Abstract
Peroxisome proliferator-activated receptor γ (PPARγ), a member of the nuclear receptor superfamily, is expressed predominantly in adipose tissue. Forced expression of the two isoforms of this receptor, PPARγ1 and PPARγ2, in fibroblasts initiates a transcriptional cascade that leads to the development of adipocyte phenotype. Using the yeast two-hybrid system and GAL4-PPARγ as bait to screen mouse liver cDNA library, we isolated a mouse steroid receptor coactivator (mSRC-1) involved in nuclear hormone receptor transcriptional activity as a mPPARγ interactive protein. mSRC-1 cDNA we isolated contains an open reading frame of 1447 amino acids and encodes a new member of the basic helix-loop-helix-PAS domain family. We show that the binding of mSRC-1 to mPPARγ is ligand independent and coexpression of mSRC-1 with mPPARγ increases the transcriptional activity of mPPARγ in the presence of mPPARγ ligand. We have identified the presence of two putative mPPARγ binding sites in the mSRC-1, one between residues 620 and 789, and the second between residues 1231 and 1447. These two regions exhibit different degrees of binding affinity for mPPARγ. We also show that mSRC-1 exhibits its own constitutive transcriptional activity in the yeast as well as in mammalian cells. These results suggest that mSRC-1 interacts with PPARγ and plays a role in the PPARγ-mediated signaling pathway.
Keywords: Mouse steroid receptor coactivator-1, Peroxisome proliferator-activated receptor γ, Transcriptional activity
PEROXISOME proliferators constitute a broad spectrum of synthetic and naturally occurring compounds which includes certain hypolipidemic drugs, phthalate ester plasticizers, industrial solvents, herbicides, and the adrenal steroid dehydroepiandrosterone (29,30). When these structurally diverse compounds are administered to rats, mice, and certain nonrodent species, including primates, they induce predictably similar pleiotropic responses, the hallmark of which is profound proliferation of peroxisomes in liver parenchymal cells (29,30). Peroxisome proliferator-induced pleiotropic responses are characterized as immediate and chronic, with the immediate pleiotropic responses comprised of significant hepatomegaly, marked increase in peroxisome volume density in liver cells, enhancement of transcriptional activity of the three genes encoding the peroxisomal fatty acid β-oxidation enzyme system, and increases in the activities of certain nonperoxisomal enzymes (27,29,30). The delayed pleiotropic response manifesting as a result of chronic exposure to peroxisome proliferators is the development of liver tumor (29). Because peroxisome proliferators are nongenotoxic in nature, the carcinogenicity of these agents has been attributed to induction of H2O2-generating peroxisomal β-oxidation enzyme system and the resultant oxidative stress in the liver (7,27,28).
The liver-specific induction of peroxisome proliferation vis-à-vis the transcriptional activation of the β-oxidation enzyme system genes by peroxisome proliferators is mediated by members of the nuclear receptor superfamily, termed peroxisome proliferator-activated receptors (PPARs), which are closely related to the TR and retinoid receptors (18,37,38). To date, three isoforms of PPARs have been identified in amphibians, rodents, and humans: PPARα, PPARδ (also called β or NUC-1), and PPARγ (9,20,42). PPAR isoforms exhibit distinct patterns of tissue distribution, and differ considerably in their ligand binding domains, suggesting that they perform different functions in different cell types (2). PPARα is highly expressed in hepatocytes, enterocytes, and the proximal tubular epithelium of kidney, and plays an essential role in the peroxisome proliferator-induced pleiotropic responses (2). Corroborative evidence for the functional role of PPARα in peroxisome proliferator-induced signal transduction and transcriptional activation of genes comes from the observation that disruption of PPARα gene results in the abolishment of the pleiotropic effects of peroxisome proliferators in mice (23). PPARγ has been shown to play an important role in the adipocyte differentiation, and the actions of this receptor are regulated by thiazolidinediones, a class of antidiabetic drugs, and the fatty acid derivative 15-deoxy-Δ12,4-prostaglandin-J2, which bind PPARγ and promote adipogenesis (10,21,35,36).
To understand the role of PPARs as mediators of cell- and species-specific effects of peroxisome proliferators, it is essential to gain insight into the mechanism underlying transcriptional activation of responsive genes. It has been postulated that nuclear receptors activate their target gene transcription by directly or indirectly promoting assembly of basal transcription factors into the preinitiation complex through the recruitment of cofactors. In support of this concept, several potential nuclear receptor cofactors have been identified in recent years. Some examples include: Tripl interaction with the AF-2 domain of TR, RAR, VDR, and ER (23); TIF1 interaction with AF-2 domains of RAR, RXR, and ER (22); and interaction of RIP140 and RIP160 with AF-2 region of ER, RAR, and TR in a hormone-dependent fashion (4,13). Recently, two structurally related proteins, designated as SMRT (silencing mediator for retinoid and thyroid receptors) and N-CoR (nuclear receptor corepressor), have been shown to interact with RAR and TR and repress basal transcription in the absence of ligand (5,15). In an attempt to identify coactivators or corepressors for PPARs, we utilized a yeast two-hybrid system to isolate the possible cofactor(s) of mPPARγ that may be required for its transcriptional activation function. We cloned a cDNA from mouse liver cDNA library, which encodes a protein that interacts with AF-2 domain of PPARγ. The amino acid sequence comparison indicates that this protein is the homologue of human SRC-1, which interacts with and enhances the PR transcriptional activity (26). mSRC-1 has two mPPARγ binding sites, which show varying degrees of affinity to bind mPPARγ. Although both these binding sites interact with mPPARγ in a ligand-independent manner, mSRC-1 stimulates mPPARγ transcriptional activity in a ligand-dependent manner.
MATERIALS AND METHODS
Plasmids
The vector GAL4-PPARγ for expressing the fusion protein of GAL4DBD (DNA binding domain) and mPPARγ LBD (ligand binding domain) in yeast was constructed by inserting PCR-amplified cDNA fragment coding for mPPARγ1 ligand binding region (174–475 amino acids) into EcoRI/SalI site of PGBT9 (Clontech). PCMV-mSRC-1 cDNA, amplified by PCR, was generated by inserting the PCR-amplified mSRC-1 cDNA with the full length of coding region into BamHI/Xhol site of PCMV-Amp (Invitrogen). PCMV-mPPARγ was made by inserting mPPARγ2 cDNA into HindIII/Xhol site of PCMV-Amp (Invitrogen). To construct PCMV-mSRC-1 (1231–1447 amino acids), the partial mSRC-1 cDNA fragment encoding amino acids 1231 to 1447 was obtained by PCR and subcloned into HindIII/SalI site of PCMV-FLAG (Kodak). PCMV-GAL was made by inserting the GAL4 DNA binding domain sequence into the HindIII/XbaI site of PCMV-Amp. Plasmids encoding fusion proteins between different truncated mSRC-1 and GAL-AD were made by PCR amplification of mSRC-1 fragment encoding the amino acids indicated, then insertion into the appropriate restriction site of PGAD424. We constructed GAL-mSRC-1 by the insertion of mSRC-1 coding region into BamHI/ SalI site of PCMV-GAL, and glutathione-S-transferase (GST)-mPPARγ expression vector was generated by inserting a EcoRI fragment from mPPARγ cDNA encoding a 251–475 amino acid region into PGEX-3X. The PPRE-TK-LUC was constructed by inserting three copies of PPRE into HindIII/Sal I site of TK-LUC. The GAL-TK-LUC was produced by insertion of three copies of the GAL4 binding element into the BamHI/SalI site of TK-LUC.
Yeast Two-Hybrid System
To isolate cDNAs encoding proteins that specifically interact with PPARγ, the yeast two-hybrid screening (6) was performed by using the matchmaker two-hybrid system kit (Clontech). Briefly, the Saccharomyces cerevisiae strain HF7C [MATa, ura3–52, his3–200,lys2–801,ade2–101, trpl–901,leu2–3,112,gal4–542,gal80–538, LYS2:: GAL1-HIS3, URA3:: (GAL4 17-mers)-CYC1-lacZ] was cotransformed with a mouse matchmaker liver cDNA expression library and GAL4-PPARγ. The positive clones were selected by their growth in medium lacking histidine, and expression of β-galactosidase. Screening was performed in the presence or absence of 1 × 10–6 M of mPPARγ ligand, thiazolidinedione (BRL 49653). To localize the binding sites on the mSRC-1, plasmids encoding fusion protein between different truncated mSRC-1 GAL4-AD were cotransformed into HF7C with GAL4-PPARγ. The β-galactosidase activity was examined by the filter lift method or quantitatively by CPRG method (6).
Race PCR
After identifying the positive clones that contain the partial mSRC-1, the remaining 5′ end sequence was cloned by 5′ RACE PCR (11) amplification from the mouse liver marathon ready cDNA (Clontech) using rTth DNA polymerase. Briefly, the first amplification was performed using the adapter primer 1 and the gene-specific primer (5′-TGTCCCATCATTCAATATGAATCT-3′) for 20 cycles. Each cycle includes 20 s at 94°C, 30 s at 60°C, and 4 min at 68°C; 1 μl of the PCR product was used as the template for the second amplification with the adapter primer 2 and the nested gene-specific primer (5′-GGATGGGCTGGAGGCAGTGC-3′) for 20 cycles, each cycle consisting of 20 s at 94°C, 30 s at 65°C, and 4 min at 68°C. The PCR products were cloned into pGEM-T (Promega) and sequenced.
Interaction of mSRC-1 and mPPARγ In Vitro
mSRC-1 in PCMV-Amp was transcribed and translated in rabbit reticulocyte lysates (Promega) and labeled with [35S]methionine. GST and GST-mPPARγ in PGEX-3X were produced in E. coli and bound to glutathione-sepharose beads according to the manufacturer’s instructions (Pharmacia LKB). A 10 μl aliquot of GST-mPPARγ fusion protein, loaded on glutathione-sepharose beads, was incubated with 2 μl of [35S]methionine-labeled mSRC protein for 2 h in 600 μl of NETN (20 mM Tris-HCl, pH 7.5, 100 mM KCl, 0.7 mM EDTA, 0.5% Nonidet p-40, 1 mM phenylmethylsulfonyl fluoride), and the beads were washed three times with NETN. BRL 49653 was added to NETN at a final concentration of 1 × 10–6 M when required. The protein(s) bound to the beads was eluted by boiling in 20 μl of 1 × SDS sample buffer and analyzed by SDS/PAGE and autoradiographed.
Cell Culture and Transfection
HeLa or CV-1 cells, 5.7 × 105, were plated in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum and cultured for 24 h before transfection. Cells were transfected for 20 h with 5 μg of luciferase reporter plasmid DNA, 4 μg of appropriate expression plasmid DNA, and 2 μg of β-galactosidase expression vector pCMVB DNA (Clontech), using calcium phosphate precipitation technique (1). Cell extracts were prepared 36 h after transfection by three cycles of freezing and thawing and assayed for luciferase and β-galactosidase activities (1).
RESULTS
Cloning of the mSRC-1 cDNA Encoding the PPARγ Interacting Protein
To isolate cDNA clone(s) capable of interacting with PPARγ, we used a yeast two-hybrid screening system. This screening system used GAL4-PPARγ (expressing the GAL4 DNA-binding domain and PPARγ-LBD fusion protein), which was cotransformed into yeast with a second vector that expressed fusion proteins between GAL4-activating domain and mouse liver cDNA. By screening about 4 × 106 yeast transfectants in the presence or absence of 1 × 10–6 M of the PPARγ ligand, BRL-49653, we identified 13 clones that exhibited positive interaction with PPARγ; these clones failed to interact with other unrelated proteins such as p53 and lamin, used as controls in this screening system (6). Partial nucleotide sequence analysis revealed that two of the clones represented different lengths of the same mRNA. Both these clones interacted with mPPARγ LBD in yeast cells in the absence of the ligand but with different affinity, and the addition of ligand, BRL 49653, did not influence the β-galactosidase reporter gene activity (data not shown), indicating that the interaction between mPPARγ and these two clones is largely ligand independent. These two clones were selected for further analysis.
The cDNA fragments recovered from the above two clones were 3.5 and 1 kb in size, respectively. Nucleotide sequence analysis revealed that they represented different lengths of the same mRNA and each of them contained a stop codon. We used RACE PCR to clone the remaining 5′ end sequence of the cDNA; the entire coding region was obtained and sequenced. This cDNA encompassed a 4341-nucleotide open reading frame that predicted a 1447-amino acid protein, with an estimated molecular mass of 157 kDa (Fig. 1). The clone with the 3.5-kb insert encoded amino acid residues 313–1447, whereas the clone with the 1.0-kb insert encoded amino acid residues 1231–1447. GenBank comparison revealed that the C-terminal portion of this protein (residues 381–1447) corresponds to the human SRC-1, and shares 89% identity with human SRC-1 (26). Human SRC-1, isolated from human B lymphocyte cDNA library, has been shown to interact with and enhance the PR transcriptional activity in a hormone-dependent fashion (26). The N-terminal 380-amino acid residue segment of this mouse PPAR-interacting protein described here revealed that the amino acid sequence from 32 to 86 (Fig. 2) is a basic helix–loop–helix (bHLH) domain most homologous to Drosophila Hairy, Her-1, and Hes-1 (32,33). The amino acid residues 113 to 165 of the PPAR-interacting protein also showed homology to the ‘A’ subdomain of the PAS domain (25). Proteins with the PAS domain include Drosophila circadian rhythm protein, Per (41), “Single-minded” protein Siml (25), and “trachealess” TCL, which is the essential gene for the tracheal development (17), and the mammalian aryl hydrocarbon receptor, AHR (3), aryl hydrocarbon receptor nuclear translocator, ARNT (31), and the hypoxia-inducible factor 1α, HIF-1α (39). As there is great similarity of the C-terminal portion of this protein to human SRC-1 (26), we designate this PPARγ-interacting protein as mouse steroid receptor coactivator (mSRC-l). During the preparation of this manuscript, Kamei et al. (19) reported the sequence of variants of the mouse SRC-1. Comparison with the published sequence indicated that they represent the same cDNA except for the difference of a few amino acids (Fig. 1).
FIG. 1.

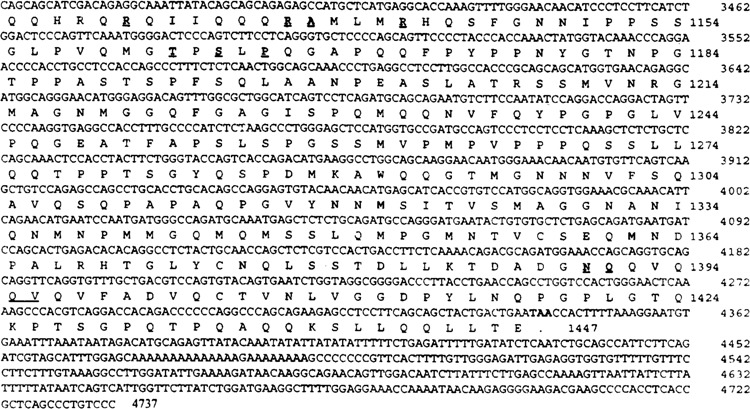
Amino acid sequence of the mSRC-1 deduced from cDNA nucleotide sequence. The positive numbers of the nucleotide sequence start at the first ATG codon. The amino acids for the single open reading frame are numbered starting with the first methionine. The initiation codon ATG and the termination codon TAA are shown in bold face. The different amino acids compared with the published sequence (19) are in bold and underlined. Their corresponding amino acids from the published sequence are: G for amino acid number 45 E, C for 223 R, E for 234 G, T for 465 S, T for 466 S, Q for 699 P, K for 1130 R, K for 1137 R, P for 1138 A, K for 1142 R, D for 1162 T, R for 1164 S, L for 1166 P, T for 1390 N, L for 1391 Q. There is an additional amino acid M between amino acid 1395 Q and 1396 V in the published sequence.
FIG. 2.

N-terminal regions of mSRC-l contain bHLH motif and the A region of the PAS domain. (A) Homology between an N-terminal region (amino acid 32-84) of mSRC-l and basic helix–loop–helix domains of Drosophila HAIRY, HER-1, and HES-1 proteins. (B) Homology between an N-terminal region (amino acid 113–167) and the A region of the PAS domain of AHR, ARNT, HIF-lα, TCL, and SIM1 proteins.
The expression of mSRC-l was analyzed by Northern blotting. The results showed that mSRC-l is ubiquitously expressed as a 7.5-kb transcript in all tissues examined (Fig. 3). The levels of mSRC-l mRNA appeared abundant in the brain, kidney, skeletal muscle, and liver. In the spleen, it is expressed in low quantities (Fig. 3). A less abundant 6-kb transcript is also observed in all tissues, but it appears most prominent in the testis and liver and may represent an alternate splicing form of the mRNA transcript (Fig. 3).
FIG. 3.

Tissue distribution of mSRC-l mRNA. A mouse multiple tissue Northern blot (Clontech) containing 2 μg poly(A) RNA for each tissue was probed with 32P-labeled mSRC-l cDNA. Heart (lane 1), brain (lane 2), spleen (lane 3), lung (lane 4), liver (lane 5), skeletal muscle (lane 6), kidney (lane 7), and testis (lane 8).
mSRC-1 Interacts With Mouse PPARγ In Vitro
To further confirm the specificity of interaction between mPPARγ and mSRC-l, we first expressed the mPPARγ ligand binding domain as GST fusion protein and linked this GST-mPPARγ fusion protein to GST-Sepharose beads. Incubation of these preloaded beads with the full-length, in vitro-synthesized [35S]methionine-labeled mSRC-1 revealed that the GST-Sepharose beads with preloaded fusion protein can pull out [35S]methioine-labeled mSRC-l (Fig. 4, see GST-mPPARγ). No interaction was seen when only the GST-Sepharose beads (without the GST-mPPARγ fusion protein) were incubated with mSRC-l (Fig. 4, see GST), indicating that mSRC binding to GST-mPPARγ is due to the interaction of mSRC with mPPARγ. Addition of 1 × 10–6 M of mPPARγ ligand, BRL 49653, did not influence the interaction between mPPARγ and mSRC-1 (Fig. 4). These in vitro assays further confirm the strong interaction observed in the yeast two-hybrid assay.
FIG. 4.

Interaction of mSRC-1 with mPPARγ in vitro. [35S]Methionine-labeled full-length mSRC-1 generated by in vitro transcription and translation was incubated with GST-Sepharose beads bound with purified E. coli expressed GST-mPPARγ or with GST, in the presence (+) or absence (–) of ligand BRL 49653. The bound proteins were eluted and analyzed by SDS-PAGE and autoradiographed. Note that raSRC-1 binds to GST-mPPARγ with or without the ligand. No binding is seen to GST alone.
mSRC-1 Contains Two Binding Sites for mPPARγ
Because the two clones recovered from two hybrid system interacted with mPPARγ with different affinity, it appeared possible the clone with longer insert has additional binding sites for the mPPARγ. This two-hybrid system was used to delineate the mSRC-1 structural requirements for PPARγ binding. The yeast expression vector with different lengths of partial mSRC-1 cDNAs (Fig. 5A) fused inframe to the GAL4 activation domain were cotransformed into yeast with the vector expressing GAL4-DB-PPARγ. These studies led to the identification of two binding sites. The one, which included a small polypeptide (residues 620–789), displayed much stronger interaction with mPPARγ; the second site localized at the carboxyl-terminus (residues 1231–1447) also interacted with mPPARγ, but the interaction was weaker than the first one. The presence or absence of the PPARγ ligand, BRL49653, exerted no influence on the interaction of these sites with mPPARγ (Fig. 5B).
FIG. 5.

(A) mSRC-1 contains two mPPARγ binding domains. Deletions of mSRC-1 with different remaining amino acids (indicated on the left) were inserted into yeast expression vecter PGAD424, then checked with its interaction with mPPARγ by two-hybrid system. The two identified sites are shown. (B) The two binding sites have different affinity for mPPARγ. The β-galactosidase activity was measured after growth for 12 h in the presence or absence of 10–6 M of BRL-46593.
mSRC-1 Coactivates the Transcriptional Activity of mPPARγ
To investigate the consequences of the interaction between the mSRC-1 and mPPARγ on the mPPARγ transcriptional activity in vivo, transient transfection assays were performed. HeLa cells were transfected with expression vector encoding full-length mPPARγ either alone or with mSRC-1 (cotransfection) along with a reporter plasmid containing three copies of peroxisome proliferator-response elements (PPREs) upstream of the thymidine kinase (TK) promoter driving the luciferase gene, and a β-galactosidase expression vector as a control. As expected, transfection with mPPARγ without mSRC-1 resulted in ∼18-fold increase in the luciferase reporter activity in the presence of the ligand, BRL-49653 (Fig. 6). When mSRC-1 was coexpressed with mPPARγ, a threefold increase in the ability of mPPARγ to transactivate the reporter gene in the presence of the ligand was observed (Fig. 6). mSRC-1 did not affect the mPPARγ activity in the absence of its ligand (Fig. 6). mSRC also coactivated the ligand-mediated transactivation capacity of PPARα, another member of PPAR subfamily (data not shown). To clarify that the mSRC-1 acts by interacting with the receptor instead of the general promoter, PPRE-TK-LUC was replaced with GAL-TK-LUC, which contained three copies of GAL4 DNA binding site upstream of the TK promoter driving the luciferase gene in the cotransfection assay. As illustrated in Fig. 6, mSRC-1 did not influence the expression of GAL-TK-LUC.
FIG. 6.
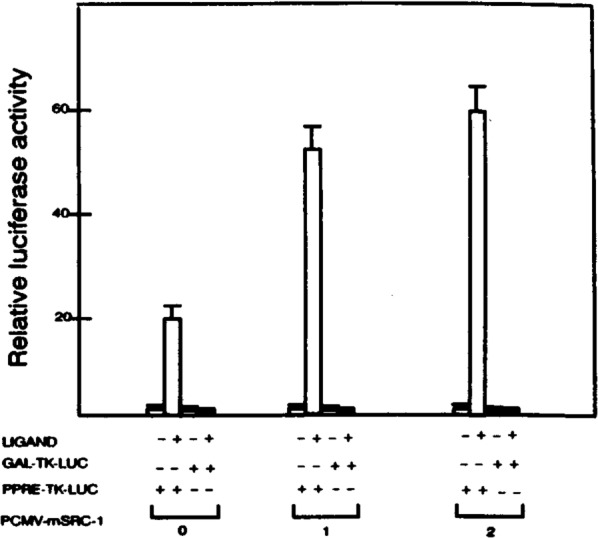
Enhancement of mPPARγ-mediated transactivation of reporter expression by mSRC-1. HeLa cells were cotransfected with 5 μg reporter construct PPRE-TK-LUC or GAL-TK-LUC, with different amounts of PCMV-mSRC-1, 2 μg PCMV-mPPARγ, and 2 μg PCMVB in the absence (–) or presence (+) of 10–6 M BRL 49653. Transfections with less PCMV-mSRC-1 were compensated by adding appropriate amounts of PCMV-AMP. The activity obtained on transfection of the PPRE-TK-LUC without exogenous mSRC-1 in the absence of ligand was taken as 1. Results are the mean of three independent transfections normalized to the internal controls of β-galactosidase expression.
The coactivator function of mSRC-1 was further confirmed by using its truncated form to act as a dominant-negative inhibitor for the endogenous mSRC-1 function. Partial mSRC-1 peptide (residues 1231–1447), which contains one of the mPPARγ binding sites, when cotransfected with mPPARγ into CV-1 cells, inhibited the ligand-induced mPPARγ transcriptional activity in a concentration-dependent manner (Fig. 7). This mSRC-1 peptide (residues 1231–1447) also showed similar inhibitory effect on mPPARγ activity in HeLa cells (data not shown). Thus, the ability of the truncated mSRC-1 to act as dominant-negative repressor in both CV-1 and HeLa cells strongly indicates that mSRC-1 is a true coactivator for mPPARγ.
FIG. 7.

Repression of mPPARγ-mediated transactivation by truncated mSRC-1. Reporter construct or PPRE-TK-LUC was cotransfected with PCMV-mPPARγ along with different amounts of PCMV-mSRC-1 (amino acid 1231 C-terminus), and PCMVB into CV-1 cells in the presence of 10–6 M BRL 49653. The transfections with less PCMV-mSRC-1 (1231 C-terminus) were compensated with appropriate amounts of PCMV-Flag. Luciferase activity is presented as percent where induced mPPARγ activity in the presence of BRL 49653 is arbitrarily set at 100%.
mSRC-1 Exhibits Constitutive Transcriptional Activation Capacity in Yeast and Mammalian Cells
As mSRC-1 exhibits the coactivator property, we investigated whether mSRC-1 contains the transcriptional activity itself, because it contains bHLH-PAS domain. GAL-mSRC-1 expressing the GAL4 DNA binding domain and mSRC-1 fusion protein were cotransfected into CV-1 cells with GAL-TK-LUC. The expression of luciferase gene was increased by GAL-mSRC-1 but not by GAL4 DNA binding domain alone or mSRC-1 (Fig. 8A), indicating mSRC-1 possesses the transcriptional activation function. The transcriptional activation function is also seen in the yeast as the GAL4-mSRC-l can activate the expression of the β-galactosidase gene with the promoter containing the GAL4 binding elements (Fig. 8B).
FIG. 8.
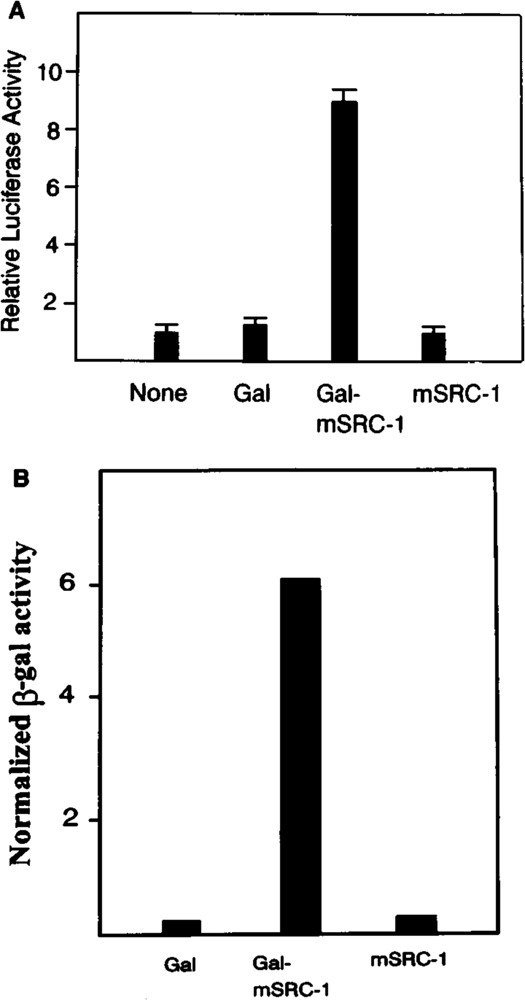
(A) Transcriptional activation by mSRC-1 in mammalian cell. CV-1 cells were cotransfected with 5 μg reporter construct GAL-TK-LUC, 2 μg PCMVB as internal control, 4 μg expression plasmid. The luciferase activity from the transfection of GAL-TK-LUC without expression plasmid was taken as 1. (B) Transcriptional activation by mSRC-1 in yeast. The yeast with β-galactosidase reporter gene directed by GAL4 binding sites was transformed with the yeast expression vector encoding GAL4 DB alone (GAL) or fusion protein between GAL4 DB and mSRC-1(GAL-mSRC-1) or mSRC-1 alone. The β-galactosidase activity was measured after growth for 12 h.
DISCUSSION
The role of PPARs in the physiologic modulation of lipid metabolism is increasingly being recognized. Several genes involved in lipid metabolism that respond to peroxisome proliferators have regulatory regions designated as PPRE, which bind PPAR/RXR heterodimers, and this binding initiates gene transcription (20,37). Evidence also indicates that mice with PPARα gene disruption fail to respond to the peroxisome proliferators (23). Furthermore, PPARγ isoform, which is abundantly expressed in adipocytes, functions as a key regulator of adipocyte differentiation (35,36). We now present evidence that mSRC-1, which we isolated from the mouse cDNA library using the yeast two-hybrid screen, interacts with and enhances the transcriptional activity of mPPARγ. The mPPARγ binding protein we cloned represents the full-length mouse homologue of the human SRC-1 (26). The C-terminal 1067-amino acid sequence of this protein (amino acid 381 to the C-terminus) shares high homology to the human steroid hormone receptor coactivator-1 (SRC-1) identified recently as a transcriptional coactivator of steroid receptors (26). Full-length mPPARγ-interacting protein cDNA encodes a polypeptide of 1447 amino acids with an estimated molecular mass of 157 kDa. This protein shows strong interaction with mPPARγ both in vivo and in vitro (Fig. 3) and stimulates the transactivation capacity of both mPPARγ (Fig. 4) and PPARα (data not shown). The N-terminal truncated form of this protein functions as a dominant-negative repressor, further indicating that the mPPARγ-interacting protein is indeed a coactivator required for the full transcriptional activity of PPARγ and possibly other PPAR isoforms. The N-terminal 380-amino acid region of mSRC-1 contains a bHLH motif. Such motifs are found in mammalian transcription factors AHR (3), and ARNT (14,31), and also in Drosophila Hairy, Hes-1, and Her-1 genes (32,33), implying that the mouse PPARγ coactivator has the potential to function as a transcriptional factor by itself. This protein also possesses a domain homologous to the subdomain “A” of PAS domain (14,25), which is implicated in the protein–protein heterodimerization as well as homodimerization (16). In particular, this protein may provide important insights into the interrelationships between peroxisome proliferator-induced pleiotropic responses, the biological/toxicological effects of TCDD and other halogenated hydrocarbons, and signal transductions mediated by different steroid hormones.
Two mPPARγ binding sites are found in mSRC-1 using the truncational analysis. It would be interesting to ascertain if these two sites bind to the same domain on mPPARγ. Because mPPARγ binds to PPRE as a heterodimer with RXR, it is possible that one mSRC-1 molecule may interact with both mPPARγ and RXR with these two binding regions. Additional information about the interactions between mSRC-1, mPPARγ, and RXR is necessary to understand the molecular aspects of cell- and tissue-specific aspects of mPPARγ-mediated transcriptional activation.
Interaction between mPPARγ hormone binding domain and mSRC-1 in the yeast two-hybrid system is not influenced by the presence or absence of mPPARγ ligand, BRL-49653. The addition of BRL-49653 did not affect the binding of mSRC-1 to mPPARγ in vitro. In this context, mSRC-1 binding to mPPARγ differs when compared to human SRC-1 binding to PR, which is ligand dependent both in the yeast two-hybrid system and in vitro assays (26). During the preparation of this manuscript, Kamei et al. (19) and Yao et al. (40) reported the isolation of mSRC-1, and in these studies mSRC-1 has been shown to interact with CBP and RAR. mSRC-1 binding to RAR has been shown to be ligand dependent (40). This suggests that the interactions between mSRC-1 and various receptors are highly complex and that receptor-dependent differences in interaction may dictate the transcriptional activity. Although the interaction between mSRC-1 and mPPARγ is ligand independent, cotransfection experiments demonstrated clearly that the mSRC-1 can only fully increase the mPPARγ transcriptional activity in the presence of ligand. Considering that mSRC-1 and other transcriptional activators (12) and repressors (5,15) interact with multiple target proteins to exert their function, it is reasonable to assume the mPPARγ must bind to other proteins in addition to mSRC-1 to exert maximum transcriptional activity. In a related investigation, using the yeast two-hybrid screen with PPARα as the bait, we have identified dUTPase as an inhibitor of rat PPARα (8). Full-length dUTPase prevented PPAR-RXR heterodimerization, resulting in an inhibition of PPAR activity in a ligand-independent manner (8). It is possible that other proteins may bind to PPARα or other PPAR isoforms in a ligand-dependent fashion. The mechanism by which coactivators and corepressors mediate the transactivation remains unknown. It has been proposed that coactivators may act as physical bridges between the DNA-bound transcriptional activator protein and the basal transcription complex (34). We anticipate that mSRC-1 may provide some clues or serve as a tool to detect this multistep complex pathway.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grant R37 GM23750 and by the Joseph L. Mayberry, Sr. Endowment Fund.
REFERENCES
- 1. Ausubel F. M.; Brent R.; Kingston R. E.; Moore D. D.; Seidman J. G.; Smith J A.; Struhl K. Current protocols in molecular biology. New York: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2. Braissant O.; Foufelle F.; Scotto C.; Dauca M.; Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology 137:354–366; 1996. [DOI] [PubMed] [Google Scholar]
- 3. Burbach K. M.; Poland A.; Bradfield C. A. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc. Natl. Acad. Sci. USA 89:8185–8189; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cavailles V.; Dauvois S.; L’Horset F.; Lopez G.; Hoare S.; Kushner P. J.; Parker M. G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 14:3741–3751; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J. D.; Evans R. M. A transcriptional compressor that interacts with nuclear hormone receptors. Nature 377:454–457; 1995. [DOI] [PubMed] [Google Scholar]
- 6. Chien C. T.; Bartel P. L.; Sternglanz R.; Fields S. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578–9582; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chu S.; Huang Q.; Alvares K.; Yeldandi A. V.; Rao M. S.; Reddy J. K. Transformation of mammalian cells by overexposing H2O2-generating peroxisomal fatty acyl-CoA oxidase. Proc. Natl. lead. Sci. USA 92:7080–7084; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu R.; Lin Y.; Rao M. S.; Reddy J. K. Cloning and identification of rat deoxyuridine triphosphatase as an inhibitor of peroxisome proliferator-activated receptor α . J. Biol. Chem. 271:27670–27676; 1996. [DOI] [PubMed] [Google Scholar]
- 9. Dreyer C.; Krey G.; Keller H.; Givel F.; Helftenbein G.; Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell 68:879–887; 1992. [DOI] [PubMed] [Google Scholar]
- 10. Forman B. M.; Tontonoz P.; Chen J.; Brun R. P.; Spiegelman B. M.; Evans R. M. 15-Deoxydelta 12, 14-prostaglandin J2 is a ligand for the adipoevte determination factor PPARγ . Cell 83:803–812; 1995. [DOI] [PubMed] [Google Scholar]
- 11. Frohman M. A.; Dush M. K.; Martin G. R. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998–9002; 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodrich J. A.; Hoey T.; Thut C. J.; Admon A.; Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75:519–530; 1995. [DOI] [PubMed] [Google Scholar]
- 13. Halachmi S.; Marden E.; Martin G.; MacKay H.; Abbondanza C.; Brown M. Estrogen receptor-associated proteins: Possible mediators of hormone-induced transcription. Science 264:1455–1458; 1994. [DOI] [PubMed] [Google Scholar]
- 14. Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35:307–340; 1995. [DOI] [PubMed] [Google Scholar]
- 15. Horlein A. J.; Naar A. M.; Heinzel T.; Torchia, J, Gloss B.; Kurokawa R.; Ryan, A, Kamei Y.; Soderstrom M.; Glass C. K.; Rosenfeld M. G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor compressor. Nature 377:397–404; 1995. [DOI] [PubMed] [Google Scholar]
- 16. Huang Z. J.; Edery I.; Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364:259–262; 1993. [DOI] [PubMed] [Google Scholar]
- 17. Isaac D. D.; Andrew D. J. Tubulogenesis in Drosophilia: A requirement for the trachealess gene product. Genes Dev. 10:103–117; 1996. [DOI] [PubMed] [Google Scholar]
- 18. Issemann I.; Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 347:645–650; 1990. [DOI] [PubMed] [Google Scholar]
- 19. Kamei Y.; Xu L.; Heinzel T.; Torchia J.; Kuro-kawa R.; Gloss B.; Lin S. C.; Heyman R. A.; Rose D. W.; Glass C. K.; Rosenfeld M. G. ACBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403–414; 1996. [DOI] [PubMed] [Google Scholar]
- 20. Kliewer S. A.; Forman B. M.; Blumberg B.; Ong E. S.; Borgmeyer U.; Mangelsdorf D J.; Umesono K.; Evans R. M. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA 91:7355–7359; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kliewer S. A.; Lenhard J. M.; Willson T. M.; Patel I.; Morris D. C.; Lehmann J. M. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor γ and promotes adipocyte differentiation. Cell 83:813–819; 1995. [DOI] [PubMed] [Google Scholar]
- 22. LeDouarin B.; Zechel C.; Garnier J. M.; Lutz Y.; Tora L.; Pierrat P.; Heery D.; Gronemeyer H.; Chambon P.; Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 14:2020–2033; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee J. W.; Ryan F.; Swaffield J. C.; Johnston S. A.; Moore D D. Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature 374:91–94; 1995. [DOI] [PubMed] [Google Scholar]
- 24. Lee S. S.; Pineau T.; Drago J.; Lee E. J.; Owens J. W.; Kroetz D. L.; Fernandex-Salguero P. M.; Westphal H.; Gonzalez F. J. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15:3012–3022; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nambu J. R.; Lewis J. O.; Wharton K. A. Jr.; Crews S. T. The Drosophila single-minded gene encodes a helix–loop–helix protein that acts as a master regulator of CNS midline development. Cell 67:1157–1167; 1991. [DOI] [PubMed] [Google Scholar]
- 26. Onate S. A.; Tsai S. Y.; Tsai M. J.; O’Malley B. W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270;1354–1357; 1995. [DOI] [PubMed] [Google Scholar]
- 27. Rao M. S.; Reddy J. K. Peroxisome proliferation and hepatocarcinogenesis. Carcinogenesis 8:631–636; 1987. [DOI] [PubMed] [Google Scholar]
- 28. Reddy J. K.; Goel S. K.; Nemali M. R.; Carrino J. J.; Laffer T. G.; Reddy M. K.; Sperbeck S. J.; Osumi T.; Hashimoto T.; Lalwani N. D.; Rao M. S. Transcription regulation of peroxisomal fatty acyl-CoA oxidase and enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase in rat liver by peroxisome proliferators. Proc. Natl. Acad. Sci. USA 83:1747–1751; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reddy J. K.; Lalwani N. D. Carcinogenesis by hepatic peroxisome proliferators: Evaluation of the risk of hypolipidemic drugs and industrial plasticizers to humans. Crit. Rev. Toxicol. 12:1–58; 1983. [DOI] [PubMed] [Google Scholar]
- 30. Reddy J. K.; Mannaerts G. P. Peroxisomal lipid metabolism. Annu. Rev. Nutr. 14:343–370; 1994. [DOI] [PubMed] [Google Scholar]
- 31. Reisz-Porszasz S.; Probst M. R.; Fukunaga B. N.; Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT). Mol. Cell. Biol. 14:6075–6086; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rushlow C. A.; Hogan A.; Pinchin S. M.; Howe K. M.; Lardelli M.; Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 8:3095–3103; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sasai Y.; Kageyama R.; Tagawa Y.; Shigemoto R.; Nakanishi S. Two mammalian helix–loop–helix factors structurally related to Drosophila hairy and enhancer of split. Genes Dev. 6:2620–2634; 1992. [DOI] [PubMed] [Google Scholar]
- 34. Tjian R.; Maniatis T. Transcriptional activation: A complex puzzle with few easy pieces. Cell 77:5–8; 1994. [DOI] [PubMed] [Google Scholar]
- 35. Tontonoz P.; Hu E.; Graves R. A.; Budavari A. I.; Spiegelman B. M. mPPARγ2: Tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8:1224–1234; 1994. [DOI] [PubMed] [Google Scholar]
- 36. Tontonoz P.; Hu E.; Spiegelman B. M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79:1147–1156; 1994. [DOI] [PubMed] [Google Scholar]
- 37. Tugwood J. D.; Issemann I.; Anderson R. G.; Bundell K. R.; Mcpheat W. L.; Green S. The peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 11:433–439; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varanasi U.; Chu R.; Huang Q.; Castellon R.; Yeldandi A. V.; Reddy J. K. Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. J. Biol. Chem. 271:2147–2155; 1996. [DOI] [PubMed] [Google Scholar]
- 39. Wang G. L.; Jiang B. H.; Rue E. A.; Semenza G. L. Hypoxia-inducible factor 1 is a basic-helix–loop–helix–PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510–5514; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yao T.-P.; Ku G.; Zhou N.; Scully R.; Livingston D. M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. USA 93:10626–10631; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zehring W. A.; Wheeler D. A.; Reddy P.; Konopka R. J.; Kyriacou C. P.; Rosbash M.; Hall J. C. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster . Cell 39:369–376; 1984. [DOI] [PubMed] [Google Scholar]
- 42. Zhu Y.; Alvares K.; Huang Q.; Rao M. S.; Reddy J. K. Cloning of a new member of the peroxisome proliferator-activated receptor gene family from mouse liver. J. Biol. Chem. 268:26817–26820; 1993. [PubMed] [Google Scholar]


