Abstract
The role of Hedgehog signaling in normal and malignant T-cell development is controversial. Recently, Hedgehog pathway mutations have been described in T-ALL, but whether mutational activation of Hedgehog signaling drives T-cell transformation is unknown, hindering the rationale for therapeutic intervention. Here, we show that Hedgehog pathway mutations predict chemotherapy resistance in human T-ALL, and drive oncogenic transformation in a zebrafish model of the disease. We found Hedgehog pathway mutations in 16% of 109 childhood T-ALL cases, most commonly affecting its negative regulator PTCH1. Hedgehog mutations were associated with resistance to induction chemotherapy (P = 0.009). Transduction of wild-type PTCH1 into PTCH1-mutant T-ALL cells induced apoptosis (P = 0.005), a phenotype that was reversed by downstream Hedgehog pathway activation (P = 0.007). Transduction of most mutant PTCH1, SUFU and GLI alleles into mammalian cells induced aberrant regulation of Hedgehog signaling, indicating that these mutations are pathogenic. Using a CRISPR/Cas9 system for lineage-restricted gene disruption in transgenic zebrafish, we found that ptch1 mutations accelerated the onset of notch1-induced T-ALL (P = 0.0001), and pharmacologic Hedgehog pathway inhibition had therapeutic activity. Thus, Hedgehog-activating mutations are driver oncogenic alterations in high-risk T-ALL, providing a molecular rationale for targeted therapy in this disease.
INTRODUCTION
The Hedgehog signal transduction pathway stimulates growth and proliferation in multiple cell types during embryonic development (1–3). Loss-of-function mutations of PTCH1, a negative regulator of Hedgehog signaling, result in aberrant Hedgehog pathway activation, and are driver oncogenic mutations in some tumor types, including basal cell carcinoma, medulloblastoma and rhabdomyosarcoma (4–9). However, the role of Hedgehog signaling in normal and malignant T-cell development is controversial. In normal T-cell development, several lines of evidence suggest that Hedgehog signaling regulates proliferation or differentiation at multiple stages of T-cell development (10–14). However, the deletion of Smo in prethymic hematopoietic progenitors, which completely abolishes Hedgehog signaling, has no detectable effect on T-cell development (15, 16). Taken together, these findings suggest that acute changes in Hedgehog signaling can modulate T-cell output, but T-cell progenitors can adapt to chronic Hedgehog pathway inactivation, presumably due to compensation by alternative signals.
Hedgehog pathway mutations have recently been described in T-cell acute lymphoblastic leukemia (T-ALL) patient samples (17), and pharmacologic Hedgehog inhibitors demonstrate toxicity to T-ALL cases with autocrine Hedgehog activation (18). However, inactivating Hedgehog signaling has no effect on the onset or maintenance of Notch1-induced T-ALL (15), indicating that autocrine or paracrine Hedgehog signaling is not necessary for T-cell leukemogenesis. Further, despite the remarkable clinical activity of SMO inhibitors in tumors where Hedgehog-activating mutations are driver oncogenic lesions (19–22), clinical trials in tumor types with autocrine or paracrine Hedgehog activation have revealed little clinical benefit (23–26). Whether mutations that activate Hedgehog signaling drive T-cell transformation is unknown, hindering the rationale for therapeutic intervention.
Here, we report a high frequency of Hedgehog pathway mutations in T-ALL cases that are resistant to induction chemotherapy, and we show that Hedgehog signaling represses apoptosis in PTCH1-mutant T-ALL cells. We further demonstrate that ptch1 mutations accelerate the onset of notch1-induced T-ALL in transgenic zebrafish, demonstrating that mutational Hedgehog pathway activation is a driver oncogenic lesion in the molecular pathogenesis of T-ALL.
MATERIALS AND METHODS
T-ALL clinical specimens were collected from children with newly diagnosed T-ALL enrolled on Dana-Farber Cancer Institute Study 05-001 (27), or Children’s Oncology Group Study AALL0434 (28) clinical trials, with informed consent and institutional review board approval in accordance with the Declaration of Helsinki.
Complete methods are included in the SI Appendix.
RESULTS
Hedgehog pathway mutations in human T-ALL
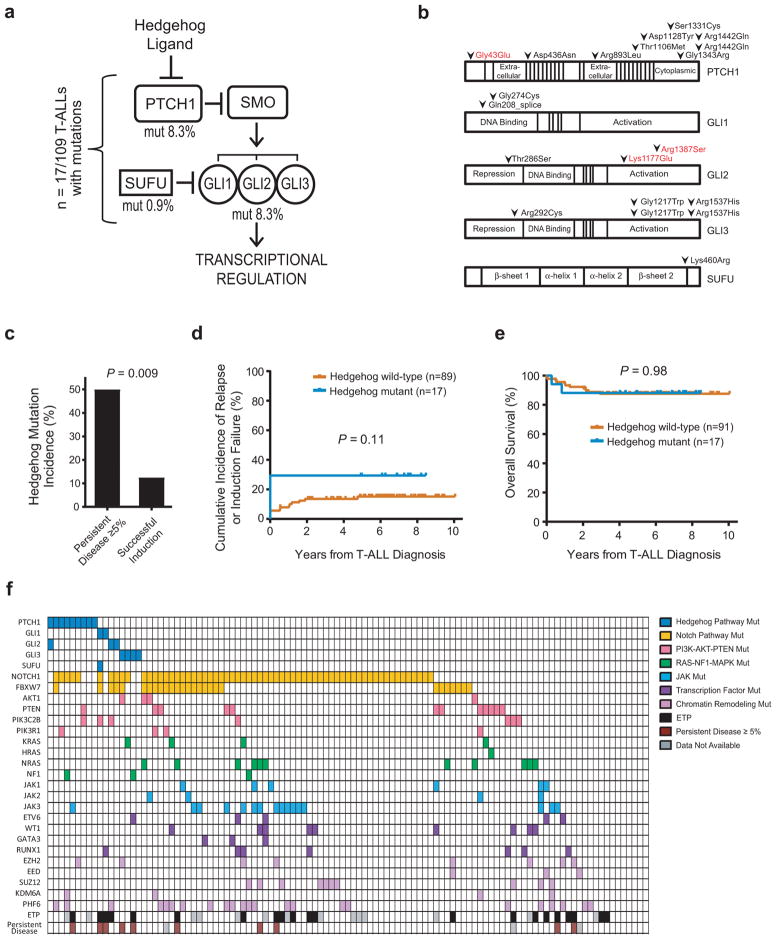
Using targeted exon sequencing of T-ALL diagnostic clinical specimens (Supplementary Tables 1 and 2), we found 20 mutations of genes encoding canonical Hedgehog pathway components in 16% (n = 17 of 109) of patient samples analyzed (Figure 1a). With the exception of one case with low mutant allele frequencies (MAF) (<0.1), the MAFs (0.34–0.65) were consistent with monoallelic mutations in the bulk tumor population (Supplementary Table 1). Two of the T-ALL cases harbored monoallelic mutations of two Hedgehog pathway genes, PTCH1/GLI2 and GLI1/SUFU, and a third case harbored two distinct mutations of GLI3 at low allele frequencies. The remaining 14 T-ALL cases had a single heterozygous mutation of the pathway (Supplementary Table 1). PTCH1 was the single most commonly mutated gene, with 9 cases harboring heterozygous missense PTCH1 mutations (Figure 1b), all of which were validated by Sanger sequencing (Supplementary Figure 1a). Sequencing of remission specimens, which were available in 5 of the 9 cases with PTCH1 missense mutations, revealed that 3 of these mutations were absent at remission, indicating a somatic origin, whereas 2 were detected in the remission specimen (Supplementary Figure 1b). We lacked a non-hematopoietic germline specimen to distinguish whether these were germline mutations, possibly indicating an underlying cancer predisposition syndrome, or somatic mutations associated with clonal hematopoiesis.
Figure 1. Hedgehog pathway mutations in childhood T-ALL.
(a) Hedgehog pathway genes mutated in T-ALL. (b) Amino acid alterations predicted to result from each of the mutations identified. Amino acid numbering based on the following transcripts: PTCH1, NM_000264 (mutations in black font), ENST00000375274.2 (red font). GLI1, NM_005269. GLI2, NM_005270 (black), ENST00000452319.1 (red). GLI3, NM_000168. SUFU, NM_016169. Note that mutations were annotated on the transcriptional variant predicted to be most significantly affected, with detailed annotation results shown in Supplementary Table 2. (c) Association of Hedgehog pathway mutations and resistance to induction chemotherapy, defined as ≥ 5% leukemic blasts in the bone marrow at end-induction by morphology (DFCI samples) or flow cytometric analysis (COG samples). Number of cases of persistent disease = 10; number of cases of successful induction = 96. A two-sided Fisher’s exact test was used to assess significance. (d) Analysis of cumulative incidence of relapse or induction failure in Hedgehog mutant (n = 17) or Hedgehog wild-type (n = 89) T-ALL patients. Note that induction deaths, which occurred in 2 Hedgehog wild-type cases, were defined as competing events in the cumulative incidence calculation. (e) Kaplan-Meier analysis of overall survival in Hedgehog mutant (n = 17) or Hedgehog wild-type (n = 91) T-ALL patients. Note that all early deaths in Hedgehog-mutant T-ALL were caused by leukemic progression. A two-sided log rank (Mantel-Cox) test was used to test for differences in survival between groups. (f) Summary of gene mutations identified in the T-ALL samples analyzed. “Persistent Disease ≥ 5%” indicates patients with induction failure or ≥ 5% end-induction minimal residual disease.
Hedgehog pathway mutations were associated with resistance to the initial induction cycle of intensive combination chemotherapy (Figure 1c), which suggests primary, or pre-existing, chemotherapy resistance. Hedgehog mutations were also associated with a trend towards an increased incidence of relapse or induction failure, which did not reach statistical significance (Figure 1d), but there was no difference in overall survival (Figure 1e). These findings suggest that Hedgehog-mutant T-ALL cases with a poor response to induction chemotherapy respond favorably to subsequent intensification of treatment, which is applied in response to high levels of residual leukemia. Hedgehog mutations were not significantly associated with other described biomarkers of early treatment failure, such as early T-cell precursor (ETP) phenotype or absence of biallelic TCRγ deletion (Supplementary Figure 2), suggesting that these markers identify largely non-overlapping groups of T-ALL cases at increased risk of early treatment failure. Hedgehog pathway mutations co-occurred with a number of oncogenic lesions typical of T-ALL, such as NOTCH1 and PRC2 mutations, and deletions of CDKN2A and TAL1 (Figure 1f and Supplementary Figure 3), although the incidence of these cooperating mutations was similar in Hedgehog-mutant versus wild-type T-ALL. These findings suggest that Hedgehog mutations can cooperate with a number of classical T-ALL oncogenic lesions in T-cell transformation.
Transduction of wild-type PTCH1 induces apoptosis in PTCH1-mutant T-ALL
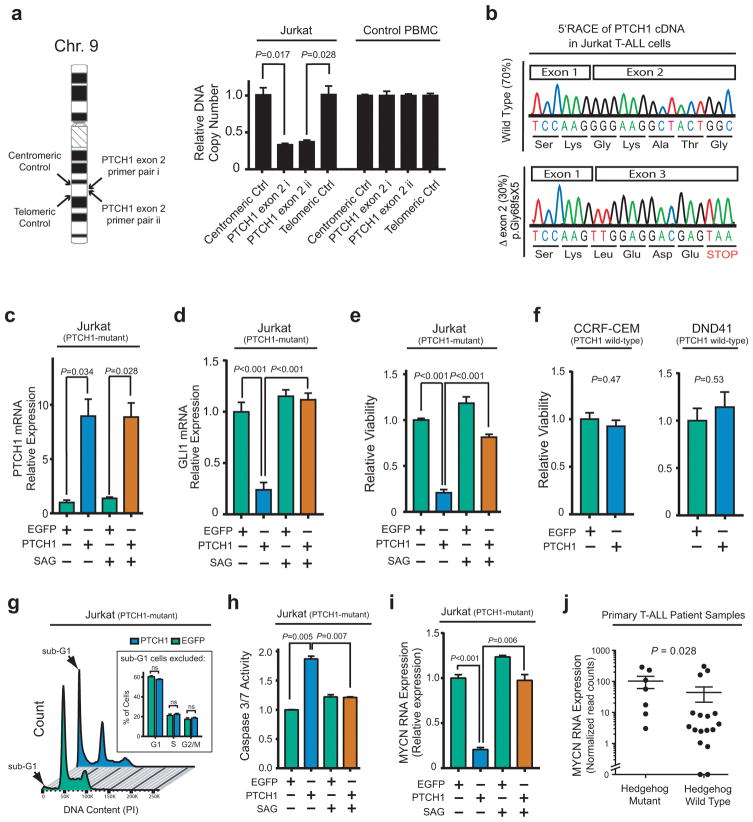
To investigate the role of Hedgehog signaling in T-ALL, we first focused on its negative regulator PTCH1, because this was the most commonly mutated Hedgehog pathway gene in this cohort. Analysis of the PTCH1 locus in a panel of human T-ALL cell lines revealed that Jurkat cells harbored a heterozygous microdeletion of exon 2 (Figure 2a), associated with production of a mutant PTCH1 mRNA predicted to encode a p.Gly68fsX5 truncated protein product (Figure 2b). We confirmed that this PTCH1 p.Gly68fsX5 mutant allele was unable to silence Hedgehog signaling, as assessed by mRNA expression of the canonical Hedgehog target Gli1, upon transduction into ASZ001 cells derived from a Ptch1 heterozygous knockout mouse (29, 30) (Supplementary Figure 4). In fact, expression of this mutant allele appeared to interfere with residual Ptch function in these cells. Thus, Jurkat T-ALL cells harbor a pathogenic PTCH1 mutation.
Figure 2. Aberrant hedgehog pathway activation is required for survival of PTCH1-mutant T-ALL cells.
(a) Quantitative DNA PCR analysis of DNA copy number using independent primer pairs located within exon 2 of PTCH1 (NM_000264), as well as centromeric and telomeric primer pair controls (location shown on left), on genomic DNA from Jurkat T-ALL cells. (b) 5′RACE analysis of PTCH1 mRNA transcripts expressed in Jurkat T-ALL cells. (c–d) Jurkat cells were transduced with wild-type PTCH1 or EGFP control, and treated with DMSO control or 100nM SAG to activate the Hedgehog pathway downstream of PTCH1. Quantitative reverse transcriptase PCR analysis was performed to assess mRNA expression of PTCH1 (c) or GLI1 (d), a canonical reporter of Hedgehog pathway activity. Results were normalized to β-Actin. A two-sided Welch t-test was used for statistical analysis of PTCH1 mRNA overexpression (c), and an ANOVA with Tukey adjustment for multiple comparisons was performed to assess differences in GLI1 mRNA expression (d). (e–f) Viability of the indicated T-ALL cells transduced with EGFP control or wild-type PTCH1, assessed 48 hours after selection in puromycin using trypan blue staining. A two-sided Welch t-test was used for statistical analysis. A Bonferroni correction was applied to adjust for multiple hypotheses testing (e). (g) Jurkat cells transduced with PTCH1 or EGFP control were harvested 48 hours after selection in puromycin and stained with PI for cell cycle analysis. A two-way ANOVA with Tukey adjustment was applied to assess differences in G1, S, and G2/M populations; p-values for each comparison in these three populations were not significant. (h) Caspase 3/7 activity in Jurkat cells transduced with PTCH1 versus EGFP control, and treated with 100 nM SAG or vehicle control (DMSO); results were normalized to DMSO-treated EGFP control. A two-sided Welch t-test with Bonferroni correction was applied for statistical analysis. (i) Relative MYCN mRNA expression (normalized to β-Actin control) was assessed by RT-PCR in Jurkat cells transduced with either EGFP or wild-type PTCH1, and treated with DMSO control or 100nM SAG to activate the Hedgehog pathway downstream of PTCH1. A two-sided Welch t-test with Bonferroni correction was used to assess significance. (j) Association of MYCN mRNA expression assessed using RNA sequencing analysis in 24 of the 109 patients in this study, with Hedgehog pathway mutations. A Wilcoxon sum-rank test was used to assess significance.
We then asked whether transducing wild-type PTCH1 inhibits Hedgehog signaling in PTCH1-mutant Jurkat cells. Transduction of wild-type PTCH1 downregulated Hedgehog signaling, an effect that was reversed by downstream Hedgehog pathway activation using SAG, a small molecule Smoothened agonist (Figure 2c–d). Transduction of PTCH1 impaired the viability of PTCH1-mutant Jurkat cells (Figure 2e), but had no toxicity to PTCH1 wild-type CCRF-CEM or DND41 T-ALL cells (Figure 2f). Importantly, the toxicity of PTCH1 transduction into PTCH1-mutant Jurkat cells was reversed by downstream Hedgehog pathway activation with SAG (Figure 2e), indicating on-target, Hedgehog-dependent toxicity. Cell cycle analysis revealed that transduction of wild-type PTCH1 induced formation of a large sub-G1 peak, suggesting induction of apoptosis, with little effect on the cell cycle distribution of live cells (Figure 2g). To confirm that PTCH1 transduction induced Hedgehog-dependent apoptosis in PTCH1-mutant T-ALL cells, we assessed effector caspase activity following transduction of PTCH1 or EGFP in Jurkat cells, alone or in combination with SAG treatment. This revealed that PTCH1 induced caspase activation, an effect that was reversed by downstream Hedgehog pathway activation using SAG (Figure 2h). Taken together, these findings demonstrate that Hedgehog signaling represses apoptosis in PTCH1-mutant T-ALL.
Hedgehog signaling positively regulates MYCN expression
The MYCN proto-oncogene is a transcriptional target of Hedgehog signaling in neuronal progenitors (31). MYCN is overexpressed in approximately 20% of human T-ALL via unknown mechanisms (32), and its overexpression drives T-cell transformation (33). Moreover, we have previously shown that overexpression of its paralog, MYC, represses mitochondrial apoptosis in T-ALL (34, 35). To test whether Hedgehog signaling regulates MYCN transcription in T-ALL, we transduced PTCH1-mutant Jurkat T-ALL cells with wild-type PTCH1 or EGFP control. We found that PTCH1 transduction downregulated MYCN mRNA expression, an effect that was rescued by downstream Hedgehog pathway activation (Figure 2i). Analysis of MYCN RNA expression in the subset of primary T-ALL patient samples on which RNA sequencing was available revealed that Hedgehog mutations were associated with increased MYCN expression (Figure 2j), but MYCN RNA expression was not associated with resistance to induction chemotherapy (P = 1.00 by Wilcoxon rank-sum test; data not shown). These findings indicate that MYCN is a transcriptional target of Hedgehog signaling in human T-ALL, but suggest that other Hedgehog targets may underlie chemotherapy resistance.
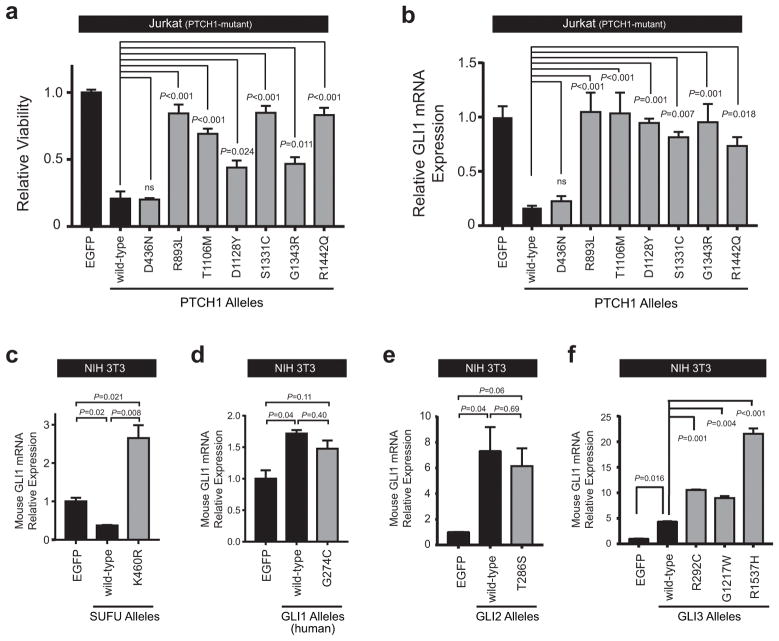
Most of the identified Hedgehog pathway mutations encode pathogenic alleles
Numerous missense mutations throughout the PTCH1 gene have been described in the cancer predisposition syndrome Gorlin syndrome (36, 37). While most of the PTCH1 mutations we identified in T-ALL were predicted to mutate evolutionarily conserved amino acid residues (Supplementary Figure 5), none of these are known to be pathogenic. To test whether the PTCH1 mutations identified are pathogenic, we transduced mutant alleles identified into the PTCH1-mutant T-ALL cell line Jurkat, and assessed their effects on leukemic cell viability and Hedgehog signaling in these cells. We found that 6 of the 7 mutant PTCH1 alleles tested had impaired growth-suppressive activity when compared to wild-type PTCH1 (Figure 3a), with a concordant impairment in their ability to suppress Hedgehog signaling, as assessed by GLI1 mRNA expression (Figure 3b). Transduction of wild-type or mutant PTCH1 alleles had no effect on the viability of PTCH1 wild-type DND41 cells (Supplementary Figure 6). Taken together, these data indicate that most of the PTCH1 mutations identified in T-ALL encode functionally impaired alleles.
Figure 3. Most Hedgehog pathway mutations encode pathogenic alleles.
(a) Jurkat cells were transduced with EGFP control, wild-type PTCH1, or PTCH1 mutant alleles identified within our patient cohort, and viability was assessed following selection in puromycin on day 5 using trypan blue staining. Results were normalized to EGFP control. A two-sided, one-way ANOVA with Dunnett’s adjustment for multiple comparisons was used for statistical analysis. (b) Relative GLI1 mRNA expression (normalized to β-Actin control) was assessed in Jurkat cells transduced with EGFP, wild-type PTCH1, or mutant PTCH1 alleles. A one-way ANOVA with Dunnett’s adjustment for multiple comparisons was used for statistical analysis. NIH 3T3 mouse fibroblast cells were transfected with EGFP control, (c) wild-type SUFU or K460R mutant SUFU allele, (d) wild-type GLI1 or G274C mutant GLI1 allele, (e) wild-type GLI2 or the T286S mutant GLI2 allele, or (f) wild-type GLI3, or one of GLI3 mutant alleles (R292C, G1217W, R1537H) identified within our T-ALL patient cohort, and relative mRNA expression of mouse Gli1 (normalized to mouse Gapdh control) was assessed using RT-PCR. Results were normalized to EGFP control. A one-way ANOVA with Dunnett’s adjustment for multiple comparisons was used for statistical analysis.
To test whether mutations of other Hedgehog pathway genes identified induce aberrant pathway activation, we turned to NIH 3T3 cells, a mouse cell line with low basal Hedgehog activity that is competent for Hedgehog pathway activation (38). SUFU encodes a negative regulator of Hedgehog signaling, and transduction of NIH 3T3 cells with wild-type SUFU further suppressed basal Hedgehog signaling in these cells (Figure 3c). By contrast, transduction of the SUFU K460R mutant led to a significant increase in Hedgehog pathway activity above that in basal conditions (Figure 3c), suggesting that this allele has dominant-negative activity and can interfere with function of the wild-type protein. To test the effect of wild-type or mutant GLI1 on Hedgehog pathway activity, we first confirmed that our RT-PCR assay for expression of endogenous mouse Gli1 mRNA (the marker for Hedgehog pathway activation) did not cross-react with the human GLI1 expression construct utilized (Supplementary Figure 7). Transduction of NIH 3T3 cells with the human GLI1 G274C mutant allele did not activate Hedgehog signaling beyond the degree of activation induced by wild-type GLI1 overexpression, suggesting that this mutation is not activating (Figure 3d). Similarly, transduction of the GLI2 T286S mutant into NIH 3T3 cells did not activate the Hedgehog pathway beyond that induced by wild-type GLI2 overexpression (Figure 3e). By contrast, all three GLI3 mutant alleles identified (R292C, G1217W and R1537H) induced significantly more Hedgehog pathway activation than wild-type GLI3, indicating that expression of these mutations is sufficient to induce aberrant Hedgehog pathway activation (Figure 3f). Taken together, these findings indicate that most of the Hedgehog pathway mutations identified in our T-ALL patient cohort are pathogenic. Indeed, of the 15 T-ALL cases harboring Hedgehog pathway mutations that were tested, 12 harbored mutations that were clearly pathogenic. We confirmed that the association of Hedgehog mutations with resistance to induction chemotherapy remains statistically significant if this analysis is limited to cases with known pathway activating mutations (Supplementary Figure 8; P = 0.011).
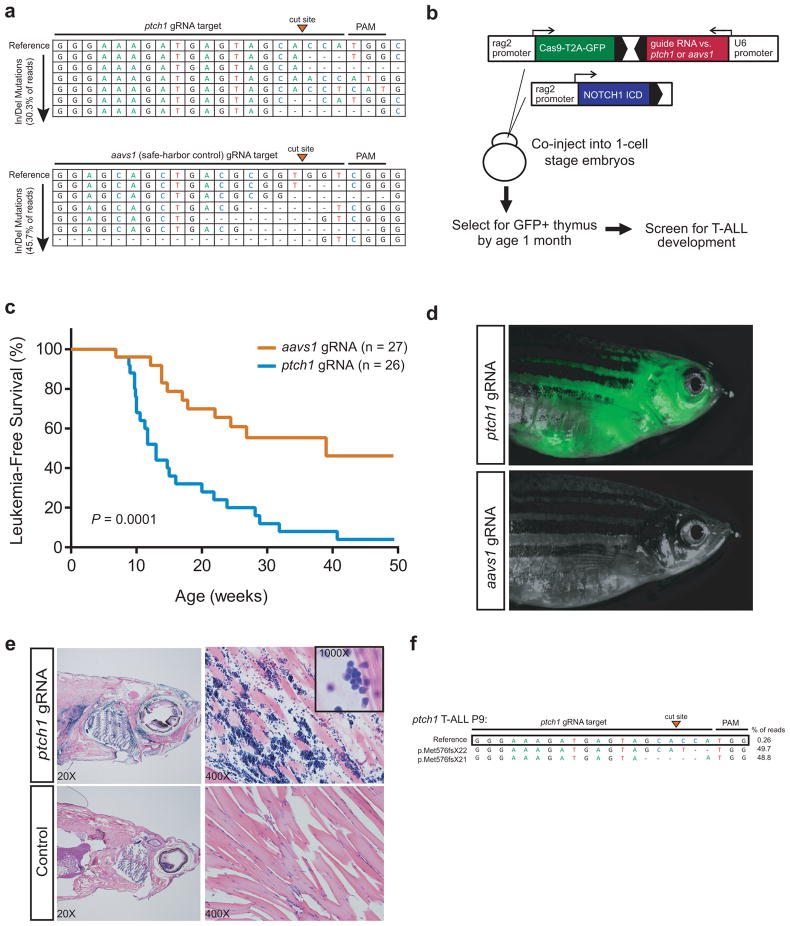
ptch1 mutations accelerate the onset of notch1-induced T-ALL
If Hedgehog-activating mutations promote T-cell transformation in part via MYCN upregulation, we reasoned that this effect would be best detected in collaboration with an oncogenic lesion that promotes MYC-independent transformation. We thus turned to the zebrafish model, where notch1a promotes T-cell transformation without upregulating myc (39, 40). To generate a CRISPR/Cas9 system for lineage-restricted gene disruption, we first designed a panel of guide RNAs (gRNAs) targeting exon 13 of zebrafish ptch1, because mutation of this exon is known to induce aberrant Hedgehog pathway activation (41). As a control for CRISPR/Cas9-induced DNA damage, we also designed gRNAs targeting the locus syntenic to the human safe-harbor AAVS1 locus, which is located within intron 1 of the ppp1r12c gene (42). To identify gRNAs that efficiently mutagenize each of these loci, individual gRNAs were co-injected with Cas9 mRNA into zebrafish embryos at the 1-cell stage, and cutting efficiency was assessed by next-generation sequencing of the target locus. This revealed gRNAs that induced insertion/deletion mutations in at least 30% of each target locus (Figure 4a). These gRNAs were then cloned into an expression vector in which the U6 promoter drives RNA polymerase III-mediated expression of the gRNA. Lineage-restricted gene disruption was achieved via a separate expression cassette within the same vector that drives expression of a Cas9-T2A-GFP self-cleaving protein under the control of the rag2 promoter (Figure 4b, top), which drives expression in immature T-cell progenitors.
Figure 4. Mutations of ptch1 accelerate the onset of notch1-induced T-ALL.
(a) Guide RNAs targeting ptch1 or the zebrafish locus syntenic to the human aavs1 safe-harbor locus were co-injected with Cas9 mRNA into zebrafish embryos at the 1-cell stage, genomic DNA was harvested 48h later, and cutting efficiency was assessed by next-generation sequencing. Results are shown for the most efficient gRNAs targeting each locus, which were used in the subsequent generation of transgenic zebrafish. Top line indicates the reference sequence, and individual reads harboring insertion or deletion (In/Del) mutations are shown in decreasing order of abundance. The total fraction of reads with In/Del mutations is shown in parentheses. (b) Schema of experimental design. (c) Kaplan-Meier analysis of zebrafish injected with the CRISPR/Cas9 construct targeting ptch1 or aavs1 (safe harbor control), together with rag2-notch1aICD (aavs1, n=27; ptch1, n=26). A two-sided Log-rank (Mantel-Cox) test was used to assess differences in survival between these two groups. (d) Representative ptch1 (top panel) or aavs1 (control) (bottom panel);rag2- notch1aICD double transgenic zebrafish at 20 weeks of age. (e) H&E staining of sagittal cross-section (left) and muscle (right) from ptch1 (top) and wild-type (bottom) age-matched zebrafish. (f) Next-generation sequencing of the locus targeted by ptch1 gRNA in a ptch1-mutant rag2- notch1aICD double transgenic zebrafish. Note that ptch1 is transcribed from the strand complementary to the gRNA target sequence shown here.
To test whether ptch1 mutations accelerate the onset of notch1-induced T-ALL, we co-injected wild-type zebrafish embryos at the 1-cell stage with the CRISPR/Cas9 expression vector targeting either ptch1 or aavs1, together with a rag2 promoter-driven activated notch1a allele (Figure 4b). Zebrafish with successful transgene integration and expression were identified by thymic GFP fluorescence, and all fish demonstrating thymic GFP fluorescence by 4 weeks of age were screened for T-ALL onset by weekly fluorescence microscopy. Expression of the ptch1-targeting CRISPR/Cas9 cassette significantly accelerated the onset and increased the penetrance of notch1-induced T-ALL, with a median time to T-ALL onset of 13 weeks in ptch1 fish, versus 39 weeks in the aavs1 controls (P=0.0001; Figure 4c–d). Histologic analysis of leukemic zebrafish revealed diffuse infiltration of normal tissues by small cells with round to slightly irregular nuclear contours, homogeneous chromatin, and scant cytoplasm (Figure 4e), consistent with leukemic T-lymphoblasts. Next-generation sequencing of the gRNA target locus in a subset of T-ALL cases revealed the presence of dominant mutations, confirming effective mutagenesis of the intended locus and indicating a clonal origin of these leukemias (Figure 4f).
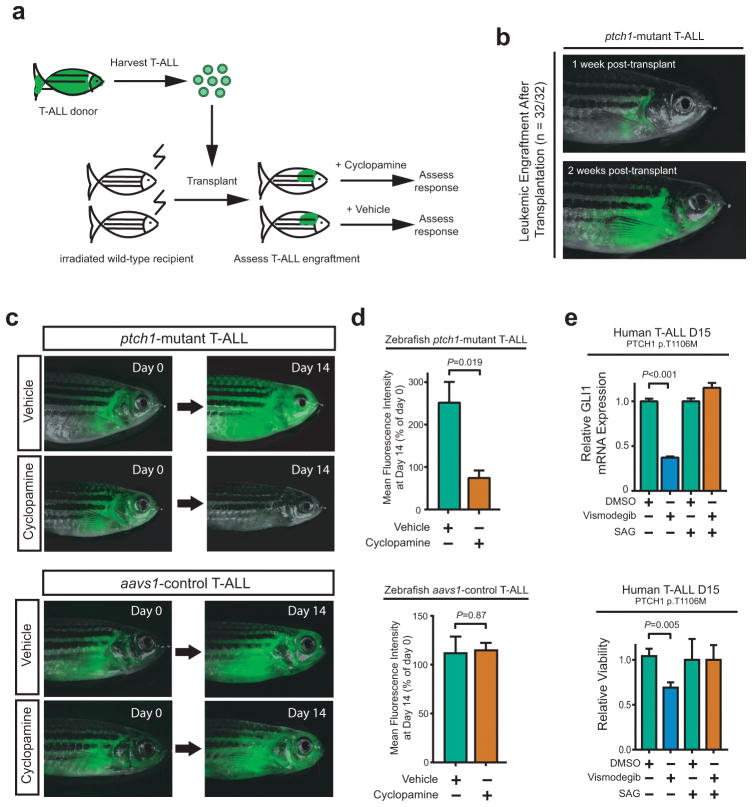
To assess the transplantability of zebrafish ptch1-mutant leukemias, T-ALL blasts were harvested from zebrafish with ptch1-mutant T-ALL, and injected into irradiated wild-type recipients (Figure 5a). We observed successful leukemic engraftment in 32 of 32 injected recipients, which rapidly progressed in all cases (Figure 5b), indicating that the aberrant growth and proliferation of these cells is cell-autonomous. Taken together, these findings demonstrate that mutational activation of Hedgehog signaling via disruption of its negative regulator ptch1 functions as an oncogenic driver in T-ALL.
Figure 5. Transplantability of ptch1-mutant T-ALL and therapeutic activity of cyclopamine.
(a) Schema of experimental design. (b) A representative adult ptch1-mutant recipient fish shown at day 7 (top) and 14 (bottom) following transplantation with ptch1; rag2-notch1aICD T-ALL. All 32 transplanted ptch1-mutant zebrafish demonstrated similar leukemic engraftment and rapid disease progression. Following leukemic engraftment, fish were treated with 1μM cyclopamine or vehicle control (ethanol) for 14 days. (c) Representative zebrafish in cyclopamine-treated and vehicle control (ethanol) for ptch1-mutant (top panel) and aavs1-control zebrafish (bottom panel) at the indicated time points. (d) Leukemic burden was assessed by mean fluorescence intensity (MFI), with MFI at day 14 normalized to day 0 for each individual fish. Of note, 5 ptch1-mutant fish from each group (vehicle and cyclopamine) met criteria for euthanasia due to radiation toxicity by day 14, and most others became ill soon thereafter, thus the study was terminated at this time point. A two-sided Welch t-test was used to assess differences among treatment groups. (Number of fish per group: ptch1-mutant recipient fish - Day 0, vehicle=10, cyclopamine=11; Day 14, vehicle=5, cyclopamine=5; aavs1-control recipient fish - Day 0, vehicle=9, cyclopamine=11; Day 14, vehicle=8, cyclopamine=9). (e) Cells from a primary T-ALL patient sample that harbored a p.T1106M PTCH1 mutation, which had been engrafted and expanded in immunodeficient mice, were treated with DMSO control, 10nM vismodegib, or 100nM SAG. After 48 hours, (top panel) relative GLI1 mRNA expression (to β-Actin control) and (bottom panel) viability using trypan blue staining were assessed. Results were normalized to DMSO control. Differences were assessed using a Welch t-test.
ptch1-mutant T-ALLs are responsive to Hedgehog pathway inhibition
We then asked whether pharmacologic Hedgehog pathway inhibition has in vivo therapeutic activity in ptch1-mutant T-ALL. Vismodegib is a potent and specific Smoothened inhibitor with clinical activity in Hedgehog-driven human cancers (19, 43), but this drug failed to inhibit zebrafish Smoothened, as evidenced by its inability to induce cyclopia in zebrafish embryos (data not shown). Thus, we leveraged the structurally unrelated Smoothened inhibitor cyclopamine, which effectively inhibits Hedgehog signaling in the zebrafish [(44) and data not shown]. T-ALL blasts were harvested from ptch1-mutant or aavs1-control fish, and transplanted into a cohort of irradiated recipients. After T-ALL engraftment, fish were treated with either cyclopamine or vehicle control. Assessment of tumor response by fluorescence microscopy after 2 weeks of treatment revealed tumor regression in most cyclopamine-treated ptch1-mutant zebrafish, versus marked tumor progression in vehicle controls (Figure 5c–d, top panels). By contrast, cyclopamine had no therapeutic activity against T-ALLs expressing the aavs1 control gRNA (Figure 5c–d, bottom panels), indicating that the effect of the ptch1-targeting gRNA on T-ALL is Hedgehog-dependent. Moreover, treatment of leukemic cells from patient T-ALL D15, which harbor a pathogenic PTCH1 T1106M mutation, with the FDA-approved Smoothened inhibitor vismodegib revealed that vismodegib inhibits Hedgehog signaling and impairs viability in these cells (Figure 5e). Thus, pharmacologic Hedgehog pathway inhibition has therapeutic activity against ptch1-mutant T-ALL.
DISCUSSION
Despite improvements achieved through the intensification of conventional cytotoxic chemotherapy, front-line therapy for T-ALL fails in 15–20% of children and 50–70% of adults (45–48), and these patients face a very poor prognosis (49–51). To date, no targeted therapies have been shown to improve clinical outcomes in this disease, highlighting the urgent clinical need for novel effective therapeutic approaches. We have identified a high frequency of Hedgehog pathway mutations in T-ALL cases with primary resistance to induction chemotherapy, and have demonstrated that mutations that activate Hedgehog signaling function as driver oncogenic events in the molecular pathogenesis of T-ALL. Given the remarkable clinical benefit of SMO inhibitors in tumors in which Hedgehog-activating mutations are driver oncogenic alterations (19–22), our findings thus provide a compelling rationale for clinical trials testing Hedgehog pathway inhibitors in patients with PTCH1-mutant T-ALL.
The 16% incidence of Hedgehog mutations we identified in childhood T-ALL is in line with 13% incidence reported by Dagklis and colleagues in a cohort of 67 cases of childhood and adult T-ALL (17). By contrast, a recent whole-genome sequencing (WGS) study revealed Hedgehog pathway mutations in only 4 of 264 T-ALL cases analyzed (33). One factor that likely contributed to this apparent discrepancy is the association of Hedgehog mutations with induction failure, whereas such cases appear to have been excluded from WGS analysis due to lack of a remission specimen with which to distinguish germline versus somatic variants. Additionally, accurately distinguishing missense substitutions that are pathogenic from those that are non-pathogenic passengers by informatics alone remains a major challenge, and all of the pathogenic Hedgehog mutations we identified were missense substitutions not previously known to be pathogenic. Thus, we speculate that strict algorithms for mutation calling, which are appropriately applied to WGS data to avoid the risk that the list of mutation calls will be dominated by non-pathogenic variants, may have missed pathogenic Hedgehog pathway mutations. These findings highlight the ability of functional genetics to complement genomic investigation of human disease.
Independent validation of the association of Hedgehog pathway mutations with early treatment resistance will be important before this biomarker is incorporated into clinical decision-making. Nevertheless, this finding suggests that Hedgehog signaling may directly mediate chemotherapy resistance. In acute myeloid leukemia, Hedgehog pathway activation mediates chemotherapy resistance by inducing glucuronidation of nucleoside analogs (52). Induction of apoptosis resistance provides an additional potential mechanism linking Hedgehog pathway activation to chemotherapy resistance, because mitochondrial apoptosis resistance provides one cellular mechanism for induction of resistance to conventional chemotherapy [(53–55) and unpublished observations]. However, the fact that Hedgehog mutations did not predict inferior overall survival suggests that Hedgehog-mutant T-ALLs respond favorably to the intensification of therapy applied in reaction to a poor response to induction chemotherapy. The treatment intensification strategies differed between the two clinical trials on which patients in this study were treated. On DFCI 05-001, patients with ≥5% leukemic blasts in the bone marrow at end-induction were deemed to have failed front-line therapy and taken off-study. While subsequent treatment was at the discretion of the treating physician, most of these patients likely underwent intensive re-induction chemotherapy followed by allogeneic hematopoietic stem cell transplantation after myeloablative doses of cyclophosphamide and total body irradiation. By contrast, on COG AALL0434, patients with 1–25% leukemic blasts at end-induction remained on study, but most were eligible for randomization to additional intensification of therapy with Nelarabine. While we remain blinded to outcomes of this randomization, it will be of considerable interest to determine whether the addition of nelarabine or allogeneic stem cell transplantation improves outcomes for patients with Hedgehog-mutant T-ALL. Even so, Hedgehog pathway inhibitors have a much more favorable toxicity profile than conventional therapy for high-risk T-ALL (19, 23, 24). Given the clear clinical activity of these drugs in tumors driven by Hedgehog-activating mutations (19–22), our findings provide a compelling rationale for clinical trials testing whether combining Hedgehog pathway inhibitors with conventional chemotherapy will improve clinical outcomes for patients with high-risk, PTCH1-mutant T-ALL.
Supplementary Material
Acknowledgments
We thank Julien Ablain for the pCS2-Cas9, pDest-Tol2-CG2-U6-gRNA, pME-MCS-zfCas9-T2A-GFP, and p3E-polyA vectors. We thank Christine Reynolds and Oscar Calzada for technical assistance; and Sofie Peirs, Bjorn Menten and Pieter Van Vlierberghe for the array CGH analysis. We thank Jessica Blackburn for the rag2-notch1aICD vector; and Jae Cho for the basal cell carcinoma cell line, ASZ001. We are grateful to the patients and families who provided samples for these studies. This work was supported by NIH R01 CA193651 (A.G.) and the Boston Children’s Hospital Translational Research Program (A.G.). The Children’s Oncology Group work was supported by U10 CA98543 (COG Chair’s grant), U10 CA98413 (COG Statistical Center), U24 CA114766 (COG Specimen Banking), U10 CA 180886 (COG Operations Center), and U10 CA 180899 (COG Statistics and Data Center). M.B. is supported by the Children’s Leukemia Research Association, Inc.. S.P.H. is the Jeffrey E. Perelman Distinguished Chair in the Department of Pediatrics, Children’s Hospital of Philadelphia. A.G. is supported by an Investigatorship at Boston Children’s Hospital.
Footnotes
CONFLICT OF INTEREST
The authors have no competing interests to declare.
AUTHOR CONTRIBUTIONS
M.A.B., Z.W.L, and A.G. conceived the project, designed and performed experiments, interpreted data, and wrote the manuscript. A.R.T. and M.D. performed the targeted exon sequencing and analysis. M.A.B., G.P.P., and A.G. analyzed targeted exon sequencing, RNA sequencing and array CGH data. M.A.B., E.A.S., and N.Y. performed and interpreted experiments. K.E.S. and D.S.N. performed statistical analyses and interpreted data. K.E.S., D.S.N., S.P.H., M.L.L., S.S.W., K.P.D., M.D., L.B.S., and S.E.S. aided in data collection of primary patient samples, and in analysis and interpretation of data. All authors aided in critical revision and approval of the submitted manuscript.
References
- 1.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews Molecular cell biology. 2013;14(7):416–29. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 2.Sharpe HJ, Wang W, Hannoush RN, de Sauvage FJ. Regulation of the oncoprotein Smoothened by small molecules. Nature chemical biology. 2015;11(4):246–55. doi: 10.1038/nchembio.1776. [DOI] [PubMed] [Google Scholar]
- 3.Duman-Scheel M, Weng L, Xin S, Du W. Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature. 2002;417(6886):299–304. doi: 10.1038/417299a. [DOI] [PubMed] [Google Scholar]
- 4.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272(5268):1668–71. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 5.Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–51. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 6.Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384(6605):129–34. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 7.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277(5329):1109–13. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 8.Hahn H, Wojnowski L, Zimmer AM, Hall J, Miller G, Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat Med. 1998;4(5):619–22. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- 9.Hettmer S, Teot LA, Kozakewich H, Werger AM, Davies KJ, Fletcher CD, et al. Myogenic tumors in nevoid Basal cell carcinoma syndrome. J Pediatr Hematol Oncol. 2015;37(2):147–9. doi: 10.1097/MPH.0000000000000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drakopoulou E, Outram SV, Rowbotham NJ, Ross SE, Furmanski AL, Saldana JI, et al. Non-redundant role for the transcription factor Gli1 at multiple stages of thymocyte development. Cell Cycle. 2010;9(20):4144–52. doi: 10.4161/cc.9.20.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Andaloussi A, Graves S, Meng F, Mandal M, Mashayekhi M, Aifantis I. Hedgehog signaling controls thymocyte progenitor homeostasis and differentiation in the thymus. Nature immunology. 2006;7(4):418–26. doi: 10.1038/ni1313. [DOI] [PubMed] [Google Scholar]
- 12.Furmanski AL, Saldana JI, Rowbotham NJ, Ross SE, Crompton T. Role of Hedgehog signalling at the transition from double-positive to single-positive thymocyte. European journal of immunology. 2012;42(2):489–99. doi: 10.1002/eji.201141758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Outram SV, Hager-Theodorides AL, Shah DK, Rowbotham NJ, Drakopoulou E, Ross SE, et al. Indian hedgehog (Ihh) both promotes and restricts thymocyte differentiation. Blood. 2009;113(10):2217–28. doi: 10.1182/blood-2008-03-144840. [DOI] [PubMed] [Google Scholar]
- 14.Rowbotham NJ, Hager-Theodorides AL, Furmanski AL, Ross SE, Outram SV, Dessens JT, et al. Sonic hedgehog negatively regulates pre-TCR-induced differentiation by a Gli2-dependent mechanism. Blood. 2009;113(21):5144–56. doi: 10.1182/blood-2008-10-185751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Graves S, Koch U, Liu S, Jankovic V, Buonamici S, et al. Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell stem cell. 2009;4(6):548–58. doi: 10.1016/j.stem.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann I, Stover EH, Cullen DE, Mao J, Morgan KJ, Lee BH, et al. Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell stem cell. 2009;4(6):559–67. doi: 10.1016/j.stem.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagklis A, Pauwels D, Lahortiga I, Geerdens E, Bittoun E, Cauwelier B, et al. Hedgehog pathway mutations in T-cell acute lymphoblastic leukemia. Haematologica. 2015;100(3):e102–5. doi: 10.3324/haematol.2014.119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dagklis A, Demeyer S, De Bie J, Radaelli E, Pauwels D, Degryse S, et al. Hedgehog pathway activation in T-cell acute lymphoblastic leukemia predicts response to SMO and GLI1 inhibitors. Blood. 2016;128(23):2642–54. doi: 10.1182/blood-2016-03-703454. [DOI] [PubMed] [Google Scholar]
- 19.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366(23):2171–9. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Von Hoff DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. The New England journal of medicine. 2009;361(12):1164–72. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 21.Robinson GW, Orr BA, Wu G, Gururangan S, Lin T, Qaddoumi I, et al. Vismodegib Exerts Targeted Efficacy Against Recurrent Sonic Hedgehog-Subgroup Medulloblastoma: Results From Phase II Pediatric Brain Tumor Consortium Studies PBTC-025B and PBTC-032. J Clin Oncol. 2015;33(24):2646–54. doi: 10.1200/JCO.2014.60.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang JY, Ally MS, Chanana AM, Mackay-Wiggan JM, Aszterbaum M, Lindgren JA, et al. Inhibition of the hedgehog pathway in patients with basal-cell nevus syndrome: final results from the multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2016;17(12):1720–31. doi: 10.1016/S1470-2045(16)30566-6. [DOI] [PubMed] [Google Scholar]
- 23.LoRusso PM, Rudin CM, Reddy JC, Tibes R, Weiss GJ, Borad MJ, et al. Phase I trial of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with refractory, locally advanced or metastatic solid tumors. Clin Cancer Res. 2011;17(8):2502–11. doi: 10.1158/1078-0432.CCR-10-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catenacci DV, Junttila MR, Karrison T, Bahary N, Horiba MN, Nattam SR, et al. Randomized Phase Ib/II Study of Gemcitabine Plus Placebo or Vismodegib, a Hedgehog Pathway Inhibitor, in Patients With Metastatic Pancreatic Cancer. J Clin Oncol. 2015;33(36):4284–92. doi: 10.1200/JCO.2015.62.8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaye SB, Fehrenbacher L, Holloway R, Amit A, Karlan B, Slomovitz B, et al. A phase II, randomized, placebo-controlled study of vismodegib as maintenance therapy in patients with ovarian cancer in second or third complete remission. Clin Cancer Res. 2012;18(23):6509–18. doi: 10.1158/1078-0432.CCR-12-1796. [DOI] [PubMed] [Google Scholar]
- 26.Rodon J, Tawbi HA, Thomas AL, Stoller RG, Turtschi CP, Baselga J, et al. A phase I, multicenter, open-label, first-in-human, dose-escalation study of the oral smoothened inhibitor Sonidegib (LDE225) in patients with advanced solid tumors. Clin Cancer Res. 2014;20(7):1900–9. doi: 10.1158/1078-0432.CCR-13-1710. [DOI] [PubMed] [Google Scholar]
- 27.Place AE, Stevenson KE, Vrooman LM, Harris MH, Hunt SK, O’Brien JE, et al. Intravenous pegylated asparaginase versus intramuscular native Escherichia coli L-asparaginase in newly diagnosed childhood acute lymphoblastic leukaemia (DFCI 05-001): a randomised, open-label phase 3 trial. Lancet Oncol. 2015;16(16):1677–90. doi: 10.1016/S1470-2045(15)00363-0. [DOI] [PubMed] [Google Scholar]
- 28.Winter SS, Dunsmore KP, Devidas M, Eisenberg N, Asselin BL, Wood BL, et al. Safe integration of nelarabine into intensive chemotherapy in newly diagnosed T-cell acute lymphoblastic leukemia: Children’s Oncology Group Study AALL0434. Pediatr Blood Cancer. 2015;62(7):1176–83. doi: 10.1002/pbc.25470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.So PL, Langston AW, Daniallinia N, Hebert JL, Fujimoto MA, Khaimskiy Y, et al. Long-term establishment, characterization and manipulation of cell lines from mouse basal cell carcinoma tumors. Experimental dermatology. 2006;15(9):742–50. doi: 10.1111/j.1600-0625.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 30.Aszterbaum M, Epstein J, Oro A, Douglas V, LeBoit PE, Scott MP, et al. Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat Med. 1999;5(11):1285–91. doi: 10.1038/15242. [DOI] [PubMed] [Google Scholar]
- 31.Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130(1):15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 32.Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017 doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutierrez A, Grebliunaite R, Feng H, Kozakewich E, Zhu S, Guo F, et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J Exp Med. 2011;208(8):1595–603. doi: 10.1084/jem.20101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds C, Roderick JE, LaBelle JL, Bird G, Mathieu R, Bodaar K, et al. Repression of BIM mediates survival signaling by MYC and AKT in high-risk T-cell acute lymphoblastic leukemia. Leukemia. 2014;28(9):1819–27. doi: 10.1038/leu.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindstrom E, Shimokawa T, Toftgard R, Zaphiropoulos PG. PTCH mutations: distribution and analyses. Hum Mutat. 2006;27(3):215–9. doi: 10.1002/humu.20296. [DOI] [PubMed] [Google Scholar]
- 37.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Human genetics. 2017;136(6):665–77. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406(6799):1005–9. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007;21(3):462–71. doi: 10.1038/sj.leu.2404546. [DOI] [PubMed] [Google Scholar]
- 40.Blackburn JS, Liu S, Raiser DM, Martinez SA, Feng H, Meeker ND, et al. Notch signaling expands a pre-malignant pool of T-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia. 2012;26(9):2069–78. doi: 10.1038/leu.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koudijs MJ, den Broeder MJ, Keijser A, Wienholds E, Houwing S, van Rooijen EM, et al. The zebrafish mutants dre, uki, and lep encode negative regulators of the hedgehog signaling pathway. PLoS Genet. 2005;1(2):e19. doi: 10.1371/journal.pgen.0010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2011;12(1):51–8. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 43.Robarge KD, Brunton SA, Castanedo GM, Cui Y, Dina MS, Goldsmith R, et al. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg Med Chem Lett. 2009;19(19):5576–81. doi: 10.1016/j.bmcl.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 44.Neumann CJ, Grandel H, Gaffield W, Schulte-Merker S, Nusslein-Volhard C. Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development. 1999;126(21):4817–26. doi: 10.1242/dev.126.21.4817. [DOI] [PubMed] [Google Scholar]
- 45.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21(19):3616–22. doi: 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 46.Marks DI, Paietta EM, Moorman AV, Richards SM, Buck G, DeWald G, et al. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993) Blood. 2009;114(25):5136–45. doi: 10.1182/blood-2009-08-231217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children’s oncology group. J Clin Oncol. 2012;30(14):1663–9. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360(26):2730–41. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko RH, Ji L, Barnette P, Bostrom B, Hutchinson R, Raetz E, et al. Outcome of patients treated for relapsed or refractory acute lymphoblastic leukemia: a Therapeutic Advances in Childhood Leukemia Consortium study. J Clin Oncol. 2010;28(4):648–54. doi: 10.1200/JCO.2009.22.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrappe M, Hunger SP, Pui CH, Saha V, Gaynon PS, Baruchel A, et al. Outcomes after induction failure in childhood acute lymphoblastic leukemia. The New England journal of medicine. 2012;366(15):1371–81. doi: 10.1056/NEJMoa1110169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fielding AK, Richards SM, Chopra R, Lazarus HM, Litzow MR, Buck G, et al. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood. 2007;109(3):944–50. doi: 10.1182/blood-2006-05-018192. [DOI] [PubMed] [Google Scholar]
- 52.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511(7507):90–3. doi: 10.1038/nature13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vo TT, Ryan J, Carrasco R, Neuberg D, Rossi DJ, Stone RM, et al. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151(2):344–55. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhola PD, Mar BG, Lindsley RC, Ryan JA, Hogdal LJ, Vo TT, et al. Functionally identifiable apoptosis-insensitive subpopulations determine chemoresistance in acute myeloid leukemia. The Journal of clinical investigation. 2016;126(10):3827–36. doi: 10.1172/JCI82908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, del Moore VG, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334(6059):1129–33. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.