Abstract
“Hemoglobin switching,” or the sequential expression of globin genes in erythroid cells during development, has provided an important paradigm for tissue- and stage-specific gene regulation. Over the past decade, regulatory DNA sequences and transcription factors involved in controlling the expression of individual globin genes in erythroid cells have been identified. The picture that has emerged indicates that gene proximal control elements collaborate with a “locus control region” located far upstream — probably via a DNA looping mechanism — to ensure that each gene is turned on only in erythroid cells and at the appropriate time during development. Interactions among the various regulatory sequences are thought to be mediated and stabilized by an array of tissue-specific and ubiquitous proteins. Chromatin structure plays a critical but still poorly understood role in this process.
Keywords: Hemoglobin switching, Gene regulation, Erythroid transcription factors, Locus control region
THE development of multicellular organisms is directed by the spatial and temporal regulation of specific sets of genes. An important model for understanding the molecular basis for such differential gene expression has been provided by the vertebrate globin gene family. As development progesses, distinct members of each of the two globin gene subfamilies (α and β) are expressed sequentially in red blood cells (erythroid cells). In humans, this “hemoglobin switching” occurs at two developmental stages: a) the embryonic-to-fetal switch occurs during the first few weeks of gestation and involves a change in expression of both the α- and β-globin clusters; b) the fetal-to-adult switch, involving only the β cluster, occurs around the time of birth (59). These changes in globin gene transcription (and production of hemoglobin proteins) are associated with changes in red blood cell morphology and site of production (Fig. 1). Embryonic globin genes are expressed in the “primitive” erythroblasts of the yolk sac blood islands, whereas the fetal and adult globin genes are active in the fetal liver and adult bone marrow.
FIG. 1.
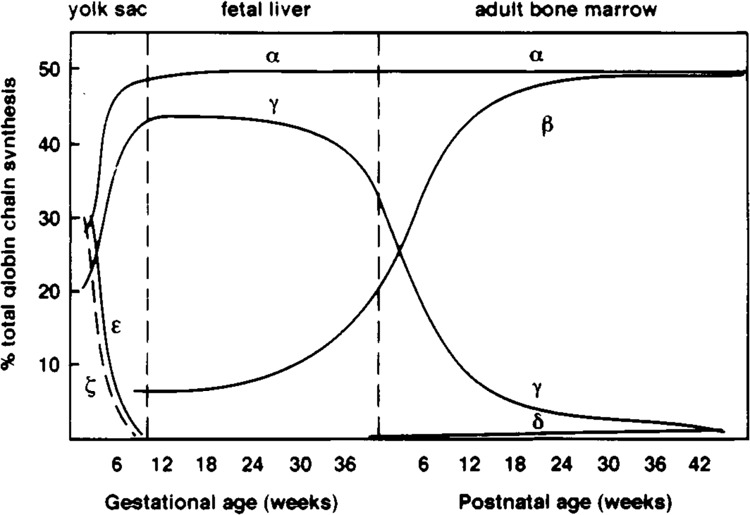
Stage-specific expression of α- and β-like globin genes. Changes in globin gene expression during development are associated with changes in the site of production (yolk sac, fetal liver, adult bone marrow) and morphology (size, presence or absence of nucleus) of erythroid cells. The α-like globin genes are ζ (embryonic) and α (fetal, adult); the β-like globin genes are ϵ (embryonic), γ (fetal), and β, δ (adult). Their protein products assemble into heterotetramers to form functional hemoglobin protein.
Hemoglobin contains two types of polypeptide, α-like and β-like, that are encoded by two gene clusters located within distinct chromosomal loci in mammals and birds. The α- and β-globin gene families apparently evolved by duplication and sequence divergence from a single ancestral gene common to all vertebrates (24). When the linkage relationships among the various globin genes were first defined, the intriguing observation was made that they are organized on the chromosome in order of expression during development: 5′-embryonic-fetal-adult-3′ (shown for human β-globin locus in Fig. 2). This gene order is conserved among nearly all species and appears to be of functional significance.
FIG. 2.
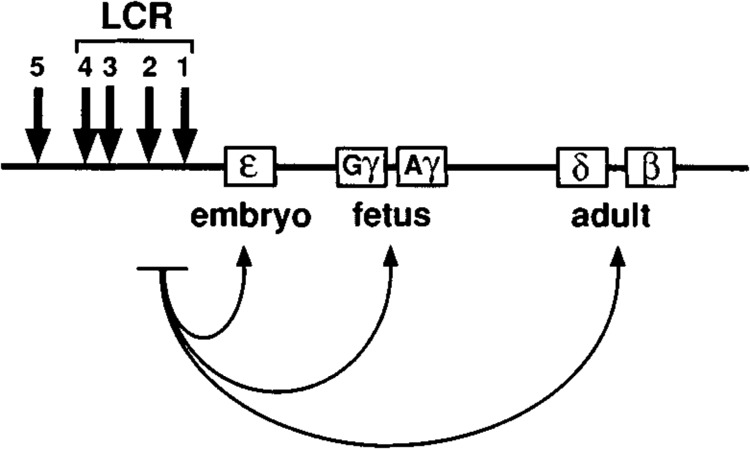
Structure of the human β-globin locus. The β-LCR extends from 6 to 18 kb upstream of the ϵ-globin gene and contains four strong erythroid-specific DNAse I-hypersensitive sites (represented by arrows) that are present at all stages of development (HS1-4). HS5 is constitutively sensitive to nuclease digestion and may function as an insulator or boundary element. Interactions between the LCR and the promoters of individual globin genes (indicated by curved arrows) are thought to be mediated by stage-specific regulatory proteins that stabilize the formation of DNA loops. The figure is not drawn to scale.
In this review I will highlight some of the key regulatory problems presented by these complex gene clusters. For the most part, examples will be drawn from studies on the α-like globin genes, but the α-like globin genes will also be used to emphasize certain points. Although the expression of the genes within these two loci is coordinated, important differences in their control mechanisms have been defined.
CONTROL SEQUENCES WITHIN GLOBIN GENE LOCI
Despite the apparent similarities between the two globin gene families, their chromosomal environments are quite different. In nonerythroid cells, the chromatin domain of the β-globin cluster is late-replicating, insensitive to DNase I (closed chromatin), and transcriptionally silent [discussed in (33)]. Tissue-specific changes in chromatin structure (from closed to open) and replication of the locus (from late to early) in erythroid cells are thought to be controlled by the locus control region (LCR) (39) located far upstream (Fig. 2). The a-globin cluster, in contrast, is contained within an early replicating domain that is constitutively open (DNase I sensitive). A dominant control element (HS-40) (42) located far upstream of the α-globin structural genes is superficially analogous to the β-LCR. However, unlike the β-LCR, HS-40 appears to have no effect on long-range chromatin structure and does not influence formation of DNase I-hypersensitive sites within the a-globin gene promoters; its major role is thought to be enhancement of a-globm transcription within a constitutively open chromatin domain (17). It is conceivable that some of the other properties associated with the β-LCR (such as a domain-opening function) might reside in as yet unidentified sequences located at a distance from HS-40 (5).
Both the β-LCR (39) and HS-40 (42) contain very strong enhancer activities. When introduced into the germline of mice, β-LCR-linked globin genes show high-level, erythroid-specific expression that is copy number dependent but relatively position independent. Although still not well understood, the absence of “position effects” in transgenic mice containing LCR sequences is thought to result from inhibition of heterochro-matin spreading (64).
The β-LCR contains four strong erythroid-specific DNAse I-hypersensitive sites (5′-HSl, 2, 3, and 4) found at all stages of development [reviewed in (40); see Fig. 2]. Only one such site is present in HS-40 (42). Nuclease sensitivity is thought to reflect nucleosome-free regions of DNA that are accessible for binding by transcription factors (25). Nucleosome disruption has recently been demonstrated for HS4 of the β-LCR (60) and presumably is associated with the formation of other HS sites as well.
Most of the activity of the β-LCR is associated with 5′-HS2, HS3, and HS4; naturally occurring deletions of HS1 are not associated with obvious abnormalities in globin gene expression [reviewed in (40)]. A detailed discussion of the properties of the β-LCR nuclease sensitive sites can be found in several reviews (5,40,52).
Globin gene promoters contain a number of control elements found within the promoters of many other genes. These include a TATA-like, CCAAT, and CACCC motifs. Characteristically, they also contain one or more GATA binding sites that are recognized by the lineage-specific tran-scriptional regulator, GATA-1 (see below). Other positive and negative control sequences have been identified further upstream, downstream, and within the transcribed regions of the structural α- and β-like globin genes (5,59). The upstream region of the human embryonic β-like globin gene (e), for example, contains clusters of positive and negative regulatory sequences (62). At least some of these regions interact synergistically, in different combinations, to drive tissue- and stage-specific expression of this gene (62). Interestingly, the regulatory specificity observed when these elements are combined is not overshadowed by the presence of LCR sequences. Thus, the correct pattern of expression may be determined largely by sequences located in the neighborhood of an individual globin gene, whereas high-level expression shows a critical dependence on the dominant control region located far upstream.
THE PROTEIN PLAYERS
Interactions between gene-proximal regulatory sequences and the LCR are essential for the developmental transitions in globin gene expression to occur normally [reviewed in (40)]. Although not well understood, they most likely are mediated by distinct combinations of lineage-specific, ubiquitous, and stage-specific regulatory proteins. Some of these proteins presumably contribute to the erythroid- and stage-specific globin gene activation observed when erythroid cells and nonerythroid cells are fused to form heterokaryons (2,3). According to a current model for globin gene switching, direct or indirect interactions among proteins bound at sites within the LCR and at sites located in the vicinity of each globin gene result in the stable formation of DNA loops (see below).
A handful of erythroid lineage-restricted transcription factors have now been identified and cloned and represent a number of different protein families [for recent reviews, see (5,52)]. Several of these (notably the zinc finger proteins GATA-1 and EKLF and the leucine zipper protein NF-E2) have been implicated in erythroid development through gene disruption experiments in embryonic stem cells. At least one (EKLF) plays a direct role in globin gene activation and possibly in switching. Ubiquitous (or at least widely expressed) proteins also serve important functions in erythroid development and/or regulation of globin genes (5,52).
It has been known for some time that proteins play a role in formation of nuclease-hypersensitive sites (27,28,61). In an in vitro chromatin-assembled system, binding by NF-E2 facilitates binding by GATA-1 to its nearby sites (1), induces formation of HS2 (6,7), and is associated with nucleosome disruption that can be detected at a distance of at least 200 bp (1). The probability of hypersensitive site formation within a β-globin gene enhancer was shown to increase in additive fashion with increasing density of transcription factor binding sites (11). However, as binding sites were mutated, nuclease sensitivity did not decrease uniformly within all copies of the enhancer, but were simply either accessible or not accessible. Formation of HS sites therefore resembles an on/off switch (11,30).
The HS core regions of the LCR and HS-40 each contain various combinations and arrangements of three sites: GATA, NF-E2/AP-1, and CACC/GGTG (40,52,60). The grouping [NF-E2-(50 bp spacer)-(inverted GATA repeat)] is conserved in HS2, HS3, and HS4 and, at least for HS4, the spacing between these sites is critical (60). GATA factor binding is required for position-independent expression of an LCR-linked promoter, whereas binding by NF-E2 is required for the enhancer activity of HS2 [reviewed in (40,52)].
It is not yet known which protein(s) interact with the CACC/GGTG sites in vivo. Because null mutations in EKLF (Erythroid Krüppel-like factor) (50) affect only the adult β-globin gene (51,53), it seems unlikely that binding by this protein is required for LCR function, unless the pattern of occupation of CACC/GGTG sites in the LCR is stage specific. These sites could be bound by one of the widely expressed members of the Kruppel family of transcription factors (e.g., Sp1 or BKLF) or by another (currently unknown) erythroid-specific member of the family.
Several of the proteins that have been implicated in erythroid differentiation and/or globin gene regulation are known to interact physically in vitro and in vivo and can synergistically activate expression of a linked gene. In view of the likely involvement of protein-protein interactions in LCR-dependent globin gene activation, this problem merits close attention. Globin gene promoters and the LCR contain GATA binding sites, and homotypic interactions between GATA-1 poly-peptides have been implicated in transcriptional activation (19,67). Therefore, GATA-1 might mediate loop formation between globin gene promoters and the LCR. Moreover, GATA-1 also interacts at a distance with Spl and EKLF, binding sites for which are found in the β-LCR HS sites and in globin gene promoters.
Potential binding sites for GATA-1 and YY1 are found scattered throughout the β-globin locus [e.g., see (69)]. While the significance of this observation has not been systematically examined, it suggests that GATA-1 and YY1 could play an architectural role in formation of higher order protein-DNA complexes and in opening of the locus.
CHROMATIN REMODELING AND LOCUS OPENING
The thalassemias (a severe form of anemia caused by deficiency or absence of α-like or β-like globin chain production) comprise the most common single gene disorders in the human population (65). The vast majority of mutations are found within the α- or β-globin gene clusters on human chromosomes 16 and 11, respectively. However, some forms of thalassemia or hereditary persistence of fetal hemoglobin segregate independently [e.g., see (18,36)], suggesting the existence of additional, trans-acting loci that influence the expression of one or more globin genes. To date, only one such locus has been characterized. Mutations in the XH2 gene cause a complex syndrome of a-thalassemia and mental retardation (ATR-X syndrome) (37). XH2 is homologous to the SWI/SNF superfamily of regulatory proteins, different subgroups of which are involved in DNA repair and/or transcriptional regulation (14,21). By analogy, it has been speculated that XH2 functions by altering chromatin structure (37). However, the biochemical properties of XH2 have not yet been reported, and it is not known whether this protein, like the SWI/SNF complex (55), binds DNA and/or alters helical twist. Notably, expression of the β-like globin genes is not affected by mutations in XH2. The preferential effect of XH2 mutations on the α-globin cluster may be related in some way to its distinct chromosomal environment (discussed above).
At least one SWI/SNF relative, the Drosophila protein brahma, plays a role in regulation of HOX genes, whose clustered organization is reminiscent of that of the globin gene loci. The organization of both HOX and globin genes along the chromosome is conserved and is related to their temporal and/or spatial patterns of expression. Activation of both globin (26,40) and homeobox (46) genes is thought to require stable associations between promoters and distant chromosomal elements, and these interactions are thought to be mediated by multiprotein complexes. Interestingly, it is speculated that brahma may facilitate HOX gene promoter-enhancer interactions by opposing the repressive effects of heterochromatin [reviewed in (46,49)]. Perhaps a related protein serves a similar function for vertebrate globin gene promoter-LCR interactions.
Transcription of a globin gene within a chromosomal context has been achieved in vitro, in synthetic nuclei (7). An erythroid-specific chromatin structure was reconstituted in vitro on a DNA template containing the entire chick β-globin locus using extracts from Xenopus oocytes. Enhancer-dependent activation of the adult β A-globin gene was observed in the presence of an appropriate complement of nuclear proteins, provided that DNA replication had first been allowed to proceed. The requirement for DNA replication in this system may reflect the need of differentiated cells to stably maintain their patterns of gene expression even after many rounds of cell division. Het-erokaryon experiments have suggested, however, that the differentiated phenotype depends on the relative levels of positive and negative regulators (4,10), apparently independently of DNA replication (15). At present the requirement for DNA replication for activation of a chromatin-assembled template (7) is difficult to reconcile with the results of the cell fusion studies (2,3). A simple possibility that remains to be tested is that the protein extracts used to create a nucleosome-repressed DNA template in vitro may lack certain components present in vivo. Nevertheless, these findings are very exciting because they open up the possibility of examining the activities of cloned transcription factors in a chromatin context that more closely approximates what is observed in vivo.
A ROLE FOR DNA BENDING IN GLOBIN GENE REGULATION?
Recent work in the author’s laboratory suggests that DNA bending may play a role in regulation of the human embryonic β-like globin gene (ϵ) (23). The control element PRE II (62) is bound by a nuclear phosphoprotein termed PREIIBF that is enriched in embryonic erythroid cells. PREIIBF has both embryonic and adult forms and recognizes a novel DNA sequence element within PRE II (23,62,63). Synergistic interactions between PRE II and PRE V require binding by this protein to PRE II (63). A directed bend is introduced into target DNA upon binding by PREIIBF; this bend may facilitate protein-protein interactions and may help to explain the ability of PRE II to synergize with other regulatory regions located at a distance (23).
Expression cloning has led to isolation of a cDNA that encodes an HMG domain protein (22). The biochemical properties and DNA binding specificity of the recombinant protein are identical to those of PREIIBF isolated from embryonic erythroid cell nuclear extracts. In view of the well-known ability of HMG domain proteins to bend DNA (38) and, in some cases, to function as transcriptional activators (57,58), the identification of PREIIBF as a member of the HMG domain protein family is intriguing. This work suggests that PREIIBF-induced DNA bending might be required for synergistic activation of a linked promoter (22,23).
HEMOGLOBIN SWITCHING: HOW DOES IT WORK?
Any mechanism for hemoglobin switching must somehow account for the ability of the LCR to activate globin gene promoters from a distance of many kilobases. At present, the favored model involves DNA looping in which stage-specific factors mediate stable associations between the LCR and globin gene promoters (5,40,52) (see Fig. 2). Developmental switching of globin genes probably does not involve the formation of stage-specific chromatin structures within the LCR (40). However, each HS site of the β-LCR may contribute preferentially to the developmental regulation of particular globin genes (12,34). Thus, the sequential expression of globin genes may be controlled both by sequences proximal to the genes and by elements within the LCR. However, the details of the mechanism remain unclear, and there is not yet uniform agreement about the stage preference of each HS site [for example, compare (34,43)]. Nevertheless, it is generally agreed that individual HS elements interact with one another to form a functional unit or “holocomplex” (Fig. 3) which then interacts with regulatory sequences located in the neighborhood of each globin gene (26,66).
FIG. 3.
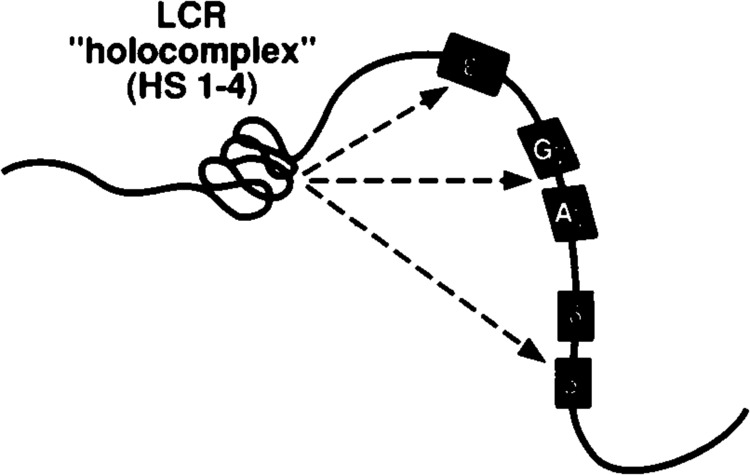
Cartoon of the LCR “holocomplex.” The hypersensitive regions of the LCR are thought to interact as a unit or “holocomplex” (see text). This figure was adapted from Wij-gerdeetal. (66).
The question of whether the LCR interacts with only one globin gene at a time on a given chromosome is a very interesting one and is central to a complete understanding of the mechanistic basis for globin gene switching. The LCR-globin gene interaction appears to be monogenic (66). Confocal microscopy and single cell in situ hybridization experiments with primary globin gene transcript (unprocessed) probes suggest that, at the time of a globin gene switch, the LCR may alternate back and forth between transcriptional activation of the earlier- and later-expressed globin genes (e.g., embryonic and fetal or fetal and adult) but does not appear to activate these genes simultaneously (66). As development progresses, changes in the complement of transcription factors are thought to modulate the stability of LCR-globin gene complexes; the earlier-expressed gene is downregulated and the more newly activated gene continues to be expressed. Thus, as suggested by the heterokaryon studies (2–4), switching is a dynamic process in which individual globin genes respond to stage-specific changes in the levels of transcriptional regulators; it does not involve irreversible changes in the expressed state of globin genes.
It seems clear that competitive interactions between the promoters of the fetal and adult β-like globin genes are a key component of the fetal-to-adult switch (8,29). Although it was initially believed that the embryonic β-like globin gene (ϵ) is regulated autonomously during development (i.e., in the absence of any other globin genes, it is expressed embryonically but not at later stages in transgenic mice), the real story may be more complex. Analysis of transgenic mice carrying the entire human β-globin locus suggests that the embryonic and fetal β-like globin genes also compete with one another for interactions with the LCR [reviewed in (5)].
Globin gene competition is polar: interaction between the LCR and a more proximally located gene is favored over that with a more distal gene (41,54). Thus, the globin gene in closest proximity with the LCR is activated earliest during development, consistent with the idea that temporal regulation reflects the relative frequency of interaction between a globin gene promoter and the LCR (41).
The first “promoter competition” model for globin gene switching emerged from transient expression studies with chick globin genes in primitive and definitive chick cells (16). That model envisioned a competition between the promoters of the chick adult (β A) and embryonic (ϵ) β-like globin genes for interactions with an enhancer located between them on the chromosome. Silencing of the ϵ-globin gene in definitive erythroid cells was shown to require the presence of a “stage-selector element” (SSE) in the promoter of the β A-globin gene (16,31). Although more recently it has been argued that promoter competition does not enter into globin gene switching in the chicken, the studies on which this conclusion is based (32) were carried out using a heterologous (transgenic mouse) system and the results remain open to a different interpretation [discussed in (5)].
A stage-selector element has also been identified in the promoter of the human fetal γ-globin gene promoter (44). Competitive silencing by the SSE is mediated in part by a protein complex (SSP) containing an adult stage-specific protein related to NF-E4 (35) and a ubiquitous protein, CP2 (45). NF-E4 binds to the chick SSE and is thought to mediate β-globin enhancer-promoter interactions in vivo (35). Together with GATA-1, it activates transcription of a chromatin-assembled β A-globin gene in vitro (6). The gene encoding NF-E4 has not yet been cloned, but biochemical characterization suggests that the protein may be related to Spl (68). The SSP complex interacts with human TAFII130 in vivo (20) and therefore may interact directly with the transcription initiation complex.
The globin gene switching mechanism involves activation of one gene and turnoff of the previously expressed gene. This negative regulation may to some degree be passive, as stage-specific factors disappear or become inactive. However, silencer elements (or at least, negative control elements) have also been identified for each of the β-like globin genes (9,13,47,56,62). Transcriptional downregulation of the human embryonic β-like globin gene appears to be mediated by several negative regulatory elements [(62), C. No-guchi, personal communication], at least one of which fulfills the operational definition of a silencer (13). Silencing of the human embryonic α like globin gene (ζ) in definitive erythroid cells requires the combined activities of the promoter, sequences within and downstream from the body of the gene, and involves both transcriptional and posttranscriptional mechanisms (48).
SUMMARY
Hemoglobin switching is a complex phenomenon in which individual members of a set of closely linked genes are sequentially activated and then turned off as development progresses. These events are associated with the differentiation of primitive erythroid cells in the embryonic yolk sac and definitive erythroid cells in the fetal liver and adult bone marrow. Whether these morphologically distinct erythroid cells arise from a common progenitor (and therefore share a common lineage) or represent different lineages has not been rigorously determined. It may well be that the embryonic-to-fetal switch results not only from changes in the transcriptional state of specific globin genes but also as a natural consequence of lineage switching.
During the past decade, many of the globin gene regulatory sequences required for tissue- and stage-specific activation or silencing have been dissected, a handful of key erythroid transcription factors have been identified and cloned, and some understanding of the mechanism by which the locus control region drives high-level expression of downstream globin genes has emerged. In some respects, these breakthroughs have raised more questions than they answered: How does the LCR influence the chromatin structure and replication pattern of the β-globin locus? What is the higher order structure of its active elements within the postulated “holocomplex,” and how do they interact with the regulatory regions associated with each globin gene? How do multiple positive and negative globin gene control elements cooperate to achieve the remarkable specificity in expression observed during development? What are the hierarchical relationships among the known erythroid transcription factors, and what other proteins are required to ensure that the correct globin gene will be expressed at the appropriate time? How do these regulatory proteins overcome the repressed state of a gene in a chromosomal context, and is protein-induced DNA bending a component of the mechanism?
Few of these questions are specific to globin gene switching: they have broad implications for the orchestration of differential gene expression in specialized cell types. The globin gene families are therefore likely to remain a subject of intense investigation for some time to come.
ACKNOWLEDGEMENTS
I am grateful to numerous colleagues for kindly providing preprints and reprints of their work. I thank Beverly Emerson, Sarah Farrington, and Michael Dyer for helpful discussions and for critical comments on this review. Research in my laboratory was supported by grants from the NIH and from the Lucille P. Markey Charitable Trust. I was a Lucille P. Markey Scholar in Biomedical Sciences.
REFERENCES
- 1. Armstrong J. A.; Emerson B. M. NF-E2 disrupts chromatin structure at the human β-globin locus control region HS2 in vitro. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- 2. Baron M. H.; Maniatis T. Rapid reprogramming of globin gene expression in transient heterokaryons. Cell 46:599–602; 1986. [DOI] [PubMed] [Google Scholar]
- 3. Baron M. H.; Maniatis T. Regulated expression of human α- and β-globin genes in transient heterokaryons. Mol. Cell. Biol. 11:1239–1247; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baron M. H. Reversibility of the differentiated state in somatic cells. Curr. Opin. Cell Biol. 5:1050–1056; 1993. [DOI] [PubMed] [Google Scholar]
- 5. Baron M. H. Transcriptional control of globin gene switching during vertebrate development. Biochim. Biophys. Acta (in press). [DOI] [PubMed]
- 6. Barton M. C.; Madani N.; Emerson B. M. The erythroid protein cGATA-1 functions with a stage-specific factor to activate transcription of chromatin-assembled β-globin genes. Genes Dev. 7:1796–1809; 1993. [DOI] [PubMed] [Google Scholar]
- 7. Barton M. C.; Emerson B. M. Regulated expression of the β-globin gene locus in synthetic nuclei. Genes Dev. 8:2453–2465; 1994. [DOI] [PubMed] [Google Scholar]
- 8. Behringer R. R.; Ryan T. M.; Palmiter R. D.; Brinster R. L.; Townes T. M. Human gamma- to beta-globin switching in transgenic mice. Genes Dev. 4:380–389; 1990. [DOI] [PubMed] [Google Scholar]
- 9. Berg P. E.; Williams D. M.; Qian R.-L.; Cohen R. B.; Cao S.-X.; Mittelman M.; Schechter A. N. A common protein binds to two silencers 5′ to the human β-globin gene. Nucleic Acids Res. 17:8833–8852; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blau H. M. Differentiation requires continuous active control. Annu. Rev. Biochem. 61:1213–1230; 1992. [DOI] [PubMed] [Google Scholar]
- 11. Boyles J.; Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. (in press). [PMC free article] [PubMed]
- 12. Bungert J.; Dave U.; Lim K. C. ; Lieuw K. H.; Shavit J. A.; Liu Q.; Engel J. D. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 9:3083–3096; 1995. [DOI] [PubMed] [Google Scholar]
- 13. Cao S.; Gutman P.; Dave H.; Schechter A. Identification of a transcriptional silencer in the 5′-flanking region of the human epsilon globin gene. Proc. Natl. Acad. Sci. USA 86:5306–5309; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carlson M.; Laurent B. C. The SNF/SWI family of global transcriptional activators. Curr. Biol. 6:396–402; 1994. [DOI] [PubMed] [Google Scholar]
- 15. Chiu C.-P.; Blau H. M. Reprogramming cell differentiation in the absence of DNA synthesis. Cell 37:879–887; 1984. [DOI] [PubMed] [Google Scholar]
- 16. Choi O.-R.; Engel J. D. Developmental regulation of β-globin gene switching. Cell 55:17–26; 1988. [DOI] [PubMed] [Google Scholar]
- 17. Craddock C. F.; Vyas P.; Sharpe J. A.; Ayyub H.; Wood W. G.; Higgs D. R. Contrasting effects of α and β globin regulatory elements on chromatin structure may be related to their different chromosomal environments. EMBO J. 14:1718–1726; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Craig J. E.; Rochette J.; Fisher C. A.; Weather-all D. J.; Marc S.; Lathrop G. M.; Demenais F.; Thein S. Dissecting the loci controlling fetal haemoglobin production on chromosomes 11p and 6q by the regressive approach. Nat. Genet. 12:58–64; 1996. [DOI] [PubMed] [Google Scholar]
- 19. Crossley M.; Merika M.; Orkin S. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448–2456; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cunningham J. M.; Amrolia P. J.; Jane S. M. Human CP2, a component of the stage selector protein, interacts with the transcription initiation complex [abstract]. Blood 86:248a; 1995. [Google Scholar]
- 21. Drapkin R.; Sancar A.; Reinberg D. Where transcription meets repair. Cell 77:9–12; 1994. [DOI] [PubMed] [Google Scholar]
- 22. Dyer M. A.; Hayes P.; Wattanga H.; Baron M. H. unpublished data.
- 23. Dyer M. A.; Naidoo R.; Hayes P.; Larson C. J.; Verdine G. L.; Baron M. H. A DNA bending protein interacts with an essential upstream regulatory element of the human embryonic β-like globin gene. Mol. Cell. Biol. 16:829–838; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Efstratiadis A.; Posakony J. W.; Maniatis T.; Lawn R. M.; O’Connell C.; Spritz R.; DeRiel J.; Forget B. G.; Weissman S. M.; Slightom J. L.; Blechl A. E.; Smithies O.; Baralle F. E.; Shoulders C. C.; Proudfoot N. J. The structure and evolution of the human beta-globin gene family. Cell 21:653–668; 1980. [DOI] [PubMed] [Google Scholar]
- 25. Elgin S. C. R. The formation and function of DNase I hypersensitive sites in the process of gene activation. J. Biol. Chem. 263:19259–19262; 1995. [PubMed] [Google Scholar]
- 26. Ellis J.; Tan-Un K.; Harper A.; Michalovich D.; Yannoutsos N.; Philipsen S.; Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 15:562–568; 1996. [PMC free article] [PubMed] [Google Scholar]
- 27. Emerson B.; Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5′ end of the chicken adult beta-globin gene. Proc. Natl. Acad. Sci. USA 81:95; 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emerson B.; Lewis C.; Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: Nature of the binding domain. Cell 41:21–30; 1985. [DOI] [PubMed] [Google Scholar]
- 29. Enver T.; Raich N.; Ebens A. J.; Papayannopoulou T.; Costantini F.; Stamatoyannopoulos G. Developmental regulation of human fetal-to-adult globin gene switching in transgenic mice. Nature 344:309–313; 1990. [DOI] [PubMed] [Google Scholar]
- 30. Felsenfeld G. Chromatin unfolds. Cell 86:13–19; 1996. [DOI] [PubMed] [Google Scholar]
- 31. Foley K. P.; Engel J. D. Individual stage selector element mutations lead to reciprocal changes in β-vs. ϵ-globin gene transcription: Genetic confirmation of promoter competition during globin gene switching. Genes Dev. 6:730–744; 1992. [DOI] [PubMed] [Google Scholar]
- 32. Foley K. P.; Pruzina S.; Winick J. D.; Engel J. D.; Grosveld F.; Fraser P. The chicken β/ϵ-globin enhancer directs autonomously regulated, high-level expression of the chicken ϵ-globin gene in transgenic mice. Proc. Natl. Acad. Sci. USA 91:7252–7256; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Forrester W. C.; Epner E.; Driscoll M. C.; Enver T.; Brice M.; Papayannopoulou T.; Groudine M. A deletion of the human β-globin locus activation region causes a major alternation in chromatin structure and replication across the entire β-globin locus. Genes Dev. 4:1637–1649; 1990. [DOI] [PubMed] [Google Scholar]
- 34. Fraser P.; Pruzina S.; Antoniou M.; Grosveld F. Each hypersensitive site of the human β-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes Dev. 7:106–113; 1993. [DOI] [PubMed] [Google Scholar]
- 35. Gallarda J. L.; Foley K. P.; Yang Z.; Engel J. D. The beta-globin stage selector element (SSE) factor is erythroid specific promoter/enhancer binding protein NF-E4. Genes Dev. 3:1845–1859; 1989. [DOI] [PubMed] [Google Scholar]
- 36. Gibbons R. J.; Suthers G. K.; Wilkie A. O. M.; Buckle V. J.; Higgs D. R. X-linked α thalassemia/mental retardation (ATR-X) syndrome: Localisation to Xq12-21.31 by X-inactivation and linkage analysis. Am. J. Hum. Genet. 51:1136–1149; 1992. [PMC free article] [PubMed] [Google Scholar]
- 37. Gibbons R. J.; Picketts D. J.; Villard L.; Higgs D. R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 80:837–845; 1995. [DOI] [PubMed] [Google Scholar]
- 38. Grosschedl R.; Giese K.; Pagel J. HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10:94–100; 1994. [DOI] [PubMed] [Google Scholar]
- 39. Grosveld F.; Assendelft G. B. V.; Greaves D.; Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51:975–985; 1987. [DOI] [PubMed] [Google Scholar]
- 40. Grosveld F.; Antoniou M.; Berry M.; deBoer E.; Dillon N.; Ellis J.; Fraser P.; Hurst J.; Imam A.; Meijer D.; Philipsen S.; Pruzina S.; Strouboulis J.; Whyatt D. Regulation of human globin gene switching. Cold Spring Harbor Symp. Quant. Biol. 58:7–13; 1993. [DOI] [PubMed] [Google Scholar]
- 41. Hanscombe O.; Whyatt D.; Fraser P.; Yannoutsos N.; Greaves D.; Dillon N.; Grosveld F. Importance of globin gene order for correct developmental expression. Genes Dev. 5:1387–1394; 1991. [DOI] [PubMed] [Google Scholar]
- 42. Higgs D. R.; Wood W. G.; Jarman A. P.; Sharpe J.; Lida J.; Pretorius I.-M.; Ayyub H. A major positive regulatory region located far upstream of the human α-globin gene locus. Genes Dev. 4:1588–1601; 1990. [DOI] [PubMed] [Google Scholar]
- 43. Hug B. A.; Wesselschmidt R. L.; Fiering S.; Bender M. A.; Epner E.; Groudine M.; Ley T. J. Analysis of mice containing a targeted deletion of β-globin locus control region 5′-hypersensitive site 3. Mol. Cell. Biol. 16:2906–2912; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jane S. M.; Ney P. A.; Vanin E. F.; Gumucio D. L.; Nienhuis A. W. Identification of a stage selector element in the human γ-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the β-promoter. EMBO J. 11:2961–2969; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jane S. M.; Nienhuis A. W.; Cunningham J. M. Hemoglobin switching in man and chicken is mediated by a heteromeric complex between the ubiquitous transcription factor CP2 and a developmentally specific protein. EMBO J. 14:97–105; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kennison J. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet. 9:75–79; 1993. [DOI] [PubMed] [Google Scholar]
- 47. Li Q.; Stamatoyannopoulos J. Position independence and proper developmental control of γ-globin gene expression require both a 5′ locus control region and a downstream sequence element. Mol. Cell. Biol. 14:6087–6096; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liebhaber S. A.; Wang Z.; Cash F. E.; Monks B.; Russell J. E. Developmental silencing of the embryonic ζ-globin gene: Concerted action of the promoter and the 3′-flanking region combined with stage-specific silencing by the transcribed segment. Mol. Cell. Biol. 16:2637–2646; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Manak J. R.; Scott M. P. A class act: Conservation of homeodomain protein functions. Development Suppl. 61–71; 1994. [PubMed] [Google Scholar]
- 50. Miller I. J.; Bieker J. J. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell. Biol. 13:2776–2786; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nuez B.; Michalovich D.; Bygrave A.; Ploe-macher R.; Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316–318; 1995. [DOI] [PubMed] [Google Scholar]
- 52. Orkin S. H. Regulation of globin gene expression in erythroid cells. Eur. J. Biochem. 231:271–281; 1995. [DOI] [PubMed] [Google Scholar]
- 53. Perkins A. C.; Sharpe A. H.; Orkin S. H. Lethal β-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318–322; 1995. [DOI] [PubMed] [Google Scholar]
- 54. Peterson K.; Stamatoyannopoulos G. Role of gene order in developmental control of human γ- and β-globin gene expression. Mol. Cell. Biol. 13:4836–4843; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Quinn J.; Fyrberg A. M.; Ganster R. W.; Schmidt M. C.; Peterson C. L. DNA-binding properties of the yeast SWI/SNF complex. Nature 379:844–847; 1996. [DOI] [PubMed] [Google Scholar]
- 56. Raich N.; Papayannopoulou T.; Stamatoyannopoulos G.; Enver T. Demonstration of a human ϵ-globin gene silencer with studies in transgenic mice. Blood 79:861–864; 1992. [PubMed] [Google Scholar]
- 57. Sheridan P. L.; Sheline C.; Cannon K.; Voz M.; Pazin M.; Kadonaga J.; Jones K. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 9:2090–2104; 1995. [DOI] [PubMed] [Google Scholar]
- 58. Shykind B.; Kim J.; Sharp P. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev. 9:1354–1365; 1995. [DOI] [PubMed] [Google Scholar]
- 59. Stamatoyannopoulos G.; Nienhuis A. Hemoglobin switching. In: Stamatoyannopoulos G.; Nienhuis A. W.; Leder P.; Majerus P. W., eds. The molecular basis of blood diseases. Philadelphia: W. B. Saunders Co.; 1994:107–155. [Google Scholar]
- 60. Stamatoyannopoulos J. A.; Goodwin A.; Joyce T.; Lowrey C. H. NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human β-globin locus control region. EMBO J. 14:106–116; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Strauss E. C.; Orkin S. H. In vivo protein-DNA interactions at hypersensitive site 3 of the human β-globin locus control region. Proc. Natl. Acad. Sci. USA 89:5809–5813; 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trepicchio W. L.; Dyer M. A.; Baron M. H. Developmental regulation of the human embryonic β-globin gene is mediated by synergistic interactions among multiple tissue- and stage-specific elements. Mol. Cell. Biol. 13:7457–7468.; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Trepicchio W. L.; Dyer M. A.; Baron M. H. A novel developmental regulatory motif required for stage-specific ϵ-globin gene activation and nuclear factor binding in embryonic erythroid cells. Mol. Cell. Biol. 14:3763–3771; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Walters M. C. ; Magis W.; Fiering S.; Eidemiller J.; Scalzo D.; Groudine M.; Martin D. I. K. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 10:185–195; 1996. [DOI] [PubMed] [Google Scholar]
- 65. Weatherall D. J. The thalassemias. In: Stamatoyannopoulos G.; Nienhuis A. W.; Leder P.; Majerus P. W., eds. Molecular basis of blood diseases. Philadelphia: Saunders; 1994:157–205. [Google Scholar]
- 66. Wijgerde M.; Grosveld F.; Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature 377:209–213; 1995. [DOI] [PubMed] [Google Scholar]
- 67. Yang H.; Evans T. Homotypic interactions of chicken GATA-1 can mediate transcriptional activation. Mol. Cell. Biol. 15:1353–1363; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang Z.; Engel J. D. Biochemical characterization of the developmental stage- and tissue-specific erythroid transcription factor, NF-E4. J. Biol. Chem. 269:10079–10087; 1994. [PubMed] [Google Scholar]
- 69. Yant S. R.; Zhu W.; Millinoff D.; Slightom J. L.; Goodman M.; Gumucio D. L. High affinity YY1 binding motifs: Identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 23:4353–4362; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


