The NONOCTMUT clustered point mutant was described previously (6). Random mutations were introduced using a degenerate primer by PCR as described in the Materials and Methods section. Two degenerate primers, RM1 and RM2, were used in separate PCR preparations, and the region of degeneracy for each primer is delineated by the lines. All random mutation (RM) plasmids also contained a clustered point mutation to disrupt the consensus octamer motif [OCTCONMUT in (6)]. (B) Transient expression of U6 maxigene templates containing random mutations in the region of the NONOCT(SPH) element after transfection of human 293 cells. U6 maxigene expression in transfected cells was detected by primer extension analysis and electrophoresis on 10% polyacrylamide/8.3 M urea gels. “MaxiU6” represents the primer extension product from U6 maxigene RNA, and “cβ3” represents the major primer extension product from transcripts initiated from a chicken 0-tubulin gene contained in a cotransfected plasmid used as a control to normalize for variable transfection efficiency and RNA recovery. The lane marked “M” contained radiolabeled DNA fragments from Mspl digestion of pGEMl plasmid DNA. The “dl-148” plasmid used to generate the sample electrophoresed in lane 2 was a U6 maxigene template lacking 5′-flanking sequence upstream of –148, and, hence, missing the distal region. (C) Quantitation of transfection experiments. Primer extension products that had been electrophoresed on polyacrylamide gels were quantitated with a Fujix BAS2000 Phosphorimager (Fuji). After background subtraction, the level of each maxiU6 band was normalized according to the cβ3 band intensity in that lane and compared with that from a wild-type (w.t.) U6 maxigene template included in each experiment. The height of each bar represents the average value from at least four separate experiments (actual number of samples tested given in parentheses above each bar), and the height from the midpoint of the error bar shows one standard deviation from the mean. All mutant templates contained a disrupted consensus octamer motif. The “NONOCT-/OCTCONMUT” template contained both disrupted octamer and NONOCT(SPH) motifs [NONOCTMUT in (A)]. (D) Electrophoretic mobility shift assays using NONOCT(SPH) region random mutant templates as competitors. A radiolabeled probe containing both the NONOCT(SPH) and octamer motifs from the human U6 distal region was prepared and used for gel shift assays as described in the Materials and Methods section except all binding reactions contained 20 μg/ml poly(dl-dC)·poly(dl-dC) and 20 μg/ml plasmid DNA. Binding reactions contained 2 μg of protein from a HeLa SI00 extract. To effect specific competition of the hSBF/DNA complex, 50, 300, or 1000 ng of plasmid DNAs containing a normal NONOCT(SPH) motif (RM2-13) or mutant templates were substituted for the same amount of pGEM3Zf(–) DNA in binding reactions. Radioactivity in hSBF/DNA complexes separated on native polyacrylamide gels was determined using a Fujix BAS2000 Phosphorimager (Fuji). After background subtraction, the amount of radioactivity at each titration point was compared as a percentage to the signal from the sample where only 1 μg, pGEM3Zf(–) vector, and no specific competitor, was used as the plasmid DNA. Each data point is the average of two experiments.
A radiolabeled DNA fragment containing the human U6 NONOCT(SPH) and octamer motifs was prepared by PCR using pGEM/NPLUSO plasmid DNA, SP6 promoter primer (Promega) radiola-beled with [γ-32P]ATP and polynucleotide kinase, and T7 promoter primer (Promega). The 140-bp PCR product was purified by agarose gel electrophoresis, incubated with proteins, and electrophoresed on 4% polyacrylamide gels as described previously (6), except for variations described in the figure legends. All binding reactions contained MgCl2 at a final concentration of 2 mM. The sequences of unlabeled competitor oligonucleotides used for experiments in Fig. 3 are listed in Table 1.
DNase I Footprinting
For footprinting the human U6 distal region sequence, the singly end-labeled DNA fragment was the same one as described above for gel shift reactions. Approximately 3 fmol (20,000 dpm) was mixed with or without protein in a total volume of 25 μ1 containing 40 μg of poly(dI-dC) poly(dl-dC) per ml, 2 mM MgCl2, 16 mM 4 - (2 - hydroxyethyl) -1 - piperazineethanesulfonic acid (HEPES) (pH 7.9), 0.16 mM EDTA, 16% glycerol, 80 mM KC1, and 0.8 mM dithiothreitol for 30 min at 30°C. Then CaCl2 was added to a final concentration of 0.1 mM, followed by 1 pi of DNase I (15 ng/μl). After incubation for 2 min at room temperature the reaction was quenched by adding EDTA, and DNA was extracted and electrophoresed on a 10% polyacrylamide/8.3 M urea gel as described previously (6).
Transfection Experiments
Transient transfection assays using human 293 cells and primer extension to quantitate RNA levels were performed exactly as described previously (6). Relative radioactivity in specific bands on polyacrylamide gels was quantitated using a Fujix BAS2000 Phosphorimager (Fuji).
UV Cross-Linking
Internally radiolabeled double-stranded oligonucleotides containing the linked human U6 NONOCT(SPH) and octamer motifs or the chicken U4B SPH element were prepared essentially by a previously described technique (24). For the human U6 probe, oligonucleotides NPLU-SOBT: 5′ -GATCATATGCAAATATGAAGGA ATCATGGGAAATAG-3′ and UV1: 5′-CTAT TTCCCA-3′ were annealed and filled in with Klenow DNA polymerase, [α-32P]dATP, 5-bromo-2′ -deoxyuridine-5′ -triphosphate (Sigma), dCTP, and dGTP. For the chicken U4 SPH probe, the two oligonucleotides 5′-GCATA GCGCgaaCCAGCATGCaTaGCGGCCGCCCA -3′ and 5′-TGGGCGGCCG-3′ were annealed and filled in with Klenow DNA polymerase and dN-TPs, including 5-bromo-2′-deoxyuridine-5′-triphosphate and [α-32P]dCTP. This probe was based upon the chicken U4B gene SPH motif sequence, but contains several substitutions shown in lower case letters to maximize the incorporation of BrdU and radiolabel. These substitutions did not significantly affect binding of SBF in competitive mobility shift assays (data not shown).
For the experiment shown in Fig. 7A, approximately 40 fmol (100,000 dpm) of U6 probe was mixed with 11 μg HeLa phosphocellulose extract in a solution with a total volume of 20 μ1 containing 100 μg of poly(dI-dC)-poly(dI-dC) per ml, 2 mM MgCl2, 50 mM KC1, 10 mM HEPES (pH 7.9), 10% glycerol, 0.1 mM EDTA, 0.5 mM dithio-threitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg/m1 of pepstatin A, and 0.5 μg/ml of leupeptin. Each mixture contained unlabeled oligonucleotide competitors as detailed in the figure legend. Samples were incubated at 30°C for 30 min and irradiated for 15 min with 254 nm ultraviolet light in a Stratalinker UV Crosslinker (Strata-gene). During irradiation the samples were held in ice at a distance of approximately 11 cm from the bulbs to the bottom of the tube. Immediately following irradiation, CaCl2 was added to a final concentration of 10 mM and DNA was digested with a mixture of micrococcal nuclease (Sigma) at 2.8 units/ml and RQ1 DNase I (Promega) at 83 units/ml at 37 °C for 30 min. Samples were then diluted twofold with SDS loading buffer and elec-trophoresed on 10% polyacrylamide Laemmli gels. Dried gels were autoradiographed with an intensifying screen.
For the experiment shown in Fig. 7B, approximately 50 fmol of U4B SPH probe (150,000 cpm) was incubated with 5 μg chicken SBF heparin agarose fraction or 7 μg HeLa phosphocellulose fraction in a total volume of 24 μ1 containing 2 μg poly (dI-dC)poly(dl-dC), 20 mM HEPES (pH 7.9), 5 mM MgCl2, 0.01 mM ZnCl2, 0.2 mM EDTA, 16% glycerol, and 2 mM dithiothreitol. Some reactions contained 3 pmol of competitor oligonucleotide as described in the legend. After incubation at 20°C for 30 min, the open tubes were irradiated with 312 nm ultraviolet light for 45 min at a distance of 10 cm. CaCl2 was added to a final concentration of 10 mM, followed by 0.5 units of DNase I and 0.02 units of micrococcal nuclease. Samples were incubated at 37°C for 1 h and analyzed by electrophoresis in 8% polyacryl-amide Laemmli gels.
RESULTS
A Mg2+-Dependent DNA Binding Activity in a HeLa Cell Extract Has Properties Similar to Chicken SBF
A 147-bp DNA fragment containing the octamer and SPH motifs of the chicken U1 snRNA gene enhancer (sequence shown in Fig. 1A) was incubated with increasing amounts of HeLa cell extract, and protein-bound DNA was separated from free DNA by native gel electrophoresis. In the absence of MgCl2, two complexes of retarded mobility were apparent (labeled Oct-1 and X in Fig. 1B, lanes 1–5). The band labeled Oct-1 was previously shown to be due to an activity that footprints on the octamer sequence in the U1 enhancer (20), undoubtedly the ubiquitous Oct-1 factor originally identified in HeLa extract. In the presence of MgCl2 (5 mM), two additional complexes (labeled SBF and Y) could be observed (Fig. 1B, lanes 6–10), although the Y complex was poorly resolved from the X complex in the experiment shown in Fig. 1B.
It seemed possible that the Mg2+-dependent DNA binding activity present in HeLa cell extracts could be due to a human homolog of chicken SBF. Therefore, we next investigated the Mg2+ dependence of chicken SBF DNA binding activity by using SBF prepared from chicken embryo nuclear extracts and purified by sequence-specific DNA affinity chromatography (Fig. 1C, lanes 2, 3, 5, and 6). In the absence of MgCl2, only a low level of binding of SBF to the U1 enhancer was observed (lanes 2 and 6). In contrast, inclusion of 5 mM MgCl2 resulted in strong SBF c2 and cl complex formation (lanes 3 and 5). Thus, the specific binding of chicken SBF to DNA is highly dependent upon the presence of Mg2+ in the binding reaction. The ability of SBF to bind DNA increased sharply between 0.5 and 1.5 mM MgCl2, but decreased above 5 mM MgCl2 (results not shown). Moreover, it was notable that the chicken SBF c2 complex and the Mg2+-dependent HeLa complex migrated with nearly identical mobilities. The similarity of these properties provided initial evidence that the HeLa extract contains a factor homologous to chicken SBF. [It is worth noting that the SBF c2 complex is believed to represent the DNA/protein complex that contains full-length chicken SBF, whereas cl is likely due to a specific degradation product of SBF produced during purification (our unpublished results).]
To confirm the identities of the complexes formed on the chicken U1 enhancer, the competition assays shown in Fig. 2 were performed. Increasing amounts of unlabeled octamer motif oli-gonucleotide specifically competed away the Mg2+-independent band labeled Oct-1 (lanes 1-5), but not the Mg2+-dependent band labeled SBF. Conversely, unlabeled SPH motif oligonucleotide specifically competed away the SBF band (lanes 6–9), but not the Oct-1 band. These competition experiments provided strong evidence that the designated bands are due, respectively, to human Oct-1 and to a second human protein with DNA binding properties very similar or identical to chicken SBF.
The Human U6 NONOCT Element Is Similar to a Small RNA Gene SPH Element
In previous work we identified a distinct DNA sequence immediately upstream of the consensus octamer motif of a human U6 snRNA gene that was required for efficient template activity in transient transfection experiments and was bound by a factor in crude HeLa cell extracts (6). It was important to determine whether this transcriptional control element was novel or another representative of a previously identified family of elements. Although the extent of the U6 NONOCT sequence was not known, we noticed some sequence similarity within this region to motifs located adjacent to octamers of other small RNA gene distal promoter regions. For example, within the NONOCT region, 7 out of 10 bp were identical with a bovine tRNA(Ser)Sec SPH motif [CCCA GAATGC (16)], 6 out of 10 bp were identical with a chicken U1 snRNA gene SPH motif [CCCGG CATGC (19,20)], 6 out of 8 bp matched an AP-2 consensus that is found in the human U4 snRNA gene 5′-flanking region [CCC(A/C)N(G/C)(G/ C)(G/C) (23)], 5 out of 9 bp matched a sequence including the CACCC motif from a human 7SK gene [CACCCATCT (11)], and 7 out of 10 bp were identical with the sequence surrounding the CCAAT motif from a rat U3 snRNA gene distal region [CCAATCATAC (1)].
Complementary, 20-mer oligonucleotides including the human 7SK CACCC, rat U3 CCAAT, and bovine SPH motifs (Table 1) were synthesized and annealed to use as competitors in a gel mobility shift assay. In addition, double-stranded Spl and AP-2 oligonucleotides were purchased from Promega and used as competitors (Table 1). The gel mobility shift assay employed a probe containing both the NONOCT and consensus octamer (OCTCON) motifs of the human U6 distal region and a crude HeLa cell S100 extract containing both binding activities (Fig. 3). The upper band of the major doublet corresponds to an octamer complex because it was completely eliminated by an excess of OCTCON oligonucleotide (Fig. 3A, lanes 2–4). The lower band of the doublet corresponds to a Nonoct complex, because it was competed by the NONOCT oligonucleotide (Fig. 3A, lanes 5, 6), but not a similar oligonucleotide containing a set of clustered point mutations (NON-OCTMUT; Fig. 3A, lanes 7, 8). Of the other competitor DNAs, only the bovine tRNA(Ser)Sec SPH motif decreased the amount of Nonoct complex (tRNASERSEC; Fig. 3A, lanes 9, 10). Therefore, it is possible that the human U6 NONOCT motif is a representative of the small RNA SPH elements.
To further test the specificity of the NONOCT-(SPH) binding factor we used an oligonucleotide corresponding to the chicken U1 snRNA gene SPH motif in a competition gel mobility shift assay. This oligonucleotide served as a very efficient competitor in our reactions with the human U6 distal region probe and a HeLa cell S100 extract (Fig. 3B, lanes 6-8). Binding to the chicken U1 SPH DNA was at least as good as to the human U6 NONOCT DNA (Fig. 3B, lanes 3–5).
The converse experiment was performed also, in which a protein preparation containing chicken SPH binding factor (SBF) was used for gel mobility shift experiments. The human U6 NONOCT oligonucleotide was a specific competitor for binding of SBF to a chicken U1 SPH motif (Fig. 4, compare lanes 5–7 with lanes 8–11), although not as effective as the homologous chicken U1 SPH competitor (lanes 2–4). Furthermore, specific chicken SBF complexes were formed on a radiolabeled human U6 NONOCT probe (lane 12), and these were competed with either a chicken U1 SPH or U6 NONOCT oligonucleotide, but not with a nonspecific DNA oligonucleotide (Fig. 4, lanes 13–21).
These results confirm that the human U6 NONOCT element is a bona fide small RNA gene promoter SPH motif. A comparison of the human U6 sequence with several functional SPH motifs, as well as a consensus sequence derived from them, is shown in Fig. 5. Of the six sequences, the human U6 is the most divergent, and this probably accounts for it being a less effective competitor in the band shift assays than the chicken U1 sequence (Figs. 3B and 4). The left, or distal, portion of the U6 SPH motif is the most conserved relative to the consensus sequence. The right end, on the other hand, is more divergent. Particularly striking is the absence of the GCG triplet in the last three positions that are perfectly conserved in the other examples. Perhaps this divergence is related to the fact that the right end of the SPH motif in the human U6 distal region is directly abutted to the adjacent octamer motif with no intervening nucleotides.
FIG. 5.

Comparison of sequences of snRNA distal region SPH motifs and spacing to octamer motifs. The sequences of several SPH motifs and of a consensus derived from them are shown. Also noted are the distance to and the orientation of the octamer motif that is nearby the SPH element for all but the Xenopus tRNA(Ser)Sec promoter. The consensus sequence of OCTforw is ATGCAAAT and OCTrev is ATTTGCAT. The sequences listed are from the following references: chicken U1 (20), chicken U4B and U4X (9), X. laevis tRNA(Ser)Sec (16), bovine tRNA(Ser)Sec (7), and human U6 (13).
Characterization of Human SPH Binding Factor
To further analyze the binding of the HeLa cell factor to the SPH motif of the human U6 distal region, we performed DNase I footprinting experiments using partially purified transcription factor. Two steps of fractionation, using phosphocellulose and DEAE-cellulose chromatography, yielded a fraction that protected only the U6 SPH motif sequence (Fig. 6, compare lanes 2 and 3).
FIG. 6.
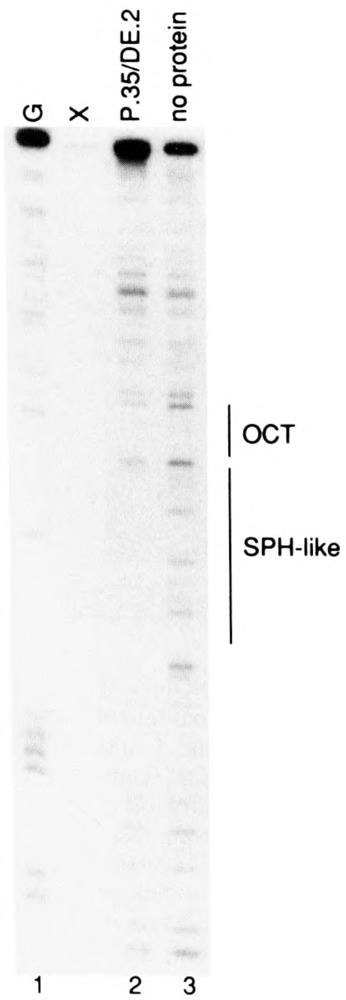
DNase I footprint over the NONOCT(SPH) region of the human U6 distal region. A singly end-labeled DNA fragment containing the human U6 NONOCT(SPH) and octamer motifs was prepared, incubated with fractionated HeLa cell extract, treated with DNase I, and electrophoresed as described in the Materials and Methods section. The human SBF fraction (approximately 15 μg) that was incubated with the probe to generate the sample in lane 2 was HeLa SI00 extract fractionated in series over phosphocellulose and DEAE-cellulose as described previously (6). Lane 1 displays the result of Maxam-Gilbert G-reaction cleavage of the probe that was used as a marker, and lane 3 shows DNase I cleavage of the probe carried out in the absence of protein. The lane marked “X” was a misloaded sample irrelevant to the results presented here.
In addition, we used an ultraviolet light-mediated DNA/protein cross-linking assay to estimate the molecular weight of both the HeLa and chicken SPH binding factors. Internally labeled probes containing either the human U6 SPH and octamer motifs (Fig. 7A) or chicken U4 SPH motif (Fig. 7B) were prepared by filling in a partially duplex oligodeoxynucleotide with bromodeoxyur-idine triphosphate and deoxynucleoside triphosphates, one of which was radiolabeled with 32P. After incubation with partially fractionated HeLa extract (P.35) or a chicken SBF fraction, followed by cross-linking with UV light, the samples were digested with nucleases and electrophoresed on an SDS gel.
Several polypeptides were detected by autoradiography after irradiating the HeLa extract that had been incubated with a human U6 distal region probe (Fig. 7A). One polypeptide of approximately 85 kDa was specifically correlated with SBF binding because it was competed by addition of unlabeled U6 NONOCT oligonucleotide during the reaction (Fig. 7A, lane 3), but not by inclusion of an oligonucleotide containing a “nonspecific” sequence (Fig. 7A, lane 1), nor by one containing the consensus octamer motif (Fig. 7A, lane 2). Because the phosphocellulose-fractionated HeLa extract contains very little Oct-1, the absence of a radiolabeled polypeptide competed by the octamer oligonucleotide was not surprising.
We extended these results by irradiating a chicken U4B SPH probe bound by either a chicken SBF preparation or the same HeLa phosphocellulose fraction used in the previous experiment. A specific 85-kDa hSBF polypeptide was cross-linked to the chicken SPH probe (Fig. 7B, lane 6) that was competed by addition of excess unlabeled U6 NONOCT or chicken U1 SPH oligonucleotides (Fig. 7B, lanes 7 and 8), but not by the same amount of a nonspecific oligonucleotide (Fig. 7B, lane 9). When a chicken SBF preparation was used for cross-linking, two specific polypeptides were detected as evidenced by competition with both unlabeled U6 NONOCT and chicken U1 SPH oligonucleotides but not by a nonspecific oligonucleotide (Fig. 7B, lanes 1-4). Significantly, the largest polypeptide of 85 kDa comigrated with the HeLa cross-linked polypeptide (compare lanes 1 and 6), a result consistent with the nearly identical mobilities of chicken and HeLa SBF-DNA complexes in the electrophoretic mobility shift assay (Fig. 1C). A second specific polypeptide of approximately 65 kDa was also detected after cross-linking, which could correspond to the higher mobility SBF c1/DNA complex (Fig. 1C, lanes 3, 5, 6).
Correlation of Transcription With SBF Binding Activity
Using plasmid templates containing random mutations introduced into the region of the U6 SPH element, we determined the importance of individual base pairs on U6 expression and transcription factor binding. These mutants were constructed in a U6 maxigene plasmid that contained a clustered set of point mutations to inactivate the consensus octamer motif, because previous results demonstrated that mutation either of the U6 SPH element (NONOCTMUT in Fig. 8A) or of the consensus octamer (OCTMUT), alone, caused only a minor reduction in U6 expression, but when combined caused an 8-10-fold decrease (6). The specific mutants isolated and used for these studies are listed in Fig. 8A. The three templates, RM1-1, RM2-7, and RM2-C, each contained mutations in the human U6 SPH motif that decreased its similarity to the consensus sequence.
Each mutant plasmid template was introduced into separate dishes of cultured human 293 cells along with a plasmid containing a chicken β-tubulin gene as a control for variation in transfec-tion efficiency. U6 maxigene transcription from each mutant plasmid was measured using a primer extension assay, normalized to the β-tubulin signal, and compared to that from the wild-type. Representative results are shown in Fig. 8B and summarized graphically in Fig. 8C. In these experiments, the distal region accounted for only a fourfold effect on U6 expression (dl-148 in Figs. 8B, C). As found previously, expression from a template containing substitutions only in the octamer motif was reduced only slightly, to approximately 75% of wild-type (RM2-13 in Figs. 8B, C). Thus, the range of expression of these mutants was expected to vary between 25% and 75% of wild-type. Each of the mutant templates exhibited a reduced level of U6 maxigene expression. Significantly, the cluster of three point mutations in RM2-C, or even a single C to T transition in RM2-7, inactivated the ability of the U6 SPH motif to stimulate transcription. The single C to A transversion in RM1-1 was not quite as severe but also reduced expression markedly.
We used the same mutant plasmids as competitors in gel mobility shift reactions to examine the efficacy of binding of human SPH binding factor (SBF) to the substituted DNA elements. Preliminary experiments demonstrated that the SBF/ DNA complex could be competed by the addition of 50–1000 ng of supercoiled plasmid DNA containing a normal SPH-like sequence. These amounts corresponded to approximately a 7–130-fold molar excess of competitor plasmid DNA over the radiolabeled probe. The average of results from two experiments for each competitor is graphed in Fig. 8D. All of the mutants were much less efficient competitors than templates containing normal SPH motifs (w.t. and RM2-13). A good correspondence existed between competitive ability in the gel mobility shift reaction and level of expression in the transfection assay; that is, poor competitors exhibited low levels of expression.
DISCUSSION
Many vertebrate snRNA gene distal sequence elements contain two adjacent motifs, one of which binds Oct-1 transcription factor. We demonstrated that the distal region of the pol III-transcribed human U6 gene is composed of a SPH element immediately abutted to an octamer motif. This composition is common to some pol II snRNA gene enhancer regions such as those associated with chicken U1 and U4B promoters (19,27). Thus, both pol II-and pol III-transcribed, naturally occurring snRNA gene promoters employ highly similar activating regions. Homologous SPH elements have also been identified in the 5′-flanking region of pol III-transcribed selenocysteine tRNA genes from several vertebrate species, although no octamer motif is present in the Xenopus tRNA(Ser)Sec promoter (16,17).
The interelement spacing and orientations of the SPH and octamer motifs is quite variable among promoters that contain both elements (Fig. 5). Indeed, we have found that U6 maxigene expression in transfected human 293 cells was not significantly affected when insertions of 5 or 10 bp were added between the SPH and OCT elements (G. Kunkel, unpublished results). In contrast, the normal spacing of the SPH and octamer elements was necessary for highest transcriptional levels of the chicken U1 snRNA gene injected into Xenopus oocytes and when linked to a Xenopus U6 proximal promoter (3,17). Nevertheless, elec-trophoretic mobility shift experiments provided no evidence for cooperative binding of chicken SBF and octamer protein to chicken U1 enhancer DNA (T. Cheung and W. Stumph, unpublished results). A synergistic role for chicken SBF and Oct-1 is apparent from transcriptional studies (3,19,20,27), but the mechanism for this syner-gism is not understood, and its elucidation will require use of purified transcription factors.
The activities of several mutant forms of the U6 SPH motif were analyzed in U6 transcription assays in transfected cells and as competitors in protein/DNA binding assays. Each of these mutants demonstrated a direct relationship between DNA binding and transcriptional stimulation by an activator protein. Although our collection of mutants is limited, the results point to the special significance of C residues at positions –7 and –6 (RM1-1 and RM2-7). Previous mutagenesis of the chicken U4B SPH motif demonstrated the importance of C residues at positions –6 and –5 (27). Unfortunately, we did not recover any mutants at nucleotide –5 of the U6 SPH element. The RM2-C triple point mutant was especially deleterious, and this is not unexpected because it differs from the consensus sequence at all three mutant positions.
We have also made the interesting observation that SBF requires MgCl2 to bind to DNA. CaCl2 can substitute for this divalent cation requirement (results not shown). However, neither ZnCl2 nor CuCl2 was able to substitute for MgCl2 in the DNA binding assay, eliminating the possibility that trace contamination of zinc or copper in the MgCl2 solution actually mediated the positive effect. A strong dependence on Mg2+ concentration has also been observed for the sequence-specific binding of TFIIIA to the internal control region of 5S RNA genes (4), although to our knowledge the mechanism by which Mg2+ exerts this effect is not known.
The presence of homologous SPH motifs in small RNA gene promoters from diverse vertebrate species (human, bovine, chicken, amphibian) and the cross-species binding specificity exhibited by chicken and human SBF demonstrates a conserved role for this protein in transcriptional activation. Our laboratories have also detected SBF activity in extracts from Xenopus oocytes and bovine liver (results not shown). Recently, a cDNA clone was isolated that encodes a protein from Xenopus laevis that specifically recognizes the selenocysteine tRNA gene SPH motif (Staf) (21). Interestingly, amphibian Staf, a zinc finger protein, was found to be highly homologous to a human protein of unknown function, ZNF76. However, it is not likely that the hSBF we detect in electrophoretic mobility shift and UV-mediated protein/DNA cross-linking experiments is ZNF76 because no ZNF76 transcripts were detected in HeLa cells (18). Instead, the ZNF76 transcript is highly enriched in testis. Furthermore, our estimate of the size of a DNA binding hSBF polypeptide (∼85 kDa) is larger than the predicted size of the 514-amino acid ZNF76 polypeptide (∼57 kDa). However, we cannot be certain in our experiments that the cross-linked nucleotides do not affect the mobility of the hSBF polypeptide in SDS gels. Whether human and/or chicken SBF are related to amphibian Staf will require molecular characterization of their cDNAs.
ACKNOWLEDGEMENTS
We thank Melissa McClung for purification of many plasmid DNAs used in these experiments. This research was supported by grants from NSF (MCB-9304799) (G.R.K.), NIH (GM33512) (W.E.S.), and the California Metabolic Research Foundation (W.E.S).
REFERENCES
- 1. Ach R. A.; Weiner A. M. Cooperation between CCAAT and octamer motifs in the distal sequence element of the rat U3 small nucleolar RNA promoter. Nucleic Acids Res. 19:4209–4218; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ares M. Jr.; Chung J.-S.; Giglio L.; Weiner A. M. Distinct factors with Spl and NF-A specificities bind to adjacent functional elements of the human U2 snRNA gene enhancer. Genes Dev. 1:808–817; 1987. [DOI] [PubMed] [Google Scholar]
- 3. Cheung C. H.; Fan Q. N.; Stumph W. E. Structural requirements for the functional activity of a U1 snRNA gene enhancer. Nucleic Acids Res. 21:281–287; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christensen J. H.; Hansen P. K.; Lillelund O.; Thogersen H. C. Sequence-specific binding of the N-terminal three-finger fragment of Xenopus transcription factor IIIA to the internal control region of a 5S RNA gene. FEBS Lett. 281:181–184; 1991. [DOI] [PubMed] [Google Scholar]
- 5. Dahlberg J. E.; Lund E. How does III × II make U6. Science 254:1462–1463; 1991. [DOI] [PubMed] [Google Scholar]
- 6. Danzeiser D. A.; Urso O.; Kunkel G. R. Functional characterization of elements in a human U6 small nuclear RNA gene distal control region. Mol. Cell. Biol. 13:4670–4678; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diamond A. M.; Montero-Puerner Y.; Lee B. J.; Hatfield D. Selenocysteine inserting tRNAs are likely generated by tRNA editing. Nucleic Acids Res. 18:6727; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herr W.; Cleary M. A. The POU domain: Versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 9:1679–1693; 1995. [DOI] [PubMed] [Google Scholar]
- 9. Hoffman M. L.; Korf G. M.; McNamara K. J.; Stumph W. E. Structural and functional analysis of chicken U4 small nuclear RNA genes. Mol. Cell. Biol. 6:3910–3919; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Janson L.; Bark C.; Pettersson U. Identification of proteins interacting with the enhancer of human U2 small nuclear RNA genes. Nucleic Acids Res. 15:4997–5016; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kleinert H.; Bredow S.; Benecke B.-J. Expression of a human 7SK RNA gene in vivo requires a novel pol III upstream element. EMBO J. 9:711–718; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kunkel G. R. RNA polymerase III transcription of genes that lack internal control regions. Biochim. Biophys. Acta 1088:1–9; 1991. [DOI] [PubMed] [Google Scholar]
- 13. Kunkel G. R.; Pederson T. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev. 2:196–204; 1988. [DOI] [PubMed] [Google Scholar]
- 14. Lobo S. M.; Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell 58:55–67; 1989. [DOI] [PubMed] [Google Scholar]
- 15. Mattaj I. W.; Dathan N. A.; Parry H. D.; Carbon P.; Krol A. Changing the RNA polymerase specificity of U snRNA gene promoters. Cell 55:435–442; 1988. [DOI] [PubMed] [Google Scholar]
- 16. Myslinski E.; Krol A.; Carbon P. Optimal tRNA(Ser)Sec gene activity requires an upstream SPH motif. Nucleic Acids Res. 20:203–209; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Myslinski E.; Schuster C.; Krol A.; Carbon P. Promoter strength and structure dictate module composition in RNA polymerase III transcriptional activator elements. J. Mol. Biol. 234:311–318; 1993. [DOI] [PubMed] [Google Scholar]
- 18. Ragoussis J.; Senger G.; Mockridge I.; et al. A testis-expressed Zn finger gene (ZNF76) in human 6p21.3 centromeric to the MHC is closely linked to the human homolog of the t-complex gene tcp-11. Genomics 14:673–679; 1992. [DOI] [PubMed] [Google Scholar]
- 19. Roebuck K. A.; Szeto D. P.; Green K. P.; Fan Q. N.; Stumph W. E. Octamer and SPH mottfs in the U1 enhancer cooperate to activate U1 RNA gene expression. Mol. Cell. Biol. 10:341–352; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roebuck K. A.; Walker R. J.; Stumph W. E. Multiple functional motifs in the chicken U1 RNA gene enhancer. Mol. Cell. Biol. 7:4185–4193; 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schuster, C; Myslinski E.; Krol A.; Carbon P. Staf, a novel zinc finger protein that activates the RNA polymerase III promoter of the selenocysteine tRNA gene. EMBO J. 14:3777–3787; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tebb G.; Mattaj I. W. The Xenopus laevis U2 gene distal sequence element (enhancer) is composed of four subdomains that can act independently and are partly functionally redundant. Mol. Cell. Biol. 9:1682–1690; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weller P.; Bark, C; Janson L.; Pettersson U. Transcription analysis of a human U4C gene: Involvement of transcription factors novel to snRNA gene expression. Genes Dev. 2:1389–1399; 1988. [DOI] [PubMed] [Google Scholar]
- 24. Williams M.; Brys A.; Weiner A. M.; Maizels N. A rapid method for determining the molecular weight of a protein bound to nucleic acid in a mobility shift assay. Nucleic Acids Res. 20:4935–4936; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Willis I. M. RNA polymerase III: Genes, factors and transcriptional specificity. Eur. J. Biochem. 212:1–11; 1993. [DOI] [PubMed] [Google Scholar]
- 26. Wu, C; Tsai, C; Wilson S. Affinity chromatography of sequence-specific DNA-binding proteins. Genet. Eng. 10:67–74; 1988. [Google Scholar]
- 27. Zamrod Z.; Stumph W. E. U4B snRNA gene enhancer activity requires functional octamer and SPH motifs. Nucleic Acids Res. 18:7323–7330; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]