Abstract
Aims
Long noncoding RNAs (lncRNAs) are critical regulators of cardiovascular lineage commitment and heart wall development, but their roles in regulating endogenous cardiac regeneration are unclear. The present study investigated the role of human-derived lncRNA in regulating endogenous cardiac regeneration as well as the underlying mechanisms.
Methods and results
We compared RNA sequencing data from human foetal and adult hearts and identified a novel lncRNA that was upregulated in adult hearts (Genesymbol NONHSAG000971/NONHSAT002258 or ENST00000497710.5), which was a splice variant of the AZIN2 gene (AZIN2-sv). We used quantitative PCR to confirm the increased expression of AZIN2-sv in adult rat hearts. Coexpression network analysis indicated that AZIN2-sv could regulate proliferation. Loss- and gain-of-function approaches demonstrated that AZIN2-sv negatively regulated endogenous cardiomyocyte proliferation in vitro and in vivo. Knockdown of AZIN2-sv attenuated ventricular remodelling and improved cardiac function after myocardial infarction. Phosphatase and tensin homolog (PTEN) was identified as a target of AZIN2-sv, their direct binding increased PTEN stability. Furthermore, AZIN2-sv acted as a microRNA-214 sponge to release PTEN, which blocked activation of the PI3 kinase/Akt pathway to inhibit cardiomyocyte proliferation.
Conclusions
The newly discovered AZIN2-sv suppressed endogenous cardiac regeneration by targeting the PTEN/Akt pathway. Thus, AZIN2-sv may be a novel therapeutic target for preventing and treating heart failure.
Keywords: lncRNA, AZIN2-sv, Cardiac regeneration, Heart failure
1. Introduction
Heart failure is a global public health problem, largely due to the irreversible loss of cardiomyocytes (CMs).1 During the past decade, research has focused on the development of novel therapeutic strategies to induce cardiac regeneration.2 Exogenous stem cell transplantation has emerged as a potential new approach for regenerative therapy, but the results from clinical trials have not been encouraging.3 Currently, endogenous regenerative mechanisms have been studied for cardiac tissue regeneration,4 which hold promise for regeneration during normal mammalian myocardial homeostasis and after ischemia or injury.5 Thus, enhancements of these mechanisms might facilitate regeneration of the injured myocardium. Gene-targeted therapies such as RNA interference and small interfering RNA-mediated gene silencing are effective and safe strategies for endogenous cardiac regeneration.6,7 However, limited gene targets exist for true cardiac regeneration and adequate improvement of heart function, so more effective targets need to be identified.
Long noncoding RNAs (lncRNAs), a class of RNA transcripts longer than 200 nucleotides with weak protein-coding potential, regulate mRNA and microRNA (miRNA) expression via multiple methods,8,9 and as such, may be novel targets for inducing cardiac regeneration. Base complementary pairing between lncRNAs and miRNA seed sequences competitively trigger their combination and restore transcription of downstream target mRNA. Previous studies have indicated that lncRNAs can affect mRNA by acting as an endogenous miRNA sponge (ceRNA).10 For example, lncRNA-cardiac hypertrophy-related factor (CHRF) acts as an miR-489 sponge to affect the downstream target, myeloid differentiation primary response 88 (Myd88), which regulates cardiac hypertrophy in mouse myoblasts.11 In addition, lncRNA-myocardial infarction (MI)-associated transcript affects the miR-150-5p target gene, vascular endothelial growth factor, to regulate endothelial cell function.12 Additional studies have indicated that lncRNAs can interact with miRNAs involved in myocardial repair after MI.13,14 Thus, lncRNAs may have important roles in the regulation of cardiac regeneration; however, the mechanisms underlying their roles in endogenous cardiac regeneration remain unclear.
In this study, high-throughput sequencing of foetal and adult human hearts was used to identify a novel lncRNA that was upregulated in the adult heart (Genesymbol NONHSAG000971/NONHSAT002258 or ENST00000497710.5), which was a splice variant of the AZIN2 gene (AZIN2-sv). AZIN2-sv suppressed endogenous cardiac regeneration in vivo and in vitro. Loss of AZIN2-sv attenuated adverse remodelling and improved cardiac function after MI. AZIN2-sv mediated these effects by acting as a ceRNA of miR-214 and regulating the phosphatase and tensin homolog (PTEN)-Akt pathway. Our results shed light on the roles of lncRNA during the molecular regulation of endogenous cardiac regeneration.
2. Methods
The animal experiments in this study were approved by the Animal Research Committee at Southern Medical University, and all procedures were in accordance with the Institutional Guidelines for Animal Research and complied with the Guide for the Care and Use of Laboratory Animals published by the US NIH (2011).15
2.1 Analysis of RNA-seq data
RNA-seq datasets were acquired from the European Nucleotide Archive (http://www.ebi.ac.uk/ena) (also see details in the Supplementary material online).
2.2 Analysis of the lncRNA-mRNA regulatory network
The lncRNA-mRNA regulatory network was constructed as described previously (also see details in the Supplementary material online).16,17
2.3 Correlation between lncRNA and nearby coding gene expression
Coding gene data were referenced to analyse the co-ordinates of the lncRNAs (also see details in the Supplementary material online).
2.4 Cell isolation, culture, and transfection
CMs for the in vitro experiments were isolated from D1 and D7 Sprague-Dawley (SD) neonatal rats, obtained from the Laboratory Animal Center of the Southern Medical University. Neonatal rats were anesthetized using 2% inhaled isoflurane. Isolation and culture of ventricular CMs were performed as previously described.18 CMs and cardiac fibroblasts (CFs) were cultured via differential adhesion. The AC16 human CM-like cells19 and human cardiac fibroblasts (HCFBs) were donated by Mingkong Bio Co. Ltd. (Guangzhou, China). Cells were incubated in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) with 10% foetal bovine serum (Gibco, Life Technologies, Australia) at 37°C with 5% CO2. Adenovirus vectors harbouring plasmid or 4-in-1 shRNA (Vigene Biosciences, Shandong, China) were added to overexpress or inhibit AZIN2-sv with a multiplicity of infection (MOI) = 100. The AZIN2-sv RNA interference target sequences are shown in Supplementary material online, Table S1. A scrambled form was the control. After 24 h, cells were maintained in normal fresh medium for 48 h before the experiments and subsequent analysis. Small interfering RNA of AZIN2 and vector harbouring PTEN were generated by Kidan Bio Co. Ltd. (Guangzhou, China). Transfections were performed with a Lipofectamine 2000 kit (Invitrogen, ThermoFisher Scientific, Bridgewater, NJ) according to the manufacturer’s instructions. miR-214 mimic and inhibitor were transfected at a final concentration of 50 nM for 6 h. Actinomycin D (Sigma, St. Louis, MO) was added at a concentration of 50 μM.
2.5 Quantitative real-time polymerase chain reaction
Total RNA was extracted from the CMs using E.Z.N.A.R Total RNA Kit II (Norcross, GA). Related mRNAs were quantified with the SYBR Green PCR Kit (Takara) and the Light Cycler 480 II (Roche Diagnostics, Basel, Switzerland). The primers are shown in Supplementary material online, Table S2. The qPCR reactions were performed as described previously.18 A stem-loop kit (Sangon Biotech, Shanghai, China) was applied to qPCR reactions of miRNA according to the instructions. The RT primers used for reverse transcription and the PCR primers are shown in Supplementary material online, Table S2. PCR was performed as described above.
2.6 5′- and 3′-rapid amplification of cDNA ends
The transcriptional initiation and termination sites of AZIN2-sv were determined using the 5′- and 3′-rapid amplification of cDNA ends (RACE) System for Rapid Amplification of cDNA Ends (Version 2.0, 18374058, ThermoFisher Scientific) according to the manufacturer’s instructions. The gene-specific primers used for PCR for RACE analysis were as follows: 5′-CAGGTAGCCAGCCATGCCTTGC-3′ (5′-RACE) and 5′-GGCATGGCTGGCT ACCTG-3′ (3′-RACE).
2.7 RNA FISH analysis
RNA FISH was used to identify the location of AZIN2-sv (also see details in the Supplementary material online). Slides were mounted and analysed using the Carl Zess LSM880 confocal microscope (Zess Corporation, Germany).
2.8 Cytoplasmic and nuclear fraction separation
Subcellular fractions were isolated using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher Scientific) according to the manufacturer’s instructions (also see details in the Supplementary material online).
2.9 Luciferase reporter assay
AZIN2-sv-wt and AZIN2-sv-mut were amplified using qPCR and cloned into the luciferase vector psiCHECK-2 (Saicheng Bio Co. Ltd., Guangzhou, China). For luciferase reporter assays, the miR-214 mimic was co-transfected into HEK293 with the luciferase constructs described above. Transfection was performed using Lipofectamine 2000 (Invitrogen, ThermoFisher Scientific) according to the manufacturer’s instructions. Firefly and Renilla luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega, Madison, WI) and each well had three replicates.
2.10 Pull-down assay and mass spectrometry
lncRNA-AZIN2-sv and its antisense RNA were transcribed in vitro from the pGEM-T vector, biotin-labelled with the Biotin RNA Labeling Mix (Roche Diagnostics, Indianapolis, IN) and T7 RNA polymerase (Roche), and purified using the RNeasy Mini Kit (Qiagen, Valencia, CA). Then, biotinylated probes were mixed with proteins or RNA obtained from P7 CMs, and incubated with streptavidin-coated magnetic beads (SA10004; Invitrogen). RNase-free BSA and yeast tRNA (Sigma, Shanghai, China) were used to prevent the non-specific binding of RNA and protein complexes. Finally, RNA complexes bound to beads were purified by Trizol for RT-PCR analysis, and proteins were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. The specific bands were extracted followed by mass spectrometry analysis (Supplementary material online, Table S3) (Luming Bio Co. Ltd., Shanghai, China).
2.11 RNA immunoprecipitation
The RNA immunoprecipitation (RIP) experiment was performed with the Magna RIP™ RNA-binding protein Immunoprecipitation Kit (Millipore, Stafford, VA) according to the protocol. The PTEN antibody (1:20, ab79156; Abcam) was used to co-precipitate RNA. RNA quantity was detected by qPCR.
2.12 Western blot analysis
Western blot analysis was performed as previously reported (also see details in the Supplementary material online).20 Protein expression was measured using Image J Analysis software (NIH, Bethesda, MD).
2.13 Immunofluorescence
Immunofluorescence staining was performed as previously described (also see details in the Supplementary material online).20,21 Positive cells were counted from five randomly selected fields in non-consecutive sections using confocal microscopy (CarlZess, LSM880). The CM area was quantified using Image J Analysis software (NIH).
2.14 MI model establishment
Rat MI was induced by ligation of the left anterior descending coronary artery as previously described (also see details in the Supplementary material online).20 Adult SD rats (180–200 g) were intraperitoneally anesthetized with 3% pentobarbital sodium (40 mg/kg). The left coronary artery was ligated with silk suture 2 mm distal from the ascending aorta. For sham-operated animals, an analogous surgical operation was performed without occlusion of the coronary artery. Immediately post-ligation, adenovirus vectors (1 × 1011 viral genome particles per animal) containing shAZIN2-sv or control vector were injected into the myocardium bordering the infarct zone (single injection), using an insulin syringe with a 30-gauge needle. Animal chests were closed and rats were warmed for several minutes until recovery. EdU was administered intraperitoneally every 2 days for 10 days. All rats were sacrificed by an over dose of pentobarbital sodium anaesthesia (150 mg/kg intraperitoneal injection) or cervical dislocation at different time points.
2.15 Measurement of lung weight
Lungs were weighed to evaluate the degree of heart failure as previously described.22 Thirty days after MI, lungs were removed and weights were normalized to body weight. Then, the lungs were soaked in 4% paraformaldehyde in a fixed shape.
2.16 Evaluation of infarct and fibrosis area
The infarct and fibrotic area was evaluated as previously described.18,20,23 After MI (14 and 60 days), hearts were cut into six sections and incubated in 1% triphenyl tetrazolium chloride (TTC) for 10 min at room temperature (white indicates infarct area; red indicates normal myocardium). Infarct areas were measured digitally using Image-Pro Plus 6.0 (Media Cybernetics, Bethesda, MD). The Masson trichrome stain was used to evaluate cardiac fibrosis as described elsewhere (blue indicates fibrosis area; red indicates normal myocardium).23 The percent area of cardiac fibrosis was measured in five random images from each animal, and calculated as the ratio of the Masson trichrome-stained area to the total optical field using Image J Analysis software (NIH).
2.17 Assessment of capillary density
The capillary density was assessed as previously described (also see details in the Supplementary material online).20
2.18 Cardiac function evaluation
Cardiac dimensions and function were evaluated by echocardiography with the Sequoia 512 system and a 17 L-5 probe (Siemens, Berlin, Germany) before infarction (pre-MI), and at 1, 14, 30, and 60 days post-MI (also see details in the Supplementary material online).
2.19 Statistical analysis
Quantitative data are expressed as the means ± SEM. The differences between two groups were evaluated using the two-tailed unpaired Student’s t-test, and multiple comparisons were analysed by one-way analysis of variance (ANOVA), followed by Bonferroni tests with equal variances and Dunnett’s C test with unequal variances (SPSS 16.0; SPSS Inc., Chicago, IL). For all tests, P < 0.05 was considered statistically significant.
3. Results
3.1 A human-derived lncRNA is upregulated in the adult heart
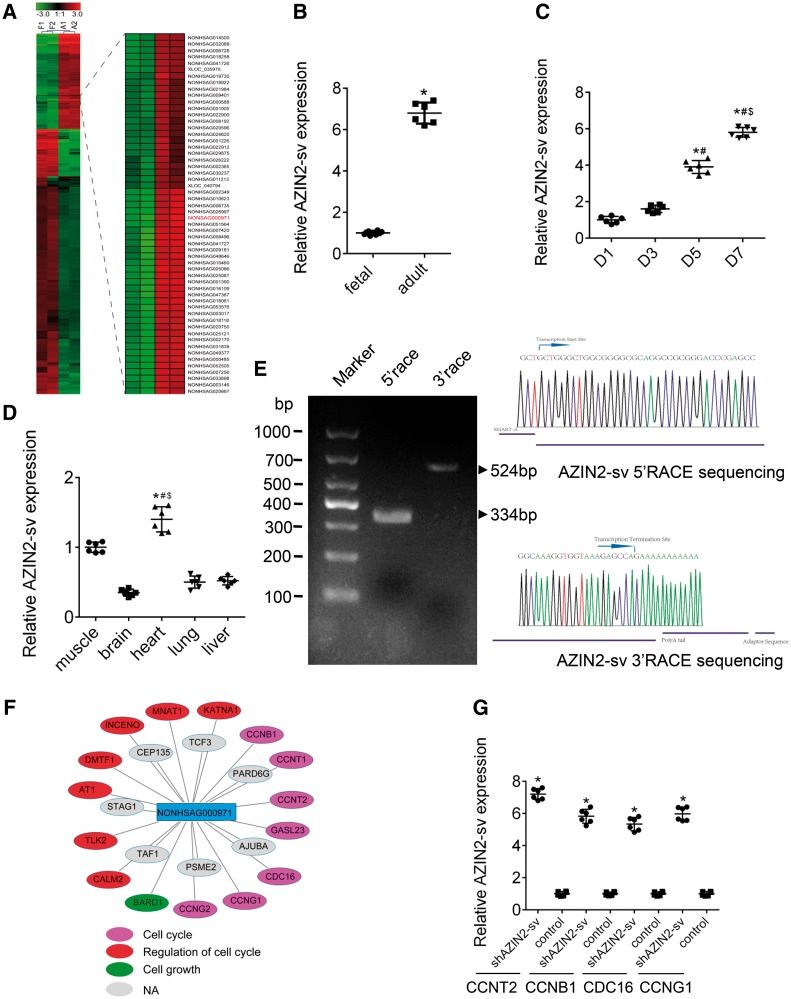
After comparing RNA sequencing (RNA-seq) data from foetal and adult hearts, we selected five differentially expressed lncRNAs that correlated with cell cycle-related protein-coding genes (r > 0.9) (Supplementary material online, Figure S1A). Gene expression was confirmed by quantitative PCR (qPCR) in foetal and adult heart specimens (Supplementary material online, Figure S1B). Of these lncRNAs, NONHSAG000971, which is located on chromosome 1 near the coding gene AZIN2 and has six exons, was the most significantly differentially expressed (Figure 1A and Supplementary material online, Figure S1C). We evaluated the expression of NONHSAG000971 in AC16 human CMs and HCFBs, the qPCR results suggested that expression in AC16 cells was higher (Supplementary material online, Figure S1D). NONHSAG000971 had weak protein-coding capability based on online coding probability prediction (Supplementary material online, Figure S1E). Next, conservation analysis suggested that the exons were highly conserved across different mammalian species such as the chimp, rhesus, mouse, and rat (Supplementary material online, Figure S1F). In SD rats, NONHSAG000971 expression was greater in the adult heart than in the foetal heart (P < 0.05; Figure 1B) and progressively increased from Day 1 (D1) to D7 after birth (P < 0.05; Figure 1C). Moreover, NONHSAG000971 expression was higher in CMs than in CFs (Supplementary material online, Figure S1G). NONHSAG000971 was detected in various tissues and was enriched in the heart, followed by the kidney, liver, and lung (P < 0.05; Figure 1D). Full-length of NONHSAG000971, which was amplified by 5′- and 3′-RACE, was 839 base pairs (Figure 1E and Supplementary material online, Table S1). We also identified the gene sequence in rats (Supplementary material online, Figure S1H and Table S1). Coexpression network analysis indicated that NONHSAG000971 was associated with many cell cycle–related mRNAs such as CCNT2, PDS5A, CDKN1B, and ANAPC1 (Figure 1F). The qPCR results suggested that several cell cycle genes increased after NONHSAG000971 knockdown (Figure 1G). This novel gene was named AZIN2-sv.
Figure 1.
AZIN2-sv is upregulated in adult hearts. (A) Cluster analysis identified upregulated AZIN2-sv in adult hearts. (B) qPCR in foetal and adult rats. *P < 0.05 vs. foetal rat group; n = 6 per group (Student’s t-test). (C) Gene expression of AZIN2-sv from D1 to D7 neonatal rats by qPCR. *P < 0.05 vs. D1 group; #P < 0.05 vs. D3 group; $P < 0.05 vs. D5 group; n = 6 per group (one-way ANOVA analysis with Dunnett C’s multiple comparison test). (D) Expression of AZIN2-sv in different tissues of D7 rats using qPCR. *P < 0.05 vs. brain; #P < 0.05 vs. lung; $P < 0.05 vs. liver; n = 6 per group (one-way ANOVA analysis with Dunnett C’s multiple comparison test). (E) A representative image of PCR products from 5′- and 3′-RACE. The major PCR product is indicated by arrow. (F) Coexpression network analysis of AZIN2-sv. Pink represents cell cycle-related mRNAs; red represents mRNAs regulating the cell cycle; green represents cell growth-related mRNAs; grey represents other unrelated mRNAs. (G) qPCR for several cell cycle-related mRNAs from coexpression analysis. *P < 0.05 vs. control; n = 6 per group (Student’s t-test).
3.2 AZIN2-sv suppresses CM proliferation in vitro
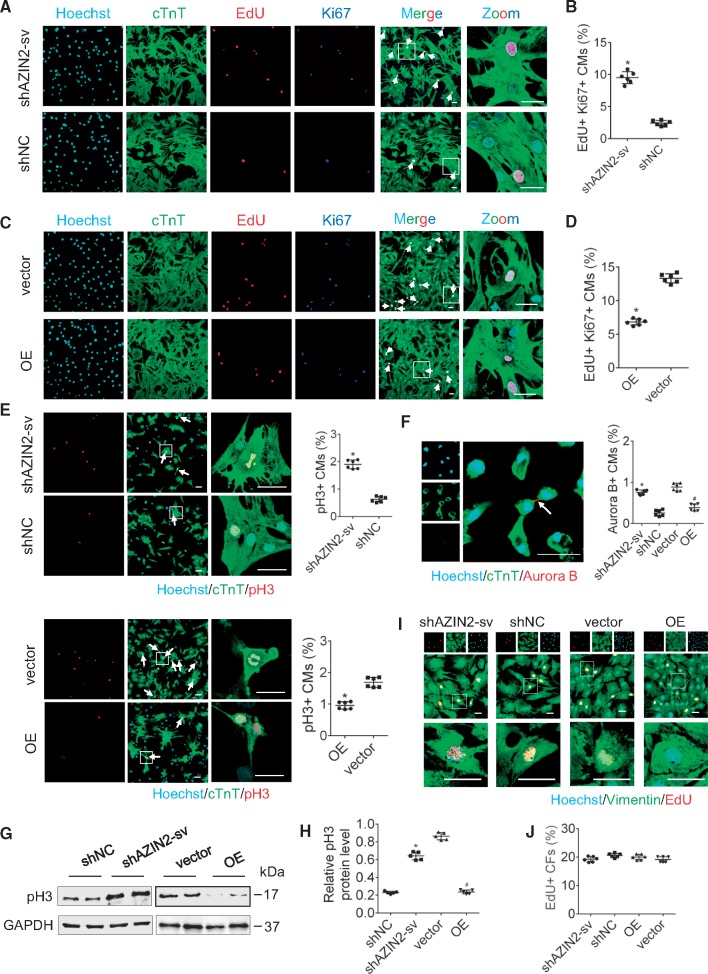
To explore the role of AZIN2-sv in CM proliferation, we reduced AZIN2-sv in D7 CMs and overexpressed AZIN2-sv (OE) in D1 CMs using adenovirus transfection technology (Supplementary material online, Figure S2A–D). In D7 CMs, AZIN2-sv reduction increased the ratio of 5-ethynyl-2′-deoxyuridine (EdU) and Ki-67-positive CMs from 2.3% to 9.1% (P < 0.05; Figure 2A and B), which indicated their proliferation potential. In D1 CMs, AZIN2-sv overexpression reduced the ratio of EdU and Ki-67-positive CMs from 13.3% to 6.4% (P < 0.05; Figure 2C and D). The mitosis markers, phosphorylated histone 3 (pH3) and Aurora B, were used to evaluate the effects of AZIN2-sv on proliferation. Compared to the controls, pH3- and Aurora B-positive CMs increased in D7 CMs when AZIN2-sv was knocked down, and decreased in D1 CMs when AZIN2-sv was overexpressed (P < 0.05; Figure 2E and F). In addition, loss of AZIN2-sv caused an increase in pH3 protein in D7 CMs, whereas the enrichment of AZIN2-sv decreased pH3 protein expression in D1 CMs (P < 0.05; Figure 2G and H). Finally, immunofluorescence data showed that the reduction or enrichment of AZIN2-sv did not increase the proliferation of CFs (Figure 2I and J). Thus, AZIN2-sv inhibited CM but not CF proliferation in vitro.
Figure 2.
AZIN2-sv reduces CM proliferation in vitro. (A) Immunostaining of D7 CMs for EdU and Ki-67 (bars, 40 μm). White arrows refer to EdU-positive and Ki-67-positive myocytes. (B) Quantification of EdU-positive and Ki-67-positive myocytes. *P < 0.05 vs. shNC; n = 6 per group (Student’s t-test). (C) Immunostaining of D1 CMs for EdU and Ki-67 (bars, 40 μm). White arrows refer to EdU-positive and Ki-67-positive myocytes. (D) Quantification of EdU-positive and Ki-67-positive myocytes. *P < 0.05 vs. vector; n = 6 per group (Student’s t-test). (E) Immunostaining of D7 and D1 CMs for pH3 (bars, 40 μm). White arrows refer to pH3-positive myocytes. *P < 0.05 vs. shNC or vector; n = 6 per group (Student’s t-test). (F) Representative fluorescent staining against Aurora B (bars, 40 μm). White arrows refer to Aurora B-positive myocytes. *P < 0.05 vs. shNC, #P < 0.05 vs. vector; n = 6 per group (Student’s t-test). (G) Protein level of pH3 (western blot) (GAPDH internal reference). (H) Quantification of pH3 protein level. *P < 0.05 vs. shNC, #P < 0.05 vs. vector; n = 5 per group (Student’s t-test). (I) Immunostaining of CFs for EdU (bars, 40 μm). (J) Quantification of EdU-positive CFs; n = 6 per group (one-way ANOVA analysis with Bonferroni’s multiple comparison test).
3.3 AZIN2-sv suppresses CM proliferation in vivo
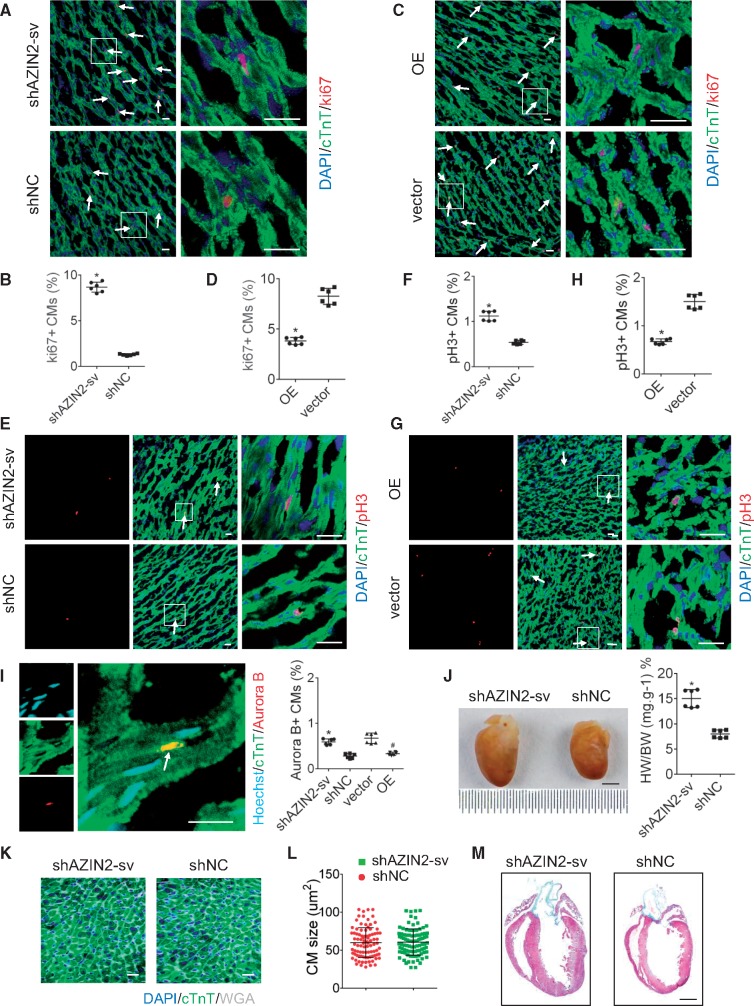
To assess the effects of AZIN2-sv on CM proliferation in vivo, adenovirus vectors containing single hairpin AZIN2-sv (shAZIN2-sv) or AZIN2-sv (OE) were injected into the hearts of D7 or D1 neonatal rats, respectively (Supplementary material online, Figure S3A and B). Three days after injection, cells were labelled with Ki-67 (Supplementary material online, Figure S3C). Compared to the control, Ki-67-positive CMs were significantly increased in D7 rats injected with shAZIN2-sv (from 1.3% to 8.67%, P < 0.05; Figure 3A and B) and were decreased in D1 rats injected with AZIN2-sv (from 8.25% to 3.82%, P < 0.05; Figure 3C and D). Similarly, the ratio of pH3- and Aurora B-positive CMs was greater in D7 rats with AZIN2-sv knockdown and lower in D1 rats with AZIN2-sv overexpression compared to their respective controls (P < 0.05; Figure 3E–I). Western blot analysis indicated that pH3 protein increased in D7 rats injected with shAZIN2-sv and decreased in D1 rats injected with AZIN2-sv (P < 0.05; Supplementary material online, Figure S3D and E). After 12 days, hearts injected with shAZIN2-sv were morphologically normal, but were significantly enlarged compared to those that received single hairpin negative control (shNC) (P < 0.05; Figure 3J). Wheat germ agglutinin (WGA) staining showed that CMs were not enlarged in the shAZIN2-sv group compared to the shNC group (Figure 3K and L). Moreover, no difference in collagen content was observed between rats injected with shAZIN2-sv or shNC (Figure 3M). These data show that AZIN2-sv repressed CM proliferation in vivo without causing myocardial hypertrophy and fibrosis, in accordance with the in vitro results.
Figure 3.
AZIN2-sv reduces myocardial proliferation in vivo. (A) Immunostaining of myocardial fibres in D7 rats for Ki-67 (bars, 40 μm). White arrows refer to Ki-67-positive myocytes. (B) Quantification of Ki-67-positive myocytes. *P < 0.05 vs. shNC; n = 6 per group (Student’s t-test). (C) Immunostaining of myocardial fibres in D1 rats for Ki-67 (bars, 40 μm). White arrows refer to Ki-67-positive myocytes. (D) Quantification of Ki-67-positive myocytes. *P < 0.05 vs. vector. n = 6 per group (Student’s t-test). (E) Immunostaining of myocardial fibres in D7 rats for pH3 (bars, 40 μm). White arrows refer to pH3-positive myocytes. (F) Quantification of pH3-positive myocytes. *P < 0.05 vs. shNC; n = 6 per group (Student’s t-test). (G) Immunostaining of myocardial fibres in D1 rats for pH3 (bars, 40 μm). White arrows refer to pH3-positive myocytes. (H) Quantification of pH3-positive myocytes. *P < 0.05 vs. vector; n = 6 per group (Student’s t-test). (I) Representative fluorescent staining against Aurora B (bars, 40 μm). White arrows refer to Aurora B-positive myocytes. *P < 0.05 vs. shNC, #P < 0.05 vs. vector; n = 6 per group (Student’s t-test). (J) Rat hearts transfected with shAZIN2-sv and shNC (12 days post-transduction) (bars, 2mm). *P < 0.05 vs. shNC; n = 6 per group (Student’s t-test). (K) WGA staining of left ventricular (LV) heart section (bars, 40 μm). (L) Quantitative analyses represent the counting of fields from six independent samples per group (n = 6) (Student’s t-test). (M) Representative masson trichrome staining of rat hearts 12 days injected with shAZIN2-sv or shNC to reveal muscle fibres (red) and collagen (blue) (bars, 500 μm); n = 6 per group.
3.4 Reduction of AZIN2-sv attenuates cardiac remodelling after MI
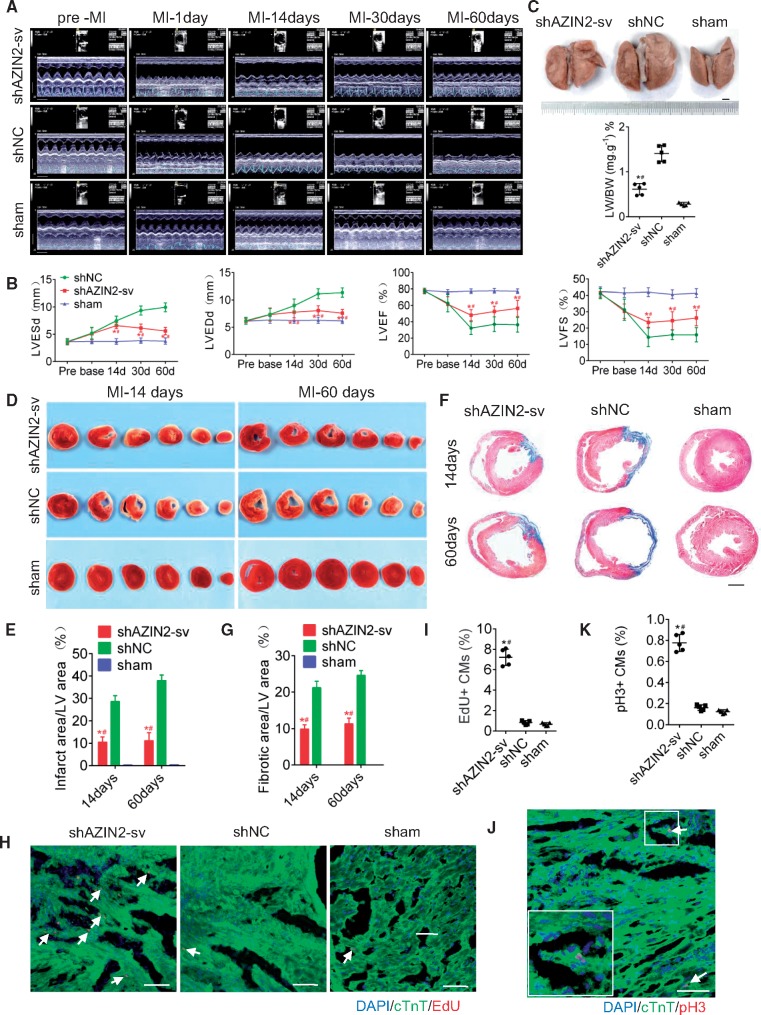
Next, we assessed the effects of AZIN2-sv on cardiac remodelling after MI (Supplementary material online, Figure S4A). AZIN2-sv knockdown was performed in the MI model (Supplementary material online, Figure S4B and C). From days 14 to 60 after MI, the left ventricular ejection fraction (LVEF), left ventricular fractional shortening (LVFS), left ventricular end systolic diameter (LVESd), and left ventricular end diastolic diameter (LVEDd) were significantly preserved in rats injected with shAZIN2-sv compared to those treated with shNC (P < 0.05; Figure 4A and B). At 6 weeks post-MI, lung weights were significantly decreased in the knockdown group compared to the shNC group (P < 0.05; Figure 4C), indicating improved cardiac function. At 14 and 60 days after MI, TTC and masson trichrome staining showed that infarct areas (white) and fibrotic scars (blue) were significantly reduced in rats treated with shAZIN2-sv compared to those treated with shNC (P < 0.05; Figure 4D–G). Furthermore, CM proliferation around the infarct area was confirmed. At 7 days after vector delivery, EdU-positive and pH3-positive CMs increased in the peri-infarct and remote areas in the shAZIN2-sv group (P < 0.05; Figure 4H–K, Supplementary material online, Figure S4D–G). The effects on myocardial angiogenesis were investigated. CD31 and CD34 were used to stain the blood vessels. At 7 days after MI, rats injected with shAZIN2-sv had more CD31 and CD34 staining than those injected with shNC in the infarct and remote areas (P < 0.05; Supplementary material online, Figure S4H and I), indicating higher vascular density. Thus, loss of AZIN2-sv after MI exerts protective effects on cardiac function possibly by stimulating endogenous CM proliferation and angiogenesis.
Figure 4.
Knockdown of AZIN2-sv reduces ventricular remodelling post-MI. (A) Evaluation of cardiac function by echocardiography at pre-MI and 1, 14, 30, and 60 days after MI (time stamps, 33 ms) (x axis); (bars, 5 mm) (y axis). (B) Quantitative analysis of LVEF, LVFS, LVEDd, LVESd. *P < 0.05 vs. shNC, #P < 0.05 vs. sham; n = 5 per group (one-way ANOVA analysis with Bonferroni’s multiple comparison test). (C) Representative images of lung from rats injected with shAZIN2-sv or shNC 6 weeks after MI (bars, 2 mm). *P < 0.05 vs. shNC, #P < 0.05 vs. sham; n = 5 per group (one-way ANOVA analysis with Dunnett C’s multiple comparison test). (D) TTC staining of rat ventricular cross sections at 14 and 60 days after MI. (E) Quantitative analysis of the infarcted area. *P < 0.05 vs. shNC, #P < 0.05 vs. sham; n = 5 per group (one-way ANOVA analysis with Bonferroni’s multiple comparison test). (F) Masson staining of rat mid-ventricular cross sections at 14 and 60 days after MI (bars, 500 μm). (G) Quantitative analysis of the fibrotic area. *P < 0.05 vs. shNC, #P < 0.05 vs. sham; n = 5 per group (one-way ANOVA analysis with Bonferroni’s multiple comparison test). (H) Immunostaining of myocardial fibres for EdU 7 days after MI in the peri-infarct area (bars, 40 μm). White arrows refer to EdU-positive myocytes. (I) Quantification of EdU-positive myocytes. *P < 0.05 vs. shNC, #P < 0.05 vs. sham; n = 5 per group (one-way ANOVA analysis with Dunnett C’s multiple comparison test). (J) Representative fluorescent staining against pH3 in the peri-infarct area (bars, 200 μm). White arrows refer to pH3-positive myocytes. (K) Quantification of pH3-positive myocytes. *P < 0.05 vs. shNC, #P < 0.05 vs. sham; n = 5 per group (one-way ANOVA analysis with Dunnett C’s multiple comparison test).
3.5 AZIN2-sv directly and specifically binds to miR-214
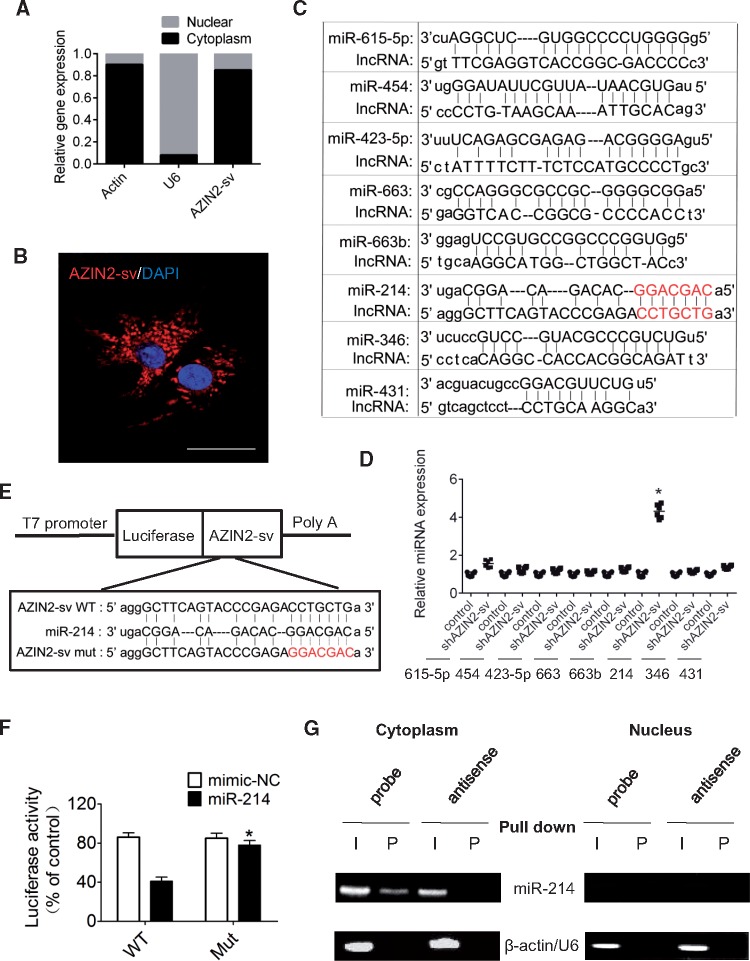
We detected subcellular AZIN2-sv and confirmed that it was localized in the cytoplasm (P < 0.05; Figure 5A and B). Using the RNAhybrid bioinformatics program, we screened miRNAs that may bind to AZIN2-sv and found that the seed sequence of miR-214 was complementary to that of AZIN2-sv (Figure 5C). In further, only miR-214 expression significantly increased when AZIN2-sv expression was reduced (Figure 5D). A luciferase construction of AZIN2-sv (Luc-AZIN2-sv-wt) and a mutated form with a derivative devoid of the miR-214 binding site (Luc-AZIN2-sv-mut) were produced (Figure 5E). The luciferase assay revealed that miR-214 could suppress the luciferase activity of Luc-AZIN2-sv-wt, but had less effect on the Luc-AZIN2-sv-mut (P < 0.05; Figure 5F), indicating that AZIN2-sv directly bound to miR-214. Moreover, a biotin–avidin pull-down assay showed that miR-214 could be co-precipitated from the cytoplasm, but not from the nucleus by AZIN2-sv (Figure 5G). In addition, the binding sites of AZIN2-sv with miR-214 were conserved between human and rats (Supplementary material online, Figure S1F). These data show that AZIN2-sv directly binds to miR-214 at specific sites in the cytoplasm.
Figure 5.
Interaction of AZIN2-sv and miR-214. (A) Nuclear and cytoplasmic distribution of AZIN2-sv in CMs by qPCR (U6 or actin internal reference). (B) Representative image of RNA FISH to confirm AZIN2-sv location in myocytes (bars, 40 μm). (C) miRNA binding with AZIN2-sv predicted by RNAhybrid. AZIN2-sv contains a site complementary to the seed sequence of miR-214. (D) Expression of miRNAs after knockdown of AZIN2-sv by qPCR. *P < 0.05 vs. control; n = 6 per group (Student’s t-test). (E) Construction of Luc-AZIN2-sv-wt and Luc-AZIN2-sv-mut vector. (F) Luciferase activity of CMs transfected with Luc-AZIN2-sv-wt or Luc-AZIN2-sv-mut. *P < 0.05 vs. WT with miR-214; n = 6 per group (two-way ANOVA analysis with Bonferroni’s multiple comparison test). (G) Co-precipitation between AZIN2-sv and miR-214 by pull-down assay. I, input (10% samples were loaded); P, pellet (100% samples were loaded).
3.6 miR-214 participates in the regulation of CM proliferation
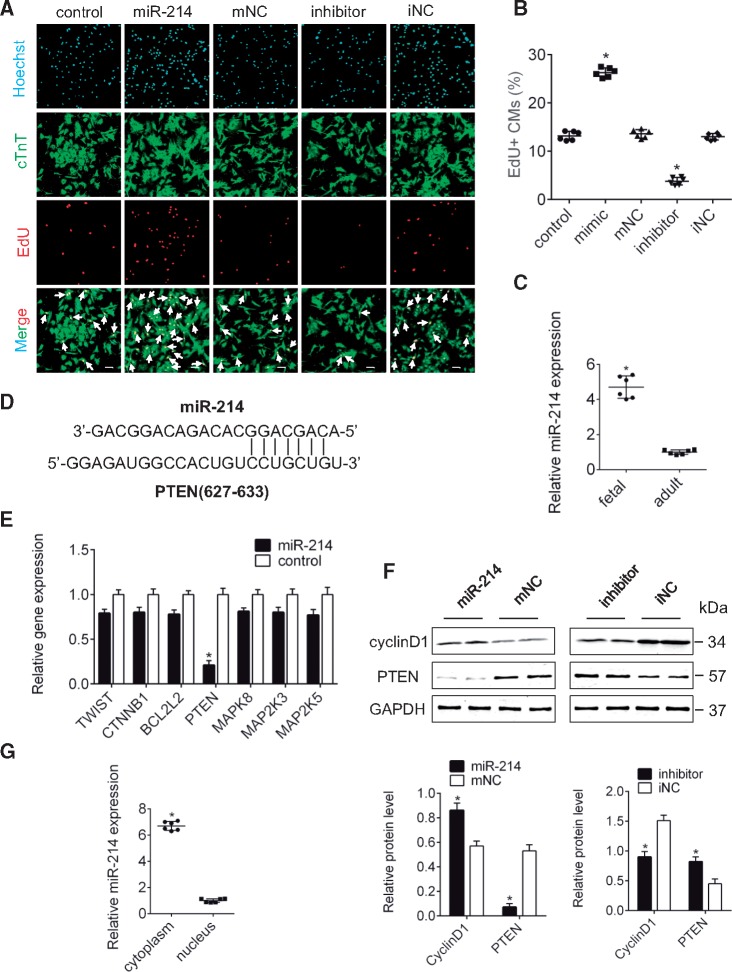
The effect of miR-214 on CM proliferation was measured using a mimic and an inhibitor. The results showed that enhanced miR-214 promoted CM proliferation and reduced miR-214 had inhibitory effects compared to their respective controls (P < 0.05; Figure 6A and B). Moreover, miR-214 was higher in foetal hearts than in adult hearts and no change in AZIN2 was observed (P < 0.05; Figure 6C, Supplementary material online, Figure S5A). In addition, miR-214 expression was higher in CMs than in CFs, with no difference in AZIN2 expression between them (Supplementary material online, Figure S5B and C). We predicted several downstream target genes of miR-214 using miRTarBase and found that PTEN had complementary pairing with the seed sequence of miR-214 (Figure 6D). qPCR showed that PTEN expression significantly decreased when miR-214 expression increased (P < 0.05; Figure 6E). Western blot analysis also confirmed that miR-214 could suppress PTEN protein levels and that cyclin D1 positively correlated with miR-214 (P < 0.05; Figure 6F). Finally, subcellular localization data indicated that miR-214 was primarily located in the cytoplasm, and no difference was for AZIN2 between cytoplasm and nucleus (P < 0.05; Figure 6G, Supplementary material online, Figure S5D). These results suggest that cytoplasmic miR-214 regulates CM proliferation in vitro and that PTEN is a potential target.
Figure 6.
miR-214 regulates PTEN level and of CMs proliferation. (A) Immunostaining for EdU in myocytes transfected with miR-214, inhibitor or control (bars, 40 μm). White arrows refer to EdU-positive myocytes. (B) Quantification of EdU-positive myocytes. *P < 0.05 vs. control; n = 6 per group (one-way ANOVA analysis with Bonferroni’s multiple comparison test). (C) qPCR for miR-214 in foetal and adult rats. *P < 0.05 vs. adult rat; n = 6 per group (Student’s t-test). (D) PTEN contains a site complementary to the seed sequence of miR-214. (E) Expression of target genes after overexpression of miR-214 according to qPCR. *P < 0.05 vs. control; n = 6 per group (Student’s t-test). (F) PTEN and cyclin D1 protein level when miR-214 was inhibited and overexpressed (western blot) (GAPDH internal reference). *P < 0.05 vs. mNC or iNC; n = 6 per group (Student’s t-test). (G) Location of miR-214 in CMs by qPCR. *P < 0.05 vs. nucleus; n = 6 per group (Student’s t-test).
3.7 AZIN2-sv regulates CM proliferation through the miR-214/PTEN/Akt pathway
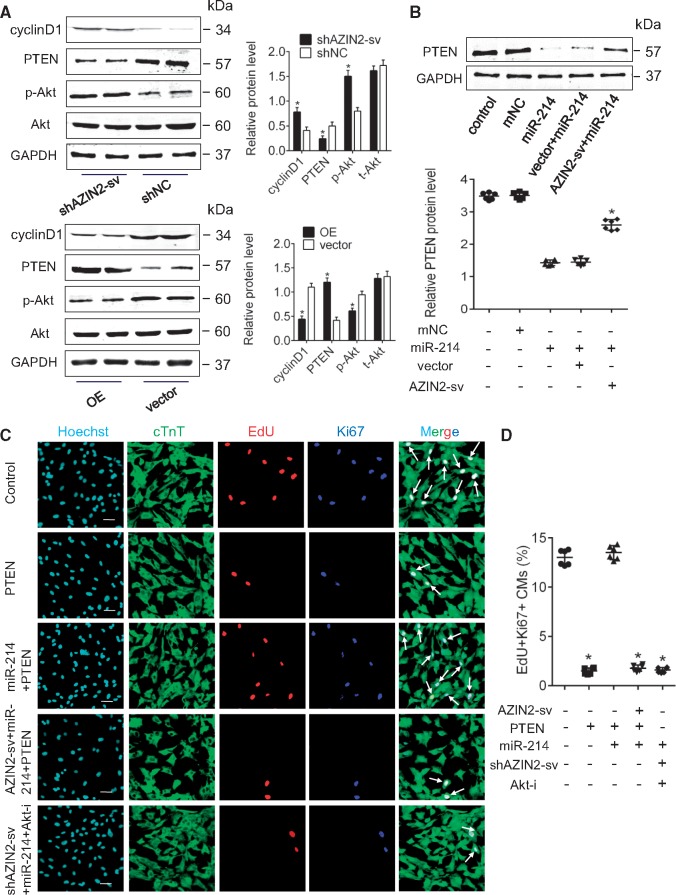
Next, we determined if AZIN2-sv regulates the PTEN/Akt pathway. Cyclin D1 was used to reflect cell cycle activity. When AZIN2-sv was inhibited, PTEN levels decreased, and phosphorylated Akt (p-Akt) and cyclin D1 levels significantly increased compared with the respective control, whereas overexpressing AZIN2-sv had the opposite effect (P < 0.05; Figure 7A). Moreover, overexpressing AZIN2-sv counteracted the inhibitory effects of miR-214 on PTEN (P < 0.05; Figure 7B). In addition, loss of AZIN2-sv had no effect on AZIN2 and inhibition of AZIN2 had no effect on miR-214 and PTEN levels (P < 0.05; Supplementary material online, Figure S5E–I). Furthermore, enforced miR-214 released the inhibitory effects of PTEN on CM proliferation, and overexpression of AZIN2-sv reduced CM proliferation, which was also blocked by an Akt inhibitor (P < 0.05; Figure 7C and D). Therefore, the regulatory effects of AZIN2-sv on CM proliferation are achieved by regulating miR-214, PTEN, and its downstream target Akt.
Figure 7.
AZIN2-sv regulates CM proliferation via miR-214/PTEN/Akt pathway. (A) The protein levels of cyclin D1, PTEN, Akt, and pAkt when AZIN2-sv was knocked down or overexpressed (western blot) (GAPDH internal reference). *P < 0.05 vs. shNC group or vector group; n = 6 per group (Student’s t-test). (B) PTEN protein level after AZIN2-sv and miR-214 interference (western blot) (GAPDH internal reference). AZIN2-sv offset the inhibitory effect of miR-214 on PTEN expression. *P < 0.05 vs. miR-214 group; n = 6 per group (one-way ANOVA analysis with Bonferroni’s multiple comparison test). (C) Immunostaining of EdU and Ki-67 in myocytes after PTEN, AZIN2-sv, and miR-214 interference (bars, 40 μm). White arrows refer to EdU-positive and Ki-67-positive myocytes. Enhanced miR-214 offset the inhibitory effects of PTEN on CM proliferation, and AZIN2-sv reduced CM proliferation, which was blocked by an Akt inhibitor. (D) Quantification of EdU-positive and Ki-67-positive myocytes. *P < 0.05 vs. control; n = 6 per group (one-way ANOVA analysis with Bonferroni’s multiple comparison test).
3.8 AZIN2-sv binds to PTEN directly and increases its stability
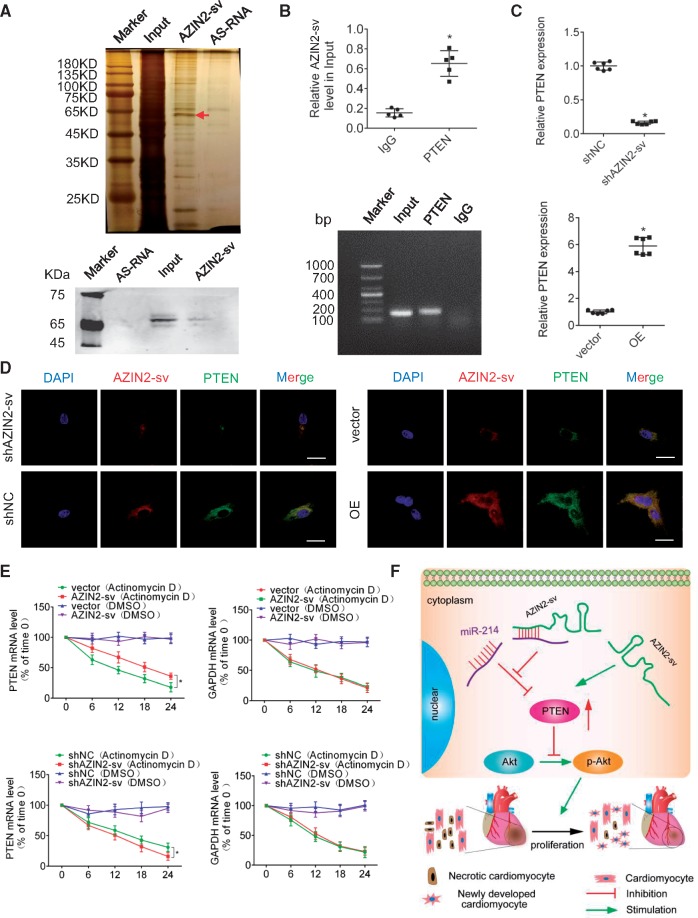
Previous studies have suggested that cytoplasmic lncRNAs can regulate signalling pathways through interactions with specific proteins. Therefore, RNA pull-down assays were performed to identify AZIN2-sv-interacting proteins in D7 CMs. The bands specific to AZIN2-sv were excised and subjected to mass spectrometry. PTEN could be detected by mass spectrometry and western blotting confirmed expression (Figure 8A). We performed RIP to demonstrate that AZIN2-sv could bind to PTEN in vivo. AZIN2-sv was enriched by the PTEN antibody compared to a non-specific IgG antibody (Figure 8B). The RNA level of PTEN could be positively regulated by AZIN2-sv (Figure 8C). In addition, fluorescence in situ hybridization (FISH) and immunofluorescence staining of PTEN showed that they were both located in the cytoplasm (Figure 8D). The forecast results showed that binding sites of AZIN2-sv with PTEN were conserved between human and rats (Supplementary material online, Figures S1F and S5J). These data suggest that AZIN2-sv may regulate the stability of PTEN. To further determine the effects of AZIN2-sv on the synthesis or degradation of PTEN mRNA, AZIN2-sv was overexpressed or reduced in CMs treated with actinomycin D or dimethylsulfoxide (DMSO) over a period of 24 h to block new RNA synthesis, after which the loss of PTEN, GAPDH, and 18sRNA were measured at different time points. The results showed that AZIN2-sv was positive correlated with PTEN stability (Figure 8E). These data suggest that AZIN2-sv can increase PTEN levels through direct binding (Figure 8F).
Figure 8.
AZIN2-sv binds to PTEN and increase its stability. (A) Silver-stained SDS-PAGE gel of proteins immunoprecipitated by AZIN2-sv and its antisense RNA. The arrow indicates the region of the gel excised for mass spectrum determination. PTEN protein was assayed by western blotting. (B) RIP experiments were performed using an antibody against PTEN or negative IgG. The purified RNA was used for qPCR analysis, and enrichment of the lncRNA-AZIN2-sv was normalized to the input. *P < 0.05 vs. IgG; n = 5 per group (Student’s t-test). (C) PTEN mRNA level after reducing or overexpressing AZIN2-sv. *P < 0.05 vs. shNC or vector; n = 6 per group (Student’s t-test). (D) FISH and immunofluorescence showed colocalization of AZIN2-sv and PTEN when AZIN2-sv was knocked down and overexpressed (bars, 40 μm). (E) PTEN and GAPDH mRNA levels measured by qPCR when AZIN2-sv was overexpressed and knocked down. Actinomycin (50 μM) or DMSO (negative control) was used to block new RNA synthesis. *P < 0.05 vs. vector (Actinomycin D) or shNC (Actinomycin D); n = 6 per group (one-way ANOVA analysis with Bonferroni’s multiple comparison test). (F) Illustration of the mechanism underlying AZIN2-sv regulation of CM proliferation through the PTEN/Akt pathway.
4. Discussion
We identified a novel upregulated lncRNA in rat hearts, namely AZIN2-sv, and found that its expression progressively increased after birth. Loss of AZIN2-sv promoted rat cardiac regeneration in vitro and in vivo and attenuated adverse ventricular remodelling post-MI. Also AZIN2-sv acted as a ceRNA of miR-214 to increase its target PTEN, which inhibited the phosphorylation of Akt and reduced CM proliferation. Moreover, AZIN2-sv directly bound to PTEN and increased its stability. Thus, our newly discovered AZIN2-sv may serve as a novel target for preventing and treating heart failure.
Numerous lncRNAs have important functions in developmental processes such as chromatin modification,24 RNA processing,25 structural scaffolds,26 and modulation of proliferation and apoptosis.27 Here, we show that lncRNAs can also participate in cardiac regeneration as AZIN2-sv negatively regulated rat CM proliferation in vitro and in vivo. AZIN2-sv knockdown significantly promoted CM proliferation in rats 7 days after birth, a time at which most cells have exited the cell cycle. This suggests a solution for non-regeneration after myocardial necrosis. Then, DNA synthesis and the appearance of mitotic feature in CMs were enhanced, which not only demonstrated the reliability of proliferation but also indicated that proliferation occurred in partially differentiated CMs, an event that occurs in zebrafish and neonatal mice. In addition, AZIN2-sv knockdown did not induce the proliferation of CFs, which may be due to participation in different signalling pathways. Furthermore, the repair capacity of AZIN2-sv in MI was proved and loss of AZIN2-sv improved ventricular remodelling after MI. CM proliferation and angiogenesis in the infarct zone are critical for the rescue of necrotic myocardium and improvements in post-MI prognosis.28 We observed that with AZIN2-sv loss, EdU- and pH3-labeled CMs increased in the peri-infarct and remote areas, higher vascular density was observed, infarct size was reduced, and cardiac function was improved. Thus, AZIN2-sv may be a key regulator of regeneration/repair capacity in the heart, and as such, may be harnessed for enhancing endogenous repair mechanisms in the adult heart after MI.
lncRNAs exert their various effects by interacting with miRNA as endogenous sponge RNAs, which is the most common interaction described.29,30 To determine if AZIN2-sv functioned through this model, we assessed its cytoplasmic localization, expression abundance, binding sites, and free energy. It has been reported that miRNA target accessibility is a critical factor in determining miRNA function.31 AZIN2-sv and miR-214 were mainly located in the cytoplasm according to FISH and RNA pull-down assays, and RNA hybrid analysis showed that AZIN2-sv had paired seven consecutive bases with the seed sequence of miR-214 and had a minimum free energy of –15.8 kcal mol−1, which made accessibility to miRNA and binding force more reliable. RNA crosstalk between AZIN2-sv and miR-214 was validated with luciferase and pull-down assays. Luciferase activity significantly increased after a mutation at the binding site of AZIN2-sv and miR-214, and the pull-down experiment confirmed their direct combination. Thus, we confirmed the miRNA sponge adsorption effect of lncRNA.10,13,29 We also showed that miR-214 was downstream of AZIN2-sv and participated in the regulation of CM proliferation. To date, more miRNAs, such as miR-199a and miR-34a, have been reported to be involved in myocardial regeneration.7,32 Previous studies have shown that miR-214 can serve as a cardioprotective molecule by attenuating CM death and promote proliferation in various cancers.33,34 Moreover, miR-214 plays an important role in myogenesis35 and muscle fate.36 Consistent with these conclusions, our results showed that miR-214 could promote CMs proliferation in vitro. Moreover, its expression was related to cyclin D1, which indicated its effects on cell cycle activity. Thus, we confirmed the interaction between AZIN2-sv and miR-214 and expanded the role of miR-214 in CM proliferation.
miRNAs inhibit the transcription of target mRNAs. PTEN, a documented target of miR-214, regulates cell proliferation and survival through inhibition of PI3K/Akt pathway.37,38 Activation of the PI3K/Akt signalling pathway is essential for cardiac cell proliferation and survival.39 Previous works have indicated that PTEN blocks activation of the downstream PI3K/Akt signalling pathway to promote CM death during ischemic heart disease.40,41 Our study confirmed that PTEN is a negative regulator of CM proliferation. Transfection of plasmids containing PTEN significantly reduced CM proliferation. More importantly, AZIN2-sv modified PTEN abundance and regulated CM proliferation via the PTEN-Akt pathway. An Akt inhibitor offset the proliferation effects caused by AZIN2-sv loss. Many lncRNAs are reported to absorb miRNA and release its targeted mRNA by competitive binding with miRNA seed sequences.42 We found that AZIN2-sv regulated PTEN by binding to miR-214. Overexpression of miR-214 reduced PTEN, which was increased by overexpressed AZIN2-sv. In addition, our results indicated that AZIN2-sv also bound directly to PTEN and increased its stability, which is a common mechanism underlying the roles of cytoplasmic lncRNAs. Thus, the effects of AZIN2-sv on PTEN occur through its absorption of miR-214 and interaction with PTEN.
Our study had some limitations. The open reading frame of AZIN2-sv partially overlaps with AZIN2, so it is possible that a short AZIN2 peptide was produced. However, there are currently no specific antibodies against this short peptide. In the future, a specific antibody should be customized to evaluate this possibility. Then, we found that AZIN2-sv could directly bind to miR-214, but in the future, AGO2 immunoprecipitation is needed to confirm the presence of miR-214, AZIN2-sv, and PTEN in the complex. In addition, the detailed mechanism underlying the role of AZIN2-sv in increasing PTEN stability should be examined. Finally, although we evaluated the expression of AZIN2-sv in CMs and CFs, further studies are needed to measure AZIN2-sv levels in endothelial cells and to explore its role in blood vessels.
In conclusion, loss of human AZIN2-sv can induce cardiac regeneration and attenuate cardiac adverse remodelling after MI by targeting the PTEN/Akt pathway. Thus, this newly discovered AZIN2-sv may be a potential therapeutic target for promoting endogenous cardiac regeneration and preventing heart failure.
Conflict of interest: none declared.
Funding
This work was supported by grants to J.B. from the National Natural Science Foundation of China (grant numbers 81771857, 81571698, 81227801, and 81271640), National Basic Research Program of China (973 Program) (grant number 2013CB733804), and the Team Program of Natural Science Foundation of Guangdong Province, China (grant number S2011030003134).
Supplementary Material
Time for primary review: 4 days
References
- 1. Karra R, Poss KD.. Redirecting cardiac growth mechanisms for therapeutic regeneration. J Clin Invest 2017;127:427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Laflamme MA, Murry CE.. Heart regeneration. Nature 2011;473:326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Afzal MR, Samanta A, Shah ZI, Jeevanantham V, Abdel-Latif A, Zuba-Surma EK, Dawn B.. Adult bone marrow cell therapy for ischemic heart disease: evidence and insights from randomized controlled trials. Circ Res 2015;117:558–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT.. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013;493:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisén J.. Evidence for cardiomyocyte renewal in humans. Science (New York, NY) 2009;324:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, Porrello ER, Sadek HA.. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 2013;497:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M.. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012;492:376–381. [DOI] [PubMed] [Google Scholar]
- 8. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG.. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013;24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA.. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013;152:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomson DW, Dinger ME.. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272–283. [DOI] [PubMed] [Google Scholar]
- 11. Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF.. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res 2014;114:1377–1388. [DOI] [PubMed] [Google Scholar]
- 12. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q.. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res 2015;116:1143–1156. [DOI] [PubMed] [Google Scholar]
- 13. Wang H, Niu L, Jiang S, Zhai J, Wang P, Kong F, Jin X.. Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget 2016;7:86174–86185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong R, Liu GB, Liu BH, Chen G, Li K, Zheng S, Dong KR.. Targeting long non-coding RNA-TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis 2016;7:e2278.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. National Research Council Committee for the Update of the Guide for the C and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. Washington, DC: National Academies Press (US) National Academy of Sciences; 2011. The National Academies Collection: reports funded by National Institutes of Health. [Google Scholar]
- 16. Dong R, Jia D, Xue P, Cui X, Li K, Zheng S, He X, Dong K.. Genome-wide analysis of long noncoding RNA (lncRNA) expression in hepatoblastoma tissues. PLoS One 2014;9:e85599.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Su Y, Li L, Zhao S, Yue Y, Yang S.. The long noncoding RNA expression profiles of paroxysmal atrial fibrillation identified by microarray analysis. Gene 2018;642:125–134. [DOI] [PubMed] [Google Scholar]
- 18. Wang Y, Chen Y, Yan Y, Li X, Chen G, He N, Shen S, Chen G, Zhang C, Liao W, Liao Y, Bin J.. Loss of CEACAM1, a tumor-associated factor, attenuates post-infarction cardiac remodeling by inhibiting apoptosis. Sci Rep 2016;6:21972.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davidson MM, Nesti C, Palenzuela L, Walker WF, Hernandez E, Protas L, Hirano M, Isaac ND.. Novel cell lines derived from adult human ventricular cardiomyocytes. J Mol Cell Cardiol 2005;39:133–147. [DOI] [PubMed] [Google Scholar]
- 20. Chen Y, Zhang C, Shen S, Guo S, Zhong L, Li X, Chen G, Chen G, He X, Huang C, He N, Liao W, Liao Y, Bin J.. Ultrasound-targeted microbubble destruction enhances delayed BMC delivery and attenuates post-infarction cardiac remodelling by inducing engraftment signals. Clin Sci (Lond) 2016;130:2105–2120. [DOI] [PubMed] [Google Scholar]
- 21. Guo S, Shen S, Wang J, Wang H, Li M, Liu Y, Hou F, Liao Y, Bin J.. Detection of high-risk atherosclerotic plaques with ultrasound molecular imaging of glycoprotein IIb/IIIa receptor on activated platelets. Theranostics 2015;5:418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liao Y, Bin J, Asakura M, Xuan W, Chen B, Huang Q, Xu D, Ledent C, Takashima S, Kitakaze M.. Deficiency of type 1 cannabinoid receptors worsens acute heart failure induced by pressure overload in mice. Eur Heart J 2012;33:3124–3133. [DOI] [PubMed] [Google Scholar]
- 23. Chen G, Yang L, Zhong L, Kutty S, Wang Y, Cui K, Xiu J, Cao S, Huang Q, Liao W, Liao Y, Wu J, Zhang W, Bin J.. Delivery of hydrogen sulfide by ultrasound targeted microbubble destruction. Sci Rep 2016;6:30643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gartler SM, Riggs AD.. Mammalian X-chromosome inactivation. Annu Rev Genet 1983;17:155–190. [DOI] [PubMed] [Google Scholar]
- 25. Gong C, Maquat LE.. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 2011;470:284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB.. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009;33:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X.. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science (New York, NY) 2014;344:310–313. [DOI] [PubMed] [Google Scholar]
- 28. Shapiro SD, Ranjan AK, Kawase Y, Cheng RK, Kara RJ, Bhattacharya R, Guzman-Martinez G, Sanz J, Garcia MJ, Chaudhry HW.. Cyclin A2 induces cardiac regeneration after myocardial infarction through cytokinesis of adult cardiomyocytes. Sci Transl Med 2014;6:224ra27.. [DOI] [PubMed] [Google Scholar]
- 29. Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, Feng C, Wang CQ, Zhao YF, Li PF.. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun 2015;6:6779.. [DOI] [PubMed] [Google Scholar]
- 30. Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, Fan J, Liu L, Sun SH, Zhou WP.. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016;63:499–511. [DOI] [PubMed] [Google Scholar]
- 31. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E.. The role of site accessibility in microRNA target recognition. Nat Genet 2007;39:1278–1284. [DOI] [PubMed] [Google Scholar]
- 32. Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, Fisch S, Unno K, Sereti KI, Liao R.. MicroRNA-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res 2015;117:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dong H, Dong S, Zhang L, Gao X, Lv G, Chen W, Shao S.. MicroRNA-214 exerts a cardio-protective effect by inhibition of fibrosis. Anat Rec (Hoboken) 2016;299:1348–1357. [DOI] [PubMed] [Google Scholar]
- 34. Yang X, Qin Y, Shao S, Yu Y, Zhang C, Dong H, Lv G, Dong S.. MicroRNA-214 inhibits left ventricular remodeling in an acute myocardial infarction rat model by suppressing cellular apoptosis via the phosphatase and tensin homolog (PTEN). Int Heart J 2016;57:247–250. [DOI] [PubMed] [Google Scholar]
- 35. Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V.. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell 2009;36:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG.. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet 2007;39:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS.. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int 2013;13:68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsai C-Y, Wu JCC, Fang C, Chang AYW.. PTEN, a negative regulator of PI3K/Akt signaling, sustains brain stem. Neuropharmacology 2017;123:175–185. [DOI] [PubMed] [Google Scholar]
- 39. Cantley LC. The phosphoinositide 3-kinase pathway. Science 2002;296:1655–1657. [DOI] [PubMed] [Google Scholar]
- 40. Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, Pu WT, Liao R, Wang DZ.. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res 2013;112:1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosivatz E. Inhibiting PTEN. Biochem Soc Trans 2007;35:257–259. [DOI] [PubMed] [Google Scholar]
- 42. Xue Y, Ni T, Jiang Y, Li Y.. LncRNA GAS5 inhibits tumorigenesis and enhances radiosensitivity by suppressing miR-135b expression in non-small cell lung cancer. Oncol Res 2017;25:1305.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.