Abstract
Mutations that disrupt the inwardly rectifying potassium channel Kir2.1 lead to Andersen-Tawil syndrome that includes periodic paralysis, cardiac arrhythmia, cognitive deficits, craniofacial dysmorphologies and limb defects. The molecular mechanism that underlies the developmental consequences of inhibition of these channels has remained a mystery. We show that while loss of Kir2.1 function does not affect expression of several early facial patterning genes, the domain in which Pou3f3 is expressed in the maxillary arch is reduced. Pou3f3 is important for development of the jugal and squamosal bones. The reduced expression domain of Pou3f3 is consistent with the reduction in the size of the squamosal and jugal bones in Kcnj2KO/KO animals, however it does not account for the diverse craniofacial defects observed in Kcnj2KO/KO animals. We show that Kir2.1 function is required in the cranial neural crest for morphogenesis of several craniofacial structures including palate closure. We find that while the palatal shelves of Kir2.1-null embryos elevate properly, they are reduced in size due to decreased proliferation of the palatal mesenchyme. While we find no reduction in expression of BMP ligands, receptors, and associated Smads in this setting, loss of Kir2.1 reduces the efficacy of BMP signaling as shown by the reduction of phosphorylated Smad 1/5/8 and reduced expression of BMP targets Smad6 and Satb2.
Introduction
Inwardly rectifying potassium (Kir/Irk) channels are best known for their role in setting resting membrane potential in neurons, muscle, and pancreatic beta cells[1–3]. Through their effects on membrane potential, Kir channels regulate neuron firing, muscle contraction/relaxation, and insulin release. Mutations that disrupt several different ion channels lead to syndromes of birth defects in humans and in several other organisms, but the molecular mechanism that underlies developmental consequences of these defects has remained a mystery. For example, Andersen-Tawil Syndrome is caused by mutations in KCNJ2, which encodes Kir2.1, with patients exhibiting periodic paralysis, cardiac arrhythmia, and cognitive deficits, dental defects, cleft lip and palate, micrognathia, hypertelorism, low set ears, and limb patterning defects including syndactyly, clinodacyly and brachydactyly [4–7]. Disruption of another member of the Kir/Irk superfamily, Kir7.1 (encoded by Kcnj13) also leads to cleft palate and other craniofacial defects suggesting that multiple Kir channels play a significant role in palate closure and craniofacial development [8]. While the periodic paralysis and cardiac arrhythmia both are logical consequences of a mutation in a potassium channel, it is less apparent why mutations that disrupt Kir2.1 and Kir7.1 would cause defects in morphological development. This requirement for Kir channels in development is conserved across organisms. For example, disruption of Kir2.1 also leads to limb defects, hypoplastic craniofacial structures, and cleft palate in mice, craniofacial defects in frogs, and morphological defects in flies [9–11].
Genetic models provide insight into potential mechanisms by which ion channels may influence morphological development. For example, the Kcnj2KO/KO mouse has the same polydactyly phenotype as limb-specific removal of BMP4 or BMP2 [10, 12]. The complete cleft of the secondary palate is similar to the defects that occur when the BMP2/4 receptor, BMPR1A, is deleted from the cranial neural crest (CNC) that will make up the palate [10, 13]. Deletion of BMP2 and 4 from the CNC cells also results in similar phenotypes including an enlarged fontanella and a smaller mandible [14]. In frogs, injection of a dominant negative Kir2.1 subunit disrupts expression of early craniofacial patterning genes [9]. In flies, disruption of Irk channels leads to wing defects reminiscent of those that occur when fly BMP ligands (Dpp and Gbb) or receptors (Punt and Thick veins) are disrupted [10, 15–20]. The similarities between phenotypes when Kir channels are disrupted and BMP signaling is disrupted suggest that Kir channels could act in the same pathway as BMP signaling.
BMP signaling is initiated when BMP binds a complex of type 1 and type 2 serine-threonine kinase receptors, the type 1 receptor phosphorylates receptor associated Smads 1, 5, or 8. The phosphorylated Smad (p-Smad) can then associate with Smad 4 and translocate to the nucleus where they can bind BMP response elements to activate transcription. Kir channels are important for BMP signaling in Drosophila [10, 21]. Specifically, Kir/Irk channels regulate the secretion of BMP [21]. Reduction of Kir/Irk channel function reduced phosphorylation of Mad, a Drosophila receptor associated Smad, and expression of Dpp transcriptional targets [10, 21]. Mammalian Kir2.1 can substitute/rescue developmental consequences of loss of native Drosophila Kir/Irk channels [21].
The goal of this work was to determine how Kir2.1 contributes to mammalian craniofacial development. Here we show that Kir2.1 is important for morphogenesis of several craniofacial structures. We find that loss of Kir2.1 function reduces proliferation of the palatal shelf mesenchyme to result in cleft of the secondary palate. We show that Kir2.1 is required in the cranial neural crest and not in the ectoderm for its developmental role. Lastly, we show that BMP signaling is reduced in developing craniofacial structures of Kcnj2KO/KO embryos compared to their wild type and heterozygous littermates. These data show that similar to flies, Kir2.1 is important for BMP signaling in mice.
Results
Kir2.1 is important for development of several craniofacial structures
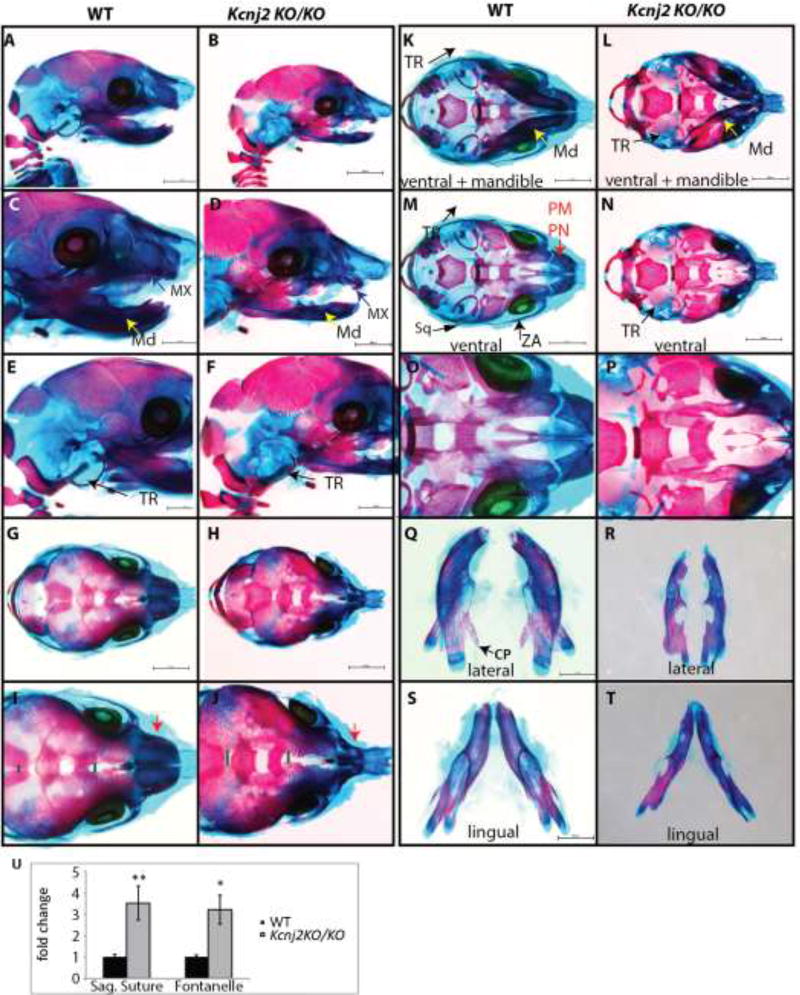
Several structures that derive from cranial neural crest cells develop abnormally in Kcnj2KO/KO animals. For example, embryonic day (E)18.5 Kcnj2KO/KO embryos have hypoplastic pre-nasal, pre-maxilla, nasal bones, and maxilla. The majority of the tympanic ring does not form in Kcnj2KO/KO animals (Fig. 1A–F). Components of the zygomatic arch (the maxillary and squamosal zygomatic processes and the basispenoid bone), and the squamosal and jugal bones are reduced in size in Kcnj2KO/KO mice compared to their wild type siblings (Fig. 1C,D). Kcnj2KO/KO mandibles are reduced in size and lack the coronoid process (Fig. 1Q–T). Kcnj2KO/KO have an enlarged anterior fontanelle compared to their wild type siblings (Fig.1G–J, U). Furthermore, the sagittal suture is wider in Kcnj2KO/KO embryos than their wild type siblings (Fig.1G–J, U). Kcnj2KO/KO mice also have a well-documented cleft of the secondary palate and limb patterning defects [10, 11]. All of these phenotypes were fully penetrant among all of the analyzed Kcnj2KO/KO embryos including males and females (n=8 of each genotype).
Figure 1.
Alizarin red and Alcian blue skeletal stains of E18.5 WT (A, C, E, G, I, K, M, O, Q, S, U) and Kcnj2KO/KO (B, D, F, H, J, L, N, P, R, T, V) mice shows that deletion of Kcnj2 causes hypoplastic mandible (MD, yellow arrow), maxilla (MX, purple arrow), premaxilla and prenasal bones (PM, PN red arrow), components of the zygomatic arch (ZA, green arrow), and tympanic ring (TR, black arrow). Skeletons are represented in lateral views (A–F), dorsal views (G–J), ventral views with mandible attached (K, L), and without mandible attached (M, N). Mandibles are compared in lingual (O, P), and lateral (Q, R), and lingual (S, T) views. Seven Kcnj2KO/KO skeletons were analyzed and compared to six wild type skeletons. The sagittal sutures and fontanelles are significantly larger in Kcnj2KO/KO compared to WT siblings, p= 0.006 and p= 0.03 by T-test (U).
Early patterning of the mouse face is grossly normal in Kcnj2KO/KO embryos
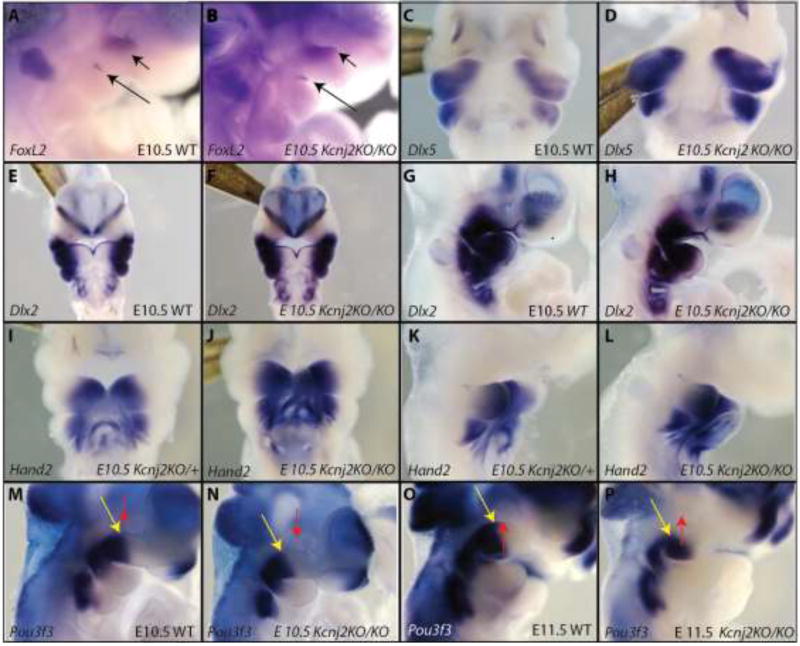
To determine whether Kir2.1 functions in early patterning of the embryonic face, we performed RNA in situ hybridization to detect transcripts of several craniofacial patterning genes in Kcnj2KO/KO and control littermates. FoxL2 is expressed just below the eye in the dorsal maxillary first pharyngeal arch and at the mandible maxillary junction [22]. We found that loss of Kir2.1 function did not affect expression of FoxL2 at E10.5 (Fig. 2A, B). Dlx5 marks the mandibular process and was unchanged in Kcnj2KO/KO embryos at E 10.5 (Fig.2C, D). Similarly, loss of Kir2.1 did not affect expression domains of two wide spread craniofacial markers Hand2 and Dlx2 (Fig. 2E–L). Pou3f3 is broadly expressed in the mesenchyme of the maxillary portion of the first pharyngeal arch[22]. Deletion of Kcnj2 resulted in a posterior and ventral shift of the Pou3f3 boundary in the maxillary processes of E10.5 and E11.5 embryos (Fig. 2M–P). These data show that loss of Kir2.1 causes a specific domain change in gene expression rather than a general change in several different patterning genes.
Figure 2.
RNA in situ hybridization shows early craniofacial patterning genes in kcnj2KO/KO mice compared to WT controls. Lateral views of WT (A) and Kcnj2KO/KO (B) show similar expression of FoxL2 (black arrows). Ventral views of WT (C) and Kcnj2KO/KO (D) show similar expression of Dlx5. Ventral views of WT (E) and Kcnj2KO/KO (F) and lateral views of E10.5 WT (G) and Kcnj2KO/KO (H) show that loss of Kir2.1 function does not affect Dlx2 expression. Ventral views of WT (I) and Kcnj2KO/KO (J) E10.5 embryos show similar expression of Hand2. Lateral views of WT (K) and Kcnj2KO/KO (L) E10.5 embryos show similar expression of Hand2. Lateral views of WT (M) and Kcnj2KO/KO (N) E10.5 embryos show a slightly reduced domain of Pou3f3 expression (yellow arrow, compared to red arrow marking the eye) so that it does not extend to under the eye. Lateral views of WT (O) and Kcnj2KO/KO (P) show posterior shift in the domain of Pou3f3 expression (yellow arrow compared to red arrow marking the eye) in E11.5 Kcnj2KO/KO embryos. N=3 embryos of each genotype for each probe.
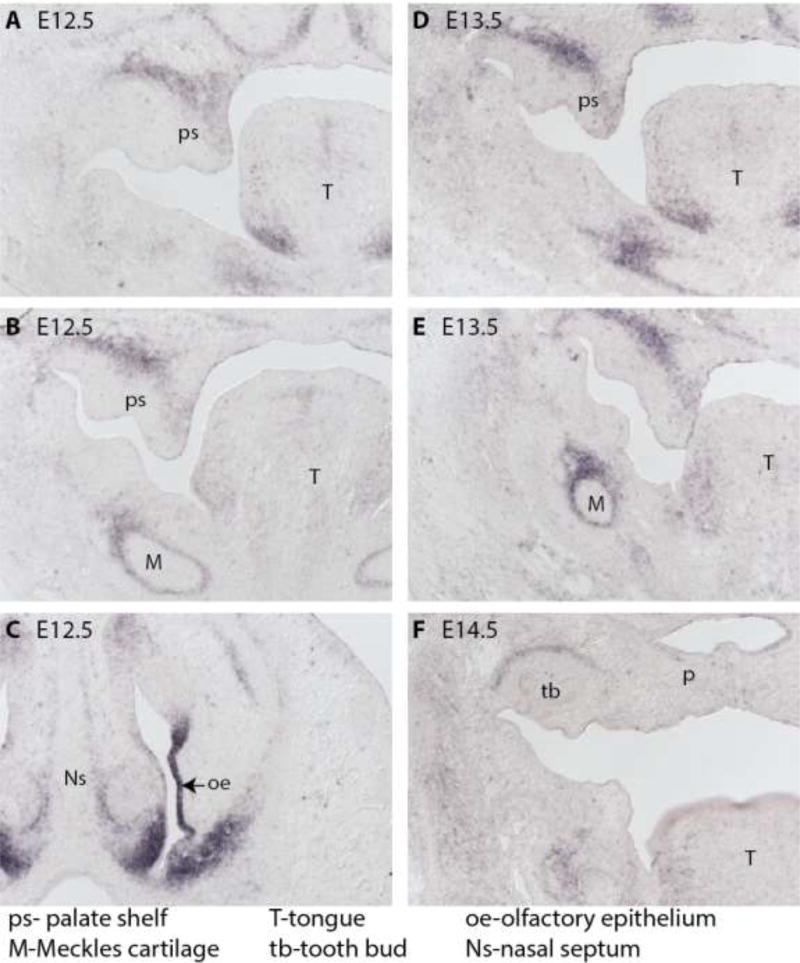
Kcnj2 is expressed in cells along the neural folds in the mouse at E8.5 [9]. Kcnj2 mRNA is detected in the frontal nasal process in E9.5 and E10.5, pharyngeal arches 1 and 2 in E9.5, and in the mandibular process at E10.5 [9]. Kcnj2 mRNA is detected in the mesenchyme of the palatal shelves, Meckel’s cartilage, and the olfactory epithelium at E12.5 (Figure 3A, B, C and [9]). At E13.5, Kcnj2 mRNA is apparent in the mesenchyme of the palatal shelves, the Meckel’s cartilage, and tongue, but by E14.5 Kcnj2 is no longer apparent in the palate, but is detected surrounding the tooth bud ([9] and Figure 3 D–F).
Figure 3.
RNA in situ hybridization reveals Kcnj2 mRNA is expressed in mesenchyme of the palate shelves and in the Meckel’s cartilage at E12.5 (A and B) and in the olfactory epithelium (C). At E13.5, Kcnj2 is still apparent in the mesenchyme of the palate shelves and also in the Meckel’s cartilage (D and E). By E14.5, Kcnj2 mRNA is not apparent in the palate, but can be detected surrounding the tooth bud in wild type mouse embryos (F).
Kir2.1 is required cell-autonomously in the cranial neural crest
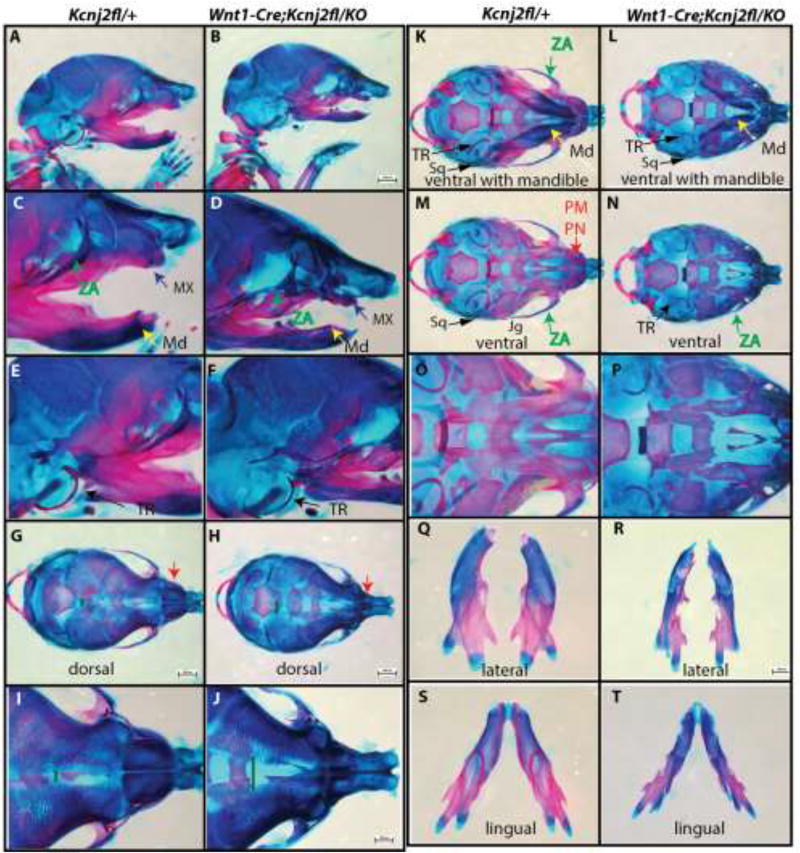
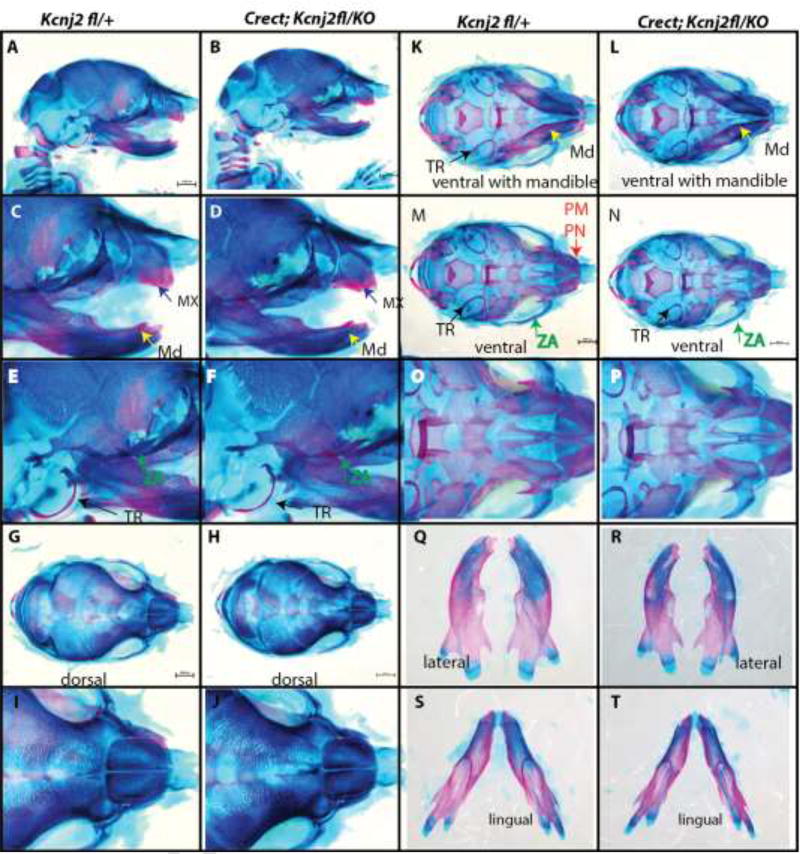
We asked whether Kir2.1 is required in cranial neural crest cells by specifically deleting Kcnj2 in the cranial neural crest using the Wnt1-Cre transgene. We found that the craniofacial structures of Wnt1-Cre+/Tg; Kcnj2KO/fl embryos were indistinguishable from Kcnj2KO/KO animals (Fig. 4) including hypoplastic mandibles, maxilla, components of the zygomatic arches, tympanic rings, and prenasal bones (Fig. 4 B, D, F, H, J, L, N, P, R, T). The anterior frontanelle and the sagittal suture are enlarged in Wnt1-Cre+/Tg; Kcnj2KO/fl E18.5 embryos compared to controls that lack Cre recombinase or the Kcnj2KO allele (Fig. 4 G–J). Furthermore, Wnt1-Cre+/Tg; Kcnj2KO/fl palatal shelves are reduced in size leading ultimately to complete cleft of the secondary palate (Fig. 6D). We used Crect to remove Kcnj2 from the ectoderm [23]. Crect+/Tg; Kcnj2KO/fl animals are indistinguishable from wild type have grossly normal development of craniofacial structures (Fig. 5) including the palate (Fig. 6F). Therefore, our data indicate that Kir2.1 is required in the cranial neural crest and not in the overlying ectoderm.
Figure 4.
Alizarin red and Alcian blue skeletal stains of Kcnj2fl/+ control (A, C, E, G, I, K, M, O, Q, S, U) compared to Wnt1-Cre; Kir2.1fl/KO (B, D, F, H, J, L, N, P, R, T, V) E18.5 mice show that deletion of Kcnj2 from the cranial neural crest cells causes hypoplastic mandible (MD, yellow arrow), maxilla (MX, purple arrow), premaxilla and prenasal bones (PM, PN red arrow), components of the zygomatic arch (ZA, green arrow), and tympanic ring (TR, black arrow). Skeletons are represented in lateral views (A–F), dorsal views (G–J), ventral views with mandible attached (K, L), and without mandible attached (M, N). Mandibles are compared in lingual (O, P), and lateral (Q, R), and lingual (S, T) views. Six Wnt1-Cre; Kcnj2fl/KO skeletons were stained and compared to four control siblings.
Figure 6.
H&E staining of E14.5 WT (A) and Kcnj2KO/KO (B) shows that the palate has fused at this stage for WT, but for Kcnj2KO/KO mice, palate shelves (PS) have elevated, but remain small and have not fused. H&E staining of E14.5 Wnt1-Cre; Kcnj2fl/+ (C) and Wnt1-Cre; Kcnj2fl/KO (D) shows that the palate shelves are small and elevated and fail to fuse in Wnt1-Cre; Kcnj2fl/KO when it has fused in Wnt1-Cre; Kcnj2fl/+ siblings at the same stage. H&E staining of E14.5 Crect; Kcnj2fl/+ (E) and Crect; Kcnj2fl/KO (F) shows that the palate fuses for both genotypes by E14.5. EdU staining of proliferating cells in E13.5 WT (G), Kcnj2KO/+ (H), Kcnj2KO/KO show that there is less proliferation in palate mesenchyme in Kcnj2KO/KO animals compared to WT and heterozygous siblings. A graph quantifies the percentage of cells marked with EdU in WT, Kcnj2KO/+, and Kcnj2KO/KO (J), p< 0.05 by two-tailed T-test comparing each genotype. EdU was quantified in 3 sections for 3 animals of each genotype. TUNEL staining marks apoptotic cells in E13.5 WT (K), Kcnj2KO/+ (L), Kcnj2KO/KO (M) palate shelves. The number of TUNEL positive cells out of 75 mesenchymal and 75 ectodermal DAPI stained nuclei were quantified in 3 sections for 3 animals of each genotype. There was no significant difference in TUNEL positive cells between Kcnj2KO/KO compared to WT siblings and heterozygous siblings, p>0.05, two tailed T-test.
Figure 5.
Alizarin red and Alcian blue skeletal stains of Kcnj2fl/+ control (A) compared to Crect; Kir2.1fl/KO (B) E18.5 mice show that deletion of Kcnj2 from the cranial neural crest cells does not significantly affect mandible (MD, yellow arrow), maxilla (MX, purple arrow), premaxilla and prenasal bones (PM, PN red arrow), components of the zygomatic arch (ZA, green arrow), and tympanic ring (TR, black arrow). Skeletons are represented in lateral views (A–F), dorsal views (G–J), ventral views with mandible attached (K, L), and without mandible attached (M, N). Mandibles are compared in lingual (O, P), and lateral (Q, R), lingual (S, T) views. Five Crect; Kcnj2fl/KO skeletons were stained and compared to five Kcnj2fl/+ control sibling skeletons.
Disruption of Kir2.1 hinders proliferation of the palatal mesenchyme
By E14.5, Kcnj2KO/KO palate shelves remain small and separate in contrast to wild type (WT) and heterozygous siblings that have a completely fused palate at this stage (Fig. 6A,B). Many steps are required for the palate to develop correctly. First, the CNC cells need to proliferate, migrate, and differentiate to form palatal shelves. Palatal shelves then extend, elevate and fuse along the midline to form a complete palate [24, 25]. Lastly, some of the cells of the palate differentiate into osteoblasts that will uptake calcium for ossification [26–30]. We found that Kcnj2KO/KO palate shelves form and elevate at all axial levels examined (anterior, middle, and posterior), but are reduced in size (medial palatal tissue shown in Fig. 6A, B). Palatal shelves that fail to extend to the midline or fuse were observed in Wnt1Cre+/Tg; Kcnj2KO/fl, but not in control littermates (Fig. 6C, D) or Crect+/Tg; Kcnj2KO/fl (Fig. 6E, F). To determine whether reduced proliferation of palatal mesenchyme could account for reduced palatal shelf size in Kcnj2KO/KO animals, we injected 5-ethynyl-2’deoxyuridine (EdU) into pregnant dams to mark proliferating cells 40 minutes before harvesting E13.5 embryos. We found 58% less proliferating palatal mesenchymal cells in Kcnj2KO/KO animals compared to wild type siblings at the same stage and same axial plane ((Fig. 6G–J, p> 0.05 T-test). Apoptosis in the palate mesenchyme as detected by TUNEL staining was insignificant in all genotypes including less than an average of 7 cells out of 150 in counted in all genotypes (Fig. 6K–N).
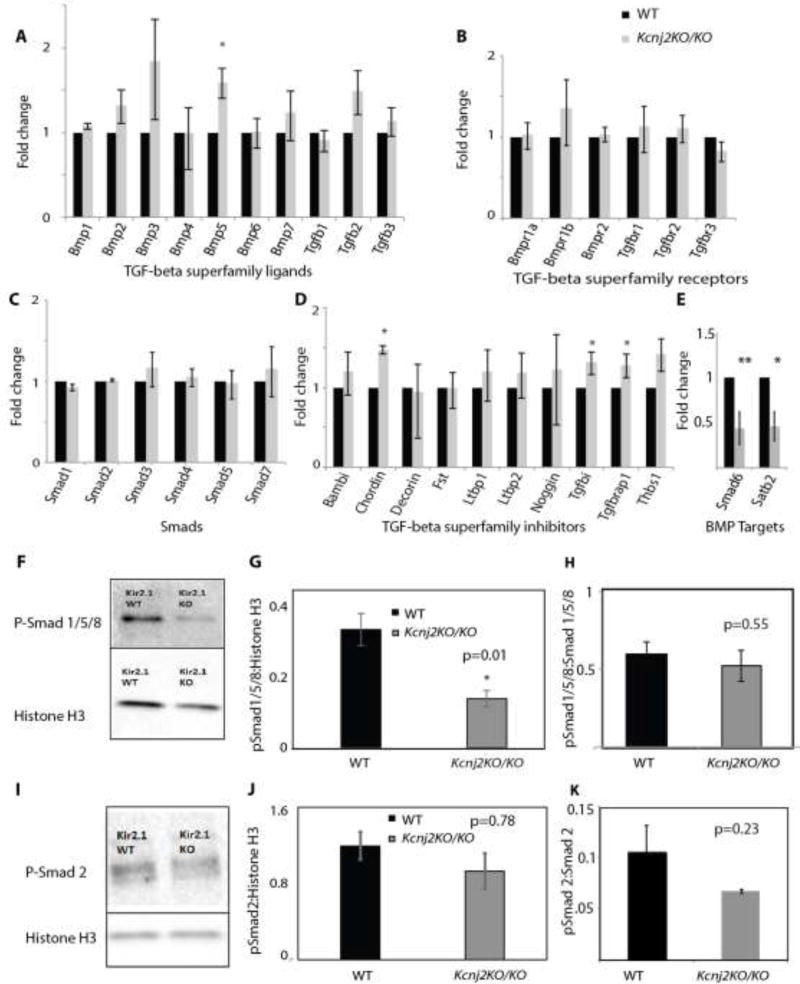
Proliferation and differentiation of cranial neural crest–derived mesenchyme of the palate shelves requires BMP signaling [31] and function of Kir2.1, supporting a role for both in the same cells at the same time. We asked whether loss of Kir2.1 function affected the BMP signaling pathway in two ways: by examining expression of BMP pathway genes and by quantifying phosphorylation of Smads. To determine if loss of Kir2.1 affected expression of BMP ligands, receptors, and Smads, we quantified their expression in total RNA from Kcnj2KO/KO and WT sibling E13.5 whole palatal shelves. We found that loss of Kcnj2 caused no significant reduction in expression of BMP ligands, receptors, or Smads (Fig. 7A–C). However, we found Kcnj2KO/KO caused a significant increase in expression of BMP5 (Fig. 6A, p=0.007, T-test). We found no significant difference in expression of BMP or TGF-β receptors or associated Smads (Fig. 7B, C). In addition, we found that expression of a BMP inhibitor called Chordin was significantly increased as were two TGF-β inhibitors: TGFbi and TGFBrap1 (Fig. 7D). We quantified expression of two established BMP target genes (Smad6 and Satb2 [14]) in Kcnj2KO/KO and WT palate shelf lysates and found a significant decrease in expression of Smad6 and Satb2 in Kcnj2KO/KO (p=0.007 and p=0.03 respectively by T-test). While the components of the BMP signaling pathway were expressed, two different BMP target genes were not efficiently transcribed without functional Kir2.1.
Figure 7.
Quantification of expression of BMP and TGF-β ligands shows that expression of BMP5 is significantly increased in Kcnj2KO/KO E13.5 palatal shelves compared to WT controls (p=0.007, T-Test). (A). Quantification of expression of BMP and TGF-β receptors shows that expression of receptors in E13.5 palatal shelves is not significantly different between Kcnj2KO/KO compared to WT controls (B). Expression of SMADs in E13.5 palatal shelves is not significantly different between Kcnj2KO/KO compared to WT controls (C). Quantification of mRNA levels encoding BMP and TGF-β inhibitors shows that Chordin, Tgfbi and Tgfbrap1 are significantly increased in Kcnj2KO/KO compared to WT controls (D). Expression of BMP target genes Smad6 and Satb2 are significantly reduced in E13.5 Kcnj2KO/KO palatal shelves compared to WT, n= 6 WT and 4 Kcnj2KO/KO, T-test p= 0.007 and 0.01 respectively (E). A western blot shows that phosphorylated Smad 1/5/8 is reduced in Kcnj2KO/KO E13.5 craniofacial structures compared to WT sibling controls (F). Phosphorylated Smad 1/5/8 is quantified and normalized to Histone H3 (G) Phosphorylated Smad 1/5/8 is quantified and normalized to total Smad 1/5 (H). Protein was quantified from lysates from 3 animals of each genotype. A western blot shows that phosphorylated Smad 2 is not significantly different in Kcnj2KO/KO E13.5 palate shelves compared to WT sibling controls (I). Phosphorylated Smad 2 is quantified and normalized to Histone H3 and is not significantly different between Kcnj2KO/KO and WT controls. (J). Phosphorylated Smad 2 is quantified and normalized to total Smad 2 (K).
To determine if Kir2.1 is important for BMP signaling, we quantified p-Smad 1/5/8 in the facial lysates (including prenasal, premaxilla, nasal, maxilla, and mandible, but excluding the brain) from E13.5 Kcnj2KO/KO and wild-type siblings. We found that phosphorylated Smad 1/5/8 was significantly reduced in Kcnj2KO/KO nuclear enriched lysates compared to WT siblings when we normalize to Histone H3 (Fig. 7E,F), indicating that Kir2.1 promotes effective BMP signaling. However, when we normalize to total Smad 1/5/8, there is not a significant decrease in Smad phosphorylation indicating that Smad protein levels are also reduced in these lysates. BMPs are members of the transforming growth factor beta (TGF-β) superfamily. To determine if Kir2.1 plays a role in TGF-β signaling like it does in BMP signaling, we quantified phosphorylation of Smad 2/3 (p-Smad 2/3) in craniofacial lysates from E13.5 Kcnj2KO/KO and wild-type siblings. We found no significant difference between phosphorylated Smad 2/3 in Kcnj2KO/KO and WT siblings when we normalize to Histone H3 or total Smad 2 (Fig. 7G,H), indicating that the requirement of Kir2.1 is specific to BMP signaling.
Materials and Methods
Mouse strains
Mouse colonies were maintained in the vivarium at the University of Colorado Anschutz Medical Campus according to IACUC protocol # B-104216(03)1E. Kcnj2KO/+ mice [11] were purchased from Jackson Laboratory (FVB.129-Kcnj2tm1Swz/J Stock No. 005057/Kir2.1). Kcnj2fl/+ mice were a generous gift from Dr. Mark T. Nelson at the University of Vermont [32]. Wnt1-Cre mice were a gift from Dr. Jenna Galloway at Harvard Medical School. Crect mice [23] were a gift from Dr. Trevor Williams at the University of Colorado Anschutz Medical Campus. Tail snips were used to isolate DNA for genotyping by PCR. The following primers were used to determine genotypes. For Kcnj2KO we use primers CACGAGACTAGTGAGACGTG, GGAGGGAGCTGGAAGCTACT, and CCTCCGCTAGTCATTCCACT in a multiplex reaction. For Kcnj2flox we use primers GGACTCTCCGATGACACTGAGAACC and TGGTCAGTTCCACATCGAAACCAACA. For detection of the gene encoding Cre recombinase, we use primers GCAGAACCTGAAGATGTTCGC and ACACCAGAGACGGAATCCATC.
Ages of mice are provided as embryonic day (E) and the day that vaginal plugs were observed is considered E0.5. When embryos were harvested, pregnant dams were euthanized by isofluorane followed by cervical dislocation.
Whole mount RNA in situ
Embryos were harvested at E10.5 (for Dlx2, Dlx5, Hand2, FoxL2, and Pou3f3) and E11.5 (for Pou3f3). Whole-mount in situ hybridization was performed using a protocol described by the Clouthier lab [33]. Probes were detected using 4-nitro blue tetrazolium chloride and 5romo-4-chloro-3-indolylphophate (Roche). Probes for whole mount RNA in situ were provided by Dr. David Clouthier at the University of Colorado Anschutz Medical Campus. For sections, OCT-embedded, developmentally staged specimens were serially cryosectioned at 10 µm intervals. E12.5, E13.5, and E14.5 mouse heads were sectioned coronally. The mouse KCNJ2 cDNA clone was purchased from Open Biosystems (Lafayette CO, USA; clone 8860860). A 518 bp fragment of the 5′ untranslated region of the KCNJ2 cDNA clone was cloned into PCRII vector (Life Technologies, Grand Island, NY, USA). DIG- labelled antisense riboprobe was generated using the DIG RNA Labelling Kit (Roche Applied Science, Indianapolis, IN, USA).
Skeletal staining
Mouse embryos were harvested at E18.5. Following removal of the skin and internal organs, skeletons were fixed in 95% ethanol for three days and acetone for two days. Skeletons were incubated in 0.3% Alcian Blue and and 0.1% Alizarin Red for five days at 37°C. Skeletons were cleared with 2% KOH for two days at room temperature. Skeletons were stored with successively more concentrated glycerol 50% and 80% in 1% KOH for 24 hours each. Skeletons were stored in 100% glycerol. Images were acquired with a Nikon SM218 microscope.
H&E
Mouse embryos were harvested at E14.5 and fixed in 10% Formalin. Heads were embedded in paraffin. Sections are 5 um thick. Hematoxylin and Eosin staining was performed with a Hematoxylin and Eosin stain kit (ScyTek Laboratories, Inc.) according to the protocol that accompanies the reagents.
Quantification of proliferation/EdU
To quantify proliferation of cells in the palate of E13.5 embryos, 50ug per gram of body weight 5 ethynyl-2’ deoxyuridine (EdU) by intraperitoneal injection into pregnant dams 40 minutes before euthanasia. Embryos were harvested and hind paws were removed for genotyping. The head of the embryo was fixed in 10% buffered formalin before embedding in paraffin. Tissue sections (5um thick) were mounted. Sections were deparafinized and rehydrated. Subsequently, EdU was detected using the Invitrogen Click-iT® EdU Imaging Kit (#C10337). Vectasheild® mounting medium with DAPI (#H1200) identified nuclei. Slides were kept in 4° C overnight before imaging. Imaging was performed on a Nikon eclipse 80 microscope at 10×. Analysis consisted of determining the ratio of EdU positive to DAPI positive nuclei in a 120 by 120 pixel region of interest.
TUNEL
TUNEL staining was conducted using the DEADEnd TUNEL staining kit (Promega Cat# G3250). Sections were deparaffinized in a graded ethanol series, re-fixed in 10% Formalin, and permeabilized with 20 ug/ml of Proteinase K. All other steps were followed according to the manufacturer’s instructions. Sections were counterstained with DAPI (Vector Laboratories, Inc, Burlingame, CA Cat # H-1200). Mesenchymal and ectodermal TUNEL positive cells were quantified independently.
Western blot
Kcnj2KO/KO and WT littermates were harvested at E13.5 in ice cold PBS. Red blood organs were collected for genotyping. Craniofacial sections including the prenasal, premaxilla, nasal, maxilla, mandible, and excluding the posterior and dorsal regions of the head and the brain were flash frozen and stored at −80°C. Tissue was homogenized in 20 mM Tris-HCL pH 7.4, 10mM NaCl, and 3mM MgCl2 using a motorized pestle on ice for 1 minute. An aliquot of 10% NP-40 was added to the homogenate, vortexed, and centrifuged for 10 minutes at 3,000 rpm at 4°C. The supernatant was transferred to a new tube and the pellet was washed with 1× PBS, re-centrifuged for 10 minutes at 3,000 rpm at 4°C and the supernatant was discarded. 100 uL of 100 mM Tris-HCL pH 7.4, 100 mM NaCl, 1% Triton X-100, 1mM EDTA, 1mM EGTA, 10% glycerol, 0.1% SDS, and 0.5% sodium deoxycholate was added to the pellet, mixed by vortexing, and incubated on ice for 2 hours. The homogenate was centrifuged at 12,000 rpm at 4°C for 20 minutes. The supernatant was transferred to a new tube and quantified using a 1:10 dilution with a PierceTM BCA Protein assay kit with BSA used as a protein standard (Thermo Scientific Prod # 23227). All buffers had Halt Protease Inhibitor and PhosStop phosphatase inhibitor cocktails added at 1× immediately before use (Thermo Scientific #87786 and Roche Cat #04906845001).
A total of 15ug of protein lysate was loaded onto 4–20% polyacrylamide gels (Bio Rad Cat #456-1095) electrophoresed and transferred onto 0.2 um nitrocellulose membranes (Bio Rad Cat #1704158) using a Trans Blot Turbo transfer system (BioRad). Membranes were blocked in 5% milk/1× TBS-T and incubated with primary antibodies overnight at 4°C. The blots were probed using primary antibodies against pSmad 1/5/9 (1:500, Cell Signaling #13820), pSmad 2 (1:500, Cell Signaling #3108), Smad1 (1:1000, Cell Signaling #6944), Smad2 (1:1000, Cell Signaling #5339), and Histone H3 (1:2000, Cell Signaling #4499). Western blots were analyzed with ProSignal Femto (Prometheus) or Clarity Western ECL substrate (BioRad).
RNA isolation and real-time PCR
Embryos were harvested at E13.5 and whole palate shelves were dissected in ice cold PBS and flash frozen. Total RNA was extracted from frozen Kcnj2KO/KO and WT palate shelves using an RNeasy Plus Mini kit (Qiagen). cDNA synthesis was carried out using the RT2 First Strand Kit with 25 ng/uL of total RNA from each sample. RT-qPCR was performed using SYBR Green and a RT2 Profiler PCR Mouse TGFb/ BMP signaling pathway array (Qiagen) on the LightCycler 480 (Roche).
BMP target genes that are expressed in craniofacial tissue during E13.5 were assessed separately using 100 ng/ul of cDNA per reaction. Smad6 (reference sequence NM_008542) and Satb2 (reference sequence NM_139146) expression was assessed using KiCqStart SYBR Green Primers (Sigma-Aldrich) and normalized to endogenous mB2m (primer sequences ACTGACCGGCCTGTATGCTA and TGAAGGACATATCTGACATCTCA). PCR was performed on a bioRad cFX Connect Real time system with 10 minutes at 95 °C followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute followed by a melting curve. Fold changes were calculated according to the ΔΔCt method [34].
Discussion
Here we show that Kir2.1 is required for development of several craniofacial structures that derive from the cranial neural crest including the mandible, the maxilla, the prenasal bones, the premaxilla, the zygomatic arch, the tympanic ring, and the palate. The hypoplastic craniofacial structures and reduction in the size of palate shelves of the Kcnj2KO/KO demonstrate that Kir2.1 is required for proper development of all of these structures. We find that Kir2.1 is important in the cranial neural crest and not in the overlying ectoderm for its developmental role. Finally, we find that BMP signaling is reduced in Kcnj2KO/KO embryos compared to wild type sibling controls. In contrast, TGF-β signaling is not significantly affected by the loss of Kir2.1 function.
To determine how early changes in craniofacial patterning in Kcnj2KO/KO embryos could be detected, we performed RNA in situ to reveal expression of established craniofacial patterning genes. Kcnj2KO/KO embryos did not display obvious shifts in the expression of several early patterning genes. In contrast, in Xenopus, RNAi reducing and overexpression of Kcnj2 affected several early neural crest markers [9]. We did detect a specific shift both posteriorly and dorsally in expression of Pou3f3 in Kcnj2KO/KO embryos in the maxillary domain (Fig. 2). Interestingly, Dlx1 and 2 positively regulate Pou3f3 and Dlx1 is a BMP4 transcriptional target gene [22]. However, Dlx5 and 6 repress expression of Pou3f3 in the mandibular arch [22] and Dlx5 is also a BMP4 target [35–37]. By RNA in situ, it is difficult to assess levels of gene expression, and quantitative PCR would be needed to assess whether there are significant changes in expression levels of any of these genes at E10.5. Pou3f3 mutant animals lack squamosal bones and jugal bones, but do not have cleft palate, smaller mandibles, smaller maxilla, or frontal bone defects that are characteristic of Kcnj2KO/KO mice [22]. Thus, the reduced expression domain of Pou3f3 is consistent with the reduction in the size of the squamosal and jugal bones in Kcnj2KO/KO animals. However, the reduction in the Pou3f3 expressing domain does not explain the wide spread craniofacial phenotypes of Kcnj2KO/KO animals and thus these defects likely arise at a later time point and/or may be do to more subtle changes in expression of other genes.
Kir2.1 and BMP signaling are required for development of the same structures. Deletion of BMP receptors and ligands from specific populations of cells reveal where BMP signaling is required. Deletion of one BMP receptor, Alk2, from the cranial neural crest results in cleft secondary palate and hypotrophic mandible, in addition to loss of the posterior bones of the zygomatic arch, hypoplastic tympanic ring, and defective squamosal bone formation [38]. Conditional deletion of another BMP receptor, Bmpr1a (Alk3), decreases proliferation of the maxillary and palatal mesenchyme and results in cleft of the secondary palate [39, 40]. BMP5 and BMP7 ligands have redundant functions in modulation of proliferation of cells of the pharyngeal arch [41]. Deletion of BMP2, BMP4, and BMP7 from the cranial neural crest enlarges the anterior fontanelle in a dose dependent manner [14]. Cranial neural crest specific deletion of the combination of BMP2, BMP4, and BMP7 results in severe mandibular defects, while deletion of only one of the ligands or heterozygous deletion of a combination of BMP ligands causes less severe mandibular defects [14]. For example, deletion of only BMP2 from the CNC impacts the formation of the coronoid process of the mandible, but loss of BMP4 or 7 does not impact the coronoid process[14]. Interestingly, deletion of any of the single BMP ligands examined, did not cause defects in prenasal or nasal bone formation [14], but deletion of BMP2 and 4 from the cranial neural crest resulted in very similar defects in nasal and prenasal bone formation to deletion of Kcnj2. Loss of Kir2.1 function impacts each of the structures that are impacted by loss of BMP2, 4, or 7, but to a lesser extent than complete deletion of all three BMP2, BMP4, and BMP7 from the cranial neural crest ([14] and Fig. 2). This comparison suggests that while Kir2.1 may contribute to BMP signaling, its loss does not completely abolish BMP signaling ([14] and Fig. 2). While deletion of Kcnj2 does not completely phenocopy deletion of any one of the specific BMP signaling components, the overlap between affected structures may suggest that Kir2.1 may be important for the efficient BMP signaling for all BMP ligands.
We assessed whether BMP signaling is reduced by measuring transcription of BMP target genes and quantifying phosphorylated Smad 1/5/8. BMP signaling is significantly reduced in Kcnj2KO/KO embryos by both measures demonstrating that Kir2.1 is necessary for efficiently transmitting the BMP signal. However, loss of Kir2.1 function does not decrease expression of BMP ligands, receptors, or Smads. In fact, BMP5 transcripts are significantly increased in Kcnj2KO/KO palatal shelves compared to wild type siblings. These data show that while components of the BMP signaling pathway are present when Kcnj2 is deleted, the BMP signal is somehow not efficiently transmitted without the function of this channel.
How can we explain how expressed BMP ligands and receptors are not able to transmit a signal? We may be able to take hints from the BMP signaling in Drosophila. In the Drosophila wing disc, inhibition or loss of inwardly rectifying K+ (Irk2) channel function increases the total Dpp (BMP homolog) and does not significantly change receptor levels, but decreases phosphorylation of Mad (Smad homolog) [10, 21]. Loss of Irk2 function caused Dpp loss-of-function phenotypes and down-regulation of Dpp target genes in the wing primordium [10]. In Drosophila, inhibition of Irk channels changed the dynamics of Dpp release [21] suggesting that the timing of presentation of the BMP ligand matters for it downstream consequences. Human Kir2.1 was able to rescue the developmental role of native Drosophila inwardly rectifying K+ channels for wing development [21]. It is intriguing to speculate that the role of Irk/Kir channels in BMP signaling may be evolutionarily conserved from flies to mammals, but further studies are needed to draw that conclusion.
Kir channels dictate when secretion occurs by regulating membrane potential in different cellular environments. For example, Kir6.1 and Kir6.2 are ATP-sensitive and couple the availability of energy (ATP/ADP ratio) to the need to secrete insulin from pancreatic beta cells [42–44]. Thus Kir6.1 and Kir6.2 regulate release of insulin through their role in regulating membrane potential. Mutations that disrupt the function of Kir6.1 or Kir6.2 depolarize beta cells resulting in excessive unregulated secretion of insulin otherwise known as hyperinsulinemia [45, 46]. Mutations that activate Kir6.1 or Kir6.2 cause neonatal diabetes [2, 47–51]. Kir6.2 is also essential in the ventromedial hypothalamus glucose response in neurons for the secretion of glucagon in hypoglycemic conditions [52]. Kir4.1 is expressed in the parietal cells and regulates the secretion of acid in the stomach [53]. Kir channels that are expressed in dopamine (D2) neurons regulate dopamine release [54–57]. We do not generally think of cranial neural crest or its derivatives as excitable cells. However, no one has tested this hypothesis. It could be that these cells regulate the secretion of BMP via a similar mechanism as pancreatic beta cells regulate the secretion of insulin. In support of this hypothesis, we found that Irk channels regulate the release of Dpp, a BMP homolog for the development of the Drosophila wing [21]. While expression of Dpp and its receptors was not reduced, phosphorylation of Mad (Smad homolog) was significantly reduced as a result of the change in the temporal pattern of exposure to the Dpp ligand [21].
If Kir channels regulate release of BMP by modulating changes in membrane potential, it is likely that other channels control different currents to affect membrane potential for the same process. Human channelopathies that include morphological defects reveal likely candidates for channels that participate in modulating membrane potential of cranial neural crest cells. Mutations in several ion channels lead to human syndromes of morphological abnormalities. Craniofacial abnormalities associated with mutations in KCNJ2 (Andersen-Tawil syndrome) commonly include cleft or high-arched palate, hypertelorism, low set ears, micrognathia, and dental defects. Mutations in KCNJ6, which encodes another inwardly rectifying K+ channel called GIRK2, also cause cleft or high-arched palate and micrognathia in Keppen-Lubinsky syndrome [58, 59]. Patients with Keppen-Lubinsky syndrome also have microcephaly, large protruding eyes, a narrow nasal bridge, and lipodystrophy [58, 59]. Mutations in KCNH1, a voltage gated potassium channel lead to Temple-Baraitser (also known as Zimmermann- Laband) syndrome have craniofacial defects: cleft or high arched palate, hypertelorism, dysmorphic ears, dysmorphic nose, gingival hypertrophy, an abnormal number of teeth, and limb defects such as absent or hypoplastic phalanges and nails [60–66]. Morphological defects are not limited to channelopathies that disrupt potassium channels. Timothy Syndrome is caused by mutations that disrupt CaV1.2, a voltage gated calcium channel [67]. Disruption of CaV1.2 leads to high-arched or cleft palate, low set ears, flattened nasal bridge, thin or cleft upper lip, and syndactyly (fusion of digits) [67, 68].
Alternatively, it could be that Kir channels impact BMP signaling via a different mechanism than has been demonstrated in flies. Inhibitors of BMP (Chordin) and TGF-β (Tgfbi and Tgfbrap1) are significantly up-regulated in Kcnj2KO/KO palate shelves. There could be a biochemical signaling cascade that impacts transcription of BMP and TGF-beta inhibitors that account for the reduction in phosphorylated Smad 1/5/8. Further studies exploring the mechanism that connects Kir2.1 function to BMP signaling are warranted.
HIGHLIGHTS.
-
*
Kcnj2KO/KO mice phenocopy cranial neural crest specific deletion of BMPs or receptors
-
*
Kir 2.1 is required in the cranial neural crest for craniofacial development.
-
*
Loss of Kir2.1 decreases expression of BMP target genes in E13.5 palate shelves.
-
*
Loss of Kir2.1 reduces phosphorylated Smad 1/5/8
Acknowledgments
We would like to thank Dr. Trevor Williams (University of Colorado Anschutz Medical Campus) for the Crect strain, Dr. Jenna Galloway (Harvard Medical School) for the Wnt1-Cre strain, and Dr. Mark T. Nelson at the University of Vermont for the floxed Kcnj2 mouse line. We would like to thank Dr. David Clouthier, Dr. Katherine Fantauzzo, and Dr. Trevor Williams for helpful insights. We would like to thank NIH-NIDCR -1R56DE025311-01 for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hibino H, et al. Expression of an inwardly rectifying K+ channel, Kir5.1, in specific types of fibrocytes in the cochlear lateral wall suggests its functional importance in the establishment of endocochlear potential. Eur J Neurosci. 2004;19(1):76–84. doi: 10.1111/j.1460-9568.2004.03092.x. [DOI] [PubMed] [Google Scholar]
- 2.Massa O, et al. KCNJ11 activating mutations in Italian patients with permanent neonatal diabetes. Hum Mutat. 2005;25(1):22–7. doi: 10.1002/humu.20124. [DOI] [PubMed] [Google Scholar]
- 3.Vaca L, Licea A, Possani LD. Modulation of cell membrane potential in cultured vascular endothelium. Am J Physiol. 1996;270(3 Pt 1):C819–24. doi: 10.1152/ajpcell.1996.270.3.C819. [DOI] [PubMed] [Google Scholar]
- 4.Plaster NM, et al. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen's syndrome. Cell. 2001;105(4):511–9. doi: 10.1016/s0092-8674(01)00342-7. [DOI] [PubMed] [Google Scholar]
- 5.Tawil R, et al. Andersen's syndrome: potassium-sensitive periodic paralysis, ventricular ectopy, and dysmorphic features. Ann Neurol. 1994;35(3):326–30. doi: 10.1002/ana.410350313. [DOI] [PubMed] [Google Scholar]
- 6.Tristani-Firouzi M, et al. Functional and clinical characterization of KCNJ2 mutations associated with LQT7 (Andersen syndrome) J Clin Invest. 2002;110(3):381–8. doi: 10.1172/JCI15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon G, et al. Andersen-Tawil syndrome: prospective cohort analysis and expansion of the phenotype. Am J Med Genet A. 2006;140(4):312–21. doi: 10.1002/ajmg.a.31092. [DOI] [PubMed] [Google Scholar]
- 8.Villanueva S, et al. Cleft Palate, Moderate Lung Developmental Retardation and Early Postnatal Lethality in Mice Deficient in the Kir7.1 Inwardly Rectifying K+ Channel. PLoS One. 2015;10(9):e0139284. doi: 10.1371/journal.pone.0139284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams DS, et al. Bioelectric signalling via potassium channels: a mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen-Tawil Syndrome. J Physiol. 2016;594(12):3245–70. doi: 10.1113/JP271930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahal GR, et al. An inwardly rectifying K+ channel is required for patterning. Development. 2012;139(19):3653–64. doi: 10.1242/dev.078592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaritsky JJ, et al. Targeted disruption of Kir2.1 and Kir2.2 genes reveals the essential role of the inwardly rectifying K(+) current in K(+)-mediated vasodilation. Circ Res. 2000;87(2):160–6. doi: 10.1161/01.res.87.2.160. [DOI] [PubMed] [Google Scholar]
- 12.Bandyopadhyay A, et al. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2(12):e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, et al. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132(6):1453–61. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- 14.Bonilla-Claudio M, et al. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012;139(4):709–19. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adachi-Yamada T, et al. Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature. 1999;400(6740):166–9. doi: 10.1038/22112. [DOI] [PubMed] [Google Scholar]
- 16.de Celis JF. Expression and function of decapentaplegic and thick veins during the differentiation of the veins in the Drosophila wing. Development. 1997;124(5):1007–18. doi: 10.1242/dev.124.5.1007. [DOI] [PubMed] [Google Scholar]
- 17.De Celis JF. Pattern formation in the Drosophila wing: The development of the veins. Bioessays. 2003;25(5):443–51. doi: 10.1002/bies.10258. [DOI] [PubMed] [Google Scholar]
- 18.Letsou A, et al. Drosophila Dpp signaling is mediated by the punt gene product: a dual ligand-binding type II receptor of the TGF beta receptor family. Cell. 1995;80(6):899–908. doi: 10.1016/0092-8674(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 19.Zecca M, Basler K, Struhl G. Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development. 1995;121(8):2265–78. doi: 10.1242/dev.121.8.2265. [DOI] [PubMed] [Google Scholar]
- 20.Zeng YA, et al. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development. 2007;134(11):2061–71. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
- 21.Dahal GR, Pradhan SJ, Bates EA. Inwardly rectifying potassium channels regulate Dpp release in the Drosophila wing disc. Development. 2017 doi: 10.1242/dev.146647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong J, et al. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development. 2008;135(17):2905–16. doi: 10.1242/dev.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bingsi Li ML, Risolino Maurizio, Ferretti Elisabetta, Hart James, Quintana Laura, Grishina Irina, Yang Hui, Choi Irene, Lewicki Patrick, Khan Sameer, Aho Robert, Feenstra Jennifer, Vincent Theresa, Brown Anthony MC, Williams Trevor, Selleri Licia. EMT Promotes Fusion of the Frontonasal Prominences in Facial Morphogenesis by Pbx-dependent regulation of Snail1. 2017 doi: 10.1242/dev.157628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301(2):309–26. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 25.Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235(9):2353–75. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- 26.Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120(8):2213–24. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- 27.Graham A, Lumsden A. The role of segmentation in the development of the branchial region of higher vertebrate embryos. Birth Defects Orig Artic Ser. 1993;29(1):103–12. [PubMed] [Google Scholar]
- 28.Imai H, et al. Contribution of early-emigrating midbrain crest cells to the dental mesenchyme of mandibular molar teeth in rat embryos. Dev Biol. 1996;176(2):151–65. doi: 10.1006/dbio.1996.9985. [DOI] [PubMed] [Google Scholar]
- 29.Le Douarin NM, Ziller C. Plasticity in neural crest cell differentiation. Curr Opin Cell Biol. 1993;5(6):1036–43. doi: 10.1016/0955-0674(93)90089-9. [DOI] [PubMed] [Google Scholar]
- 30.Noden DM. Vertebrate craniofacial development: the relation between ontogenetic process and morphological outcome. Brain Behav Evol. 1991;38(4–5):190–225. doi: 10.1159/000114388. [DOI] [PubMed] [Google Scholar]
- 31.Parada C, Chai Y. Roles of BMP signaling pathway in lip and palate development. Front Oral Biol. 2012;16:60–70. doi: 10.1159/000337617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longden TA, et al. Capillary K+-sensing initiates retrograde hyperpolarization to increase local cerebral blood flow. Nat Neurosci. 2017;20(5):717–726. doi: 10.1038/nn.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clouthier DE, et al. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125(5):813–24. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Miyama K, et al. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev Biol. 1999;208(1):123–33. doi: 10.1006/dbio.1998.9197. [DOI] [PubMed] [Google Scholar]
- 36.Ulsamer A, et al. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem. 2008;283(7):3816–26. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 37.Lee MH, et al. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J Biol Chem. 2003;278(36):34387–94. doi: 10.1074/jbc.M211386200. [DOI] [PubMed] [Google Scholar]
- 38.Dudas M, et al. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech Dev. 2004;121(2):173–82. doi: 10.1016/j.mod.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Baek JA, et al. Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev Biol. 2011;350(2):520–31. doi: 10.1016/j.ydbio.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu W, et al. Threshold-specific requirements for Bmp4 in mandibular development. Dev Biol. 2005;283(2):282–93. doi: 10.1016/j.ydbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 41.Solloway MJ, Robertson EJ. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126(8):1753–68. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- 42.Yokoshiki H, et al. ATP-sensitive K+ channels in pancreatic, cardiac, and vascular smooth muscle cells. Am J Physiol. 1998;274(1 Pt 1):C25–37. doi: 10.1152/ajpcell.1998.274.1.C25. [DOI] [PubMed] [Google Scholar]
- 43.Ashcroft SJ. The beta-cell K(ATP) channel. J Membr Biol. 2000;176(3):187–206. doi: 10.1007/s00232001095. [DOI] [PubMed] [Google Scholar]
- 44.Mikhailov MV, Ashcroft SJ. Interactions of the sulfonylurea receptor 1 subunit in the molecular assembly of beta-cell K(ATP) channels. J Biol Chem. 2000;275(5):3360–4. doi: 10.1074/jbc.275.5.3360. [DOI] [PubMed] [Google Scholar]
- 45.Sharma N, et al. Familial hyperinsulinism and pancreatic beta-cell ATP-sensitive potassium channels. Kidney Int. 2000;57(3):803–8. doi: 10.1046/j.1523-1755.2000.00918.x. [DOI] [PubMed] [Google Scholar]
- 46.Tornovsky S, et al. Hyperinsulinism of infancy: novel ABCC8 and KCNJ11 mutations and evidence for additional locus heterogeneity. J Clin Endocrinol Metab. 2004;89(12):6224–34. doi: 10.1210/jc.2004-1233. [DOI] [PubMed] [Google Scholar]
- 47.Koster JC, Permutt MA, Nichols CG. Diabetes and insulin secretion: the ATP-sensitive K+ channel (K ATP) connection. Diabetes. 2005;54(11):3065–72. doi: 10.2337/diabetes.54.11.3065. [DOI] [PubMed] [Google Scholar]
- 48.Hattersley AT, Ashcroft FM. Activating mutations in Kir6.2 and neonatal diabetes: new clinical syndromes, new scientific insights, and new therapy. Diabetes. 2005;54(9):2503–13. doi: 10.2337/diabetes.54.9.2503. [DOI] [PubMed] [Google Scholar]
- 49.Gloyn AL, et al. KCNJ11 activating mutations are associated with developmental delay, epilepsy and neonatal diabetes syndrome and other neurological features. Eur J Hum Genet. 2006;14(7):824–30. doi: 10.1038/sj.ejhg.5201629. [DOI] [PubMed] [Google Scholar]
- 50.Edghill EL, et al. Activating mutations in the KCNJ11 gene encoding the ATP-sensitive K+ channel subunit Kir6.2 are rare in clinically defined type 1 diabetes diagnosed before 2 years. Diabetes. 2004;53(11):2998–3001. doi: 10.2337/diabetes.53.11.2998. [DOI] [PubMed] [Google Scholar]
- 51.Gloyn AL, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350(18):1838–49. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 52.Miki T, et al. ATP-sensitive K+ channels in the hypothalamus are essential for the maintenance of glucose homeostasis. Nat Neurosci. 2001;4(5):507–12. doi: 10.1038/87455. [DOI] [PubMed] [Google Scholar]
- 53.Song P, et al. Kir4.1 channel expression is essential for parietal cell control of acid secretion. J Biol Chem. 2011;286(16):14120–8. doi: 10.1074/jbc.M110.151191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka T, et al. The role of ATP-sensitive potassium channels in striatal dopamine release: an in vivo microdialysis study. Pharmacol Biochem Behav. 1995;52(4):831–5. doi: 10.1016/0091-3057(95)00151-l. [DOI] [PubMed] [Google Scholar]
- 55.Neusch C, Schnierle S, Moser A. Selegiline induces dopamine release through ATP-sensitive potassium channels in the rat caudate-putamen in vitro. Neurochem Int. 1997;31(2):307–11. doi: 10.1016/s0197-0186(96)00132-5. [DOI] [PubMed] [Google Scholar]
- 56.Neusch C, Runde D, Moser A. G proteins modulate D2 receptor-coupled K(ATP) channels in rat dopaminergic terminals. Neurochem Res. 2000;25(12):1521–6. doi: 10.1023/a:1026620316090. [DOI] [PubMed] [Google Scholar]
- 57.Neusch C, et al. The dopamine D2 receptor agonist alpha-dihydroergocryptine modulates voltage-gated sodium channels in the rat caudate-putamen. J Neural Transm (Vienna) 2000;107(5):531–41. doi: 10.1007/s007020070076. [DOI] [PubMed] [Google Scholar]
- 58.Masotti A, et al. Keppen-Lubinsky syndrome is caused by mutations in the inwardly rectifying K+ channel encoded by KCNJ6. Am J Hum Genet. 2015;96(2):295–300. doi: 10.1016/j.ajhg.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basel-Vanagaite L, Shaffer L, Chitayat D. Keppen-Lubinsky syndrome: Expanding the phenotype. Am J Med Genet A. 2009;149A(8):1827–9. doi: 10.1002/ajmg.a.32975. [DOI] [PubMed] [Google Scholar]
- 60.Megarbane A, et al. Temple-Baraitser Syndrome and Zimmermann-Laband Syndrome: one clinical entity? BMC Med Genet. 2016;17(1):42. doi: 10.1186/s12881-016-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mastrangelo M, et al. Epilepsy in KCNH1-related syndromes. Epileptic Disord. 2016;18(2):123–36. doi: 10.1684/epd.2016.0830. [DOI] [PubMed] [Google Scholar]
- 62.Bramswig NC, et al. 'Splitting versus lumping': Temple-Baraitser and Zimmermann-Laband Syndromes. Hum Genet. 2015;134(10):1089–97. doi: 10.1007/s00439-015-1590-1. [DOI] [PubMed] [Google Scholar]
- 63.Kortum F, et al. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat Genet. 2015;47(6):661–7. doi: 10.1038/ng.3282. [DOI] [PubMed] [Google Scholar]
- 64.Castori M, et al. Clinical and genetic study of two patients with Zimmermann-Laband syndrome and literature review. Eur J Med Genet. 2013;56(10):570–6. doi: 10.1016/j.ejmg.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 65.Perks T, et al. The orthodontic and surgical management of Zimmerman-Laband syndrome. Orthodontics (Chic.) 2013;14(1):e168–76. doi: 10.11607/ortho.897. [DOI] [PubMed] [Google Scholar]
- 66.Sawaki K, et al. Zimmermann-Laband syndrome: a case report. J Clin Pediatr Dent. 2012;36(3):297–300. doi: 10.17796/jcpd.36.3.k854128176u764l8. [DOI] [PubMed] [Google Scholar]
- 67.Splawski I, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 68.Diep V, Seaver LH. Long QT syndrome with craniofacial, digital, and neurologic features: Is it useful to distinguish between Timothy syndrome types 1 and 2? Am J Med Genet A. 2015;167A(11):2780–5. doi: 10.1002/ajmg.a.37258. [DOI] [PubMed] [Google Scholar]