Abstract
Expression of the human ALDH3 gene is regulated in a tissue-dependent constitutive as well as drug-inducible manner. We identified a new 5 ′-noncoding exon (exon 1) existing at about 3 kilobase pairs (kb) upstream from the first coding exon (exon 2) of the human ALDH3 gene. Analysis of ALDH3 mRNA revealed the existence of several isoforms with different 5′ regions resulting from i) usage of multiple transcriptional initiation sites of the new exon 1, ii) usage of alternative splice acceptor sites at the 3′ end of the new intron 1, and iii) alternative splicing out of exon 2. Usage of alternative splice acceptor sites was only found in tissues expressing ALDH3 constitutively, but not in Hep G2 induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Nucleotide sequence analysis and chloramphenicol acetyltransferase (CAT) expression studies showed that a strong promoter region exists at nucleotide (nt) positions −216 to +54 of the gene. Repression activities were found upstream of the −216/+54 region. Several putative drug-inducible elements exist in the regulatory region. A possible regulatory mechanism for tissue-specific constitutive and inducible expression of the human ALDH3 gene is discussed.
Keywords: Gene structure, Gene regulation, ALDH3
THE mammalian aldehyde dehydrogenases (ALDH: aldehyde:NAD+ oxidoreductase, EC 1.2.1.3) are a famiiy of enzymes wiih broad substrate specificity, which catalyze the irreversible oxidation of various aliphatic or aromatic aldehydes to their corresponding acids (31). They have been frequently considered “detoxifying” enzymes that eliminate highly reactive aldehydes, including ethanol-derived acetaldehyde, medium-chain aliphatic aldehydes generated during membrane lipid peroxidation, aldehyde derived from metabolism of xenobiotics, and toxic aldehydes in the foodstuff (17,20,24,27,29). Several investigators have demonstrated that the ALDH isozymes also play crucial roles in the metabolism of important physiological molecules, including retinoic acid, bio-genic amines, and neurotransmitters (1,2,47,48). In addition, mammalian ALDH isozymes have been shown to be recruited as major lens or cor-neal proteins (e.g., η-crystallin in the lenses of elephant shrews or BCP54 in bovine cornea) (9,23) and may serve structural and/or enzymatic roles. Proteins associated with other functional activities have been shown to be aldehyde dehydrogenase (e.g., androgen-binding protein, daunorubicin-binding protein) and 11-hydroxythromboxane B2 dehydrogenase (4,30,44).
The ALDH3 isozyme utilizes benzaldehyde and medium-chain aliphatic aldehydes as optimal substrates. The expression of ALDH3 is tissue specific, being at a high constitutive level in the stomach, cornea, esophagus, and lung, but generally at a low or nondetectable level in the liver, kidney, and many other normal human and rat tissues (11–13,18). However, the repressed ALDH3 gene can be activated by stable genetic changes in neo-plastic liver nodules during hepatocarcinogenesis (8,24), and can be transiently induced in vitro both in carcinoma cells and in normal human hepatocytes by aromatic hydrocarbon xenobiotics (e.g., 3-methylcholanthrene and TCDD) or phenolic antioxidant (e.g., catechol) (19,25,26,36, 39). The inducible expression of the ALDH3 gene by TCDD is regulated at the transcriptional level in the rodent system (39,41). It is also interesting to note that the ALDH3 isozyme purified from tumors and induced human tissues exhibits higher activity in oxidation of aldophosphamide, an anti-neoplastic drug, than ALDH3 purified from normal stomach tissue (37). It is not known whether carcinogenesis and xenobiotic induction have mutagenic and/or selection effects in the coding region of ALDH3, causing changes in its catalytic activity.
Recently, a cDNA of the fatty ALDH gene, a homologue of the human ALDH3 gene, has been cloned and mapped (10). This gene has been indicated as the gene responsible for Sjögren-Larsson syndrome, an autosomal recessive disorder. The fatty ALDH gene mapped to the same chromosomal band location as the human ALDH3 gene at 17p (32).
ALDH3 cDNAs and/or genes from three different species, human stomach/cornea (18), rat tu-mor/TCDD-induced (3,21), and mouse dioxin-induced ALDH3 (42), have been cloned and characterized. The cDNAs encode 453 amino acid (aa) residues and share greater than 80% aa positional identity. The human and rat ALDH3 genes show a structural similarity except in their 5′ end region. In our previous study, the human ALDH3 gene was identified by a human stomach cDNA probe and was found to span about 8 kb in length, consisting of 10 coding exons. The 5′-untranslated region (UTR) (50 nt) of the human stomach ALDH3 cDNA was shown to be contiguous with the beginning of the coding region in a single exon. In contrast, the rat ALDH3 gene was identified by a rat hepatoma HTC cDNA probe (3). The rat gene spans approximately 9 kb in length and consists of 11 exons, including 10 coding exons and one 5′-noncoding exon. The 5′ UTRs (167 and 45 nt) of the liver-tumor/TCDD-induced ALDH3 transcripts were interrupted by a 3-kb intron 5 base pairs (bp) upstream from the coding initiator ATG. The rat ALDH3 promoter region, about 5.5 kb, upstream from the first non-coding exon was partially characterized and three functional regions were assigned. The region contains a strong TATA-less promoter, upstream inhibitory regions, and TCDD-responsive elements (38).
The discrepancy at the 5′ end of the human and rat ALDH3 gene raises interesting questions concerning the regulation of the ALDH3 gene expression. Does the difference represent different mRNA isoforms? Are there alternative promoters involved in the regulation of the synthesis of ALDH3 transcript isoforms? It has been shown that alternative promoters are a regulatory mechanism in tissue- and development-specific gene expression (22,45). Furthermore, it is known that the ALDH3 gene is a single copy gene in the human genome (18) and, yet, the ALDH3 enzyme has isoforms with multiple ALDH3 activity bands on isoelectric focusing (IEF) gels. It is not known whether this is due to various ALDH3 mRNA isoforms or due to the posttranslational modification of the ALDH3 isozyme. The aim of the present study was to identify and characterize the mRNA isoforms and promoters of the human ALDH3 gene.
MATERIALS AND METHODS
DNA Sequencing
DNA sequence determination of double-stranded DNA was performed by thermocycle sequencing with Taq polymerase (Applied Biosys-tems, model 373A). To sequence the promoter region, overlapping subclones derived from an ALDH3 genomic clone (λDASH-4) were generated in pBluescript KS( + ) vectors using various restriction enzymes.
Tissue Culture and TCDD Induction
Human Hep G2 (hepatocellular carcinoma) and MCF7 (breast adenocarcinoma) cells were maintained in Dulbecco’s modified Eagle’s medium containing 2 nM l-glutamine and supplemented with 10% fetal bovine serum (GIBCO/ BRL), penicillin, and streptomycin at 37°C and 5% CO2. The cultured cells at 50% confluence were treated for 16 h with 20 nM TCDD (> 99%, Cambridge Isotope Laboratory) in dimethyl sulfoxide to give a maximal induction effect. The final concentration of dimethyl sulfoxide was 0.1% in the culture medium. KATO III (human gastric carcinoma) and HT-29 (human colon adenocarcinoma) cells were maintained in RPMI 1640 medium with 20% and 10% FCS, respectively. AGS (human gastric adenocarcinoma) cells were maintained in Ham’s F-12 medium with 10% FCS.
RNA Preparation
Total RNA from different tissues and cultured cells used in this study was prepared by an acid guanidium thiocyanate-phenol-chloroform extraction method (RNAzol™ B, CINNA/BIOTECX Lab).
Characterization of the 5′ UTRof TCDD-induced ALDH3 Transcript
The 5′ UTR of the TCDD-induced ALDH3 mRNA was amplified by a rapid-amplification-cDNA-end (RACE) method using a commercial kit (5′-AmpiFINDER™ RACE Kit, Clontech). In brief, 50 μg of total RNA was reverse transcribed at 52°C for 30 min with 12.5 U of AMV reverse transcriptase using 77 ng of primer 12 (5′-GG GAGCTTCTGGATCATGTACTC-3′, complementary to positions +330 to +308, Fig. 2). First strand cDNA was purified with GENO-BIND™, ligated at its 3′ end with an anchor oligomer and T4 RNA ligase, and amplified by PCR using specific primer 7 (5′-CTTGTGCAGGTCTGCG-3′, complementary to nucleotide positions +256 to + 241, Fig. 2) and an anchor primer. The thermal profile used was 94°C for 1 min, 52°C for 1 min, then 72°C for 2 min. After 30 cycles, the products were separated on a 2% agarose gel. Bands larger than 230 nt were eluted and subcloned into EcoRV-treated/ddT-tailed pBluescript KS( +) vectors. The clones that showed a positive hybridization signal with radioactive-labeled ALDH3 cDNA probe were selected for nucleotide sequencing.
FIG. 2.
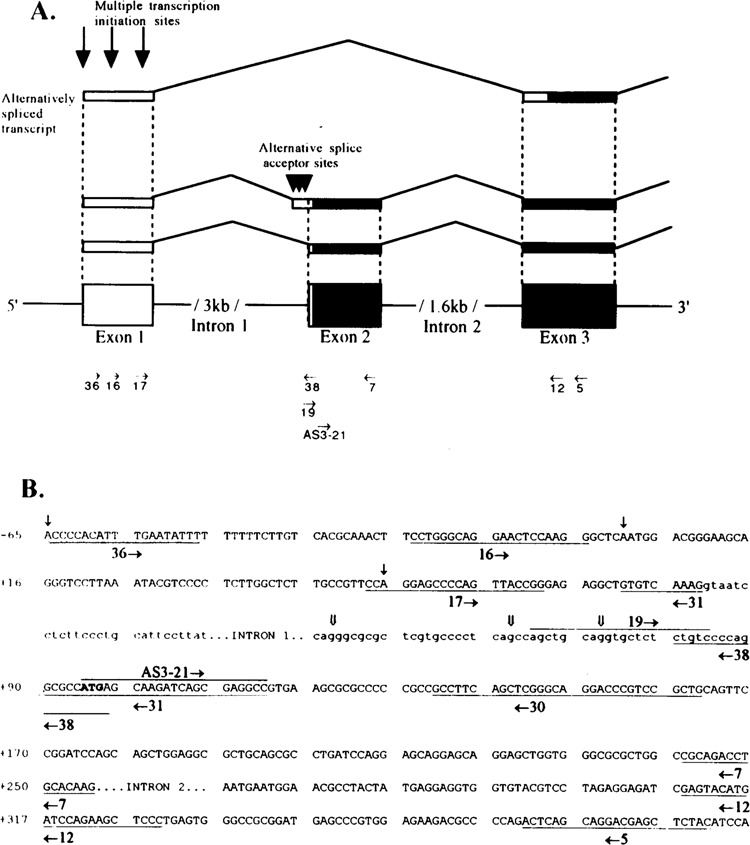
(A) Schematic diagram of the revised ALDH3 genomic structure and the 5′ ends of diverse ALDH3 transcript isoforms. Coding and noncoding regions in the exons are indicated as solid and open bars, respectively. A new noncoding exon (exon 1) is identified at 3 kb upstream from the first coding exon (exon 2). The ALDH3 transcripts have multiple initiation sites as shown by the downward arrows at the exon 1 (↓). The alternatively spliced ALDH3 transcript isoform skipping exon 2 is shown at the top. Multiple splice acceptor sites are indicated by the downward triangles at the 3′ end of intron 1 (▼). The corresponding locations of the RT-PCR primers and hybridization probes used in this study are indicated as horizontal arrows in the directions of their orientations. (B) The locations and nucleotide sequences of the primers used in this study are underlined. The horizontal arrows indicate the direction of the primers’ orientation. The nucleotide sequence in upper and lower case represent the exon and intron sequence, respectively. (↓) Transcription initiation sites, ( ↓ ) alternative splice acceptor sites.
RT-PCR Analysis of ALDH3 Transcripts
To analyze the isoforms of the ALDH3 transcripts in human stomach tissue, hepatocellular carcinoma (HCC) tissue, and cultured cells, three different reverse transcriptase-polymerase chain reaction (RT-PCR) experiments were performed. The condition of the PCR cycles was 94 °C for 45 s, 60°C for 45 s, and 72°C for 1 min. The positions of the primers are shown in the Fig. 2. i) To identify ALDH3 transcript isoform using alternative splice acceptor sites at the new intron 1, we used primer 16 (5′-−24CCTGGGCAGGAACTC CAAGG−5-3′) and primer 38 (5′-TCATGG CGCCTGGGGACAG-3′, complementary to positions at the junctions of the 3′ end of the new intron 1 and 5′ end of the exon 2) for RT-PCR, and primer 19 (5′-AGCTGCAGGTGCTCTCT GTC-3′, at 3′ end of the new intron 1) for hybridization. ii) For the study on the usage of the alternative splice acceptor sites in ALDH3-positive tissues, we used two sets of primers, primer 12 and primer 17 (5′ -+53CCAGGAGCCCCAGTTAC CGG + 72-3′) and primer 12 and primer 19. Human phosphoglycerate kinase (PGK)-specific PCR primers, 5′-AGGGATCCTTAGAGCCAGTTGC TGT-3′ and 5′-CCAGAATTCTGTGGCAGATT GACTCC-3′, were used as primer pairs for an internal standard. The Southern blot of the PCR products was hybridized with the, 4Z,ZW3-specific oligomer AS3-21 (5′-+ 95ATGAGCAAGATCA GCGAGGCC + 115-3 ’) and, then, with the PGK cDNA probe, iii) For the study of alternative splicing, we used primer 36 (5′-−65ACCCCA CATTTGAATATTT"− 5-3′) and primer 5 (5′-GTAGAGCTCGTCCTGCTGAGT-3′, complementary to positions +391 to +371) as first RT-PCR primers, primer 16 and primer 12 as second PCR primers, and primer 17 as the hybridization probe. Double PCR enhanced the amplification specificity.
Primer Extension
The 5′ end of the ALDH3 mRNA was mapped by primer extension analysis using standard techniques (34). The oligonucleotide primers were 5′ end labeled with polynucleotide kinase and [γ-32P]dATP and purified on a 20% urea-acrylamide gel. Primer 31 (5′-GGCCTCGCTGATCTTGCT CATGGCGCCTTTGACAC-3′) and primer 30 (5′ - CAGCGGACGGGTCCTGCCCGAGCTGA AGGC-3′) are complementary to positions +115 to +81 and +163 to +134, respectively (Fig. 2). The specific activity of each individual primer was about 2-4 × 108cpm/μg.
Promoter/Reporter Gene Constructs
A low background promoterless CAT expression vector pUMSVOCAT (33) was modified at its unique Smal cloning site, located at the 5′ end of the CAT gene, to create 5′-HindIII and 3′-Xbal sites as described (46). The 2.55-kb ALDH3 genomic Pstl/BamHI fragment, containing the 2.3-kb promoter region, first noncoding exon, and the 5′ end 0.2 kb of the first intron, from λDASH-4 was subcloned into the corresponding sites of the pBluescript vector (clone 96#7). To delete the first exon and part of the first intron, the 96#7 clone was digested with SstI and replaced with a Sstl-treated PCR fragment amplified with primers 24 (5′-−262GAAGAGCTCCATGCCAG GCT−243-3′) and 25 (5′-GTAGAGCTCTCTA GA +54GGAACGGCAAGAGCCAAGAG + 35-3′) on the 96#7 template. The orientation and sequence of the resulting plasmid, 3P-1 (-2263/ + 54), was confirmed by restriction endonuclease mapping and by sequencing. Plasmid 3P-1 was simultaneously digested at a number of selected restriction endonuclease sites, blunt ended, digested at a vector-encoded EcoRV site, recircular-ized, digested at the vector-encoded Hindlll and Xbal sites, and subcloned into the corresponding sites of the modified CAT expression vector pUM-SVOCAT to generate a set of 5′-nested deletions. For the downstream PstI site, steps of blunt-ending and digestion with EcoRV were skipped. For the SstI fragment, subcloning into pUC-13 was performed to obtain the vector encoded by Hindlll and Xbal sites for subsequent subcloning into modified pUMSVOCAT. The end points of the deletion constructs and the enzyme used were as follows: -2263 (PstI), -1752 (Ncol), -1337 (StuI), -726 (PstI), -259 (SstI), and -120 (Smal). For other 5′ and/or 3′ deletion constructs, sets of 5′ and 3′ promoter-specific primers flanking the designated region with respective artificial 5′-Hindlll and 3′-Xbal sites were used in the PCR amplification. The PCR products were restricted with Hindlll and Xbal, purified on the 1.5% agarose gel, and subcloned into the modified pUMSVOCAT.
pSV40CAT, which served as a positive control, was derived by inserting a SV40 promoter sequence fragment into the 5′ end of the CAT gene of pUMSVOCAT. All plasmids were purified with a QIAGEN kit according to the kit manual.
Transient Expression
Human Hep G2 cells were plated at 2 × 106 cells/60 mm culture dish the day before transfection. Lipofectin (20 μg) (GIBCO/BRL) was mixed with 10 μg of the test construct and 2 pg of pCMVβ-gal DNA in 3 ml Optimem medium to form a DNA/lipofectin complex. pCMVβ-gal was used to control for variations in transfection efficiencies. Cells were transfected by the precipitated complex for 16 h, refed with DMEM containing 10% calf serum for 8 h, and treated with or without TCDD for 15 h. Following washing and harvesting, the cells were lysed in 0.25 M Tris-HCl (pH 8) by four cycles of freezing and thawing and microfuged at 12,000 × g for 15 min. Aliquots of the cell extract were removed to measure β-galactosidase activity as described by Sambrook et al. (34). The remaining cell extract was heat inactivated at 60°C for 10 min to inactivate deace-tylase and again microfuged. This supernatant was used to measure CAT activity.
CAT Activity Assay
CAT activity was measured by the phase-extraction method as described (35). The assay condition was in a linear range as determined by preliminary experiments. In brief, CAT-containing extracts were incubated with butyrylcoenzyme A (Sigma) and [14C]chloramphenicol (Amersham). The butyrylated chloramphenicol was extracted with xylene (Aldrich). The xylene phase was back-extracted twice with 0.25 M Tris-HCl (pH 8) and was counted in a liquid scintillation counter. The CAT activity of individual construct was normalized with respect to β-galactosidase activity and compared to that of the pSV40CAT control.
RESULTS
Identification of a New Noncoding Exon of the ALDH3 Gene
An anchored RT-PCR study was performed with total human RNA isolated from TCDD-treated Hep G2 cells to obtain 5′-RACE cDNA clones containing 5′-UTR sequence of the ALDH3 transcript. Among 30 isolated 5′-RACE cDNA clones, 18 clones showed positive hybridization signals with the labeled ALDH3 cDNA probe. These positive 5′-RACE clones contained inserts with two different sizes: one with a 94-nt 5′-UTR region and the other with a 40-nt 5′-UTR region.
To localize the new 5′-UTR sequence in the human ALDH3 gene, we sequenced a 6.0-kb genomic fragment isolated from a previously characterized ALDH3 genomic clone (XDash-4) (Fig. 1). A new ALDH3 noncoding exon (exon 1) was then identified about 3 kb upstream from the previously identified first coding exon (18). The new intron 1 conforms to the consensus “GT-AG” rule for RNA splicing (5). Exon 2, the first coding exon, contains 5 bp of the 5′-UTR and 162 bp of the 5′-coding region.
FIG. 1.
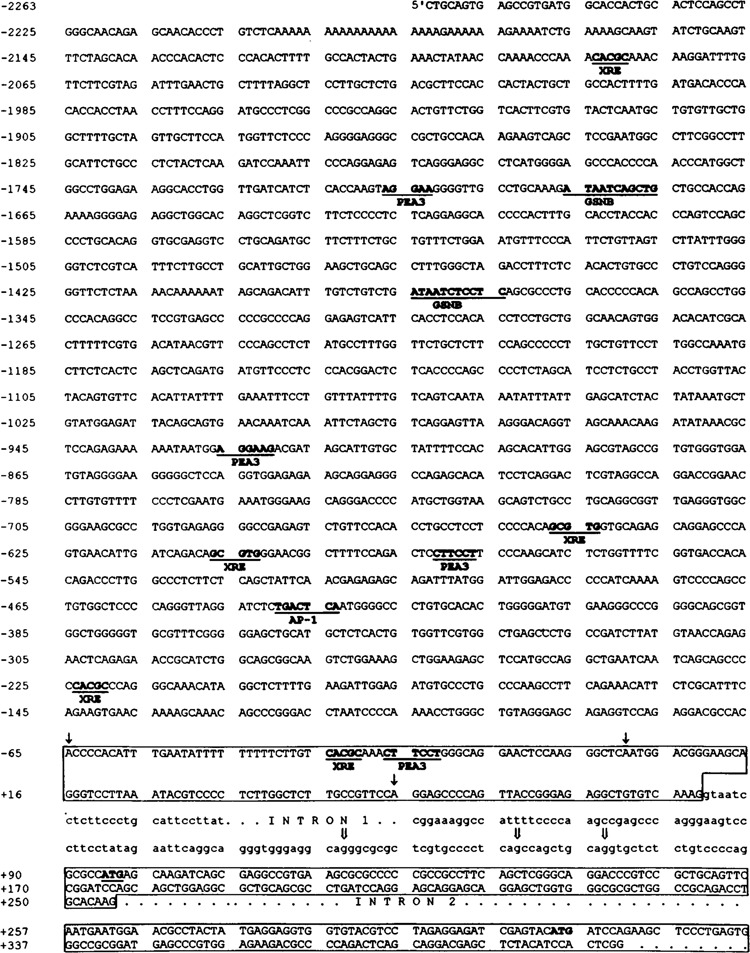
Nucleotide sequences of the 5′ regulatory region and first three exons of the human ALDH3 gene. The nucleotide sequences for putative cis-acting elements discussed in the text are in bold, underlined, and labeled (14,40). The first three exons (1, 2, and 3) are boxed. In the new exon 1, the three identified transcription initiation sites are indicated as downward single arrows (↓). The adenine nucleotide of the second transcription initiation site is assigned as nucleotide 1. In the new intron 1, alternative splice acceptor sites are indicated as downward double arrows ( ⇓). The translational initiation codon, ATG, is in bold and underlined in exon 2. The ATG codon in exon 3 at the +314 position is in bold. Abbreviations for potential binding factors or cis elements are as follows: AP-1, activator protein 1; GSNB, gastric-specific nuclear binding factor; PEA3, polyoma enhancer activator 3; XRE, xenobiotic response element.
Identification of Alternative Splice Acceptor Sites at the New Intron 1
The 3′ end of the new intron 1 contains the 5′-UTR sequence of the previously identified stomach ALDH3 cDNA (18), which indicates the possibility of the existence of alternative splice acceptor sites at the new intron 1 of the human ALDH3 gene. To examine this possibility, we performed RT-PCR analysis using total stomach and KATO III RNA, respectively, with two primers: 16 and 38. Primer 38 covers the junction region of the new intron 1 and exon 2 and thus is specific for detecting alternative splice acceptor sites (Fig. 2 for the locations of the primers). The RT-PCR products were then subjected to the sequence analysis. The sequence result indicates that in human stomach tissue the new intron 1 of the human ALDH3 gene has at its 3′ end at least three alternative splice acceptor sites (Fig. 2A, downward triangles at intron 1). These sites are 17, 27, and 47 bp upstream from the 3′ splice junction site of the new intron 1 (Figs. 1 and 2B, downward double arrows at intron 1).
To examine whether the usage of these splice acceptor sites is tissue specific, we performed RT-PCR with total RNA from other ALDH3-positive and -negative tissues using primer pairs: 12/17 and 12/19. Primer 12/19 can only amplify the transcripts using alternative splice acceptor sites (Fig. 2 for the locations of the primers). The hybridization of the RT-PCR products with exon 2-specific probe AS-3 is shown in Fig. 3. The usage of alternative splice acceptor sites, as indicated by positive bands with primer 12/19, was only found in the stomach and in ALDH3-positive HCC, but not in TCDD-treated Hep G2 cells. These positive 12/19 bands were much weaker compared to the corresponding 12/17 bands of the same tissue, probably indicating a different preference in the usage of the intron 1 splice acceptor sites or different stability of the ALDH3 transcript isoforms. Consistent with the previous observation, ALDH3 transcripts were not detected in the normal liver tissue and ALDH3-negative HCC cells.
FIG. 3.
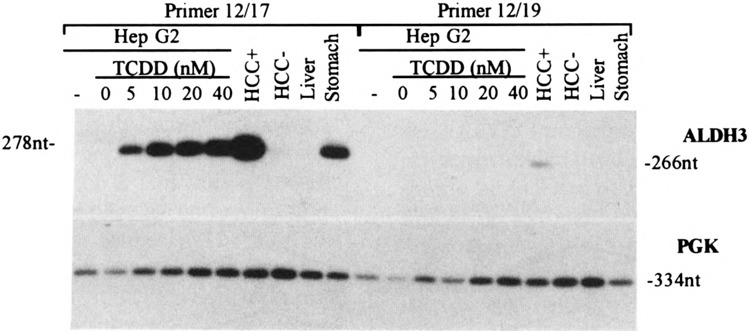
Tissue-specific usage of alternative splice acceptor sites. The RT-PCR products were amplified from 0.1 μg of total RNA from various human tissues (stomach, HCC +, HCC-, and normal liver) and Hep G2 cultured cells treated with various concentrations of TCDD, and analyzed by electrophoresis on a 2% agarose gel. Human PGK primers are used as an internal standard. The sizes of the PCR products are indicated.
Identification of an Alternatively Spliced ALDH3 Transcript
In the course of this work, we also identified alternatively spliced transcripts skipping the exon 2 sequence. When the RT-PCR products amplified with primers specific for the new exon 1 and exon 3 were subjected to probing with ALDH3 cDNA, in addition to the expected band, a shorter band was observed. Sequencing of this extra band revealed an alternatively spliced product skipping the exon 2. All the ALDH3-positive tissues and culture cells examined produced such an alternatively spliced transcript (Fig. 4).
FIG. 4.
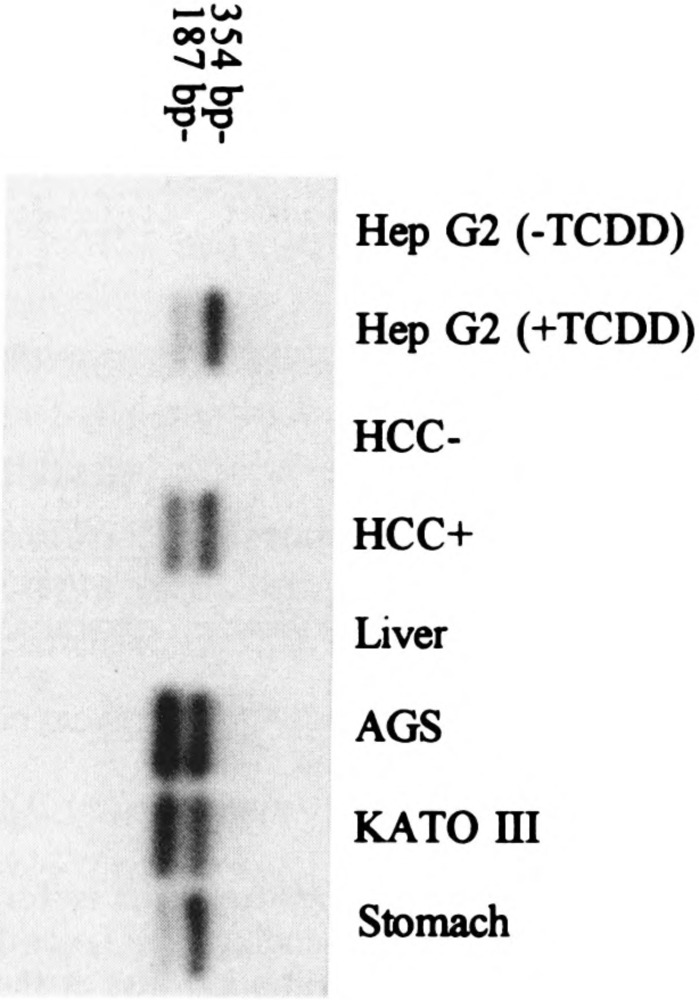
Identification of an alternatively spliced ALDH3 transcript. The PCR products were amplified from 0.1 fig of total RNAs from various human tissues and cultured cells and analyzed by electrophoresis on a 2% agarose gel. The positive bands were sliced, subcloned, and sequenced. The sizes of the expected PCR products are indicated. Top and bottom bands are the regular and alternatively spliced transcript skipping exon 2, respectively. Minor 381-bp products derived from the transcripts using alternative splice acceptor site at the 5′ end of exon 2 were also detected in the top bands from RNAs of ALDH3-posittive tissues except the TCDD-treated Hep G2 cells.
Mapping of the Transcription Initiation Site
If the positions of the most 5′-UTR nucleotides found in the anchored 5′-RACE cDNA clones correspond to the transcription initiation sites of the ALDH3 transcript, the expected lengths of the primer-extended products obtained with primer 31 should be 115 and 61 nt long, and with primer 30 should be 163 and 109 nt long, respectively (Fig. 2 for the locations of the primers). Indeed, the primer-extended products with these expected lengths are visualized as major signals together with other bands when RNA isolated from ALDH3-positive HCC cells was used as the extension template (Fig. 5). Other bands could be due to other multiple initiation sites, alternative splicing acceptor sites at the new intron 1, or mRNA degradation.
FIG. 5.
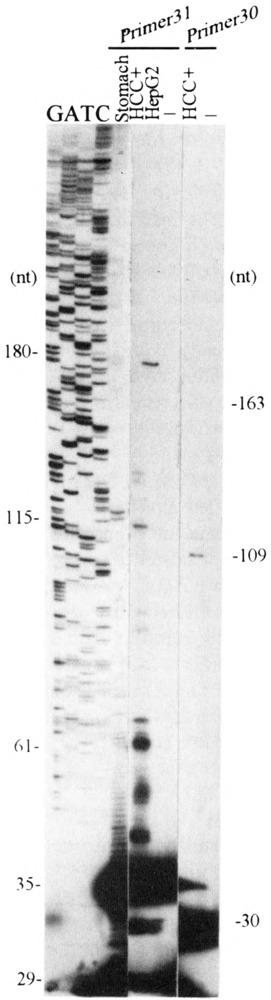
Mapping of the transcription initiation sites for the human ALDH3 gene by primer extension analysis. The sizes of the oligomer primers and the extended bands corresponding to the 5′-RACE and other characterized cDNA clones are indicated at both sides. Total RNA (80 μg) was primed and reverse transcribed with 200 units of Superscriptll reverse transcriptase for 1 h at 42 °C. The primer extension products were analyzed on a 6% sequencing gel.
When total RNA isolated from TCDD-treated Hep G2 cells was extended with primer 31, a strong signal at the 180 nt position was detected (Fig. 5). The expected 115- and 61-nt products were weak bands. The existence of transcripts corresponding to the 180-nt extension product in all the ALDH3-positive tissues was confirmed by sequence analysis of the RT-PCR products, which were amplified with primer 36 and 30. Primer 36 contains the ALDH3 genomic sequence 180 nt upstream from the extension primer 31. However, our anchored 5′-RACE protocol failed to obtained the clones corresponding to the 180-nt primer-extended fragment, possibly due to different RNA preparations used in the primer extension and 5 ’-RACE studies and/or selection of certain DNA fragments during the cloning procedure of the 5′-RACE product.
Primer extension using primer 31 and human stomach RNA demonstrated major bands at 116 and 118 nt, which were within a few bases of the expected 115-nt band. Other multiple smaller primer-extended bands might be derived from mRNA degradation (Fig. 5).
The initiation adenine nucleotides of the 61-, 115-, and 180-nt primer-extended products are located 40, 94, and 159 bp upstream from the ATG translation initiation site (Figs. 1 and 2B, downward single arrows in exon 1). The adenine nucleotide of the second transcription initiation site was assigned as nt +1 in this study partly due to its sequence identity with the conserved cap signal motif from the −2 to + 3 nt positions (14).
Tissue-Specific Expression Directed by the Human ALDH3 Gene Promoter
To test whether the 5′-flanking sequence of the human ALDH3 gene is capable of directing the tissue-specific gene expression and conferring TCDD induction, a series of deletion constructs were made and transiently transfected into human Hep G2 and MCF7 cell lines. As shown in Fig. 6A, the relative CAT activities in the human Hep G2 cells increased with increasing deletion of the 5’-flanking region up to −216. The CAT activity of this p−216/ + 54CAT construct was 11 times greater than the positive control promoter SV40 and 160 times greater than the promoterless pUM-SVOCAT vector. This region, −216/+ 54, corresponds to the minimal ALDH3 promoter, because the construct deleted further to −170 retained only 3% of the maximal activity obtained with p−216/ + 54CAT. No significant CAT activity was observed when −170 was deleted further to −120. In addition, the 3′-deletion construct, p−259/−1 CAT, retained only 17% of the maximal activity obtained with p−216/ + 54CAT and 19% with p−259/+ 54CAT. Constructs with further deletions from the 3′ end up to −120 demonstrated a further drop in the relative CAT activity. These results indicate that the −216/+ 54 region is essential for obtaining the maximal ALDH3 promoter activity in Hep G2 cells.
FIG. 6.
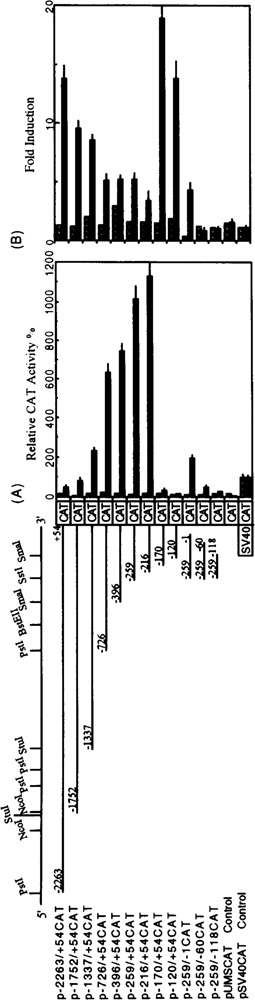
Transient expression of human ALDH3 gene promoter activity in Hep G2 and MCF7. Depicted on the left side of the figure are the ALDH3 promoter/CAT deletion constructs. The relative CAT activity was derived by normalizing the CAT activity with β-gal activity (to minimize the errors due to the differences in the transfection efficiency) and compared with that of the SV40 promoter. (A) Histogram representing the average relative CAT activities of three independent experiments carried out in duplicate of each of the constructs in Hep G2 (solid bars) and MCF7 (hatched bars). The values are referred to that obtained with the pSV40CAT control. (B) Histogram representing the fold induction of each of the constructs in the Hep G2 (solid bars) and MCF7 (hatched bars) treated with and without TCDD.
MCF7 cells, which do not express detectable levels of ALDH3 activity and whose ALDH3 gene expression cannot be induced by TCDD (data not shown), could not support the promoter activity of the transfected constructs (Fig. 6). The activity of p−216/ + 54CAT in MCF7 cells was similar to that of the promoterless control vector.
TCDD Induction of the Human ALDH3 Promoter Activity
The 5′-flanking region (2.3 kb) of the human ALDH3 gene contains five TCDD-responsive XRE sequences (5′-GCGTG-3′) as determined from a computer search (Fig. 1). To investigate whether the DNA segment containing these elements could confer TCDD induction, we performed the promoter activity analysis with transfected cells treated with TCDD. Figure 6B demonstrates the level of TCDD induction with various deletion constructs. Plasmid −2263/ + 54CAT exhibited approximately 14-fold induction of CAT expression in response to TCDD. The level of induction decreased with increasing deletion of the 5′-flanking region up to −216. However, induction increased to the original level when the 5′-flanking region was further deleted to −170 and −120.
DISCUSSION
Identification of a New Exon in the Human ALDH3 Gene
In this study, the exon/intron organization of the human ALDH3 gene was revised (Figs. 1 and 2). A new 5′-noncoding exon (exon 1) was identified. Primer extension analysis also supported the existence of such an exon. The new intron 1 follows the GT-AG rule and exon 2 is the first coding exon. The human ALDH3 gene is thus organized into 11 exons and spans about 11 kb in length, which is similar to the gene organization of the rat ALDH3 gene (3). In addition, the two ALDH3 genes share 80% sequence identity at the first non-coding exon and 60% about 700 bp upstream from the first noncoding exon, indicating the possibility of conservation of the gene regulation mechanism. This finding is consistent with similar tissue distribution of ALDH3 expression, and expression response to TCDD induction of the human and rat ALDH3 gene.
Different Isoforms of the Human ALDH3 Transcripts
Alternative splice acceptor sites
The 3′ end of the new intron 1 contains the 5′-UTR sequence of a previously identified stomach ALDH3 cDNA (18), which may be due to the following possibilities: i) The new intron 1 has at its 3′ end other alternative splice acceptor sites, which causes base insertion in some cDNA clones. Therefore, the stomach cDNA isolated previously was thus a truncated cDNA, which did not extend far enough at the 5′ end to include the new exon 1 sequence, ii) An alternative promoter located in the new intron 1 to direct the synthesis of the ALDH3 transcript starting from the 3′ end of the new intron 1 and continues downstream through exon 2, the first coding exon.
A series of deletion constructs of the 3-kb intron region were tested for the CAT activity in three ALDH3-positive cell lines [the gastric (AGS and KATO III) and colon (HT-29) carcinoma culture cells] to test whether an alternative promoter exists in the new intron 1. No promoter activity was found with any of the CAT constructs in any of the cell lines used (data not shown). Therefore, the CAT analysis did not support the existence of an alternative promoter in the new intron 1 of the human ALDH3 gene.
Sequencing analysis directly identified three alternative splice acceptor sites at the 3′ end of the new intron 1. The usage of these alternative splice sites during transcription generates nucleotide insertions in the 5′-UTR of the ALDH3 transcripts and does not affect the coding region. It is not known whether the insertions in the 5′-UTR affect the stability of the ALDH3 transcript and/ or the efficiency of the ALDH3 translation. It is interesting to note that, in our study, the usage of alternative splice acceptor sites was found only in tissues constitutively expressing ALDH3, but not in the TCDD-induced tissue (Fig. 3). Further study is necessary to reveal the biological significance of the insertions.
Alternatively spliced ALDH3 transcripts
We have also identified alternatively spliced ALDH3 transcripts skipping the exon 2 sequence. The significance of the existence of the alternatively spliced mRNA is not known. However, the alternatively spliced mRNA must use the internal in-frame methionine codon in exon 3 as the initiator to direct the synthesis of a shorter ALDH3 poly-peptide, because the skipped exon 2 contains the translational initiator Met (Fig. 1). The use of an alternative initiation codon to generate functionally distinct forms of several transcription factors has been reported (7).
Multiple transcription initiation sites
The primer extension analysis and 5′ end sequencing analysis of the 5′-RACE products from various tissues identified multiple transcription initiation sites. All three transcription initiation sites of the ALDH3 gene start at an adenine nucleotide preceded by a cytosine, which is consistent with the initiation site for RNA-polymerase II-dependent transcription (6). It is unknown whether different tissues prefer different initiation site(s).
The Human ALDH3 Gene Promoter
Human ALDH3 promoter region
A strong promoter region, −216 to +54, was localized in the human ALDH3 gene. In this region, three ATA sequences (at nt positions −208, −52, and + 26) and an inverted CCAAT box (at nt position −188) were found. The significance of these sequences is unknown. However, the transcriptional initiation sites of the ALDH3 gene, especially the one at site −65, demonstrates a high degree of sequence identity with the initiator (Inr) of the DHFR gene. The DHFR initiator is critical in positioning RNA polymerase II of TATA-less promoter (43). The expression exhibited by the p−259/−1 CAT and p−259/-00CAT constructs (Fig. 6A) might indicate the utilization of the putative initiation site at −65.
TCDD induction
The TCDD-induced transcriptional response has been studied extensively with the CYPIA1 gene (28). The induction effects are thought to be mediated by the interaction of the TCDD-Ah receptor with the xenobiotic response element (XRE, 5′-TNGCGTG-3′). Sequencing of the 5′-flanking region (2.3 kb) of the human ALDH3 gene revealed five potential XRE sequences at nt positions −2084, -648, −607, −224, and −35, whereas nt positions −648, −607, and −224 had only the core sequence of the XRE, 5′-GCGTG-3′ (Fig. 1).
The deletion of any or all of the putative XREs at nt positions −2084, −224, and -35 resulted in decreased induction by TCDD (Fig. 6B). The deletion of the other two putative XREs at nt positions −648 and −607 seemed to have no significant effect. It is worth mentioning that the latter two XRE sequences are in the same orientation as the XRE consensus sequence, whereas the other three are in the reverse orientation. It has been demonstrated in the rat ALDH3 gene, the regulatory regions in different orientation with respect to the promoter showed a significant difference in the level of induction by TCDD (38).
The deletion of the region between nt −1337 and −726, which contained no potential canonical XREs, also resulted in decreased induction, indicating the possibility of the involvement of other regulatory elements in the TCDD induction. In the rat ALDH3 gene system, a complex array of regulatory elements involved in TCDD induction was also observed (38). In our study, the original level of induction was restored when the deletion extended into the essential promoter region, −216/+ 54 (Fig. 6B, clones −170 and −120). In the presence of TCDD, the promoter activities of these two deletion clones might be driven by other regulatory elements located in the promoter inserts. It is unlikely that the vector sequence can account for the induction because the vector does not have any promoter.
Repressor activity
Repressor activities were found in many regions upstream from the ALDH3 promoter region, −216/+ 54. The relative CAT activity in transfected Hep G2 cells dropped to approximately half when −216/+ 54 region was extended 5′ upstream to - 396. A further decrease in relative CAT activity was observed with further upstream extension to -2.3 kb. A similar result was also observed in the rat ALDH3 gene (38).
The repressor regions may be responsible for repression of the ALDH3 gene in ALDH3-negative tissues such as normal liver. Mutations in the repressor region may result in the activation of the gene in transformed ALDH3-positive HCC cells. However, elements like AP-1 and PEA3 motifs may also be involved in the regulation of the ALDH3 expression in HCC cells. The combination of AP-1 and PEA3 elements has been referred to as tumor promoter and oncogene-responsive units as well as functional regulatory elements in several genes (15,16).
It is also very interesting to note that two sequence motifs recognized by the gastric-specific nuclear protein (5′-G/CPuPuG/CNGATA/ TPuPy-3′) (40) were identified in the repressor region at nt −1686 and −1385 positions of the human ALDH3 gene, implying that tissue-specific positive trans-acting factors upregulate the ALDH3 gene in the stomach. In the tissue expressing Ah receptor, it is likely that the binding of TCDD-Ah receptors to the XREs releases the repressor and activates the ALDH3 gene transcription.
The existence of both positive and negative regulatory mechanisms provides more extensive transcriptional regulation of the human ALDH3 gene. DNase I footprinting and mobility shift assays are currently being conducted to confirm the potential regulatory factor binding sites.
ACKNOWLEDGMENTS
We thank Dr. Gerald Forrest for helpful discussions and Vibha Dave for editing the manuscript.
REFERENCES
- 1. Ambroziak W.; Pietruszko R. Human aldehyde dehydrogenase: Activity with aldehyde metabolites of monoamines, diamines and polyamines. J. Biol. Chem. 266:13011–13018; 1991. [PubMed] [Google Scholar]
- 2. Ambroziak W.; Pietruszko R. Metabolic role of aldehyde dehydrogenase. Enzymol. Mol. Biol. Carbonyl Met. 4:5–15; 1993. [DOI] [PubMed] [Google Scholar]
- 3. Asman D. C.; Takimoto K.; Pitot H. C.; Dunn T. J.; Lindahl R. Organization and characterization of the rat class 3 aldehyde dehydrogenase gene. J. Biol. Chem. 268:12530–12536; 1993. [PubMed] [Google Scholar]
- 4. Banfi P.; Lanzi C. ; Falvella F. S.; Gariboldi M.; Gambetta R. A.; Dragani T. A. The daunorubicin-binding protein of Mr. 54,000 is an aldehyde dehydrogenase and is down-regulated in mouse liver tumors and in tumor cell lines. Mol. Pharmacol. 46:896–900; 1994. [PubMed] [Google Scholar]
- 5. Breathnach R.; Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu. Rev. Biochem. 50:349–383; 1981. [DOI] [PubMed] [Google Scholar]
- 6. Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212:563–578; 1990. [DOI] [PubMed] [Google Scholar]
- 7. Calligaris R.; Bottardi S.; Cogoi S.; Apezteguia I.; Santoro C. Alternative translation initiation site usage results in two functionally distinct forms of the GATA-1 transcription factor. Proc. Natl. Acad. Sci. USA 92:11598–11602; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chieco P.; Stecca B.; Bolondi L.; Melchiorri C. ; Gaiani S.; Barbara L. Cytochemical detection of a class 3 aldehyde dehydrogenase in human hepatocellular carcinoma. Liver 15:87–92; 1995. [DOI] [PubMed] [Google Scholar]
- 9. Cooper D. L.; Baptist E. W.; Enghild J. J.; Isola N. R.; Klintworth G. K. Bovine corneal protein 54K (BCP54) is a homologue of the tumor-associated (class 3) rat aldehyde dehydrogenase (RATALD). Gene 98:201–207; 1991. [DOI] [PubMed] [Google Scholar]
- 10. De Laurenzi V.; Rogers G. R.; Hamrock D. J.; Marekov L. N.; Steinert P. M.; Compton J. G.; Markova N.; Rizzo W. B. Sjögren-Larsson syndrome is caused by mutations in the fatty aldehyde dehydrogenase gene. Nature Genet. 12:52–57; 1996. [DOI] [PubMed] [Google Scholar]
- 11. Duley J.; Harris O.; Holmes R. S. Analysis of human alcohol- and aldehyde-metabolizing isozymes by electrophoresis and isoelectric focusing. Alcohol. Clin. Exp. Res. 9:263–271; 1985. [DOI] [PubMed] [Google Scholar]
- 12. Dunn T. J.; Lindahl R.; Pitot H. C. Differential gene expression in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-noncoordinate regulation of a TCDD-induced aldehyde dehydrogenase and cytochrome p-450c in the rat. J. Biol. Chem. 263:10878–10886; 1988. [PubMed] [Google Scholar]
- 13. Evces S.; Lindahl R. Characterization of rat cornea aldehyde dehydrogenase. Arch. Biochem. Biophys. 274:518–524; 1989. [DOI] [PubMed] [Google Scholar]
- 14. Faisst S.; Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 20:3–26; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gutman A.; Wasylyk B. The collagenase gene promoter contains a TPA and oncogene-responsive unit encompassing the PEA3 and AP-1 binding sites. EMBO J. 9:2241–2246; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gutman A.; Wasylyk B. Nuclear targets for transcription regulation by oncogenes. Trends Genet. 7:49–54; 1991. [DOI] [PubMed] [Google Scholar]
- 17. Harrington M. C.; Henehan G. T. M.; Tipton K. F. The roles of human aldehyde dehydrogenase isozymes in ethanol metabolism. Enzymol. Mol. Biol. Carbonyl Met. 1:111–125; 1987. [PubMed] [Google Scholar]
- 18. Hsu L. C.; Chang W-C.; Shibuya A.; Yoshida A. Human stomach aldehyde dehydrogenase cDNA and genomic cloning, primary structure, and expression in Escherichia coli . J. Biol. Chem. 267:3030–3037; 1992. [PubMed] [Google Scholar]
- 19. Huang M.; Lindahl R. Effects of hepatocarcinogenic initiators on aldehyde dehydrogenase gene expression in cultured rat hepatic cells. Carcinogenesis 11:1059–1065; 1990. [DOI] [PubMed] [Google Scholar]
- 20. Jakoby W. B.; Ziegler D. M. The enzymes of detoxication. J. Biol. Chem. 265:20715–20718; 1990. [PubMed] [Google Scholar]
- 21. Jones D. E. Jr.; Brennan M. D.; Hempel J.; Lindahl R. Cloning and complete nucleotide sequence of a full-length cDNA encoding a catalytically functional tumor-associated aldehyde dehydrogenase. Proc. Natl. Acad. Sci. USA 85:1782–1786; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lacronique V.; Lopez S.; Miquerol L.; Porteu A.; Kahn A.; Raymondjean M. Identification and functional characterization of an erythroid-specific enhancer in the L-type pyruvate kinase gene. J. Biol. Chem. 270:14989–14997; 1995. [DOI] [PubMed] [Google Scholar]
- 23. Lee D. C.; Gonzalez P.; Rao P. V.; Zigler J. S. Jr.; Wistow G. J. Carbonyl-metabolizing enzymes and their relatives recruited as structural proteins in the eye lens. Enzymol. Mol. Biol. Carbonyl Met. 4:159–168; 1993. [DOI] [PubMed] [Google Scholar]
- 24. Lindahl R. Aldehyde dehydrogenases and their role in carcinogenesis. Crit. Rev. Biochem. Mol. Biol. 27:283–335; 1992. [DOI] [PubMed] [Google Scholar]
- 25. Marselos M.; Strom S. C.; Michalopoulus G. Enhancement of aldehyde dehydrogenase activity in human and rat hepatocyte cultures by 3-methylcholanthrene. Cell. Biol. Toxicol. 2:257–269; 1986. [DOI] [PubMed] [Google Scholar]
- 26. Marselos M.; Strom S. C.; Michalopoulus G. Effect of phenobarbital and 3-methylcholanthrene on aldehyde dehydrogenase activity in cultures of HepG2 cells and normal human hepatocytes. Chem. Biol. Interact. 62:75–88; 1987. [DOI] [PubMed] [Google Scholar]
- 27. Mitchell D. Y.; Petersen D. R. The oxidation of α,β-unsaturated aldehydic products of lipid peroxidation by rat liver aldehyde dehydrogenases. Toxicol. Appl. Pharmacol. 87:403–410; 1987. [DOI] [PubMed] [Google Scholar]
- 28. Nebert D. W.; Puga A.; Vasiliou V. Role of the Ah receptor and the dioxin-inducible [Ah] gene battery in toxicity, cancer, and signal transduction. Ann. NY Acad. Sci. 685:624–640; 1993. [DOI] [PubMed] [Google Scholar]
- 29. Parrilla R.; Ohkawa K.; Lindros K. O.; Zimmerman U.-J. P.; Kobayashi K.; Williamson J. R. Functional compartmentation of acetaldehyde oxidation in rat liver. J. Biol. Chem. 249:4926–4933; 1974. [PubMed] [Google Scholar]
- 30. Pereira F.; Rosenmann E.; Nylen E.; Kaufman M.; Pinsky L.; Wrogemann K. The 56K androgen binding protein is an aldehyde dehydrogenase. Biochem. Biophy. Res. Commun. 175:831–838, 1991. [DOI] [PubMed] [Google Scholar]
- 31. Pietruszko R. Aldehyde dehydrogenase isozymes. Isozymes Curr. Top. Biol. Med. Res. 8:195–217; 1983. [PubMed] [Google Scholar]
- 32. Rogers G. R.; Rizzo W. B.; Zlotogorski A.; Hashem N.; Lee M.; Compton J. G.; Bale S. J. Genetic homogeneity in Sjögren-Larsson syndrome: Linkage to chromosome 17p in families of different non-Swedish ethnic origins. Am. J. Hum. Genet. 57:1123–1129; 1995. [PMC free article] [PubMed] [Google Scholar]
- 33. Salier J.-P.; Hirosawa S.; Kurachi K. Functional characterization of the 5′-regulatory region of human factor IX gene. J. Biol. Chem. 265:7062–7068; 1990. [PubMed] [Google Scholar]
- 34. Sambrook J.; Fritsch E. F.; Maniatis T. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989:7.79–7.83. [Google Scholar]
- 35. Seed B.; Sheen J.-Y. A simple phase-extraction assay for chloramphenicol acetyltransferase activity. Gene 67:271–277; 1988. [DOI] [PubMed] [Google Scholar]
- 36. Sreerama L.; Rekha G. K.; Sladek N. E. Phenolic antioxidant-induced overexpression of class-3 aldehyde dehydrogenase and oxazaphosphorine-specific resistance. Biochem. Pharmacol. 49:669–675; 1995. [DOI] [PubMed] [Google Scholar]
- 37. Sreerama L.; Sladek N. E. Identification of the class-3 aldehyde dehydrogenases present in human MCF-7/0 breast adenocarcinoma cells and normal human breast tissue. Biochem. Pharmacol. 48:617–620; 1994. [DOI] [PubMed] [Google Scholar]
- 38. Takimoto K.; Lindahl R.; Dunn T. J.; Pitot H. C. Structure of the 5′ flanking region of class 3 aldehyde dehydrogenase in the rat. Arch. Biochem. Biophys. 312:539–546; 1994. [DOI] [PubMed] [Google Scholar]
- 39. Takimoto K.; Lindahl R.; Pitot H. C. Regulation of 2,3,7,8-tetrachlorodibenzo-p-dioxin-inducible expression of aldehyde dehydrogenase in hepatoma cells. Arch. Biochem. Biophys. 298:492–497; 1992. [DOI] [PubMed] [Google Scholar]
- 40. Tamura S.; Oshiman K.-L. ; Nishi T.; Mori M.; Maeda M.; Futai M. Sequence motif in control regions of the H +/K + ATPase alpha and beta subunit genes recognized by gastric specific nuclear protein(s). FEBS Lett. 298:137–141; 1992. [DOI] [PubMed] [Google Scholar]
- 41. Vasiliou V.; Puga A.; Nebert D. W. Negative regulation of the murine cytosolic aldehyde dehydrogenase-3 (Aldh-3c) gene by functional CYP1A1 and CYP1A2 proteins. Biochem. Biophys. Res. Commun. 187:413–419; 1992. [DOI] [PubMed] [Google Scholar]
- 42. Vasiliou V.; Reuter S. F.; Kozak C. A.; Nebert D. W. Mouse dioxin-inducible cytosolic aldehyde dehydrogenase-3: AHD4 cDNA sequence, genetic mapping, and differences in mRNA levels. Pharma-cogenetics 3:281–290; 1993. [DOI] [PubMed] [Google Scholar]
- 43. Weis L.; Reinberg D. Transcription by RNA polymerase II: Initiator-directed formation of transcription-competent complexes. FASEB J. 6:3300–3309; 1992. [DOI] [PubMed] [Google Scholar]
- 44. Westlund P.; Fylling A. C.; Cederlund E.; Jörnvall H. 11-Hydroxythromboxane B2 dehydrogenase is identical to cytosolic aldehyde dehydrogenase. FEBS Lett. 345:99–103; 1994. [DOI] [PubMed] [Google Scholar]
- 45. Xie J.; Roddy P.; Rife T. K.; Murad F.; Young A. P. Two closely linked but separable promoters for human neuronal nitric oxide synthase gene transcription. Proc. Natl. Acad. Sci. USA 92:1242–1246; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yanagawa Y.; Chen J. C.; Hsu L. C.; Yoshida A. The transcriptional regulation of human aldehyde dehydrogenase 1 gene: The structural and functional analysis of the promoter. J. Biol. Chem. 270:17521–17527; 1995. [DOI] [PubMed] [Google Scholar]
- 47. Yoshida A.; Hsu L. C.; Davé V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme 46:239–244; 1992. [DOI] [PubMed] [Google Scholar]
- 48. Yoshida A.; Hsu L. C.; Yanagawa Y. Biological role of human cytosolic aldehyde dehydrogenase 1: Hormonal response, retinal oxidation and implication in testicular feminization. Enzymol. Mol. Biol. Carbonyl Met. 4:37–44; 1993. [DOI] [PubMed] [Google Scholar]


