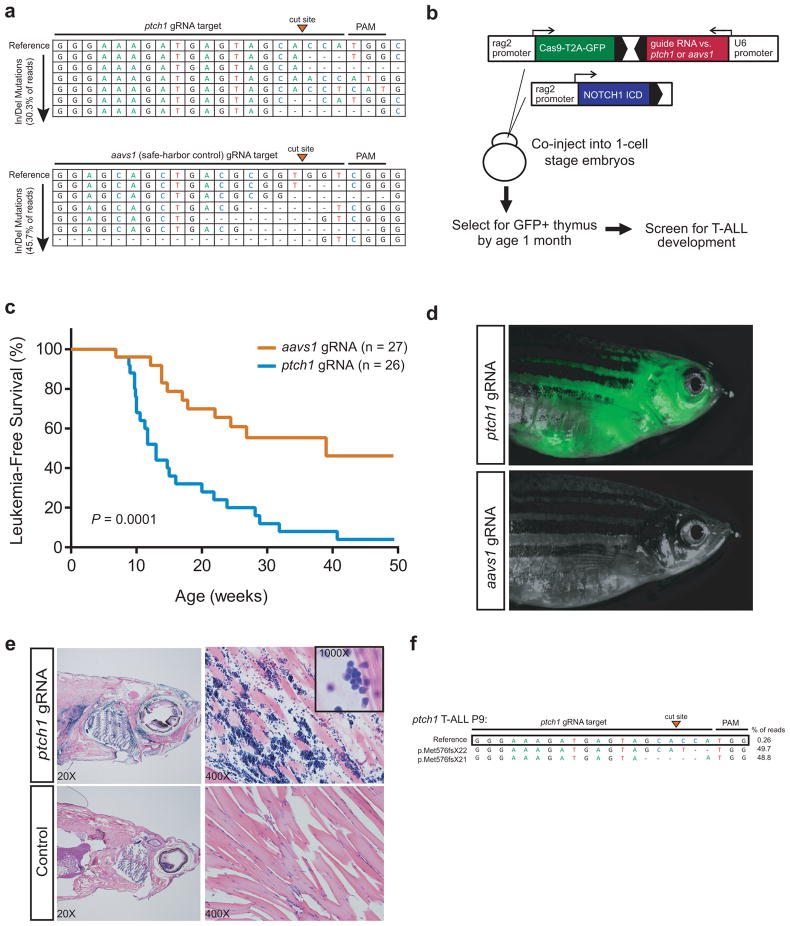
Figure 4. Mutations of ptch1 accelerate the onset of notch1-induced T-ALL.
(a) Guide RNAs targeting ptch1 or the zebrafish locus syntenic to the human aavs1 safe-harbor locus were co-injected with Cas9 mRNA into zebrafish embryos at the 1-cell stage, genomic DNA was harvested 48h later, and cutting efficiency was assessed by next-generation sequencing. Results are shown for the most efficient gRNAs targeting each locus, which were used in the subsequent generation of transgenic zebrafish. Top line indicates the reference sequence, and individual reads harboring insertion or deletion (In/Del) mutations are shown in decreasing order of abundance. The total fraction of reads with In/Del mutations is shown in parentheses. (b) Schema of experimental design. (c) Kaplan-Meier analysis of zebrafish injected with the CRISPR/Cas9 construct targeting ptch1 or aavs1 (safe harbor control), together with rag2-notch1aICD (aavs1, n=27; ptch1, n=26). A two-sided Log-rank (Mantel-Cox) test was used to assess differences in survival between these two groups. (d) Representative ptch1 (top panel) or aavs1 (control) (bottom panel);rag2- notch1aICD double transgenic zebrafish at 20 weeks of age. (e) H&E staining of sagittal cross-section (left) and muscle (right) from ptch1 (top) and wild-type (bottom) age-matched zebrafish. (f) Next-generation sequencing of the locus targeted by ptch1 gRNA in a ptch1-mutant rag2- notch1aICD double transgenic zebrafish. Note that ptch1 is transcribed from the strand complementary to the gRNA target sequence shown here.