Abstract
The role of maternal factors in the evolution of development is poorly understood. Here we describe the use of reciprocal hybridization between the surface dwelling (surface fish, SF) and cave dwelling (cavefish, CF) morphs of the teleost Astyanax mexicanus to investigate the roles of maternal genetic effects in cavefish development. Reciprocal hybridization, a procedure in which F1 hybrids are generated by fertilizing SF eggs with CF sperm (SF X CF hybrids) and CF eggs with SF sperm (CF X SF hybrids), revealed that the CF degenerative eye phenotype showed maternal genetic effects. The eyes of CF X SF hybrids resembled the degenerate eyes of CF in showing ventral reduction of the retina and corresponding displacement of the lens within the optic cup, a smaller lens and eyeball, more lens apoptosis, a smaller cartilaginous sclera, and lens-specific gene expression characteristics compared to SF X CF hybrids, which showed eye and lens gene expression phenotypes resembling SF. In contrast, reciprocal hybridization failed to support roles for maternal genetic effects in the CF regressive pigmentation phenotype or in CF constructive changes related to enhanced jaw development. Maternal transcripts encoded by the pou2f1b, runx2b, and axin1 genes, which are involved in determining ventral embryonic fates, were increased in unfertilized CF eggs. In contrast, maternal mRNAs encoded by the ß-catenin and syntabulin genes, which control dorsal embryonic fates, showed similar expression levels in unfertilized SF and CF eggs. Furthermore, maternal transcripts of a sonic hedgehog gene were detected in SF and CF eggs and early cleaving embryos. This study reveals that CF eye degeneration is controlled by changes in maternal factors produced during oogenesis and introduces A. mexicanus as a model system for studying the role of maternal changes in the evolution of development.
Keywords: Astyanax mexicanus, cavefish, maternal genetic effects, optic regression, dorsoventral pattern formation, sonic hedgehog mRNA, evolution of development
Introduction
Maternal genetic effects occur when the phenotype of an organism is controlled by the genotype of its mother. A classic example is the direction of shell coiling in snails, which depends on dextral or sinistral orientation of spiral cleavages (Boycott et al., 1930; Freeman and Lundelius, 1982). Dextral oriented development is genetically dominant, but the cleavage and shell coiling patterns of a sinistral-coiling female snail can be imposed on her offspring by depositing maternal information in eggs. During oogenesis, maternal mRNAs are transcribed and stored in a translationally inactive state or translated into stable maternal proteins. The stored maternal mRNAs and/or proteins function to regulate cleavage patterns, embryonic axis specification, and cell fate determination during embryonic development. For example, in Drosophila maternal mRNAs transcribed from the maternal effect genes bicoid and nanos are localized at the poles of the egg; where they function, along with the products of other maternal effect genes, to specify anteroposterior fates by regulating zygotic gene transcription (Nasiadka et al., 2002). Likewise, maternal mRNAs and proteins are employed to organize the first steps of embryonic development, including cleavage patterns and dorsoventral axis formation, in many other animals (Davidson, 1986; Heasman, 2006; Jeffery, 2001a; Abrams and Mullins, 2009; Landon and Mullins, 2011).
Although the importance of maternal genetic effects in evolution is recognized (Badyaev, 2008; Wolf and Wade, 2016), whether novel phenotypes can be generated by changing maternal factors is still an open question. A problem encountered in studying the evolution of maternal genetic effects is that they are usually defined by mutations, which can be difficult to interpret in the context of evolution. The existence of naturally occurring morphs in the same species provides an opportunity to study maternal genetic effects in an evolutionary context, but examples such as the dextral and sinistral coiling snails mentioned above are uncommon. An exception is the teleost Astyanax mexicanus, in which two different morphs, an eyed and pigmented surface dwelling form (surface fish, SF) and an eyeless and depigmented cave dwelling form (cavefish, CF), have evolved in the same species (Jeffery, 2001b; 2009).
The polarity of evolutionary changes is known in A. mexicanus: CF arose from SF ancestors less than a few million years ago (Gross, 2012). During this short interval, CF evolved a suite of traits associated with adaptation to dark caves (Jeffery, 2001b). They have reduced or eliminated their eyes by an unusual process in which the lens and retina are initially formed during embryogenesis and then regress during larval development (Jeffery, 2009; Strickler et al., 2007a; Yamamoto, 2016; Krishnan and Rohner, 2017). The CF eye degeneration phenotype is also defined by changes in the sclera, the outer coating of the eye, and in the craniofacial bones surrounding the orbit (Yamamoto et al., 2003; O’Quin et al., 2015; Powers et al., 2018). CF embryos have also evolved the reduction or complete loss of melanin pigmentation (Jeffery et al., 2016). These and other regressive changes are accompanied by constructive changes, such as the development of larger oral jaws with more taste buds (Yamamoto et al., 2009; Atukorata and Franz-Odendaal, 2016), improved structures involved in tactile sensations and feeding (Yoshizawa et al., 2010; 2012), and modified behaviors for adaptation to the cave environment (Elipot et al., 2014). Some of these changes may be under the control of events related to zygotic gene transcription, while others may be regulated by differences in the transcription and deposition of maternal mRNAs during oogenesis, but such distinctions have not been previously investigated in A. mexicanus.
The ability to generate viable offspring by crossing SF and CF (Wilkens, 1988; Yoshizawa et al., 2012) allows reciprocal hybridization to be used for investigating the role of maternal genetic effects in CF development. Reciprocal hybridization involves comparing the offspring generated by inseminating CF eggs with SF sperm to the progeny generated by inseminating SF eggs with CF sperm. Because virtually all of the zygote cytoplasm is contributed by the egg, the differences between reciprocal hybrids can be used to identify CF phenotypes that develop under the influence of maternal factors. Although often employed to study phenotypic differences in plants (Erickson et al., 1990; Song et al., 1993; Stanton, 2005, Lain et al., 2016) or genetic imprinting (Feil and Berger, 2007; Wolf et al., 2014), reciprocal hybridization has only rarely been used to study the evolutionary origin of novelties within a single species (Marshak, 1936; Zakas and Rockman, 2014).
In this investigation, reciprocal hybridization has revealed that the evolution of degenerative CF phenotypic changes in optic polarity, the lens, and the sclera are influenced by maternal genetic effects. Consistent with these maternal effects, unfertilized cavefish eggs were discovered to contain higher levels of maternal transcripts encoded by some of the genes responsible for determining ventral embryonic fates. These findings highlight the importance of A. mexicanus as a novel model system for studying the roles of maternal changes in the evolution of development.
Materials and Methods
Biological Materials
Surface fish (SF) and cavefish (CF, Pachón) were obtained from Jeffery laboratory stocks originally collected from Nacimiento del Rio Choy, San Luis Potosi, Mexico and Balmorhea Springs State Park, Texas or Cueva de Pachón, Tamaulipas, Mexico respectively. Adult fish were maintained in a running water system at 23ºC on a 14-hr light and 10-hr dark photoperiod as described previously (Jeffery et al., 2000). Embryos were obtained by natural spawning or by in vitro fertilization, as described below, and raised at 23–25ºC. Larvae were fed brine shrimp beginning at 5–6 days post-fertilization (dpf), and adults were fed TetraMin Pro flakes (Tetra Holding Inc, Blacksburg VA). Methods for fish handling and care were approved by the University of Maryland Animal Care and Use Committee and conformed to National Institutes of Health guidelines.
Collection of Unfertilized Eggs and Generation of Reciprocal Hybrids
Unfertilized eggs were collected by applying gentle pressure to the abdomen of gravid adults and used to extract RNA for quantitative real time RT-PCR experiments (see below) or to produce reciprocal hybrids. Sperm were obtained from males by gently squeezing the area around the cloaca. In vitro fertilization was carried out according to Borowsky (2008). Diluted sperm were added to a suspension of unfertilized eggs in fish system water. SF eggs were inseminated with CF sperm to generate SF (female) X CF (male) F1 hybrids, and CF eggs were inseminated with SF sperm to obtain CF (female) x SF (male) F1 hybrids. The reciprocal hybrids were raised under the conditions described above.
Optic Phenotype Quantification
Optic phenotypes were determined in specimens anesthetized with tricaine (2 μg/ml; Western Chemical Inc., Ferndale, WA) and viewed under a compound microscope. Optic vesicles were measured along the anterior-posterior axis in 20 somite embryos and categorized into individuals with SF-like, CF-like, or intermediate phenotypes (see Fig. 2F). Retinal and lens areas were determined in 2 dpf embryos from diameters measured using Image J software or AxioVision software (Carl Zeiss Microscopy GmbH, Jena, Germany). Eye and lens (or pupil, as a proxy) sizes were determined in 3 and 10-day post-fertilization (dpf) larvae from Image J measurements and standardized by measuring body lengths from the anterior tip of the larval rostrum to the posterior tip of the tail.
Figure 2.
Optic phenotypes in embryos and hatched larvae of reciprocal hybrids. A, B. Surface fish (SF) x cavefish (CF) (A) and CF X SF (B) hybrid embryos at the 20-somite stage. OV: optic vesicle. Scale bar: 40 μm. C. Histograms comparing optic vesicle lengths in SF X CF and CF X SF hybrids at the 20-somite stage. The number of embryos is shown at the base of each column. A total of 38 hybrids from 2 reciprocal hybridizations were analyzed. ns: no significance. Error bars: standard deviations of the means. D, E. SF X CF hybrids (D) and CF X SF hybrids (E) at 24-hours post-fertilization (hpf). Scale bar: 40 μm. F. Horizontal bar graphs showing the percentages of different of optic phenotypes in SF, SF X CF hybrids, CF X SF hybrids, and CF at 24 hpf. The number (N) of larvae is shown at the bottom of each row. A total of 314 hybrids from 4 reciprocal hybridizations were analyzed. A key of the different optic phenotypes is shown above the bars. IN: intermediate phenotype.
Melanophore Quantification
In larvae and juveniles, melanophores are concentrated over the dorsal regions of the body. Because the dorsal cranium is flattened relative to the dorsal trunk, which is oval shaped and punctuated with fins, we selected the dorsal cranium as the best place to count melanophores by microscopy without changing the plane of focus. Melanophores were counted in areas of dorsal skin centered over the hindbrain in anesthetized 10 dpf larvae or 30 dpf juveniles respectively (see Fig. 6A, D).
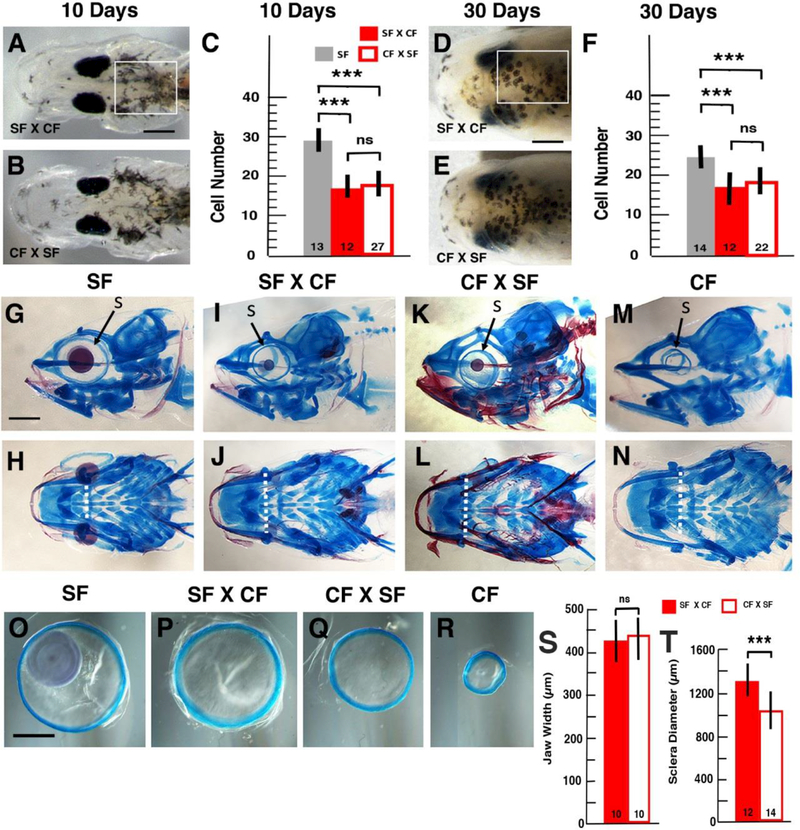
Figure 6.
Melanin pigmentation and craniofacial phenotypes in reciprocal hybrids. A-C. Larval pigmentation. Dorsal views of surface fish x cavefish (SF X CF) hybrid (A) and cavefish x surface fish (CF X SF) hybrid (B) larvae at 10-days post-fertilization (dpf) showing cranial melanophores. Scale bar: 100 μm. C. Histograms showing the number of melanophores in the dorsal skin of surface fish (SF), SF X CF hybrids, and CF X SF hybrids. D-F. Juvenile pigmentation. Dorsal images of SF X CF (A) and CF X SF hybrids (B) at 30 dpf showing cranial melanophores. Scale bar: 50 μm. C. Histograms showing the number of melanophores in the dorsal skin of SF, SF X CF hybrids, and CF X SF hybrids. The regions in which melanophores were quantified are shown as boxes in A and D. The number of individuals is shown at the base of each histogram. A total of 75 hybrids from 3 reciprocal hybridizations were analyzed. Error bars: standard deviations of the means. ***denotes significance at < 0.001. ns: no significance. G-N. Craniofacial structure in juveniles at 30 dpf with cartilage stained blue and bone stained red. Lateral (G, I, K, M) and ventral (H, J, L, N) views of SF (G, H), SF X CF hybrids (I, J), CF X SF hybrids (K, L), and CF (M, N). Scale bar: 250 μm. Horizontal dotted lines show mandible spans. S: sclera. O-R. Sclera isolated from juveniles at 60 dpf. Frontal views of scleral cartilage rings dissected from SF (O), SF X CF hybrids (P), CF X SF hybrids (Q), and CF (R) of equivalent body lengths. Scale bar: 500 μm. S, T. Histograms comparing (S) the width of the lower jaw measured at the level of the hinge (dashed lines in J, L) and (T) the diameter of the sclera in SF X CF and CF X SF hybrids. The number of individuals is shown at the base of each histogram. A total of 46 hybrids from 2 reciprocal hybridizations were analyzed. Error bars: standard deviations of the means. ***denotes significance at < 0.001. ns: no significance.
Alcian Blue/Alizarin Red Staining of Cartilage and Bone
Larvae at 10 dpf and juveniles at 30 dpf were double stained for cartilage and bone using the Alcian blue/Alizarin Red method (Wassersug, 1976) with the following modifications: 10% formalin fixation, Alcian blue staining, trypsin treatment, and alizarin red staining were each carried out at room temperature for 12 hr. Stained specimens were imaged by light microscopy and photographed.
Histology
For comparison of eye morphology, 10 dpf SF, reciprocal hybrids, and CF were fixed overnight in 4% paraformaldehyde in PBS (PFA), dehydrated through an increasing ethanol series, embedded in Paraplast (Polysciences Inc, Warrington, PA, USA), and sectioned at 8 μm. The sections were mounted on glass slides, stained with Harris hematoxylin, imaged by light microscopy, and photographed.
Apoptosis Detection
Apoptotic cells were detected in the lens of 2 dpf reciprocal hybrids by vital staining with 5 μg/ml LysoTracker Red DND-99 (Invitrogen, Carlsbad, CA) at room temperature for 30 minutes in darkness, as described by Alunni et al. (2007). The vital stained embryos were anesthetized with tricaine and imaged by fluorescence microscopy. The number of apoptotic cells in the lens was quantified from photographs of imaged specimens.
In situ Hybridization
In situ hybridization was carried out according to Yamamoto et al. (2004) for gammaM2 crystallin or according to Ma et al. (2014) for all other genes. The RNA probes used for in situ hybridization were prepared using the oligonucleotide primers shown in Table S1. After hybridization the specimens were incubated in BM Purple AP Substrate (Roche) at room temperature in the dark. Whole mount stained specimens were cleared through an increasing glycerol series in PBS, then imaged by light microscopy and photographed. Some of the stained specimens were embedded in Paraplast, and sectioned at 10 μm. The sections were mounted on glass slides, imaged by light microscopy, and photographed.
RNA Isolation and Quantitative RT-PCR
Unfertilized SF and CF eggs were washed several times in distilled water, viewed under a compound microscope to assess purity, and concentrated for RNA extraction. Total RNA was extracted using TRI Reagent Solution (Life Technologies, Grand Island NY, USA), treated with RNase-free DNase (Qiagen) to remove traces of genomic DNA, and cDNA was synthesized using the SuperScriptTM III First-Strand Synthesis SuperMix Kit and oligo (dT)20 primers (Life Technologies).
Quantitative real time RT-PCR (qPCR) was done with cDNA using oligonucleotide primers designed to amplify the A. mexicanus orthologs of Danio rerio maternal effect genes (Table S2). To confirm the specificity of the primers, BLAST searches were done against the Ensemble cavefish genome database. Dissociation curves were used to confirm that single PCR products were amplified. Real-time PCR was performed using SYBR PremixExTag (Tli RNaseH Plus) PCR Master Mix (Takara Bio USA, Mountain View, CA) Master Mix and a LightCycler 480 System using StepOnePlus System software according to the manufacturer’s protocol (Roche). All reactions were replicated at least three times. For relative quantification, the ΔCt values for each gene were normalized to ΔΔCt values against a GAPDH reference gene and converted to 2^-△△Ct values, which represent the mean fold change of cavefish mRNA compared to surface fish mRNA.
Statistics
Statistical analysis of the qPCR results and data shown in Figures 2, 3, and 6 was done using Student’s two tailed t-test. The data shown in Figures 4 and 5 were analyzed by nonparametric statistical methods due to the comparison between reciprocal hybrids, SF, and CF (Tables S3, S4). Briefly, differences of the variance among the different groups were calculated by the Kruskal-Wallis test followed by two-independent sample Mann-Whitney tests between SF and SF X CF hybrids, and between SF X CF and CF X SF hybrids. Mann-Whitney tests were corrected using the Bonferroni method. The non-parametric tests were performed in IBM SPSS version 21 (IBM Corp. North Castle, NY, USA).
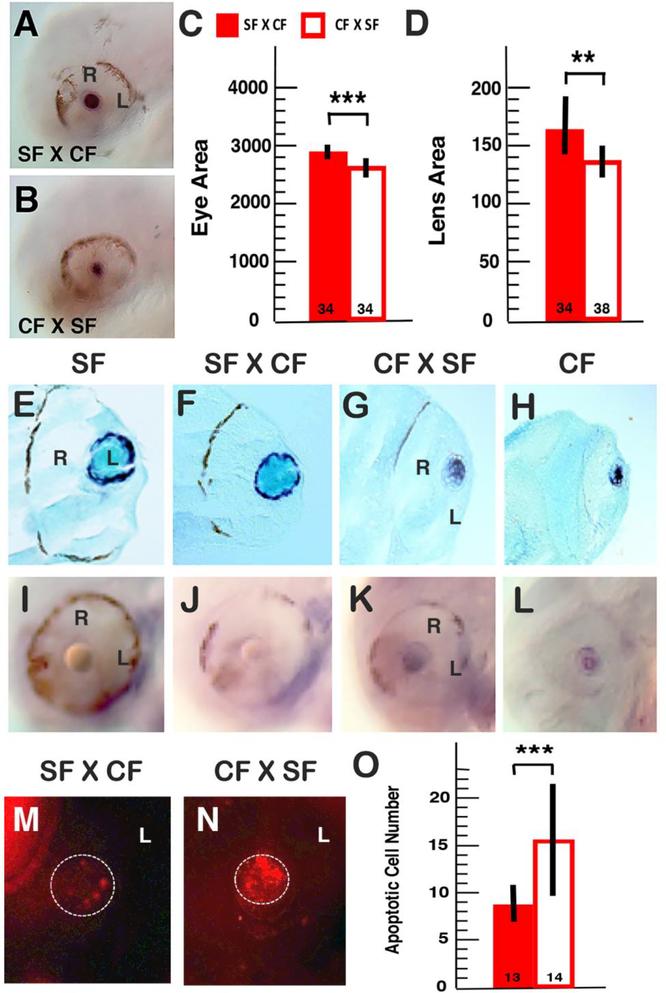
Figure 3.
Eye and lens phenotypes in reciprocal hybrids at 48 hours post-fertilization. A-B. In situ hybridization showing cryaa expression in surface fish x cavefish (SF X CF) hybrids (A) and cavefish x surface fish (CF X SF) hybrids (B). Scale bar: 20 μm. C, D. Histograms showing eye and lens sizes in SF x CF hybrids (C) and CF X SF hybrids (D). A total of 72 hybrids from 2 reciprocal hybridizations were analyzed. E-L. In situ hybridization showing gammaM2-crystallin (in sections E-H) and hsp90alpha (in whole mounted embryos I-L) expression in the lens of SF (E, I), SF X CF hybrids (F, J), CF x SF hybrids (G, K), and CF (H, L). Scale bars: 30μm in E and I. M-O. Comparison of apoptosis in the lens of SF X CF hybrids (M) and CF X SF hybrids (N). Scale bar: 30 μm. O. Histograms comparing the number of apoptotic cells in the lens of SF X CF hybrids and CF X SF hybrids. The number of individuals is shown at the base of each histogram. A total of 27 hybrids from 3 reciprocal hybridizations were analyzed. Error bars: standard deviations of the means. ** denotes significance at p < 0.01. *** denotes significance at p < 0.001. L: lens. R: retina.
Figure 4.
Eye and lens phenotypes in reciprocal hybrids at 72-hours post-fertilization (hpf). A-D. Surface fish (SF) (A), surface fish x cavefish (SF X CF) hybrid (B), cavefish x surface fish (CF X SF) hybrid (C), and cavefish (CF) (D) larvae. Lens and retina are outlined by yellow dashes in A-D. Scale bar: 30 μm. E, F. Histograms comparing eye (E) and lens (F) sizes in SF, SF X CF hybrids, CF X SF hybrids, and CF. The number of larvae is shown at the base of each histogram. A total of 63 hybrids from 3 reciprocal hybridizations were analyzed. Error bars: standard errors of the means. * denotes significance at p < 0.05. *** denotes significance at p < 0.001. n.s.: no significance. See Table S3 for statistical scores.
Figure 5.
Eye and lens phenotypes in 10-day post-fertilization (dpf) reciprocal hybrids. A-D. Dorsal views of the cranial regions of surface fish (SF) (A), surface fish x cavefish (SF X CF) hybrids (B), cavefish x surface fish (CF X SF) hybrids (C), and cavefish (CF) (D) showing differences in eye sizes. Scale bar: 200 μm. E-H. Sections through SF (E), SF X CF hybrids (F), CF X SF hybrids (G), and CF (H) eyes showing differences in lens sizes. Scale bar: 50 μm. I, J. Histograms comparing eye and pupil (as a lens proxy) sizes in SF, SF X CF hybrids, CF X SF hybrids, an CF at 10 dpf. Left axis: pupil or eye diameter divided by standard length (SL). L: lens. The number of larvae is shown at the base of each histogram. A total of 54 hybrids from 3 reciprocal hybridizations were analyzed. Error bars: standard errors of the means. n. s.: no significance. **denotes significance at < 0.01. ***denotes significance at < 0.001. See Table S4 for statistical scores.
Results
Juvenile and Adult Reciprocal Hybrid Phenotypes
Reciprocal hybrids were generated by inseminating CF eggs with SF sperm (CF X SF hybrids) and SF eggs with CF sperm (SF X CF hybrids), and their phenotypes were compared to those of SF and CF. Similar to SF, and in contrast to CF, both types of reciprocal hybrid adults exhibited external eyes (Fig. 1). However, most of the CF X SF hybrids developed slightly smaller eyes and pupils than their SF X CF counterparts. Moreover, in contrast to CF, which are albino, both types of hybrids were pigmented but pale compared to SF (Fig. 1). These observations encouraged further study of reciprocal hybrid phenotypes during embryonic and larval development with a focus on evaluating the possibility of maternal genetic effects.
Figure 1.
The phenotypes of juvenile (A) and adult (B) surface fish, reciprocal hybrids, and cavefish. Top row: surface fish. Second to top row: surface fish (female) x cavefish (male) hybrid. Second to bottom row: cavefish (female) x surface fish (male) hybrid. Bottom row: cavefish. Scale bars: 100 μm in A and 400 μm in B.
Optic Polarity and Size in Reciprocal Hybrids
SF, CF and reciprocal hybrids develop synchronously during embryonic and early larval development, allowing their phenotypes to be directly compared. We first examined the optic phenotypes of reciprocal hybrids at the 20-somite stage, about 16–20 hours post-fertilization (hpf). The optic vesicles of SF and CF embryos differ in size at this stage of development: the SF optic vesicle is larger with a longer anterior-posterior axis than the CF optic vesicle (Yamamoto et al., 2004). In hybrid embryos of both types, the length of the optic vesicle was intermediate between SF and CF, but no significant differences were found in optic vesicle lengths between SF x CF and CF x SF hybrids (Fig. 2A-C).
We next examined the optic phenotypes of reciprocal hybrids shortly after larval hatching at about 24-hours post-fertilization (hpf), when the lens and optic cup are clearly discernable. At this stage of development, most SF and CF embryos differ in the position of the lens within the optic cup (Fig. 2D, E). When viewed laterally the lens is central in the SF optic cup, whereas it is dislocated toward the ventral side of the optic cup in most CF embryos (Yamamoto et al., 2004; Alunni et al., 2007). The difference in lens placement is considered to be a consequence of the reduction or loss of the ventral retina. We found a mixture of SF, CF, and intermediate optic polarity phenotypes in reciprocal hybrids (Fig. 2D-F). The predominant phenotype of both types of reciprocal hybrids was intermediate between the SF and CF phenotypes, but some hybrid embryos also showed the typical SF and CF phenotypes (Fig. 2F). CF X SF hybrids exhibited a higher proportion of eye primordia with typical CF phenotypes (about 30%) than SF X CF hybrids (less than 10%) (Fig. 2F), consistent with a role for maternal genetic effects in the development of optic polarity.
To determine whether the differences in optic phenotypes also occur during larval development, we compared lens and eye size in reciprocal hybrids at 48 and 72 hpf (Figs. 3, 4). At both stages the eyeball and lens were smaller in CF X SF hybrids than in SF X CF hybrids (Figs. 3C, D and 4E, F). During this period, the CF lens is undergoing apoptosis (Jeffery and Martasian, 1998; Alunni et al., 2007; Strickler et al. 2007a). Apoptotic cells were observed in the lens of both types of hybrids (Fig. 3M, N), however, the number of dying cells was significantly higher in CF X SF hybrids than in SF x CF hybrids (Fig. 3O), a sign of maternal genetic effects on lens survival.
We determined the expression of cryaa, gammaM2-crystallin, and hsp90alpha genes in the lens of reciprocal hybrids by in situ hybridization. The cryaa gene is strongly expressed in the SF lens but downregulated in the CF lens (Strickler et al. 2007b, Ma et al., 2014). We found that cryaa was expressed in the lens of both types of reciprocal hybrids, but expression was weaker in most CF X SF hybrids compared to SF X CF hybrids (Fig. 3A, B), and was not detectible in the lens of some CF X SF hybrids (data not shown). The gammaM2-crystallin gene is expressed in epithelial cells around the margin of the SF lens (Fig. 3E), whereas it is expressed in primary fiber cells located the center of the CF lens (Fig. 3H). We detected gammaM2crystallin expression the lens epithelial cells of SF X CF hybrids (Fig. 3F), just as in SF, but in the central lens fiber cells in CF X SF hybrids (Fig. 3G), similar to the CF lens expression pattern. The hsp90alpha gene is expressed in the CF lens (Fig. 3L) but not in the SF lens (Fig. 3I) (Hooven et al., 2004). We found that hsp90alpha was expressed in the lens of CF X SF hybrids (Fig. 3K) but not in the lens of SF X CF hybrids (Fig. 3J). Therefore, CF X SF hybrids resemble CF in the expression patterns of lens genes, harkening a role for maternal genetic effects in zygotic gene expression.
To determine whether maternal influences on optic phenotypes persist through later development, we compared the optic phenotypes of reciprocal hybrids at 10 dpf (Fig. 5). At this stage of larval development, both types of hybrids showed smaller eyes than SF (Fig. 5A-C, E-G, I). Although the mean eye size of CF X SF hybrids was smaller than SF X CF hybrids (Fig. 5B, C), the difference was not significant (Fig. 5I). In contrast, CF X SF hybrids showed a significantly smaller lens compared to SF X CF hybrids (Fig. 5F, G, J), indicating that maternal influences on lens size persist through larval development.
In summary, the results show that CF X SF hybrids, in contrast to SF X CF hybrids, have morphological, physiological, and molecular optic phenotypes resembling CF, indicating an influence of maternal genetic effects on the regressive CF eye phenotype.
Melanin Pigmentation, Craniofacial, and Scleral Development in Reciprocal Hybrids
In addition to eyes, CF differ from SF in the extent of melanin pigmentation (Jeffery et al., 2016), jaw span (Yamamoto et al., 2009; Atukorata and Franz-Odendaal, 2016), and scleral morphology (Yamamoto et al., 2003, O’ Quin et al., 2015). To determine whether these phenotypes are controlled by maternal genetic effects, we compared melanin pigment cells, craniofacial structure, and sclera development in reciprocal hybrids.
Melanophores were quantified at 10 and 30 dpf in SF, SF X CF hybrids, and CF X SF hybrids (Fig. 6A-F). Although both classes of hybrids showed fewer melanophores than SF, no significant differences in melanophores were apparent between reciprocal hybrids at either developmental stage (Fig. 6C, F), suggesting that zygotic, rather than maternal, genetic effects control the regressive CF melanin pigmentation phenotype.
CF have wider lower jaw (mandible) spans than SF (Yamamoto et al., 2009; Atukorata and Franz-Odendaal, 2016). To compare jaw and cranial morphology in reciprocal hybrids, we stained cartilage and bone with Alcian Blue/Alizarin Red in 30 dpf juveniles (Fig. 6G-N). The overall cranial shape was similar in SF, CF, and reciprocal hybrids (Fig. 6G-N). The mandibles of reciprocal hybrids were intermediate in size between those of SF and CF, and no significant differences in lower jaw span were detected in CF X SF relative to SF X CF hybrids (Fig. 6H-N, S). We noted that all of the 23 CF x SF hybrids analyzed showed more extensive ventral cranial (and vertebral) ossification than SF X CF hybrids (Fig. 6G-N). Because hyper-ossification was not shared with CF, however, it is unlikely to be due to maternal genetic effects, but instead may be related to a difference in growth rate peculiar to CF X SF hybrids.
CF have a smaller sclera than SF, which does not develop scleral ossicles (Yamamoto et al., 2003; O’Quin et al., 2015). To determine if maternal genetic effects are involved in sclera formation, scleral rings were dissected from 60 dpf Alcian Blue/Alizarin Red stained SF, reciprocal hybrids, and CF juveniles of the same body length (Fig. 6O-R). Although CF X SF hybrid sclera were larger than CF sclera, they were significantly smaller than SF X CF hybrid (Fig. 6T) sclera, suggesting that maternal genetic effects have an impact on sclera development.
The differences in scleral ossification and orbital bone morphology previously reported between CF and SF do not appear until later in development (Yamamoto et al., 2003), and thus could not be evaluated in 30 dpf reciprocal hybrid juveniles.
Maternal mRNA Differences in Cavefish Eggs
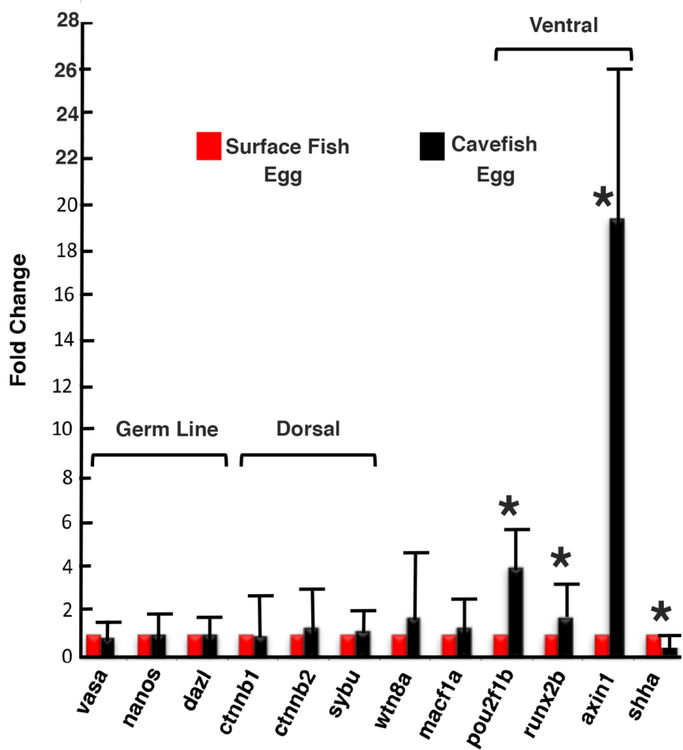
The evidence for maternal effects revealed by reciprocal hybridizations prompted an investigation of possible differences in maternal mRNAs in unfertilized SF and CF eggs. Since maternal effect genes have not been identified in A. mexicanus, the approach was to compare mRNA corresponding to the SF and CF orthologs of zebrafish maternal effect genes (Landon and Mullins, 2011). We focused on maternal effect genes involved in embryonic dorsoventral axis determination because of changes in CF and CF X SF hybrid optic polarity. Accordingly, ß-catenin (ctnnb1 and ctnnb2) and syntabulin (sybu) were selected as examples of dorsal fate determination genes, and axin1, pou2f1b, and runx2b were compared as ventral fate determination genes. We also surveyed macf1a, wtn8a, vasa, nanos, and dazl as additional maternal effect genes, and included sonic hedgehog (shha) in the analysis as a typical zygotically expressed gene. The Astyanax orthologs of the corresponding zebrafish genes were identified in the CF genome database (McGaugh et al., 2014), gene specific primers were designed, and transcript levels were compared by qPCR using cDNA templates prepared from unfertilized egg RNA of at least two different SF and CF individuals (Fig. 7). No significant differences were found in the levels of vasa, dazl, and nanos mRNAs, which are involved in germ line determination and unexpected to differ in SF and CF eggs, macf1a and wtn8a mRNAs, or ctnnb1, ctnnb2, and sybu mRNAs, which are involved in dorsal determination (Fig. 7). In contrast, the maternal transcripts of the runx2b, pou2f1b, and axin1genes, which are involved in ventral determination, were significantly increased in CF relative to SF eggs. The increases in ventralizing mRNAs in CF relative to SF eggs ranged from about 2 to 20 fold (Fig. 7). Maternal sonic hedgehog (shh) transcripts were also detected in unfertilized eggs, and were significantly increased in SF relative to CF eggs (Fig. 7).
Figure 7.
Quantitative real-time RT-PCR analysis of transcripts encoded by maternal effect genes and the shha gene in surface fish and cavefish unfertilized eggs. Brackets show maternal transcripts encoded by classes of genes involved in germ line determination, dorsal embryonic fate determination, and ventral embryonic fate determination. Error bars: Standard derivation of the mean fold change. * denotes significance at p < 0.05 calculated from ΔΔCt values.
Since the detection of maternal shh mRNA was unexpected, we conducted in situ hybridization to confirm that shh transcripts were present in eggs, rather than in contaminating follicle cells or somatic cells. In these experiments, the distribution of shh mRNA was compared with dazl mRNA, whose localization is well characterized in zebrafish oocytes and cleaving embryos (Maegara et al., 1999; Howley and Ho, 2000). Maternal dazl mRNA was detected in both the vegetal cortical and animal pole regions of SF and CF eggs and embryos (Fig. 8A-H), as described in zebrafish. In contrast, shh mRNA was restricted to the animal pole region and excluded from other parts of unfertilized eggs and cleaving blastomeres throughout the early cleavage stages (Fig. 8I-P). Consistent with the qPCR results (Fig. 7), the shh signal was stronger in SF than in CF eggs and cleaving embryos (Fig. 8I-P). The results confirm the existence of maternal shh mRNA in A. mexicanus.
Figure 8.
In situ hybridization showing the distribution of dazl (A-H) and shh (I-P) mRNA in surface fish (A-D, I-L) and cavefish (E-H, M-P) eggs and early cleavage stage embryos. Eggs and embryos are oriented with the animal pole at the top except for B, F, J, H in which the animal pole is facing the viewer. A, B, E, F, I, J, M, N: One cell stage. C, G, K, O: Two cell stage. D, H: Four cell stage. L-P: Eight cell stage. Arrows indicate vegetal cortical localization of dazl mRNA. Scale bar: 300 μm.
Discussion
This study has revealed maternal genetic effects during development of the teleost Astyanax mexicanus, a single species consisting of two morphs with dramatically different phenotypes. For detecting maternal genetic effects, we capitalized on the inter-fertility of Astyanax morphs and compared the phenotypes of reciprocal hybrids: F1 hybrids produced by inseminating SF eggs with CF sperm and CF eggs with SF sperm. We expected that most of the phenotypic differences between CF and SF would be caused by mutations in genes that are expressed after the initiation of zygotic transcription and therefore result in the development of intermediate and/or equivalent phenotypes in reciprocal hybrids. However, we were also cognizant of the inheritance of vibration attraction behavior (VAB) and its underlying neuromast sensory system in A. mexicanus, which show parental genetic effects controlled either by the sperm (a paternal genetic effect) or the egg (a maternal genetic effect) in various CF populations (Yoshizawa et al., 2012). The major finding of the present investigation is that some of the phenotypic differences between CF and SF are overexpressed in CF X SF hybrids relative to SF X CF hybrids and therefore reveal an influence of maternal genetic effects in the evolution of CF development.
Maternal Genetic Effects Revealed by Reciprocal Hybridization
The role of maternal genetic effects in different aspects of the degenerative CF eye phenotype is strongly supported by several lines of evidence from reciprocal hybridization experiments. First, the reduction of the ventral retina and displacement of the lens to the ventral side of the optic cup was observed frequently in CF X SF hybrids but not in SF X CF hybrids. The optic ventralization phenotype exhibited by CF X SF hybrids is a characteristic of CF embryonic eye primordia (Yamamoto et al., 2004; Jeffery, 2009; Yamamoto, 2016). Second, the lens and eyeball were significantly smaller in CF X SF hybrids than in SF X CF hybrids, a difference that persists into the late larval stages and is still apparent in some adult hybrids. Small lenses and eyeballs are another characteristic of CF larvae (Jeffery, 2009; Yamamoto, 2016). Third, apoptosis is more prevalent in the developing lens of CF X SF hybrids than SF X CF hybrids. Lens apoptosis is rarely seen in SF embryos and considered to be an important regulatory step in the control of CF eye degeneration (Yamamoto and Jeffery, 2000; Strickler et al. 2007a; Hinaux et al., 2014, 2017; Ma et al., 2014). Fourth, the lens of CF X SF hybrids shows striking differences in gene expression that are absent in SF X CF hybrids. One difference is that gammaM2-crystallin expression occurs primarily in primary fiber cells located in the center of the lens in CF X SF hybrids, a typical feature of the CF lens, rather than in the epithelial layer of the lens, a hallmark of the SF lens (Strickler et al., 2007b). Another difference is the restriction of hsp90alpha expression to the lens of CF X SF hybrids. Expression of the hsp90alpha gene is unique to the CF lens (Hooven et al., 2004). Furthermore, the cryaa gene, which is strongly expressed in the SF lens but downregulated in the CF lens (Strickler et al., 2007b; Ma et al., 2014; Hinaux et al., 2015), was expressed more weakly, or in some cases not detectable, in the lens of CF X SF hybrids compared to SF X CF hybrids. Lastly, the sclera of CF X SF hybrids was smaller than SF X CF counterparts. The sclera grows in concert with the eyeball, possibly under direct or indirect control by the lens (Yamamoto et al., 2003), and size differences of the sclera in reciprocal hybrid juveniles are indicative of long lasting maternal effects on optic development. In summary, morphological, physiological, and molecular differences all support the role of maternal genetic effects in the regressive CF eye phenotype, and particularly in lens degeneration.
A reciprocal hybridization experiment was recently conducted in A. mexicnaus to investigate the relationship between lens and retinal development (Hinaux et al., 2017). This study reported that the lens and eyeball were slightly smaller in CF X SF hybrids than in SF X CF hybrids at 36-hours post-fertilization, but the differences were not significant (Fig. 1 in Hinaux et al., 2017), possibly due to the relatively small number of hybrids measured and their probable origin from a single cross. The significant differences between eye phenotypes reported in the present investigation were determined in 26 reciprocal crosses resulting a total of 689 hybrid progeny generated from 15 different parental families. Thus, the use of multiple parents from different families can increase the power of reciprocal hybridization for the detection of parental genetic effects.
In contrast to CF optic degeneration, some constructive and regressive CF phenotypes did not appear to be regulated by maternal genetic effects. Similar numbers of melanin containing pigment cells were detected in larvae and juveniles of reciprocal hybrids, suggesting control by zygotic effects that are dependent on contributions of both sperm and eggs. This conclusion is supported by earlier studies in which mutations in oca2, a gene exhibiting a zygotic expression pattern in SF (Bilandzija et al., 2013; Ma et al., 2015), was demonstrated to be responsible for CF albinism (Protas et al., 2006). In addition, the mandibles, which are enlarged in CF oral jaws (Yamamoto et al., 2009), showed intermediate and equivalent phenotypes in reciprocal hybrids, suggesting that this constructive CF phenotype is also under zygotic control.
The results suggest that some of the evolutionary changes involved CF eye regression occur in the genome of the mother during oogenesis, whereas others are controlled by zygotic transcription initiated after the mid-blastula transition. In future studies, it will be important to determine whether other typical CF phenotypes, such as regional changes in the brain (Menuet et al., 2007; Rétaux et al., 2016), differences in olfactory sensitivity (Hinaux et al., 2016), and behavioral modifications (Duboué et al., 2011; Kowalko et al., 2013; 2014; Yoshizawa et al. 2015), are controlled by maternal or zygotic effects. The study of parental genetic effects in these and other phenotypes will be possible using the reciprocal hybridization approach described here.
Maternal mRNA and Ventralizing Factors in Cavefish Eggs
We reasoned that if maternal genetic effects play an important role in the degenerative CF phenotype they could be reflected by different levels of maternal transcripts. Since no maternal effect genes have been identified in Astyanax, we turned to zebrafish (Pelegrl, 2003; Harvey et al., 2013) to address this question. Accordingly, we compared transcripts encoded by the orthologous A. mexicanus maternal effect genes that regulate dorsal, ventral, and germ line determination in zebrafish. Most of the maternal mRNAs we surveyed, including transcripts of the germ line vasa, nanos, and dazl genes, the wnt8 and macf1a genes, and the ß-catenin (ctnnb1 and ctnnb2) and sybu dorsalizing genes (Kelly et al., 2000; Bellipanni et al., 2006; Nojima et al., 2010), showed no differences in abundance between CF and SF eggs. In contrast, the maternal mRNAs encoded by the ventralizing genes axin1, pou2f1b and runx2b were more abundant in CF eggs. Although the conserved function of the maternal genes involved in embryonic dorsoventral axis formation will require confirmation in Astyanax, we consider it unlikely that the reproducible differences discovered here could be coincidental. Therefore, based on the current information, we conclude that changes in dorsoventral polarity may have occurred during CF evolution.
The zebrafish ventralizing genes pou2f1b and runx2b encode transcription factors that function upstream of the bmp2b/4/7 genes (Reim and Brand, 2006), which are responsible for establishing the embryonic ventral gradient, or are direct transcriptional activators of the vox/vent/ved genes, which promote ventral fates by inhibiting dorsal fates in the zebrafish embryo (Flores, et al., 2008), respectively. Likewise, the axin1 gene, which showed the largest fold difference in transcripts in CF relative to SF eggs, encodes a factor that interferes with ßcatenin/Wnt mediated specification of dorsal fates by promoting ß-catenin degradation (Schneider et al., 2012). The increased titers of pou2f1b, runx2b, and axin1 mRNAs in CF eggs brings up the possibility that ventral fates may be expanded at the expense of dorsal fates in CF embryos by a concerted exacerbation of the BMP morphogen gradient and interference with the function of the dorsalizing system. If so, an intriguing prediction of this hypothesis would be a change in the activity of the CF organizer.
Maternal Sonic Hedgehog mRNA
The shh gene is expressed zygotically during development of zebrafish (Krause et al. 1993) and other vertebrates (Ingham and McMahon, 2001). Thus, the discovery of maternal shh transcripts in Astyanax was unexpected. Maternal shh mRNA was concentrated in the animal pole region of unfertilized eggs and cleaving embryos. We note that maternal shh mRNA has also been found in the animal pole region of another cyprinid teleost, the carp Cyprinus carpo (Wang et al., 2007). Therefore, maternal expression of shh may differ among teleosts, with maternal shh transcripts present in carp and A. mexicanus, but apparently absent in zebrafish.
The current work does not provide clues about the function of maternal shh mRNA in A. mexicanus, or why it is present at higher levels in SF eggs than in CF eggs. We speculate that the Shh morphogen could have an unknown signaling function during oogenesis; for example, during the development of oocytes and the surrounding follicle cells, and that this function has regressed in CF oocytes. It is also possible is that maternal shh transcripts are masked in unfertilized eggs, actively translated after fertilization, and have an unknown role in embryonic development, which could have been lost or modified in CF. Finally, we cannot exclude the possibility that maternal shh transcripts are the result of low levels of background gene activity during oogenesis. To resolve these issues, it will be important to determine whether Shh protein is present and localized in A. mexicanus oocytes and cleaving embryos.
Concluding Remarks
Multiple genes are involved in CF eye loss (Borowsky and Wilkens, 2002), and attention has recently focused on identifying the mutated genes responsible for CF lens and eye regression (Protas et al, 2007; O’ Quin et al., 2013; McGaugh et al., 2014; Ma et al. 2014). Based on aligning QTL with the CF genome, numerous candidates for mutated eye loss genes have now been identified (McGaugh et al., 2014). However, these candidate genes were recognized based on zygotic functions in eye development. The present results suggest that the search for mutated eye genes might benefit from expansion to include maternal effect genes.
The increase in maternal mRNAs in CF eggs is likely due to differential transcriptional regulation during oogenesis, although other possibilities such as differential mRNA processing or stability cannot be excluded. We note that epigenetic control based on gene imprinting would also be consistent with maternal gene expression changes and the phenotypic differences between reciprocal hybrids (Wolf et al., 2014). It has recently been discovered that DNA methylation is used widely for epigenetic gene modification in CF, particularly to silence the genes governing eye development (Gore et al., 2018).
Our demonstration of the importance of maternal genetic effects implies that some of the cave adapted phenotypes of A. mexicanus may be controlled by evolutionary changes during oogenesis. Differences in gene expression during oogenesis may reflect some of the first steps of a process that eventually leads to the regressive eye phenotype in CF embryos. Maternal genetic effects could broadly impact many different events that occur downstream during embryonic development and thus coordinate multiple changes in CF phenotypic evolution.
Supplementary Material
Highlights.
Cavefish regressive optic phenotypes show maternal genetic effects
Cavefish regressive pigmentation and constructive cranial skeletal phenotypes do not show maternal genetic effects
Maternal transcripts of ventral patterning genes are increased in unfertilized cavefish eggs
Maternal sonic hedgehog transcripts are present in unfertilized Astyanax eggs
Acknowledgements
This research was supported by National Institutes of Health grant EY024941 to WRJ. We are grateful to Deidre Heyser, Craig Foote, and Ruby Dessiatoun for fish maintenance and husbandry and to Mandy Ng for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams EW, Mullins MC. Early zebrafish development: It’s in the maternal genes. Cur Opin Genet Dev. 2009; 19:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alunni A, Meneut A, Candal E, Penigault J-B, Jeffery WR, Rétaux S. Developmental mechanisms for retinal degeneration in the blind cavefish Astyanax mexicanus. J Comp Neurol. 2007; 505:221–233. [DOI] [PubMed] [Google Scholar]
- Atukorata ADS, Franz-Odendaal TA. Biology and Evolution of the Mexican Cavefish Academic Press, New York, USA: 2016. Chapter 11. Evolution and development of the cavefish oral jaws: Adaptations for feeding. Pp 209–222. [Google Scholar]
- Badyaev AV. Maternal effects as generators of evolutionary change. A reassessment. Ann NY Acad Sci. 2008; 1133:151–161. [DOI] [PubMed] [Google Scholar]
- Bellipanni G, Varga M, Margawa S, Imai S, Kelly C, Pomrehn Meyers A, Chu F, Talbot WS, Weinberg ES. Essential and opposing roles of zebrafish ß-catenins in the formation of dorsal axial structures and neuroectoderm. Development 2006; 133:298–309. [DOI] [PubMed] [Google Scholar]
- Bilandzija H, Ma L, Parkhurst A, Jeffery WR. A potential benefit of albinism in Astyanax cavefish: Downregulation of the oca2 gene increases L-tyrosine and catecholamine levels as an alternative to melanin synthesis. PLoS One 2013; 8 (11):e80823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky R In vitro fertilization of Astyanax mexicanus. Cold Spr Harb Protoc. 2008; doi; 10.1101/pdb.prot5092 [DOI] [PubMed] [Google Scholar]
- Borowsky R, Wilkens H. Mapping a cavefish genome: polygenic systems and regressive evolution. J Hered. 2002; 93: 19–21. [DOI] [PubMed] [Google Scholar]
- Boycott AE, Diver C, Garstang SL, Turner FM. The inheritance of sinistrality in Limnaea pergra (Mollusca, Pulmonata). Philos Trans R Soc Lond B Biol Sci. 1930; 219:51–130. [Google Scholar]
- Davidson EH. Gene Activity in Early Development. Third Edition, Academic Press; New York, USA: 1968 [Google Scholar]
- Duboué ER, Keene AC, Borowsky RL. Evolutionary convergence on sleep loss in cavefish populations. Curr Biol. 2011; 8:671–676. [DOI] [PubMed] [Google Scholar]
- Elipot Y, Hinaux H, Callebert J, Launay J-M, Blin M, Rétaux S. A mutation in the enzyme monoamine oxidase explains part of the Astyanax cavefish behavioral syndrome. Nature Commun. 2014; 5:3647 doi: 10.1038/ncomms4647 [DOI] [PubMed] [Google Scholar]
- Erickson L, Kemble R. Paternal inheritance of mitochrondria in rapeseed (Brassica napus). Mol Gen Genet. 1990; 22:135–139. [DOI] [PubMed] [Google Scholar]
- Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007; 23:192–199. [DOI] [PubMed] [Google Scholar]
- Flores MV, Lam EY, Crosier KE, Crosier PS. Osteogenic transcription factor Runx2 is a maternal determinant of dorsoventral patterning in zebrafish. Nat Cell Biol. 2008; 10:346352. [DOI] [PubMed] [Google Scholar]
- Freeman G, Lundelius JW. The developmental genetics of dextrality and sinistrality in the gastropod Lymnaea peregra. Roux Arch Dev Biol. 1982; 191:69–83. [DOI] [PubMed] [Google Scholar]
- Gore AV, Tomins KA, Iben J, Ma L, Castranova D, Davis AE, Parkhurst A, Jeffery WR, Weinstein BM. An epigenetic mechanism for cavefish eye degeneration. Nat Eco Evol. 2018; doi: 10.1038/s441559-018-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB. The complex origin of Astyanax cavefish. BMC Evol Biol. 2012; 12:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SA, Sealy I, Kettleboroug R, Fenyes F, White R, Stemple D, Smith JC. Identification of the zebrafish maternal and paternal transcriptomes. Development 2013; 27032710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J Maternal determinants of embryonic cell fate. Sem Cell Dev Biol. 2006; 17:93–98. [DOI] [PubMed] [Google Scholar]
- Hinaux H, Blin M, Fumey J, Legendre L, Casane D, Rétaux S. Lens defects in Astyanax mexicanus cavefish: evolution of crystallins and a role for alphaAcrystallin. Dev Neurobiol. 2015; 75:505–521. [DOI] [PubMed] [Google Scholar]
- Hinaux H, Devos L, Blin M, Elipot Y, Alié A, Rétaux S. Sensory evolution in blind cavefish is driven by early embryonic events during gastrulation and neurulation. Development 2016; 143:4521–4532. [DOI] [PubMed] [Google Scholar]
- Hooven TA, Yamamoto Y, Jeffery WR. Blind cavefish and heat shock protein chaperones: a novel role for hsp90α in lens apoptosis. Int J Dev Biol. 2004; 48:731–738. [DOI] [PubMed] [Google Scholar]
- Howley C, Ho RK. mRNA localization patterns in zebrafish oocytes. Mech Dev. 2000; 92:305–309. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001; 15:3059–3087. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Determinants of cell and positional fate in ascidian embryos. Int Rev Cytol. 2001a; 203:3–62. [DOI] [PubMed] [Google Scholar]
- Jeffery WR Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001b; 231:1–12. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Evolution and development in the cavefish Astyanax. Curr Top Dev Biol. 2009; 86:191–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, Martasian D. Evolution of eye regression in the cavefish Astyanax: Apoptosis and the Pax-6 gene. Amer Zool. 1998; 38:685–696. [Google Scholar]
- Jeffery WR, Strickler AG, Guiney S, Heyser DG, Tomarev SI. Prox 1 in eye degeneration and sensory organ compensation during development and evolution of the cavefish Astyanax. Dev Genes Evol. 2000; 210:223–230. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Ma L, Parkhurst A, Bilandzija H. Biology and Evolution of the Mexican Cavefish Academic Press, New York, USA: 2016. Chapter 8. Pigment regression and albinism in Astyanax cavefish. Pp 155–173. [Google Scholar]
- Kelly C, Chin AJ, Leatherman JL, Koslowsky DJ, Weinberg ES. Maternally controlled (beta)-catenin-mediated signaling is required for organizer formation in the zebrafish. Development. 2000; 127: 3899–3911. [DOI] [PubMed] [Google Scholar]
- Kowalko JE, Rohner N, Linden TA, Rompani SB, Warren WC, Borowsky R, Tabin CJ, Jeffery WR, Yoshizawa M Convergence in feeding posture occurs through different genetic loci in independently evolved cave populations of Astyanax mexicanus. Proc Natl Acad Sci USA 2013; 110: 16933–16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalko JE, Rohner N, Rompani SB, Peterson BK, Linden T, Yoshizawa M, Kay EH, Hoekstra HE, Jeffery WR, Borowsky R, Tabin CJ. Genetic analysis of the loss of schooling behavior in cavefish reveals both sight-dependent and independent mechanisms. Curr Biol. 2013; 23:1874–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hedgehog is expressed in tissues with polarizing activity in zebrafish embryos. Cell 1993; 75:1432–1444. [DOI] [PubMed] [Google Scholar]
- Krishnan J, Rohner N. Cavefish and the basis for eye loss. Philos Trans R Soc B. 2017; 372:20150487 10.1098/rstb.2015.0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lain X, Luo G, Hui L, Xu W, Xiao Y, Bi X. Reciprocal difference of interspecific hybridization between three different colours of Iris dichotoma and I. domestica. J. Hort Sci Biotech. 2016; 91:483–490. [Google Scholar]
- Landon YG, Mullins MG (2011). Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annu Rev Genet. 2011; 45:357–377. [DOI] [PubMed] [Google Scholar]
- Ma L, Parkhurst A, Jeffery WR. The role of a lens survival pathway including sox2 and αA-crystallin in the evolution of cavefish eye degeneration. EvoDevo 2014; 5:28 doi: 10.1186/2041-9139-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Essner JJ, Jeffery WR, Kowalko JE. Genome editing using TALENS in blind Mexican cavefish, Astyanax mexicanus. 2015; PLoS One 16;10:e0119370. doi: 10.1371/journal.pone.0119370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Yasuda K, Inoue K. Maternal mRNA localization of zebrafish DAZ-like gene. Mech Dev. 1999; 81:223–236. [DOI] [PubMed] [Google Scholar]
- Marshak A Growth differences in reciprocal hybrids and cytoplasmic influence on growth in mice. J Exp Zool. 1936; 72:497–510. [Google Scholar]
- McGaugh S, Gross J, Aken B, Blin M, Borowsky R, Chaopin D, Hinaux H, Jeffery WR, Keene A, Ma L, Minx P, Murphy D, O’Quin KE, Rétaux S, Rohner N, Searle SMJ, Stahl BA, Tabin C, Volff J-N, Yoshizawa M, Warren WC. The cavefish genome reveals candidate genes for eye loss. Nature Commun. 2014; 5:5307 doi: 10.1038/ncomms6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuet A, Alunni A, Joly J-S, Jeffery WR, Rétaux S. Shh overexpression in Astyanax cavefish: multiple consequences on forebrain development and evolution. Development 2007; 134:845–855. [DOI] [PubMed] [Google Scholar]
- Nasiadka A, Dietrich BH, Krause HM. Anterior-posterior patterning in Drosophila embryos. Adv Dev Biol Biochem. 2002; 12:155–204. [Google Scholar]
- Nojima H, Rothhämel S, Shimizu T, Kim CH, Yonemura S, Marlow FL, Hibi M. Syntabulin, a motor protein linker, controls dorsal determination. Development 2010; 137:923–933. [DOI] [PubMed] [Google Scholar]
- O’ Quin KE, Yoshizawa M, Doshi P, Jeffery WR. Quantitative genetic analysis of retinal degeneration in the blind cavefish. PLoS One 2013; 8(2): e57281. doi: 10.1371/journal.pone.0057281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Quin KE, Doshi P, Lyon A, Hoenemeyer E, Yoshizawa M, Jeffery WR. Complex evolutionary and genetic patterns characterize the loss of scleral ossification in the blind cavefish Astyanax mexicanus. PLoS One. 2015; 10(12): e0142208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrl F, Maternal factors in zebrafish development. Dev Dyn. 2003; 228:535–554. [DOI] [PubMed] [Google Scholar]
- Powers AK, Kaplan SA, Boggs TE, Gross JB. Facial bone fragmentation in blind cavefish arises through two unusual ossification processes. Sci Rep. 2018; 8:7015 DOI: 10.1038/s41598-018-25107-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, Borowsky R, Tabin CJ. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006; 38:107–111. [DOI] [PubMed] [Google Scholar]
- Protas M, Conrad M, Gross JB, Tabin C, Borowsky R Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007: 17:452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim G, Brand M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POE domain protein Spg/Pou2/Oct4. Development. 2006; 133:2757–2770. [DOI] [PubMed] [Google Scholar]
- Rétaux S, Alié A, Blin M, Devos L, Elipot Y, Hinaux H. Biology and Evolution of the Mexican Cavefish Academic Press, New York, USA: 2016; Chapter 12. Neural development in Astyanax mexicanus: Comparing cavefish and surface fish brains. Pp. 227–241. [Google Scholar]
- Schneider PN, Slusarski DC, Houston DW. Differential role of axin RGS domain function in Wnt signaling during anteroposterior patterning and maternal axis formation. PLoS One 2012; 7(9): e44096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Tang K, Osborn TC. Development of synthetic Brassica amphidiploids by reciprocal hybridization and comparison to natural amphidiploids. Theoret Appl Genet. 1993; 86:811–821. [DOI] [PubMed] [Google Scholar]
- Stanton BJ. The effect of reciprocal hybridization on reproduction of the intersectional cross, Populus x Generosa. For Genet. 2005; 12:131–140. [Google Scholar]
- Strickler AG, Yamamoto Y, Jeffery WR. The lens controls cell survival in the retina. Evidence from the blind cavefish Astyanax. Dev Biol. 2007a; 311, 512–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickler AG, Byerly MS, Jeffery WR. Lens gene expression analysis reveals downregulation of the anti-apoptotic chaperone A crystallin during cavefish eye degeneration. Dev. Genes Evol. 2007b; 217:771–782. [DOI] [PubMed] [Google Scholar]
- Wang N, Sun Y-H, Liu J, Zuo Z-Y. Molecular characterization of common carp (Cyprinus carpio) Sonic Hedgehog and discovery of its maternal expression. Dev Genes Evol. 2007; 217:299–305. [DOI] [PubMed] [Google Scholar]
- Wassersug RJ. A procedure for differential staining of cartilage and bone in whole formalin-fixed vertebrates. Stain Technol. 1983; 51:131–134. [DOI] [PubMed] [Google Scholar]
- Wilkens H. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces)-Support for the neutral mutation theory. Evol Biol. 1988; 23:271–367. [Google Scholar]
- Wolf JB, Oakey RJ, Fell R. Imprinted gene expression in hybrids: perturbed mechanism and evolutionary implications. Heredity 2014: 113:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Wade MJ. Evolutionary genetics of maternal effects. Evolution 2016; 70:827839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y. Biology and Evolution of the Mexican Cavefish Academic Press, New York, USA: 2016. Chapter 9. Molecular mechanisms of eye degeneration in cavefish. Pp 175–189. [Google Scholar]
- Yamamoto Y, Espinasa L, Stock DW, Jeffery WR. Development and evolution of craniofacial patterning is mediated by eye-dependent and –independent processes in the cavefish Astyanax. 2003; Evol Dev. 5:435–446. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Stock DW, Jeffery WR. Hedgehog signaling controls eye degeneration in blind cavefish. Nature 2004; 431:844–847. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Byerly MS, Jackman WR, Jeffery WR. Pleiotropic functions of embryonic sonic hedgehog expression link jaw and taste bud amplification with eye loss during cavefish evolution. Dev Biol. 2009; 330:200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Goricki S, Soares D, Jeffery WR. Evolution of a behavioral shift mediated by superficial neuromasts helps cavefish find food in darkness. Curr Biol . 2010; 20:1631–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Ashida G, Jeffery WR. Parental genetic effects in a cavefish adaptive behavior explain disparity between nuclear and mitochondrial DNA. Evolution 2012; 66: 2975–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Yamamoto Y, O’Quin KE, Jeffery WR. Evolution of an adaptive behavior and its sensory receptors promotes eye regression in blind cavefish. BMC Biol. 2012; 10:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Robinson BG, Duboué ER, Masek P, Jaggard JB, O’Quin KE, Borowsky RL, Jeffery WR, Keene A. Distinct genetic architecture underlies the emergence of sleep loss and prey-seeking behavior in the Mexican cavefish. BMC Biol. 2015; 13:15 doi 10.1186/s12915-015-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakas C, Rockman MV. Dimorphic development in Streblospino benedicti: genetic analysis of morphological differences between larval types. Int J Dev Biol. 2014; 58:593–599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.