To adapt to ever-changing environments, pathogens quickly alter gene expression. This can occur through transcriptional, posttranscriptional, or posttranslational regulation.
KEYWORDS: EHEC, enterohemorrhagic E. coli, pathogenesis, posttranscriptional regulation, regulation of gene expression, sRNA, virulence
ABSTRACT
To adapt to ever-changing environments, pathogens quickly alter gene expression. This can occur through transcriptional, posttranscriptional, or posttranslational regulation. Historically, transcriptional regulation has been thoroughly studied to understand pathogen niche adaptation, whereas posttranscriptional and posttranslational gene regulation has only relatively recently been appreciated to play a central role in bacterial pathogenesis. Posttranscriptional regulation may involve chaperones, nucleases, and/or noncoding small RNAs (sRNAs) and typically controls gene expression by altering the stability and/or translation of the target mRNA. In this review, we highlight the global importance of posttranscriptional regulation to enterohemorrhagic Escherichia coli (EHEC) gene expression and discuss specific mechanisms of how EHEC regulates expression of virulence factors critical to host colonization and disease progression. The low infectious dose of this intestinal pathogen suggests that EHEC is particularly well adapted to respond to the host environment.
INTRODUCTION
Upon infecting a host, bacterial pathogens encounter many unique environments. To survive in each of these environments, pathogens must quickly alter gene expression. A major mechanism by which this occurs is through transcriptional regulation, during which transcription factors bind promoter sequences to recruit or occlude RNA polymerase to/from target promoters (1, 2). Because of the importance in global gene regulation, transcriptional regulation has been studied extensively in a variety of bacterial pathogens (see, e.g., references 3 to 7). However, transcriptional regulation is not the whole picture. Bacteria also coordinate gene expression at the posttranscriptional and posttranslational levels. These mechanisms of gene regulation provide distinct advantages to bacterial fitness by enabling bacterial pathogens to rapidly alter gene expression. For example, immediately after a stimulus is sensed by a bacterium, posttranscriptional regulation enables fast responses to adjust the expression and/or activities of proteins (8). Moreover, posttranscriptional regulation enables bacterial pathogens to fine-tune gene expression by uncoupling transcription and translation. This is beneficial because functionally related genes may be organized within an operon and thus be cotranscribed. However, a bacterium may require different levels of the resulting proteins, which can be achieved by selectively altering translation (9). Posttranslational regulation occurs mainly through protein-protein interactions and modifies the activity or amount of protein in the cell. Although posttranslational regulation may be energetically costly (by consuming ATP to degrade proteins), this type of regulation is rapid, robust, and sometimes reversible, thus allowing cells to quickly respond to stimuli (8).
Enterohemorrhagic Escherichia coli O157:H7 (EHEC) causes major outbreaks of foodborne illness in developed nations. The primary reservoir of EHEC is the gastrointestinal tract of ruminant animals, and people become ill following ingestion of contaminated food or water (10). Disease consists largely of hemorrhagic colitis, but infection can lead to the development of the potentially fatal complication hemolytic-uremic syndrome (HUS) (11). HUS complicates 6 to 9% of EHEC infections overall and approximately 15% of EHEC infections in children under age 10 (12–15). Antibiotics are thought to promote expression of Shiga toxin and thereby worsen clinical manifestations of EHEC disease (16, 17). Therefore, treatment for EHEC is currently limited to supportive care, i.e., rehydration therapy (18, 19). In addition to causing severe illness, EHEC outbreaks place a heavy economic burden on the agricultural industry due to recalls of contaminated food products (20).
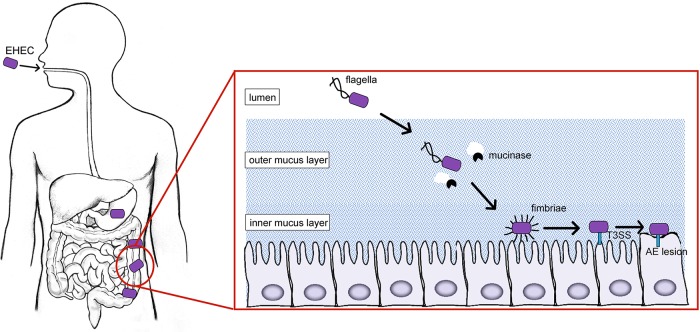
Following ingestion, EHEC survives and/or thrives within different host niches. For example, EHEC survives passage through the acidic stomach by expressing acid stress proteins (21–23). Subsequently, EHEC travels through the small intestine to reach the major site of colonization, the colon (24). Here, EHEC expresses distinct virulence factors that enable EHEC to move through the intestine and attach to enterocytes (Fig. 1). For example, EHEC produces mucinases and flagella to break down and penetrate through the mucus layer, respectively, and move to the epithelium (25–27). At the epithelial border, EHEC is thought to express adhesins that mediate initial adherence, which is then followed by expression of a type three secretion system (T3SS) and effectors that result in the formation of attaching-and-effacing (AE) lesions. AE lesions are characterized by effacement of microvilli and actin cytoskeletal rearrangement that results in the formation of a pedestal-like structure beneath the bacterium and intimate attachment of EHEC to enterocytes (28–30). Most of the genes involved in AE lesion formation are carried within a chromosomal pathogenicity island called the locus of enterocyte effacement (LEE) (31). The LEE contains five major operons (LEE1 to -5) that encode a T3SS and effector proteins (31). EHEC's arsenal of virulence factors also includes non-LEE-encoded effector proteins that influence colonization or modulate immune responses (32–39). Finally, during infection, EHEC also produces Shiga toxin, which is responsible for the severe morbidity and mortality associated with EHEC disease (11). Significantly, ingestion of as few as 10 to 100 cells is sufficient for EHEC to establish infection (40), suggesting that EHEC employs mechanisms to quickly and precisely coordinate gene expression to survive passage through the stomach and the small intestine and then mediate attachment to the colonic epithelium. Indeed, various environmental stimuli alter the transcriptional regulation of virulence traits (25, 41, 42). Increasing evidence reveals that posttranscriptional gene regulation occurs at every stage of EHEC infection. In this review, we provide an overview of key factors involved in posttranscriptional regulation in EHEC and include relevant details of posttranslational regulation when appropriate. When they are known, we also summarize mechanisms of how expression of EHEC virulence traits is controlled via these regulatory factors.
FIG 1.
EHEC colonization of the colon requires precise spatiotemporal expression of genes required to move through the lumen and attach to enterocytes (see the text for details).
POSTTRANSCRIPTIONAL REGULATION PLAYS A GLOBAL ROLE IN EHEC GENE EXPRESSION
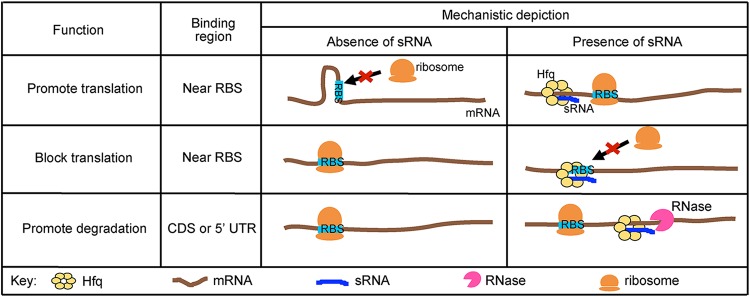
Small RNAs (sRNAs) are global mediators of posttranscriptional regulation. sRNAs are typically noncoding RNAs that are 50 to 300 nucleotides in length and regulate expression of mRNAs through base pairing (43–46). sRNAs have previously been classified as cis or trans encoded; however, the term cis encoded can lead to some confusion, as sRNAs can be encoded within the 5′ and 3′ untranslated regions (UTRs) as well as on the noncoding DNA strand. Therefore, in this review we refer to the two classes as antisense sRNAs and trans-acting sRNAs. Antisense sRNAs are carried on the DNA strand opposite from the regulated gene and bind with extensive perfect complementarity. In contrast, trans-acting sRNAs are located in a distal location from the target mRNA(s) and usually bind to target mRNAs over short regions with imperfect complementarity, which enables a trans-acting sRNA to regulate multiple genes (47, 48). However, this feature frequently requires an RNA chaperone to facilitate binding interactions (43–45). In bacteria, the canonical mechanism of sRNA-mediated regulation occurs through interaction with the 5′ UTR of the target mRNA (Fig. 2), in which the sRNA sequesters the ribosome binding site (RBS) and inhibits translation (49, 50). Conversely, an sRNA may disrupt mRNA structures that occlude the RBS and thereby promote translation (51–53). Significantly, a few recent studies revealed that sRNAs can directly affect stability of a target mRNA, independently of affecting translation initiation (54–59) (Fig. 2). In these examples, the sRNA binds upstream, to the 5′ UTR or within the coding sequence (CDS) of the target mRNA (54, 57, 59).
FIG 2.
Canonical mechanisms of posttranscriptional regulation by trans-acting sRNAs. mRNAs can have secondary structures that occlude the ribosome binding site (RBS). With the help of the RNA chaperone Hfq, a small RNA (sRNA) promotes translation by binding to the target mRNA, thereby relieving secondary structures that may occlude the RBS. Alternatively, an mRNA may be efficiently translated in the absence of an sRNA, but the presence of an sRNA can inhibit translation by binding to or near the RBS and blocking ribosome access. Furthermore, an sRNA may promote degradation by recruiting RNases to the target mRNA.
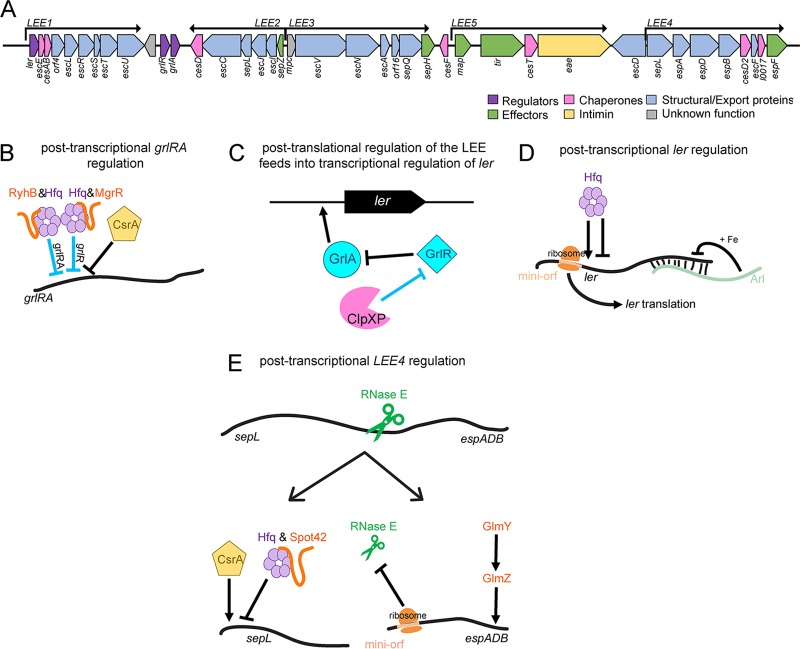
Comparative genome analysis of 17 E. coli genomes (including two lab-adapted strains, one human commensal strain, and 14 pathovars [two of which were EHEC]) revealed that of approximately 4,000 to 5,000 genes per genome, only about 2,200 genes are conserved among all strains (60). These shared genes are classified as belonging to the core genome. To date, most knowledge of sRNAs in EHEC is derived from studies that were performed using nonpathogenic E. coli as the model organism and thus were based on core sRNAs. These studies have provided invaluable insights in understanding the extent and mechanisms of posttranscriptional regulation in the Enterobacteriaceae; however, the EHEC genome contains approximately 1.34 Mb of unique DNA sequences that are carried in pathogenicity islands (also referred to as O-islands) compared to nonpathogenic E. coli (61, 62). Therefore, it is likely that EHEC contains sRNAs that are not present in nonpathogenic E. coli and/or that the regulon of core sRNAs is more expansive in EHEC and includes regulation of virulence factors. In support of this, Tree et al. reported that the density of sRNAs encoded in O-islands is 39 sRNAs per Mb of DNA, compared to 23 sRNAs per Mb of DNA in the core genome (63). Furthermore, transcriptome sequencing (RNA-seq) experiments revealed 35 to 55 sRNAs unique to EHEC (63–65), of which only a handful have been characterized (Table 1). Additionally, RNA-seq experiments showed that core genome-encoded sRNAs regulate EHEC-specific genes, perhaps through mechanisms that are distinct from those characterized in nonpathogenic E. coli (Table 1). For example, the Hfq-dependent sRNAs GlmY and GlmZ provide feedback control synthesis of GlmS, an enzyme required for cell envelope synthesis (66). In EHEC, GlmY and GlmZ also regulate LEE expression as well as expression of the non-LEE-encoded effector NleA (64) (Fig. 3A). Moreover, in nonpathogenic E. coli, these sRNAs are characterized to work in concert to regulate gene expression (53). However, microarray analyses showed that a deletion of glmY affects EHEC gene expression differently from a deletion of glmZ (64), suggesting that these sRNAs may also have independent functions.
TABLE 1.
Core and EHEC-specific sRNAs that contribute to EHEC pathogenesis
| General process targeted | sRNA | Target RNA | Type of regulation | Direct interaction | Reference(s) |
|---|---|---|---|---|---|
| Adherence | sRNA103 | fimZ | Positive | Predicted | 64 |
| Global regulation | AgvB | gcvB | Negative | Yes | 63 |
| Iron homeostasis | AsxR | fnsR | Negative | Yes | 63 |
| Esr41 | Bfr | Negative | Yes | 87 | |
| chuA | Negative | Yes | 87 | ||
| cirA | Negative | Yes | 87 | ||
| LEE and effectors | GlmY/Z | LEE4 | Negative | Yes | 130 |
| LEE5 | Negative | Unknown | 130 | ||
| nleA | Positive | Predicted no | 64 | ||
| espFu | Positive | Predicted direct | 130 | ||
| sRNA56 | espA | Positive | Predicted no | 64 | |
| sRNA103 | espA | Positive | Predicted no | 64 | |
| sRNA350 | Unknown | Positive | Unknown | 64 | |
| Arl | ler | Negative | Antisense sRNA | 119 | |
| Esr055 | Unknown | Positive | Unknown | 147 | |
| Spot42 | sepL | Negative | Yes | 87, 111 | |
| RyhB | grlRA | Negative | Yes | 112 | |
| MgrR | grlR | Negative | Yes | 112 | |
| Metabolism | sRNA56 | ureG | Positive | Predicted direct | 64 |
| Motility | Esr41 | fliC | Positive | Unknown | 175 |
FIG 3.
Comparison of the regulation and expression of core sRNAs in nonpathogenic E. coli and EHEC. (A) The GlmY/GlmZ regulon in nonpathogenic E. coli versus EHEC. (B) The EHEC-specific sRNA AgvB regulates expression of the core sRNA GcvB. Black lines indicate regulation that is conserved between nonpathogenic E. coli and EHEC, whereas red lines indicate regulation unique to EHEC. Arrows indicate positive regulation, and blunted lines indicate negative regulation.
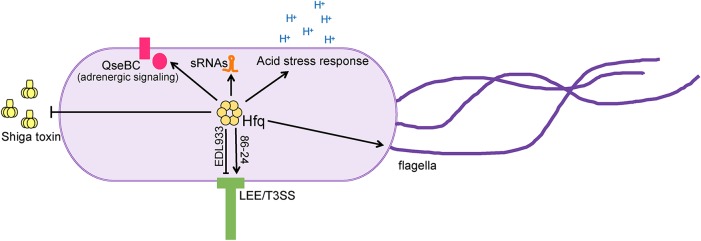
The chaperone Hfq associates with many trans-encoded sRNAs to modulate gene expression (67, 68) (Fig. 2). Hfq is a hexameric protein that binds both sRNAs and mRNAs to facilitate interactions as well as to promote sRNA stability (69, 70). Initial, unbiased studies to examine posttranscriptional regulation in EHEC investigated Hfq-dependent changes in gene expression using microarray analyses (71–73). A deletion of hfq caused extensive changes in gene expression (Fig. 4), including expression of genes encoding iron acquisition, acid stress responses, regulatory proteins, flagella and motility, and sRNAs. The hfq deletion also resulted in increased expression of Shiga toxin (72). The importance of Hfq in EHEC virulence gene expression is further underscored in that these studies each demonstrated that Hfq affects LEE expression (71–73). Interestingly, these data demonstrated that Hfq influences LEE expression differently depending on the strain of EHEC (negatively in strain EDL933 versus positively in strain 86-24) (71–73). Strains of EHEC studied in the lab vary considerably, and the most commonly studied EHEC strains are 86-24, Sakai, and EDL933, which were isolated from patients presenting with hemorrhagic colitis (86-24 and Sakai) (61, 74) or from meat associated with an EHEC outbreak (EDL933) (75). The genomes of the EDL933, Sakai, and 86-24 EHEC strains exhibit significant variation in 27 regions of the genome in addition to differences in prophage content (76). Therefore, it is likely that these variable regions and prophages of the EHEC genomes vary in sRNA content and contribute to differences in subsequent regulation of virulence factors.
FIG 4.
Hfq influences expression of virulence and regulatory factors in EHEC. Strain-dependent regulation of the LEE is indicated. Arrows indicate positive regulation, and blunted lines indicated negative regulation.
Although these studies provided an unbiased characterization of the Hfq regulon, a limitation was that the association of Hfq with a particular sRNA or specific target could not be determined (solely using microarrays). This issue is beginning to be resolved with the use of a technique called UV-induced RNA-protein cross-linking and analysis of cDNA by high-throughput sequencing (CRAC) (77). Generally, in these experiments, Hfq (or another protein) is chromosomally tagged and used as bait to capture interacting RNAs which are then cross-linked. Following Hfq purification and then subsequent digestion, the RNAs can be identified by RNA-seq. Using this method, Tree et al. identified 55 EHEC-specific sRNAs that interact with Hfq (63). Many of these EHEC-specific sRNAs are encoded within prophages and antagonize the function of other sRNAs, and thus they were termed anti-sRNAs (63). For example, one anti-sRNA, named AgvB, targets the core genome-encoded sRNA GcvB, which controls expression of genes encoding diverse functions (63, 78–81) (Fig. 3B). GcvB controls expression of the response regulator PhoP, which regulates acid resistance, magnesium homeostasis, and cell envelope homeostasis (82), as well as the sigma factor RpoS, which is involved in various stress response pathways, including acid resistance (83). GcvB also modulates expression of peptide transport systems, including decreasing expression of the dipeptide transporter DppA (63, 78–81). Based on data generated using a three-plasmid coexpression system, the anti-sRNA AgvB relieves GcvB repression of DppA. These data were substantiated by subsequent experiments in which DppA expression was measured in agvB-null strain of EHEC. Finally, the agvB-null strain was outcompeted by wild-type (WT) EHEC specifically when grown in mucus obtained from the terminal rectum of the bovine gastrointestinal (GI) tract (63), suggesting that anti-sRNAs are important for EHEC host colonization.
sRNAs may recruit RNases to destabilize target mRNA, and the RNase RNase E is responsible for the majority of RNA processing and turnover in E. coli (84–86). Based on this information, Waters et al. tagged RNase E and performed UV-cross-linking, ligation, and sequencing of hybrids (CLASH) to characterize the sRNA interactome in EHEC (87). CLASH is similar to CRAC but includes an RNA ligation step following cross-linking to purify sRNAs and the corresponding mRNA target. Using this method, Waters et al. identified nearly 2,000 sRNA-mRNA interactions, 152 unique sRNA-sRNA interactions (or “anti-sRNA”-sRNA interactions), and 320 unique sRNA-tRNA interactions (87). Whereas sRNA-mRNA and sRNA-sRNA interactions regulate gene expression, sRNA-tRNA interactions are hypothesized to serve as sponges against sRNA activity (87). As sponges, tRNAs interact with sRNAs, preventing sRNA-mRNA and sRNA-sRNA interactions. In this manner, the tRNA-sRNA interactions act to dampen sRNA activity. Although the importance of these RNA-RNA interactions in EHEC pathogenesis is not fully understood, the idea that tRNAs antagonize sRNA activity adds a new level of complexity to RNA-based regulation.
RNase E can act independently to cleave RNA molecules or in a complex called the degradosome (86). The C terminus of RNase E is a scaffold that recruits the other components of the degradosome, including the RNA helicase RhlB, the glycolytic enzyme enolase, and the polynucleotide phosphorylase (PNPase) (88–90). RhlB relieves secondary structures in mRNA (88), enolase influences RNase E localization in the cell and thus modifies RNase E activity (91), and PNPase degrades cleaved mRNA molecules (86, 92). A deletion of pnp (which encodes the PNPase enzyme) caused various effects on virulence gene expression in EHEC, including a decrease in expression of Shiga toxin and adherence to colonic epithelial cells in vitro as well as an increase in expression of some of the LEE-carried genes (93). Although this study underscored a role for PNPase in EHEC pathogenesis, the mechanism of PNPase-dependent regulation of the LEE and Shiga toxin has not yet been reported.
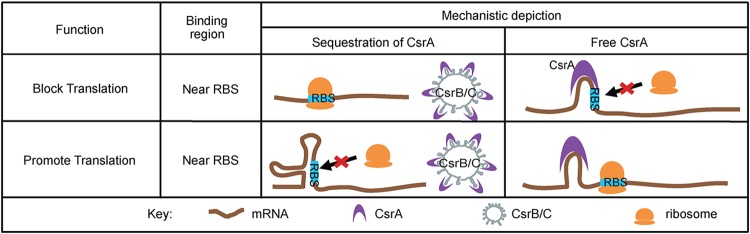
The CsrA (carbon storage regulatory) protein binds RNA and plays an extensive role in gene regulation in a myriad of bacteria (94). CsrA typically represses translation initiation by binding to the Shine-Dalgarno (SD) sequence and blocking ribosomal binding (95–97); however, CsrA has also been shown to activate translation and promote RNA stability (98, 99) (Fig. 5). CsrA activity is regulated by the sRNAs CsrB and CsrC. CsrB and CsrC bind multiple molecules of CsrA with a relatively high affinity and thus prevent CsrA from interacting with regulatory targets (94) (Fig. 5). The CsrA regulon in EHEC was recently elucidated by RNA-seq (100). Similar to the case for other bacteria, CsrA plays a global role in EHEC gene expression and affects expression of the LEE as well as genes encoding motility, transport, metabolism, and signal transduction proteins (100). Mechanistic insights concerning CsrA-dependent regulation of T3S are detailed in the next section.
FIG 5.
Canonical mechanisms of posttranscriptional regulation by CsrA. The substrate-mimetic sRNAs CsrB and CsrC sequester CsrA and limit CsrA activity. When unbound (free), CsrA binds mRNA and relieves secondary structures to promote translation. Conversely, CsrA may bind to or near the ribosome binding site (RBS) to block ribosome access and inhibit translation.
SPECIFIC MECHANISMS OF VIRULENCE FACTOR REGULATION
T3SS and effectors.
The LEE is a horizontally acquired pathogenicity island that is perhaps the most canonical virulence factor of EHEC (101). The importance of the LEE to pathogenesis is exemplified by the requirement for colonization and pathogenesis in human in vitro organ culture as well as in a variety of animal models, including mice, infant rabbits, gnotobiotic pigs, and calves (24, 102–107). Proper spatiotemporal control of LEE expression is imperative to prevent energy from being wasted on premature T3SS formation as well as to evade host immune detection. The LEE carries two transcriptional regulators, Ler and GrlA. Ler is encoded by ler, the first gene in LEE1, and is the master regulator of the LEE (108). grlR and grlA are carried within an operon between LEE1 and LEE2 (Fig. 6A). GrlA directly binds the ler promoter to activate LEE expression (33, 109). However, GrlR sequesters GrlA and prevents its interaction with the ler promoter (109, 110). The expression and activity of Ler, GrlA, and GrlR are a hub of regulation at all levels within the cell. A recent study provided evidence that Hfq binds multiple RNA transcripts throughout the LEE, including LEE1 and grlRA (111). Although the authors extensively characterized the involvement of Hfq in expression of LEE4 (detailed below), no data describing the specific mechanism of Hfq-dependent regulation of Ler or GrlA expression were reported in that study. However, the Hfq-dependent sRNAs MgrR and RyhB directly influence LEE expression in enteropathogenic E. coli (112). These sRNAs were identified following a screen that assessed the impact of core sRNAs on the activity of grlR′-′lacZ reporter fusions. Further characterization demonstrated that MgrR base pairs to the 5′ UTR of grlR to repress grlR expression and to activate expression of grlA and the LEE, whereas RyhB binds to the grlRA mRNA to repress translation (112) (Fig. 6B). CsrA directly binds to the grlRA transcript, repressing translation of grlA in enteropathogenic E. coli (EPEC) and corresponding expression of ler (113) (Fig. 6B). This mechanism is likely conserved in EHEC because the predicted CsrA binding site is identical (113). Finally, ClpXP, an AAA+ protease that hydrolyzes ATP to degrade substrates (114), degrades GrlR to relieve GrlA, which ultimately results in activation of LEE transcription (110, 115) (Fig. 6C).
FIG 6.
Posttranscriptional and posttranslational regulation of the LEE. (A) Schematic of the LEE pathogenicity island. Functions of LEE-carried genes are indicated. (B to E) Posttranscriptional or posttranslational regulation of LEE-carried genes. Black lines represent interactions shown through genetic and biochemical data, whereas blue lines represent interactions relying solely on genetic data. Arrows indicate positive regulation, and blunted lines indicate negative regulation. For details, see the associated text.
Expression of ler is also controlled by cis-acting factors. For example, a 2-amino-acid-encoding open reading frame (ORF), termed a “minigene,” promotes ler translation and corresponding AE lesion formation (116) (Fig. 6D). The minigene is located upstream of the ler start codon and affects ler expression in a manner that is dependent on the spacing between the minigene and the ler start codon (116). This is an example of translational coupling, in which translation of the minigene may increase the local concentration of ribosomes available to initiate ler translation or remove secondary structures that may be prohibitive for translation initiation of ler (117, 118); the specific mechanism of how the minigene promotes Ler expression remains to be established. Furthermore, transcription of the antisense sRNA arl results in repression of ler specifically under high-iron conditions (119) (Fig. 6D).
In addition to LEE-encoded regulatory factors, LEE genes encoding T3SS structural and functional components are posttranscriptionally regulated. For example, expression of most genes in LEE3 is translationally coupled with the first gene in the operon, mpc. Disrupting mpc translation results in decreased expression of the entire operon to various degrees (120–122). The authors progressively mutated every SD sequence in LEE3 and determined that of the six genes downstream of mpc, translation of five of the genes was dependent on translation of the gene immediately upstream (120). Mpc forms a complex with SepL and SepD. SepL and SepD are cytoplasmic proteins required for T3SS formation and function (34, 121, 123). Additionally, Mpc interacts with and promotes expression of EscA, which is required for effector secretion (33, 121). Therefore, this translational coupling may be important to ensure coordinated expression of these essential components of the basal T3SS machinery.
The LEE4 operon encodes proteins that comprise the T3SS filament (EspA) and translocon (EspB and -D) in addition to SepL, which controls protein translocation and the chaperones CesD2 and L0017. Initial work demonstrated that different EHEC isolates expressed various levels of EspA even though levels of LEE4 transcription in the strains were equivalent (124), which suggested a posttranscriptional mechanism of control. Indeed, subsequent work has revealed a sophisticated mechanism of LEE4 regulation in EHEC (Fig. 6E). sepL is the first gene in LEE4, and several studies indicated that expression of the entire operon is dependent on its transcription and translation (111, 124–126) (Fig. 6E). The nascent sepL 5′ UTR forms a structure that sequesters the SD sequence and thereby prevents translation (111). CsrA binds the sepL leader sequence to expose the SD sequence and promote translation of the entire operon (111, 113). Hfq and the sRNA Spot42 also bind sepL mRNA and repress translation (111). The CsrA and Hfq interaction sequences on sepL mRNA overlap, suggesting that either CsrA or Hfq binds exclusively the transcript (111). Future work is necessary to understand whether CsrA binding to sepL causes additional changes in the mRNA structure (beyond exposing the RBS) that reduce CsrA binding affinity and/or enhance Hfq binding. Additionally, levels of CsrA or Hfq in the cell may affect regulation via direct competition for binding.
Additional studies have shown that RNase E also contributes to LEE4 regulation (127, 128) (Fig. 6E). Specifically, RNase E cleaves LEE4 transcripts between sepL and espA, which ultimately yields larger amounts of EspA than of SepL (127). RNase E degrades transcripts containing a 5′ monophosphate (86), and thus the newly cleaved espADB transcript should be vulnerable to degradation. Significantly, the leader region of the espA mRNA carries a 6-codon “mini-ORF” that prevents further degradation of the espADB transcript (129). The mini-ORF contains a strong SD element that recruits ribosomes to the espADB leader sequence and ultimately occludes RNase E from accessing the transcript. Thus, RNase E processing of LEE4 provides a means to differentially produce amounts of SepL and T3SS structural proteins, which are needed in smaller and larger amounts, respectively.
After reaching the required levels, expression of EspA-CesD2 needs to be turned off to allow redirection of the cell's energy to producing effector proteins. Recently, the sRNAs GlmY and GlmZ were shown to specifically destabilize the espB-cesD2 transcript to limit further protein synthesis, without affecting stability of the sepL transcript (130) (Fig. 6E). Importantly, GlmY and GlmZ modulate expression of additional components important for AE lesion formation, including destabilizing the LEE5 transcript and potentially promoting translation of the non-LEE-encoded effector EspFu/TccP (130). In conjunction with the effector Tir, EspFu/TccP recruits N-WASP and Arp2/3 to the bacterial site of attachment, which promotes effector translocation, actin rearrangement, and pedestal formation (32, 33, 35, 37, 131).
The T3SS-dependent effector NleA is an important virulence factor in EHEC. NleA contributes to host colonization and modulates host processes, including protein secretion, maintenance of the epithelial barrier, and immune responses (36, 132–134). Importantly, translation of nleA is precisely tuned so that NleA is not expressed until the bacterium attaches to the host cell (135). The nleA 5′ UTR contains a CsrA recognition sequence to which CsrA binds and inhibits translation. Significantly, the T3SS chaperone CesT antagonizes CsrA to promote NleA expression (135). Specifically, CesT binds to CsrA on a motif that overlaps the CsrA RNA binding site, which ultimately results in inhibition of CsrA-RNA interactions (136). Therefore, CesT interaction with CsrA not only promotes NleA expression but also relieves other CsrA-dependent repression within the cell to redirect gene expression upon host cell contact (135). Altogether, these findings reveal that EHEC has evolved complex and precise mechanisms to ensure proper and coordinated expression of the T3SS and effectors.
Shiga toxin.
Shiga toxin (Stx) is a an AB subunit toxin that is comprised of a catalytic A subunit and a pentamer of B subunits (137, 138). The B subunits bind globotriaosylceramide (Gb3) on the surfaces of Paneth cells in the intestinal tract as well as renal epithelial cells, which induces endocytosis (139, 140). The A subunit cleaves an adenosine residue from the 28S rRNA of the 60S ribosomal subunit, inhibits protein synthesis, and ultimately triggers apoptotic host cell death (141, 142). The translocation of Stx to the kidneys is responsible for the development of HUS. Stx is encoded within a prophage, and phage-mediated cell lysis releases Stx (143). The phage-encoded sRNA 24B_1 and the EHEC-encoded enzyme PAPI influence phage entry into the lytic cycle (144, 145). These studies suggest that both phage-encoded and EHEC-encoded factors contribute to posttranscriptional regulation of the phage life cycle, which impacts Stx production and potentially disease severity. However, the implications of phage life cycle regulation for EHEC pathogenesis are unclear because these studies were performed in nonpathogenic E. coli strains transduced with Stx-encoding phages. Moreover, examples of direct regulation of stx/Stx by any posttranscriptional regulator have yet to be reported.
Adhesion and motility.
EHEC carries 16 fimbrial loci in addition to other adhesins that enable attachment to abiotic and biotic surfaces and that may influence tissue tropism. The EHEC-specific sRNA Esr055 was identified in a screen for upregulated genes/sRNAs from EHEC attached to HeLa cells compared to EHEC grown in Dulbecco modified Eagle medium (DMEM) (146). Further characterization revealed that a deletion of the sRNA esr055 increased EHEC adherence to HeLa cells. Additionally, greater numbers of the esr055 deletion strain were recovered from the colons than from the ceca of infected mice (147). The latter findings correlate with decreased expression of esr055 in the colon compared to in the ileum, which suggests that Esr055 contributes to tissue tropism (147). Finally, RNA-seq experiments comparing expression of WT EHEC and the Δesr055 strains grown in vitro revealed differential expression of over 400 genes, including genes carried within fimbrial loci (147); however, direct versus indirect targets were not determined in that study.
Flagella are long whiplike surface structures that are important for motility and that also promote attachment. Flagellar expression is controlled by the master transcriptional regulator encoded by flhD and flhC (148, 149) and is also subject to complex posttranscriptional regulation (Fig. 7). In nonpathogenic E. coli, OxyS, OmrA, OmrB, and ArcZ negatively regulate flagellar expression by direct interaction with flhD (150). Furthermore, McaS positively regulates flagellar expression by binding to flhD (150, 151). Additionally, a deletion of the z5898 gene, which is predicted to encode an EHEC-specific RNA helicase, resulted in decreased levels of flagellar expression and corresponding motility (152). Finally, GrlA negatively regulates transcription of flhD and flhC (110). Thus, ClpXP-dependent degradation of GrlR leads to downregulation of flagellar expression (110, 115).
FIG 7.
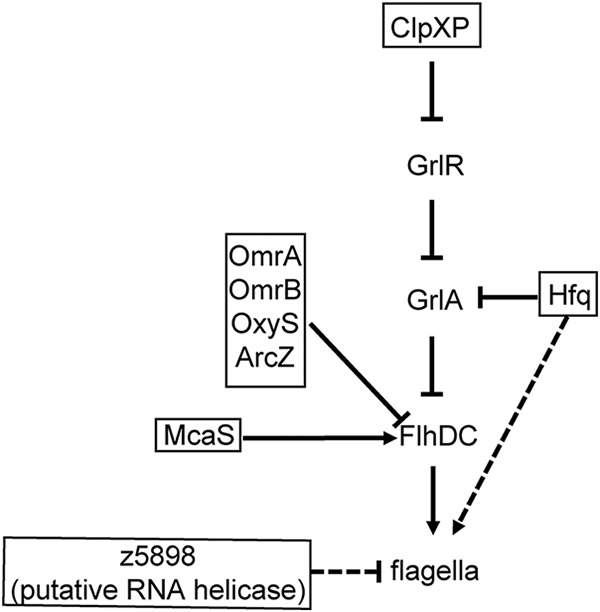
Posttranscriptional and posttranslational regulation of flagella. Solid lines indicate direct interactions, and dashed lines indicated indirect regulation or regulation based on genetic data. Arrows indicate positive regulation, and blunted lines indicate negative regulation.
Acid resistance.
Although E. coli strains in general encode acid resistance pathways, the regulation of these pathways varies between nonpathogenic E. coli and EHEC (and even among EHEC isolates) (153–155). To reach the colon, EHEC must survive the acidity of the stomach. Therefore, acid resistance is considered a critical virulence factor and is thought to contribute to the low infectious dose of EHEC (23). In addition to the stomach, EHEC is exposed to weak acids in the intestine. EHEC encodes three acid resistance mechanisms to counteract these acidic environments: an oxidative system, an arginine-dependent system, and a glutamate-dependent system (21, 22). The protease ClpXP indirectly promotes expression of the glutamate-dependent acid resistance genes gadE and gadX. ClpXP rapidly degrades the sigma factor RpoS, which negatively regulates gadE and gadX transcription (156). Furthermore, the core genome-encoded antisense sRNA GadY binds to the 3′ UTR of gadX and thereby promotes expression (157).
Iron homeostasis.
Iron is critical for the survival of most pathogens, including EHEC, because many enzymes are dependent on iron redox reactions (158). EHEC encodes several iron transporters and siderophores to acquire iron from the iron-limited intestinal environment (159). Fur (ferric uptake regulator) is an iron-sensing transcription factor and master regulator of genes encoding iron homeostasis proteins (160). As part of this, Fur activates expression of the core genome sRNA RyhB under iron-limited conditions (161). RyhB represses expression of genes encoding iron-utilizing proteins, including sodB, which encodes an Fe-superoxide dismutase (161, 162). Furthermore, RyhB promotes siderophore production by positively regulating expression of shiA, which encodes a shikimate permease. Under iron-limited conditions, the production of shikimate allows for the synthesis of siderophores and therefore the acquisition of iron (52). RyhB and the EHEC-specific sRNA Esr41 modulate expression of the iron transport and storage proteins CirA, ChuA, and Bfr (161). RyhB negatively influences chuA and bfr expression but positively affects expression of cirA (161), whereas Esr41 represses translation of chuA, bfr, and cirA (87). Moreover, in support of the genetic data, deletion of esr41 resulted in a growth advantage when EHEC was cultured in an iron-depleted medium (87).
SUMMARY AND OUTLOOK
Clearly, posttranscriptional regulation plays a key role in regulating traits essential for EHEC pathogenesis. The vast amount of data produced by unbiased techniques indicates that EHEC has coopted core sRNAs as well as evolved EHEC-specific sRNAs to regulate virulence. These studies have demonstrated a global role for posttranscriptional regulation; however, for the majority of sRNAs identified in EHEC, the mRNA targets and mechanisms of regulation remain to be uncovered. Moreover, most of the targeted studies have focused on regulation of the LEE, with fewer detailed reports regarding posttranscriptional regulation of other virulence factors. Posttranscriptional regulation of non-LEE virulence factors, especially Stx, warrants further study, as these pathways may represent therapeutic targets.
Furthermore, recent work has provided important insights regarding new roles for known regulatory factors, new players in posttranscriptional regulation, and novel mechanisms important for gene expression and protein function. For example, although the canonical function of Hfq is to interact with sRNAs, recent studies have shown that Hfq also functions independently of sRNAs to inhibit translation in nonpathogenic E. coli (163, 164). Moreover, the FinO domain protein ProQ was recently found to bind a large repertoire of sRNAs in Salmonella enterica serovar Typhimurium (68). In S. Typhimurium, ProQ not only promotes stability of a target sRNA, RaiZ, but is a component of the ternary complex that forms between the sRNA RaiZ and the 5′ UTR of the target mRNA hupA. The formation of the ProQ-RaiZ-hupA complex inhibits HU-α protein synthesis (165). Interestingly, in vitro biochemical data indicate that ProQ remains associated with RaiZ at the target mRNA binding site (165), whereas Hfq rapidly disassociates following sRNA-mRNA association (166). Moreover, in addition to ProQ, RaiZ also interacted with Hfq (165), suggesting that RaiZ may regulate gene expression differently depending on the associated chaperone. ProQ is conserved in EHEC, and thus similar interwoven webs of regulation will likely be uncovered in this pathogen.
RNA nucleotide modifications have also been shown to influence gene expression. For example, similar to the decapping of eukaryotic mRNAs, in bacterial transcripts, the conversion of the 5′-terminal triphosphate to a monophosphate initiates RNA decay by exposing the transcript to attack by 5′-monophosphate-dependent RNase. RppH (Nud/YgdP) is a member of the Nudix hydrolase family of enzymes. Specifically, RppH is an RNA pyrophosphohydrolase that removes 5′ phosphate from the mRNA, which results in rapid decay of the mRNA (167, 168). RppH has been shown to play an extensive role in Helicobacter pylori gene expression (169) and to contribute to the pathogenesis of diverse bacteria, including E. coli K1, Legionella pneumophila, and S. Typhimurium (170–172). Other types of mRNA modifications, such as incorporation of methylated nucleotides and/or pseudouridines, have been shown to result in premature translation termination (173).
Finally, growing evidence indicates that subcellular localization of RNAs impacts distribution of the corresponding protein products (174). This is a relatively nascent field in bacterial RNA biology, in general, and will likely have implications for understanding the function and activity of virulence factors, such as the membrane-associated T3SS or adhesins, in EHEC and other pathogens.
In summary, posttranscriptional gene regulation functions at a global level in bacteria and provides rapid and coordinated gene expression, which is essential for a pathogen to evade and/or overcome host defenses to establish infection and cause disease. Current data underscore the importance of posttranscriptional gene regulation to spatiotemporal control of EHEC virulence factors at all stages of infection. Despite the increasing number of recent reports, it is clear that these findings are just a drop in the bucket and that there is much more to investigate. Future studies will reveal new strategies that EHEC uses to cause disease and will likely be applicable to understanding bacterial physiology and host-pathogen interactions in general.
ACKNOWLEDGMENTS
We thank members of the Kendall lab for thoughtful discussions and feedback. We apologize to colleagues whose work was not included in this review due to space constraints.
Work in the Kendall lab is supported by National Institutes of Health National Institute for Allergy and Infectious Diseases grants R01AI118732 and R21AI130439. A.B.S. also received support from NIH training grant 5 T32 AI055432.
REFERENCES
- 1.Lee DJ, Minchin SD, Busby SJ. 2012. Activating transcription in bacteria. Annu Rev Microbiol 66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 2.Balleza E, López-Bojorquez LN, Martínez-Antonio A, Resendis-Antonio O, Lozada-Chávez I, Balderas-Martínez YI, Encarnación S, Collado-Vides J. 2009. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol Rev 33:133–151. doi: 10.1111/j.1574-6976.2008.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorman CJ, Dorman MJ. 2017. Control of virulence gene transcription by indirect readout in Vibrio cholerae and Salmonella enterica serovar Typhimurium. Environ Microbiol 19:3834–3845. doi: 10.1111/1462-2920.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erhardt M, Dersch P. 2015. Regulatory principles governing Salmonella and Yersinia virulence. Front Microbiol 6:949. doi: 10.3389/fmicb.2015.00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harel J, Martin C. 1999. Virulence gene regulation in pathogenic Escherichia coli. Vet Res 30:131–155. [PubMed] [Google Scholar]
- 6.Marceau M. 2005. Transcriptional regulation in Yersinia: an update. Curr Issues Mol Biol 7:151–177. [PubMed] [Google Scholar]
- 7.Mellies JL, Barron AM, Carmona AM. 2007. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun 75:4199–4210. doi: 10.1128/IAI.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, Margalit H. 2007. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol 3:138. doi: 10.1038/msb4100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt S, Romeo T, Kalman D. 2011. Honing the message: post-transcriptional and post-translational control in attaching and effacing pathogens. Trends Microbiol 19:217–224. doi: 10.1016/j.tim.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferens WA, Hovde CJ. 2011. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog Dis 8:465–487. doi: 10.1089/fpd.2010.0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karmali MA, Petric M, Lim C, Fleming PC, Steele BT. 1983. Escherichia coli cytotoxin, haemolytic-uraemic syndrome, and haemorrhagic colitis. Lancet ii:1299. [DOI] [PubMed] [Google Scholar]
- 12.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. [DOI] [PubMed] [Google Scholar]
- 13.Bell BP, Goldoft M, Griffin PM, Davis MA, Gordon DC, Tarr PI, Bartleson CA, Lewis JH, Barrett TJ, Wells JG, Baron R, Kobayashi J. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA 272:1349–1353. [PubMed] [Google Scholar]
- 14.Boyce TG, Swerdlow DL, Griffin PM. 1995. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N Engl J Med 333:364–368. doi: 10.1056/NEJM199508103330608. [DOI] [PubMed] [Google Scholar]
- 15.Bender J, Hedberg C, Besser J, Boxrud D, MacDonald K, Osterholm M. 1997. Surveillance for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N Engl J Med 337:388–394. doi: 10.1056/NEJM199708073370604. [DOI] [PubMed] [Google Scholar]
- 16.Walterspiel J, Ashkenazi S, Morrow A, Cleary T. 1992. Effect of subinhibitory concentrations of antibiotics on extracellular Shiga-like toxin I. Infection 20:25–29. doi: 10.1007/BF01704889. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, McDaniel A, Wolf L, Keusch G, Waldor M, Acheson D. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis 181:664–670. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]
- 18.Wong CS, Jelacic S, Habeeb RL, Watkins SL, Tarr PI. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med 342:1930. doi: 10.1056/NEJM200006293422601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grif K, Dierich MP, Karch H, Allerberger F. 1998. Strain-specific differences in the amount of Shiga toxin released from enterohemorrhagic Escherichia coli O157 following exposure to subinhibitory concentrations of antimicrobial agents. Eur J Clin Microbiol Infect Dis 17:761. doi: 10.1007/s100960050181. [DOI] [PubMed] [Google Scholar]
- 20.Pennington H. 2010. Escherichia coli O157. Lancet 376:1428. doi: 10.1016/S0140-6736(10)60963-4. [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol 177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol 62:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamin MM, Datta AR. 1995. Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol 61:1669–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis SB, Cook V, Tighe R, Schüller S. 2015. Enterohemorrhagic Escherichia coli colonization of human colonic epithelium in vitro and ex vivo. Infect Immun 83:942–949. doi: 10.1128/IAI.02928-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnett Foster D. 2013. Modulation of the enterohemorrhagic E. coli virulence program through the human gastrointestinal tract. Virulence 4:315–323. doi: 10.4161/viru.24318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lackraj T, Kim JI, Tran SL, Barnett Foster DE. 2016. Differential modulation of flagella expression in enterohaemorrhagic Escherichia coli O157: H7 by intestinal short-chain fatty acid mixes. Microbiology 162:1761–1772. doi: 10.1099/mic.0.000357. [DOI] [PubMed] [Google Scholar]
- 27.Bäumler AJ, Sperandio V. 2016. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci U S A 92:7996–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A 87:7839–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520. doi: 10.1016/S0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 31.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92:1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campellone KG, Robbins D, Leong JM. 2004. EspFU Is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev Cell 7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A 101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garmendia J, Frankel G, Crepin VF. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect Immun 73:2573–2585. doi: 10.1128/IAI.73.5.2573-2585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garmendia J, Phillips AD, Carlier MF, Chong Y, Schüller S, Marches O, Dahan S, Oswald E, Shaw RK, Knutton S, Frankel G. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol 6:1167–1183. doi: 10.1111/j.1462-5822.2004.00459.x. [DOI] [PubMed] [Google Scholar]
- 36.Gruenheid S, Sekirov I, Thomas NA, Deng W, O'Donnell P, Goode D, Li Y, Frey EA, Brown NF, Metalnikov P, Pawson T, Ashman K, Finlay BB. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- 37.Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, Younis R, Mattthews S, Marches O, Frankel G, Hayashi T, Pallen MJ. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A 103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torres AG, Giron JA, Perna NT, Burland V, Blattner FR, Avelino-Flores F, Kaper JB. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect Immun 70:5416–5427. doi: 10.1128/IAI.70.10.5416-5427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres AG, Kanack KJ, Tutt CB, Popov V, Kaper JB. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol Lett 238:333–344. [DOI] [PubMed] [Google Scholar]
- 40.Tuttle J, Gomez T, Doyle MP, Wells JG, Zhao T, Tauxe RV, Griffin PM. 1999. Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol Infect 122:185. doi: 10.1017/S0950268898001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luzader DH, Kendall MM. 2016. Commensal ‘trail of bread crumbs’ provide pathogens with a map to the intestinal landscape. Curr Opin Microbiol 29:68–73. doi: 10.1016/j.mib.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connolly JP, Finlay BB, Roe AJ. 2015. From ingestion to colonization: the influence of the host environment on regulation of the LEE encoded type III secretion system in enterohaemorrhagic Escherichia coli. Front Microbiol 6:568. doi: 10.3389/fmicb.2015.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43:880. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narberhaus F, Vogel J. 2009. Regulatory RNAs in prokaryotes: here, there and everywhere. Mol Microbiol 74:261. doi: 10.1111/j.1365-2958.2009.06869.x. [DOI] [PubMed] [Google Scholar]
- 46.Bossi L, Figueroa-Bossi N. 2016. Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat Rev Microbiol 14:775–784. doi: 10.1038/nrmicro.2016.129. [DOI] [PubMed] [Google Scholar]
- 47.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Wagner EGH. 2003. RNomics in Eshcerichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner EGH, Romby P. 2015. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet 90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Darfeuille F, Unoson C, Vogel J, Wagner EG. 2007. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell 26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Morita T, Mochizuki Y, Aiba H. 2006. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci U S A 103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammer BK, Bassler BL. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 104:11145–11149. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E. 2007. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 53.Urban JH, Vogel J. 2008. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol 6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corcoran CP, Podkaminski D, Papenfort K, Urban JH, Hinton JCD, Vogel J. 2012. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol 84:428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- 55.Durand S, Braun F, Helfer AC, Romby P, Condon C. 2017. sRNA-mediated activation of gene expression by inhibition of 5′-3′ exonucleolytic mRNA degradation. Elife 6:e23602. doi: 10.7554/Elife.23602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frohlich KS, Papenfort K, Fekete A, Vogel J. 2013. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J 32:2963–2979. doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lalaouna D, Morissette A, Carrier MC, Massé E. 2015. DsrA regulatory RNA represses both hns and rbsD mRNAs through distinct mechanisms in Escherichia coli. Mol Microbiol 98:357–369. doi: 10.1111/mmi.13129. [DOI] [PubMed] [Google Scholar]
- 58.Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. 2013. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153:426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfeiffer V, Papenfort K, Lucchini S, Hinton JCD, Vogel J. 2009. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16:840–847. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 60.Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C-G, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 62.Perna NT, Plunkett Gr Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 63.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. 2014. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell 55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gruber CC, Sperandio V. 2015. Global analysis of posttranscriptional regulation by GlmY and GlmZ in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 83:1286–1295. doi: 10.1128/IAI.02918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neuhaus K, Landstorfer R, Simon S, Schober S, Wright PR, Smith C, Backofen R, Wecko R, Keim DA, Scherer S. 2017. Differentiation of ncRNAs from small mRNAs in Escherichia coli O157:H7 EDL933 (EHEC) by combined RNAseq and RIBOseq—ryhB encodes the regulatory RNA RyhB and a peptide, RyhP. BMC Genomics 18:216. doi: 10.1186/s12864-017-3586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gopel Y, Khan MA, Gorke B. 2014. Ménage à trois: post-transcriptional control of the key enzyme for cell envelope synthesis by a base-pairing small RNA, an RNase adaptor protein, and a small RNA mimic. RNA Biol 11:433–442. doi: 10.4161/rna.28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, Becher D, Reinhardt R, Vogel J. 2016. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci U S A 113:11591–11596. doi: 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Updegrove TB, Zhang A, Storz G. 2016. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 30:133–138. doi: 10.1016/j.mib.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hansen A-M, Kaper JB. 2009. Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli. Mol Microbiol 73:446–465. doi: 10.1111/j.1365-2958.2009.06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kendall MM, Gruber CC, Rasko DA, Hughes DT, Sperandio V. 2011. Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86-24. J Bacteriol 193:6843–6851. doi: 10.1128/JB.06141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shakhnovich EA, Davis BM, Waldor MK. 2009. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol Microbiol 74:347–363. doi: 10.1111/j.1365-2958.2009.06856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. 1988. Illnesses associated with Escherichia coli O157:H7. Ann Intern Med 109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 75.Wells JG, Davis BR, Wachsmuth IK, Riley LW, Remis RS, Sokolow R, Morris GK. 1983. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol 18:512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wick LM, Qi W, Lacher DW, Whittam TS. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol 187:1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smirnov A, Schneider C, Hor J, Vogel J. 2017. Discovery of new RNA classes and global RNA-binding proteins. Curr Opin Microbiol 39:152–160. doi: 10.1016/j.mib.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 78.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. 2007. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev 21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Urbanowski ML, Stauffer LT, Stauffer GV. 2000. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol 37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 80.Jin Y, Watt RM, Danchin A, JDH . 2009. Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics 10:165. doi: 10.1186/1471-2164-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coornaert A, Chiaruttini C, Springer M, Guillier M. 2013. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet 9:e1003156. doi: 10.1371/journal.pgen.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Monsieurs P, De Keersmaecker S, Navarre WW, Bader MW, De Smet F, McClelland M, Fang Y, De Moor B, Vanderleyden J, Marchal K. 2005. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J Mol Evol 60:462–474. doi: 10.1007/s00239-004-0212-7. [DOI] [PubMed] [Google Scholar]
- 83.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laalami S, Zig L, Putzer H. 2014. Initiation of mRNA decay in bacteria. Cell Mol Life Sci 71:1799–1828. doi: 10.1007/s00018-013-1472-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohanty BK, Kushner SR. 2016. Regulation of mRNA decay in bacteria. Annu Rev Microbiol 70:25–44. doi: 10.1146/annurev-micro-091014-104515. [DOI] [PubMed] [Google Scholar]
- 86.Mackie GA. 2013. RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11:45–57. doi: 10.1038/nrmicro2930. [DOI] [PubMed] [Google Scholar]
- 87.Waters SA, McAteer SP, Kudla G, Pang I, Deshpande NP, Amos TG, Leong KW, Wilkins MR, Strugnell R, Gally DL, Tollervey D, Tree JJ. 2017. Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J 36:374–387. doi: 10.15252/embj.201694639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 89.Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci U S A 93:3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. 1994. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 91.Murashko ON, Lin-Chao S. 2017. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc Natl Acad Sci U S A 114:E8025–E8034. doi: 10.1073/pnas.1703731114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carpousis AJ. 2007. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol 61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 93.Hu J, Zhu MJ. 2015. Defects in polynucleotide phosphorylase impairs virulence in Escherichia coli O157:H7. Front Microbiol 6:806. doi: 10.3389/fmicb.2015.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kusmierek M, Dersch P. 2018. Regulation of host-pathogen interactions via the post-transcriptional Csr/Rsm system. Curr Opin Microbiol 41:58–67. doi: 10.1016/j.mib.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 95.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol 44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 96.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol 56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 98.Ren B, Shen H, Lu ZJ, Liu H, Xu Y. 2014. The phzA2-G2 transcript exhibits direct RsmA-mediated activation in Pseudomonas aeruginosa M18. PLoS One 9:e89653. doi: 10.1371/journal.pone.0089653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Patterson-Fortin LM, Vakulskas CA, Yakhnin H, Babitzke P, Romeo T. 2013. Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. J Mol Biol 425:3662–3677. doi: 10.1016/j.jmb.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang S, Yang F, Yang B. 2017. Global effect of CsrA on gene expression in enterohemorrhagic Escherichia coli O157:H7. Res Microbiol 168:700–709. doi: 10.1016/j.resmic.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 101.Law D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J Appl Microbiol 88:729–745. doi: 10.1046/j.1365-2672.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- 102.Dean-Nystrom EA, Bosworth BT, Moon HW, O'Brien AD. 1998. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun 66:4560–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deng W, VAllance BA, Li Y, Puente JL, Finlay BB. 2003. Citrobacter rodentium translocated intimin receptor (Tir) is an essential virulence factor needed for actin condensation, intestinal colonization and colonic hyperplasia in mice. Mol Microbiol 48:95–115. doi: 10.1046/j.1365-2958.2003.03429.x. [DOI] [PubMed] [Google Scholar]
- 104.Donnenberg MS, Tzipori S, McKee ML, O'Brien AD, Alroy J, Kaper JB. 1993. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Invest 92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McKee ML, Melton-Celsa AR, Moxley RA, Francis DH, O'Brien AD. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun 63:3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ritchie JM, Thorpe CM, Rogers AB, Waldor MK. 2003. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect Immun 71:7129–7139. doi: 10.1128/IAI.71.12.7129-7139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ritchie JM, Waldor MK. 2005. The locus of enterocyte effacement-encoded effector proteins all promote enterohemorrhagic Escherichia coli pathogenicity in infant rabbits. Infect Immun 73:1466–1474. doi: 10.1128/IAI.73.3.1466-1474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Elliott SJ, Sperandio V, Girón JA, Shin S, Mellies JL, Wainwright L, Hutcheson SW, McDaniel TK, Kaper JB. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun 68:6115–6126. doi: 10.1128/IAI.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Russell R, Sharp F, Rasko DA, Sperandio V. 2007. QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J Bacteriol 189:5387–5392. doi: 10.1128/JB.00553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iyoda S, Koizumi N, Satou H, Lu Y, Saitoh T, Ohnishi M, Watanabe H. 2006. The GrlR-GrlA regulatory system coordinately controls the expression of flagellar and LEE-encoded type III protein secretion systems in enterohemorrhagic Escherichia coli. J Bacteriol 188:5682–5692. doi: 10.1128/JB.00352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang D, McAteer SP, Wawszczyk AG, Russell CD, Tahoun A, Elmi A, Cockroft SL, Tollervey D, Granneman S, Tree JJ, Gally DL. 2018. An RNA-dependent mechanism for transient expression of bacterial translocation filaments. Nucleic Acids Res doi: 10.1093/nar/gky096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhatt S, Egan M, Ramirez J, Xander C, Jenkins V, Muche S, El-Fenej J, Palmer J, Mason E, Storm E, Beurkert T. 2017. Hfq and three Hfq-dependent small regulatory RNAs—MgrR, RyhB and McaS—coregulate the locus of enterocyte effacement in enteropathogenic Escherichia coli. Pathog Dis 75:ftw113. doi: 10.1093/femspd/ftw113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bhatt S, Edwards AN, Nguyen HTT, Merlin D, Romeo T, Kalman D. 2009. The RNA binding protein CsrA is a pleiotropic regulator of the locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli. Infect Immun 77:3552–3568. doi: 10.1128/IAI.00418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Olivares A, Baker T, Sauer R. 2016. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat Rev Microbiol 14:33–44. doi: 10.1038/nrmicro.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iyoda S, Watanabe H. 2005. ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J Bacteriol 187:4086–4094. doi: 10.1128/JB.187.12.4086-4094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Islam MS, Shaw RK, Frankel G, Pallen MJ, Busby SJ. 2012. Translation of a minigene in the 5′ leader sequence of the enterohaemorrhagic Escherichia coli LEE1 transcription unit affects expression of the neighbouring downstream gene. Biochem J 441:247–253. doi: 10.1042/BJ20110912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCarthy JE, Gualerzi C. 1990. Translational control of prokaryotic gene expression. Trends Genet 6:78–85. doi: 10.1016/0168-9525(90)90098-Q. [DOI] [PubMed] [Google Scholar]
- 118.Levin-Karp A, Barenholz U, Bareia T, Dayagi M, Zelcbuch L, Antonovsky N, Noor E, Milo R. 2013. Quantifying translational coupling in E. coli synthetic operons using RBS modulation and fluorescent reporters. ACS Synth Biol 2:327–336. doi: 10.1021/sb400002n. [DOI] [PubMed] [Google Scholar]
- 119.Tobe T, Yen H, Takahashi H, Kagayama Y, Ogasawara N, Oshima T. 2014. Antisense transcription regulates the expression of the enterohemorrhagic Escherichia coli virulence regulatory gene ler in response to the intracellular iron concentration. PLoS One 9:e101582. doi: 10.1371/journal.pone.0101582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sun W-SW, Chen J-W, Wu Y-C, Tsai H-Y, Kuo Y-L, Syu W Jr.. 2016. Expression regulation of polycistronic lee3 genes of enterohaemorrhagic Escherichia coli. PLoS One 11:e0155578. doi: 10.1371/journal.pone.0155578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lin CN, Sun WS, Lu HY, Ng SC, Liao YS, Syu WJ. 2014. Protein interactions and regulation of EscA in enterohemorrhagic E. coli. PLoS One 9:e85354. doi: 10.1371/journal.pone.0085354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tsai NP, Wu YC, Chen JW, Wu CF, Tzeng CM, Syu WJ. 2006. Multiple functions of l0036 in the regulation of the pathogenicity island of enterohaemorrhagic Escherichia coli O157:H7. Biochem J 393:591–599. doi: 10.1042/BJ20051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Younis R, Bingle LE, Rollauer S, Munera D, Busby SJ, Johnson S, Deane JE, Lea SM, Frankel G, Pallen MJ. 2010. SepL resembles an aberrant effector in binding to a class 1 type III secretion chaperone and carrying an N-terminal secretion signal. J Bacteriol 192:6093–6098. doi: 10.1128/JB.00760-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Roe AJ, Yull H, Naylor SW, Woodward MJ, Smith DGE, Gally DL. 2003. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect Immun 71:5900–5909. doi: 10.1128/IAI.71.10.5900-5909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kresse A, Beltrametti F, Müller A, Ebel F, Guzmán CA. 2000. Characterization of SepL of enterohemorrhagic Escherichia coli. J Bacteriol 182:6490–6498. doi: 10.1128/JB.182.22.6490-6498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang D, Roe AJ, McAteer SP, Shipston MJ, Gally DL. 2008. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol Microbiol 69:1499–1512. doi: 10.1111/j.1365-2958.2008.06377.x. [DOI] [PubMed] [Google Scholar]
- 127.Lodato PB, Kaper JB. 2009. Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol Microbiol 71:273–290. doi: 10.1111/j.1365-2958.2008.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lodato PB, Thuraisamy T, Richards J, Belasco J. 2017. Effect of RNase E deficiency on translocon protein synthesis in an RNase E-inducible strain of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol Lett 364. doi: 10.1093/femsle/fnx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lodato PB, Hsieh P, Belasco J, Kaper JB. 2012. The ribosome binding site of a mini-ORF protects a T3SS mRNA from degradation by RNase E. Mol Microbiol 86:1167–1182. doi: 10.1111/mmi.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gruber CC, Sperandio V. 2014. Posttranscriptional control of microbe-induced rearrangement of host cell actin. mBio 5:e01025-. doi: 10.1128/mBio.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vingadassalom D, Campellone KG, Brady MJ, Skehan B, Battle SE, Robbins D, Kapoor A, Hecht G, Snapper SB, Leong JM. 2010. Enterohemorrhagic E. coli requires N-WASP for efficient type III translocation but not for EspFU-mediated actin pedestal formation. PLoS Pathog 6:e1001056. doi: 10.1371/journal.ppat.1001056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim J, Thanabalasuriar A, Chaworth-Musters T, Fromme JC, Frey EA, Lario PI, Metalnikov P, Rizg K, Thomas NA, Lee SF, Hartland EL, Hardwidge PR, Pawson T, Strynadka NC, Finlay BB, Schekman R, Gruenheid S. 2007. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe 2:160–171. doi: 10.1016/j.chom.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 133.Mundy R, Petrovska L, Smollett K, Simpson N, Wilson RK, Yu J, Tu X, Rosenshine I, Clare S, Dougan G, Frankel G. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect Immun 72:2288–2302. doi: 10.1128/IAI.72.4.2288-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yen H, Sugimoto N, Tobe T. 2015. Enteropathogenic Escherichia coli uses NleA to inhibit NLRP3 inflammasome activation. PLoS Pathog 11:e1005121. doi: 10.1371/journal.ppat.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Katsowich N, Elbaz N, Pal RR, Mills E, Kobi S, Kahan T, Rosenshine I. 2017. Host cell attachment elicits posttranscriptional regulation in infecting enteropathogenic bacteria. Science 355:735–739. doi: 10.1126/science.aah4886. [DOI] [PubMed] [Google Scholar]
- 136.Ye F, Yang F, Yu R, Lin X, Qi J, Chen Z, Cao Y, Wei Y, Gao G, Lu G. 2018. Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT. Nat Commun 9:1196. doi: 10.1038/s41467-018-03625-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fraser ME, Chernaia MM, Kozlov YV, James MN. 1994. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 A resolution. Nat Struct Biol 1:59–64. doi: 10.1038/nsb0194-59. [DOI] [PubMed] [Google Scholar]
- 138.Stein PE, Boodhoo A, Tyrrell GJ, Brunton JL, Read RJ. 1992. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature 355:748–750. doi: 10.1038/355748a0. [DOI] [PubMed] [Google Scholar]
- 139.Jacewicz M, Clausen H, Nudelman E, Donohue-Rolfe A, Keusch GT. 1986. Pathogenesis of Shigella diarrhea. XI. Isolation of a Shigella toxin-binding glycolipid from rabbit jejunum and HeLa cells and its identification as globotriaosylceramide. J Exp Med 163:1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lindberg AA, Brown JE, Stromberg N, Westling-Ryd M, Schultz JE, Karlsson KA. 1987. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J Biol Chem 262:1779–1785. [PubMed] [Google Scholar]
- 141.Pacheco AR, Sperandio V. 2012. Shiga toxin in enterohemorrhagic E. coli regulation and novel anti-virulence strategies. Front Cell Infect Microbiol 2:81. doi: 10.3389/fcimb.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Romer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D, Aly MR, Fraisier V, Florent JC, Perrais D, Lamaze C, Raposo G, Steinem C, Sens P, Bassereau P, Johannes L. 2007. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450:670–675. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- 143.Shimizu T, Ohta Y, Nota M. 2009. Shiga toxin 2 is specifically released from bacterial cells by two different mechanisms. Infect Immun 77:2813–2823. doi: 10.1128/IAI.00060-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nejman-Faleńczyk B, Bloch S, Licznerska K, Dydecka A, Felczykowska A, Topka G, Węgrzyn A, Węgrzyn G. 2015. A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage Φ24Β. Sci Rep 5:10080. doi: 10.1038/srep10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nowicki D, Bloch S, Nejman-Faleńczyk B, Szalewska-Pałasz A, Węgrzyn A, Węgrzyn G. 2015. Defects in RNA polyadenylation impair both lysogenization by and lytic development of Shiga toxin-converting bacteriophages. J Gen Virol 96:1957–1968. doi: 10.1099/vir.0.000102. [DOI] [PubMed] [Google Scholar]
- 146.Yang B, Feng L, Wang F, Wang L. 2015. Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat Commun 6:6592. doi: 10.1038/ncomms7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Han R, Xu L, Wang T, Liu B, Wang L. 2017. A small regulatory RNA contributes to the preferential colonization of Escherichia coli O157:H7 in the large intestine in response to a low DNA concentration. Front Microbiol 8:274. doi: 10.3389/fmicb.2017.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kutsukake K, Ohya Y, Iino T. 1990. Transcriptional analysis of the flageelar regulon of Salmonella typhimurium. J Bacteriol 172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu X, Matsumura P. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol 176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De Lay N, Gottesman S. 2012. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol 86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Thomason MK, Fontaine F, De Lay N, Storz G. 2012. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu Y, Xu X, Lan R, Xiong Y, Ye C, Ren Z, Liu L, Zhao A, Wu LF, Xu J. 2013. An O island 172 encoded RNA helicase regulates the motility of Escherichia coli O157:H7. PLoS One 8:e64211. doi: 10.1371/journal.pone.0064211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang J, Russell TW, Hocking DM, Bender JK, Srikhanta YN, Tauschek M, Robins-Browne RM. 2015. Control of acid resistance pathways of enterohemorrhagic Escherichia coli strain EDL933 by PsrB, a prophage-encoded AraC-like regulator. Infect Immun 83:346–353. doi: 10.1128/IAI.02758-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bergholz TM, Whittam TS. 2007. Variation in acid resistance among enterohaemorrhagic Escherichia coli in a simulated gastric environment. J Appl Microbiol 102:352–362. doi: 10.1111/j.1365-2672.2006.03099.x. [DOI] [PubMed] [Google Scholar]
- 155.Arnold KW, Kaspar CW. 1995. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol 61:2037–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tomoyasu T, Takaya A, Handa Y, Karata K, Yamamoto T. 2005. ClpXP controls the expression of LEE genes in enterohaemorrhagic Escherichia coli. FEMS Microbiology Lett 253:59–66. doi: 10.1016/j.femsle.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 157.Opdyke JA, Kang JG, Storz G. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol 186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Doherty CP. 2007. Host-pathogen interactions: the role of iron. J Nutr 137:1341–1344. doi: 10.1093/jn/137.5.1341. [DOI] [PubMed] [Google Scholar]
- 159.Torres AG, Payne SM. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 160.Ernst JF, Bennett RL, Rothfield LI. 1978. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J Bacteriol 135:928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Masse E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Blasi U. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol Microbiol 50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 163.Ellis MJ, Trussler RS, Haniford DB. 2015. Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Mol Microbiol 96:633–650. doi: 10.1111/mmi.12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen J, Gottesman S. 2017. Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev doi: 10.1101/gad.302547.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Smirnov A, Wang C, Drewry LL, Vogel J. 2017. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J 36:1029–1045. doi: 10.15252/embj.201696127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hopkins J, Panja S, Woodson SA. 2011. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res 39:5193–5202. doi: 10.1093/nar/gkr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Luciano D, Vasilyev N, Richards J, Serganov A, Belasco J. 2017. A novel RNA phosphorylation state enables 5′ end-dependent degradation in Escherichia coli. Mol Cell 67:44–54. doi: 10.1016/j.molcel.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Deana A, Celesnik H, Belasco J. 2008. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 169.Bischler T, Hsieh PK, Resch M, Liu Q, Tan HS, Foley PL, Hartleib A, Sharma CM, Belasco JG. 2017. Identification of the RNA pyrophosphohydrolase RppH of Helicobacter pylori and global analysis of its RNA targets. J Biol Chem 292:1934–1950. doi: 10.1074/jbc.M116.761171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Badger JL, Wass CA, Kim KS. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol Microbiol 36:174–182. doi: 10.1046/j.1365-2958.2000.01840.x. [DOI] [PubMed] [Google Scholar]
- 171.Edelstein P, Hu B, Shinzato T, Edelstein M, Xu W, Bessman M. 2005. Legionella pneumophila NudA Is a Nudix hydrolase and virulence factor. Infect Immun 73:6567–6576. doi: 10.1128/IAI.73.10.6567-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ismail T, Hart C, McLennan A. 2003. Regulation of dinucleoside polyphosphate pools by the YgdP and ApaH hydrolases is essential for the ability of Salmonella enterica serovar typhimurium to invade cultured mammalian cells. J Biol Chem 278:32602–32607. doi: 10.1074/jbc.M305994200. [DOI] [PubMed] [Google Scholar]
- 173.Hoernes T, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, Huttenhofer A, Erlacher M. 2016. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res 44:852–862. doi: 10.1093/nar/gkv1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Buskila A, Kannaiah S, Amster-Choder O. 2014. RNA localization in bacteria. RNA Biol 11:1051–1060. doi: 10.4161/rna.36135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sudo N, Soma A, Muto A, Iyoda S, Suh M, Kurihara N, Abe H, Tobe T, Ogura T, Hayashi H, Kurokawa K, Ohnishi M, Sekine Y. 2014. A novel small regulatory RNA enhances cell motility in enterohemorrhagic Escherichia coli. J Gen Appl Microbiol 60:44–50. doi: 10.2323/jgam.60.44. [DOI] [PubMed] [Google Scholar]